Abstract
The IL1A and IL1B genes lie in close proximity on chromosome 2 near the gene for their natural inhibitor, IL1RN. In spite of diverse functions, they are all three inducible through TLR4 signaling but with distinct kinetics. This study analyzed transcriptional induction kinetics, chromosome looping, and enhancer RNA production to understand the distinct regulation of these three genes in human cells. IL1A, IL1B and IL1RN were rapidly induced after LPS, however, IL1B mRNA production was less inhibitable by iBET151, suggesting it does not use pause-release regulation. Surprisingly, chromatin looping contacts between IL1A and IL1B were highly intermingled although those of IL1RN were distinct and we focused on comparing IL1A and IL1B transcriptional pathways. Our studies demonstrated that enhancer RNAs were produced from a subset of the regulatory regions, that they were critical for production of the mRNAs and that they bound a diverse array of RNA-binding proteins, including p300 but not CBP. We furthermore demonstrated that recruitment of p300 was dependent on MAP kinases. Integrator is another RNA-binding protein recruited to the promoters and enhancers and its recruitment was more dependent on NFκB than MAP kinases. We found that Integrator and NELF, an RNA polymerase II pausing protein, were associated with RNA in a manner that facilitated interaction. We conclude that IL1A and IL1B share many regulatory contacts, signaling pathways, and interactions with enhancer RNAs. A complex of protein interactions with enhancer RNAs emphasize the role of enhancer RNAs and the overall structural aspects of transcriptional regulation.
Introduction
The IL-1 family of cytokines is comprised of 11 members that all function in the regulation of inflammation (1). Among the IL-1 family members are IL-1α and IL-1β, the genes for which are located on chromosome 2q14 in close apposition. IL-1α is a dual function cytokine regulating transcription directly through a DNA-binding function as well as regulating the inflammatory response through binding of a cell membrane receptor (2). IL-1α in the nucleus binds chromatin and is generally induced through pro-apoptotic signals. Necrotic signals, in contrast, cause IL-1α to distribute primarily in the cytoplasm (3). When released from the cell, IL-1α stimulates a robust inflammatory response, driving chemokine expression that regulates the influx of neutrophils and monocytes.
IL-1β has similar but non-overlapping functions and it functions as a cytokine. It was discovered as a key inflammatory agent driving fever in rabbits (4). IL-1β is transcriptionally and post-translationally regulated and is primarily induced after recognition of microbial patterns (5, 6). Monocytes appear to be the main cell that produces IL-1β following stimulation with LPS. The transcriptional induction of IL-1β leads to the accumulation of an inactive protein precursor in the cytoplasm (pro- IL-1β that is processed by the activation of nucleotide-binding domain and leucine-rich repeat pyrin containing protein-3 (NLRP3)) to active IL-1β (7).
Near the IL1B gene, encoding IL-1β, lies the gene IL1RN that encodes the IL-1 receptor antagonist (IL-1ra). IL-1ra is a potent inhibitor of both IL-1α and IL-1β (8) by blocking the IL-1 receptor 1. This agent is utilized to treat inflammatory conditions such as arthritis and disorders of the inflammasome (9). In spite of the fact that IL-1ra functions as an antagonist of IL-1α and IL-1β, it too is induced primarily by microbial byproducts. The close approximation of these three genes with similar regulatory patterns offers an opportunity to dissect commonalities and differences in the transcriptional regulatory pattern of these three genes. Understanding the regulation of these three genes is of critical importance in chronic inflammatory conditions where increased expression of the antagonist might provide clinical benefit. We noted increased expression of all three mRNAs in cells from patients with systemic lupus erythematosus (SLE) and the use of the therapeutic IL-1ra antagonist has shown promise in SLE (10). Thus, there is a clinical rationale for the examination of transcriptional pathways regulating this locus.
There is also a strong scientific rationale for the study of enhancer-promoter interactions at this locus. Much of what is understood regarding the mechanism by which enhancers regulate transcription has come from genome-wide studies of developmentally-regulated systems (11, 12). The IL-1 locus, in contrast, exhibits a poised status and the mechanisms of enhancer function are not well understood in this setting (13). In developmental systems, enhancers are active in a cell-specific and development-specific fashion. Upon activation driven by developmental signals, most enhancers are marked by the histone modification H3K27ac and they produce regulatory RNAs (14, 15). The enhancer-derived RNA (eRNA) is important for bridging the enhancer to the proximity of the promoter and plays a role in elongation by RNA polymerase II (RNAPII) (16, 17). The contacts are often stabilized by cohesin and CTCF and loops are not only critical for activation of transcription but CTCF can also act to insulate promoters from enhancer effects, contributing to the specificity of the enhancer effect. 3-dimensional chromatin loops actively regulate the transcriptional program (18–20) and the final transcriptional state of any given promoter integrates signals from multiple enhancers (whose effects are additive) (21), promoter-proximal transcription factors, and insulators and other negative regulators. Very few studies have examined enhancer effects in settings where the induction of expression is rapid and promoters are already poised (22). These genes are of critical importance in inflammatory settings, yet the rules governing enhancer activation in such a setting are largely unexplored. Another compelling reason to better understand the dynamics and mechanisms of this process is the recent recognition that most common variants implicated in disease susceptibility lie in regulatory regions and impact level of transcription, functioning as expression quantitative trait loci (eQTL) (23–25).
Our data demonstrate that these three genes in the IL-1 locus (IL1A, IL1B, IL1RN) lie within a chromosome region with pre-established chromatin loops and a poised transcriptional status. LPS treatment appears to drive eRNA production and nucleation of RNA-binding proteins that collectively regulate expression. In spite of similar kinetics and signaling pathways, transcriptional mechanisms are distinct at the three genes.
Methods
Cells
Primary monocytes were obtained from a campus core facility under an IRB-approved protocol. Subjects gave informed consent. The SLE samples were previously reported and all subjects gave informed consent (26–28), although these analyses are new. The MonoMac6 cell line is a human monocyte line maintained in RPMI with 10% cosmic calf serum with OPI supplement (Sigma-Aldrich, St. Louis, MO). MonoMac6 cells were obtained from the German source and thawed from primary vials every 2–3 months. LPS treatment utilized 1μg.
Transfection of cells
Transfection of cells was performed by electroporation with the Amaxa Cell Line Lonza Nucleofector Kit (Amaxa Biosystems, Gaithersburg, MD) or Lipofectamine 2000 (ThermoFisher, Waltham, MA). Two million cells were transfected with 500nM antisense oligonucleotide using nucleofection. Anti-sense oligonucleotides utilized:
IL1–299ASO1: 5’- G*G*A* A*G*T* T*C*T* T*G*C* T*G*T* G*C*A-3’
IL1–299ASO2: 5’-A*G*A* A*A*T* A*T*C* C*A*C* A*T*T* T*T*T-3’
IL1ALNC327_ASO2: 5’- C*C*C* T*T*T* C*T*G* T*A*C* T*C*T* T*C*C* C*T*C-3’
IL1ALNC327_ASO7: 5’-G*T*T* C*T*C* T*C*C* T*T*C* C*T*C* C*T*T* T-3’
Overexpression primers for vectors:
CD831A327- Fwd: 5’-TAGAGCTAGCGAATTATACAAAATTAGCTGGGCATGG-3’
CD831A299-Fwd: 5’-TAGAGCTAGCGAATTCCCAAAGCTGGACGTGCTG-3’
CD831A299- Rev: 5’-ACGCGGCCGCGGATCAATTGCAGTGTAAAGTGTAAAACAC-3’
CD831A327- Rev: 5’-ACGCGGCCGCGGATCGATGAACTTTCCAATAAGACAAACC-3’
Inhibitors and stimuli
The inhibitors included 20μM C646 (Santa Cruz Biotechnology, Dallas, Texas). It is an inhibitor of p300 induced histone acetylation (29, 30). 20μM iBET151 (Life science Research, Billerica, Massachusetts) was used at as a specific inhibitor of BRD3/4, important for transcriptional activation and elongation at paused genes (31). 40μM DRB (Sigma-Aldrich, Allentown, PA ) was used as an inhibitor of elongation regulating eviction of NELF via P-TEFb (32). 20μM JSH-23 (Santa Cruz Biotechnology) was used as a specific inhibitor of NFκB translocation (33) and 10μM IT901 (Bio-Techne, Minneapolis, MN) was used as a specific antagonist of p65 NFκB (34). 10μM SP600125 (Calbiochem, Darmstadt, Germany) was used as an inhibitor of the JNK MAP kinase. Toxicity analyses confirmed minimal cell death at these concentrations. HPLC-purified LPS was purchased from Sigma-Aldrich (St. Louis, MO).
Transcriptional analysis
Total RNA was extracted by TRI Reagent® (Sigma-Aldrich, St. Louis, MO, USA) and the Direct-zol™ RNA MiniPrep Kit (Zymo Research Irvine, CA) was used for clean-up. The Clontech Advantage RT for PCR kit (Clontech, Mountain View, CA) was used to make cDNA. Transcript levels were detected by real-time PCR using the TaqMan 7900 normalizing to the 18S signal. ncRNAs were detected with custom SYBR green primers and normalized to actin. Primer sequences are listed below.
