Abstract
De novo mutations (DNMs) arising on the paternal chromosome make the largest known contribution to autism risk, and correlate with paternal age at the time of conception. The recurrence risk for autism spectrum disorders (ASD) is substantial, leading many families to decline future pregnancies, but the potential impact of assessing parental gonadal mosaicism has not been considered. We measured sperm mosaicism using deep whole genome sequencing, both for variants present in an offspring and evident only in father’s sperm, and identified single nucleotide, structural, and short tandem repeat variants. We found that mosaicism quantification can stratify ASD recurrence risk due to DNMs into the vast majority with near 0% recurrence and a small fraction with a substantially higher and quantifiable risk, and we identify novel mosaic variants at risk for transmission to a future offspring. Thus, this suggests that genetic counseling would benefit from the addition of sperm mosaicism assessment.
Introduction
Clinicians are facing an ever increasing incidence of ASD in the population, without effective strategies to prevent disease or counsel families. Recent studies have identified gene-damaging DNMs in at least 10– 30% of simplex ASD cases1–4, along with the realization that the number of DNMs increase as a function of paternal age at the time of conception, doubling in DNM number in an offspring every 16.5 years of father’s age at the time of conception5,6. A DNM, defined as a genetic variant present in an offspring but not detectable in either parent, can have any of several different origins7,8. While classically considered to occur in the fertilized egg at the one-cell stage, most probably occur either post-zygotically in the offspring, or in a parent, either in the gonads or broadly in a mosaic pattern9. DNMs that occur during embryogenesis of a parent cause mosaicism in the soma, the gonads, or both, and remain throughout life, yet may be undetectable or barely detectable in blood10. Parental age is the single largest risk factor for the number of DNMs in a child, with a doubling in DNM number in an offspring every 16.5 years of father’s age at the time of conception5,6. However, the balance of gonadal-specific compared to broadly distributed DNMs in the father has not been carefully assessed, and thus the role of gonadal mosaicism on DNM recurrence risk remains uncertain.
Knowledge of the rates and mechanisms by which gonadal mutations arise has been advanced through assessment of multiple transmissions of DNMs within families, where approximately 1.3% of DNMs are shared by siblings11. Although only 3.8% of offspring DNMs are detectably mosaic in parental blood, this increases to 57.2% if shared by two or more offspring 10,11. Counterintuitively, DNM recurrence risk decreases by 1.8–2.3% per year of parent age, due to an increase in aging-associated DNMs10,11, thereby decreasing the relative contribution of parental mosaic variants to mutation burden.
Results
Sperm sequencing allows stratification of variants into low and high recurrence risk
We recruited eight families from our autism spectrum disorder (ASD) cohort12,13 where each father agreed to submit a sperm sample for sequencing (Supplementary Data 1). Employing 30× whole genome sequencing (WGS) from blood 12,13, we defined 912 de novo single nucleotide variants (dSNVs) in the 14 offspring (Fig. 1a, Methods). We then isolated sperm from the ejaculates and performed 200× WGS on paternal blood and sperm cells to determine which dSNVs were detectable in sperm based on three or more mutant reads (Extended Data Fig.1, Methods). We found 23 (2.5%) de novo single nucleotide variants (dSNVs) were also detected in paternal blood or sperm, leaving 889 (97.5%) dSNVs undetectable (Fig. 1b, Supplementary Data 2). Orthogonal validation of a subset with ultra-deep target amplicon sequencing (TAS) showed ~83% validation rate, 15/18 (Extended Data Fig. 2, Supplementary Data 3, 4, see Methods). All 3 non-validated variants were at allelic fractions (AFs) below 3% and located within repetitive elements (SINE or LINE).
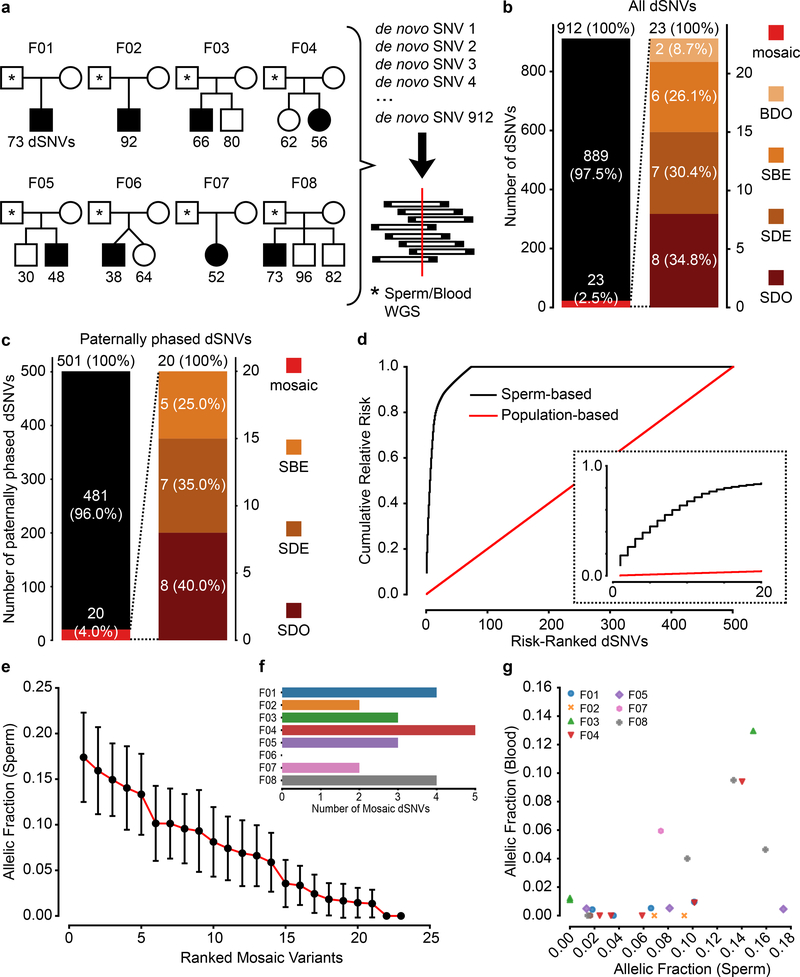
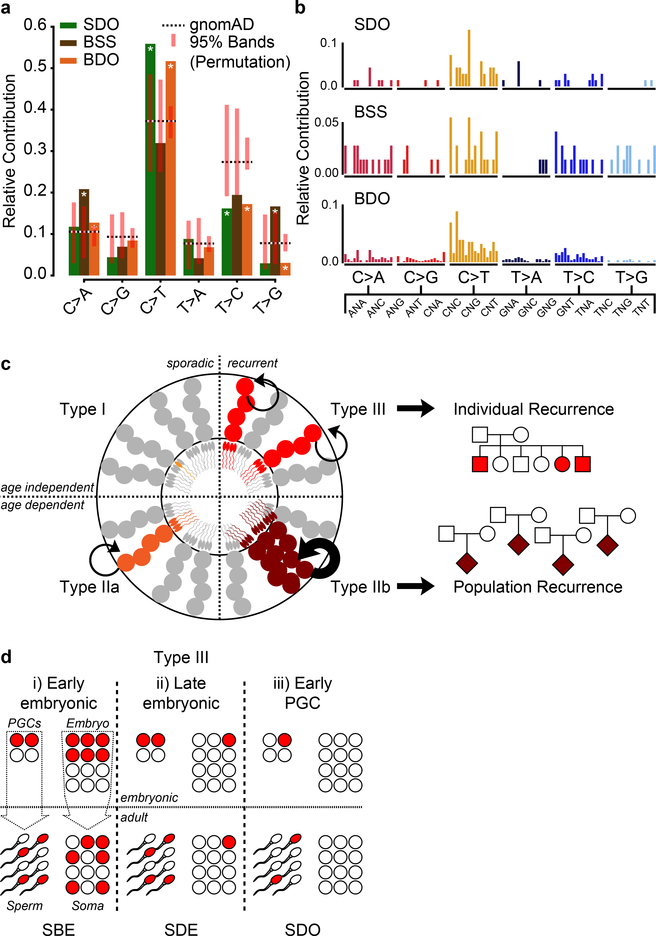
Figure 1. Recurrence risk stratification and mosaicism rates of 912 dSNVs is different in sperm compared with blood.
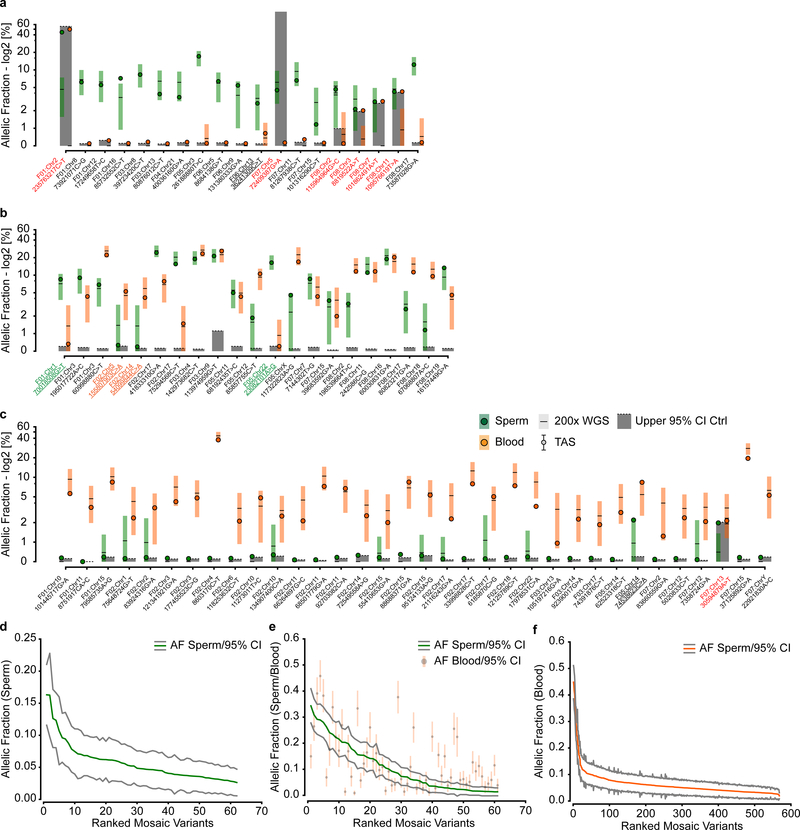
a, 8 Nuclear families used for 200× WGS analysis of father’s sperm and blood. dSNVs in offspring were evaluated in paternal sperm and blood using WGS data. Filled symbols: autism spectrum disorder (ASD) diagnosis. b, dSNV assessment in 8 families identified 912 dSNVs, of which 23 (2.5%) were detected in father’s sperm or blood with ≥3 mutant reads; 34.8% of these were sperm detectable only (SDO), 30.4% were sperm detectable enriched (SDE; α<3), 26.1% were present at equal AF in sperm and blood (sperm blood equal; SBE), and 8.7% were blood detectable only (BDO). c, Relative number of paternal dSNVs that showed evidence (≥3 reads) of mosaicism in blood, sperm, or both. d, Contribution to the cumulative relative recurrence risk for all dSNVs. Risk derived from sperm mosaicism (≥1 alternate read; black), assuming equal risk for all variants (red). Dashed box shows only the first 20 identified paternally phased mosaic variants. e, Ranked plot of the estimated sperm AF (estimated fraction ± binomial 95% CI; based on the fraction of mutant reads, see Supplementary Data 2) for all mosaic variants. f, Number of mosaic variants found in each father’s sperm. F04 had the most at 5 and F06 had none detected. g, Sperm vs. blood AF for all detected mosaic variant coded by family. Most sperm mosaic AFs <8% were either SDO or SDE, whereas most mosaic variants >8% were also detected in blood at similar AFs.
Using the ratio of mutant to reference reads in blood and sperm, we defined four dSNV classes: Sperm Detectable Only (SDO); Sperm Detectable Enriched (SDE) for which the AF was >3-fold higher in sperm than in blood; Sperm Blood Equal (SBE, enrichment <3-fold); and Blood Detectable Only (BDO) (Table 1). Of the 23 variants, 34.8% were SDO, 30.4% were SDE, 26.1% were SBE, and 8.7% were BDO. Nanopore long read sequencing of the children allowed phasing of 501 of the 912 dSNVs to the paternal haplotype. Of the 23 mosaic variants, 20 resided on the paternal chromosome (40% as SDO, 35.0% as SDE, 25.0% as SBE, and none were BDO) (Fig. 1c). Thus, assessment of blood underestimates paternal gonadal mosaicism (PGM) for most dSNVs (Fig. 1d). Further, most dSNVs are not present in paternal sperm at this sensitivity level, and thus have little measurable chance to recur.
Table 1.
