Abstract
Perfluorooctane sulfonate (PFOS) is a persistent environmental chemical whose biological effects are mediated by multiple mechanisms. Recent evidence suggests that the gut microbiome may be directly impacted by and/or alter the fate and effects of environmental chemicals in the host. Thus, the aim of this study was to determine whether PFOS influences the gut microbiome and its metabolism, and the host metabolome. Four groups of male C57BL/6J mice were fed a diet with or without 0.003%, 0.006%, or 0.012% PFOS, respectively. 16S rRNA gene sequencing, metabolomic, and molecular analyses were used to examine the gut microbiota of mice after dietary PFOS exposure. Dietary PFOS exposure caused a marked change in the gut microbiome compared to controls. Dietary PFOS also caused dose-dependent changes in hepatic metabolic pathways including those involved in lipid metabolism, oxidative stress, inflammation, TCA cycle, glucose, and amino acid metabolism. Changes in the metabolome correlated with changes in genes that regulate these pathways. Integrative analyses also demonstrated a strong correlation between the alterations in microbiota composition and host metabolic profiles induced by PFOS. Further, using isolated mouse cecal contents, PFOS exposure directly affected the gut microbiota metabolism. Results from these studies demonstrate that the molecular and biochemical changes induced by PFOS are mediated in part by the gut microbiome, which alters gene expression and the host metabolome in mice.
Keywords: Perfluorooctane sulfonate, microbiome, metabolism, metabolome
1. Introduction
Recent studies show that the gut microbiome, which is composed of different types of bacteria, viruses and fungi, can significantly impact the effect of chemical exposure in the host (Snedeker and Hay 2012, Claus, Guillou and Ellero-Simatos 2016, Atlas et al. 2018). For example, there is clear evidence that the gut microbiome can markedly influence the outcome of acetaminophen metabolism in humans (Clayton et al. 2009). There are several levels of regulation by which the gut microbiome may influence the effect of a chemical. Gut bacteria may: 1) modulate the absorption of chemicals; 2) metabolize the chemical to various metabolites; and, 3) be directly affected by the chemical (e.g., antibiotics kill many types of gut bacteria). Given potential mechanisms by which gut bacteria can influence the effects and disposition of chemicals, there remains a need to study the impact of the gut microbiome on the outcome of chemical exposures.
Perfluorooctane sulfonate (PFOS) is a persistent environmental chemical with distinct chemical and physical properties that has been used directly as a surfactant in various industrial and consumer applications or may be generated by degradation of certain fluorochemical materials (Lau et al. 2007). The biological effects induced by PFOS can be mediated in part by activation of the peroxisome proliferator activated receptor α (PPARα), a ligand-activated transcription factor, but there is also evidence that PFOS may activate other nuclear receptors such as the constitutive androstane receptor (CAR) or pregnane X receptor (PXR) (Bijland et al. 2011, Elcombe et al. 2012, Lau et al. 2007). However, the tumorigenic effect mediated by activation of PPARα, CAR, or PXR by PFOS in liver is unlikely to be relevant to humans (Corton, Peters and Klaunig 2018, Hirose et al. 2009, Lake 2009, Ross et al. 2010) because significant differences exist between the response of human liver versus rodent liver by activation of these receptors as the responses in human liver is greatly attenuated or absent as compared to rodent liver (Bjork, Butenhoff and Wallace 2011, Bjork and Wallace 2009, Wolf et al. 2008). Most studies examining the effects of PFOS in animal and human models focus on exploring the host biochemical and molecular events that result from PFOS exposure. Whether the gut microbiota can impact the effect of PFOS on host metabolism is not known. This is important because epidemiological studies suggest that chronic exposure to some environmental chemicals may contribute to the development of metabolic disorders including obesity and diabetes, which may be associated with disruption of gut microbiota community structure and function (Lee et al. 2014, Myre and Imbeault 2014). For these reasons, the present study examined whether exposure to PFOS in mice caused alterations in the gut microbiome and/or the metabolome, and whether these changes were associated with altered host gene expression.
2. MATERIALS AND METHODS
2.1. Materials and Chemicals
Potassium PFOS (87% purity) was provided by the 3M Company. NaCl, K2HPO4, and NaH2PO4 (analytical grade) were purchased from Sigma-Aldrich Chemical Co. Ltd. (St Louis, MO, USA). Sodium 3-trimethylsilyl propionate (TSP-d4) and D2O (99.9% in D) were purchased from Cambridge Isotope Laboratories (Miami, FL, USA). Dyes for flow cytometry, SYBR green and carboxyfluorescein diacetate (CFDA), were purchased from Invitrogen (Carlsbad, CA). Reduced (1x) phosphate-buffered saline (PBS) solution containing 1 g/l l-cysteine was prepared and stored anaerobically for use in flow cytometry.
2.2. Animal and diets
Animal experimental procedures were performed according to the National Institutes of Health (NIH) guidelines. The samples used for these studies were obtained from a previously published study (Zhang et al. 2016). The animal use protocol was reviewed and approved by the Pennsylvania State University Institutional Animal Care and Use Committee. Stocks of potassium PFOS were prepared by Dyets, Inc. (Bethlehem, PA, USA) by adding potassium PFOS to small volumes of sucrose, which was then added to the diet formulation to maintain a constant amount of sucrose but increasing amount of potassium PFOS to obtain a final dietary concentration of 0.003%, 0.006% or 0.012% PFOS in pelleted form. The dietary content of PFOS was considered as low dose exposure and confirmed as previously described (Chang et al. 2017). Twenty male C57BL/6J mice (8–10 weeks old) were housed in an Association for Assessment and Accreditation of Laboratory Animal Care accredited facility, with a temperature (25 °C) and light (12 h light/12 h dark) controlled environment and provided water and AIN-93G diet ad libitum. After acclimatization for two weeks, mice were randomly divided into four groups of five and subjected to treatments for three weeks; controls were fed with control AIN-93G diet and the treatment groups were fed AIN-93G diet containing 0.003%, 0.006% or 0.012% PFOS. Liver and cecal content samples were collected immediately following asphyxiation with carbon dioxide and stored at −80 °C until analysis.
2.3. RNA isolation and quantitative real-time PCR (qPCR)
RNA was extracted from frozen liver tissues (~50 mg) using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized from 1 μg of total RNA using qScript cDNA SuperMix (Quanta Biosciences, Beverly, MA, USA) and the products were diluted to 1:10 before use in subsequent reactions. Gene-specific primers were designed with qPrimerDepot and used in each reaction and all results were normalized to mRNA encoding β-actin (primer sequences are listed in Table S1). qPCR assays were performed by using SYBR green qPCR master mix on an ABI Prism 7900HT Fast Real-Time PCR sequence detection system (Applied Biosystems, Foster City, CA, USA). The reactions were analyzed with the ΔΔCT method.
2.4. 16S rRNA gene sequencing of the gut microbiome
The bacteria in the cecal content were extracted using the E.Z.N.A.® Stool DNA Kit (Omega Bio-Tek) based on the manufacturers’ instructions. PCR amplification was performed on the bacteria genomic DNA samples using V4-V4 primer set. PCR reactions were initially heated to 98°C for 2 min, followed by 25 cycles of 98°C for 10 s, 56.5°C for 20 s, 72°C for 15 s. Reactions were completed at 72°C for 5 min. The amplicon products were checked on an agarose gel to confirm the correct amplicon size (350 bp). The amplicons were quantified by Qubit (Lifetech, Carlsbad, CA, USA) and were submitted for 250×250 paired end sequencing on the Illumina Miseq (Illumina, San Diego, CA, USA) at the Pennsylvania State University Genomics Core Facility. Raw sequence data were analyzed with the mothur software package (Kozich et al. 2013), resulting in a taxonomic distribution of the bacteria. Additionally, a generalized UniFrac analysis (Chen et al. 2012) was completed on the sequencing data, which combines both weighted and unweighted UniFrac distance matrices and uses them to compare the bacterial populations.
