Abstract
Upon activation by CD40 or TLR signaling, B lymphocytes activate NF-κB to induce AID and, therefore, immunoglobulin class switch DNA recombination, as central to the maturation of the antibody and autoantibody responses. Here we show that NF-κB activation is boosted by co-localization of engaged immune receptors, such as CD40, with RAB7 small GTPase on mature endosomes, in addition to signals emanating from the receptors localized on the plasma membrane, in mouse B cells. In mature endosomes, RAB7 directly interacts with TRAF6 E3 ubiquitin ligase, which catalyzes K63 polyubiquitination for NF-κB activation. RAB7 overexpression in Cd19+/creRosa26fl-STOP-fl-Rab7 mouse B cells upregulates K63 polyubiquitination activity of TRAF6, enhances NF-κB activation and AID induction, and boosts IgG antibody and autoantibody levels. This, together with the extensive intracellular localization of CD40 and strong correlation of RAB7 expression with NF-κB activation in mouse lupus B cells, shows that RAB7 is an integral component of the B cell NF-κB activation machinery, likely through interaction with TRAF6 for the assembly of “intracellular membrane signalosomes”.
Keywords: intracellular membrane signalosomes, NF-κB, RAB7, receptor internalization, TRAF6
Introduction
B cells are critical for host immunity by producing antibodies to infectious and tumoral antigens as well as by regulating other immune elements. They can be activated directly by complex antigens that engage both a Toll-like receptor (TLR) and the B cell receptor (BCR) or, in a T cell-dependent manner, upon CD40 engagement by CD154 (CD40 ligand) expressed on activated cognate T cells (1). These stimuli trigger activation of NF-κB, which directly binds to the Aicda gene to induce the expression of activation-induced cytidine deaminase (AID), as essential for class switch DNA recombination (CSR) in the immunoglobulin heavy chain (IgH) locus (2,3). CSR substitutes the constant Cμ region with Cγ, Cα or Cε to give rise to class-switched IgG, IgA or IgE antibodies, which display wide tissue distributions and possess diverse biological effector functions (4). CSR of autoreactive B cells, however, leads to production of pathogenic IgG autoantibodies that mediate tissue/organ damages in systemic lupus (5).
An important pathway in NF-κB activation is through TRAF6, an E3 ubiquitin ligase that is recruited to CD40 or TLR-activated IRAK-1 and then catalyzes polyubiquitination via the Lys63 (K63) residue of ubiquitin (6,7). The K63 polyubiquitination of TRAF6 itself and other molecules would provide a “platform” for activation of TAK1 and IKKγ kinases, both of which can phosphorylate IKKβ in the IKKα/IKKβ complex (8), leading to IκB phosphorylation and subsequent IκB ubiquitination/degradation to relieve inhibition of NF-κB, which in turn induces gene transcription. Unlike immune cell functions requiring only a burst of NF-κB activation (e.g., cytokine release by macrophages), induction of AID in B cells, as well as 14–3-3 proteins and histone modifying enzymes for the AID targeting in the IgH locus (9,10), would entail sustained NF-κB activation (11). In B cells, the AID/CSR induction by TLRs and CD40 also depends on RAB7 (12), a small GTPase that, by replacing RAB5 on immature endosomes through a “GTPase switch” process, mediates maturation of endosomes and is localized exclusively on mature endosomes (13). B cell expression of RAB7 is upregulated by the same NF-κB- and AID-inducing stimuli. Mice that are conditional knockout (KO) in Rab7 in activated B cells (IghCγ1-creRab7fl/fl) cannot mount IgG responses despite normal IgM production and plasma cell differentiation (12). Further, NF-κB activation and AID/CSR induction are impaired, albeit not abolished, in IghCγ1-creRab7fl/fl B cells stimulated in vitro (12) and in B cells treated with CID 1067700, the only known small molecule RAB7 inhibitor (14). Such impairment can be rescued by the enforced expression of a constitutively active mutant of IKKβ, suggesting a role of RAB7 in NF-κB activation (12,15). The crucial role of RAB7 in mature endosome formation also implies the involvement of such intracellular membrane structures in signal transduction from immune receptor TLRs and CD40 to transcription factors, particularly NF-κB.
Here, to address how endosomal RAB7 mediates NF-κB activation for B cell differentiation, we reasoned that since surface CD40 or TLR4 can be internalized upon engagement in several immune cells (16–18), a subset of such endocytosed receptors would traffick through the vesicle system to reach RAB7-marked endosomes and transduce signals from there. We further reasoned that the rest of endocytosed receptor molecules could remain associated with the plasma membrane by anchoring on lipid rafts and initiate signal transduction, as prompted by findings showing an important role of lipid rafts in the CD40 signaling in B lymphoma cells and other immune cell types (19–22) – whether CD40 is endocytosed or in the original surface location in those studies, however, is unclear. Finally, we reasoned that the lipid raft-dependent pathway would supplement or backup the endosomal RAB7-depdent pathway in activated primary B cells, and would mediate residual NF-κB activation and AID/CSR induction when the expression or activity of RAB7 is inhibited. With these considerations, we aimed to explore the mechanistic basis of RAB7-dependent pathway in NF-κB activation by increasing the contribution of this pathway through two complementary strategies: by boosting RAB7 expression, i.e., through generation of mice with B cell-specific RAB7 overexpression, and by inhibiting the lipid raft-dependent pathway, in wildtype B cells as well as B cells in which the RAB7 expression/activity were impaired or enhanced. These two strategies, as supported by our comprehensive approaches in molecule biology, cell biology, imaging and in silico docking in the analyses of receptor internalization, their co-localization with RAB7 and TRAF6, NF-κB activation in single cells and AID/CSR induction, have revealed an important role of RAB7-dependent “intracellular membrane signalosomes” in promoting TRAF6 K63 polyubiquitination and NF-κB activation during B cell differentiation.
Materials and Methods
Mice and immunization
Cd19+/creRosa26+/fl-STOP-fl-Rab7 (Rab7 B-Tg) mice and their “wildtype” littermate controls were generated by crossing Cd19+/cre mice (JAX, stock number 006785) with Rosa26+/fl-STOP-fl-Rab7 mice, as generated by a customized TurboKnockout® approach (Cyagen Biosciences). Briefly, the targeting vector contains two arms homologous to the first and second exon, respectively, of the Rosa26 locus. In between the arms lies the CMV promoter-driven Rab7 cDNA, which is preceded by a loxP site-flank “STOP” cassette sequence, as followed by a neomycin resistance gene (Neor) in the inverse direction (Fig. 1A). After injection of the targeting vector into ES cells from C57 mice and screening by Southern Blotting, selected ES clones showing correct targeting were used for blastocyst microinjection to produce chimeric mice. These were bred with a C57 mouse strain that expressed Flipase to screen for mice with germline-transmission as founders – as Flipase also deleted Neor in the offspring, the entire process to generate founders on the C57 background with no Neor took only 6 months. Among the five founders (one female and four male heterozygous mice), two were randomly selected for breeding with Cd19+/cre mice to establish independent lines. As progenies from these two lines displayed no significant difference in the parameters we measured, data from one line are depicted here. In Rab7 B-Tg mice, Cre expression driven by the Cd19 promoter in the Cd19Cre allele mediated deletion of the STOP cassette in the Rosa26 locus, thereby allowing for the CMV promoter-driven expression of untagged RAB7 in CD19+ B cells.
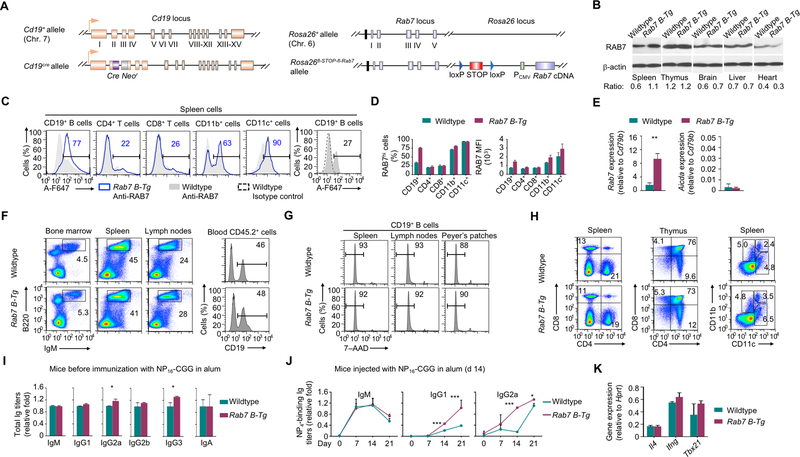
FIGURE 1.
B cell-specific RAB7 overexpression enhances the antibody response. (A) Illustration of Cd19 and Rosa26 alleles in Cd19+/creRosa26+/fl-STOP-fl-Rab7 (Rab7 B-Tg) mice. (B) Immunoblotting analysis of RAB7 expression in different organs. The signal intensity of each band was quantified by ImageJ (NIH) and the ratio of the value of RAB7 signals to that of β-actin signals was depicted below. (C) Flow cytometry analysis of RAB7 expression in immune cells. Numbers indicate the proportion of RAB7hi cells. Representative of three independent experiments. (D) Proportions of RAB7hi cells (top) and mean fluorescence intensities (MFIs) of RAB7 (bottom) in each immune cell type in Rab7 B-Tg mice (mean and s.e.m., n=3). (E) Levels of Rab7 transcripts (left) in B cells isolated from non-immunized Rab7 B-Tg mice. Data were expressed as the ratio to that of Cd79b levels (mean and s.d., triplicates). Also depicted were Aicda expression levels, which were low in non-immunized mice (right). (F) Flow cytometry analysis of B cell development and homeostasis in the bone marrow and the periphery. Numbers indicate the proportion of B220+IgMhi B cells. Representative of three independent experiments. (G) Flow cytometry analysis of B cell survival. Numbers indicate the proportion of live (7–AAD–) B cells. Representative of three independent experiments. (H) Flow cytometry analysis of T cell subsets (CD4+CD8– and CD4–CD8+) and myeloid cell types (CD11b+CD11c– macrophages, CD11b–CD11c+ DCs and CD11bhiCD11c+ DCs), as indicated. Numbers indicate the proportions of each cell types. Representative of three independent experiments. (I) Titers of serum Ig isotypes in Rab7 B-Tg mice before immunization with NP-CGG and their wildtype littermate counterparts (normalized to the values in wildtype mice; mean and s.e.m., n=4). (J) Titers of NP4-binding IgM, IgG1 and IgG2a in the serum of mouse littermates after immunization with NP-CGG (normalized to the values in wildtype mice; mean and s.e.m., n=4). (K) Levels of different transcripts, as indicated, in B cells from immunized mice, as in (J). Data were normalized to the values in wildtype mice (mean and s.d., triplicates). *, p < 0.05; ***, p < 0.005.
