Abstract
Leveraging emerging opportunities in data science to open new frontiers in heart, lung, blood, and sleep (HLBS) research is one of the major strategic objectives of the National Heart, Lung, and Blood Institute (NHLBI), one of the 27 Institutes/Centers (ICs) within the National Institutes of Health (NIH). To assess NHLBI’s recent funding of research grants in data science and to identify its relative areas of focus within data science, a portfolio analysis from fiscal year (FY) 2008 to FY 2017 was performed. In this portfolio analysis, an efficient and reliable methodology was used to identify data science research grants by utilizing several NIH databases and search technologies (iSearch, Query View Reporting system, and IN-SPIRE™). Six-hundred thirty data science-focused extramural research grants supported by NHLBI were identified using keyword searches based primarily on NIH’s working definitions of bioinformatics and computational biology. Further analysis characterized the distribution of these grants among the HLBS disease areas as well as the sub-types of data science projects funded by NHLBI. Information was also collected for data science research grants funded by other NIH ICs using the same search and analysis methodology. The funding comparison among different NIH ICs highlighted relative data science areas of emphasis and further identified opportunities for potential data science areas in which NHLBI could foster research advances.
Keywords: bioinformatics, data science, research grant, portfolio analysis, NHLBI
Journal Subject Terms: Basic Science Research, Computational Biology
Introduction
Data science approaches and methodologies are very dynamic and have expanded in almost all aspects of scientific research, including biomedical science. Thanks to the automation of data collection procedures, coupled with the development of vast capacity for data storage and the creation of highly sophisticated tools for analyzing and processing data, advances in data science are changing the way research is conducted.
In recent years, NIH-funded researchers have generated substantial quantities of biomedical research information, which includes omics data (e.g., genomic, transcriptomic, proteomic, glycomic, metabolomic, etc.), data from clinical, observational, and epidemiological studies, and data from basic research using model organisms, among other data types. For example, with the advance of the Next-Generation Sequencing technology and dramatic cost reductions, a vast amount of genomic sequencing data has been generated. The total amount of genomics data alone is expected to equal or exceed the data from three other major data producers: astronomy, Twitter, and YouTube, by 2025 1. Moreover, there is no sign of slowing down of this exponential data growth trend. Both hardware and software are becoming more sophisticated and cheaper every day, and these trends support the growth of data science.
NHLBI is supporting substantial data generation through its Trans-Omics for Precision Medicine (TOPMed) program 2, that has generated about 150,000 human whole genome sequences. This vast trove of data has the potential to provide important new insights into the preemption and precise treatment of many HLBS diseases. Achieving this potential will require use of cutting-edge data science techniques and tools. In an effort to understand the current status of application of data science to HLBS conditions, a portfolio analysis was conducted with the specific goals of providing an overview of NHLBI’s data science research grant awards and to identify its relative areas of focus within data science. This portfolio analysis focused on extramural research grants, including original and independent research grants, cooperative agreements and program projects. It encompasses both investigator-initiated research grants, as well as those submitted and funded in response to NHLBI Funding Opportunity Announcements (FOA). Given that collectively, extramural research grants account for more than 70% of NHLBI’s budget spending 3, findings from this analysis should offer insight into the current interests and potential needs, challenges, and future trends of the extramural research community with respect to data science. Understanding these NHLBI-supported data science awards in the past ten years will also help to identify opportunities to guide future NHLBI efforts to foster data science research.
Methods
Definition of data science
The term “data science” has many definitions and spans multiple scientific disciplines, including applied mathematics, statistics, and computer science. The use of this term has evolved over time. To guide our analysis, various definitions for data science were reviewed before an internal consensus working definition was established. For example, the Wikipedia definition states “Data science is a ‘concept to unify statistics, data analysis, machine learning and their related methods in order to understand and analyze actual phenomena’ with data. It employs techniques and theories drawn from many fields within the context of mathematics, statistics, information science, and computer science” 4. The Techopedia version states “data science is a broad field that refers to the collective processes, theories, concepts, tools and technologies that enable the review, analysis and extraction of valuable knowledge and information from raw data. It is geared toward helping individuals and organizations make better decisions from stored, consumed and managed data” 5.
The following working definition of bioinformatics and computational biology was developed by the NIH Biomedical Information Science and Technology Initiative Consortium (BISTIC) Definition Committee and released on July 17, 2000 6.
