Abstract
It is generally regarded that the progression of an infection within host macrophages is the consequence of a failed immune response. However, recent appreciation of macrophage heterogeneity, with respect to both development and metabolism, indicates that the reality is more complex. Different lineages of tissue-resident macrophages respond divergently to microbial, environmental and immunological stimuli. The emerging picture that the developmental origin of macrophages determines their responses to immune stimulation and to infection stresses the importance of in vivo infection models. Recent investigations into the metabolism of infecting microorganisms and host macrophages indicate that their metabolic interface can be a major determinant of pathogen growth or containment. This Review focuses on the integration of data from existing studies, the identification of challenges in generating and interpreting data from ongoing studies, and a discussion of the technologies and tools that are required to best address future questions in the field.
Subject categories: Biological sciences, Microbiology, Bacteria, Bacterial host response, Physiology, Metabolism, Immunology, Innate immune cells, Monocytes and macrophages, Phagocytes, Antimicrobial responses
Table of Contents
Different lineages of macrophages respond divergently to immune stimuli and microbial infection. This review explores our current knowledge of how the different metabolic states of macrophage lineages impacts the control or progression of intracellular bacterial infections.
Introduction
It has long been known that macrophage metabolism is extremely plastic and frequently reflects the pathology that is associated with specific states of disease1–3. For example, atherosclerosis is associated with the accumulation of foamy macrophages [G], increased lipid metabolism and fatty acid oxidation (FAO). By contrast, tumour-associated macrophages have a marked metabolic shift to glycolysis, fatty acid synthesis and altered nitrogen cycle metabolism. Until recently, these divergent metabolic responses of macrophages were thought to be controlled by environmental cues and the local cytokine milieu, partly as a result of the long-held belief that all macrophages had a common origin in the blood4,5. This concept was extended by the paradigm of M1 or M2 macrophages [G], which proposed that macrophages in a ground-state of M0 can be driven to either M1 or M2 states of activation following exposure to defined cytokines6. Although this model has generated robust in vitro data, the in vivo reality is of much greater nuance and complexity than can be accommodated by a simple model with only two polarized states of activation7–9.
Different types of macrophage are now known to arise from distinct cell lineages emerging at different stages in embryonic development10–12. Blood monocytes are replenished from the bone marrow throughout life, and these cells can also invade and populate various organs as monocyte-derived macrophages upon specific recruitment. By contrast, tissue-resident macrophages are specific lineages derived from fetal stem cells. Finally, data are accumulating to indicate that these different macrophage lineages are programmed to respond divergently to both tissue injury and disease.
Much of the current literature on the metabolism of microorganisms within their host cells has been generated in tissue culture models using cell lines; however, as researchers have started to investigate the metabolism of host macrophages and pathogens in vivo it is becoming clear that the simplicity of these models has become a limitation. The development of dual RNA-sequencing [G] (dual RNA-seq) of both host and pathogen at the single cell level and the use of fluorescent bacterial fitness reporters are enabling the in vivo heterogeneity of responses to be investigated in a more holistic manner. In this Review, we summarize our existing knowledge of the metabolism of macrophage sub-populations and the metabolism of intracellular microorganisms. We focus predominantly on bacterial pathogens because they represent the more complete body of available literature, and we highlight the impact of host cell metabolism on pathogen growth or restriction. Finally, we discuss those tools and technologies that are enabling a greater understanding of the immunometabolic interplay between host macrophages and pathogens in in vivo infection models.
Macrophage immunometabolism
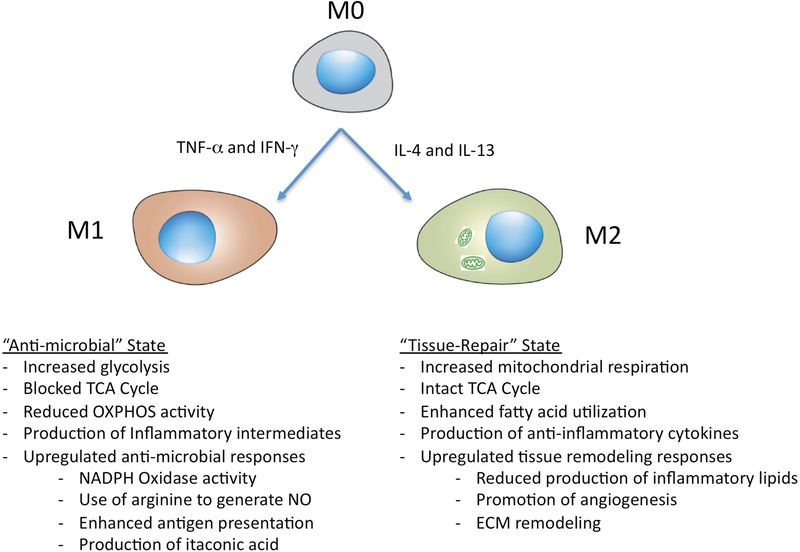
Immunometabolism — in particular, the metabolic reprogramming of macrophages — is now established as a rapidly growing field in immunology2,13–15. The key metabolic and functional differences between the M1 and M2 macrophage polarization states in vitro are summarized briefly in FIG. 1.
Figure 1 |. Phenotypic characteristics of the M1-like and M2-like macrophage polarization states.
The M1-like and M2-like macrophage polarization states are an in vitro paradigm that highlights the pro-inflammatory or anti-inflammatory phenotype of these cells but oversimplifies the highly specialized, transcriptomically dynamic and extremely heterogeneous nature of macrophages in vivo. In brief, M0 macrophages are activated to an M1-like macrophage status by exposure to IFNγ and/or tumour necrosis factor (TNF), or LPS, whereas M2-like macrophages are induced through treatment with IL-4 and/or IL-13. M1-like macrophages are more glycolytically active and markedly upregulate activities that are associated with microbicidal responses. By contrast, M2-like macrophages have increased mitochondrial respiration and enhanced fatty acid metabolism and upregulate activities that are associated with tissue remodeling or wound healing. ECM, extracellular matrix; NO, nitric oxide; OXPHOS, oxidative phosphorylation; TCA, tricarboxylic acid cycle.
M1 macrophages, which are induced by stimulation with tumour necrosis factor (TNF) or IFNγ, have marked upregulation of glycolytic metabolism for generating ATP to support phagocytic and microbicidal functions while feeding an interrupted mitochondrial tricarboxylic acid cycle [G] (TCA cycle). This blocked TCA cycle drives the accumulation of citrate16, itaconate17 and succinate16. Lipopolysaccharide (LPS) stimulation drives the downregulation of expression of the TCA enzyme isocitrate dehydrogenase (IDH), which leads to the production of excess citrate to support fatty acid synthesis and the generation of pro-inflammatory lipid intermediates. Downregulation of IDH also promotes the accumulation of citrate-based precursors to fuel the production of itaconate, which ultimately inhibits the succinate dehydrogenase complex [G] (SDH complex) in mitochondria. To compensate for the decreased expression of IDH, LPS-activated host cells increase glutamine metabolism for the production of the key metabolic intermediates, α-ketoglutarate and succinate. α-ketoglutarate sustains cellular amino acid synthesis and the excess succinate perturbs immune function by stabilizing hypoxia-inducible factor 1α (HIF1α) to promote the production of the pro-inflammatory cytokine IL-1β. In addition, these cells also have increased activity of the pentose phosphate pathway that is needed to convert glucose-6-phosphate into nucleotides and proteins while generating NADPH, which supports lipid biosynthesis as well as the production of reactive oxygen species (ROS) and reactive nitrogen intermediates. These macrophages have an enhanced antimicrobial response, including up-regulation of the superoxide burst generated by the NADPH oxidase complex3, the generation of reactive nitrogen intermediates through inducible nitric oxide synthase (NOS2)18 and increased production of antimicrobial effectors such as cationic peptides, lysosomal hydrolases and metabolic inhibitors19, including itaconate (antibacterial)17 and 25-hydroxycholesterol (antiviral)20.
By contrast, M2 macrophages, which are induced by stimulation with IL-4 and IL-13, have an intact TCA cycle and enhanced mitochondrial oxidative phosphorylation (OXPHOS) activity. Whereas it was previously thought that this metabolic process was mainly fueled by fatty acid oxidation (FAO), it is now understood that glucose also fuels OXPHOS in M2 macrophages21. Multiple studies have shown that the perturbation of key metabolic enzymes in these pathways by either small molecule inhibitors or genetic manipulation can markedly affect the major immune functions of macrophages and other myeloid cells (recently reviewed in REF.15). The M2 macrophage state is frequently associated with the generation of anti-inflammatory products such as glucocorticoids, IL-10, IL-13 and colony-stimulating factor 1 (CSF1; also known as M-CSF), but it is an oversimplification of their complex biological properties to view these cells as a polar counterpart of the M1 macrophage activation state22.
The emerging field of immunometabolism has particular relevance to the study of intracellular infections. Many microbial pathogens are phagocytosed by macrophages early in the infection cycle. Although most microorganisms are killed by phagocytosis and fusion with acidic lysosomes in host cells, some professional pathogens have evolved to survive and even grow in the apparently hostile environment within phagocytes. For these microorganisms, particularly those that maintain a stable relationship with host cells, the coupled metabolism between the host and pathogen is of considerable importance. Thus, the metabolic divergence of the different macrophage lineages will affect pathogen survival and growth, and is likely to be a key factor in determining disease control or progression.
Pathogen metabolism in macrophages
The greatest body of data available with regards to host-pathogen metabolism refers to bacterial pathogens, so although we include discussion of eukaryotic pathogens where appropriate, this article emphasizes bacteria as illustrative examples. It also focuses predominantly on the central carbon metabolism of host and pathogen because the relationship between immunometabolism and macrophage ontogeny is fundamental to a full appreciation of how pathogens maintain their infection within heterogeneous macrophage populations in vivo. The major metabolic pathways that are favored by a range of eukaryotic and prokaryotic pathogens of macrophages are summarized in TABLE 1. Disease progression is invariably linked to growth of the pathogen population, but the persistence of microorganisms in a non-replicative, but metabolically active, state is also required for the maintenance of chronic infections by many intracellular pathogens. Therefore, it should be noted that the optimal conditions for intracellular growth of a pathogen detailed in TABLE 1 do not necessarily describe the only metabolic state that is relevant to infection and disease.
Table 1.
