Abstract
Caged morpholino oligonucleotides (cMOs) are useful research tools in developmental biology because they allow spatiotemporal control of gene expression in whole organisms. While cMOs are usually triggered by light of a single wavelength, the introduction of spectrally distinct chromophores can enable combinatorial regulation of multiple genes. This chapter describes the general principles and methods of wavelength-selective cMO design and synthesis from commercially available reagents. Synthetic protocols for the linkers and the two-step cMO assembly are described in detail, as well as the microinjection and photoactivation techniques. Following these protocols, spectrally separated cyclic cMOs for multiple genes of interest can be prepared, enabling their inhibition in zebrafish embryos and other animal models.
Keywords: Antisense oligonucleotides, Caged morpholinos, Zebrafish, Gene silencing, Wavelength-selective activation, Photolabile linkers, Developmental Biology
1. Introduction
Embryonic development is controlled by a complex network of genetic programs that are precisely regulated to create tissues and organs (Perrimon, Pitsouli, & Shilo, 2012). One of the major objectives of modern developmental biology is to understand how these genes act in space and time to coordinate embryonic patterning. In recent years, genetic technologies such as antisense oligonucleotides, RNA interference, and genome-editing tools have enabled new ways to dissect embryonic pathways at the transcriptional or translational level (Housden et al., 2017). In particular, morpholino antisense oligonucleotides (MOs) have been widely used to inhibit gene expression in a variety of model systems including zebrafish, Xenopus, and chick embryos(Ekker & Larson, 2001; Nasevicius & Ekker, 2000). MOs are chemically modified DNA derivatives that are composed of nucleobase-appended morpholine rings connected with neutral phosphorodiamidate linkages (Figure 1A) (Bhadra, Pattanayak, & Sinha, 2015; Summerton & Weller, 1997). These oligonucleotides are resistant to cellular nucleases due to their unnatural backbones and persist in vivo for up to 4 days (Bill, Petzold, Clark, Schimmenti, & Ekker, 2009). MOs are typically 25-base sequences, designed to block translation start sites or RNA splice junctions, and the resulting loss-of-function phenotypes can be rapidly generated in a non-Mendelian manner (Corey & Abrams, 2001). While it may be required to alter the MO length for a specific application, shorter sequences can have diminished efficacy and longer sequences can have decreased specificity. When MOs are microinjected into early-stage embryos (typically before 4-cell stage in zebrafish) they uniformly distribute throughout the organism and constitutively inhibit protein synthesis.
Figure 1. Morpholinos and their caged versions.
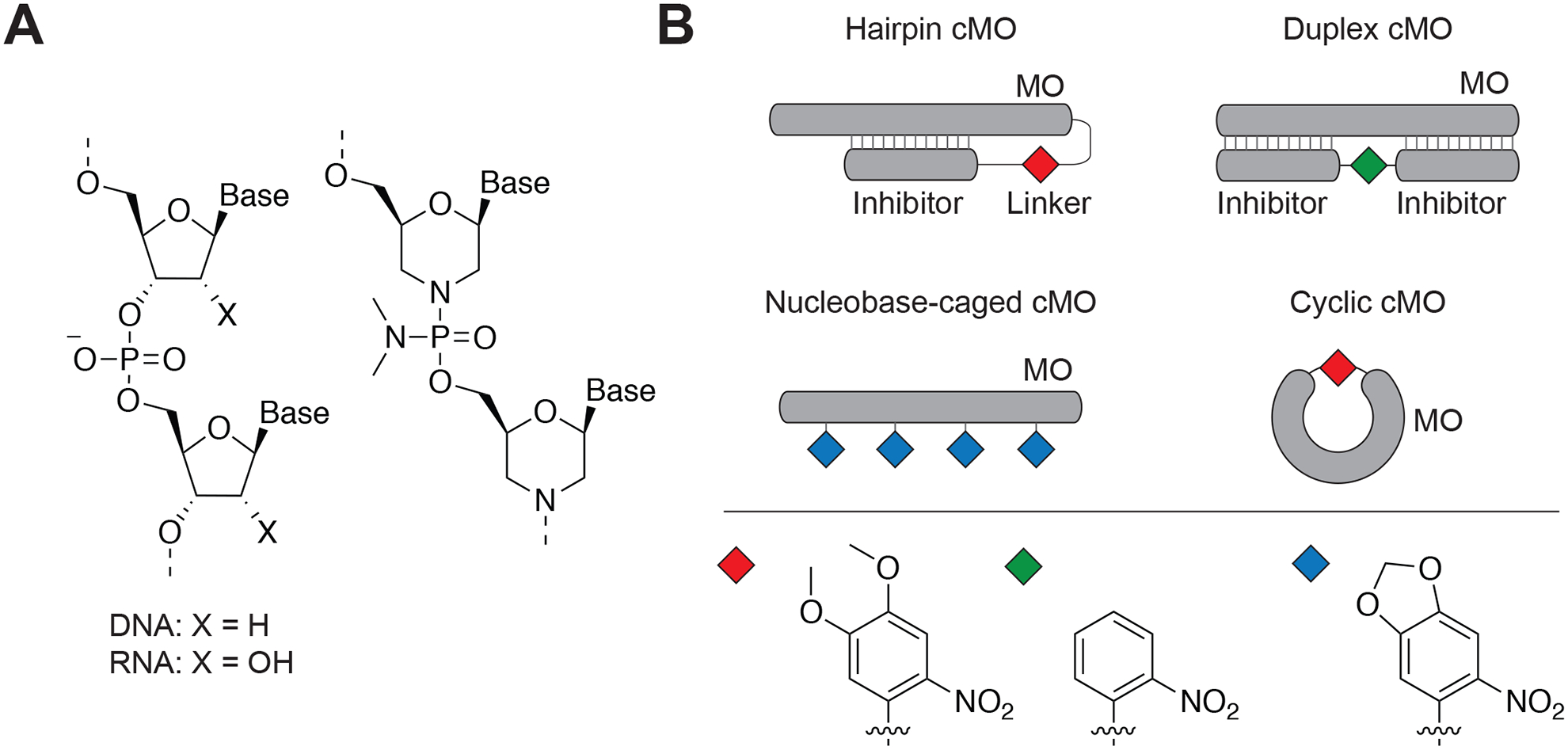
A) DNA/RNA and MO chemical structures. B) cMO design strategies showing hairpin, duplex, nucleobase-caged, and cyclic cMOs. The photolabile moiety has been indicated as diamond shapes. Adapted with permission from Yamazoe et al., 2012.
While MOs are routinely used to interrogate gene functions in zebrafish and other organisms, their constitutive activity renders these reagents inadequate for studying genes that have pleiotropic functions. Since gene expression is inhibited immediately after microinjection, conventional MOs do not allow any spatiotemporal control. Moreover, silencing essential genes in early development can cause embryonic lethality, making them challenging to study(Timme-Laragy, Karchner, & Hahn, 2012). To overcome these limitations, we and others have recently developed caged morpholinos (cMOs) that remain inactive in vivo until activated by light of the appropriate wavelength. The first generation of cMOs were composed of an RNA-targeting MO, tethered with a short complementary MO inhibitor through a photocleavable linker, thereby forming a hairpin structure (Figure 1B). The hairpin MOs contained dimethoxynitrobenzyl (DMNB)- or bromohydroxyquinoline (BHQ)- based linkers, which can be cleaved with ultraviolet (365 nm) or two-photon irradiation, respectively (Ouyang et al., 2009; Shestopalov, Sinha, & Chen, 2007). Several other MO-caging strategies also have been developed. MOs have been caged by duplexing the antisense oligonucleotide with an inhibitory strand (Figure 1B) that contains an internal photolabile nitrobenzyl-based linker (Tallafuss et al., 2012; Tomasini, Schuler, Zebala, & Mayer, 2009). Alternatively, cMOs were generated by incorporating multiple nucleobase-caged morpholino monomers during solid-phase synthesis (Figure 1B) (Deiters et al., 2010; Liu & Deiters, 2014). The most recent strategy to generate cMOs utilizes restricting linear morpholinos into a closed circular configuration, thereby preventing target mRNA hybridizations (Figure 1B) (Yamazoe, Shestopalov, Provost, Leach, & Chen, 2012). The cyclic cMOs provide several advantages over previous hairpin, duplex, and nucleobase-caged versions because they are modular in linker design, can be synthesized and purified easily, do not use auxiliary nucleotides, and utilize a single photolabile group. The first generation of cyclic cMOs utilized DMNB - based linkers that are readily cleaved by ultraviolet light (Yamazoe et al., 2012).
