Abstract
Background
Recent emergence of Zika virus (ZIKV), and the global spread of dengue (DENV), chikungunya (CHIKV) and West Nile viruses (WNV) raised urgent need of accurate and affordable molecular diagnosis of these clinically indistinguishable arboviral infections.
Objectives
We established a pentaplex real-time reverse transcription PCR (rRT-PCR) assay (CII-ArboViroPlex rRT-PCR) for specific and sensitive detection of the African and American genotypes of ZIKV, all four serotypes of DENV, CHIKV, WNV and a housekeeping gene as internal control in single reaction.
Study design
Specific primers and probe sets were designed for ZIKV, DENV, CHIKV, WNV and RNase P (housekeeping gene) and tested for in-vitro transcribed RNA standards, virus cultures, clinical samples positive for ZIKV, DENV, CHIKV and WNV and limit of detection (LOD) were determined for each.
Results
Using ten-fold serially diluted in-vitro transcribed RNA, CII- ArboViroPlex rRT-PCR assay has LOD of 100 RNA copies/reaction (Rn) for ZIKV in serum or urine, 100 RNA copies/Rn for DENV in serum, and 10 RNA copies/Rn for CHIKV and WNV in serum. LODs from sera spiked with quantitated viral stocks were 2.6 × 102 GEQ/Rn for ZIKV, 2.2 × 101 GEQ/Rn for DENV-1, 9.4 × 100 GEQ/Rn for DENV-2, 2.3 × 102 GEQ/Rn for DENV-3, 1.4 × 103 GEQ/Rn for DENV-4, 2.7 × 102 GEQ/Rn for CHIKV, and 1.05 × 101 GEQ/Rn for WNV.
Conclusions
The CII-ArboViroPlex rRT-PCR assay is a quantitative one-step pentaplex rRT-PCR assay for the molecular detection and differential diagnosis of ZIKV, DENV, CHIKV, WNV and a human housekeeping gene control in a single- PCR reaction.
Keywords: qPCR, Zika virus, Dengue virus, Chikungunya virus, West Nile virus, Arboviruses, CII-ArboViroPlex rRT PCR
1. Background
ZIKV infection is typically asymptomatic or associated with a mild febrile illness but may result in fetal abnormalities during pregnancy or Guillain-Barré syndrome in adults (1, 2). Infection with DENV results in a broad spectrum of symptoms that range from mild fever to dengue hemorrhagic fever and dengue shock syndrome (3). Although the majority of WNV infections are asymptomatic, approximately 1% of cases result in neuroinvasive disease that manifests as aseptic meningitis, encephalitis or flaccid paralysis (4) (5). CHIKV frequently causes a febrile illness associated with arthralgia and skin rash. Like ZIKV, CHIKV has also been associated with Guillain-Barre syndrome (6, 7).
Through January 2019, more than two million cases of Zika virus (ZIKV) infection have been reported globally, including the Americas, Asia, and Oceania (8). Recent outbreaks in Central and South America, particularly in Brazil, have been associated with neurodevelopmental disorders, including microcephaly in infants born to mothers with a history of gestational infection, as well as Guillain Barre syndrome in infected adults (1, 2). Regions where ZIKV infections are reported are frequently endemic for dengue virus (DENV), chikungunya virus (CHIKV), or West Nile virus (WNV). ZIKV, DENV, and CHIKV are carried by same mosquito vector, Aedes aegypti and Aedes albopictus (9). Early clinical manifestations of infection with these viruses are similar and typically comprise a mild febrile illness. Thus, differential diagnosis requires molecular or serological assays. During the acute phase of infection when patients are viremic, real time reverse transcription PCR (rRT-PCR) assays are the mainstay of molecular diagnostics. Many singleplex to triplex PCR assays have been developed in the last few years for detection of ZIKV, DENV, CHIKV and WNV (10–13) but to our knowledge none of the current molecular assays can simultaneously detect both African and American ZIKV strains, all four DENV serotypes, CHIKV, WNV and a human housekeeping control gene for assay validity.
2. Objectives
Our main objective focused to establish an affordable assay to diiferentially diagnose both African and American ZIKV strains, all four DENV serotypes, CHIKV, WNV and also detect a human housekeeping control simultaneously in a single PCR reaction.
3. Study design
Primer and probe selection
We aligned 40 complete genomic sequences of ZIKV (available at the time of study) from Africa, Asia and Americas, 37 representative complete genomic sequences of DENV (representing all four serotypes), 20 complete genomic sequences of CHIKV from different geographical locations, and 32 complete genomic sequences of WNV from GenBank using Geneious 10.1.2 software (https://www.geneious.com, Biomatters, Auckland, New Zealand) (Supplementary table 1). Primers and probes for specific detection of viral RNAs were designed and synthesized (Biosearch Technologies, Novato, CA) in the 3’ UTR regions of ZIKV and DENV, NSP2 region of CHIKV and NS5 region of WNV genomes (Table 1). Primers and a probe for the detection of a human housekeeping gene, RNase P, were also selected.