Modulation of host macrophage metabolism through pathogen-associated molecular patterns and bacterial effector proteins
| Pathogen | Intracellular niche | Primary intracellular carbon source | Modulation of host cell metabolism | Differential survival in macrophage lineages | Refs |
|---|---|---|---|---|---|
| Protozoa | |||||
| Leishmania spp. | Vacuolar | Glucose essential in vivo but can also use amino acids | Arginine deprivation response in host macrophages | Preferential growth in tissue-resident macrophages | 113,139–141 |
| Trypanosoma cruzi | Cytosolic | Strongly glucose dependent | Enhanced glycolysis | ND | 138,142,143 |
| Toxoplasma gondii | Vacuolar | Flexible, co-metabolizes glucose and glutamine | Innate immune activation through mTOR | Differential growth linked to macrophage polarization | 144–147 |
| Fungi | |||||
| Candida albicans | Vacuolar | Glucose, through glycolysis and gluconeogenesis | Innate immune activation | ND | 148,149 |
| Bacteria | |||||
| Listeria monocytogenes | Cytosolic | Glucose and glucose-6-phosphate | Innate immune activation and induction of specific effectors to reduce mitochondrial function | Type I interferon enhances pathogen spread | 46,75,80,150 |
| Shigella flexneri | Cytosolic | Pyruvate and glucose | Innate immune activation and induction of specific effectors to enhance glycolysis | ND | 151–153 |
| Salmonella enterica serovar Typhimurium | Vacuolar | Generalist | Innate immune activation and induction of specific effectors | Persister phenotype enhanced by type 1 immune responses | 27,85,89,92 |
| Coxiella burnetti | Vacuolar | Bipartite metabolism | Innate immune activation and induction of specific effectors to modulate lipid homeostasis | Tissue-resident macrophages highly susceptible to infection | 154–157 |
| Brucella spp. | Vacuolar | Glucose, through glycolysis | Enhanced glycolysis | ND | 76 |
| Legionella pneumophila | Vacuolar | Bipartite metabolism with serine preference | Innate immune activation and induction of specific effectors to reduce mitochondrial function | ND | 54,79,158 |
| Mycobacterium tuberculosis | Vacuolar | Cholesterol and fatty acids | Innate immune activation and induction of specific effectors to modulate host cell response | Growth sustained best in resident alveolar macrophages | 40,42,107 |
mTOR, mechanistic target of rapamycin; ND, not determined.
Many enteric bacteria exhibit carbon catabolite repression, which is a regulatory mechanism to restrict metabolism to a single, preferred carbon source, such as glucose. It is suggested that this enables bacteria living in a mixed population to outcompete their neighbors23. However, intracellular bacteria are not under the same evolutionary pressure as enteric bacteria owing to their relatively sterile intracellular environment. As shown by the following examples, these intracellular pathogens usually have an increased capacity to co-metabolize different carbon sources, which provides greater metabolic flexibility24.
This section provides a brief insight into the diverse metabolic solutions that have evolved amongst pathogenic bacteria to enable them to survive and multiply within host macrophages. The solutions are complex and some bacteria rigorously compartmentalize their use of nutrients, whereas other bacteria retain extensive metabolic flexibility. An understanding of the metabolic state of the pathogen is required to appreciate the complexity of the interplay with different macrophage subsets and with an evolving immune environment. We believe that the challenges facing host cells in terms of limiting the growth of these pathogens reflect the diverse metabolism of these infectious microorganisms.
Salmonella Typhimurium.
Salmonella enterica serovar Typhimurium can invade and disseminate systemically in its hosts. In tissues, these bacteria tend to reside and proliferate in tissue-resident macrophages, although they can invade and grow in many different cell types25. Inside the cell, S. Typhimurium remains predominantly intravacuolar but remodels the intracellular compartment into an extensive, filamentous network26. Metabolically, intracellular S. Typhimurium seems to be a ‘generalist’ with an extremely flexible metabolic capacity, such that few single mutations in metabolic pathways have a major effect on bacterial survival27,28.
Legionella pneumophila.
L. pneumophila also remains within a membrane-bound compartment in its host cell29, and, similarly to S. Typhimurium, it can infect several different cell types in vivo. Different isotope and carbon flux analyses indicate that L. pneumophila has a bipartite metabolism [G] using serine as a major carbon source, while using glycerol and glucose for anabolic processes30,31. In tissue culture models, L. pneumophila has two differential growth phases, with an exponential growth phase that depends on serine32 and a post-exponential phase that relies more heavily on glucose and glycerol33.
Mycobacterium tuberculosis.
Similarly to S. Typhimurium and L. pneumophila, M. tuberculosis also remains predominantly intravacuolar. The bacterium can escape from the vacuole, but this transition culminates with host cell death in tissue culture model systems34–36. Access to the cytosol depends on the ESX-1 bacterial secretion system and induces a type I interferon response in the host cell37. Early genetic studies identified the glyoxylate shunt enzyme isocitrate lyase38 and the bacterial cholesterol transporter Mce439 as being important for the establishment and maintenance of intracellular M. tuberculosis infection. An extensive empirical screen for compounds that blocked the growth of intracellular M. tuberculosis identified inhibitors of the bacterial cholesterol degradation pathway40. Recent data indicate that cholesterol and fatty acid breakdown are balanced, which suggests that there is complex regulation of how the bacterium uses these substrates within the host cell environment41,42. Finally, M. tuberculosis has also been shown to use glucose43 and C3 glycolytic substrates44 to sustain in vivo and intracellular infections.
Listeria monocytogenes.
L. monocytogenes, which is the canonical ‘cytosolic’ pathogen, escapes from its vacuole rapidly following uptake by phagocytosis. Studies with L. monocytogenes have shown that the host cell cytosol can be nutritionally restrictive and that the bacterium requires specific co-factors, such as lipoate, for a functional pyruvate dehydrogenase complex45. In this cellular compartment, L. monocytogenes also seems to operate a bipartite metabolism, using glycerol predominantly for energy generation and glucose-6-phosphate for anabolic functions46. The bacteria metabolize pyruvate and lactate poorly, despite their apparent abundance in the cytosol.
Metabolic restriction of pathogens
In addition to the direct antimicrobial mechanisms of macrophages, such as the superoxide burst, generation of ROS, autophagy and production of antimicrobial peptides (not discussed here), macrophages also control microbial growth through nutrient limitation. The concept of nutritional immunity was first proposed in 1973 in reference to iron sequestration as a means of restricting bacterial growth in vivo47. Since then, this mechanism has been found to be relevant in many different microbial infections. In addition to iron sequestration, restricted access to other metal ions such as Zn2+, Cu2+and Mn2+ can also reduce microbial fitness and growth48–51. Most intracellular bacteria are auxotrophic [G] for purines, pyridines and/or amino acids52–55, which —coupled with the known metabolic restrictions of immune cells56 — indicates an obvious link between host cell metabolism and the growth limitation of intracellular pathogens. In pivotal early experiments, IFNγ-induced degradation of host cell tryptophan was shown to restrict the growth of Toxoplasma gondii57. The enzyme involved, indoleamine 2,3-dioxygenase (IDO), has been shown to have a role in inhibiting growth of several intracellular pathogens, including M. tuberculosis and Chlamydia trachomatis 58.
One of the more intriguing pathways that controls the growth of intracellular pathogens involves the production of itaconate. Immunoresponsive gene 1 (Irg1) encodes an enzyme that produces itaconate from cis-aconitate, a citrate-derived intermediate in the TCA cycle17 (the biology of which is discussed extensively in a companion Review59). In addition to its activities as a mitochondrial SDH inhibitor and an immunomodulator60–62, itaconate is a potent inhibitor of bacterial isocitrate lyase activity. Isocitrate lyase is required to facilitate the retention of carbon from fatty acids through the glyoxylate shunt38,63 in many Gram-negative bacteria. In M. tuberculosis, the enzyme also functions as a methyl-isocitrate lyase, which is required for processing propionyl-CoA generated from cholesterol breakdown, and inhibition of the enzyme leads to intoxication of the bacteria64,65. Several intracellular microorganisms, including M. tuberculosis, S. Typhimurium17 and L. pneumophila66 have reduced virulence and growth when exposed to itaconate, and IRG1-deficient mice have markedly increased susceptibility to M. tuberculosis infection67. Moreover, several pathogenic bacteria encode a pathway for the detoxification of itaconate through transfer of a CoA group from a succinyl-CoA donor, which indicates that at least some pathogens can counteract this host-derived metabolic poison68.
Metabolic reprogramming by pathogens
Infection of macrophages by microbial pathogens results in marked changes to the physiology and metabolism of host cells. Some of these changes are mediated by the innate immune response of host cells, which detects and responds to pathogen-associated molecular patterns (PAMPs) through pattern-recognition receptors (PRRs) and generates an inflammatory response against infection (FIG. 2). However, other changes to host cell metabolism are mediated by pathogen-derived effectors, or ‘virulence factors’, that have evolved to modulate host cell function in a targeted, specific manner. Both types of change affect the outcome of the infection process.
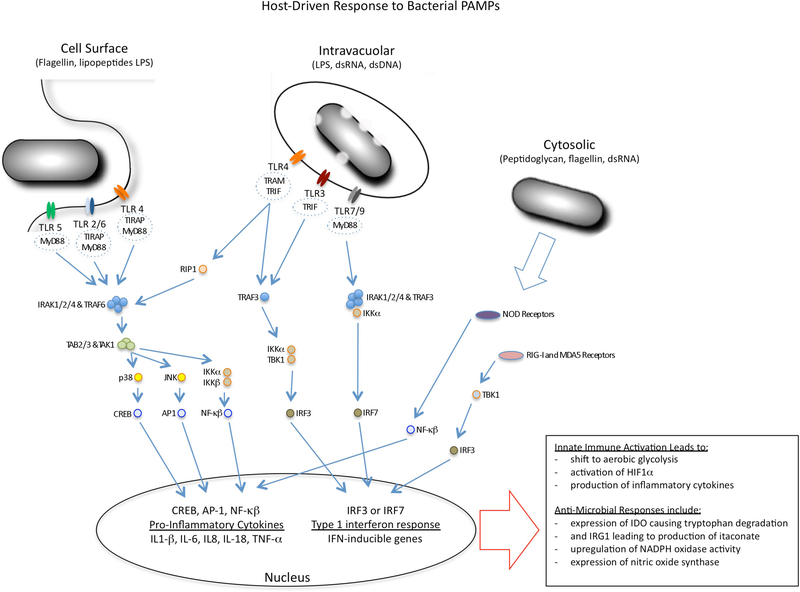
Figure 2 |. Modulation of macrophage metabolism by microbial mediators.