The application of cMOs in optically transparent model organisms like zebrafish has provided new insights into gene functions during embryonic development. For example, we have used hairpin cMOs to optically inhibit the expression of mesodermal genes in zebrafish, including no tail-a (tbxta), heart of glass (heg), and floating head (flh) (Ouyang et al., 2009). The nucleobase-caged cMOs have been used inhibit zebrafish chordin expression (Deiters et al., 2010). Similarly, the cyclic cMOs have been used to perturb genes like tbxta, t-box 16/spadetail (tbx16), pancreas transcription factor 1 alpha (ptf1a), ets variant gene 2 (etv2), and focal adhesion kinase (Fak) (Gutzman et al., 2018; Yamazoe et al., 2012). We also have integrated cMOs with photoactivatable lineage tracers, fluorescence-activated cell sorting and mRNA profiling to dynamically elucidate Tbxta- and Tbx16-regulated transcriptomes at different stages of mesoderm development(Payumo, McQuade, Walker, Yamazoe, & Chen, 2016; Shestopalov, Pitt, & Chen, 2012).
In these examples, cMOs were used to inhibit individual genes since their reliance on ultraviolet light-sensitive caging groups precludes sequential control of multiple genes. However, such a capability is highly desirable since cMOs can then be used to decipher the combinatorial activities of two or more genes in a more complex biological network. To achieve this goal, we recently reported the development of spectrally separated cMOs that can be activated by illumination with distinct wavelengths of light. In this chapter, we describe the detailed methods for cyclic cMO preparation based on our published article (Yamazoe, Liu, McQuade, Deiters, & Chen, 2014).
2. Design and synthesis of cyclic cMOs
2.1. Design of cyclic cMOs
As described in the introduction, the general strategy to design cMOs is to restrict their hybridization with the target mRNA until being activated by light. The basis of cyclic cMOs is the concept that oligonucleotide duplex formation has limited tolerance to local curvature of the hybridizing strands (Ramstein & Lavery, 1988). Our first generation of cyclic cMOs were prepared by connecting the two ends of a linear MO 1 using a DMNB-based photolabile linker 2 (Figure 2) (Yamazoe et al., 2012). The bifunctional linker was synthesized with an amine-reactive N-hydroxysuccinimide (NHS) ester functionality and a thiol-reactive chloroacetamide group. The 25-base MO was designed to have corresponding 5’-amine and 3’-alkyl disulfide end-modifications. To prepare the cyclic cMO, the linear MO was first completely converted to the linker-MO conjugate through the NHS ester-amine reaction using an excess amount of the linker, while keeping the thiol-functionality protected as the disulfide group. Subsequent disulfide reduction generates the free thiol that reacts spontaneously with the chloroacetamide to give the desired macrocycle in high yields (Figure 2).
Figure 2. Synthesis of cyclic cMOs.
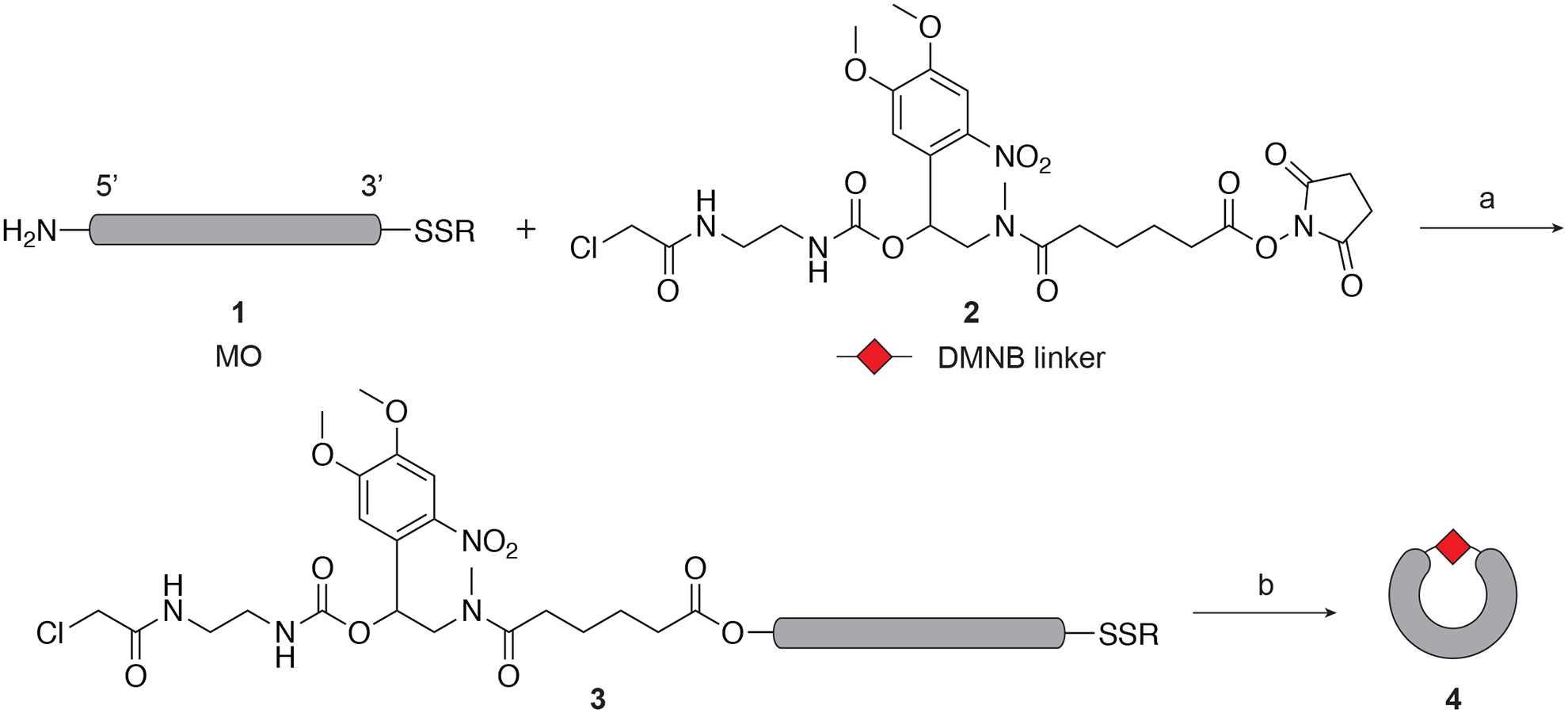
The DMNB linker is shown as an example. a) 0.1 M Na2B4O7, pH 8.5, DMSO, 69–95%; b) tris(2-carboxyethyl)phosphine, 0.1 M Tris-HCl buffer, pH 8.4, 84–92%. Adapted with permission from Yamazoe et al., 2012.
We tested the strategy by targeting Tbxta, the zebrafish ortholog of mammalian Brachyury (Yamazoe et al., 2012). Tbxta deficiency is characterized by loss of notochord and posterior mesoderm, abnormal medial floor plate and somite mispatterning. When the cyclic tbxta cMOs were injected into zebrafish zygotes at a dose of 1 ng/embryo and cultured in the dark, 75% of the embryos exhibited no developmental defects at 24 hours post fertilization (hpf). However, when the cyclic cMO was uncaged through illumination with ultraviolet light at 3.5 hpf, 90% of the embryos showed complete ta-mutant phenotype.
2.2. Design of wavelength-selective cMOs
Since the design of cMOs is modular, they can be synthesized with a variety of linkers and chromophores. To prepare spectrally differentiated cMOs, we chose 2-nitrobenzyl (NB) and [7-(diethylamino)coumarin-4-yl]methyl (DEACM) chromophore–based linkers (Figure 3A) because of their photo-deprotection efficiencies and minimal spectral overlap. Swapping the DMNB moiety with the NB chromophores blue-shifts the absorption maximum and makes it less susceptible to photolysis at wavelengths greater than 400 nm. In contrast, the DEACM chromophore efficiently photolyzes at wavelengths over 400 nm, thus would allow us to orthogonally control the cleavage of these two linkers (Figure 3B)(Yamazoe et al., 2014).
Figure 3. Sequential and wavelength-activation of cyclic cMOs.
