Abstract
Spatial positioning is a fundamental principle governing nuclear processes. Chromatin is organized as a hierarchy from nucleosomes to Mbp chromatin domains (CD) or topologically associating domains (TADs) to higher level compartments culminating in chromosome territories (CT). Microscopic and sequencing techniques have substantiated chromatin organization as a critical factor regulating gene expression. For example, enhancers loop back to interact with their target genes almost exclusively within TADs, distally located coregulated genes reposition into common transcription factories upon activation, and Mbp CDs exhibit dynamic motion and configurational changes in vivo. A longstanding question in the nucleus field is whether an interactive nuclear matrix provides a direct link between structure and function. The findings of nonrandom radial positioning of CT within the nucleus suggest the possibility of preferential interaction patterns among populations of CT. Sequential labeling up to 10 CT followed by application of computer imaging and geometric graph mining algorithms revealed cell-type specific interchromosomal networks (ICN) of CT that are altered during the cell cycle, differentiation, and cancer progression. It is proposed that the ICN correlate with the global level of genome regulation. These approaches also demonstrated that the large scale 3-D topology of CT is specific for each CT. The cell-type specific proximity of certain chromosomal regions in normal cells may explain the propensity of distinct translocations in cancer subtypes. Understanding how genes are dysregulated upon disruption of the normal “wiring” of the nucleus by translocations, deletions, and amplifications that are hallmarks of cancer, should enable more targeted therapeutic strategies.
1 |. INTRODUCTION
Spatial positioning has emerged as a fundamental principle governing nuclear processes and, together with the field of genomics, has led to a paradigm shift in the study of gene regulation.1–17 Rather than studying individual genes and their regulation, the emphasis is now on understanding the regulation and coordination of up to 1000s of genes at any given time. An even more daunting challenge is deciphering how these large numbers of genes are spatially arranged, expressed, and regulated within the three-dimensional (3-D) context of the cell nucleus.
Key to this understanding is the realization that the chromatin in the cell nucleus is arranged as a hierarchy (Figure 1A) ranging from the DNA double helix organized into chromatin to the arrangement of chromatin into increasingly higher levels of organization culminating with the entire chromosome as a 3-D entity termed the chromosome territory (CT).1,2,18,19 In this manner, the CT acts as an epigenetic feedback system where events and modifications occurring at all levels of chromatin organization affect the global expression and regulation of the genome. For example, at the molecular level, histones are dynamically regulated through a diverse array of modifications defining the histone code and leading to alterations in chromatin organization and function.20–23,25,28,30,31 Targeted alterations in DNA methylation along with a variety of other factors, such as chromatin remodelers, have further provided the foundation for studying chromatin as an integral factor driving the epigenetic regulation of the genome.22,26,28–30 Analyses of these epigenetic markers enable the distinction of euchromatin or “open” chromatin—where active genes are predominantly located—from the gene poor heterochromatic or “closed” chromatin regions.22,30,31 In higher order chromatin domains (CD) up to entire chromosome territories (CT), nuclear architecture coupled with genome organization have been implicated in the regulation of genomic functions such as DNA replication, transcription, and RNA processing.1–4,8,12–15,17,18,23,24,27,32–40 With this in mind, our review is focused on the 3-D architecture of CT and their potential role in the global orchestration of genomic expression and regulation within the functional milieu of the cell nucleus.
FIGURE 1.
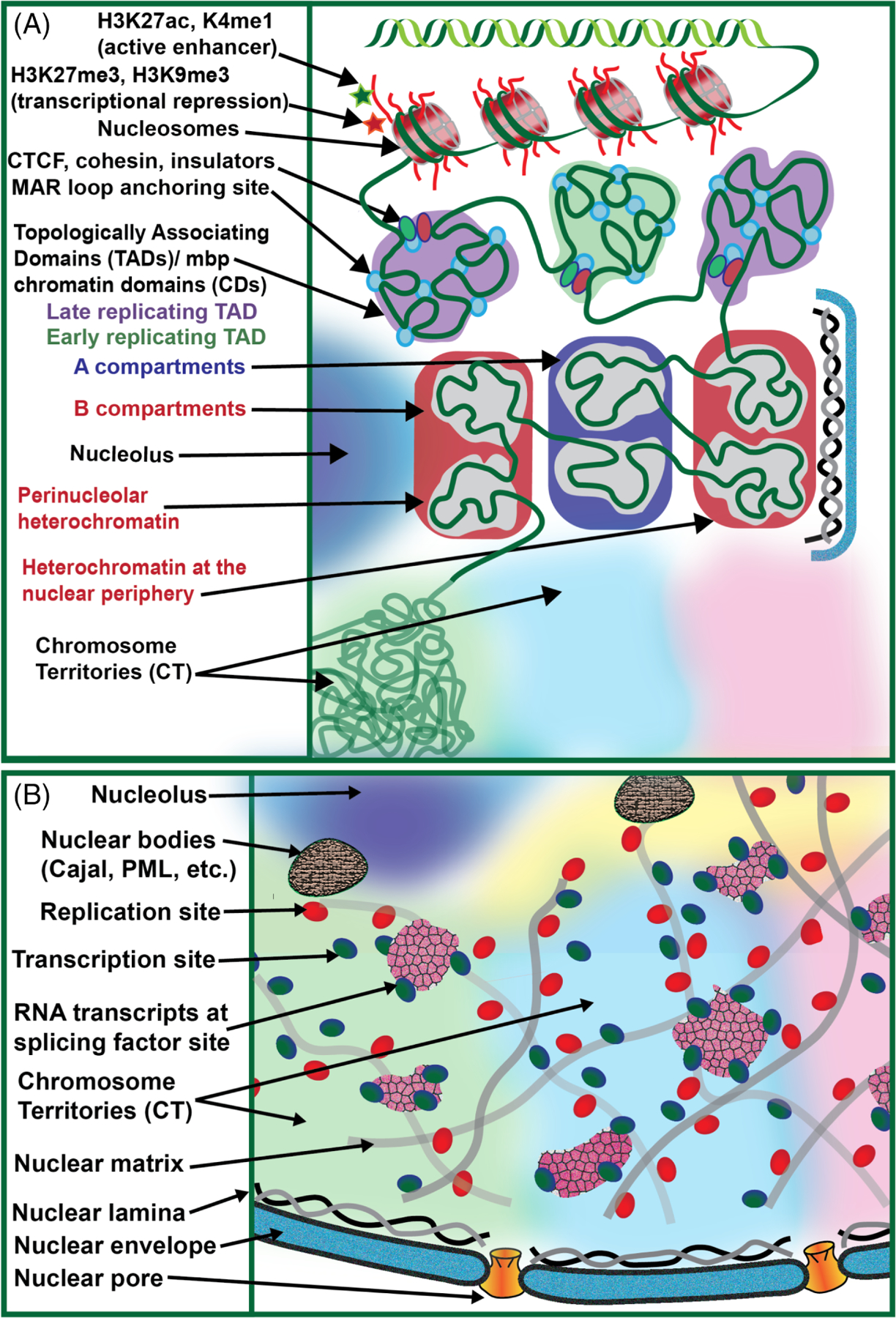
Higher order chromatin organization and functional nuclear architecture. A, Hierarchal levels of chromatin organization are shown including: nucleosomes, chromatin fibers, chromatin loops, mbp CDs (Chromatin Domains)/TADs (Topologically Associating Domains), A and B compartments, and CT (chromosome territories). B, The functional nuclear architecture is depicted. Replication sites and nuclear speckles are associated with the nuclear matrix. Transcription sites associate with the nuclear matrix or nuclear speckles. Other nuclear bodies and compartments are also shown such as the nuclear envelope, nuclear lamina, nuclear pore complex, and the nucleolus
1.1 |. The Concept of CT
It is now well established that in the nucleus the chromosomes are present as discrete entities. Carl Rabl in 188541 first suggested a territorial organization of interphase chromosomes in animal cell nuclei. Later, Theodor Boveri coined the term chromosome territory (CT) during his study of roundworm blastomere stage.42,43 Despite its early discovery, the existence of CT was not well accepted, and in the 1950s to 1970s, it was generally believed that the chromatin intermingled in the cell nucleus “like a bowl of spaghetti”. The idea was still tested, and the first substantial experimental evidence was provided in the 1970s. In these experiments, squashed Chinese hamster ovary cells were treated with acetic acid, air dried, and subjected to Giemsa staining. Upon visualizing these nuclei, large patches of chromatin were observed and interpreted as CT.44 While some of these treatments may have artificially “induced” territorial-like arrangements of the chromatin, other evidence avoided this caveat. Using a laser micro beam, DNA damage was inflicted to a small region in the nucleus. The cells were then immediately pulse labeled with3H-thymidine, which marked sites of DNA repair. Observing the cells during mitosis revealed that the labeled DNA was restricted to a region on one of the chromosomes rather than being present in smaller amounts on several chromosomes, which would be expected if the chromatin was freely intermingling.45,46
Development of in situ hybridization and chromosome-specific painting probes enabled the first direct visualization of CT in the nuclei of fixed cells.47–49 Later techniques were developed to visualize CT in live cells. This was achieved by incorporating fluorescent nucleotide analogs into replicating DNA in cells, followed by several cell divisions. Through independent assortment after several generations, individually labeled CT could be seen in the live cells.50–52 While these studies indicated that the intranuclear position of CT are relatively stable throughout the interphase, further investigations in living cells revealed that discrete replication labeled CD in the Mbp size range undergo significant oscillatory motion and frequent shape transitions between condensed and open configurations.52–55 Remarkably, chromatin dynamics in live cells appears to correlate with gene activity. Chromatin enriched in active genes had higher levels of oscillatory motion and configurational changes than gene poor chromatin regions.53,54 Furthermore, chromatin motion is significantly reduced by inhibitors of RNA synthesis, such as actinomycin D.53,54
1.2 |. Interactions of CT Within the Nuclear Milieu
Behind the complex 3-D structural organization of the cell nucleus is a striking degree of functional order (Figure 1B) wherein specific genomic functions such as DNA replication, transcription, and RNA splicing are compartmentalized within discrete sites along with a host of nuclear bodies such as the PML, Cajal, and histone loci.1,6,8,39,56–63 While our understanding of the biogenesis and assembly of various nuclear bodies are still in their infancy, insight has been gained from physiochemical considerations. For example, it has been proposed that nuclear bodies form when proteins bind and cross-link the chromatin as polymer segments thereby collapsing the chromatin into globule-like configurations. Another proposed mechanism is that proteins binding to the chromatin induce liquid-liquid phase separations whereby “liquid-like” nuclear bodies form around the chromatin globules.64,65
A longstanding question in the field is whether the plethora of functional domains in the cell nucleus are present as separated entities or, alternatively, are they linked into an interactive network as studies into the nuclear matrix suggest (Figure 1B).4,12,37,39,66–72 The nuclear matrix was first identified by treating cells with high salt and nucleases.73 Under these conditions, the nucleus retains its overall architectural features such as the nuclear lamina, nucleoli, and a fibrogranular matrix network.35,73–75 Detailed electron microscopic studies revealed a close resemblance of this fibogranular matrix network to nonchromatin structures visible under certain staining conditions in the interchromatinic regions of intact cells.35,68,71,72,74–76 The retention of functionally related properties associated with the isolated nuclear matrix (eg, DNA replication, repair, transcription, RNA splicing, steroid hormone receptors, viral associations, protein kinases, oncogenes, tumor suppressors, and many other regulatory factors) is consistent with the nuclear matrix as a structural framework for nuclear functions. In this view, the in vivo matrix is a milieu in the nucleus where domains of chromatin and ribonucleoproteins dynamically interact to form higher order nuclear architecture undergirding genomic functions.1,35,68,70,71,75,76 Indeed most of these functional associations are maintained in a structural organization that is indistinguishable from those in the nuclei of untreated cells. This is most strikingly illustrated in studies of DNA replication and transcription sites that remain bound to the isolated nuclear matrix.39,71,77 Moreover, DNA replication and transcriptional activities are still present in these preparations and function on the matrix-attached DNA fragments.39,71,77,90
Despite this progress, it is important to realize that isolated nuclear matrix systems are likely static and highly stabilized versions of a dynamic nuclear architecture in living cells.13 Moreover, the precise relationships of the isolated matrices to in vivo nuclear architecture and function remain to be defined. In one previously stated view, the “in vivo nuclear matrix” is considered as the milieu in the nucleus where domains of chromatin and ribonucleoprotein complexes dynamically interact to form modules of higher order architecture and function.1,13,35,68–71,75,76 To further extend this model, interactions among individual modules may occur resulting in many local network-like connections dispersed throughout the nucleus.13,69 Isolation conditions for the nuclear matrix may then result in one stable network termed the nuclear matrix. In this view, a continuous nuclear matrix structure is not present in the in vivo nucleus. Instead, higher order discontinuous architectural elements may provide the basis for nuclear matrix isolation. More research on many levels is needed to resolve these issues.
The architectural role of the nuclear matrix likely extends to the CT themselves. Previous studies have indicated the association of telomeres,78,80,81 centromeres,82,83 chromatin loops and the attachment sites for chromatin loops termed S/MARs (matrix or scaffold attachment regions)79,84–93 on the nuclear matrix. S/MARs DNA binding elements have been identified in silico94,95 and recently analyzed on a genomic scale, which should open additional doors to the understanding of these critical but enigmatic regions of the genome.96,97 Moreover, experiments on whole cells to isolate nuclear matrix demonstrated that despite the removal of up to 90% of the chromosomal histones, the chromosomal DNA in the remaining nuclear matrix structures are still folded into territorial arrays resembling intact CT.98 Subsequent disruption and extraction of nuclear matrix proteins, led to a corresponding disruption of territorial organization.98
Several proteins associated with isolated nuclear matrix have been identified as potential factors involved at chromatin loop S/MAR attachment sites and/or higher order CD. These include topoisomerase II,99–102 CTCF,103 cohesin,104–106 matrin 3 (also called as P-130 nuclear scaffold protein107,108), HnRNP-U (also called as SAF-A, for Scaffold/Matrix attachment region factor109–113), SAF-B,114,115 ARBP (Attachment Region Binding Protein116), SAT-B,117–119 and nuclear lamins.120,121 Recent proteomic approaches have further confirmed the S/MAR binding activities of SAF-A, matrin 3, and nuclear lamins.122–124 CTCF and cohesin have been identified as key players in the arrangement of chromatin into higher order domains termed Topologically Associated Domains (TADs), which mediate long range intrachromosomal and interchromosomal interactions within the nucleus.15,106,125–134 Recently, HnRNP-U (SAF-A) has been demonstrated to be integral to maintaining genome organization at TADs and higher levels of chromatin.113
Matrin 3 is an acidic ~120 kDa nuclear matrix protein35,135 that is among the major proteins of isolated nuclear matrix based on two-dimensional (2-D) gel separations.136 It has both DNA and RNA binding properties and is involved in a wide range of processes ranging from cell survival to association at S/MAR chromatin loop anchor sites to a variety of functions involved in RNA metabolism including RNA splicing, mRNA assembly/stabilization, RNA export/retention, and viral RNA regulation.107,108,123,137–145 Consistent with these findings, yeast two hybrid screening identified interactions with a constellation of proteins involved in DNA replication/repair, transcription, and RNA splicing.66 A recent transcriptome and interactome analysis revealed over 100 proteins that bind matrin 3 (prominently including proteins associated with RNA processing/splicing146). Deletion of a RNA recognition motif (RRM1) or Zn finger motifs (ZnF1 or ZnF2) diminished the binding of a subset of matrin 3 interacting proteins and altered its intranuclear organization.124 This study also uncovered a crucial role of the RRM2 motif of matrin 3 for regulating interactions with other nuclear proteins and maintaining its characteristic intranuclear localization.
