SUMMARY
The somatotopic motor-neuron projections onto their cognate target muscles are essential for coordinated movement, but how that occurs for facial motor circuits, which have critical roles in respiratory and interactive behaviors, is poorly understood. We report extensive molecular heterogeneity in developing facial motor neurons in the mouse and identify markers of subnuclei and the motor pools innervating specific facial muscles. Facial subnuclei differentiate during migration to the ventral hindbrain, where neurons with progressively later birth dates—and evolutionarily more recent functions—settle in more-lateral positions. One subpopulation marker, ETV1, determines both positional and target muscle identity for neurons of the dorsolateral (DL) subnucleus. In Etv1 mutants, many markers of DL differentiation are lost, and individual motor pools project indifferently to their own and neighboring muscle targets. The resulting aberrant activation patterns are reminiscent of the facial synkinesis observed in humans after facial nerve injury.
Graphical Abstract
In Brief
Tenney et al. demonstrate that embryonic facial motor neurons are transcriptionally diverse as they establish somatotopic innervation of the facial muscles, a process that requires the transcription factor ETV1. Facial-motor axon-targeting errors in Etv1 mutants cause coordination of whisking and eyeblink evocative of human blepharospasm.
INTRODUCTION
The development of neuronal circuits requires highly organized connections between discrete neuronal populations and their synaptic targets. The functional consequences of developmental or maladaptive miswiring can be severe. However, despite much interest in this question, there are few cases in vertebrates for which we understand how cell identity drives innervation of the correct target and avoidance of neighboring alternatives (Bonanomi, 2019).
Facial muscles have a vital role in many complex behaviors. Distributed across the surface of the skull and neck in ~30 pairs, their nuanced activation by branches of the facial motor nerve (cranial nerve VII) underlies respiration and feeding in fish (Gorlick, 1989), rhythmic whisking of sensory vibrissae in rodents (Hill et al., 2008), and communication through facial expression and spoken language in humans. Loss of facial nerve function because of injury or congenital conditions, such as Moebius syndrome (Terzis and Anesti, 2011), can lead to socially debilitating facial paralysis. Moreover, facial motor fibers regenerating after nerve injury can innervate incorrect muscles, resulting in facial synkinesis, an inappropriate coordination of facial movement, such as eye closure, triggered by volitional movement of the mouth. Although some cases can be treated by selective chemodenervation (Husseman and Mehta, 2008), synkinesis remains a significant clinical challenge, and the molecular mechanisms underlying these wiring abnormalities have been little studied.
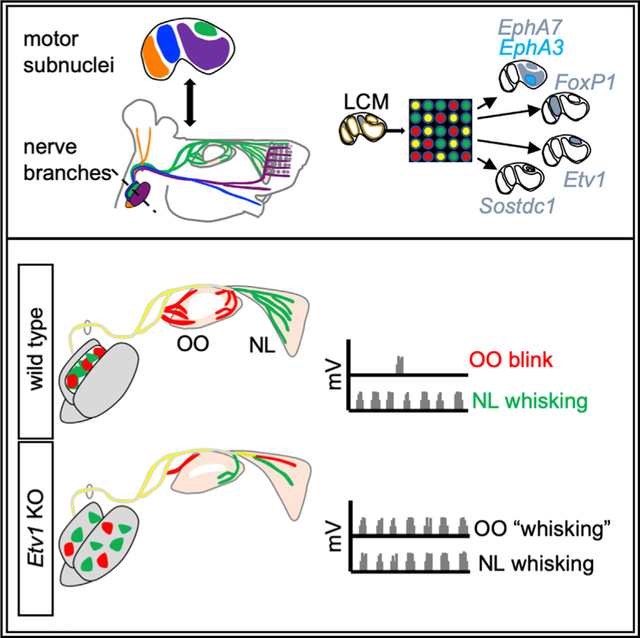
The muscles of the facial expression are controlled by a population of hindbrain branchiomotor neurons forming the facial motor nucleus, which is structured according to the diversity of its muscle targets (Cattaneo and Pavesi, 2014). Mammalian facial motor neuron (FMN) cell bodies are organized into distinct subnuclei: medial (M), intermediate (I), dorsolateral (DL), and lateral (L) (Figures 1A and 1B) (Baisden et al., 1987; Papez, 1927; Semba and Egger, 1986). FMN axons form five distinct branches of the facial nerve that innervate separate subsets of muscles. In the mouse, the posterior auricular (PA) and anterior auricular (AA) branches supply the muscles that rotate the ear pinnae (Ashwell and Watson, 1983), the zygomatic/temporal (Z/T) branch controls the eyelid-closing orbicularis oculi (OO) and extrinsic whisking nasolabialis (NL) muscles (Shaw and Baker, 1985), the buccolabial (BL) branch innervates intrinsic muscles of the lip and sensory vibrissae (Baisden et al., 1987; Hinrichsen and Watson, 1984), the marginal mandibular (MM) branch contacts the muscles that move the lower lip (Semba and Egger, 1986), and the cervical (C) branch innervates muscles of the lower jaw (Martin and Lodge, 1977) (Figure 1C). This musculature is thought to have evolved from the jaw- and gill slit-opening muscles of primitive aquatic tetrapods, with sometimes extensive remodeling (Baisden et al., 1987; Guest et al., 2018; Hinrichsen and Watson, 1984) to support adaptations, such as eyelid-closing muscles in terrestrial animals, somatosensory whisking in mammals, and facial expression in humans (Diogo et al., 2008; Grant et al., 2012). The adaptation of the relative sizes of facial subnuclei to specific evolutionary needs in different mammalian species (Furutani and Sugita, 2008) suggests they may serve as organizing centers for facial nerve branches, and multiple studies indicate that individual facial branches map to specific subnuclei to varying extents (Baisden et al., 1987; Courville, 1966; Han et al., 2018; Martin and Lodge, 1977; Papez, 1927; Semba and Egger, 1986; Uemura-Sumi et al., 1986; Wang-Bennett and Coker, 1990). However, a comprehensive map of facial nucleus musculotopic organization in mouse is lacking.
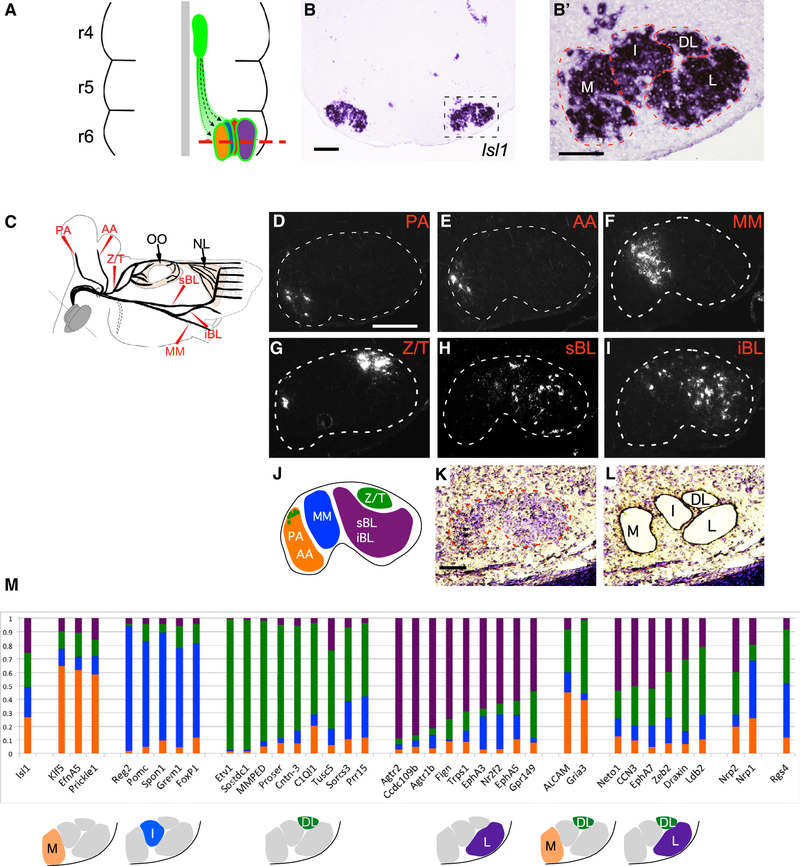
Figure 1. Molecular Heterogeneity among Somatotopic Facial Motor Subnuclei.
(A) Schematic of facial-motor nucleus development in the embryonic mouse hindbrain. Facial motor neurons (FMN) emerge from the rhombomere 4 (r4) midline germinal zone (green oval) and migrate caudally and laterally (dotted lines) to form somatotopic subnuclei in ventral r6. Red dashed line marks the plane of section for (B) and (B′).
(B and B′) In situ hybridization (ISH) for Isl1 in a transverse-hindbrain section through the facial nucleus at E16.5. Dashed box denotes position of high-power image (B′). Dashed lines in (B′) outline approximate anatomic borders of the medial (M), intermediate (I), dorsolateral (DL), and lateral (L) facial motor subnuclei.
(C) Schematic indicating the positions at which the posterior auricular (PA) and anterior auricular (AA), marginal mandibular (MM), zygomatic/temporal (Z/T), superior buccolabial (sBL) and inferior buccolabial (iBL) nerves were transected and marked with the retrograde rhodamine-dextran (Rh-Dex) tracer. Eyelid-closing orbicularis oculi (OO) and extrinsic whisking nasolabialis (NL) muscles are indicated. Dotted region represents the cervical (C) facial-nerve branch that was not included in the study
(D–I) Coronal sections through P2 mouse hindbrains after retrograde Rh-Dx labeling of the PA (D), AA (E), MM (F), Z/T (G), sBL (H), and iBL (I) facial nerve branches as described in (C). Dotted line denotes facial nucleus border.
(J) Schematic summary of the distribution of FMNs contributing to the indicated facial nerve branches.
(K and L) Cresyl violet-stained coronal section through the E16.5 wild-type facial nucleus (dashed line) before (K) and after (L) laser-capture microdissection (LCM) to isolate M, I, DL, and L facial subnuclei.
(M) Predicted subnucleus-specific enrichment of select markers identified in microarray screen. (Top) Histograms show genes for which a microarray probe showed approximately 40% or more total normalized expression enriched in a single subnucleus, color coded to represent the normalized expression in each of the subnuclei. (Bottom) Diagrams show predicted gene-expression patterns.
Scale bars: 200 μm in (B) and (D)–(I) and 100 μm in (B’) and (K).
See also Table S1 and Figures S1 and S6.
To identify candidate regulators of the late stages of facial motor map development, we transcriptionally profiled individual facial subnuclei from 16.5 embryonic days (E16.5) mice and found that each subpopulation was defined by a unique combinatorial gene-expression program. The molecular identity of a given subnucleus correlates with neuronal birth date, whereas spatial aggregation occurs through stereotyped patterns of migration toward, and within, the developing facial nucleus. We studied in more detail the role of the Ets transcription factor ETV1—already shown to label a subset of FMNs (Alfonsi et al., 2008)—in the development of FMNs of the DL subnucleus and the motor pools innervating the OO and NL muscles. ETV1 is required for correct positioning of each DL motor pool and for complete innervation of its target muscle. Moreover, it activates downstream expression of a series of candidate effectors. As a striking functional correlate of this role, we show that loss of Etv1 generates synkinesis between eyelid and nasal muscles. We, therefore, propose that ETV1 acts as a regulator of late aspects of facial motor-neuron development, driving both the growth of their axons and the segregation of their cell bodies in distinct subnuclei, required for non-overlapping innervation of their targets.
RESULTS
Somatotopic Projections of Facial Subnuclei in Mouse
We used retrograde labeling in post-natal day (P)-zero mice to map the subnuclei of origin for each facial nerve branch (Figure 1C). We observed that the PA and AA branches originated exclusively in the ventral and dorsal regions of the M subnucleus, respectively (Figures 1D, 1E, S1A, and S1B). The MM branch arose solely from the I subnucleus (Figures 1F and S1C). The Z/T branch was formed by FMNs of the DL subnucleus as well as a few in the dorsal M subnucleus (Figures 1G and S1D) (Semba and Egger, 1986). Lastly, the buccolabial branch arose primarily from the L subnucleus, with a smaller contribution from the I subnucleus (Figures 1H, 1I, S1E, and S1F). These observations, summarized in Figure 1J, provide a firm basis on which to link FMN identity to somatotopic innervation of target muscles.