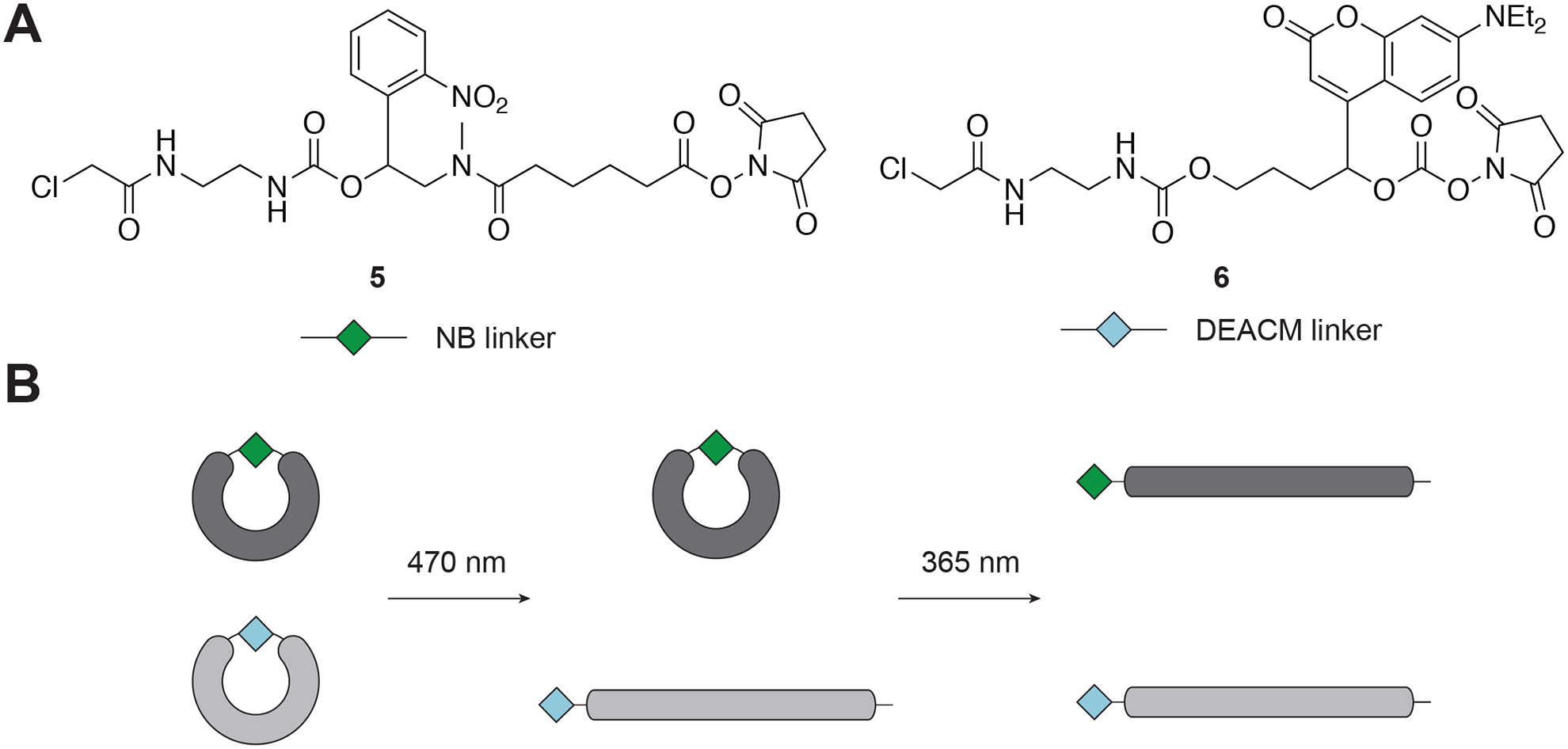
A) Chemical structures of the NB linker and the DEACM linker. B) Concept of sequential activation of cMOs, prepared by using these two linkers. Adapted with permission from Yamazoe et al., 2014.
The cyclic cMOs were prepared by using procedures analogous to our first generation of DMNB-containing linkers (Figure 2) (Yamazoe et al., 2012). We have systematically evaluated the validity of this approach in zebrafish embryos using the tbxta cMO as a test system (Yamazoe et al., 2014). We found that when the embryos were illuminated with 365-nm light at 3.5 hpf, 86% of NB cMO-injected embryos exhibited strong tbxta mutant phenotypes, whereas DEACM cMO-injected embryos exhibited only variable defects. Conversely, upon 470-nm light illumination, 95% of DEACM cMO-injected embryos showed complete loss of tbxta functions, while 91% of NB cMO-injected embryos developed normally.
We further tested the applicability of the approach in two additional regulators of zebrafish mesoderm development, the Tbx16 and Flh (Yamazoe et al., 2014). Our results showed that NB and DEACM cyclic cMOs could be used in combination to sequentially silence genes.
2.3. Methods
2.3.1. Synthesis of the photocleavable linkers
Materials and instruments
All reactions were carried out in flame-dried glassware and stirred magnetically under a nitrogen atmosphere, unless stated otherwise.
Reactions were monitored by thin layer chromatography (TLC), using glass-backed silica gel 60F254 (Merck or Sorbent Technologies, 250-μm thickness), and a compact ultraviolet lamp.
Solvents: tetrahydrofuran (THF), dioxane, and toluene were distilled from sodium/benzophenone ketyl prior to use; dichloromethane (DCM), dimethylformamide (DMF), acetonitrile, and methanol were distilled from calcium hydride and stored over 4 Å molecular sieves.
Reagents obtained from commercial sources were stored under nitrogen and used directly without further purification.
Yields refer to chromatographically and spectroscopically pure compounds.
Flash chromatography was performed with silica gel (EM Science silica gel 60 Å, 70–230 mesh; Sorbtech silica gel 60 Å, 230–400 mesh) as a stationary phase.
The 1H NMR and 13C NMR spectra were recorded on a 300 MHz or 400 MHz Varian NMR spectrometer, and chemical shifts are in δ units (ppm).
Electrospray ionization (ESI) mass spectra were obtained using a Micromass ZQ single quadrupole liquid chromatography-mass spectrometer (LC-MS) or a Micromass Q-TOF hybrid quadrupole LC-MS.
2.3.1.1. Preparation of the DEACM Linker 6.
The DEACM linker 6 can be synthesized from commercially-available 7-(diethylamino)-4-methyl-2H-chromen-2-one (7) in 10 steps (Figure 4). Briefly, coumarin 7 was oxidized with selenium dioxide and subsequently allylated to produce the alcohol 9, which was protected with a tert-butyldimethylsilyl (TBS) group to provide the alkene 10. The alkene 10 was subjected to hydroboration to give the alcohol 11. The chloroacetamide group was then installed in three steps to obtain 12, followed by the removal of the silyl group with tetra-n-butylammonium fluoride (TBAF), delivering the secondary alcohol 13. Finally, the hydroxyl group of 13 was activated by disuccinimidyl carbonate (DSC) to yield the linker 6. A detailed protocol for the synthesis of 6 can be found below.
Figure 4. Synthesis of the DEACM linker.