Microorganisms modulate the metabolic status of macrophages through the activation of innate immune signalling pathways or through the use of specific effectors, known as ‘virulence’ factors. The figure summarizes the major pattern recognition receptors (PRRs) found at the cell surface of macrophages and inside the cells, together with key components of their signalling pathways. Microorganisms activate Toll-like receptor (TLR) signalling pathways in host cells through their nucleic acids and cell wall constituents — known as pathogen-associated molecular patterns (PAMPs) — which drive macrophages into a state of enhanced glycolysis and increased production of lactate. The TLRs sense their ligands within the endosomal network as well as at the cell surface. They use different adaptor proteins that can modulate the outcome of TLR ligation, although the dominant responses are linked to either the production of pro-inflammatory cytokines through the activation of nuclear factor-κB (NF-κB) or the induction of a type I interferon response through interferon regulatory factor 3 (IRF3) and IRF7. PAMPs that are delivered into the cell cytosol by bacterial secretion systems activate signalling pathways through NOD-like receptors (NLRs), retinoic acid inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), with similar metabolic and cellular outcomes. The metabolic shift in the macrophages is accompanied by the upregulation of antimicrobial responses, including expression of indoleamine 2,3-dioxygenase (IDO) to limit tryptophan availability, production of itaconate through the activity of immunoresponsive gene 1 (IRG1) to intoxicate bacterial metabolism, and enhanced production of reactive oxygen and nitrogen species. ds, double-stranded; HIF1α, hypoxia inducible factor 1α; IKK, inhibitor of NF-κB kinase; IRAK, IL-1 receptor-associated kinase; LPS, lipopolysaccharide; MYD88, myeloid differentiation primary-response gene 88; NOS2, inducible nitric oxide synthase; TBK1, TANK-binding kinase 1; TIRAP, Toll/IL-1R (TIR)-domain-containing adaptor protein; TRAF, TNF receptor associated factor; TRAM, TRIF-related adaptor molecule; TRIF, TIR-domain-containing adaptor protein inducing IFNβ.
Mediated by PAMPs.
Mammalian cells in resting state generally have low levels of catabolic and anabolic activity and efficient generation of ATP through mitochondrial respiration (OXPHOS)69, by engaging a complete TCA cycle. By contrast, proliferating cells, including cancer cells, exhibit a shift to aerobic glycolysis, known as the Warburg effect, which involves increased uptake of glucose, the energetically inefficient generation of ATP and accumulation of by-products of glycolysis and fermentation70.
Mammalian cells have both surface and cytosolic sensors — for example, PRRs such as Toll-like receptors (TLRs) and NOD-like receptors (NLRs) — that respond to a range of microbial products from cell wall components (LPS and peptidoglycan) to modified DNA and RNA (dsRNA and CpG), and the stimulation of these sensors drive cells into a Warburg-like response16,71–73. The Warburg shift is closely associated with fueling many of the antimicrobial responses that are generated following the acquisition of an M1-like macrophage phenotype. However, there are significant differences in metabolism between proliferating cancer cells and LPS-stimulated macrophages14. Cancer cells use glucose-6-phosphate to fuel both lipid and nucleotide synthesis, and they metabolize pyruvate to support amino acid and fatty acid synthesis for cell growth. By contrast, the inflammatory response induced by TLR ligands such as LPS yields lactate and results in the generation of nitric oxide, ROS, prostaglandins and itaconate, all of which are linked to the antimicrobial activities of phagocytes (as detailed above for itaconate). So, although cancer cells and infected macrophages are superficially similar with respect to increased glycolysis, the downstream consequences of their metabolic reprogramming are disparate.
In addition, a recent study used human monocytes to show that the metabolic switch induced by stimulation with whole-pathogen lysates from Escherichia coli, Staphylococcus aureus and M. tuberculosis, and with the TLR2 ligand Pam3CysSK4, was not comparable to that induced by stimulation with the TLR4 ligand LPS74. Measurements of the oxygen consumption rate [G] (OCR) and the spare respiratory capacity [G] (SRC) indicated that whereas all stimuli induced an increased OCR and therefore increased glycolysis, only LPS triggered a reduction in the SRC and therefore OXPHOS. These data indicate that different stimuli that induce an M1-like macrophage phenotype are not necessarily equivalent and that the metabolic response(s) to such stimuli need to be assessed individually.
Glycolytically active macrophages differentially support pathogen growth. M1-like macrophages have increased microbicidal capacity, but some pathogens benefit from the enhanced glycolytic activity. For example, replication of intracellular L. monocytogenes is enhanced by the increased glucose uptake in primary macrophages that is triggered upon infection. Interestingly, this increased growth rate of L. monocytogenes was not observed in J774A.1 cells. However, J774A.1 cells are a macrophage-like neoplastic cell line that constitutively expresses MYC and, therefore, these cells are already shifted to a Warburg-like metabolism prior to infection75. These data raise a note of caution for investigators using transformed cell lines to study the metabolism of intracellular pathogens. Similarly, infection of human macrophage-like THP-1 cells with Brucella abortus also triggers a shift to aerobic glycolysis [G] and increased lactate production76. B. abortus can use lactate as a sole carbon and energy source, and the bacterium grows robustly in THP-1 cells; blocking host glycolysis and lactate dehydrogenase using chemical inhibitors resulted in a marked decrease in pathogen number76. In addition, infection with B. abortus leads to a reduction in mitochondrial function and a metabolic shift away from OXPHOS towards aerobic glycolysis, which presumably increases lactate production in host cells.
An emerging concept of direct relevance to the metabolic reprogramming of macrophage lineages by microorganisms in vivo is that of ‘trained immunity’ (BOX 1). The induction of a trained immune response by the fungal cell wall component p-glucan involves a shift from OXPHOS to glycolysis mediated through the AKT-mechanistic target of rapamycin (mTOR) signalling pathway and HIF1α77. Moreover, the epigenetic programming of these cells is modulated by the metabolic intermediates acetyl-CoA, NAD, α-ketoglutarate and succinate, all of which are extensively altered by macrophage lineage and polarization, and are directly relevant to immune-mediated and vaccine-induced pathways of control78. The major themes and salient points of PAMP-induced conditioning of phagocyte populations are summarized in BOX 1.
Box 1 |. Trained immunity.
Recent data indicate that innate immune cells can develop memory-type responses that intensify their recall response following an initial exposure to a pathogen. This phenomenon, termed ‘trained immunity’, has been reported in myeloid cell lineages and is thought to result from epigenetic modification and cellular metabolic rewiring. Compared with the classical immunological memory that is a feature of the adaptive immune system, trained immunity has reduced specificity as re-stimulated cells respond more robustly to a broader range of secondary stimuli. Two well-documented stimuli that trigger a trained immune response in macrophages are β-glucan of Candida albicans and Mycobacterium bovis bacillus Calmette-Guérin (BCG). The trained immune response induced by both agonists activates the AKT-mTOR (mechanistic target of rapamycin) pathway, triggers a shift of cellular metabolism towards glycolysis and promotes genome-wide changes in histone modification77,127. More recently, mevalonate, which is an intermediate of the cholesterol synthesis pathway, has been shown to promote trained immunity through mTOR activation128,129. It is clear that both intrinsic cellular metabolic alteration and sustained epigenetic reprogramming are required to establish trained immunity. Furthermore, β-glucan and BCG both also stimulate myelopoiesis at the level of haematopoietic stem cells129,130. These results suggest that trained immune reprogramming endures and therefore has considerable potential as a novel route to the correction or realignment of ineffective immune responses. A more comprehensive understanding of trained immunity as the product of a complex interaction between epigenetics, cellular metabolism and signalling networks, may provide a novel avenue to improve disease therapeutics and vaccines.
Mediated by specific microbial effectors.
In addition to the modulation of macrophage metabolism through the activation of mammalian PRRs, some pathogens have evolved more direct mechanisms to modulate the metabolism of host cells.
L. pneumophila causes pulmonary infections and replicates within human lung macrophages. The bacterium encodes a type IV secretion system [G] that can inject more than 300 bacterial effector proteins into host cells29, which carry out a wide range of overlapping and apparently redundant activities. One feature of infected macrophages is the close apposition between L. pneumophila-containing vacuoles and mitochondria. A recent study reports that the subsequent fragmentation of these vacuole-associated mitochondria depends on the bacterial effector protein MitF, which is released into host cells79. Mitochondrial fusion and fission [G] are tightly regulated processes in mammalian cells that are mediated in part through the activity of the GTPase dynamin 1-like protein (DNM1L; also known as DRP1), and the activity of this host enzyme is increased during infection with L. pneumophila. The resulting increased fragmentation of mitochondria leads to reduced mitochondrial respiration and increased host cell glycolysis, and, as a result, increased growth of bacteria within the host cell.
The specific targeting of mitochondrial function is not unique to L. pneumophila. For example, L. monocytogenes uses its toxin Listeriolysin O (LLO) to escape from the phagosome and access the host cell cytosol, where LLO targets host cell mitochondria and induces their fragmentation80. Interestingly, in contrast to the mitochondrial fragmentation induced by L. pneumophila, this phenomenon is transient in L. monocytogenes-infected cells, leading to decreased cellular respiration without the activation of cell death pathways. It is thought that mitochondrial fragmentation is required for optimal early infection with L. monocytogenes as infection is impaired in cells with previously fissioned mitochondria and is enhanced in cells with mitochondria that were already fused80.
Macrophage heterogeneity in vivo
The data discussed so far were generated predominantly using in vitro infection of tissue cultures consisting of relatively homogeneous cell populations, comprising either macrophage-like cell lines or primary cells such as human monocyte-derived macrophages matured from peripheral blood monocytes, or murine bone marrow-derived macrophages. Although it is understandable and necessary to have started with such homogeneous challenge models to establish what is possible, the extrapolation of in vitro data to the heterogeneous nature of the in vivo environment presents many challenges. To do so requires a capacity to detect and assess the functional relevance of heterogeneity, and there are two main routes to this that researchers have explored so far. The first is through the generation of fitness reporter organisms [G] (BOX 2) that provide information regarding the metabolic and replicative state of the pathogen in a specific host cell or host tissue environment81–88. The second is single cell transcriptional profiling, specifically dual RNA-seq methodology, that enables simultaneous transcriptional profiling of host and pathogen89–91. These two, mutually informative methods are still in development and, so far, they have only been applied in combination to models of infection with S. Typhimurium and M. tuberculosis. Both of these pathogens can result in persistent infections that are thought to be maintained by bacteria that are live but non-replicative, with reduced metabolic activity86,92,93. Furthermore, the acquisition of this ‘persister’ phenotype is closely associated with the development of drug tolerance, resulting in the reduced effectiveness of antibiotics in vivo83,94–96. Therefore, understanding the physiological states of host macrophages and the infecting bacteria during persistence and disease progression is key to the development of new effective strategies for treatment. The different biological phenotypes that have been documented in in vivo infection models for S. Typhimurium and M. tuberculosis are illustrated in FIG. 3 and FIG. 4, respectively.
Box 2 |. Tools to understand the metabolic crosstalk between host and pathogen.
The integration of these tools and technical platforms is presented graphically in FIG. 5.
Microbial fitness reporter strains
There are many types of fluorescent fitness reporter that have been generated in bacterial strains. These include ribosomal RNA correlates [G]96, inducible GFP dilution85,131 TIMER fluorescence83,132, chromosomal replication complex reporters 88,107,133 environmentally responsive promoter reporters88,107,109,134 and pH-sensitive GFP [G]135,136. The value of these reagents is that they enable the status of a pathogen to be linked spatially to the host macrophage population. This allows analysis and quantitation by confocal microscopy, phenotypic analysis of surface markers on host cells and cell sorting by flow cytometry for RNA-sequencing (RNA-seq) analysis or metabolic profiling.