List of acronyms used in this study.
| List of acronyms | |
|---|---|
| DNM | De Novo Mutation |
| AF | Allelic Fraction |
| dSNV | De Novo Single Nucleotide Variant |
| dSV | De Novo Structural Variant |
| dDel | De Novo Deletion |
| dDup | De Novo Duplication |
| dSTRΔ | De Novo Short Tandem Repeat Variant |
| SDO | Sperm Detectable Only |
| BDO | Blood Detectable Only |
| SDE | Sperm Detectable Enriched |
| BDE | Blood Detectable Enriched |
| SBE | Sperm Blood Equal |
| BSS | Blood Sperm Shared (includes SDE, BDE, and SBE) |
| ASD | Autism Spectrum Disorder |
| PGM | Paternal Gonadal Mosaicism |
| WGS | Whole Genome Sequencing |
| TAS | Targeted Amplicon Sequencing |
| PGC | Primordial Germ Cell |
| SSC | Spermatogonial Stem Cell |
| CH | Clonal Hematopoiesis |
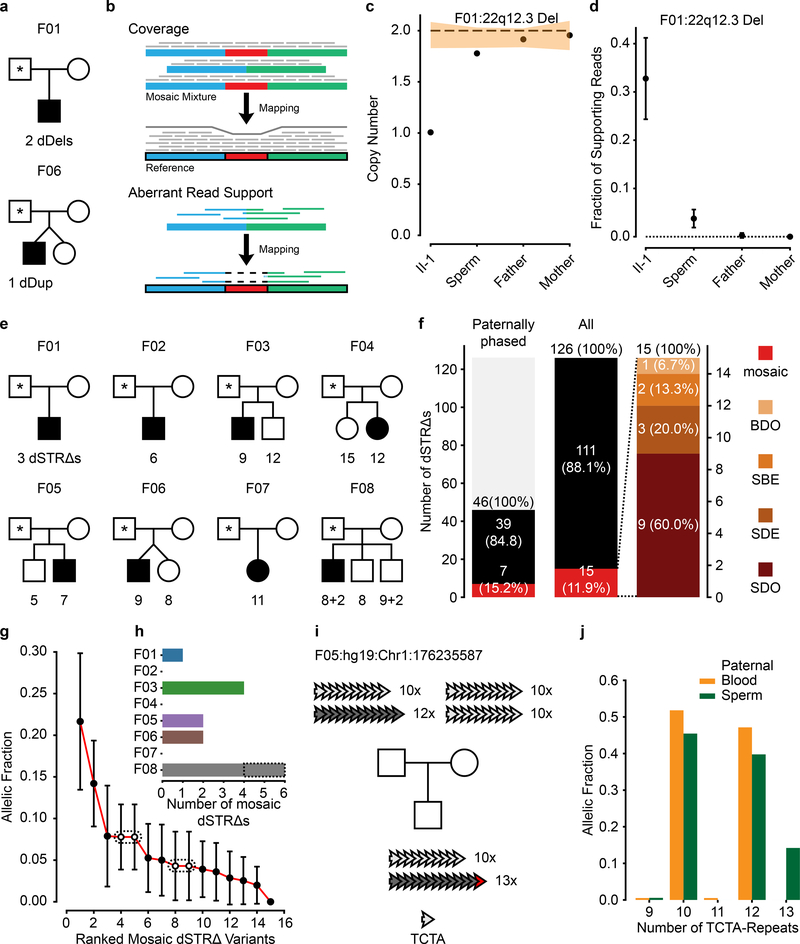
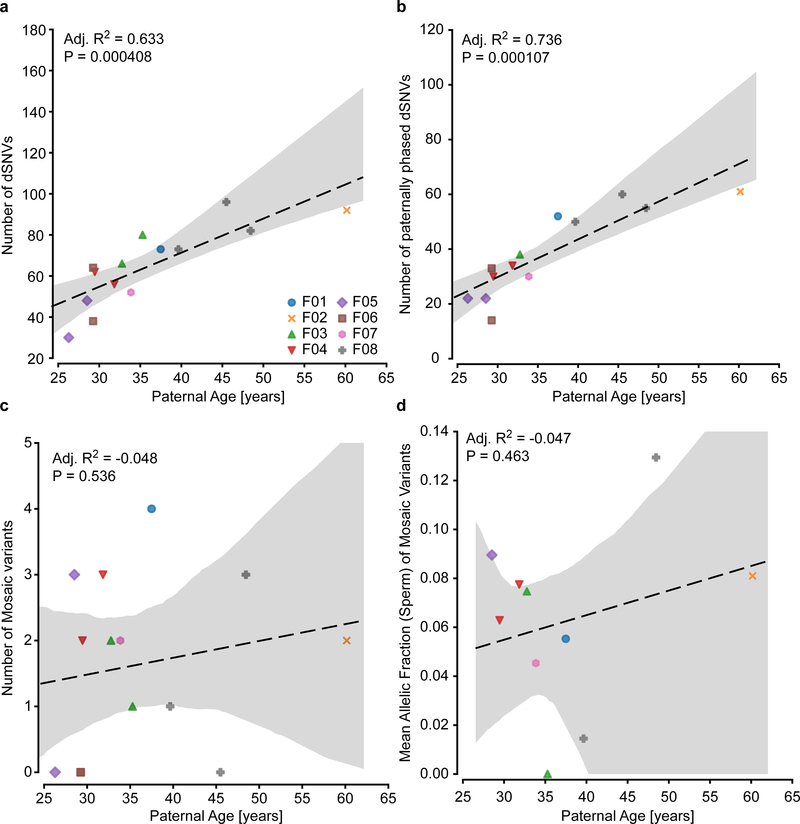
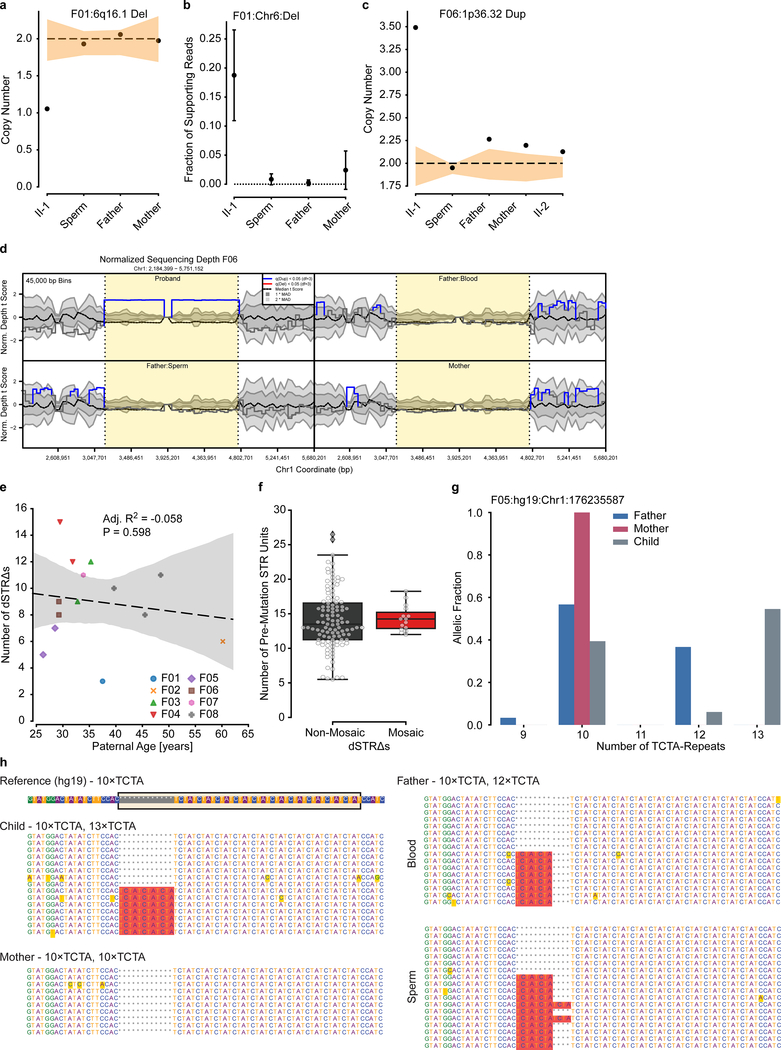
The PGM burden was roughly equally distributed among the eight families (0–5 PGM variants/male), with AFs varying between 17% to the lower detection limit of 1.3% (Fig. 1e). Neither number of mosaic variants nor their AF correlated with paternal age (Extended Data Fig. 3). We observed a mutational signature for PGM variants consistent with a developmental origin, not observed for the non PGM DNMs (e.g. relative decrease in T>C variants6,10) (Extended Data Fig. 4). While PGM variants above 7% AF were often also detectable in blood (10/11 SBE or SDE), variants below this level were typically restricted to sperm (7/12 SDO). Together, these data are consistent with an origin of PGM during embryonic development of the father, with those occurring earlier showing broader tissue distributions and higher AFs14.We next assessed the potential of sperm/blood sequencing to measure PGM for de novo structural variants (dSVs) and de novo short tandem repeats (dSTRΔs) (see Methods). Among the eight families, F01 had two de novo deletions (dDels) and F06 had one de novo duplication (dDup) (Fig. 2a). One of these variants was detectably mosaic in paternal sperm with an AF of 2–6% (Fig. 2b–d, Extended Data Fig. 5a–d). Among the eight families we identified 126 different dSTRΔs; 15 (11.9%) were mosaic (Fig. 2e–j, Extended Data Fig. 5e–h, Supplementary Data 5). Because 12 of 15 variants were SDO or SDE, recurrence risk assessment from blood only would be erroneous for 80%.
Figure 2. Risk stratification can be applied to other classes of DNMs.
a, Pedigrees for F01 and F06 and detected dSVs. b, Approaches to detect dSV gonadal mosaicism: coverage and aberrant read support. c, Calculated copy number (CN) for the 22q12.3 deletion in F01. Dashed line: expected CN (2 copies). Orange band: ±1 SD of the CN using similarly sized regions across the genome (n=1,000 random regions, see Methods). d, Estimated fraction of supporting reads (estimated fraction ± binomial 95% CI; based on the fraction of mutant reads, see Supplementary Data 7), for the 22q12.3 deletion in F01. e, 8 nuclear families and the detected de novo short tandem repeat changes (dSTRΔs) for each child (total of 126 variants, two of which are recurrent in F08). f, Gonadal mosaicism assessment for 126 dSTRΔs in father’s sperm from 8 families. g, Ranked plot of the estimated sperm AF and 95% confidence intervals (estimated fraction ± binomial CI; based on the fraction of mutant reads, see Supplementary Data 7) for all mosaic variants. Dashes: recurrent variants that suggest parental mosaicism. h, Number of mosaic dSTRΔs found in each father. i, Exemplary dSTRΔ in F05, where the child had an expansion of a tetranucleotide repeat (TCTA) on the paternal haplotype (12x to 13x) based upon bulk sequencing. j, TCTA repeat AFs from 200× WGS data for paternal blood and sperm demonstrated MGM for the 13x variant.