2.5. 1H NMR-based metabolomics analysis
Liver tissue (~50 mg) and cecal content were extracted three times and two times, respectively, with 1 mL of a pre-cooled methanol-water mixture (2/1, v/v) using PreCellys Tissue Homogenizer (Bertin Technologies, Rockville, MD, USA). After centrifugation for 10 min (11,180 X g, 4°C for liver tissue and 3200 X g, 4°C for cecal content), the supernatants were dried by vacuum. The aqueous extracts were separately reconstituted into 600 μL phosphate buffer (K2HPO4/NaH2PO4, 0.1 M, pH 7.4, 50% v/v D2O) containing 0.005% TSP-d4 as a chemical shift reference. 550 μL of the extracts were transferred into 5 mm NMR tube for NMR analysis after centrifugation.
1H NMR spectra were acquired at 298 K on a Bruker Avance III 600 MHz spectrometer (for aqueous liver extracts) and Bruker Avance Neo (for cecal content) (operating at 600.08 MHz for 1H and at 150.93 MHz for 13C) equipped with a Bruker inverse cryogenic probe (Bruker Biospin, Germany). A standard one-dimensional NMR spectrum was recorded for each of all samples using the first increment of NOESY pulse sequence (NOESYPR1D). 90° pulse was set to approximately 10 μs with water signal suppression by a weak continuous wave irradiation. A total of 64 scans were collected into 32K data points for each spectrum with a spectral width of 20 ppm and recycle delay of 2 s. For the purposes of NMR signal assignments, a range of 2D NMR spectra were acquired and processed for selected samples of liver extracts including 1H-1H correlation spectroscopy (COSY), 1H-1H total correlation spectroscopy (TOCSY), 1H-13C heteronuclear single quantum correlation (HSQC), and 1H-13C heteronuclear multiple bond correlation (HMBC) spectra.
All free induction decays were multiplied by an exponential function with a 1 Hz line broadening factor prior to Fourier transformation. Each 1H NMR spectrum (δ 0.5–9.5) was manually corrected for phase and baseline distortions by use of Topspin 3.0 (Bruker, BioSpin, Germany). The spectra were calibrated to TSP-d4 at δ 0.00 and then divided into regions with equal width of 0.004 ppm (2.4 Hz) using AMIX software package (V3.8, Bruker Biospin, Germany). Region δ 4.65–5.12 was removed for imperfect water suppression. Each bucketed region was then normalized to the total sum of the spectral integrals.
Multivariate data analysis was carried out with SIMCAP+ software (version 13.0, Umetrics, Sweden). Briefly, orthogonal projection to latent structures with discriminant analysis (OPLS-DA) was performed on the normalized NMR data. The OPLS-DA models were validated using a 7-fold cross validation method and the quality of the model was described by the parameters R2X and Q2 values. After back-transformation of the loadings generated from the OPLS-DA, color-coded correlation coefficient loading plots (MATLAB, The Mathworks Inc.; Natick, MA, USA) were used to determine the significance of the metabolite contributing to the class discrimination with a “hot” color (e.g., red) being more significant than a “cold” color (e.g., blue). In this study, a cutoff value of |r| > 0.755 (r > 0.755 and r < −0.755) was chosen for correlation coefficient as significant based on the discrimination significance (p < 0.05).
2.6. Correlation analysis of gut microbiome and liver metabolome
Pearson correlation analysis was used to examine the relationships between bacterial composition and metabolite levels after low dose PFOS exposure. Statistical significance was found by transforming the Pearson r-value into t-value and then using the t distribution to find the p-value. Correlation values above 0.63 or below −0.63 were considered statistically significant.
2.7. GC-MS analysis of fatty acid composition
Sample preparation and total fatty acid compositional measurements were conducted as described previously (Zhang et al. 2015a). Briefly, liver tissue (~50 mg) was mixed with 1 mL of methanol-chloroform mixture (2/1, v/v) with the addition of 5 μL internal standards (50 μM C15:0 free fatty acid and the methyl ester of C17:0) and then homogenized with a tissue homogenizer. 500 μL 0.9 % saline solution was added into the supernatant after centrifugation (20,187 X g, 4 °C) for 15 min. After vortexing for 5 min and centrifugation (20,187 X g, 4 °C) for 15 min, the organic layer was transferred into 10 mL glass tube and dried down with nitrogen. 1 mL methanol/HCl was added and incubated overnight at 60°C. The resultant mixture was combined with 5 mL hexane and 5 mL 0.9% saline. After vortexing for 5 min, the top layer was collected and dried down with nitrogen. The resultant residues were re-dissolved in 200 μL hexane and then transferred to an autosampler vial for GC-MS analysis. Fatty acid composition was measured on an Agilent 7890A-5975C GC-MS system (Agilent Technologies, Santa Clara, CA, USA). An HP-5MS (Agilent Technologies, Santa Clara, CA, USA) capillary column (30 m, 0.25 mm ID, 0.25 μm film thickness) was used with helium as a carrier gas at flow rate of 1 mL/min. Sample injection volume was 0.5 μL with a pressure pulsed split ratio (1:10 split, 10 p.s.i.). The injection port and detector temperatures were 230 and 250 °C, respectively. The initial column temperature was 80 °C, which was maintained for 1 min, then increased to 205 °C at a rate of 20 °C/min, then increased to 220 °C at a rate of 2 °C/min, and finally increased to 310 °C at a rate of 15 °C/min, where it was maintained for 2 min. Fatty acids were relatively quantified by comparing integrated peak areas following normalization to the internal standards.
2.8. LC-Orbitrap MS analysis of GSH/GSSG
Liver extracts were analyzed by LC-Orbitrap MS using an ion pairing reversed phase negative ion electrospray ionization method. Sample preparation and LC-Orbitrap MS conditions are described in the supplemental materials.
2.9. In vitro gut microbiota culture and flow cytometry
Isolation, incubation of gut microbiota, and flow cytometry preparation procedures were derived from a previously published protocol (Cai et al. 2018) with slight modifications. Briefly, cecal contents from male 8–12 weeks old wild-type C57BL/6J mice were collected following asphyxiation with carbon dioxide. The cecum was tied by silk suture (Fine Science Tools, Foster City, CA), removed from the mouse, and emersed in 70% ethanol for 10 s before transferring into the anaerobic chamber with monitored oxygen levels below 20 ppm. The cecal content was diluted 1:10 (1 g in 10 mL) with pre-reduced brain heart infusion (BHI) broth (Sigma, St. Louis, MO) and then divided into 5 replicates of a 1.7 mL suspension into four groups. Three groups were treated with PFOS at final concentrations of 5 μg/g (low dose), 25 μg/g (medium dose) and 50 μg/g (high dose) of cecal content, vortexed and then incubated at 37 °C for 4 h in the dark. These doses are lower than the doses of dietary exposure accounting for the high absorption of PFOS following ingestion (Cui et al. 2010) which limits the concentration in the gut. After incubation, the samples were centrifuged (700 X g, 4 °C for 1 min). Six hundred microliters of microbial supernatant were transferred to new tubes and stained for flow cytometry analysis. The remaining supernatants were stored at −80 °C for 1H NMR metabolomics profiling.
