Abstract
Background
Schizophrenia is a multigene disease with a complex etiology and different clinical manifestations. It is of great significance to understand the etiology and pathogenesis of schizophrenia patients from different clinical dimensions and to interpret the potential molecular changes of schizophrenia patients from different clinical dimensions.
Material/Methods
RNA-Seq was performed on peripheral blood leukocytes of 50 patients with schizophrenia and 50 healthy controls. Phenotypic information of patients with schizophrenia was collected during blood sampling. Differentially expressed genes (DEGs) were screened by the edgeR package of R software. To better analyze the correlation between DEG expression values, explore the potential association between differential genes and clinical dimensions of schizophrenia, and identify hub genes, we constructed a DEG co-expression network using weighted gene co-expression network analysis (WGCNA).
Results
We provide the transcription profiles of peripheral blood leukocytes in patients with schizophrenia and found a gene module (including 89 genes) closely related to the clinical dimension of abnormal psychomotor behavior in schizophrenia.
Conclusions
The findings enhance our understanding of the biological processes of schizophrenia, enabling us to identify specific clinical dimensions of genes for diagnosis and prognostic markers and possibly for targeted therapy.
MeSH Keywords: Gene Expression Profiling, Leukocytes, Schizophrenia
Background
Schizophrenia is a common and difficult disease to treat clinically. Genetic factors play an important role in the pathogenesis of schizophrenia [1–3]. It is generally accepted that schizophrenia is a polygenic disease [4–7]. A single gene or SNP mutation site has little effect on schizophrenia [6]. Previous large-scale genome-wide association studies (GWAS) have found that multiple genetic loci are closely related to the occurrence of schizophrenia [8–10], however, identifying the risk chromosomes in schizophrenia is difficult. Great advancements have been made in genetic research on schizophrenia in recent years, with more than 100 genome-wide significant risk variations identified [10], but how reported risk variations affect the pathogenesis of schizophrenia remains elusive. Because schizophrenia is a mixed clinical syndrome, the clinical manifestations of schizophrenia vary. Some patients mainly have positive symptoms such as delusions and hallucinations [11,12]. Whereas, some patients mainly have negative symptoms such as apathy and decreased willpower [13,14]. Other patients have emotional symptoms such as mania or depression [15]. Schizophrenia is a heterogeneous aggregation of different clinical phenotypes. At present, clinical and basic research mostly focuses on schizophrenia itself. There are few studies on the relationship between different clinical manifestations and heredity. Defining the specific clinical dimensions of schizophrenia is a great challenge for psychiatrists.
In the past, the traditional clinical classification system classified schizophrenia as paranoid, catatonic, simple, undifferentiated, disorganized, and residual [16]. However, due to the heterogeneity of the disease, the stability of clinical diagnosis is poor, as is the standardization of treatment. Therefore, the 5th edition of the U.S. Diagnostic and Statistical Manual on Mental Illness (DSM-5; published in 2013) excludes these traditional subtypes of schizophrenia [17,18]. It is recommended that the psychiatric symptom severity rating scale be used to evaluate symptoms in different dimensions. In recent years, some scholars have divided the clinical manifestations of schizophrenia into the following 8 symptom groups (8-dimensional symptoms): abnormal psychomotor behavior, disorganized speech, hallucination, delusion, negative symptom, impaired cognition, depression, and mania [17,19,20]. This approach can deepen psychiatrists’ further understanding of schizophrenia and guide clinical treatment and scientific research. Compared with the use of subtypes, the use of psychopathological dimensions in DSM-5 significantly improves the ability to describe the heterogeneity of the disease in a more effective and clinically useful way, and this approach can be targeted to different dimensions of symptoms for research and treatment [19–22].
To further study the relationship between different dimensions of schizophrenia and heredity, we collected clinical information from schizophrenia patients and used the dimension method in DSM-5 (Clinician-Rated Dimensions of Psychosis Symptom Severity scale) to assess the severity of the core symptoms of schizophrenia [19,20]. If the score of the core symptoms in a dimension was 2 or greater than 2, we considered the symptoms in this dimension to be positive. If the score of the core symptoms in a dimension was less than 2, we considered the symptoms in this dimension to be negative [17,19]. The total RNA of peripheral blood leukocytes from study participants was collected at the same time as the dimension score assessment. After removing ribosomal RNA (rRNA), the DNA library was constructed. Illumina HiSeqTM 4000 sequencing technology was used to obtain information on the transcription groups and to explore the differences in genes between 2 groups: schizophrenia patients and healthy controls who were roughly matched for sex and age. Weighted gene co-expression network analysis (WGCNA) was used to analyze the relationship between differential genes and 8 clinical dimensions. Interestingly, we found that Turquoise module was positively correlated with abnormal psychomotor behavior, and the difference was statistically significant. We randomly selected 5 hub genes (CXCL8, EGR1, EGR3, IL1B, and PTGS2) for quantitative analysis. Quantitative real-time polymerase chain reaction (qRT-PCR) validation showed that the results were consistent with those of messenger RNA (mRNA) sequencing. In our study, the Turquoise module (89 genes) was identified as new risk genes for the abnormal psychomotor behavior dimension of schizophrenia, and we speculate that risk variation might affect the expression of these genes. This leads to the susceptibility of patients with schizophrenia to the abnormal psychomotor behavior dimension.
Material and Methods
Study patients
A total of 50 patients with schizophrenia were enrolled from the Honghe Second People’s Hospital and Yuxi Second People’s Hospital from May 2018 to July 2018. During the same period, 50 healthy volunteers with similar sex and age were recruited into the normal control group at the Sixth Affiliated Hospital of Kunming Medical University. After the patients with schizophrenia and healthy controls provided informed consent, we used anticoagulant extraction to obtain 5 mL of peripheral blood.
Inclusion criteria for the patient case group
Inclusion criteria for the patient case group were as follows: 1) meet the criteria for diagnosis of schizophrenia in DSM-5; 2) first onset schizophrenia or no antipsychotic drugs for 5 weeks before enrollment; 3) Han Chinese aged 18 to 60 years; 4) no significant RNA degradation in the sample; and 5) signed informed consent and voluntary participation in this study.
Exclusion criteria for the patient case group
Exclusion criteria for the patient case group were as follows: 1) combination of mental disorders caused by other mental illnesses such as depression or physical and physical diseases; 2) serious neurological diseases or severe physical diseases such as the combination of liver and kidney insufficiency; 3) a medical history of alcohol dependence, substance abuse and addiction; 4) a history of blood transfusion within 1 month before admission; 5) treatment with electroconvulsive therapy (ECT) within 3 months; and 6) severe audio-visual disorders and intellectual development disorders.
Inclusion criteria in the healthy control group
Inclusion criteria for the health control group were as follows: 1) a normal physical examination of adults without abnormalities, without any family history of mental illness; 2) no history of severe traumatic brain injury, no neurological diseases, and no major physical diseases and trauma history; and 3) of Chinese Han nationality roughly matched with the sex and age of the case group.
Ethical approval and informed consent of patients
All experimental schemes involving human participants in this study were approved by the Ethics Committee of the Sixth Affiliated Hospital of Kunming Medical University. All participants in the study provided written informed consent, and all participants agreed to use their clinical and RNA-Seq data for research and publication. The research methods adopted were based on the Helsinki Declaration and the guidelines of the Ethics Committee of the Sixth Affiliated Hospital of Kunming Medical University. Patients were told that refusing to participate in the study would not affect future treatment.
Peripheral blood leukocyte (PBL) collection and total RNA extraction
Collection of fresh whole blood samples
At approximately 7: 00 a.m., 5 mL of whole blood was extracted by venipuncture from the study participants. An EDTA anticoagulant tube was used. The samples used to extract leukocyte RNA were centrifuged within 60 minutes and transferred to a 1.5 mL EP tube. The leukocytes were mixed with 1 mL TRIzol and then transferred into a 1.5 mL freezer tube (which was tightly sealed with sealing film and stored in liquid nitrogen or −80°C freezer.
Total RNA extraction
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract leukocytes from peripheral blood. A Nanodrop ND200 (Thermo Scientific Inc., Waltham, MA, USA) was used to determine the concentration and purity of RNA. According to OD260, the concentration of the RNA sample was calculated as follows: RNA (mg/mL) = 40×OD260×dilution multiple/1000. The value of OD260/OD280 of pure RNA is generally 1.8–2.0, so the purity of RNA can be estimated according to the value of OD260/OD280. If the ratio is low, there are residual proteins. It is important to note that the value of RNA OD260/OD280 extracted by this method was between 1.8 and 2.0 (hands are the main source of RNase).
Library construction sequencing
After extracting the total RNA, the rRNA was removed using the Ribo-ZeroTM Magnetic Kit to enrich the mRNA. The enriched mRNA fragments were then converted into short fragments with fragmentation buffer, and the cDNA was reversed transcribed with a random primer. Second chain cDNA was synthesized with DNA polymerases I, RNase H, DNTP, and buffers. The samples were then amplified using QIAquick-PCR to purify the cDNA fragments, repair the ends, add polyadenine (a), and connect the Illumina sequencing adaptor. The product was sequenced with the Illumina HeSeqTM 4000, and underwent agarose gel electrophoresis and PCR amplification at Gidio Biotechnology (Guangzhou, China).
Analysis of differential expression transcription products (DEGS) of coding RNA
To identify differentially expressed transcripts between the schizophrenia patients and the healthy controls, the edgeR package (http://www.r-project.org/) was used [23] and Benjamini-Hochberg (B-H) correction for multiple comparisons. We categorized the transcripts with multiple fold changes (>2) and false discovery rate (FDR) <0.05 as indicting significant differentially expressed genes (DEGs). Then, enrichment analysis of Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was carried out for differentially expressed coding RNA.
Comparison of our RNA sequencing of differential genes with genes identified by GWAS identification and genes identified by gene set enrichment (GSE) data set
We downloaded the GWAS identification gene on SZDB data [24] (http://www.szdb.org/SZDB/gwas.php) and several data sets from the GEO public data set (GEO, http://www.ncbi.nlm.nih.gov/geo), including GSE62191 (comprising 29 patients with schizophrenia and 30 healthy controls) [25,26], GSE17612 (including 28 patients with schizophrenia and 23 cases of healthy controls) [27], and GSE12649 (including 35 patients with schizophrenia and 34 cases of healthy control) [28]. First, GEO2R was used to identify the differential genes between the schizophrenia group and the healthy control group in the GSE dataset. With P less than 0.05 indicating statistical significance (GSE dataset differential gene results are detailed in Supplementary Document 1), a Venn diagram was drawn using the Omicshare tool, a free online data analysis platform (www.omicshare.com/tools).
WGCNA analysis
The WGCNA algorithm is a common algorithm for building gene co-expression networks. The WGCNA algorithm first assumes that the gene network obeys the scale less distribution, defines the adjoining function of the gene co-expression correlation matrix and gene network formation, calculates the difference coefficient of different nodes, and constructs a hierarchical cluster tree accordingly. Different branches of the cluster tree represent different genetic modules. The degree of gene co-expression is high, while the degree of gene co-expression is low in different modules. Finally, the relationship between the module and the specific phenotype was explored, and the target gene and gene network of the disease-specific phenotype were finally obtained. We extracted 559 upregulated and downregulated DEGs from edgeR analysis and performed WGCNA with the expressed sequencing data [29]. The R-package “WGCNA” was used to search for related modules and hub genes of clinical traits.
Analysis of trait-related characteristics module
The main goal of network co-expression analysis is to identify the modules and genes most relevant to biological significance or clinically relevant. Hence, the modules most relevant to the traits for schizophrenia were identified by analyzing the correlation between the modules and specific phenotypes or traits. By calculating the correlation between each module’s characteristic values and traits, we identified the modules and genes most relevant to the traits so that the corresponding modules could be selected for further study. In general, if a module has a significantly higher correlation with the selected trait than the other modules, it indicates that the module may have the strongest correlation with the trait. The grouping relationship is considered a trait used to determine the relationship between modules and groupings. Then, the gene function of a clinical dimension-related (and statistically significant) module was analyzed, and the GO and KEGG pathways were enriched to analyze the biological function of the module.
Protein–protein interaction (PPI) network analysis
Protein–protein interaction (PPI) was used to find the biologically or clinically meaningful modules and analyze the gene expression profiles of the study patients. Each gene in the module was calculated; the highly connected genes are often hub genes, which may have important functions. The network was visualized using String database (http://www.string-db.org/).
qPCR verification
Illumina offers a wide range of powerful library preparation kits that provide fast and easy library preparation workflow, and the Illumina HiSeqTM 4000 sequencing results are highly technically repeatable. However, to further verify the accuracy of our peripheral white blood cell sample sequencing results, we used qRT-PCR to validate 5 hub genes (CXCL8, EGR1, EGR3, IL1B, and PTGS2) in the Turquoise module to evaluate whether the results were consistent with mRNA sequencing results and to verify the accuracy of the Illumina HiSeqTM 4000 sequencing results. Extracted RNA was synthesized with complementary DNA (cDNA) using a reverse transcription kit (Takara). Real-time PCR was carried out with TB Green series kit (Takara) and monitored with the Bio-RAD (CFX96 real-time) system. β-actin for mRNA was applied to normalize the result. All reactions were repeated 3 times and relative gene expressions were evaluated by the 2−ΔΔCt method. Primer sequences are shown in Table 1.
Table 1.
Primer sequences.
| Genes | Forward | Reverse |
|---|---|---|
| EGR1 | GGTCAGTGGCCTAGTGAGC | GTGCCGCTGAGTAAATGGGA |
| EGR3 | GACATCGGTCTGACCAACGAG | GGCGAACTTTCCCAAGTAGGT |
| IL1B | ATGATGGCTTATTACAGTGGCAA | GTCGGAGATTCGTAGCTGGA |
| CXCL8 | AAGACATACTCCAAACCTTTCCACC | CTTCAAAAACTTCTCCACAACCCTC |
| PTGS2 | CTGGCGCTCAGCCATACAG | CGCACTTATACTGGTCAAATCCC |
| ACTB | CATGTACGTTGCTATCCAGGC | CTCCTTAATGTCACGCACGAT |
Statistical methods
All transcript sequencing data were analyzed by R software. GEO data sets were analyzed by GEO2R tools, and qRT-PCR validation data were tested by 2 independent samples t-test. The test level a=0.05, P<0.05 was considered statistically significant.
Results
Analysis of the difference in the expression of mRNA between the 2 groups
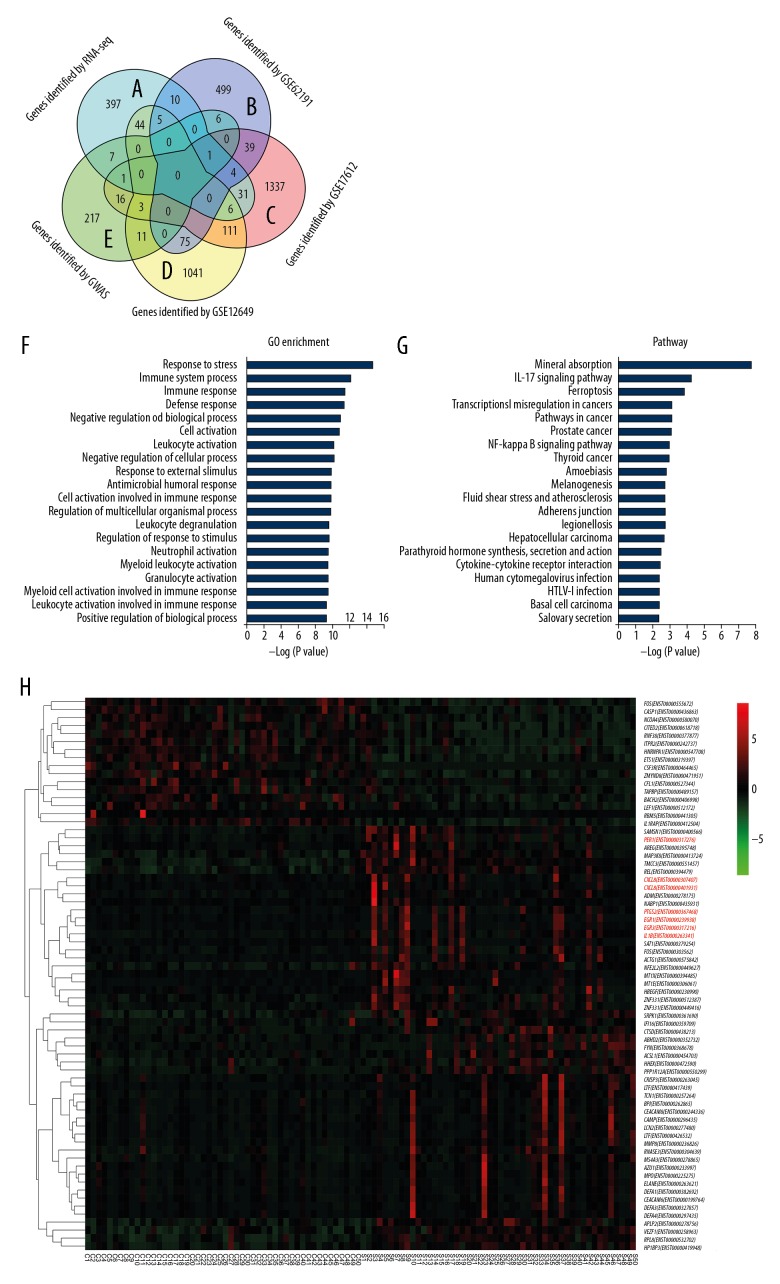
We used edgeR software to analyze the difference in FPKM (fragments per kilobase of transcript per million) values between the healthy control group and schizophrenia patients. FDR and log2FC were used to screen differential transcripts. The screening conditions were FDR <0.05 and |log2FC| >1. A total of 559 differentially expressed transcripts were screened, including 206 downregulated transcripts and 353 upregulated transcripts (see Supplementary Table 1 for details). Also, we compared with GWAS and other brain GSE datasets and used a Venn map (Figure 1A–1E). The comparisons between GWAS identifying genes and GSE dataset identifying genes in brain regions can be found in the Supplementary Document 1; a Venn chart compared GWAS and GSE data.
Figure 1.
(A–E) Comparisons between genome-wide association studies (GWAS) and gene set enrichment (GSE) dataset. Identifying genes in the brain regions can be found in Supplementary Document 1 Venn chart that compares GWAS and GSE data. (F) Gene Ontology (GO) item in differentially expressed genes (DEGs). (G) The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis chart. (H) Expression patterns of differentially expressed transcripts (69 transcripts) with an average value of FPKM (fragments per kilobase of transcript per million) greater than 1 in the healthy control group and schizophrenia group and constructed a heat map.
Differential mRNA GO/pathway enrichment analysis
After obtaining differentially expressed transcripts, GO functional analysis and KEGG pathway analysis were performed on the differentially expressed transcripts. Taking FDR <0.05 as the threshold, we calculated the P values. Meeting this condition was defined as a significant enrichment GO item in DEGs. This analysis can identify the main biological functions performed by different genes. GO analysis indicated that DEGs were mainly concentrated in response to stress, immune system process, and immune response, as shown in Figure 1F. The KEGG pathway analysis showed that DEGs are mainly involved in mineral absorption, IL-17 signaling pathways, etc. The KEGG enrichment analysis chart is shown in Figure 1G.
Cluster analysis of expression patterns
Based on the expression of transcripts, the relationship between transcripts was hierarchically clustered, and the clustering results were presented using a heat map. We analyzed the expression patterns of differentially expressed transcripts (69 transcripts) with an average value of FPKM greater than 1 in the healthy control group and schizophrenia group, and we constructed a heat map (Figure 1H). The rows were normalized (z-score), and hierarchical clustering analysis was carried out for different transcripts. Each column in the graph represents one sample: C1–C50 are the 50 samples of the healthy control group, and S1–S50 are the 50 samples of the schizophrenia group. Each row represents one transcript, and the expression of transcripts in different samples was expressed in different colors. The redder the color, the higher the expression level, and the greener the color, the lower the expression level.
WGCNA module division
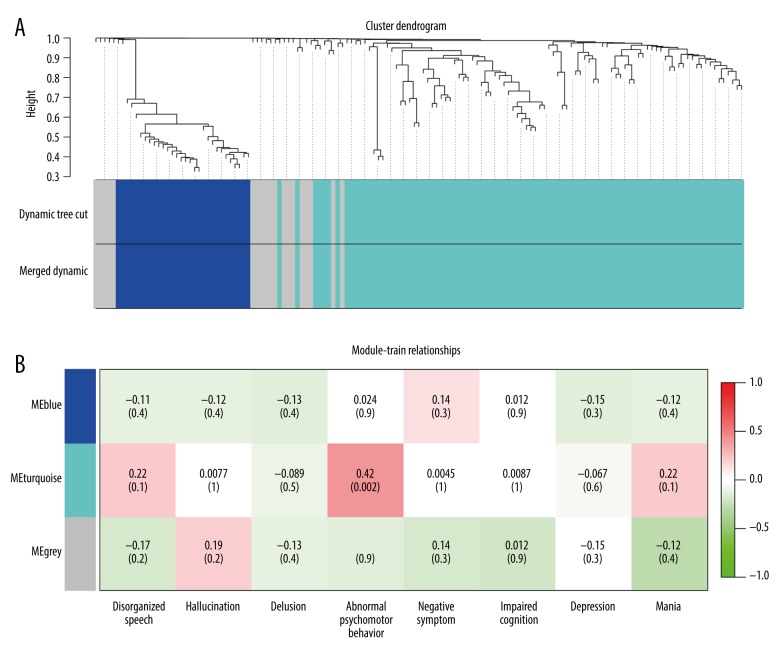
The adjacency matrix was transformed into a topological overlap matrix. According to the TOM-based difference measure, genes are divided into different gene modules. According to the principle of a scale-free network, the power value was determined. The power value selected in this analysis was 5. The parameters (similarity) of the merging module were 0.75, and the number of genes needed to be included in each module was at least 30. The hierarchical clustering tree of modules is as follows: 1) Dynamic Tree Cut is a module divided according to clustering results, 2) Dynamics is a combined module division of modules that express similar patterns based on module similarity, followed by analysis according to the merged module, and 3) for Tree Diagrams, vertical distance represents the distance between 2 nodes (between genes), and the horizontal distance is meaningless. See Figure 2.
Figure 2.
(A) The hierarchical clustering tree of modules. (B) The results of the trait-module correlation are shown in the Module-Trait Relationships chart.
Character module analysis
The main goal of the network co-expression analysis was to identify the most relevant modules and genes of biological of clinical significance. Therefore, the most relevant modules can be found by analyzing the correlation between modules and specific phenotypes or traits. By calculating the correlation between each module’s characteristic values and traits, we can identify the modules and genes most relevant to the traits, so that the corresponding modules can be selected for further study. In general, if one module has a significantly higher correlation with the trait than the other modules, this indicates the module may have the strongest correlation with the trait. The 8-dimensional characteristics of schizophrenia samples are detailed in Supplementary Table 2: Schizophrenia Sample Trait. The grouping relationship was used to find the relationship between the module and the grouping. The results of the trait-module correlation are shown in the Module-Trait Relationships chart (Figure 2B) which contains the correlation coefficients and P values of each trait and module characteristic values.
Each column of the Profile-Related Characteristics Module (Figure 2B) represents a personality, and each row represents a genetic module. The number in each box represents the correlation between the module and the characteristic. The closer the number is to 1, the stronger the positive correlation between the module and the characteristic. The closer the module to −1, the stronger the module’s negative correlation with the trait. The numbers in parentheses represent the significance of the P value, and the smaller the number, the greater the significance. The P value was calculated using the Student’s t-test, the smaller the P value, the more significant the correlation between the representative form and the module. The results suggest that the Turquoise module is positively correlated with abnormal psychomotor behavior (P=0.002).
Turquoise module gene expression pattern and functional enrichment
We show the expression patterns of the genes contained in the Turquoise module using thermal maps and changes in the module’s eigenvalues in different samples (equivalent to the module expression pattern) by histogram, as shown in Figure 3A. The calorimetric expression of genes in different samples is shown. Red indicates upregulated genes, and green indicates downregulated genes. The expression pattern of module eigenvalues in different samples is shown. GO analysis reveals that the Turquoise module genes are mainly concentrated in biological processes, such as response, locomotion, immune system process, cell motility, and localization of cells. The GO enrichment analysis is shown in Figure 3B. The KEGG pathway analysis (Figure 3C) shows that the Turquoise module genes are mainly involved in malaria, mineral absorption, the IL-17 signaling pathway, Salmonella infection, the NF-κB signaling pathway, and the tumor necrosis factor (TNF) signaling pathway.
Figure 3.
(A) Expression patterns of the genes contained in the Turquoise module. (B) The Gene Ontology (GO) enrichment analysis of the genes contained in the Turquoise module. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the genes contained in the Turquoise module.
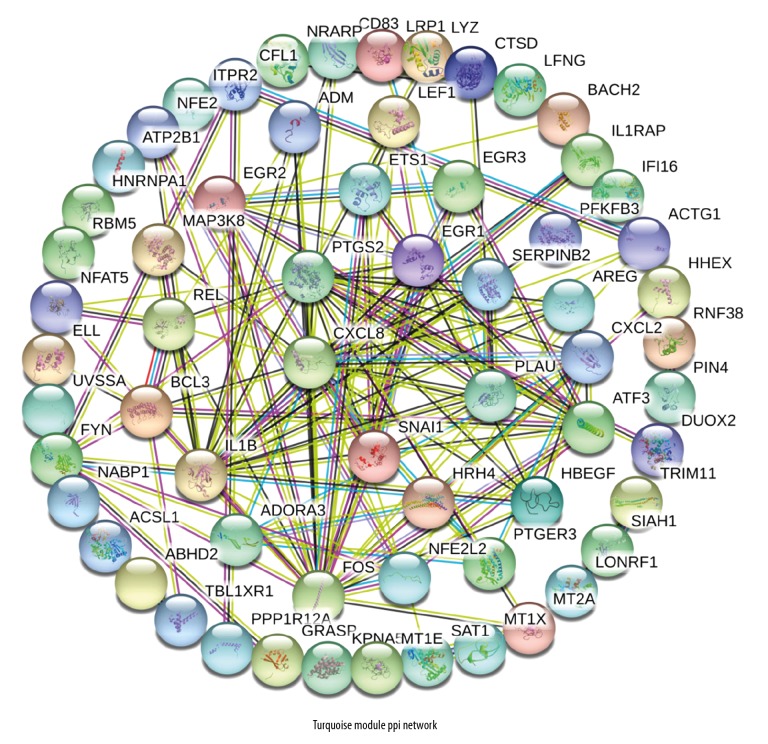
PPI network construction and hub gene analysis
After mapping the genetic symbols of all the Turquoise modules to String, a PPI network (Figure 4) was generated. PPI statistical analysis indicates 89 nodes with 141 edges; the average node degree was 3.17, the expected number was 57, and PPI p-enrichment was 1.0e-16. At the same time, we selected the hub gene using the CytoHubba plug-in in Cytoscape to determine the top 20 hub genes with high connectivity in turn: CXCL8, FOS, PTGS2, IL1B, ATF3, EGR1, CXCL2, REL, NFE2L2, SNAI1, PLAU, HBEGF, AREG, EGR2, ETS1, PTGER3, LEF1, ADM, SERPINB2, and EGR3.
Figure 4.
Turquoise modules to string, a protein–protein interaction (PPI) network was generated.
Gene validation
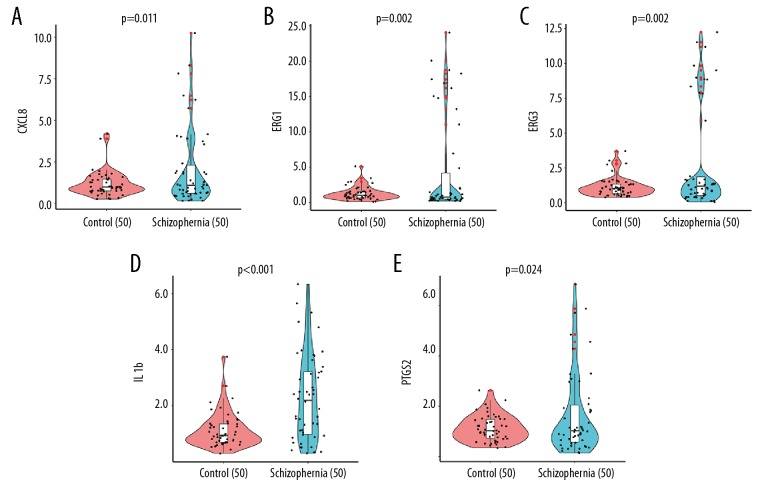
To further verify the accuracy of the sequencing results of our peripheral blood leukocyte samples, we randomly selected 5 hub genes (CXCL8, EGR1, EGR3, IL1B, and PTGS2) in used the Turquoise module for qRT-PCR verification. Two independent sample t-tests showed that the expression of CXCL8, EGR1, EGR3, IL1B, and PTGS2 in the schizophrenia group was higher than that of the healthy control group (P<0.05). The qRT-PCR results were consistent with those of mRNA sequencing (Figure 5).
Figure 5.
(A–E) Quantitative Real-Time polymerase chain rection results of 5 hub genes in the Turquoise module.
Discussion
Information regarding mRNA expression in the brain of patients after death is very important to elucidate the molecular genetic mechanism of schizophrenia and other neuropsychiatric diseases. However, brain tissue specimens from living patients are difficult to obtain. Recent studies have found that the immune system plays a vital role in the repair, maintenance, and brain reserves of the central nervous system [30,31], that immune cells help maintain neurogenesis and spatial learning in adulthood [32], and that age-related spatial memory loss can be partially recovered through immune activation [33]. The interaction between neurons, glial cells, and the immune system contributes to the healthy functioning of the brain [34]. The occurrence of schizophrenia is closely related to immunity [35]. Peripheral blood macrophage/monocyte inflammation patterns may indicate the activation of small glial cells, which, in the brain, are macrophages, and are considered the main immune cells [36]. Neuroimmune interactions allow the nerves and immune systems to regulate each other: immune cells play an important role as peripheral neurotransmitters, peripheral T cells exhibit similar characteristics as dopamine-activated cells, and the brain can regulate immune cells [37]. The cells that compose the peripheral immune system are mainly white blood cells, which are easy to collect in a clinical setting and can be used as an alternative to brain tissue for biomarker screening studies.
The results of this study can help us understand the relationship between different dimensions of schizophrenia and genetics. In the past, genetic studies of schizophrenia have paid little attention to its heterogeneity (and little attention to symptom dimensions). In this study, we describe a large transcription dataset of blood cells in a patient with schizophrenia (detailing their symptom dimensions) as well as gender and age-matching control samples of white blood cells.