PGM extends to ASD pathogenic variants
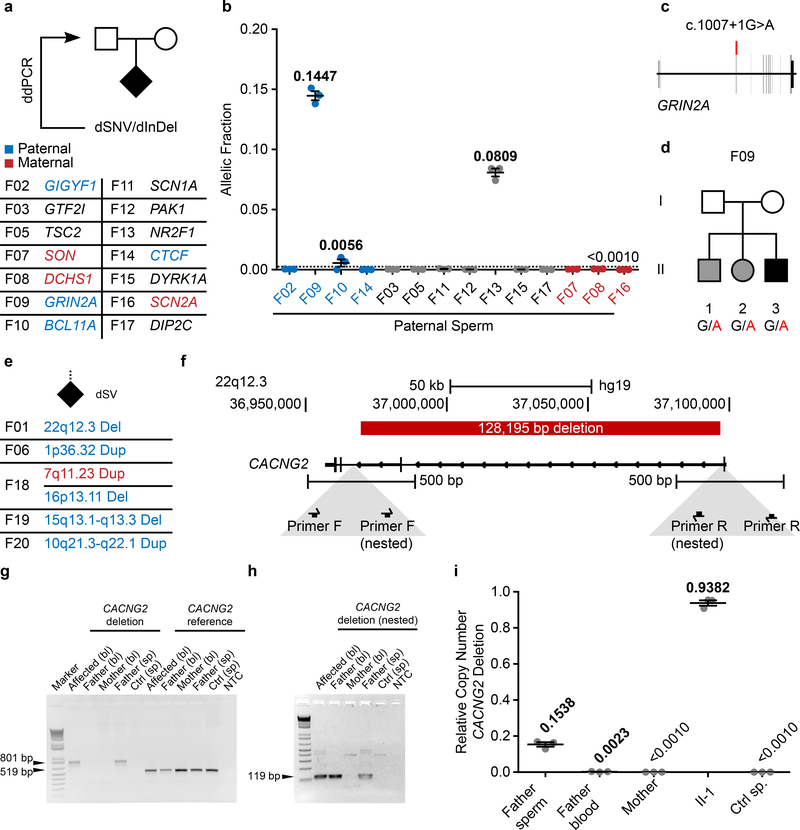
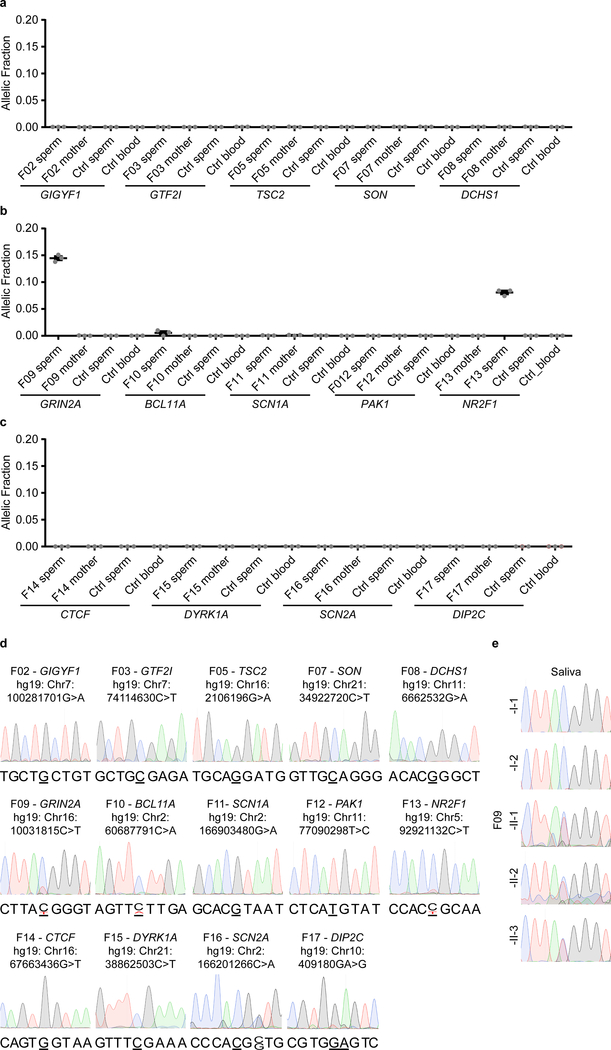
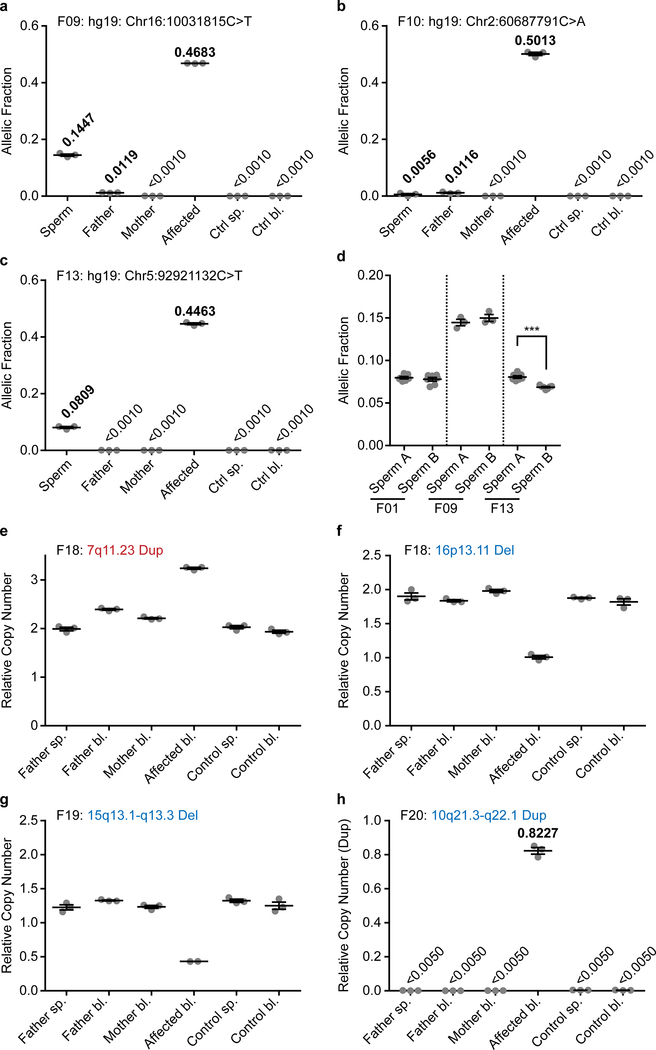
We next assessed whether clinically significant DNMs could be detected in parental sperm, which could impact clinical decision-making. We assessed a cohort of 14 families in which an offspring had ASD attributed to a dSNV or 1 basepair dDel based upon ACMG guidelines (Fig. 3a, Supplementary Text, Supplementary Table 1, Supplementary Data 1). Using ddPCR, three of 14 (21.4%) DNMs were detected as mosaic in sperm, with AFs of 14.47% (F09), 0.56% (F10), and 8.09% (F13) (Fig. 3b, Extended Data Fig. 6a–d, Supplementary Data 3). We were successful in phasing the 14.47% and the 0.56% AF variants to the paternal haplotype (Supplementary Data 2 and 6). Three variants phased to the maternal haplotype posed no risk of PGM; seven variants could not be phased, including the 8.09% AF variant. The F13 variant was absent in paternal blood (SDO), and the F09 variant was substantially reduced (SDE) (Extended Data Fig. 7a–c). These results, while representing a small number of DNMs, suggest that a substantial fraction of paternally phased as well as unphased disease-related DNMs are detectable as PGM, and thus recurrence risk can be estimated directly.
Figure 3. Pathogenic ASD DNMs benefit from risk stratification through sperm sequencing.
a, 14 ASD families with a causative dSNV/de novo insertion/deletion (dInDel) in the child and phased, where possible, to the parental haplotype (blue: paternal; red: maternal). Gonadal mosaicism was assessed by ddPCR for each dSNV/dInDel. b, AF (determined by ddPCR) of the mutant alleles in paternal sperm (n=3 experimental replicates for each sample, shown are mean ± SEM and individual values). c, Schematic of GRIN2A and the PGM variant found in F09. d, Pedigree of family F09. Black: ASD; grey: epilepsy with ADHD symptoms. All three children shared the GRIN2A G>A conversion. e, 5 ASD families with a causative dSV. Haplotype was determined from the WGS data as paternal (blue) or maternal (red). Only the 22q12.3 deletion in F01 showed gonadal mosaicism, as also described in Fig. 2c–d. f, Genomic CACNG2 locus (22q12.3) and the pathogenic 128,195 bp deletion in F01. Below: primers for nested PCR for deletion detection. g, Agarose gel for the primary PCR products from blood (bl) and sperm (sp). CACNG2 deletion: 801 bp band detected in DNA from affected and paternal sperm; 519 bp reference band detected in all samples as a control. h, Agarose gel for nested PCR products (arranged as in g). g and h show representative gels from two independent replicates. i, ddPCR showed CN mosaicism at 0.1538 copies or~7.5% AF in sperm and 0.0023 copies or ~0.1% in blood from father, 0.9382 copies or ~47.0% AF in blood from the affected individual, undetectable in samples from mother and control (n=3 experimental replicates for each sample, shown are mean ± SEM and individual values).
Two variants showed sperm AF that predict substantially elevated recurrence above the basal 1% risk in families (F09 at 14.47% AF and F13 at 8.09% AF). While F13 had a single child, F09, with a c.1007+1G>A known pathogenic variant in GRIN2A15,16 (Fig. 3c), had two older siblings lacking criteria for ASD, but deeper questioning revealed that both siblings showed neurodevelopmental abnormalities without a known cause (Fig. 3d, Supplementary Table 2). The middle child showed ADHD and speech impairment, and the oldest child had ADHD and seizures, all consistent with GRIN2A haploinsufficiency. We collected DNA samples from the whole family and found that the GRIN2A c.1007+1G>A variant was heterozygous in all three children (Extended Data Fig. 6e). Thus, the mosaic variant in father’s sperm at 14.47% was transmitted to all three offspring, an unlikely but confirmed event, resulting in pleiotropic clinical features.
In our larger ASD cohort five families had dSVs detected with standard WGS that were considered as risk alleles (Supplementary Text)12,13. These included the de novo 22q12.3 dDel in F01 and the 1p36.32 dDup in F06 (Fig. 2a), as well as F18 with a 7q11.23 dDup and a 16p13.11 dDel, F19 with a 15q13.1-q13.3 dDel and F20 with a 10q21.3-q22.1 dDup (Fig. 3e). Several of the CNVs (e.g. 15q13.1-q13.3) were flanked by directly oriented segmental duplications, suggesting that they may have arisen during meiosis through nonallelic homologous recombination (NHR)17,18. A meiotic origin of these variants would preclude any possibility of mosaicism; however, as NHR may also occur during mitosis, these were still included for this analysis19.
We phased all of these variants and found that all except the 7q11.23 dDup phased to the paternal haplotype. Probe sets were designed to interrogate these variants from sperm using PCR and ddPCR copy number assessment (Fig. 3f, Supplementary Data 7). These assays confirmed the presence of the dSV in all tested probands, but did not reveal sperm mosaicism in any additional cases beyond F01 (Fig. 3g–i,Extended Data Fig.7e–h). The 22q12.3 variant in F01 was mosaic in father’s sperm sample based upon the presence of a junction fragment matching the band in the proband and assessment of the deletion by nested PCR (Fig. 3g–h). ddPCR quantification showed 0.9382 mutant allele abundance in the proband (i.e. heterozygous), whereas father sperm showed a 0.1538 abundance and father’s blood showed 0.0023 abundance (Fig. 3i), suggesting that 7.69% of sperm carry the deletion. Thus, one of five dSVs was detectably mosaic in parental sperm at AF that could be considered clinical significant since it would increase recurrence risk by over 7-fold (Extended Data Fig. 7e–h). The specificity of these assays precluded the confident exclusion of mosaicism in paternal sperm except for one additional variant (F20 with a 10q21.3-q22.1 Dup) and thus a negative predictive value is more difficult to calculate for most dSVs.
For three of the four pathogenic variants that were mosaic in sperm, a second semen sample, collected 1–4 months apart, was subjected to mosaicism analysis by ddPCR (Extended Data Fig.7d). While all three tested variants were detected at similar AF in both, the NR2F1 mutation exhibited a slight, but significant difference between the two samples (P<0.001). This suggests that mosaicism at these higher AFs is relatively stable over time.
Unbiased analysis of sperm mosaicism detects 9 to 23 mosaic variants in sperm
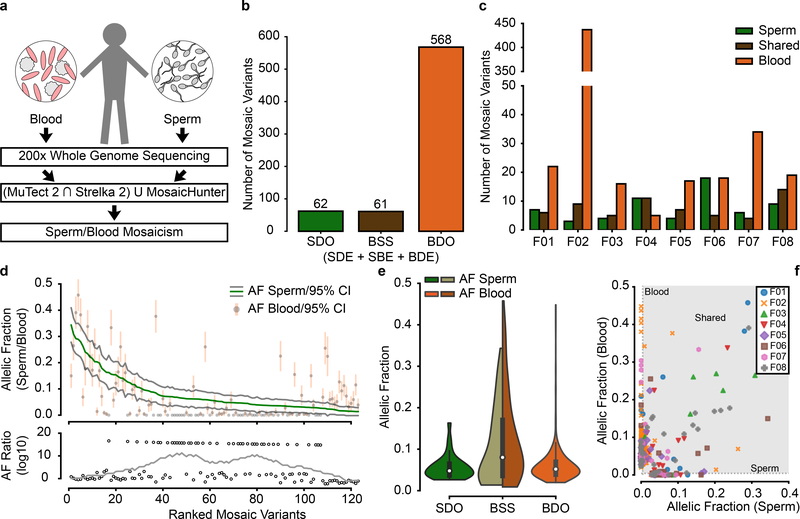
While our data demonstrate the value of sperm sequencing to determine if DNMs are mosaic in sperm, we also wanted to assess its value in identifying gonadal mosaicism for variants not yet observed in children. Using the 200× sperm WGS on the eight fathers, we identified mosaic variants using the intersection of variants of MutTect2 and Strelka220,21, both optimized for mosaic variant detection in one tissue compared with another, as well as MosaicHunter22, optimized for mosaic variant detection shared between two tissues (Fig. 4a,Extended Data Fig. 8, Supplementary Data 8). This method identified 6/23 DNMs (from Fig. 1b) as PGM, since many of the DNMs occurred in repetitive sequences that were masked by these callers. This low recall rate was partially due to optimization of the pipeline for specificity (TAS: ~90% validation rate) (Extended Data Fig. 9). To increase power for subsequent analyses on variants detected in blood and sperm, we defined three major groups of mosaic mutations SDO, BSS (consisting of SDE, SBE and blood detectable enriched - BDE) and BDO (Fig. 4b). We identified 62 SDO, 61 BSS and 568 BDO, the latter likely reflecting clonal hematopoiesis23 (CH) primarily arising from the father of F02 (Fig. 4c). There were 9 to 23 variants in the sperm of each father, each with the potential to transmit to an offspring.
Figure 4. Unbiased analysis of sperm mosaicism reveals that sperm sequencing reclassifies risk for ~50% of mosaic variants.
a, Blood and sperm from 8 fathers was subjected to 200× whole genome sequencing (WGS) followed by detection of mosaicism using the intersection of MuTect 2 and Strelka 2 and union with MosaicHunter. b-c, Total number of mosaic variants (b) and those found in each father (c) that were SDO, blood/sperm shared (BSS; includes SDE, SBE, and blood detectable enriched - BDE), or BDO. F02 showed a substantially increased number of BDO variants, most likely related to clonal hematopoiesis/collapse due to his advanced age at sampling (70 years). d, Ranked plot of the estimated sperm and blood AF with 95% confidence intervals (estimated fraction ± binomial CI; based on the fraction of mutant reads, see Supplementary Data 8) for all 123 gonadal mosaic variants that were detected as mosaic in sperm. Lower plot shows the log10 transformed ratio of sperm and blood AFs (0 replaced with 1*10−8) and the rolling average over 20 data points to display the local trend. e, Violin plots with inner box plots (showing median and quartiles) of AFs of all three types of variants as indicated (n=62 SDO variants, 61 BSS, and 568 BDO). f, Sperm vs. blood AF for all detected mosaic variant coded by individual. BDO and BSS mosaic variants reached higher AF than those that were SDO. Grey area denotes region of variants that are detectable in both sperm and blood.