C57BL/6 and lupus-prone MRL/Lpr mice were from the Jackson Laboratory (JAX, stock number 000664 and 000585, respectively). All mice were maintained in a pathogen-free vivarium of the University of Texas Health Science Center at San Antonio (UTHSCSA) and were used for experiments at 8–12 weeks of age, without any apparent infection or disease. For immunization, mice were injected intraperitoneally (i.p.) with 100 μg of NP-CGG (in average 16 molecules of 4-hydroxy-3-nitrophenyl acetyl coupled to 1 molecule of chicken γ-globulin; Biosearch Technologies) in alum (Imject® Alum, Pierce). All protocols were in accordance with the rules and regulations of the Institutional Animal Care and Use Committee (IACUC) of UTHSCSA.
Mouse B cell preparation and purification
Mouse immune cells were isolated from single cell suspensions prepared from spleen, lymph node or blood samples. Spleen cells were resuspended in ACK Lysis Buffer (Lonza) to lyse red blood cells. After quenching with FCS-RPMI (RPMI-1640 medium (Invitrogen) supplemented with FCS (10% v/v, Hyclone), penicillin-streptomycin (1% v/v, Invitrogen) and amphotericin B (1% v/v, Invitrogen)) and centrifugation, cells were resuspended in FCS-RPMI for further preparation/analysis. Whole blood samples (250 μl) were collected by submandibular bleeding into Eppendorf tubes containing heparin (25 μl) and then subjected to ACK lysis three times. Cells were resuspended in FCS-RPMI for further preparation/analysis. To isolate B cells, spleen or lymph node cells were subjected to negative selection (against CD43, CD4, CD8, CD11b, CD49b, CD90.2, Gr-1 or Ter-119) using the EasySep™ Mouse B-Cell Isolation Kit (StemCell™ Technologies) following the manufacturer’s instructions, resulting in preparations of more than 98% IgM+IgDhi B cells. After pelleting, B cells were resuspended in FCS-RPMI for analysis or culturing.
Mouse B cell stimulation and drug treatment
For CSR induction, spleen B cells were cultured (3 × 105 cell/ml) in FCS-RPMI and stimulated for 96 h (or 48 h for transcript and protein analyses) by: CD154 (3 U/ml), LPS (3 μg/ml, from E. coli, serotype 055:B5, deproteinized by chloroform extraction; Sigma-Aldrich), TLR1/2 ligand Pam3CSK4 (100 ng/ml, Invivogen), TLR4 ligand monophosphoryl lipid A (lipid A, 1 μg/ml, Sigma-Aldrich; TLR7 ligand R-848 (30 ng/ml; Invivogen) or TLR9 ligand CpG ODN1826 with a phosphorothioate backbone (CpG, 1 μM, sequence 5’–TCCATGACGTTCCTGACGTT–3’; Operon). Recombinant IL-4 (3 ng/ml; R&D Systems) was added for CSR to IgG1 and IgE, IFN-γ (50 ng/ml; R&D Systems) for CSR to IgG2a, and TGF-β (2 ng/ml; R&D Systems) and anti–Igδ mAb (clone 11–26c) conjugated to dextran (anti–δ mAb/dex; 100 ng/ml; Fina Biosolutions), which crosslinks BCR in IgD+ B cells, for CSR to IgA. CSR to IgG3 and IgG2b was induced by LPS.
To treat mouse B cells in vitro with the cell permeable RAB7 inhibitor, CID 1067700 (Glixx Laboratories) dissolved in DMSO (stock concentration 40 mM, 16 mg/ml) was further diluted in DMSO and added to cell cultures to a final concentration of 40 μM. To treat B cells with methyl-β-cyclodextrin (MβCD, Sigma Aldrich), which solubilizes lipid rafts, MβCD was freshly dissolved in FCS-RPMI to make a stock solution of 16 mM (21 mg/ml) and then further diluted with FCS-RPMI before adding to cell cultures.
To treat B cells with dynasore (Selleckchem), a cell permeable inhibitor of dynamin, the stock solution (10mM in DMSO) was added to the B cell culture medium to a final concentration of 15 μM (or different doses as indicated) when stimulation started. In CD40 internalization assays, purified B cells were stimulated with LPS for 24 h to induce CD40 expression, washed three times to completely remove LPS, and cultured for an additional 6 h to allow intracellular LPS signals to subside. B cells were then stimulated with CD154 plus IL-4 for 24 h and treated with DMSO (nil) or dynasore in the presence of serum-free medium before being harvested for flow cytometry analysis.
Total and NP-specific antibody titers
To determine titers of total or NP-specific IgM, IgG subclasses and IgA, mouse sera were first diluted 1 to 100-fold with PBS (pH 7.4) plus 0.05% (v/v) Tween-20 (PBST). Two-fold serially diluted samples and standards for each Ig isotypes were incubated in 96-well plates coated with pre-adsorbed goat anti–IgM, anti–IgA or anti–IgG Abs (all 1mg/mL) in sodium carbonate/bicarbonate buffer (pH 9.6). After washing with PBST, captured Igs were detected with biotinylated anti–IgM, –IgA or –IgG isotypes Abs, followed by reaction with horseradish peroxidase (HRP)-labeled streptavidin (Sigma-Aldrich), development with o-phenylenediamine, and measurement of absorbance at 492 nm. Ig concentrations were determined using Prism® (GraphPad Software) or Excel® (Microsoft) software. To analyze titers of antigen-specific antibodies (high-affinity NP-binding Abs), sera were diluted 1,000-fold in PBST. Two-fold serially diluted samples were incubated in a 96-well plate pre-blocked with BSA and coated with NP4-BSA (4 NP molecules on one BSA molecule). Captured Igs were detected with biotinylated mAbs. Data are relative values based on end-point dilution factors.
Flow cytometry
To analyze B cells stimulated in vitro, cells were harvested, stained with fluorochrome-conjugated mAbs in Hank’s Buffered Salt Solution plus 0.1% BSA (BSA-HBSS) for 20 m and analyzed by flow cytometry for Igγ3, Igγ1, Igγ2b and Igγ2a expression (CSR to IgG3, IgG1, IgG2b and IgG2a), CD138 (plasma cell marker) and other B cell surface molecules. For intracellular Igε analysis, B cells (2 × 106) were treated with 200 μl of trypsin for 2 m to cleave off FcεRI, followed by quenching with 1 ml FCS-RMPI. Cells were then stained with fixable viability dye (FVD) in HBSS-BSA. After fixing in 200 μl of formaldehyde (3.7% v/v) at 25°C for 10 m and quenching with 22 μl 1 M glycine (pH 7.0), cells were permeablized in 500 μl of 90% cold methanol on ice for 30 m and, after washing, stained with fluorochrome-conjugated Abs. Dead (FVD+) cells were excluded from analysis. To analyze B cells and other immune cells in vivo, spleen or blood cells (2 × 106) were first stained with fluorophore-labeled mAbs to surface markers in the presence of mAb Clone 2.4G2, which blocks FcγIII and FcγII receptors, and FVD-506. After washing, cells were resuspended in HBSS for FACS or in the BD Cytofix/Cytoperm™ buffer (250 μl, BD Biosciences) for intracellular staining (see below) before FACS analysis. FACS data were analyzed by the FlowJo® software (BD).
For flow cytometry analysis of B cell proliferation in vivo, mice were injected twice i.p. with 2 mg of bromodeoxyuridine (BrdU) in 200 μl PBS, with the first and second injection at 24 h and 20 h prior to sacrificing, respectively. Spleen B cells were analyzed for BrdU incorporation (by anti–BrdU mAb staining) and DNA contents (by 7–AAD staining after cells were permeabilized) using an APC BrdU Flow kit® (BD Biosciences) following the manufacturer’s instructions. For B cell proliferation analysis in vitro, B cells were labeled with CellTrace™ CFSE or CellTrace™ Yellow (ThermoFisher) following the manufacturer’s instructions and then cultured for 96 h. Cells were analyzed by flow cytometry for CellTrace™ CFSE or CellTrace™ Yellow intensity, which was reduced by half after completion of each cell division, as CellTrace™ CFSE or CellTrace™ Yellow-labeled cell constituents were equally distributed between daughter cells. The number of B cell divisions was determined by the FlowJo® software to calculate the average cell divisions that B cells had completed.
Intracellular staining
Freshly isolated lymphocytes from the spleen, lymph node or blood samples, or B cells after stimulation were washed with BSA-HBSS and stained with fluorochrome-conjugated mAbs specific for surface markers in the presence of 2.4G2 and FVD. After washing, cells were resuspended in Cytofix/Cytoperm™ buffer (250 μl) and incubated at 4°C for 20 m. After washing twice with BD Perm/Wash™ buffer, cells were stained in the same buffer with fluorochrome-labeled mAbs.