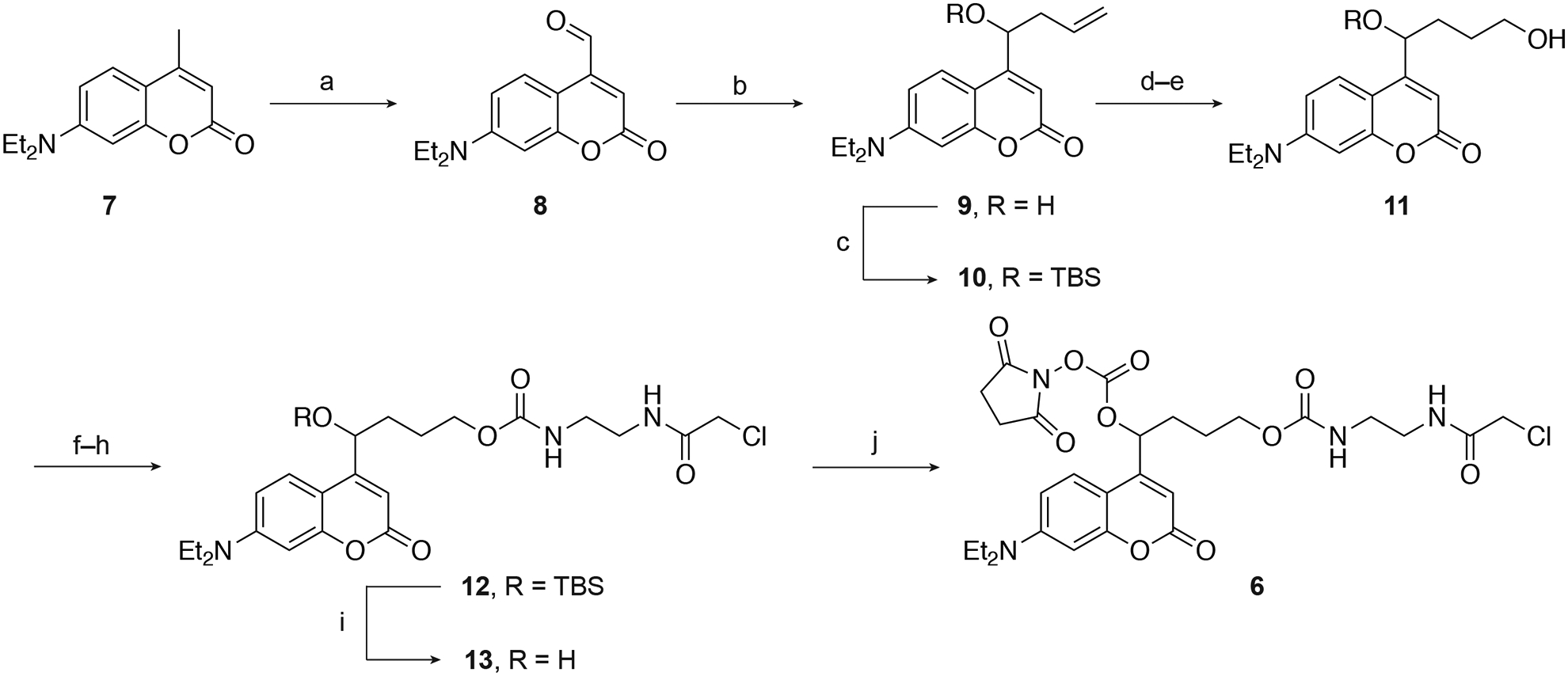
a) SeO2, dioxane, reflux, 42%; b) allyltributyltin, ZnCl2, CH3CN/H2O, 83%; c) TBSCl, imidazole, DMF, 97%; d) BH3-Me2S, THF, 0°C; e) NaOH, H2O2, 0°C – RT, 49% over 2 steps; f) CDI, DCM; g) ethylenediamine, DCM; h) 2-chloroacetyl chloride, DIPEA, DCM, 0°C – RT, 65% over 3 steps; i) TBAF, THF, 0°C, 44%; (j) DSC, DMAP, DCM, 82%. Adapted with permission from Yamazoe et al., 2014.
7-(Diethylamino)-2-oxo-2H-chromene-4-carbaldehyde (2)
-
1
Add selenium dioxide (3.32 g, 30.0 mmol) to commercially-available 7-(diethylamino)-4-methyl-2H-chromen-2-one (7, 4.64 g, 20.0 mmol) dissolved in anhydrous dioxane (120 mL) in a round-bottom flask.
-
2
Attach a Liebig condenser to the round-bottom flask.
-
3
Reflux the mixture at 60°C using an oil bath, while stirring overnight.
-
4
Let the reaction mixture cool to room temperature, then filter through Celite (25 g) and concentrate the filtrate under reduced pressure.
-
5
Purify by silica gel chromatography eluting with hexanes/ethyl acetate (4:1) to afford 7-(diethylamino)-2-oxo-2H-chromene-4-carbaldehyde (8) as a red solid (2.06 g, 42%). Characterization properties: 1H NMR (300 MHz, CDCl3): δ = 1.12–1.20 (t, J = 7.2 Hz, 6H), 3.33–3.40 (q, J = 7.2 Hz, 4H), 6.36 (s, 1H), 6.42–6.43 (d, J = 2.7 Hz, 1H), 6.53–6.57 (dd, Ja = 9.0 Hz, Jb = 2.7 Hz, 1H), 8.19–8.22 (d, J = 2.7 Hz, 1H), 10.95 (s, 1H). 13C NMR (400 MHz, CDCl3): δ = 12.4, 44.7, 97.4, 103.6, 109.4, 117.1, 126.9, 143.7, 150.9, 157.3, 161.7, 192.5. HRMS-ESI: m/z calculated for C14H15NO3 [M + H]+: 246.1130; observed: 246.1130.
7-(Diethylamino)-4-(1-hydroxybut-3-en-1-yl)-2H-chromen-2-one (9)
-
6
Stir a mixture of 7-(diethylamino)-2-oxo-2H-chromene-4-carbaldehyde (8, 500 mg, 2.0 mmol), allyltri-n-butyltin (0.99 mL, 3.0 mmol), and zinc chloride (409 mg, 3.0 mmol), dissolved in a 4:1 mixture of acetonitrile and water (10 mL) at room temperature overnight.
-
7
Concentrate the mixture under reduced pressure.
-
8
Add water (10 mL) to the residue and extract with dichloromethane (DCM, 3 × 10 mL).
-
9
Wash the combined organic layers with brine (10 mL).
-
10
Dry the organic layer over anhydrous sodium sulfate (10 g), and filter.
-
11
Concentrate the filtrate under reduced pressure.
-
12
Purify the crude product by silica gel chromatography eluting with hexanes/ethyl acetate (1:1) to afford 7-(diethylamino)-4-(1-hydroxybut-3-en-1-yl)-2H-chromen-2-one (9) as a green solid (477 mg, 83%). Characterization properties: 1H NMR (400 MHz, CDCl3): δ = 1.14–1.18 (t, J = 7.2 Hz, 6H), 2.38–2.45 (m, 1H), 2.57–2.63 (m, 1H), 3.33–3.38 (q, J = 7.2 Hz, 4H), 4.97–5.00 (dd, Ja = 8.0 Hz, Jb = 4.0 Hz, 1H), 5.10–5.17 (m, 2H), 5.81–5.90 (m, 1H), 6.22 (s, 1H), 6.40–6.41 (d, J = 2.4 Hz, 1H), 6.52–6.55 (dd, Ja = 9.2 Hz, Jb = 2.4 Hz, 1H), 7.36–7.38 (d, J = 9.2 Hz, 1H). 13C NMR (400 MHz, CDCl3): δ = 12.5, 41.6, 44.7, 68.7, 97.8, 105.1, 106.2, 108.6, 118.8, 124.9, 133.6, 150.3, 156.4, 158.2, 163.0. HRMS-ESI: m/z calculated for C17H21NO3 [M + H]+: 288.1660; observed: 288.1602.
4-(1-((tert-Butyldimethylsilyl)oxy)but-3-en-1-yl)-7-(diethylamino)-2H-chromen-2-one (10)
-
13
To a solution of 7-(diethylamino)-4-(1-hydroxybut-3-en-1-yl)-2H-chromen-2-one (9, 862 mg, 3.0 mmol) in dimethylformamide (DMF, 3.4 mL), add imidazole (612 mg, 9.0 mmol) and tert-butyldimethylsilyl chloride (904 mg, 6.0 mmol).
-
14
Stir the solution at room temperature overnight.
-
15
Quench the reaction with saturated aqueous sodium bicarbonate (15 mL).
-
16
Extract the resulting aqueous phase with ethyl acetate (3 × 10 mL).
-
17
Wash the combined organic layers with water (15 mL) and then brine (10 mL).
-
18
Dry the organic layer over anhydrous sodium sulfate (5 g), and filter.
-
19
Concentrate the filtrate under reduced pressure.
-
20
Purify the crude product by silica gel chromatography eluting with hexanes/ethyl acetate (4:1) to afford 4-(1-((tert-butyldimethylsilyl)oxy)but-3-en-1-yl)-7-(diethylamino)-2H-chromen-2-one (10) as a yellow solid (1.16 g, 97%). Characterization properties: 1H NMR (400 MHz, CDCl3): δ = 0.03 (s, 6H), 0.86 (s, 9H), 1.13–1.16 (t, J = 7.2 Hz, 6H), 2.38–2.48 (m, 2H), 3.32–3.37 (q, J = 7.2 Hz, 4H), 4.85–4.88 (m, 1H), 4.97–5.02 (m, 2H), 6.13 (s, 1H), 6.13–6.14 (s, 1H) 6.456.46 (d, J = 2.8 Hz, 1H), 6.52–6.55 (dd, Ja = 9.2 Hz, Jb = 2.8 Hz, 1H), 7.43–7.45 (d, J = 9.2 Hz, 1H). 13C NMR (400 MHz, CDCl3): δ = −5.1, −4.8, −3.6, 12.4, 18.0, 25.7, 42.9, 44.6, 71.0, 97.8, 105.7, 105.9, 108.4, 117.8, 125.2, 133.8, 150.2, 156.5, 158.3, 162.7. HRMS-ESI: m/z calculated for C23H35NO3Si [M + H]+: 402.2464; observed: 402.2468.