Detailed visualization studies including computer-assisted analysis demonstrated that matrin 3 forms a network-like structure in the nucleus, which permeates the interior of CT and is in close proximity to functional domains containing active sites of transcription, RNA polymerase II, RNA splicing, and other functionally related factors.4,8,12,66 While the potential role of matrin 3 as a major component of an overall scaffolding network in the cell nucleus remains to be resolved, it is interesting to note that deletion mutations in the RRM2 domain resulted in major alterations of the network-like organization of matrin 3 into large nuclear body-like structures and protein-protein interactions.124 Moreover, colocalization studies demonstrated a similar network organization and close 3-D proximity of the S/MAR binding protein SAF-A (hnRNP-U) with matrin 3 including penetration into the interior of CT.8,12,66 Based on these findings and similar properties of these two proteins, it was proposed that SAF-A and matrin 3 are major components of a nuclear scaffolding structure that serve as dynamic assembly centers for the organization of functional neighborhoods of genomic function in the cell nucleus.4,12 The potential physiological roles that matrin 3 plays in nuclear organization and function are further emphasized by the recent findings connecting mutations in the matrin 3 gene (MATR3) to the diseases ALS, distal myopathy, and left ventricular outflow tract obstruction (LVOTO), a common congenital heart defect.124,141,146–149
Within the nuclear matrix, the nuclear lamina—the network of peripheral proteins surrounding the inner nuclear membrane—has also been posited as a major factor in the organization of the genome.61,150–156 The major proteins of the nuclear lamina are lamins A, B, and C. These bind to a myriad of other proteins involved in many nuclear functions. Among these proteins are histone H2A or H2B dimers, actin, retinoblastoma protein, components of the RNA-polymerase-II-dependent transcription, and replication complexes.150,151 Lamins also bind to integral membrane proteins such as emerin, lamin-B receptor (LBR), and MAN1.157 Through these many interactions, it is proposed that specific regions within the genome are tethered to the nuclear periphery.152,153,158 Lamins are also MAR binding proteins and are generally believed to be involved in the binding of the characteristic heterochromatin along the nuclear periphery120,121 and domains of chromatin loops termed LADs, which are enriched in inactive genes and serve a regulatory role in gene repression along the nuclear periphery.11,153,158,159
The important role of nuclear lamin proteins in genomic organization and function is further highlighted by the linkage of mutant forms of the lamins to disease states termed laminopathies.154–156,160 Over 400 mutations throughout the LMNA gene, which codes for both lamins A and C, have been linked to various forms of laminopathies such as muscle dystrophies, cardiomyopathies, and partial lipodystrophies.154 Laminopathies typically lead to nuclear blebbing and striking alterations in both nuclear shape and chromatin morphology in the nuclear interior.161,162 These alterations are consistent with a fundamental role of the nuclear lamina in genomic organization and function.
In considering one mode in which the lamins on the nuclear periphery might influence chromatin in the nuclear interior, a recent study demonstrated a specific binding site in the C-terminal tail of lamin A for matrin 3.163 This was corroborated by pull-down experiments and the close proximity of a small portion of matrin 3 with the nuclear lamins on the nuclear periphery by immunofluorescence. A cell line, containing a C-terminal mutant lamin A that leads to characteristic forms of distal myopathy and lacks the matrin 3 binding domain, was devoid of close proximity with matrin 3 along the nuclear periphery.163 It is tempting to speculate that this particular laminopathy involves disruption of a normal linkage between lamin A and matrin 3 on the nuclear periphery. This in turn may be linked to the elaborate network structure that matrin 3 forms in the nuclear interior including localization within the interior of CT.66
While substantial findings indicate that nuclear lamin A is also present throughout the nuclear interior, its exact role in this noncanonical location remains unknown.164,165 In this regard, striking increases were reported in overall chromatin motion in a lamin A deficient double null cell line. Appropriate experiments suggested that this increase was a result of changes in lamin A interactions within the nuclear interior where it might serve as part of a scaffolding system for chromatin.166 Recently, it was determined that lamin A is responsible for the viscoelasticity of the nuclear interior.167 It would be interesting to determine if this interior lamin A population is influencing chromatin motion through its association with matrin 3 and/or other components of the fibrogranular internal nuclear matrix structure. Compatible with a scaffolding role of interior lamin A, increases in the intranuclear movement of PML bodies was also reported in a lamin A double null cell line.168
Chromatin loops within LADs may also play a role in the overall anchoring of CT within the nuclear interior through association with the nuclear lamina. Early studies of DNA attached to the nuclear matrix suggested that while the bulk of the DNA loop attachment sites are located within the nuclear matrix interior, a small but reproducible proportion (<20%) are present within nuclear lamin preparations.90 While removal of the internal components of the nuclear matrix leads to the disruption of the characteristic territorial organization of CT in the cell nucleus, the DNA remains tightly packaged inside the cell nucleus.98 Because the nuclear lamina was maintained in those preparations, it is conceivable that the DNA is anchored in the nucleus via the persistence of LAD DNA loops. Further studies are necessary to resolve this issue and the potential architectural role of the nuclear lamin-associated LADs and chromatin loops in CT organization.
The third major architectural component of the isolated nuclear matrix is the residual nucleolar structure.35,74,75 A large body of studies demonstrate the association of a subset of CT termed the nucleolar organizing region (NOR)- chromosomes (CT13, 14, 15, 21, 22) with the nucleolus.169 The NOR-CT contain the amplified arrays of rDNA genes, which provide the basis for ribosomal RNA production and underlies the formation of the nucleolus as a ribosome assembly factory.169–171 The nucleolus is also associated with a population of chromatin loop domains termed nucleolar-associated domains, which like the LADs are enriched in inactive genes and are believed to be involved in the repressive state of the nucleolar-associated heterochromatin.172,173
1.3 |. The Hierarchy of Chromatin Organization and Function
The organization of chromatin and the impact of nuclear structure on the regulation of the cell have fascinated biologists because the nucleus was first visualized by microscopy. The eukaryotic genome is hierarchically organized across a spectrum of levels (see Figure 1A). At the lowest scale, chromatin is packaged by histone proteins into repeating structures known as nucleosomes. These consist of 147 base pairs of DNA, core histone subunits, and tails that extend out and are subject to posttranslational modifications such as acetylation, phosphorylation, and methylation. At this level, nucleosomes on the DNA have been described as “beads on a string” 10 nm fibers.174,175 Upon methylation and the addition of histone H1, chromatin is believed to be condensed into a 30 nm fiber. However, the exact structure of the 30 nm fiber and whether it exists in vivo is still controversial and unresolved.176
The next level of organization involves the folding of the chromatin into loops anchored to the nuclear matrix or chromosome scaffold1,19,177 Chromatin loops were first identified in human cells as DNA supercoiled domains with an average size of ~ 200 kbp.178 Electron microscopic analysis further revealed that the DNA halos, which appear following relaxation of the supercoiled domains,88 consisted of tightly packed and highly convoluted DNA loops anchored to the chromosome scaffold and nuclear matrix, respectively.75,179,180 Following the initial finding that newly replicating DNA is associated with the nuclear matrix,181 it was demonstrated that replication occurs in a bidirectional movement of the DNA loops through fixed replication complexes present at the base of nuclear matrix attached DNA loops. In this way, postreplicated DNA accumulates progressively in the distal portions of the loops at a rate consistent with the bidirectional replication fork rate.88,182–184 Analysis of gene positioning along the DNA loops further indicated that inactive genes positioned in the distal regions of DNA loops are repositioned near the nuclear matrix attached sites for the DNA loops in correlation with gene activation.185–187 This suggests the intriguing idea of a dynamic chromatin loop program corresponding to the overall transcriptional program of a cell.75,76 Consistent with these results, biochemical and genomic approaches have revealed that S/MAR attachment sites preferentially associate with actively transcribed genes.96,188
Further studies of DNA replication sites using 3-D microscopy and quantitative computer image analysis resulted in an important breakthrough in our understanding of higher order CD beyond the chromatin loops. Appropriate pulse-chase experiments revealed that replication is organized at discrete replication sites (RS) in the nucleus with each RS containing an average of ~1 Mbp DNA.58,189 Based on the average lifetime of each RS or replication factory, this corresponded to ~ 5 to 6 replicons or chromatin loops with an average size of ~ 180 kbp.58 These results were consistent with earlier biochemical findings demonstrating the organization of DNA polymerases and primase into large megacomplexes bound to the isolated nuclear matrix from regenerating rat liver.190 Chase studies extending hours to days revealed that the originally labeled RS persist as similarly sized CD throughout the cell cycle and into subsequent cell generations.58,189 Similar CD have also been visualized several generations after longterm labeling of DNA.52 These Mbp CDs are, therefore, a fundamental unit for both the higher order assembly of chromatin and it replication at replication factories.1 More recent studies of higher order CD using the high throughput chromosome conformation capture (Hi-C) approach (described below) have led to the identification of similarly sized domains termed TADs.127,128,132 It is now generally accepted that TADs and Mbp CDs represent the same higher order CD identified by two different approaches.125,128
The findings of discrete transcription factories where multiple genes can be transcribed in close proximity may represent another functional aspect of the Mbp CDs.10,191–194 Genes from different regions of individual CDs could loop out to form a cluster of active genes under common transcriptional regulation within the transcription factory.8,12
Understanding the arrangement of chromatin beyond the Mbp CDs is a challenging area of research. In one direction, studies of the arrangement of RS and transcription sites (TS) in 3-D led to the finding of discrete higher order zones of replication and transcription that appeared as separate network structures in the cell nucleus.1,36 These higher order zones of replication and transcription are maintained following extraction of whole cells for nuclear matrix further implicating a critical role of this overall network structure in the structural compartmentalization of these two fundamental genomic processes.39
In recent years, a compendium of chromosome conformation capture (3C) approaches have provided biochemical methods for determining genome organization.133,195 These techniques rely on the ligation of DNA fragments that are in close proximity in fixed nuclei. The advancement of next generation sequencing technologies has allowed for high throughput unbiased investigation of genome organization in the technique termed Hi-C132,133 (Figure 2). In brief, Hi-C involves the 3C technique followed by adapter ligation and results in a library of chimeric DNA fragments, each end of which are subsequently mapped to the genome. The pairwise frequency of interactions between regions of the genome is then represented as a matrix. The diagonal of this matrix denotes local interactions, whereas distal interactions are represented farther off the diagonal (see Figure 2).
FIGURE 2.
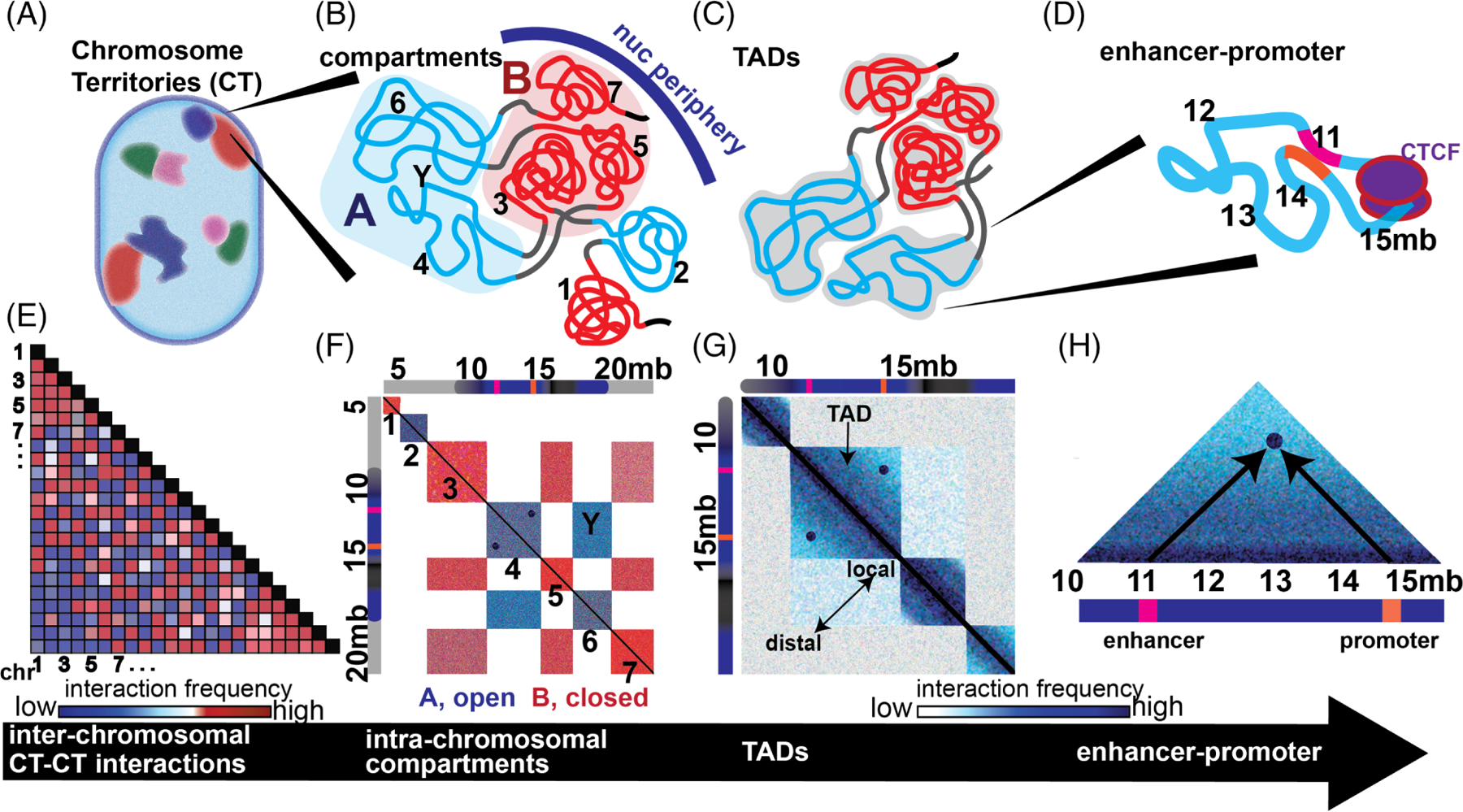
The hierarchy of higher order chromatin organization based on high throughput chromatin conformation capture (Hi-C). Diagrams are shown of higher order chromatin organization in a hierarchy from the whole CT to A and B compartments to topologically associating domains (TADs)/mbp chromatin domains (CDs) to specific enhancer-promoter interactions A-D, with corresponding schematics of Hi-C visualizations representing interchromosomal interactions, A and B compartmentalization, TADs/CDs, and intra-TAD interactions E-H. The Hi-C technique enables determination of pairwise interaction frequencies between regions of chromatin across the genome (represented as a matrix). A,E, At the highest level, total pairwise interaction frequencies between chromosomes can be determined. A simulated matrix of pairwise CT–CT interactions is shown (red = high frequency, blue = low, and white = intermediate). BF, Within CT, chromatin is organized into open, A, euchromatic or closed, B, heterochromatic, compartments. These compartments are in turn composed of TADs exhibiting a triangular pattern of higher frequency interactions along the diagonal. Seven TADs numbered from 1 to 7 are colored red or blue with respect to whether they are contained within A (blue) or B (red) compartments. An interaction between two TADs within a compartment is demarcated with a Y. C,G, A section of this matrix encompassing TADs 3 to 6 is displayed enlarged (darker blue - high frequency, light blue - low). Local interactions are closer to the diagonal while distal interactions are farther from the diagonal. D,H. Enhancer-promoter interactions are generally contained within TADs. A simulated enhancer-promoter interaction is displayed within TAD number 4. This TAD is enlarged to demonstrate the enhancer at ~11 mbp (pink) interacting with the promoter at ~15 mbp (orange)
Using these techniques, investigators have confirmed and extended most of the findings that were based upon microscopy and increased the resolution at which these fundamental principles of nuclear organization are measured (Figure 2). For example, the previously discussed microscopic visualization experiments that identified ~ 1 Mbp CDs,58,189 were confirmed by Hi-C to be TADs and can be seen in interaction matrices as increased local frequencies of contact125,128,132 (ie, looping interactions, see Figure 2).TADs are largely invariable across cell types and conserved across evolution.125 Identification of replication domain boundaries intersect with boundaries of TADs.196 Thus TADs define modules of early or late replicating CD and are dynamic domains that correlate with genomic function197 just as proposed for the Mbp CDs analyzed by microscopic and computer imaging approaches.58,189
Hi-C experiments have also resolved the genome into two major compartments, A and B, representing euchromatic vs heterochromatic microenvironments, respectively.198 While constitutive heterochromatin is typically composed of repetitive DNA and is involved in structures such as centromeres and telomeres, facultative heterochromatin is the result of gene silencing via histone deacetylation.199 Thus, facultative heterochromatin is a more dynamic and reversible structure that can decondense and become transcriptionally active.199
In Hi-C matrices, compartments are seen as blocks of interaction that are off-diagonal and therefore represent coalescing TAD regions of heterochromatin or euchromatin (Figure 2). These compartments are enriched in histone marks indicative of their chromatin state. As expected, A compartments are generally more gene rich and have higher levels of expression than B. This demarcation of chromatin states has become more complex because heterochromatin has been subdivided into four or five distinct states that range from constitutive to facultative and are marked by different epigenetic modifications.200 Similarly, using a higher resolution Hi-C technique, the A and B compartments can be subdivided into distinct subcompartments.201 More recently, however, single cell Hi-C approaches have determined that rather than distinct subcompartments TADs exist within a spectrum of compartments that range from A to B.202
Heterochromatin is preferentially located at the nuclear periphery.203,204 As such inactive TADs associated with B compartments are preferentially localized at the nuclear periphery, while active TADs in A compartments are generally more interior.201 A role for the nuclear lamins in this localization is supported by the finding that lamin A knock-out mice lose this peripheral organization of heterochromatin.205 This is also the case in cells of progeria patients with mutated lamin A.206 In fact, the nuclear compartmentalization is lost in late passages of cells with progeria.207 While still under investigation, the specific association of lamin proteins with heterochromatin likely occurs through the interactions of lamins with the lamin receptor LBR which in turn binds to the Heterochromatin Protein 1 (HP1),208 which plays a vital role in heterochromatin formation.209–211 The gene repressive microenvironment of heterochromatin has functional implications for the regulation of replication, transcription, and DNA repair. Interior “nucleoplasmic” lamin A likely also plays a role in these processes and should also be considered.165
1.4 |. Organization of CT
Following the establishment of CT as the basis for the organization of the genome in the cell nucleus, investigators focused on understanding how the individual territories were arranged within the 3-D architecture of the cell nucleus. As a first step, the relative radial arrangement of chromosomes was examined with respect to the nuclear periphery and center. A fundamental question was whether CT are randomly arranged or whether there was a level of nonrandomness in their positions within the cell nucleus. These studies clearly established a high level of order in the radial positioning of individual CT in the cell nucleus.9,11,18,212–217,219
Because the transcriptionally inactive heterochromatin in the nucleus is preferentially localized along the nuclear periphery, the relationship of peripheral position to the gene density of individual chromosomes was analyzed in detail. In an initial study, the inactive X (Xi) CT had a more peripheral location within the nucleus than its active (Xa) counterpart.212,213,219
The relationship between radial positioning and gene activity can also be demonstrated for the somatic CT because gene density generally correlates with gene activity.218 Thus CT that are gene poor may be positioned more peripheral than those that are gene rich because they are less transcriptionally active. For this reason, several studies have concluded that gene density is the major contributing factor to the radial positioning of CT.212,213,219 For example, the gene poor CT18 is more peripheral than the gene rich CT19.9,212,220 This paradigm can be extended to individual genes or gene complexes within the chromosomes. Individual genes that are expressed are typically found more interior than those that are not.10,221–227 Interestingly, aberrant localization to the nuclear periphery by tethering regions to the nuclear lamina results in the inhibition of transcription.223 Moreover, the radial positioning of CT in the same cell type are conserved throughout primate evolution.214
Studies demonstrating a close correlation between gene density and radial positioning were performed predominantly in the relatively spherical nuclei of lymphocytes. Analysis of radial positioning in flatter cells such as fibroblasts, however, have shown a more complex situation where chromosome size becomes an important factor. In these cases, the radial positioning generally correlates with chromosome size and not gene density.228–232,246
It was recently reported that the radial positions of a subset of seven chromosomes were not altered during the cell cycle but had limited alterations between fibroblast and epithelial cell types.229 In addition, five of nine CT examined showed significant differences in radial position between normal breast epithelial cells and a corresponding malignant cell line.230 Even more striking, it was reported that the radial positioning of seven CT in undifferentiated human keratinocytes strongly correlated with gene density, while the positioning in the corresponding CT from late differentiating keratinocytes were highly altered with no relationship to gene density or chromosome size.233 These findings are consistent with the probabilistic radial positioning code proposed by Cremer and associates213 and support the view that the probabilistic positioning code is cell-type-specific and can undergo alterations during both malignant progression and cell differentiation where massive changes in the genomic expression programs generally occur.