The AF of PGM variants ranged from a maximum of ~35% to the lower limits of detection of ~1.5% (Fig. 4d). Compared with the sperm AF, blood AF showed two trends: At higher sperm AFs, blood AFs were similar to sperm AF; at lower sperm AFs, the blood AFs were very low or undetectable (Extended Data Fig. 9d–f). This suggested two separate origins of PGM during paternal embryogenesis: the former occurring before and the latter after germ cell specification. The AF distributions of SDO, BSS and BDO was consistent with this model, where most SDO variants occurred at AFs<10%, whereas BSSs showed an AF range up to 35% (Fig. 4e–f). BDO AFs tended to mimic SDOs, but there was a distribution tail with higher AFs likely reflecting CH. These BDO variants, while numerous, have little chance to transmit to an offspring because they were absent in sperm. Therefore, sequencing only blood to identify potentially transmissible variants would not distinguish BDO from BSS and would miss SDO variants completely.
Mutational signatures suggest an embryonic origin of PGM
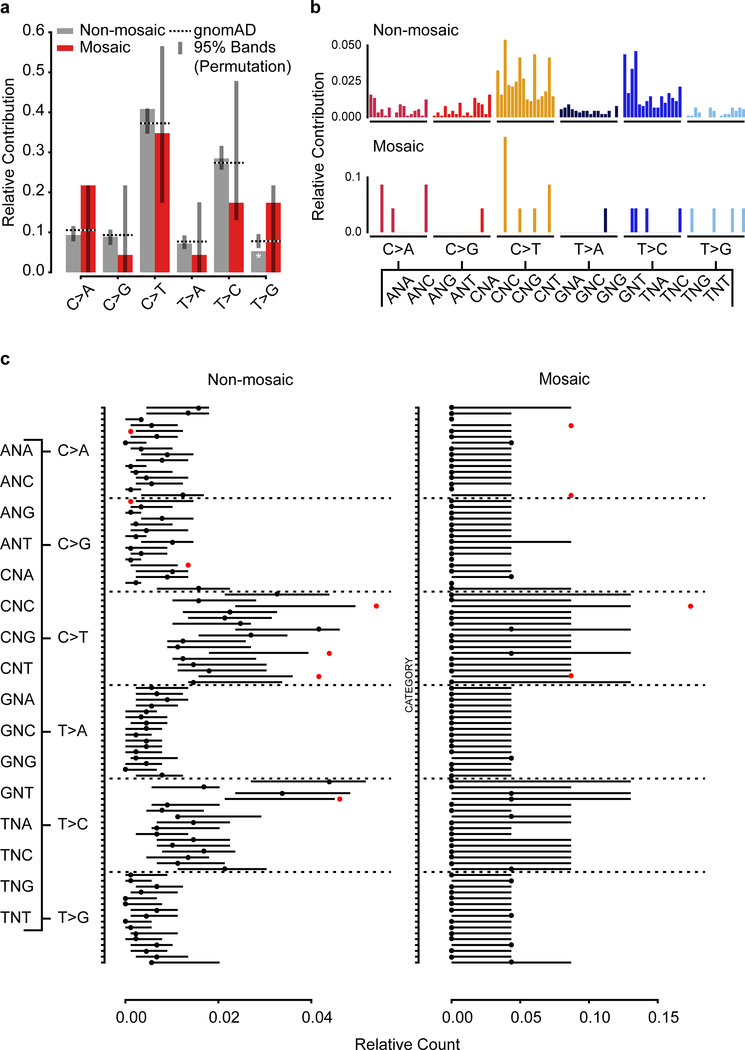
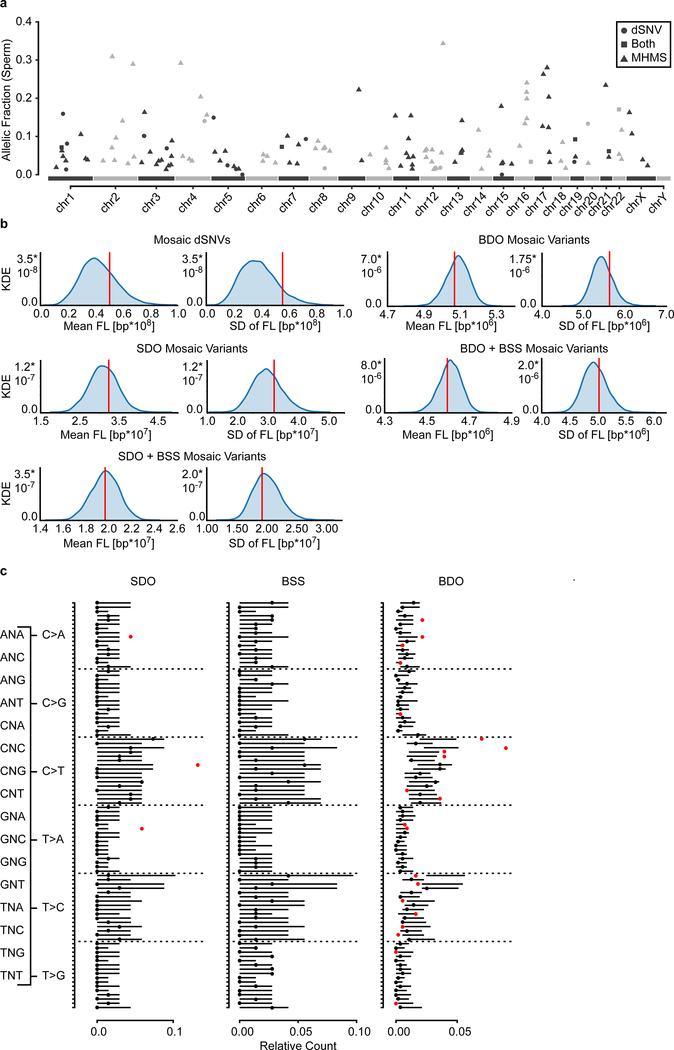
We then combined all mosaic SNVs detected in both approaches (Fig. 1 and Fig. 4) to observe common patterns for these variants. While there was no clustering along or enrichment across chromosomes (Extended Data Fig. 10a–b, Supplementary Data 9), we observed distinct mutational signatures differentiating variant classes. Assessing the relative contribution of each of the six possible base substitutions, mosaic variants differed from the background of gnomAD variants in several categories (Fig. 5a–b, Extended Data Fig. 10c).The early shared BSS mosaics differed from SDO and BDO variants that were similar to each other. Supporting an embryonic origin of these variants, they were all depleted in T>C variants, a class that was correlated with environmental damage and aging gonads and depleted in variants that were shared among siblings6,10. The differential signals for BSS variants enriched in C>A and T>G mutations relative to gnomAD and SDO and BDO mosaics are consistent with distinct mutational mechanisms in early embryonic development compared to later stages14,24.
Figure 5. Sperm mosaic variant mutation patterns support a developmental origin.
a, Mutational signatures (6 categories) for the three classes of mosaicism, compared to the overall gnomAD signature and a permuted subset (n=1,000 permutations for n=68 (SDO), 72 (BSS), and 568 (BDO) gnomAD SNVs; shown is the 95% band). Asterisks indicate observed signatures that lie outside the 95% band of the permuted variants. SDO and BDO showed signatures that differed from gnomAD and the BSS variants; BSS variants likewise showed a mutational signature that was distinct from the gnomAD population. b, Mutational signatures (96 categories; trinucleotide environment) of the three classes of mosaicism. c, Model for four types of PGM from testis tubule cross-section, with spermatogonial stem cells (SSC) at perimeter, and mature sperm at lumen. Type I and IIa PGM occurs in a single sperm (I) or SSC (IIa), thus contributing to only a fraction of total sperm and associate with non-recurrent disease. Type IIb and III mutations lead to selective growth advantage and elevate population-level recurrence risk (IIb), or occur during paternal embryogenesis, leading to multiple independent mutant SSCs and associate with mutational recurrence (III). d, Type III PGM mosaicism occur (i) during early paternal embryogenesis, seeding sperm and somal progenitors at equally high AFs, (ii) during late embryogenesis, seeding stochastically at variable AFs between tissues, or (iii) in early primordial germ cell (PGC) differentiation, seeding only gonads. Note: PGCs are the early embryonic progenitors of SSCs.
Discussion
Our results represent a significant improvement over previous strategies, where only assessment of parental blood mosaicism was used to predict future offspring in combination with advanced population statistics7,10. The role of sperm mosaicism has been increasingly recognized in single gene disorders25–28 and our work complements these efforts by providing a more general assessment of sperm mosaicism. Our data suggest a model of three major types of PGM (Fig. 4c): type I arise during the terminal differentiation of sperm and never recur. Type II arise in proliferating spermatogonial stem cells (SCCs) and include those that are extant clonally (IIa) or those under positive selection (IIb), akin to the ‘selfish sperm’ hypothesis 29. Type IIa likely represent mutations accumulating in individual SSCs and proposed to underlie the increased mutational load with age10,11, although their importance in this process is controversial24. Multiple inheritance is rare for IIa, whereas IIb are similar to IIa, as they have the same origin, but their selective advantage results in over-proliferation of the SSC clone and the potential for population-wide recurrence.
Type III arise during paternal embryonic development, prior to primordial germ cell (PGC; the early embryonic progenitors of gonadal stem cells) specification or within the PGC population, and may be detectably mosaic in sperm, resulting in the potential for recurrence. The timing of a mutation likely determines its abundance and patterns of mosaicism between sperm and somatic tissue, and our data suggest distinct mutational mechanisms between BSS and SDO variants.
Employing our methods, a distinction between the contribution of Type I and Type II mosaicism to the male-specific mutational burden is not possible, as both are below the detection limits; similarly, Type III PGM occurring after PGC specification is probably not possible, unless it is positively selected27. In contrast, our work focuses on the detection of Type III mosaicism that can stratify risk of recurrence. Considering the fraction of mosaic variants detected for each father, we estimate that on average 2.9% (95% CI: 1.4–4.4%) of variants fall within this category; and this number increases to 4.3% (95% CI: 1.6%−7.1%) if a variant can be phased to the paternal haplotype. Yet, based on our data and those of previous, gene-centric studies on sperm mosaicism, even within this group, risk can vary by an order of magnitude25,27,28.
Thus, the patterns of sperm mosaicism and the resulting framework for its detection that we present have the potential to impact clinical testing in two ways. First, direct assessment of previously transmitted pathogenic variants in paternal sperm allows for the stratification of fathers with low and high recurrence risk through TAS or ddPCR analysis. Second, even without any prior risk or family history, prospective fathers who may want to know their risk of transmitting a high-impact variant to their child could undergo deep sequencing of their sperm, followed by mosaic analysis of these data. This potential is highlighted by our finding that one of the SDO variants (F06:chr9:g.131380333G>A; NP_001123910.1:p.Arg1849Gln; 3.7% AF) was located in SPTAN1, a known gene causing infantile epileptic encephalopathy (MIM: 613477)30. While this specific variant has not been previously reported, it was predicted to be “potentially disease-causing” by MutationTaster31, had a MutPred2 score of 0.68732, and a different non-synonymous change in this same amino acid residue has been reported in affected children in ClinVar (SCV000243194.10, SCV000553140.2; p.Arg1849Trp). Based on our results, we would predict that this variant, which has the potential to be pathogenic, has a 3.7% inheritance risk for any subsequent child of the father in F06.
There are still several limitations and impediments for the application of sperm mosaicism testing. First, both approaches require the assessment of suspected high-penetrance variants and currently ignore modifiers and polygenic risk scores. This limitation is exemplified by the GRIN2A variant in family F09, where it is unclear whether the variability in expressivity is due to environment, genetic modifiers, or stochasticity15,16,33. Second, the absence of detectable mosaicism in paternal sperm can only stratify the family into low risk if the mutation of interest has been phased to the paternal haplotype. While phasing can be achieved through several experimental approaches34,35, including the nanopore sequencing we present in this manuscript, its implementation into clinical practice is still uncommon. Third, while we show examples of resampling for three of the pathogenic variants and the relative stability of mosaicism between samples, it is unclear whether this is true across all mosaic variants and should be studied systematically in future. Yet, it is a problem that would be less relevant when testing sperm samples that are directly used for in vitro fertilization. Finally, our framework for the unbiased detection of mosaicism is tuned for specificity and may therefore miss clinically relevant variants. Similarly to mosaic analysis of cancer, implementation of sperm analysis for mosaic risk mutation has to be tuned for clinical application and may require large-scale secondary validation by methods such as TAS.
Methods
Life Sciences Reporting Summary
A Reporting Summary document is published alongside this manuscript.