4-(1-((tert-Butyldimethylsilyl)oxy)-4-hydroxybutyl)-7-(diethylamino)-2H-chromen-2-one (11)
-
21
Dissolve 4-(1-((tert-butyldimethylsilyl)oxy)but-3-en-1-yl)-7-(diethylamino)-2H-chromen-2-one (10, 80 mg, 0.20 mmol) in tetrahydrofuran (THF, 700 μL).
-
22
Cool down to 0°C.
-
23
Add a 2 M solution of borane–dimethyl sulfide complex in THF (500 μL, 1.0 mmol) dropwise over 5 minutes.
-
24
Stir at 0°C for 3 h.
-
25
Still at this temperature, slowly add a 3 M aqueous solution of sodium hydroxide (500 μL) and 30% hydrogen peroxide in water (400 μL).
-
26
Allow the mixture to warm up to room temperature for 2 h while stirring.
-
27
Extract the reaction mixture with ethyl acetate (3 × 2 mL).
-
28
Wash the combined organic layers with water (2 mL) and brine (2 mL).
-
29
Dry the organic layer over anhydrous sodium sulfate (5 g), and filter.
-
30
Concentrate the filtrate under reduced pressure.
-
31
Purify the crude product by silica gel chromatography eluting with hexanes/ethyl acetate (1:1) to afford 4-(1-((tert-butyldimethylsilyl)oxy)-4-hydroxybutyl)-7-(diethylamino)-2H-chromen-2-one (11) as a yellow solid (41 mg, 49%). Characterization properties: 1H NMR (400 MHz, CDCl3): δ = −0.04 (s, 3H) 0.08 (s, 3H), 0.91 (s, 9H), 1.18–1.21 (t, J = 7.2 Hz, 6H), 1.62–1.90 (m, 5H), 3.37–3.42 (q, J = 7.2 Hz, 3H), 3.62–3.67 (m, 2H), 4.91–4.93 (dd, Ja = 6.4 Hz, Jb = 4.0 Hz, 1H), 6.17 (s, 1H), 6.49–6.50 (d, J = 2.8 Hz, 1H), 6.54–6.57 (dd, Ja = 9.2 Hz, Jb = 2.8 Hz, 1H), 7.47–7.49 (d, J = 9.2 Hz, 1H). 13C NMR (400 MHz, CDCl3): δ = −5.0, −4.6, 12.6, 18.3, 25.9, 28.5, 34.5, 44.8, 62.7, 71.0, 97.9, 105.9, 106.1, 108.5, 125.4, 150.3, 156.6, 158.7, 162.7. HRMS-ESI: m/z calculated for C23H37NO4Si [M + H]+: 420.2570; observed: 408.4193.
4-((tert-Butyldimethylsilyl)oxy)-4-(7-diethylamino-2-oxo-2H-chromen-4-yl)butyl (2-(2-chloroacetamido)ethyl)carbamate (12)
-
32
To a solution of 4-(1-((tert-butyldimethylsilyl)oxy)-4-hydroxybutyl)-7-(diethylamino)-2H-chromen-2-one (11, 47 mg, 0.11 mmol) in DCM (3 mL), add 1,1’-cabonyldiimidazole (45 mg, 0.28 mmol).
-
33
Stir at room temperature for 3 h.
-
34
Dilute the reaction mixture by adding more DCM (2 mL).
-
35
Wash the solution with water (5 mL) and brine (5 mL).
-
36
Dry the organic layer over anhydrous sodium sulfate (2 g), and filter.
-
37
Concentrate the filtrate under reduced pressure.
-
38
Confirm the crude product by 1H NMR and use in the next step without further purification.
-
39
Dissolve the activated alcohol in DCM (2.5 mL).
-
40
Add ethylenediamine (16 μL, 0.25 mmol).
-
41
Stir at room temperature for 2 h.
-
42
Remove the solvent and excess ethylenediamine under reduced pressure.
-
43
Confirm the crude product by 1H NMR and use in the next step without further purification.
-
44
Dissolve the amine residue in DCM (2.0 mL).
-
45
Add N,N-diisopropylethyl amine (8 μL, 0.05 mmol) and stir at room temperature for 10 minutes.
-
46
Cool down to 0°C.
-
47
Add 2-chloroacetyl chloride (16 μL, 0.20 mmol) dropwise.
-
48
Stir at 0°C for 10 minutes.
-
49
Quench the reaction with saturated aqueous sodium bicarbonate (2 mL).
-
50
Extract the aqueous layer with DCM (3 × 2 mL).
-
51
Wash the combined organic layers with brine (2 mL).
-
52
Dry the organic layer over anhydrous sodium sulfate (3 g), and filter.
-
53
Concentrate the filtrate under reduced pressure.
-
54
Purify the crude product by silica gel chromatography eluting with hexanes/ethyl acetate (1:1) to afford 4-((tert-Butyldimethylsilyl)oxy)-4-(7-diethylamino-2-oxo-2H-chromen-4-yl)butyl (2-(2-chloroacetamido)ethyl)carbamate (12) as a yellow foam (42 mg, 65%). Characterization properties: 1H NMR (400 MHz, CDCl3): δ = −0.04 (s, 3H) 0.08 (s, 3H), 0.91 (s, 9H), 1.18–1.22 (t, J = 7.2 Hz, 6H), 1.71–1.80 (m, 4H), 3.35–3.43 (m, 8H), 4.03 (s, 2H), 4.05–4.06 (m, 2H), 4.89 (m, 1H), 5.04 (m, 1H), 6.17 (s, 1H), 6.50–6.51 (d, J = 2.8 Hz, 1H), 6.54–6.57 (dd, Ja = 8.8 Hz, Jb = 2.4 Hz, 1H), 7.13 (br, 1H), 7.44–7.47 (d, J = 8.8 Hz, 1H). 13C NMR (400 MHz, CDCl3): δ = −5.0, −4.5, 12.6, 18.3, 24.9, 25.9, 29.8, 34.6, 40.5, 40.8, 42.6, 44.8, 64.9, 70.8, 97.9, 105.9, 106.1, 108.4, 125.3, 150.4, 156.7, 157.4, 158.4, 162.7, 166.9. HRMS-ESI: m/z calculated for C28H44ClN3O6Si [M + H]+: 582.2766; observed: 582.2761.
4-(7-Diethylamino-2-oxo-2H-chromen-4-yl)-4-hydroxybutyl (2-(2-chloroacetamido)ethyl) carbamate (13)
-
55
To a solution of 4-((tert-butyldimethylsilyl)oxy)-4-(7-diethylamino-2-oxo-2H-chromen-4-yl)butyl (2-(2-chloroacetamido)ethyl)carbamate (12, 38 mg, 0.06 mmol) in THF (1.5 mL), add TBAF (1 M in THF, 0.1 mL, 0.1 mmol) dropwise.
-
56
Stir at room temperature for 1 h.
-
57
Concentrate under reduced pressure.
-
58
Purify the crude product by silica gel chromatography eluting with chloroform/acetone (1:1) to afford 4-(7-diethylamino-2-oxo-2H-chromen-4-yl)-4-hydroxybutyl (2-(2-chloroacetamido)ethyl) carbamate (13) as a yellow foam (13 mg, 44%). Characterization properties: 1H NMR (400 MHz, CDCl3): δ = 1.17–1.21 (t, J = 7.2 Hz, 6H), 1.80–1.90 (m, 4H), 3.34–3.45 (m, 9H), 4.03 (s, 2H), 4.10–4.13 (m, 2H), 5.01 (m, 1H), 5.44 (m, 1H), 6.25 (s, 1H), 6.45–6.46 (d, J = 2.0 Hz, 1H), 6.54–6.57 (dd, Ja = 9.2 Hz, Jb = 2.4 Hz, 1H), 7.21 (br, 1H), 7.37–7.39 (d, J = 9.2 Hz, 1H). 13C NMR (400 MHz, CDCl3): δ = 12.6, 25.3, 33.7, 40.6, 40.7, 42.7, 44.8, 64.9, 69.3, 97.9, 105.1, 106.2, 108.7, 125.1, 150.5, 156.5, 157.5, 158.9, 163.1, 167.2. HRMS-ESI: m/z calculated for C22H30ClN3O6 [M + H]+: 468.1901; observed: 468.1870.
4-(7-Diethylamino-2-oxo-2H-chromen-4-yl)-4-((((2,5-dioxopyrrolidin-1-yl)oxy)carbonyl) oxy)butyl (2-(2-chloroacetamido)ethyl)carbamate (6)
-
59
Dissolve 4-(7-Diethylamino-2-oxo-2H-chromen-4-yl)-4-hydroxybutyl (2-(2-chloroacetamido)ethyl) carbamate (13, 13 mg, 0.03 mmol) in DCM (500 μL).