First, we analyzed the differential expression patterns of transcripts between the schizophrenia group and the healthy control group. A total of 559 differential transcripts (506 genes) were found. Functional enrichment analysis of differential genes indicated that 139 transcripts were involved in response to stress biological processes (GO: 0006950), and 114 transcripts were involved in immune system processes (GO: 0002376). Enrichment analysis indicated that 93 transcripts were involved in immune response processes (GO: 0006955). DEGs were mainly involved in the immune response, IL-17 signaling pathway, and NF-κB signaling pathway. Dimitrov et al. showed that the IL-17 signaling pathway may play an important role in schizophrenia [38]. Numerous studies have shown that the NF-κB signaling pathway plays an important role in nerve protrusion growth, activity-dependent plasticity, and cognitive function, and that abnormal changes in the NF-κB signaling pathways in patients with schizophrenia can occur [39,40].
A Venn diagram was used to analyze where the DEGs identified by RNA-Seq and the genes identified by GWAS overlapped. A total of 9 overlapping genes (LRP1, ABCB9, NISCH, EGR1, FES, DGKZ, FYN, CTND1, and ALDOA) were found. GSE62191 had 20 overlapping genes, GSE12649 had 55 overlapping genes, and GSE17612 had 43 overlapping genes. Our study found that FOS, IL1B, CXCL8, CASP1, CFL1, CAMP, ITPR2, ACTG1 and other inflammation and immunity related genes were differently expressed between the schizophrenia patient group and the healthy control group, These findings were consistent with the neuroinflammatory hypothesis of schizophrenia. Previous studies have shown that the loss of mTOR inhibitor TSC1 or PTEN can increase the growth of somatic cells and dendrites, and that altered TSC/mTOR or PTEN/mTOR signal can play an important role in the development of autism spectrum disorders [41–44]. In our study, we found that the ASD related genes PTEN, mTOR, TSC1, and FMR1 expression in peripheral blood of patients with schizophrenia had no significant change when compared with healthy controls. This may indicate that these 2 common psychiatric diseases have different pathogenesis. According to the differential gene thermogram, the expression of CASP1 and other genes in schizophrenia patients decreased significantly. CASP1 is a risk gene for depression [45,46]. At present, there are no studies on the relationship between CASP1 and schizophrenia. Some studies found that the depression-like symptoms of CAPS1 knockout rats were alleviated, while pleasure-like behaviors occurred, and the speed of exercise was increased in rats [45]. The decreased expression of CASP1 in patients with schizophrenia may be related to the positive symptoms of patients with schizophrenia, and the specific role of CASP1 in schizophrenia is unclear. We need to conduct more research on CASP1 in the future.
To find the key module most relevant to the clinical dimension of schizophrenia, we carried out WGCNA on DEGs using edgeR analysis. Clinical sample information was collected from patients, such as abnormal psychomotor behavior, disorganized speech, hallucination, delusion, negative symptom, and 8 other dimensions. Through WGCNA of differential transcripts, we determined 3 modules (nonclustering degree of gray display). From the thermogram of the module-feature correlation, we found that the Turquoise module was the module that was most correlated with the clinical features of abnormal psychomotor behavior, and the correlation coefficient was 0.42, P=0.002. The Turquoise module contains 96 transcripts (89 genes). We used the CytoHubba plug-in in Cytoscape to select hub genes and identify the top 20 hub genes with high connectivity: CXCL8, FOS, PTGS2, IL1B, ATF3, EGR1, CXCL2, REL, NFE2L2, SNAI1, PLAU, HBEGF, AREG, EGR2, ETS1, PTGER3, LEF1, ADM, SERPINB2, and EGR3. To reveal the potential biological functions of genes in the Turquoise module, we performed GO and KEGG analysis. This analysis shows that the genes in the Turquoise module are mainly involved in the response to stress, inflammatory response, locomotion, immune system processes and other biological processes. ADORA3, IL1B, ADM, FOS, CXCL8, PER1, ETS1, LRP1, IFI16, PTGS2, PTGER3, REL, IL1RAP, HRH4, and CXCL2 were involved in inflammatory response (GO: 0006954), This indicated that inflammation related genes play an important role in the abnormal psychomotor behavior characteristics in patients with schizophrenia, These results provide a new clue for further study of the mechanism of abnormal psychomotor behavior in schizophrenia. The Turquoise module contains 5 genes (CXCL8, FOS, PTGS2, IL1B, and CXCL2) enriched in the IL-17 signaling pathway, 5 genes (PLAU, IL1B, CXCL2, CXCL8, and PTGS2) enriched in the NF-κB signaling pathway, 3 genes (IL1B, FYN, and EGR1) enriched in prion diseases, 6 genes (FOS, MAP3K8, IL1B, CXCL2, BCL3, and PTGS2) enriched in the TNF signaling pathway, and 4 genes (FOS, MAP3K8, IL1B, and CXCL8) enriched in the Toll-like receptor signaling pathway. Previous studies have found that the TNF signaling pathway and Toll-like receptor signaling pathway are closely related to the onset of schizophrenia [47–49].
WGCNA identified 89 risk genes for schizophrenia (Supplementary Table 3), and their expression changes were positively correlated with abnormal psychomotor behavior in schizophrenia. The Turquoise module contains 3 genes of the EGR family (EGR1, EGR2, and EGR3). Hub gene EGR1 plays an important role in the occurrence of schizophrenia. Kurian et al. [50] suggested that EGR1 gene expression is increased in the blood of patients with schizophrenia who have a high illusion state. Other studies have shown that EGR1 is downregulated in the prefrontal cortex of postmortem brain samples from patients with schizophrenia [51,52]. Ramaker et al. [53] performed transcriptome analysis of 24 patients with schizophrenia and 24 bipolar disorder patients and 24 control brain tissues. The results showed that the expression of EGR1 in the anterior cingulate cortex of patients with schizophrenia was significantly lower than that of the control group. Studies have found that EGR3 is considered a potential susceptibility gene for schizophrenia [54–56]. CXCL8 is a risk gene for depression [57], but recent studies have shown that chemokine CXCL8 is involved in the neurobiological process associated with schizophrenia. The increase in maternal CXCL8 levels is also related to the increased risk of mental illness in offspring [58–60]. The PTGS2 gene, also known as the COX2 gene, is closely related to schizophrenia. The PTGS2-specific inhibitor has a curative effect on schizophrenia [61,62].
Our data showed that there was a significant difference in gene expression between the schizophrenia group and the healthy control group. Further analysis of the gene and clinical phenotype by WGCNA suggested that the Turquoise module in patients with schizophrenia was significantly related to abnormal psychomotor behavior, among which key genes were involved. IL1B participates in 5 pathways, thus, we speculate that IL1B in core genes may lead to abnormal psychomotor behavior in schizophrenia through the IL-17 signaling pathway, NF-κB signaling pathway, TNF signaling pathway, and Toll-like receptor signaling pathway. In addition, these data provide useful resources for future research and help to test preliminary hypotheses or verify important findings.
Conclusions
In the present study, compared with the healthy control group, 206 downregulated transcripts and 353 upregulated transcripts were found in the schizophrenia group samples. Functional analysis of GO showed that DEGs were mainly enriched in response to stress, immune system process, and immune response. The KEGG pathway analysis showed that DEGs mainly participated in the TNF signaling pathway and the IL-17 signaling pathway. WGCNA divided DEGs into 3 co-expression modules, which indicated that the Turquoise module was positively correlated with abnormal psychomotor behavior (P=0.002). We randomly selected 5 hub genes (CXCL8, EGR1, EGR3, IL1B, and PTGS2) of the Turquoise module for qRT-PCR verification. The results of qRT-PCR were consistent with those of mRNA sequencing. We provided the transcription profiles of peripheral blood leukocytes in patients with schizophrenia and found a gene module (including 89 genes) closely related to the clinical dimension of abnormal psychomotor behavior in schizophrenia. We also speculate that IL1B, a hub gene, may play an important role in the abnormal psychomotor behavior of schizophrenia through multiple pathways. These findings enhance our understanding of the biological processes of schizophrenia, enabling us to identify specific clinical dimensions of genes for diagnosis and prognostic markers and possibly for targeted therapy
Supplementary Data
Supplementary Document 1
1. Genes identified by GSE62191
KRT6A, TNFSF10, GBP4, SLC14A1, SCGB1D2, DUSP1, FAM86B3P, ETV5, ARC, KIF19, DUSP6, PLIN1, TTC7A, KDF1, USP45, ZMYM5, ELK3, SPAG5-AS1, PLPP5, SCGB1D1, FKBP5, CBX3P2, HSPA8, CCT6P1, C1orf74, EGR2, HES4, DYRK1A, PGBD1, PROCR, DYNLL2, RHCE, EIF3F, SERPIND1, CRTAM, EPM2A, CLEC18A, CASP14, NHLRC2, LIF, VEGFC, DUSP5P1, ATP13A4, DUS4L, GLI2, TEX2, SPRR2C, ZNF615, LOC158435, EGR1, DCAKD, CD52, EGR4, PER2, TTC13, HLA-DRB3, TTC33, GPR149, MRVI1, HRAS, SAMHD1, MDC1, GPR20, ALDH1L1, TRPC3, GNAZ, USF2, CCND1, TRIM24, PDZD2, MOCOS, PTPN7, PAPSS1, SMCHD1, BBS2, TSACC, PRR15, CD69, CCL4, EGFR, PIGA, ATP1B2, F7, SDC4, TRPV2, TECPR2, MCM6, LOC100129397, MFGE8, ZNF442, SLC25A21-AS1, LINC00469, GALNT4, CD1B, LOC101927598, OR7G3, C2CD4B, SLC15A2, WNT7A, CALML4, RRN3, SOD3, AQP1, PGM5, IRX6, CPZ, ITPKB, DGKE, WHSC1L1, HIF3A, DNAJB6, CMYA5, SERBP1, FAM161A, TMPRSS5, NR3C1, PLAU, MLXIPL, SNX2, AOC2, TUBA3D, GGACT, LINC00671, TCP11, MXRA5, SBK1, TAF15, POU2F1, TRIB2, MTSS1L, DSCC1, FAM117B, FGD3, FKTN, NIM1K, LBH, CLC, MEGF11, NR4A1, SLC16A9, MON1B, TRIOBP, EIF4E3, ZNF223, NOP14-AS1, ECM1, SLC22A6, ZFAND2B, PLK4, THBS1, SOCS6, FOS, PRKCA, NGF, FERMT2, PIP5K1C, CISD3, ST8SIA2, NAAA, PAX4, ARHGAP9, LMNTD2, CITED4, LOC100652930, CNNM2, FAM81B, ROBO3, BORCS8-MEF2B, LOC100507663, GLTSCR1L, SLC36A1, ZBTB39, C4BPB, GPIHBP1, IRX5, LOC285957, NLRP6, LCK, POLR3H, CRABP1, YY1AP1, CTNNB1, DDIT4L, ETS1, TEX40, ABL1, USP22, PPID, ARHGEF10, SLC35F6, ENTPD5, CFAP70, DPYSL3, MYBPC1, DLGAP1-AS4, LOC100288254, TMC2, FKBP4, UTP11, DZIP1L, SOHLH1, VGF, PMFBP1, MT1X, AGRN, SMOC1, COL4A1, CHRD, CEP95, ZFHX4-AS1, NOS1, C4B, GUCA1B, RARA, ZNF443, LOC105378499, B3GNT9, BCL6B, RHOBTB3, SLC30A7, DDRGK1, ARV1, CD300LG, TP53TG5, CLASP2, PGLYRP2, CEBPB, TTC29, PLIN3, GABBR1, POU5F1P4, SPATA24, KCNQ1DN, VILL, CCL19, DUSP8, R3HDM2, NPBWR1, ITGB3, LPIN1, RHO, ADGRG6, TXNRD2, FBXW11, POLR2A, HSPBP1, GRIN2C, LOC153546, ALB, AMT, C6orf141, OAS1, DLX1, FAM189A2, PCDHB9, UCA1, ZNF256, CREM, FOXA3, KNCN, CMC2, TXNDC16, ZBTB6, KIF13A, GZMH, CT45A1, C12orf49, APOC1, DPYSL2, C3AR1, AGXT, MAOB, NPBWR2, CASP8, TUSC5, APC2, MASP2, VWF, CH25H, HLA-DRB1, CD48, NXF1, CDC5L, NFATC1, CPA5, SPON1, ETNPPL, LRAT, PHLDB2, GGA1, ANKRD11, CCIN, PGAM1, GINS2, GIMAP2, SMIM6, RAB34, GDF10, ZNF738, CCL3L3, UCKL1, NOTCH2NL, NKX2-1, EXO1, B4GALT2, DOCK3, KRTAP19-1, SCGN, TCP10, GRAMD1C, TNFRSF11B, SPRY4, KAT2B, FBXO21, PPFIBP1, VRTN, MYF6, PSIP1, ABCC3, SYT6, CSNK1G2, TTC32, CDKN2AIPNL, INTS10, CHKB, P2RY13, TDGF1, COL24A1, KAZALD1, ZC3H10, PRAP1, ARID1B, ABHD12, TGFB3, MGST1, RFX4, ECHDC2, RMND5B, METTL7A, PLEC, PAPLN, MYOD1, CTBS, SMARCA5, KNSTRN, ABL2, ARMC6, IGF2BP1, TMEM56, NBPF3, SLU7, IFI44L, CFAP157, XIRP1, CEACAM1, EPSTI1, GABPA, USP47, FOSL2, ENDOU, TTC30A, PLD6, COLCA2, UTP15, ZNF382, LINC01554, GPSM3, PTGER2, MAFA, C3orf70, SPACA6, IL10, H3F3B, SH3KBP1, CCDC122, BCL2L11, RUFY3, CALCA, ATG4D, LOC100506548, ARX, PYGB, FAM197Y2P, CD79A, ENPP1, GJA1, AVP, RRM1, TTC21A, PDE1A, HES1, PBX4, NID1, ZC4H2, NOS3, ITGAX, C6orf52, LOC439938, SLC2A4RG, SIRPB1, LPAR6, PQLC3, MUC1, KLK1, PIGQ, FRZB, IRAK1, PBXIP1, GPAT3, LOC105369230///HLA-DRB5///HLA-DRB4///HLA-DRB3///HLA-DRB1, SERPINA3, LENG8, KLHL28, SRGAP2C, PTCH1, SLC52A3, ENOX1, ZNF416, TXNIP, FN1, ST3GAL2, DHRS3, CCDC88C, ORAI2, ALDH1A2, VSIG4, FAM198A, APOL5, TMPRSS15, GLIS3-AS1, NPNT, ZCCHC7, ENPEP, LOC100133091, A2M, PSPH, GAA, SLC25A5, CELF4, SLC39A3, HFM1, DUSP2, CNGB1, AQP4, LOC101927402, ETV4, CLN3, HS6ST3, UBA6, EDN1, WFS1, PTPRS, SIK1, OSGEP, GPR45, CHRNA4, ERVH-4, ERVH-6, MCPH1, FGL1, SGSM3, TDRD9, ZNF433, MCM9, AASDH, SLC44A5, LOC101927502, PROK2, ARMC3, MATN3, PKM, RBM23, GLIS3, ARMC12, SRSF10, CD3G, GALNT3, DCN, ABCG2, MYCBPAP, IRAK4, INO80D, RCC2, RNF157, RANBP17, GMNN, KMT5B, GPCPD1, NTRK2, SLC2A4, ISYNA1, KIF1A, HLA-H, CWF19L2, AGFG2, TMCO3, HSPA12B, DAZL, ZNF563, TET1, ARPP19, MTA1, PDK4, PPAT, GIMAP8, NUDT14, ZBTB40, ZNF77, CYB561D1, PSMB7, MYOC, MPP3, TNXB, NUTF2, ZSCAN25, ADARB2, BMP8B, PON2, TPM4, PON3, SHMT1, RDH8, TPCN1, NEK9, ZNF8, TLK2, SARNP, RBBP8NL, IFT88, RB1, VCAM1, ASB4, TYMSOS, RASL11B, NETO2, ZNF595, PARD6A, ITGB1BP1, SMCR5, EMX2, MAGEA5, SLC12A7, RALGDS, AARS2, VPS13C, TYW5, ZNF182, ZNF214, GH1, GOT1L1, RPGR, SEMA3A, SYNPO, GTF2H5, HLA-DMA, ZNF175, PDZRN3, OR2H2, NIP7, TMEM254, ABCC12, GPA33, ING1, HSPB1, MRPL30, ERBB2, BCAT2, MKNK2, IL17RB, FLJ13224, ACTR5, EIF2AK3, ZNF84, LINC00602, CSDC2, COX18, MAP3K2, RHPN1, GRAMD2, ZNF250, MGAT1, APOL3, BIRC3, TWIST2, BTN1A1, MOB3B, PIM3, MAN2A2, HSD3B7, C1orf226, ZNF713, HAUS5, TRMT6, CRH, G0S2, LMX1B, GAL3ST2, ILF2, CACNA1H, SOX18, ZNF146, MRS2, KDM1B, LOC284561, MT1B, JMJD1C-AS1, PPFIA1, LEF1, KIF20B, XYLT1, OSBPL11, RAB24, FAM110B, SDHB, ALDH1L2, PITPNA, TAT, NUFIP2, LGALS3, GPC3, UTY, LOC100506544, BBOX1, SLC10A7, DBF4, ADCY1, PLK1, CD70, ZNF799, DGAT1, NUDT22, FNDC4, BBIP1, ELOVL2-AS1, HRK, CYP51A1, ZNF578, LYPD6B, RIC1, THNSL2, CD320, ALDH1A3, AKAP1, DPYD, MIAT, SERTAD4, RPS6KA3.
2. Genes identified by GSE17612
ZNF578, PMAIP1, LOC100506446, FAM204A, FRZB, S100A9, ZNF775, OGN, ALS2CR11, SERP2, FHAD1, LAT2, S100A8, PXDN, WARS2, GSDMB, PEX7, GSTM3, KDM3A, KIAA0408, PTRH1, SVOPL, SCARA5, LOC101928973, DNAJC7, MPV17, FMOD, DKK1, LOC101928238, MROH1, HK2, MLLT3, LOC101927783, TBC1D2B, LINC01361, LINC00838, AKAP12, LINC01020, DSG2, CHD2, CTTNBP2NL, TMEM218, PTPN14, GLRX3, ABCC13, LOC101928409, STK4, MAX, SLC28A1, EPB41L4A, OR10D3, SLC13A3, ATF6, USP37, CTD-2194D22.4, GPR132, GRAMD2, SEC23IP, AGBL3, LACTB, LOC101929757, TIMP4, WIPF3, ST20-AS1, TGFBR1, SATB2-AS1, LOC728613, LINC00900, FBXW2, ZNF577, IL1RN, IFT140, LOC374443, FLJ41455, SAMHD1, ACVRL1, LOC101930112///SPG7, FRY, CYP26B1, SLC13A4, LOC728613///PDCD6, STYX, SLC25A36, LOC101929607, RORA-AS1, CDH6, LOC286154, TRIB2, ATP2A1, DNAJC5G, TREML3P, FMNL1, SPAG11A///SPAG11B, RERGL, CCDC80, ARMCX4, ZDHHC3, GTSF1L, NRCAM, ITGBL1, COLEC10, SCIN, PYGL, SMYD2, TRAF3IP2, PDGFD, GPBP1L1, ZNF518B, HP11014, FUZ, ALOX5AP, LOC100996579, LOC100653005, ASXL3, RPF1, TCP11, CIRBP-AS1, PCBP1-AS1, OSR1, DOCK2, ZNF764, UGGT2, TRAT1, CDHR1, LZTS1, GHR, C19orf43, SAMSN1, MALT1, COL6A2, AGA, IPMK, CHST2, MFAP3L, CDADC1, HLA-DOA, ZNF599, JMJD6, FGF2, SPAG5, SHANK3, KREMEN2, MTHFD1L, SMIM21, VPS13B, PDPN, ATG10, SOAT1, COL3A1, FLYWCH1, LINC01118, SPRNP1, ATP23, CREBBP, DUSP9, RAPGEF1, HGD, HSBP1, C8orf34, EWSR1, TRPC3, CFAP206, COL23A1, BAG3, MLNR, HRH3, LOC440028, UPP2, GSTA3, ANAPC7, AMELY, CD59, FCGR3B///FCGR3A, SEC14L1, C10orf71, CRYBG3, TEKT5, NTS, PIGO, LOC101927021, SRSF5, SMCHD1, CHCHD5, KIF1C, GSS, RNA45S5, LOXL2, CRABP2, MOB1A, MGP, FGD2, PRADC1, TPCN2, COLEC11, LOC730098, HIST1H2APS5, PRKY///PRKX, LCOR, GIMAP1-GIMAP5///GIMAP5, ATPIF1, LOC105371967, P4HA1, NARF, ATOH7, SERP1, KANSL3, IGFN1, LTB, CCL3L3///CCL3L1///CCL3, SUPT20H, CDC42-IT1, ZNRF4, SPCS1, GPRIN2, C12orf54, TNC, GRTP1, MPPED2, SLC25A25-AS1, LOC101929036///PAH, EFHC2, DCAF12L1, NT5DC3, ATP11B, RASGRP1, PNN, HDAC4, SRSF8, UNC5C, TMEM184C, ETV1, GMPR, RHBDF2, SAMD8, F11, EIF3M, CECR5-AS1, CNOT6, SYT5, PTGER3, SYK, TMEM256, GYG2, EIF4A2, C11orf1, LOC101929248, TTTY7, SSH1, IMPAD1, LOC107985971, COX4I1, IRGM, MIR6787///SLC16A3, RBM48, LINC00507, SLCO4A1, LOC101927907, PTK7, CEP131, IKZF1, ADAMTS2, TMEM51-AS1, C1QC, HNF4A, NABP1, FAM193B, ARHGAP45, WDR86, ETV5, AHSA2, ARHGAP30, GSTCD, TLR10, SRSF10, BMP5, ZDHHC2, IQCH, SCGB1C2///SCGB1C1, BANP, MSR1, SLC22A6, IFT27, SLC35E3, CHID1, ZNF350, RNASET2, SFTPC, CREB1, POLR2F, LOC101928879///ZNF250///COMMD5, LOC101927843, DSP, LOC102724884///LOC728613///PDCD6, CACNA1I, ACTR3BP2, RABL3, ADTRP, PITPNC1, CACNA1C, PITRM1, TAF12, CYB5D2, THUMPD3, ARL4D, PIEZO1, LINC01270, COMTD1, SLC15A4, GPR161, LPCAT2, DNAJB4, IGHM, SRSF3, ZFYVE28, FGFRL1, GJB5, FCGR3B, BTG1, ZNF17, TBC1D23, AGK, AADACP1, ABCB5, TRAF6, MKS1, SRMS, PAPOLA, ALKBH1, SPG20, SLC25A24, NCAPH2, NDUFA10, ZC2HC1A, ABI3BP, NCR3, TUBB4A, MIR210HG, TYROBP, PEPD, TRPT1, C1QB, LOC338620, KISS1R, CHRDL2, PLOD2, LOC728485, JMJD4, RGS16, LOC100505585///ARHGEF1, ETV2, ZNF451, KIAA2022, IGK///IGKC, TCL6, PDLIM2, RCOR3, USP22, TM9SF3, CCDC82, SLC2A5, ZNF837, EPS8L1, BCAT1, CYAT1///IGLV1-44///IGLC1, KLHL21, PLA2G4E-AS1, GGA2, CNNM4, ZNF595, ITGB3BP, SERGEF, UBE2J1, COMMD4, ZNF395, CNTNAP3B///CNTNAP3, ADGRE2, NR4A3, LAPTM5, LOC101927787, LINC01267, CEACAM1, KATNAL2, SMAD7, LOC254896///TNFRSF10C, FUBP1, PLCXD2, JAKMIP2, PCYOX1L, SMIM8, EPB41, GATA3, TMLHE, EPHA5-AS1, TOM1, GPR137, NUP160, EPPK1, FER1L4, NR4A1, AMFR, ALDH1A1, CEP128, STXBP5-AS1, CYBA, CPXM2, TRUB1, S100A12, MTF1, HEBP2, ZNF215, ZDHHC22, LOC100505716, IGLC1, PDP2, CEACAM21, SCAMP2, CASP10, TEX9, ADSS, CHEK1, HNMT, RPS6KB1, CCDC69, WDR97, COL1A1, SNORA65, ATXN2L, DLX5, LOC101060363///PPIA, MEIG1, ABCC6P1, ITGB2, SNRPE, LOC101928314, TTLL11, ZNF493, LIN9, NDUFA7, RHBDL3, C14orf105, CCDC117, GRIN2D, CXADR, IGF2BP1, IMMP1L, MRPS6, TMF1, KANK2, FIGNL1, TMEM208, CDK5RAP1, MARCO, RPS6KA2, DNAJC22, KIZ, CR2, FUCA1, ADAMTS19, TMED3, SNX6, FCGR1CP///FCGR1B///FCGR1A, FBXO16///ZNF395, CDC14A, TFDP3, FAM181A-AS1, FBXO9, CALCA, GPR156, RPTOR, SPRTN, PRRC2B, GLP1R, SERPINB9, CWF19L1, IL1R2, LOC441178, FOXC2, NOV, NENF, LOC101928401, EMILIN2, EBF3, SLC38A5, ZNF616, EGR1, RPS6KA5, CA4, FKBP4, C11orf94, CLEC18A///CLEC18C, APOA2, UBE2G2, C11orf80, TMEM222, NPSR1-AS1, TOR1A, SIRT2, ATP5C1, HRASLS2, SDHB, RHOBTB1, RBBP9, SEMA5A, HCFC2, NSUN6, ARPP19, ACAD9, MUC6, RAP1GDS1, HBZ, TAB2, ARID5B, HSD17B1, CASZ1, MAPT-AS1, ABCC5, RPS8, ZNF271P, RNF126, HAVCR2, FAM174B, GOLM1, EFNA4, MOB2, PSTK, HCG26, SLC6A13, GPNMB, PPFIBP1, ADGRA1, SCAF4, LINC00396, PBRM1, PARVG, FBXO10, PSMD6-AS2, EXOSC10, PARL, KCTD12, PNKD, CLU, HAX1, AMIGO3///GMPPB, DNAJB1, NPTXR, GART, PRKXP1, MIA2, ST3GAL3, LINC01120, PHLDB2, CD22, C1orf162, OFCC1, DENND1B, LOC101928505, ARHGEF28, PCDHB18P, HSPB1, NAA30, DSCR8, BCL6, APOC4-APOC2///APOC4///APOC2, PCGF6, NFYC, PAPOLG, HACD4, DYX1C1, TSPAN18, GRP, ANXA3, NKX2-3, CCDC169-SOHLH2///SOHLH2, C2orf54, GH1, CCDC116, SARS2, OOEP, ADAMTS8, LOC730268///ANAPC1, LOC100130872///SPON2, LAMA3, PCAT19, C6orf120, ZNF226, MGST1, PSME1, CCR1, LINC01553, GALNT16, RABIF, MRPL16, LOC100134317///LOC284412, GPATCH2, TMEM184A, TSPY26P, ZNF91, KAZALD1, LOC100130502, SERPINA1, MIR6789///PLEKHJ1, SNRPB2, ATP5D, GUCY2F, SLC35F6, OR5AK4P, ST3GAL1, SDF2L1, C21orf59, SLITRK2, RSBN1, IDUA, GPER1, LINC00936, TAF2, CKS2, ZNF582-AS1, ATG12, PRO2958, EXOC4, PRR5L, WDR11-AS1, SLC30A7, NEDD4L, NRSN2, C10orf10, KLHL5, NRG2, ELOVL2-AS1, MFAP4, MUC16, LOC647070, LIPG, ROPN1B, SLC12A3, TDRD5, SYNPO2, LINC00545, ORAI2, ANP32A-IT1, IQCF5, MTMR1, MEIOC, RGS18, PHOSPHO2-KLHL23///KLHL23, CTTN, PARD6B, LOC102724312, FILIP1L, ALDH1A3, C2CD2, FOXE1, MLX, HHEX, ENPP3, FXR1, LOC101926921///DAB2, FNIP2, UPK3A, ALMS1, PPP5D1, XXYLT1, RSPH1, NOP14-AS1, ALPK1, ACTG2, AP1S3, MIR924HG, LOC100996246, MIR6805///RPL28, COL22A1, KLHL31, MSLN, LOC102723927, CHRNA4, NPAS3, DOCK8, C10orf25, SSH2, DCAKD, LOC105375773, LOC101927604, OGDH, UBASH3B, VWA7, RBM47, CARD6, COL6A5, COX19, ZNF654, PTGS1, C7, ZNF138, LINC00167, LOC101928747///RBMX///SNORD61, SYNE3///LINC00341, MRPL44, RXRG, LOC100421494, NIPAL3, MFSD9, HCK, AHI1, FAM53B-AS1, CCDC169-SOHLH2///CCDC169, GAS2L1, DCAF1, CSF2RA, RGS19, CEP104, ZNF217, MCM4, BID, PLSCR4, ARRDC1, PTGDR, VPS37A, RERE, CDC20, LOC100130285, ARG1, FAM27B///FAM27C, AGR3, LOC101927705///P4HA2, CRYGB, LOC101930370, EHD3, SH3GL1P2, LOC100506526, ARVCF, LOC283745, FGB, LGALS13, FER, SERPINB13, EXOSC2, CNIH3, DCLRE1C, YIPF1, CDC42EP4, KIR3DL3, HFE, LOC101060553, CD14, LOC101927450, DGCR9, CHMP6, LHFPL3, HBE1, C11orf31, ABCB9, ANKRD44, NET1, TRIM52, SGK494, ACO1, FCGR1B, ANKRD37, BSND, CYP4Z2P, C17orf62, DSEL, EIF4E, PRCAT47, AMBP, C8orf33, AP3D1, SHISA6, MMAB, LRRC1, MIR100HG, CP, PRKAR2A, PAPD5, LOC101928161, LOC105371038, N6AMT1, LOC101929050, DESI2, NHLRC2, CD37, HMCN2, PTCD2, IER2, GPCPD1, LOC105376689, FCER1G, MKLN1, CFAP97, UBN2, ENTPD5, TBC1D7, CELF4, SCARA3, HMGA2, SPATA41, GIMAP5, POLR3D, TRMT61A, PAXIP1-AS1, DTX4, TINCR, COPS6, ATIC, AVPR1A, HILPDA, WDTC1, NLRP13, DOK3, LOC340107, MTOR, SAMM50, FGFR2, FAM3D, PEX12, SFT2D2, ZBTB20, PCDHA1///PCDHA2///PCDHA3///PCDHA4///PCDHA5///PCDHA6///PCDHA7///PCDHA8///PCDHA10///PCDHA11///PCDHA12///PCDHA13///PCDHAC1///PCDHAC2///PCDHA9, ASH2L, ZC3H12C, LOC286437, MAGEA4, FAM226B///FAM226A, ZNF45, CFLAR, LRRC17, ZBTB40, PATL1, PTGIR, MAP3K3, KIAA1644, MFSD3, CFAP126, ARID1B, R3HDM4, EPB41L5, ACAP1, LST1, MORC3, RIC1, TARBP1, ZNF391, MGC4859, MCTP2, PSORS1C3, HDHD2, OAS2, PAX8-AS1, CLPTM1, CSNK1A1, PTH2, ZNF136, FGD4, DSC1, EDN3, CBX4, LRRC31, BACE2, HNRNPA1, BCL2A1, ZC3HAV1, TTC9C, MYNN, FAM63B, MAP7D3, NPR2, HECTD1, ZBTB46, PSMD10, CPNE5, LINC00521, ZBTB10, ADAMTS6, MTDH, C14orf28, DNAJC21, CACHD1, DBF4B, MAP4K4, CSN1S2AP, GPRIN3, TNRC6C, EPS8L3, LOC157273, LINC00896, ZNF670-ZNF695///ZNF670///ZNF695, GRM3, RTN4, TSPAN2, OSMR, RALGAPA1, FAM192A, LOC105369507, MRE11A, LOC101060604///NPIPA5///SMG1P3///NPIPB5///SMG1P1///LOC613037///NPIPB4///SLC7A5P1///NPIPB3, ACP6, WEE1, DNAJC10, TRPM6, HCLS1, GAS8, AHSG, FPR1, PLEKHA6, MYL9, LYN, AGTR2, TRIM4, C1orf21, EP400, TIGIT, TBX3, PRKD3, PALLD, DCAF13, RAB5C, ZNF678, IL17RA, B4GALT3, AJAP1, ACP1, MTHFSD, PXT1, FBLN5, PTGDS, SSFA2, CNPY2, METTL8, IZUMO1, DIS3L2, MFI2-AS1, MSH5-SAPCD1///SAPCD1, DNAH5, MLF1, C1QA, LINC01089, ALOX12B, MARVELD1, PGRMC1, ENDOD1, TMEM132C, PHYH, DHX9, CHP1, VGLL2, COPS9, LINC00839, SORBS1, RDH11, DTNB, SPATA6L, REM2, WIZ, LINC01085, CUL7, FAM118A, CLIP1, IL13RA1, PADI4, WHAMMP2///WHAMMP3, ELK1, TOR1AIP1, LOC100507033, LINC00629, KLF14, TMEM156, STK31, AAK1, TSSK2///DGCR14, ZC3H7A, PTAFR, CEP57L1, COLGALT1, KCTD7///RABGEF1, RSPH3, ZNF354A, LOC339803, SUSD5, FZR1, MPZL3, LOC100996542, TP53RK, RNF103, GPAA1, ORAOV1, COL14A1, KCNG1, LOXL1, CDH4, KRT12, IRF8, LINC00382, KDM5B, LGALS8, TCF3, SGSH, PPIL2, WLS, BAG6, SP100, ESR1, LOC101928631///ZNF77, HMGB2, TMEM119, MIR6132///ST7-OT3///ST7, CASP1, LMLN, MRPS2, LOC101060521///POLR3E, IL1RL2, CXCL14, SLC47A1, FARSA, ZER1, ISLR, MIR7111///RPL10A, ADAM7, CAST, RNF130, ISM1, C2orf82, HMX1, SAP30, NAA50, DYNC2LI1, INS-IGF2///IGF2, KLHL34, HAGHL, PDK1, HEXA, GTF3C3, TRIAP1, LOC728743///ZNF775, VWCE, ZNF541, VGLL3, LINC01587, MRPS18A, SLC6A5, DNMT1, ANGPT2, LOC106146153, KLC1, NT5E, PPP1R16B, NEU3, LOC101927272, SYCE2, LOC101929709, LOC100129924///C1orf50, NFIA-AS2, PLA2G4C, ZMYND10, TAF9B, ATP5F1, XRCC4, ZNF407, OK/SW-CL.58, CLDN15, DAD1, SH3TC1, SNHG16///SNORD1C///SNORD1A, VEGFA, ANKRD9, RNF185-AS1, UNC119B, AKAP13, LPAR5, LOC102723886, LOC101929373///SLC9B1, PM20D2, UNC93B1, RRP12, GPX8, SMIM10L2B///SMIM10L2A, HMGB1, ADGRG3, CHD7, PODXL, PTP4A3, SCEL, PDCD2, PTBP3, MIR7-3HG, LOC285692, DEPDC1, INTS4, KITLG, ZNRF2P1, TMPRSS3, PI15, RFXAP, SCAF11, ZNHIT2, SCOC-AS1, XBP1, TFE3, CSPG4P5, REG4, ARHGEF12, LOC101929622, TRIO, ATP2B2, AKAP14, RAP1A, STMN1, MSANTD1, ACER2, ARF5, CTSC, GATA2, OR1D2, PITX2, FOXP1, LOC441204, MTMR11, JAK3, TRIM17, APBB1IP, LOC105372881, ZNF506, GRK4, SLC25A5-AS1, ZNF408, UBE2I, P2RX2, PCGF5, INSL3, THOC7, ZNF765, ERCC6, LOC101927933///LRRC8C, TRIM16, IL1RAP, ST20, AP1G2, ALOX5, ORC5, TMEM176B, TMEM44, LPAR2, PLN, DPM2, COA5, USPL1, DIEXF, GOLPH3L, ANAPC13, DSG2-AS1, KANSL1L, ZBTB32, ARIH2OS, LINC01350, BRINP2, DYNC2H1, TAPT1, SERPINB3, GDPGP1, IRAK2, C2orf83, CANT1, SLC10A7, ADPRHL1, FLJ41170, BBS9, PRR34-AS1, DNAH17, LYPD3, MAML2, SESN2, KCNH6, LOC101927157, SNAPC5, SNRNP48, CYB5RL, AIF1, RPP30, TRIM72, ERAP1, ERI2, CTXN1, MLXIP, TMEM161A, DPYSL5, NUDT22, STUM, AQR, PKD1P1, CIRBP, RNASEK-C17orf49///C17orf49, HDGFRP3, AADAC, ALKBH6, IQGAP3, SERPINB1, CST3, ZSCAN20, PYGO1, DIO3OS, APOM, ZNF169, KLF16, TRMO, LOC105274304, LOC102800447///LOC101930566, CSF2, ZNF396, WDR11, DAPP1, MS4A4A, TTLL10, THBS1, TRA2A, ST6GAL2, SLC2A11, GPC6, IDH1, SEPSECS, TAAR3, PTPA, RABEP2, ITGA4, TMEM221, CARD10, PCMTD1, ATP4A, MGARP, ZNF823, HYKK, MCM10, ADRB1, UQCRC1, C9orf84, RPUSD3, ASTE1, PTPRC, TACR3, KCNMA1, POU2F2, COL1A2, PPARA, FCAR, RHEBL1, SLC16A1-AS1, NDC1, LOXL4, SIRPD, LINC01007, DCN, MYH11, LINC00112, B3GALNT2, OSBPL2, TSEN15, ESYT2, PPP1R11, ARHGAP27, GTF2A2, PLAA, LOC101929450, FLCN, PEBP4, PCYT2, LINC00910, SOCS2, LOC100507855///AK4, HKDC1, WBP4, RPE, PARD6G, C14orf39, CCDC177, P2RY8, PGAM5, EED, LOC100506351, CDK9, SMAD4, SPATA5, SEC23B, ZNF347, RNFT2, TTTY2B///TTTY2, PAG1, RDH12, CASP8, IKZF4, LOC100129476, GPAT2, ZNF521, LOC101930006///FRMPD2B///FRMPD2, LINC00858, LINC01366, ACTA2, PIK3IP1, RNF139-AS1, LATS1, LOC102724275, CAMK2G, TPD52L2, GNL1, STK38L, KLHL2, WDR66, SPRY4-IT1, ATP4B, ARID4B///RBM34, ECM2, TRPM1, SLC25A51, YPEL2, ZSCAN30, SCAND1, CCDC13, SPATA13, BMT2, CNOT3, CTGF, COQ9, ATP2A3, ZNF587, DKFZP586I1420, TSR2, MAPKAPK5-AS1, ASIP, IPO7, KIAA1109, FAM50B, PTPN18, LOC285889, EXOSC3, KLB, SMURF2, SBSPON, CYB5D1, C16orf86, LINC00116, SHMT2, GPRC5A, CELF2, QRSL1, HS2ST1, E2F3, MALAT1, LINC01278, PTPRCAP, PIK3CG, SLC25A30, PIAS2, DNAH1, NTPCR, RAB29, OLAH, USP42, OR51E2, LOX, RNH1, NFATC3, NFATC2IP, MYH14, STOM, LINC00565, PIK3AP1, SASH3, EFCAB1, KBTBD3, SUPT7L, TGFB2, METTL21A, EPHA8, ATP5S, SP3, AKAP10, LOC153546, IL18, AP3B1, LYSMD4, AGAP6, SEC11C, PDXDC1, ACTL8, OSTM1, XKRY2///XKRY, NUP188, RCL1, ANKIB1, SEC14L3, METTL22, TXNL4B, SZRD1, MAPK14, MND1, EMILIN1, SLC39A14, ZBTB25, LOC101929774, SERPIND1, C18orf25, WASF2, NCF1, DALRD3, FABP3P2, DANCR, MACC1, CDC42EP5, GBP5, CAPNS1, LINC01255, ZNF750, LOC101928269///LOC100506403///RUNX1, FAM214A, FBXO22, NCBP1, FLT1, NT5C, C15orf57, PDCD6, TRPM4, CMTM1, HOXB-AS3, ANKRD11, CLUL1, LDAH, GSDMD, RS1, DENND5B, ZNF821, TIMM10, LOC729732, SWAP70, LZTS2, MIR3190///MIR3191///BBC3, LOC101927164, TFR2, TEP1, VMO1, CECR2, PTPN2, ZNF789, HAND2-AS1, ZNF625-ZNF20///ZNF625, HTR3A, HGF, CD6, C3, HIVEP3, KSR1, C9orf66, LINC00313, LOC153682, LOC400768, SPATA17, CFI, PTPN21, MMP24-AS1, SIGIRR, DHX30, WWP2, ASB1, CHORDC1, SYNCRIP, IGF1R, NAGLU, RASSF7, SLC45A2, SMARCA4, LOC102724532///SP2-AS1, NUP93, GIN1, PDZD11, ATP6V0E2-AS1, ZNF26, ING5, NPTX2, FAM166A, TGFB1, LIN7C, DDI1, QKI, FGFR1, KLRD1, LLPH, STXBP6, C4B_2///C4B///C4A, C1orf87, NDUFA6, LTC4S, CABP7, LINC00895, ADAMTS7, GNRH1, TP53TG1, PTCD3, LOC101927616, GZF1, SLF1, PSMA3-AS1, ACSL6, C21orf91-OT1, ICAM1, DIAPH3, GPM6A, GOLGA1, CENPN, ARHGAP18, LHX9, WDR13, IFT22, C3orf80, PPP4R3CP, PCDHB3, HRG, GALNT5, TRA2B, SLC26A7, RASSF1-AS1, PCDH18, MSTN, LOC101927044, PEX16, LOC102724250///NBPF26///NBPF10///NBPF8///NBPF9///NBPF15///NBPF11///NBPF12///NBPF1///NBPF14, SNX18, BORCS6, LOC253805, IRAK3, ZKSCAN3, SPRED1, PTN, PDE7B, EVI5, ZNF24, IRX5, CHMP1B, TMED5, NSL1, ZNF780B, VWA1, DOK1, HPSE, LY86, LAMTOR1, GUCA1B, JUND, LOC101929084, ALOXE3, LOC101927446///GTPBP10, LOC100506548///RPL37, SEBOX///VTN, SMOX, UFSP1, RNASEH2A, ZBED5, NDUFB10, CHFR, SETDB2, PSRC1, IGHV3-23///IGHV4-31///IGHM///IGHG3///IGHG1///IGHA2///IGHA1.
3. Genes identified by GSE12649
ST8SIA4, IGFBP6, LOC101930400///AKR1C2, NOX5, TAF2, TRIM24, FUT7, HSD11B1, ABCG2, BID, CTBP2, MBIP, MGMT, CP, NXPH4, DOPEY2, TRIB2, LST1, DIAPH2, RAD52, ATP6V0E1, TAPT1, NR4A3, AAAS, RSBN1, HNRNPDL, IL6ST, AKR1C1, DST, PHACTR2, COL6A2, DZIP1, PDZD2, TMEM97, HLA-C, SLC27A3, NR3C1, PLBD1, PRRG3, ATF6B, DHX15, ASXL1, ENTPD1-AS1, GNS, TUBB1, FAM189A2, TNFSF10, PPP6C, RPS6KA5, PAPOLG, FYB, GPR137, UBR5, PSME3, SLC39A8, NOL12, SMR3A///SMR3B, UBASH3A, A2M, HLA-F, SMARCA1, ATG5, FXYD3, LAMTOR5, PPFIA1, DR1, KRT6B, ZFP2, MBNL1, RBMS1, PALB2, MPZL1, TIMM8B, TGFBR2, GNG12, DDX27, UBE2A, DNAJB6, PIAS1, ADD3-AS1, GIP, SLC25A38, SCAF11, LRRC31, MKL2, ERAP1, SF3A1, PEG10, RAB9BP1, LBH, ZNF26, LGI2, CRHBP, SDF4, TRMT61A, MGEA5, CPSF6, CEP350, RBM12, CCL3L3///CCL3L1///CCL3, GAS2L1, SVIL, CA4, CAPN9, LOC145783///ZNF280D, CPVL, EIF2S2, RRP12, CRYBA2, UCP3, DCTD, FOXD4L1///FOXD4, RRN3, PLOD2, CRB1, ZNF771, CYTL1, MAGEA6, TCF7L2, ST6GALNAC2, SUCLG2, PMS2P1, MAP2K6, PLEKHA5, ZNF611, U2SURP, MT1X, PIH1D1, ECM1, FAM13A, OLFML1, NFATC2IP, APLP2, AIP, SLC29A1, C1orf27, AQP4, GAB1, CHKB-CPT1B///CPT1B, VAMP4, FGF2, VPS13A, TMEM147, ACACB, DCX, KPNA3, ASPN, PTGS1, HAX1, ZBTB22, SSTR4, SURF1, OVCA2///DPH1, PDX1, SFPQ, CDKN2C, NKG7, TAF13, ISLR, CYP2W1, HLA-B, CSNK1A1, NT5DC2, THNSL2, GLI2, MICAL1, DNAJB5, CCL27, TTC17, SNTG1, ALDH1L1, HMGB2, INS-IGF2///IGF2, SNORD14D///SNORD14C///HSPA8, EPN3, RLF, TNFSF12-TNFSF13///TNFSF13, ETV5, SNAPC3, PLN, CDC25B, NDUFB2, SPHAR///RAB4A, MYO7A, ADAM22, ZYX, SRSF11, SLC7A8, TACC1, TRPM8, NOLC1, DNAJB12, CBR4, PRKCQ, PPT2-EGFL8///EGFL8///PPT2, CCL18, RBBP5, ROCK2, JARID2, S100A9, IRAK1, CDK19, NMB, EIF2B2, NR2C1, RAPGEF1, COL3A1, TFDP1, JTB, SENP6, RASL10A, SORBS1, IMPA1, CASP5, GREB1, CENPC, YME1L1, CFLAR, HMCES, DHX16, MTCH2, PSMB10, ITPR2, TIMM10B, WDR76, DUS2, ATG12, FERMT2, MST1R, DNAJC22, EDRF1, SLC20A2, POU2F2, DBT, MRPS7, VEGFC, SLC6A11, FOLR2, HIST1H4H, IMPACT, GATC///SRSF9, GSR, ERLIN1, IGHV4-31///IGHM///IGHG3///IGHG2///IGHG1///IGHD///IGHA2///IGHA1///IGH, RARB, MLYCD, SP100, RNF43, MYBPC1, ETNPPL, UGGT2, FLI1, OSBP2, KCNE2, LY6E, NIPAL3, CTDSPL, ROBO3, IGF1, FLII, RYR3, MED13L, RAMP3, LOC101060835///LOC100996809///HLA-DRB5///HLA-DRB4///HLA-DRB1///HLA-DQB1, UPF3A, RRM2, PPP6R2, SLC2A4RG, SDC4, CKS1B, CD52, MAP4K1, AMBRA1, GINS1, LANCL2, USP24, LOC100996809///HLA-DRB4///HLA-DRB1, ABI2, ZC3H7B, CHST1, FBXO38, PANX1, MAOB, CAMSAP1, MYBL2, CDV3, PINK1, EFNA4, CST6, DNMT3L, PTBP3, UXT, FRY, ZNF148, EIF3I, SMR3A, ELOVL2, PCLO, NTSR1, GJA9-MYCBP///MYCBP, SYT2, ANP32E, CELP, TBC1D29, SIK1, MIR6789///PLEKHJ1, HEPH, PHF20, PMPCA, BMP8A, CNOT4, SLC25A5, KATNBL1, IL23A///TRBV19, APCS, FRZB, FBXO42, LOC100507855///AK4, LAT2, MTHFD1, LOC101926921///DAB2, ZNF266, BMP1, SDF2, NAALADL1, MID1, LRIG1, GHRHR, TIMELESS, PIGA, NR4A2, PRIM2B///PRIM2, MICALL2, C12orf49, MINOS1-NBL1///NBL1, SLCO2B1, EBAG9, CRH, TARP, VRK3, CASP1, MYF6, PSAP, IPCEF1, BUD31, POT1, HCCS, TLE3, MCL1, SLC1A4, SOX9, SLC8B1, GRB10, HLA-G, TRIM38, DUSP6, RAB15, TUBB2B, DUSP1, ARG2, PLGLB1///PLGLB2, BHLHE40, MYH2, HBQ1, RAP2B, MRS2, MAGI2, FBN2, TERF1, FGL2, CENPJ, SLC14A1, BCKDK, ERLIN2, ID4, UFL1, ZNF267, RABL2A///RABL2B, HIF1A, SRSF7, BAG1, RAPSN, TRBC1, FGF5, HERPUD1, DNAJB14, SCGB1A1, APBB2, KIF13A, FBXO9, HIST1H2BJ///HIST1H2BG, EDNRB, RYR2, FKBP4, COPZ2, KCNJ8, ZBTB7A, TUSC2, SPTSSA, OSBPL3, ASGR2, IDH3B, AQP1, MIOS, PLPP3, LOC101930405, RREB1, RGL3///EPOR, PENK, SERTAD2, S100A8, SETSIP///SET, SEC16A, KPNA6, ACP1, PROC, THAP9-AS1, ZNF304, PBX2, RNF6, TAS2R10, DYX1C1-CCPG1///CCPG1, TMEM131, ARHGAP33, GK, SNRPF, RNF216, HSPB2, NOTCH2, RYBP, RCOR1, GSTA3, ZCCHC8, CENPI, NPY, C21orf2, ACOT1///ACOT2, TESC, DHX34, SRSF3, BCAT1, SLC4A4, NEUROD1, C19orf66, LOC102724229///RASA4B///RASA4, SIAH1, GPR88, GTF2H1, PAM16, KLHL35, LOC101060835///LOC100996809///HLA-DRB4///HLA-DRB3///HLA-DRB1///HLA-DQB1, ARMC9, PCF11, DDX24, RRP8, NUP54, CYLD, SIPA1L1, IFI27, MYO1E, IFNA16, NDP, SVEP1, RGS5, EXOSC2, METTL7A, LIN37, RAB20, NCOA1, INHBA, RRP1B, RAB5C, SHQ1, ABCC4, N4BP2L2, PTGIS, PEX6, CLIC5, NPAS3, TMEM100, ABCE1, NFKB2, COQ7, PEX7, CTC-338M12.4, PAF1, GCC2, BATF3, EML2, SPON1, TBC1D19, CCS, DPYSL3, CALM3///CALM2///CALM1, SH3BP5, MYRIP, DBF4, AFDN, HTR2B, RHOBTB3, PUS7, TMEM231, ABI1, HLA-DPA1, MYDGF, MT1HL1, HOXD4, C9orf16, ANK1, ARFGAP3, KRT10, ZCCHC14, PCDH7, MRPL48, ZNF443, PIGV, SRI, NOL9, MTSS1, UFM1, SREBF2, ATG16L1, RPS6KA3, TMEM109, TLN1, APEH, FAM76A, EZH1, MYOC, MAGEB3, SCAMP1, GABRA4, DUSP4, GGA3, MMP14, NPEPPS, POLQ, LOC101927792, MIR15A///DLEU2L///DLEU2, ALMS1, TAF1B, PELI1, GJA1, PMS2P9///PMS2P5, TRANK1, AP4E1, RAB40C, RNF115, WTAP, EMX2, FAM206A, EFR3B, GPX4, CEP57, SST, DND1, AATF, FAT4, CDYL, GALNS, NBN, MEIS2, FZD7, ANGEL2, CCKAR, ANK2, BOLA2B///BOLA2, CRK, ANGPTL2, TSC22D3, MFSD1, F3, OAZ2, LGALS1, TUBGCP2, CINP, UGT2A2///UGT2A1, RNASEH2B, LPAR3, GID8, ZNF729///ZNF43, TOX3, PEX26, STX6, LOC100293211, ANAPC5, SPRY4, SERPINA2, TCF3, EDN3, PPARA, ZFR, UBAC1, ASAP2, GUK1, PLG///LPA, USP7, SMC5, NTRK2, VPS54, SDC2, SATB2, AFF1, TNIK, IGF2BP2, PDE12, BRCC3, CDK5R1, WAPL, ERGIC3, SFTPB, GJB5, CDKN2B, CHAC1, CDK1, NNAT, STX4, ZNF667, SNCA, PACSIN3, ATP2B4, RMI1, ADH5, VPS37C, APOC1, LOC100420758, PAIP1, RNF128, NCOA2, ART3, NLE1, PPIG, PRPF6, MTO1, TNRC6B, BTBD7, FAM66D, GCK, AVL9, CCDC181, ZNF419, EPB41L4A-AS2, TMEM47, HSDL2, MDM2, CPD, AK2, SART3, KDM4B, HFE, MEFV, SMARCA2, APRT, EGFL7, RCAN1, PDPN, ANKRD36B, B2M, PLAU, ORAI2, PPP1R11, EIF4E2, ATP7A, ALG13, FKTN, THUMPD1, FABP7, WWC2///CLDN22, GLUL, NEDD9, IL17RB, AKAP8, HMGN5, HHEX, ANK3, HNRNPA3///HNRNPA3P1, HSBP1, SESN1, CHERP, ALDH6A1, ZNF292, GNA11, LEF1, RPP25, KDM5A, SEC24B, PTCH1, WBP4, WLS, MCM6, PSMA5, ENOX2, FAM169A, DKK3, ZC3H14, BAK1, PCDHB3, ADGRL3, CHKB, GHR, MCF2, IRF4, HLA-DRA, ACO1, ARC, LSAMP, UCP2, IL33, MMP15, TTK, GUCA2B, ZBTB20, MXD1, RABEPK, RPS9, PRKAA1, ITGB2, HPN, PLK3, LRRC42, TBL1X, ABCF1, NOS1AP, CSHL1, AMIGO3///GMPPB, ZCCHC2, HLCS, HNRNPH3, NR4A1, LOC101928635///ALDH1A2, METAP2, C18orf25, ZDHHC4, ACSS3, ALDOB, NFATC1, FCGR2C, ARHGEF2, NXF1, FUT6, MGAT2, RRAS2, GPM6A, IGHM///IGHG1///IGHD, AMMECR1, EDA, USH1C, FAM173A, PITPNM1, SPHK2, UBE2H, RGS20, ARHGAP35, TMEM259, MPRIP, HTN1, C22orf24, SOCS5, SNCG, MINA, CARTPT, DNAI2, UBE2E1, GLE1, DGKG, GRPEL1, HOXB9, FKBP2, NTRK1, SERP1, NR0B2, TM2D1, PDE3B, LEFTY1///LEFTY2, ASMT, MUS81, LPP, FBXO40, LSM1, FBXO11, SOSTDC1, LOC101927051, CA12, RAB3GAP2///AURKAPS1, ZBTB3, HNRNPU, PTPRZ1, UBQLN2, SYDE1, BTNL8, GNAL, NR2F2, ABCC9, NOTCH2NL, ASAH1, RASL11B, PLIN1, ALPL, ALAD, VPS4B, BET1, FN1, PRKAR2B, KCNJ5, NDUFB11, CTAGE5, CAV2, PALLD, KHSRP, PUS1, KCNN3, EIF1, MYO1C, SNX4, ZNF16, PHF3, PPARGC1A, NUDCD3, SLK, ACVR1B, GLRX3, WDR43, LAPTM4B, VAMP1, PPP2R2A, KCNAB1, LAIR1, ZBTB43, CES3, EPHX1, TFRC, SLC16A1, POLR3C, PPP4R3B, PLSCR4, EHD2, RGPD6///RGPD8///RGPD3///RGPD4///RGPD5, SNF8, HCG4B, LOC101929479///LOC727751///LOC642423///GOLGA2P7///GOLGA2P10, SCRT1, PRUNE2, LOC102724200///TRAPPC10, TATDN2, SEMA3G, UHRF1BP1L, WNT5A, SDHC, FOXJ3, NPY1R, INS, IGLC1, THEMIS2, GRSF1, FOXG1, MAGOHB///MAGOH, SFI1, PLEKHF2, MLH3, PPM1F, UBE2Z, TMC6, CD86, HNRNPD, CPS1-IT1, FGF18, TPD52, SLC1A2, NR1I3, FCF1, GAD1, CALU, CHST5, SLC25A30, SPAG1, ARHGEF12, TTN, OTUD4, FANCF, OCLN, NCOA3, CUL5, CPNE3, GPR182, SLC22A13, MEX3C, SKP2, DCAF11, TMPRSS6, TGS1, USP21, CSGALNACT1, DNAJC8, OPHN1, TBX3, ADGRA3, ZSCAN32, LANCL1, ZNF562, CDH6, RABGAP1, TEAD3, TP53BP2, GYG1, PANK3, THAP9, SOS1, THBD, CYP2C18, NUDT11, CREBZF, AVEN, ZNF3, ADORA2B, CXCR4, TBL2, ZNF189, ZNF696, STS, DRC3, TRPV2, ZBP1, CSNK1D, RPAP1, PXMP2, PIKFYVE, MT2A, HIST1H2AH///HIST1H2AG///HIST1H2AM///HIST1H2AL///HIST1H2AK///HIST1H2AI, MKRN2, MPO, MIR6125///USP15, KCNK12, HERC1, AHCYL1, BLOC1S1, SIGMAR1, URB1, CDK17, NCR1, KRBOX4, PRPF40A, TREM2, SMCHD1, FBXO2, SLC24A1, LOC101929500///CRIM1, PIR, SNX1, RXRB, FOXC1, DCN, STON1, TMEM63A, RBM23, TSPAN6, HMG20A, PCNP, ST8SIA5, HEMK1, KNG1, SNX3, DET1, TSC22D2, PDLIM4, NOX3, MBNL3, LSM14A, TPSAB1, LARP4, SMPD2, ALOX12, ATP8A1, SMG7-AS1, SNRNP25, DSC2, SLC39A7, GSPT1, CACNA1C, ATMIN, SPATA2, BTD, PAFAH1B1, ARPC1B, PRKY///PRKX, STC1, UBXN7, ATF4, MIR6756///MCAM, PSCA, SRPK1, CBS, H2AFV, SENP3, TM4SF1, ITIH5, TYROBP, PAK5, TRIP13, ZMPSTE24, SND1, SNORD19B///GNL3, HLA-DRB1, SGSM2, PPP1R12A, B3GALT4, SYNPO2L, ST6GAL1, ZMYM2, CLCN7, GIPR, SH3D21, GNAQ, HLA-DPB1, RAB5A, FMOD, PRKCG, IL11, PHLDA2, SAMM50, ELF2, USP34, HIST1H1A, SPTBN1, RPL17-C18orf32///SNORD58C///SNORD58A///SNORD58B///RPL17, L3MBTL1, UPK1B, MECR, CAMK1D, ECE1, RCL1, EIF3K, RAB2A, MET, ADRA1A, TENM3, DICER1, KLK2, SNU13///ANXA2, PRKCDBP, PPP1R2, CRYBB3, LAMB4, AGT, TMEM11, CORT, NDUFS7, B3GNT2, SF3B5, DIS3, TAT, CTSS, DNAJB9, DMD, LMO1, EPHB2, SF1, TRIM33, STAG2, DIAPH3, SHANK2, NUDT13, CC2D1A, CETN3, COA1, MED8, RBM10, CD22, EMC2, MPP3, DTNA, TRAPPC2, IDS, ANKRD6, CD36, ITSN2, OVOL2, FTH1, BCL11B, SLC35C1, MIR4680///PDCD4, GPC5, EGR4, MICALL1, RGS14, SNORD18C///SNORD18B///SNORD18A///SNORD16///RPL4, WDR78, PAPOLA, MSH6, LOC101929540///LOC101928670///LOC101928344///LOC100996442///LOC100288069///LOC100134822, N4BP2L1, SCP2, WASF1, ASMTL-AS1, VGLL3, RAD51, CASC1, FMNL1, TMEM223, GABRB1, RPS10-NUDT3///NUDT3, POLR1B, LOC101930363///LOC101928349///LOC100507387///FAM153C///FAM153A///FAM153B, ZNF83, SSR1, INPPL1, MRPS18B, CDC42EP3, KAT2B, MAGEB2, SRP9, DPP3, NR1H4, BBOX1, MUL1, ST3GAL6, MIS12, PTGER1, TTC28, TTC30A, SPAG9, RAPGEF3, GCN1, PLIN2, MTF2, INPP5K, KLHL21, THRA, NUP155, GYG2, CFDP1, KIF1C, SERHL2, WDR48, CHN2, FPR1, ZNF747, CASKIN2, NUP107, HTR7, REV1, PRPF4B, NEUROD4, COL8A2, PRKAR1A, TAOK2, HPD, GUCA1A, S1PR1, CD2, PRICKLE3, FAM30A, USE1, APOBEC3G, RORA, PKN2, XK, WISP3, KLHL2, GDF10, OPN1SW, ANP32D, EIF4H, MAOA, MFGE8, PNN, ARHGAP19, FANCA, ORC4, CCL21, CRLF3, ATP2C1, PHLPP2, PDZRN4, BNIP1, EXTL3, CLTA, AP5M1, BTRC, NCBP2, KSR1, TIMP3, ILF3, CSN3, NDST1, RNASEH1, OXA1L, HAUS7///TREX2, HSP90AA1, POM121C, FBXO21, IST1, PDE4B, GJA4, B4GALT1, NDUFC2-KCTD14///KCTD14, ARFIP1, MYO9B, IPO7, ANP32A, MRPS10, FUZ, EMC6, ANOS1, ELL3, PRSS3P2, SEL1L, C1orf112, GJA8, THTPA, CSRNP3, ZNF140, ITGA4, MAP3K11, CNPPD1, USP12, SPAG7, PDCD6, ARPC2, PTCH2, PPP5C, LOX, USH2A, PI4K2A, CNOT8, PKMYT1, ARHGAP5, VDR, HSF2, MIR6516///SCARNA16///SNHG20, RPRM, NNT, ZNF702P, ZBTB44, ADAM10, CD164, SSNA1, HSD17B6, HIPK1, CPT2, BMPR1A, UPF3B, KLF6, BRWD1, GON4L, DCAF16, TMCO3, EPAS1, PON2, CLCN4, LIMK2, ALDH7A1, POFUT1, LRRD1///CYP51A1, FBXL5, KLF12, LOC101927562///DUSP8, MPHOSPH6, GPAA1, ACTR1B, FGFR4, DOPEY1, PDSS2, ZNF516, ATF6, MAPK13, ACACA, PSME1, RASSF7, SLC52A1, ACTR6, MLF1, DRICH1, SYNE1, STEAP1B, CLN5, TUG1, GMCL1, TBC1D30, RBCK1, DNAJA1, TLK1, PLCB1, DTNB, OPA1, SLC1A3, PLAA, LBR, TWF1, SLITRK5, IGHV4-31///IGHM///IGHG3///IGHG1///IGHD///IGHA2///IGHA1///IGH, ANXA7, CCL24, MTMR10, SLC6A7, ZNF160, TSPYL1, IDI1, MBD4, RPS6KA2, TAL1, TAF10, FIS1, LOC101929726, SP3.