Several factors other than gene density or chromosome size have been implicated in the radial arrangement of CT. Interactions with other nuclear compartments are proposed as one mechanism by which radial positioning is established. For example, the interaction with nucleoli is implicated in the radial positioning of the nucleolar NOR-CT.169,228,234 The nuclear lamina has also been demonstrated to be important for the radial organization of CT.235 Specifically, cells that express the aberrant truncated lamin-A protein (progerin) have an altered radial arrangement of CT.236
In addition to interactions with other nuclear compartments, several protein factors or protein modifications may have a role in the radial positioning of CT. Knocking down the actin family member Arp6,237 a histone variant H2A.Z,237 myosin,235 or inhibiting histone deacetylation238 all lead to abnormal radial positioning of CT.
1.5 |. Interactions Between CT
The discovery of nonrandom radial positioning of CT suggested the intriguing possibility that there might be nonrandom interactions patterns among the population of CT. Initial studies using multifluorescence in situ hybridization (FISH) techniques were consistent with this view and indicated nonrandom albeit probabilistic interchromosomal associations.228,239,240 Alterations in these associations in different cell and tissue types,66,231,241,242 during cell differentiation243 and in cancer215,231,244 further supported a relationship to genomic function.
One limitation of most of these studies, however, is that they evaluated interchromosomal associations based on the distances between the centers of gravity of the CT. Thus specific interactions at the borders or interfaces between neighboring CT are not captured. Studies measuring CT-interactions based on distances between their nearest border sites, revealed that the corresponding distances based on CT centers are highly variable and often do not correspond to the positive associations measured by nearest border-to-border interactions.230,245 Furthermore, due to technical limitations of multi-FISH labeling and analysis, most of these studies analyzed up to only three chromosome pairs per nucleus.
To circumvent these difficulties, a 3-D re-FISH approach228 was adapted to simultaneously analyze large subsets of CT in individual nuclei. Figure 3 depicts typical CT multicolor labeled images from re-FISH experiments with MCF10 normal human breast and malignant breast cancer cells (Figure 3A,B), undifferentiated and differentiated human keratinocytes (Figure 3C,D), WI38 normal human fibroblasts (Figure 3E), and the NOR-CT in WI38 cells (Figure 3F). An integrated computer imaging program was then applied to segment the individual CT and determine a large battery of measurements among the total population of labeled CT in the individual nuclei.229,230,233,245,246 These measurements included the 3-D nearest neighbor distances between the borders of all labeled CT. The latter computations resulted in a listing of the 3-D nearest neighbor distances for every possible combination of chromosome pairs. In studies involving nine CT pairs, a total of 153 pairwise CT interactions were determined.230 From this data, the percentages of pairwise associations (interactions) were calculated using a threshold distance of ≤4 pixels, which corresponded to ≤0.28 μm or the approximate resolution limitation of the fluorescence microscopy used for collecting the images.229,230,233,246
FIGURE 3.
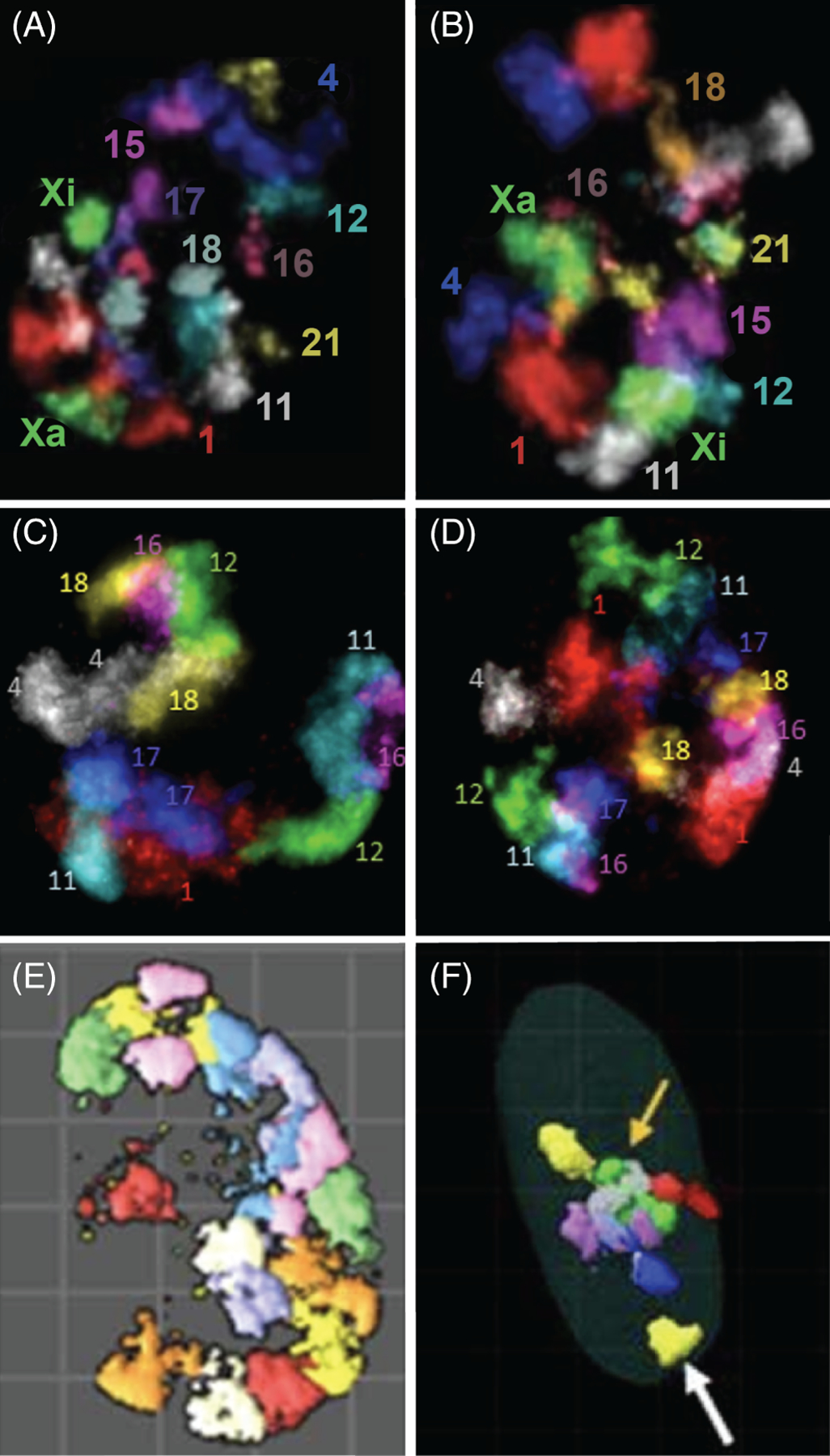
CT multicolor images from re-FISH experiments. Typical CT multicolor labeled images from re-FISH experiments are depicted in nine CT pairs (1, 4, 11, 12, 15, 16, 18, 21, X) from MCF10 normal human breast A, and malignant breast cancer cells B230; seven CT pairs (1, 4, 11, 12, 16–18) from undifferentiated C, and differentiated D human keratinocytes233; eight CT pairs (1–4, 6–9) from WI38 normal human fibroblasts E 247; and the NOR-CT pairs (13–15, 21, 22) in WI38 cells F.245 Note the NOR-CT distal to any visible nucleolus denoted by a white arrow and the dominant nucleolus with the many associated NOR-CT denoted by an orange arrow in F.245 A to D are 2-D projection images while E and F are 3-D surface renderings. An integrated computer imaging program was then applied to segment the individual CT images followed by a large battery of measurements among the total population of labeled CT in the individual nuclei229,230,233,245,246
Using these techniques, major differences were reported in the total pairwise interchromosomal interaction profiles between normal luminal MCF10A epithelial cells and the corresponding malignant MCF10Ca1a breast cells derived from the MCF10 normal cells.230 Nineteen of the 36 heterologous CT pairs changed by >10% association, and these alterations were even more pronounced (26 of 36 CT pairs) in profiles showing multiple pairwise interactions. Analysis of the total number of heterologous CT interacting with each CT demonstrated a similar average of ~ 3.5 interactions of the possible 16 heterologs in both 10A and Ca1a cells with the exception of CTX where both Xi and Xa had much lower values in both cell lines.230 Assuming these interactions are representative of the entire genome, each CT interacts on average with 9 to 10 other heterologs at any given moment or nearly one-fourth of the genome. These findings predict a high level of interactions among the individual CT in the genome. Similar results in CT from human WI38 fibroblasts support this conclusion.229
Major differences in the pairwise interactive CT profiles for WI38 and MCF10A, point to cell-type specificity in interchromosomal interactions.229 In addition, striking changes in the CT interactive profiles involving multiple interactions (>2) were detected between undifferentiated vs highly differentiated human keratinocytes.233 These overall findings of major alterations in the interchromosomal interactions relating to cell type, malignant progression, and epidermal cell differentiation are consistent with the view that the probabilistic patterns of CT interchromosomal interactions in the genome are dynamic and correlate with corresponding alterations in the global gene expression programs.
The interchromosomal positioning and interactions of the subset of human NOR chromosomes (CT13, 14, 15, 21, 22)—which participate in the formation of nucleoli and contribute rDNA for transcription and ribosome biogenesis—were investigated in WI38 human diploid fibroblast cells.245 The radial positioning of NOR-CT in the nucleus correlated with their size wherein the smaller NOR-CT (CT 21, 22) were more interior than the larger ones (CT 13–15). High levels of pairwise CT interactions ranging from 52% (CT13–21) to 82% (CT15–21) were detected as well as a triplet arrangement of CT 15-21-22 (72%). In cells with multiple nucleoli, one of the nucleoli (termed “dominant”) always associated with a higher number of NOR-bearing CT. Also, certain CT pairs more frequently contributed to the same nucleolus than to others.245 This nonrandom pattern suggested that a large number of NOR-chromosomes are poised in close proximity during the postmitotic nuclear recovery and through their NORs may contribute to the formation of the same nucleolus.
1.6 |. Dynamics of Interchromosomal Networks
The nonrandom profiles of pairwise interchromosomal associations, which demonstrate cell-type specificity and are altered during the cell cycle, malignant transformation, and keratinocyte differentiation, raise a fundamental question: Is there an overall arrangement of the individual CT into a global interactive network that might in turn drive genomic function and regulation? A major limitation in addressing this issue is that the demonstrated pairwise CT interactions, while preferred, are also probabilistic. This is most evident when one examines individual nuclei labeled with a series of CT. No two overall patterns are precisely alike and many appear very different. Thus, if a preferred overall pattern of CT associations exists in the nucleus, it will by nature be probabilistic and difficult to identify. Another difficulty is the enormous number of potential CT interactions in any given nucleus. With nine sets of labeled CT, there are 153 potential pairwise interactions among the nine homologous pairs.230 There is also the problem of properly distinguishing the two copies of each CT and their individual interchromosomal associations.
To solve these problems, computational geometry algorithms were specifically developed to determine the possible presence of probabilistic interchromosomal arrangements of an entire population of labeled CT. In an investigation involving eight chromosome pairs in WI38 lung fibroblast cells, a geometric graph mining approach termed the Generalized Median Graph or GMG247 was applied to determine the probabilistic best fit for the spatial arrangement of the entire set of eight CT pairs (see Figure 3E for multicolor CT FISH image).246 The GMG has been widely used to model the best representation for a set of structural associations in computer vision and pattern recognition research. For every image set under analysis, each of the 16 CT in the eight labeled chromosomes is segmented, masked, and represented as a simplified graph called an “association graph” containing 16 nodes corresponding to each CT. The individual association graphs are first compared with an initially unknown GMG to obtain its parameterization. A nonconvex optimization procedure is then applied to determine the GMG, which is the “best fit” match graph to all the individual association graphs. While this network of CT associations was probabilistic, other computational algorithms determined that the association graphs for each image set in the analysis contained a minimum of 40% of the connecting associations found in the final GMG interchromosomal network (ICN).246
One limitation of the GMG method was that it identified each homologous CT of a pair based on a slight difference in the apparent volumes of each homologous CT rather than their interchromosomal association differences. This problem was solved with the introduction of a geometric technique termed the Chromatic Median or CM, which uses combinatorial optimization to infer the common chromosome interaction pattern or network for the overall cell population.248 While the GMG used integer linear programming and rounding techniques, the CM is more accurate and robust. It is based on a number of newer techniques, such as semidefinite programming, multilevel rounding, geometric peeling, and adaptive sampling.249,250 The CM technique results in much better approximation ratios and yields near optimal solutions in all tested random or real data sets.248 Figure 4 illustrates the major steps of the CM to generate the most probable network of CT interchromosomal interactions.
FIGURE 4.