Table 1.
Primer and probe sets and linked reporter fluorophores and quenchers used for detection of ZIKV, DENV, CHIKV, WNV and RNase P in the CII-ArboViroPlex rRT-PCR assay.
| Prime and Probe name | Gene traget | “Sequence (5’ to 3’) | Length (bp) | Probe modifications | |
|---|---|---|---|---|---|
| 5’-modification (dye) | 3’-modification (quencher) | ||||
| ZIKA.3P.QF | 3’ UTR | ARTGTTGTCAGGCCTGCTAG | 20 | ||
| ZIKA.3P.QR | 3’ UTR | CTTGRTTTCCCAGCKTCTCCT | 21 | ||
| ZIKA.3P.Probe.R610 | 3’ UTR | GGGGAAAGCTGTGCAGCCTGT | 22 | CAL Fluor Red 610 | BHQ-2 |
| DENV.3P.QF | 3’ UTR | ACTAGAGGTTAGAGGAGACCCCC | 23 | ||
| DENV.3P.QRA | 3’ UTR | GGCGCTCTGTGCCTGGATT | 19 | ||
| DENV.3P.QRB | 3’ UTR | TGGCGTTCTGTGCCTGGAAT | 20 | ||
| DENV.3P.Probe.O560 | 3’ UTR | CCCAGCGTCAATATGCTGT | 19 | CAL Fluor Orange 560 | BHQ-1 plus |
| CHIK.NSP2.NQF | NSP2 | CATCTGCACYCAAGTGTACCA | 21 | ||
| CHIK.NSP2.NQR | NSP2 | GCGCATTTTGCCTTCGTAATG | 21 | ||
| CHIK.NSP2.Probe.FAM | NSP2 | AAAAGTATCTCCAGGCGG | 18 | FAM | BHQ-1 plus |
| WNV.NS5.NQF | NS5 | CATTTGCTCCGCTGTCCCTGTGAA | 24 | ||
| WNV.NS5.NQR | NS5 | CCACTCTCCTCCTGCATGGATGGAC | 25 | ||
| WNV.NS5.Probe.Q670 | NS5 | TGGGTCCCTACCGGAAGAACCACGT | 25 | Quasar 670 | BHQ-2 |
| RNASE-P-QF | RPP30 | AGATTT GGACCTGCGAGCG | 19 | ||
| RNASE-P-QR | RPP30 | GAGCGGCTGTCTCCACAAGT | 20 | ||
| RNASE-P-Probe.Q705 | RPP30 | TTCTGACCTGAAGGCTCTGCGCG | 23 | Quasar 705 | BHQ-3 |
Primer and probe sequences from the targeted viruses were mapped amongst each other to test homology and avoid non-specific detection (Supplementary table 1). All primer and probe sequences showed 100% sequence identity with their expected targets. Furthermore, specificity for the cognate templates was also assessed in silico comparison to other agents that may cause febrile illness but were not available for laboratory testing (Supplementary table 2). All primers and probes were evaluated using the BLASTn algorithm (NCBI). We did not identify >80% nucleotide homology with any of our primer or probe sequences. Empirical testing of cross-reactivity of the primers and probes was performed by testing additional viruses and bacteria (Supplementary table 3) which may cause febrile illness. No amplification was observed with any of the templates, providing no evidence of potential false positive results due to closely related organisms.
Plasmid synthesis and generation of in vitro transcribed RNA standard curves
The 249 bp long 3’ UTR region of ZIKV (ArD165531), the 284 bp long 3’ UTR region of DENV-3 (449335_Boyaca_CO_2015), the 245 bp long NSP2 region of CHIKV (S27-African prototype), and the 174 bp long NS5 region of WNV (HNY1999) were commercially synthesized and cloned into pUC57 plasmid DNA Vector (GenScript, USA). Plasmids were digested using BamHI and XbaI restriction enzymes to release target segments (New England Biolabs) which included a T7 promoter site on 5’ end after digestion (Supplementary figure 1). Gel purified target inserts were quantified by NanoDrop (ThermoFisher) and used as templates for in vitro transcription with RiboMAX Large Scale RNA Production Systems (Promega, USA). In vitro transcribed RNA was DNase-treated, purified with Trizol (ThermoFisher), and quantitated by NanoDrop. Based on optical density and insert size, RNA copy numbers (RNA copies) were calculated and serially diluted (109 - 100 RNA copies / μL) in distilled water with a background of yeast tRNA (50 ng/uL).