Transcriptional Profiling Identifies Markers of Developing Facial Motor Subnuclei and Motor Pools
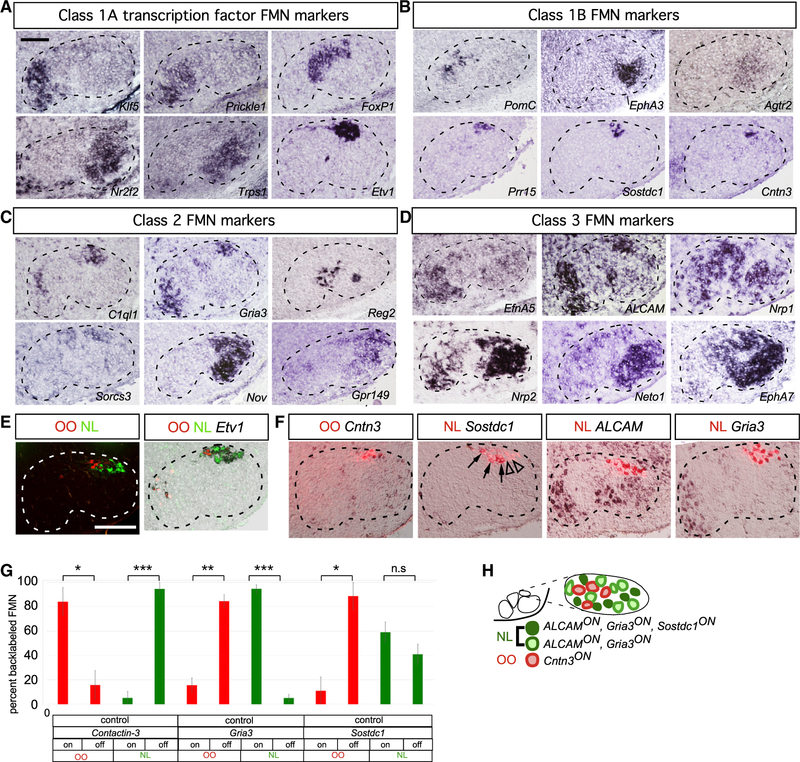
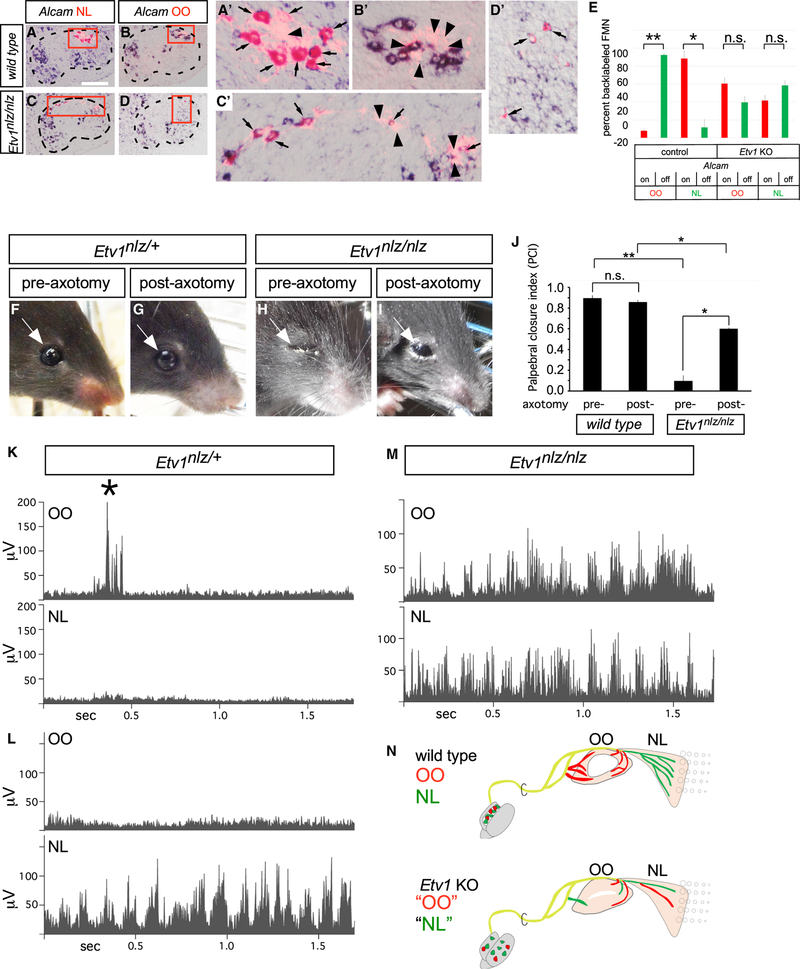
To identify potential molecular correlates of FMN cell-body position, we used laser-capture microdissection to isolate M, I, DL, and L subnuclei from mouse hindbrain cryosections at embryonic day E16.5 (Figures 1K and 1L), a time point at which facial subnuclei can be distinguished anatomically (Figure 1B’) but may still retain expression of genes regulating FMN migration and target innervation (Song et al., 2006) (Figures 3A–3F and 6A). Microarray analysis of the individual facial motor subnuclei revealed a striking level of molecular heterogeneity among the four subpopulations, with 58–172 transcripts showing >4-fold enrichment in pairwise comparisons among these populations (Figure 1M; Table S1). We used in situ hybridization (ISH) to confirm the microarray data and map the markers with increased resolution (Figures 2 and S1G–S1P). We identified three groups of expression patterns of graded selectivity (Figures 2A–2D): class 3 genes, expressed in three or more FMN subnuclei; class 2 genes, which marked two different FMN subnuclei; and class 1 genes, whose expression was restricted to a single FMN subnucleus. We classified the last group into two subgroups. Class 1A genes labeled entire subnuclei (Figure 2A), suggesting a potential role determining subnucleus identity. In accordance with that, several class 1A genes are transcription factors whose expression is most pronounced in the M (Klf5, Prickle1), I (FoxP1), and L (Nr2f2, Trps1) or DL (Etv1) subnucleus. Class 1B genes showed even more restricted expression, limited to a subset of neurons within a single subnucleus only (Figure 2B), raising the possibility that they might be markers for specific facial motor pools. To test that idea, we injected the two muscles controlled by the DL facial subnucleus, the OO and NL, with Alexa 488- and Alexa 597-conjugated cholera toxin B subunit (A488CTB, A597CTB), respectively. Retrogradely labeled OO and NL FMNs formed two distinct groups within the Etv1ON DL subnucleus, partially segregated along the mediolateral axis (Figure 2E). Single cholera toxin B (CTB) injections, followed by ISH, showed that the class 1B DL marker Cntn3 defined the OO motor pool, whereas Sostdc1 specifically marked a subset of the NL motor-pool neurons. Within the DL, class 2 marker Gria3 and class 3 marker Alcam were also restricted to the NL pool (Figures 2F, 2G, and 7E). Therefore, similar to their spinal counterparts (Dalla Torre di Sanguinetto et al., 2008), FMNs are organized into distinct motor pools defined by combinatorial expression of marker genes (Figure 2H).
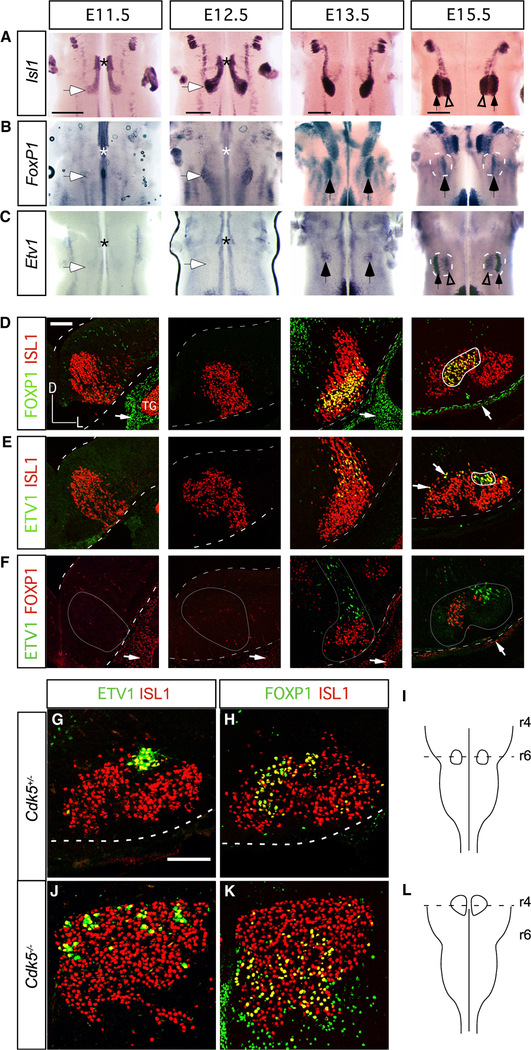
Figure 3. FMN Molecular Identity and Nerve-Branching Pattern Are Maintained in the Absence of Caudal Migration.
(A–C) Time course of Isl1 (A), FoxP1 (B), and Etv1 (C) expression in the developing hindbrain. Whole mount ISH, hindbrain ventral views. Asterisks indicate the r4 FMN germinal zone; white-filled arrows mark the final FMN settling position at E11.5 and E12.5; black-filled arrows indicate FoxP1-expressing I (B) and Etv1-expressing DL (C) subnuclei at E13.5 and E15.5; open arrowheads in (A) and (C) mark a subpopulation of Etv1-expressing FMNs occupying M positions within the E15.5 facial subnucleus.
(D–F) Immunofluorescence staining for FOXP1/ISLl (D), ETV1/ISL1 (E), and ETV1/FOXP1 (F) on coronal cryosections through r5/6 (E11.5 and E12.5) and r6 (E13.5 and E15.5). Dashed white lines outline the dorsal and ventral surfaces of the hindbrain; thick solid white lines in (D) and (E) outline DL and I facial motor subnuclei, respectively; thin gray lines in (F) outline the FMN migratory stream and nascent facial nucleus. Representative images, n = 3 embryos per time point.
(G–L) ETV1 (G and J) and FOXP1 (H and K) expression in control (G and H) and Cdk5−/−(J and K) FMNs at E16.5. Coronal planes of section indicated in (I) and (L), n = 1 Cdk5−/−, n = 1 wild-type littermate control.
Scale bars: 500 μm in (A)–(C) and 100 μm in (D)–(F), (G), (H), (J), and (K).
See also Figure S2.
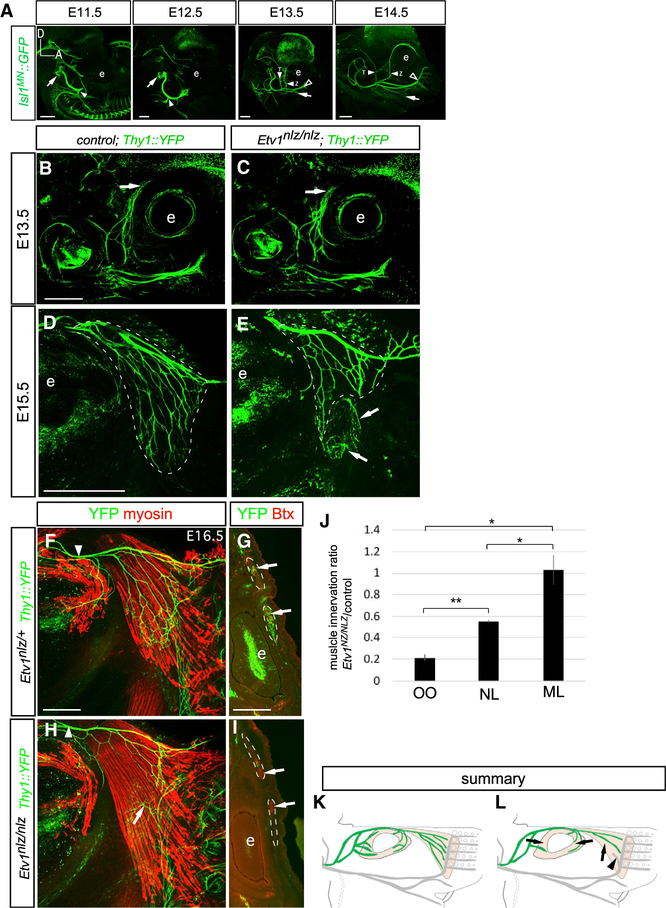
Figure 6. Attenuation of DL Facial Motor Subnucleus Target Muscle Innervation in Etv1-Mutant Mice.
