Abstract
Pancreatic ductal adenocarcinoma (PDAC) remains one of the deadliest malignancies. It is phenotypically heterogeneous with a highly unstable genome and provides few common therapeutic targets. We found that MCL1, Cofilin1 (CFL1) and SRC mRNA were highly expressed by a wide range of these cancers, suggesting that a strategy of dual MCL-1 and SRC inhibition might be efficacious for many patients. Immunohistochemistry revealed that MCL-1 protein was present at high levels in 94.7% of patients in a cohort of PDACs from Australian Pancreatic Genome Initiative (APGI). High MCL1 and Cofilin1 mRNA expression was also strongly predictive of poor outcome in the TCGA dataset and in the APGI cohort. In culture, MCL-1 antagonism reduced the level of the cytoskeletal remodeling protein Cofilin1 and phosphorylated SRC on the active Y416 residue, suggestive of reduced invasive capacity. The MCL-1 antagonist S63845 synergized with the SRC kinase inhibitor dasatinib to reduce cell viability and invasiveness through 3D-organotypic matrices. In preclinical murine models, this combination reduced primary tumor growth and liver metastasis of pancreatic cancer xenografts. These data suggest that MCL-1 antagonism, while reducing cell viability, may have an additional benefit in increasing the antimetastatic efficacy of dasatinib for the treatment of PDAC.
Subject terms: Pancreatic cancer, Targeted therapies, Apoptosis
PDAC is the 8th most common cause of cancer death worldwide accounting for approximately 430,000 deaths in 2018, being one of the most lethal cancers and exhibiting an mortality to incidence ratio of 94% [1]. An in-depth characterization of the pancreatic cancer genomic landscape [2–4] has revealed great heterogeneity among PDACs where highly penetrant variants are rare. The translation of this genomic information into clinical benefit remains a significant challenge [5] and there is desperate need to identify new treatments that improve the outcomes of patients suffering PDAC. In spite of the genomic heterogeneity observed in PDAC, the nonreceptor tyrosine kinase SRC is present at high levels in most PDAC specimens and pancreatic cancer cell lines. A high level of its activated form (phosphorylated on Y416) is predictive of poor outcome among low-grade pancreatic tumors [6, 7]. SRC is a member of the SRC family kinases (SFK) with pleotropic roles in the growth, survival, and invasion of pancreatic cancer [8] and suppression of SRC activity by dasatinib slows the growth of PDAC models in vitro and in vivo [9, 10]. Unfortunately the promise of these preclinical models has not been realized in clinical trials of metastatic PDAC, where single agent SFK inhibitors alone or in combination with gemcitabine showed no clinical benefit in the adjuvant setting [11–13]. Other combinatorial approaches show better activity with the triple combination of dasatinib, erlotinib (an EGFR inhibitor) and gemcitabine resulting in stable disease in ~70% of patients with tolerable safety profiles [14]. Thus the activity of agents targeting SRC may be improved with other targeted therapies that enhance its activity.
Antagonizing Myeloid cell leukemia 1 (MCL-1) in triple negative breast cancer (TNBC) can enhance the efficacy of SFK inhibitors [15]. MCL-1 is a member of the BCL-2 family of proteins that regulate the intrinsic (mitochondrial) apoptotic cascade, and a mediator of survival in both healthy and cancerous tissues [16]. MCL-1 protein levels correlate with outcome, tumor grade and therapeutic resistance in many cancers including those of the hematopoietic system, breast, lung, and pancreas [17–21]. In preclinical models of TNBC, we showed that MCL-1 modulated metastatic progression via two possible mechanisms; firstly via modulating the output of SFKs and the secondly via direct regulation of Cofilin. Cofilin is a cytoskeletal remodeling protein that is regulated by SRC activity [22, 23] and essential for actin remodeling during cellular invasion [24, 25]. As MCL-1 regulated the activity of Cofilin and the output of the SFKs in breast cancer cells, this led us to discover that drugs that antagonize MCL-1 can sensitize TNBC cells to dasatinib and suppress metastatic progression [15].