-
60
Add a catalytic amount of 4-dimethylaminopyridine (10 mol%, 0.3 mg).
-
61
Add N,N’-disuccinimidyl carbonate (36 mg, 0.14 mmol).
-
62
Stir reaction mixture at room temperature overnight.
-
63
Concentrate under reduced pressure.
-
64
Purify the crude product by silica gel chromatography eluting with DCM/acetone (2:1) to afford 4-(7-diethylamino-2-oxo-2H-chromen-4-yl)-4-((((2,5-dioxopyrrolidin-1-yl)oxy)carbonyl) oxy)butyl (2-(2-chloroacetamido)ethyl)carbamate (6) as a yellow solid (15 mg, 82%). Characterization properties: 1H NMR (400 MHz, CDCl3): δ = 1.19–1.22 (t, J = 7.2 Hz, 6H), 1.72–1.86 (m, 2H), 2.10–2.14 (m, 2H), 2.84 (s, 4H), 3.35–3.42 (m, 8H), 4.03 (s, 2H), 4.15–4.18 (t, J = 7.2 Hz, 2H), 5.43 (m, 1H), 5.96–5.99 (t, J = 6.0 Hz, 1H), 6.15 (s, 1H), 6.51–6.52 (d, J = 2.8 Hz, 1H), 6.59–6.63 (dd, Ja = 9.2 Hz, Jb = 2.8 Hz, 1H), 7.18 (br, 1H), 7.33–7.35 (d, J = 9.2 Hz, 1H). 13C NMR (400 MHz, CDCl3): δ = 12.5, 14.3, 24.3, 25.5, 25.6, 31.9, 40.3, 42.6, 44.9, 64.5, 78.3, 98.2, 105.1, 109.4, 124.6, 150.9, 151.2, 151.6, 156.7, 157.4, 161.8, 167.3, 172.2. HRMS-ESI: m/z calculated for C27H33ClN4O10 [M + H]+: 609.1963; observed: 609.1990.
2.3.1.2. Preparation of the NB Linker 5.
The photocleavable NB-linker can be synthesized from commercially available 1-(2-nitrophenyl)ethane-1,2-diol (14) in eight steps (Figure 5), and detailed procedures for the synthetic steps have been previously reported (Yamazoe et al., 2012). Briefly, the primary hydroxyl of 14 was tosylated and substituted with methylamine to obtain the amino alcohol 15. It was then reacted with methyl adipoyl chloride to get the methyl ester 16. The secondary alcohol of 16 was subsequently activated with 1,1-carbonyldiimidazole and converted to a primary amine with the treatment of ethylenediamine. The chloroacetamide moiety was then installed by capping this amine with 2-chloroacetyl chloride to yield 17. Finally, base-catalyzed hydrolysis and activation with N,N’-disuccinimidyl carbonate provided the fully functionalized NB linker 5.
Figure 5. Synthesis of the NB linker.
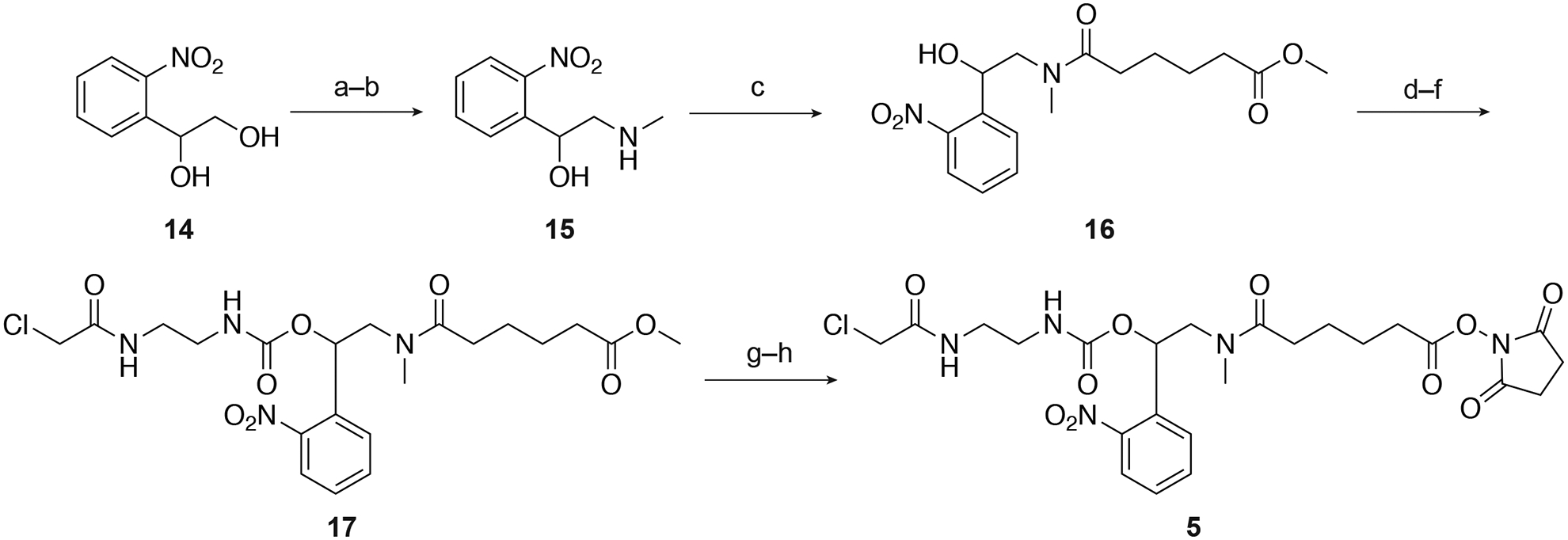
a) TsCl, pyridine, 99%; b) methylamine,THF, 91%; c) methyladipoyl chloride, DIPEA, CH2Cl2, 41%; d) CDI,CH2Cl2; e) ethylenediamine, CH2Cl2; f) 2-chloroacetyl chloride, Et3N,CH2Cl2, 65% over 3 steps; g) LiOH, THF, H2O; h) DSC, pyridine,CH3CN, 62% over 2 steps. Ts = toluene-sulfonyl, DIPEA = N,N-diisopropylethylamine. Adapted with permission from Yamazoe et al., 2014.
2.3.2. Synthesis and purification of cMOs
Materials and instruments
Custom MO oligomers with 5’-primary amine and 3’-disulfide amide modifications were purchased from Gene Tools, LLC.
Reaction buffers: 0.1 M Na2B4O7 (pH 8.5), 0.1 M Tris-HCl buffer (pH 8.4).
A vortex mixer, a Speedvac, a sonicator, and a benchtop centrifuge.
Illustra NAP-5 columns (GE Healthcare, Catalog number – 17-0853-01).
Pierce™ Filter Spin Cups (ThermoFisher, Catalog number – 69700).
Pierce™ Immobilized TCEP Disulfide Reducing Gel (ThermoFisher, Catalog number – 77712).
SulfoLink™ Coupling Resin (ThermoFisher, Catalog number – 20401).
A Nanodrop™ UV-Vis spectrophotometer.
2.3.2.1. General procedure for the cMO preparation
Dissolve the lyophilized, 25-base MO oligomer with 5’-primary amine and 3’-disulfide amide modifications (50.0 nmol) in 0.1 M Na2B4O7 (pH 8.5, 100 μL) buffer in an Eppendorf tube. Sonicate to dissolve if required.
Add a solution of the photocleavable linker 5 or 6 (172 nmol) in 15 uL DMSO to the above MO solution.
Cover with aluminum foil and shake overnight using a vortex mixer.
Purify the linker-MO conjugate from the unreacted linker and salts using a NAP-5 column. To do so, equilibrate the NAP-5 column three times with 5 mL water each time. Dilute the reaction mixture to 500 μL with water and load onto the column. Elute the MO with water and collect 500 μL fractions in Eppendorf tubes. Check fractions to have absorbance at 260 nm and combine. Generally, the first 1 mL contains the MO.
Concentrate to 200 μL using a Speedvac.
It’s important to eliminate all the unreacted linker impurities to avoid side reaction in the subsequent step. Since Step - 4 may not remove the linker completely, acidify with 2 μL AcOH to protonate any hydrolyzed linker. Then wash the aqueous layer with CHCl3 (3 × 200 μL) and EtOAc (3 × 200 μL) to remove the linker. Care should be taken not to remove the opaque interface of the solvents.