The bacterial mixtures were centrifuged, washed and diluted 120 times with reduced PBS. Diluted samples were stained with carboxyfluorescein diacetate (CFDA) fluorescent dye to assess metabolic activity. SYBR green dye, which stains all cells irrespective of whether the cells are dead or alive, was used to normalize the CFDA counts. Analysis was performed on a BD Accuri C6 flow cytometer (BD Biosciences, Franklin Lakes, NJ) and data were analyzed using FlowJo software (version 10; Tree Star, Ashland, OR).
2.10. Statistical Data Analysis
All the experimental values are presented as the mean ± standard deviation (s.d.). Statistical analyses were performed with GraphPad Prism version 6.0 (GraphPad). Multiple group comparisons were performed by One-way ANOVA with Tukey’s correction and P-values < 0.05 were considered significant.
3. RESULTS
3.1. Dietary PFOS alters the composition of the gut microbiota
Exposure to environmental chemicals such as dioxins and arsenic through the diet or drinking water can alter the gut microbiome (Zhang et al. 2015b, Lu et al. 2014). In this study, generalized UniFrac analysis of 16S rRNA gene sequencing data revealed clear separation of microbial communities between control and PFOS-treated mice (Fig. 1a). Indeed, administration of 0.003% dietary PFOS caused a marked change in the gut microbiota composition at both phylum and genus levels as compared to control mice as especially characterized by decreased Firmicutes and increased Bacteroidetes, respectively (Fig. 1b; Fig. S1).
Fig. 1.

PFOS exposure alters gut microbiota community. a) Generalized UniFrac analysis of the total population of gut microbiome of the cecal contents from control or mice fed 0.003% PFOS in the diet. P value was obtained via permutational multivariate analysis of variance using the Adonis.2 package in Rstudio. b) PFOS exposure alters the composition of gut microbiota at a phylum level. 16S rRNA gene sequencing results for the Firmicutes and Bacteroidetes phyla. Data were analyzed using one-way ANOVA with Tukey’s correction and are presented as means ± s.d., (n = 5 per group); *p < 0.05; **p < 0.01; ***p < 0.001 compared to control.
3.2. PFOS exposure dose-dependently alters the host metabolome
Untargeted 1H NMR-based metabolomic analyses were used to investigate potential changes in the liver metabolome of mice in response to PFOS administration. 1H NMR spectra of liver extracts were determined and the NMR resonances were assigned to specific metabolites as described previously (Huang et al. 2013, Zhao et al. 2015). Multivariate data analysis using OPLS-DA was used to determine variations in metabolite levels in response to exposure to PFOS. The changes in metabolites levels between control and PFOS-treated mice were identified with the corresponding color-coded coefficient plots (Fig. S2). PFOS-treated mice exhibited higher hepatic levels of lipid moieties, unsaturated fatty acids (UFA), 3-HB, lactate, acetate, glutamine, branched chain amino acids (BCAAs), histidine, phenylalanine, choline, TMAO (Trimethylamine N-oxide) +betaine, oxidized glutathione (GSSG), and lower glucose and glycogen (Fig. 2; Fig. S2).
Fig. 2.

PFOS dose dependently impacts mouse liver metabolomic profile. Relative abundance of the significantly changed metabolites in the liver obtained from vehicle and PFOS-treated mice. n = 5 mice per group. Data were analyzed using one-way ANOVA with Tukey’s correction and are presented as mean ± s. d., (n = 5 per group); *p < 0.05, **p < 0.01, ***p < 0.001 compared with vehicle treatment.
LC-Orbitrap MS measurements of the ratio of reduced glutathione (GSH) to oxidized glutathione (GSSG) in the livers serve as a good indicator of the cellular redox status and relative oxidative stress (Rebrin and Sohal 2004). A dose-dependent decrease in the GSH/GSSG ratio was observed in mouse liver after administration of low dose dietary PFOS as compared to controls (Fig. 3a). These data are consistent with the changes in relative GSSG content in the liver of mice fed the low dose PFOS diet as compared to control as revealed by 1H NMR metabolomic analysis (Fig. S2). Further, LC-orbitrap-MS analyses showed that low dose dietary PFOS exposure caused marked reductions of the NAD+/NADH and the NADP+/NADPH ratios in the livers (Fig. 3b and 3c).
Fig. 3.

PFOS dose dependently induces oxidative stress. Quantification of the ratios of (A) GSH/GSSG, (B) NAD+/NADH, (C) NADP+/NADPH in the liver obtained from vehicle and PFOS-treated mice. n = 5 mice per group. Data were analyzed using one-way ANOVA with Tukey’s correction and are presented as mean ± s. d., (n = 5 per group); *p < 0.05, **p < 0.01, ***p < 0.001 compared with vehicle treatment.
Targeted analyses of total fatty acid composition by GC-MS showed that low dose dietary PFOS exposure caused an increase in the levels of hepatic saturated fatty acids (SFA: C16:0, C18:0), mono-saturated fatty acids (MUFA: C16:1n7, C18:1n9) and polyunsaturated fatty acids (PUFA: C18:2n6) (Fig. 4). Combined, these observations suggest that exposure to low dose dietary PFOS impacts the host metabolome and these changes are reflected by changes in intracellular redox state/oxidative stress and altered lipid homeostasis in the liver.
Fig. 4.