As both SRC and MCL-1 are important in the etiology of multiple cancers [26, 27], we used publicly available data to identify additional cancer contexts where a combined SRC and MCL-1 inhibitor strategy may be effective, identifying PDAC as possibly responsive to a dual SRC and MCL-1 inhibitor therapeutic strategy. We then utilized patient-derived pancreatic cell lines and orthotopic xenografts from the APGI to examine whether a dual MCL-1 and SRC inhibitor strategy was an effective antimetastatic in PDAC.
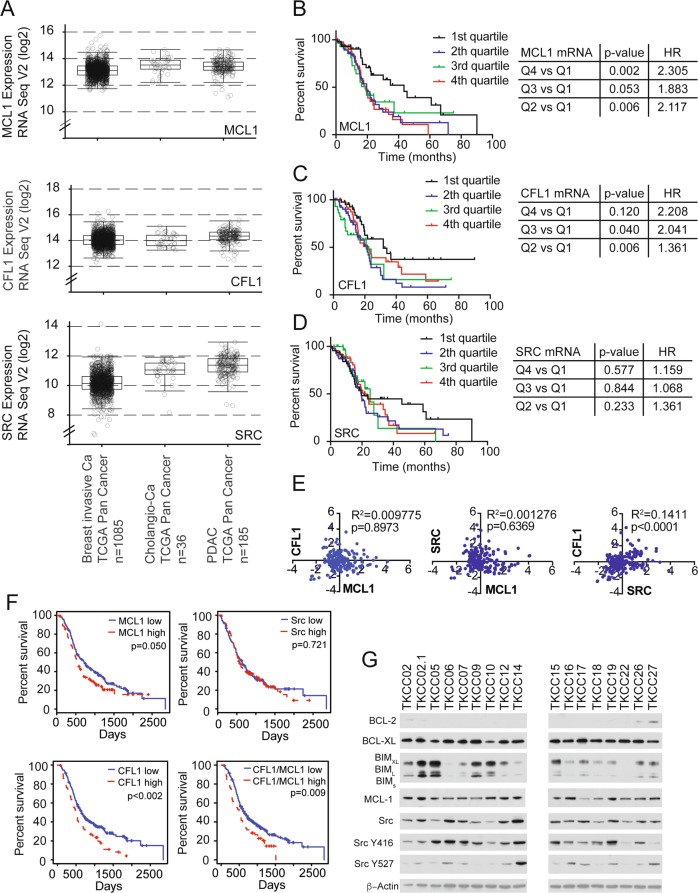
We first explored the mRNA expression of MCL1, SRC, and Cofilin1 (CFL1) across cancers in the TCGA and Australian Pancreatic Genome Initiative (APGI) to identify contexts where a dual MCL-1, and SRC inhibitor strategy may be effective. Interrogation of the TCGA datasets using cBioPortal indicated that MCL1, SRC and CFL1 are expressed among cholangiocarcinomas and PDACs to a similar extent to that of invasive breast carcinomas (Fig. 1a). Immunohistochemistry using an antibody to human MCL-1 on a tissue microarray cohort of 228 pancreatic cancers (including 188 PDACs, 20 intraductal papillary mucinous neoplasms with invasion and other mixed subtypes) from the APGI revealed a large proportion (94.7%) of PDACs and (90%) of intraductal papillary mucinous neoplasms with invasion expressed high levels of MCL-1 by IHC consistent with previous reports [28] (Supplementary Table 1 and Supplementary Fig. 1).