4. Genes identified by GWAS
HSPE1-MOB4, RLTPR, YJEFN3, NDST3, ADGRV1, AKAP10, AKAP6, AP3B2, BNIP3L, C6orf118, CDH11, CDIP1, CENPE, CISD2, CORO7, CPEB1, DNAJA3, EMB, EMX1, EYS, FLRT2, FYN, GPD1L, HACE1, HYAL3, JMJD1C, KCNG2, LIN28B, MAGI2, MSI2, MTMR7, NMRAL1, NOS1, NPAS3, NRBF2, PHF3, POLG, PPP1R3A, PPP2R2A, PTP4A1, RASSF1, RLBP1, SLC9B1, SPECC1, VPS37A, YWHAE, ZDHHC2, ABCB9, ACD, ACTR5, ADAMTSL3, ADRBK2, AGPHD1, AKT3, ALDOA, AMBRA1, ANKRD44, ANKRD63, ANP32E, APH1A, APOPT1, ARHGAP1, ARL3, ARL6IP4, AS3MT, ASPHD1, ATG13, ATP2A2, ATPAF2, ATXN7, BAG5, BCL11B, BCL9, BOLL, BRP44, BTBD18, C10orf32, C11orf31, C11orf87, C12orf42, C12orf65, C16orf86, C16orf92, C1orf51, C1orf54, C2orf47, C2orf69, C2orf82, C4orf27, CA14, CA8, CACNA1C, CACNA1I, CACNB2, CCDC39, CCDC68, CD14, CD46, CDC25C, CDK2AP1, CENPM, CENPT, CHADL, CHRM4, CHRNA3, CHRNA5, CHRNB4, CILP2, CKAP5, CKB, CLCN3, CLP1, CLU, CNKSR2, CNNM2, CNOT1, CNTN4, COQ10B, CR1L, CREB3L1, CSMD1, CTNNA1, CTNND1, CTRL, CUL3, CYP17A1, CYP26B1, CYP2D6, DDX28, DFNA5, DGKI, DGKZ, DNAJC19, DND1, DOC2A, DPEP2, DPEP3, DPP4, DPYD, DRD2, DRG2, DUS2L, EDC4, EFHD1, EGR1, ENKD1, EP300, EPC2, EPHX2, ERCC4, ESAM, ESRP2, ETF1, F2, FAM109B, FAM53C, FAM57B, FAM5B, FANCL, FES, FONG, FURIN, FUT9, FXR1, GALNT10, GATAD2A, GDPD3, GFOD2, GFRA3, GID4, GIGYF2, GLT8D1, GNL3, GOLGA6L4, GPM6A, GPX5, GPX6, GRAMD1B, GRIA1, GRIN2A, GRM3, HAPLN4, HARBI1, HARS2, HARS, HCN1, HIRIP3, HIST1H2BJ, HIST1H2BL, HSPA9, HSPD1, HSPE1, IFT74, IGSF9B, IK, IMMP2L, INA, INO80E, IREB2, ITIH1, ITIH3, ITIH4, KCNB1, KCNJ13, KCNV1, KCTD13, KDM3B, KDM4A, KLC1, L3MBTL2, LCAT, LRP1, LRRC48, LRRIQ3, LSM1, LUZP2, MAD1L1, MAN2A1, MAN2A2, MAPK3, MARS2, MAU2, MDK, MED19, MEF2C, MKL1, MLL5, MMP16, MPHOSPH9, MPP6, MSANTD2, MSL2, MUSTN1, MYO15A, MYO18B, MYO1A, NAB2, NAGA, NCAN, NCK1, NDUFA13, NDUFA2, NDUFA4L2, NDUFA6, NEK1, NEK4, NFATC3, NGEF, NISCH, NKAPL, NLGN4X, NOSIP, NOTCH4, NRGN, NRN1L, NT5C2, NT5DC2, NUTF2, NXPH4, OGFOD2, OSBPL3, OTUD7B.
Supplementary Table 1.
Differentially expressed mRNAs in peripheral blood Leukocyte from Schizophrenia patients versus healthy controls by RNA-seq.
| ID | Control_fpkm | SZ_fpkm | log2(FC) | P value | Symbol |
|---|---|---|---|---|---|
| ENST00000010404 | 0.0006 | 0.067 | 6.803055 | 0.000242 | MGST1 |
| ENST00000064780 | 0.343 | 0.9742 | 1.506009 | 5.61E-05 | RELT |
| ENST00000164227 | 0.001 | 0.081 | 6.33985 | 3.93E-05 | BCL3 |
| ENST00000199764 | 1.007 | 2.7254 | 1.436404 | 0.00132 | CEACAM6 |
| ENST00000204726 | 0.001 | 0.0072 | 2.847997 | 0.000598 | GOLGA3 |
| ENST00000216127 | 0.0028 | 0.027 | 3.269461 | 0.000102 | RASD2 |
| ENST00000218432 | 0.1038 | 0.2788 | 1.425424 | 0.000598 | PIN4 |
| ENST00000221327 | 0.0432 | 0.3458 | 3.000835 | 1.81E-05 | ZNF180 |
| ENST00000224862 | 0.0234 | 0.001 | −4.54844 | 0.000707 | FBXL15 |
| ENST00000225275 | 1.8012 | 3.7582 | 1.061083 | 4.22E-05 | MPO |
| ENST00000230173 | 0.001 | 0.0062 | 2.632268 | 0.00066 | ADGRG6 |
| ENST00000230990 | 2.3934 | 5.1126 | 1.094996 | 0.000229 | HBEGF |
| ENST00000231173 | 0.0012 | 0.009 | 2.906891 | 0.000221 | PCDHB15 |
| ENST00000231751 | 0.2456 | 0.623 | 1.342922 | 0.001308 | LTF |
| ENST00000232424 | 0.1168 | 0.4342 | 1.894319 | 2.58E-08 | HES1 |
| ENST00000233714 | 0.001 | 0.0276 | 4.786596 | 0.001496 | LANCL1 |
| ENST00000233997 | 1.5778 | 3.3686 | 1.094235 | 0.000295 | AZU1 |
| ENST00000236826 | 1.054 | 3.4208 | 1.698459 | 3.56E-07 | MMP8 |
| ENST00000237264 | 0.001 | 0.0462 | 5.529821 | 2.69E-07 | TBPL1 |
| ENST00000238508 | 0.1864 | 0.5244 | 1.492266 | 0.000341 | SERPINB10 |
| ENST00000239316 | 0.0026 | 0.0172 | 2.725825 | 0.000322 | INSL4 |
| ENST00000239938 | 35.003 | 138.1178 | 1.980349 | 1.06E-08 | EGR1 |
| ENST00000241356 | 0.8688 | 1.8864 | 1.11854 | 9.38E-06 | ADORA3 |
| ENST00000242208 | 0.082 | 0.3154 | 1.943487 | 2.64E-10 | INHBA |
| ENST00000242480 | 0.8694 | 2.736 | 1.653976 | 7.11E-08 | EGR2 |
| ENST00000242737 | 2.8472 | 1.0502 | −1.43888 | 0.000686 | ITPR2 |
| ENST00000244050 | 0.3902 | 0.9026 | 1.209873 | 0.000287 | SNAI1 |
| ENST00000244336 | 2.7612 | 7.5754 | 1.456027 | 2.50E-06 | CEACAM8 |
| ENST00000244364 | 0.0102 | 0.001 | −3.3505 | 2.04E-06 | DST |
| ENST00000252675 | 0.0456 | 0.1404 | 1.622437 | 3.31E-05 | FUT5 |
| ENST00000253408 | 0.0088 | 0.0636 | 2.853451 | 0.000537 | GFAP |
| ENST00000255427 | 0.0126 | 0.0916 | 2.861924 | 0.000663 | CHIT1 |
| ENST00000257264 | 2.2706 | 5.1756 | 1.188653 | 1.84E-10 | TCN1 |
| ENST00000258400 | 0.001 | 0.0108 | 3.432959 | 0.000351 | HTR2B |
| ENST00000258494 | 0.001 | 0.0076 | 2.925999 | 7.91E-05 | ALDH1L2 |
| ENST00000258963 | 3.2004 | 6.5878 | 1.041545 | 0.000937 | VEZF1 |
| ENST00000259206 | 0.0306 | 0.001 | −4.93546 | 0.000568 | IL1RN |
| ENST00000259951 | 0.0736 | 0.0034 | −4.4361 | 0.000871 | HLA-F |
| ENST00000261944 | 0.001 | 0.0094 | 3.232661 | 0.000368 | CDHR2 |
| ENST00000262304 | 0.013 | 0.001 | −3.70044 | 7.49E-06 | PKD1 |
| ENST00000262809 | 0.7448 | 1.837 | 1.302427 | 0.000152 | ELL |
| ENST00000262865 | 3.837 | 7.9572 | 1.052282 | 5.32E-05 | BPI |
| ENST00000263045 | 1.6118 | 4.4596 | 1.468242 | 3.68E-09 | CRISP3 |
| ENST00000263341 | 16.5414 | 43.6896 | 1.401209 | 2.67E-12 | IL1B |
| ENST00000263545 | 0.001 | 0.0358 | 5.161888 | 8.82E-05 | LUC7L2 |
| ENST00000263621 | 1.157 | 2.7144 | 1.230244 | 5.73E-05 | ELANE |
| ENST00000263707 | 0.067 | 0.002 | −5.06609 | 4.20E-05 | TFCP2L1 |
| ENST00000264156 | 0.0116 | 0.3634 | 4.969362 | 5.28E-07 | MCM6 |
| ENST00000264498 | 0.001 | 0.0062 | 2.632268 | 0.000744 | FGF2 |
| ENST00000264956 | 0.0154 | 0.001 | −3.94486 | 4.17E-05 | EVC |
| ENST00000265881 | 0.1112 | 0.001 | −6.79701 | 3.41E-05 | REXO2 |
| ENST00000265990 | 0.096 | 0.0124 | −2.95269 | 1.45E-09 | BTAF1 |
| ENST00000273063 | 0.0136 | 0.001 | −3.76553 | 7.83E-05 | SLC4A3 |
| ENST00000273077 | 0.001 | 0.0144 | 3.847997 | 0.001435 | PNKD |
| ENST00000273347 | 0.1102 | 0.2382 | 1.112049 | 0.000991 | NXPE3 |
| ENST00000274710 | 0.0096 | 0.001 | −3.26303 | 0.000112 | PSD2 |
| ENST00000277480 | 6.1284 | 15.075 | 1.298576 | 0.000139 | LCN2 |
| ENST00000278175 | 4.0576 | 9.7064 | 1.25831 | 9.36E-07 | ADM |
| ENST00000278590 | 0.005 | 0.001 | −2.32193 | 0.001398 | ZC3H12C |
| ENST00000278756 | 4.8168 | 10.7066 | 1.152353 | 0.001103 | APLP2 |
| ENST00000278865 | 2.041 | 4.6692 | 1.193899 | 1.80E-07 | MS4A3 |
| ENST00000282120 | 0.0408 | 0.001 | −5.3505 | 3.96E-06 | TGOLN2 |
| ENST00000282282 | 0.1732 | 0.3508 | 1.01821 | 5.87E-06 | ZNF547 |
| ENST00000283426 | 0.0212 | 0.0044 | −2.26849 | 0.001026 | PLEKHG4B |
| ENST00000284551 | 0.9296 | 2.254 | 1.277806 | 1.59E-06 | TRIM11 |
| ENST00000286627 | 0.0096 | 0.001 | −3.26303 | 7.95E-05 | KCNMA1 |
| ENST00000292169 | 0.3618 | 0.0032 | −6.82098 | 6.20E-05 | S100A1 |
| ENST00000293662 | 0.331 | 0.8206 | 1.309848 | 4.16E-06 | GRASP |
| ENST00000293677 | 0.001 | 0.0918 | 6.520422 | 3.14E-06 | RAVER1 |
| ENST00000294702 | 0.0106 | 0.001 | −3.40599 | 0.001431 | GFI1 |
| ENST00000295992 | 0.0286 | 0.1144 | 2 | 0.000404 | PCOLCE2 |
| ENST00000296435 | 8.3026 | 17.1748 | 1.048658 | 5.01E-06 | CAMP |
| ENST00000297239 | 0.0388 | 0.001 | −5.27798 | 0.000112 | SYTL3 |
| ENST00000297435 | 2.3918 | 6.9666 | 1.542358 | 5.58E-06 | DEFA4 |
| ENST00000299502 | 0.3266 | 1.0236 | 1.648055 | 1.48E-06 | SERPINB2 |
| ENST00000299969 | 0.0292 | 0.008 | −1.8679 | 0.000283 | SERINC4 |
| ENST00000300027 | 0.001 | 0.0152 | 3.925999 | 3.53E-05 | FANCI |
| ENST00000301624 | 0.0006 | 0.119 | 7.631783 | 1.71E-06 | TNRC6C |
| ENST00000301698 | 0.019 | 0.0024 | −2.98489 | 0.000768 | PRR25 |
| ENST00000302035 | 0.0116 | 0.1026 | 3.144834 | 0.001254 | SLAMF1 |
| ENST00000302291 | 0.001 | 0.01 | 3.321928 | 0.000215 | LUZP1 |
| ENST00000302326 | 0.0614 | 0.1664 | 1.438345 | 0.000946 | MN1 |
| ENST00000302328 | 0.0024 | 0.0244 | 3.345775 | 0.000313 | SCN11A |
| ENST00000302779 | 0.0414 | 0.001 | −5.37156 | 0.000216 | PXK |
| ENST00000303391 | 0.001 | 0.597 | 9.221587 | 1.02E-13 | MECP2 |
| ENST00000303562 | 122.7578 | 289.7258 | 1.238873 | 1.13E-07 | FOS |
| ENST00000304639 | 1.7764 | 4.1508 | 1.224433 | 0.000958 | RNASE3 |
| ENST00000306061 | 1.0988 | 6.8182 | 2.633462 | 4.06E-17 | MT1E |
| ENST00000306090 | 0.0032 | 0.2008 | 5.971544 | 0.000344 | GNAS |
| ENST00000306151 | 0.0008 | 0.0346 | 5.434628 | 4.01E-05 | MUC17 |
| ENST00000306585 | 0.001 | 0.016 | 4 | 0.001358 | CHTF8 |
| ENST00000307407 | 260.8124 | 576.8646 | 1.14522 | 1.53E-06 | CXCL8 |
| ENST00000307885 | 0.001 | 0.0096 | 3.263034 | 0.000155 | ADCY6 |
| ENST00000309539 | 0.2124 | 0.8638 | 2.023914 | 9.00E-06 | OLR1 |
| ENST00000309902 | 0.0422 | 0.001 | −5.39917 | 8.46E-06 | ZNF407 |
| ENST00000312943 | 0.001 | 0.0882 | 6.462707 | 3.62E-06 | DOK3 |
| ENST00000317216 | 6.415 | 22.1836 | 1.789972 | 2.73E-09 | EGR3 |
| ENST00000317276 | 3.6124 | 8.6866 | 1.265834 | 2.47E-05 | PER1 |
| ENST00000317721 | 0.0048 | 0.001 | −2.26303 | 0.000439 | NCKAP5 |
| ENST00000319397 | 18.132 | 8.0092 | −1.17881 | 5.15E-09 | ETS1 |
| ENST00000322748 | 0.0032 | 0.0552 | 4.108524 | 0.000709 | ZNF18 |
| ENST00000323938 | 0.011 | 0.1666 | 3.920813 | 0.000919 | LGALS8 |
| ENST00000325094 | 0.0258 | 0.001 | −4.6893 | 1.80E-05 | TMEM33 |
| ENST00000327857 | 31.7274 | 106.71 | 1.749894 | 9.79E-06 | DEFA3 |
| ENST00000330439 | 0.1554 | 2.5418 | 4.031792 | 8.71E-18 | MT1E |
| ENST00000330862 | 0.0046 | 0.0522 | 3.504344 | 5.35E-06 | TMEM89 |
| ENST00000331224 | 0.008 | 0.0796 | 3.314697 | 0.000587 | DLK1 |
| ENST00000332904 | 0.207 | 0.0994 | −1.05831 | 0.001385 | CDHR1 |
| ENST00000337532 | 0.084 | 0.001 | −6.39232 | 1.85E-05 | MPP7 |
| ENST00000337843 | 0.0172 | 0.001 | −4.10434 | 0.000386 | C1QTNF6 |
| ENST00000338183 | 0.001 | 0.023 | 4.523562 | 0.000193 | C15orf41 |
| ENST00000338962 | 0.3224 | 0.7634 | 1.243587 | 6.55E-11 | LRP1 |
| ENST00000339861 | 0.001 | 0.0292 | 4.867896 | 1.99E-06 | SEMA4D |
| ENST00000340552 | 0.5436 | 0.024 | −4.50144 | 0.000124 | LIMK2 |
| ENST00000340800 | 0.0516 | 0.001 | −5.6893 | 4.78E-07 | ACSL4 |
| ENST00000340855 | 0.001 | 0.0092 | 3.201634 | 0.000583 | IDS |
| ENST00000341671 | 0.0252 | 0.001 | −4.65535 | 1.97E-05 | LRRC37B |
| ENST00000341852 | 0.1028 | 0.0064 | −4.00562 | 0.000818 | TNIK |
| ENST00000341911 | 0.0206 | 0.0008 | −4.6865 | 0.001394 | MYB |
| ENST00000342579 | 0.1454 | 1.0296 | 2.823985 | 0.000996 | BPTF |
| ENST00000343195 | 0.0122 | 0.086 | 2.817456 | 0.000125 | KCNIP2 |
| ENST00000344722 | 0.001 | 0.0224 | 4.485427 | 2.76E-05 | SMC4 |
| ENST00000344774 | 0.0256 | 0.0022 | −3.54057 | 0.000298 | FAM166A |
| ENST00000346330 | 0.0532 | 0.0018 | −4.88536 | 0.000158 | UBE2A |
| ENST00000346964 | 0.001 | 0.0464 | 5.536053 | 5.51E-06 | DLG1 |
| ENST00000347401 | 0.001 | 0.0044 | 2.137504 | 0.001491 | COL6A3 |
| ENST00000347471 | 0.0006 | 0.0272 | 5.5025 | 0.001259 | SMARCC2 |
| ENST00000350199 | 0.8082 | 1.771 | 1.13178 | 0.000665 | RNF38 |
| ENST00000350773 | 0.001 | 0.0092 | 3.201634 | 0.000568 | TSC2 |
| ENST00000352551 | 0.0008 | 0.0414 | 5.693487 | 0.000166 | UBE2C |
| ENST00000352732 | 5.5954 | 13.9578 | 1.318758 | 3.88E-09 | ABHD2 |
| ENST00000353660 | 0.001 | 0.027 | 4.754888 | 0.000636 | RBCK1 |
| ENST00000355667 | 0.0576 | 0.001 | −5.848 | 1.38E-05 | DNM2 |
| ENST00000355946 | 0.0062 | 0.001 | −2.63227 | 0.000318 | SH3PXD2A |
| ENST00000356249 | 0.001 | 0.008 | 3 | 0.000446 | CEMIP |
| ENST00000356348 | 0.3538 | 0.7496 | 1.083187 | 0.000111 | KPNA5 |
| ENST00000356628 | 0.05 | 0.1298 | 1.37629 | 0.000832 | NRARP |
| ENST00000357008 | 0.001 | 0.0166 | 4.053111 | 0.000103 | CHD4 |
| ENST00000357570 | 0.0246 | 0.001 | −4.62059 | 6.71E-07 | CACNA1E |
| ENST00000357702 | 0.0094 | 0.001 | −3.23266 | 0.00129 | SAP130 |
| ENST00000357736 | 0.001 | 0.2582 | 8.012345 | 4.95E-10 | MAFG |
| ENST00000358077 | 0.0016 | 0.0698 | 5.447083 | 0.000602 | DAPK1 |
| ENST00000358768 | 0.0136 | 0.0008 | −4.08746 | 0.000292 | PARD3B |
| ENST00000358791 | 0.0354 | 0.0002 | −7.46761 | 4.40E-05 | BCAS4 |
| ENST00000358825 | 0.0144 | 0.001 | −3.848 | 8.97E-05 | PRDM10 |
| ENST00000359576 | 0.001 | 0.011 | 3.459432 | 0.000434 | CLASP2 |
| ENST00000359645 | 0.0418 | 0.0012 | −5.1224 | 0.00031 | MBP |
| ENST00000359671 | 0.001 | 0.008 | 3 | 0.000112 | FN1 |
| ENST00000359709 | 3.1242 | 6.2692 | 1.004795 | 0.000505 | IFI16 |
| ENST00000359972 | 0.001 | 0.0506 | 5.661065 | 2.45E-06 | APAF1 |
| ENST00000360289 | 0.0104 | 0.001 | −3.37851 | 0.00091 | HK1 |
| ENST00000360997 | 0.0004 | 0.0084 | 4.392317 | 0.00041 | FAM107A |
| ENST00000361690 | 3.569 | 7.8604 | 1.139083 | 0.000707 | SRPK1 |
| ENST00000366837 | 0.0202 | 0.001 | −4.33628 | 0.001503 | EPHX1 |
| ENST00000367030 | 0.001 | 0.0098 | 3.292782 | 0.000144 | LAMB3 |
| ENST00000367084 | 0.001 | 0.029 | 4.857981 | 4.48E-06 | YOD1 |
| ENST00000367468 | 69.4708 | 156.7534 | 1.174018 | 2.66E-07 | PTGS2 |
| ENST00000367949 | 0.0294 | 0.001 | −4.87774 | 5.37E-05 | FCRLA |
| ENST00000368114 | 0.022 | 0.2212 | 3.329776 | 0.000973 | FCER1A |
| ENST00000368285 | 0.033 | 0.001 | −5.04439 | 2.58E-05 | SEMA4A |
| ENST00000368286 | 0.0338 | 0.001 | −5.07895 | 8.70E-05 | SEMA4A |
| ENST00000368678 | 1.792 | 4.7582 | 1.408845 | 0.001159 | FYN |
| ENST00000368682 | 0.001 | 0.2784 | 8.121015 | 3.77E-10 | FYN |
| ENST00000368932 | 0.001 | 0.1176 | 6.877744 | 7.10E-07 | CDC40 |
| ENST00000369040 | 0.0284 | 0.001 | −4.82782 | 3.96E-06 | ATE1 |
| ENST00000369298 | 0.0348 | 0.001 | −5.12102 | 9.27E-05 | PIAS3 |
| ENST00000369733 | 0.0408 | 0.1026 | 1.33039 | 0.000542 | COL17A1 |
| ENST00000369915 | 0.001 | 0.0146 | 3.867896 | 0.001168 | TKTL1 |
| ENST00000370401 | 0.0088 | 0.001 | −3.1375 | 0.000488 | MAMLD1 |
| ENST00000370924 | 0.098 | 0.2312 | 1.238288 | 0.000224 | PTGER3 |
| ENST00000371160 | 0.001 | 0.0166 | 4.053111 | 0.000108 | STAG2 |
| ENST00000371489 | 0.001 | 0.0184 | 4.201634 | 0.000805 | MYOF |
| ENST00000372764 | 0.2062 | 0.969 | 2.232452 | 5.01E-11 | PLAU |
| ENST00000373525 | 0.001 | 0.0146 | 3.867896 | 0.000491 | ATG16L1 |
| ENST00000374024 | 0.0044 | 0.0294 | 2.740241 | 0.000318 | GPR3 |
| ENST00000374848 | 0.0012 | 0.0436 | 5.183222 | 0.000259 | TMEM246 |
| ENST00000375053 | 0.001 | 0.0516 | 5.689299 | 7.01E-05 | MAGED2 |
| ENST00000375840 | 0.0446 | 0.0012 | −5.21594 | 0.000996 | STRADA |
| ENST00000377081 | 0.0082 | 0.001 | −3.03562 | 0.000361 | KIF1B |
| ENST00000377390 | 0.001 | 0.2122 | 7.729281 | 7.99E-07 | SF1 |
| ENST00000377877 | 2.8064 | 1.2408 | −1.17745 | 0.001502 | RNF38 |
| ENST00000378239 | 0.001 | 0.1114 | 6.799605 | 5.65E-06 | ERLEC1 |
| ENST00000378569 | 0.001 | 0.1352 | 7.078951 | 3.51E-06 | CCL7 |
| ENST00000379016 | 0.001 | 0.0082 | 3.035624 | 0.00074 | HIPK3 |
| ENST00000379153 | 0.271 | 0.8604 | 1.666715 | 1.04E-05 | CD83 |
| ENST00000379254 | 3.0092 | 10.6272 | 1.82031 | 1.27E-06 | SAT1 |
| ENST00000379333 | 0.02 | 0.001 | −4.32193 | 4.02E-05 | SLC23A2 |
| ENST00000379651 | 0.0454 | 0.0042 | −3.43423 | 0.000604 | MAP7D2 |
| ENST00000379811 | 0.0298 | 0.926 | 4.957628 | 3.54E-07 | MT1G |
| ENST00000380877 | 0.0876 | 0.201 | 1.198193 | 1.77E-05 | GABPB1 |
| ENST00000381309 | 0.0856 | 0.001 | −6.41954 | 7.44E-07 | KDM4C |
| ENST00000381497 | 0.001 | 0.0362 | 5.177918 | 0.001034 | AC138969.1 |
| ENST00000381573 | 0.0132 | 0.001 | −3.72247 | 0.000699 | CD274 |
| ENST00000381923 | 0.027 | 0.0608 | 1.171112 | 0.001012 | TMEM52B |
| ENST00000382111 | 0.0144 | 0.001 | −3.848 | 0.000207 | DEPDC5 |
| ENST00000382542 | 0.0472 | 0.001 | −5.56071 | 0.001266 | GZMB |
| ENST00000382692 | 122.2304 | 289.618 | 1.244548 | 8.64E-05 | DEFA1 |
| ENST00000389169 | 0.001 | 0.0626 | 5.968091 | 1.75E-06 | FLCN |
| ENST00000389282 | 0.001 | 0.022 | 4.459432 | 7.64E-05 | WIZ |
| ENST00000389851 | 0.089 | 0.2336 | 1.392163 | 0.001061 | UVSSA |
| ENST00000391834 | 0.0216 | 0.001 | −4.43296 | 0.000892 | AKT1S1 |
| ENST00000392746 | 0.037 | 0.0018 | −4.36146 | 0.000621 | RFX5 |
| ENST00000392975 | 0.0282 | 0.001 | −4.81762 | 0.000127 | CYB561 |
| ENST00000393001 | 0.0002 | 0.0748 | 8.546894 | 7.87E-06 | AMMECR1L |
| ENST00000393041 | 0.0206 | 0.001 | −4.36457 | 0.000322 | BIN1 |
| ENST00000393307 | 0.087 | 0.001 | −6.44294 | 7.80E-07 | RAB43 |
| ENST00000393386 | 0.0002 | 0.0072 | 5.169925 | 6.39E-05 | PTPRZ1 |
| ENST00000393484 | 0.0116 | 0.196 | 4.078657 | 0.000943 | TES |
| ENST00000394132 | 0.0068 | 0.001 | −2.76553 | 0.001094 | TTC23 |
| ENST00000394479 | 2.4894 | 5.0416 | 1.018084 | 7.68E-07 | REL |
| ENST00000394485 | 7.5228 | 16.8156 | 1.160459 | 2.99E-08 | MT1X |
| ENST00000394611 | 0.001 | 0.0316 | 4.981853 | 2.04E-05 | RTN4 |
| ENST00000394649 | 0.0232 | 0.001 | −4.53605 | 1.15E-05 | MYO1D |
| ENST00000394713 | 0.0412 | 0.001 | −5.36457 | 8.29E-07 | LRRC37B |
| ENST00000394725 | 0.4972 | 1.123 | 1.17546 | 5.21E-07 | SIAH1 |
| ENST00000394904 | 0.001 | 0.024 | 4.584963 | 1.82E-05 | SLC11A2 |
| ENST00000395032 | 0.5426 | 1.2672 | 1.223683 | 0.00021 | MS4A3 |
| ENST00000395097 | 0.0016 | 0.3804 | 7.893302 | 5.05E-09 | NR4A3 |
| ENST00000395287 | 0.0424 | 0.001 | −5.40599 | 1.35E-05 | BRD2 |
| ENST00000395392 | 0.0004 | 0.0296 | 6.209453 | 2.12E-06 | KIF23 |
| ENST00000395467 | 0.001 | 0.0236 | 4.560715 | 0.000405 | ADAMTSL5 |
| ENST00000395748 | 4.005 | 8.6518 | 1.111198 | 0.000558 | AREG |
| ENST00000395989 | 0.0216 | 0.1538 | 2.831952 | 0.000414 | ZFP64 |
| ENST00000396200 | 0.0106 | 0.001 | −3.40599 | 0.000676 | PDP1 |
| ENST00000396650 | 0.0622 | 0.2466 | 1.987186 | 0.001487 | C15orf48 |
| ENST00000397049 | 0.0574 | 0.001 | −5.84298 | 0.000248 | NUDT1 |
| ENST00000397121 | 0.0008 | 0.0118 | 3.882643 | 0.00084 | ZNF676 |
| ENST00000397283 | 0.001 | 0.0324 | 5.017922 | 1.12E-06 | ERMN |
| ENST00000398042 | 0.001 | 0.0252 | 4.655352 | 0.00024 | TANGO2 |
| ENST00000398246 | 0.2174 | 0.5198 | 1.257605 | 8.55E-08 | LONRF1 |
| ENST00000398499 | 0.001 | 0.049 | 5.61471 | 3.92E-07 | ZBTB21 |
| ENST00000398905 | 0.031 | 0.1174 | 1.921092 | 0.001283 | ERG |
| ENST00000398907 | 0.006 | 0.001 | −2.58496 | 0.00126 | ERG |
| ENST00000399169 | 0.0228 | 0.001 | −4.51096 | 6.10E-05 | STOX1 |
| ENST00000399839 | 0.001 | 0.0168 | 4.070389 | 0.000379 | ADA2 |
| ENST00000399975 | 0.001 | 0.0944 | 6.560715 | 2.72E-07 | USP16 |
| ENST00000400202 | 0.0072 | 0.001 | −2.848 | 0.000623 | NRIP1 |
| ENST00000400566 | 13.5744 | 28.7188 | 1.081107 | 2.36E-14 | SAMSN1 |
| ENST00000401931 | 11.6478 | 36.344 | 1.64166 | 4.94E-07 | CXCL8 |
| ENST00000403870 | 0.001 | 0.0168 | 4.070389 | 0.000606 | CCNJ |
| ENST00000405636 | 0.001 | 0.1964 | 7.617651 | 4.58E-06 | TOMM40 |
| ENST00000406998 | 2.4486 | 1.1584 | −1.07982 | 6.03E-05 | BACH2 |
| ENST00000407218 | 0.0008 | 0.0462 | 5.851749 | 0.00017 | MECP2 |
| ENST00000407322 | 0.031 | 0.001 | −4.9542 | 0.000925 | RGS6 |
| ENST00000409198 | 0.001 | 0.0022 | 1.137504 | 0.001363 | NEB |
| ENST00000409417 | 0.001 | 0.0494 | 5.626439 | 0.000358 | PDLIM2 |
| ENST00000409588 | 0.0002 | 0.027 | 7.076816 | 7.12E-05 | NIF3L1 |
| ENST00000409787 | 0.001 | 0.0578 | 5.852998 | 0.000436 | NT5C3A |
| ENST00000409917 | 0.1768 | 0.0146 | −3.59808 | 0.000336 | INO80B |
| ENST00000410034 | 0.001 | 0.0836 | 6.385431 | 1.45E-05 | ANKZF1 |
| ENST00000411732 | 0.0066 | 0.0812 | 3.620942 | 0.000329 | EGR2 |
| ENST00000412359 | 0.0148 | 0.001 | −3.88753 | 0.000514 | BUB1B |
| ENST00000412504 | 2.2868 | 1.0012 | −1.1916 | 1.16E-10 | IL1RAP |
| ENST00000412734 | 0.1042 | 0.007 | −3.89586 | 0.000954 | GET4 |
| ENST00000413393 | 0.001 | 0.0732 | 6.193772 | 4.20E-06 | MAPRE2 |
| ENST00000413636 | 0.7148 | 3.4676 | 2.278326 | 0.000724 | CIRBP |
| ENST00000413724 | 1.8446 | 3.7196 | 1.011839 | 2.21E-06 | MAP3K8 |
| ENST00000414217 | 0.011 | 0.001 | −3.45943 | 0.000328 | PPIP5K2 |
| ENST00000414248 | 0.001 | 0.0742 | 6.213347 | 1.40E-05 | FES |
| ENST00000415603 | 0.8546 | 0.2186 | −1.96696 | 0.00021 | DHX16 |
| ENST00000416658 | 0.033 | 0.357 | 3.435386 | 0.001058 | SLMAP |
| ENST00000417439 | 1.0032 | 5.0328 | 2.326752 | 6.76E-05 | LTF |
| ENST00000418259 | 0.001 | 0.016 | 4 | 0.000205 | ATG2A |
| ENST00000418359 | 0.0004 | 0.328 | 9.67948 | 1.04E-08 | CNOT2 |