(A) Time course of embryonic facial nerve development from E11.5 to E15.5. GFP immunostaining in embryonic Isl1MN::GFP mice. Arrows at E11.5 and E12.5 indicate the exit of the facial nerve from the pons. The ramification of the auricular (filled arrowheads at E11.5 and E12.5), zygomatic/temporal (filled arrowheads at E13.5 and E14.5), buccolabial (open arrowheads at E13.5 and E14.5), and marginal mandibular branches (arrows at E13.5 and E14.5) are indicated. Eye is marked with “e”; dorsal (D) and anterior (A) directions are indicated.
(B–E) Z/T axon growth in wild-type (B and D) and Etv1 mutant (C and E) embryos at E13.5 (B and C) and E15.5 (D and E). Z/T branch growth is unchanged in Etv1 mutants at E13.5 (B and C), but attenuation of NL innervation is apparent by E15.5 (dotted lines in D and E). Arrows in (E) mark ectopic innervation of the NL by BL nerve-branch fibers.
(F and H) GFP (green) and myosin (red) immunostaining on whole-mount NL muscles from control (F) and Etv1−/−(H) E16.5 Thy1::YFP embryos. NL innervation remains attenuated, arrow in (H) marks the ectopic invasion of the ventral NL by the BL nerve-branch fibers. No obvious change in Z/T facial nerve-branch caliber was observed in Etv1-mutant embryos, arrows in (B) and (C) and arrowheads in (F) and (H).
(G and I) GFP immunofluorescence (green) and Alexa555 bungarotoxin (Btx, red) staining on coronal cross sections through nasal OO from control (G) and Etv1-mutant (I) Thy1::YFP mice at E16.5. OO innervation is reduced in Etv1 mutants; dotted lines denote OO muscle, arrows label regions of OO innervation in the control muscle that is absent from the Etv1 mutant.
(J) NL and OO muscle innervation is reduced in Etv1-mutant mice, represented as the means of the ratio of muscle innervation detected in Etv1nlz/nlz compared with littermate controls. Innervation of the maxillolabialis (ML) muscle supplied by the BL nerve branch is unchanged. n = 3 embryos per genotype (unpaired t test, *p < 0.05, **p < 0.005). Samples were cleared for imaging using iDISCO (A) or BABB (B–I); see Method Details.
(K and L) Summary of reduced innervation of the NL and OO muscles (shaded tan) by Z/T facial-nerve fibers (green) in Etv1 mutants. Arrowhead in (L) indicates BL fibers from the L subnucleus mistargeted to the NL.
Scale bars: 500 μm in (A)–(E), 250 μm in (F) and (H), and 500 μm in (G) and (I).
See also Figure S5.
Figure 2. Unique, Combinatorial Gene Expression Programs Define Facial Motor Subnuclei and Pools.
(A–F) ISH for facial subnucleus markers in coronal cryosections at E16.5 (A–D) and P3 (E and F). Dashed lines in (A)–(F) outline the approximate boundary of the facial nucleus.
(A) Class 1A microarray hits defining distinct facial subnuclei.
(B) Class 1B microarray hits marking subpopulations within a single subnucleus.
(C) Class 2 microarray hits defining two subnuclei or subpopulations therein.
(D) Class 3 microarray hits marking FMNs of three or more facial subnuclei.
(E) Simultaneous injection of retrograde Alexa 555- and Alexa 647-conjugated cholera toxin B (A555CTB and A647CTB) labels into OO and NL muscles, respectively, marking distinct motor pools within the Etv1ON DL facial-motor subnucleus. Representative images.
(F) Single CTB injection followed by ISH identifies Cntn3 as an OO motor pool marker, whereas Sostdc1, ALCAM, and Gria3 define the NL pool. Sostdc1ON (arrows) and Sostdc1OFF (arrowheads) NL FMNs are indicated.
(G) Specificity of motor pool markers in (F) represented as mean ± SE percentage of FMNs marked by retrograde muscle labeling expressing the indicated motor-pool markers. n = 3 mice per muscle injection, (*p < 0.05, **p < 0.005, ***p < 0.0005, unpaired t test).
(H) Summary of OO and NL motor-pool molecular identities.
Scale bars: 100 μm in (A)–(D) and 200 μm (E) and (F).
See also Figures S1, S2, and S6.
Figure 7. Near Randomization of DL Facial Motor Axon Targeting in Etv1 Mutant Mice.
(A–D) Facial motor nucleus cryosections from neonatal wild-type (A and B) and Etv1 mutant (C and D) mice showing retrograde Rh-Dx labeling from NL and OO motor fibers (red) overlaid with Alcam expression (blue). Boxes denote positions of high-power images (A’–D’). Selectivity of Alcam expression in NL (A’, arrows) and its exclusion from OO (B’, arrowheads) FMNs is lost in Etv1 mutants, with many AlcamOFF FMNs innervating the NL (arrowheads in C’) and AlcamON FMNs innervating the OO (arrows in D’). Arrow head in (A’) marks a likely AlcamOFF NL FMN.
(E) Coincidence of Alcam NL pool-marker expression in OO or NL FMNs, as represented by the percentage (± SE) of FMNs marked by retrograde muscle labeling that express Alcam. The specificity of NL innervation by AlcamON FMNs in control mice is lost in Etv1 mutants. FMNs marked by OO and NL retrograde labeling were scored for colocalization of Alcam in control and age-matched Etv1nlz/nlz mice. Data represented as the percentage (± SE) of FMNs marked by retrograde label and Alcam expression, n = 3 mice per genotype, 52 NL and 47 OO FMNs from control mice and 98 NL and 47 OO FMNs from Etv1-mutant mice (unpaired t test, *p < 0.05, **p < 0.0005). Scale bar: 200 μm.
(F–I) Eyelid behavior in control (F and G) and Etv1-mutant (H and I) mice at P21 before (F and H) and after (G and I) Z/T facial nerve-branch axotomy. OO axotomy partially rescued tonic eyelid closure in Etv1 mutants, whereas the dimensions of the palpebral fissure remained unchanged in controls.
(J) Quantitation of axotomy-induced changes in palpebral fissure dimensions. Values represented as PCI means ± SE, n = 4 mice per genotype (unpaired t test, *p < 0.001, **p = 0.0001).
(K–M) Electromyographic (EMG) recordings from the OO and NL muscles of control (K and L) and Etv1-mutant (M) mice at P20. OO muscle did not contract during exploratory whisking in control mice (K) but in Etv1-mutant mice, the OO was activated simultaneously and synchronously with the NL during bouts of exploratory whisking (M). n = 8 control, 6 mutant mice.
(N) Diagram of changes in peripheral facial-motor circuitry leading to OO/NL synkinesis in Etv1 mutants.
See also Figure S7.
ETV1 and FOXP1 Predefine Specific Subnuclei during Ventral Migration and Segregation of FMNs
We next studied the developmental steps that lead to precise segregation of FMN subsets. As visualized by whole-mount ISH for Isl1 between E11.5 and E15.5 (Figure 3A), the developmental migration of FMNs encompasses three main phases: caudal migration from rhombomere (r) 4 to r6, ventral (radial) migration within r6 to form the facial nucleus (Garel et al., 2000; Rossel et al., 2005), and segregation within the facial nucleus to form the subnuclei. Differential gene expression within the caudal migratory stream has been reported (Song et al., 2006), but the stage at which the distinct, terminal identities of FMNs are determined, and how they segregate on the way to their final location, has not, to our knowledge, been studied. We used FoxP1 and Etv1 as markers of specific FMN populations to address those questions, applying ISH on intact brainstems to visualize their entire expression pattern at once. Neither gene was expressed in FMNs during the initial caudal phase of migration (Figures 3B and 3C). A subset of FoxP1-expressing (FoxP1ON) FMNs was first detected at E12.5 during ventral migration and by E15.5 had congregated into an anatomically distinct I subnucleus (Figure 3B). Etv1 expression was not apparent until E13.5, when it also marked a scattered subpopulation of FMNs (Figure 3C). By E15.5, most Etv1ON FMNs had coalesced into a distinct DL subnucleus (Figure 3C, arrows) with some Etv1ON FMNs forming a sparse, more-M band of cells that occupy dorsal positions in the M and I subnuclei (Figures 2A and arrowheads in 3C). Expression of the class 3 marker Nrp2 and the class 2 marker Zeb2 also displayed subpopulation specificity within the migratory stream at E13.5, which presaged the distribution of expression observed when FMNs reached their final somatotopic positions at E16 (Figures S1G–S1J).
We used immunolabeling on E11.5–E15.5 sections to determine how the ETV1ON and FOXP1ON populations emerged and segregated (Figures 3D–3F). No FMNs co-expressed ETV1 and FOXP1, and at no point did we observe intermingling (Figure 3F). Moreover, each group projected only to its cognate target; 94.0 ± 2.5% of FMNs labeled by Z/T branch retrograde tracing at P1 were ETV1-immunopositive and located within the DL subnucleus (Figure S2A; n = 3 control mice, 143 FMNs). By early postnatal stages, FOXP1 expression expanded to FMNs beyond the I subnucleus (Figure S2B), but FMNs labeled from the MM branch remained, nonetheless, positioned as the FOXP1ON I population observed at E15.5–E16.5 (Figures 2A and 3F). To determine whether migration itself is required for the onset of expression of each marker, we analyzed cyclin-dependent kinase 5 (Cdk5) knockout mice, in which FMNs are born normally but fail to migrate caudally from r4 (Ohshima et al., 2002). In E16.5 Cdk5 mutants, the ETV1ON DL and FOXP1ON I FMNs did not form subnuclei and, instead, were dispersed within the dorsal and ventral compartments, respectively, of the r4 FMN population (Figures 3G–3L), but nevertheless, the facial nerve exited the skull and formed branches (Figures S2F–S2H). Thus, the migration of FMNs from r4 to r6 does not appear to be required for expression of Etv1 and FoxP1 in FMN subpopulations at later stages.
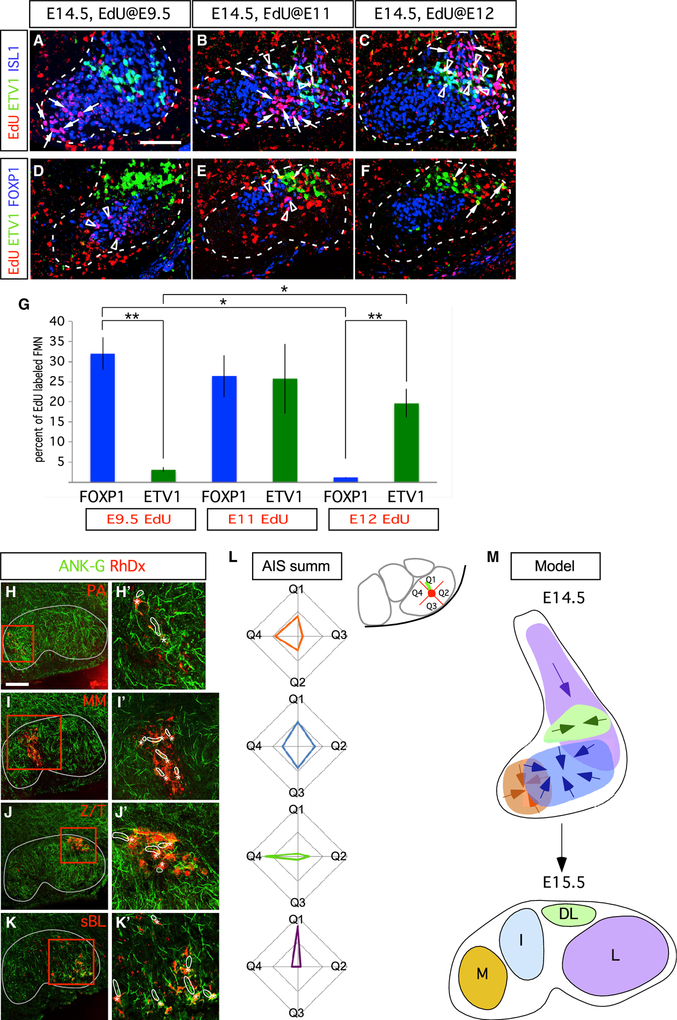
To determine the relation of FMN birth date to terminal identity, we labeled mouse embryos in utero at E9.5, E11, and E12 with the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU). The later the time of injection, the more lateral were the FMN populations labeled at E14.5 (Figures 4A–4F). FOXP1ON I FMNs were born early, between E9.5 and E11, whereas most ETV1ON DL FMNs were born later (E11–E12; Figure 4G).