Assay performance controls
In-vitro transcribed synthetic RNA standards for ZIKV, DENV, CHIKV and WNV (serial dilutions of 105 - 100 RNA copies/Rn) were used as virus positive controls. Sterile, nuclease-free water was used as a no template control (NTC). Extracted total nucleic acid from a human cell culture preparation known to contain RNase P was used as RNase P control/rRT-PCR control. Each of the rRT-PCR plate (experiment) contained positive control, negative control and rRT-PCR control.
Clinical specimens
Viral RNA positive clinical specimens (serum or urine) were obtained from patients who were ZIKV, DENV, CHIKV or WNV RNA positive using eastablished rRT-PCRs (14–17). These samples included sera from 41 patients with ZIKV, 24 patients with DENV (4 DENV-1, 4 DENV-2, 6 DENV-3, 3 DENV-4 and 7 DENV of unknown serotype), 20 patients with CHIKV, and 19 patients with WNV infection. We also obtained urine from 24 patients with ZIKV infection. Sixty one sera and 50 urine samples from patients with a febrile illness not attributed to infection with ZIKV, DENV, CHIKV or WNV were used as negative clinical controls.
Nucleic acid extraction
Total nucleic acid (TNA) was extracted from 250 μL of tissue culture supernatant, serum or urine by using the NucliSENS easyMAG automated extraction platform (bioMérieux). TNA was eluted in 50 μL of elution buffer and stored at −80°C until further use.
Multiplex one-step real-time RT-PCR assay
The CII-ArboViroPlex rRT-PCR assay includes primers and dual-labeled BHQ probes (Biosearch Technologies, Novato, CA) to detect RNA from ZIKV, DENV, CHIKV, WNV and RNase P. Dual-labeled BHQ probes are linear, incorporating a fluorophore and quencher covalently attached to the 5′ and 3′ ends of the oligonucleotide, respectively. One-step rRT-PCR reactions were performed in 25 μL volumes containing all primers and probes as described in the Supplementary table 4. Ten microliters of purified in-vitro transcribed RNA or extracted TNA from specimens were reverse transcribed and amplified using RNA UltraSense One-Step Quantitative RT-PCR System (ThermoFisher) with thermal cycling and detection on the CFX96 Real-Time PCR Detection System (Bio-Rad). The thermal cycling conditions were used as follows; a) reverse transcription step at 50°C for 30 min, b) reverse transcription inactivation and Taq polymerase enzyme activation at 95°C for 2 min, c) followed by 45 cycles of qPCR at 95°C for 15 s (denaturation) and, d) 60°C for 1 min (annealing and extension). Fluorescence signal intensity was detected after completion of each qPCR cycle and data was collected using CFX Manager Software.
Virus strains, cell culture, virus amplification, and concentration
ZIKV (strain PRVABC59), DENV type 1 (strain Hawaii), DENV type 2 (strain New Guinea C), DENV type 3 (strain H87), DENV type 4 (Strain H241), CHIKV (strain R80422) and WNV (strain HNY1999) were cultured for this study (Supplementary methods, Supplementary table 5). Genomic equivalent quantities (GEQ) were estimated as viral copy number/Rn of each viral supernatant and quantified using serial dilutions of quantitated in vitro T7-transcribed RNA standards diluted in yeast t-RNA (50 ng/μL) as background.
4. Results
Assay establishment
The assay comprises positive, negative, and rRT-PCR controls. The assay runs were considered valid only when amplification products were obtained for the relevant positive controls and not for irrelevant viral template targets. The sample was considered positive If the rRT-PCR amplification curve, based on increasing fluorescence for a primer/probe set, crossed the threshold within (≤) 38.00 cycles. If the amplification curve for a primer probe set crossed the threshold above (>) 38 cycles, the sample result was considered negative. The Ct value ‘38’ cutoff was selected because lowest dilution of in-vitro transcribed RNA standards for ZIKV, DENV, CHIKV and WNV were consistently positive in duplicate runs with similar Ct values and proper sigmoid amplification curve.