To analyze CD40 distribution, same B cells were equally divided into two groups, with one group undergoing surface staining of CD40 and the other group undergoing intracellular staining using anti-CD40 mAb, which recognized CD40 localized both on the surface and inside the cell (total). The ratio of the proportion of CD40hi cells in the surface staining group to that in the total CD40 staining group would be inversely correlating with CD40 internalization. To analyze active internalization of CD40, purified B cells were stimulated with LPS for 24 h to upregulate CD40 expression and then stimulated with CD154 for 0, 10, 20, 30 and 60 m. Total and surface CD40 levels were analyzed as above.
Flow imaging
Cells after surface CD19 staining and intracellular CD40 staining were resuspended in BSA-HBSS at the concentration of 2×107 cells/ml and analyzed using an ImageStream X multispectral imaging flow cytometer (Amnis®), with >10,000 events collected in each sample. Images were analyzed using IDEAS image-analysis software (Amnis®), with single-stained samples being used for compensation of fluorescence signals among different channels. CD40 distribution was assessed by the “Intracellular localization” score, which was defined as the ratio of the intensity “inside the cell” to the intensity of the entire cell, with the “inside of the cell” determined by the “erosion of a mask” function of the software. For each sample, at least 4,000 cells were scored, as positive for CD40 intracellular localization when displaying primarily internal fluorescence and negative when showing little intracellular localization (cells with intensive “nuclear” staining of CD40 were excluded from the analysis). The intracellular localization score, which would be independent of cell size and signal intensities of a wide range, is depicted by values within – 3 ~ + 3 and the proportion of cells with the score >0 was quantified.
Immunofluorescence microscopy
To visualize RAB7 and CD40 expression and co-localization, B cells were spun onto cover slips pre-coated with poly-D-lysine (10 μg/ml, Sigma-Aldrich) and fixed with 2% paraformaldehyde for 10 m. After washing with HBSS and permeabilization with 0.5% Triton X-100 for 10 m, cells were stained with a rabbit anti–Rab7 mAb and a mouse anti–CD40 mAb at 25°C for 1 h, followed by staining with fluorescence-labeled goat anti–rabbit and goat anti–mouse secondary Abs (Supplemental Table 1). After washing, cells were mounted using ProLong® Gold with DAPI for analysis under a Zeiss LM710 confocal microscope.
Intracellular membrane fractionation
B cells (2 × 107, freshly isolated or stimulated with CD154 plus IL-4 for 48 h) were harvested and washed twice with cold PBS. They were lysed with 500 μl homogenization buffer (250 mM sucrose, 3 mM imidazole and 1 mM EDTA, pH 7.4) supplemented with the protease inhibitor cocktail, as above, and 10 mM cycloheximide. After microcentrifugation at full speed for 15 m at 4°C, the post-nuclear fraction (PNF) was layered onto a 10%−40% continuous sucrose gradient, as prepared in the same lysis buffer (3.5 ml), and subjected to ultracentrifugation at 100,000 g for 16 h at 4°C. Two bands of membrane fractions corresponding to the mature and immature endosomes appearing at the position of 25% and 35% sucrose, respectively, were collected, through holes on the side wall of the ultra-centrifugation tube punctured by a 26-gauge needle, and then analyzed by SDS-PAGE and immunoblotting.
Immunoblotting and Co-IP
B cells (107) were harvested by centrifugation at 500 g for 5 m, resuspended in 0.5 ml of lysis buffer (20 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1% (v/v) NP-40) supplemented with Halt™ Protease & Phosphatase Inhibitors Cocktail (#1861281, ThermoScientific). Cell lysates were separated by SDS–PAGE and transferred onto PVDF membranes for immunoblotting involving specific Abs. Membranes were then stripped with Restore™ PLUS Western Blot Stripping Buffer (#46430, ThermoScientific) for re-immunoblotting with an anti–β-actin mAb.
For Co-IP, spleen B cells (107) stimulated with CD154 plus IL-4 for 48 h were collected and resuspended in lysis buffer (20 mM Tris-Cl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.5% (v/v) NP-40) supplemented with Halt™ Protease & Phosphatase Inhibitors Cocktail. After sonication and centrifugation, protein lysates were precleared with equilibrated Protein G-conjugated Sepharose™ 4B beads (ThermoFisher, 50 μl) and then incubated, in 500 μl of lysis buffer, with anti–TRAF6 mouse mAb or a mouse IgG mAb with irrelevant specificities at 4°C for 4 h in the presence of Protein G/Sepharose beads. After three times of washing with the lysis buffer, immunoprecipitates were eluted in the SDS sample buffer and fractionated by SDS-PAGE for immunoblotting.
Bi-fluorescence complementation (BiFC) assay
EYFP was split into two complementary moieties: the N-terminal 154 amino acids (EYFP1–154) and the C-terminal 84 amino acids (EYFP155–238). EYFP1–154 was fused with Flag–tagged RAB7, RAB5A, AID or UNG, and EYFP155–238 was fused with different domains of TRAF6 (RING, zinc finger and C-terminal domain TRAF-C) tagged with influenza hemagglutinin (HA). HeLa cells (5 × 105) cultured in DMEM (Invitrogen) supplemented with FBS, were transfected with 1 μg of plasmid using Lipofectamine™ (Life Technologies). After 24 hours, cells were analyzed for cell viability (7–AAD–) and for EYPF intensity by flow cytometry.
Real-time qRT-PCR transcript analysis
RNA was extracted from 5 × 106 B cells using the RNeasy Mini Kit (Qiagen). First-strand complementary DNA was synthesized from equal amounts of total RNA (4 μg) using the SuperScript III System (Invitrogen) and analyzed by the SYBR Green dye (Bio-Rad) incorporation in PCR reactions involving specific primers and performed in an iCycler and MyIQ real-time qPCR system (Bio-Rad). For qRT-PCR, the ΔΔCt method was used to determine levels of transcripts and data were normalized to levels of Cd79b (mouse), which encodes the BCR Igβ chain that is constitutively expressed in B cells.
Modeling
Modeling studies were performed using the Schrodinger 2015–3 software suite. Protein-protein docking was performed with the Piper algorithm, using the GTP-bound Rab7 (PDB ID: 1yhn derived from a complex with RILP) as the ligand and the complex containing TRAF6 a.a. 55–160 (the RING domain and the first zinc finger, ZF1), Ubc13 and ubiquitin (PDB ID 5vo0) as the receptor. All the protein structures were pre-processed with a protein preparation routine. A number of iterations of the ligand (30,000) were analyzed, and the top 30 poses were saved. These poses were visually analyzed and those showing binding of RAB7 to the TRAF6 RING dimerization interface and forming contacts with ubiquitin were depicted. The RAB7 structure (PDB ID: 1t91) was visualized by Swiss-PdbViewer.
Statistical analysis
Statistical analysis was performed by either GraphPad (Prism) or Excel (Microsoft) software to determine p values by paired student t-test or F test.
Results
B cell-specific RAB7 overexpression enhances the antibody response
In Cd19+/CreRosa26+/fl-STOP-fl-Rab7 (Rab7 B-Tg) mice, homeostatic expression of untagged RAB7 from the Rosa26 locus (which is in close proximity to the endogenous Rab7 locus, Fig. 1A) was largely restricted to lymphoid organs, resulting in increased Rab7 transcript levels and RAB7 overexpression in about 75% of CD19+ B cells in the spleen (Fig. 1B–E). The remaining B cells expressed RAB7 at levels comparable to those in the majority of B cells from Cd19+/creRosa26+/+ and Cd19+/+Rosa26+/fl-STOP-fl-Rab7 littermates, which were collectively referred to as “wildtype” mice (a proportion of B cells from wildtype mice expressed high RAB7 levels, likely reflecting their activated state). The smaller difference in RAB7 protein expression (2-fold in Rab7 B-Tg mice) than that in Rab7 transcript levels (5-fold) likely due to post-transcriptional regulation mechanisms that would limit RAB7 overexpression.
In Rab7 B-Tg mice, B cell-specific RAB7 overexpression did not affect B cell development in the bone marrow or maturation and survival in the periphery; nor did it affect the development of other immune cells (Fig. 1F–H). These mice displayed serum levels of IgM and switched IgG1, IgG2b and IgA isotypes comparable to those in their wildtype littermates, but higher IgG2a and IgG3 titers, with IgE undetectable (Fig. 1I). Upon immunization with NP-CGG in alum, they produced more high-affinity NP4-binding IgG1 and IgG2a, the two main IgG isotypes induced by NP-CGG, with normal expression of the genes encoding IL-4 and IFNγ, which direct CSR to IgG1 and IgG2, respectively (Fig. 1J, 1K). This and the previous findings showing reduced IgG responses in B cell-specific KO IghCγ1-creRab7fl/fl mice provide two-sided evidence for the B cell-autonomous role of RAB7 in the antibody response.