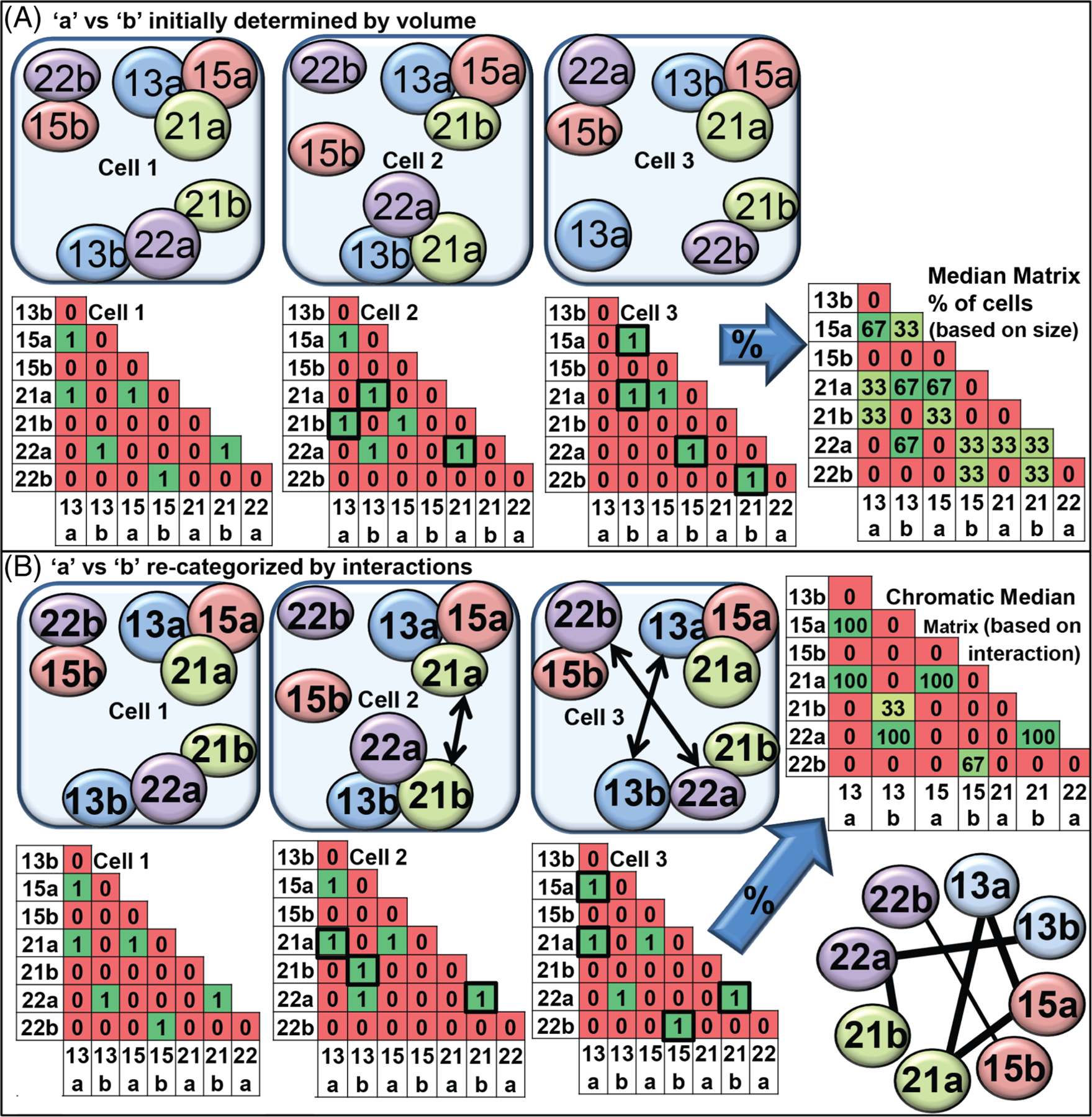
Diagram illustrating application of the chromatic median (CM) analysis to determine the best fit probabilistic arrangement of a set of CT into an interchromosomal network (ICM). A, Three schematic drawings of CT associations in nuclei are shown with the larger homolog defined as copy ‘a’ and the smaller one as copy ‘b’. The associations are represented in binary matrices wherein a 1 indicates an interaction and a 0 the absence of an interaction; B, the chromatic median program determines which homolog is “copy a” vs “copy b” based upon which other CT are associated and switches “a” for “b” to match the best fit model for the population. The percent of cells with an interaction at any given position within the matrix is calculated. Using a threshold, a chromatic median graph enriches for those connections, which are greater than randomizations of the input matrices245
Using the CM graph mining algorithm, a network was reported of 20 interchromosomal associations among a subset of nine labeled chromosomes in both normal human epithelial breast cells (MCF10A) and a malignant breast cancer line (MCF10CA1a) derived from MCF10A.230 The ICN was nearly completely altered in the malignant CA1a with only one of twenty connections in common (Figure 5A,B). Similarly, the preferred probabilistic ICN were nearly completely different between human WI38 lung fibroblasts and MCF10 human breast epithelial cells, strikingly altered during the cell cycle in MCF10 cells229 as well as undergoing major alterations (~ 70% differences in interchromosomal connections) following differentiation of human keratinocytes (Figure 5C,D).233 An ICN was also determined among the entire population of NOR-CT.245 Because NOR-CT are also found in other CT ICNs, they are likely part of an overall genomic network.
FIGURE 5.
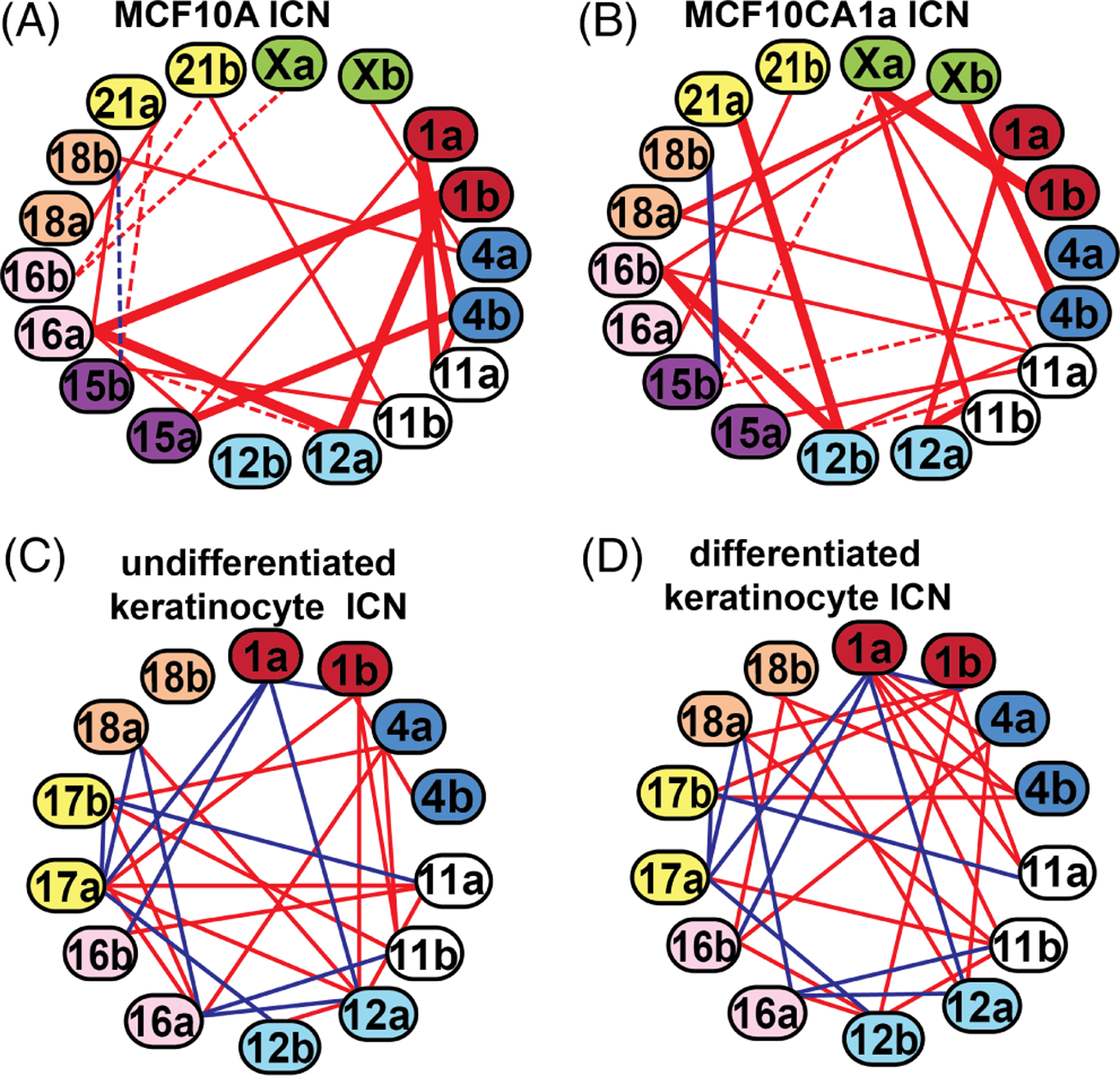
Global nonrandom interchromosomal network (ICN) models. Chromatic median (CM) analysis of the interchromosomal association data generated from re-FISH experiments involving subsets of CT have enabled the generation of the most probable interactive network for all the CT in a particular subset. The general method involved is summarized in Figure 3. The connections between CT pairs depicted in the model were all well above random simulation values.229,230 ICNs of CT 1, 4, 11, 12, 15, 16, 18, 21, X in MCF10A normal human luminal breast cells A, and MCF10CA1a malignant breast cells derived from the MCF10A cells B. There are massive differences in the ICNs of 10A and CA1a. Nineteen of the twenty preferred nonrandom pairwise associations were unique to both 10A and CA1a (red lines) with only one in common (CT 15b–18b, blue line). Thick solid connections scored the highest in pairwise association levels, thin solid lines are at moderate levels of association, while dashed connections are among the lowest of those above randomization. ICNs of CT 1, 4, 11, 12, 16, 17, 18 in undifferentiated human keratinocytes C, and differentiated keratinocytes derived from the same undifferentiated cells D. Red connecting lines are unique to each cell type while blue lines depict common interchromosomal interactions common between undifferentiated and differentiated keratinocytes. Approximately two-thirds of the pairwise associations were unique to each cell differentiation state
In conclusion, CT investigations combining the tools of re-FISH, computer imaging algorithms, and geometric graph mining approaches have revealed that large subsets of CT form ICNs with cell-type specificity. Alterations in these interactive networks during malignant cancer progression, cell differentiation, and the cell cycle illustrate their dynamic nature and suggest their possible role in the corresponding genomic expression programs of cells. Moreover, the discovery of ICNs in several different cell lines under different conditions and involving over 50% of the total human chromosomes provide support for a probabilistic “chromosome positional code” wherein the overall interactive networks are different between cell types and may contribute to the global regulation of gene expression.231,246 It is further speculated that the preferred network defined by this positional code is maintained epigenetically and facilitates specific genomic expression programs characteristic of the particular cell and its differentiation state. The probabilistic nature of the overall interactive network could in turn provide the flexibility for alterations and contribute to corresponding changes in the overall genomic program.
1.7 |. Interchromosomal Interactions and Translocations
A characteristic feature of cancer cells are the large number of chromosomal rearrangements. More than 600 recurrent balanced chromosomal rearrangements have been documented in human cancers.251 This strikingly illustrates the key role that interchromosomal interactions and translocations likely play in tumorigenesis. In order for translocations to occur, the CT involved in the translocations should be in close proximity.252,253 Indeed, the close associations of certain CT may be characteristic of the normal cell/tissue types that progress into tumorigenesis. For example, in a T-cell lymphoma mouse cell line that has a reoccurring 12:14 translocation, a close proximity of CT 12 and 14 were found in both the lymphoma cells and normal splenocytes.215 In differentiated adipocytes, CT 12 and 16 are closely associated enabling the (12, 16) translocation that is characteristic of human liposarcoma.243 Importantly, this association is not observed in preadipocytes indicating that a dynamic rearrangement of interacting CT during adipocyte differentiation plays a crucial role in human liposarcoma tumorigenesis.243 Tissue specificity of interchromosomal interactions and translocations have also been reported.242 Proximity of CT 12 and 15 but not CT 5 and 6 were observed in mouse lymphocytes where 12/15 translocations occur in lymphomas.242 Conversely, close proximity of CT 5 and 6 but not CT 12 and 15 were detected in mouse hepatocytes where 5/6 translocations are common in hepatomas.242
Studies have also documented the proximity of genes involved at the breakpoints for translocations. A t(9;22) translocation involving the ABL and BCR genes is a recurrent primary abnormality in chronic myeloid leukemia (CML) and adult acute lymphoblastic leukemia (ALL). A generally accepted hypothesis is that the physical proximity of the involved chromosomal regions may be one important factor in the genesis of these phenomena. In landmark studies, a close proximity was discovered in hematopoietic cells belonging to different cell lineages for the ABL and BCR genes of t(9;22)32,254 and the PML and RAR genes of t(15;17).32 A high level of proximity of the BCR (CT 22) and ABL (CT 9) genes involved in this translocation was also detected in B-lymphocytes.255 It was, therefore, concluded that this characteristic translocation is mediated by the close association of the participating CT in the normal cell type that give rise to these leukemia cells.255 Similarly, MYC, BCL, and immunoglobulin loci, which are recurrently translocated in various B-cell lymphomas, are preferentially positioned in close spatial proximity relative to each other in normal B cells.256
The findings that translocations are more prevalent between sites of higher interaction frequency were confirmed via the Hi-C approach.257 As anchor sites on chromatin loops are fragile sites for double strand breaks,258,259 double strand breaks are enriched at TAD boundaries.258,259 Overall, these studies implicate cell- and tissue type-specific interchromosomal interactions and higher order chromatin organization as the epigenetic basis for the generation of characteristic chromosome translocations during cancer progression.
1.8 |. Gene Positioning and Dynamics Within CT
An earlier report concluded that active genes are found at the CT boundaries.260 Other studies concluded that genes are distributed throughout the CT regardless of their level of expression.261,262 More recently single-cell Hi-C studies suggested that domains of active gene expression are enriched at the interface of CT.263,264 These findings, however, were based on a very limited number of individual cells and will require further analysis. Several studies have demonstrated that upon very active expression, genes are found within chromatin loops that project out of the CT. This has been reported for the major histocompatibility complex on CT6,24 HOX genes on CT11,23 and the epidermal differentiation complex on CT1.265 In a groundbreaking study, it was discovered that during murine erythroid differentiation, several genes involved in hemoglobin synthesis (eg, Hbb-b, Eraf, and Uros) are repositioned in close proximity to the same transcription factories for co-regulated gene expression.266 This occurs despite the large Mbp distances between these genes on CT7.
Repositioning of actively transcribed genes on different chromosomes to the same transcription factory, which must be mediated by specific interchromosomal interactions, has also been reported between Myc and Igh267 and olfactory genes.268 Another study found that several housekeeping genes colocalize into an active hub.269 Repositioning of genes to sites of high proximity may also play a role in gene silencing. Clusters of inactive genes were detected at the nuclear periphery,159,172,173 perinucleolar regions,172,173 and within polycomb bodies in Drosophila.270
The repositioning of distally located genes into common transcriptional factories is indicative of a major role that the 3-D topology of CT and their interchromosomal interactions may play in regulating gene expression. Consistent with this view, studies have found that enhancers loop back to interact with their target genes.271 These contacts have been observed across a myriad of physiological conditions and can occur over vast distances of the DNA sequence with some enhancers even looping back over megabases.272 For example, upon transcription, the β-globin locus, α-globin gene cluster, TH2, IFNG, MHC class II, CF, IgH and their respective enhancers are in close physical proximity.271,273–277 The interactions between the beta-globin and its locus of control were shown to require specific transcription factors, including EKLF1 and GATA1.276,278 It was further reported that not only do the regulatory regions of the locus control region (LCR) and TH2 interact on CT11, but they also form an interchromosomal cluster with the IFN-γ gene on CT10.279
A subset of these enhancers called super-enhancers (SEs) are clustered together and occupied by a large number of factors that function in a synergistic cooperative manner to increase expression of their cognate genes.280 SEs have been implicated in cell-type specificity281,282 and driving expression of oncogenes in cancer.281,283 The advancement of chromosome capture techniques has allowed for unbiased and global determination of enhancer-promoter interactions within the context of the global genomic organization.284 Investigations into the higher order chromatin organization around enhancers has provided the insight that the concentrated activity around SEs induce phase separation of nearby chromatin64 and that enhancers-promoter interactions are generally constrained within the TADs.128,285,286
1.9 |. 3-D Topology of Chromosomes and Gene Regulation
It is widely assumed that the 3-D arrangement of CT and the spatial positioning of genes within the CT are linked to genomic function and regulation.9,18,19,287,288 While elucidating the 3-D arrangement of individual CT and their relationships to gene activity is a challenging endeavor,214,216,228,242,289,290 progress has been made in understanding the overall shape and 3-D organization of chromatin within individual CT.239,291–299 CT display a wide range of 3-D shapes from regular ellipsoid-like to highly irregular.300,301 Properties that could influence the global organization of individual CT include heterochromatin/euchromatin levels and arrangements,302 gene density,301,303 RIDGES/ANTI-RIDGES,296 and overall levels of gene activity.304,305 A higher degree of irregularity in CT shape is found with increasing gene density.301 For example, despite being similar in sequence length, the gene-rich CT 17 is less compact and much more irregular in shape than the gene-poor CT 18.301 The potential influence of gene activity on shape irregularity is demonstrated in female cells, where one homolog of the X chromosome is inactivated (Xi) and more regular in comparison to its active counterpart Xa.304,305
Gene-rich CT have a greater proportion of regions that are sensitive to micrococcal nuclease than gene-poor CT and thus have more open chromatin fibers.302 Moreover, the CT regions that are most sensitive to micrococcal nuclease were among the most gene dense.302 By mapping the transcriptome, it was shown that these gene dense regions were more active in gene expression (RIDGES) than those that are gene poor (ANTI-RIDGES).295,306 Chromatin within RIDGES is much less compact and more irregular in shape than the chromatin in ANTI-RIDGES.295,296
If there are distinct differences among CT in overall shape, how is the chromatin arranged three dimensionally at the global level of the entire CT? Limited studies using multi-FISH and computer analysis have revealed distinct 3-D organization and specificity for relatively short regions (<5 Mbp) within CT.296,299,307 A morphometric geometrical approach and statistical shape theory for 3-D reconstruction and visualization of the mean positions of five consecutive probes on a 3.7 Mbp region of chromosome X, provided evidence for a nonrandom organization that differed between Xa and Xi.307 Similarly, a nonrandom organization in a 4.3 Mbp region of CT14 in mice was detected299 and significant differences in organization in RIDGE and anti-RIDGE regions were demonstrated for chromosomes 1 and 11 in six different cell lines.296 More recently, integrated yeast 3C data were used to model 3-D chromatin structures based on a Bayesian inference framework.308 This approach, however, is designed to model chromatin structure at a level ≤ 1Mbp.