Figure 4. FMN Transcriptional and Positional Identities Are Linked to Birth Date and Secondary Migration within the Developing Facial Nucleus.
(A–F) FMNs settle in the ventral hindbrain along a mediolateral developmental axis. Time course of EdU birthdate labeling (red) combined with ETV1 (green) and ISL1 (blue) immunofluorescence in (A)–(C), and ETV1 (green) and FOXP1 (blue) immunofluorescence in (D)–(F). EdU administration at E9.5 marks M FMNs, whereas administration at E11 and E12 mark progressively more lateral FMN populations. Arrows indicate EdU+/ISL1ON in (A)–(C) and EdU+/ETV1ON in (D)–(F) cells; triangles show EdU+/ETV1ON/ISL1ON in (A)–(C) and EdU+/FOXP1ON in (D)–(F) cells.
(G) Percentage of EdU+/FOXP1ON and EdU+/ETV1ON detected at E14.5 after EdU delivery at the indicated time points represented as means ± SE. n = 3 embryos per injection time points E9.5 (290 ETV1ON and 1,013 FOXP1ON FMNs), E11 (639 ETV1ON and 859 FOXP1ON FMNs), and E12 (690 ETV1ON and 876 FOXP1ON). Unpaired t test, *p < 0.05, **p < 0.005.
(H–K) Ankyrin-G immunostaining (ANK-G, green) reveals variable axon initial segment (AIS) orientation among perinatal mouse facial subnuclei defined by retrograde Rh-Dx facial-nerve branch labeling (red); thin gray lines outline facial nucleus border. (H’–K’) High-power images of boxed regions; Rh-Dx labeled FMNs (asterisks) and corresponding AISs (outlined by thick white lines) are indicated. n = 3 mice per nerve branch.
(L) Radar plots indicating, for each subnucleus, the proportion of AISs assigned to the indicated directional quadrant bins (Q1–Q4); inset shows example diagram with an L FMN AIS (green) assigned to the Q1 bin. n = 3 mice each for each subnucleus, with 42 M, 65 I, 75 DL, and 80 L FMNs marked by retrograde labeling of the PA/AA, MM, Z/T, and BL nerve branches, respectively.
(M) Model for facial subnucleus formation through secondary migration based on marker expression and AIS orientation vectors. Directional vectors for final steps of secondary migration through which FMNs coalesce into discrete subnuclei are indicated.
Scale bars: 50 μm in (A)–(F) and 200 μm in (H)–(K).
See also Figure S6.
Although it is possible that the shifts in ETV1 and FOXP1 expression reflect temporal changes of gene regulation sweeping through the FMN population, the position of FOXP1ON FMNs ahead of the ETV1ON population in the migratory stream at E13.5 (Figure 3F, third column) suggests that the facial nucleus is built from waves of FMNs with successively later birth dates. In this model, later-born FMNs occupy progressively more-lateral settling positions, consistent with recent analysis of zebrafish FMN development (McArthur and Fetcho, 2017).
The final stage of migration is segregation within the developing facial nucleus. Because the trailing axon deposited by migrating FMNs provides a record of their migratory path (Shaw and Baker, 1985), we used immunostaining for the axon initial segment (AIS) marker ANKYRIN-G to assess proximal axon orientation in facial subnuclei in neonatal control mice (Figures 4H–4K). For a series of neurons in each subnucleus, we plotted the deduced direction of recent migration as a vector and quantified their orientations (Figure 4L). M and I subnucleus axonal vectors displayed a radial distribution, indicating that these structures formed from the clustering of initially dispersed subpopulations, whereas mediolateral and dorsoventral biases were observed for DL and L subnuclei, respectively, implying that they migrate to their final positions via single, but distinct, migratory paths.
Our findings, thus far, indicate that subnucleus-specific gene-expression patterns are activated during migration of FMNs toward the site of the presumptive facial nucleus. Initially, FMNs settle along the mediolateral axis as a function of birth date. Nevertheless, the congregation of these FMNs into coherent subnuclei requires a fine-grained, secondary migration, which leads to precise sorting within the nascent nucleus (Figures 4H–4M). However, it remained to be determined whether any of the class 1 genes used as markers have a functional role in that migration.
Etv1 Assigns Transcriptional and Positional Identity to DL FMNs
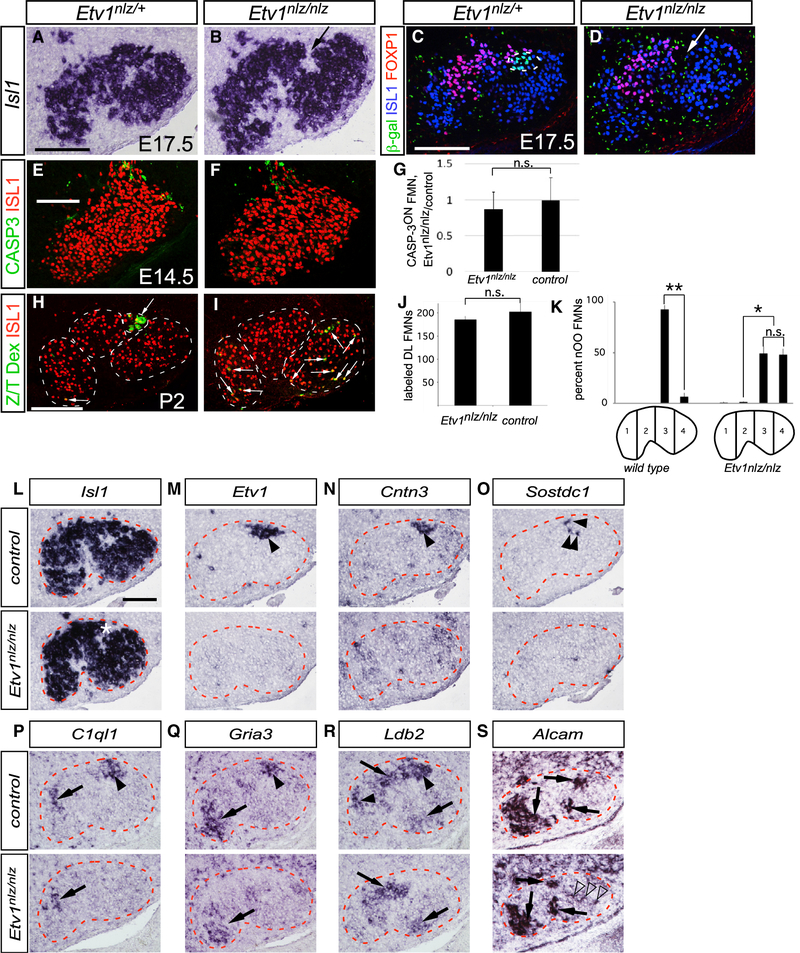
Because Etv1 expression appears in DL FMNs during the final stages of their migration to somatotopic positions and target-muscle selection by the corresponding axons, we focused our attention on its potential role in orchestrating those processes. In Etv1nls-LacZ/nls-LacZ (referred to here as Etvnlz/nlz) mutants (Arber et al., 2000), an anatomically distinct DL subnucleus failed to form and was, instead, replaced by a large cleft devoid of FMNs flanked by apparently enlarged I and L subnuclei (Figures 5B and 5D; arrows). We attempted to visualize the positions of DL FMNs using the β-galactosidase (β-gal) reporter encoded by the Etv1nlz mutant allele. In heterozygous Etv1nlz/+ facial nuclei at E16.5, β-gal expression was coincident with ETV1 immunoreactivity (Figure S3A). However, no β-galON FMNs were detected in Etv1nlz/nlz knockouts (Figure 5D), whereas β-gal expression in ETV1ON dorsal root ganglion and inferior olive neurons was maintained (Figures S3B–S3E). Therefore, either DL FMNs are lost in the absence of Etv1 or ETV1 is required to maintain its own transcription through an autoregulatory module.
Figure 5. Loss of DL Subnucleus FMNs Positional and Transcriptional Identities in Etv1 Mutant Mice.
(A–D) Facial nucleus cytoarchitecture in E17.5 Etv1nlz/+ (A and C) and Etv1nlz/nlz (B and D) embryos as defined by Isl1 ISH (A and B) and ISL1 immunostaining, blue in (C) and (D). An anatomically distinct DL subnucleus is absent from Etv1 mutant embryos, arrows in (B) and (D); and the β-gal reporter marking DL FMNs in Etv1nlz/+ embryos, green in (C), is absent from Etv1-mutant FMNs (D). The location of FOXP1ON I FMNs (red in C and D) is unchanged in Etv1 mutants. Green puncta, in (C) and (D), are non-specific background binding of the anti β-gal antibody.
(E and F) Immunofluorescent detection of cleaved caspase-3 apoptosis marker (CASP3, green) in control (E) and Etv1 mutant (F) ISL1ON (red) FMNs at E14.5.
(G) Ratio of CASP-3ON, ISL1ON to CASP-3OFF, ISL1ON FMNs detected in Etv1nlz/nlz and control E14.5 embryos in (E) and (F) represented as means ± SE, n = 3 Etv1 mutant (208 FMNs) and control (238 FMNs) littermate pairs (unpaired t test, p = 0.7639).
(H and I) Retrograde labeling (green cells, white arrows) of DL FMNs in neonatal control (H) and Etv1nlz/nlz (I) mice with Rh-Dx application to the transected Z/T nerve branch.
(J) Similar numbers of DL FMNs are marked in neonatal control and Etv1nlz/nlz mice, represented as ISL1ON FMNs and Rh-Dx-labeled FMNs means ± standard error. n = 3 mice for each genotype: 139 Etv1 mutant and 152 control DL FMNs (unpaired t test, p = 0.4769).
(K) Rh-Dx-labeled OO FMNs assigned to one of four mediolateral positional bins. OO FMNs are more broadly distributed in Etv1 mutant mice than they are in littermate controls, represented as the percentage of OO FMNs assigned to each bin ± SEM. n = 3 Etv1nlz/nlz and control littermate pairs, 121 and 114 OO FMNs analyzed, respectively, from the two genotypes (unpaired t test, *p < 0.005, **p < 0.0005).
(L–S) Expression of the motor neuron marker Isl1 (L); the class 1 DL markers Etv1 (M), Cntn3 (N), and Sostdc1 (O); class 2 DL markers C1ql1 (P), and Gria3 (Q); and the class 3 DL markers Ldb2 and Alcam in control (upper panels) and Etv1nlz/nlz (lower panels) facial nuclei at E16.5. Black arrowheads in (M)–(R) indicate DL motor-pool markers present in wild type and their absence from Etv1 mutant, facial motor nuclei; arrows in (P)–(S) indicate FMN marker expression maintained in Etv1 mutants. Open arrowheads in (S) mark AlcamON FMNs ectopically occupying the L subnucleus.
Scale bars: 100 μm in (A–D) and (L–S); 50 μm in (E) and (F); and 200 μm in (H) and (I), n ≥ 3 mice for each gene.
See also Figures S3 and S4.
We found no overall change in the total number of FMNs (Figures S4A–S4C) and no increase in programmed cell death as determined by caspase-3 cleavage at E14.5 (Figures 5E–5G) in Etv1 mutants, indicating that DL FMNs are not lost. Furthermore, retrograde labeling from the Z/T nerve at P2 (Figures 5H and 5I) marked similar numbers of FMNs in controls and Etv1 mutants (Figure 5J). However, in mutants, some of these neurons occupied ectopic positions deep within the L subnucleus (Figures 5I, arrows, and 5K). They also showed striking modifications in gene expression patterns: the OO motor-pool marker Contactin-3 and the NL motor-pool marker Sostdc1 were barely detectable at E16.5 and were excluded from OO and NL FMNs in Etv1-mutant neonatal mice (Figures 5N, 5O, S4H, and S4I). Similarly, the class 2 NL motor-pool markers C1ql1 and Gria3 and the class 3 DL marker Ldb2 were absent from the DL and L regions of the mutant facial nucleus but were maintained as expected in the other subnuclei (Figures 5P–5R). By contrast, the class 3 DL marker Alcam was detected in ectopic positions in the L subnucleus of the Etv1 mutants, where it may label mislocalized DL FMNs in an ETV1-independent manner (Figure 5S). The persistence of DL FMNs in mice lacking Etv1 and the requirement of ETV1 for the expression of a battery of secreted molecules and transcription factors—including itself and with the exception of Alcam—underscore the essential role ETV1 has in establishing DL FMNs mature transcriptional identity.