RAB7 overexpression upregulates CSR in vivo and in vitro
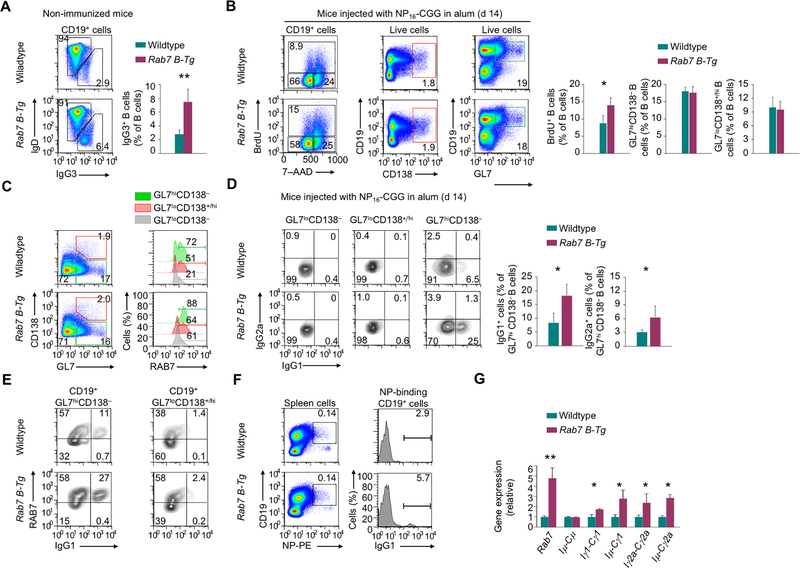
Non-immunized Rab7 B-Tg mice had increased frequencies of IgG3+ B cells in the spleen (Fig. 2A). Upon immunization, they displayed enhanced B cell proliferation, but largely normal B cell differentiation into (GL7hiCD138–) germinal center B cells and (GL7loCD138+/hi) plasma cells (Fig. 2B). While (GL7loCD138–) naïve Rab7 B-Tg B cells remained IgG-negative despite their high RAB7 expression, germinal center B cells from the same mice underwent CSR to IgG1 and IgG2a, and did so at higher levels than their wildtype counterparts in spite of the reduced difference in RAB7 expression due to induction of endogenous RAB7 (Fig. 2C, 2D). Irrespective of the endogenous or exogenous source of RAB7, IgG1+ B cells segregated within RAB7hi cells (Fig. 2E). RAB7 overexpression also resulted in elevated CSR in antigen-specific NP-binding B cells (Fig. 2F) and increased levels of IgH post-recombination transcripts Iμ-Cγ1 and Iμ-Cγ2a (Fig. 2G).
FIGURE 2.
RAB7 overexpression upregulates CSR in vivo. (A) Flow cytometry analysis of surface IgD and IgG3 expression in B cells from the spleen in mice before NP-CGG/alum immunization (n=3, mean and s.e.m. in the right panel). (B) Flow cytometry analysis of spleen B cell proliferation (left) and differentiation into plasma cells (middle) and germinal center B cells (right) in mice immunized with NP-CGG in alum for 14 d (n=3, mean and s.e.m.). (C) Flow cytometry analysis of B cell differentiation into germinal center B cells and plasma cells (left), and RAB7 expression (right) in immunized mice, as in (B). Representative of three independent experiments. (D) IgG1 and IgG2a expression in different mouse spleen B cell subsets, as in (C; mean and s.e.m., n=3 in the histogram, right). (E) Surface expression of IgG1 and intracellular expression of RAB7 in germinal center B cells and plasmablasts in immunized mice, as in (C). Representative of three independent experiments. (F) Surface expression of NP-binding BCR in B cells (left; numbers indicate the proportions of NP-specific B cells) and IgG1 expression within NP-specific B cells (right; numbers indicate the proportions of IgG1+ cells) in immunized mice, as in (B). Representative of three independent experiments. (G) Levels of different transcripts, as indicated, in B cells from immunized mice, as in (B). Data were normalized to the values in wildtype mice (mean and s.d., triplicates). *, p < 0.05; **, p < 0.01.
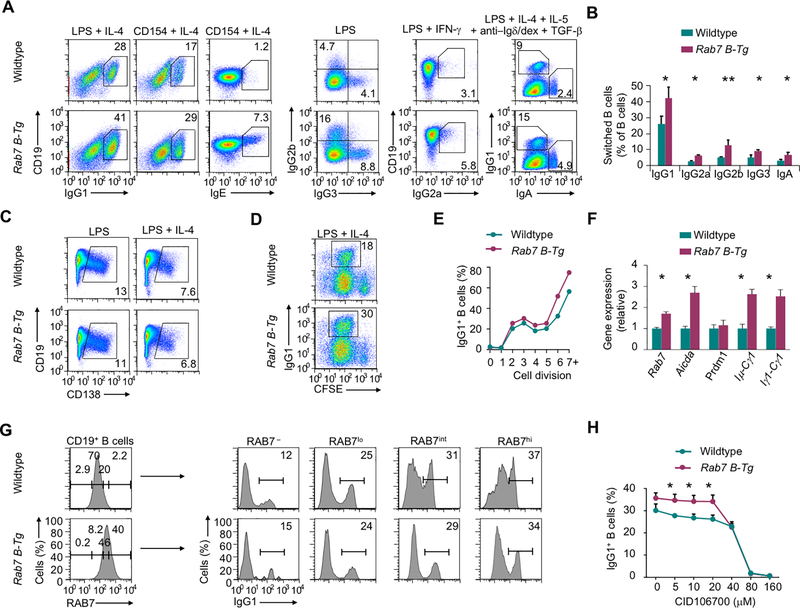
Like germinal center B cells, Rab7 B-Tg B cells stimulated in vitro by LPS, which engaged both TLR4 and BCR (1,23), or CD154 plus appropriate secondary stimuli showed higher CSR, but normal plasma cell differentiation, than their wildtype counterparts (Fig. 3A–C). The increase in CSR was across all cell divisions and associated with upregulation of IgH germline transcription and Aicda expression, but normal expression of Prdm1, which encodes Blimp-1 (Fig. 3D–F). Like in vivo, IgG1+ B cells generated in vitro preferentially segregated within RAB7hi/RAB7int cells in both Rab7 B-Tg and wildtype B cells (Fig. 3G). Finally, Rab7 B-Tg B cells showed higher CSR in the presence of CID 1067700, consistent with the specific targeting of RAB7 by CID 1067700 (Fig. 3H).
FIGURE 3.
RAB7 overexpression upregulates CSR in vitro. (A) Expression of class-switched Ig isotypes in B cells stimulated in vitro, as indicated. (B) Proportion of class-switched Ig isotypes in B cells stimulated in vitro by LPS plus appropriate cytokines (n=5 in IgG1, n=3 in other IgG isotypes and IgA; mean and s.e.m.). (C) Flow cytometry analysis of plasma cells after B cells were stimulated with LPS or LPS plus IL-4 for 2 d. Numbers indicate the proportions of CD19loCD138hi plasma cells. Representative of three independent experiments. (D, E) Flow cytometry analysis of IgG1+ B cells after stimulation of CFSE-labeled B cells by LPS plus IL-4 (D). Proportions of IgG1+ cells within B cells that had completed each cell division, as assessed by CFSE intensities, were depicted in (E). Representative of two independent experiments. (F) Levels of different transcripts in B cells stimulated with LPS plus IL-4. Data were normalized to the values in wildtype B cells (mean and s.d., triplicates). (G) Intracellular expression of RAB7 in B cells stimulated with CD154 plus IL-4 for 96 h (left) and surface expression of IgG1 in B cells with different levels of RAB7 expression. Representative of two independent experiments. (H) Proportions of IgG1+ B cells stimulated with LPS plus IL-4 in the presence of different doses of CID 1067700 (mean and s.d., triplicates). *, p < 0.05; **, p < 0.01.
RAB7 potentiates NF-κB activation and interacts with TRAF6 to enhance TRAF6 activity
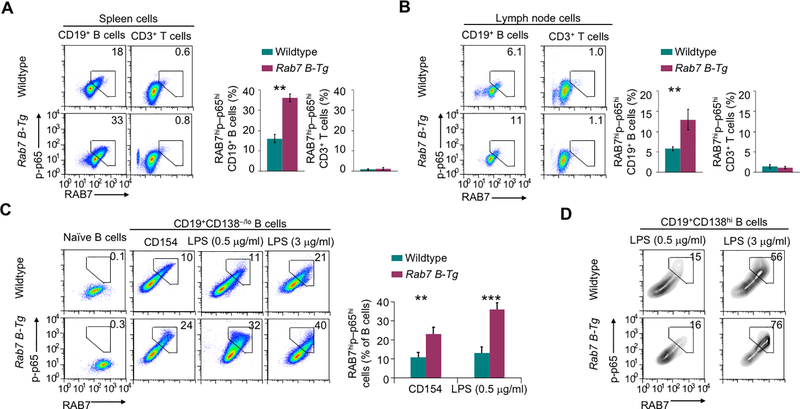
The dependency of AID/CSR induction on NF-κB prompted us to analyze NF-κB activation in B cells from immunized Rab7 B-Tg mice. Only B cells that expressed high levels of RAB7 displayed phosphorylation of the NF-κB p65 (RELA) subunit at Ser532, a hallmark of NF-κB activation through the canonical pathway, with Rab7 B-Tg B cells showing a higher degree of RAB7 expression and p65 phosphorylation than wildtype B cells (Fig. 4A, 4B). A B cell-intrinsic role of RAB7 in NF-κB activation was further confirmed in vitro in highly purified Rab7 B-Tg B cells, which displayed little p65 phosphorylation when freshly isolated despite the high basal levels of RAB7 expression, but upregulated it upon CD154 or LPS stimulation (Fig. 4C). In these cells, p65 phosphorylation was restricted in RAB7hi cells and occurred at a level higher than that in wildtype B cells. Plasma cells generated from stimulated Rab7 B-Tg and wildtype B cells also showed p65 phosphorylation in RAB7hi cells (Fig. 4D).
FIGURE 4.
RAB7 potentiates NF-κB activation. (A,B) Flow cytometry analysis of intracellular levels of RAB7 expression and NF-κB p65 phosphorylation in B and T cells in the spleen (A) and pooled lymph nodes (B) of immunized mice. Numbers indicate the proportions of B cells showing high RAB7 expression and p65 phosphorylation (mean and s.e.m., n=3 in the histograms). (C) Flow cytometry analysis of RAB7 expression and p65 phosphorylation in freshly isolated naïve B cells and B cells stimulated in vitro with CD154 or LPS for 48 h (mean and s.e.m., n=4). (D) Flow cytometry analysis of RAB7 expression and p65 phosphorylation in plasma cells generated after in vitro B cell stimulation by LPS, as indicated, for 48 h (CD154 stimulation did not generate enough plasma cells for analysis). Representative of three independent experiments. **, p < 0.01; ***, p < 0.005.