Estimation of the limit of detection (LOD)
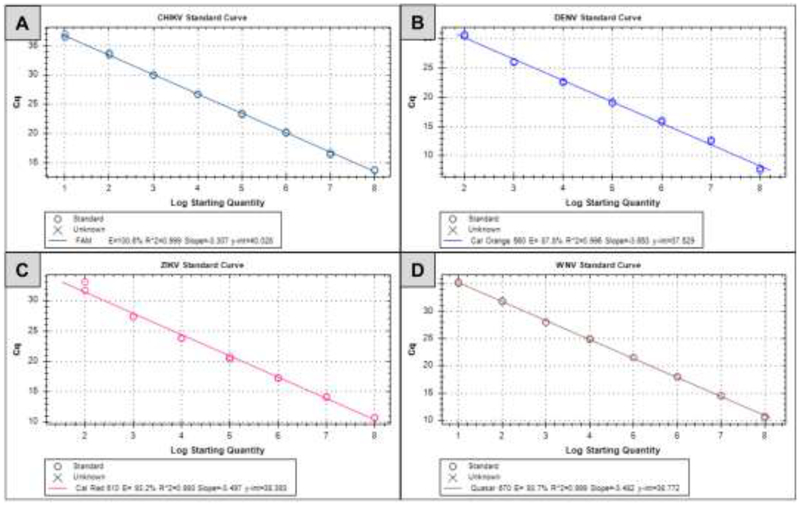
Analytical sensitivities were determined by testing serial dilutions (ten-folds) of RNA standards for all four viruses. Ten microliters of serially diluted (107 - 100 copies) RNA standards for each virus were tested in duplicate with the rRT-PCR reaction mix (Supplementary table 4). The highest RNA dilution positive in duplicates was considered as limit of detection (LOD) for that virus. The LOD for CHIKV and WNV RNA standards was 10 copies/Rn. The LOD for DENV and ZIKV RNA standards was 100 copies/Rn. The coefficient of determination (R2 values) for each assay was greater than 0.99 in all cases (0.999 for CHIKV, 0.996 for DENV, 0.993 for ZIKV, and 0.999 for WNV; Figure 1).
Figure 1.
Standard curves for CHIKV (A), DENV (B), ZIKV (C) and WNV (D) were generated from Ct values obtained for serial 10-fold dilutions plotted against the amounts of standard RNA copy number. All the assays were performed in duplicate.
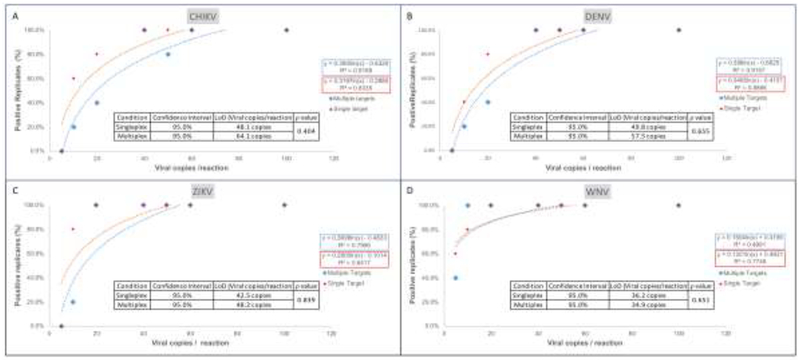
To estimate assay performance in the window between 10 and 100 copies/Rn and in the presence of competing templates, assays with one template (singleplex) and all four viral templates present (multiplex) were performed at 5, 10, 20, 40, 60, 100, 250, 500, 1,000, 5,000 and 10,000 copies/Rn with 10 replicates each. Probit regression analysis (18, 19) indicated comparable performance for all four viruses (ZIKV, DENV, CHIKV and WNV) in singleplex and multiplex as indicated by closely matched LoD values at 95% confidence interval (CI) detection (Figure 2). Since cases of infection with two or more viruses have been reported (20–23), these results suggest sensitive detection of simultaneous infection even with multiple viruses.
Figure 2.
Probit regression analysis of singleplex versus multiplex CII-ArboViroPlex rRT-PCR. Ten replicates of 10,000, 5,000, 1,000, 500, 250, 100, 60, 50, 40, 20, 10 and 5 of in vitro transcribed viral RNA copies were run in CII-ArboViroPlex assays in singleplex format (one template per assay; red symbols) or in multiplex format (all four viral templates per assay; blue symbols). Probit analysis was performed by MATLAB v12 and trendline curves were calculated in Microsoft Excel v16.16.12 for CHIKV (A), DENV (B), ZIKV (C) and WNV (D). LoDs at 95% CI and p-values (MATLAB) were calculated comparing singleplex and multiplex results. Curves are shown for the range of 5 to 100 copies/Rn; for 250 to 10,000 copies/Rn all replicates (100%) were positive with comparable Ct values in all cases.
Serially diluted (105 - 102 copies/Rn) synthetic RNAs were tested in triplicate and compared with extracted TNA from virus culture supernatants using similar viral concentrations (GEQ/Rn). Synthetic in vitro transcribed RNA and TNA extracts from cultured virus generated comparable Ct values for the matching dilutions (Supplementary figure 2).