The organization of larger regions has been until recently largely limited to investigations of chromatin folding by the method of polymer modeling and mean squared distances (MSDs)309,310 with only one previous study analyzing entire human chromosomes (CT 4, 5, and 19).310 These investigations have led to general models of chromatin loops and higher level folding that are of potentially great significance,17,309–313 but do not directly address the precise organization of chromatin within individual CT. In this regard, studies involving both FISH and chromosome capture (3C and Hi-C) techniques have been performed to fit chromosomes or regions of chromosomes to the proposed polymer models.312,314,315
While the Hi-C approach has been instrumental for understanding higher order CD of 1 to 10 Mbp across the entire chromosome and defining specific sequences within these domains such as the TADs,125,128,316,317 a multi-FISH approach is necessary for analyzing sequences separated by the much larger genomic distances that range up to the full length of the chromosome (80–240 Mbp). As valuable as 3C, Hi-C and other chromatin conformation capture techniques are for elucidation of the 3-D organization of CT,318,319 the findings of these approaches may not always correlate precisely with the actual spatial distances between DNA sites in the 3-D microenvironment of the cell nucleus.318 A more reliable approach would be to perform both multi-FISH and chromatin capture approaches.
In earlier whole chromosome studies using microscopy, CT 4, 5, and 19 were shown to behave according to a “random walk or giant loop” based on MSD analysis.310,313 These investigations demonstrated a large increase in MSD within 2 Mbp followed by a much more gradual increase in MSD extending the length of each chromosome.310 A more recent study combined the tools of 3-D microscopy, multi-FISH, computer imaging, and computational geometric analysis to analyze six labeled regions spanning each CT in the G1 and S phase of WI38 normal diploid human fibroblasts.320 The six CT investigated (1, 4, 12, 17, 18, X) are representative of both the range of gene densities and CT size in the genome. The average distance between labeled DNA probes ranged from ~15 to 60 Mbp. At these large Mbp distances between probes, MSD analysis along with studies of the folding ratios (FRs) across the CT demonstrated that each CT had a specific folding pattern with some limited alterations across the cell cycle.320 Random simulation plots of all the CT except CT 17 were very different from the actual CT determinations and uniformly showed little or no changes in MSD or FRs across the entire simulated CT. Thus, when measured at large genomic separations, each CT displays a different profile of genomic to spatial distances that are nonrandom. An exception was CT 17 whose MSD/FR plots were similar to its random simulations with only a small portion displaying nonrandom folding.320
A classic data-mining algorithm termed the k-means321–324 was then applied to determine the best fit probabilistic 3-D topology of the labeled probes across each CT320 Consistent with the MSD analysis, the 3-D topological models derived from this data mining approach were specific for each CT (Figure 6) and had a high degree of nonrandomness compared with models generated from random simulations. CT 17, however, was again characterized by a high level of randomness.320 Alterations were detected in G1 vs S phase and it was concluded that each CT except CT 17 had a specific probabilistic 3-D topology at the global level spanning the whole chromosome (Figure 6). This analysis further revealed that at least one of the telomeres of each chromosome was located at or near the CT periphery. The most striking examples of this were the telomere regions of 17-q during S phase and Xi-p, which were positioned on projections extending from the main CT body.320 Interestingly, the labeled q-telomeric region of CT 17 contains the gene for tubulin cofactor D, which is a cell cycle regulated protein, and plays a role in cell division.325 Similarly, the pseudoautosomal region on chromosome X, which is homologous to a region on the Y chromosome and escapes X-inactivation,326 was found to be the most peripheral in the inactive X in comparison with all other regions in Xi or its Xa counterpart.320
FIGURE 6.
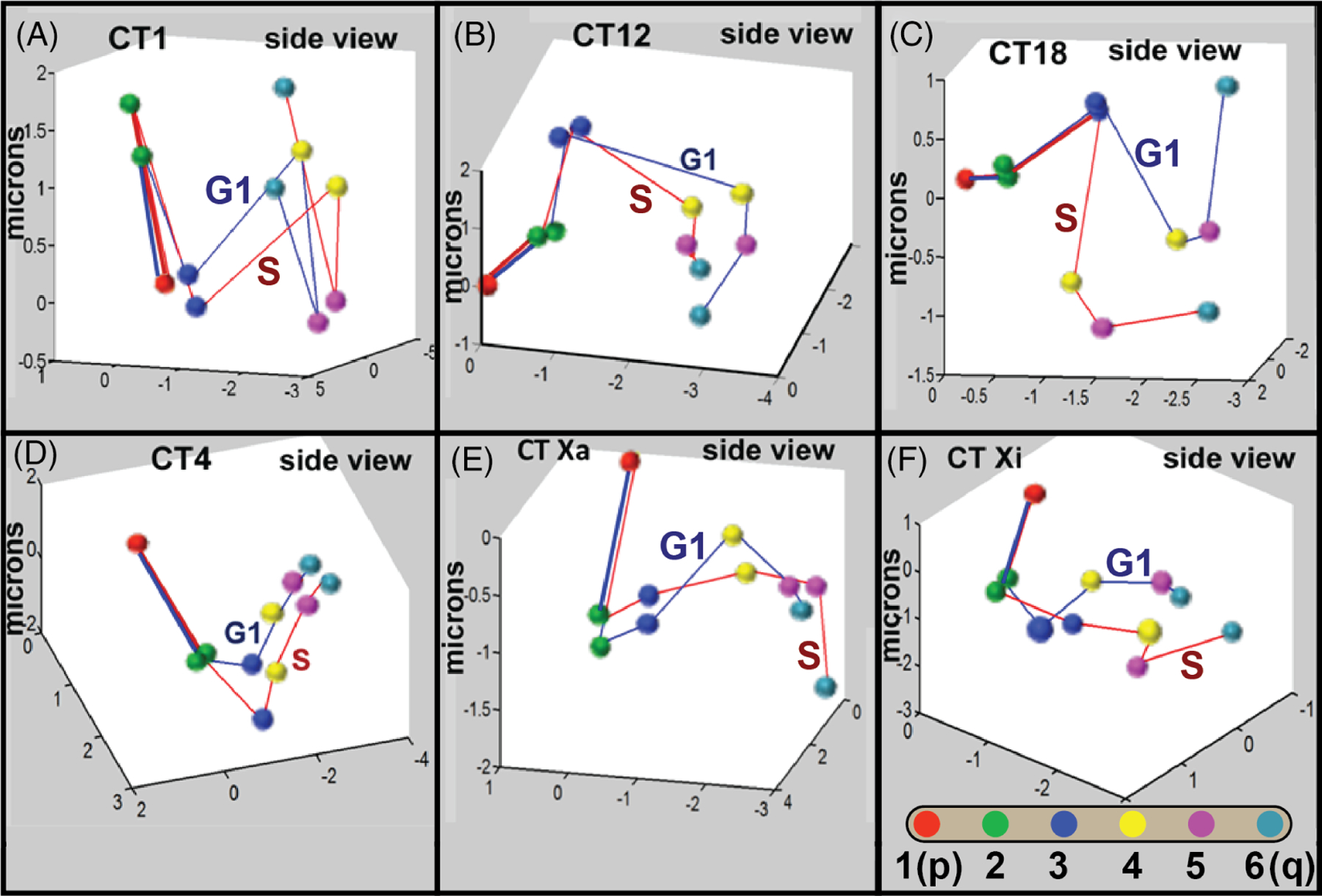
Large scale 3-D topology of CT. The large scale 3-D topology of six CT ranging in size and gene density (CT 1, 4, 12, 17, 18, X) was determined in the G1 and S-phases of human WI-38 diploid fibroblasts. As a first step, multicolor FISH was performed using six BAC probes extending across each chromosome. In-house computational geometric algorithms were applied to measure the 3-D distances between every combination of probes and to elucidate data-mined structural patterns. These findings demonstrated a high degree of nonrandom arrangement of individual CT that were specific for each CT and displayed distinct changes during the cell cycle.320 A classic clustering and pattern recognition algorithm (k means) was then applied to determine the best fit probabilistic arrangement (topology) in the 3-D positioning of the six BAC probe positions within each CT.320 The analysis successfully defined a single most probable arrangement and specificity in the 3-D topology of each CT. Comparisons with random simulations demonstrated that all the CT except CT 17 showed significant levels of nonrandomness in the preferred 3-D models with some significant differences between the G1 and S-phases. Side views of these 3-D models are shown in panels A-F with connecting blue lines for CT in G1 and red lines for CT in S-phase. Position 1 and the trajectory to position 2 are overlaid in order to compare topology between individual CT in G1 and S phase. The positional six color scheme from the p(1) to q(6) ends of each CT are shown in panel F
In conclusion, recent advancements in chromosome capture techniques such as Hi-C are enabling analysis of the intricacies of chromatin looping and folding and the identification of specific DNA interactions within CD ≤10 Mbp.9,125,128,316,319 Several physical models have been proposed to explain the organization of chromatin at even higher levels of organization but their application to resolving the global 3-D topology of individual chromosomes has been limited.309 By combining multi-FISH 3-D imaging with computational and pattern recognition algorithms, progress has been made in understanding both specificity and uniqueness in the overall global folding of each chromosome as well as some cell cycle-related alterations.320 Chromosome specific differences in structural organization and changes during the cell cycle may provide topological signatures that contribute to the global expression programs of individual chromosomes.
Overall, studies of CT organization and their alterations have potentially important implications for understanding cancer progression and deciphering optimal treatment strategies. Cancer is characterized by compromised global interactions and regulation of the genome.327 A major challenge in cancer screening is the determination of which patients require aggressive treatment or only to be monitored. Determining whether particular alterations in epigenetics and/or chromatin organization are indicative of poor outcomes could aid in defining tumors that are particularly deleterious. Furthermore, understanding how genes are dysregulated upon disruption of the normal “wiring” of the nucleus by translocations, deletions, and amplifications that are hallmarks of cancer should also enable more targeted therapeutic strategies.
Funding information
Division of Information and Intelligent Systems, Grant/Award Numbers: IIIS1422591, IIS-0713489, IIS-1115220; NIH Clinical Center, Grant/Award Numbers: GM072131, GM-23922; University at Buffalo Foundation, Grant/Award Number: 9351115726; National Cancer Institute, Grant/Award Number: F32-CA220935; University at Buffalo; National Science Foundation; National Institutes of Health
ABBREVIATIONS:
- CT
Chromosome territory
- S/MARs
Matrix or scaffold attachment regions
- CTCF
CCCTC-Binding Factor
- MATR3
Matrin 3
- LBR
Lamin-B receptor
- LADs
Lamin associated domains
- NOR
Nucleolar organizer region
- NADs
Nucleolar-associated domains
- RS
Replication sites
- TS
Transcription site
- CD
Chromatin domain
- TAD
Topologically associating domain
- 3C
Chromosome conformation capture
- Hi-C
High throughput chromosome conformation capture
- HP1
Heterochromatin protein 1
- SEs
Superenhancers
- RIDGES
regions of increased gene expression
- MSD
Mean squared distance
- FRs
Folding ratios
- GMG
Generalized Median Graph
- CM
Chromatic Median
REFERENCES
- 1.Berezney R Regulating the mammalian genome: the role of nuclear architecture. Adv Enzyme Regul. 2002;42:39–52. [DOI] [PubMed] [Google Scholar]
- 2.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8(2):104–115. [DOI] [PubMed] [Google Scholar]
- 3.Stein GS, Zaidi SK, Stein JL, et al. Genetic and epigenetic regulation in nuclear microenvironments for biological control in cancer. J Cell Biochem. 2008;104(6):2016–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malyavantham KS, Bhattacharya S, Barbeitos M, et al. Identifying functional neighborhoods within the cell nucleus: Proximity analysis of early S-phase replicating chromatin domains to sites of transcription, RNA polymerase II, HP1gamma, matrin 3 and SAF-A. J Cell Biochem. 2008;105(2):391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaidi SK, Young DW, Javed A, et al. Nuclear microenvironments in biological control and cancer. Nat Rev Cancer. 2007;7(6):454–463. [DOI] [PubMed] [Google Scholar]
- 6.Misteli T Beyond the sequence: cellular organization of genome function. Cell. 2007;128(4):787–800. [DOI] [PubMed] [Google Scholar]
- 7.Misteli T Concepts in nuclear architecture. Bioessays. 2005;27(5): 477–487. [DOI] [PubMed] [Google Scholar]
- 8.Malyavantham KS, Bhattacharya S, Alonso WD, Acharya R, Berezney R. Spatio-temporal dynamics of replication and transcription sites in the mammalian cell nucleus. Chromosoma. 2008;117(6):553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bickmore WA. The spatial organization of the human genome. Annu Rev Genomics Hum Genet, Vol 14. 2013;14:67–84. [DOI] [PubMed] [Google Scholar]
- 10.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447(7143):413–417. [DOI] [PubMed] [Google Scholar]
- 11.Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135(1):9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malyavantham KS, Bhattacharya S, Berezney R. The architecture of functional neighborhoods within the mammalian cell nucleus. Adv Enzyme Regul, Vol 50. 2010;50:126–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cremer T, Cremer M, Hubner B, et al. The 4D nucleome: evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett. 2015;589(20 Pt A):2931–2943. [DOI] [PubMed] [Google Scholar]
- 14.Hubner MR, Eckersley-Maslin MA, Spector DL. Chromatin organization and transcriptional regulation. Curr Opin Genet Dev. 2013;23(2): 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonev B, Cavalli G. Organization and function of the 3D genome. Nat Rev Genet. 2016;17(12):772. [DOI] [PubMed] [Google Scholar]
- 16.Kumaran RI, Thakar R, Spector DL. Chromatin dynamics and gene positioning. Cell. 2008;132(6):929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pombo A, Dillon N. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol. 2015;16(4):245–257. [DOI] [PubMed] [Google Scholar]
- 18.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2(3):a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories - a functional nuclear landscape. Curr Opin Cell Biol. 2006;18(3):307–316. [DOI] [PubMed] [Google Scholar]
- 20.Grant PA. A tale of histone modifications. Genome Biol. 2001;2(4): REVIEWS0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schubeler D, MacAlpine DM, Scalzo D, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18(11):1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18(10):1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volpi EV, Chevret E, Jones T, et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci. 2000;113(Pt 9):1565–1576. [DOI] [PubMed] [Google Scholar]
- 25.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. [DOI] [PubMed] [Google Scholar]
- 26.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. [DOI] [PubMed] [Google Scholar]
- 27.Pombo A, Cuello P, Schul W, et al. Regional and temporal specialization in the nucleus: a transcriptionally-active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J. 1998;17(6):1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geiman TM, Robertson KD. Chromatin remodeling, histone modifications, and DNA methylation-how does it all fit together? J Cell Biochem. 2002;87(2):117–125. [DOI] [PubMed] [Google Scholar]
- 29.Hashimshony T, Zhang J, Keshet I, Bustin M, Cedar H. The role of DNA methylation in setting up chromatin structure during development. Nat Genet. 2003;34(2):187–192. [DOI] [PubMed] [Google Scholar]
- 30.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. [DOI] [PubMed] [Google Scholar]
- 31.Jenuwein T, Allis CD. Translating the histone code. Science. 2001; 293(5532):1074–1080. [DOI] [PubMed] [Google Scholar]
- 32.Neves H, Ramos C, da Silva MG, Parreira A, Parreira L. The nuclear topography of ABL, BCR, PML, and RARalpha genes: evidence for gene proximity in specific phases of the cell cycle and stages of hematopoietic differentiation. Blood. 1999;93(4):1197–1207. [PubMed] [Google Scholar]
- 33.Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007; 21(23):3027–3043. [DOI] [PubMed] [Google Scholar]
- 34.Razin SV, Iarovaia OV, Sjakste N, et al. Chromatin domains and regulation of transcription. J Mol Biol. 2007;369(3):597–607. [DOI] [PubMed] [Google Scholar]
- 35.Berezney R, Mortillaro MJ, Ma H, Wei X, Samarabandu J. The nuclear matrix: a structural milieu for genomic function. Int Rev Cytol. 1995; 162A:1–65. [DOI] [PubMed] [Google Scholar]
- 36.Wei X, Samarabandu J, Devdhar RS, Siegel AJ, Acharya R, Berezney R. Segregation of transcription and replication sites into higher order domains. Science. 1998;281(5382):1502–1506. [DOI] [PubMed] [Google Scholar]
- 37.Markaki Y, Gunkel M, Schermelleh L, et al. Functional nuclear organization of transcription and DNA replication: a topographical marriage between chromatin domains and the interchromatin compartment. Cold Spring Harb Symp Quant Biol. 2010;75:475–492. [DOI] [PubMed] [Google Scholar]
- 38.Fritz AJ, Barutcu AR, Martin-Buley L, et al. Chromosomes at work: organization of chromosome territories in the interphase nucleus. J Cell Biochem. 2016;117(1):9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei X, Somanathan S, Samarabandu J, Berezney R. Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. J Cell Biol. 1999;146(3):543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matharu NK, Ahanger SH. Chromatin insulators and topological domains: adding new dimensions to 3D genome architecture. Genes (Basel). 2015;6(3):790–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rabl C Über Zelltheilung. Morph Jb. 1885;10:214–330. [Google Scholar]
- 42.Boveri T Die Blastomerenkerne von Ascaris megalocephala und die Theorie der Chromosomenindividualität. Arch Zellforsch. 1909;3:181–268. [Google Scholar]
- 43.Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective. Part I. The rise of chromosome territories. Eur J Histochem. 2006;50(3):161–176. [PubMed] [Google Scholar]
- 44.Stack SM, Brown DB, Dewey WC. Visualization of interphase chromosomes. J Cell Sci. 1977;26:281–299. [DOI] [PubMed] [Google Scholar]
- 45.Zorn C, Cremer C, Cremer T, Zimmer J. Unscheduled DNA synthesis after partial UV irradiation of the cell nucleus. Distribution in interphase and metaphase. Exp Cell Res. 1979;124(1):111–119. [DOI] [PubMed] [Google Scholar]
- 46.Cremer T, Peterson SP, Cremer C, Berns MW. Laser microirradiation of Chinese hamster cells at wavelength 365 nm: effects of psoralen and caffeine. Radiat Res. 1981;85(3):529–543. [PubMed] [Google Scholar]
- 47.Lichter P, Cremer T, Borden J, Manuelidis L, Ward DC. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum Genet. 1988;80(3):224–234. [DOI] [PubMed] [Google Scholar]
- 48.Manuelidis L Individual interphase chromosome domains revealed by in situ hybridization. Hum Genet. 1985;71(4):288–293. [DOI] [PubMed] [Google Scholar]