Bioinformatics:
Research, development, or application of computational tools and approaches for expanding the use of biological, medical, behavioral or health data, including those to acquire, store, organize, archive, analyze, or visualize such data.
Computational Biology:
The development and application of data-analytical and theoretical methods, mathematical modeling and computational simulation techniques to the study of biological, behavioral, and social systems.
The NIH BISTIC committee recognized that no definition could completely eliminate overlap with other activities or preclude variations in interpretation by different individuals and organizations. The BISTIC definitions served as the foundation for the NIH Research, Condition, and Disease Categorization (RCDC) report terms in the data science and informatics research category 7. The NIH RCDC provides consistent and transparent information to the public about NIH-funded research, providing a complete list of all NIH-funded projects related to each category. However, due to the evolving nature in the field of data science, the NIH RCDC for data science is still under development and hence not available for use in this portfolio analysis.
In June, 2018, NIH released a strategic plan for data science 8, which defines data science as “the interdisciplinary field of inquiry in which quantitative and analytical approaches, processes, and systems are developed and used to extract knowledge and insights from increasingly large and/or complex sets of data.” The lexicon developed for the described portfolio analysis is based on this definition. For this analysis, the working definition for data science comprises two interrelated and overlapping areas: bioinformatics and computational biology. Both of these interdisciplinary approaches are drawn from specific disciplines such as mathematics, statistics, physics, computer science and engineering, biology, and behavioral science. Bioinformatics applies principles of information sciences and technologies to make the vast, diverse, and complex life sciences data more understandable and useful. Computational biology uses mathematical and computational approaches to address theoretical and experimental questions in biology.
Term search
A portfolio analysis on NHLBI-funded research grants from FY 2008–2017 was conducted. The NIH internal data platforms, Query View Report (QVR) 9 and iSearch (NIH Office of Portfolio Analysis’s next-generation portfolio analysis platform) (v2.0) 10 were used for the search to retrieve the targeted grants using 22 lexicon terms related to data science. As described above, the lexicon was developed largely based on the NIH working definitions of bioinformatics and computational biology and NIH RCDC terms. The lexicon terms used for searches in both QVR and iSearch are summarized in Table 1. After a free text search in the title and abstracts using both QVR and iSearch, and removing all training grants, a total of 1224 unique grants were retrieved after combining the search results from QVR and iSearch (Figure 1). The grant information is accessible using the publicly available NIH Research Portfolio Online Reporting Tools (NIH RePORT) system 11. Since both iSearch and QVR are “keyword-based searches, it is anticipated that the initial search would require further refinement to remove those grants that were not closely related to the working definition of data science. Thus, as a next step, a text-mining and clustering tool called IN-SPIRE™ was used 12 to filter the initial search results based on grant specific aims. As long as one specific aim contains data science component, the grant is classified as the data science grant. IN-SPIRE™ is useful for content analysis, and it analyzes a multitude of text files and determines key topics or themes to create a signature for each document in the collection. The key topics or themes are based on the frequency of lexicon terms (keywords) and co-occurrences of those lexicon terms. The lexicon terms were derived for the IN-SPIRE™ search using terms from the QVR and iSearch searches. After working with different lexicon terms, a more refined search was conducted with IN-SPIRE™ (5 Update 9.4.1) using the following terms: “algorithm” or “Bayesian modeling” or “bioinformatics” or “computational” or “computer” or “data modeling” or “decision modeling” or “systems biology” or “data analysis” or “computerized modeling” (Table 1). Using IN-SPIRE™, the initial QVR and iSearch results of 1224 grants were further refined. IN-SPIRE™ generated 669 “positive” grants with a relevance score between 0.23 to 1, which are considered as highly relevant to data science. IN-SPIRE™ also generated 555 “negative” grants with a relevance score of 0 which are considered not relevant to data science. There was no grant with a relevance score between 0 to 0.23.
Table 1.