LODs of viral RNA in clinical samples were estimated by serial dilutions of virus stocks (Supplementary table 5) spiked in single donor human serum (SDHS). For TNA extraction, 225 μL of SDHS aliquots were spiked individually with 25 μL of each serially diluted virus stock. The spiked serum mixes (250 μL in total) were extracted in triplicate and eluted in 50-μL of elution buffer. Ten μL of TNA was tested in triplicate and the lowest concentration at which all replicates were positive was determined as the tentative LOD (GEQ/Rn) for that virus. Only results with a positive RNase P signal were included. The LODs were 2.6 × 102 GEQ/Rn for ZIKV, 2.2 × 101 GEQ/Rn for DENV-1, 9.4 × 100 GEQ/Rn for DENV-2, 2.3 × 102 GEQ/Rn for DENV-3, 1.4 × 103 GEQ/Rn for DENV-4, 2.7 × 102 GEQ/Rn for CHIKV, and 1.05 × 101 GEQ/Rn for WNV (Table 2). Based on the tentative LODs, 20 replicates of viral stock spiked into SDHS at the LOD were processed. The LOD from urine samples spiked with quantitated ZIKV viral stock was 5.6 × 102 GEQ/Rn. Based on the tentative LOD 20 replicates of viral stock spiked into single donor urine sample at LOD were processed. All samples were positive (Ct 31.01 −33.51), yielding a >95% confidence for detection (Table 2).
Table 2.
Determination of the presumptive and final limit of detection (LOD) of the ZIKV, DENV 1, DENV 2, DENV 3, DENV 4, CHIKV and WNV for the CII-ArboViroPlex rRT-PCR assay with serum specimens and ZIKV with urine specimens.
| ZIKV | DENV 1 | DENV 2 | DENV 3 | DENV 4 | CHIKV | WNV | |
|---|---|---|---|---|---|---|---|
| LoD | |||||||
| Virus concn (Copies/reaction) | 2.58E+02 | 2.19E+01 | 9.35E+00 | 2.32E+02 | 1.44E+03 | 2.70E+02 | 1.05E+01 |
| Dilution factor from stock | 1.00E−05 | 1.00E−05 | 1.00E−05 | 1.00E−05 | 1.00E−05 | 1.00E−05 | 1.00E−06 |
| Confirmation of the LoD | |||||||
| Average Ct (20 replicates) | 31.81 | 32.87 | 32.43 | 31.01 | 31.2 | 31.52 | 33.51 |
| standard deviation | 0.76 | 0.8 | 0.93 | 0.81 | 0.52 | 0.28 | 0.48 |
| Call rate | 20/20 | 20/20 | 20/20 | 20/20 | 20/20 | 20/20 | 20/20 |
| Call rate % | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
Assay assessment with clinical samples
Negative specimens:
All negative specimens were tested with the CII-ArboViroPlex rRT-PCR assay, the comparator CDC-Trioplex (ThermoFisher) assay, and a published WNV qPCR (16) assay. None of these assays showed signal for ZIKV, DENV, CHIKV or WNV (Table 3). RNase P was positive for all the negative specimens by CII-ArboViroPlex rRT-PCR and CDC-Trioplex assays.
Table 3.
Clinical Performance of CII-ArboViroPlex rRT-PCR assay for ZIKV, DENV, CHIKV and WNV in Serum and for ZIKV in urine
| Category | ZIKV Positive | DENV Positive | CHIKV Positive | WNV Positive |
|---|---|---|---|---|
| Positive Controls | ||||
| ZIKV Positive (Serum) n=41 | ||||
| CII ArboViroPlex | 39/41* | 0/41 | 0/41 | 0/41 |
| Comparator assays# | 41/41 | 0/41 | 0/41 | 0/41 |
| ZIKV Positive (Urine) n=26 | ||||
| CII ArboViroPlex | 25/26* | 0/26 | 0/26 | 0/26 |
| Comparator assays# | 26/26 | 0/26 | 0/26 | 0/26 |
| DENV Positive (n=24 serum) | ||||
| CII ArboViroPlex | 0/24 | 24/24 | 0/24 | 0/24 |
| Comparator assays# | 0/24 | 24/24 | 0/24 | 0/24 |
| CHIKV Positive (n=20 serum) | ||||
| CII ArboViroPlex | 0/20 | 0/20 | 20/20 | 0/20 |
| Comparator assays# | 0/20 | 0/20 | 20/20 | 0/20 |
| WNV Positive (n=19 serum) | ||||
| CII ArboViroPlex | 0/19 | 0/19 | 0/19 | 19/19 |
| Comparator assays# | 0/19 | 0/19 | 0/19 | 19/19 |
| Negative Controls | ||||
| Negative Controls(n=61 serum) | ||||
| CII ArboViroPlex | 0/61 | 0/61 | 0/61 | 0/61 |
| Comparator assays# | 0/61 | 0/61 | 0/61 | 0/61 |
| Negative (n=50 urine) | ||||
| CII ArboViroPlex | 0/50 | 0/50 | 0/50 | 0/50 |
| Comparator assays# | 0/50 | 0/50 | 0/50 | 0/50 |
| Positive agreement (Serum) | (39/41) | (24/24) | (20/20) | (19/19) |
| Positive % agreement | 95.10% | 100.00% | 100.00% | 100.00% |
| 95% CI | 89.8–99.2% | 93.5–100.0% | 92.3–100.0% | 92.1–100.0% |
| Negative agreement (Serum) | (124/124)❖ | (141/141)❖ | (145/145)❖ | (146/146)❖ |
| Negative % agreement | 100.00% | 100.00% | 100.00% | 100.00% |
| 95% CI | 97.1–100.0% | 96.1–99.9% | 97.5–100.0% | 97.5–100.0% |
| Positive agreement (Urine) | (25/26) | NA | NA | NA |
| Positive percent agreement | 96.20% | |||
| 95% CI | 89.9–99.7% | |||
| Neative agreement (Urine) | 100% | NA | NA | NA |
| Negative % agreement | (50/50) | |||
| 95% CI | 92.9–100.0% | |||
Includes clinical positive specimens for other viral targets as additional negatives, e.g. for Zika virus the CHIKV, DENV and WNV positive specimens were included as negatives in the Zika virus performance.