Binomial modeling of the detection threshold
Depicted curves were based on a classic binomial model assuming that the AF of a mutation represents the probability of encountering a mutant read. The cumulative probability was calculated using the integrate.quad function from the scipy module from python.
Simulation and analysis
To determine our sensitivity to detect mosaic variants, we created simulated datasets that contained known mosaic variants at low frequencies. We first randomly generated 10,000 variants from chromosome 22 as our set of mosaic variants. We then used Pysim36 to simulate Illumina paired-end sequencing reads from a reference chromosome 22 and a version of chromosome 22 that contained the alternate alleles from our 10,000 mosaic variants. These two sets of reads were then combined to create a series of datasets with mosaic variants at 1, 2, 3, 4, 5, 10, 15, 20, 25, and 50% AF. The coverage of these datasets was 200×. We processed these reads through our standard mapping and somatic variant calling pipelines (see below), and calculated sensitivity to detect mosaic variants at each AF as the fraction of simulated variants called by our dSNV pipeline or both MuTect 2/Strelka 2 and MosaicHunter.
Patient recruitment
Patients were enrolled according to approved human subjects protocols at the University of California for blood, saliva, and semen sampling. Semen was collected for all fathers of families F01-F20. For F09–012, saliva from the fathers and their family members was obtained, for F01-F08 and F13–20, DNA from blood was extracted. WES trio analysis for F09-F12 was performed on DNA extracted from lymphocyte cell lines (generated by the NIMH Repository) and results were confirmed in saliva samples, WGS trio analysis for F01–08 and F13–20 was performed on DNA derived from blood. Each father provided a single sperm samples, with the exception of F01, F09, and F13, where a second sample was obtained 1, 3.5, and 4 moths, respectively, after the first. Patients were all part of two independent cohorts, assembled to identify dSNVs and dSVs through trio sequencing1,4: the REACH cohort12,13, consisting of 265 families with a proband with general features of ASD, recruited at Rady Children’s Hospital San Diego (J.S.) and at Mutua Terrassa Hospital Barcelona (M.A., H.A., and J.S.), and one focusing on 98 probands with ASD and an additional diagnosis of epilepsy, recruited at NYU Medical School (O.D. and J.G.G; unpublished). The REACH cohort has been described before12,13. The cohort assembled by J.G.G. and O.D. represents a new recruitment effort that focused on patients with a diagnosis of ASD with associated epilepsy. Patients were evaluated by a child neurologist and a clinical geneticist for general and neurological assessment after referral from their primary care physician for concern about developmental delay and autism. Intellectual function was assessed by IQ score. Speech was assessed by a speech therapist fluent in the child’s native language. Brief videos of each affected member were collected during the examination as part of the clinical assessment. Autism was assessed by a clinical psychologist using ADIR, ADOS, and CARS, developmental milestones were assessed with the Vineland and hyperactivity with the Conners Parent/Teacher Scale, all administered in the child’s native language by a trained psychologist. Epilepsy was assessed by a trained specialist and included history of daytime and nighttime seizures, seizure types, length, onset, and resolutions, and treatment history. EEG was assessed awake and asleep using minimum 21 electrodes and “10 to 20” system placements recommended by the IFCN. In specific subjects, a 24 hour EEG was recorded to evaluate the possibility of nighttime seizures and to identify seizure foci. All subjects were recruited between the ages of 3–8 years. Patients were followed longitudinally to assess response to anticonvulsant therapy and behavioral therapy. All patients were seen at NYU School of Medicine and were recruited through the ethical framework at the University of California, San Diego.
Blood and saliva extraction
DNA was extracted on an Autopure LS instrument (Qiagen, Valencia, CA).
WES and WGS trio analysis
Exome capture and sequencing of F09–012 was performed at the New York Genome Center (Agilent Human All Exon 50 Mb kit, Illumina HiSeq 2000, paired-end: 2×100) and the Broad Institute (Agilent Sure-Select Human All Exon v2.0, 44 Mb baited target, Illumina HiSeq 2000, paired-end:2×76). Sequencing reads were aligned to the hg19 reference genome using BWA (v0.7.8). Duplicates were marked using Picard’s MarkDuplicates (v1.83, http://broadinstitute.github.io/picard) and reads were re-aligned around insertion/deletions (InDels) with GATK’s IndelRealigner. Variant calling for SNVs and InDels was according to GATK’s best practices by first calling variants in each sample with HaplotypeCaller and then jointly genotyping them across the entire cohort using CombineGVCFs and GenotypeGVCFs. Variants were annotated with SnpEff (v4.2) and SnpSift (v4.2) and allele frequencies from the 1000 Genomes Project and the Exome Aggregation Consortium (ExAC)37. De novo variants were called for probands using Triodenovo (v0.06) with a minimum de novo quality score (minDQ) of 2.0 and subjected to manual inspection. WGS sequencing and analysis for F01–08 and F13–20 were performed as described previously13,38. Variants from F01-F08 were further interrogated for postzygotic mosaic variants (PMV) that might be present in the children9,39. Among all 912 variants only 4 showed a significant deviation from an expected 0.5 AF using a binomial model; this effect was prior to multiple testing and disappeared following a Bonferroni correction. This lack of PMVs in our data is most likely a reflection of limited sequencing depth (~40×) and cannot conclusively exclude the existence of PMVs in our data. Nevertheless, conservatively, we assumed that all 912 dSNVs were true DNMs. We further interrogated F01–08 for possible paternally mosaic variants that might have been erroneously reported as inherited heterozygous variants; such artifacts might result in an underestimation of mosaicism and overestimation of SDO and SDE variants. However, multiple filtering approaches did not result in the identification of any such variants. While we cannot exclude their existence, we believe that their contribution to mosaicism – if any – is minor in our dataset.
Sperm extraction
Extraction of sperm cell DNA from fresh ejaculates was performed as previously described40. In short, sperm cells were isolated by centrifugation of the fresh (up to 2 days) ejaculate over an isotonic solution (90%) (Sage/Origio, ART-2100; Sage/Origio, ART-1006) using up to 2 mL of the sample. Following a washing step, quantity and quality were assessed using a cell counting chamber (Sigma-Aldrich, BR717805–1EA). Cells were pelleted and lysis was performed by addition of RLT lysis buffer (Qiagen, 79216), Bond-Breaker TCEP solution (Pierce, 77720), and 0.2 mm stainless steel beads (Next Advance, SSB02) on a Disruptor Genie (Scientific Industries, SI-238I). The lysate was processed using reagents and columns from an AllPrep DNA/RNA Mini Kit (Qiagen, 80204). Concentration of the final eluate was assessed employing standard methods. Concentrations ranged from ~0.5–300 ng/μl.
WGS of matched sperm and blood samples
WGS was performed using an Illumina TrueSeq PCR-free kit (350 bp insertion) or a TrueSeq Nano kit (350 bp insertion) on an Illumina HiSeqX. Paired-end FASTQ files of deeply (~200×) sequenced blood and sperm samples from fathers were aligned to the hg19 reference genome (1000Genomes version 37) with BWA mem (version 0.7.15-r1140), specifying the –M option that tags chimeric reads as secondary, required for some downstream applications that implement this legacy option. The resulting average mean coverage was 227× for blood samples and 222× for sperm samples with an average read length of 150bp for both sets. Duplicates were removed with the markdup command from sambamba (version 0.6.6), and base quality scores were recalibrated with the Genome Analysis ToolKit (GATK version 3.5–0-g36282e4). SNPs and InDels were called with HaplotypeCaller jointly genotyping within pedigrees, consisting of the deep coverage (~200×) genomes from father’s blood and sperm and ~40× coverage genomes derived from blood of the parents, and the children.
Oxford Nanopore sequencing (ONP) and analysis
Whole genome sequencing libraries were generated with Oxford Nanopore 1D long reads for all children (except for F03-II-2 due to lack of sufficient DNA) in deep whole genome families (F01-F08) and a subset of families with pathogenic variants (F13-F15) according to manufacturer’s recommendations. FASTQs were aligned to the hg19 reference genome with BWA mem with the ‘-x ont2d’ option for ONP reads. Coverage of proband samples ranged from 3× to 15× (average 8.6×) with an average read length of 5,349 bp.
Haplotype phasing
To phase dSNVs, a set of phase-informative single nucleotide polymorphisms (SNPs) from the WGS germline variant calls or from an assembly of the local area using Nextera sequencing (see below) of a 20 kb region around the dSNV was determined. Phase-informative SNPs were those where the child was heterozygous and either 1) one parent was heterozygous or homozygous for the alternate allele while the other parent was homozygous for the reference allele, or 2) one parent was heterozygous while the other parent was homozygous for the alternate allele. Second, where applicable, long-reads (Oxford Nanopore reads, average length 5,349 bp) were identified that contained both a dSNV and one or more phase-informative SNPs. The number of dSNV and phase-informative SNP combinations that were present in reads and consistent with the dSNV occurring on a maternal or paternal haplotype were counted. Reads containing an InDel flanking either the dSNV or the phase-informative SNP were excluded from the analysis. Finally, the dSNVs were assigned to maternal and paternal haplotypes if there were: 1) a minimum of two counts, and 2) the haplotype with the majority of counts had at least 2/3 of total counts. For F09-F12, F16, and F17, we attempted phasing using a Drop-Phase approach41. In short, a complementary assay to the mutant allele at the dSNV position was designed for both the wild-type and the variant allele (see ddPCR methods). Then the co-occurrence of the mutant dSNV was assessed for both genotypes and quantified as described before41 (Data S8).
Sanger sequencing of SNVs
PCR and Sanger sequencing were performed according to standard methods. Primer sequences can be found in Supplementary Data 10. Validated mutations and surrounding SNPs were also used as basis for the design of ddPCR assays where applicable.
ddPCR design, validation, and setup of experiments for SNV analysis
Using the Primer3Plus web interface42–44, the amplicon and probes for wild-type and mutant were designed to distinguish reference and alternate allele (settings in Supplementary Information under Additional Information). Probes were required to be located within 15 bp up- and 15 bp downstream of the mutation and adjusted, so melting temperatures (Tm) were matched between reference and alternate probe. In addition, if possible, amplicons were kept at 100 bp or shorter and probes at 20 bp or shorter. Specificity of the primers was assessed using Primer-BLAST. Custom primer and probe mixes (primer to probe ratio of 3.6) were ordered from IDT with FAM-labeled probes for the alternate, and HEX-labeled probes for the reference allele (Supplementary Data 10). Optimal annealing temperature, specificity, and efficiency were tested using custom gblocks (IDT) or patient DNA at a range of dilutions. ddPCR was performed on a BioRad platform, using a QX200 droplet generator, a C1000 touch cycler, a PX1 PCR Plate Sealer, and a QX200 droplet reader with the following reagents: ddPCR Supermix (BioRad, 1863024), droplet generation oil (BioRad, 1863005), cartridge (BioRad, 1864008), and PCR plates (Eppendorf, 951020346). Aiming for 30–60 ng per reaction, up to 8 μl of DNA solution were used in a single reaction. Data analysis was performed using the software packages QuantaSoft and QuantaSoft Analysis Pro (BioRad). Each run included technical duplicates or triplicates (as indicated in figure legends). For direct comparison of sperm samples we used seven technical replicates, except for F09, where the total amount of sperm DNA was limiting. Across all ddPCR reactions that were designed for SNV detection, we determined that the minimum AF that we could reliably detect was 0.1%. Therefore, we set this as threshold of detection. Raw data for ddPCR experiments can be found in Data S3.
Targeted amplicon sequencing (TAS)
PCR products for sequencing were designed with a target length of 160–190 bp with primers being at least 60 bp away from the base of interest. Primers were designed using the command-line tool of Primer3 with a python wrapper (Supplementary Data 10). PCR was performed according to standard procedures using GoTaq Colorless Master Mix (Promega, M7832) on sperm, blood, and an unrelated control. Amplicons were either enzymatically cleaned with ExoI (NEB, M0293S) and SAP (NEB, M0371S) treatment or gel extraction where necessary (Zymo Research, D4007). Following normalization with the Qubit HS Kit (ThermFisher Scientific, Q33231), amplification products were processed according to the manufacturer’s protocol with SureSelect SPRI Beads (Beckman Colture, A63881) at a ratio of 1.2x. Library preparation was performed according to the manufacturer’s protocol using a Kapa Hyper Prep Kit (Kapa Biosystems, KK8501) and barcoded independently with unique dual indexes (IDT for Illumina, 20022370). After sequencing on an Illumina HiSeq 4000 with 100 bp paired-end reads, reads were mapped to the hg19 reference genome (1000Genomes version 37) and processed according to GATK v3.8 best practices. Across all amplicons, read numbers (mean ± SD) were 636,636 ± 382,226 in sperm, 831,556 ± 530,332 in blood, and 857,289 ± 570,612 in control. Overall, read depth reached between 93–3,138,968x, with 99% >261x and 95% >1809x. Putative mosaic sites were retrieved using samtools mpileup and pileup filtering scripts described in previous TAS pipelines28,45. Variants were considered validated if 1) their lower 95% CI boundary was above the upper 95% CI boundary of the control; 2) their AF was >0.5%.