IL1–327F: 5’- GCATGGAACTGTGCTCTACAGGAA −3’
IL1–327R: 5’- TTCTATCCCATGCAAAGTTGCTCA −3’
IL1–288 FWD : 5’- GCA AGC CGA CTA ATG TGT TAT TT −3’
IL1–288 REV : 5’- GGT CCT TCT AAT CCC GTT GTG −3’
IL1–299 FWD: 5’- CTTGTGTCCTGTTGAGAGAGAG-3’
IL1–299 REV: 5’- CTTGAGTCCCTGCTTCCATATT −3’
IL1–338 FWD: 5’- AGGCTCAGGAATTGGGTATTT-3’
IL1–338REV: 5”-GGGTGGAAAGGTAGAGAAAGAG-3’
896_FWD: ATCCCACCTCTTCCGACT
896_REV: CAGTGACGCTGAATGAATGAATAC
477 FWD: 5’-TGCTCTTCACTATGGCTCTTTAC-3’
477 REV: 5’-GTCAGCGTTGTTGTCAGTTTAC-3’
Capture-C
Promoter connectome analysis was performed essentially as in Chesi et al. (35). Briefly, 1×107 primary monocytes were fixed with 1% formaldehyde. After 10 minutes, 1M glycine was added to stop the process. Cells were multiply washed before ultimately being snap frozen in lysis buffer (10mM Tris, pH8.0, 10mM NaCl, 0.2% NP40 with protease inhibitor) and stored in −80°C. The 3C process was followed as described (35). The pellet was centrifuged and lysis buffer was removed. Protease inhibitor cocktail was added and the resuspended samples were divided into 6 tubes with a predigestion control reserved (control 1). Digestion of DNA utilized DpnII (NEB) overnight at 37°C. Before starting the ligation reaction, 100μL of each digestion was removed and combined to create the digest control (control 2). DpnII was inactivated at 65°C. T4 DNA Ligase (HC ThermoFisher) was added. Proteinase K (Invitrogen) was used to de-crosslink at 65°C and RNAse A (Millipore) was added. The efficiency of the digestion process was tested through quantitative PCR and a 0.9% agarose gel. A 10μg aliquot of each 3C library was then sonicated using a QSonica Q800R. Once the DNA was sheared AMPure XP beads (Agencourt) were used to clean the samples and size was assessed on a Qubit fluorometer (Life Technologies) and a Bioanalyzer 2100. Capture C libraries were made using the Agilent SureSelect XT Library Prep Kit (Agilent). After the pre-capture PCR, the concentration and size of the sample were measured. 1μg of each library was hybridized and captured following the SureSelect XT method. The concentration was tested on the Qubit and the size was measured by BioAnalyzer using a DNA High Sensitivity Chip. Five replicate libraries were barcoded using Nextera Index and free adaptor blocking kits, and sequenced to 1.2 billion 50 bp reads per library on the Illumina NovaSeq 6000 platform. Paired-end reads from five biological replicates for both LPS-treated and untreated Monocytes were pre-processed using the HICUP pipeline (v0.5.9) (36), with bowtie2 as aligner and hg19 as the reference genome. Significant promoter interactions at 1-DpnII fragment resolution were called using CHiCAGO (v1.1.8) (37) with default parameters except for binsize set to 2500. Significant interactions at 4-DpnII fragment resolution were also called at artificial 4 fragment resolution in which DpnII fragments were concatenated in silico into 4 consecutive fragments using default parameters except for remove Adjacent set to False. The significant interactions (CHiCAGO score > 5) from both 1-fragment and 4-fragment resolutions were exported in .ibed format and merged into a single file using custom a PERL script to remove redundant interactions and to keep the max CHiCAGO score for each interaction.
ATAC-seq and data analysis
Chromatin profiling was performed by ATAC-Seq as described previously (38). Briefly, 12,000 to 50,000 cells were washed in cold PBS and lysed in cold lysis buffer (10mM Tris-HCl, pH 7.4, 10mM NaCl, 3mM MgCl2, 0.1% IGEPAL). Transposition was performed at 42°C for 45 min with Nextera DNA library Prep kit (illumnia, San Diego, CA). DNA was purified with the MinElute PCR purification kit (Qiagen, Hilden, Germany), and was amplified for 12 cycles with the primers from Nextera Index kit ( Illumina). Additional PCR cycles were evaluated by real time PCR. Final product was cleaned by Ampure Beads. Libraries were sequenced on a Hiseq 2500 1T in a 50bp/50bp paired end run, using the TruSeq SBS Kit v3 (Illumina). LPS-treated and control Monocyte ATAC-seq peaks were called independently using the ENCODE ATAC-seq pipeline. Briefly, paired-end reads from four biological replicates for each condition were aligned to hg19 genome using bowtie2, and duplicate reads were removed from the alignment. Narrow peaks were called independently for each replicate using macs2 (-p 0.01 --nomodel --shift −75 --extsize 150 -B --SPMR --keep-dup all --call-summits) and ENCODE blacklist regions (ENCSR636HFF) were removed from peaks in individual replicates. Peaks from all replicates were merged by bedtools (v2.25.0) within each cell type and the merged peaks present in less than two biological replicates were removed from further analysis. Finally, ATAC-seq peaks from both conditions were merged to obtain reference open chromatin regions. To determine whether an open coding region (OCR) is present in LPS or control cells, we first intersected peaks identified from individual replicates in each condition with reference OCRs. If any peaks from at least one replicate overlapped with a given reference OCR, we considered the region as open.
ChIP assays
5–10 × 106 cells in each condition were treated with 1 % formaldehyde for 10 min at room temperature to crosslink. Glycine was used to quench the cross-linking, as previously described (39, 40). Lysed cells were sonicated and immunoprecipitated overnight at 4°C. Immune complexes were collected with protein A (Invitrogen, Carlsbad, CA), washed extensively and eluted. DNA was extracted by phenol-chloroform and treated with proteinase K (200 μg/ml, Roche) and RNase (40 μg/ml, Roche). Antibodies utilized included those to: CDK9 (a component of P-TEFb), NELF-A, c-JUN, NFκB p65, CBP and p300 antibodies (all from Santa Cruz Biotechnology, Santa Cruz, CA); H3K4me3 antibody (Active Motif, Carlsbad, CA) and H3K27ac antibody (Active Motif) were used in ChIP assays and RNA-immunoprecipitation. The GST antibody (Invitrogen, Camarillo, CA) was used as a negative control for all ChIP assays. Cells were harvested at 30 minutes after LPS stimulation. Signals are reported as units normalized to 10% input according to the formula 2^(10%input CT-Sample CT). The primers for the ChIP assays are listed below:
IL1–299 Enhancer FWD: 5’-GCATGTTACAGCTGTGGATAAC-3’
IL1–299 Enhancer REV : 5’-GACATGGGTGAAGGGAAAGA-3’
IL1–299 Enhancer PRB: 5’- /56-FAM/AGCATCCTATTGTGTTTAGATGAGCCTGG/3IABkFQ/−3’
IL1– 338 enhancer FWD: 5’-TTTAGATGAGCCTGGTCTTTCC-3’
IL1– 338 enhancer Probe : 5’- /56-FAM/ ATGCACATTAAAGGCCTCCCTGGA (Sense) /3IABkFQ-3’
IL1–338 enhancer REV 5’-TGCTATTGTCTGCCTCACTTC-3’
IL1–327 enhancer FWD: 5’-TTTCCTGAAATCTAGAGAGCATACA-3’
IL1–327 enhancer PRB: 5’−56-FAM/ TAGGCAGTGCTCACCAAATACCTGC/3IABkFQ-3’
IL1–327 enhancer REV: 5’-GCAAGGGAGACCAGGAAATTA-3’
IL1A promoter FWD: 5’-GACTCAAACGCCAATGAAATGA-3’
IL1A promoter PRB: 5’- /56-FAM/ AAAGGCGAAGAAGACTGACTCAGGC /3IABkFQ-3’
IL1A promoter REV: 5’-GTAGCCACGCCTACTTAAGAC-3’
IL1B promoter FWD: 5’-GCCATGCACTGGATGCTG-3’
IL1B promoter REV: 5’-TGGCTGCTTCAGACACCT-3’
IL1B promoter PRB: 5’-/56-FAM/ACACATGAACGTAGCCGTCATGGG/36-/3IABkFQ-3’
GAPDH FWD: 5’-CGGTGCGTGCCCAGTT-3’
GAPDH PRB: 5’-/56-FAM/ACCAGGCGGCTGCGGAAAAAA/36–3IABkFQ-3’
GAPDH REV: 5’-CCCTACTTTCTCCCCGCTTT-3’
RNA-Protein Analyses
The RNA immunoprecipitation (RNA-IP) assay was modified as described (41). Briefly, 20–40 million cells were crosslinked using 1% formaldehyde for 10 min and quenched with glycine. The fixed cells were lysed in RIPA buffer (1%NP40, 0.5% sodium deoxycholate, 0.05%SDS, 1mM EDTA, 150mM NaCl) and sonicated. The precleared lysates were incubated with antibodies and protein A agarose at 4°C. Immune complexes were washed and reverse cross-linked. Direct-zol™ RNA MiniPrep Kit was used to extract the immunoprecipitated RNA.