Positive Specimens:
Thirty-nine of 41 sera and 25/26 ZIKV positive urine samples were positive for ZIKV in the CII-ArboViroPlex rRT-PCR assay. All 67 were negative for DENV, CHIKV and WNV. All 24 DENV positive sera were positive for DENV in the CII-ArboViroPlex rRT-PCR assay. All 24 were negative for ZIKV, CHIKV and WNV. All 20 sera positive for CHIKV were found positive for CHIKV in the CII-ArboVirolPlex rRT-PCR assay. All 20 were negative for ZIKV, DENV and WNV. All 19 sera that were WNV positive were positive for WNV in the CII-ArboViroPlex rRT-PCR assay. All 19 were negative for ZIKV, DENV and CHIKV (Table 3).
Potential cross binding of individual primers or probes that share nucleotide identity to other agent’s sequences may occur in some instances and affect the sensitivity of multiplex PCR assays (20). To address this concern, we performed an interference experiment for ZIKV and DENV targets. DENV-3P.F primer, DENV.3P.PROBE had ~80% identity to ZIKV sequence and ZIKA.3P.PROBE had ~70% identity to DENV sequence (Supplementary table 1). Two different concentrations of ZIKV and DENV (high concn=1000×LoD and low concn=3×LoD) were spiked in SDHS and tested in triplicate after extractions with CII-ArboViroPlex rRT-PCR assay as single agent and also as co-infection in same sample well. No noticeable difference in Ct values were recorded despite the high sequence identity with individual primers/probes and in the presence of the low and high backgrounds of non-cognate viral templates (Table 4). We did not detect any signal for CHIKV or WNV. Based on our experiment we do not anticipate sensitivity of the assay will be compromised in presence of more than one agent in the same sample.
Table 4.
Interference experiment between ZIKV and DENV primer/probe sets of the CII-ArboViroPlex rRT-PCR assay (in vitro testing)
| Ct values in ZIKV (Cal fluor red 610) channel | Ct values in DENV (Cal fluor orange 560) channel | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| ZIKV 1 Ct | ZIKV 2 Ct | ZIKV 3 Ct | Average ZIKV Ct | DENV 1 Ct | DENV 2 Ct | DENV 3 Ct | Average DENV Ct | ||
| High ZIKV concn (1000 × LoD) | 18.67 | 18.75 | 18.15 | 18.52 | ND | ND | ND | ND | |
| Low ZIKV concn (3 × LoD) | 30.32 | 30.55 | 30.20 | 30.36 | ND | ND | ND | ND | |
| High DENV concn (1000 × LoD) | ND | ND | ND | ND | 18.17 | 18.56 | 17.65 | 18.13 | |
| Low DENV concn (3 × LoD) | ND | ND | ND | ND | 29.50 | 30.10 | 30.50 | 30.03 | |
| High ZIKV (1000 × LoD) + Low DENV (3 × LoD) | 19.13 | 18.87 | 18.85 | 18.95 | 29.80 | 30.10 | 29.95 | 29.95 | |
| Low ZIKV (3 × LoD) + High DENV (1000 × LoD) | 30.15 | 30.66 | 31.44 | 30.75 | 18.20 | 18.65 | 18.50 | 18.45 | |
The concordance for results obtained using the CII-ArboViroPlex rRT-PCR assay and other established assays including the LightMix Zika rRT-PCR test (Roche), Trioplex Assay (Thermofisher), NYS-approved LDT assays (14), CDC-Dengue virus real-time RT-PCR assay (15), and WNV-NS3 RT-PCR Assay (16) is shown in supplementary table 6. All of the samples were also tested with the CDC-Trioplex-Assay (Thermofisher) and a WNV-NS3 RT-PCR Assay (16). The positive percentage agreement was 95.1% for ZIKV serum assays, 96.2% for ZIKV urine assays, and 100% for DENV, CHIKV and WNV serum assays.