Fig. 1.
a Box and whisker graphs of MCL1, SRC, and Cofilin1 (CFL1) mRNA expression across breast invasive carcinoma (n = 1085), cholangiocarcinoma (cholangio-Ca) (n = 36), pancreatic adenocarcinoma (n = 185) among the TCGA cohort. b Kaplan Meier survival curves of MCL1 c CFL1, d SRC mRNA expression split by quartiles in the TCGA PDAC cohorts (n = 185). e mRNA correlation of MCL1 mRNA vs. CFL1 (left panel) and SRC (middle panel) as well as CFL1 vs. SRC (right panel). f Kaplan Meier survival curves of MCL1 (top left panel), CFL1 (bottom left panel), SRC (top right panel) and combined MCL1 and CFL1 mRNA expression split by quartiles in the APGI cohort (n = 247). Log Rank-p-value and hazard ratios indicated. g Western blots of BCL-2, BCL-XL, BIM, MCL-1, total SRC, Y416 SRC, Y527 SRC, and beta ACTIN among pancreatic cancer cells derived from the APGI cohort
To explore the clinical significance of MCL1, CFL1, and SRC in PDAC, Kaplan Meier survival analysis was performed using the mRNA expression quartiles of each gene from a total of 185 PDAC patients in the TCGA dataset. This analysis revealed that, although widely expressed among PDACs, when compared to the lowest levels of MCL1 in quartile 1, the quartiles with higher MCL1 mRNA expression were associated with worse overall survival in PDAC (Fig. 1b). A similar and significant pattern was observed using CFL1 mRNA expression quartiles (Fig. 1c), although the highest compared to the lowest quartiles failed to reach significance. SRC mRNA expression quartiles were not predictive of outcome in this cohort (Fig. 1d). There was no association of MCL1 mRNA expression with either CFL1 or SRC but we observed a significant positive correlation of SRC mRNA with CFL1 mRNA (Fig. 1e). We confirmed the observations made in the TCGA databases using data obtained from 247 PDAC patients with gene expression data from the APGI. Clinicopathological information for this cohort is provided Supplementary Table 2 and in Bailey et al. [4]. This analysis showed that the highest levels (top 25% vs. lowest 25%) of both MCL1 and CFL mRNA correlated with worse overall survival (Fig. 1f, left panels). The mRNA expression of SRC showed no prognostic power (Fig. 1f top right panel). When used together, top quartile levels of both MCL1 and CFL1 were predictive of worse outcome when compared to lower quartile levels in the APGI (Fig. 1f bottom right panel). Western blotting showed that activated SRC (Y416) was a feature among a panel of patient-derived pancreatic cancer cell lines (Fig. 1g). The BH3 only pro-apoptotic and MCL-1 interacting protein BIM was variable across each line. Furthermore the majority of PDACs were MCL-1 and BCL-XL positive but BCL-2 negative potentially indicating a preference on either MCL-1 or BCL-XL for survival.
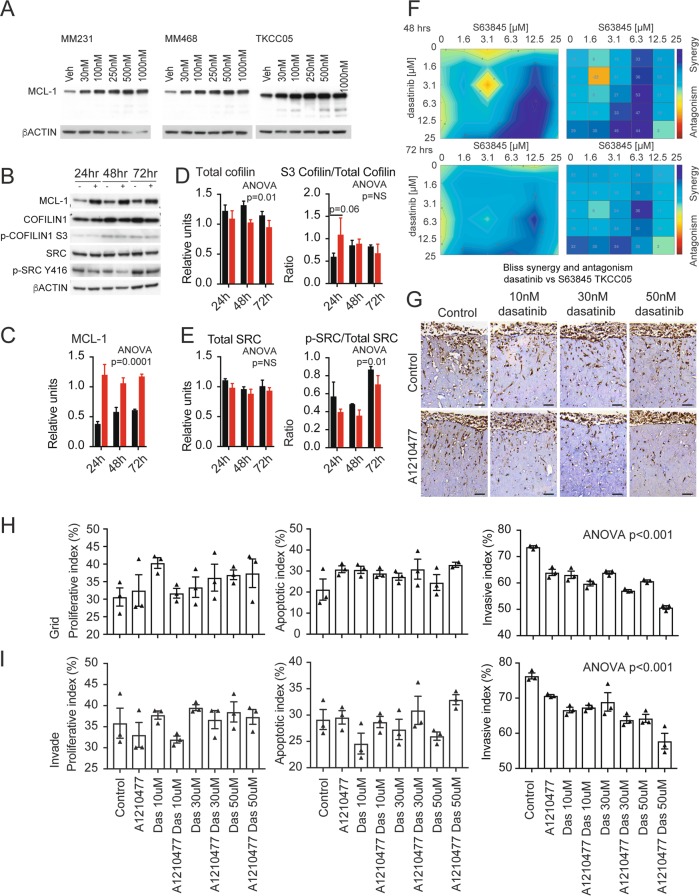
As the TKCC05 PDAC patient-derived cell line showed high levels of MCL-1, BIM and total and pSRC levels, this line was selected to examine the efficacy of a dual MCL-1 and SRC inhibitor strategy. This line can also invade into 3-dimensional collagen I matrices and successfully engraft as orthotopic xenografts in immune-compromised mice, spread to the liver and other organs providing a useful model of pancreatic metastasis [29]. Increasing concentrations of the MCL-1 antagonist S63845 resulted in elevated levels of MCL-1 similar to what was observed when human breast cancer cell lines MDA-MB-231 and MDA-MB-468 were