| ENST00000418561 | 0.001 | 0.1072 | 6.744161 | 7.83E-05 | TTC17 |
| ENST00000418622 | 0.0222 | 0.001 | −4.47249 | 3.63E-05 | RIC1 |
| ENST00000419436 | 0.0006 | 0.0404 | 6.073249 | 0.000492 | PAXIP1 |
| ENST00000419948 | 1.8154 | 4.1536 | 1.194075 | 0.0007 | HP1BP3 |
| ENST00000420808 | 0.001 | 0.0644 | 6.008989 | 1.43E-05 | NISCH |
| ENST00000421217 | 0.125 | 0.0006 | −7.70275 | 0.000118 | BCAP29 |
| ENST00000422754 | 0.064 | 0.8278 | 3.693138 | 0.000281 | IVNS1ABP |
| ENST00000424802 | 0.001 | 0.0416 | 5.378512 | 0.001222 | QKI |
| ENST00000424834 | 0.001 | 0.0052 | 2.378512 | 0.000931 | SPATA13 |
| ENST00000425413 | 0.0388 | 0.001 | −5.27798 | 8.54E-06 | UBR4 |
| ENST00000426455 | 0.001 | 0.0076 | 2.925999 | 0.00146 | STAG3 |
| ENST00000426532 | 8.2416 | 24.4034 | 1.566086 | 2.71E-06 | LTF |
| ENST00000426880 | 0.2412 | 0.532 | 1.141196 | 0.001505 | HRH4 |
| ENST00000428637 | 0.023 | 0.1354 | 2.557522 | 0.000271 | RAPH1 |
| ENST00000428982 | 0.0128 | 0.001 | −3.67807 | 0.000724 | SYNGAP1 |
| ENST00000429492 | 0.0026 | 0.043 | 4.047753 | 0.000353 | OSBPL10 |
| ENST00000430436 | 0.0324 | 0.001 | −5.01792 | 2.25E-06 | RALGAPA2 |
| ENST00000431281 | 0.0176 | 0.001 | −4.1375 | 0.00076 | ACBD4 |
| ENST00000431380 | 0.001 | 0.0286 | 4.837943 | 0.000645 | SPON2 |
| ENST00000431674 | 0.1392 | 0.4262 | 1.614371 | 2.60E-05 | TBL1XR1 |
| ENST00000432018 | 0.5166 | 1.365 | 1.401781 | 5.40E-05 | IL1B |
| ENST00000432753 | 0.0752 | 0.001 | −6.23266 | 0.000907 | POR |
| ENST00000433368 | 0.004 | 0.0992 | 4.632268 | 0.000405 | CRISP3 |
| ENST00000435761 | 0.001 | 0.078 | 6.285402 | 1.39E-05 | PSMD2 |
| ENST00000435931 | 1.5656 | 3.9292 | 1.32752 | 7.71E-05 | NABP1 |
| ENST00000436757 | 0.001 | 0.2208 | 7.786596 | 1.11E-09 | PITPNM1 |
| ENST00000436863 | 12.846 | 5.579 | −1.20324 | 2.46E-06 | CASP1 |
| ENST00000437626 | 0.0104 | 0.0002 | −5.70044 | 0.000491 | ADARB1 |
| ENST00000437822 | 0.001 | 0.0266 | 4.733354 | 5.31E-05 | TAF6 |
| ENST00000438213 | 1.036 | 3.1064 | 1.58422 | 0.000695 | CTSD |
| ENST00000439889 | 0.0364 | 0.001 | −5.18587 | 0.000166 | SIGLEC10 |
| ENST00000441305 | 2.4346 | 1.1666 | −1.06137 | 6.07E-05 | RBM5 |
| ENST00000442028 | 0.0118 | 0.001 | −3.56071 | 0.000276 | ABCB9 |
| ENST00000442366 | 0.0826 | 0.001 | −6.36807 | 1.53E-06 | AIMP1 |
| ENST00000442506 | 0.052 | 0.001 | −5.70044 | 2.80E-06 | NBAS |
| ENST00000443183 | 0.001 | 0.0384 | 5.263034 | 6.72E-06 | DLG1 |
| ENST00000444415 | 0.0048 | 0.0568 | 3.564785 | 0.000898 | TRAF4 |
| ENST00000444487 | 0.7756 | 2.0002 | 1.36676 | 9.40E-06 | BCL3 |
| ENST00000444533 | 1.3736 | 0.6516 | −1.0759 | 6.11E-07 | SSBP3 |
| ENST00000444837 | 0.0386 | 2.2802 | 5.884416 | 1.91E-08 | MT1G |
| ENST00000447379 | 0.001 | 0.0592 | 5.887525 | 0.000291 | NEK6 |
| ENST00000447485 | 0.0484 | 0.001 | −5.59694 | 0.000584 | ATP6AP2 |
| ENST00000448077 | 0.003 | 0.0454 | 3.919658 | 0.001184 | PPARD |
| ENST00000448100 | 0.1936 | 0.016 | −3.59694 | 0.001476 | ZGPAT |
| ENST00000449182 | 0.001 | 0.0136 | 3.765535 | 1.17E-05 | PTPRZ1 |
| ENST00000449406 | 0.001 | 0.0506 | 5.661065 | 0.000179 | ST3GAL4 |
| ENST00000449416 | 1.2372 | 3.1574 | 1.351658 | 2.17E-10 | ZNF331 |
| ENST00000449627 | 2.5712 | 6.812 | 1.405637 | 0.000886 | NFE2L2 |
| ENST00000450232 | 0.2532 | 0.586 | 1.210623 | 7.48E-05 | PFKFB3 |
| ENST00000450926 | 0.0216 | 0.001 | −4.43296 | 0.000979 | INTS11 |
| ENST00000451619 | 0.1024 | 0.2836 | 1.469642 | 0.000141 | F13A1 |
| ENST00000451956 | 0.0178 | 1.7578 | 6.62575 | 2.77E-05 | GNAI2 |
| ENST00000452463 | 0.001 | 0.0502 | 5.649615 | 0.000419 | CREB3L2 |
| ENST00000452696 | 0.0568 | 0.2382 | 2.068211 | 0.001317 | DOT1L |
| ENST00000453873 | 0.0002 | 0.0174 | 6.442943 | 0.000669 | KIAA1841 |
| ENST00000453960 | 0.8178 | 0.2442 | −1.74368 | 0.000906 | MECP2 |
| ENST00000454703 | 3.2506 | 7.0162 | 1.109984 | 5.54E-06 | ACSL1 |
| ENST00000455263 | 0.0166 | 0.001 | −4.05311 | 0.000429 | TP53 |
| ENST00000456224 | 0.0216 | 0.0072 | −1.58496 | 0.000857 | SCN11A |
| ENST00000456990 | 0.001 | 0.1192 | 6.89724 | 7.58E-06 | THEMIS2 |
| ENST00000457296 | 0.001 | 0.0432 | 5.432959 | 0.001313 | FGR |
| ENST00000457416 | 0.037 | 0.001 | −5.20945 | 9.15E-05 | FGFR2 |
| ENST00000457692 | 0.0054 | 0.1254 | 4.537434 | 0.000295 | SERPINB2 |
| ENST00000457765 | 0.012 | 0.0014 | −3.09954 | 0.000924 | PLPPR4 |
| ENST00000458278 | 0.071 | 0.0166 | −2.09664 | 0.000839 | ANKRD54 |
| ENST00000458358 | 0.001 | 0.0186 | 4.217231 | 0.0003 | ITGA6 |
| ENST00000464465 | 12.5894 | 0.9218 | −3.77161 | 0.000105 | CSF3R |
| ENST00000464818 | 0.0458 | 0.001 | −5.51728 | 0.000475 | PRKCD |
| ENST00000465903 | 0.0544 | 0.0038 | −3.83954 | 8.98E-05 | SMC4 |
| ENST00000466255 | 0.001 | 0.0442 | 5.465974 | 0.001456 | SLMAP |
| ENST00000466335 | 0.0884 | 0.2028 | 1.197939 | 0.000394 | HACD1 |
| ENST00000466983 | 0.0734 | 0.1758 | 1.260083 | 0.001065 | SLC25A25 |
| ENST00000467517 | 0.0366 | 0.0072 | −2.34577 | 0.00081 | NOS3 |
| ENST00000469075 | 0.001 | 0.0134 | 3.744161 | 0.001462 | NVL |
| ENST00000470821 | 0.0912 | 0.0004 | −7.83289 | 1.41E-05 | ARMC8 |
| ENST00000471298 | 0.001 | 0.0982 | 6.617651 | 6.22E-05 | CEACAM1 |
| ENST00000471855 | 0.119 | 3.9434 | 5.050407 | 0.000186 | RPL36A |
| ENST00000471858 | 0.001 | 0.0334 | 5.061776 | 8.77E-06 | CD200R1 |
| ENST00000471951 | 3.2666 | 1.5126 | −1.11076 | 7.45E-06 | ZMYND8 |
| ENST00000472590 | 1.5542 | 3.3256 | 1.097442 | 6.03E-05 | HHEX |
| ENST00000489157 | 8.3084 | 4.1198 | −1.012 | 0.000851 | TAPBP |
| ENST00000490703 | 0.1034 | 0.0066 | −3.96963 | 0.00065 | TBC1D10B |
| ENST00000492950 | 0.0294 | 0.001 | −4.87774 | 0.00102 | USP21 |
| ENST00000494383 | 0.02 | 0.001 | −4.32193 | 0.000266 | AC097637.1 |
| ENST00000503683 | 0.2528 | 0.0056 | −5.49643 | 1.45E-06 | SEC24D |
| ENST00000503860 | 0.0376 | 0.0054 | −2.7997 | 0.000712 | CXCL11 |
| ENST00000504148 | 0.114 | 0.001 | −6.83289 | 7.13E-05 | TIMM8B |
| ENST00000504228 | 0.024 | 0.002 | −3.58496 | 0.001061 | KIAA1211 |
| ENST00000504342 | 0.001 | 0.0252 | 4.655352 | 0.000247 | ACSL1 |
| ENST00000507661 | 0.1578 | 0.0454 | −1.79733 | 0.000477 | ARHGAP10 |
| ENST00000508487 | 0.1572 | 0.532 | 1.758825 | 1.38E-05 | CXCL2 |
| ENST00000509047 | 0.0426 | 0.2188 | 2.360687 | 0.00013 | ZNF331 |
| ENST00000509570 | 0.0788 | 0.001 | −6.30012 | 0.000236 | SELENOW |
| ENST00000510624 | 0.001 | 0.0742 | 6.213347 | 0.000147 | LEF1 |
| ENST00000510911 | 0.037 | 0.0042 | −3.13906 | 0.001388 | FGFR4 |
| ENST00000511481 | 0.068 | 0.5106 | 2.908587 | 0.001063 | SEC24D |
| ENST00000512172 | 2.2486 | 1.1202 | −1.00527 | 0.000636 | LEF1 |
| ENST00000512387 | 2.2552 | 5.118 | 1.182325 | 0.000219 | ZNF331 |
| ENST00000512470 | 0.0342 | 0.001 | −5.09592 | 0.001384 | SLC41A3 |
| ENST00000512778 | 0.001 | 0.0886 | 6.469235 | 0.000221 | SEPTIN11 |
| ENST00000513042 | 0.001 | 0.0132 | 3.722466 | 0.000103 | ARHGEF28 |
| ENST00000513137 | 0.0036 | 0.0174 | 2.273018 | 0.000978 | SEMA6A |
| ENST00000513795 | 0.0022 | 0.1 | 5.506353 | 0.000119 | SPDL1 |
| ENST00000513826 | 0.001 | 0.0618 | 5.949535 | 3.42E-06 | FBXO38 |
| ENST00000514970 | 0.001 | 0.236 | 7.882643 | 8.76E-06 | AFF1 |
| ENST00000515359 | 0.0134 | 0.001 | −3.74416 | 0.00024 | DAG1 |
| ENST00000515524 | 0.0034 | 0.0796 | 4.549162 | 0.000818 | NSA2 |
| ENST00000515735 | 0.1588 | 0.0366 | −2.1173 | 0.000405 | NSD1 |
| ENST00000517649 | 0.001 | 0.0306 | 4.93546 | 2.48E-05 | NAIP |
| ENST00000518977 | 0.8398 | 0.0586 | −3.84107 | 0.000743 | TNIP1 |
| ENST00000519940 | 0.001 | 0.056 | 5.807355 | 2.40E-05 | CARD8 |
| ENST00000520223 | 0.001 | 0.0388 | 5.277985 | 0.001191 | GPAT4 |
| ENST00000520309 | 0.0154 | 0.001 | −3.94486 | 0.000304 | ANKAR |
| ENST00000520356 | 0.001 | 0.0486 | 5.602884 | 1.06E-06 | ZFAT |
| ENST00000520986 | 0.0412 | 0.6888 | 4.063369 | 0.000338 | DENND3 |
| ENST00000522100 | 0.001 | 0.4874 | 8.928962 | 1.98E-07 | TNIP1 |
| ENST00000523149 | 1.197 | 0.4168 | −1.522 | 0.001372 | EXTL3 |
| ENST00000525042 | 0.0452 | 0.001 | −5.49825 | 0.000753 | LGALS8 |
| ENST00000525809 | 0.001 | 0.0368 | 5.201634 | 0.000876 | BBS1 |
| ENST00000525919 | 2.132 | 0.9952 | −1.09915 | 0.000944 | EMSY |
| ENST00000526014 | 0.001 | 0.0166 | 4.053111 | 0.00133 | FXYD6 |
| ENST00000526285 | 0.001 | 0.0404 | 5.336283 | 0.000729 | GRK2 |
| ENST00000527344 | 12.5394 | 5.6772 | −1.14322 | 6.96E-06 | CFL1 |
| ENST00000527659 | 0.001 | 0.0218 | 4.446256 | 0.000322 | EPB41L2 |
| ENST00000528615 | 0.044 | 0.001 | −5.45943 | 1.48E-05 | DGKZ |
| ENST00000529318 | 0.0228 | 0.001 | −4.51096 | 0.000514 | SAAL1 |
| ENST00000529447 | 0.5138 | 0.2184 | −1.23423 | 0.000414 | ZDHHC5 |
| ENST00000531445 | 0.0096 | 0.0004 | −4.58496 | 0.000749 | C4orf50 |
| ENST00000532057 | 0.098 | 0.627 | 2.677612 | 0.000144 | FBXO3 |
| ENST00000532463 | 0.001 | 0.0212 | 4.405992 | 7.03E-06 | CTNND1 |
| ENST00000532702 | 29.7676 | 65.1756 | 1.130589 | 8.77E-09 | RPL8 |
| ENST00000532703 | 0.001 | 0.0704 | 6.137504 | 0.000488 | PITPNM1 |
| ENST00000532846 | 0.0106 | 0.0492 | 2.214594 | 0.001415 | RECQL4 |
| ENST00000533129 | 0.001 | 0.158 | 7.303781 | 1.43E-06 | CAPN1 |
| ENST00000534247 | 0.2926 | 0.0252 | −3.53743 | 8.49E-05 | ZHX2 |
| ENST00000536108 | 0.014 | 0.001 | −3.80735 | 0.000533 | KDM4C |
| ENST00000537194 | 0.0394 | 0.4072 | 3.36947 | 0.000208 | SMAD3 |
| ENST00000537561 | 0.001 | 0.0134 | 3.744161 | 0.000225 | NUDT16 |
| ENST00000537562 | 0.0004 | 0.0152 | 5.247928 | 0.000579 | AMN1 |
| ENST00000537750 | 0.0072 | 0.137 | 4.250035 | 0.001144 | SYNE1 |
| ENST00000537817 | 0.2318 | 0.0948 | −1.28992 | 0.001488 | C12orf43 |
| ENST00000538384 | 1.4048 | 0.5874 | −1.25795 | 0.00016 | NOL10 |
| ENST00000538577 | 0.001 | 0.0372 | 5.217231 | 0.00029 | SLC35B2 |
| ENST00000539661 | 0.001 | 0.03 | 4.906891 | 0.000294 | RAB40C |
| ENST00000540264 | 0.534 | 1.1212 | 1.070132 | 0.000316 | NFE2 |
| ENST00000540338 | 0.001 | 0.0204 | 4.350497 | 7.49E-05 | CLIP1 |
| ENST00000540510 | 0.001 | 0.0348 | 5.121015 | 4.19E-05 | GPR19 |
| ENST00000540933 | 0.0224 | 0.001 | −4.48543 | 4.08E-05 | GANAB |
| ENST00000541043 | 0.001 | 0.0336 | 5.070389 | 4.85E-05 | ACO1 |
| ENST00000541102 | 0.165 | 0.002 | −6.36632 | 4.83E-06 | LTBR |
| ENST00000541951 | 0.0324 | 0.001 | −5.01792 | 0.000645 | PAAF1 |
| ENST00000542120 | 0.0274 | 0.001 | −4.7761 | 4.72E-06 | TBC1D30 |
| ENST00000544017 | 0.001 | 0.0202 | 4.336283 | 0.00028 | TIMM50 |
| ENST00000544327 | 0.0254 | 0.1286 | 2.33999 | 0.000982 | SCARB1 |
| ENST00000544484 | 0.001 | 0.0126 | 3.655352 | 6.22E-05 | CHD4 |
| ENST00000544971 | 0.0272 | 0.0016 | −4.08746 | 7.14E-05 | TRPV4 |
| ENST00000545872 | 0.119 | 0.001 | −6.89482 | 2.49E-07 | ZNF83 |
| ENST00000547708 | 37.8826 | 18.019 | −1.07202 | 0.000121 | HNRNPA1 |
| ENST00000548839 | 0.5064 | 1.824 | 1.848756 | 2.49E-09 | LYZ |
| ENST00000550299 | 1.36 | 6.5598 | 2.270045 | 2.28E-05 | PPP1R12A |
| ENST00000550716 | 0.7612 | 1.676 | 1.138675 | 0.000165 | ATP2B1 |
| ENST00000551457 | 3.2292 | 9.1232 | 1.498363 | 8.11E-10 | TMCC3 |
| ENST00000552244 | 0.05 | 0.359 | 2.843984 | 0.000937 | RNF41 |
| ENST00000552496 | 0.001 | 0.1098 | 6.778734 | 0.000361 | CDK17 |
| ENST00000554144 | 0.001 | 0.0132 | 3.722466 | 0.001031 | HOPX |
| ENST00000554174 | 0.1876 | 0.4072 | 1.118078 | 4.44E-06 | LRP1 |
| ENST00000555672 | 83.829 | 38.2274 | −1.13284 | 0.000337 | FOS |
| ENST00000556134 | 0.001 | 0.018 | 4.169925 | 7.57E-06 | KIAA0586 |
| ENST00000556994 | 0.0208 | 0.001 | −4.37851 | 0.000901 | SRP54 |
| ENST00000558373 | 0.001 | 0.0208 | 4.378512 | 0.000187 | CTDSPL2 |
| ENST00000558968 | 0.696 | 0.336 | −1.05063 | 0.000949 | CTDSPL2 |
| ENST00000560177 | 0.001 | 0.0378 | 5.240314 | 8.70E-07 | NUSAP1 |
| ENST00000560338 | 0.397 | 1.4782 | 1.896631 | 0.001011 | EIF5 |
| ENST00000560520 | 0.0446 | 0.0008 | −5.8009 | 0.000433 | ICE2 |
| ENST00000560521 | 0.1708 | 0.6464 | 1.920119 | 0.000464 | RABGGTA |
| ENST00000561212 | 0.001 | 0.0308 | 4.944858 | 0.000581 | ZEB1 |
| ENST00000561491 | 0.8696 | 7.0584 | 3.020917 | 4.50E-12 | MT2A |
| ENST00000562939 | 0.2528 | 2.0322 | 3.006974 | 1.16E-13 | MT1X |
| ENST00000563402 | 0.2108 | 0.001 | −7.71973 | 2.13E-06 | MAZ |
| ENST00000563987 | 0.001 | 0.5306 | 9.051481 | 1.91E-06 | ALDOA |
| ENST00000564003 | 0.001 | 0.054 | 5.754888 | 5.63E-05 | MPI |
| ENST00000566419 | 0.0514 | 0.001 | −5.6837 | 0.001194 | NTAN1 |
| ENST00000566742 | 0.2954 | 0.0668 | −2.14475 | 0.000702 | JPT2 |
| ENST00000566946 | 0.0678 | 0.001 | −6.08321 | 2.87E-05 | ATXN2L |
| ENST00000567481 | 0.1732 | 0.0652 | −1.4095 | 0.00095 | ZFP1 |
| ENST00000567887 | 0.001 | 0.0028 | 1.485427 | 0.00048 | MACF1 |
| ENST00000568293 | 0.0752 | 0.9304 | 3.629047 | 3.34E-07 | MT1E |
| ENST00000568675 | 0.001 | 0.106 | 6.72792 | 1.85E-06 | MT1G |
| ENST00000569155 | 0.001 | 0.0356 | 5.153805 | 0.000262 | MT1H |
| ENST00000569500 | 0.0042 | 0.3174 | 6.239769 | 1.56E-06 | MT1G |
| ENST00000575842 | 2.7222 | 8.4102 | 1.627367 | 9.08E-12 | ACTG1 |
| ENST00000576965 | 0.001 | 0.1454 | 7.183883 | 0.00012 | RNF167 |
| ENST00000577040 | 0.2602 | 0.0266 | −3.29012 | 0.000167 | GPS2 |
| ENST00000579624 | 0.0492 | 0.001 | −5.62059 | 0.000554 | CNDP2 |
| ENST00000580070 | 26.092 | 13.0098 | −1.00401 | 0.001288 | NCOA4 |
| ENST00000581552 | 0.001 | 0.027 | 4.754888 | 0.000297 | PIK3R5 |
| ENST00000582117 | 0.0238 | 0.2492 | 3.388271 | 0.000107 | LRRC37B |
| ENST00000584114 | 0.0028 | 0.1546 | 5.78697 | 0.000904 | WSB1 |
| ENST00000584650 | 0.02 | 0.001 | −4.32193 | 0.001088 | ELAC2 |
| ENST00000585194 | 0.0862 | 0.001 | −6.42962 | 7.64E-06 | ZNF286A |
| ENST00000587615 | 0.0982 | 0.0118 | −3.05694 | 0.001018 | UBALD1 |
| ENST00000588735 | 0.0014 | 0.0086 | 2.61891 | 0.00112 | GFAP |
| ENST00000588982 | 0.0088 | 0.001 | −3.1375 | 0.000927 | ZBTB7C |
| ENST00000589377 | 0.0004 | 0.0092 | 4.523562 | 0.000909 | KCNJ16 |
| ENST00000589629 | 0.0012 | 0.1034 | 6.429058 | 0.000392 | CDC37 |
| ENST00000589640 | 0.3442 | 0.0236 | −3.86639 | 0.000498 | GPI |
| ENST00000589881 | 0.001 | 0.0154 | 3.944858 | 9.67E-05 | ZNF24 |
| ENST00000590414 | 0.001 | 0.0302 | 4.916477 | 0.000643 | ZNF573 |
| ENST00000590935 | 0.0178 | 0.001 | −4.15381 | 0.000365 | DUSP3 |
| ENST00000591768 | 0.2142 | 0.0168 | −3.67243 | 0.000201 | USP32 |
| ENST00000591811 | 0.404 | 0.1552 | −1.38023 | 5.67E-05 | CANT1 |
| ENST00000592282 | 0.0056 | 0.001 | −2.48543 | 0.001412 | ZNF260 |
| ENST00000593591 | 0.0726 | 0.0016 | −5.50383 | 0.000306 | SH3GL1 |
| ENST00000594442 | 0.001 | 0.1102 | 6.78398 | 2.60E-05 | ZFP36 |
| ENST00000594517 | 0.2572 | 0.1012 | −1.34568 | 0.000295 | ZNF677 |
| ENST00000595618 | 0.001 | 0.0382 | 5.255501 | 7.91E-06 | MYO9B |
| ENST00000596544 | 0.2146 | 1.6754 | 2.964784 | 0.001286 | CEACAM3 |
| ENST00000596853 | 0.319 | 2.1026 | 2.720546 | 0.000554 | DNAJB1 |
| ENST00000596894 | 0.0004 | 0.0306 | 6.257388 | 0.000248 | ZNF546 |
| ENST00000598915 | 0.0198 | 0.0002 | −6.62936 | 0.000546 | NAPSA |
| ENST00000599359 | 0.001 | 0.0826 | 6.36807 | 3.77E-06 | TNFSF14 |
| ENST00000599806 | 0.001 | 0.0284 | 4.827819 | 0.000404 | VAV1 |
| ENST00000600909 | 0.305 | 0.6274 | 1.040576 | 1.00E-05 | KCNN4 |
| ENST00000603300 | 0.0062 | 0.0206 | 1.732304 | 0.000727 | DUOX2 |
| ENST00000606731 | 0.8274 | 0.3442 | −1.26534 | 1.50E-05 | HOMEZ |
| ENST00000610538 | 0.0166 | 0.001 | −4.05311 | 0.000429 | TP53 |
| ENST00000610640 | 0.001 | 0.0672 | 6.070389 | 0.000107 | BTNL8 |
| ENST00000610724 | 0.0248 | 0.0004 | −5.9542 | 4.87E-05 | TBL2 |
| ENST00000610831 | 0.001 | 0.061 | 5.930737 | 5.27E-06 | CARHSP1 |
| ENST00000611646 | 0.016 | 0.001 | −4 | 1.77E-05 | RNF38 |
| ENST00000611707 | 0.032 | 0.001 | −5 | 0.000163 | GPAT3 |
| ENST00000612895 | 0.001 | 0.0086 | 3.104337 | 8.80E-05 | ACACA |
| ENST00000613058 | 0.025 | 0.001 | −4.64386 | 0.000301 | SLC4A1AP |
| ENST00000613071 | 0.001 | 0.0122 | 3.608809 | 0.000425 | TDRP |
| ENST00000613104 | 0.1144 | 0.5906 | 2.368094 | 3.64E-05 | ATF3 |
| ENST00000613273 | 0.0612 | 0.001 | −5.93546 | 2.13E-05 | TMEM185A |
| ENST00000613315 | 0.0042 | 0.1454 | 5.113494 | 0.000215 | UBAP2L |
| ENST00000614035 | 0.001 | 0.5596 | 9.128252 | 2.46E-08 | TNFAIP3 |
| ENST00000614134 | 0.001 | 0.0068 | 2.765535 | 0.000529 | JRK |
| ENST00000614382 | 0.1376 | 0.0166 | −3.05123 | 3.84E-08 | LFNG |
| ENST00000616144 | 0.2214 | 0.082 | −1.43296 | 0.001005 | BCL2L12 |
| ENST00000616923 | 0.0424 | 0.001 | −5.40599 | 2.70E-05 | GCLC |
| ENST00000617249 | 0.001 | 0.022 | 4.459432 | 0.00107 | PDIA6 |
| ENST00000617706 | 0.026 | 0.001 | −4.70044 | 0.000131 | CBSL |
| ENST00000617859 | 0.0114 | 0.001 | −3.51096 | 0.000702 | IKZF5 |
| ENST00000618040 | 0.001 | 0.0032 | 1.678072 | 0.000229 | KCNN3 |
| ENST00000618515 | 0.0286 | 0.001 | −4.83794 | 1.67E-05 | KIF3A |
| ENST00000618718 | 4.2948 | 1.8492 | −1.21569 | 0.00099 | CITED2 |
| ENST00000618741 | 0.001 | 0.0524 | 5.711495 | 7.82E-06 | LYPLA1 |
| ENST00000618765 | 1.698 | 0.583 | −1.54227 | 0.000248 | RRP12 |
| ENST00000619321 | 0.001 | 0.0438 | 5.452859 | 0.000148 | SLC16A3 |
| ENST00000619572 | 0.0024 | 0.1014 | 5.400879 | 0.000254 | FAM214A |
| ENST00000619580 | 0.001 | 0.0456 | 5.510962 | 0.001259 | PTMS |
| ENST00000619595 | 0.3864 | 0.8364 | 1.114098 | 0.000668 | SERPINB10 |
| ENST00000620047 | 0.0652 | 0.001 | −6.0268 | 0.000619 | PPIA |
| ENST00000620100 | 0.001 | 0.0172 | 4.104337 | 0.000131 | STAG3 |
| ENST00000620374 | 0.001 | 0.0224 | 4.485427 | 0.000106 | ZNF195 |
| ENST00000620571 | 0.001 | 0.015 | 3.906891 | 0.000113 | MGAM |
| ENST00000621226 | 0.0002 | 0.004 | 4.321928 | 0.000803 | MUC5AC |
| ENST00000621537 | 0.0664 | 0.001 | −6.05311 | 7.61E-07 | CMIP |
| ENST00000622396 | 0.0402 | 0.2478 | 2.623909 | 0.001249 | TBKBP1 |
| ENST00000625924 | 0.038 | 0.001 | −5.24793 | 0.000827 | RUNX2 |
| ENST00000627059 | 0.0996 | 0.0116 | −3.10202 | 0.001199 | ALDOA |
| ENST00000627621 | 0.0966 | 0.8118 | 3.071029 | 0.000682 | NFAT5 |
| ENST00000629272 | 0.0252 | 0.001 | −4.65535 | 0.001174 | ATL2 |
| ENST00000629371 | 0.001 | 0.0194 | 4.277985 | 8.43E-05 | AK2 |
| ENST00000629688 | 0.0008 | 0.092 | 6.84549 | 0.000992 | DRG1 |
| ENST00000635833 | 0.001 | 0.036 | 5.169925 | 0.000121 | RGS9 |
| ENST00000636347 | 0.0182 | 0.001 | −4.18587 | 9.83E-05 | SLC9A6 |
| ENST00000636941 | 0.001 | 0.013 | 3.70044 | 0.001408 | TCAF2C |
| ENST00000637581 | 0.0148 | 0.001 | −3.88753 | 0.000361 | SLC9A6 |
| ENST00000638375 | 0.001 | 0.0694 | 6.116864 | 0.000359 | HLA-A |
| ENST00000638451 | 0.0364 | 0.001 | −5.18587 | 0.000245 | LIAS |
| ENST00000638972 | 0.0138 | 0.0002 | −6.10852 | 0.000663 | ADAMTS19 |
| ENST00000639132 | 0.01 | 0.001 | −3.32193 | 0.000903 | BIVM-ERCC5 |
| ENST00000639145 | 0.0158 | 0.001 | −3.98185 | 0.000143 | SCARB2 |
| ENST00000640254 | 0.001 | 0.0134 | 3.744161 | 0.000711 | STAT6 |
| ENST00000642044 | 0.0258 | 0.001 | −4.6893 | 0.000118 | MICU1 |
| ENST00000642248 | 0.001 | 0.029 | 4.857981 | 0.000295 | CTNNB1 |
| ENST00000643242 | 0.0094 | 0.001 | −3.23266 | 0.001346 | TCAF2 |
| ENST00000643927 | 0.028 | 0.001 | −4.80735 | 9.01E-06 | PRKCB |
| ENST00000643977 | 0.001 | 0.0866 | 6.436295 | 2.38E-06 | CTNNB1 |
| ENST00000644774 | 0.138 | 0.001 | −7.10852 | 3.03E-06 | ACTG1 |
| ENST00000644913 | 0.2388 | 0.065 | −1.87729 | 0.001064 | PLEKHG1 |
| ENST00000644983 | 0.0974 | 0.3932 | 2.01327 | 0.000987 | ALAS2 |
| ENST00000645095 | 0.001 | 0.018 | 4.169925 | 0.000121 | CHD4 |
| ENST00000645632 | 0.001 | 0.08 | 6.321928 | 4.39E-08 | VPS13A |
| ENST00000645988 | 0.0764 | 0.0088 | −3.118 | 0.001439 | DSE |
| ENST00000646168 | 0.001 | 0.0268 | 4.744161 | 0.00036 | PRKCB |
| ENST00000646303 | 0.0236 | 0.001 | −4.56071 | 9.59E-05 | SPG7 |
| ENST00000646369 | 0.001 | 0.0922 | 6.526695 | 1.62E-06 | CTNNB1 |
| ENST00000646646 | 0.0014 | 0.059 | 5.397216 | 0.00048 | HSD17B4 |
| ENST00000646649 | 0.001 | 0.0134 | 3.744161 | 1.31E-05 | CUX1 |
| ENST00000646673 | 0.0546 | 3.5362 | 6.017155 | 4.26E-07 | SAMHD1 |
| ENST00000647508 | 0.001 | 0.0226 | 4.498251 | 0.000398 | SMARCE1 |
Supplementary Table 2.