Neutralize with 10% NH4OH and confirm the pH ≥ 8.0.
Speedvac to dryness and dissolve in 200 μL water. To calculate the yield using a Nanodrop spectrophotometer, dilute 1 μL of the MO solution in 9 μL 0.1 N HCl and measure the absorption at 260 nm, following MO-manufacturer’s instructions. Typical amide-coupling yields for the MO-linker intermediates are 71–78%.
Use mass spectrometry to confirm the product identity and quality. Dissolve 0.5 ng of the MO-conjugate in 10 μL HPLC grade water and record the mass spec on an ESI LC-MS instrument. The multicharged peaks can be deconvoluted using MaxEnt 1™ software (Waters Corporation).
Remove water using a Speedvac and dissolve in 0.1 M Tris-HCl buffer (pH 8.4, 100 μL).
Wash the TCEP reducing gel (100 μL of the 50% slurry) with the same Tris-HCl buffer (3 × 300 μL) using a Pierce™ Filter Spin Cup.
Add the MO-linker intermediate solution to the TCEP-resin and shake overnight in dark.
Centrifuge at 13000 g for 30 seconds to collect the filtrate.
Wash the gel slurry with 0.1 M Tris-HCl buffer (pH 8.4, 3 × 100 μL), collect by centrifugation and combine with the filtrate.
Concentrate to 200 μL using a Speedvac.
Wash Sulfolink Coupling Resin (200 μL of the 50% slurry) with 0.1 M Tris-HCl buffer (pH 8.4, 400 μL) in a Pierce™ Filter Spin Cup.
Add the MO solution to the resin and shake for 15 min and then left upright without shaking for 30 min.
Centrifuge at 13000 g for 30 seconds to collect the filtrate.
Wash the gel slurry with 0.1 M Tris-HCl buffer (pH 8.4, 3 × 100 μL), collect by centrifugation and combine with the filtrate.
Remove the salts from the cyclic cMO using a NAP-5 column following procedures described above.
Speedvac to dryness, dissolve in 200 μL water, quantify and calculate the yield using a Nanodrop spectrophotometer. Typical thiol-halogen reaction yields are 39–71%.
Use mass spectrometry to confirm the final cyclic cMO identity and quality as described before.
3. Microinjection of cMOs into zebrafish embryos
Microinjection is an effective and common way to introduce reagents into zebrafish embryos. The MOs or cMOs were microinjected either into the animal cell or the yolk, following previously described standard procedures (Ekker, 2004). Injection solutions were prepared in cell-culture-grade water containing 100 mM KCl and 0.1% (w/v) phenol red. Prior to injection, the solutions were heated to 100°C for 30 seconds to dissociate any MO aggregates, and were centrifuged (13000 g, 1 min) to remove undissolved particles. One- to four-cell stage zebrafish embryos were arrayed in an agarose template and submerged in E3 medium for microinjection. Typically, 1–2 nL solutions were injected either into the animal cell or the yolk prior to the 4-cell stage, and the embryos were subsequently cultured in E3 medium at 28.5°C.
Accurate microinjection dosing of cMOs is essential to achieve their optimum dynamic ranges. We recommend performing a careful titration with the 25-base linear MO to find the minimum required dose, as described for the tbxta hairpin cMO in our published paper (Ouyang et al., 2009). The cMO injection solutions should be stored in air-tight O-ring-capped microcentrifuge tubes to avoid concentration changes due to evaporation. We also recommend careful quantification of the injection volume by the mineral oil droplet method (Nüsslein-Volhard C, 2002). Care should be taken to minimize exposure to ambient light or light from the microinjection stereoscope, especially while working with the DEACM linker. To minimize photobleaching, the stereoscope light in the bottom can be covered with UV or blue absorbing plastic such as 3M™ Sun Control window film (3M USA Ltd. Catalog number – PR50).
4. Photoactivation of cMOs
The cMOs can be uncaged with the irradiation of appropriate wavelengths of light. The light should also have enough intensity to effectively complete the photochemical reaction within seconds with minimal photodamage. As described earlier, to uncage NB cMOs we illuminate with 365-nm light, which in our hands only partially uncage DEACM cMOs. In contrast, 470-nm light illumination is used to uncage DEACM cMOs. Our set-up for the light irradiation is shown in Figure 6. For global uncaging, zebrafish embryos were arranged in an agarose microinjection template (560 μm × 960 μm) that contain evenly spaced individual wells (Figure 6A–B). The illumination was performed with a mercury short arc lamp (Osram HBO™ 103 W) using a Leica DM4500B upright compound epifluorescence microscope equipped with a 20X water-immersion objective (Leica 506147, HCX APO L 20X/0.5 NA). To activate NB-cMOs, individual embryos were irradiated with 365-nm light for 10 seconds using a narrow-band DAPI filter cube (Ex: 365 nm, 10-nm bandpass; Chroma). Similarly, DEACM-cMOs were activated with 470-nm light using a GFP filter cube (Ex: 470 nm, 40-nm bandpass; Leica) for 30 seconds. The light intensities were measured using a digital energy meter (Thor Labs PM100D) and microscope slide sensor (Thor Labs S170C). Both light beams create a spot with 2-mm diameter and the light intensities were as follows: 365-nm filter, 41 mW/cm2; 470 nm filter, 64 mW/cm2.
Figure 6. Photoactivation of cMOs.
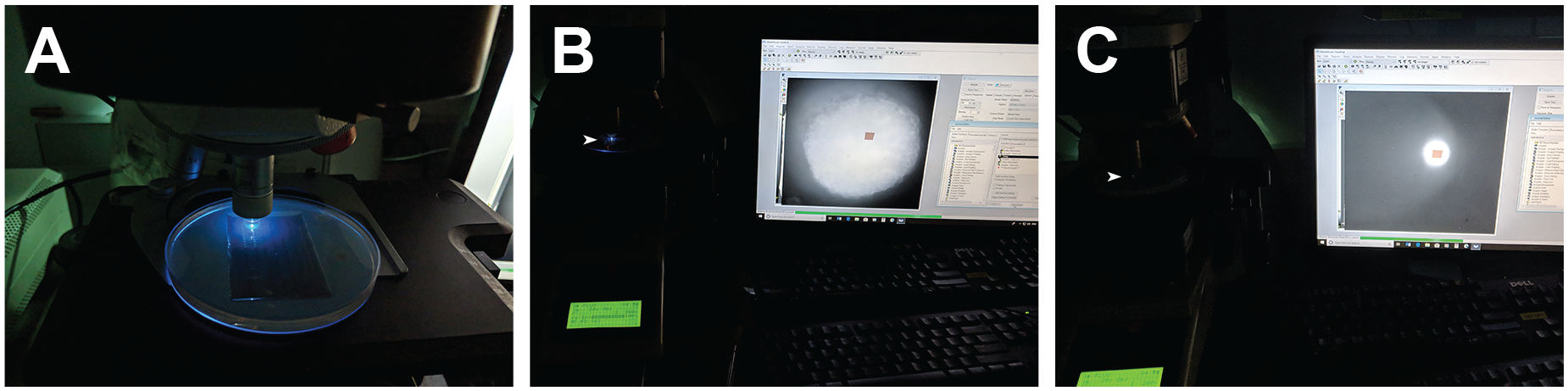
A) Irradiation set-up for zebrafish embryos using an agarose template in a Leica DM4500B upright fluorescence microscope equipped with a 20×, 0.5 NA water-immersion objective. B) Global irradiation of an embryo controlled by Metamorph™ software. Full illumination of animal cells is shown. C) Local irradiation of an embryo using similar set-up. Localized illumination of animal cells is shown. The illumination light beam has been indicated by the arrow-head.
In the microinjection template, we recommend orienting the chorionated embryos such that the animal cells face upward and remain directly in the path of the light beam. While unlikely, in mis-oriented embryos, the light (especially UV) may not pass the top cell layers/yolk to reach the bottom cells, resulting inefficient uncaging. To avoid exposure to scattered light from another irradiation, the embryos can be placed in alternate wells.
The above set-up can also be used for localized activation of cMOs by reducing the size of the illumination field diaphragm (Figure 6C). To further trace the uncaged cells, a photoactivatable fluorescent tracer such as caged fluorescein dextran can be co-injected with cMOs, and the targeted cells can be isolated with fluorescence activated cell sorting for further analysis (Payumo et al., 2016; Shestopalov et al., 2012).