treated with S63845 for 48 h (Fig. 2a) [15, 30]. Treatment with 500 nM S63845 produced a significant suppression of total Cofilin, which was maintained over a 72-h period (Fig. 1b, c) and also resulted in a trend towards an increased ratio of serine 3 (S3) phosphorylated (inactivated) Cofilin to total Cofilin at 24 h post treatment (Fig. 2d). MCL-1 antagonism did not alter the levels of total SRC but decreased the ratio of Y416 phosphorylated (activated) SRC to total SRC over the entire 72 h period suggestive of reduced activity (Fig. 2e). Bliss synergy analysis showed that the combination of S63845 and dasatinib (0–25 µM) was synergistic across a wide range of concentrations at 48 h and 72 h post treatment (Fig. 2f).
Fig. 2.
a Western blots of MCL-1 and beta ACTIN from MDA-MB-231, MDA-MB-468 breast cancer and TKCC05 pancreatic cancer cells treated with increasing concentrations of S63845. b Western blots and densitometry quantification of MCL-1 (c), total Cofilin (d, left panel), ratio of S3 phosphorylated Cofilin/total Cofilin (d, right panel), total SRC (e, left panel), the ration of Y416 phosphorylated SRC to total SRC (e, right panel) from TKCC05 pancreatic cancer cells treated with 250 nM S63845 over a 72 h period and normalized to beta ACTIN. N = 4 independent experiments, error bars, unpaired t-tests between groups and two-way ANOVA for treatments (vehicle vs. S63845) indicated. f Bliss synergy contour plot (left panels) and synergy matrix (right plots) of TKCC05 pancreatic cancer cells treated with increasing concentrations (0–25 µM) of S63845 and dasatinib at 48 h (upper panels) and 72 h (lower panels). g Representative immunohistochemistry using an antibody to human Vimentin on TKCC05 pancreatic cancer cells invading into fibrillar Collagen I organotypic matrices and treated with the indicated concentrations of A1210477 and dasatinib (h, i). Bar graphs showing the quantification of Ki67 (proliferating cells, left panels), cleaved caspase 3 (apoptotic cells, middle panels) and Vimentin (invasion index, right panels) of TKCC05 pancreatic cancer cells treated with the indicated concentrations of A1210477 and dasatinib at seeding (upper panels, grid) or 5 days after seeding (lower panels, invade). Error bars and two-way ANOVA p-value between treatments indicated
We then examined the effects of MCL-1 or SFK antagonism alone and in combination in three-dimensional fibrillar Collagen I matrices in vitro (Fig. 2g [31]). There were no significant effects of SRC inhibition by dasatinib or MCL-1 antagonism by A1210477 alone or in combination on proliferation or apoptosis as measured by Ki67 and cleaved caspase 3 immunohistochemistry respectively (Fig. 2h, i). However, there was a trend towards enhanced apoptosis when S63845 was combined with dasatinib when administered 5 days post exposure to an air-liquid interface. This was after when they had begun to invade, mimicking the clinical presentation of this disease, which often is associated with local invasion. In contrast dasatinib treatment resulted in a significant and dose dependent decrease in the ability of TKCC05 cells to invade through the organotypic matrix. Treatment with A1210477 similarly reduced their invasive capacity and significantly enhanced the effects of dasatinib across the dosage range equally when the drugs were administered just after seeding (Fig. 2h right panel) and after when they had begun to invade (Fig. 2i, right panel).