Animal models
An animal model should recapitulate as much of the biology of the infection site of human disease as is possible. All models have caveats but we believe that a model that generates functionally relevant heterogeneity of host cell populations in vivo has considerably greater biological relevance than the in vitro infection of homogeneous transformed mammalian cell lines.
Flow-sorting capacity
To interrogate specific host–pathogen pairings with respect to infection control and progression requires sufficient flow-sorting capacity under appropriate biosafety conditions to discriminate macrophage lineages and microbial fitness phenotypes.
Single-cell RNA-seq and dual RNA-seq
The limited coverage of single-cell sequencing can be experimentally challenging, but this is an advancing field and the technologies are improving with extraordinary rapidity. Some platforms, such as the Seq-Well platform, can readily be used under biosafety level 3 (BSL3) conditions and require no specialized equipment137. Dual RNA-seq has the added advantage that the transcriptional profiles of host cells and bacteria can be directly integrated, and this has been carried out at both population level and for single cells89,90.
Metabolic profiling
Metabolic profiling through the quantitation of key intermediates is readily achievable, and live profiling of metabolic activity can be assessed using Agilent’s Seahorse platform138.
Chemical inhibitors
There are several chemical inhibitors of metabolic pathways of immune cells such as macrophages that have been invaluable in the manipulation of cell physiology to probe immune phenotype15. These compounds, together with new inhibitors of specific bacterial pathways40, provide new tools for modulation of the host–pathogen interface.
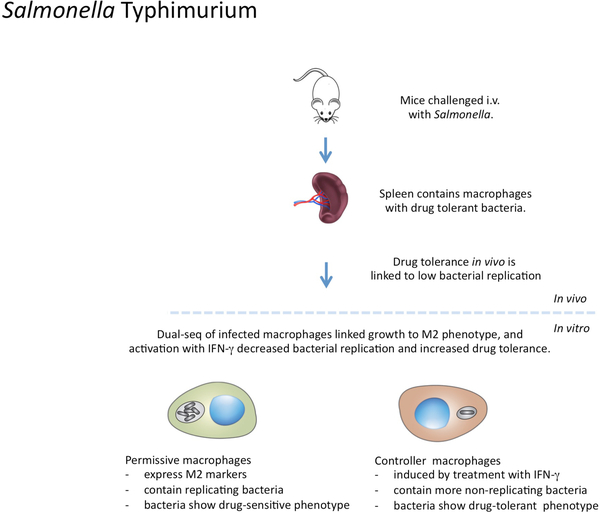
Figure 3 |. The links between macrophage phenotype and Salmonella Typhimurium physiology.
In mice infected with Salmonella enterica serovar Typhimurium, the presence of ‘persister’ bacteria was shown in splenic macrophages83,85. These bacteria were non-replicating and had strong antibiotic tolerance. In vitro experiments established that the induction of a drug-tolerant phenotype was greatly enhanced in bacteria present in macrophages activated by IFNγ85. Single-cell RNA-sequencing of in vitro-infected, bone marrow-derived macrophages indicated that rapidly growing bacteria were found mainly in macrophages expressing markers of M2-like polarization89. These data suggested that the in vivo induction of drug tolerance in S. Typhimurium was most likely due to M1-like macrophages, and that support of bacterial growth was most likely due to macrophages with an M2-like phenotype.
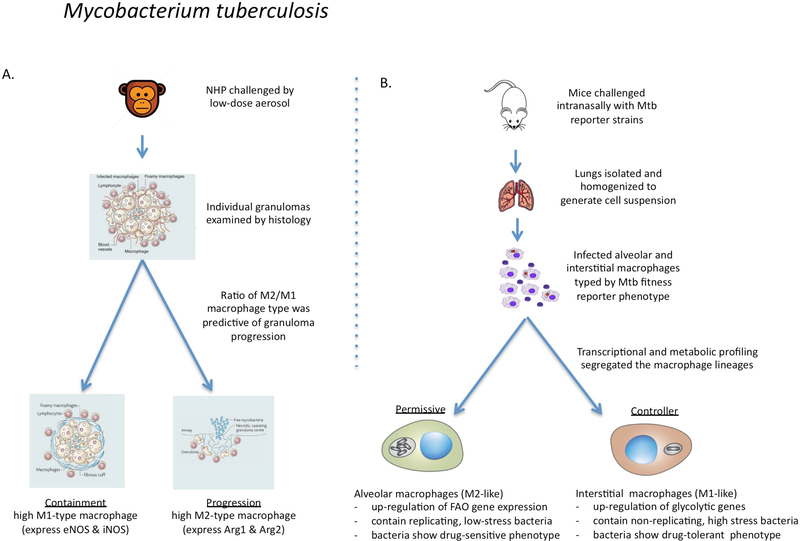
Figure 4 |. The links between macrophage phenotype and the progression of Mycobacterium tuberculosis infection in non-human primates and mice.
a | In macaque monkeys infected with Mycobacterium tuberculosis, the bacterial burden in individual granulomas and progression to active disease were found to correlate directly with an increased ratio of macrophages expressing M2 markers over Ml markers105,106. Progression was determined at the level of the individual granuloma without any consensus at the level of the host. b | In mice infected with fitness and replication reporter strains of M. tuberculosis, resident alveolar macrophages were more permissive of bacterial replication than were recruited interstitial macrophages107. RNA-sequencing showed that the alveolar macrophages were more committed to fatty acid oxidation (FAO) and oxidative phosphorylation, whereas the interstitial macrophages had higher levels of glycolysis. Selective depletion of the alveolar macrophage population reduced the bacterial burden, whereas depletion of the interstitial macrophage population led to an increase in bacterial burden. Therefore, in agreement with the data from non-human primates105,106, the ratio of alveolar (M2-like) macrophages to interstitial (M1-like) macrophages affects bacterial growth and disease progression in mice. In addition, drug-tolerant M. tuberculosis were more abundant in vivo in macrophages expressing M1-like activation markers95.
During S. Typhimurium infection.
S. Typhimurium causes a systemic infection in mice, infecting macrophages in multiple tissues including the Peyer’s patches, mesenteric lymph nodes (MLNs), spleen and liver. A recently developed fluorescence dilution reporter strain [G] to quantify relative rates of bacterial replication has been used to determine the relative replication rates of S. Typhimurium in different tissue environments85 (FIG. 3). This study reported that the proportion of non-replicating bacteria isolated over the first 24 hours post challenge was markedly lower in macrophages isolated from the Peyer’s patches than in those isolated from the MLNs. The viability of these non-replicating bacteria was shown by subsequent in vitro outgrowth. Activation of infected macrophages in vitro with IFNγ led to the generation of a non-replicative bacterial phenotype and to the acquisition of drug tolerance in S. Typhimurium. Moreover, treatment of macrophages with inhibitors of the vacuolar ATPase that is responsible for phagosomal acidification caused a marked decrease in the frequency of the ‘persister’ bacterial phenotype, which indicates that pH-mediated stress is a primary driver of reduced bacterial replication and increased drug tolerance in intracellular bacteria.
Subsequent publications have connected macrophage polarization in vitro to the growth rates of intracellular S. Typhimurium. Mouse bone-marrow-derived macrophages were infected with the fluorescence dilution reporter strain of S. Typhimurium and sorted into populations at the extremes of fluorescence, which represent ‘controller’ host cell populations and ‘permissive’ host cell populations, for single-cell sequencing analysis89. All infected macrophages showed down-regulation of replication markers, which is consistent with previous reports that PAMP-mediated activation of macrophages drives differentiation into a non-replicative state. Infected macrophages containing non-growing bacteria were, by principle component analysis (PCA), most similar to uninfected bystander macrophages. By contrast, macrophages with growing populations of S. Typhimurium had increased expression of M2 macrophage-associated transcripts, such as Arg1, Cd206 and Il4ra, although the analysis was not extended to cellular metabolism. Recent dual-RNA-seq analysis of the splenic macrophages from S. Typhimurium-infected mice confirmed the permissiveness of the M2 host macrophage phenotype in vivo and demonstrated that this correlated with the increased expression of SPI2 effectors in the bacteria in these cells suggesting that the bacteria actively modulate host macrophage polarization97.
Another study used a fluorescent protein variant known as TIMER [G], which spontaneously changes its emission fluorescence from green to orange as a function of age83. By quantifying the fluorescence ratio of the green to orange signal, one can estimate the division rate of individual bacteria in culture. Analysis of the S. Typhimurium fluorescence signal from infected macrophages and in mice confirmed the presence of bacterial populations with differing growth rates that tended to vary between host macrophages, although this was not correlated with the activation state of the host cells. Proteomic analysis showed that more actively replicating bacteria contained higher levels of ribosomal proteins, whereas slower replicating bacteria had higher levels of expression of the transcription factor cAMP receptor protein (CRP), the expression of which is linked to metabolic regulation under starvation conditions98. As reported in other studies, slower replicating bacteria had higher levels of phenotypic tolerance to antibacterial drugs83.
These data suggest that the phenotype of the infecting bacteria, with respect to both growth state and drug sensitivity, is determined predominantly by the polarized state of their host macrophage. Macrophages expressing M1 macrophage-associated markers control bacterial growth and induce drug tolerance, whereas macrophages expressing M2 macrophage-associated markers are more permissive for bacterial growth.
During M. tuberculosis infection.
Researchers have long been aware of the heterogeneity within M. tuberculosis populations during infection (reviewed recently in REF.93), and it is known that M. tuberculosis has evolved hard-wired genetic mechanisms to generate inequality between its daughter cells to maximize the benefits of phenotypic heterogeneity96,99,100. It is thought that this bacterial heterogeneity better equips M. tuberculosis to exploit the environmental diversity found in host cells and host tissues.
Recent studies of early low-dose infection in mice show that M. tuberculosis is internalized by alveolar macrophages that subsequently invade the underlying tissue101. Immunohistology and flow cytometric analysis of infected tissues from mice and non-human primates (NHPs) have shown that M. tuberculosis becomes established in several types of host phagocyte, primarily alveolar macrophages, recruited interstitial macrophages and neutrophils102. The bacteria are also found in dendritic cells, which are known to have a role in the development of an adaptive immune response through the presentation of mycobacterial antigens103 but are not thought to be an important host cell population for bacterial replication104. Extensive analysis of macrophage subsets in individual granulomas of NHPs infected with M. tuberculosis has documented different populations of macrophages that express M1 macrophage-associated markers, such as endothelial nitric oxide synthase (NOS3) and inducible nitric oxide synthase (NOS2), or M2 macrophage-associated markers, such as arginase (ARG1 and ARG2)105 (FIG. 4a). In a disease outcome model based on these data, the authors suggest that an increased ratio of M2 to M1 macrophage subsets is an accurate predictor that an individual granuloma is likely to progress to active disease106. Similarly, when M. tuberculosis-infected macrophages were isolated from mouse lung tissue and flow-sorted into activated (CD80hi) versus resting (CD80low) cells, the bacteria in the activated host macrophages had much higher levels of phenotypic drug tolerance95, which suggests that these bacteria were in a state of low-level replication.