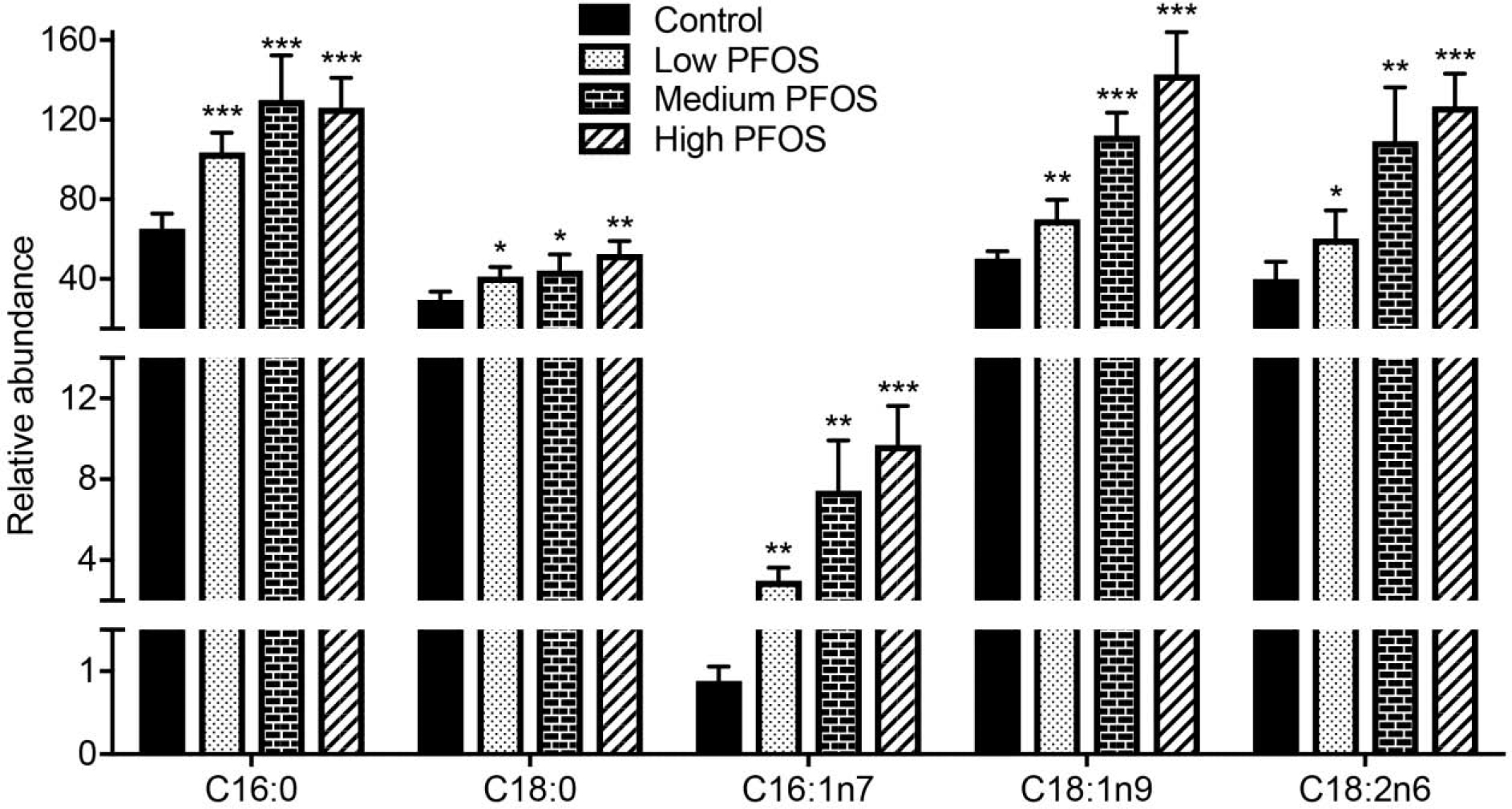
PFOS exposure impacts total fatty acid composition. Total fatty acids were quantified in liver from vehicle-treated and PFOS-treated mice by GC-MS. Data were analyzed using a two-tailed Student’s t-test and are presented as mean ± s. d., n = 5 per group; *p < 0.05, **p < 0.01, ***p < 0.001.
3.3. PFOS alters expression of genes that modulate intracellular metabolism
It is well known that PPARα plays an important role in the regulation of a variety of processes such as inflammation, lipid metabolism, and energy homeostasis (Kersten 2014, Peters, Cheung and Gonzalez 2005), however, there are other mechanisms by which PFOS could alter expression of genes. Dietary PFOS administration increased hepatic expression of mRNAs encoding PPARα target genes, including fatty acid translocase (Cd36), acyl-CoA oxidase 1 (Acox1) and carnitine palmitoyltransferase 2 (Cpt2) as compared to controls (Fig. 5), which are involved in fatty acid β-oxidation. Interestingly, dietary PFOS administration also increased hepatic expression of stearoyl coenzyme A desaturase-1 (Scd1), fatty acid synthase (Fasn) and acetyl-CoA carboxylase 1 (Acaca) as compared to controls (Fig. 5). The latter is of interest because these gene products are involved in lipogenic pathways rather than catabolic pathways mediated by CD36, ACOX and CPT2. Previous studies have also shown that PPARα regulates mitochondrial oxidative stress (Le May et al. 2000). Expression of superoxide dismutase 1 (Sod1) mRNA that encodes an enzyme that modulates mitochondria-derived reactive oxygen species was markedly increased in the liver of high dose of PFOS-treated mice (Fig. 5). Interestingly, PPARα is also known to regulate inflammation (Youssef and Badr 2004). Thus, it is of interest to note that results from the present study show that dietary PFOS exposure reduced hepatic expression of mRNAs encoding proteins involved in pro-inflammatory signaling, such as Lcn-2, Il-1β and Tnf-α (Fig. 5).
Fig. 5.

PFOS dose dependently impacts PPARα signaling and gene expression. qPCR analyses of mRNA levels of Pparα, Cd36, Acox1, Cpt1a, Cpt2, Srebp1, Cidea, Fasn, Scd1, Acaca, Hmgcs2, Acaa1b, G6pase, Glut2, Pepck, Ucp2, and Sod1 associated with lipid, fatty acids, glucose metabolism and oxidative stress and inflammatory cytokines (Lcn-2, IL-1β, and Tnf-α) in the liver of mice exposed to different dosage of PFOS. Data were analyzed using one-way ANOVA with Tukey’s correction and are presented as mean ± s. d., (n = 5 per group); *p < 0.05, **p < 0.01, ***p < 0.01 compared with vehicle treatment.
3.4. Relationship between the gut microbiome and host metabolome after PFOS exposure
Recent evidence suggests that the gut microbiota can indirectly influence liver metabolism and thereby impact a number of metabolic diseases, such as obesity and fatty liver disease (Miele et al. 2013, Vajro, Paolella and Fasano 2013). It is also known that PFOS can induce fatty liver (Zhang et al. 2016). To begin to determine whether the gut microbiome is associated with the host metabolic alterations induced by PFOS (0.003%), a correlation matrix was generated by calculating the Pearson’s correlation coefficient. Clear correlations could be identified between changes in the gut microbiome and altered metabolic profiles (r > 0.63 or < −0.63). The resulting association map indicated positive (red) and negative (blue) correlations between the levels of host liver metabolites and the gut microbiome of PFOS (0.003%)-treated mice as compared with controls (Fig. 6). Each metabolite in Fig. 6 has at least one significant relationship with a particular genus. These metabolites were used to bisect results shown in Fig. 6. The first half (Proteus – Clostridium XI) includes direct relationships between total hepatic lipid, GSSG, TMAO/betaine, BCAAs, acetate, succinate, nicotinamide, lactate, and UFA, with genera predominately belonging to the Firmicutes phyla. By contrast, some metabolites including glycogen and glucose had inverse relationships with the aforementioned genera. The second half (Unclassified Desulfovibrionales–Anarosporobacter) have the opposite relationships as the first half (Fig. 6). These results indicated that the significant modulation of the gut microbiota was highly correlated with the host metabolic disorder induced by PFOS treatment.
Fig. 6.