To test the role of RNA in protein-protein interactions, we treated lysates with RNases before co-immunoprecipitation. MonoMac6 cells were stimulated with HPLC-purified LPS (1μg/ml, Sigma) for 0, 10 and 30 minutes. Cells were lysed and precleared with protein A agarose beads (ThermoFisher). 1500μg of protein was used in each immunoprecipitation. 2μg of antibody (NELF-A, Santa Cruz) was added to the protein and incubated overnight at 4 degrees. 30μl of protein A agarose beads were added for 60 minutes the next day and the beads were washed. Beads were resuspended in 30μl of lysis buffer with 50μg/ml of RNase A (Roche) and incubated at 37 degrees for 15 minutes. After the incubation, they were spun down and both the supernatant and beads were collected. The beads were washed twice in PBS and resuspended in lysis buffer and both supernatant and beads were run on a 4–12% Bis Tris Novex gel (ThermoFisher). The membrane was probed with antibody to the Integrator 11 component (INTS11) (Bethyl Lab). To test the ability of RNA to increase Integrator 11-NELF-A interactions, both proteins were produced as GST-tagged proteins and purified using glutathione beads. In vitro production of eRNAs utilized SP6 or T7. Proteins were mixed 1:1 at 10−8M with a 3X molar excess of eRNAs and incubated at 37° for 15 minutes. Anti-NELF-A was used for recovery of the complexes on protein A beads overnight. Blotting with antibody to Integrator 11 was performed as above.
Statistical analyses
The results represent a minimum of three separate biological experiments. Each figure legend describes the number of individual experiments. Comparisons across treatments were performed using Student’s t test.
Results
Increased NLRP3 activation leading to IL-1β production has been observed in SLE patients, and is thought to be critically important in lupus nephritis (42–45). In our previously published RNA-seq analysis (27), we noted that the IL-1 family genes were up-regulated in monocytes from patients with SLE (Supplemental Table 1) (27). IL1RN, encoding IL-1ra, was increased approximately two-fold over control monocytes, while the IL1A RNA abundance was also significantly increased in monocytes from patients with SLE. IL1B was increased but the difference did not reach statistical significance, and the nearby IL37 gene was not expressed in monocytes. Based on these data, we hypothesized that there may be abnormal transcriptional induction of the IL-1 gene family in SLE and designed our studies to understand commonalities and discordant mechanisms of gene regulation in the IL-1 family at this locus.
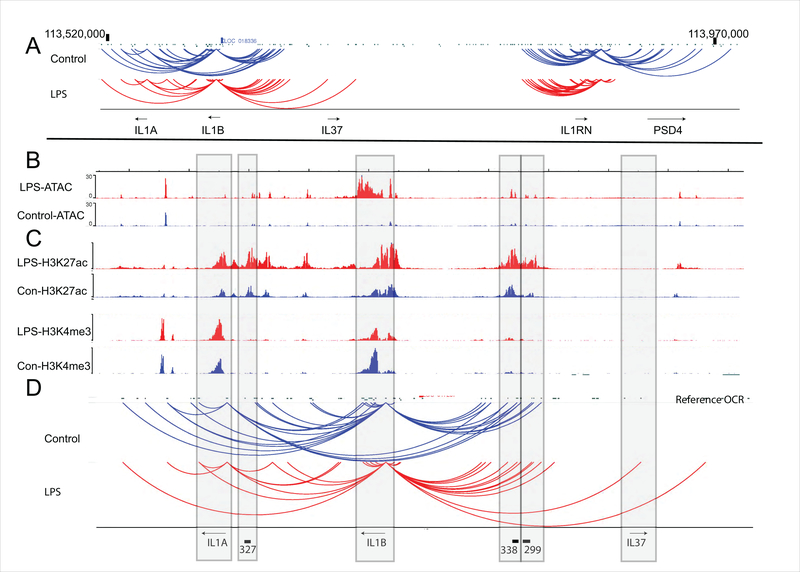
We first established the three dimensional architecture of the IL-1 locus in primary monocytes. The arrangement of the genes in the context of the one-dimensional sequence of the human IL-1 family locus is depicted in Figure 1A, To better understand the transcriptional regulation at this locus, we defined the relationship of the enhancers to the target promoter using a genome-wide promoter-focused chromatin conformation capture approach we recently developed (35). The resulting Capture-C data were utilized to specifically understand the local chromatin folding at this locus. Interactions between regulatory elements and the IL1B, IL1A, and IL1RN promoters were identified in cells stimulated with LPS, and unstimulated cells. Two distinct three-dimensional regulatory domains were apparent (Figure 1A). The first notable finding is that IL1A and IL1B share numerous contacts with putative regulatory elements while IL1RN, in spite of similar transcriptional kinetics, utilizes its own unique set of regulatory contacts. IL37, which is not expressed in these cells, has no loops. The second notable finding is that the looping pattern is more focused after LPS. IL1RN loses all of its downstream contacts after LPS and IL1A loses a major set of loops from the promoter to a cluster of regulatory sites that lie between IL1B and IL37. Loss of loops could imply release from a negative regulatory effect or simply refocusing of loops on the strongest positive regulatory effect. Also noted were loops from each promoter to the 3’ end of the gene. These were stable after stimulation. For better visualization of IL1A and IL1B, expanded views are shown in Figure 1B, 1C, and 1D. ATAC-seq tracks from the same cells as the Capture-C studies are shown in Figure 1B and ChIP-seq tracks from published data are shown in Figure 1C. Figure 1D is an expanded view of the data on Figure 1A. We selected regulatory elements with diverse behaviors: stable contacts (299 and 338 for IL1B), or contacts lost with LPS (327 for IL1B, 299 and 338 for IL1A) for further study based on these findings. All three regulatory elements are embedded in H3K27ac peaks that increase after LPS (Figure 1C). All three have detectable ATAC-seq peaks at baseline that increase after LPS (Figure 1B). 338 and 299 lie in a complex web of contacts upstream of IL1B and have baseline contacts with IL1A and IL1B that simplify to IL1B alone after LPS. The 327 eRNA, in contrast, has contacts with IL1B at baseline which are lost after LPS.
Figure 1. Long-range chromatin interactions at the IL1 region.
Primary monocytes were mock treated (blue) or treated with 1μg/ml LPS (red) for 30 minutes. A) Promoter contacts from IL1A, IL1B, IL37, IL1RN were analyzed by Capture-C in five separate biological replicates. Interactions were designated based on the geometric mean of the replicates. B) ATAC-Seq signal profile across the IL1A, IL1B, and IL37. This was performed on the same cells. C) H3K27ac, H3K4me1 and H3K4me3 peaks from primary monocytes treated with or without LPS for 1 hour. The ChIP-seq data were extracted from a published study (80). D) The long-range chromatin contacts defined by Capture-C and eRNA locations are displayed as an expanded view for IL1A, IL1B, and IL37.
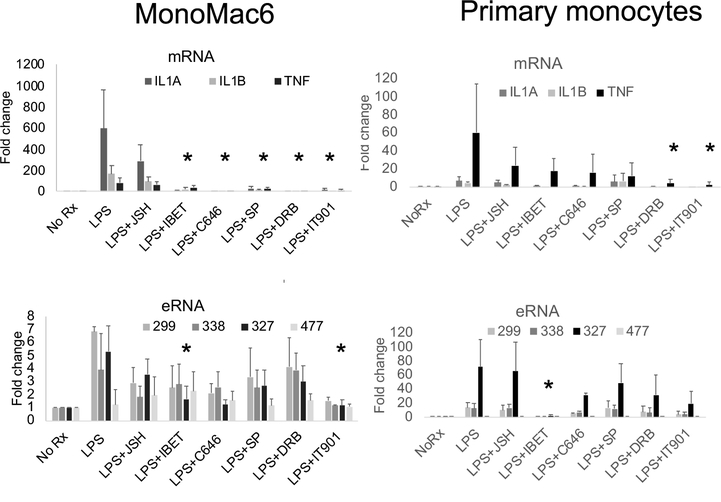
In monocytes, both IL1A and IL1B mRNAs were induced by LPS. We compared responses in a cell line model of peripheral blood monocytes and primary monocytes from healthy donors (Figure 2). Kinetics for the effect of the inhibitors in MonoMac6 cells are shown in Supplemental Figure 1. One key finding is that in the kinetic analysis IL-1B is less inhibitable by iBET151 compared to IL1A and IL1RN, an effect previously described for IL1B, which does not utilize pause release in the standard fashion for transcriptional regulation (46, 47). Both primary monocytes and MonoMac6 cells exhibited induction of IL1A and IL1B after 90 minutes of LPS. The eRNAs were similarly induced after LPS with the exception of 477 which we use as a negative off-target control. 477 is an enhancer RNA located on chromosome 1 but not induced by LPS and was used as a negative control. TNF was included as a positive control for LPS induction (40, 48). There were differences between cell types and between inhibitors, The NFκB inhibitor, IT901, and the BRD4 inhibitor, iBET, had the most consistent inhibitory effects across mRNAs and eRNAs and both cell types. The magnitude of the LPS effect and the inhibitors was greater in MonoMac6 cells, and the need for high cell numbers for the ChIP studies, led us to utilize this model cell line for subsequent studies. In MonoMac6 cells, the 327 RNA species was more inhibitable by iBET and C646 than the other eRNAs. The 299 and 338 eRNAs behaved similarly with respect to inhibitor effects and they lie in a single complex regulatory region upstream of IL1B.