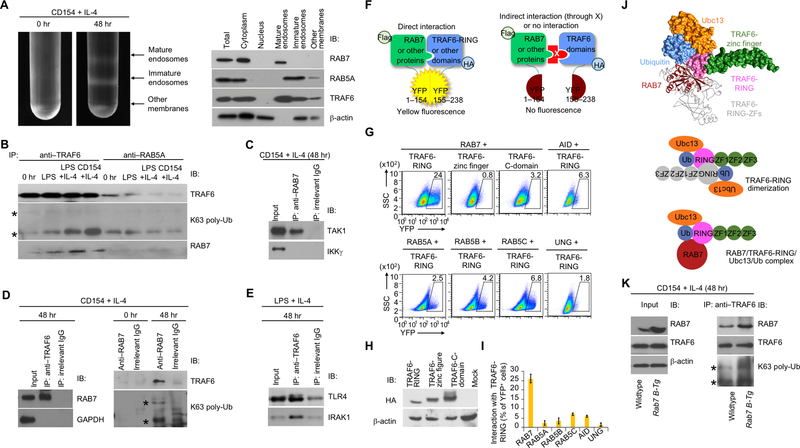
RAB7 interacts with TRAF6 and enhances TRAF6 activity
The enhancement of p65 phosphorylation by overexpression of RAB7, which is the marker of mature endosomes, suggested the involvement of these intracellular membrane structures in NF-κB activation in B cells. While virtually absent in naïve B cells, immature and mature endosomes formed in CD154-activated B cells (Fig. 5A). In these cells, TRAF6 was present mainly in (RAB7-marked) mature endosomes and co-immunoprecipitated with RAB7, together with its K63 polyubiquitination form and in association with RAB7 and TAK1, but not IKKγ (Fig. 5B–D). Likewise, TRAF6 co-immunoprecipitated with RAB7 as well as TLR4 receptor and signal adaptor IRAK1 in LPS-stimulated B cells (Fig. 5B, 5E). By contrast, TRAF6 showed only weaker association with RAB5 and much less localization in RAB5-marked immature endosomes or other membranes, which would include the plasma membrane, where CD40 and TLR4 were located prior to the engagement (Fig. 5A, 5B). The specific association of TRAF6 with RAB7 was consistent with its direct interaction with RAB7 (but not RAB5) through the N-terminal RING domain, which did not interact with AID or UNG, an enzyme involved in the processing of AID-generated DNA lesions (Fig. 5F–I). Finally, consistent with the elevated NF-κB activation and AID/CSR induction in Rab7 B-Tg B cells, RAB7 overexpression upregulated the RAB7/TRAF6 interaction and TRAF6 K63 polyubiquitination (Fig. 5J).
FIGURE 5.
RAB7 interacts with TRAF6 and enhances TRAF6 activity. (A) Intracellular membrane fractionation of B cells before or after CD154 plus IL-4 stimulation for 48 h (left) and detection of protein molecules in different member fractions by immunoblotting (right). (B) Immunoblotting analysis of proteins immunoprecipitated by anti-TRAF6 or anti-RAB5A IgG from B cells prior to stimulation (0 hr) or stimulated as indicated for 48 h (K63 poly-Ub, K63 polyubiquitination). Two bands denoted by “*” indicated K63 polyubiquitinated TRAF6. (C) Immunoblotting of proteins immunoprecipitated by anti-RAB7 IgG or by an irrelevant IgG from B cells stimulated with CD154 plus IL-4 for 48 h. (D) Immunoblotting analysis of proteins immunoprecipitated by anti-TRAF6 (left panels), anti-RAB7 (right panels) or an irrelevant IgG from B cells prior to stimulation (0 hr) or stimulated as indicated for 48 h. (E) Immunoblotting of proteins immunoprecipitated by anti-TRAF6 IgG or by an irrelevant IgG from B cells stimulated with LPS plus IL-4 for 48 h. (F) Illustration of the principle of the BiFC assay. (G) BiFC analysis of direct interaction of TRAF6-RING domain with RAB7, but not other proteins, as indicated. (H) Immunoblotting of expression of HA-tagged different TRAF6 domains. (I) BiFC analysis and quantified interaction of TRAF6-RING with different protein factors, as indicated (n=3, mean and s.e.m. in the histogram). (J) Immunoblotting analysis of total proteins (input) and proteins immunoprecipitated by anti-TRAF6 in stimulated B cells. (K) The interaction of RAB7 (red) with the TRAF6-RING domain (pink) in complex with ubiquitin (blue) and Ubc13 (orange), as modeled by docking analysis (top panel; PDB ID: RAB7 1yhn; TRAF6-RING/Ubc13/ubiquitin: 5vo0). In general, TRAF6 needs to be dimerized to catalyze K63 polyubiquitination, with the two RING domains in an anti-parallel conformation to stabilize the interaction with ubiquitin through reciprocal binding to the first zinc finger (ZF1, green; bottom panel). RAB7 can substitute the RING-domain and ZF1 of one TRAF6 monomer, as depicted by its overlaying of a TRAF6 monomer (grey, top panel), in stabilizing the interaction of TRAF6-RING and ubiquitin (middle panel). A-E, H, J, representative of two independent experiments.
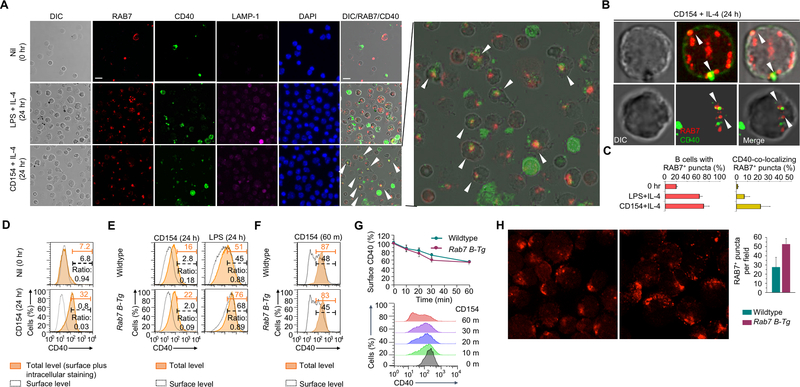
CD40 is localized in the cytoplasm upon engagement and co-localizes with RAB7 in activated B cells
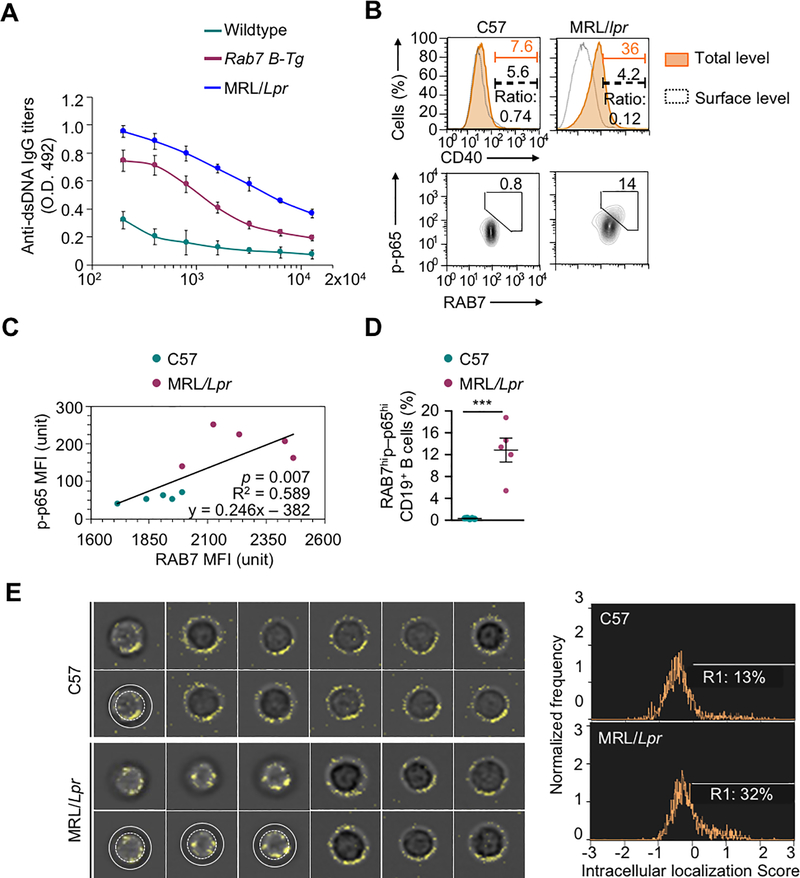
Engagement of CD40 and TLR4 triggers the recruitment of TRAF6 through binding of its C-terminal (TRAF-C) domain to CD40 and IRAK1, respectively. As TRAF6 directly interacted with RAB7 through its N-terminal RING domain, we reasoned that TRAF6 and RAB7 would form a complex with CD40 (or TLR4 together with IRAK1) in mature endosomes. Supporting this notion, RAB7, upon induction by CD154, readily formed multi-molecule puncta, in contrast to LAMP-1, which showed a largely diffused pattern with a small proportion forming puncta (Fig. 6A). RAB7 also co-localized with CD40 in the cytoplasm (Fig. 6A–C), consistent with an increased proportion (e.g., 80%) of B cells showing higher overall CD40 expression but little surface CD40 expression (Fig. 6D, 6E), likely due to internalization of CD40 upon engagement (Fig. 6F, 6G). Overexpression of RAB7 in activated Rab7 B-Tg cells increased the number of RAB7+ puncta (Fig. 6H), but did not accelerate CD40 internalization (Fig. 6G). Like B cells from immunized Rab7 B-Tg mice, which generated anti-dsDNA IgG autoantibodies (Fig. 7A), B cells from MRL/Faslpr/lpr lupus mice displayed high levels of RAB7 expression and NF-κB activation that were correlated (Fig. 7B–D). Accordingly, they showed more CD40 intracellular localization (Fig. 7E).