- 49.Schardin M, Cremer T, Hager HD, Lang M. Specific staining of human chromosomes in Chinese hamster x man hybrid cell lines demonstrates interphase chromosome territories. Hum Genet. 1985;71(4):281–287. [DOI] [PubMed] [Google Scholar]
- 50.Zink D, Cremer T. Cell nucleus: chromosome dynamics in nuclei of living cells. Curr Biol. 1998;8(9):R321–R324. [DOI] [PubMed] [Google Scholar]
- 51.Visser AE, Aten JA. Chromosomes as well as chromosomal subdomains constitute distinct units in interphase nuclei. J Cell Sci. 1999; 112(Pt 19):3353–3360. [DOI] [PubMed] [Google Scholar]
- 52.Zink D, Cremer T, Saffrich R, et al. Structure and dynamics of human interphase chromosome territories in vivo. Hum Genet. 1998;102(2):241–251. [DOI] [PubMed] [Google Scholar]
- 53.Pliss A, Malyavantham K, Bhattacharya S, Zeitz M, Berezney R. Chromatin dynamics is correlated with replication timing. Chromosoma. 2009;118(4):459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pliss A, Malyavantham KS, Bhattacharya S, Berezney R. Chromatin dynamics in living cells: Identification of oscillatory motion. J Cell Physiol. 2013;228(3):609–616. [DOI] [PubMed] [Google Scholar]
- 55.Gasser SM. Nuclear architecture - visualizing chromatin dynamics in interphase nuclei. Science. 2002;296(5572):1412–1416. [DOI] [PubMed] [Google Scholar]
- 56.Stein GS, Zaidi SK, Braastad CD, et al. Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. Trends Cell Biol. 2003;13(11):584–592. [DOI] [PubMed] [Google Scholar]
- 57.Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. [DOI] [PubMed] [Google Scholar]
- 58.Ma H, Samarabandu J, Devdhar RS, et al. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J Cell Biol. 1998;143(6):1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20(3):290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shopland LS, Johnson CV, Byron M, McNeil J, Lawrence JB. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J Cell Biol. 2003;162(6):981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mehta IS, Figgitt M, Clements CS, Kill IR, Bridger JM. Alterations to nuclear architecture and genome behavior in senescent cells. Ann N Y Acad Sci. 2007;1100:250–263. [DOI] [PubMed] [Google Scholar]
- 62.Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19(2):172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghule PN, Seward DJ, Fritz AJ, et al. Higher order genomic organization and regulatory compartmentalization for cell cycle control at the G1/S-phase transition. J Cell Physiol. 2018;233(10):6406–6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A phase separation model for transcriptional control. Cell. 2017;169(1):13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Erdel F, Rippe K. Formation of chromatin subcompartments by phase separation. Biophys J. 2018;114(10):2262–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeitz MJ, Malyavantham KS, Seifert B, Berezney R. Matrin 3: chromosomal distribution and protein interactions. J Cell Biochem. 2009; 108(1):125–133. [DOI] [PubMed] [Google Scholar]
- 67.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4(9):677–687. [DOI] [PubMed] [Google Scholar]
- 68.Nickerson J Experimental observations of a nuclear matrix. J Cell Sci. 2001;114(Pt 3):463–474. [DOI] [PubMed] [Google Scholar]
- 69.Albiez H, Cremer M, Tiberi C, et al. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 2006;14(7):707–733. [DOI] [PubMed] [Google Scholar]
- 70.Nickerson JA, Blencowe BJ, Penman S. The architectural organization of nuclear metabolism. Int Rev Cytol. 1995;162A:67–123. [DOI] [PubMed] [Google Scholar]
- 71.Berezney R The nuclear matrix: structure, function and DNA replication In: Bittar EE, ed. Advances in Molecular and Cell Biology. Vol 4 Connecticut. Stamford, CT: Jai Press Inc; 1992:37–73. [Google Scholar]
- 72.Berezney R Dynamic properties of the nuclear matrix In: Busch H, ed. The Cell Nucleus Chromatin Part D. Vol 7 New York, NY: Academic Press; 1979:413–456. [Google Scholar]
- 73.Berezney R, Coffey DS. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974;60(4):1410–1417. [DOI] [PubMed] [Google Scholar]
- 74.Berezney R, Coffey DS. Nuclear matrix. Isolation and characterization of a framework structure from rat liver nuclei. J Cell Biol. 1977;73(3): 616–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berezney R. Organization and functions of the nuclear matric In: Hnilica LS, ed. Chromosomal Nonhistone Proteins - Structural Associations. Boca Raton, FL: CRC Press; 1984:119–180. [Google Scholar]
- 76.Berezney R, Basler J, Buccholtz LA, Smith HC, Siegel AJ. Nuclear matrix organization and DNA replication In: Maul GG, ed. The Nuclear Envelope and Nuclear Matrix. Vol 1982 New York: Alan R. Liss Inc.; 1982:183–197. [Google Scholar]
- 77.Nakayasu H, Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol. 1989;108(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luderus ME, van Steensel B, Chong L, Sibon OC, Cremers FF, de Lange T. Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J Cell Biol. 1996;135(4): 867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gasser SM, Laemmli UK. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986;46(4):521–530. [DOI] [PubMed] [Google Scholar]
- 80.de Lange T Human telomeres are attached to the nuclear matrix. EMBO J. 1992;11(2):717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang GS, Luo WJ, Pan WJ, Ding MX, Zhai ZH. Association of chromosomal telomere DNA with nuclear matrix in HeLa cell. Sci China B. 1994;37(6):691–700. [PubMed] [Google Scholar]
- 82.Chaly N, Little JE, Brown DL. Localization of nuclear antigens during preparation of nuclear matrices in situ. Can J Biochem Cell Biol. 1985; 63(6):644–653. [DOI] [PubMed] [Google Scholar]
- 83.Petrova NV, Iarovaia OV, Verbovoy VA, Razin SV. Specific radial positions of centromeres of human chromosomes X, 1, and 19 remain unchanged in chromatin-depleted nuclei of primary human fibroblasts: evidence for the organizing role of the nuclear matrix. J Cell Biochem. 2005;96(4):850–857. [DOI] [PubMed] [Google Scholar]
- 84.Mirkovitch J, Mirault ME, Laemmli UK. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984;39(1):223–232. [DOI] [PubMed] [Google Scholar]
- 85.Dijkwel PA, Hamlin JL. Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol Cell Biol. 1988;8(12):5398–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wilson RHC, Coverley D. Transformation-induced changes in the DNA-nuclear matrix interface, revealed by high-throughput analysis of DNA halos. Sci Rep. 2017;7(1):6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heng HH, Goetze S, Ye CJ, et al. Chromatin loops are selectively anchored using scaffold/matrix-attachment regions. J Cell Sci. 2004; 117(Pt 7):999–1008. [DOI] [PubMed] [Google Scholar]
- 88.Vogelstein B, Pardoll DM, Coffey DS. Supercoiled loops and eucaryotic DNA replicaton. Cell. 1980;22(1 Pt 1):79–85. [DOI] [PubMed] [Google Scholar]
- 89.Cockerill PN, Garrard WT. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44(2):273–282. [DOI] [PubMed] [Google Scholar]
- 90.Smith HC, Puvion E, Buchholtz LA, Berezney R. Spatial distribution of DNA loop attachment and replicational sites in the nuclear matrix. J Cell Biol. 1984;99(5):1794–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schubeler D, Mielke C, Maass K, Bode J. Scaffold/matrix-attached regions act upon transcription in a context-dependent manner. Biochemistry. 1996;35(34):11160–11169. [DOI] [PubMed] [Google Scholar]
- 92.Cockerill PN, Yuen MH, Garrard WT. The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements. J Biol Chem. 1987;262(11):5394–5397. [PubMed] [Google Scholar]
- 93.Razin SV, Mantieva VL, Georgiev GP. DNA adjacent to attachment points of deoxyribonucleoprotein fibril to chromosomal axial structure is enriched in reiterated base sequences. Nucleic Acids Res. 1978;5(12):4737–4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Frisch M, Frech K, Klingenhoff A, Cartharius K, Liebich I, Werner T. In silico prediction of scaffold/matrix attachment regions in large genomic sequences. Genome Res. 2002;12(2):349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Platts AE, Quayle AK, Krawetz SA. In-silico prediction and observations of nuclear matrix attachment. Cell Mol Biol Lett. 2006;11(2):191–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Keaton MA, Taylor CM, Layer RM, Dutta A. Nuclear scaffold attachment sites within ENCODE regions associate with actively transcribed genes. PLoS One. 2011;6(3):e17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dobson JR, Hong D, Barutcu AR, et al. Identifying nuclear matrix-attached DNA across the genome. J Cell Physiol. 2017;232(6):12951305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ma H, Siegel AJ, Berezney R. Association of chromosome territories with the nuclear matrix: disruption of human chromosome territories correlates with the release of a subset of nuclear matrix proteins. J Cell Biol. 1999;146(3):531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Berrios M, Osheroff N, Fisher PA. In situ localization of DNA topoisomerase II, a major polypeptide component of the Drosophila nuclear matrix fraction. Proc Natl Acad Sci U S A. 1985;82(12):4142–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gasser SM, Laroche T, Falquet J, Boy de la Tour E, Laemmli UK. Metaphase chromosome structure. Involvement of topoisomerase II. J Mol Biol. 1986;188(4):613–629. [DOI] [PubMed] [Google Scholar]
- 101.Adachi Y, Kas E, Laemmli UK. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 1989; 8(13):3997–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Miyaji M, Furuta R, Sano K, Tsutsui KM, Tsutsui K. Genomic regions targeted by DNA topoisomerase IIbeta frequently interact with a nuclear scaffold/matrix protein hnRNP U/SAF-A/SP120. J Cell Biochem. 2015;116(4):677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dunn KL, Zhao H, Davie JR. The insulator binding protein CTCF associates with the nuclear matrix. Exp Cell Res. 2003;288(1):218–223. [DOI] [PubMed] [Google Scholar]
- 104.Gregson HC, Schmiesing JA, Kim JS, Kobayashi T, Zhou S, Yokomori K. A potential role for human cohesin in mitotic spindle aster assembly. J Biol Chem. 2001;276(50):47575–47582. [DOI] [PubMed] [Google Scholar]
- 105.McCracken S, Longman D, Marcon E, et al. Proteomic analysis of SRm160-containing complexes reveals a conserved association with cohesin. J Biol Chem. 2005;280(51):42227–42236. [DOI] [PubMed] [Google Scholar]
- 106.Guillou E, Ibarra A, Coulon V, et al. Cohesin organizes chromatin loops at DNA replication factories. Genes Dev. 2010;24(24):2812–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hibino Y, Usui T, Morita Y, et al. Molecular properties and intracellular localization of rat liver nuclear scaffold protein P130. Biochim Biophys Acta. 2006;1759(5):195–207. [DOI] [PubMed] [Google Scholar]
- 108.Hibino Y, Ohzeki H, Sugano N, Hiraga K. Transcription modulation by a rat nuclear scaffold protein, P130, and a rat highly repetitive DNA component or various types of animal and plant matrix or scaffold attachment regions. Biochem Biophys Res Commun. 2000;279(1): 282–287. [DOI] [PubMed] [Google Scholar]
- 109.Romig H, Fackelmayer FO, Renz A, Ramsperger U, Richter A. Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J. 1992;11(9):3431–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fackelmayer FO, Dahm K, Renz A, Ramsperger U, Richter A. Nucleic-acid-binding properties of hnRNP-U/SAF-A, a nuclear-matrix protein which binds DNA and RNA in vivo and in vitro. Eur J Biochem. 1994; 221(2):749–757. [DOI] [PubMed] [Google Scholar]
- 111.Gohring F, Fackelmayer FO. The scaffold/matrix attachment region binding protein hnRNP-U (SAF-A) is directly bound to chromosomal DNA in vivo: a chemical cross-linking study. Biochemistry. 1997;36(27):8276–8283. [DOI] [PubMed] [Google Scholar]
- 112.Kipp M, Gohring F, Ostendorp T, et al. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol Cell Biol. 2000;20(20):7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fan H, Lv P, Huo X, et al. The nuclear matrix protein HNRNPU maintains 3D genome architecture globally in mouse hepatocytes. Genome Res. 2018;28(2):192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Renz A, Fackelmayer FO. Purification and molecular cloning of the scaffold attachment factor B (SAF-B), a novel human nuclear protein that specifically binds to S/MAR-DNA. Nucleic Acids Res. 1996;24(5):843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nayler O, Stratling W, Bourquin JP, et al. SAF-B protein couples transcription and pre-mRNA splicing to SAR/MAR elements. Nucleic Acids Res. 1998;26(15):3542–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.von Kries JP, Buhrmester H, Stratling WH. A matrix/scaffold attachment region binding protein: identification, purification, and mode of binding. Cell. 1991;64(1):123–135. [DOI] [PubMed] [Google Scholar]
- 117.Cai S, Han HJ, Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat Genet. 2003;34(1):42–51. [DOI] [PubMed] [Google Scholar]
- 118.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38(11):1278–1288. [DOI] [PubMed] [Google Scholar]
- 119.Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452(7184):187–193. [DOI] [PubMed] [Google Scholar]
- 120.Luderus ME, de Graaf A, Mattia E, et al. Binding of matrix attachment regions to lamin B1. Cell. 1992;70(6):949–959. [DOI] [PubMed] [Google Scholar]
- 121.Luderus ME, den Blaauwen JL, de Smit OJ, Compton DA, van Driel R. Binding of matrix attachment regions to lamin polymers involves single-stranded regions and the minor groove. Mol Cell Biol. 1994;14(9):6297–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Albrethsen J, Knol JC, Jimenez CR. Unravelling the nuclear matrix proteome. J Proteomics. 2009;72(1):71–81. [DOI] [PubMed] [Google Scholar]
- 123.Barboro P, Repaci E, D’Arrigo C, Balbi C. The role of nuclear matrix proteins binding to matrix attachment regions (Mars) in prostate cancer cell differentiation. PLoS One. 2012;7(7):e40617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Iradi MCG, Triplett JC, Thomas JD, et al. Characterization of gene regulation and protein interaction networks for Matrin 3 encoding mutations linked to amyotrophic lateral sclerosis and myopathy. Sci Rep. 2018;8(1):4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dixon JR, Gorkin DU, Ren B. Chromatin domains: the unit of chromosome organization. Mol Cell. 2016;62(5):668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Williams A, Flavell RA. The role of CTCF in regulating nuclear organization. J Exp Med. 2008;205(4):747–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xu Y, Guo W, Li P, et al. Long-range chromosome interactions mediated by cohesin shape circadian gene expression. PLoS Genet. 2016; 12(5):e1005992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19(1):24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Holwerda SJ, de Laat W. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos Trans R Soc Lond B Biol Sci. 2013;368(1620):20120369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tang Z, Luo OJ, Li X, et al. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell. 2015;163 (7):1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lieberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14(6):390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Seitan VC, Faure AJ, Zhan Y, et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013;23(12):2066–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Belgrader P, Dey R, Berezney R. Molecular cloning of matrin 3. A 125-kilodalton protein of the nuclear matrix contains an extensive acidic domain. J Biol Chem. 1991;266(15):9893–9899. [PubMed] [Google Scholar]
- 136.Nakayasu H, Berezney R. Nuclear matrins: identification of the major nuclear matrix proteins. Proc Natl Acad Sci U S A. 1991;88(22): 10312–10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Salton M, Elkon R, Borodina T, et al. Matrin 3 binds and stabilizes mRNA. PLoS One. 2011;6(8):e23882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54 (nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106(4):465–475. [DOI] [PubMed] [Google Scholar]