Figure 2. Analysis of LPS induction of RNAs.
MonoMac6 cells and primary monocytes were treated with 1μg/ml LPS for 90 minutes. Total RNA was extracted and qRT-PCR was performed. IL1A, IL1B and TNF were examined as mRNAs. The inhibitors JSH-23, iBET151, C646, SP600125, DRB, and IT901 were tested for effects on RNA abundance. These graphs represent the mean of n=3 biological replicates (error bars represent SD). The asterisks indicate a consistent statistical significance across all measured RNA species (except TNF for mRNAs and 477 eRNAs which represent controls). Supplemental Figure 1 provides additional detailed views of the data in a time course.
Although eRNA production is not a universal requirement for enhancer activation, the majority of active enhancers are actively transcribed in either a unidirectional or bidirectional pattern (49, 50). We defined the location of the RNA transcripts associated with these putative regulatory contact regions using MonoMac6 cells. The RNAs were found in chromatin and they increased after LPS stimulation (Supplemental Figure 2). Chromatin localization supports a regulatory role for these RNA species. Actin (ACTB) is shown as a typical mRNA localized to cytoplasm and XIST is shown as a non-coding regulatory RNA localized to chromatin as controls for cell fraction purity. These results indicate that these RNA species associated with the loop contacts are chromatin-associated and embedded in chromatin with regulatory characteristics. Thus, we will refer to these RNAs as eRNAs.
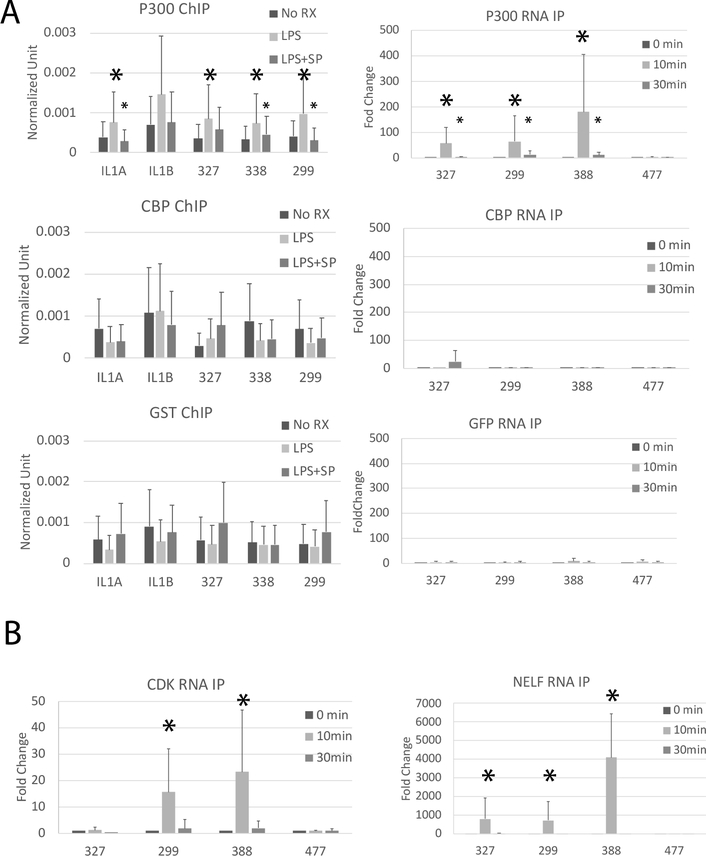
A recognized function of eRNAs is to activate enzymatic activity of the RNA-binding histone acetyltransferases CBP and p300 (51). We hypothesized that one or both of these proteins play a role in enhancer activation at LPS stimulation. To determine if CBP and p300 are localized to the enhancer or promoter of these genes, we performed a ChIP analysis for CBP and p300 using anti-GST as a control (Figure 3A). We observed increased binding of p300, but not CBP, to the enhancers and promoters of IL1A and IL1B following LPS treatment in MonoMac6 cells. SP600125 (JNK inhibitor) inhibited p300 binding to chromatin, indicating that LPS-driven recruitment of p300 to chromatin is JNK-dependent. p300 is directly phosphorylated by MAP kinases (52–54) and it is known to interact with a variety of transcription factors (55–57). Our data place p300 recruitment and activation at an early point in transcriptional activation. CBP surprisingly does not appear to be involved.
Figure 3. LPS induces p300 but not CBP binding to eRNAs and chromatin.
ChIP analysis of p300 and CBP binding. MonoMac6 cells were pretreated with JNK inhibitor SP600125 (SP) for 30 minutes and then stimulated with 1μg/ml LPS for 15 minutes. ChIP assays were performed using p300, CBP or GST (negative control) antibodies. Bound promoter (IL1B and IL1A) and enhancers (327, 338, 299) were detected by qPCR. Results are the means plus standard deviation (n=4, * p<0.05 comparing LPS treatment to unstimulated [large asterisks], and inhibitor plus LPS to LPS alone [small asterisks]). RNA-immunoprecipitation (RNA IP) was used to identify RNA binding to proteins. MonoMac6 cells were treated with 1μg/ml LPS for 0, 10 or 30 minutes. RNA IPs were performed using p300, CBP, or GFP (negative control) antibodies. Bound RNAs were detected by qRT-PCR. 477 is an enhancer RNA located on chromosome 1 but not induced by LPS and was used as a negative control. Results displayed represent the means plus standard deviation (n=4, asterisks indicate the same as for ChIP assays). B) MonoMac6 cells were treated with 1μg/ml LPS for 0, 10, or 30 minutes. RNA immunoprecipitation was performed using NELF-A or CDK9. Bound RNAs were detected by qRT-PCR. The eRNAs were observed to significantly bind to CDK9 and NELF upon LPS treatment, peaking at 10 minutes. 477 is a control eRNA that is not LPS inducible. Results displayed represent the means with standard deviations (n=4, * p<0.05).
To further test whether CBP and/or p300 regulate IL-1 family members via eRNA binding, we performed RNA-immunoprecipitation by pulling down CBP and p300 and analyzing the bound RNA by qRT-PCR (Figure 3A). GFP is a negative control antibody. Consistent with the chromatin binding results, eRNA binding to p300 was significantly increased after LPS treatment, while eRNA binding to CBP was not detected. These results together suggest that LPS-driven enhancer activation of IL-1 family genes is mediated by JNK-dependent p300 binding to chromatin and rapid eRNA interaction with p300.
To additionally understand whether other RNA-binding proteins nucleate at the promoter-enhancer interface, we examined binding of the CDK9 component of P-TEFb and the E component of NELF, two proteins involved in pause-release (Figure 3B). We had previously shown that the SERPINB2 promoter was regulated in a pause-release manner (58). The IL-1 family is similarly induced rapidly after stimulation and we tested whether NELF and the CDK9 component of P-TEFb bind to these eRNAs in a manner similar to that observed at the SERPINB2 locus. Recent data support a role for enhancers at pause-release promoters (59–61). NELF rapidly bound to all the eRNAs whereas CDK9 bound only 299 and 338 after LPS. The kinetics of binding were very rapid, and were selective for eRNAs induced by LPS. 327 did not bind CDK9 and had a distinct pattern of inhibition in Figure 2 compared to 338 and 299 which share a single complex regulatory region. These data suggest that a complex of interactions occurs in a stimulus-inducible manner, centered on RNA, uniquely for each region; a concept with growing support (62–64).
To test whether the eRNAs recruit additional protein complexes, we analyzed the binding pattern of other known RNA-binding proteins to the eRNAs using RNA-IP (Supplemental Figure 3). J2 is an antibody that recognizes dsRNA. S9.6 recognizes RNA:DNA hybrids. RBFOX bridges PRC2 to chromatin and is required for transcriptional repression and binding is RNA-dependent, similar to CBP and p300. SUZ12 is a Zn finger protein component of PRC2 and it binds nascent transcripts. EZH2 is a methyltransferase component of PRC2 and binding RNA inhibits enzymatic activity. TIP60 recognizes R loops (RNA:DNA at transcription bubbles). TIP60 binding was not induced by LPS. EZH2 binding went down with LPS and SUZ12 showed increased binding to the eRNAs after LPS. Therefore, there are considerable protein-RNA interactions after LPS even if there are no major changes in chromatin loops. We concluded that the eRNAs participate in multiple protein complexes at the enhancer-promoter interface.