Terms used for the search.
| Tool | Terms used for search |
|---|---|
| QVR | Algorithm // Bayesian Modeling // Bioinformatics // Computational Algorithm // Computational Analyses // Computational Biology // Computational Model // Computational Modeling // Computational Molecular Biology // Computational Network Modeling // Computational Tools // Computer Aided OR Computer Algorithm // Computer Analyses // Computer Based Biological Model // Computer Based Models // Computer Data Analysis // Computer Data Processing // Computerized Modeling // Data Modeling // Decision Modeling // Systems Biology Modeling |
| iSearch | “Algorithm” OR “Bayesian Modeling” OR “Bioinformatics” OR “Computational Algorithm” OR “Computational Analyses” OR “Computational Biology” OR “Computational Model” OR “Computational Modeling” OR “Computational Molecular Biology” OR “Computational Network Modeling” OR “Computational Tools” OR “Computer Aided” OR “Computer Algorithm” OR “Computer Analyses” OR “Computer Based Biological Model” OR “Computer Based Models” OR “Computer Data Analysis” OR “Computer Data Processing” OR “Computerized Modeling” OR “Data Modeling” OR “Decision Modeling” OR “Systems Biology Modeling” |
| IN-SPIRE™ | “algorithm” or “bayesian modeling” or “bioinformatics” or “computational” or “computer” or “data modeling” or “decision modeling” or “systems biology” or “data analysis” or “computerized modeling” |
Figure 1.
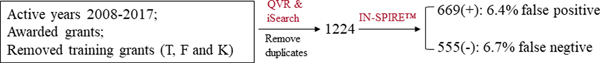
Key term search in both QVR and iSearch (22 terms) and refined by IN-SPIRE™ Key terms on data science were used to search NHLBI awarded research grants and projects active during 2008–2017 in both QVR and iSearch (v2.0), 1224 were retrieved after removing duplicates and training grants; after a filter through IN-SPIRE™ , 669 showed “positive” and 555 showed “negative”, the manual curation showed that 6.4% of them were false positive, and 6.7% were false negative.
To determine the accuracy and consistency of the data-science relevance scores provided by IN-SPIRE™ (5 Update 9.4.1), the data was manually reviewed by analyzing the abstracts of the top 70 and the bottom 70 relevance-scoring grants of the total 669 “positive” grants (21%). All 70 grants from the highest relevance group were determined to be data science related, and 9 out of the bottom 70 as not data science related. Thus out of the total of 140 assessed, there were only 9 false positives, yielding a rate of false positives at 6.4%. The results confirmed that the matching score generated by IN-SPIRE™ reflected sufficient relevance of the grants to our data science definition. A similar manual review of 60 grants (11%) among the 555 “negative” grants (i.e., those with a relevance score of 0 for data science generated by IN-SPIRE™ ) was performed. After the review and adjudication, 4 out of the 60 were found to be, in fact, data science related, which reflected a false negative rate of 6.7% (Figure 1). These results support the accuracy of our data collection and analysis methods of using a combination of QVR and iSearch with IN-SPIRE™ to identify data science research grants.
It should be noted that the manual review is only to assess the dependability and consistency of the methods we used for our study. To make the data and analysis reproducible and avoid human bias, the manually identified false positive and false negatives were not removed nor added to the final analytic set of research grants. The same validated search method and process were used to retrieve and analyze the research data science grants of other ICs in this study. It should also be noted that among the 669 “positive” grants, 39 projects were excluded from subsequent analyses because they were actually research conducted within the internal research program of the NHLBI, known as the NHLBI intramural program 13. In our primary analyses, only the NHLBI grants awarded to the extramural research community were included. The same inclusion (of extramural grants only) was applied for the subsequent analyses of other ICs’ data science portfolios.
Results
Data science research grant distribution among different disease areas within NHLBI
The distribution of the 630 extramural research grants among heart, lung, blood and sleep disease areas was assessed using Program Classification Codes (PCCs), which is an NIH internal grant coding system, assigned by the NHLBI when those grant applications were initially received to denote specific scientific or disease areas. Figure 2 shows the final distribution of these 630 grants across specific disease areas: 399 grants were related to cardiovascular sciences, 140 grants were related to lung and airway related diseases, 78 grants were related to blood diseases, and 13 grants were related to sleep.
Figure 2.
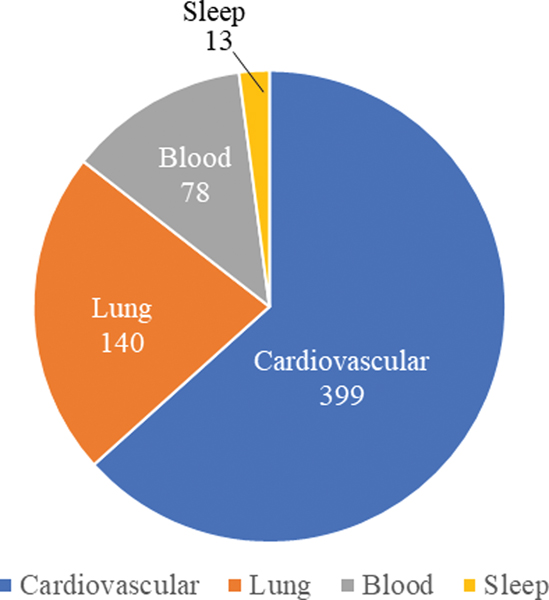
Data science research grant distribution among different disease areas within NHLBI. Among the 630 NHLBI extramural research grants on data science, 399 grants were related to cardiovascular sciences, 140 grants were related to lung and airway related diseases, 78 grants were related to blood diseases, 13 grants were related to sleep.