- 139.Yedavalli VS, Jeang KT. Matrin 3 is a co-factor for HIV-1 Rev in regulating post-transcriptional viral gene expression. Retrovirology. 2011; 8:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Coelho MB, Attig J, Bellora N, et al. Nuclear matrix protein Matrin 3 regulates alternative splicing and forms overlapping regulatory networks with PTB. EMBO J. 2015;34(5):653–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Boehringer A, Garcia-Mansfield K, Singh G, Bakkar N, Pirrotte P, Bowser R. ALS associated mutations in Matrin 3 alter protein-protein interactions and impede mRNA nuclear export. Sci Rep. 2017;7(1):14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Erazo A, Goff SP. Nuclear matrix protein Matrin 3 is a regulator of ZAP-mediated retroviral restriction. Retrovirology. 2015;12:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Giordano G, Sanchez-Perez AM, Montoliu C, et al. Activation of NMDA receptors induces protein kinase A-mediated phosphorylation and degradation of matrin 3. Blocking these effects prevents NMDA-induced neuronal death. J Neurochem. 2005;94(3):808–818. [DOI] [PubMed] [Google Scholar]
- 144.Przygodzka P, Boncela J, Cierniewski CS. Matrin 3 as a key regulator of endothelial cell survival. Exp Cell Res. 2011;317(6):802–811. [DOI] [PubMed] [Google Scholar]
- 145.Rayaprolu S, D’Alton S, Crosby K, et al. Heterogeneity of Matrin 3 in the developing and aging murine central nervous system. J Comp Neurol. 2016;524(14):2740–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gallego-Iradi MC, Clare AM, Brown HH, Janus C, Lewis J, Borchelt DR. Subcellular localization of Matrin 3 containing mutations associated with ALS and distal myopathy. PLoS One. 2015;10 (11):e0142144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Johnson JO, Pioro EP, Boehringer A, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17(5):664–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Quintero-Rivera F, Xi QJ, Keppler-Noreuil KM, et al. MATR3 disruption in human and mouse associated with bicuspid aortic valve, aortic coarctation and patent ductus arteriosus. Hum Mol Genet. 2015;24 (8):2375–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Senderek J, Garvey SM, Krieger M, et al. Autosomal-dominant distal myopathy associated with a recurrent missense mutation in the gene encoding the nuclear matrix protein, matrin 3. Am J Hum Genet. 2009;84(4):511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gruenbaum Y, Goldman RD, Meyuhas R, et al. The nuclear lamina and its functions in the nucleus. Int Rev Cytol. 2003;226:1–62. [DOI] [PubMed] [Google Scholar]
- 151.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6(1):21–31. [DOI] [PubMed] [Google Scholar]
- 152.Bridger JM, Foeger N, Kill IR, Herrmann H. The nuclear lamina. Both a structural framework and a platform for genome organization. FEBS J. 2007;274(6):1354–1361. [DOI] [PubMed] [Google Scholar]
- 153.Amendola M, van Steensel B. Mechanisms and dynamics of nuclear lamina-genome interactions. Curr Opin Cell Biol. 2014;28:61–68. [DOI] [PubMed] [Google Scholar]
- 154.Briand N, Collas P. Laminopathy-causing lamin A mutations reconfigure lamina-associated domains and local spatial chromatin conformation. Nucleus. 2018;9(1):216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Burke B, Stewart CL. The laminopathies: the functional architecture of the nucleus and its contribution to disease. Annu Rev Genomics Hum Genet. 2006;7:369–405. [DOI] [PubMed] [Google Scholar]
- 156.Dobrzynska A, Gonzalo S, Shanahan C, Askjaer P. The nuclear lamina in health and disease. Nucleus. 2016;7(3):233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lin F, Morrison JM, Wu W, Worman HJ. MAN1, an integral protein of the inner nuclear membrane, binds Smad2 and Smad3 and antagonizes transforming growth factor-beta signaling. Hum Mol Genet. 2005;14(3):437–445. [DOI] [PubMed] [Google Scholar]
- 158.van Steensel B, Belmont AS. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell. 2017;169(5):780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Guelen L, Pagie L, Brasset E, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453(7197):948–951. [DOI] [PubMed] [Google Scholar]
- 160.Butin-Israeli V, Adam SA, Goldman AE, Goldman RD. Nuclear lamin functions and disease. Trends Genet. 2012;28(9):464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Maraldi NM, Lattanzi G, Capanni C, et al. Laminopathies: a chromatin affair. Adv Enzyme Regul. 2006;46:33–49. [DOI] [PubMed] [Google Scholar]
- 162.Maraldi NM, Lattanzi G, Squarzoni S, Capanni C, Cenni V, Manzoli FA. Implications for nuclear organization and gene transcription of lamin A/C specific mutations. Adv Enzyme Regul. 2005;45:1–16. [DOI] [PubMed] [Google Scholar]
- 163.Depreux FF, Puckelwartz MJ, Augustynowicz A, et al. Disruption of the lamin A and matrin-3 interaction by myopathic LMNA mutations. Hum Mol Genet. 2015;24(15):4284–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. Nuclear lamins. Cold Spring Harb Perspect Biol. 2010;2(11):a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Moir RD, Yoon M, Khuon S, Goldman RD. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J Cell Biol. 2000;151(6):1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bronshtein I, Kepten E, Kanter I, et al. Loss of lamin A function increases chromatin dynamics in the nuclear interior. Nat Commun. 2015;6:8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Taheri F, Isbilir B, Muller G, et al. Random motion of chromatin is influenced by Lamin A interconnections. Biophys J. 2018;114(10):2465–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stixova L, Matula P, Kozubek S, et al. Trajectories and nuclear arrangement of PML bodies are influenced by A-type lamin deficiency. Biol Cell. 2012;104(7):418–432. [DOI] [PubMed] [Google Scholar]
- 169.Raska I, Shaw PJ, Cmarko D. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol. 2006;18(3):325–334. [DOI] [PubMed] [Google Scholar]
- 170.Pliss A, Koberna K, Vecerova J, et al. Spatio-temporal dynamics at rDNA foci: global switching between DNA replication and transcription. J Cell Biochem. 2005;94(3):554–565. [DOI] [PubMed] [Google Scholar]
- 171.Koberna K, Malinsky J, Pliss A, et al. Ribosomal genes in focus: new transcripts label the dense fibrillar components and form clusters indicative of “Christmas trees” in situ. J Cell Biol. 2002;157(5):743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nemeth A, Conesa A, Santoyo-Lopez J, et al. Initial genomics of the human nucleolus. PLoS Genet. 2010;6(3):e1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.van Koningsbruggen S, Gierlinski M, Schofield P, et al. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell. 2010;21(21):3735–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98(3):285–294. [DOI] [PubMed] [Google Scholar]
- 175.Oudet P, Gross-Bellard M, Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975;4(4):281–300. [DOI] [PubMed] [Google Scholar]
- 176.Hansen JC, Connolly M, McDonald CJ, et al. The 10-nm chromatin fiber and its relationship to interphase chromosome organization. Biochem Soc Trans. 2018;46(1):67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Demeret C, Vassetzky Y, Mechali M. Chromatin remodelling and DNA replication: from nucleosomes to loop domains. Oncogene. 2001;20(24):3086–3093. [DOI] [PubMed] [Google Scholar]
- 178.Cook PR, Brazell IA. Supercoils in human DNA. J Cell Sci. 1975;19(2): 261–279. [DOI] [PubMed] [Google Scholar]
- 179.McCready SJ, Akrigg A, Cook PR. Electron-microscopy of intact nuclear DNA from human cells. J Cell Sci. 1979;39:53–62. [DOI] [PubMed] [Google Scholar]
- 180.Paulson JR, Laemmli UK. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12(3):817–828. [DOI] [PubMed] [Google Scholar]
- 181.Berezney R, Coffey DS. Nuclear protein matrix: association with newly synthesized DNA. Science. 1975;189(4199):291–293. [DOI] [PubMed] [Google Scholar]
- 182.Berezney R, Buchholtz LA. Dynamic association of replicating DNA fragments with the nuclear matrix of regenerating liver. Exp Cell Res. 1981;132(1):1–13. [DOI] [PubMed] [Google Scholar]
- 183.Dijkwel PA, Mullenders LH, Wanka F. Analysis of the attachment of replicating DNA to a nuclear matrix in mammalian interphase nuclei. Nucleic Acids Res. 1979;6(1):219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pardoll DM, Vogelstein B, Coffey DS. A fixed site of DNA replication in eucaryotic cells. Cell. 1980;19(2):527–536. [DOI] [PubMed] [Google Scholar]
- 185.Gerdes MG, Carter KC, Moen PT Jr, Lawrence JB. Dynamic changes in the higher-level chromatin organization of specific sequences revealed by in situ hybridization to nuclear halos. J Cell Biol. 1994; 126(2):289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Robinson SI, Nelkin BD, Vogelstein B. The ovalbumin gene is associated with the nuclear matrix of chicken oviduct cells. Cell. 1982;28 (1):99–106. [DOI] [PubMed] [Google Scholar]
- 187.Robinson SI, Small D, Idzerda R, McKnight GS, Vogelstein B. The association of transcriptionally active genes with the nuclear matrix of the chicken oviduct. Nucleic Acids Res. 1983;11(15):5113–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bode J, Schlake T, Rios-Ramirez M, et al. Scaffold/matrix-attached regions: structural properties creating transcriptionally active loci. Int Rev Cytol. 1995;162A:389–454. [DOI] [PubMed] [Google Scholar]
- 189.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140(6):1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tubo RA, Berezney R. Identification of 100 and 150 S DNA polymerase alpha-primase megacomplexes solubilized from the nuclear matrix of regenerating rat liver. J Biol Chem. 1987;262(12):5857–5865. [PubMed] [Google Scholar]
- 191.Carter DR, Eskiw C, Cook PR. Transcription factories. Biochem Soc Trans. 2008;36(Pt 4):585–589. [DOI] [PubMed] [Google Scholar]
- 192.Cook PR. The organization of replication and transcription. Science. 1999;284(5421):1790–1795. [DOI] [PubMed] [Google Scholar]
- 193.Papantonis A, Cook PR. Transcription factories: genome organization and gene regulation. Chem Rev. 2013;113(11):8683–8705. [DOI] [PubMed] [Google Scholar]
- 194.Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10(7):457–466. [DOI] [PubMed] [Google Scholar]
- 195.Barutcu AR, Fritz AJ, Zaidi SK, et al. C-ing the genome: a compendium of chromosome conformation capture methods to study higher-order chromatin organization. J Cell Physiol. 2016;231(1):31–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Pope BD, Ryba T, Dileep V, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515 (7527):402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hansen AS, Cattoglio C, Darzacq X, Tjian R. Recent evidence that TADs and chromatin loops are dynamic structures. Nucleus. 2018;9(1):20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fortin JP, Hansen KD. Reconstructing A/B compartments as revealed by Hi-C using long-range correlations in epigenetic data. Genome Biol. 2015;16:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Oberdoerffer P, Sinclair DA. The role of nuclear architecture in genomic instability and ageing. Nat Rev Mol Cell Biol. 2007;8(9):692–702. [DOI] [PubMed] [Google Scholar]
- 200.van Steensel B Chromatin: constructing the big picture. EMBO J. 2011;30(10):1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Rao SS, Huntley MH, Durand NC, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nagano T, Lubling Y, Varnai C, et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature. 2017;547(7661): 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Belmont AS, Zhai Y, Thilenius A. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J Cell Biol. 1993;123(6 Pt 2):1671–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Belmont AS, Bignone F, Ts’o PO. The relative intranuclear positions of Barr bodies in XXX non-transformed human fibroblasts. Exp Cell Res. 1986;165(1):165–179. [DOI] [PubMed] [Google Scholar]
- 205.Nikolova V, Leimena C, McMahon AC, et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J Clin Invest. 2004;113(3):357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Goldman RD, Shumaker DK, Erdos MR, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2004;101(24):8963–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.McCord RP, Nazario-Toole A, Zhang H, et al. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 2013;23(2):260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Holmer L, Worman HJ. Inner nuclear membrane proteins: functions and targeting. Cell Mol Life Sci. 2001;58(12–13):1741–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Aagaard L, Laible G, Selenko P, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3–9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18(7):1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wreggett KA, Hill F, James PS, Hutchings A, Butcher GW, Singh PB. A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet Cell Genet. 1994;66(2):99–103. [DOI] [PubMed] [Google Scholar]
- 211.Sharma GG, Hwang KK, Pandita RK, et al. Human heterochromatin protein 1 isoforms HP1(Hsalpha) and HP1(Hsbeta) interfere with hTERT-telomere interactions and correlate with changes in cell growth and response to ionizing radiation. Mol Cell Biol. 2003;23(22): 8363–8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10(3):211–219. [DOI] [PubMed] [Google Scholar]
- 213.Kreth G, Finsterle J, von Hase J, Cremer M, Cremer C. Radial arrangement of chromosome territories in human cell nuclei: a computer model approach based on gene density indicates a probabilistic global positioning code. Biophys J. 2004;86(5):2803–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tanabe H, Muller S, Neusser M, et al. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc Natl Acad Sci U S A. 2002;99(7):4424–4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Parada LA, McQueen PG, Munson PJ, Misteli T. Conservation of relative chromosome positioning in normal and cancer cells. Curr Biol. 2002;12(19):1692–1697. [DOI] [PubMed] [Google Scholar]
- 216.Cremer M, von Hase J, Volm T, et al. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9(7):541–567. [DOI] [PubMed] [Google Scholar]
- 217.Parada LA, Sotiriou S, Misteli T. Spatial genome organization. Exp Cell Res. 2004;296(1):64–70. [DOI] [PubMed] [Google Scholar]
- 218.Versteeg R, van Schaik BD, van Batenburg MF, et al. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 2003;13(9):1998–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Tanabe H, Habermann FA, Solovei I, Cremer M, Cremer T. Non-random radial arrangements of interphase chromosome territories: evolutionary considerations and functional implications. Mutat Res. 2002;504(1–2):37–45. [DOI] [PubMed] [Google Scholar]
- 220.Marella NV, Seifert B, Nagarajan P, Sinha S, Berezney R. Chromosomal rearrangements during human epidermal keratinocyte differentiation. J Cell Physiol. 2009;221(1):139–146. [DOI] [PubMed] [Google Scholar]
- 221.Dietzel S, Zolghadr K, Hepperger C, Belmont AS. Differential large-scale chromatin compaction and intranuclear positioning of transcribed versus non-transcribed transgene arrays containing beta-globin regulatory sequences. J Cell Sci. 2004;117(Pt 19):4603–4614. [DOI] [PubMed] [Google Scholar]
- 222.Fedorova E, Zink D. Nuclear genome organization: common themes and individual patterns. Curr Opin Genet Dev. 2009;19(2):166–171. [DOI] [PubMed] [Google Scholar]
- 223.Finlan LE, Sproul D, Thomson I, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4(3):e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180(1):51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38(9):1005–1014. [DOI] [PubMed] [Google Scholar]
- 226.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452(7184):243–247. [DOI] [PubMed] [Google Scholar]
- 227.Zink D, Amaral MD, Englmann A, et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;166(6):815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bolzer A, Kreth G, Solovei I, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3(5):826–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Fritz AJ, Stojkovic B, Ding H, Xu J, Bhattacharya S, Berezney R. Cell type specific alterations in interchromosomal networks across the cell cycle. PLoS Comput Biol. 2014;10(10):e1003857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Fritz AJ, Stojkovic B, Ding H, et al. Wide-scale alterations in interchromosomal organization in breast cancer cells: defining a network of interacting chromosomes. Hum Mol Genet. 2014;23(19):5133–5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Marella NV, Bhattacharya S, Mukherjee L, Xu J, Berezney R. Cell type specific chromosome territory organization in the interphase nucleus of normal and cancer cells. J Cell Physiol. 2009;221(1):130–138. [DOI] [PubMed] [Google Scholar]
- 232.Sun HB, Shen J, Yokota H. Size-dependent positioning of human chromosomes in interphase nuclei. Biophys J. 2000;79(1):184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sehgal N, Seifert B, Ding H, et al. Reorganization of the interchromosomal network during keratinocyte differentiation. Chromosoma. 2016;125(3):389–403. [DOI] [PubMed] [Google Scholar]