Mosaic dSNV analysis
Using the read depth information generated by HaplotypeCaller, the AF for previously called dSNVs was determined. Additionally, dSNVs that fell in repetitive regions of the human genome were annotated using the repeatMasker (rmsk.txt) file from UCSC. Variants that were homozygous alternate in the father and heterozygous in the proband, as well as variants that were present in both blood and sperm at AFs that suggested an inherited heterozygous SNP (i.e. AF>35% in both blood and sperm) were removed. Variants were further filtered to include only those with a gnomAD frequencies <0.0146. Mosaic variants were categorized based on their presence or absence in sperm and blood (≥3 reads minimum requirement in one of them; if ≥3 reads were present for one, the other only had to show ≥1 reads). The 3 read minimum was based on an expected Illumina per base error rate of Q30/0.1% (i.e. ~0.033% error rate to substitute to the expected dSNV). Given a read depth of 200×, a minimum of 1 read as evidence would result in a falsely assigned mosaic variant with ~6.5% probability, with 2 reads this drops to ~0.2%, with 3 reads to ~0.005%. Given the number of interrogated dSNVs is ~1000, this would result in ~60, ~2, and ~0.05 false positive variants. To be called sperm enriched, a variant’s AF had to be three times higher in sperm than in blood (α>3), which was an arbitrarily determined threshold, mainly based on the size of the 95% CI at ~200× at low AF. To assess the sensitivity of 200× WGS, in one family we also performed Multiplex Accurate Sensitive Quantitation (MASQ) on sperm DNA, which is capable of detecting AFs as low as 10−4-10−6 (A.B.M., Z.W., unpublished). We selected dSNVs where sperm WGS did not suggest gonadal mosaicism for the majority of variants, in order to assess whether additional mosaics might be identified. From 73 such dSNVs in F01, MASQ assay design was successful for 23, two of which were already detected as mosaic from 200× WGS of sperm. MASQ confirmed these two, but did not detect any additional variants that were mosaic in sperm. Only 1 of these variants remained unphased, confirming that the remaining 20 paternally derived dSNVs even at this level of detection were not found to be mosaic in sperm, and that they likely arose either later in the sperm lineage, zygotically, or post-zygotically.
MuTect 2/Strelka 2 and MosaicHunter mosaic variant calling
Sperm- and blood-specific SNVs were called in the 200× WGS data using two somatic variant callers with default parameters, MuTect 2 (v2.1)20 and Strelka 2 (v2.9.2)21, setting the sperm sample as “tumor” and the blood sample as “normal” and vice versa. High confidence calls for somatic mosaicism for each sperm-blood and blood-sperm comparison was performed by taking the intersection of variants identified by both callers (MS). These candidates were further filtered to reduce potential false positives as follows: we removed those that fell into repetitive regions, that fell within 5 bp of a germline InDel, that were part of a homopolymer or dinucletide repeat, or that were present in gnomAD at allele frequencies >0.01. The latter filter was employed as common variants that appear to be mosaic are most often artifacts. Shared mosaic variants (and some tissue-specific variants missed by MS) were called using the whole genome single mode provided by MosaicHunter v1.022 as previously described14 (MH). Additionally, variants had to be within the 5th and 95th percentile (76<read depth<280) for sequencing depth across all variants to control for artifacts, had to be absent or at an allele frequency <0.01 in gnomAD, could not be recurrent in our data set, had to have a major allele consistent with the reference allele in hg19, and had to have an AF below 30% in at least one tissue (to remove likely heterozygous calls). If a mosaic variant was only found in one tissue by MH, the variant was only determined to be shared mosaic if there were ≥3 reads supporting the alternative allele in the second tissue. Calls from both methods (MS and MH) were then combined to obtain tissue-specific and shared mosaicism.
Assessment of the location of the genome-wide distribution of mosaic variants
In order to assess the distribution of mosaic variants along the chromosomes, an equal number of variants (for mosaic dSNVs and unbiased calls that were sperm-specific, sperm mosaic, blood mosaic, or blood-specific) was randomly generated with BEDTools from the called region from Strelka 2 with or without subtraction of the repeatMasker (rmsk.txt) file from UCSC as appropriate. This process was repeated 10,000 times to generate a distribution of the mean and standard deviation of the distance of neighboring variants according to a broken stick model47.
Mutational signatures
Mutational signatures were determined for each variant by retrieving the tri-nucleotide sequence context using Python with pysam and plotting the trans- or conversion based on the pyrimidine base of the original pair similar to previous studies48. gnomAD mutational signatures were obtained by retrieving SNVs present in the publicly available VCF. In order to obtain a 95% band of expectation, an equivalent number of variants was randomly chosen from the gnomAD VCF. This process was performed for a total of 1,000 times to obtain a distribution and the 2.5th and 97.5th percentile of the simulated mutational signatures. Significance was reported if a mutational signature was outside the permuted 95% bands.
Mosaic dSV analysis of WGS data
We searched for evidence for mosaicism of structural variants in the fathers using depth of coverage, split-reads, discordant paired-ends, and B-allele frequency in deeply sequenced paired-end genomes. Depth of coverage was estimated as the median per base-pair coverage within the SV locus, while omitting positions that overlapped assembly gaps, RepeatMasker elements, short tandem repeats, and segmental duplications. We estimated copy number by dividing the median depth of coverage by the median coverage of the chromosome and multiplying by the ploidy number (2 for autosomes). Standard deviation of copy number was calculated by sampling the estimated copy number of 1,000 random non-overlapping regions of the same length of the dSV, ensuring each region did not overlap more than 50% to exclude elements listed above. Since the reported copy number of the de novo duplication appears to be elevated in related non-carriers, we opted to estimate the standard deviation of copy number while controlling for repeat content. Hence, sampled regions contained a RepeatMasker content in the range of 30–35% (dSV=33%). Split-reads (also known as chimeric reads) are those with multiple alignments to the genome. Generally, if a read spanned a deletion or tandem duplication breakpoint, two alignments were generated with each segment mapping to opposite ends of the breakpoint. Similar to split-reads, discordant paired-ends had read fragments that span the SV breakpoint, but the SV breakpoint resided in the unsequenced insert of the fragment. Consequently, the paired-ends mapped to opposite ends of the breakpoint producing an insert size approaching the size of the SV. We searched ±250 bp from the predicted breakpoint for SV supporting reads, which were unique reads that were either split or contained discordant paired-ends with breakpoints that overlap at least 95% reciprocally to the SV. We reported the proportion of supporting reads to non-informative reads (those that do not support the SV) within the ±250bp windows, which roughly estimates proportion of mosaicism. Additionally for the de novo duplication SV, we searched for deviations in B-allele frequency defined as the proportion of reads that support the alternate variant to all reads covering the variant in question. Normalized sequencing depth calculations generated by CNView49 was derived from binned coverages in 45Kb non-overlapping windows.
dSTRΔ calling and mosaicism detection
Analysis of STR expansions and contractions were performed using HipSTR50 (version v0.6) jointly on all BAM files (40× trios and >200× blood and sperm of fathers). The reference STR set provided by HipSTR for GRCh37 (GRCh37.hipstr_reference.bed) and default options were used except for: --def-stutter-model and --output-gls. Furthermore, a modified version of HipSTR’s denovofinder tool was run on each of the 40× trios. The posterior probability of a de novo mutation was calculated using HipSTR genotype likelihood and STR loci mutation rates as priors. Strict quality filters to detect de novo STRs were applied within trios. STR loci were excluded from analysis if they were in segmentally duplicated (UCSC hg19.genomicSuperDups table)51,52 regions. Genotype STR calls in all family members were required to have a minimum genotype quality of 0.9, a maximum of 15% of reads with stutter or InDel, at least ten spanning reads, and at least 20% of reads to support each allele. STR loci were excluded if homozygous in the child or if they contained homopolymers and dinucleotide repeat motifs. De novo STR mutations were further required to have a posterior probability of de novo mutation ≥0.8. Mutations were excluded if they were not a multiple of the repeat motif unit, or if the de novo allele was found in one of the parents at >0.1 allele frequency. STR mutations were further only considered if the repeat unit was ≥3 as homopolymers and dinucleotide repeats were enriched for false-positive calls. The remaining loci were annotated with their phase where possible and de novo allele frequencies in the >200× sperm and blood samples. dSTRΔs were qualified as inconclusive if mosaicism was detected in mother and father, as true de novo if no mosaicism was detected in the parents, as maternal if mosaicism was only detected in the mother, and as paternal if mosaicism was detected in blood, sperm, or both. As for dSNVs, sperm-enriched variants were annotated as such if the AF was more than 3 times higher in sperm than blood. Phase of STR was inferred from genotypes: if a unique allele was inherited from one of the parents, the STRΔ was assumed to be derived from the other.
Nextera sequencing to identify informative SNPs
PCR products for sequencing were designed to encompass 1kbp for the assembly of the local region around the mutation for phasing of F09-F12 (Supplementary Data 10). Parallelized primer design was achieved using the web interface of PCRTiler53. PCR was performed according to standard procedures using GoTaq Colorless Master Mix (Promega, M7832). Successful amplification products were processed according to the manufacturer’s protocol with SureSelect SPRI Beads (Beckman Colture, A63881) at a ratio of 0.8–1.0x, a Nextera DNA Library Preparation Kit (Illumina, FC-121–1031), and a Nextera Index Kit (Illumina, FC-121–1011). After sequencing on an Illumina MiSeq, reads were mapped and processed according to GATK best practices. Variants were called using GATK’s HaplotypeCaller.
Mosaic SV analysis using PCR and ddPCR
Nested PCR was performed using blood DNA extracted from the F01 trio (proband, mother, and father), as well as sperm from the F01 father and a non-related male. Primers were designed using Primer3Plus online software to span the deletion breakpoints within CACNG2 determined by WGS analysis within 500 bp windows up- and down-stream of the predicted deletion. Additionally, a reverse primer was designed to be used with the nested forward primer as an amplification control (Supplementary Data 10). All PCR reactions were 25 μl volumes and included 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 2 mM MgCl2, 1 U of Taq (Thermo Fisher Scientific, Waltham, MA), and 300 nM of each appropriate primer. DNA template was 50 ng of DNA from blood or sperm for the initial PCR (using the external set of primers), or μl of the initial PCR product for the nested (internal) PCR. PCR reactions were run following a standard ramp speed protocol using a C1000 Touch Thermal Cycler (Bio Rad, Hercules, CA) with cycling consisting of a 2 min initiation at 95°C, 35 cycles of 95°C for 30 s, 55°C anneal for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 3 min. Products were resolved on 2% agarose gels. For ddPCR analysis, primer and probe sets for the SVs (copy number and break point analysis) were designed using Primer3Plus (Supplementary Text, Supplementary Data 10). Primers were designed to span the deletion breakpoints within the region or to lie within an intron of a centrally located gene within the deleted or duplicated region. Custom primer and FAM-labeled probe mix at a primer:probe ratio of 750 nM:250 nM was ordered from Bio Rad (Hercules, CA) or from IDT as described above, as well as a HEX-labeled pre-validated copy number variation assay specific for RPP30 as an internal control (assay ID: dHsaCP2500350). ddPCR was performed and analyzed as described above. Raw data for ddPCR experiments can be found in Data S3.
Data processing
Data analysis and plotting were performed using GraphPad Prism, R, and Python (pandas, matplotlib, and seaborn modules).
Statistics
Statistical analyses and fitting were performed using GraphPad Prism or Python with the SciPy, Astropy, or StatsModels modules, or calculated directly using Pandas for percentiles. Regression analysis was performed using a simple ordinary least squares model with StatsModels. 95% confidence intervals around the estimated fraction (Allelic Fraction) were calculated as binomial confidence intervals based on the estimated fraction and read number for each data point. Mean and standard error of the mean (SEM) were calculated using GraphPad Prism’s integrated function. GraphPad Prism was also used to perform the unpaired, two-tailed t-test, and the two-tailed Mann-Whitney test. Permutation analyses are described above in the respective methods sections. To calculate the mean population fraction of mosaic variants from the dSNV data, the mean and 95% confidence intervals (CI) were calculated from the 8 fathers, for both all variants and those that were paternally phased in each individual. 95% confidence intervals were calculated using the statsmodels.stats.api.DescrStatsW() and the associated .tconfint_mean() functions.
Data availability statement
Aligned BAM files generated for this study through deep WGS or TAS are available on SRA (accession number: PRJNA588332). WGS data used for de novo calling are available through the NIMH Data Archive (NDA; collection ID: 2019). Long read sequencing data are likewise available on NDA (collection ID: 2795). NDA access is regulated by the standard organizational process and is subject to review by NDA. Data are also available through the corresponding authors on reasonable request. Additionally, summary tables of the data are included as supplementary information.
Code availability statement
Algorithms used for mosaic variant detection were published before. Any custom code is available through the corresponding authors upon reasonable request.