These findings were extended recently to examine the metabolic state induced by M. tuberculosis infection in different mouse macrophage lineages in vivo107 (FIG. 4b). Mice were infected with a panel of M. tuberculosis fitness reporters, including a ‘clock’ plasmid [G]84,108, a chromosomal replication complex reporter [G] and an environmentally responsive promoter reporter [G]84 sensitive to nitric oxide88,109, which showed that alveolar macrophages were more permissive for bacterial replication than monocyte-derived interstitial macrophages. Selective depletion of the alveolar macrophage population by clodronate liposomes delivered to the airways led to a decreased bacterial burden in the lung, whereas depletion of the recruited interstitial macrophages, through intravenous administration of clodronate liposomes or the use of CC-chemokine ligand 2 (CCL2)-deficient mice, led to an increased bacterial burden in the lung. RNA-seq analysis of the infected populations of alveolar macrophages and interstitial macrophages indicated that the two host cell populations had acquired metabolically distinct states. Alveolar macrophages were enriched for transcripts associated with OXPHOS and fatty acid metabolism, whereas interstitial macrophages exhibited a marked Warburg shift. Treatment of M. tuberculosis-infected mice with the non-hydrolysable glucose analogue, 2-deoxyglucose, selectively decreased the size of the interstitial macrophage population, resulting in an increase in the bacterial burden. These findings were verified by in vitro experiments showing that treatment of M. tuberculosis-infected bone marrow-derived macrophages with 2-deoxyglucose promoted bacterial growth, whereas the FAO inhibitor etomoxir restricted bacterial growth. These data indicate that the different host macrophage lineages respond divergently to infection with M. tuberculosis with respect to their metabolic commitment, and that this divergence is relevant to infection control. These data are consistent with the recent report that the reduced number of recruited interstitial macrophages in CC-chemokine receptor 2 (Ccr2)−/− mice upon infection with the highly virulent HN878 strain of M. tuberculosis explains the increased susceptibility of these mice110. The data are also supported by metabolic flux analysis of human macrophages, which shows that cigarette smoking reduces the glycolytic response of macrophages upon M. tuberculosis infection and that this increases bacterial survival111.
The development of vaccines against tuberculosis has been hampered by the lack of biomarkers for immune protection112 and the realization that IFNγ production, while clearly required for protection, is not sufficient to prevent disease progression. It has been observed that M. tuberculosis infection drives bystander macrophage populations at the site of infection into cell division107. This provides a possible alternative explanation for disease development and posits that, even in the face of an effective, controlling T helper 1 (TH1) cell-mediated immune response, the selective expansion of alveolar macrophage populations, given their apparent commitment to OXPHOS, could support bacterial growth.
The observation that tissue-resident macrophages can be driven into replication by infection or inflammatory insult is not unique to M. tuberculosis infection. Similar observations have been made for skin macrophages in a murine cutaneous leishmaniasis model113, whereby infection with Leishmania parasites was maintained by the presence of replicating, M2 macrophage-like resident dermal macrophages, even in the presence of a robust local TH1 cell-mediated response.
These studies indicate that the M2 macrophage-like characteristics of alveolar macrophages provide an environment that is more permissive for the growth of M. tuberculosis, directly associated with the metabolic state of these macrophages and their bias towards FAO. By contrast, and comparable to S. Typhimurium, the M1 macrophage-like characteristics of interstitial macrophages are associated with control of M. tuberculosis growth, but also with the induction of a drug-tolerant phenotype.
Macrophage ontogeny and metabolism
The data from the mouse M. tuberculosis challenge model have broader implications with respect to macrophage biology in infectious disease control. Transcriptional profiling has shown that there are major differences between the embryonically derived, resident alveolar macrophages and the haematopoietically derived interstitial macrophages in the mouse lung114,115. All interstitial macrophages expressed transcripts encoding the tyrosine kinase MERTK, CD64, CD11b and CX3CR1, whereas all alveolar macrophages expressed the macrophage receptor MARCO114,115. The differences between the macrophage lineages were increased markedly in an acute lung injury model116. After treatment of the lungs with LPS to induce inflammation, functional analysis of the resident and recruited lung macrophages was carried out, looking at both transcript abundance and the concentrations of key metabolic intermediates. The resident alveolar macrophages proliferated locally and showed commitment to increased TCA cycle and OXPHOS. By contrast, the recruited macrophages were more inflammatory in phenotype and had increased glycolytic activity and arginine metabolism. These data are consistent with the murine tuberculosis model and indicate that the developmental origin of macrophages rather than the inflammatory immune environment is the dominant factor in determining macrophage phenotype and metabolism.
Helminth infections are known to strongly induce alternatively activated macrophages, and they are recognized drivers of the localized proliferation of resident macrophages117,118. The functional plasticity of macrophage populations has been studied in a co-infection model combining helminth challenge (with Heligmosomoides polygyrus) with S. Typhimurium infection. H. polygyrus is a naturally occurring gastrointestinal nematode of mice that induces a strong type 2 immune response that drives the proliferative expansion and M2-type activation of resident peritoneal macrophages119. Upon subsequent intraperitoneal injection of S. Typhimurium, there was an influx of monocyte-derived macrophages and neutrophils to the peritoneal cavity. This led to a reduction in size of the F4/80hi nematode-expanded, resident macrophage population. The F4/80low inflammatory macrophages recruited by S. Typhimurium were exclusively haematopoietic in origin and were not derived through conversion of the F4/80hi macrophages. The decreased numbers of F4/80hi resident macrophages that remained were reprogrammed by S. Typhimurium to acquire antimicrobial properties, such as expression of NOS2 and increased MHC class II expression, although the extent of macrophage re-polarization was restricted and these cells retained many characteristics that reflected their resident macrophage origin.
Finally, it was interesting to note that the recruitment of the inflammatory monocyte-derived macrophages seemed to be linked to the disappearance of the F4/80hi resident macrophages119. It is thought that this disappearance occurred as a result of induced apoptosis and could be linked to niche competition as a means of regulating the phenotypic characteristics of phagocyte populations in the presence of infectious stimuli.
In addition to the traditional cytokines that are associated with the polarization of macrophages and with driving functional phenotypes (FIG. 1), the transcription factor peroxisome proliferator-activated receptor-γ (PPARγ) is being increasingly recognized as a modulator of lipid metabolism linked to inflammation and immunity, particularly in tissue-resident macrophages. Early evidence indicated that PPARs inhibit proinflammatory responses in macrophages120, but the role of PPARγ seems to be much more nuanced and is influenced by the target macrophage lineage121. PPARγ is required for tissue macrophage-mediated clearance of S. aureus skin infections and for control of Pseudomonas aeruginosa in the lung122, and it is implicated in both the control and progression of M. tuberculosis infection, dependent on the model123–126. Further study of the role of PPARγ in macrophage polarization is required; however, it is clear that this nuclear receptor protein has an important role in the modulation of tissue-resident macrophage function.
Conclusions and future directions
Most of the studies that have been published on the control of microbial infections by macrophages have focused on classical killing pathways, such as the production of ROS and reactive nitrogen intermediates, the up-regulation of expression of antimicrobial peptides, the triggering of autophagy and the sequestration of co-factors. All of these mechanisms are highly relevant to infection control but we believe that these pathways overlay a heterogeneity in macrophage lineages that is developmentally defined, and that these macrophage lineages, through their metabolic commitment, determine how the different phagocyte populations respond to activating cytokines. Understanding how macrophage metabolism interfaces with microbial metabolism, and how this affects the control or progression of infection promises to be fascinating. Existing data from in vitro and in vivo infections with S. Typhimurium and M. tuberculosis link the phenotypes of tissue resident (M2-like) macrophages with bacterial growth, and the phenotypes of recruited inflammatory (M1-like) macrophages with control of bacterial growth, but also with the induction of a drug tolerance programme in the bacteria that is linked to bacterial persistence.
Currently, there is a disconnect between the research community studying the metabolism of intracellular pathogens and those immunologists who appreciate the relevance of macrophage ontogeny. It is important to understand the metabolic status of host cells and how this can be manipulated by pathogens trying to establish their optimum growth conditions. However, the homogeneous populations of transformed mammalian cell lines that are used routinely for these studies are not an accurate reflection of the in vivo situation. Given the central role of macrophages in most microbial infections, it is important to appreciate the functions of the specific macrophage lineages and populations that determine the outcome of infection in vivo.
Our research on M. tuberculosis infections in tissue culture models and in mice has increased our appreciation of the value of certain tools (BOX 2) and our awareness of the information that needs to be generated to interrogate the macrophage-pathogen interface effectively. The iterative interactions of these tools and technical platforms are illustrated in FIG. 5. Integration of these tools will be required to cement the emerging links between host macrophage ontogeny and metabolism and the growth and replication states of intracellular pathogens. Tools and techniques — such as fluorescent fitness reporters, dual RNA-seq of host and pathogen mRNA and appropriate animal models — are already available for several microbial pathogens, and it is likely that data from these infection models will continue to accumulate. Such an abundance of tools and models has never before been available to study biochemistry and cellular metabolism, nor with such intriguing problems to address. It is likely that an increased appreciation of macrophage diversity will be crucial to the development of more effective vaccines, and more efficacious drug combinations that might target both host and pathogen.
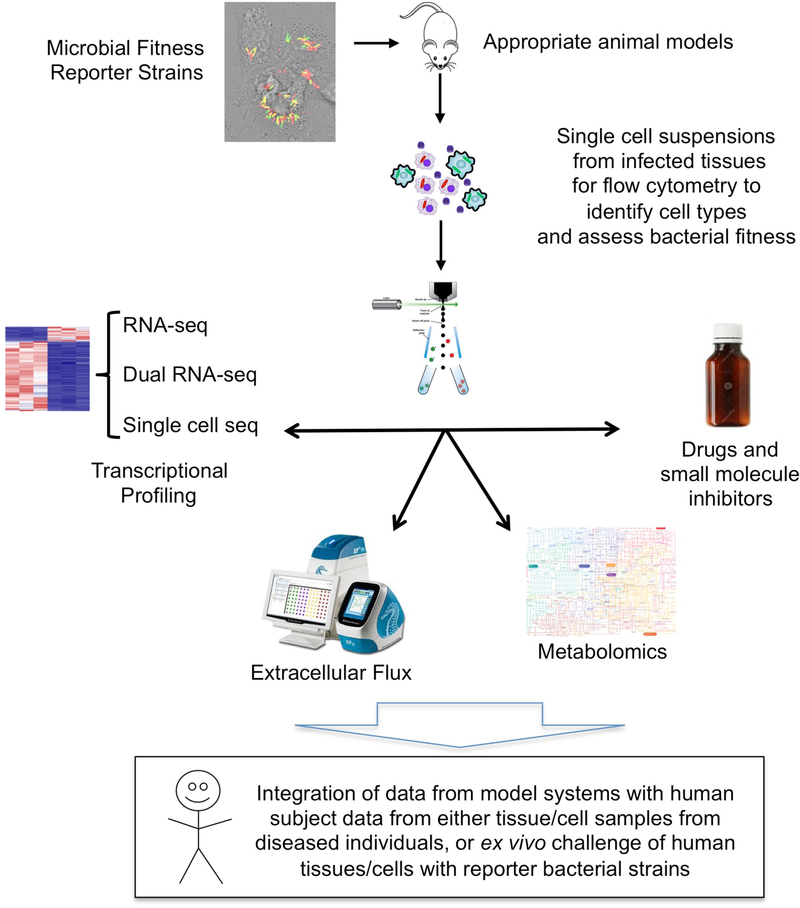
Figure 5 |. Technologies and tools for investigating the metabolic interplay between host and pathogen.