Etv1 Controls the Innervation of Select Facial Muscles
As FMNs migrate, their axons are simultaneously projecting toward their target muscle anlagen. The facial nerve emerges from the developing skull by E11.5 and, over the subsequent 2 days, facial nerve branches become apparent. At E15.5, the pattern of facial nerve-branch formation is established, and the innervation of the facial musculature is underway (Figure 6A; Dörfl, 1985). We asked whether ETV1 has a role in the innervation of DL target muscles. The overall trajectories of the Z/T and other facial nerve branches were unchanged in Etv1 mutants at E13.5 (Figures 6B and 6C). By E15.5, Z/T axons of control mice have grown dorsally over the eye to the nasal side of the eyelid, where a subset of DL axons defasciculate to innervate the OO muscle and the remainder continue to grow into the NL and the smaller nasolabialis superficialis muscle (Figure 6D) (Haidarliu et al., 2010). Strikingly, in Etv1 mutants, the extent of NL innervation by the Z/T was reduced, reaching 50% loss at E16.5 (Figures 6E, 6H, and 6J). The few Z/T fibers that did enter the NL muscle were restricted to its dorsal half, leaving space for ectopic invasion by neighboring buccolabial axons (Figures 6E, 6H, arrows, and 6L, arrowhead). Innervation of the OO muscles in Etv1 mutants was reduced by ~85% compared with controls (Figures 6G, 6I, and 6J). The trajectory of the Z/T branch and its caliber at a point proximal to the OO muscle were unchanged in Etv1 mutants (Figures 6B, 6C, arrows, 6F, and 6H, arrowheads), indicating that the failure to completely innervate the OO and NL is a distal defect. Moreover, innervation of the ML muscle, an extrinsic whisking muscle lying deep to the NL—and innervated by Etv1OFF L subnucleus FMNs (Figure 2B) (Dörfl, 1982; Hill et al., 2008)—was unchanged (Figure 6J). Etv1 expression was largely absent from the facial periphery and sensory ganglia as FMN axons grow toward, and invade, their muscle targets (Figure S5), consistent with ETV1 having a selective and likely cell-autonomous role in driving the full innervation of the DL target muscles.
Loss of Etv1 Function Leads to Facial Synkinesis
Although the innervation of OO and NL muscles in Etv1 mutants is reduced, each muscle nonetheless still appears to receive a detectable level of motor input. We asked to what degree selective projections by each pool onto their appropriate muscle target were maintained in the absence of Etv1, using retrograde labeling in conjunction with ISH for the ETV1-independent NL motor-pool marker Alcam. In control mice, we observed the expected close correlation between Alcam expression and NL muscle innervation (Figures 7A and 7E), whereas almost no Alcam was detected in the OO motor pool (Figures 7B and 7E). By contrast, in Etv1 mutants, ~60% of OO-innervating FMNs expressed Alcam, whereas a similar proportion of NL-innervating FMNs were Alcam-negative as expected for OO FMNs (Figures 7C–7E). Thus, the specificity of axonal projection is severely eroded in Etv1 mutants, in which a large fraction of motor inputs onto the OO muscle originate from FMNs that display the molecular identity of the NL motor pool and vice versa.
A potential behavioral correlate for these axon-targeting errors came from the observation that the eyes of Etv1 mutants remained completely or partially closed at P18 (Figures 7F and 7H). We found an 80% reduction in eye opening in Etv1 mutants compared with control mice (Figure 7J). Despite that, the overall structure of mutant eyelids—including the separation of the upper and lower lids into distinct structures—matched that of controls (Figures 7H and 7I).
Abnormal eyelid closure could result from tonic activation of the OO muscle, as seen in the human disorder of blepharospasm, or from impaired activation of the levator palpebrae superioris (LPS) muscle, the OO antagonist that opens the eye (Boghen, 1996). To directly test the former hypothesis, we unilaterally severed the Z/T branch to denervate the OO. In control mice, this intervention did not affect the palpebral closure index (PCI; palpebral fissure height/width) (Figures 7F, 7G, and 7J). In striking contrast, OO denervation in Etv1 mutants led to a >4-fold improvement in palpebral closure index (Figures 7H–7J), likely reflecting preserved tonic activation of the LPS muscle relieved of aberrant OO antagonism.
To determine directly whether the OO muscle had been aberrantly recruited into the whisking circuit in Etv1 mutants, we recorded electromyographic (EMG) activity from the OO and NL muscles in awake mice at P19 (Pearson et al., 2005). In control animals, infrequent (~0.01 Hz), single bursts of OO activity, coincident with eyelid blinks, were detected (Figure 7K, asterisk) (Blount, 1927), with no corresponding activation of the NL. Conversely, OO activation during whisking bouts was very rare (1/99 epochs; Figure 7L, bottom panel; n = 8 mice). In Etv1 mutants, despite the reduced motor input, the frequency of NL activation during whisking was similar to that of controls (Figure 7M, bottom panel). In striking contrast, Etv1 mutant OO activity was often modulated during epochs of whisking, in phase with the high-frequency (~8 Hz) NL activation (28/39 epochs, 72%, n = 6 mice; Figure 7M, top panel). The high-frequency motor drive ectopically directed onto the OO muscle in Etv1 mutants is, therefore, likely to originate from the NL motor pool (Figures 7N and S7).
These observations, together with misprojection by OO and NL motor pools, support a model in which aberrant targeting of NL motor axons onto the OO muscle causes it to contract improperly at the consistently rhythmic firing rates of whisking (8–15 Hz) and respiration (~3 Hz) (Hill et al., 2008; Moore et al., 2013), leading to an eyelid-closure phenotype reminiscent of synkinesis in human patients.
DISCUSSION
The coordinated activation of facial muscles—which underlies multiple essential behaviors—requires somatotopic connections between FMNs and specific muscle targets with diverse functions. We report extensive molecular heterogeneity among FMNs, which can be mapped to the level of single facial motor pools. We used marker genes to show that birth date, together with stereotyped patterns of caudal migration and cell sorting, helps determine the segregation of FMNs into subnuclei with discrete peripheral-projection areas. One such marker, the Ets transcription factor ETV1, acts as an essential regulator of the specific molecular and positional identity of DL FMNs, as well as the innervation of their muscle targets. In its absence, motor pools that normally project to neighboring muscles with no overlap show randomization of their projections, resulting in aberrant patterns of muscle activation reminiscent of the human condition of facial synkinesis.
Molecular Diversity of Facial Subnuclei and Motor Pools
The patterns of gene expression we found to define FMN subpopulations fell into four main classes. Class 1A genes, such as transcription factors Klf5, FoxP1, and Etv1, are expressed by all neurons in a single subnucleus, suggesting that they have a major role in coordinating the different projections within the corresponding nerve branch (Figures 1J and 2A). In this study, we have begun to characterize the role of Etv1, but other class 1A genes may similarly assign facial motor subnucleus identity. Class 1B genes that label subpopulations within a single subnucleus include markers for specific facial motor pools: Cntn3, expressed only in OO FMNs, and Sostdc1, restricted to a subpopulation of NL FMNs. Many class 2 and class 3 genes also labeled small clusters of FMNs, including Alcam and Gria3, which mark the NL motor pool within the DL subnucleus, suggesting that, like their spinal counterparts (Kanning et al., 2010), each FMN pool has a unique combinatorial, transcriptional identity.
Many of the genes identified by our screen encode cell-surface and secreted molecules that have the potential to guide migration of FMN cell bodies and growth cones (Figure 2). For example, the complementary expression of EfnA5 and of multiple EphA receptors by M and L facial nuclei (Figures 2B and 2D) would allow bidirectional, repulsive signaling to cluster the corresponding axon fibers within the common nerve in a manner that anticipates their branch-specific ramifications (Lee et al., 1991) as found in the optic nerve (Suetterlin and Drescher, 2014). At later stages, highly restricted expression of EphA3 in a ventral group of L FMNs (Figure 2B) could sensitize those fibers to Efna5-repellent ligands expressed in the developing mystacial pad (Visel et al., 2004), thereby attenuating their dorsal growth and guiding them to their ventral target muscles. This would be reminiscent of topographic maps in the visual system in which Eph/ephrin signaling has a central role (Triplett et al., 2012).
Similarities and Differences Compared with Spinal Motor Neuron Development
In the ventral spinal cord, motor neurons (MNs) congregate into discrete clusters, termed motor pools, each of which innervates a single muscle (Romanes, 1964; Shaw and Baker, 1985). A comparable organization has been proposed in the simian facial nucleus (Horta-Júnior et al., 2004). The molecular characterization we report demonstrates the existence of such FMN pools in the mouse (Figure 2F). At limb levels, spinal motor pools are grouped into columels with fixed spatial coordinates (Sürmeli et al., 2011), which may bear similarities to facial subnuclei. Late stages of spinal MN differentiation are controlled by ETV5 in specific brachial motor pools, whereas ETV1 controls targeting of proprioceptive circuits onto skeletal muscle (Arber et al., 2000; Haase et al., 2002; Livet et al., 2002). ETV1 acts at relatively earlier stages in the facial nucleus than it does in the spinal cord, guiding DL subnucleus positioning and projection in a manner reminiscent to that of ETV5 in the brachial spinal cord (Livet et al., 2002). Etv1 expression is also absent from the trigeminal ganglia that supply sensory input to facial tissues, indicating that its role in spinal proprioceptive circuit formation is not recapitulated in the facial motor system (Figures S5I and S5J). However, although the underlying mechanisms of its action may differ, ETV1 is important in establishing motor circuits at both hindbrain and spinal levels. Therefore, there are strong parallels, despite clear differences, between branchiomotor and somatic motor neurons in the mechanisms controlling somatotopic innervation of their target muscles.
Evolutionary Influences on FMN Migration, Subnucleus Formation, and Somatotopy
The developmental logic of the caudal migration of FMNs that occurs in embryonic mammals and fish, but not in embryonic birds and reptiles, has eluded explanation (Gilland and Baker, 2005). Caudal migration may bring the FMNs that mediate respiratory behaviors—gill opening in fish and airway patency in mammals (Gorlick, 1989; Moore et al., 2014)—in closer proximity to caudal hindbrain respiratory centers (McArthur and Fetcho, 2017). However, FMNs in fish and mammals do control facial motor behaviors beyond respiration (Diogo et al., 2008; Gorlick, 1989; Stiassny, 2013). Cell migration could also provide the time and space needed for those cells to “sample” the repertoires of cell-surface receptors displayed by their neighbors and aggregate into somatotopic subnuclei, based on shared receptor expression, a mechanism used in the assembly of spinal motor pools (Demireva et al., 2011). The failure of ETV1ON DL and FOXP1ON I FMNs to cluster into subnuclei when caudal migration is interrupted in Cdk5 mutants is consistent with that model, although the effects of CDK5 loss are likely pleiotropic and could cause an overall, non-specific impairment of cell migratory behavior.
The mediolateral progression we report in the settling of FMN in the developing nucleus may reflect the evolution of tetrapod facial motor specialization (Figure S6). The jaw-opening muscles of primitive aquatic tetrapods are thought to have given rise to mammalian proximal ear, jaw, and neck muscles innervated by the M and I FMNs (Lazar et al., 1992; Northcutt and Bemis, 1993), whereas muscles that open the gill slits are ancestral to more distal mammalian eyelid, nose, and lip muscles innervated by DL and L FMNs (Diogo et al., 2008). Early born mammalian M and I subnuclei may, therefore, represent the vestige of an ancient-core facial nucleus that supported aquatic feeding and respiratory behaviors. DL and L subnuclei would have subsequently arisen to innervate new facial muscles, mediating behaviors essential for terrestrial life (e.g., eyelid closure and somatosensory whisking) by deploying complex, combinatorial, transcriptional, and axon-targeting schemes to accurately innervate increasingly complex anterior facial musculature (Figure S6C) (Grant et al., 2012; Sherwood, 2005).