Extended Data
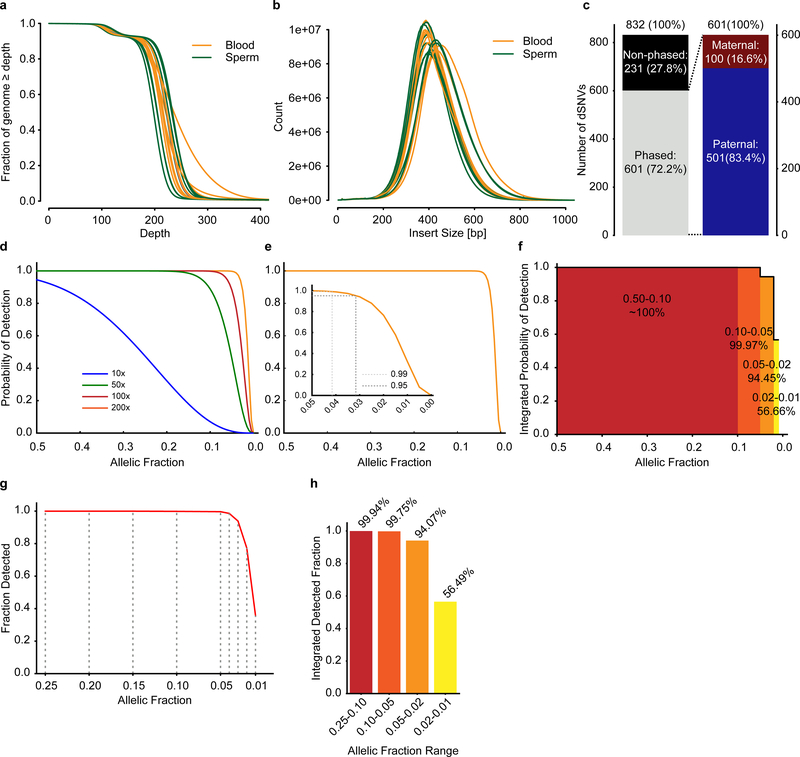
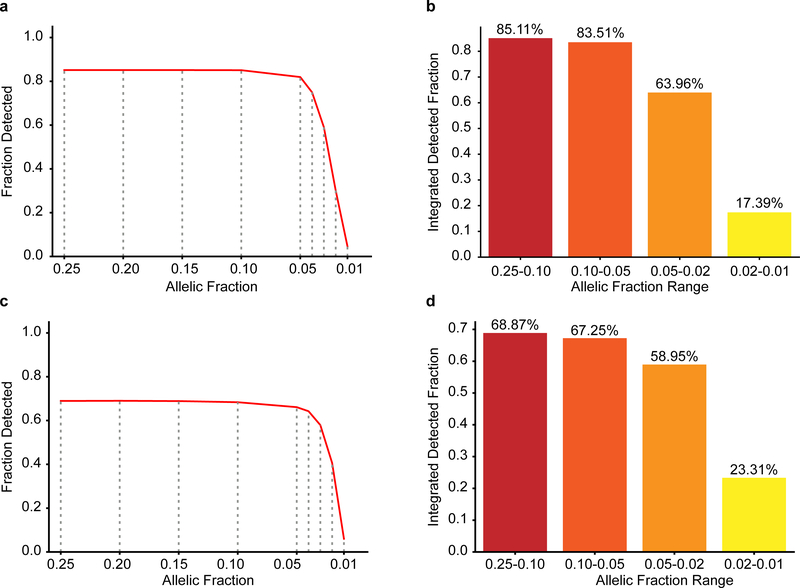
Extended Data Fig. 1. 200× WGS allows detection of mosaic variants down to 1% sensitivity.
a, Plot showing the fraction of the genome that is covered at a given depth for blood and sperm following WGS with a target coverage of 200×. b, Plot showing the insert size of the reads for blood and sperm. c, Nanopore long-read technology (average read length 5,349 bp) was able to assign parental haplotype to 601/832 dSNVs in 13 children. Out of these, 501 were paternal, resulting in α~4 as reported previously. d-e, Binomial models for the detection limit of mosaic variants. Plots show the probability of detecting a given variant at a specific allelic fraction (AF) when requiring at 3 alternate reads at different read-depths (d) or including a magnified inset for AF between 0.05 and 0 at 200× (e). f, Analysis of the power of detection assuming a minimum requirement of 3 reads at 200× sequencing. Plot shows the integrated probability of detection for the indicated tiers based on the curve seen in e. g-h, Plot of the fraction of detected variants (g) and the integrated detected fraction for the indicated AF ranges (h) of simulated data using Pysim. Results are from 10,000 variants simulated at 0.25, 0.20, 0.15, 0.10, 0.05, 0.02, and 0.01 AF. HaplotypeCaller was employed to detect variants as for data in Figure 1.
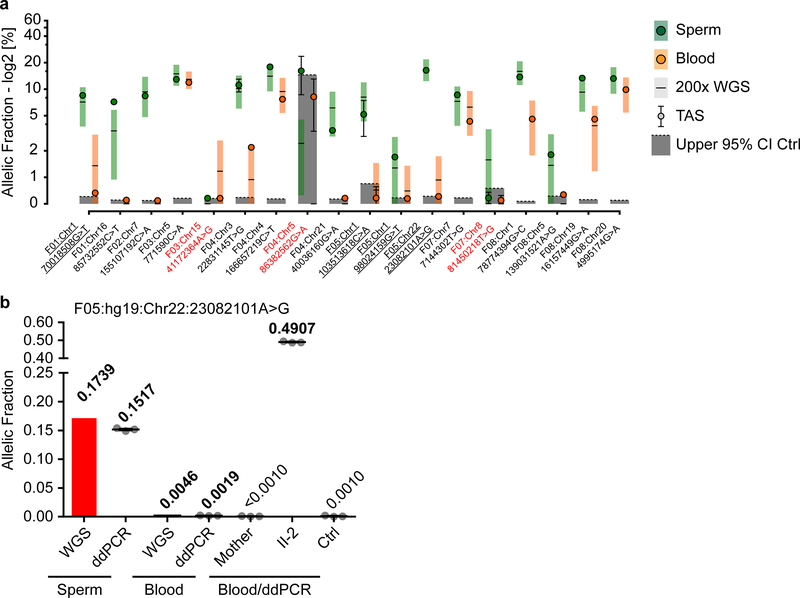
Extended Data Fig. 2. Orthogonal validation of a subset of mosaic dSNVs.
a, 18 variants that could be assessed by ultra-deep target amplicon sequencing (TAS): shown are the reported 200× WGS results (square with horizontal line) and the results from TAS (closed circle) (shown are estimated fraction ± binomial 95% CI). Sperm (left, green) and blood (right, orange). Dashed line and grey box: upper 95% CI of an unrelated control and the area beneath to visualize likely false positive variants. y-axis: allelic fraction (%) for a log2 transformation of the data. Red text: variants that were considered to have failed orthogonal validation: 15/18 variants were successfully confirmed. Underlined variants were confirmed, but likely annotated as the wrong class (all 5 are probably SDO rather than SDE). For all data points, the estimated fraction and CI are based on the fraction of mutant reads, see Supplementary Data 2 and 4. b, Allelic fraction (determined by ddPCR or WGS read counts) of the mutant allele with the highest allelic fraction in sperm (F05: Chr22:23082101A>G). Sperm and Blood indicate samples from the father, other samples (Blood/ddPCR) were derived from the mother, the child harboring the dSNV (II-2), or control (Ctrl) blood. Graph shows individual data points (experimental triplicates) and mean ± SEM for the ddPCR data.
Extended Data Fig. 3. Age correlation of all and mosaic dSNVs.
a, Plot showing the increase in dSNV number with paternal age at birth, as described previously1,5. Dashed line shows a regression curve demonstrating this dependence (n=14 trios, adjusted R2=0.526, P=0.0020). b, Plot showing the increase in dSNV number with paternal age at birth for paternal variants only. As expected, this correlation was stronger than for non-phased variants (n=13 trios, adjusted R2=0.736, P=0.000107). c-d, Plots showing correlation for paternal age and the number of mosaic variants or the mean AF in sperm. Paternal age/the number of mosaic variants (c; n=14 trios, adjusted R2=−0.048, P=0.536) and paternal age/mean AF in sperm (d; n=14 trios, adjusted R2=−0.047, P=0.463) did not show any significant correlation. Adjusted R2, coefficient of determination, and F-statistic nominal P-values are derived from a linear regression model through ordinary least squares. All graphs show individual data points, a regression line, and the 95% CI.
Extended Data Fig. 4. Mutational signature for non-mosaic and mosaic dSNVs.
a, Mutational signatures (6 categories) for non-mosaic and mosaic dSNVs, compared to the overall gnomAD signature and a permuted subset (n=1,000 permutations for n=889 (non-mosaic) and n=23 (mosaic) dSNVs; shown is the 95% band). Asterisks indicate observed signatures that lie outside the 95% band of the permuted variants.. Non-mosaic variants are largely reminiscent of the gnomAD signature (with the exception of a significant depletion of T>G). Mosaic variants exhibit some differences, but none reach significance due to the low number of available mutations. b, Mutational signatures (96 categories; trinucleotide environment for non-mosaic and mosaic dSNVs. c, Detailed view of the 96 mutational categories for non-mosaic and mosaic dSNVs, compared to the overall gnomAD signature and a permuted subset (n=1,000 permutations for n=889 (non-mosaic) and n=23 (mosaic) dSNVs; shown is the 95% band). Dots indicate the observed mutational signature (black: within 95% band; red: outside the 95% band).
Extended Data Fig. 5. Sperm mosaicism stratifies recurrence risk for dSV and dSTRΔ variants.
a-c,, Calculated copy number (a, c) and fraction of supporting reads (b) for the 6q16.1 deletion in F01 and The 1p36.32 duplication as indicated. Orange band in a and c: ±1 SD of the CN using similarly sized regions across the genome (n=1,000 random regions, see Methods). Plot in d shows the estimated fraction of supporting reads (estimated fraction ± binomial 95% CI; based on the fraction of mutant reads, see Supplementary Data 7). Together, these approaches suggest that these dSVs are not mosaic in paternal sperm. Note that the fraction of supporting reads could not be used for the duplication due to the repetitive elements flanking this SV. d, Copy number variant plot for the duplication in F06 for the Proband (40×), Father (200× both), and the mother (40×). Visualization was performed with the CNView36 tool (see Methods). e, Correlation of the number of dSTRΔs with paternal age at birth. Dashed line shows a regression curve (n=14 trios, adjusted R2=−0.058, P=0.598). Adjusted R2, coefficient of determination, and F-statistic nominal P-value are derived from a linear regression model through ordinary least squares. Graph shows individual data points, a regression line, and the 95% CI. f, Number of STR repeat units for non-mosaic dSTRΔs or those that are mosaic. No significant difference can be observed between the two groups (n=111 non-mosaic variants and n=15 mosaic variants; two-tailed Mann Whitney test; nominal P=0.5490). Boxplots show median and quartiles with outliers as well as individual values. g, Detailed analysis of the TCTA repeat numbers in paternal, maternal, and child’s blood at low sequencing depth. Results show a de novo 13× repeat in the child that is neither present in the father nor the mother. h, Sample reads showing the presence of a 10× and 13× allele in the child, a homozygous 10× allele in the mother, a 10× and a 12× allele in the father, and the presence of a mosaic 13× allele exclusively in paternal sperm.
Extended Data Fig. 6. Sperm mosaicism stratifies risk for pathogenic ASD mutations.
a-c AF (determined by ddPCR) of the mutant allele in paternal sperm (sperm) and maternal blood (mother) for the relevant dSNV in the 14 families. Part of this panel is also presented in Figure 3. Ctrl –an unrelated sperm or blood sample, as indicated, acting as control. Graphs show individual data points (experimental triplicates) and mean ± SEM. d, Sanger sequencing results of paternal sperm for the locus harboring the dSNV for each family. Confirming the ddPCR results, F09, F10, and F13 showed mosaicism at their respective positions. e, Sanger sequencing results showing the C>T conversion locus in GRIN2A in F09 for all family members. The mutation was absent in the saliva of both parents, but present as a heterozygous allele in all 3 children.
Extended Data Fig. 7. ddPCR assessment of pathogenic structural variants and recurrent sampling of pathogenic DNMs in F01, F09, and F13.
a-c, AF (determined by ddPCR) of the mutant alleles in F09 (a), F10 (b), and F13 (c). DNA tested was derived from paternal sperm (indicated as sp.) and the saliva (a and b; sal.) or blood (c, bl.) of the father, mother, or affected child. In addition, controls for sperm (sp) and blood (bl) are provided. d, AF (determined by ddPCR) comparing two biological replicates of paternal sperm for F01, F09, and F13. The samples showed comparable levels of AF over time for all three samples, however, F13 exhibited a minor, but statistically significant difference. ***P<0.001 (unpaired t-test, two-tailed, degrees of freedom=12). e-g, Relative copy number (determined by ddPCR) for the three indicated dSVs for blood-derived samples, labeled as SNV assays in Extended Data Figure 6. Note that there is no detectable abnormality in the paternal sperm copy number above noise level, suggesting absence of sperm mosaicism in these samples. h, Direct copy number quantification of the duplication by ddPCR. Samples as before. All graphs show individual data points (experimental triplicates except for Affected in g [experimental duplicate], and F01 and F13 in d [7 experimental replicates]) and mean ± SEM.
Extended Data Fig. 8. Limit of detection analysis for the unbiased analysis of gonadal mosaic SNVs.
a-d, Plots of the fraction of detected variants (a, c) and the integrated detected fraction for the indicated AF ranges (b, d) of simulated data using Pysim for the intersection of MuTect 2/Strelka 2 (a, b) and MosaicHunter (c, d). Results were from 10,000 variants simulated at 0.25, 0.20, 0.15, 0.10, 0.05, 0.02, and 0.01 AF. This was the same data set as used in Extended Data Figure 1. The MuTect 2/Strelka 2 and MosaicHunter pipelines were employed with the same filters as for the data in Figure 4.