Given the large number of cardiovascular awards, the disease distribution within cardiovascular sciences was further interrogated with a “Term Search” in IN-SPIRE™ using key terms that represent various major cardiovascular disease categories. The results showed that funding of data science research grants was well distributed among several major cardiovascular diseases (Table 2). Lung disease and sleep disorder projects were manually examined and showed that developmental and pediatric diseases had the greatest number of research grants, followed by common and chronic diseases, chronic obstructive pulmonary disease (COPD), asthma, and sleep disorders (Table 3). Blood disease projects were also manually examined (Table 4). Both hemoglobinopathies and hemostasis-thrombosis comprised more than half of the data science awards. Hemoglobinopathies predominantly captures sickle cell disease (SCD) and other globin related disorders, while hemostasis and thrombosis includes platelet biology, blood’s coagulable states, as well as their associated bleeding and thromboembolic disorders. Also reflected in the table are cell therapies that represent well defined erythropoiesis cascades to model red cell development. Under special programs, glycobiology has focused on understanding the complex sugar nomenclatures and the structure, function, and biology of carbohydrates (glycans) and their stochastic distribution to provide important post-translational modifications.
Table 2.
Research grant distribution across different cardiovascular disease areas.
| Type of CVD | Terms used for search in IN-SPIRE™ | No. out of 399 DCVS grants |
|---|---|---|
| Heart Failure | [heart failure] | 81 |
| Vascular Diseases | [vascular disease*] OR aortic OR hypertension | 70 |
| Arrhythmia | [arrhythmia] OR rhythm* | 53 |
| Coronary Artery Disease | [coronary artery] | 40 |
| Heart Attack, Myocardial Infarction | [heart attack] OR infarction | 38 |
| Congenital Heart Disease | congenital | 32 |
| Heart Valve Disease | valve | 21 |
| Cardiomyopathies | [cardiomyopathies] | 14 |
Table 3.
Research grant distribution across lung diseases and sleep disorders.
| Disease | No. out of 140 DLD grants |
|---|---|
| Developmental and Pediatric Diseases | 30 |
| COPD | 28 |
| Asthma | 19 |
| Sleep disorders | 19 |
| Pulmonary Vascular Diseases | 14 |
| Infectious Disease | 11 |
| Immunology/Fibrosis Diseases | 11 |
| Lung Diseases | 10 |
| Critical Care/Acute Lung Injury | 10 |
Table 4.
Research grant distribution across different blood disease areas.
| Disease | No. out of 78 DBDR grants |
|---|---|
| Hemostasis and Thrombosis | 24 |
| Hemoglobinopathies and Genetics | 23 |
| Systems Biology | 19 |
| Transfusion Medicine and Cell Therapies | 13 |
| Special Programs | 8 |
Data science investment vs. total NHLBI funding
To gain additional perspective regarding the NHLBI data science portfolio, we retrieved additional data from QVR to compare: a) the total number of NHLBI research grants to the total number of data science research grants (Table 5 and Figure 3A and 3B), and b) the total annual funding for all NHLBI research grants to the total annual funding for data science research grants (Table 6 and Figure 3B). The trend of NHLBI data science investment during FY 2008–2017, was calculated as the ratio of NHLBI data science research grants vs. all NHLBI grants by both number (Table 5 and Figure 3C) and level of funding (Table 6 and Figure 3D). NHLBI’s investment in data science research grants averaged about 1% of its overall research grant investment, which has remained relatively constant over the time period in this analysis. Given the focus of this portfolio analysis on research grants only, it should be noted that NHLBI has also made substantive investments in generating big data and building a data science platform, funded primarily using mechanisms other than grants – namely through Other Transaction Authority (OTAs) and contracts, both of which were not accounted for in this analysis. The main reason OTAs and contracts were not included is because they go beyond the intended scope of this paper. This paper focuses on extramural research grants, which account for more than 70% of NHLBI research funding 3. Furthermore, OTA and contracts data are not fully available in the current NIH RePORT system which if used for comparison purposes could lead to inaccurate conclusions.