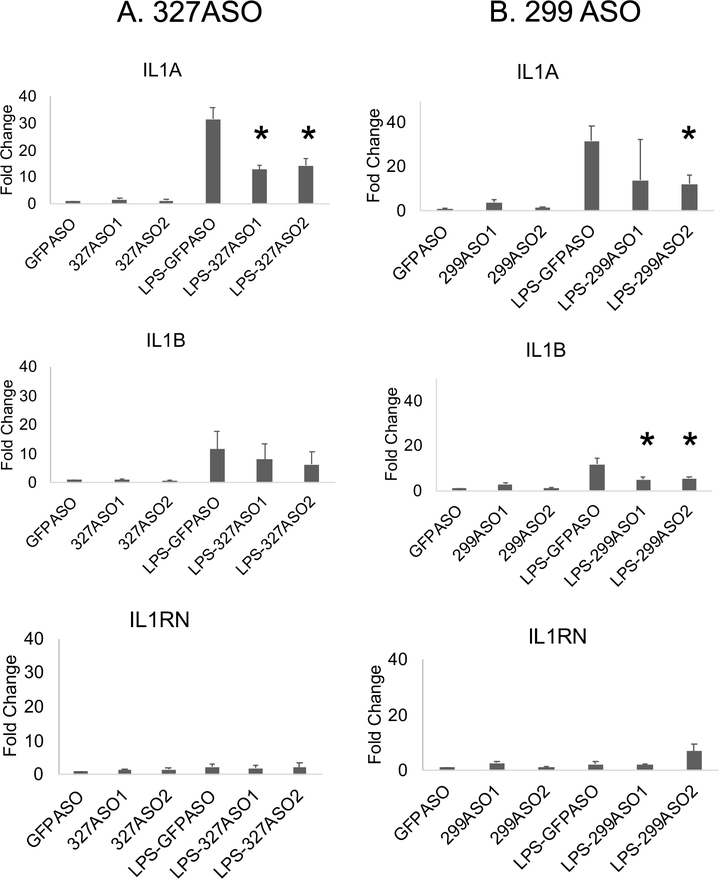
We hypothesized that eRNAs, produced before mRNA transcription, bind p300, driving increased histone acetylation. This is a recognized model for eRNA function but this setting is conceptually different than that seen in development where activation of enhancers and eRNA production underlies chromatin loop formation to bring the enhancer into proximity of the regulated promoter(s) (16, 65, 66). Our Capture-C data suggested that the loops are largely pre-established at these loci. To test whether the eRNAs participate in mRNA regulation, we utilized an anti-sense oligonucleotide (ASO) knockdown approach to examine the role of the eRNAs on mRNA expression. We first confirmed that each ASO led to diminished levels of the cognate eRNA (not shown). The knockdown of eRNA 327, which contacts the IL1B promoter in unstimulated monocytes and gains a connection to IL1A after LPS, resulted in diminished IL1A mRNA after LPS treatment, with less of an effect on IL1B transcript levels (Figure 4A). In contrast, the knockdown of eRNA 299, which is part of a large enhancer complex that loops to both IL1B and IL1A promoters in unstimulated monocytes, significantly impaired expression of both IL1B and IL1A after LPS induction (Figure 4B). In addition, IL1RN mRNA levels remained unchanged when knocking down these two eRNAs, as anticipated since IL1RN does not loop to these two regions. This study demonstrates that the eRNAs are required for full transcriptional induction after LPS even without large changes in chromatin loops and suggests a hierarchy of effects of the eRNAs on mRNA regulation.
Figure 4. IL1 eRNAs regulate transcription.
MonoMac6 cells were transfected with the indicated 20-bp phosphorothioate antisense oligonucleotides (ASO) designed to knock down eRNAs (two different ASOs each) or an ASO to GFP for 3 hours and stimulated with or without 1μg/ml LPS for 60 minutes. Total RNA was isolated and qRT-PCR was performed to measure levels of eRNAs and mRNAs. A) Knockdown of 327 eRNA significantly decreased the LPS-induction of IL1A but had less effect on IL1B and no effect on IL1RN. B) Knockdown of 299 eRNA significantly decreased the LPS-induction of IL1A and IL1B mRNAs but did not affect the IL1RN transcript levels. Fold change indicates levels compared to the GFP ASO in the unstimulated cells. (n =3, * p<0.05). PCRs were normalized to Actin.
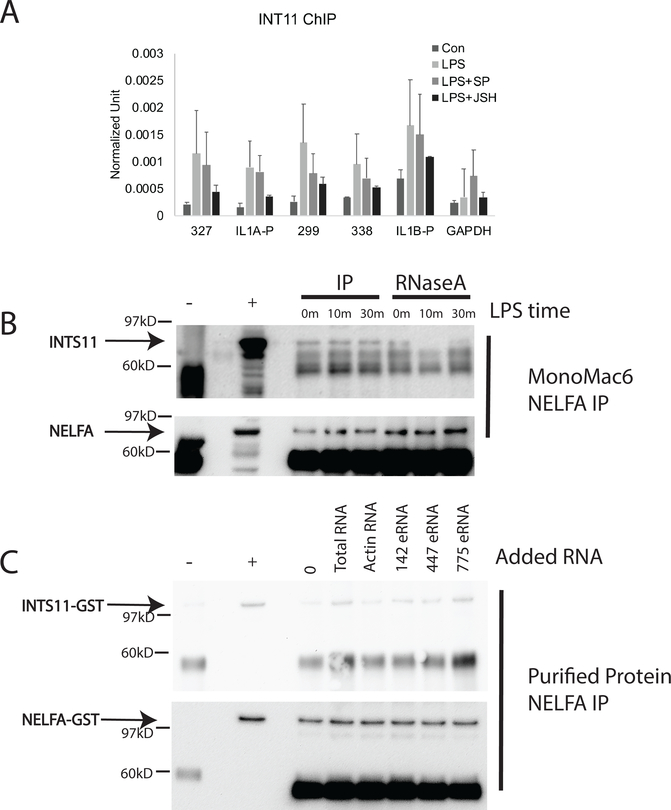
Finding that the eRNAs are structurally important for mRNA transcription at the IL-1 locus and that they bound to pause-release proteins suggested to us that Integrator may be involved in IL1A and IL1B transcriptional regulation. Integrator is known to regulate NELF eviction from the paused RNA polymerase complex (67). We first performed a ChIP assay for Integrator 11 and found that Integrator was bound to both promoters and enhancers after LPS stimulation. Furthermore, blockade of NFκB with JSH-23 led to markedly compromised recruitment of Integrator at all sites, whereas JNK inhibition had a more modest effect, the opposite of what was observed for p300 recruitment to chromatin (Figure 5A). We observed greater Integrator occupancy at the IL1B promoter at baseline compared to the other targets, and recruitment after LPS was less susceptible to inhibition. This is again consistent with the low level baseline transcription of IL1B and less dependence on pause-release. We hypothesized that such a large complex including enhancer DNA, eRNA, promoter DNA and at least five proteins besides the RNA polymerase complex might require a scaffold to maintain structural integrity. To test whether the protein complexes might be held by a structural aspect of the eRNAs, we digested cell lysates with RNase A and then immunoprecipitated with NELF and blotted with an antibody to Integrator 11 (Figure 5B). While Integrator 11 was easily co-immunoprecipitated in the absence of RNase treatment, RNase treatment led to release of Integrator from the complex. These data suggest that the eRNAs may act as scaffolding for some of the proteins complexed at the enhancer-promoter interface. To further test this, we produced NELF and Integrator as GST-linked proteins and incubated them with various RNA species (Figure 5C). All of the RNA species increased the protein-protein interaction although the eRNAs appeared to have a greater effect than actin.
Figure 5. Integrator interactions.
(A) An Integrator 11 (INT11) ChIP assay was performed to analyze Integrator 11 binding to the promoters and enhancers. Inhibition of the NFκB pathway by JSH-23 (JSH) and the JNK pathway with SP600125 (SP) led to diminished binding with greater effects observed with JSH-23. (n=4) B) Co-immunoprecipitation was used to show the interaction of Integrator 11 (INTS11) and NELF-A. MonoMac6 cells were treated with LPS (1μg/ml) for 0, 10, or 30 minutes. Lysates were collected and treated with RNase A as indicated and anti-NELFA used for immunoprecipitation. After RNase treatment, Integrator 11 interactions with NELF were decreased. The NELF blot of the same gel is shown below for quantitative comparison. C) Purified proteins were incubated with various types of RNAs in an effort to test the ability of RNAs to facilitate interactions between Integrator and NELF. Various RNAs increased the interaction between the two proteins. (There were at least five biological replicates of these analyses). + represents the lysate, - represents the negative control of mouse IgG.