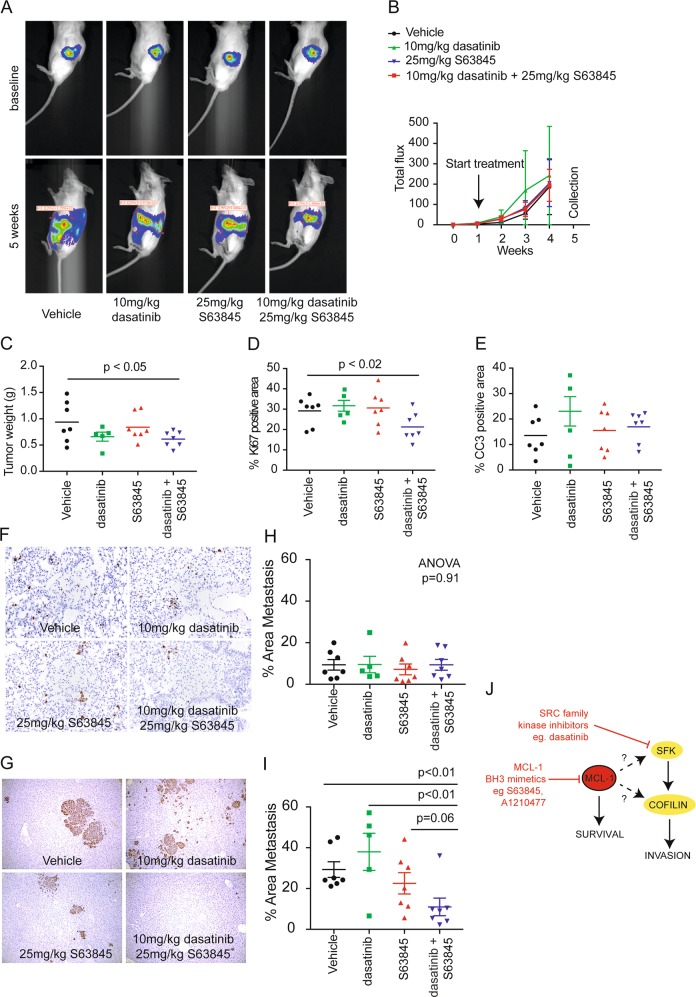
We next investigated whether dual inhibition of MCL-1 and SRC would be effective in the treatment of PDACs in vivo (Fig. 3). TKCC05 patient-derived pancreatic cells were implanted directly in the pancreas of immune-compromised NODScidIL2gamma–/– mice and bioluminescent imaging was used confirm successful engraftment and monitor the growth and spread of TKCC05 patient-derived pancreatic xenografts over 5 weeks (Fig. 3a). The rate of expansion of primary pancreatic tumors was not significantly different between mice treated with vehicle, S63845, dasatinib or a combination (Fig. 3b) but we observed a small but significant reduction in the weight of the primary tumor at 5 weeks post implantation (Fig. 3c). There were no effects of the single agents on primary tumor proliferation and apoptosis as measured by Ki67 and cleaved caspase 3 immunohistochemistry respectively, but a small and significant decrease in proliferation was observed in response to combination treatment (Fig. 3d, e). Bioluminescent imaging at 5 weeks post surgery suggested that the combination with S63845 and dasatinib reduced the spread of the TKCC05 patient-derived pancreatic xenografts (Fig. 3a). Immunohistochemistry using an antibody to human MCL-1 in resected PDAC tumors from this model revealed both nuclear and cytoplasmic staining (Supplementary Fig. 2A). Treatment with S63845 produced a significant increase in MCL-1 intensity (Supplementary Fig. 2B) consistent with S63845 extending MCL-1 protein half-life levels and providing a biomarker of response [30]. Both the lungs and livers of mice bearing TKCC05 patient-derived pancreatic xenografts were collected at 5 weeks and stained with an antibody against human vimentin to highlight disseminated PDAC cells [32] (Fig. 3f–i). We observed far fewer metastases in the lungs compared to the livers at this time point. While no effect of any treatment was detected in the lungs of these mice (Fig. 3g), the combination of S63845 and dasatinib produced a significant reduction in liver metastasis compared to vehicle and single agent therapy (Fig. 3i).