The score of Clinician-Rated Dimensions of Psychosis Symptom Severity scale
| Sample | Disorganized Speech | Hallucination | Delusion | Abnormal psychomotor behavior | Negative symptom | Impaired cognition | Depression | Mania |
|---|---|---|---|---|---|---|---|---|
| Js 78 | 3 | 0 | 1 | 3 | 3 | 4 | 3 | 0 |
| Js 70 | 2 | 1 | 3 | 3 | 0 | 3 | 0 | 2 |
| Yx33 | 2 | 1 | 0 | 3 | 2 | 1 | 0 | 2 |
| Yx23 | 2 | 2 | 3 | 2 | 0 | 3 | 0 | 2 |
| Yx40 | 2 | 0 | 2 | 2 | 2 | 1 | 0 | 2 |
| YX1 | 2 | 0 | 3 | 3 | 2 | 4 | 1 | 3 |
| Yx34 | 2 | 2 | 2 | 2 | 0 | 1 | 0 | 2 |
| Yx39 | 2 | 2 | 3 | 3 | 0 | 3 | 0 | 3 |
| Yx47 | 2 | 0 | 1 | 3 | 2 | 3 | 0 | 0 |
| Js 86 | 2 | 0 | 3 | 4 | 3 | 3 | 0 | 0 |
| YX18 | 2 | 3 | 3 | 4 | 0 | 4 | 0 | 3 |
| Js 81 | 2 | 2 | 2 | 2 | 0 | 1 | 0 | 0 |
| JS31 | 0 | 1 | 3 | 3 | 3 | 3 | 0 | 0 |
| Yx36 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 0 |
| Yx20 | 2 | 2 | 2 | 2 | 0 | 1 | 0 | 2 |
| Js 67 | 2 | 3 | 3 | 0 | 0 | 4 | 0 | 3 |
| Yx37 | 2 | 2 | 2 | 2 | 2 | 1 | 0 | 0 |
| JS48 | 2 | 1 | 1 | 0 | 3 | 1 | 0 | 0 |
| Yx25 | 2 | 2 | 2 | 3 | 0 | 1 | 2 | 0 |
| JS33 | 3 | 0 | 1 | 3 | 3 | 4 | 1 | 0 |
| JS19 | 2 | 1 | 3 | 0 | 0 | 1 | 0 | 0 |
| YX2 | 3 | 0 | 3 | 3 | 0 | 4 | 0 | 3 |
| YX4 | 0 | 3 | 1 | 2 | 2 | 1 | 0 | 0 |
| JS40 | 2 | 2 | 2 | 2 | 0 | 1 | 2 | 2 |
| YX15 | 3 | 2 | 4 | 3 | 0 | 4 | 0 | 3 |
| YX8 | 2 | 3 | 3 | 3 | 2 | 4 | 0 | 0 |
| Yx28 | 2 | 3 | 2 | 3 | 0 | 3 | 0 | 2 |
| JS35 | 2 | 0 | 3 | 2 | 2 | 4 | 0 | 0 |
| Yx21 | 2 | 0 | 1 | 2 | 2 | 1 | 0 | 2 |
| JS55 | 0 | 2 | 3 | 2 | 0 | 1 | 0 | 0 |
| JS32 | 2 | 2 | 2 | 0 | 0 | 1 | 0 | 0 |
| JS2 | 2 | 0 | 1 | 2 | 2 | 0 | 1 | 0 |
| JS4 | 2 | 0 | 3 | 0 | 3 | 4 | 0 | 0 |
| Yx26 | 3 | 3 | 3 | 3 | 0 | 4 | 0 | 2 |
| JS39 | 0 | 3 | 1 | 0 | 2 | 1 | 2 | 0 |
| Yx42 | 2 | 0 | 3 | 2 | 2 | 4 | 0 | 1 |
| Yx45 | 2 | 1 | 1 | 2 | 2 | 1 | 0 | 0 |
| JS52 | 2 | 3 | 2 | 2 | 0 | 3 | 0 | 0 |
| JS53 | 2 | 2 | 2 | 0 | 0 | 1 | 0 | 0 |
| Js 66 | 2 | 2 | 2 | 2 | 0 | 1 | 0 | 0 |
| JS25 | 2 | 2 | 1 | 0 | 2 | 1 | 0 | 0 |
| Yx46 | 3 | 1 | 1 | 3 | 2 | 4 | 0 | 0 |
| JS42 | 2 | 0 | 2 | 2 | 2 | 1 | 0 | 2 |
| Yx49 | 3 | 3 | 3 | 4 | 0 | 4 | 0 | 0 |
| JS37 | 2 | 2 | 2 | 0 | 0 | 1 | 2 | 0 |
| JS22 | 2 | 1 | 2 | 0 | 0 | 1 | 0 | 0 |
| JS41 | 2 | 2 | 3 | 0 | 2 | 3 | 1 | 0 |
| JS38 | 2 | 0 | 2 | 2 | 2 | 3 | 1 | 0 |
| JS50 | 2 | 1 | 4 | 3 | 0 | 4 | 0 | 3 |
| JS27 | 2 | 0 | 3 | 3 | 2 | 3 | 1 | 0 |
Supplementary Table 3.
WGCNA identified 89 risk genes related to abnormal psychomotor behavior characteristics in patients with schizophrenia.
| IFI16, ETS1, BTAF1, LFNG, HHEX, ITPR2, MT1X, REL, RELT, NCOA4, SAMSN1, SIAH1, FBXO3, TAPBP, CTSD, GRASP, CXCL8, PTGS2, ACTG1, RNF38, HES1, ADM, PPP1R12A, FOS, SAT1, MAP3K8, FYN, NFE2L2, NRARP, DNAJB1, SNAI1, TBL1XR1, MN1, BCL3, CFL1, GABPB1, LONRF1, PLAU, ADORA3, SSBP3, F13A1, CD83, TRIM11, CITED2, IL1B, DUOX2, AREG, SERPINB2, CXCL2, VEZF1, PTGER3, FUT5, ABHD2, LEF1, TMCC3, HP1BP3, ZNF547, ATP2B1, NFE2, HBEGF, IL1RAP, ZNF331, PFKFB3, KPNA5, MT1E, LYZ, UVSSA, NXPE3, MT2A, PER1, EGR2, RBM5, HOMEZ, EGR3, BACH2, ZNF180, HRH4, EGR1, ATF3, NFAT5, PIN4, LRP1, ELL, ACSL1, EIF5, HNRNPA1, NABP1, CTDSPL2, CIRBP |
Acknowledgements
The authors thank all the patients and healthy volunteers who participated in the study. We thank Professor Xiongjian Luo for his assistance in data statistics.
Footnotes
Source of support: This work was supported by the National Natural Science Foundation of China (81760253), Yunnan Application Research Plan (2018fe001-009)
Conflict of interest
None.
References
- 1.Wang D, Liu S, Warrell J, et al. Comprehensive functional genomic resource and integrative model for the human brain. Science. 2018;362(6420) doi: 10.1126/science.aat8464. pii: eaat8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Girdhar K, Hoffman GE, Jiang Y, et al. Cell-specific histone modification maps in the human frontal lobe link schizophrenia risk to the neuronal epigenome. Nat Neurosci. 2018;21(8):1126–36. doi: 10.1038/s41593-018-0187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huckins LM, Dobbyn A, Ruderfer DM, et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat Genet. 2019;51(4):649–54. doi: 10.1038/s41588-019-0364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schrode N, Ho SM, Yamamuro K, et al. Synergistic effects of common schizophrenia risk variants. Nat Genet. 2019;51(10):1475–85. doi: 10.1038/s41588-019-0497-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Donovan MC, Owen MJ. The implications of the shared genetics of psychiatric disorders. Nat Med. 2016;22(11):1214–19. doi: 10.1038/nm.4196. [DOI] [PubMed] [Google Scholar]
- 6.Fromer M, Roussos P, Sieberts SK, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19(11):1442–53. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium. Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell. 2018;173(7):1705–15. doi: 10.1016/j.cell.2018.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pardiñas AF, Holmans P, Pocklington AJ, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381–89. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Chen J, Yu H, et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet. 2017;49(11):1576–83. doi: 10.1038/ng.3973. [DOI] [PubMed] [Google Scholar]
- 10.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–27. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bighelli I, Salanti G, Huhn M, et al. Psychological interventions to reduce positive symptoms in schizophrenia: Systematic review and network meta-analysis. World Psychiatry. 2018;17(3):316–29. doi: 10.1002/wps.20577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avram M, Brandl F, Cabello J, et al. Reduced striatal dopamine synthesis capacity in patients with schizophrenia during remission of positive symptoms. Brain. 2019;142(6):1813–26. doi: 10.1093/brain/awz093. [DOI] [PubMed] [Google Scholar]
- 13.Szulc A, Samochowiec J, Gałecki P, et al. Recommendations for the treatment of schizophrenia with negative symptoms. Standards of pharmacotherapy by the Polish Psychiatric Association (Polskie Towarzystwo Psychiatryczne), Part 1. Psychiatr Pol. 2019;53(3):497–524. doi: 10.12740/PP/OnlineFirst/100698. [DOI] [PubMed] [Google Scholar]
- 14.Németh G, Laszlovszky I, Czobor P, et al. Cariprazine versus risperidone monotherapy for treatment of predominant negative symptoms in patients with schizophrenia: A randomised, double-blind, controlled trial. Lancet. 2017;389(10074):1103–13. doi: 10.1016/S0140-6736(17)30060-0. [DOI] [PubMed] [Google Scholar]
- 15.Musliner KL, Mortensen PB, McGrath JJ, et al. Association of polygenic liabilities for major depression, bipolar disorder, and schizophrenia with risk for depression in the Danish Population. JAMA Psychiatry. 2019;76(5):516–25. doi: 10.1001/jamapsychiatry.2018.4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGlashan TH, Fenton WS. Classical subtypes for schizophrenia: literature review for DSM-IV. Schizophr Bull. 1991;17(4):609–32. doi: 10.1093/schbul/17.4.609. [DOI] [PubMed] [Google Scholar]
- 17.Barch DM, Bustillo J, Gaebel W, et al. Logic and justification for dimensional assessment of symptoms and related clinical phenomena in psychosis: Relevance to DSM-5. Schizophr Res. 2013;150(1):15–20. doi: 10.1016/j.schres.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 18.Carpenter WT, Tandon R. Psychotic disorders in DSM-5: Summary of changes. Asian J Psychiatr. 2013;6(3):266–68. doi: 10.1016/j.ajp.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Heckers S, Barch DM, Bustillo J, et al. Structure of the psychotic disorders classification in DSM-5. Schizophr Res. 2013;150(1):11–14. doi: 10.1016/j.schres.2013.04.039. [DOI] [PubMed] [Google Scholar]
- 20.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorder. 5th ed. American Psychiatric Publishing; Arlington, VA, USA: 2013. [Google Scholar]
- 21.Tandon R, Gaebel W, Barch DM, et al. Definition and description of schizophrenia in the DSM-5. Schizophr Res. 2013;150(1):3–10. doi: 10.1016/j.schres.2013.05.028. [DOI] [PubMed] [Google Scholar]
- 22.Mattila T, Koeter M, Wohlfarth T, et al. Impact of DSM-5 changes on the diagnosis and acute treatment of schizophrenia. Schizophr Bull. 2015;41(3):637–43. doi: 10.1093/schbul/sbu172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.RoBinSon MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardiñas AF, Holmans P, Pocklington AJ, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381–89. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Baumont A, Maschietto M, Lima L, et al. Innate immune response is differentially dysregulated between bipolar disease and schizophrenia. Schizophr Res. 2015;161(2–3):215–21. doi: 10.1016/j.schres.2014.10.055. [DOI] [PubMed] [Google Scholar]
- 26.Maschietto M, Tahira AC, Puga R, et al. Co-expression network of neural-differentiation genes shows specific pattern in schizophrenia. BMC Med Genomics. 2015;8:23. doi: 10.1186/s12920-015-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maycox PR, Kelly F, Taylor A, et al. Analysis of gene expression in two large schizophrenia cohorts identifies multiple changes associated with nerve terminal function. Mol Psychiatry. 2009;14(12):1083–94. doi: 10.1038/mp.2009.18. [DOI] [PubMed] [Google Scholar]
- 28.Iwamoto K, Bundo M, Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum Mol Genet. 2005;14(2):241–53. doi: 10.1093/hmg/ddi022. [DOI] [PubMed] [Google Scholar]
- 29.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9(1):559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ron-Harel N, Schwartz M. Immune senescence and brain aging: can rejuvenation of immunity reverse memory loss? Trends Neurosci. 2009;32(7):367–75. doi: 10.1016/j.tins.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Tsyglakova M, McDaniel D, Hodes GE. Immune mechanisms of stress susceptibility and resilience: lessons from animal models. Front Neuroendocrinol. 2019;54:100771. doi: 10.1016/j.yfrne.2019.100771. [DOI] [PubMed] [Google Scholar]
- 32.Ziv Y, Ron N, Butovsky O, et al. Immune cells contribute to the maintenance of neurogenesis and spatial learning abilities in adulthood. Nat Neurosci. 2006;9:268–75. doi: 10.1038/nn1629. [DOI] [PubMed] [Google Scholar]
- 33.Ron-Harel N, Segev Y, Lewitus GM, et al. Age-dependent spatial memory loss can be partially restored by immune activation. Rejuvenation Res. 2008;11:903–13. doi: 10.1089/rej.2008.0755. [DOI] [PubMed] [Google Scholar]
- 34.Pape K, Tamouza R, Leboyer M, et al. Immunoneuropsychiatry – novel perspectives on brain disorders. Nat Rev Neurol. 2019;15(6):317–28. doi: 10.1038/s41582-019-0174-4. [DOI] [PubMed] [Google Scholar]
- 35.Müller N, Schwarz MJ. Immune system and schizophrenia. Curr Immunol Rev. 2010;6(3):213–20. doi: 10.2174/157339510791823673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergink V, Gibney SM, Drexhage HA. Autoimmunity, inflammation, and psychosis: A search for peripheral markers. Biol Psychiatry. 2014;75(4):324–31. doi: 10.1016/j.biopsych.2013.09.037. [DOI] [PubMed] [Google Scholar]
- 37.Ilani T, Strous RD, Fuchs S, et al. Dopaminergic regulation of immune cells via D3 dopamine receptor: A pathway mediated by activated T cells. FASEB J. 2004;18(13):1600–2. doi: 10.1096/fj.04-1652fje. [DOI] [PubMed] [Google Scholar]
- 38.Dimitrov DH, Lee S, Yantis J, et al. Differential correlations between inflammatory cytokines and psychopathology in veterans with schizophrenia: potential role for IL-17 pathway. Schizophr Res. 2013;151(1–3):29–35. doi: 10.1016/j.schres.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 39.Roussos P, Katsel P, Davis KL, et al. Convergent findings for abnormalities of the NF-κB signaling pathway in schizophrenia. Neuropsychopharmacology. 2013;38(3):533–39. doi: 10.1038/npp.2012.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto S, Ohta N, Matsumoto A, et al. Haloperidol suppresses NF-kappaB to inhibit lipopolysaccharide-induced pro-inflammatory response in RAW 264 cells. Med Sci Monit. 2016;22:367–72. doi: 10.12659/MSM.895739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weston MC, Chen H, Swann JW. Loss of mTOR repressors Tsc1 or Pten has divergent effects on excitatory and inhibitory synaptic transmission in single hippocampal neuron cultures. Front Mol Neurosci. 2014;7:1. doi: 10.3389/fnmol.2014.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Vries PJ. Targeted treatments for cognitive and neurodevelopmental disorders in tuberous sclerosis complex. Neurotherapeutics. 2010;7(3):275–82. doi: 10.1016/j.nurt.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato A. mTOR, a potential target to treat autism spectrum disorder. CNS Neurol Disord Drug Targets. 2016;15(5):533–43. doi: 10.2174/1871527315666160413120638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ehninger D, Silva AJ. Rapamycin for treating tuberous sclerosis and autism spectrum disorders. Trends Mol Med. 2011;17(2):78–87. doi: 10.1016/j.molmed.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inserra A, Choo JM, Lewis MD, et al. Mice lacking Casp1, Ifngr and Nos2 genes exhibit altered depressive- and anxiety-like behaviour, and gut microbiome composition. Sci Rep. 2019;9(1):6456. doi: 10.1038/s41598-018-38055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li MX, Zheng HL, Luo Y, et al. Gene deficiency and pharmacological inhibition of caspase-1 confers resilience to chronic social defeat stress via regulating the stability of surface AMPARs. Mol Psychiatry. 2018;23(3):556–68. doi: 10.1038/mp.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoseth EZ, Ueland T, Dieset I, et al. A study of TNF pathway activation in schizophrenia and bipolar disorder in plasma and brain tissue. Schizophr Bull. 2017;43(4):881–90. doi: 10.1093/schbul/sbw183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morgan JA, Singhal G, Corrigan F, et al. TNF signalling via the TNF receptors mediates the effects of exercise on cognition-like behaviours. Behav Brain Res. 2018;353:74–82. doi: 10.1016/j.bbr.2018.06.036. [DOI] [PubMed] [Google Scholar]
- 49.Yu H, Bi W, Liu C, et al. Protein-interaction-network-based analysis for genome-wide association analysis of schizophrenia in Han Chinese population. J Psychiatr Res. 2014;50:73–78. doi: 10.1016/j.jpsychires.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 50.Kurian SM, Le-Niculescu H, Patel SD, et al. Identification of blood biomarkers for psychosis using convergent functional genomics. Mol Psychiatry. 2011;16(1):37–58. doi: 10.1038/mp.2009.117. [DOI] [PubMed] [Google Scholar]
- 51.Yamada K, Gerber DJ, Iwayama Y, et al. Genetic analysis of the calcineurin pathway identifies members of the EGR gene family, specifically EGR3, as potential susceptibility candidates in schizophrenia. Proc Natl Acad Sci USA. 2007;104:2815–20. doi: 10.1073/pnas.0610765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pérez-Santiago J, Diez-Alarcia R, Callado LF, et al. A combined analysis of microarray gene expression studies of the human prefrontal cortex identifies genes implicated in schizophrenia. J Psychiatr Res. 2012;46:1464–74. doi: 10.1016/j.jpsychires.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 53.Ramaker RC, Bowling KM, Lasseigne BN, et al. Post-mortem molecular profiling of three psychiatric disorders. Genome Med. 2017;9(1):72. doi: 10.1186/s13073-017-0458-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim SH, Song JY, Joo EJ, et al. EGR3 as a potential susceptibility gene for schizophrenia in Korea. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(7):1355–60. doi: 10.1002/ajmg.b.31115. [DOI] [PubMed] [Google Scholar]
- 55.Nishimura Y, Takizawa R, Koike S, et al. Association of decreased prefrontal hemodynamic response during a verbal fluency task with EGR3 gene polymorphism in patients with schizophrenia and in healthy individuals. Neuroimage. 2014;85(Pt 1):527–34. doi: 10.1016/j.neuroimage.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 56.Williams AA, Ingram WM, Levine S, et al. Reduced levels of serotonin 2A receptors underlie resistance of Egr3-deficient mice to locomotor suppression by clozapine. Neuropsychopharmacology. 2012;37(10):2285–98. doi: 10.1038/npp.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stuart MJ, Baune BT. Chemokines and chemokine receptors in mood disorders, schizophrenia, and cognitive impairment: A systematic review of biomarker studies. Neurosci Biobehav Rev. 2014;42:93–115. doi: 10.1016/j.neubiorev.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 58.Brown AS, Hooton J, Schaefer CA, et al. Elevated maternal interleukin-8 levels and risk of schizophrenia in adult offspring. Am J Psychiatry. 2004;161:889–95. doi: 10.1176/appi.ajp.161.5.889. [DOI] [PubMed] [Google Scholar]
- 59.Kelland EE, Gilmore W, Weiner LP, Lund BT. The dual role of CXCL8 in human CNS stem cell function: Multipotent neural stem cell death and oligodendrocyte progenitor cell chemotaxis. Glia. 2011;59:1864–78. doi: 10.1002/glia.21230. [DOI] [PubMed] [Google Scholar]
- 60.Ellman LM, Deicken RF, Vinogradov S, et al. Structural brain alterations in schizophrenia following fetal exposure to the inflammatory cytokine interleukin-8. Schizophr Res. 2010;121(1–3):46–54. doi: 10.1016/j.schres.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fond G, Hamdani N, Kapczinski F, et al. Effectiveness and tolerance of anti-inflammatory drugs’ add-on therapy in major mental disorders: A systematic qualitative review. Acta Psychiatr Scand. 2014;129(3):163–79. doi: 10.1111/acps.12211. [DOI] [PubMed] [Google Scholar]
- 62.Müller N, Myint AM, Krause D, et al. Anti-inflammatory treatment in schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2013;42:146–53. doi: 10.1016/j.pnpbp.2012.11.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Differentially expressed mRNAs in peripheral blood Leukocyte from Schizophrenia patients versus healthy controls by RNA-seq.