Table 5.
Data science research grants as a percentage of all NHLBI-funded research grants, FY 2008–2017.
| Year | Total # of Grants | # of Data Science Grants | Data Science vs. NHLBI total (%) |
|---|---|---|---|
| 2008 | 5471 | 52 | 0.95% |
| 2009 | 7113 | 69 | 0.97% |
| 2010 | 6754 | 58 | 0.86% |
| 2011 | 6086 | 60 | 0.99% |
| 2012 | 6119 | 42 | 0.69% |
| 2013 | 5801 | 39 | 0.67% |
| 2014 | 5617 | 61 | 1.09% |
| 2015 | 5909 | 56 | 0.95% |
| 2016 | 6274 | 61 | 0.97% |
| 2017 | 6883 | 72 | 1.05% |
Figure 3.
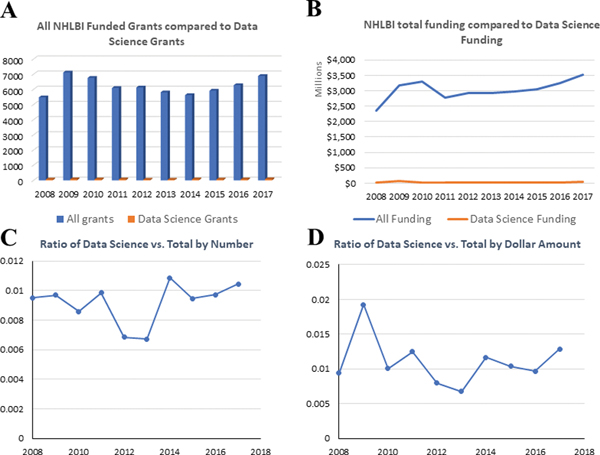
All NHLBI funded research grants compared to data science research grants. A. The comparison in research grant number by year during 2008–2017; B. The comparison in dollar amount by year during 2008–2017; C. The ratio of data science funding number vs. total NHLBI funding number by year during 2008–2017; D. The ratio of data science funding dollar amount vs. total NHLBI funding dollar amount by year during 2008–2017.
Table 6.
Data science research grant funding as a percentage of all NHLBI research grant funding, FY 2008–2017.
| Year | Total dollars all grants | Total dollars data science grants | Data Science funding vs. NHLBI total funding (%) |
|---|---|---|---|
| 2008 | $2,357,508,494 | $22,210,328 | 0.94% |
| 2009 | $3,174,256,040 | $61,067,424 | 1.92% |
| 2010 | $3,310,824,719 | $33,294,598 | 1.01% |
| 2011 | $2,772,455,495 | $34,682,373 | 1.25% |
| 2012 | $2,919,095,986 | $23,201,331 | 0.79% |
| 2013 | $2,927,694,300 | $19,723,599 | 0.67% |
| 2014 | $2,983,759,460 | $34,788,610 | 1.17% |
| 2015 | $3,045,975,305 | $31,610,686 | 1.04% |
| 2016 | $3,256,281,277 | $31,533,038 | 0.97% |
| 2017 | $3,522,794,122 | $45,413,861 | 1.29% |
Identification of data science areas of relative focus
The distribution of the 630 NHLBI data science research grants across different data science categories was also studied (Figure 4 and Table 7). These categories cover nine common data science domains including: “Modeling”, “Genetics, Genomics, Proteomic and other Omics”, “Precision Medicine”, “Big Data”, “Electronic Medical Record/Electronic Health Record (EMR/EHR)”, “Clinical Decision Support”, “Image Processing/Image Analysis”, “Computational Tools” and “Systems Biology/Synthetic Biology.” Some of these areas include sub areas (Table 7). Also employed was the “Term Search” function in IN-SPIRE™ using key terms that represent each area (Table 7). Among nine data science categories, omics, modeling, and systems biology are the top three data science sub-fields supported by NHLBI research grants in recent years (Figure 4 and Table 7).
Figure 4.