5. Discussion
Differentially diagnoising ZIKV, DENV, CHIKV, and WNV infection is important for epidemiology and risk assessment. As additional antiviral interventions are developed, we anticipate that differential diagnosis may also become important for clinical management. Molecular methods are rapidly replacing culture in viral diagnostics due to advantages in cost, sensitivity, ease of use and time to result. Virus isolation from a clinical sample is the gold standard for detection, but is a laborious and time-consuming method. Virus culture at reference labs is also not clinically feasible because it requires extensive laboratory setup and these methods are time consuming and exhaustive. There are several singleplex and multiplex RT-PCR assays for detection of arboviruses (24); however, the CII-ArboViroPlex rRT-PCR assay has the advantage of allowing simultaneous detection and quantitation of ZIKV, DENV (all 4 serotypes), CHIKV, and WNV. In addition, it detects a human housekeeping gene (RNase P) as an internal control for ensuring the integrity of RNA template and overall assay performance. Out of 67 ZIKV postive clinical samples, 2 sera and 1 urine sample that were negative at 38 cycles in the CII-ArboViroPlex rRT-PCR assay were found to be positive with comparator assays (1 sample each in LightMix Zika rRT-PCR test, Wadsworth Center New York State-approved ZIKV assay, and the Trioplex assay). These samples were positive at 38.63, 38.40 and 38.05 cycles respectively in the CII-ArboViroPlex rRT-PCR assay. We speculate that freeze-thaw cycles may have caused degredation of template. Based on its performance characteristics the CII-ArboViroPlex rRT-PCR assay received emergency use authorization by the FDA for detection of ZIKV, DENV, WNV and CHIKV in serum and for ZIKV in urine (25).
6. Conclusions
We developed a quantitative one-step pentaplex rRT-PCR assay “CII-ArboViroPlex rRT-PCR” for the molecular detection and differential diagnosis of ZIKV, DENV, CHIKV and WNV and a human housekeeping gene (RNase P) which ensures assay validity in single-PCR reaction.
Supplementary Material
Highlights.
Accurate diagnosis is needed for clinically indistinguishable arboviral infections
Establishment of differential and rapid diagnosis of ZIKV, DENV, CHIKV, and WNV
Simultaneous detection of all ZIKV strains, all four DENV serotypes, CHIKV, WNV
Acknowledgments:
The authors are grateful to the patients, clinicians, Boca Biolistics [FL, USA], and Creative Testing Solutions [AZ, USA] who provided samples without which this work could not have been done. We also thank Adam Nitodo [CII], and Dakai Liu, Taryn Burke and Aaron Olsen [New York City Department of Health and Mental Hygiene] for technical assistance, and Sydney Silverman and Gregory J. Keough [CII] for organizing sample transport and project logistics. Financial support for this study was provided by National Institutes of Health award U19AI109761 [CETR: Center for Research in Diagnostics and Discovery].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: NM, WIL, RT and TB are listed as inventors in patent application No. PCT/US2017/023012 that covers the CII-ArboViroPlex rRT-PCR assay. The remaining authors declare no competing interests.
References
- 1.Krauer F, Riesen M, Reveiz L, Oladapo OT, Martinez-Vega R, Porgo TV, Haefliger A, Broutet NJ, Low N, Group WHOZCW. 2017. Zika Virus Infection as a Cause of Congenital Brain Abnormalities and Guillain-Barre Syndrome: Systematic Review. PLoS Med 14:e1002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. 2016. Zika Virus and Birth Defects--Reviewing the Evidence for Causality. N Engl J Med 374:1981–1987. [DOI] [PubMed] [Google Scholar]
- 3.Slon Campos JL, Mongkolsapaya J, Screaton GR. 2018. The immune response against flaviviruses. Nat Immunol 19:1189–1198. [DOI] [PubMed] [Google Scholar]
- 4.Briese T, Bernard KA. 2005. West Nile virus--an old virus learning new tricks? J Neurovirol 11:469–475. [DOI] [PubMed] [Google Scholar]
- 5.Sejvar JJ. 2016. West Nile Virus Infection. Microbiol Spectr 4. [DOI] [PubMed] [Google Scholar]
- 6.Pinheiro TJ, Guimaraes LF, Silva MT, Soares CN. 2016. Neurological manifestations of Chikungunya and Zika infections. Arq Neuropsiquiatr 74:937–943. [DOI] [PubMed] [Google Scholar]
- 7.Pialoux G, Gauzere BA, Jaureguiberry S, Strobel M. 2007. Chikungunya, an epidemic arbovirosis. Lancet Infect Dis 7:319–327. [DOI] [PubMed] [Google Scholar]