ETV1 as a Determinant of DL FMN Fate
Facial muscles with widely divergent firing patterns lie in close apposition (Diogo et al., 2008). Therefore, even a low level of miswiring could cause debilitating synkinesis among these small, but functionally vital, muscles. ETV1 is a likely mediator of the gene-expression program required to prevent inaccuracies in DL axon targeting. Surprisingly, we found that the loss of ETV1 did not lead to the constitutively open eyelid associated with OO paresis but, instead, to a closed-eye phenotype caused by tonic OO muscle contraction (Figure 7), a condition that was relieved by OO denervation. We demonstrate that there is near-randomization of the synaptic targeting of DL motor axons in the absence of ETV1 (Figure 7) and, as a result, synkinetic activation of the OO coordinated with high-frequency NL contraction during bouts of active whisking. Together, these observations suggest that a single determinant of facial subnucleus identity may establish specificity even within the groups of neurons whose phenotypes it defines. How may that be controlled? The ectopic activation of OO-innervating FMNs during whisking in Etv1 mutants may reflect the loss of pool specific factors that allow whisking afferents to distinguish NL and OO FMNs. It may also be a secondary effect of the displacement of some OO FMNs into the L subnucleus territory normally occupied by FMNs that innervate intrinsic vibrissal muscles (Figures 5I, 5K, 7D, and S4E) (Klein and Rhoades, 1985; Takatoh et al., 2013). In the spinal cord, MN soma position determines afferent input (Sürmeli et al., 2011), and displaced OO neurons might be the target for inappropriate innervation from nearby relays of cortical whisking command (Takatoh et al., 2013).
“Terminal selector” transcription factors determine the anatomic and functional properties of mature neuronal subpopulations by regulating “terminal effector” genes that, in turn, define the cytoarchitectural, electro-physiological, and functional properties of postmitotic, mature neurons (Hobert, 2008; Serrano-Saiz et al., 2018; Spencer and Deneris, 2017). The expression of terminal selectors is sustained over time by feed-forward, autoregulatory loops, through which, they activate their own promoters (Deneris and Hobert, 2014). Etv1 displays several hallmarks of a candidate terminal selector for DL FMN identity, including subpopulation-specific expression in adult stages (Figures S2C and S2E), transcriptional autoregulation (Figures 5C and 5D), and regulation of a battery of candidate effectors (Figures 5N–5R). Recent chromatin immunoprecipitation sequencing (ChIP-seq) analyses performed on non-neuronal cell types are indeed consistent with direct Etv1 autoregulation and ETV1 binding to possible regulatory regions of the candidate effectors Sostdc1 and Ldb2 (Zheng et al., 2019). In a striking parallel, ETV1 also serves as a terminal selector in mouse olfactory bulb neurons, where it regulates dopaminergic gene expression (Flames and Hobert, 2009). It remains to be determined whether the low level of ETV1 maintained in postnatal DL FMNs is required for DL functional identity at adult stages and, if so, whether any of the ETV1-dependent class 1 DL subnucleus markers identified in our screen function postnatally as terminal effectors.
The gene regulatory mechanisms that lie upstream of the initial expression of class 1A genes such as Etv1 remain to be defined. In the spinal cord, motor pool-specific expression of Etv1 and Etv5 depends on signals from the periphery (Haase et al., 2002; Livet et al., 2002; Wang and Scott, 2004). In a striking parallel, LIFRβ signaling is required for the formation of a single DL subnucleus by ETV1-expressing FMNs (Alfonsi et al., 2008). The effectors that guide the architectural and physiological characteristics of DL FMNs also remain to be identified. Our screen identified several candidates (Figures 5N–5R). Paralogs of the OO motor pool marker Cntn3 function in cell adhesion and axon guidance (Mohebiany et al., 2014), and CNTN3 would be positioned to detect eyelid-specific guidance ligands. The NL pool marker and glutamate receptor Gria3 may function to support the physiological demand of high-frequency whisking and respiratory activation of these FMNs. The Wnt signaling antagonist and NL pool marker Sostdc1 could modulate NL motor axon response to peripheral Wnt signals expressed in the region of the nasal eyelid choice point (Lintern et al., 2009; Liu et al., 2003).
In conclusion, our findings begin to assign logic to a remarkable molecular diversity of developing facial motor-neuron subpopulations. We demonstrate that ETV1 has a critical role in defining the final transcriptional and functional identity of dorsolateral facial motor neurons. The loss of facial somatotopy and coordinated motor control in the absence of ETV1 underscores how critically dependent motor function is on the neuronal positioning and axon targeting established during development. In the future, it may be possible to manipulate those same pathways for the treatment of facial nerve injury and disease.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Requests for further information should be directed to Alan P. Tenney (alantenney@alumni.harvard.edu). Reagents were obtained from commercial sources or donating laboratories as indicated. This study did not generate new unique reagents.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
The following previously published mouse strains were used in this study: Etv1 (former Er81)nlslacZ (RRID: MGI:3621019) (Arber et al., 2000), Thy1::YFP Line 16 (RRID: IMSR_JAX:003709) (Feng et al., 2000), Cdk5−/−(RRID: IMSR_JAX:003536) (Ohshima et al., 2002), Isl1MN::GFP (RRID: IMSR_JAX:017952) (Lewcock et al., 2007). Male and female mice were both used depending on availability and were maintained in microisolator cages. All comparisons were made between experimental mice and littermate controls. For timed developmental studies, detection of a vaginal plug at 9AM was considered embryonic day 0.5. All experiments and procedures were performed according to NIH guidelines and approved by the Institutional Animal Care and Use Committee of Columbia University, NIH, or Boston Children’s Hospital.
METHOD DETAILS
Retrograde tracing and axotomy
In order to label specific facial nerve branches with a retrograde tracer, facial motor nerve and intramuscular branches of cryoanesthetized neonatal Thy1::YFP mice were visualized on an Olympus dissecting fluorescent microscope, severed with fine forceps or a tungsten needle, respectively, and 0.3 μl of lysine fixable tetramethylrhodamine-dextran (Rh-Dx; 10,000MW, Thermofisher) was applied to the proximal nerve stumps. Individual NL and OO motor pools were also labeled singly and doubly through intramuscular injection of Alexa Fluor-conjugated choleratoxin B (Alexa-CTB). Mice were perfused with 4% paraformaldehyde/0.1 M phosphate buffer (4%PFA/PB) 24 h post injection, hindbrains post-fixed in 4%PFA/PB overnight, cryoprotected in 30% sucrose/PB, and embedded in optimal cutting temperature compound (OCT, Tissue-Tek) for storage and cryosectioning. For behavioral studies, the Z/T facial nerve branches of P19 Thy1::YFP mice were visualized and severed as described above under isofluorane anesthesia.
Laser Capture Microdissection and Microarray analysis
12 μm-thick cryosections were collected from E16.5 C57/Bl6 mouse hindbrains fresh frozen in OCT and mounted on RNase-free, PEN-foil covered glass slides (Zeiss), followed by dehydration and staining with 1% Cresyl Violet prior to LCM using PALM Micro-beam system (Zeiss). Every 5th section was collected on a glass reference slide and stained with guinea pig anti-ISL1/2 primary (gift from Tom Jessell, Columbia University) and horse radish peroxidase (HRP)-conjugated anti-guinea pig secondary (Sigma-Aldrich) antibodies. Antibody binding was visualized with a 3,3′-Diaminobenzidine tetrahydrochloride (DAB) chromogenic HRP stain (Sigma-Aldrich) to confirm the range of sections containing FMNs. M, I, DL and L subnuclei from sets of 3 hindbrains were collected directly into lysis buffer, pooled, processed for RNA extraction (Absolutely RNA, NanoPrep kit, Stratagene, La Jolla, CA). RNA integrity number (RIN) was assessed on the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Approximately 1.5ng of purified RNA was amplified in the WT-Ovation Pico RNA Amplification System (Nugen) and hybridized to Affymetrix GeneChip Mouse Genome 430 2.0 Arrays (Kaplan et al., 2014).
In situ Hybridization (ISH)
ISH protocol was followed as previously described on cryosections (Schaeren-Wiemers and Gerfin-Moser, 1993) and whole mount embryonic hindbrains (Garcès et al., 2000) at the indicated developmental stages. Riboprobe templates were generated from cDNA clones or through PCR amplification using excess subnucleus-specific cDNA generated for the microarray as template (see also Table S2). For gene expression studies on hindbrain cryosections, Isl1 expression in near-adjacent sections was used to determine the approximate boundaries of the facial motor nucleus.
Immunostaining
Embryonic mouse heads were prepared and sectioned as described in Retrograde tracing and axotomy above. Cryosections were blocked for immunofluorescent staining (2% goat serum in phosphate buffered saline [PBS] supplemented with 0.1% Triton X-100 [PBST]) and incubated overnight at 4°C with the following primary antibodies diluted in blocking buffer: rabbit anti-ETV1 (RRID: AB_2617167) 1:4000, guinea pig anti-FOXP1 (RRID: AB_2665444) 1:1000, guinea pig anti-ISL1/2 (RRID: AB_2631974) 1:1000 (kind gifts of Tom Jessell), rabbit anti-FOXP1 (RRID: AB_732428) 1:1000 (Abcam), rabbit anti-GFP (ThermoFisher Cat#A11122, RRID: AB_221569) 1:2000 (sections), 1:500 (whole mount), chick anti-GFP (ThermoFisher Cat#A10262, RRID: AB_2534023) 1:1000, rabbit anti-cleaved Caspase-3 (Cell Signaling Technologies Cat#9661S, RRID: AB_2341188) 1:1000, rabbit anti-Ankyrin G (Santa Cruz Cat# sc-31776, RRID: AB_633908) 1:250, mouse anti-myosin, fast (Sigma Aldrich Cat# M1570–200UL, RRID: AB_2147168) 1:500; rabbit anti-neurofilament (EMD Millipore Cat# AB1987, RRID: AB_91201), 1:500. Slides were washed in PBST and incubated for two h at room temperature with Alexa Fluor 488-, 568- and 647-conjugated secondary antibodies diluted 1:1000 in blocking buffer. Alexa Fluor 594 alpha-bungarotoxin (Life Technologies) was included in some instances to label facial muscles in cross section. Slides were coverslipped in Fluoromount G (SouthernBiotech) and imaged on a LSM 5 Pascal confocal microscope (Zeiss). To examine the overlap of FMN subpopulation identity and gene expression, facial nerve branches and muscles of neonatal mice were labeled with retrograde tracers and hindbrains processed and cryosectioned as described above. Cryosections were air dried, imaged for the accumulation of fluorescent label, and processed for chromogenic ISH detection. Images documenting the retrograde fluorescent labeling were superimposed on corresponding ISH images in Photoshop and transparency of the fluorescent retrograde label image was adjusted to approximately 50%. Size and position of the fluorescent retrograde label images were aligned with the ISH images based on common, preserved sample contours and anatomic landmarks.
Whole-mount immunostaining of wild-type E11.5-E15.5 mouse heads was performed using the iDISCO method (Renier et al., 2016). Briefly, embryos and heads were fixed overnight in 4% paraformaldehyde, dehydrated through a methanol series, bleached overnight in 5% H2O2 in methanol, rehydrated through a methanol series, and washed in PBS with 2% Triton X-100. Samples were then permeabilized and blocked (two days each process) and incubated with primary antibody diluted in a solution of 2% Tween-20 and 0.01mg/ml heparin in PBS (PTwH) supplemented with 5% DMSO and 3% donkey serum for 7 days. Following one day of washes using PTwH, samples were placed in secondary antibody diluted in a solution of PTwH with 3% donkey serum for 7 days. Stained tissue was washed in PTwH, dehydrated through a methanol series, and incubated in a solution of dichloromethane and methanol. Samples were cleared in dibenzyl ether, mounted on coverslips, and imaged on a Zeiss LSM 700 series laser scanning confocal microscope using Zen Software (Carl Zeiss MicroImaging GmbH, Gottingen, Germany) and manipulated in three dimensions using Imaris software (Bitplane, Zurich, Switzerland).
Examination of proximal axon orientation
Facial motor subnuclei of wild-type Thy1::YFP mice were marked with retrograde Rh-Dx tracer as described above. Coronal facial motor nucleus cryosections (50 μm thick) were stained for Ankyrin G immunofluorescence. Quantitation was restricted to FMNs for which Ankyrin G labeling terminated unambiguously in a discrete neuron marked by dextran labeling and GFP expression. AIS directional vectors were determined, superimposed on a common origin in PowerPoint, and assigned to one of four bins corresponding to dorsal, lateral, ventral, and medial quadrants (Molofsky et al., 2014).