The figure illustrates how fluorescent microbial fitness and replication reporter strains (BOX 2) can be used to infect appropriate animal models to generate in vivo heterogeneity of relevance to disease progression or control. Infected tissues are analyzed by the generation of single cell suspensions for flow cytometry to yield cell populations defined by immune markers and bacterial fitness profiles. These cell suspensions can be sorted according to the host or microbial readout of relevance and then assessed further by various RNA-sequencing protocols to define the transcriptomes of both host and pathogen, and by metabolic profiling through the analysis of intermediates or through metabolic flux studies. Small molecule inhibitors or drugs specific for host or microbial targets can be used to perturb the system and test hypotheses relevant to the metabolic interface between host cells and pathogens. Ultimately the data generated in the model systems need to be integrated with data from actual human disease to test its validity.
Acknowledgements
D.G.R., L.H. AND B.C.V. WOULD LIKE TO GRATEFULLY ACKNOWLEDGE THE SUPPORT OF THE NATIONAL INSTITUTES OF HEALTH, USA, AND THE BILL AND MeLinda Gates Foundation.
Glossary
- Foamy macrophages
Macrophages with cytosolic lipid droplets containing cholesterol, cholesterol ester and triacylgycerol, which are frequently induced by chronic pro-inflammatory stimuli
- Ml or M2 macrophages
Ml’ and ‘M2’ are classifications historically used to define macrophages activated in vitro as pro-inflammatory (when ‘classically’ activated with IFNγ and LPS) or anti-inflammatory (when ‘alternatively’ activated with IL-4 or IL-10), respectively. However, in vivo, macrophages are highly specialized, transcriptomically dynamic and extremely heterogeneous with regards to their phenotypes and functions, which are continuously shaped by their tissue microenvironment. Therefore, the Ml or M2 classification is too simplistic to explain the true nature of in vivo macrophages, although these terms are still often used to indicate whether the macrophages in question are more pro- or anti-inflammatory
- Dual RNA-sequencing
(Dual RNA-seq). A transcriptional profiling technique that enables simultaneous acquisition of the expression levels of mRNA transcripts in both the host cell and the pathogen
- Tricarboxylic acid cycle
(TCA cycle). Also known as the citric acid cycle or Krebs cycle. This is a series of enzymatic reactions used in aerobic metabolism to release energy through the oxidation of acetyl-CoA to yield ATP and carbon dioxide
- Succinate dehydrogenase complex
(SDH complex). An enzyme complex found in bacterial cells and in the inner mitochondrial membrane of eukaryotic mitochondria that is active in both the TCA cycle and the electron transport chain
- Bipartite metabolism
A metabolic programme whereby one carbon source is used exclusively as an energy supply, while another carbon source, or sources, are used for anabolic processes
- Auxotrophic
Organisms that are unable to synthesis all of the compounds required for growth are auxotrophic, meaning that they are dependent on their hosts to supply those compounds they cannot synthesize
- Oxygen consumption rate
The total oxygen utilization capacity of a biological system under examination against time
- Spare respiratory capacity
(SRC). The difference in the amount of ATP generated by oxidative phosphorylation at basal rate and at maximal respiratory capacity
- Aerobic glycolysis
The conversion of glucose to lactate under conditions where oxygen is present at nonlimiting concentrations
- Type IV secretion system
An ATP-dependent bacterial transporter complex that is frequently used to inject bacterial effector proteins or bacterial DNA into eukaryotic and prokaryotic target cells
- Mitochondrial fusion and fission
The OXPHOS capacity of eukaryotic cells can be regulated in part through the fusion or fragmentation of mitochondria in a highly controlled manner
- Fitness reporter organisms
Bacterial reporter strains, usually encoding a fluorescent protein-based readout, that are used to assess bacterial fitness with respect to responsiveness to noxious stimuli and replicative capacity
- Fluorescence dilution reporter strain
A bacterial strain that can be induced to transiently express a fluorescent protein, which can then be quantified as it becomes diluted when the bacteria divide to infer bacterial replication rates
- TIMER
A re-engineered red fluorescent protein that undergoes a conformational shift, and hence a change in fluorescence emission wavelength, as it ages thus providing a correlate of replication rates
- ‘Clock’ plasmid
An episomal plasmid encoding an antibiotic-resistance marker that is lost from a bacterial population at a fixed rate directly proportional to the rate of replication
- Chromosomal replication complex reporter
A single-stranded DNA-binding protein-GFP fusion complex that persists for the duration of chromosomal replication and can be used to assess the replication status of a bacterial population
- Environmentally responsive promoter reporter
A dual-fluorescent bacterial reporter strain that expresses one fluorescent protein constitutively and the other fluorescent protein under the control of promoters that are responsive to specific environmental stimuli
- Ribosomal RNA correlates
Sequences encoding a destabilized or short-lived GFP are inserted into ribosomal RNA loci to provide a correlate of ribosomal RNA activity and bacterial replication
- pH-sensitive GFP
A green fluorescent protein derivative that exhibits a shift in fluorescence emission wavelength in a pH-dependent manner
Footnotes
Competing interests
THE AUTHORS DECLARE NO COMPETING INTERESTS.
Publisher’s note
SPRINGER NATURE REMAINS NEUTRAL WITH REGARD TO JURISDICTIONAL CLAIMS IN PUBLISHED MAPS AND INSTITUTIONAL AFFILIATIONS.
Reviewer information
NATURE REVIEWS IMMUNOLOGY THANKS K. FITZGERALD, C. SASSETTI AND OTHER ANONYMOUS REVIEWERCS) FOR THEIR CONTRIBUTION TO THE PEER REVIEW OF THIS WORK.
References
- 1.O’Neill LA & Pearce EJ Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med 213, 15–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van den Bossche J, O’Neill LA & Menon D Macrophage Immunometabolism: Where Are We (Going)? Trends. Immunol 38, 395–406 (2017) [DOI] [PubMed] [Google Scholar]
- 3.VanderVen BC, Yates RM & Russell DG Intraphagosomal measurement of the magnitude and duration of the oxidative burst. Traffic. 10, 372–378 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Furth R & Cohn ZA The origin and kinetics of mononuclear phagocytes. J. Exp. Med 128, 415–435 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Furth R et al. The mononuclear phagocyte system: a new classification of macrophages, monocytes, and their precursor cells. Bull. World Health Organ 46, 845–852 (1972). [PMC free article] [PubMed] [Google Scholar]
- 6.Munder M, Eichmann K & Modolell M Alternative metabolic states in murine macrophages reflected by the nitric oxide synthase/arginase balance: competitive regulation by CD4+ T cells correlates with Th1/Th2 phenotype. J. Immunol 160, 5347–5354 (1998). [PubMed] [Google Scholar]
- 7.Gordon S & Martinez-Pomares L Physiological roles of macrophages. Pflugers Arch. 469, 365–374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sica A & Mantovani A Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest 122, 787–795 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue J et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 40, 274–288 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ginhoux F & Guilliams M Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity. 44, 439–449 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Ginhoux F & Jung S Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol 14, 392–404 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Gordon S & Pluddemann A Tissue macrophages: heterogeneity and functions. BMC Biol. 15, 53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diskin C & Palsson-McDermott EM Metabolic Modulation in Macrophage Effector Function. Front. Immunol 9, 270 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escoll P & Buchrieser C Metabolic reprogramming of host cells upon bacterial infection: Why shift to a Warburg-like metabolism? FEBS J. 285, 2146–2160 2018 [DOI] [PubMed] [Google Scholar]
- 15.O’Neill LA, Kishton RJ & Rathmell J A guide to immunometabolism for immunologists. Nat. Rev. Immunol 16, 553–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jha AK et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 42, 419–430 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Michelucci A et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. U S A 110, 7820–7825 (2013).This study reports the identification of Irgl as encoding an enzyme that generates itaconate, and the finding that itaconate is a potent antimicrobial that can block the growth of intracellular M. tuberculosis and Salmonella spp.
- 18.MacMicking JD et al. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc. Natl. Acad. Sci. USA 94, 5243–5248 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss G & Schaible UE Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev 264, 182–203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cyster JG, Dang EV, Reboldi A & Yi T 25-Hydroxycholesterols in innate and adaptive immunity. Nat. Rev. Immunol 14, 731–743 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Tan Z et al. Pyruvate dehydrogenase kinase 1 participates in macrophage polarization via regulating glucose metabolism. J. Immunol 194, 6082–6089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosser DM & Edwards JP Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol 8, 958–969 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorke B & Stulke J Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol 6, 613–624 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Olive AJ & Sassetti CM Metabolic crosstalk between host and pathogen: sensing, adapting and competing. Nat. Rev. Microbiol 14, 221–234 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Burton NA et al. Disparate impact of oxidative host defenses determines the fate of Salmonella during systemic infection in mice. Cell Host Microbe. 15, 72–83 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Figueira R & Holden DW Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology. 158, 1147–1161 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Bumann D & Schothorst J Intracellular Salmonella metabolism. Cell. Microbiol 19, e12766 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Steeb B et al. Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS. Pathog 9, e1003301 (2013).An extremely comprehensive analysis of the major metabolic pathways that are required to support the survival and growth of Salmonella spp. inside mammalian cells using proteomics, genetics and computational modelling.