Extended Data Fig. 9. Mosaic SNVs identified by unbiased analysis have a high validation rate and their AF differs depending on their origin.
a-c, 74 variants that could be assessed by ultra-deep target amplicon sequencing (TAS): shown are the reported 200× WGS results (square with horizontal line) and the results from TAS (closed circle) (shown are estimated fraction ± binomial 95% CI). Sperm (left, green) and blood (right, orange). Dashed line and grey box: upper 95% CI of an unrelated control and the area beneath to visualize likely false positive variants. y-axis: allelic fraction (%) for a log2 transformation of the data. Plots are split by the three categories: SDO (a), BSS (b), and BDO (c). Red text denotes variants that were considered to have failed orthogonal validation: 13/19 (a), 21/21 (b), and 33/34 (c) were successfully confirmed. Underlined variants were confirmed, but likely annotated as the wrong class (i.e. they are actually BSS for SDO and BDO variants in a and c, or are SDO (green text) or BDO (orange text) for BSS variants in c). For all data points, the estimated fraction and CI are based on the fraction of mutant reads, see Supplementary Data 2 and 8. d-f, Ranked plot of the estimated sperm and blood AF with 95% confidence intervals (estimated fraction ± binomial CI; based on the fraction of mutant reads, see Supplementary Data 8) for all variants detected in the three categories. SDO (d) and BDO (f) variants both show curves that are reminiscent of exponential decay, consistent with an increase of the number of mutations with expansion of the progenitor pool at a constant mutational rate. However, BSS (e) mosaicism for the first 40 variants appears to be more linear, suggesting that mutation rates for early division might be higher than those for later. This is consistent with previous models that estimated an elevated mutation rate in early embryonic development14.
Extended Data Fig. 10. Mosaic variants do not exhibit clustering but differ in their mutational signatures depending on their origin.
a, Plot of the chromosomal location for each of the mosaic variants and their allelic fraction found in sperm from F01–08. Circles, triangles, and squares denote variants found to be mosaic by the dSNV approach, by the unbiased approach, or by both, respectively. b, Permutation simulations (n=10,000 simulations of n=23 mosaic dSNVs, n=62 SDO mosaics, n=123 SDO+BSS mosaics, n=568 BDO mosaics, and n=629 BDO+BSS mosaics) of variant locations to obtain mean and SD of broken stick fragment lengths. Vertical lines mark the observed value from mosaic dSNVs and mosaic variants from the indicated classes. These simulations illustrate that the observed distributions of variants along the chromosomes (as visualized in A for those that were mosaic in sperm) were within expectation. c, Detailed view of the 96 mutational categories for SDO, shared, and BDO mosaic variants,, compared to the overall gnomAD signature and a permuted subset (n=1,000 permutations for n=68 (SDO), 72 (BSS), and 568 (BDO) gnomAD SNVs; shown is the 95% band). Dots indicate the observed mutational signature (black: within 95% band; red: outside the 95% band).
Supplementary Material
Acknowledgements
We thank the participants in this study for their contribution. M.W.B. was supported by an EMBO Long-Term Fellowship (ALTF 174-2015), which is co-funded by the Marie Curie Actions of the European Commission (LTFCOFUND2013, GA-2013-609409), and an Erwin Schrödinger Fellowship by the Austrian Science Fund (FWF): J 4197-B30. This study was supported by grants to J.G.G. from the NIH (U01MH108898, R01NS083823), Simons Foundation Autism Research Initiative (SFARI) to J.G.G., J.S., and Michael Wigler, NIH (MH076431, MH113715) to J.S., and the Howard Hughes Medical Institute to J.G.G. Sequencing support was provided by the Rady Children’s Institute for Genomic Medicine and Oxford Nanopore. O.D. acknowledges support from the Silverman Family Foundation and Finding A Cure for Epilepsy and Seizures. We thank Bruce Hamilton, Neil Chi, Valentina Stanley, Ashley Marsh, Michael Wigler, and Ludmil Alexandrov for suggestions. We thank R. Sinkovits, A. Majumdar, S. Strande, and the San Diego Supercomputer Center for hosting the computing infrastructure necessary for completing this project.
Footnotes
Competing Interests Statement
M.W.B., D.A., M.K., K.N.J., W.M.B., J.S., and J.G.G. are inventors on a provisional patent (PCT ref. no. SD2017–181-2PCT) filed by UC, San Diego that is titled “Assessing risk of de novo mutations in males”.
References
- 1.Iossifov I et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221, doi: 10.1038/nature13908 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner TN et al. Genomic Patterns of De Novo Mutation in Simplex Autism. Cell 171, 710–722 e712, doi: 10.1016/j.cell.2017.08.047 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Roak BJ et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250, doi: 10.1038/nature10989 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neale BM et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245, doi: 10.1038/nature11011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kong A et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488, 471–475, doi: 10.1038/nature11396 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jonsson H et al. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature 549, 519–522, doi: 10.1038/nature24018 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Campbell IM et al. Parent of origin, mosaicism, and recurrence risk: probabilistic modeling explains the broken symmetry of transmission genetics. Am. J. Hum. Genet 95, 345–359, doi: 10.1016/j.ajhg.2014.08.010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acuna-Hidalgo R, Veltman JA & Hoischen A New insights into the generation and role of de novo mutations in health and disease. Genome Biol 17, 241, doi: 10.1186/s13059-016-1110-1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freed D, Stevens EL & Pevsner J Somatic mosaicism in the human genome. Genes (Basel) 5, 1064–1094, doi: 10.3390/genes5041064 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonsson H et al. Multiple transmissions of de novo mutations in families. Nat. Genet 50, 1674–1680, doi: 10.1038/s41588-018-0259-9 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Rahbari R et al. Timing, rates and spectra of human germline mutation. Nat. Genet 48, 126–133, doi: 10.1038/ng.3469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandler WM et al. Paternally inherited cis-regulatory structural variants are associated with autism. Science 360, 327–331, doi: 10.1126/science.aan2261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brandler WM et al. Frequency and Complexity of De Novo Structural Mutation in Autism. Am. J. Hum. Genet 98, 667–679, doi: 10.1016/j.ajhg.2016.02.018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang AY et al. Distinctive types of postzygotic single-nucleotide mosaicisms in healthy individuals revealed by genome-wide profiling of multiple organs. PLoS Genet. 14, e1007395, doi: 10.1371/journal.pgen.1007395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carvill GL et al. GRIN2A mutations cause epilepsy-aphasia spectrum disorders. Nat. Genet 45, 1073–1076, doi: 10.1038/ng.2727 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemke JR et al. Mutations in GRIN2A cause idiopathic focal epilepsy with rolandic spikes. Nat. Genet 45, 1067–1072, doi: 10.1038/ng.2728 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Turner DJ et al. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat. Genet 40, 90–95, doi: 10.1038/ng.2007.40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hehir-Kwa JY et al. De novo copy number variants associated with intellectual disability have a paternal origin and age bias. J. Med. Genet 48, 776–778, doi: 10.1136/jmedgenet-2011-100147 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Escaramis G, Docampo E & Rabionet R A decade of structural variants: description, history and methods to detect structural variation. Brief Funct Genomics 14, 305–314, doi: 10.1093/bfgp/elv014 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Cibulskis K et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31, 213–219, doi: 10.1038/nbt.2514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594, doi: 10.1038/s41592-018-0051-x (2018). [DOI] [PubMed] [Google Scholar]
- 22.Huang AY et al. MosaicHunter: accurate detection of postzygotic single-nucleotide mosaicism through next-generation sequencing of unpaired, trio, and paired samples. Nucleic Acids Res. 45, e76, doi: 10.1093/nar/gkx024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaiswal S et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med 371, 2488–2498, doi: 10.1056/NEJMoa1408617 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Z et al. Overlooked roles of DNA damage and maternal age in generating human germline mutations. Proc. Natl. Acad. Sci 116, 9491–9500, doi: 10.1073/pnas.1901259116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernkopf M et al. Quantification of transmission risk in a male patient with a FLNB mosaic mutation causing Larsen syndrome: Implications for genetic counseling in postzygotic mosaicism cases. Hum Mutat 38, 1360–1364, doi: 10.1002/humu.23281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hancarova M et al. Parental gonadal but not somatic mosaicism leading to de novo NFIX variants shared by two brothers with Malan syndrome. Am J Med Genet A, doi: 10.1002/ajmg.a.61302 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Wilbe M et al. A novel approach using long-read sequencing and ddPCR to investigate gonadal mosaicism and estimate recurrence risk in two families with developmental disorders. Prenat Diagn 37, 1146–1154, doi: 10.1002/pd.5156 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X et al. Genomic mosaicism in paternal sperm and multiple parental tissues in a Dravet syndrome cohort. Sci Rep 7, 15677, doi: 10.1038/s41598-017-15814-7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goriely A & Wilkie AO Paternal age effect mutations and selfish spermatogonial selection: causes and consequences for human disease. Am. J. Hum. Genet 90, 175–200, doi: 10.1016/j.ajhg.2011.12.017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamdan FF et al. Identification of a novel in-frame de novo mutation in SPTAN1 in intellectual disability and pontocerebellar atrophy. Eur. J. Hum. Genet 20, 796–800, doi: 10.1038/ejhg.2011.271 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarz JM, Rodelsperger C, Schuelke M & Seelow D MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 7, 575–576, doi: 10.1038/nmeth0810-575 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Pejaver V et al. MutPred2: inferring the molecular and phenotypic impact of amino acid variants. bioRxiv 10.1101/134981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C & Kehrer-Sawatzki H Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum Genet 132, 1077–1130, doi: 10.1007/s00439-013-1331-2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snyder MW, Adey A, Kitzman JO & Shendure J Haplotype-resolved genome sequencing: experimental methods and applications. Nat Rev Genet 16, 344–358, doi: 10.1038/nrg3903 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Browning SR & Browning BL Haplotype phasing: existing methods and new developments. Nat Rev Genet 12, 703–714, doi: 10.1038/nrg3054 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia Y, Liu Y, Deng M & Xi R Pysim-sv: a package for simulating structural variation data with GC-biases. BMC Bioinformatics 18, 53, doi: 10.1186/s12859-017-1464-8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291, doi: 10.1038/nature19057 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michaelson JJ et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151, 1431–1442, doi: 10.1016/j.cell.2012.11.019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krupp DR et al. Exonic Mosaic Mutations Contribute Risk for Autism Spectrum Disorder. Am. J. Hum. Genet 101, 369–390, doi: 10.1016/j.ajhg.2017.07.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu H, de Gannes MK, Luchetti G & Pilsner JR Rapid method for the isolation of mammalian sperm DNA. Biotechniques 58, 293–300, doi: 10.2144/000114280 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Regan JF et al. A rapid molecular approach for chromosomal phasing. PloS one 10, e0118270, doi: 10.1371/journal.pone.0118270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Untergasser A et al. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, W71–74, doi: 10.1093/nar/gkm306 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Untergasser A et al. Primer3--new capabilities and interfaces. Nucleic Acids Res. 40, e115, doi: 10.1093/nar/gks596 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koressaar T & Remm M Enhancements and modifications of primer design program Primer3. Bioinformatics 23, 1289–1291, doi: 10.1093/bioinformatics/btm091 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Xu X et al. Amplicon Resequencing Identified Parental Mosaicism for Approximately 10% of “de novo” SCN1A Mutations in Children with Dravet Syndrome. Hum Mutat 36, 861–872, doi: 10.1002/humu.22819 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karczewski KJ et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 10.1101/531210 (2019). [DOI] [Google Scholar]
- 47.Goss PJ & Lewontin RC Detecting heterogeneity of substitution along DNA and protein sequences. Genetics 143, 589–602 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexandrov LB et al. Signatures of mutational processes in human cancer. Nature 500, 415–421, doi: 10.1038/nature12477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins RL, Stone MR, Brand H, Glessner JT & Talkowski ME CNView: a visualization and annotation tool for copy number variation from whole-genome sequencing. bioRxiv 10.1101/049536 (2016). [DOI] [Google Scholar]
- 50.Willems T et al. Genome-wide profiling of heritable and de novo STR variations. Nat. Methods 14, 590–592, doi: 10.1038/nmeth.4267 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailey JA et al. Recent segmental duplications in the human genome. Science 297, 1003–1007, doi: 10.1126/science.1072047 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Karolchik D et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–496, doi: 10.1093/nar/gkh103 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gervais AL, Marques M & Gaudreau L PCRTiler: automated design of tiled and specific PCR primer pairs. Nucleic Acids Res. 38, W308–312, doi: 10.1093/nar/gkq485 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Aligned BAM files generated for this study through deep WGS or TAS are available on SRA (accession number: PRJNA588332). WGS data used for de novo calling are available through the NIMH Data Archive (NDA; collection ID: 2019). Long read sequencing data are likewise available on NDA (collection ID: 2795). NDA access is regulated by the standard organizational process and is subject to review by NDA. Data are also available through the corresponding authors on reasonable request. Additionally, summary tables of the data are included as supplementary information.