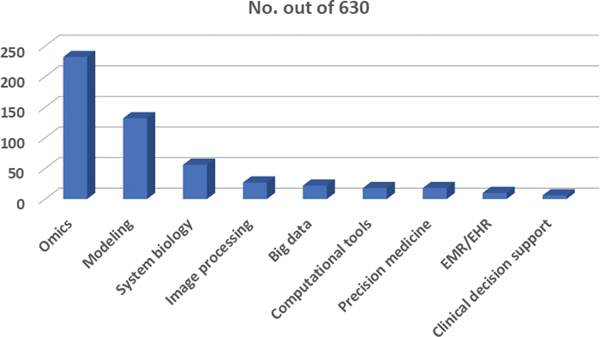
Research grant distribution in different data science areas among NHLBI. The number of NHLBI data science research grants in each common data science area out of a total of 630.
Table 7.
Research grant distribution across different data science areas among selected Institutes.
| Data Science areas | Terms used for search in IN-SPIRE™ | No. out of 630 NHLBI grants | No. out of 1584 NCI grants | No. out of 1483 NIAID grants | No. out of 1007 NINDS+NIMH+NIAAA grants | No. out of 408 NHGRI grants | No. out of 151 NLM grants |
|---|---|---|---|---|---|---|---|
| Genetics/Genomics/Proteomic/-omics | [genetics] OR [genom*] OR [gene expression] OR [genetic variant] OR [whole genome sequencing] OR [genome wide association] OR [proteom*] OR [pharmacogenom*] OR [epigenome*] OR [imaging genomi*] | 232 (36.8%) | 923 (58.3%) | 999 (67.4%) | 323 (32.1%) | 405 (99.3%) | 83 (55.0%) |
| Modeling | [algorithm] OR [Bayesian modeling] | 132 (21.0%) | 419 (26.5%) | 203 (13.7%) | 189 (18.8%) | 49 (12.0%) | 44 (29.1%) |
| Systems Biology/Synthetic Biology | [Systems biology] OR [Synthetic biology] | 56 (8.9%) | 95 (6.0%) | 133 (9.0%) | 29 (2.9%) | 15 (3.7%) | 11 (7.3%) |
| Image Processing/Image Analysis | [Image process*] OR [Image analysis] | 27 (4.3%) | 110 (6.9%) | 18 (1.2%) | 38 (3.8%) | 3 (0.7%) | 7 (4.6%) |
| Big Data | [deep learning] OR [machine learning] OR [artificial intelligence] OR [AI] | 22 (3.5%) | 94 (5.9%) | *47 (3.2%) | 49 (4.9%) | 16 (4.0%) | 43 (28.5%) |
| Computational Tools | [Computational tools] | 18 (2.9%) | 97 (6.1%) | 86 (5.8%) | 45 (4.5%) | 59 (14.5%) | 16 (10.6%) |
| Precision Medicine | [Precision medicine] | 18 (2.9%) | 99 (6.3%) | 11 (0.7%) | 9 (0.9%) | 17 (4.2%) | 5 (3.3%) |
| EMR/EHR | “EMR” OR “EHR” | 10 (1.6%) | 12 (0.8%) | 5 (0.3%) | 4 (0.4%) | 11 (2.7%) | 23 (15.2%) |
| Clinical Decision Support | [Clinical decision support] | 6 (1.0%) | 8 (0.5%) | 0 | 0 | 6 (1.5%) | 11 (7.3%) |
“[deep learning] OR [machine learning] OR [artificial intelligence]”, “AI” cannot be used because it’s also the institute code for NIAID.
To gain insight regarding potential areas of relative focus, NHLBI funding patterns were compared to other ICs within NIH on different data science disciplines. The comparator ICs included those that traditionally have substantial data science programs, including National Cancer Institute (NCI), National Institute of Allergy and Infectious Diseases (NIAID), National Library of Medicine (NLM), National Human Genome Research Institute (NHGRI), National Institute of Neurological Disorders and Stroke (NINDS), National Institute of Mental Health (NIMH) and National Institute on Alcohol Abuse and Alcoholism (NIAAA). Funding by NINDS, NIMH and NIAAA was combined for this analysis due to neuroscience synergies within their missions. We applied the same data science research grants retrieval method we used for the NHLBI portfolio (Figure 1). Similarly, intramural projects were also excluded from the analyses of all ICs. The results are shown in both Table 7 and Figure 5. The total number of data science research grants for each IC or IC groups are listed in the column heading of Table 7.
Figure 5.