FIGURE 6.
CD40 intracellular localization in activated normal B cells. (A) Immunofluorescence staining and differential interference contrast (DIC) microscopy of RAB7, CD40 and LAMP-1 in B cells prior to stimulation or stimulated with LPS plus IL-4 or CD154 plus IL-4 for 24 h. The panel showing RAB7/CD40 co-localization in CD154-stimulated B cells was enlarged (right). The white arrowheads indicate co-localization of RAB7 and CD40 (original magnification: 1,000X; scale bar, 5 μM). (B) Confocal microscopy of CD40 and RAB7 in two individual B cells stimulated with CD154 plus IL-4. The white arrowheads in the B cell indicate co-localization of CD40 with RAB7-containing puncta, as shown in yellow. The co-localizing areas in the first cell (left) were dense in molecules, as shown by DIC microscopy, and close to the plasma membrane. (C) Quantification of RAB7+ puncta in B cells before or after stimulation, as indicated, and co-localization of RAB7 foci with CD40 only in B cells stimulated with CD154 plus IL-4 (n=3, mean and s.e.m.). (D) Flow cytometry analysis of surface CD40 expression (dashed black lines) and total CD40 expression (surface plus intracellular, orange shaded areas) in B cells before or after stimulation by CD154 plus IL-4. Numbers colored in black and orange indicate the proportions of B cells showing high surface and overall CD40 expression, respectively, and were used to calculate the ratio of cells displaying only surface CD40 expression, as the readout of the inverse of CD40 intracellular localization. (E) Total (orange shaded area) and surface (dashed black lines) CD40 expression, as in (D), in wildtype and Rab7 B-Tg B cells stimulated with CD154 or LPS for 24 h. The ratio of cells displaying only surface CD40 expression, as the readout of the inverse of CD40 intracellular localization. (F,G) Surface and total CD40 levels in wildtype and Rab7 B-Tg B cells primed with LPS for 24 h and then stimulated with CD154 for different time. The numbers below gates indicated the proportion of B cells showing high total or surface CD40 levels (F) and were used to calculate the proportion of CD40 molecules on the B cell surface (G, top, n=3). The time-dependent loss of surface CD40 expression, e.g., about 50% within 60 min (G, bottom), reflected CD154-triggered CD40 internalization. (H) Immunofluorescence staining of RAB7 in wildtype and Rab7 B-Tg B cells stimulated with LPS plus IL-4 for 48 h. Quantification of the number of RAB7+ puncta in B cells was also depicted (right). D-G, representative of two independent experiments.
FIGURE 7.
CD40 intracellular localization in lupus B cells. (A) ELISA of titers of circulating anti-dsDNA autoantibodies in Rab7 B-Tg mice and their wildtype counterparts (n=3; mean and s.d.). (B) Flow cytometry analysis of total (orange shaded area) and surface (dashed black lines) CD40 expression in B cells from PBMCs of MRL/Lpr and C57 mice (top panels). The ratio of cells displaying only surface CD40 expression, as the readout of the inverse of CD40 internalization. Also depicted is flow cytometry analysis of B cells showing high RAB7 expression and NF-κB p65 phosphorylation in such B cells (bottom panels (bottom panels). (C) Linear regression of intracellular RAB7 expression and levels of phosphorylated p65 in B cells from PBMCs of MRL/Lpr and C57 mice, as determined by MFIs in flow cytometry (right; n=5; p value, F test). (D) RAB7 expression and p65 phosphorylation in B cells from PBMCs of MRL/Lpr and C57 mice (mean and s.d., n=5 in the histogram). (E) Flow imaging analysis of CD40 intracellular localization in CD19+ spleen B cells. The images of B cells showing CD40 intracellular localization, i.e., 1 out of 11 representative C57 B cells and 3 out of 9 representative MRL/Lpr B cells, were duplicated beneath for the annotation by white concentric circles of the peripheral dark ring, which indicated the cell surface and resulted from lack of surface staining of CD40. Also depicted is the proportion of B cells showing CD40 intracellular localization score of >0 (histogram, right). B, E, representative of at least two independent experiments). ***, p < 0.005.
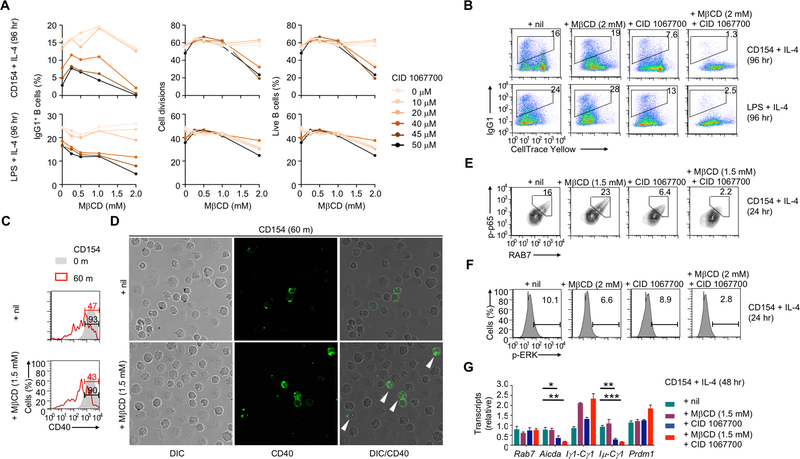
Lipid raft disruption and RAB7 inhibition synergize to abrogate NF-κB activation
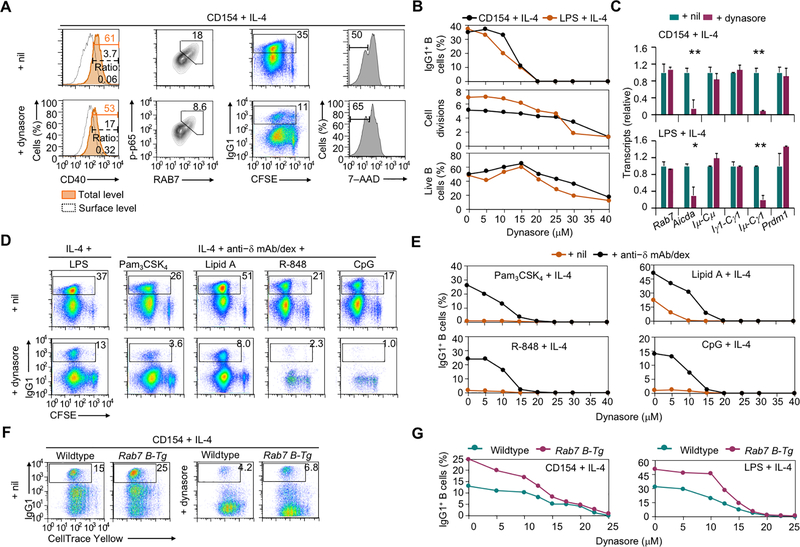
Blocking receptor internalization in CD154- and LPS-stimulated B cells by dynasore (15 μM), a small molecule inhibitor of dynamin, impaired NF-κB activation, Aicda expression and CSR (e.g., decreased IgG levels and Iμ-Cγ post-recombination transcripts) with marginal changes in B cell proliferation, survival and expression of Rab7 and Prdm1 (Fig. 8A–D). The dynasore treatment also reduced CSR in B cells activated by TLR engagement and BCR crosslinking and, in the case of TLR7/8 and TLR9, even B cell proliferation, likely reflecting the key role of trafficking of these endosomal TLRs during their maturation (Fig. 8D, 8E). The CSR enhancement in Rab7 B-Tg B cells was largely abrogated by the dynasore treatment (Fig. 8F, 8G), showing that RAB7 overexpression promoted CSR in a manner dependent on receptor internalization.
FIGURE 8.
CD40 internalization in activated B cells promotes NF-κB activation and CSR. (A) Flow cytometry analysis of CD40 internalization, intracellular RAB7 expression and levels of phosphorylated p65 in B cells stimulated with CD154 plus IL-4 for 24 h and treated with nil (DMSO) or dynasore (left two columns), and CSR to IgG1 and viability (7–AAD–) in CFSE-labeled B cells stimulated with CD154 plus IL-4 for 96 h and treated with nil or dynasore (right two columns). Representative of three independent experiments. (B) Flow cytometry analysis of IgG1+ cells (top), average divisions completed by B cells (middle) and proportion of 7–AAD– live B cells (bottom) after CFSE-labeled B cells were stimulated with CD154 or LPS plus IL-4 and treated with dynasore at different doses for 4 d. Representative of two independent experiments. (C) Levels of different transcripts in B cells stimulated with CD154 (top) or LPS (bottom) plus IL-4 and treated with nil (DMSO) or dynasore. Data were normalized to the values in nil-treated B cells (mean and s.d., triplicates). Representative of two independent experiments. (D, E) Flow cytometry analysis of IgG1+ B cells after CFSE-labeled B cells were stimulated with LPS (left) or different TLR ligands together with anti–IgD mAb/dex (right) plus IL-4 and treated with nil (DMSO) or dynasore at 15 μM (D) or different doses (E) for 4 d. Proportions of IgG1+ B cells were depicted in the histograms (E). Representative of two independent experiments. (F,G) Flow cytometry analysis of IgG1+ B cells after CellTrace Yellow-labeled wildtype and Rab7 B-Tg B cells were stimulated with CD154 plus IL-4 and treated with nil (DMSO) or dynasore at 15 μM (F) or different doses (G) for 4 d. Representative of three independent experiments. *, p < 0.05; **, p < 0.01.