- 29.Isaac DT & Isberg R Master manipulators: an update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Future Microbiol. 9, 343–359 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eylert E et al. Isotopologue profiling of Legionella pneumophila: role of serine and glucose as carbon substrates. J. Biol. Chem 285, 22232–22243 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliva G, Sahr T & Buchrieser C The Life Cycle of L. pneumophila: Cellular Differentiation Is Linked to Virulence and Metabolism. Front. Cell. Infect. Microbiol 8, 3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hauslein I et al. Legionella pneumophila CsrA regulates a metabolic switch from amino acid to glycerolipid metabolism. Open Biol. 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang C & Flieger A Characterisation of Legionella pneumophila phospholipases and their impact on host cells. Eur. J. Cell. Biol 90, 903–912 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Lerner TR et al. Mycobacterium tuberculosis replicates within necrotic human macrophages. J. Cell Biol 216, 583–594 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahamed D et al. Intracellular growth of Mycobacterium tuberculosis after macrophage cell death leads to serial killing of host cells. Elife. 6, e28205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Wel N et al. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 129, 1287–1298 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Stanley SA, Johndrow JE, Manzanillo P & Cox JS The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J. Immunol 178, 3143–3152 (2007). [DOI] [PubMed] [Google Scholar]
- 38.McKinney JD et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 406, 735–738 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Pandey AK & Sassetti CM Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. USA 105, 4376–4380 (2008).This study identifies the Mce4 membrane protein complex as the primary transporter of cholesterol in M. tuberculosis, and shows that the activity of this transporter is necessary for maintenance of M. tuberculosis infection in vivo.
- 40.VanderVen BC et al. Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium’s metabolism is constrained by the intracellular environment. PLoS. Pathog 11, e1004679 (2015).The first identification of chemical inhibitors of M. tuberculosis enzymes that are involved in the degradation of host-derived cholesterol. It shows that chemical inhibition of this pathway in the bacteria limits their intracellular growth.
- 41.Lee W, VanderVen BC, Fahey RJ & Russell DG Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J. Biol. Chem 288, 6788–6800 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nazarova EV et al. Rv3723/LucA coordinates fatty acid and cholesterol uptake in Mycobacterium tuberculosis. Elife. 6, e26969 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marrero J, Trujillo C, Rhee KY & Ehrt S Glucose phosphorylation is required for Mycobacterium tuberculosis persistence in mice. PLoS. Pathog 9, e1003116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beste DJ et al. 13C-flux spectral analysis of host-pathogen metabolism reveals a mixed diet for intracellular Mycobacterium tuberculosis. Chem. Biol 20, 1012–1021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Riordan M, Moors MA & Portnoy DA Listeria intracellular growth and virulence require host-derived lipoic acid. Science. 302, 462–464 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Grubmuller S, Schauer K, Goebel W, Fuchs TM & Eisenreich W Analysis of carbon substrates used by Listeria monocytogenes during growth in J774A.1 macrophages suggests a bipartite intracellular metabolism. Front. Cell. Infect. Microbiol 4, 156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kochan I The role of iron in bacterial infections, with special consideration of host-tubercle bacillus interaction. Curr. Top. Microbiol. Immunol 60, 1–30 (1973). [DOI] [PubMed] [Google Scholar]
- 48.Botella H et al. Mycobacterial p(1)-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe. 10, 248–259 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crouch ML, Castor M, Karlinsey JE, Kalhorn T & Fang FC Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol. Microbiol 67, 971–983 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Liu JZ et al. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe. 11, 227–239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolschendorf F et al. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 108, 1621–1626 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bange FC, Brown AM & Jacobs WR Jr. Leucine auxotrophy restricts growth of Mycobacterium bovis BCG in macrophages. Infect. Immun 64, 1794–1799 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clark-Curtiss JE & Curtiss R 3rd. Salmonella Vaccines: Conduits for Protective Antigens. J. Immunol 200, 39–48 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Ensminger AW, Yassin Y, Miron A & Isberg RR Experimental evolution of Legionella pneumophila in mouse macrophages leads to strains with altered determinants of environmental survival. PLo. SPathog 8, e1002731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vilcheze C et al. Rational Design of Biosafety Level 2-Approved, Multidrug-Resistant Strains of Mycobacterium tuberculosis through Nutrient Auxotrophy. MBio. 9, e00938 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murray PJ Amino acid auxotrophy as a system of immunological control nodes. Nat. Immunol 17, 132–139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfefferkorn ER Interferon gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proc. Natl. Acad. Sci. USA 81, 908–912 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt SV & Schultze JL New Insights into IDO Biology in Bacterial and Viral Infections. Front. Immunol 5, 384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Neill LAJ & Artyomov MN Itaconate: the poster child of metabolic reprogramming in macrophage function. Nat. Rev. Immunol in press (2019). [DOI] [PubMed] [Google Scholar]
- 60.Lampropoulou V et al. Itaconate Links Inhibition of Succinate Dehydrogenase with Macrophage Metabolic Remodeling and Regulation of Inflammation. Cell. Metab 24, 158–166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mills EL et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 556, 113–117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weiss JM et al. Itaconic acid mediates crosstalk between macrophage metabolism and peritoneal tumors. J. Clin. Invest 128, 3794–3805 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Honer Zu Bentrup K, Miczak A, Swenson DL & Russell DG Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J. Bacteriol 181, 7161–7167 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Munoz-Elias EJ & McKinney JD Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med 11, 638–644 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Savvi S et al. Functional characterization of a vitamin B12-dependent methylmalonyl pathway in Mycobacterium tuberculosis: implications for propionate metabolism during growth on fatty acids. J. Bacteriol 190, 3886–3895 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naujoks J et al. IFNs Modify the Proteome of Legionella-Containing Vacuoles and Restrict Infection Via IRGl-Derived Itaconic Acid. PLoS. Pathog 12, e1005408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nair S et al. Irgl expression in myeloid cells prevents immunopathology during M. tuberculosis infection. J. Exp. Med 215, 1035–1045 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sasikaran J, Ziemski M, Zadora PK, Fleig A & Berg IA Bacterial itaconate degradation promotes pathogenicity. Nat. Chem. Biol 10, 371–377 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Ito K & Suda T Metabolic requirements for the maintenance of self-renewing stem cells. Nat. Rev. Mol. Cell. Biol 15, 243–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vander Heiden MG & DeBerardinis RJ Understanding the Intersections between Metabolism and Cancer Biology. Cell. 168, 657–669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Everts B et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKvarepsilon supports the anabolic demands of dendritic cell activation. Nat. Immunol 15, 323–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodriguez-Prados JC et al. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J. Immunol 185, 605–614 (2010). [DOI] [PubMed] [Google Scholar]
- 73.Tan Z et al. The monocarboxylate transporter 4 is required for glycolytic reprogramming and inflammatory response in macrophages. J. Bio.l Chem 290, 46–55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lachmandas E et al. Microbial stimulation of different Toll-like receptor signalling pathways induces diverse metabolic programmes in human monocytes. Nat. Microbiol 2, 16246 (2016). [DOI] [PubMed] [Google Scholar]
- 75.Gillmaier N, Gotz A, Schulz A, Eisenreich W & Goebel W Metabolic responses of primary and transformed cells to intracellular Listeria monocytogenes. PLoS. One 7, e52378 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Czyz DM, Willett JW & Crosson S Brucella abortus Induces a Warburg Shift in Host Metabolism That Is Linked to Enhanced Intracellular Survival of the Pathogen. J. Bacteriol 199, e00227 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheng SC et al. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 345, 1250684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Netea MG et al. Trained immunity: A program of innate immune memory in health and disease. Science. 352, aaf1098 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Escoll P et al. Legionella pneumophila Modulates Mitochondrial Dynamics to Trigger Metabolic Repurposing of Infected Macrophages. Cell Host Microbe. 22, 302–316 (2017).An interesting study showing that L. pneumophila modulates the metabolism of its host cell through driving fission of the mitochondria in a DNM1L-dependent manner.
- 80.Stavru F, Bouillaud F, Sartori A, Ricquier D & Cossart P Listeria monocytogenes transiently alters mitochondrial dynamics during infection. Proc. Natl. Acad. Sci. USA 108, 3612–3617 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abramovitch RB Mycobacterium tuberculosis Reporter Strains as Tools for Drug Discovery and Development. IUBMB. Life 70, 818–825 (2018). [DOI] [PubMed] [Google Scholar]
- 82.Boot M et al. Accelerating Early Antituberculosis Drug Discovery by Creating Mycobacterial Indicator Strains That Predict Mode of Action. Antimicrob. Agents Chemother 62, e00083 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Claudi B et al. Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell. 158, 722–733 (2014).This study uses fluorescent TIMER-expressing Salmonella spp. in an in vivo infection model to analyze the rate of bacterial division, identify a sub-population of non-replicating bacteria, and demonstrate that these bacilli have acquired a drug-tolerant phenotype.
- 84.Gill WP et al. A replication clock for Mycobacterium tuberculosis. Nat. Med 15, 211–214 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Helaine S et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 343, 204–208 (2014).In this study, a GFP-dilution reporter strain is used to examine non-replicating Salmonella bacteria in vivo and to show that, in vitro, IFNγ-activated macrophages induce markedly higher levels of drug tolerance in Salmonella spp. than do resting macrophages.
- 86.Helaine S & Holden DW Heterogeneity of intracellular replication of bacterial pathogens. Curr. Opin. Microbiol 16, 184–191 (2013). [DOI] [PubMed] [Google Scholar]
- 87.MacGilvary NJ & Tan S Fluorescent Mycobacterium tuberculosis reporters: illuminating host-pathogen interactions. Pathog. Dis 76 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sukumar N, Tan S, Aldridge BB & Russell DG Exploitation of Mycobacterium tuberculosis reporter strains to probe the impact of vaccination at sites of infection. PLoS. Pathog 10, e1004394 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saliba AE et al. Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella. Nat. Microbiol 2, 16206 (2016).The use of single-cell RNA-seq to show that macrophages that preferentially support the growth of Salmonella spp. in vitro express polarization markers consistent with an M2-like macrophage phenotype.