- 234.Heride C, Ricoul M, Kieu K, et al. Distance between homologous chromosomes results from chromosome positioning constraints. J Cell Sci. 2010;123(23):4063–4075. [DOI] [PubMed] [Google Scholar]
- 235.Mehta IS, Amira M, Harvey AJ, Bridger JM. Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol. 2010;11(1):R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Mehta IS, Eskiw CH, Arican HD, Kill IR, Bridger JM. Farnesyltransferase inhibitor treatment restores chromosome territory positions and active chromosome dynamics in Hutchinson-Gilford progeria syndrome cells. Genome Biol. 2011;12(8):R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Maruyama EO, Hori T, Tanabe H, et al. The actin family member Arp6 and the histone variant H2A.Z are required for spatial positioning of chromatin in chicken cell nuclei. J Cell Sci. 2012;125(Pt 16):3739–3743. [DOI] [PubMed] [Google Scholar]
- 238.Strasak L, Bartova E, Harnicarova A, Galiova G, Krejci J, Kozubek S. H3K9 acetylation and radial chromatin positioning. J Cell Physiol. 2009;220(1):91–101. [DOI] [PubMed] [Google Scholar]
- 239.Khalil A, Grant JL, Caddle LB, Atzema E, Mills KD, Arneodo A. Chromosome territories have a highly nonspherical morphology and non-random positioning. Chromosome Res. 2007;15(7):899–916. [DOI] [PubMed] [Google Scholar]
- 240.Nagele RG, Freeman T, McMorrow L, Thomson Z, Kitson-Wind K, Lee H. Chromosomes exhibit preferential positioning in nuclei of quiescent human cells. J Cell Sci. 1999;112(Pt 4):525–535. [DOI] [PubMed] [Google Scholar]
- 241.Mayer R, Brero A, von Hase J, Schroeder T, Cremer T, Dietzel S. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol. 2005;6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Parada LA, McQueen PG, Misteli T. Tissue-specific spatial organization of genomes. Genome Biol. 2004;5(7):R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Kuroda M, Tanabe H, Yoshida K, et al. Alteration of chromosome positioning during adipocyte differentiation. J Cell Sci. 2004;117(Pt 24):5897–5903. [DOI] [PubMed] [Google Scholar]
- 244.Caddle LB, Grant JL, Szatkiewicz J, et al. Chromosome neighborhood composition determines translocation outcomes after exposure to high-dose radiation in primary cells. Chromosome Res. 2007;15(8):1061–1073. [DOI] [PubMed] [Google Scholar]
- 245.Pliss A, Fritz AJ, Stojkovic B, et al. Non-random patterns in the distribution of NOR-bearing chromosome territories in human fibroblasts: a network model of interactions. J Cell Physiol. 2015;230(2):427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Zeitz MJ, Mukherjee L, Bhattacharya S, Xu J, Berezney R. A probabilistic model for the arrangement of a subset of human chromosome territories in WI38 human fibroblasts. J Cell Physiol. 2009;221(1):120–129. [DOI] [PubMed] [Google Scholar]
- 247.Mukherjee L, Singh V, Peng JM, Xu JH, Zeitz MJ, Berezney R. Generalized median graphs and applications. J. Comb. Optim 2009;17(1):21–44. [Google Scholar]
- 248.Ding H, Stojkovic B, Berezney R, Xu J. Gauging association patterns of chromosomes territories via chromatic median. CVPR. 2013;2:1296–1303. [Google Scholar]
- 249.Thompson SK, Seber GAF. Adaptive Sampling. New York: Wiley; 1996. [Google Scholar]
- 250.Vandenberghet L, Boyd S. Semidefinite programming. SIAM Rev. 1996;38:49–95. [Google Scholar]
- 251.Mitelman F Recurrent chromosome aberrations in cancer. Mutat Res. 2000;462(2–3):247–253. [DOI] [PubMed] [Google Scholar]
- 252.Parada L, Misteli T. Chromosome positioning in the interphase nucleus. Trends Cell Biol. 2002;12(9):425–432. [DOI] [PubMed] [Google Scholar]
- 253.Sachs RK, Chen AM, Brenner DJ. Review: proximity effects in the production of chromosome aberrations by ionizing radiation. Int J Radiat Biol. 1997;71(1):1–19. [DOI] [PubMed] [Google Scholar]
- 254.Lukasova E, Kozubek S, Kozubek M, et al. Localisation and distance between ABL and BCR genes in interphase nuclei of bone marrow cells of control donors and patients with chronic myeloid leukaemia. Hum Genet. 1997;100(5–6):525–535. [DOI] [PubMed] [Google Scholar]
- 255.Kozubek S, Lukasova E, Mareckova A, et al. The topological organization of chromosomes 9 and 22 in cell nuclei has a determinative role in the induction of t(9,22) translocations and in the pathogenesis of t (9,22) leukemias. Chromosoma. 1999;108(7):426–435. [DOI] [PubMed] [Google Scholar]
- 256.Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet. 2003;34(3):287–291. [DOI] [PubMed] [Google Scholar]
- 257.Engreitz JM, Agarwala V, Mirny LA. Three-dimensional genome architecture influences partner selection for chromosomal translocations in human disease. PLoS One. 2012;7(9):e44196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Canela A, Maman Y, Jung S, et al. Genome organization drives chromosome fragility. Cell. 2017;170(3):507–521 e518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Kaiser VB, Semple CA. Chromatin loop anchors are associated with genome instability in cancer and recombination hotspots in the germline. Genome Biol. 2018;19(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kurz A, Lampel S, Nickolenko JE, et al. Active and inactive genes localize preferentially in the periphery of chromosome territories. J Cell Biol. 1996;135(5):1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Kupper K, Kolbl A, Biener D, et al. Radial chromatin positioning is shaped by local gene density, not by gene expression. Chromosoma. 2007;116(3):285–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Mahy NL, Perry PE, Gilchrist S, Baldock RA, Bickmore WA. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J Cell Biol. 2002;157 (4):579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Nagano T, Lubling Y, Stevens TJ, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502(7469): 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Stevens TJ, Lando D, Basu S, et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature. 2017;544(7648): 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Williams RR, Broad S, Sheer D, Ragoussis J. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp Cell Res. 2002;272(2): 163–175. [DOI] [PubMed] [Google Scholar]
- 266.Osborne CS, Chakalova L, Brown KE, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004; 36(10):1065–1071. [DOI] [PubMed] [Google Scholar]
- 267.Osborne CS, Chakalova L, Mitchell JA, et al. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5(8):e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Clowney EJ, LeGros MA, Mosley CP, et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151(4):724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Zhou GL, Xin L, Song W, et al. Active chromatin hub of the mouse alpha-globin locus forms in a transcription factory of clustered housekeeping genes. Mol Cell Biol. 2006;26(13):5096–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Tolhuis B, Blom M, Kerkhoven RM, et al. Interactions among Polycomb domains are guided by chromosome architecture. PLoS Genet. 2011;7(3):e1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Krivega I, Dean A. Enhancer and promoter interactions-long distance calls. Curr Opin Genet Dev. 2012;22(2):79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Mora A, Sandve GK, Gabrielsen OS, Eskeland R. In the loop: promoter-enhancer interactions and bioinformatics. Brief Bioinform. 2016;17(6):980–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32(4): 623–626. [DOI] [PubMed] [Google Scholar]
- 274.Gheldof N, Smith EM, Tabuchi TM, et al. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010;38(13):4325–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kadauke S, Blobel GA. Chromatin loops in gene regulation. Biochim Biophys Acta. 2009;1789(1):17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet. 2003;35(2):190–194. [DOI] [PubMed] [Google Scholar]
- 277.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10(6):1453–1465. [DOI] [PubMed] [Google Scholar]
- 278.Drissen R, Palstra RJ, Gillemans N, et al. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18(20):2485–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435(7042):637–645. [DOI] [PubMed] [Google Scholar]
- 280.Pott S, Lieb JD. What are super-enhancers? Nat Genet. 2015;47(1):8–12. [DOI] [PubMed] [Google Scholar]
- 281.Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Loven J, Hoke HA, Lin CY, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Schoenfelder S, Javierre BM, Furlan-Magaril M, Wingett SW, Fraser P. Promoter capture Hi-C: high-resolution, genome-wide profiling of promoter interactions. J Vis Exp. 2018;(136). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Jin F, Li Y, Dixon JR, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503 (7475):290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Moore BL, Aitken S, Semple CA. Integrative modeling reveals the principles of multi-scale chromatin boundary formation in human nuclear organization. Genome Biol. 2015;16:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2(4):292–301. [DOI] [PubMed] [Google Scholar]
- 288.Meaburn KJ, Misteli T. Cell biology - chromosome territories. Nature. 2007;445(7126):379–381. [DOI] [PubMed] [Google Scholar]
- 289.Neusser M, Schubel V, Koch A, Cremer T, Muller S. Evolutionarily conserved, cell type and species-specific higher order chromatin arrangements in interphase nuclei of primates. Chromosoma. 2007; 116(3):307–320. [DOI] [PubMed] [Google Scholar]
- 290.Parada LA, Roix JJ, Misteli T. An uncertainty principle in chromosome positioning. Trends Cell Biol. 2003;13(8):393–396. [DOI] [PubMed] [Google Scholar]
- 291.Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci U S A. 2006;103(20):7688–7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Dietzel S, Jauch A, Kienle D, et al. Separate and variably shaped chromosome arm domains are disclosed by chromosome arm painting in human cell nuclei. Chromosome Res. 1998;6(1):25–33. [DOI] [PubMed] [Google Scholar]
- 293.Eils R, Dietzel S, Bertin E, et al. Three-dimensional reconstruction of painted human interphase chromosomes: active and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J Cell Biol. 1996;135(6 Pt 1):1427–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Federico C, Cantarella CD, Di Mare P, Tosi S, Saccone S. The radial arrangement of the human chromosome 7 in the lymphocyte cell nucleus is associated with chromosomal band gene density. Chromosoma. 2008;117(4):399–410. [DOI] [PubMed] [Google Scholar]
- 295.Gierman HJ, Indemans MH, Koster J, et al. Domain-wide regulation of gene expression in the human genome. Genome Res. 2007;17(9): 1286–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Goetze S, Mateos-Langerak J, Gierman HJ, et al. The three-dimensional structure of human interphase chromosomes is related to the transcriptome map. Mol Cell Biol. 2007;27(12):4475–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Munkel C, Eils R, Dietzel S, et al. Compartmentalization of interphase chromosomes observed in simulation and experiment. J Mol Biol. 1999;285(3):1053–1065. [DOI] [PubMed] [Google Scholar]
- 298.Rego A, Sinclair PB, Tao W, Kireev I, Belmont AS. The facultative heterochromatin of the inactive X chromosome has a distinctive condensed ultrastructure. J Cell Sci. 2008;121(Pt 7):1119–1127. [DOI] [PubMed] [Google Scholar]
- 299.Shopland LS, Lynch CR, Peterson KA, et al. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol. 2006;174(1):27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Muller I, Boyle S, Singer RH, Bickmore WA, Chubb JR. Stable morphology, but dynamic internal reorganisation, of interphase human chromosomes in living cells. PLoS One. 2010;5(7):e11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Sehgal N, Fritz AJ, Morris K, et al. Gene density and chromosome territory shape. Chromosoma. 2014;123(5):499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118(5):555–566. [DOI] [PubMed] [Google Scholar]
- 303.Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145(6):1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Edelmann P, Bornfleth H, Zink D, Cremer T, Cremer C. Morphology and dynamics of chromosome territories in living cells. Biochim Biophys Acta. 2001;1551(1):M29–M39. [DOI] [PubMed] [Google Scholar]
- 305.Teller K, Illner D, Thamm S, et al. A top-down analysis of Xa- and X-iterritories reveals differences of higher order structure at >/= 20 Mb genomic length scales. Nucleus. 2011;2(5):465–477. [DOI] [PubMed] [Google Scholar]
- 306.Caron H, van Schaik B, van der Mee M, et al. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291(5507):1289–1292. [DOI] [PubMed] [Google Scholar]
- 307.Yang S, Illner D, Teller K, et al. Structural analysis of interphase X-chromatin based on statistical shape theory. Biochim Biophys Acta. 2008;1783(11):2089–2099. [DOI] [PubMed] [Google Scholar]
- 308.Wang S, Xu J, Zeng J. Inferential modeling of 3D chromatin structure. Nucleic Acids Res. 2015;43(8):e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Mateos-Langerak J, Bohn M, de Leeuw W, et al. Spatially confined folding of chromatin in the interphase nucleus. Proc Natl Acad Sci U S A. 2009;106(10):3812–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Yokota H, van den Engh G, Hearst JE, Sachs RK, Trask BJ. Evidence for the organization of chromatin in megabase pair-sized loops arranged along a random walk path in the human G0/G1 interphase nucleus. J Cell Biol. 1995;130(6):1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Barbieri M, Chotalia M, Fraser J, et al. Complexity of chromatin folding is captured by the strings and binders switch model. Proc Natl Acad Sci U S A. 2012;109(40):16173–16178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Nicodemi M, Pombo A. Models of chromosome structure. Curr Opin Cell Biol. 2014;28:90–95. [DOI] [PubMed] [Google Scholar]
- 313.Sachs RK, van den Engh G, Trask B, Yokota H, Hearst JE. A random-walk/giant-loop model for interphase chromosomes. Proc Natl Acad Sci U S A. 1995;92(7):2710–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Barbieri M, Fraser J, Lavitas LM, et al. A polymer model explains the complexity of large-scale chromatin folding. Nucleus. 2013;4(4): 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Tark-Dame M, van Driel R, Heermann DW. Chromatin folding--from biology to polymer models and back. J Cell Sci. 2011;124(Pt 6): 839–845. [DOI] [PubMed] [Google Scholar]
- 316.Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49(5):773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Imakaev M, Fudenberg G, McCord RP, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9(10):999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Williamson I, Berlivet S, Eskeland R, et al. Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev. 2014;28(24):27782791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152(6):1270–1284. [DOI] [PubMed] [Google Scholar]
- 320.Sehgal N, Fritz AJ, Vecerova J, et al. Large-scale probabilistic 3D organization of human chromosome territories. Hum Mol Genet. 2016;25(3):419–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Aloise D, Deshpande A, Hansen P, Popat P. NP-hardness of Euclidean sum-of-squares clustering. Machine Learning. 2009;75:245–249. [Google Scholar]
- 322.David Arthur BM, Röglin H. k-Means has polynomial smoothed complexity. Paper presented at: Proceedings of the 50th Annual IEEE Symposium on Foundations of Computer Science (FOCS 2009) 2009. [Google Scholar]
- 323.Kanungo T, Mount DM, Netanyahu NS, Piatko CD, Silverman R, Wu AY. An efficient K-means clustering algorithm: Analysis and implementation. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2002;24(7):881–892, 10.1109/TPAMI.2002.1017616 [DOI] [Google Scholar]
- 324.MacQueen J Some methods for classification and analysis of multivariate observations Proceedings on the 5th Berkeley Symposium on Mathematical Statistics and Probability. Berkeley, CA, USA; 1967: 281–297. www.stat.ucla.edu. [Google Scholar]
- 325.Hirata D, Masuda H, Eddison M, Toda T. Essential role of tubulin-folding cofactor D in microtubule assembly and its association with microtubules in fission yeast. EMBO J. 1998;17(3):658–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434(7031): 400–404. [DOI] [PubMed] [Google Scholar]
- 327.Zaidi SK, Fritz AJ, Tracy KM, et al. Nuclear organization mediates cancer-compromised genetic and epigenetic control. Adv Biol Regul. 2018;69:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]