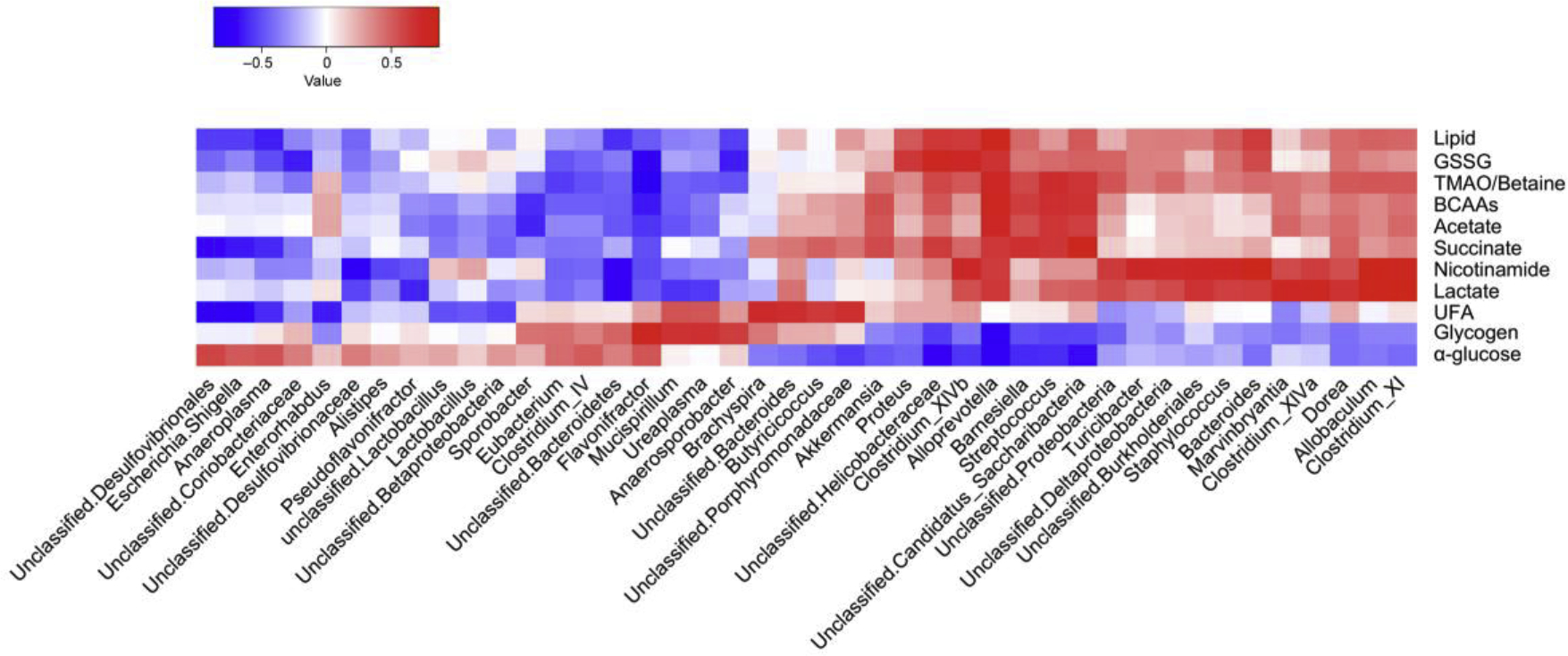
Relationship between gut microbiome and host metabolome. Heat maps indicated positive (red) and negative (blue) correlations between the levels of host metabolites and the identified gut microbiome at the genus levels of PFOS-treated mice as compared with control mice. The legend shows correlation values from −1 to 1 and assigns the appropriate color to them (Red for positive correlations and blue for negative correlations)
3.5. Low dose PFOS directly affects the metabolism of gut microbiota in vitro
An in vitro model was used to investigate the direct effect of PFOS on the metabolic activities of the gut microbiota. Two different stains were used: SYBR green which stains all cells and CFDA which is a cell permeable dye that acts as a proxy for enzymatic or metabolic activity (Cai et al, 2018). The counts of CFDA stained bacteria were normalized using the cell counts obtained using SYBR green. PFOS caused a significant dose-dependent decrease in CFDA stained bacteria (from 63.0% ± 3.3 to 26.4% ± 0.9%) indicating decreased metabolic activity of bacterial cells (Fig. 7a).
Fig. 7.
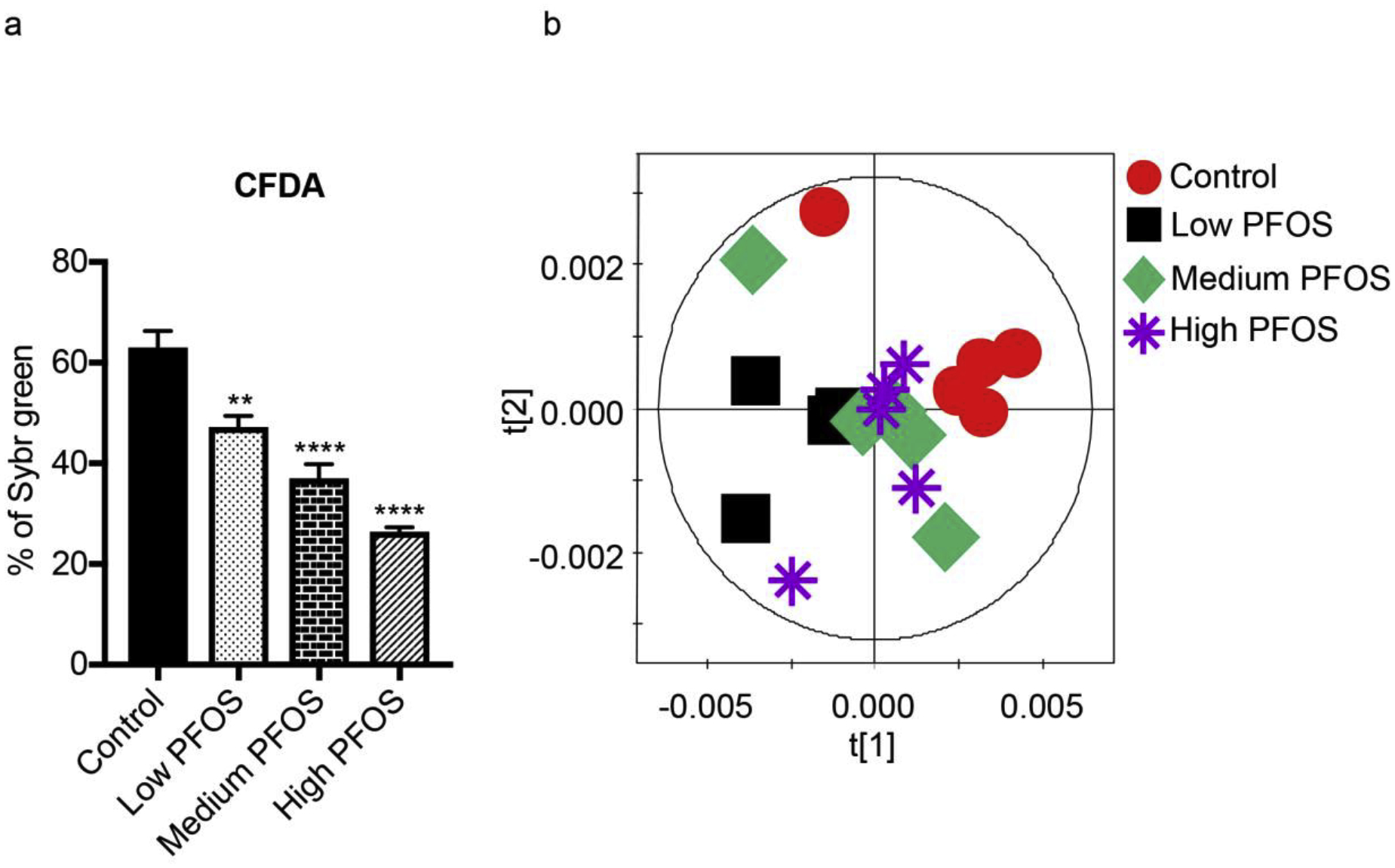
Direct PFOS exposure influences gut microbiome metabolism. a) The relative proportion of CFDA in the flow cytometric analysis of cecal content after PFOS treatment. Data were analyzed using one-way ANOVA with Tukey’s correction and are presented as means ± s. d., (n = 5 per group); *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 compared to control. b) Orthogonal projection to latent structure-discriminant (OPLS-DA) analysis scores plot derived from 1H NMR spectra of cecal content treated with control and three doses of PFOS.