Fig. 3.
a Representative bioluminescent images of mice bearing TKCC05 pancreatic cancer xenografts at surgery (baseline) or at 5 weeks after surgery (5 weeks) and treated with vehicle (n = 7), 25 mg/kg S63845 (n = 7), 10 mg/kg dasatinib (n = 5, 2 were excluded from the dasatinib cohort as they reached ethical end point one week early due to ascites) or combined S63845 and dasatinib (n = 7). b Line graphs of the average bioluminescence of mice bearing TKCC05 pancreatic cancer xenografts at surgery (baseline) over a 5 week period treated with vehicle, 25 mg/kg S63845, 10 mg/kg dasatinib or combined S63845 and dasatinib. Dot plots of c tumor weight, d tumor Ki67 positivity (e) and cleaved caspase 3 positivity in TKCC05 pancreatic cancer orthotopic primary tumors. Representative photomicrographs taken at ×20 objective of the f lungs and g livers from mice bearing TKCC05 pancreatic cancer xenografts subjected to immunohistochemistry using an antibody against vimentin and (h) dot plots showing the average area of metastasis in the lungs and (i) livers of mice bearing TKCC05 pancreatic cancer xenografts at 5 weeks post surgery (each dot is average of 15 images within one mouse). Unpaired t-tests between groups and one-way ANOVA p-value for treatments (vehicle vs. S63845) illustrated. j Model schematic of MCL-1 and SRC regulation of Cofilin. Combined inhibition of MCL-1 by BH3 mimetics such as S63845 and A1210477 can enhance the anti-invasive effects of dasatinib via a possible direct or indirect regulation of Cofilin via SRC
Here we have shown that MCL-1, Cofilin, and SRC are widely expressed among PDACs (Fig. 1) with high MCL-1 protein levels detected among 94.7% of all PDACs in the APGI tissue microarray cohort. Elevated expression of MCL1 resulted in a two-fold higher risk of death when compared to patients with the lowest quartile mRNA expression of MCL1 in the TCGA and APGI cohorts (Fig. 1a, f respectively). Similar observations were true for Cofilin in both the TCGA (Fig. 1c) and the APGI cohorts (Fig. 1f), although there was no significant difference between CFL1 low group and CFL1 high group in the TCGA cohort (Fig. 1c). Possible reasons for this discrepancy could be the methodology in assessing mRNA expression (RNAseq in TCGA vs. array based gene expression in the APGI) as well as a greater number of patients analyzed in the APGI cohort (247) vs. the TCGA cohort (185), reaching significance in the APGI cohort. As Cofilin is tightly linked to SRC activity [22, 23], and we have shown can be regulated by MCL-1, these data suggest that up to 75% of patients with PDACs may benefit from a combinatorial MCL-1 and SFK inhibitor strategy. A similar benefit could be possible for patients with cancers dependent on MCL-1 and SFK activity via Cofilin e.g. cholangiocarcinomas, but this remains to be investigated (Fig. 1a). Furthermore, as 75% of PDACs contain inactivating mutations in TP53 [33], it is accepted that these tumors are likely to have an intact apoptotic cascade and therefore sensitive to antagonism by BH3 mimetics [34]. We have shown that MCL-1 antagonism can potently sensitize PDACs to SFK inhibition by dasatinib, and that MCL-1 protein levels as measured by immunohistochemistry could be used as a biomarker for response. The importance of SFK in pancreatic cancer is widely recognized [35–37], hence there has been extensive research into the development of agents that target the SFK in the clinical setting. Unfortunately the promise of preclinical experiments has been met with disappointment in clinical trials with single agent dasatinib [11], and Phase II clinical trials of dasatinib or saracatinib in combination with gemcitabine failing to show any clinical benefit in patients with refractory PDAC [12, 13]. A more recent combination shows better activity with the triple combination of dasatinib, erlotinib (an EGFR inhibitor) and gemcitabine resulting in stable disease in ~70% of patients with tolerable safety profiles [14]. Interestingly this combination includes an agent that antagonizes EGFR, a key growth factor that controls MCL-1 transcription [38], possibly suggesting that the success of this trial could be at least, in part, due to the effects of erlotinib on EGFR driven MCL-1 transcription.