| ID | Control_fpkm | SZ_fpkm | log2(FC) | P value | Symbol |
|---|---|---|---|---|---|
| ENST00000010404 | 0.0006 | 0.067 | 6.803055 | 0.000242 | MGST1 |
| ENST00000064780 | 0.343 | 0.9742 | 1.506009 | 5.61E-05 | RELT |
| ENST00000164227 | 0.001 | 0.081 | 6.33985 | 3.93E-05 | BCL3 |
| ENST00000199764 | 1.007 | 2.7254 | 1.436404 | 0.00132 | CEACAM6 |
| ENST00000204726 | 0.001 | 0.0072 | 2.847997 | 0.000598 | GOLGA3 |
| ENST00000216127 | 0.0028 | 0.027 | 3.269461 | 0.000102 | RASD2 |
| ENST00000218432 | 0.1038 | 0.2788 | 1.425424 | 0.000598 | PIN4 |
| ENST00000221327 | 0.0432 | 0.3458 | 3.000835 | 1.81E-05 | ZNF180 |
| ENST00000224862 | 0.0234 | 0.001 | −4.54844 | 0.000707 | FBXL15 |
| ENST00000225275 | 1.8012 | 3.7582 | 1.061083 | 4.22E-05 | MPO |
| ENST00000230173 | 0.001 | 0.0062 | 2.632268 | 0.00066 | ADGRG6 |
| ENST00000230990 | 2.3934 | 5.1126 | 1.094996 | 0.000229 | HBEGF |
| ENST00000231173 | 0.0012 | 0.009 | 2.906891 | 0.000221 | PCDHB15 |
| ENST00000231751 | 0.2456 | 0.623 | 1.342922 | 0.001308 | LTF |
| ENST00000232424 | 0.1168 | 0.4342 | 1.894319 | 2.58E-08 | HES1 |
| ENST00000233714 | 0.001 | 0.0276 | 4.786596 | 0.001496 | LANCL1 |
| ENST00000233997 | 1.5778 | 3.3686 | 1.094235 | 0.000295 | AZU1 |
| ENST00000236826 | 1.054 | 3.4208 | 1.698459 | 3.56E-07 | MMP8 |
| ENST00000237264 | 0.001 | 0.0462 | 5.529821 | 2.69E-07 | TBPL1 |
| ENST00000238508 | 0.1864 | 0.5244 | 1.492266 | 0.000341 | SERPINB10 |
| ENST00000239316 | 0.0026 | 0.0172 | 2.725825 | 0.000322 | INSL4 |
| ENST00000239938 | 35.003 | 138.1178 | 1.980349 | 1.06E-08 | EGR1 |
| ENST00000241356 | 0.8688 | 1.8864 | 1.11854 | 9.38E-06 | ADORA3 |
| ENST00000242208 | 0.082 | 0.3154 | 1.943487 | 2.64E-10 | INHBA |
| ENST00000242480 | 0.8694 | 2.736 | 1.653976 | 7.11E-08 | EGR2 |
| ENST00000242737 | 2.8472 | 1.0502 | −1.43888 | 0.000686 | ITPR2 |
| ENST00000244050 | 0.3902 | 0.9026 | 1.209873 | 0.000287 | SNAI1 |
| ENST00000244336 | 2.7612 | 7.5754 | 1.456027 | 2.50E-06 | CEACAM8 |
| ENST00000244364 | 0.0102 | 0.001 | −3.3505 | 2.04E-06 | DST |
| ENST00000252675 | 0.0456 | 0.1404 | 1.622437 | 3.31E-05 | FUT5 |
| ENST00000253408 | 0.0088 | 0.0636 | 2.853451 | 0.000537 | GFAP |
| ENST00000255427 | 0.0126 | 0.0916 | 2.861924 | 0.000663 | CHIT1 |
| ENST00000257264 | 2.2706 | 5.1756 | 1.188653 | 1.84E-10 | TCN1 |
| ENST00000258400 | 0.001 | 0.0108 | 3.432959 | 0.000351 | HTR2B |
| ENST00000258494 | 0.001 | 0.0076 | 2.925999 | 7.91E-05 | ALDH1L2 |
| ENST00000258963 | 3.2004 | 6.5878 | 1.041545 | 0.000937 | VEZF1 |
| ENST00000259206 | 0.0306 | 0.001 | −4.93546 | 0.000568 | IL1RN |
| ENST00000259951 | 0.0736 | 0.0034 | −4.4361 | 0.000871 | HLA-F |
| ENST00000261944 | 0.001 | 0.0094 | 3.232661 | 0.000368 | CDHR2 |
| ENST00000262304 | 0.013 | 0.001 | −3.70044 | 7.49E-06 | PKD1 |
| ENST00000262809 | 0.7448 | 1.837 | 1.302427 | 0.000152 | ELL |
| ENST00000262865 | 3.837 | 7.9572 | 1.052282 | 5.32E-05 | BPI |
| ENST00000263045 | 1.6118 | 4.4596 | 1.468242 | 3.68E-09 | CRISP3 |
| ENST00000263341 | 16.5414 | 43.6896 | 1.401209 | 2.67E-12 | IL1B |
| ENST00000263545 | 0.001 | 0.0358 | 5.161888 | 8.82E-05 | LUC7L2 |
| ENST00000263621 | 1.157 | 2.7144 | 1.230244 | 5.73E-05 | ELANE |
| ENST00000263707 | 0.067 | 0.002 | −5.06609 | 4.20E-05 | TFCP2L1 |
| ENST00000264156 | 0.0116 | 0.3634 | 4.969362 | 5.28E-07 | MCM6 |
| ENST00000264498 | 0.001 | 0.0062 | 2.632268 | 0.000744 | FGF2 |
| ENST00000264956 | 0.0154 | 0.001 | −3.94486 | 4.17E-05 | EVC |
| ENST00000265881 | 0.1112 | 0.001 | −6.79701 | 3.41E-05 | REXO2 |
| ENST00000265990 | 0.096 | 0.0124 | −2.95269 | 1.45E-09 | BTAF1 |
| ENST00000273063 | 0.0136 | 0.001 | −3.76553 | 7.83E-05 | SLC4A3 |
| ENST00000273077 | 0.001 | 0.0144 | 3.847997 | 0.001435 | PNKD |
| ENST00000273347 | 0.1102 | 0.2382 | 1.112049 | 0.000991 | NXPE3 |
| ENST00000274710 | 0.0096 | 0.001 | −3.26303 | 0.000112 | PSD2 |
| ENST00000277480 | 6.1284 | 15.075 | 1.298576 | 0.000139 | LCN2 |
| ENST00000278175 | 4.0576 | 9.7064 | 1.25831 | 9.36E-07 | ADM |
| ENST00000278590 | 0.005 | 0.001 | −2.32193 | 0.001398 | ZC3H12C |
| ENST00000278756 | 4.8168 | 10.7066 | 1.152353 | 0.001103 | APLP2 |
| ENST00000278865 | 2.041 | 4.6692 | 1.193899 | 1.80E-07 | MS4A3 |
| ENST00000282120 | 0.0408 | 0.001 | −5.3505 | 3.96E-06 | TGOLN2 |
| ENST00000282282 | 0.1732 | 0.3508 | 1.01821 | 5.87E-06 | ZNF547 |
| ENST00000283426 | 0.0212 | 0.0044 | −2.26849 | 0.001026 | PLEKHG4B |
| ENST00000284551 | 0.9296 | 2.254 | 1.277806 | 1.59E-06 | TRIM11 |
| ENST00000286627 | 0.0096 | 0.001 | −3.26303 | 7.95E-05 | KCNMA1 |
| ENST00000292169 | 0.3618 | 0.0032 | −6.82098 | 6.20E-05 | S100A1 |
| ENST00000293662 | 0.331 | 0.8206 | 1.309848 | 4.16E-06 | GRASP |
| ENST00000293677 | 0.001 | 0.0918 | 6.520422 | 3.14E-06 | RAVER1 |
| ENST00000294702 | 0.0106 | 0.001 | −3.40599 | 0.001431 | GFI1 |
| ENST00000295992 | 0.0286 | 0.1144 | 2 | 0.000404 | PCOLCE2 |
| ENST00000296435 | 8.3026 | 17.1748 | 1.048658 | 5.01E-06 | CAMP |
| ENST00000297239 | 0.0388 | 0.001 | −5.27798 | 0.000112 | SYTL3 |
| ENST00000297435 | 2.3918 | 6.9666 | 1.542358 | 5.58E-06 | DEFA4 |
| ENST00000299502 | 0.3266 | 1.0236 | 1.648055 | 1.48E-06 | SERPINB2 |
| ENST00000299969 | 0.0292 | 0.008 | −1.8679 | 0.000283 | SERINC4 |
| ENST00000300027 | 0.001 | 0.0152 | 3.925999 | 3.53E-05 | FANCI |
| ENST00000301624 | 0.0006 | 0.119 | 7.631783 | 1.71E-06 | TNRC6C |
| ENST00000301698 | 0.019 | 0.0024 | −2.98489 | 0.000768 | PRR25 |
| ENST00000302035 | 0.0116 | 0.1026 | 3.144834 | 0.001254 | SLAMF1 |
| ENST00000302291 | 0.001 | 0.01 | 3.321928 | 0.000215 | LUZP1 |
| ENST00000302326 | 0.0614 | 0.1664 | 1.438345 | 0.000946 | MN1 |
| ENST00000302328 | 0.0024 | 0.0244 | 3.345775 | 0.000313 | SCN11A |
| ENST00000302779 | 0.0414 | 0.001 | −5.37156 | 0.000216 | PXK |
| ENST00000303391 | 0.001 | 0.597 | 9.221587 | 1.02E-13 | MECP2 |
| ENST00000303562 | 122.7578 | 289.7258 | 1.238873 | 1.13E-07 | FOS |
| ENST00000304639 | 1.7764 | 4.1508 | 1.224433 | 0.000958 | RNASE3 |
| ENST00000306061 | 1.0988 | 6.8182 | 2.633462 | 4.06E-17 | MT1E |
| ENST00000306090 | 0.0032 | 0.2008 | 5.971544 | 0.000344 | GNAS |
| ENST00000306151 | 0.0008 | 0.0346 | 5.434628 | 4.01E-05 | MUC17 |
| ENST00000306585 | 0.001 | 0.016 | 4 | 0.001358 | CHTF8 |
| ENST00000307407 | 260.8124 | 576.8646 | 1.14522 | 1.53E-06 | CXCL8 |
| ENST00000307885 | 0.001 | 0.0096 | 3.263034 | 0.000155 | ADCY6 |
| ENST00000309539 | 0.2124 | 0.8638 | 2.023914 | 9.00E-06 | OLR1 |
| ENST00000309902 | 0.0422 | 0.001 | −5.39917 | 8.46E-06 | ZNF407 |
| ENST00000312943 | 0.001 | 0.0882 | 6.462707 | 3.62E-06 | DOK3 |
| ENST00000317216 | 6.415 | 22.1836 | 1.789972 | 2.73E-09 | EGR3 |
| ENST00000317276 | 3.6124 | 8.6866 | 1.265834 | 2.47E-05 | PER1 |
| ENST00000317721 | 0.0048 | 0.001 | −2.26303 | 0.000439 | NCKAP5 |
| ENST00000319397 | 18.132 | 8.0092 | −1.17881 | 5.15E-09 | ETS1 |
| ENST00000322748 | 0.0032 | 0.0552 | 4.108524 | 0.000709 | ZNF18 |
| ENST00000323938 | 0.011 | 0.1666 | 3.920813 | 0.000919 | LGALS8 |
| ENST00000325094 | 0.0258 | 0.001 | −4.6893 | 1.80E-05 | TMEM33 |
| ENST00000327857 | 31.7274 | 106.71 | 1.749894 | 9.79E-06 | DEFA3 |
| ENST00000330439 | 0.1554 | 2.5418 | 4.031792 | 8.71E-18 | MT1E |
| ENST00000330862 | 0.0046 | 0.0522 | 3.504344 | 5.35E-06 | TMEM89 |
| ENST00000331224 | 0.008 | 0.0796 | 3.314697 | 0.000587 | DLK1 |
| ENST00000332904 | 0.207 | 0.0994 | −1.05831 | 0.001385 | CDHR1 |
| ENST00000337532 | 0.084 | 0.001 | −6.39232 | 1.85E-05 | MPP7 |
| ENST00000337843 | 0.0172 | 0.001 | −4.10434 | 0.000386 | C1QTNF6 |
| ENST00000338183 | 0.001 | 0.023 | 4.523562 | 0.000193 | C15orf41 |
| ENST00000338962 | 0.3224 | 0.7634 | 1.243587 | 6.55E-11 | LRP1 |
| ENST00000339861 | 0.001 | 0.0292 | 4.867896 | 1.99E-06 | SEMA4D |
| ENST00000340552 | 0.5436 | 0.024 | −4.50144 | 0.000124 | LIMK2 |
| ENST00000340800 | 0.0516 | 0.001 | −5.6893 | 4.78E-07 | ACSL4 |
| ENST00000340855 | 0.001 | 0.0092 | 3.201634 | 0.000583 | IDS |
| ENST00000341671 | 0.0252 | 0.001 | −4.65535 | 1.97E-05 | LRRC37B |
| ENST00000341852 | 0.1028 | 0.0064 | −4.00562 | 0.000818 | TNIK |
| ENST00000341911 | 0.0206 | 0.0008 | −4.6865 | 0.001394 | MYB |
| ENST00000342579 | 0.1454 | 1.0296 | 2.823985 | 0.000996 | BPTF |
| ENST00000343195 | 0.0122 | 0.086 | 2.817456 | 0.000125 | KCNIP2 |
| ENST00000344722 | 0.001 | 0.0224 | 4.485427 | 2.76E-05 | SMC4 |
| ENST00000344774 | 0.0256 | 0.0022 | −3.54057 | 0.000298 | FAM166A |
| ENST00000346330 | 0.0532 | 0.0018 | −4.88536 | 0.000158 | UBE2A |
| ENST00000346964 | 0.001 | 0.0464 | 5.536053 | 5.51E-06 | DLG1 |
| ENST00000347401 | 0.001 | 0.0044 | 2.137504 | 0.001491 | COL6A3 |
| ENST00000347471 | 0.0006 | 0.0272 | 5.5025 | 0.001259 | SMARCC2 |
| ENST00000350199 | 0.8082 | 1.771 | 1.13178 | 0.000665 | RNF38 |
| ENST00000350773 | 0.001 | 0.0092 | 3.201634 | 0.000568 | TSC2 |
| ENST00000352551 | 0.0008 | 0.0414 | 5.693487 | 0.000166 | UBE2C |
| ENST00000352732 | 5.5954 | 13.9578 | 1.318758 | 3.88E-09 | ABHD2 |
| ENST00000353660 | 0.001 | 0.027 | 4.754888 | 0.000636 | RBCK1 |
| ENST00000355667 | 0.0576 | 0.001 | −5.848 | 1.38E-05 | DNM2 |
| ENST00000355946 | 0.0062 | 0.001 | −2.63227 | 0.000318 | SH3PXD2A |
| ENST00000356249 | 0.001 | 0.008 | 3 | 0.000446 | CEMIP |
| ENST00000356348 | 0.3538 | 0.7496 | 1.083187 | 0.000111 | KPNA5 |
| ENST00000356628 | 0.05 | 0.1298 | 1.37629 | 0.000832 | NRARP |
| ENST00000357008 | 0.001 | 0.0166 | 4.053111 | 0.000103 | CHD4 |
| ENST00000357570 | 0.0246 | 0.001 | −4.62059 | 6.71E-07 | CACNA1E |
| ENST00000357702 | 0.0094 | 0.001 | −3.23266 | 0.00129 | SAP130 |
| ENST00000357736 | 0.001 | 0.2582 | 8.012345 | 4.95E-10 | MAFG |
| ENST00000358077 | 0.0016 | 0.0698 | 5.447083 | 0.000602 | DAPK1 |
| ENST00000358768 | 0.0136 | 0.0008 | −4.08746 | 0.000292 | PARD3B |
| ENST00000358791 | 0.0354 | 0.0002 | −7.46761 | 4.40E-05 | BCAS4 |
| ENST00000358825 | 0.0144 | 0.001 | −3.848 | 8.97E-05 | PRDM10 |
| ENST00000359576 | 0.001 | 0.011 | 3.459432 | 0.000434 | CLASP2 |
| ENST00000359645 | 0.0418 | 0.0012 | −5.1224 | 0.00031 | MBP |
| ENST00000359671 | 0.001 | 0.008 | 3 | 0.000112 | FN1 |
| ENST00000359709 | 3.1242 | 6.2692 | 1.004795 | 0.000505 | IFI16 |
| ENST00000359972 | 0.001 | 0.0506 | 5.661065 | 2.45E-06 | APAF1 |
| ENST00000360289 | 0.0104 | 0.001 | −3.37851 | 0.00091 | HK1 |
| ENST00000360997 | 0.0004 | 0.0084 | 4.392317 | 0.00041 | FAM107A |
| ENST00000361690 | 3.569 | 7.8604 | 1.139083 | 0.000707 | SRPK1 |
| ENST00000366837 | 0.0202 | 0.001 | −4.33628 | 0.001503 | EPHX1 |
| ENST00000367030 | 0.001 | 0.0098 | 3.292782 | 0.000144 | LAMB3 |
| ENST00000367084 | 0.001 | 0.029 | 4.857981 | 4.48E-06 | YOD1 |
| ENST00000367468 | 69.4708 | 156.7534 | 1.174018 | 2.66E-07 | PTGS2 |
| ENST00000367949 | 0.0294 | 0.001 | −4.87774 | 5.37E-05 | FCRLA |
| ENST00000368114 | 0.022 | 0.2212 | 3.329776 | 0.000973 | FCER1A |
| ENST00000368285 | 0.033 | 0.001 | −5.04439 | 2.58E-05 | SEMA4A |
| ENST00000368286 | 0.0338 | 0.001 | −5.07895 | 8.70E-05 | SEMA4A |
| ENST00000368678 | 1.792 | 4.7582 | 1.408845 | 0.001159 | FYN |
| ENST00000368682 | 0.001 | 0.2784 | 8.121015 | 3.77E-10 | FYN |
| ENST00000368932 | 0.001 | 0.1176 | 6.877744 | 7.10E-07 | CDC40 |
| ENST00000369040 | 0.0284 | 0.001 | −4.82782 | 3.96E-06 | ATE1 |
| ENST00000369298 | 0.0348 | 0.001 | −5.12102 | 9.27E-05 | PIAS3 |
| ENST00000369733 | 0.0408 | 0.1026 | 1.33039 | 0.000542 | COL17A1 |
| ENST00000369915 | 0.001 | 0.0146 | 3.867896 | 0.001168 | TKTL1 |
| ENST00000370401 | 0.0088 | 0.001 | −3.1375 | 0.000488 | MAMLD1 |
| ENST00000370924 | 0.098 | 0.2312 | 1.238288 | 0.000224 | PTGER3 |
| ENST00000371160 | 0.001 | 0.0166 | 4.053111 | 0.000108 | STAG2 |
| ENST00000371489 | 0.001 | 0.0184 | 4.201634 | 0.000805 | MYOF |
| ENST00000372764 | 0.2062 | 0.969 | 2.232452 | 5.01E-11 | PLAU |
| ENST00000373525 | 0.001 | 0.0146 | 3.867896 | 0.000491 | ATG16L1 |
| ENST00000374024 | 0.0044 | 0.0294 | 2.740241 | 0.000318 | GPR3 |
| ENST00000374848 | 0.0012 | 0.0436 | 5.183222 | 0.000259 | TMEM246 |
| ENST00000375053 | 0.001 | 0.0516 | 5.689299 | 7.01E-05 | MAGED2 |
| ENST00000375840 | 0.0446 | 0.0012 | −5.21594 | 0.000996 | STRADA |
| ENST00000377081 | 0.0082 | 0.001 | −3.03562 | 0.000361 | KIF1B |
| ENST00000377390 | 0.001 | 0.2122 | 7.729281 | 7.99E-07 | SF1 |
| ENST00000377877 | 2.8064 | 1.2408 | −1.17745 | 0.001502 | RNF38 |
| ENST00000378239 | 0.001 | 0.1114 | 6.799605 | 5.65E-06 | ERLEC1 |
| ENST00000378569 | 0.001 | 0.1352 | 7.078951 | 3.51E-06 | CCL7 |
| ENST00000379016 | 0.001 | 0.0082 | 3.035624 | 0.00074 | HIPK3 |
| ENST00000379153 | 0.271 | 0.8604 | 1.666715 | 1.04E-05 | CD83 |
| ENST00000379254 | 3.0092 | 10.6272 | 1.82031 | 1.27E-06 | SAT1 |
| ENST00000379333 | 0.02 | 0.001 | −4.32193 | 4.02E-05 | SLC23A2 |
| ENST00000379651 | 0.0454 | 0.0042 | −3.43423 | 0.000604 | MAP7D2 |
| ENST00000379811 | 0.0298 | 0.926 | 4.957628 | 3.54E-07 | MT1G |
| ENST00000380877 | 0.0876 | 0.201 | 1.198193 | 1.77E-05 | GABPB1 |
| ENST00000381309 | 0.0856 | 0.001 | −6.41954 | 7.44E-07 | KDM4C |
| ENST00000381497 | 0.001 | 0.0362 | 5.177918 | 0.001034 | AC138969.1 |
| ENST00000381573 | 0.0132 | 0.001 | −3.72247 | 0.000699 | CD274 |
| ENST00000381923 | 0.027 | 0.0608 | 1.171112 | 0.001012 | TMEM52B |
| ENST00000382111 | 0.0144 | 0.001 | −3.848 | 0.000207 | DEPDC5 |
| ENST00000382542 | 0.0472 | 0.001 | −5.56071 | 0.001266 | GZMB |
| ENST00000382692 | 122.2304 | 289.618 | 1.244548 | 8.64E-05 | DEFA1 |
| ENST00000389169 | 0.001 | 0.0626 | 5.968091 | 1.75E-06 | FLCN |
| ENST00000389282 | 0.001 | 0.022 | 4.459432 | 7.64E-05 | WIZ |
| ENST00000389851 | 0.089 | 0.2336 | 1.392163 | 0.001061 | UVSSA |
| ENST00000391834 | 0.0216 | 0.001 | −4.43296 | 0.000892 | AKT1S1 |
| ENST00000392746 | 0.037 | 0.0018 | −4.36146 | 0.000621 | RFX5 |
| ENST00000392975 | 0.0282 | 0.001 | −4.81762 | 0.000127 | CYB561 |
| ENST00000393001 | 0.0002 | 0.0748 | 8.546894 | 7.87E-06 | AMMECR1L |
| ENST00000393041 | 0.0206 | 0.001 | −4.36457 | 0.000322 | BIN1 |
| ENST00000393307 | 0.087 | 0.001 | −6.44294 | 7.80E-07 | RAB43 |
| ENST00000393386 | 0.0002 | 0.0072 | 5.169925 | 6.39E-05 | PTPRZ1 |
| ENST00000393484 | 0.0116 | 0.196 | 4.078657 | 0.000943 | TES |
| ENST00000394132 | 0.0068 | 0.001 | −2.76553 | 0.001094 | TTC23 |
| ENST00000394479 | 2.4894 | 5.0416 | 1.018084 | 7.68E-07 | REL |
| ENST00000394485 | 7.5228 | 16.8156 | 1.160459 | 2.99E-08 | MT1X |
| ENST00000394611 | 0.001 | 0.0316 | 4.981853 | 2.04E-05 | RTN4 |
| ENST00000394649 | 0.0232 | 0.001 | −4.53605 | 1.15E-05 | MYO1D |
| ENST00000394713 | 0.0412 | 0.001 | −5.36457 | 8.29E-07 | LRRC37B |
| ENST00000394725 | 0.4972 | 1.123 | 1.17546 | 5.21E-07 | SIAH1 |
| ENST00000394904 | 0.001 | 0.024 | 4.584963 | 1.82E-05 | SLC11A2 |
| ENST00000395032 | 0.5426 | 1.2672 | 1.223683 | 0.00021 | MS4A3 |
| ENST00000395097 | 0.0016 | 0.3804 | 7.893302 | 5.05E-09 | NR4A3 |
| ENST00000395287 | 0.0424 | 0.001 | −5.40599 | 1.35E-05 | BRD2 |
| ENST00000395392 | 0.0004 | 0.0296 | 6.209453 | 2.12E-06 | KIF23 |
| ENST00000395467 | 0.001 | 0.0236 | 4.560715 | 0.000405 | ADAMTSL5 |
| ENST00000395748 | 4.005 | 8.6518 | 1.111198 | 0.000558 | AREG |
| ENST00000395989 | 0.0216 | 0.1538 | 2.831952 | 0.000414 | ZFP64 |
| ENST00000396200 | 0.0106 | 0.001 | −3.40599 | 0.000676 | PDP1 |
| ENST00000396650 | 0.0622 | 0.2466 | 1.987186 | 0.001487 | C15orf48 |
| ENST00000397049 | 0.0574 | 0.001 | −5.84298 | 0.000248 | NUDT1 |
| ENST00000397121 | 0.0008 | 0.0118 | 3.882643 | 0.00084 | ZNF676 |
| ENST00000397283 | 0.001 | 0.0324 | 5.017922 | 1.12E-06 | ERMN |
| ENST00000398042 | 0.001 | 0.0252 | 4.655352 | 0.00024 | TANGO2 |
| ENST00000398246 | 0.2174 | 0.5198 | 1.257605 | 8.55E-08 | LONRF1 |
| ENST00000398499 | 0.001 | 0.049 | 5.61471 | 3.92E-07 | ZBTB21 |
| ENST00000398905 | 0.031 | 0.1174 | 1.921092 | 0.001283 | ERG |
| ENST00000398907 | 0.006 | 0.001 | −2.58496 | 0.00126 | ERG |
| ENST00000399169 | 0.0228 | 0.001 | −4.51096 | 6.10E-05 | STOX1 |
| ENST00000399839 | 0.001 | 0.0168 | 4.070389 | 0.000379 | ADA2 |
| ENST00000399975 | 0.001 | 0.0944 | 6.560715 | 2.72E-07 | USP16 |
| ENST00000400202 | 0.0072 | 0.001 | −2.848 | 0.000623 | NRIP1 |
| ENST00000400566 | 13.5744 | 28.7188 | 1.081107 | 2.36E-14 | SAMSN1 |
| ENST00000401931 | 11.6478 | 36.344 | 1.64166 | 4.94E-07 | CXCL8 |
| ENST00000403870 | 0.001 | 0.0168 | 4.070389 | 0.000606 | CCNJ |
| ENST00000405636 | 0.001 | 0.1964 | 7.617651 | 4.58E-06 | TOMM40 |
| ENST00000406998 | 2.4486 | 1.1584 | −1.07982 | 6.03E-05 | BACH2 |
| ENST00000407218 | 0.0008 | 0.0462 | 5.851749 | 0.00017 | MECP2 |
| ENST00000407322 | 0.031 | 0.001 | −4.9542 | 0.000925 | RGS6 |
| ENST00000409198 | 0.001 | 0.0022 | 1.137504 | 0.001363 | NEB |
| ENST00000409417 | 0.001 | 0.0494 | 5.626439 | 0.000358 | PDLIM2 |
| ENST00000409588 | 0.0002 | 0.027 | 7.076816 | 7.12E-05 | NIF3L1 |
| ENST00000409787 | 0.001 | 0.0578 | 5.852998 | 0.000436 | NT5C3A |
| ENST00000409917 | 0.1768 | 0.0146 | −3.59808 | 0.000336 | INO80B |
| ENST00000410034 | 0.001 | 0.0836 | 6.385431 | 1.45E-05 | ANKZF1 |
| ENST00000411732 | 0.0066 | 0.0812 | 3.620942 | 0.000329 | EGR2 |
| ENST00000412359 | 0.0148 | 0.001 | −3.88753 | 0.000514 | BUB1B |
| ENST00000412504 | 2.2868 | 1.0012 | −1.1916 | 1.16E-10 | IL1RAP |
| ENST00000412734 | 0.1042 | 0.007 | −3.89586 | 0.000954 | GET4 |
| ENST00000413393 | 0.001 | 0.0732 | 6.193772 | 4.20E-06 | MAPRE2 |
| ENST00000413636 | 0.7148 | 3.4676 | 2.278326 | 0.000724 | CIRBP |
| ENST00000413724 | 1.8446 | 3.7196 | 1.011839 | 2.21E-06 | MAP3K8 |
| ENST00000414217 | 0.011 | 0.001 | −3.45943 | 0.000328 | PPIP5K2 |
| ENST00000414248 | 0.001 | 0.0742 | 6.213347 | 1.40E-05 | FES |
| ENST00000415603 | 0.8546 | 0.2186 | −1.96696 | 0.00021 | DHX16 |
| ENST00000416658 | 0.033 | 0.357 | 3.435386 | 0.001058 | SLMAP |
| ENST00000417439 | 1.0032 | 5.0328 | 2.326752 | 6.76E-05 | LTF |
| ENST00000418259 | 0.001 | 0.016 | 4 | 0.000205 | ATG2A |
| ENST00000418359 | 0.0004 | 0.328 | 9.67948 | 1.04E-08 | CNOT2 |
| ENST00000418561 | 0.001 | 0.1072 | 6.744161 | 7.83E-05 | TTC17 |
| ENST00000418622 | 0.0222 | 0.001 | −4.47249 | 3.63E-05 | RIC1 |
| ENST00000419436 | 0.0006 | 0.0404 | 6.073249 | 0.000492 | PAXIP1 |
| ENST00000419948 | 1.8154 | 4.1536 | 1.194075 | 0.0007 | HP1BP3 |
| ENST00000420808 | 0.001 | 0.0644 | 6.008989 | 1.43E-05 | NISCH |
| ENST00000421217 | 0.125 | 0.0006 | −7.70275 | 0.000118 | BCAP29 |
| ENST00000422754 | 0.064 | 0.8278 | 3.693138 | 0.000281 | IVNS1ABP |
| ENST00000424802 | 0.001 | 0.0416 | 5.378512 | 0.001222 | QKI |
| ENST00000424834 | 0.001 | 0.0052 | 2.378512 | 0.000931 | SPATA13 |
| ENST00000425413 | 0.0388 | 0.001 | −5.27798 | 8.54E-06 | UBR4 |
| ENST00000426455 | 0.001 | 0.0076 | 2.925999 | 0.00146 | STAG3 |
| ENST00000426532 | 8.2416 | 24.4034 | 1.566086 | 2.71E-06 | LTF |
| ENST00000426880 | 0.2412 | 0.532 | 1.141196 | 0.001505 | HRH4 |
| ENST00000428637 | 0.023 | 0.1354 | 2.557522 | 0.000271 | RAPH1 |
| ENST00000428982 | 0.0128 | 0.001 | −3.67807 | 0.000724 | SYNGAP1 |
| ENST00000429492 | 0.0026 | 0.043 | 4.047753 | 0.000353 | OSBPL10 |
| ENST00000430436 | 0.0324 | 0.001 | −5.01792 | 2.25E-06 | RALGAPA2 |
| ENST00000431281 | 0.0176 | 0.001 | −4.1375 | 0.00076 | ACBD4 |
| ENST00000431380 | 0.001 | 0.0286 | 4.837943 | 0.000645 | SPON2 |
| ENST00000431674 | 0.1392 | 0.4262 | 1.614371 | 2.60E-05 | TBL1XR1 |
| ENST00000432018 | 0.5166 | 1.365 | 1.401781 | 5.40E-05 | IL1B |
| ENST00000432753 | 0.0752 | 0.001 | −6.23266 | 0.000907 | POR |
| ENST00000433368 | 0.004 | 0.0992 | 4.632268 | 0.000405 | CRISP3 |
| ENST00000435761 | 0.001 | 0.078 | 6.285402 | 1.39E-05 | PSMD2 |
| ENST00000435931 | 1.5656 | 3.9292 | 1.32752 | 7.71E-05 | NABP1 |
| ENST00000436757 | 0.001 | 0.2208 | 7.786596 | 1.11E-09 | PITPNM1 |
| ENST00000436863 | 12.846 | 5.579 | −1.20324 | 2.46E-06 | CASP1 |
| ENST00000437626 | 0.0104 | 0.0002 | −5.70044 | 0.000491 | ADARB1 |
| ENST00000437822 | 0.001 | 0.0266 | 4.733354 | 5.31E-05 | TAF6 |
| ENST00000438213 | 1.036 | 3.1064 | 1.58422 | 0.000695 | CTSD |
| ENST00000439889 | 0.0364 | 0.001 | −5.18587 | 0.000166 | SIGLEC10 |
| ENST00000441305 | 2.4346 | 1.1666 | −1.06137 | 6.07E-05 | RBM5 |
| ENST00000442028 | 0.0118 | 0.001 | −3.56071 | 0.000276 | ABCB9 |
| ENST00000442366 | 0.0826 | 0.001 | −6.36807 | 1.53E-06 | AIMP1 |
| ENST00000442506 | 0.052 | 0.001 | −5.70044 | 2.80E-06 | NBAS |
| ENST00000443183 | 0.001 | 0.0384 | 5.263034 | 6.72E-06 | DLG1 |
| ENST00000444415 | 0.0048 | 0.0568 | 3.564785 | 0.000898 | TRAF4 |
| ENST00000444487 | 0.7756 | 2.0002 | 1.36676 | 9.40E-06 | BCL3 |