1H NMR-based metabolomics was used to further investigate the global metabolic changes in the cecal content after PFOS treatment. Multivariate data analysis using OPLS-DA plot (Fig. 7b) shows distinct clustering of bacterial samples after PFOS treatments compared with control samples. Further, PFOS treatment resulted in a significant increase in the levels of nucleosides and nucleoside derivatives, particularly that of uracil, inosine, hypoxanthine, xanthine and uridine diphosphate (UDP) (Fig. 8). PFOS treatment at low dose was also found to significantly lower lipid levels (Fig. 8).
Fig. 8.

Direct PFOS exposure of the gut the microbiome impacts the metabolic profile. The relative abundance of cecal content metabolites measured by 1H NMR data after exposure to three doses of PFOS. Data were analyzed using one-way ANOVA with Tukey’s correction and are presented as means ± s. d., (n = 5 per group); *p < 0.05, **p < 0.01, ***p < 0.001 compared to control.
4. DISCUSSION
In the current study, a combination of 16S rRNA gene sequencing, untargeted 1H NMR-based metabolomics, targeted metabolite profiling by LC/GC-MS, and gene expression analysis were used to explore the effect of PFOS exposure on the gut microbiota and host metabolome in mice. In addition, associations between the gut microbial composition and metabolic pathways involved in lipid, glucose, and amino acid metabolism were explored. Further, flow cytometry and untargeted 1H NMR-based metabolomics were used to assess the direct effect of PFOS on microbiome using isolated mice cecal content. These findings provide new insights and understanding of the mechanisms that may modulate the effects of PFOS.
In Sprague Dawley rats, long-term dietary administration of PFOS (up to 104 weeks) did not elicit any significant toxicological changes in gastrointestinal tract histology (Butenhoff et al. 2012), however the gut microbiota was not evaluated. In the present study, short-term PFOS exposure caused a striking change in the mouse gut microbiota. Within the intestinal microbiome, at the phylum level, PFOS-treated mice exhibited a lower proportion of Firmicutes but higher proportion of Bacteroidetes than those of the vehicle-treated mice, indicating that the total population of the gut microbiota was modulated by PFOS. Similar observations have been reported in the reduction of Firmicutes/Bacteroidetes ratio in the gut microbiome at the phylum level induced by the persistent environmental contaminants TCDF and PCBs (Zhang et al, 2015; Choi et al. 2013). PFOS administration also reduced body weight of mice as compared to controls as observed in previous study (Zhang et al. 2016). Further, PFOS treatment induced elevated levels of the genera, Clostridium cluster XIVb, Clostridium cluster XI, Clostridium Cluster XIVa and Streptococcus, and depleted the genera Flavonifractor, Alistipes, and an unclassified genus of Bacteroidetes.
Increasing evidence suggests a cross-talk between the gut microbiota and liver that plays an important role in the development of liver diseases such as non-alcoholic fatty liver disease and hepatocellular carcinoma (Miele et al. 2013, Visschers et al. 2013). In the current study, a significant correlation was identified between the alterations in gut microbiota and metabolite changes in the PFOS-treated mice. For example, hepatic lipids, GSSG, UFA, branch chain amino acids, choline metabolites (TMAO+betaine) and short chain fatty acids were found to be positively correlated with genera predominantly belonging to the Firmicutes phyla in the cecal content of PFOS-treatment mice. By contrast, glucose and glycogen were negatively correlated with genera predominantly belonging to the phylum Firmicutes in the cecal content of PFOS-treatment mice. Of particular note, the genus Clostridium cluster XIVa is positively correlated with the levels of acetate and lactate. One possible explanation is that Clostridium cluster XIVa includes many Clostridium spp. that utilize acetate and lactate to create butyrate and can also increase the mucous layer of the gut, both of which may serve to protect the gut against inflammation (Van den Abbeele et al. 2013). Additionally, the genus Streptococcus was found to be positively correlated with choline metabolites including, TMAO and betaine, critical components of the cell membrane and important for regulation of osmolarity (Klein 2000, Zerbst-Boroffka et al. 2005). Streptococcus, was reported to be associated with formation of the choline metabolite, trimethylamine (TMA) (Chao et al. 1990). Interestingly, numerous reports have identified TMAO levels to be positively correlated with atherosclerosis and chronic kidney disease (Koeth et al. 2013, Tang et al. 2013, Wang et al. 2011, Tang et al. 2015). Given that PFOS is known to cause hypolipidemia in rodents and non-human primates (Bijland et al. 2011, Butenhoff et al. 2012, Chang et al. 2017, Seacat et al. 2002), further investigation into the connection between PFOS, the gut microbiome, and cardiovascular disease is warranted.
Results from these studies also show that changes in the host metabolome reflect alterations in the expression of PPARα target genes induced by PFOS. Administration of PFOS caused an increase in the expression of mRNAs encoding Cd36, Acox1 and Cpt2 that mediate fatty acid β-oxidation. This is consistent with the increase in hepatic acetate and ketone bodies such as 3-hydroxybutyrate, the catabolic byproducts of fatty acid β-oxidation, and the increase in carnitines including free carnitine, hexanoylcarnitine, octanoylcarnitine, lauroylcarnitine, and palmitoleoylcarnitine observed in the liver of PFOS-treated mice compared to controls (data not shown). Low dose PFOS administration also caused an increase in the expression of mRNAs encoding Scd1, Fasn, and Acaca that mediate lipogenesis. This is consistent with the increase in hepatic fatty acids observed in PFOS-treated mice as compared to controls, a phenotype known to occur in response to administration of a PPARα activator, which may be due in part to increased expression of SCD1 (Miller and Ntambi 1996, Yan et al. 2014). In addition to the alterations in lipid metabolism caused by PFOS exposure, it is noteworthy that PFOS treatment also caused reductions in the levels of hepatic glucose, glycogen, and succinate accompanied by elevated lactate and pyruvate. These metabolic changes likely reflect disruption of carbohydrate and energy metabolism and TCA cycle after PFOS treatment. This is similar to a previous microarray study showing that perfluorododecanoic acid affected the expression of key enzymes such as involved (solute carrier family 3 and aconitase 1) in carbohydrate metabolism and resulted in liver damage by interfering with the TCA process and glycolysis (Ding et al. 2009). PFOS exposure likely causes mitochondrial oxidative stress and lipid peroxidation as depletion in the GSH/GSSG, NAD+/NADH, and NADP+/NADPH ratios was observed along with marked up-regulation of uncoupling protein 2 (Ucp2) and superoxide dismutase 1 (Sod1) mRNAs, two important regulators of mitochondria-derived ROS production. Lastly, since PFOS can activate PPARα, it is interesting to note the decrease in the expression of mRNAs encoding Lcn2, Il1b and Tnfa, as it is well known that PPARα can indirectly mediate this effect (Delerive et al. 1999). This suggests that administration of PFOS can also cause anti-inflammatory effects in the liver.
Given the correlation between the alterations in the gut microbiota and metabolic changes in the host upon PFOS exposure, the gut microbiota could be acting in a causal way in altering host metabolism. While there have been limited studies on the effect of PFOS on the metabolism of isolated bacteria (Roberto et al. 2010, Liu et al. 2016), its direct effect on the bacterial community has not yet been explored. In the current study, PFOS was found to directly alter metabolism of bacteria obtained from isolated cecal contents. Within four hours of treatment, PFOS caused a significant decrease in the overall metabolism of the cecal bacterial community thus demonstrating that PFOS can directly impact the metabolism of the gut microbiome in vitro. The flow cytometry findings were corroborated by 1H NMR-based metabolomics which also showed distinct clustering of samples treated with PFOS. The significant increase in the relative abundance of nucleosides and nucleoside derivatives could be attributed to DNA damage elicited by PFOS which has been reported in E. coli (Liu et al. 2016). The decrease in relative abundance of lipids after PFOS exposure may be explained by lipid peroxidation because of oxidative stress elicited by PFOS treatment. Overall, these studies suggest that PFOS directly alters the metabolism of gut microbiome which may modulate the effects of PFOS.