In addition to trafficking into RAB7-marked mature endosomes, internalized CD40 or TLR4 could remain associated with the plasma membrane by localizing on cholesterol-rich lipid rafts, from which they emanate signals. We reasoned that disruption of such lipid rafts, e.g., by methyl-β-cyclodextrin (MβCD) (20,24), would divert more internalized CD40 molecules to the RAB7-dependent endosomal pathway. Indeed, treatment of CD154-stimulated B cells with MβCD at selected doses did not affect CD40 internalization (i.e., loss of surface CD40), but increased CD40 intracellular localization and enhanced NF-κB activation and CSR (Fig. 9A–D), likely by augmenting the RAB7-dependent pathway. Importantly, MβCD combined with RAB7-specific inhibitor CID 1067700 severely impaired NF-κB and ERK activation and cell proliferation/survival, and virtually abolished AID/CSR induction by CD154 or LPS (Fig. 9A–B, E–G), showing the functional redundancy of the MβCD-sensitive pathway and RAB7-dependent pathway.
FIGURE 9.
Lipid raft disruption and RAB7 inhibition synergize to abrogate NF-κB activation. (A,B) Flow cytometry analysis of CSR to IgG1, proliferation and viability (7–AAD–) of CellTrace Yellow-labeled B cells stimulated with CD154 or LPS plus IL-4 for 96 h and treated with nil, MβCD, CID 106770 or both at different doses, as indicated. Representative of three independent experiments. (C) FACS analysis of surface CD40 in B cells primed with LPS for 24 h and then stimulated with CD154 for 0 or 60 m in the presence of nil or MβCD, as indicated. The numbers indicated the proportions of B cells showing high levels of surface CD40. (D) DIC microscopy and immunofluorescence staining of CD40 in B cells primed with LPS for 24 h and then stimulated with CD154 for 60 m in the presence of nil or MβCD. The white arrowheads indicate intracellular localization of CD40 (original magnification: 1,000X). (E,F) Flow cytometry analysis of RAB7 expression and p65 phosphorylation (E) as well as of ERK phosphorylation (F) in B cells stimulated with CD154 plus IL-4 for 24 h and treated with nil, MβCD, CID 106770 or both. Representative of two independent experiments. (G) Levels of different transcripts in B cells stimulated with CD154 (top) or LPS (bottom) plus IL-4 and treated with nil (DMSO) or dynasore. Data were normalized to the values in nil-treated B cells (mean and s.d., triplicates). Representative of two independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.005.
Discussion
Here we have shown a new mechanisms of B cell NF-κB activation for AID induction, CSR (and likely AID-dependent SHM) and the maturation of the antibody response. Upon engagement, immune receptors, such as CD40, were localized in the cytoplasm, where they were associated with signal adaptor TRAF6 in RAB7-marked mature endosomes, as facilitated by direct RAB7/TRAF6 interaction, to enhance TRAF6-catalyzed K63 polyubiquitination. Such a RAB7/mature endosome-dependent pathway could be strengthened by RAB7 overexpression, as occurring in Rab7 B-Tg B cells, or the MβCD treatment due to the diversion of CD40 or TLR4 from the lipid raft-dependent pathway. When both pathways were inhibited, NF-κB activation was severely impaired, providing strong evidence for the functional redundancy of RAB7-dependent intracellular membrane signalosomes and lipid raft-anchored plasma membrane signalosomes in B cell NF-κB activation. Extending our current study on RAB7, future identification of the linchpin factors in plasma membrane signalosomes would further verify the functional redundancy of the two pathways through genetic (e.g., double KO) or pharmacological inhibitor approaches. Such factors may be shared by previously reported CD40-containing signalosomes localized in lipid rafts in B lymphoma cells and DCs (19–22). They may also mediate the crosstalk with signalosomes assembled on the plasma membrane following BCR crosslinking (25) – BCR can also be endocytosed upon crosslinking, but, instead of transducing signals, mostly recycled back to the cell surface or sorted to the lysosomal degradation pathway for antigen processing and MHC presentation (26).
Despite the redundancy by plasma membrane signalosomes, the RAB7-nucleated intracellular membrane signalosomes probably play a major pathophysiological role in B cell NF-κB activation through the canonical pathway, as indicated by the strong correlation of RAB7 expression with NF-κB p65 phosphorylation in normal and lupus B cells, likely due to the RAB7 upregulation and its enhancement of the enzymatic activities of TRAF6, also consistent with the role of TRAF6 in NF-κB activation through the canonical, but not non-canonical, pathway. The endosomal localization of TRAF6 was unexpected, as this adaptor had been thought to be localized in the cytosol and, upon CD40 engagement, recruited to lipid rafts (21). According to our in silico modeling, RAB7 could potentially form a complex with the RING domain of TRAF6 in either the monomeric or trimeric form, Ubc13 E2 ubiquitin ligase and ubiquitin, thereby stabilizing the TRAF6-RING/Ubc13/ubiquitin complex (Fig. 5K), within which RAB7/TRAF6 interaction be sufficient to stabilize the ubiquitin binding for K63 polyubiquitination without the need for RING domain dimerization (27). Alternatively, RAB7 may simply stabilize the CD40/TRAF6 interaction on endosomes. In either case, intracellular membrane signalosomes would be advantageous over plasma membrane signalosomes for heightened signal outputs and may involve different IKKβ kinases for signal transduction, as suggested by the association of RAB7 with TAK1, but not IKKγ. Within RAB7, the TRAF6-interacting interface was apparently outside the catalytic pocket (as occupied by GTP, GDP or CID 1067700) and the second binding site of CID 1067700 (our in silico modeling). The role of TRAF6/RAB7 interaction in NF-κB activation could be further verified by CSR impairment when such interaction is disrupted by point mutations at the interaction interface or small molecules identified by high-throughput screening.
As a prerequisite for the generation of the RAB7/mature endosome-dependent intracellular membrane signalosomes, endocytosed CD40 and TLR4, as originally in microdomains of the plasma membrane, need to traffick through RAB5-marked immature endosomes, possibly assisted by JLP scaffold protein, before reaching RAB7-marked mature endosomes (17). Analysis of such a dynamic process, as lacking in the current studies, would be best performed using live cell imaging involving fluorochrome-labeled recombinant CD154 (or agonistic anti-CD40 mAb) and RAB7-RFP fusion protein (28). Previous cell line-based studies have shown that CD40 internalization or localization in lipid rafts occurred in B cells and other cell types (18–22,29,30) and used TRAF2 and TRAF3 (31) for signal transduction. By contrast, in mature endosomes, CD40 would recruit TRAF6 to form macromolecular complexes through multi-valent protein interactions, as CD40 and RAB7 interacted with different TRAF6 domains. Trafficking of CD154-engaged CD40 within T cell:B cell synapses likely involves formation of mini-vesicles that enclose the CD154-CD40 conjugate and could travel in both directions, to B cells and T cells (32,33) – along this line, a subset of CD40 may “bud out” and become circulating biomarkers. In lupus B cells, CD40 is likely upregulated due to strong stimulation by TLR ligands or CD154 from hyper-activated T cells, and extensively endocytosed.
RAB7 overexpression per se in naïve Rab7 B-Tg B cells failed to trigger NF-κB activation, as other requirements of signalosome formation (e.g., CD40/TLR internalization and intracellular membrane biogenesis) had not yet been met. In activated B cells, the two spatially distinct signalosomes would show different kinetics in their formation, with plasma membrane signalosomes rapidly forming and triggering the first wave of NF-κB activation. This would feed into the second phase by initiating the upregulation of RAB7 and biogenesis of intracellular membrane structures, such as endosomes (shown here), ER and autophagosome-like double-membrane structures, all of which could support signalosome formation in immune cells (34,35). Thus, the exquisite function of RAB7-dependent intracellular membrane signalosomes would be most relevant in B cells with lengthier stimulation (e.g., 24–48 hr), and was likely missed by previous studies that employed genetics and structure biology approaches to identify components of CD40 and TLR signaling pathways using B cells stimulated for short periods of time, e.g. a few minutes.
The correlation of aberrant RAB7 expression and heightened NF-κB activation in single lupus B cells extends previous findings involving bulk lupus B cells (36,37). As NF-κB hyperactivation is also responsible for a wide range of other disease conditions, such as B cell lymphoma (38,39), RAB7 may be a common causative factor and, therefore, attractive therapeutic targets – NF-κB per se is a poor target due to the critical role of basal NF-κB activity in homeostasis and many combinations of NF-κB heterodimers. Disruption of RAB7-dependent intracellular membrane signalosomes would only block a subset of NF-κB signals, while leaving plasma membrane signalosomes intact to provide basal NF-κB activity to maintain essential functions. It can be achieved by CID 1067700 derivatives or compounds that interfere with the RAB7/TRAF6 interaction, particularly those with good PK/PD profiles and a high therapeutic index.
Supplementary Material
Key Points.
RAB7 small GTPase co-localizes with engaged receptors on mature endosomes in B cells.
RAB7 directly interacts with TRAF6 to transduce receptor signals in NF-κB activation.
RAB7 overexpression in normal and lupus B cells enhances NF-κB and AID induction.
Acknowledgements
We thank Dr. Tonika Lam, Dr. Dmitri Ivanov (UTHSCSA), Dr. William Bauta and Dr. Jonathan Bohman (Southwest Research Institute), and Dr. Aimee Edinger (UC Irvine) for help. We thank Karla Gorena of the UTHSCSA Flow Cytometry Core Facility for help.
C. E. R. was supported by NIH Research Supplement to Promote Diversity in Health Related Research grant AI 105813S-1; J. B. M. by NIH T32 AI 138944; P. C. by NIH grants AI 079705, AI 105813 and T32 AI 138944; and Z. X. by NIH grants AI 124172, AI 131034 and AI 135599. The UTHSCSA Flow Cytometry Core Facility is supported by the NIH/NCI P30 CA054174-20 and NIH UL1 TR001120 grants.