| ENST00000444533 | 1.3736 | 0.6516 | −1.0759 | 6.11E-07 | SSBP3 |
| ENST00000444837 | 0.0386 | 2.2802 | 5.884416 | 1.91E-08 | MT1G |
| ENST00000447379 | 0.001 | 0.0592 | 5.887525 | 0.000291 | NEK6 |
| ENST00000447485 | 0.0484 | 0.001 | −5.59694 | 0.000584 | ATP6AP2 |
| ENST00000448077 | 0.003 | 0.0454 | 3.919658 | 0.001184 | PPARD |
| ENST00000448100 | 0.1936 | 0.016 | −3.59694 | 0.001476 | ZGPAT |
| ENST00000449182 | 0.001 | 0.0136 | 3.765535 | 1.17E-05 | PTPRZ1 |
| ENST00000449406 | 0.001 | 0.0506 | 5.661065 | 0.000179 | ST3GAL4 |
| ENST00000449416 | 1.2372 | 3.1574 | 1.351658 | 2.17E-10 | ZNF331 |
| ENST00000449627 | 2.5712 | 6.812 | 1.405637 | 0.000886 | NFE2L2 |
| ENST00000450232 | 0.2532 | 0.586 | 1.210623 | 7.48E-05 | PFKFB3 |
| ENST00000450926 | 0.0216 | 0.001 | −4.43296 | 0.000979 | INTS11 |
| ENST00000451619 | 0.1024 | 0.2836 | 1.469642 | 0.000141 | F13A1 |
| ENST00000451956 | 0.0178 | 1.7578 | 6.62575 | 2.77E-05 | GNAI2 |
| ENST00000452463 | 0.001 | 0.0502 | 5.649615 | 0.000419 | CREB3L2 |
| ENST00000452696 | 0.0568 | 0.2382 | 2.068211 | 0.001317 | DOT1L |
| ENST00000453873 | 0.0002 | 0.0174 | 6.442943 | 0.000669 | KIAA1841 |
| ENST00000453960 | 0.8178 | 0.2442 | −1.74368 | 0.000906 | MECP2 |
| ENST00000454703 | 3.2506 | 7.0162 | 1.109984 | 5.54E-06 | ACSL1 |
| ENST00000455263 | 0.0166 | 0.001 | −4.05311 | 0.000429 | TP53 |
| ENST00000456224 | 0.0216 | 0.0072 | −1.58496 | 0.000857 | SCN11A |
| ENST00000456990 | 0.001 | 0.1192 | 6.89724 | 7.58E-06 | THEMIS2 |
| ENST00000457296 | 0.001 | 0.0432 | 5.432959 | 0.001313 | FGR |
| ENST00000457416 | 0.037 | 0.001 | −5.20945 | 9.15E-05 | FGFR2 |
| ENST00000457692 | 0.0054 | 0.1254 | 4.537434 | 0.000295 | SERPINB2 |
| ENST00000457765 | 0.012 | 0.0014 | −3.09954 | 0.000924 | PLPPR4 |
| ENST00000458278 | 0.071 | 0.0166 | −2.09664 | 0.000839 | ANKRD54 |
| ENST00000458358 | 0.001 | 0.0186 | 4.217231 | 0.0003 | ITGA6 |
| ENST00000464465 | 12.5894 | 0.9218 | −3.77161 | 0.000105 | CSF3R |
| ENST00000464818 | 0.0458 | 0.001 | −5.51728 | 0.000475 | PRKCD |
| ENST00000465903 | 0.0544 | 0.0038 | −3.83954 | 8.98E-05 | SMC4 |
| ENST00000466255 | 0.001 | 0.0442 | 5.465974 | 0.001456 | SLMAP |
| ENST00000466335 | 0.0884 | 0.2028 | 1.197939 | 0.000394 | HACD1 |
| ENST00000466983 | 0.0734 | 0.1758 | 1.260083 | 0.001065 | SLC25A25 |
| ENST00000467517 | 0.0366 | 0.0072 | −2.34577 | 0.00081 | NOS3 |
| ENST00000469075 | 0.001 | 0.0134 | 3.744161 | 0.001462 | NVL |
| ENST00000470821 | 0.0912 | 0.0004 | −7.83289 | 1.41E-05 | ARMC8 |
| ENST00000471298 | 0.001 | 0.0982 | 6.617651 | 6.22E-05 | CEACAM1 |
| ENST00000471855 | 0.119 | 3.9434 | 5.050407 | 0.000186 | RPL36A |
| ENST00000471858 | 0.001 | 0.0334 | 5.061776 | 8.77E-06 | CD200R1 |
| ENST00000471951 | 3.2666 | 1.5126 | −1.11076 | 7.45E-06 | ZMYND8 |
| ENST00000472590 | 1.5542 | 3.3256 | 1.097442 | 6.03E-05 | HHEX |
| ENST00000489157 | 8.3084 | 4.1198 | −1.012 | 0.000851 | TAPBP |
| ENST00000490703 | 0.1034 | 0.0066 | −3.96963 | 0.00065 | TBC1D10B |
| ENST00000492950 | 0.0294 | 0.001 | −4.87774 | 0.00102 | USP21 |
| ENST00000494383 | 0.02 | 0.001 | −4.32193 | 0.000266 | AC097637.1 |
| ENST00000503683 | 0.2528 | 0.0056 | −5.49643 | 1.45E-06 | SEC24D |
| ENST00000503860 | 0.0376 | 0.0054 | −2.7997 | 0.000712 | CXCL11 |
| ENST00000504148 | 0.114 | 0.001 | −6.83289 | 7.13E-05 | TIMM8B |
| ENST00000504228 | 0.024 | 0.002 | −3.58496 | 0.001061 | KIAA1211 |
| ENST00000504342 | 0.001 | 0.0252 | 4.655352 | 0.000247 | ACSL1 |
| ENST00000507661 | 0.1578 | 0.0454 | −1.79733 | 0.000477 | ARHGAP10 |
| ENST00000508487 | 0.1572 | 0.532 | 1.758825 | 1.38E-05 | CXCL2 |
| ENST00000509047 | 0.0426 | 0.2188 | 2.360687 | 0.00013 | ZNF331 |
| ENST00000509570 | 0.0788 | 0.001 | −6.30012 | 0.000236 | SELENOW |
| ENST00000510624 | 0.001 | 0.0742 | 6.213347 | 0.000147 | LEF1 |
| ENST00000510911 | 0.037 | 0.0042 | −3.13906 | 0.001388 | FGFR4 |
| ENST00000511481 | 0.068 | 0.5106 | 2.908587 | 0.001063 | SEC24D |
| ENST00000512172 | 2.2486 | 1.1202 | −1.00527 | 0.000636 | LEF1 |
| ENST00000512387 | 2.2552 | 5.118 | 1.182325 | 0.000219 | ZNF331 |
| ENST00000512470 | 0.0342 | 0.001 | −5.09592 | 0.001384 | SLC41A3 |
| ENST00000512778 | 0.001 | 0.0886 | 6.469235 | 0.000221 | SEPTIN11 |
| ENST00000513042 | 0.001 | 0.0132 | 3.722466 | 0.000103 | ARHGEF28 |
| ENST00000513137 | 0.0036 | 0.0174 | 2.273018 | 0.000978 | SEMA6A |
| ENST00000513795 | 0.0022 | 0.1 | 5.506353 | 0.000119 | SPDL1 |
| ENST00000513826 | 0.001 | 0.0618 | 5.949535 | 3.42E-06 | FBXO38 |
| ENST00000514970 | 0.001 | 0.236 | 7.882643 | 8.76E-06 | AFF1 |
| ENST00000515359 | 0.0134 | 0.001 | −3.74416 | 0.00024 | DAG1 |
| ENST00000515524 | 0.0034 | 0.0796 | 4.549162 | 0.000818 | NSA2 |
| ENST00000515735 | 0.1588 | 0.0366 | −2.1173 | 0.000405 | NSD1 |
| ENST00000517649 | 0.001 | 0.0306 | 4.93546 | 2.48E-05 | NAIP |
| ENST00000518977 | 0.8398 | 0.0586 | −3.84107 | 0.000743 | TNIP1 |
| ENST00000519940 | 0.001 | 0.056 | 5.807355 | 2.40E-05 | CARD8 |
| ENST00000520223 | 0.001 | 0.0388 | 5.277985 | 0.001191 | GPAT4 |
| ENST00000520309 | 0.0154 | 0.001 | −3.94486 | 0.000304 | ANKAR |
| ENST00000520356 | 0.001 | 0.0486 | 5.602884 | 1.06E-06 | ZFAT |
| ENST00000520986 | 0.0412 | 0.6888 | 4.063369 | 0.000338 | DENND3 |
| ENST00000522100 | 0.001 | 0.4874 | 8.928962 | 1.98E-07 | TNIP1 |
| ENST00000523149 | 1.197 | 0.4168 | −1.522 | 0.001372 | EXTL3 |
| ENST00000525042 | 0.0452 | 0.001 | −5.49825 | 0.000753 | LGALS8 |
| ENST00000525809 | 0.001 | 0.0368 | 5.201634 | 0.000876 | BBS1 |
| ENST00000525919 | 2.132 | 0.9952 | −1.09915 | 0.000944 | EMSY |
| ENST00000526014 | 0.001 | 0.0166 | 4.053111 | 0.00133 | FXYD6 |
| ENST00000526285 | 0.001 | 0.0404 | 5.336283 | 0.000729 | GRK2 |
| ENST00000527344 | 12.5394 | 5.6772 | −1.14322 | 6.96E-06 | CFL1 |
| ENST00000527659 | 0.001 | 0.0218 | 4.446256 | 0.000322 | EPB41L2 |
| ENST00000528615 | 0.044 | 0.001 | −5.45943 | 1.48E-05 | DGKZ |
| ENST00000529318 | 0.0228 | 0.001 | −4.51096 | 0.000514 | SAAL1 |
| ENST00000529447 | 0.5138 | 0.2184 | −1.23423 | 0.000414 | ZDHHC5 |
| ENST00000531445 | 0.0096 | 0.0004 | −4.58496 | 0.000749 | C4orf50 |
| ENST00000532057 | 0.098 | 0.627 | 2.677612 | 0.000144 | FBXO3 |
| ENST00000532463 | 0.001 | 0.0212 | 4.405992 | 7.03E-06 | CTNND1 |
| ENST00000532702 | 29.7676 | 65.1756 | 1.130589 | 8.77E-09 | RPL8 |
| ENST00000532703 | 0.001 | 0.0704 | 6.137504 | 0.000488 | PITPNM1 |
| ENST00000532846 | 0.0106 | 0.0492 | 2.214594 | 0.001415 | RECQL4 |
| ENST00000533129 | 0.001 | 0.158 | 7.303781 | 1.43E-06 | CAPN1 |
| ENST00000534247 | 0.2926 | 0.0252 | −3.53743 | 8.49E-05 | ZHX2 |
| ENST00000536108 | 0.014 | 0.001 | −3.80735 | 0.000533 | KDM4C |
| ENST00000537194 | 0.0394 | 0.4072 | 3.36947 | 0.000208 | SMAD3 |
| ENST00000537561 | 0.001 | 0.0134 | 3.744161 | 0.000225 | NUDT16 |
| ENST00000537562 | 0.0004 | 0.0152 | 5.247928 | 0.000579 | AMN1 |
| ENST00000537750 | 0.0072 | 0.137 | 4.250035 | 0.001144 | SYNE1 |
| ENST00000537817 | 0.2318 | 0.0948 | −1.28992 | 0.001488 | C12orf43 |
| ENST00000538384 | 1.4048 | 0.5874 | −1.25795 | 0.00016 | NOL10 |
| ENST00000538577 | 0.001 | 0.0372 | 5.217231 | 0.00029 | SLC35B2 |
| ENST00000539661 | 0.001 | 0.03 | 4.906891 | 0.000294 | RAB40C |
| ENST00000540264 | 0.534 | 1.1212 | 1.070132 | 0.000316 | NFE2 |
| ENST00000540338 | 0.001 | 0.0204 | 4.350497 | 7.49E-05 | CLIP1 |
| ENST00000540510 | 0.001 | 0.0348 | 5.121015 | 4.19E-05 | GPR19 |
| ENST00000540933 | 0.0224 | 0.001 | −4.48543 | 4.08E-05 | GANAB |
| ENST00000541043 | 0.001 | 0.0336 | 5.070389 | 4.85E-05 | ACO1 |
| ENST00000541102 | 0.165 | 0.002 | −6.36632 | 4.83E-06 | LTBR |
| ENST00000541951 | 0.0324 | 0.001 | −5.01792 | 0.000645 | PAAF1 |
| ENST00000542120 | 0.0274 | 0.001 | −4.7761 | 4.72E-06 | TBC1D30 |
| ENST00000544017 | 0.001 | 0.0202 | 4.336283 | 0.00028 | TIMM50 |
| ENST00000544327 | 0.0254 | 0.1286 | 2.33999 | 0.000982 | SCARB1 |
| ENST00000544484 | 0.001 | 0.0126 | 3.655352 | 6.22E-05 | CHD4 |
| ENST00000544971 | 0.0272 | 0.0016 | −4.08746 | 7.14E-05 | TRPV4 |
| ENST00000545872 | 0.119 | 0.001 | −6.89482 | 2.49E-07 | ZNF83 |
| ENST00000547708 | 37.8826 | 18.019 | −1.07202 | 0.000121 | HNRNPA1 |
| ENST00000548839 | 0.5064 | 1.824 | 1.848756 | 2.49E-09 | LYZ |
| ENST00000550299 | 1.36 | 6.5598 | 2.270045 | 2.28E-05 | PPP1R12A |
| ENST00000550716 | 0.7612 | 1.676 | 1.138675 | 0.000165 | ATP2B1 |
| ENST00000551457 | 3.2292 | 9.1232 | 1.498363 | 8.11E-10 | TMCC3 |
| ENST00000552244 | 0.05 | 0.359 | 2.843984 | 0.000937 | RNF41 |
| ENST00000552496 | 0.001 | 0.1098 | 6.778734 | 0.000361 | CDK17 |
| ENST00000554144 | 0.001 | 0.0132 | 3.722466 | 0.001031 | HOPX |
| ENST00000554174 | 0.1876 | 0.4072 | 1.118078 | 4.44E-06 | LRP1 |
| ENST00000555672 | 83.829 | 38.2274 | −1.13284 | 0.000337 | FOS |
| ENST00000556134 | 0.001 | 0.018 | 4.169925 | 7.57E-06 | KIAA0586 |
| ENST00000556994 | 0.0208 | 0.001 | −4.37851 | 0.000901 | SRP54 |
| ENST00000558373 | 0.001 | 0.0208 | 4.378512 | 0.000187 | CTDSPL2 |
| ENST00000558968 | 0.696 | 0.336 | −1.05063 | 0.000949 | CTDSPL2 |
| ENST00000560177 | 0.001 | 0.0378 | 5.240314 | 8.70E-07 | NUSAP1 |
| ENST00000560338 | 0.397 | 1.4782 | 1.896631 | 0.001011 | EIF5 |
| ENST00000560520 | 0.0446 | 0.0008 | −5.8009 | 0.000433 | ICE2 |
| ENST00000560521 | 0.1708 | 0.6464 | 1.920119 | 0.000464 | RABGGTA |
| ENST00000561212 | 0.001 | 0.0308 | 4.944858 | 0.000581 | ZEB1 |
| ENST00000561491 | 0.8696 | 7.0584 | 3.020917 | 4.50E-12 | MT2A |
| ENST00000562939 | 0.2528 | 2.0322 | 3.006974 | 1.16E-13 | MT1X |
| ENST00000563402 | 0.2108 | 0.001 | −7.71973 | 2.13E-06 | MAZ |
| ENST00000563987 | 0.001 | 0.5306 | 9.051481 | 1.91E-06 | ALDOA |
| ENST00000564003 | 0.001 | 0.054 | 5.754888 | 5.63E-05 | MPI |
| ENST00000566419 | 0.0514 | 0.001 | −5.6837 | 0.001194 | NTAN1 |
| ENST00000566742 | 0.2954 | 0.0668 | −2.14475 | 0.000702 | JPT2 |
| ENST00000566946 | 0.0678 | 0.001 | −6.08321 | 2.87E-05 | ATXN2L |
| ENST00000567481 | 0.1732 | 0.0652 | −1.4095 | 0.00095 | ZFP1 |
| ENST00000567887 | 0.001 | 0.0028 | 1.485427 | 0.00048 | MACF1 |
| ENST00000568293 | 0.0752 | 0.9304 | 3.629047 | 3.34E-07 | MT1E |
| ENST00000568675 | 0.001 | 0.106 | 6.72792 | 1.85E-06 | MT1G |
| ENST00000569155 | 0.001 | 0.0356 | 5.153805 | 0.000262 | MT1H |
| ENST00000569500 | 0.0042 | 0.3174 | 6.239769 | 1.56E-06 | MT1G |
| ENST00000575842 | 2.7222 | 8.4102 | 1.627367 | 9.08E-12 | ACTG1 |
| ENST00000576965 | 0.001 | 0.1454 | 7.183883 | 0.00012 | RNF167 |
| ENST00000577040 | 0.2602 | 0.0266 | −3.29012 | 0.000167 | GPS2 |
| ENST00000579624 | 0.0492 | 0.001 | −5.62059 | 0.000554 | CNDP2 |
| ENST00000580070 | 26.092 | 13.0098 | −1.00401 | 0.001288 | NCOA4 |
| ENST00000581552 | 0.001 | 0.027 | 4.754888 | 0.000297 | PIK3R5 |
| ENST00000582117 | 0.0238 | 0.2492 | 3.388271 | 0.000107 | LRRC37B |
| ENST00000584114 | 0.0028 | 0.1546 | 5.78697 | 0.000904 | WSB1 |
| ENST00000584650 | 0.02 | 0.001 | −4.32193 | 0.001088 | ELAC2 |
| ENST00000585194 | 0.0862 | 0.001 | −6.42962 | 7.64E-06 | ZNF286A |
| ENST00000587615 | 0.0982 | 0.0118 | −3.05694 | 0.001018 | UBALD1 |
| ENST00000588735 | 0.0014 | 0.0086 | 2.61891 | 0.00112 | GFAP |
| ENST00000588982 | 0.0088 | 0.001 | −3.1375 | 0.000927 | ZBTB7C |
| ENST00000589377 | 0.0004 | 0.0092 | 4.523562 | 0.000909 | KCNJ16 |
| ENST00000589629 | 0.0012 | 0.1034 | 6.429058 | 0.000392 | CDC37 |
| ENST00000589640 | 0.3442 | 0.0236 | −3.86639 | 0.000498 | GPI |
| ENST00000589881 | 0.001 | 0.0154 | 3.944858 | 9.67E-05 | ZNF24 |
| ENST00000590414 | 0.001 | 0.0302 | 4.916477 | 0.000643 | ZNF573 |
| ENST00000590935 | 0.0178 | 0.001 | −4.15381 | 0.000365 | DUSP3 |
| ENST00000591768 | 0.2142 | 0.0168 | −3.67243 | 0.000201 | USP32 |
| ENST00000591811 | 0.404 | 0.1552 | −1.38023 | 5.67E-05 | CANT1 |
| ENST00000592282 | 0.0056 | 0.001 | −2.48543 | 0.001412 | ZNF260 |
| ENST00000593591 | 0.0726 | 0.0016 | −5.50383 | 0.000306 | SH3GL1 |
| ENST00000594442 | 0.001 | 0.1102 | 6.78398 | 2.60E-05 | ZFP36 |
| ENST00000594517 | 0.2572 | 0.1012 | −1.34568 | 0.000295 | ZNF677 |
| ENST00000595618 | 0.001 | 0.0382 | 5.255501 | 7.91E-06 | MYO9B |
| ENST00000596544 | 0.2146 | 1.6754 | 2.964784 | 0.001286 | CEACAM3 |
| ENST00000596853 | 0.319 | 2.1026 | 2.720546 | 0.000554 | DNAJB1 |
| ENST00000596894 | 0.0004 | 0.0306 | 6.257388 | 0.000248 | ZNF546 |
| ENST00000598915 | 0.0198 | 0.0002 | −6.62936 | 0.000546 | NAPSA |
| ENST00000599359 | 0.001 | 0.0826 | 6.36807 | 3.77E-06 | TNFSF14 |
| ENST00000599806 | 0.001 | 0.0284 | 4.827819 | 0.000404 | VAV1 |
| ENST00000600909 | 0.305 | 0.6274 | 1.040576 | 1.00E-05 | KCNN4 |
| ENST00000603300 | 0.0062 | 0.0206 | 1.732304 | 0.000727 | DUOX2 |
| ENST00000606731 | 0.8274 | 0.3442 | −1.26534 | 1.50E-05 | HOMEZ |
| ENST00000610538 | 0.0166 | 0.001 | −4.05311 | 0.000429 | TP53 |
| ENST00000610640 | 0.001 | 0.0672 | 6.070389 | 0.000107 | BTNL8 |
| ENST00000610724 | 0.0248 | 0.0004 | −5.9542 | 4.87E-05 | TBL2 |
| ENST00000610831 | 0.001 | 0.061 | 5.930737 | 5.27E-06 | CARHSP1 |
| ENST00000611646 | 0.016 | 0.001 | −4 | 1.77E-05 | RNF38 |
| ENST00000611707 | 0.032 | 0.001 | −5 | 0.000163 | GPAT3 |
| ENST00000612895 | 0.001 | 0.0086 | 3.104337 | 8.80E-05 | ACACA |
| ENST00000613058 | 0.025 | 0.001 | −4.64386 | 0.000301 | SLC4A1AP |
| ENST00000613071 | 0.001 | 0.0122 | 3.608809 | 0.000425 | TDRP |
| ENST00000613104 | 0.1144 | 0.5906 | 2.368094 | 3.64E-05 | ATF3 |
| ENST00000613273 | 0.0612 | 0.001 | −5.93546 | 2.13E-05 | TMEM185A |
| ENST00000613315 | 0.0042 | 0.1454 | 5.113494 | 0.000215 | UBAP2L |
| ENST00000614035 | 0.001 | 0.5596 | 9.128252 | 2.46E-08 | TNFAIP3 |
| ENST00000614134 | 0.001 | 0.0068 | 2.765535 | 0.000529 | JRK |
| ENST00000614382 | 0.1376 | 0.0166 | −3.05123 | 3.84E-08 | LFNG |
| ENST00000616144 | 0.2214 | 0.082 | −1.43296 | 0.001005 | BCL2L12 |
| ENST00000616923 | 0.0424 | 0.001 | −5.40599 | 2.70E-05 | GCLC |
| ENST00000617249 | 0.001 | 0.022 | 4.459432 | 0.00107 | PDIA6 |
| ENST00000617706 | 0.026 | 0.001 | −4.70044 | 0.000131 | CBSL |
| ENST00000617859 | 0.0114 | 0.001 | −3.51096 | 0.000702 | IKZF5 |
| ENST00000618040 | 0.001 | 0.0032 | 1.678072 | 0.000229 | KCNN3 |
| ENST00000618515 | 0.0286 | 0.001 | −4.83794 | 1.67E-05 | KIF3A |
| ENST00000618718 | 4.2948 | 1.8492 | −1.21569 | 0.00099 | CITED2 |
| ENST00000618741 | 0.001 | 0.0524 | 5.711495 | 7.82E-06 | LYPLA1 |
| ENST00000618765 | 1.698 | 0.583 | −1.54227 | 0.000248 | RRP12 |
| ENST00000619321 | 0.001 | 0.0438 | 5.452859 | 0.000148 | SLC16A3 |
| ENST00000619572 | 0.0024 | 0.1014 | 5.400879 | 0.000254 | FAM214A |
| ENST00000619580 | 0.001 | 0.0456 | 5.510962 | 0.001259 | PTMS |
| ENST00000619595 | 0.3864 | 0.8364 | 1.114098 | 0.000668 | SERPINB10 |
| ENST00000620047 | 0.0652 | 0.001 | −6.0268 | 0.000619 | PPIA |
| ENST00000620100 | 0.001 | 0.0172 | 4.104337 | 0.000131 | STAG3 |
| ENST00000620374 | 0.001 | 0.0224 | 4.485427 | 0.000106 | ZNF195 |
| ENST00000620571 | 0.001 | 0.015 | 3.906891 | 0.000113 | MGAM |
| ENST00000621226 | 0.0002 | 0.004 | 4.321928 | 0.000803 | MUC5AC |
| ENST00000621537 | 0.0664 | 0.001 | −6.05311 | 7.61E-07 | CMIP |
| ENST00000622396 | 0.0402 | 0.2478 | 2.623909 | 0.001249 | TBKBP1 |
| ENST00000625924 | 0.038 | 0.001 | −5.24793 | 0.000827 | RUNX2 |
| ENST00000627059 | 0.0996 | 0.0116 | −3.10202 | 0.001199 | ALDOA |
| ENST00000627621 | 0.0966 | 0.8118 | 3.071029 | 0.000682 | NFAT5 |
| ENST00000629272 | 0.0252 | 0.001 | −4.65535 | 0.001174 | ATL2 |
| ENST00000629371 | 0.001 | 0.0194 | 4.277985 | 8.43E-05 | AK2 |
| ENST00000629688 | 0.0008 | 0.092 | 6.84549 | 0.000992 | DRG1 |
| ENST00000635833 | 0.001 | 0.036 | 5.169925 | 0.000121 | RGS9 |
| ENST00000636347 | 0.0182 | 0.001 | −4.18587 | 9.83E-05 | SLC9A6 |
| ENST00000636941 | 0.001 | 0.013 | 3.70044 | 0.001408 | TCAF2C |
| ENST00000637581 | 0.0148 | 0.001 | −3.88753 | 0.000361 | SLC9A6 |
| ENST00000638375 | 0.001 | 0.0694 | 6.116864 | 0.000359 | HLA-A |
| ENST00000638451 | 0.0364 | 0.001 | −5.18587 | 0.000245 | LIAS |
| ENST00000638972 | 0.0138 | 0.0002 | −6.10852 | 0.000663 | ADAMTS19 |
| ENST00000639132 | 0.01 | 0.001 | −3.32193 | 0.000903 | BIVM-ERCC5 |
| ENST00000639145 | 0.0158 | 0.001 | −3.98185 | 0.000143 | SCARB2 |
| ENST00000640254 | 0.001 | 0.0134 | 3.744161 | 0.000711 | STAT6 |
| ENST00000642044 | 0.0258 | 0.001 | −4.6893 | 0.000118 | MICU1 |
| ENST00000642248 | 0.001 | 0.029 | 4.857981 | 0.000295 | CTNNB1 |
| ENST00000643242 | 0.0094 | 0.001 | −3.23266 | 0.001346 | TCAF2 |
| ENST00000643927 | 0.028 | 0.001 | −4.80735 | 9.01E-06 | PRKCB |
| ENST00000643977 | 0.001 | 0.0866 | 6.436295 | 2.38E-06 | CTNNB1 |
| ENST00000644774 | 0.138 | 0.001 | −7.10852 | 3.03E-06 | ACTG1 |
| ENST00000644913 | 0.2388 | 0.065 | −1.87729 | 0.001064 | PLEKHG1 |
| ENST00000644983 | 0.0974 | 0.3932 | 2.01327 | 0.000987 | ALAS2 |
| ENST00000645095 | 0.001 | 0.018 | 4.169925 | 0.000121 | CHD4 |
| ENST00000645632 | 0.001 | 0.08 | 6.321928 | 4.39E-08 | VPS13A |
| ENST00000645988 | 0.0764 | 0.0088 | −3.118 | 0.001439 | DSE |
| ENST00000646168 | 0.001 | 0.0268 | 4.744161 | 0.00036 | PRKCB |
| ENST00000646303 | 0.0236 | 0.001 | −4.56071 | 9.59E-05 | SPG7 |
| ENST00000646369 | 0.001 | 0.0922 | 6.526695 | 1.62E-06 | CTNNB1 |
| ENST00000646646 | 0.0014 | 0.059 | 5.397216 | 0.00048 | HSD17B4 |
| ENST00000646649 | 0.001 | 0.0134 | 3.744161 | 1.31E-05 | CUX1 |
| ENST00000646673 | 0.0546 | 3.5362 | 6.017155 | 4.26E-07 | SAMHD1 |
| ENST00000647508 | 0.001 | 0.0226 | 4.498251 | 0.000398 | SMARCE1 |
Supplementary Table 2.
The score of Clinician-Rated Dimensions of Psychosis Symptom Severity scale
| Sample | Disorganized Speech | Hallucination | Delusion | Abnormal psychomotor behavior | Negative symptom | Impaired cognition | Depression | Mania |
|---|---|---|---|---|---|---|---|---|
| Js 78 | 3 | 0 | 1 | 3 | 3 | 4 | 3 | 0 |
| Js 70 | 2 | 1 | 3 | 3 | 0 | 3 | 0 | 2 |
| Yx33 | 2 | 1 | 0 | 3 | 2 | 1 | 0 | 2 |
| Yx23 | 2 | 2 | 3 | 2 | 0 | 3 | 0 | 2 |
| Yx40 | 2 | 0 | 2 | 2 | 2 | 1 | 0 | 2 |
| YX1 | 2 | 0 | 3 | 3 | 2 | 4 | 1 | 3 |
| Yx34 | 2 | 2 | 2 | 2 | 0 | 1 | 0 | 2 |
| Yx39 | 2 | 2 | 3 | 3 | 0 | 3 | 0 | 3 |
| Yx47 | 2 | 0 | 1 | 3 | 2 | 3 | 0 | 0 |
| Js 86 | 2 | 0 | 3 | 4 | 3 | 3 | 0 | 0 |
| YX18 | 2 | 3 | 3 | 4 | 0 | 4 | 0 | 3 |
| Js 81 | 2 | 2 | 2 | 2 | 0 | 1 | 0 | 0 |
| JS31 | 0 | 1 | 3 | 3 | 3 | 3 | 0 | 0 |
| Yx36 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 0 |
| Yx20 | 2 | 2 | 2 | 2 | 0 | 1 | 0 | 2 |
| Js 67 | 2 | 3 | 3 | 0 | 0 | 4 | 0 | 3 |
| Yx37 | 2 | 2 | 2 | 2 | 2 | 1 | 0 | 0 |
| JS48 | 2 | 1 | 1 | 0 | 3 | 1 | 0 | 0 |
| Yx25 | 2 | 2 | 2 | 3 | 0 | 1 | 2 | 0 |
| JS33 | 3 | 0 | 1 | 3 | 3 | 4 | 1 | 0 |
| JS19 | 2 | 1 | 3 | 0 | 0 | 1 | 0 | 0 |
| YX2 | 3 | 0 | 3 | 3 | 0 | 4 | 0 | 3 |
| YX4 | 0 | 3 | 1 | 2 | 2 | 1 | 0 | 0 |
| JS40 | 2 | 2 | 2 | 2 | 0 | 1 | 2 | 2 |
| YX15 | 3 | 2 | 4 | 3 | 0 | 4 | 0 | 3 |
| YX8 | 2 | 3 | 3 | 3 | 2 | 4 | 0 | 0 |
| Yx28 | 2 | 3 | 2 | 3 | 0 | 3 | 0 | 2 |
| JS35 | 2 | 0 | 3 | 2 | 2 | 4 | 0 | 0 |
| Yx21 | 2 | 0 | 1 | 2 | 2 | 1 | 0 | 2 |
| JS55 | 0 | 2 | 3 | 2 | 0 | 1 | 0 | 0 |
| JS32 | 2 | 2 | 2 | 0 | 0 | 1 | 0 | 0 |
| JS2 | 2 | 0 | 1 | 2 | 2 | 0 | 1 | 0 |
| JS4 | 2 | 0 | 3 | 0 | 3 | 4 | 0 | 0 |
| Yx26 | 3 | 3 | 3 | 3 | 0 | 4 | 0 | 2 |
| JS39 | 0 | 3 | 1 | 0 | 2 | 1 | 2 | 0 |
| Yx42 | 2 | 0 | 3 | 2 | 2 | 4 | 0 | 1 |
| Yx45 | 2 | 1 | 1 | 2 | 2 | 1 | 0 | 0 |
| JS52 | 2 | 3 | 2 | 2 | 0 | 3 | 0 | 0 |
| JS53 | 2 | 2 | 2 | 0 | 0 | 1 | 0 | 0 |
| Js 66 | 2 | 2 | 2 | 2 | 0 | 1 | 0 | 0 |
| JS25 | 2 | 2 | 1 | 0 | 2 | 1 | 0 | 0 |
| Yx46 | 3 | 1 | 1 | 3 | 2 | 4 | 0 | 0 |
| JS42 | 2 | 0 | 2 | 2 | 2 | 1 | 0 | 2 |
| Yx49 | 3 | 3 | 3 | 4 | 0 | 4 | 0 | 0 |
| JS37 | 2 | 2 | 2 | 0 | 0 | 1 | 2 | 0 |
| JS22 | 2 | 1 | 2 | 0 | 0 | 1 | 0 | 0 |
| JS41 | 2 | 2 | 3 | 0 | 2 | 3 | 1 | 0 |
| JS38 | 2 | 0 | 2 | 2 | 2 | 3 | 1 | 0 |
| JS50 | 2 | 1 | 4 | 3 | 0 | 4 | 0 | 3 |
| JS27 | 2 | 0 | 3 | 3 | 2 | 3 | 1 | 0 |
Supplementary Table 3.
WGCNA identified 89 risk genes related to abnormal psychomotor behavior characteristics in patients with schizophrenia.
| IFI16, ETS1, BTAF1, LFNG, HHEX, ITPR2, MT1X, REL, RELT, NCOA4, SAMSN1, SIAH1, FBXO3, TAPBP, CTSD, GRASP, CXCL8, PTGS2, ACTG1, RNF38, HES1, ADM, PPP1R12A, FOS, SAT1, MAP3K8, FYN, NFE2L2, NRARP, DNAJB1, SNAI1, TBL1XR1, MN1, BCL3, CFL1, GABPB1, LONRF1, PLAU, ADORA3, SSBP3, F13A1, CD83, TRIM11, CITED2, IL1B, DUOX2, AREG, SERPINB2, CXCL2, VEZF1, PTGER3, FUT5, ABHD2, LEF1, TMCC3, HP1BP3, ZNF547, ATP2B1, NFE2, HBEGF, IL1RAP, ZNF331, PFKFB3, KPNA5, MT1E, LYZ, UVSSA, NXPE3, MT2A, PER1, EGR2, RBM5, HOMEZ, EGR3, BACH2, ZNF180, HRH4, EGR1, ATF3, NFAT5, PIN4, LRP1, ELL, ACSL1, EIF5, HNRNPA1, NABP1, CTDSPL2, CIRBP |