Discussion
These studies of acute activation of transcription define a distinct model for the role of chromatin in transcriptional regulation. In developmental systems, eRNAs are critical for loop formation and differentiation depends on the correct approximation of enhancers and promoters related to fate determination and differentiation (68, 69). Many inflammatory genes utilize a pause-release transcriptional mechanism, presumed to facilitate rapid activation of transcription in response to threat (70, 71). Our data defined a complex set of loops at baseline that focused after LPS, a conceptually different view of looping than that seen in developmental systems. This may be a systematic effort to promote the rapid transcriptional response of these critical cytokines. With enhancers already approximated to their cognate promoters, transcription of mRNAs can be very rapid. With this information, it was possible to ask about the role of the eRNAs. If the loops are established at baseline what might the role be for eRNAs in promoting transcription? To address this question, we first established the critical pathways regulating the eRNAs and mRNAs. We hoped to exploit commonalities and differences between IL1A and IL1B to dissect pathways regulating enhancer-promoter interactions.
The transcriptional patterns of IL1A and IL1B are subtly different. IL1A mRNA accumulates more slowly than that of IL1B and its final abundance is lower. IL1B mRNA level at baseline is substantially higher than that of IL1A. In accordance with the baseline transcription of IL1B, H3K4me3 is very high at the IL1B promoter at baseline and declines after LPS whereas H3K4me3 is more moderate at baseline at the IL1A promoter and increases after LPS. The IL1B gene has been described as having Spi-1/PU.1 (Spi1) bound to the promoter at baseline and then stimulus induction of NFκB to the same region of the promoter (46). Based on our data, recruitment of Integrator is an additional step downstream with PU.1 ⇒NFκB ⇒Integrator representing a central regulatory pathway that is in turn part of a larger complex of RNA-binding proteins. IL1A may require recruitment of a histone methyltransferase, usually SET1 at proinflammatory genes (72). Requirement for this extra step, compared to IL1B, may explain the delayed kinetics of IL1A mRNA accumulation. We also found that the BRD4 inhibitor, iBET151, had markedly different effects on IL1B compared to IL1A. IL1B mRNA expression was delayed but expression of IL1A was nearly completely abrogated with iBET151 treatment. The role of BRD4 is to activate P-TEFb and release RNA polymerase II from pausing. These data suggest that IL1A is more dependent on BRD4 for pause-release than IL1B and coupled with the requirement to recruit a histone methyltransferase, certainly contributes to the slower kinetics of expression. IL1B has previously been found to have less dependence of pause-release (46). Commonalities noted for IL1A and IL1B are their reliance on NFκB and MAP kinase pathways for mRNA production and their overall rapid induction. Collectively, our data identify central differences in mechanisms of regulation that may help to explain differences in their kinetics of expression and highlight the critical role of chromatin in regulating transcription.
With this detailed information on transcriptional induction of the mRNAs, we went on to similarly dissect eRNA production. In general, the eRNAs were less inhibitable than the mRNAs but were again dependent on MAP kinases and NFκB. The three eRNAs we focused on in this study arose from H3K27ac peaks that increased after LPS. In terms of expression patterns, two eRNAs behaved similarly with subtly different responses to the inhibitors compared to 327 which is located in a different region. We went on to demonstrate a key role for the eRNAs in IL1A and IL1B mRNA production by knocking them down with an antisense technology. This confirmed, as has been seen in many systems, a mechanistic effect of eRNAs, however, the precise role and sequence of events involving eRNAs is not yet defined for pause-release genes. We therefore identified key binding partners and dependencies. CBP and p300 are highly homologous histone acetyltransferases (73). The enzymatic activity is activated by RNA but the functional niches in which each is dominant remain to be defined (51). The p300 protein was inducibly recruited to both enhancers and promoters after LPS and bound eRNAs. LPS-inducible CBP binding to chromatin was not identified. These data suggest that p300 may be more critical for inflammatory gene expression than CBP and in our previous study of a different locus, we also found that the eRNAs bound to p300 after LPS and not CBP (64) supporting the inflammatory gene set as a key niche for p300. The MAP kinase pathway was critical for p300 recruitment to chromatin. Several studies identified a role for MAP kinases in activating p300 binding in very specific settings but it is not yet clear if MAP kinases represent the major pathway for p300 activation (55, 74, 75). Our study contributes to the emerging understanding of early assembly of transcriptional complexes at enhancers. These data allow the sequence of events to be established at least at the IL-1 locus. NFκB may be responsible for the initial eRNA production and Integrator recruitment to the promoter but MAP kinases appear to be important for recruitment of p300 to the enhancers. Upon recruitment of p300 and binding eRNAs, p300 acetylates H3K27ac. This is thought to act as a platform for the assembly of the transcriptional activation complex.
Integrator has been characterized as a multi-molecular complex that serves as a scaffold and has recently been implicated in the pause-release transcriptional regulation (67, 76). Both enhancers and promoters bound Integrator 11 in an LPS-inducible manner although the IL1B promoter appeared to have some Integrator 11 bound at baseline. This is consistent with the overall impression that IL1B transcription is active at baseline. Integrator 11 assembly into a higher order complex with NELF, one of the main pausing proteins, was facilitated by RNA, although not wholly dependent on RNA. Integrator 11 has been previously shown to interact with NELF using co-immunoprecipitation from nuclear extracts and it may be that unrecognized RNA was enhancing the interaction (67). These key findings lead to a model where eRNAs act in many and varied structural roles. Through binding to p300, histone acetyltransferase activity is regulated by eRNAs, and the very landscape of H3K27ac is therefore dependent on eRNA production. In addition, the higher order complex of enhancer-promoter-Integrator appears to have an RNA dependency. How the eRNAs correctly partner with the proteins in each promoter-enhancer pair is not known and the Capture-C data suggest that multiple enhancers can participate in the transcriptional regulation of a single promoter. This leads to a dynamic model in which the chromatin is variably looped and interactions may be transient with stabilization of favorable interactions after LPS.
We selected this locus specifically to compare IL1A and IL1B. In a previous study we identified that these pivotal cytokines cluster together with IL-6 in terms of their kinetics of expression and their behavior with various inhibitors (77). IL-1α and IL-1β are functionally similar although structurally very different. Surprisingly, their promoter contacts to regulatory regions were highly intermingled but completely distinct from those of IL1RN. Although we identified their co-increased expression in SLE, common features by clustering analysis, and rapid induction after LPS, there were differences in the mechanisms of transcriptional regulation. IL1B appeared to have completed more steps in the path to expression than IL1A and low levels of mRNA were seen at baseline. This is consistent with emerging models where pause-release is not a binary quality but is part of a spectrum of pausing (78, 79). According to this model, IL1B is not highly paused and is therefore less susceptible to inhibition by iBET151, as was observed. Although the enhancers we focused on also had many similarities and either at baseline or after LPS were found to interact with both promoters, there were also differences between the enhancers. 327 differed from 299 and 338 because it failed to bind CDK9, the RNA-binding component of P-TEFb. Looping to 327 was gained by the IL1A promoter after LPS but apparently P-TEFb binding to the eRNA is not critical for that to occur.
This study dissected the IL-1 locus in unprecedented detail to define key interactions between enhancers and promoters. Four key notable findings emerged. 1) The regulatory contacts for IL1A and IL1B are highly intermingled but separated from those of IL1RN. 2) Regulatory looping contacts exhibit modest changes after LPS. 3) The eRNAs interact with multiple proteins in large complexes that may be in part dependent structurally on RNA. 4) The eRNAs bound to p300 but not CBP after LPS induction of transcription. These findings highlight key differences from developmental systems where looping is dependent on eRNAs and begin to characterize the landscape for inflammatory gene expression. There are some limitations to these analyses, however. A single stimulus was utilized at early time points. This offered the advantage of being able to exploit the abundant knowledge of signaling pathways activated by TLR4 engagement. However, it is possible and even likely that not all stimuli will impact gene expression in the same manner. Another limitation is the use of a cell line for most mechanistic studies. Not all epigenetic pathways are equally recapitulated in cell lines and these studies must be extended to primary cells where possible. Finally, we selected a deep dive into a single locus to best dissect the mechanisms. Extending these analyses to a genome-wide approach will be desirable. In summary, our data show a pivotal role for eRNAs in regulating transcription of IL1A and IL1B that may include a structural role. While overall similarities were found in the transcriptional blueprint, we exploited differences to better understand key mechanisms regulating expression.
Supplementary Material
Key points.
Enhancer RNAs are functionally important for IL1A/B mRNA transcription
MAP kinases and NFκB regulate distinct transcriptional processes
Enhancer RNAs bind pausing proteins to release RNA polymerase II
Acknowledgments
This study was supported by NIH R01 ES017627 and the Wallace Chair of Pediatrics (Dr. Sullivan). Dr. Grant is funded by the Daniel B. Burke Endowed Chair for Diabetes Research, the Children’s Hospital of Philadelphia and by NIH grant R01 HG010067. Dr. Wells is supported by NIH grants R01 AI130115 and R01 AI123539.