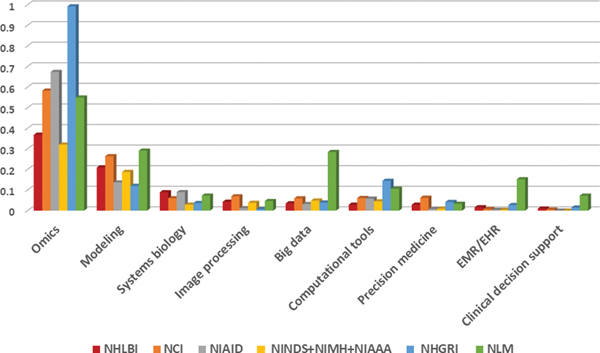
Research grant distribution in different Data Science areas among different Institutes. The relative number of each data science area to the total number of data science research grants in different institutes.
Across all institutes, most funded data science research grants were related to genomics and other -omics data. Modeling ranked second in terms of proportion of research grants funded. The graph in Figure 5 reflects the funding focus by institutes, for example, NHGRI has a focus on omics (“Genetics, Genomics, Proteomic and other Omics”), and NLM has a focus on “Big data.” This difference in funding focus is anticipated since every IC funding is mission focused. NHLBI funded fewer data science research grants in total than NCI or NIAID and also in comparison to the three neuroscience focused institutes combined (NINDS, NIMH, and NIAAA).
In general, NHLBI’s data science research project portfolio was similar to those of the other Institutes with respect to data science disciplines. Among the relative differences of potential focus, NHLBI funded a smaller proportion of “Computational Tools” research grants than the other Institutes. Furthermore, NHLBI supported a smaller proportion of “EMR/EHR” and “Clinical Decision Support” research grants compared to NLM and NHGRI, and a smaller proportion of “Genetics, Genomics, Proteomic and other Omics” research grants compared to NHGRI, NIAID, NCI, and NLM.
Discussion
The purpose of this analysis was to survey the landscape of the data science extramural research grants that were supported by NHLBI from FY 2008 to FY 2017. These research grants included awards across the spectrum of HLBS conditions and data science sub-fields. Omics, modeling, and systems biology were the top three data science sub-fields supported by NHLBI research grants in recent years. Among the different disciplines within data science, omics (including whole genome sequencing, RNA sequencing, proteomics and metabolomics) was relatively well supported across the NIH institutes studied here. The results indicate that precision medicine, clinical informatics, clinical decision support, imaging informatics, and computational tools may be areas of opportunity for fostering data science relevant to HLBS and other conditions. Over the period of the analysis, NHLBI has devoted about 1% of its extramural grant support to data science research grants. This level of support remained constant throughout the period. The NHLBI data science portfolio was similar to that of other NIH Institutes, with some variability potentially reflecting the scientific missions and priorities of the different ICs.
This analysis included only extramural research grant awards. All intramural research projects, although captured in the initial searches, were excluded in the final analyses. Awards made under other funding mechanisms, such as contracts and OTAs, were outside the scope of this analysis, particularly since they are not fully captured in the NIH RePORT system. Contracts and OTAs are typically Institute-led initiatives, while research grants predominantly reflect research initiated by the investigators. Not captured in this analysis but worth highlighting for awareness are NHLBI contracts and OTA-funded efforts, such as trans-omics data acquisition through the TOPMed program, data platform and management through the NHLBI cloud-based platform, or technical framework, for tools, applications, and workflows, , and data and biospecimens repository through the Biologic Specimen and Data Repository Information Coordinating Center (BioLINCC) 14. Despite these limitations, it is worth emphasizing that collectively, extramural research grants account for more than 70% of NHLBI’s budget spending 3; hence, this analysis provides the opportunity to explore the current work of the NHLBI-supported extramural research community with respect to data science.
Our search covered very broad aspects of data science. The lexicon was developed using the NIH working definition of Bioinformatics and Computational Biology and NIH RCDC terms, plus the keywords used in NIH data science related research grants. Since the IN-SPIRE™ search was based on both frequency and co-occurrences of the key words, it provided additional refinement of the research results obtained from QVR and iSearch. Manual curation of the research grants from IN-SPIRE™ showed both false positive and false negative ratios of about 6–7% allowing the use of IN-SPIRE™ for the identification of data science research grants to perform valid portfolio analyses.