In conclusion, dietary PFOS exposure profoundly induced alteration of the gut microbiota community structure and its function, and host metabolome in mice. Notably, PFOS dose-dependently caused induction of hepatic PPARα signaling and its target genes that was associated with disruption of related metabolic pathways, including hepatic lipogenesis, fatty acid β-oxidation, and carbohydrate and energy metabolism. Interestingly, the host metabolic dysregulation was found to be highly associated with the modulated gut microbiome by PFOS exposure. These findings suggest possible mechanisms of hepatic effects induced by PFOS and illustrate for the first time that exposure to PFOS impacts the gut microbiota-host metabolic homeostasis in mice. The direct effect of PFOS on microbiome structure and function suggests PFOS toxicity on host metabolism could be mediated by microbiome. Further investigation on the effects of PFOS on the gut microbiota and specific metabolic pathways with Pparα-null mice will increase a more detailed mechanistic understanding of the effects induced by PFOS exposure.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by an unrestricted gift from the 3M Company (J.M.P.), USDA National Institute of Food and Federal Appropriations under Project PEN04607 and Accession number 1009993 (J.M.P, A.D.P.), the National Natural Science Foundation of China (Z.L.M., 2018YFE0110800, 21577169), the National Institute of Environmental Health Sciences (A.D.P., ES022186; ES028288), and the Pennsylvania Department of Health using Tobacco C.U.R.E. Funds (J.M.P., A.D.P). The Department specifically disclaims responsibility for any analyses, interpretations or conclusions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
John L. Butenhoff, Shu-Ching Chang, were or are employees of 3M Company, a former manufacturer of PFOS.
REFERENCES
- Atlas RM, Aagaard KM, Hsiao EY, Huang Y, Huttenhower C, Krajmalnik-Brown R, Lynch S, Nazaroff WW, Patterson AD, Rawls JF, Rodricks JV, Shubat P & Thrall B (2018) Environmental Chemicals, the Human Microbiome, and Health Risk. [Google Scholar]
- Bijland S, Rensen PC, Pieterman EJ, Maas AC, van der Hoorn JW, van Erk MJ, Havekes LM, Willems van Dijk K, Chang SC, Ehresman DJ, Butenhoff JL & Princen HM (2011) Perfluoroalkyl sulfonates cause alkyl chain length-dependent hepatic steatosis and hypolipidemia mainly by impairing lipoprotein production in APOE*3-Leiden CETP mice. Toxicol Sci, 123, 290–303. [DOI] [PubMed] [Google Scholar]
- Bjork JA, Butenhoff JL & Wallace KB (2011) Multiplicity of nuclear receptor activation by PFOA and PFOS in primary human and rodent hepatocytes. Toxicology, 288, 8–17. [DOI] [PubMed] [Google Scholar]
- Bjork JA & Wallace KB (2009) Structure-activity relationships and human relevance for perfluoroalkyl acid-induced transcriptional activation of peroxisome proliferation in liver cell cultures. Toxicol Sci, 111, 89–99. [DOI] [PubMed] [Google Scholar]
- Butenhoff JL, Chang SC, Olsen GW & Thomford PJ (2012) Chronic dietary toxicity and carcinogenicity study with potassium perfluorooctanesulfonate in Sprague Dawley rats. Toxicology, 293, 1–15. [DOI] [PubMed] [Google Scholar]
- Cai J, Nichols R, Koo I, Kalikow Z, Zhang L, Tian Y, Zhang J, Smith P & Patterson A (2018) Multiplatform Physiologic and Metabolic Phenotyping Reveals Microbial Toxicity. Msystems, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Allen BC, Andres KL, Ehresman DJ, Falvo R, Provencher A, Olsen GW & Butenhoff JL (2017) Evaluation of Serum Lipid, Thyroid, and Hepatic Clinical Chemistries in Association With Serum Perfluorooctanesulfonate (PFOS) in Cynomolgus Monkeys After Oral Dosing With Potassium PFOS. Toxicol Sci, 156, 387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, Collman RG, Bushman FD & Li H (2012) Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics, 28, 2106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus SP, Guillou H & Ellero-Simatos S (2016) The gut microbiota: A major player in the toxicity of environmental pollutants? npj Biofilms and Microbiomes, 2, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton TA, Baker D, Lindon JC, Everett JR & Nicholson JK (2009) Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A, 106, 14728–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corton JC, Peters JM & Klaunig JE (2018) The PPARα-dependent rodent liver tumor response is not relevant to humans: addressing misconceptions. Arch Toxicol, 92, 83–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Liao C.-y., Zhou Q.-f., Xia T.-m., Yun Z.-j. & G.-b. Jiang (2010) Excretion of PFOA and PFOS in Male Rats During a Subchronic Exposure. Archives of Environmental Contamination and Toxicology, 58, 205–213. [DOI] [PubMed] [Google Scholar]
- Delerive P, De Bosscher K, Besnard S, Vanden Berghe W, Peters JM, Gonzalez FJ, Fruchart JC, Tedgui A, Haegeman G & Staels B (1999) Peroxisome proliferator-activated receptor alpha negatively regulates the vascular inflammatory gene response by negative cross-talk with transcription factors NF-kappaB and AP-1. J Biol Chem, 274, 32048–54. [DOI] [PubMed] [Google Scholar]
- Ding L, Hao F, Shi Z, Wang Y, Zhang H, Tang H & Dai J (2009) Systems biological responses to chronic perfluorododecanoic acid exposure by integrated metabonomic and transcriptomic studies. J Proteome Res, 8, 2882–91. [DOI] [PubMed] [Google Scholar]
- Elcombe CR, Elcombe BM, Foster JR, Chang SC, Ehresman DJ & Butenhoff JL (2012) Hepatocellular hypertrophy and cell proliferation in Sprague-Dawley rats from dietary exposure to potassium perfluorooctanesulfonate results from increased expression of xenosensor nuclear receptors PPARα and CAR/PXR. Toxicology, 293, 16–29. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Nagahori H, Yamada T, Deguchi Y, Tomigahara Y, Nishioka K, Uwagawa S, Kawamura S, Isobe N, Lake BG & Okuno Y (2009) Comparison of the effects of the synthetic pyrethroid Metofluthrin and phenobarbital on CYP2B form induction and replicative DNA synthesis in cultured rat and human hepatocytes. Toxicology, 258, 64–9. [DOI] [PubMed] [Google Scholar]
- Huang C, Lei H, Zhao X, Tang H & Wang Y (2013) Metabolic influence of acute cyadox exposure on Kunming mice. J Proteome Res, 12, 537–45. [DOI] [PubMed] [Google Scholar]
- Kersten S (2014) Integrated physiology and systems biology of PPARα. Mol Metab, 3, 354–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J (2000) Membrane breakdown in acute and chronic neurodegeneration: focus on choline-containing phospholipids. J Neural Transm (Vienna), 107, 1027–63. [DOI] [PubMed] [Google Scholar]
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ & Hazen SL (2013) Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med, 19, 576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK & Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Applied and Environmental Microbiology, 79, 5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BG (2009) Species differences in the hepatic effects of inducers of CYP2B and CYP4A subfamily forms: relationship to rodent liver tumour formation. Xenobiotica, 39, 582–96. [DOI] [PubMed] [Google Scholar]