- 90.Westermann AJ, Barquist L & Vogel J Resolving host-pathogen interactions by dual RNA-seq. PLoS. Pathog 13, e1006033 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Westermann AJ et al. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature. 529, 496–501 (2016). [DOI] [PubMed] [Google Scholar]
- 92.Bumann D & Cunrath O Heterogeneity of Salmonella-host interactions in infected host tissues. Curr. Opin. Microbiol 39, 57–63 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Cadena AM, Fortune SM & Flynn JL Heterogeneity in tuberculosis. Nat. Rev. Immunol 17, 691–702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Helaine S & Kugelberg E Bacterial persisters: formation, eradication, and experimental systems. Trends Microbiol. 22, 417–424 (2014). [DOI] [PubMed] [Google Scholar]
- 95.Liu Y et al. Immune activation of the host cell induces drug tolerance in Mycobacterium tuberculosis both in vitro and in vivo. J. Exp. Med 213, 809–825 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Manina G, Dhar N & McKinney JD Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe. 17, 32–46 (2015). [DOI] [PubMed] [Google Scholar]
- 97.Stapels DAC et al. Salmonella persisters undermine host immune defenses during antibiotic treatment. Science. 362, 1156–1160 (2018). [DOI] [PubMed] [Google Scholar]
- 98.Perez-Morales D & Bustamante VH The global regulatory system Csr senses glucose through the phosphoenolpyruvate: carbohydrate phosphotransferase system. Mol. Microbiol 99, 623–626 (2016). [DOI] [PubMed] [Google Scholar]
- 99.Aldridge BB et al. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science. 335, 100–104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rego EH, Audette RE & Rubin EJ Deletion of a mycobacterial divisome factor collapses single-cell phenotypic heterogeneity. Nature. 546, 153–157 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cohen SB et al. Alveolar Macrophages Provide an Early Mycobacterium tuberculosis Niche and Initiate Dissemination. Cell Host Microbe. 24, 439–446 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Srivastava S, Ernst JD & Desvignes L Beyond macrophages: the diversity of mononuclear cells in tuberculosis. Immunol. Rev 262, 179–192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Srivastava S, Grace PS & Ernst JD Antigen Export Reduces Antigen Presentation and Limits T Cell Control of M. tuberculosis. Cell Host Microbe. 19, 44–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bodnar KA, Serbina NV & Flynn JL Fate of Mycobacterium tuberculosis within murine dendritic cells. Infect. Immun 69, 800–809 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mattila JT et al. Microenvironments in tuberculous granulomas are delineated by distinct populations of macrophage subsets and expression of nitric oxide synthase and arginase isoforms. J. Immunol 191, 773–784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Marino S et al. Macrophage polarization drives granuloma outcome during Mycobacterium tuberculosis infection. Infect. Immun 83, 324–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang L, Nazarova EV, Tan S, Liu Y & Russell DG Growth of Mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J. Exp. Med 215, 1135–1152 (2018).A recent study showing that macrophages of different developmental lineages differentially support the growth of M. tuberculosis in vivo, and that alveolar macrophages are more permissive for bacterial growth than are interstitial macrophages through metabolism-dependent mechanisms.
- 108.Rohde KH, Veiga DF, Caldwell S, Balazsi G & Russell DG Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS. Pathog 8, e1002769 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tan S, Sukumar N, Abramovitch RB, Parish T & Russell DG Mycobacterium tuberculosis responds to chloride and pH as synergistic cues to the immune status of its host cell. PLoS. Pathog 9, e1003282 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dunlap MD et al. A novel role for C-C motif chemokine receptor 2 during infection with hypervirulent Mycobacterium tuberculosis. Mucosal Immunol. (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gleeson LE et al. Cigarette Smoking Impairs the Bioenergetic Immune Response to Mycobacterium Tuberculosis Infection. Am. J. Respir. Cell. Mol. Biol (2018). [DOI] [PubMed] [Google Scholar]
- 112.Goletti D, Petruccioli E, Joosten SA & Ottenhoff TH Tuberculosis Biomarkers: From Diagnosis to Protection. Infect. Dis. Rep 8, 6568 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee SH et al. Mannose receptor high, M2 dermal macrophages mediate nonhealing Leishmania major infection in a Th1 immune environment. J. Exp. Med 215, 357–375 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gibbings SL et al. Transcriptome analysis highlights the conserved difference between embryonic and postnatal-derived alveolar macrophages. Blood. 126, 1357–1366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gibbings SL et al. Three Unique Interstitial Macrophages in the Murine Lung at Steady State. Am. J. Respir. Cell. Mol. Biol 57, 66–76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mould KJ et al. Cell Origin Dictates Programming of Resident versus Recruited Macrophages during Acute Lung Injury. Am. J. Respir. Cell. Mol. Biol 57, 294–306 (2017).An interesting study showing that resident and recruited macrophages respond differently to LPS challenge in the lung airways, which indicates that the developmental origin of macrophages, rather than their environment, is decisive in determining their response to the same immune stimulus.
- 117.Jenkins SJ et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 332, 1284–1288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jenkins SJ et al. IL-4 directly signals tissue-resident macrophages to proliferate beyond homeostatic levels controlled by CSF-1. J. Exp. Med 210, 2477–2491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ruckerl D et al. Macrophage origin limits functional plasticity in helminth-bacterial co-infection. PLoS. Pathog 13, e1006233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Devchand PR et al. The PPARalpha-leukotriene B4 pathway to inflammation control. Nature. 384, 39–43 (1996). [DOI] [PubMed] [Google Scholar]
- 121.Nobs SP & Kopf M PPAR-gamma in innate and adaptive lung immunity. J Leukoc. Biol 104, 737–741 (2018). [DOI] [PubMed] [Google Scholar]
- 122.Bedi B et al. Enhanced Clearance of Pseudomonas aeruginosa by Peroxisome Proliferator-Activated Receptor Gamma. Infect. Immun 84, 1975–1985 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Almeida PE, Carneiro AB, Silva AR & Bozza PT PPARgamma Expression and Function in Mycobacterial Infection: Roles in Lipid Metabolism, Immunity, and Bacterial Killing. PPAR. Res 2012, 383829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guirado E et al. Deletion of PPARgamma in lung macrophages provides an immunoprotective response against M. tuberculosis infection in mice. Tuberculosis (Edinb). 111, 170–177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim YS et al. PPAR-alpha Activation Mediates Innate Host Defense through Induction of TFEB and Lipid Catabolism. J. Immunol 198, 3283–3295 (2017). [DOI] [PubMed] [Google Scholar]
- 126.Salamon H et al. Cutting edge: Vitamin D regulates lipid metabolism in Mycobacterium tuberculosis infection. J. Immunol 193, 30–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Arts RJW et al. Immunometabolic Pathways in BCG-Induced Trained Immunity. Cell Rep. 17, 2562–2571 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bekkering S et al. Metabolic Induction of Trained Immunity through the Mevalonate Pathway. Cell. 172, 135–146 (2018). [DOI] [PubMed] [Google Scholar]
- 129.Mitroulis I et al. Modulation of Myelopoiesis Progenitors Is an Integral Component of Trained Immunity. Cell. 172, 147–161e112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kaufmann E et al. BCG Educates Hematopoietic Stem Cells to Generate Protective Innate Immunity against Tuberculosis. Cell. 172, 176–190 (2018). [DOI] [PubMed] [Google Scholar]
- 131.Roostalu J, Joers A, Luidalepp H, Kaldalu N & Tenson T Cell division in Escherichia coli cultures monitored at single cell resolution. BMC Microbiol. 8, (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Terskikh A et al. “Fluorescent timer”: protein that changes color with time. Science. 290, 1585–1588 (2000). [DOI] [PubMed] [Google Scholar]
- 133.Reyes-Lamothe R, Sherratt DJ & Leake MC Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 328, 498–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Abramovitch RB, Rohde KH, Hsu FF & Russell DG aprABC: a Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol. Microbiol 80, 678–694 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Miesenbock G, De Angelis DA & Rothman JE Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 394, 192–195 (1998). [DOI] [PubMed] [Google Scholar]
- 136.Vandal OH, Pierini LM, Schnappinger D, Nathan CF & Ehrt S A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nat. Med 14, 849–854 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gierahn TM et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 14, 395–398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shah-Simpson S, Pereira CF, Dumoulin PC, Caradonna KL & Burleigh BA Bioenergetic profiling of Trypanosoma cruzi life stages using Seahorse extracellular flux technology. Mol. Biochem. Parasitol 208, 91–95 (2016). [DOI] [PubMed] [Google Scholar]
- 139.Kloehn J et al. Using metabolomics to dissect host-parasite interactions. Curr. Opin. Microbiol 32, 59–65 (2016). [DOI] [PubMed] [Google Scholar]
- 140.Saunders EC, Naderer T, Chambers J, Landfear SM & McConville MJ Leishmania mexicana can utilize amino acids as major carbon sources in macrophages but not in animal models. Mol. Microbiol 108, 143–158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Goldman-Pinkovich A et al. An Arginine Deprivation Response Pathway Is Induced in Leishmania during Macrophage Invasion. PLoS. Pathog 12, e1005494 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li Y et al. Transcriptome Remodeling in Trypanosoma cruzi and Human Cells during Intracellular Infection. PLoS. Pathog 12, e1005511 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shah-Simpson S, Lentini G, Dumoulin PC & Burleigh BA Modulation of host central carbon metabolism and in situ glucose uptake by intracellular Trypanosoma cruzi amastigotes. PLoS. Pathog 13, e1006747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Blume M et al. A Toxoplasma gondii Gluconeogenic Enzyme Contributes to Robust Central Carbon Metabolism and Is Essential for Replication and Virulence. Cell Host Microbe. 18, 210–220 (2015). [DOI] [PubMed] [Google Scholar]
- 145.Jacot D, Waller RF, Soldati-Favre D, MacPherson DA & MacRae JI Apicomplexan Energy Metabolism: Carbon Source Promiscuity and the Quiescence Hyperbole. Trends Parasitol. 32, 56–70 (2016). [DOI] [PubMed] [Google Scholar]
- 146.Jensen KD et al. Toxoplasma polymorphic effectors determine macrophage polarization and intestinal inflammation. Cell Host Microbe. 9, 472–483 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Leroux LP et al. The Protozoan Parasite Toxoplasma gondii Selectively Reprograms the Host Cell Translatome. Infect. Immun 86, e00244 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Barelle CJ et al. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell. Microbiol 8, 961–971 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lorenz MC & Fink GR The glyoxylate cycle is required for fungal virulence. Nature. 412, 83–86 (2001). [DOI] [PubMed] [Google Scholar]
- 150.Osborne SE et al. Type I interferon promotes cell-to-cell spread of Listeria monocytogenes. Cell. Microbiol 19, e12660 (2017). [DOI] [PubMed] [Google Scholar]
- 151.Harouz H et al. Shigella flexneri targets the HP1gamma subcode through the phosphothreonine lyase OspF. EMBO J. 33, 2606–2622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kentner D et al. Shigella reroutes host cell central metabolism to obtain high-flux nutrient supply for vigorous intracellular growth. Proc. Natl. Acad. Sci. U S A 111, 9929–9934 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Waligora EA et al. Role of intracellular carbon metabolism pathways in Shigella flexneri virulence. Infect. Immun 82, 2746–2755 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Calverley M, Erickson S, Read AJ & Harmsen AG Resident alveolar macrophages are susceptible to and permissive of Coxiella burnetii infection. PLoS. One 7, e51941 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Graham JG et al. Virulent Coxiella burnetii pathotypes productively infect primary human alveolar macrophages. Cell. Microbiol 15, 1012–1025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hauslein I et al. Multiple Substrate Usage of Coxiella burnetii to Feed a Bipartite Metabolic Network. Front. Cell. Infect. Microbiol 7, 285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mulye M, Zapata B & Gilk SD Altering lipid droplet homeostasis affects Coxiella burnetii intracellular growth. PLoS. One 13, e0192215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hauslein I, Manske C, Goebel W, Eisenreich W & Hilbi H Pathway analysis using (13) C-glycerol and other carbon tracers reveals a bipartite metabolism of Legionella pneumophila. Mol. Microbiol 100, 229–246 (2016). [DOI] [PubMed] [Google Scholar]