- 8.PAHO-WHO. 2018. Zika suspected and confirmed cases reported by countries and territories in the Americas Cumulative cases, 2015–2018. http://www.paho.org/zikavirus, PAHO Website. [Google Scholar]
- 9.Kauffman EB, Kramer LD. 2017. Zika Virus Mosquito Vectors: Competence, Biology, and Vector Control. J Infect Dis 216:S976–S990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waggoner JJ, Gresh L, Vargas MJ, Ballesteros G, Tellez Y, Soda KJ, Sahoo MK, Nunez A, Balmaseda A, Harris E, Pinsky BA. 2016. Viremia and Clinical Presentation in Nicaraguan Patients Infected With Zika Virus, Chikungunya Virus, and Dengue Virus. Clin Infect Dis 63:1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu W, Wang J, Yu N, Yan J, Zhuo Z, Chen M, Su X, Fang M, He S, Zhang S, Zhang Y, Ge S, Xia N. 2018. Development of multiplex real-time reverse-transcriptase polymerase chain reaction assay for simultaneous detection of Zika, dengue, yellow fever, and chikungunya viruses in a single tube. J Med Virol 90:1681–1686. [DOI] [PubMed] [Google Scholar]
- 12.Nyan DC, Swinson KL. 2015. A novel multiplex isothermal amplification method for rapid detection and identification of viruses. Sci Rep 5:17925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calvo EP, Sanchez-Quete F, Duran S, Sandoval I, Castellanos JE. 2016. Easy and inexpensive molecular detection of dengue, chikungunya and zika viruses in febrile patients. Acta Trop 163:32–37. [DOI] [PubMed] [Google Scholar]
- 14.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. 2008. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 14:1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CDC. 2013. CDC DENV-1–4 Real-Time RT-PCR Assay Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases, Division of Vector-Borne Diseases, Dengue Branch, 1324 Canada Street, San Juan, PR 00920 [Google Scholar]
- 16.Briese T, Glass WG, Lipkin WI. 2000. Detection of West Nile virus sequences in cerebrospinal fluid. Lancet 355:1614–1615. [DOI] [PubMed] [Google Scholar]
- 17.Santiago GA, Vazquez J, Courtney S, Matias KY, Andersen LE, Colon C, Butler AE, Roulo R, Bowzard J, Villanueva JM, Munoz-Jordan JL. 2018. Performance of the Trioplex real-time RT-PCR assay for detection of Zika, dengue, and chikungunya viruses. Nat Commun 9:1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jalal H, Stephen H, Curran MD, Burton J, Bradley M, Carne C. 2006. Development and validation of a rotor-gene real-time PCR assay for detection, identification, and quantification of Chlamydia trachomatis in a single reaction. J Clin Microbiol 44:206–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutfalla G, Uze G. 2006. Performing quantitative reverse-transcribed polymerase chain reaction experiments. Methods Enzymol 410:386–400. [DOI] [PubMed] [Google Scholar]
- 20.Vogels CBF, Ruckert C, Cavany SM, Perkins TA, Ebel GD, Grubaugh ND. 2019. Arbovirus coinfection and co-transmission: A neglected public health concern? PLoS Biol 17:e3000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrillo-Hernandez MY, Ruiz-Saenz J, Villamizar LJ, Gomez-Rangel SY, Martinez-Gutierrez M. 2018. Co-circulation and simultaneous co-infection of dengue, chikungunya, and zika viruses in patients with febrile syndrome at the Colombian-Venezuelan border. BMC Infect Dis 18:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acevedo N, Waggoner J, Rodriguez M, Rivera L, Landivar J, Pinsky B, Zambrano H. 2017. Zika Virus, Chikungunya Virus, and Dengue Virus in Cerebrospinal Fluid from Adults with Neurological Manifestations, Guayaquil, Ecuador. Front Microbiol 8:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villamil-Gomez WE, Rodriguez-Morales AJ, Uribe-Garcia AM, Gonzalez-Arismendy E, Castellanos JE, Calvo EP, Alvarez-Mon M, Musso D. 2016. Zika, dengue, and chikungunya co-infection in a pregnant woman from Colombia. Int J Infect Dis 51:135–138. [DOI] [PubMed] [Google Scholar]
- 24.Pabbaraju K, Wong S, Gill K, Fonseca K, Tipples GA, Tellier R. 2016. Simultaneous detection of Zika, Chikungunya and Dengue viruses by a multiplex real-time RT-PCR assay. J Clin Virol 83:66–71. [DOI] [PubMed] [Google Scholar]
- 25.Center for Infection and Immunity CU. 2017. CII-ArboViroPlex rRT-PCR assay: Authorization of the emergency use of in vitro diagnostics for detection of Zika virus and/or diagnosis of Zika virus infection., U.S. Food and Drug Administration. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.