- Lau C, Anitole K, Hodes C, Lai D, Pfahles-Hutchens A & Seed J (2007) Perfluoroalkyl acids: a review of monitoring and toxicological findings. Toxicol Sci, 99, 366–94. [DOI] [PubMed] [Google Scholar]
- Le May CL, Pineau T, Bigot K, Kohl C, Girard J & P JP (2000) Reduced hepatic fatty acid oxidation in fasting PPARα null mice is due to impaired mitochondrial hydroxymethylglutaryl-CoA synthase gene expression. FEBS Letters, 475, 163–166. [DOI] [PubMed] [Google Scholar]
- Lee DH, Porta M, Jacobs DR Jr. & Vandenberg LN (2014) Chlorinated persistent organic pollutants, obesity, and type 2 diabetes. Endocr Rev, 35, 557–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Zhang S, Yang K, Zhu L & Lin D (2016) Toxicity of perfluorooctane sulfonate and perfluorooctanoic acid to Escherichia coli: Membrane disruption, oxidative stress, and DNA damage induced cell inactivation and/or death. Environmental Pollution, 214, 806–815. [DOI] [PubMed] [Google Scholar]
- Lu K, Abo RP, Schlieper KA, Graffam ME, Levine S, Wishnok JS, Swenberg JA, Tannenbaum SR & Fox JG (2014) Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ Health Perspect, 122, 284–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele L, Marrone G, Lauritano C, Cefalo C, Gasbarrini A, Day C & Grieco A (2013) Gut-liver axis and microbiota in NAFLD: insight pathophysiology for novel therapeutic target. Curr Pharm Des, 19, 5314–24. [PubMed] [Google Scholar]
- Miller CW & Ntambi JM (1996) Peroxisome proliferators induce mouse liver stearoyl-CoA desaturase 1 gene expression. Proc Natl Acad Sci U S A, 93, 9443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myre M & Imbeault P (2014) Persistent organic pollutants meet adipose tissue hypoxia: does cross-talk contribute to inflammation during obesity? Obes Rev, 15, 19–28. [DOI] [PubMed] [Google Scholar]
- Peters JM, Cheung C & Gonzalez FJ (2005) Peroxisome proliferator-activated receptor-α and liver cancer: where do we stand? J Mol Med (Berl), 83, 774–85. [DOI] [PubMed] [Google Scholar]
- Rebrin I & Sohal RS (2004) Comparison of thiol redox state of mitochondria and homogenates of various tissues between two strains of mice with different longevities. Exp Gerontol, 39, 1513–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Plummer SM, Rode A, Scheer N, Bower CC, Vogel O, Henderson CJ, Wolf CR & Elcombe CR (2010) Human constitutive androstane receptor (CAR) and pregnane X receptor (PXR) support the hypertrophic but not the hyperplastic response to the murine nongenotoxic hepatocarcinogens phenobarbital and chlordane in vivo. Toxicol Sci, 116, 452–66. [DOI] [PubMed] [Google Scholar]
- Seacat AM, Thomford PJ, Hansen KJ, Olsen GW, Case MT & Butenhoff JL (2002) Subchronic toxicity studies on perfluorooctanesulfonate potassium salt in cynomolgus monkeys. Toxicol Sci, 68, 249–64. [DOI] [PubMed] [Google Scholar]
- Snedeker SM & Hay AG (2012) Do interactions between gut ecology and environmental chemicals contribute to obesity and diabetes? Environ Health Perspect, 120, 332–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y & Hazen SL (2013) Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med, 368, 1575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WHW, Wang Z, Kennedy DJ, Wu Y, Buffa JA, Agatisa-Boyle B, Li XS, Levison BS & Hazen SL (2015) Gut Microbiota-Dependent Trimethylamine N-oxide (TMAO) Pathway Contributes to Both Development of Renal Insufficiency and Mortality Risk in Chronic Kidney Disease. Circ Res, 116, 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajro P, Paolella G & Fasano A (2013) Microbiota and gut-liver axis: their influences on obesity and obesity-related liver disease. J Pediatr Gastroenterol Nutr, 56, 461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Abbeele P, Belzer C, Goossens M, Kleerebezem M, De Vos WM, Thas O, De Weirdt R, Kerckhof FM & Van de Wiele T (2013) Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J, 7, 949–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visschers RG, Luyer MD, Schaap FG, Olde Damink SW & Soeters PB (2013) The gut-liver axis. Curr Opin Clin Nutr Metab Care, 16, 576–81. [DOI] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ & Hazen SL (2011) Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature, 472, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf CJ, Takacs ML, Schmid JE, Lau C & Abbott BD (2008) Activation of mouse and human peroxisome proliferator-activated receptor α by perfluoroalkyl acids of different functional groups and chain lengths. Toxicol Sci, 106, 162–71. [DOI] [PubMed] [Google Scholar]
- Yan F, Wang Q, Xu C, Cao M, Zhou X, Wang T, Yu C, Jing F, Chen W, Gao L & Zhao J (2014) Peroxisome proliferator-activated receptor α activation induces hepatic steatosis, suggesting an adverse effect. PLoS One, 9, e99245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef J & Badr M (2004) Role of Peroxisome Proliferator-Activated Receptors in Inflammation Control. J Biomed Biotechnol, 2004, 156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbst-Boroffka I, Kamaltynow RM, Harjes S, Kinne-Saffran E & Gross J (2005) TMAO and other organic osmolytes in the muscles of amphipods (Crustacea) from shallow and deep water of Lake Baikal. Comp Biochem Physiol A Mol Integr Physiol, 142, 58–64. [DOI] [PubMed] [Google Scholar]
- Zhang L, Hatzakis E, Nichols RG, Hao R, Correll J, Smith PB, Chiaro CR, Perdew GH & Patterson AD (2015a) Metabolomics Reveals that Aryl Hydrocarbon Receptor Activation by Environmental Chemicals Induces Systemic Metabolic Dysfunction in Mice. Environ Sci Technol, 49, 8067–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Krishnan P, Ehresman DJ, Smith PB, Dutta M, Bagley BD, Chang SC, Butenhoff JL, Patterson AD & Peters JM (2016) Editor’s Highlight: Perfluorooctane Sulfonate-Choline Ion Pair Formation: A Potential Mechanism Modulating Hepatic Steatosis and Oxidative Stress in Mice. Toxicol Sci, 153, 186–97. [DOI] [PubMed] [Google Scholar]
- Zhang L, Nichols RG, Correll J, Murray IA, Tanaka N, Smith PB, Hubbard TD, Sebastian A, Albert I, Hatzakis E, Gonzalez FJ, Perdew GH & Patterson AD (2015b) Persistent Organic Pollutants Modify Gut Microbiota-Host Metabolic Homeostasis in Mice Through Aryl Hydrocarbon Receptor Activation. Environ Health Perspect, 123, 679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Zitzow JD, Ehresman DJ, Chang SC, Butenhoff JL, Forster J & Hagenbuch B (2015) Na+/Taurocholate Cotransporting Polypeptide and Apical Sodium-Dependent Bile Acid Transporter Are Involved in the Disposition of Perfluoroalkyl Sulfonates in Humans and Rats. Toxicol Sci, 146, 363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