Abbreviations used
- AID
activation-induced cytidine deaminase
- BCR
B cell receptor
- CSR
class switch DNA recombination
- Rab7
Ras-related in brain7
- SLE
systemic lupus erythematosus
References
- 1.Pone EJ, Zhang J, Mai T, White CA, Li G, Sakakura J, Patel PJ, Al-Qahtani A, Zan H, Xu Z, and Casali P. 2012. BCR-signaling synergizes with TLR-signaling to induce AID and immunoglobulin class-switching through the non-canonical NF-κB pathway. Nat. Commun 3, 767 (761–712). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park SR, Zan H, Pal Z, Zhang J, Al-Qahtani A, Pone EJ, Xu Z, Mai T, and Casali P. 2009. HoxC4 binds to the promoter of the cytidine deaminase AID gene to induce AID expression, class-switch DNA recombination and somatic hypermutation. Nat. Immunol 10, 540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran TH, Nakata M, Suzuki K, Begum NA, Shinkura R, Fagarasan S, Honjo T, and Nagaoka H. 2010. B cell-specific and stimulation-responsive enhancers derepress Aicda by overcoming the effects of silencers. Nat. Immunol 11, 148–154. [DOI] [PubMed] [Google Scholar]
- 4.Xu Z, Zan H, Pone EJ, Mai T, and Casali P. 2012. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat. Rev. Immunol 12, 517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsokos GC 2011. Systemic lupus erythematosus. N. Engl. J. Med 365, 2110–2121. [DOI] [PubMed] [Google Scholar]
- 6.O’Neill LA 2002. Signal transduction pathways activated by the IL-1 receptor/toll-like receptor superfamily. Curr. Top. Microbiol. Immunol 270, 47–61. [PubMed] [Google Scholar]
- 7.Chen ZJ 2012. Ubiquitination in signaling to and activation of IKK. Immunol. Rev 246, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adhikari A, Xu M, and Chen ZJ. 2007. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene 26, 3214–3226. [DOI] [PubMed] [Google Scholar]
- 9.Mai T, Pone EJ, Li G, Lam TS, Moehlman J, Xu Z, and Casali P. 2013. Induction of AID-targeting adaptor 14–3-3γ is mediated by NF-κB-dependent recruitment of CFP1 to the 5’-CpG-3’-rich 14–3-3γ promoter and is sustained by E2A. J. Immunol 191, 1895–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li G, White CA, Lam T, Pone EJ, Tran DC, Hayama KL, Zan H, Xu Z, and Casali P. 2013. Combinatorial H3K9acS10ph histone modification in IgH locus S regions targets 14–3-3 adaptors and AID to specify antibody class-switch DNA recombination. Cell Rep. 5, 702–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lou Z, Casali P, and Xu Z. 2015. Regulation of B cell differentiation by intracellular membrane-associated proteins and microRNAs: Role in the antibody response. Front. Immunol 6, 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pone EJ, Lam T, Lou Z, Wang R, Chen Y, Liu D, Edinger AL, Xu Z, and Casali P. 2015. B cell Rab7 mediates induction of activation-induced cytidine deaminase expression and class-switching in T-dependent and T-independent antibody responses. J. Immunol 194, 3065–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rink J, Ghigo E, Kalaidzidis Y, and Zerial M. 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell 122, 735–749. [DOI] [PubMed] [Google Scholar]
- 14.Agola JO, Hong L, Surviladze Z, Ursu O, Waller A, Strouse JJ, Simpson DS, Schroeder CE, Oprea TI, Golden JE, Aube J, Buranda T, Sklar LA, and Wandinger-Ness A. 2012. A competitive nucleotide binding inhibitor: in vitro characterization of Rab7 GTPase inhibition. ACS Chem. Biol 7, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam T, Kulp DV, Wang R, Lou Z, Taylor J, Rivera CE, Yan H, Zhang Q, Wang Z, Zan H, Ivanov DN, Zhong G, Casali P, and Xu Z. 2016. Small molecule inhibition of Rab7 impairs B cell class switching and plasma cell survival to dampen the autoantibody response in murine lupus. J. Immunol 197, 3792–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, Granucci F, and Kagan JC. 2011. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell 147, 868–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang HM, Yan Q, Yang T, Cheng H, Du J, Yoshioka K, Kung SK, and Ding GH. 2015. Scaffold protein JLP is critical for CD40 signaling in B lymphocytes. J. Biol. Chem 290, 5256–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menard LC, Habte S, Gonsiorek W, Lee D, Banas D, Holloway DA, Manjarrez-Orduno N, Cunningham M, Stetsko D, Casano F, Kansal S, Davis PM, Carman J, Zhang CK, Abidi F, Furie R, Nadler SG, and Suchard SJ. 2016. B cells from African American lupus patients exhibit an activated phenotype. JCI Insight 1, e87310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vidalain PO, Azocar O, Servet-Delprat C, Rabourdin-Combe C, Gerlier D, and Manie S. 2000. CD40 signaling in human dendritic cells is initiated within membrane rafts. EMBO J. 19, 3304–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hostager BS, Catlett IM, and Bishop GA. 2000. Recruitment of CD40 and tumor necrosis factor receptor-associated factors 2 and 3 to membrane microdomains during CD40 signaling. J. Biol. Chem 275, 15392–15398. [DOI] [PubMed] [Google Scholar]
- 21.Pham LV, Tamayo AT, Yoshimura LC, Lo P, Terry N, Reid PS, and Ford RJ. 2002. A CD40 Signalosome anchored in lipid rafts leads to constitutive activation of NF-kappaB and autonomous cell growth in B cell lymphomas. Immunity 16, 37–50. [DOI] [PubMed] [Google Scholar]
- 22.Manning E, Pullen SS, Souza DJ, Kehry M, and Noelle RJ. 2002. Cellular responses to murine CD40 in a mouse B cell line may be TRAF dependent or independent. Eur. J. Immunol 32, 39–49. [DOI] [PubMed] [Google Scholar]
- 23.Pone EJ, Lou Z, Lam T, Greenberg ML, Wang R, Xu Z, and Casali P. 2015. B cell TLR1/2, TLR4, TLR7 and TLR9 interact in induction of class switch DNA recombination: modulation by BCR and CD40, and relevance to T-independent antibody responses. Autoimmunity 48, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barman S, and Nayak DP. 2007. Lipid raft disruption by cholesterol depletion enhances influenza A virus budding from MDCK cells. J. Virol 81, 12169–12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fruman DA, Satterthwaite AB, and Witte ON. 2000. Xid-like phenotypes: a B cell signalosome takes shape. Immunity 13, 1–3. [DOI] [PubMed] [Google Scholar]
- 26.Hou P, Araujo E, Zhao T, Zhang M, Massenburg D, Veselits M, Doyle C, Dinner AR, and Clark MR. 2006. B cell antigen receptor signaling and internalization are mutually exclusive events. PLoS Biol. 4, e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Middleton AJ, Budhidarmo R, Das A, Zhu J, Foglizzo M, Mace PD, and Day CL. 2017. The activity of TRAF RING homo- and heterodimers is regulated by zinc finger 1. Nat. Commun 8, 1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cioni JM, Lin JQ, Holtermann AV, Koppers M, Jakobs MAH, Azizi A, Turner-Bridger B, Shigeoka T, Franze K, Harris WA, and Holt CE. 2019. Late endosomes act as mRNA translation platforms and sustain mitochondria in axons. Cell 176, 56–72 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Chen J, Xiong Y, Da Q, Xu Y, Jiang X, and Tang H. 2006. Internalization of CD40 regulates its signal transduction in vascular endothelial cells. Biochem. Biophys. Res. Commun 345, 106–117. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Chen L, Wang G, and Tang H. 2007. Cholesterol-dependent and -independent CD40 internalization and signaling activation in cardiovascular endothelial cells. Arterioscler. Thromb. Vasc. Biol 27, 2005–2013. [DOI] [PubMed] [Google Scholar]
- 31.Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, and Noelle RJ. 2009. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol. Rev 229, 152–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yellin MJ, Sippel K, Inghirami G, Covey LR, Lee JJ, Sinning J, Clark EA, Chess L, and Lederman S. 1994. CD40 molecules induce down-modulation and endocytosis of T cell surface T cell-B cell activating molecule/CD40-L. Potential role in regulating helper effector function. J. Immunol 152, 598–608. [PubMed] [Google Scholar]
- 33.Gardell JL, and Parker DC. 2017. CD40L is transferred to antigen-presenting B cells during delivery of T-cell help. Eur. J. Immunol 47, 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaturvedi A, Dorward D, and Pierce SK. 2008. The B cell receptor governs the subcellular location of Toll-like receptor 9 leading to hyperresponses to DNA-containing antigens. Immunity 28, 799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexia C, Poalas K, Carvalho G, Zemirli N, Dwyer J, Dubois SM, Hatchi EM, Cordeiro N, Smith SS, Castanier C, Le Guelte A, Wan L, Kang Y, Vazquez A, Gavard J, Arnoult D, and Bidere N. 2013. The endoplasmic reticulum acts as a platform for ubiquitylated components of nuclear factor kappaB signaling. Sci. Signal 6, ra79. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Shi Q, Xu X, Chen H, Lin W, Zhang F, Zeng X, Zhang X, Ba D, and He W. 2012. Aberrant CD40-induced NF-kappaB activation in human lupus B lymphocytes. PLoS One 7, e41644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohan C, and Putterman C. 2015. Genetics and pathogenesis of systemic lupus erythematosus and lupus nephritis. Nat. Rev. Nephrol 11, 329–341. [DOI] [PubMed] [Google Scholar]
- 38.Basso K, and Dalla-Favera R. 2015. Germinal centres and B cell lymphomagenesis. Nat. Rev. Immunol 15, 172–184. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Q, Lenardo MJ, and Baltimore D. 2017. 30 years of NF-kappaB: a blossoming of relevance to human pathobiology. Cell 168, 37–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.