FMN birth-dating
Timed pregnant mice were treated with 5-ethynyl-2′-deoxyuridine (EdU, 50 mg/kg, approximately 200 μl of 5 mg/ml EdU in phosphate buffered saline [PBS]) (Salic and Mitchison, 2008) by interperitoneal injection. Embryos were harvested at E14.5 and fixed for cryosectioning as described above. Cryosections (20 μm) were stained with antibodies against ISL1 and either ETV1 or FOXP1 and corresponding Alexa Fluor secondary antibodies as described above. Following two washes with PBS, slides were permeabilized in PBS supplemented with 0.2% Triton X-100 for 30 min and then washed twice in PBS. For each slide, 215 μL of PBS was mixed with 10 μl of 0.1M CuSO4 and 0.5 μl of Alexa 555-azide while in a second tube 22.5 μL of water was mixed with 2.5 μl ascorbic acid (0.5 M). The two components were mixed by pipetting, applied to the slide and incubated for 30 min at room temperature with light protection. Slides were washed twice in PBS, incubated with 1.4 μM 4’,6-diamidino-2-phenylindole (DAPI) to mark nuclei, washed twice in PBS, mounted, and imaged on a Zeiss confocal microscope. FMNs detected in individual optical sections were scored as EdU-positive if the Alexa-azide staining coincided with approximately 50% or more of the neuronal nucleus defined by ISL1 immunoreactivity.
FMN quantification
Hindbrain cryosections (25 μm) from E16.5 Etv1 mutant and control littermates were stained for rabbit anti-ISL1 immunofluorescence and DAPI as above. Nuclei that co-stained for ISL1 and DAPI were counted in confocal stack images with IMAGEJ (NIH). The minimum sample size required to detect the possible loss of approximately 600 DL FMNs in the Etv1 mutant mice was determined using 2 sample, 1-sided power analysis assuming 80% power and an error rate of 5% using the observed average FMN numbers and standard deviations.
Analysis of OO FMN settling position
OO FMNs were identified by retrograde Rh-Dex labeling of cut motor fibers supplying the nasal OO muscle in neonatal control and Etv1 mutant mice. Coronal cryosections through the facial nucleus were imaged for ISL1 immunofluorescence and the distribution of Rh-Dex. To quantify the mediolateral distribution of labeled OO FMNs, the facial nucleus was divided along the mediolateral axis into four bins of equal width and the percentage of labeled FMNs occupying each bin in control and Etv1 mutants was determined.
Analysis of facial muscle innervation
Heads were collected from control and Etv1 mutant E16.5 embryos (both in a Thy1::YFP Line 16 background), skinned and stained with rabbit anti-GFP and mouse anti-fast myosin (MY32) antibodies as described (Huber et al., 2005). Briefly, embryonic heads were fixed and dehydrated as described above, incubated overnight in Dent’s bleach (1 part H2O2, 2 parts Dent’s fix) and in Dent’s fix (1 part DMSO, 4 parts methanol). Samples were then incubated in primary antibodies diluted in blocking solution (5% goat serum, 20% DMSO in PBS) for 3 days at room temperature, followed by similarly diluted secondary antibodies. Samples were washed in PBS, dehydrated through a methanol series, cleared in 2:1 parts benzoic acid:benzyle benzoate (BABB), and mounted on a coverslip. Confocal Z series were collected with microscope settings that placed the anti-GFP signal from the Z/T branch dorsal to the eye at saturation. Maximum projections of the myosin immunostaining defined the region of interest (ROI) encompassing the NL muscle, and the maximum projection of the GFP immunostaining was uniformly contrast enhanced, made binary, and the GFP signal within the NL ROI was quantified using ImageJ image analysis software (NIH). To quantify OO and ML innervation, GFP immunofluorescence associated with cryosection (25 μm thick) muscle ROIs defined by diffuse Alexa 555-alpha bungarotoxin (A555-Btx) was quantified as described for whole mount NL preparations.
Determination of Palpebral Closure Index
Photographs of P19 control and Etv1 mutant faces oriented in profile centered on the eyelid were collected and the ratio of the height and width of the palpebral fissure was determined using ImageJ and averaged across five photographs per animal.
Analysis of facial muscle physiology
Bipolar, single strand steel wire EMG electrodes were fabricated and implanted as described (Pearson et al., 2005) into the belly of the nasolabialis (NL) and the dorsotemporal quadrant of the orbicularis oculi (OO) muscles of control and Etv1 mutant mice at P18, a time point necessitated by the lethality of the homozygous Etv1 mutation in the first 3–5 postnatal weeks (Arber et al., 2000). Mice were restrained but free range of motion was maintained for the head and mystacial pad. Only epochs recorded when whisking behavior was observed in the absence of locomotion or body movement were analyzed. Successful NL muscle implantation was defined by the detection of high frequency, rhythmic electromyographic (EMG) activity coincident with whisking behavior; successful OO implantation was defined by the detection of short duration, high amplitude activation coincident with blink behavior in response to noxious air puffs.
EMG signals were amplified (MA-102, Zoological Institute, University of Cologne) and recorded (1401, Cambridge Electronic Design) for offline analysis using Spike2 (CED, v.6.02) and Excel. Assays of EMG fidelity with muscle activation were initially checked against time-synched video capture (240 fps, Casio Exilim FC200) of whisking and eyelid blink behavior to eliminate artifacts, with recording of muscle-activated bursts subsequently identified manually. Behavior-related EMG activation was defined as greater than 3 SD above the average baseline value recorded 20–500 msec preceding EMG burst onset. Exploratory whisking behavior was defined as high-frequency, rhythmic EMG activation between 5–11 Hz lasting 1.9–5.9 s (n = 14 mice).
QUANTIFICATION AND STATISTICAL ANALYSIS
Averages and standard deviations were calculated using Excel; standard error was equal to (standard deviation of the average of n independent experiments/). Statistical significance was defined by p values < 0.05 from an unpaired t test (https://www.graphpad.com/quickcalcs/ttest1/?Format=SEM). Power and sample size for FMN quantification were evaluated using 2 sample, 1-sided power analysis (http://powerandsamplesize.com/Calculators/Compare-2-Means/2-Sample-1-Sided).
DATA AND CODE AVAILABILITY
The accession number for the E16.5 facial motor subnuclei microarray cel files reported in this paper is GEO: GSE134807.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Etv1 | Tom Jessell laboratory | AB_2617167 |
| Guinea pig anti-FoxP1 | Tom Jessell laboratory | AB_2665444 |
| Guinea pig anti-Isl1/2 | Tom Jessell laboratory | AB_2631974 |
| Rabbit anti-FoxP1 | Abcam | AB_732428 |
| Rabbit anti-GFP | ThermoFisher | AB_221569 |
| Chicken anti-GFP | ThermoFisher | AB_2534023 |
| Rabbit anti-cleaved Caspase-3 | Cell Signaling Technologies | AB_2341188 |
| Rabbit anti-Ankyrin G | Santa Cruz | AB_633908 |
| Mouse anti-myosin, fast | Sigma Aldrich | AB_2147168 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dextran, Tetramethylrhodamine-conjguated, 10,000MW | ThermoFisher | NA |
| Alpha bungarotoxin | Life Technologies | NA |
| Choleratoxin B subunit,Alexa-congugated | ThermoFisher | NA |
| Deposited Data | ||
| .cel data for microarray analysis of E16.5 dorsolateral (DL1–3), intermediate (I1–3), lateral (L1–3), medial (M1–3) | Gene Expression Omnibus (GEO) | GSE134807 |
| Experimental Models: Organisms/Strains | ||
| Etv1nlsLacZ/+ mice | Arber et al., 2000 | RRID:MGI:3621019 |
| B6.Cg-Tg(Thy1-YFP)16Jrs/J mice | Feng et al., 2000 | RRID:IMSR_JAX:003709 |
| B6;129-Cdk5tm1Kul/J mice | Ohshima et al., 1996 | RRID:IMSR_JAX:003536) |
| Tg(Isl1-EGFP*)1Slp/J mice | Lewcock et al., 2007 | RRID:IMSR_JAX:017952 |
| Oligonucleotides | ||
| Primers for RNA riboprobe template amplification with PCR; See Table S1 | NA | NA |
| Software and Algorithms | ||
| IMAGEJ | NIH | RRID:SCR_003070 |
| Spike2 (CED, v.6.02) | Cambridge Electronic Design | RRID:SCR_000903 |
Highlights.
Developing somatotopic facial motor neurons are transcriptionally diverse
Etv1 is required for dorsolateral facial-motor neuron migration and axon targeting
Axon mistargeting makes the eyelid muscle “whisk” in Etv1 mutants
ACKNOWLEDGMENTS
We thank Paolo Guarnieri for statistical analysis of microarray data; Florencia Marcucci for the EdU birth-dating protocol; Silvia Arber and Tom Jessell for Etv1 mutant mice; Susan Morton and Tom Jessell for anti-ETV1, anti-FOXP1, and anti-ISL1/2 antibodies; Turgay Akay for training for EMG recording; Timothy Spencer for help with laser-capture microdissection; Kevin Kanning, Rosa Polan, Ashley Juavinet, and Sarah Dogar for contributing expression studies; John Smerdon for advice on axon initial segment analysis; Dominick Papandrea for advice on surgeries; Dominick Papandrea and Nen Pagano for mouse-colony maintenance; and members of the Henderson, Wichterle, Dodd, Jessell, and Engle laboratories for helpful discussions. This work was supported by the Tow Foundation, the SMA Foundation, Project ALS, and NINDS (grant R01-NS056422). A.P.T was the recipient of a Kirschstein NRSA award F32NS060566-01A2. J.L. received support from Agence Nationale de la Recherche under contracts ANR-10-LABX-65 (LabEx LifeSenses).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.08.078.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Alfonsi F, Filippi P, Salaun D, deLapeyrière O, and Durbec P (2008). LIFRβ plays a major role in neuronal identity determination and glial differentiation in the mouse facial nucleus. Dev. Biol 313, 267–278. [DOI] [PubMed] [Google Scholar]
- Arber S, Ladle DR, Lin JH, Frank E, and Jessell TM (2000). ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell 101, 485–498. [DOI] [PubMed] [Google Scholar]
- Ashwell KW, and Watson CR (1983). The development of facial motoneurones in the mouse–neuronal death and the innervation of the facial muscles. J. Embryol. Exp. Morphol 77, 117–141. [PubMed] [Google Scholar]
- Baisden RH, Woodruff ML, Whittington DL, Baker DC, and Benson AE (1987). Cells of origin of the branches of the facial nerve: a retrograde HRP study in the rabbit. Am. J. Anat 178, 175–184. [DOI] [PubMed] [Google Scholar]
- Blount W (1927). Studies of the movements of the eyelids of animals: blinking. Q. J. Exp. Physiol 18, 16. [Google Scholar]
- Boghen DR (1996). Disorders of facial motor function. Curr. Opin. Ophthalmol 7, 48–52. [DOI] [PubMed] [Google Scholar]
- Bonanomi D (2019). Axon pathfinding for locomotion. Semin. Cell Dev. Biol 85, 26–35. [DOI] [PubMed] [Google Scholar]
- Cattaneo L, and Pavesi G (2014). The facial motor system. Neurosci. Biobehav. Rev 38, 135–159. [DOI] [PubMed] [Google Scholar]
- Courville J (1966). The nucleus of the facial nerve; the relation between cellular groups and peripheral branches of the nerve. Brain Res 1, 338–354. [DOI] [PubMed] [Google Scholar]
- Dalla Torre di Sanguinetto SA, Dasen JS, and Arber S (2008). Transcriptional mechanisms controlling motor neuron diversity and connectivity. Curr. Opin. Neurobiol 18, 36–43. [DOI] [PubMed] [Google Scholar]
- Demireva EY, Shapiro LS, Jessell TM, and Zampieri N (2011). Motor neuron position and topographic order imposed by β- and γ-catenin activities. Cell 147, 641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneris ES, and Hobert O (2014). Maintenance of postmitotic neuronal cell identity. Nat. Neurosci 17, 899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diogo R, Abdala V, Lonergan N, and Wood BA (2008). From fish to modern humans—comparative anatomy, homologies and evolution of the head and neck musculature. J. Anat 213, 391–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörfl J (1982). The musculature of the mystacial vibrissae of the white mouse. J. Anat 135, 147–154. [PMC free article] [PubMed] [Google Scholar]
- Dörfl J (1985). The innervation of the mystacial region of the white mouse: A topographical study. J. Anat 142, 173–184. [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, and Sanes JR (2000). Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28, 41–51. [DOI] [PubMed] [Google Scholar]