It should be noted that the selection of lexicon terms was not exhaustive, and perhaps reflects a stronger emphasis on capturing research grants that involve computational analytics, bioinformatics, modeling and algorithm development. Some research grants without intensive computational or modeling components that leveraged large or multi-dimensional datasets that others might still consider as “Data Science” may have been missed. There is the need for an iterative approach including identification of new terms emerging as the field advances and periodic reassessment. Furthermore, the terms used in this analysis for describing the disciplines within data science may be slightly different from those used by others, such as a recent report from Zhu and Zheng 15. However, similarities should be emphasized. For example, the use of “EMR/EHR” and “Clinical Decision Support” in the sub analysis is comparable to “Clinical informatics”, a sub-field described by Zhu and Zheng 15. Similarly, “Image Processing/Image Analysis” corresponds to “Imaging informatics”; and “Modeling”, “Genetics, Genomics, Proteomic and other Omics”, “Precision Medicine”, “Big Data”, “Computational Tools” and “Systems Biology/Synthetic Biology” correspond to “Bioinformatics” in the Zhu and Zhang paper 15.
Conclusion
In 2016, NHLBI released “The NHLBI Strategic Vision” document 16, with a key objective to leverage emerging opportunities in data science and to open new frontiers in HLBS research. The NHLBI Strategic Vision states that “It will be essential to develop innovative approaches to the integration, analysis, and interpretation of data from multiple sources so that this information can be effectively utilized to improve patient outcomes.” This portfolio analysis was conducted to understand the NHLBI data science research grants portfolio and to identify potential opportunities to foster advances in HLBS research through data science approaches.
Supplementary Material
Acknowledgments:
We thank Drs. Youngsuk Oh and Zorina Galis for their helpful comments. Disclaimer: The views expressed in this paper are those of the authors and do not necessarily represent those of the NHLBI, the National Institutes of Health, or the US Department of Health and Human Services.
Sources of Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Non-standard Abbreviations and Acronyms:
- NIH
National Institutes of Health
- ICs
Institutes/Centers
- NHLBI
National Heart, Lung, and Blood Institute
- FY
Fiscal Year
- QVR
Query View Reporting system
- TOPMed
Trans-Omics for Precision Medicine
- FOA
Funding Opportunity Announcements
- BISTIC
Biomedical Information Science and Technology Initiative Consortium
- RCDC
Research, Condition, and Disease Categorization
- NIH RePORT
NIH Research Portfolio Online Reporting Tools
- PCCs
Program Classification Codes
- COPD
Chronic Obstructive Pulmonary Disease
- SCD
Sickle Cell Disease
- OTAs
Other Transaction Authority
- EMR/EHR
Electronic Medical Record/Electronic Health Record
- NCI
National Cancer Institute
- NIAID
National Institute of Allergy and Infectious Diseases
- NLM
National Library of Medicine
- NHGRI
National Human Genome Research Institute
- NINDS
National Institute of Neurological Disorders and Stroke
- NIMH
National Institute of Mental Health
- NIAAA
National Institute on Alcohol Abuse and Alcoholism
- DataSTAGE
Storage, Toolspace, Access and analytics for biG data Empowerment
- BioLINCC
Biologic Specimen and Data Repository Information Coordinating Center
Footnotes
Disclosures: None
References:
- 1.Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, Iyer R, Schatz MC, Sinha S, Robinson GE. Big Data: Astronomical or Genomical? PLOS Biology. 2015;13:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. https://www.nhlbi.nih.gov/science/trans-omics-precision-medicine-topmed-program.
- 3.FY20 NHLBI Congressional Justification: https://www.nhlbi.nih.gov/sites/default/files/media/docs/FY_2020_NHLBI_CJ.pdf.
- 4. https://en.wikipedia.org/wiki/Data_science.
- 5. https://www.techopedia.com/definition/30202/data-science.
- 6.http://www.binf.gmu.edu/jafri/math6390-bioinformatics/workingdef.pdf.
- 7. https://report.nih.gov/rcdc/.
- 8. https://www.nih.gov/news-events/news-releases/nih-releases-strategic-plan-data-science.
- 9. https://apps.era.nih.gov/qvr/web/home_main_hmnav.cfm.
- 10. https://itools.od.nih.gov/isearch/grants/.
- 11. https://report.nih.gov/.
- 12. https://in-spire.pnnl.gov/.
- 13. https://www.nhlbi.nih.gov/about/divisions/division-intramural-research.
- 14. https://biolincc.nhlbi.nih.gov/home/.
- 15.Zhu L, Zheng WJ. Informatics, Data Science, and Artificial Intelligence. JAMA. 2018;320:1103–1104. [DOI] [PubMed] [Google Scholar]
- 16. https://www.nhlbi.nih.gov/about/strategic-vision.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