References
- 1.Dinarello CA 2018. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol. Rev 281: 8–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Werman A, Werman-Venkert R, White R, Lee JK, Werman B, Krelin Y, Voronov E, Dinarello CA, and Apte RN 2004. The precursor form of IL-1alpha is an intracrine proinflammatory activator of transcription. Proc. Natl. Acad. Sci. U. S. A 101: 2434–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen I, Rider P, Carmi Y, Braiman A, Dotan S, White MR, Voronov E, Martin MU, Dinarello CA, and Apte RN 2010. Differential release of chromatin-bound IL-1alpha discriminates between necrotic and apoptotic cell death by the ability to induce sterile inflammation. Proc. Natl. Acad. Sci. U. S. A 107: 2574–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinarello CA, Goldin NP, and Wolff SM 1974. Demonstration and characterization of two distinct human leukocytic pyrogens. J. Exp. Med 139: 1369–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schindler R, Mancilla J, Endres S, Ghorbani R, Clark SC, and Dinarello CA 1990. Correlations and interactions in the production of interleukin-6 (IL-6), IL-1, and tumor necrosis factor (TNF) in human blood mononuclear cells: IL-6 suppresses IL-1 and TNF. Blood 75: 40–47. [PubMed] [Google Scholar]
- 6.Schindler R, Ghezzi P, and Dinarello CA 1990. IL-1 induces IL-1. IV. IFN-gamma suppresses IL-1 but not lipopolysaccharide-induced transcription of IL-1. J. Immunol 144: 2216–2222. [PubMed] [Google Scholar]
- 7.Perregaux DG, McNiff P, Laliberte R, Conklyn M, and Gabel CA 2000. ATP acts as an agonist to promote stimulus-induced secretion of IL-1 beta and IL-18 in human blood. J. Immunol 165: 4615–4623. [DOI] [PubMed] [Google Scholar]
- 8.Hannum CH, Wilcox CJ, Arend WP, Joslin FG, Dripps DJ, Heimdal PL, Armes LG, Sommer A, Eisenberg SP, and Thompson RC 1990. Interleukin-1 receptor antagonist activity of a human interleukin-1 inhibitor. Nature 343: 336–340. [DOI] [PubMed] [Google Scholar]
- 9.Aksentijevich I, Nowak M, Mallah M, Chae JJ, Watford WT, Hofmann SR, Stein L, Russo R, Goldsmith D, Dent P, Rosenberg HF, Austin F, Remmers EF, Balow JE Jr., Rosenzweig S, Komarow H, Shoham NG, Wood G, Jones J, Mangra N, Carrero H, Adams BS, Moore TL, Schikler K, Hoffman H, Lovell DJ, Lipnick R, Barron K, O’Shea JJ, Kastner DL, and Goldbach-Mansky R 2002. De novo CIAS1 mutations, cytokine activation, and evidence for genetic heterogeneity in patients with neonatal-onset multisystem inflammatory disease (NOMID): a new member of the expanding family of pyrin-associated autoinflammatory diseases. Arthritis Rheum. 46: 3340–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostendorf B, Iking-Konert C, Kurz K, Jung G, Sander O, and Schneider M 2005. Preliminary results of safety and efficacy of the interleukin 1 receptor antagonist anakinra in patients with severe lupus arthritis. Ann. Rheum. Dis 64: 630–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels PA, Herman B, Happe S, Higgs A, LeProust E, Follows GA, Fraser P, Luscombe NM, and Osborne CS 2015. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet 47: 598–606. [DOI] [PubMed] [Google Scholar]
- 12.Cheng JH, Pan DZ, Tsai ZT, and Tsai HK 2015. Genome-wide analysis of enhancer RNA in gene regulation across 12 mouse tissues. Sci. Rep 5: 12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang MD, Zhang Y, McDevit D, Marecki S, and Nikolajczyk BS 2006. The interleukin-1beta gene is transcribed from a poised promoter architecture in monocytes. J. Biol. Chem 281: 9227–9237. [DOI] [PubMed] [Google Scholar]
- 14.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, and Pennacchio LA 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, and Jaenisch R 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A 107: 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang X, Sun T, Sweeney CJ, Lee GS, Chen S, Balk SP, Liu XS, Brown M, and Kantoff PW 2014. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. U. S. A 111: 7319–7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, and Fu XD 2011. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474: 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, and Young RA 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Wit E, Vos ES, Holwerda SJ, Valdes-Quezada C, Verstegen MJ, Teunissen H, Splinter E, Wijchers PJ, Krijger PH, and de Laat W 2015. CTCF Binding Polarity Determines Chromatin Looping. Mol. Cell 60: 676–684. [DOI] [PubMed] [Google Scholar]
- 20.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, Lu Y, Wu Y, Jia Z, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, and Wu Q 2015. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell 162: 900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thormann V, Rothkegel MC, Schopflin R, Glaser LV, Djuric P, Li N, Chung HR, Schwahn K, Vingron M, and Meijsing SH 2018. Genomic dissection of enhancers uncovers principles of combinatorial regulation and cell type-specific wiring of enhancer-promoter contacts. Nucleic Acids Res. 46: 2868–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, and Kim TK 2014. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell 56: 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, and Stamatoyannopoulos JA 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, Cairns J, Wingett SW, Varnai C, Thiecke MJ, Burden F, Farrow S, Cutler AJ, Rehnstrom K, Downes K, Grassi L, Kostadima M, Freire-Pritchett P, Wang F, Consortium B, Stunnenberg HG, Todd JA, Zerbino DR, Stegle O, Ouwehand WH, Frontini M, Wallace C, Spivakov M, and Fraser P 2016. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell 167: 1369–1384 e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, Li Y, Lin S, Lin Y, Barr CL, and Ren B 2016. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Rep 17: 2042–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Shi L, Song L, Maurer K, Petri M, and Sullivan K 2018. Overall Downregulation of mRNAs and Enrichment of H3K4me3 Change near Genome-Wide Association Study Signals in Systemic Lupus Erythematosus: Cell-Specific Effects. Front. Immunol 9: article 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi L, Zhang Z, Yu AM, Wang W, Wei Z, Akhter E, Maurer K, Reis PC, Song L, Petri M, and Sullivan KE 2014. The SLE Transcriptome Exhibits Evidence of Chronic Endotoxin Exposure and Has Widespread Dysregulation of Non-Coding and Coding RNAs. PLoS One 9: e93846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi L, Zhang Z, Song L, Leung YT, Petri MA, and Sullivan KE 2015. Monocyte enhancers are highly altered in systemic lupus erythematosus. Epigenomics 7: 921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, Larocca C, Saldanha SA, Abagyan R, Sun Y, Meyers DJ, Marmorstein R, Mahadevan LC, Alani RM, and Cole PA 2010. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem. Biol 17: 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dekker FJ, van den Bosch T, and Martin NI 2014. Small molecule inhibitors of histone acetyltransferases and deacetylases are potential drugs for inflammatory diseases. Drug Discov Today 19: 654–660. [DOI] [PubMed] [Google Scholar]
- 31.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, and Zhou Q 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19: 535–545. [DOI] [PubMed] [Google Scholar]
- 32.Yankulov K, Yamashita K, Roy R, Egly JM, and Bentley DL 1995. The transcriptional elongation inhibitor 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription factor IIH-associated protein kinase. J. Biol. Chem 270: 23922–23925. [DOI] [PubMed] [Google Scholar]
- 33.Shin HM, Kim MH, Kim BH, Jung SH, Kim YS, Park HJ, Hong JT, Min KR, and Kim Y 2004. Inhibitory action of novel aromatic diamine compound on lipopolysaccharide-induced nuclear translocation of NF-kappaB without affecting IkappaB degradation. FEBS Lett. 571: 50–54. [DOI] [PubMed] [Google Scholar]
- 34.Vaisitti T, Gaudino F, Ouk S, Moscvin M, Vitale N, Serra S, Arruga F, Zakrzewski JL, Liou HC, Allan JN, Furman RR, and Deaglio S 2017. Targeting metabolism and survival in chronic lymphocytic leukemia and Richter syndrome cells by a novel NF-kappaB inhibitor. Haematologica 102: 1878–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chesi A, Wagley Y, Johnson ME, Manduchi E, Su C, Lu S, Leonard ME, Hodge KM, Pippin JA, Hankenson KD, Wells AD, and Grant SFA 2019. Genome-scale Capture C promoter interactions implicate effector genes at GWAS loci for bone mineral density. Nat Commun 10: 1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wingett S, Ewels P, Furlan-Magaril M, Nagano T, Schoenfelder S, Fraser P, and Andrews S 2015. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res 4: 1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cairns J, Freire-Pritchett P, Wingett SW, Varnai C, Dimond A, Plagnol V, Zerbino D, Schoenfelder S, Javierre BM, Osborne C, Fraser P, and Spivakov M 2016. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 17: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garrett S, Dietzmann-Maurer K, Song L, and Sullivan KE 2008. Polarization of primary human monocytes by IFN-gamma induces chromatin changes and recruits RNA Pol II to the TNF-alpha promoter. J. Immunol 180: 5257–5266. [DOI] [PubMed] [Google Scholar]
- 40.Lee JY, Kim NA, Sanford A, and Sullivan KE 2003. Histone acetylation and chromatin conformation are regulated separately at the TNF alpha promoter in monocytes and macrophages. J. Leukoc. Biol 73: 862–871. [DOI] [PubMed] [Google Scholar]