- Flames N, and Hobert O (2009). Gene regulatory logic of dopamine neuron differentiation. Nature 458, 885–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani R, and Sugita S (2008). Comparative histological study of the mammalian facial nucleus. J. Vet. Med. Sci 70, 367–372. [DOI] [PubMed] [Google Scholar]
- Garcès A, Haase G, Airaksinen MS, Livet J, Filippi P, and deLapeyrière O (2000). GFRalpha 1 is required for development of distinct subpopulations of motoneuron. J. Neurosci 20, 4992–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel S, Garcia-Dominguez M, and Charnay P (2000). Control of the migratory pathway of facial branchiomotor neurones. Development 127, 5297–5307. [DOI] [PubMed] [Google Scholar]
- Gilland E, and Baker R (2005). Evolutionary patterns of cranial nerve efferent nuclei in vertebrates. Brain Behav. Evol 66, 234–254. [DOI] [PubMed] [Google Scholar]
- Gorlick DL (1989). Motor innervation of respiratory muscles and an opercular display muscle in Siamese fighting fish Betta splendens. J. Comp. Neurol 290, 412–422. [DOI] [PubMed] [Google Scholar]
- Grant RA, Mitchinson B, and Prescott TJ (2012). The development of whisker control in rats in relation to locomotion. Dev. Psychobiol 54, 151–168. [DOI] [PubMed] [Google Scholar]
- Guest JM, Seetharama MM, Wendel ES, Strick PL, and Oberlaender M (2018). 3D reconstruction and standardization of the rat facial nucleus for precise mapping of vibrissal motor networks. Neuroscience 368, 171–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase G, Dessaud E, Garcès A, de Bovis B, Birling M, Filippi P, Schmalbruch H, Arber S, and deLapeyrière O (2002). GDNF acts through PEA3 to regulate cell body positioning and muscle innervation of specific motor neuron pools. Neuron 35, 893–905. [DOI] [PubMed] [Google Scholar]
- Haidarliu S, Simony E, Golomb D, and Ahissar E (2010). Muscle architecture in the mystacial pad of the rat. Anat. Rec. (Hoboken) 293, 1192–1206. [DOI] [PubMed] [Google Scholar]
- Han AY, Gupta S, and Novitch BG (2018). Molecular specification of facial branchial motor neurons in vertebrates. Dev. Biol 436, 5–13. [DOI] [PubMed] [Google Scholar]
- Hill DN, Bermejo R, Zeigler HP, and Kleinfeld D (2008). Biomechanics of the vibrissa motor plant in rat: rhythmic whisking consists of triphasic neuromuscular activity. J. Neurosci 28, 3438–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichsen CF, and Watson CD (1984). The facial nucleus of the rat: representation of facial muscles revealed by retrograde transport of horseradish peroxidase. Anat. Rec 209, 407–415. [DOI] [PubMed] [Google Scholar]
- Hobert O (2008). Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc. Natl. Acad. Sci. USA 105, 20067–20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horta-Júnior JA, Tamega OJ, and Cruz-Rizzolo RJ (2004). Cytoarchitecture and musculotopic organization of the facial motor nucleus in Cebus apella monkey. J. Anat 204, 175–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber AB, Kania A, Tran TS, Gu C, De Marco Garcia N, Lieberam I, Johnson D, Jessell TM, Ginty DD, and Kolodkin AL (2005). Distinct roles for secreted semaphorin signaling in spinal motor axon guidance. Neuron 48, 949–964. [DOI] [PubMed] [Google Scholar]
- Husseman J, and Mehta RP (2008). Management of synkinesis. Facial Plast. Surg 24, 242–249. [DOI] [PubMed] [Google Scholar]
- Kanning KC, Kaplan A, and Henderson CE (2010). Motor neuron diversity in development and disease. Annu. Rev. Neurosci 33, 409–440. [DOI] [PubMed] [Google Scholar]
- Kaplan A, Spiller KJ, Towne C, Kanning KC, Choe GT, Geber A, Akay T, Aebischer P, and Henderson CE (2014). Neuronal matrix metalloproteinase-9 is a determinant of selective neurodegeneration. Neuron 81, 333–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BG, and Rhoades RW (1985). Representation of whisker follicle intrinsic musculature in the facial motor nucleus of the rat. J. Comp. Neurol 232, 55–69. [DOI] [PubMed] [Google Scholar]
- Lazar G, Szabo T, Libouban S, Ravaille-Veron M, Toth P, and Brändle K (1992). Central projections and motor nuclei of the facial, glossopharyngeal, and vagus nerves in the mormyrid fish Gnathonemus petersii. J. Comp. Neurol 325, 343–358. [DOI] [PubMed] [Google Scholar]
- Lee SH, Ito J, and Yamamoto E (1991). A horseradish peroxidase study of the fiber orientation in the facial nerve. Eur. Arch. Otorhinolaryngol 248, 366–369. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Genoud N, Lettieri K, and Pfaff SL (2007). The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron 56, 604–620. [DOI] [PubMed] [Google Scholar]
- Lintern KB, Guidato S, Rowe A, Saldanha JW, and Itasaki N (2009). Characterization of wise protein and its molecular mechanism to interact with both Wnt and BMP signals. J. Biol. Chem 284, 23159–23168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Mohamed O, Dufort D, and Wallace VA (2003). Characterization of Wnt signaling components and activation of the Wnt canonical pathway in the murine retina. Dev. Dyn 227, 323–334. [DOI] [PubMed] [Google Scholar]
- Livet J, Sigrist M, Stroebel S, De Paola V, Price SR, Henderson CE, Jessell TM, and Arber S (2002). ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron 35, 877–892. [DOI] [PubMed] [Google Scholar]
- Martin MR, and Lodge D (1977). Morphology of the facial nucleus of the rat. Brain Res 123, 1–12. [DOI] [PubMed] [Google Scholar]
- McArthur KL, and Fetcho JR (2017). Key features of structural and functional organization of zebrafish facial motor neurons are resilient to disruption of neuronal migration. Curr. Biol 27, 1746–1756.e1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohebiany AN, Harroch S, and Bouyain S (2014). New insights into the roles of the contactin cell adhesion molecules in neural development. Adv. Neurobiol 8, 165–194. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Kelley KW, Tsai HH, Redmond SA, Chang SM, Madireddy L, Chan JR, Baranzini SE, Ullian EM, and Rowitch DH (2014). Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature 509, 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JD, Deschênes M, Furuta T, Huber D, Smear MC, Demers M, and Kleinfeld D (2013). Hierarchy of orofacial rhythms revealed through whisking and breathing. Nature 497, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JD, Kleinfeld D, and Wang F (2014). How the brainstem controls orofacial behaviors comprised of rhythmic actions. Trends Neurosci 37, 370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcutt RG, and Bemis WE (1993). Cranial nerves of the coelacanth, Latimeria chalumnae [Osteichthyes: Sarcopterygii: Actinistia], and comparisons with other craniata. Brain Behav. Evol 42 (Suppl 1), 1–76. [DOI] [PubMed] [Google Scholar]
- Ohshima T, Ward JM, Huh CG, Longenecker G, Veeranna, Pant HC, Brady RO, Martin LJ, and Kulkarni AB (1996). Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc. Natl. Acad. Sci. USA 93, 11173–11178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima T, Ogawa M, Takeuchi K, Takahashi S, Kulkarni AB, and Mikoshiba K (2002). Cyclin-dependent kinase 5/p35 contributes synergistically with Reelin/Dab1 to the positioning of facial branchiomotor and inferior olive neurons in the developing mouse hindbrain. J. Neurosci 22, 4036–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papez J (1927). Subdivisions of the facial nucleus. J. Comp. Neurol 43, 159–191. [Google Scholar]
- Pearson KG, Acharya H, and Fouad K (2005). A new electrode configuration for recording electromyographic activity in behaving mice. J. Neurosci. Methods 148, 36–42. [DOI] [PubMed] [Google Scholar]
- Renier N, Adams EL, Kirst C, Wu Z, Azevedo R, Kohl J, Autry AE, Kadiri L, Umadevi Venkataraju K, Zhou Y, et al. (2016). Mapping of brain activity by automated volume analysis of immediate early genes. Cell 165, 1789–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanes GJ (1964). The motor pools of the spinal cord. Prog. Brain Res 11, 93–119. [DOI] [PubMed] [Google Scholar]
- Rossel M, Loulier K, Feuillet C, Alonso S, and Carroll P (2005). Reelin signaling is necessary for a specific step in the migration of hindbrain efferent neurons. Development 132, 1175–1185. [DOI] [PubMed] [Google Scholar]
- Salic A, and Mitchison TJ (2008). A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 105, 2415–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, and Gerfin-Moser A (1993). A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry 100, 431–440. [DOI] [PubMed] [Google Scholar]
- Semba K, and Egger MD (1986). The facial “motor” nerve of the rat: control of vibrissal movement and examination of motor and sensory components. J. Comp. Neurol 247, 144–158. [DOI] [PubMed] [Google Scholar]
- Serrano-Saiz E, Leyva-Diaz E, De La Cruz E, and Hobert O (2018). BRN3-type POU homeobox genes maintain the identity of mature postmitotic neurons in nematodes and mice. Curr. Biol 28, 2813–2823.e2812. [DOI] [PubMed] [Google Scholar]
- Shaw MD, and Baker R (1985). Morphology of motoneurons in a mixed motor pool of the cat facial nucleus that innervate orbicularis oculis and quadratus labii superioris, stained intracellularly with horseradish peroxidase. Neuroscience 14, 627–643. [DOI] [PubMed] [Google Scholar]
- Sherwood CC (2005). Comparative anatomy of the facial motor nucleus in mammals, with an analysis of neuron numbers in primates. Anat. Rec. A Discov. Mol. Cell. Evol. Biol 287, 1067–1079. [DOI] [PubMed] [Google Scholar]
- Song MR, Shirasaki R, Cai CL, Ruiz EC, Evans SM, Lee SK, and Pfaff SL (2006). T-Box transcription factor Tbx20 regulates a genetic program for cranial motor neuron cell body migration. Development 133, 4945–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer WC, and Deneris ES (2017). Regulatory mechanisms controlling maturation of serotonin neuron identity and function. Front. Cell. Neurosci 11, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiassny M (2013). Emotions in Animals and Humans: Comparative Perspective (Springer; ). [Google Scholar]
- Suetterlin P, and Drescher U (2014). Target-independent ephrina/EphA-mediated axon-axon repulsion as a novel element in retinocollicular mapping. Neuron 84, 740–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sürmeli G, Akay T, Ippolito GC, Tucker PW, and Jessell TM (2011). Patterns of spinal sensory-motor connectivity prescribed by a dorsoventral positional template. Cell 147, 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatoh J, Nelson A, Zhou X, Bolton MM, Ehlers MD, Arenkiel BR, Mooney R, and Wang F (2013). New modules are added to vibrissal premotor circuitry with the emergence of exploratory whisking. Neuron 77, 346–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzis JK, and Anesti K (2011). Developmental facial paralysis: a review. J. Plast. Reconstr. Aesthet. Surg 64, 1318–1333. [DOI] [PubMed] [Google Scholar]
- Triplett J, Phan A, Yamada J, and Feldheim D (2012). Alignment of multiomodal sensory input in the superior colliculus through a gradient-matching mechanism. J. Neurosci 32, 5264–5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura-Sumi M, Manabe Y, Matsushima R, and Mizuno N (1986). Correlation of the main peripheral branches of the facial nerve with the cytoarchitec-tonic subdivisions of the facial nucleus in the guinea pig. Anat. Embryol. (Berl.) 174, 161–166. [DOI] [PubMed] [Google Scholar]
- Visel A, Thaller C, and Eichele G (2004). GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res 32, D552–D556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, and Scott SA (2004). An early broad competence of motoneurons to express ER81 is later sculpted by the periphery. J. Neurosci 24, 9789–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang-Bennett LT, and Coker NJ (1990). Analysis of axonal regeneration through the silicone regeneration chamber: a retrograde tracing study in the rabbit facial nerve. Exp. Neurol 107, 222–229. [DOI] [PubMed] [Google Scholar]
- Zheng R, Wan C, Mei S, Qin Q, Wu Q, Sun H, Chen CH, Brown M, Zhang X, Meyer CA, and Liu XS (2019). Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res 47 (D1), D729–D735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the E16.5 facial motor subnuclei microarray cel files reported in this paper is GEO: GSE134807.