5. Conclusion
The protocols described in this chapter for wavelength-selective cyclic cMO design and synthesis can likely be extended to other genes and organisms. cMOs can complement genome-editing technologies like TALEN (transcription activator-like effector nucleases) and CRISPR (clustered regularly interspaced short palindromic repeats) that have been successfully used in zebrafish (Ata, Clark, & Ekker, 2016). However, these genetic technologies often require raising multiple animal generation to obtain homozygous mutants, and are subject to Mendelian phenotypic distributions. Current genome editing technologies also have limited capability in spatiotemporal control. In comparison, cyclic cMOs can be synthesized and implemented within days and can have 100% phenotypic penetration. These synthetic reagents are thus valuable research tools, allowing rapid loss-of-function studies with precise spatiotemporal control. Moreover, the wavelength-selective cMOs discussed in this chapter permit combinatorial regulation of multiple genes, extending the applicability of these tools in studying more complex biological network.
While our studies demonstrate that multiple and spectrally distinct synthetic reagents can have useful applications for in vivo studies, further advances could expand their experimental scope. Current cMO technologies suffer from narrow dynamic ranges, and the simultaneous use of multiple cMOs could be associated with significant basal activities. For example, while the tbxta and tbx16 cMOs are effective individually, their combined dark activities precluded their combinatorial use in studying medial floor plate development in zebrafish (Payumo, Walker, McQuade, Yamazoe, & Chen, 2015). This is likely because these two genes are regulated reciprocally (Griffin, Amacher, Kimmel, & Kimelman, 1998), and their mild basal activities have synergistic effects on the mesoderm development. New cMO designs may be necessary to overcome this limitation. Additionally, as described in the Section 2.2, overlapping spectral features of the NB and DEACM groups can have some influence on the non-orthogonal activation of the two cMOs, and more red-shifted chromophores maybe necessary to uncover the full potential of this approach. As novel photolabile chromophores continue to be developed(Gandioso et al., 2017; Gorka, Nani, Zhu, Mackem, & Schnermann, 2014; Hansen, Velema, Lerch, Szymanski, & Feringa, 2015; Jean-Gerard, Vasseur, Scherninski, & Andrioletti, 2018), we anticipate that wavelength-selective optochemical tools will increasingly provide insights into complex biological processes that cannot be readily achieved through other means.
Acknowledgements
We gratefully acknowledge support from NIH grants R01 GM087292 (JKC), R35 GM127030 (JKC), and R01 GM112728 (AD).
References
- Ata H, Clark KJ, & Ekker SC (2016). The zebrafish genome editing toolkit. Methods Cell Biol, 135, 149–170. [DOI] [PubMed] [Google Scholar]
- Bhadra J, Pattanayak S, & Sinha S (2015). Synthesis of Morpholino Monomers, Chlorophosphoramidate Monomers, and Solid-Phase Synthesis of Short Morpholino Oligomers. Curr Protoc Nucleic Acid Chem, 62, 4.65.61–26. [DOI] [PubMed] [Google Scholar]
- Bill BR, Petzold AM, Clark KJ, Schimmenti LA, & Ekker SC (2009). A primer for morpholino use in zebrafish. Zebrafish, 6(1), 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey DR, & Abrams JM (2001). Morpholino antisense oligonucleotides: tools for investigating vertebrate development. Genome Biol, 2(5), reviews1015.1011–reviews1015.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiters A, Garner RA, Lusic H, Govan JM, Dush M, Nascone-Yoder NM, & Yoder JA (2010). Photocaged morpholino oligomers for the light-regulation of gene function in zebrafish and Xenopus embryos. J Am Chem Soc, 132(44), 15644–15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker SC (2004). Nonconventional antisense in zebrafish for functional genomics applications. Methods Cell Biol, 77, 121–136. [DOI] [PubMed] [Google Scholar]
- Ekker SC, & Larson JD (2001). Morphant technology in model developmental systems. Genesis, 30(3), 89–93. [DOI] [PubMed] [Google Scholar]
- Gandioso A, Contreras S, Melnyk I, Oliva J, Nonell S, Velasco D, … Marchan V (2017). Development of Green/Red-Absorbing Chromophores Based on a Coumarin Scaffold That Are Useful as Caging Groups. J Org Chem, 82(10), 5398–5408. [DOI] [PubMed] [Google Scholar]
- Gorka AP, Nani RR, Zhu J, Mackem S, & Schnermann MJ (2014). A near-IR uncaging strategy based on cyanine photochemistry. J Am Chem Soc, 136(40), 14153–14159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin KJ, Amacher SL, Kimmel CB, & Kimelman D (1998). Molecular identification of spadetail: regulation of zebrafish trunk and tail mesoderm formation by T-box genes. Development, 125(17), 3379–3388. [DOI] [PubMed] [Google Scholar]
- Gutzman JH, Graeden E, Brachmann I, Yamazoe S, Chen JK, & Sive H (2018). Basal constriction during midbrain-hindbrain boundary morphogenesis is mediated by Wnt5b and focal adhesion kinase. Biol Open, 7(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen MJ, Velema WA, Lerch MM, Szymanski W, & Feringa BL (2015). Wavelength-selective cleavage of photoprotecting groups: strategies and applications in dynamic systems. Chem Soc Rev, 44(11), 3358–3377. [DOI] [PubMed] [Google Scholar]
- Housden BE, Muhar M, Gemberling M, Gersbach CA, Stainier DY, Seydoux G, … Perrimon N (2017). Loss-of-function genetic tools for animal models: cross-species and cross-platform differences. Nat Rev Genet, 18(1), 24–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean-Gerard L, Vasseur W, Scherninski F, & Andrioletti B (2018). Recent advances in the synthesis of [a]-benzo-fused BODIPY fluorophores. Chem Commun (Camb), 54(92), 12914–12929. [DOI] [PubMed] [Google Scholar]
- Liu Q, & Deiters A (2014). Optochemical control of deoxyoligonucleotide function via a nucleobase-caging approach. Acc Chem Res, 47(1), 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A, & Ekker SC (2000). Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet, 26(2), 216–220. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, D. R (2002). Zebrafish, a practical approach: Oxford University Press, Oxford. [Google Scholar]
- Ouyang X, Shestopalov IA, Sinha S, Zheng G, Pitt CL, Li WH, … Chen JK (2009). Versatile synthesis and rational design of caged morpholinos. J Am Chem Soc, 131(37), 13255–13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payumo AY, McQuade LE, Walker WJ, Yamazoe S, & Chen JK (2016). Tbx16 regulates hox gene activation in mesodermal progenitor cells. Nat Chem Biol, 12(9), 694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payumo AY, Walker WJ, McQuade LE, Yamazoe S, & Chen JK (2015). Optochemical dissection of T-box gene-dependent medial floor plate development. ACS Chem Biol, 10(6), 1466–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N, Pitsouli C, & Shilo BZ (2012). Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb Perspect Biol, 4(8), a005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramstein J, & Lavery R (1988). Energetic coupling between DNA bending and base pair opening. Proc Natl Acad Sci U S A, 85(19), 7231–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestopalov IA, Pitt CL, & Chen JK (2012). Spatiotemporal resolution of the Ntla transcriptome in axial mesoderm development. Nat Chem Biol, 8(3), 270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestopalov IA, Sinha S, & Chen JK (2007). Light-controlled gene silencing in zebrafish embryos. Nat Chem Biol, 3(10), 650–651. [DOI] [PubMed] [Google Scholar]
- Summerton J, & Weller D (1997). Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev, 7(3), 187–195. [DOI] [PubMed] [Google Scholar]
- Tallafuss A, Gibson D, Morcos P, Li Y, Seredick S, Eisen J, & Washbourne P (2012). Turning gene function ON and OFF using sense and antisense photo-morpholinos in zebrafish. Development, 139(9), 1691–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme-Laragy AR, Karchner SI, & Hahn ME (2012). Gene knockdown by morpholino-modified oligonucleotides in the zebrafish (Danio rerio) model: applications for developmental toxicology. Methods Mol Biol, 889, 51–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini AJ, Schuler AD, Zebala JA, & Mayer AN (2009). PhotoMorphs: a novel light-activated reagent for controlling gene expression in zebrafish. Genesis, 47(11), 736–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazoe S, Liu Q, McQuade LE, Deiters A, & Chen JK (2014). Sequential gene silencing using wavelength-selective caged morpholino oligonucleotides. Angew Chem Int Ed Engl, 53(38), 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazoe S, Shestopalov IA, Provost E, Leach SD, & Chen JK (2012). Cyclic caged morpholinos: conformationally gated probes of embryonic gene function. Angew Chem Int Ed Engl, 51(28), 6908–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]