- 41.Niranjanakumari S, Lasda E, Brazas R, and Garcia-Blanco MA 2002. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 26: 182–190. [DOI] [PubMed] [Google Scholar]
- 42.Kahlenberg JM, and Kaplan MJ 2014. The inflammasome and lupus: another innate immune mechanism contributing to disease pathogenesis? Curr. Opin. Rheumatol 26: 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin MS, Kang Y, Lee N, Wahl ER, Kim SH, Kang KS, Lazova R, and Kang I 2013. Self double-stranded (ds)DNA induces IL-1beta production from human monocytes by activating NLRP3 inflammasome in the presence of anti-dsDNA antibodies. J. Immunol 190: 1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cigni A, Pileri PV, Faedda R, Gallo P, Sini A, Satta AE, Marras R, Carta E, Argiolas D, Rum I, and Masala A 2014. Interleukin 1, interleukin 6, interleukin 10, and tumor necrosis factor alpha in active and quiescent systemic lupus erythematosus. J. Investig. Med 62: 825–829. [DOI] [PubMed] [Google Scholar]
- 45.Yang CA, Huang ST, and Chiang BL 2015. Sex-dependent differential activation of NLRP3 and AIM2 inflammasomes in SLE macrophages. Rheumatology (Oxford) 54: 324–331. [DOI] [PubMed] [Google Scholar]
- 46.Adamik J, Wang KZ, Unlu S, Su AJ, Tannahill GM, Galson DL, O’Neill LA, and Auron PE 2013. Distinct mechanisms for induction and tolerance regulate the immediate early genes encoding interleukin 1beta and tumor necrosis factor alpha. PLoS One 8: e70622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pulugulla SH, Packard TA, Galloway NLK, Grimmett ZW, Doitsh G, Adamik J, Galson DL, Greene WC, and Auron PE 2018. Distinct mechanisms regulate IL1B gene transcription in lymphoid CD4 T cells and monocytes. Cytokine 111: 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan KE, Suriano A, Dietzmann K, Lin J, Goldman D, and Petri MA 2007. The TNFalpha locus is altered in monocytes from patients with systemic lupus erythematosus. Clin. Immunol 123: 74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mikhaylichenko O, Bondarenko V, Harnett D, Schor IE, Males M, Viales RR, and Furlong EEM 2018. The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 32: 42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schor IE, Bussotti G, Males M, Forneris M, Viales RR, Enright AJ, and Furlong EEM 2018. Non-coding RNA Expression, Function, and Variation during Drosophila Embryogenesis. Curr. Biol 28: 3547–3561 e3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bose DA, Donahue G, Reinberg D, Shiekhattar R, Bonasio R, and Berger SL 2017. RNA Binding to CBP Stimulates Histone Acetylation and Transcription. Cell 168: 135–149 e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bratton MR, Frigo DE, Vigh-Conrad KA, Fan D, Wadsworth S, McLachlan JA, and Burow ME 2009. Organochlorine-mediated potentiation of the general coactivator p300 through p38 mitogen-activated protein kinase. Carcinogenesis 30: 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gusterson R, Brar B, Faulkes D, Giordano A, Chrivia J, and Latchman D 2002. The transcriptional co-activators CBP and p300 are activated via phenylephrine through the p42/p44 MAPK cascade. J. Biol. Chem 277: 2517–2524. [DOI] [PubMed] [Google Scholar]
- 54.Wang QE, Han C, Zhao R, Wani G, Zhu Q, Gong L, Battu A, Racoma I, Sharma N, and Wani AA 2013. p38 MAPK- and Akt-mediated p300 phosphorylation regulates its degradation to facilitate nucleotide excision repair. Nucleic Acids Res. 41: 1722–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li QJ, Yang SH, Maeda Y, Sladek FM, Sharrocks AD, and Martins-Green M 2003. MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300. EMBO J. 22: 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foulds CE, Nelson ML, Blaszczak AG, and Graves BJ 2004. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol. Cell. Biol 24: 10954–10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ortega E, Rengachari S, Ibrahim Z, Hoghoughi N, Gaucher J, Holehouse AS, Khochbin S, and Panne D 2018. Transcription factor dimerization activates the p300 acetyltransferase. Nature 562: 538–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi L, Song L, Maurer K, Zhang Z, and Sullivan K 2017. SERPINB2 is regulated by dynamic interactions with pause release proteins and enhancer RNAs. Mol. Immunol 88: 20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen FX, Xie P, Collings CK, Cao K, Aoi Y, Marshall SA, Rendleman EJ, Ugarenko M, Ozark PA, Zhang A, Shiekhattar R, Smith ER, Zhang MQ, and Shilatifard A 2017. PAF1 regulation of promoter-proximal pause release via enhancer activation. Science 357: 1294–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal AK, and Rosenfeld MG 2013. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 155: 1581–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patel MC, Debrosse M, Smith M, Dey A, Huynh W, Sarai N, Heightman TD, Tamura T, and Ozato K 2013. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol. Cell. Biol 33: 2497–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vos SM, Pollmann D, Caizzi L, Hofmann KB, Rombaut P, Zimniak T, Herzog F, and Cramer P 2016. Architecture and RNA binding of the human negative elongation factor. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pagano JM, Kwak H, Waters CT, Sprouse RO, White BS, Ozer A, Szeto K, Shalloway D, Craighead HG, and Lis JT 2014. Defining NELF-E RNA binding in HIV-1 and promoter-proximal pause regions. PLoS Genet 10: e1004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi L, Li S, Maurer K, Zhang Z, Petri M, and Sullivan KE 2018. Enhancer RNA and NFkappaB-dependent P300 regulation of ADAMDEC1. Mol. Immunol 103: 312–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, and Shiekhattar R 2013. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, and Rosenfeld MG 2013. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498: 516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stadelmayer B, Micas G, Gamot A, Martin P, Malirat N, Koval S, Raffel R, Sobhian B, Severac D, Rialle S, Parrinello H, Cuvier O, and Benkirane M 2014. Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat Commun 5: 5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim SI, Bultman SJ, Jing H, Blobel GA, and Bresnick EH 2007. Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation. Mol. Cell. Biol 27: 4551–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lai F, Gardini A, Zhang A, and Shiekhattar R 2015. Integrator mediates the biogenesis of enhancer RNAs. Nature 525: 399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diamant G, Bahat A, and Dikstein R 2016. The elongation factor Spt5 facilitates transcription initiation for rapid induction of inflammatory-response genes. Nat Commun 7: 11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, and Rogatsky I 2009. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc. Natl. Acad. Sci. U. S. A 106: 18207–18212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu L, Fang F, Dai X, Xu H, Qi X, Fang M, and Xu Y 2017. MKL1 defines the H3K4Me3 landscape for NF-kappaB dependent inflammatory response. Sci. Rep 7: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blobel GA 2000. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood 95: 745–755. [PubMed] [Google Scholar]
- 74.Jun JH, Yoon WJ, Seo SB, Woo KM, Kim GS, Ryoo HM, and Baek JH 2010. BMP2-activated Erk/MAP kinase stabilizes Runx2 by increasing p300 levels and histone acetyltransferase activity. J. Biol. Chem 285: 36410–36419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Donnell A, Yang SH, and Sharrocks AD 2008. MAP kinase-mediated c-fos regulation relies on a histone acetylation relay switch. Mol. Cell 29: 780–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gardini A, Baillat D, Cesaroni M, Hu D, Marinis JM, Wagner EJ, Lazar MA, Shilatifard A, and Shiekhattar R 2014. Integrator regulates transcriptional initiation and pause release following activation. Mol. Cell 56: 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maurer K, Ramen S, Shi L, Song L, and Sullivan KE 2018. Rapid induction of expression by LPS is accompanied by favorable chromatin and rapid binding of c-Jun. Mol. Immunol 95: 99–106. [DOI] [PubMed] [Google Scholar]
- 78.Henriques T, Scruggs BS, Inouye MO, Muse GW, Williams LH, Burkholder AB, Lavender CA, Fargo DC, and Adelman K 2018. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 32: 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Steurer B, Janssens RC, Geverts B, Geijer ME, Wienholz F, Theil AF, Chang J, Dealy S, Pothof J, van Cappellen WA, Houtsmuller AB, and Marteijn JA 2018. Live-cell analysis of endogenous GFP-RPB1 uncovers rapid turnover of initiating and promoter-paused RNA Polymerase II. Proc. Natl. Acad. Sci. U. S. A 115: E4368–E4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Novakovic B, Habibi E, Wang SY, Arts RJW, Davar R, Megchelenbrink W, Kim B, Kuznetsova T, Kox M, Zwaag J, Matarese F, van Heeringen SJ, Janssen-Megens EM, Sharifi N, Wang C, Keramati F, Schoonenberg V, Flicek P, Clarke L, Pickkers P, Heath S, Gut I, Netea MG, Martens JHA, Logie C, and Stunnenberg HG 2016. beta-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance. Cell 167: 1354–1368 e1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.