We have previously shown that in the MDA-MB-231 TNBC cell lines in culture, the effects of MCL-1 are largely limited to its anti-invasive effects possibly via its regulation of the cytoskeletal remodeling protein Cofilin and/or by the SFKs [15]. Similarly in PDAC cancer cells, S63845 also significantly modulated the expression of Cofilin and the Y416 phosphorylated and activated form of SRC. In addition, the effects of the S63845 antagonist in combination with dasatinib were predominantly restricted to outcomes of cellular invasion (Fig. 2) and metastasis (Fig. 3). These results suggest that MCL-1 modulation of metastatic progression via SRC or Cofilin may be present in multiple cancer contexts. Metastatic progression requires remodeling of the cytoskeleton, dynamic membrane changes, cellular invasion and localized tissue destruction [39], and this is regulated by the SFKs and their targets, including cSRC, FYN, YES, Paxillin, Cofilin, Cortactin, Rac and Rho [27, 40]. SRC was not predictive of outcome in the TCGA or the APGI cohorts but correlated with the expression of the cytoskeletal remodeling protein Cofilin, consistent with SRC’s known regulation of this protein [22, 23]. We have also shown that MCL-1 can modulate Cofilin expression (Fig. 2). A schematic model for these observations is provided in Fig. 3h, where MCL-1, a known pro-survival protein, may directly or indirectly regulate Cofilin expression via SRC to control invasion. We have already established that high levels of MCL-1 place it in close proximity to Cofilin in breast cancer models [15]. While the full details underlying this mechanism remain to be discovered, the data presented here provide a possible explanation as to why dual antagonism of MCL-1 and SRC is synergistic.
In conclusion we have shown MCL-1 is widely expressed by and can predict outcome in PDAC. Therapeutic targeting of MCL-1 using BH3 mimetics (e.g., S63845, A1210477, ADZ5991, MIK665/S64315 etc.) is currently being investigated in clinical trials for patients with multiple myeloma, acute myeloid leukemia, and myelodysplastic syndrome (NCT02992483, NCT02979366 and NCT03672695) and may provide a way of sensitizing these tumors to dasatinib and provide a new therapeutic strategy alone or in combination with standard of care for PDAC.
Materials and methods
All materials and methods are provided in the Supplementary information
Supplementary information
APGI membership List for Publications 2019
Acknowledgements
This work was supported by grants from the Cancer Council NSW (SRO and CJO), NHMRC Australia (CJO, DGO, PT, MP, and SRO), National Breast Cancer Foundation (ND and CEC), Cancer Institute NSW (DGO, PT, MP), Sutton Motor Pancreatic cancer funding (PT), Len Ainsworth Pancreatic Cancer Fellowship (PT), Avner Pancreatic Cancer Foundation Grant (PT) as well as philanthropic support through the Mostyn Family Foundation (SRO) and Cue Clothing Co. (SRO), Estee Lauder Australia (SRO, DGO and CJO), RT Hall Trust (CJO). We thank Gillian Lehrbach for tissue culture support and Anaiis Zaratzian and Andrew Da Silva for histology services.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Members of the Australian Pancreatic Cancer Genome Initiative (APGI) are listed in supplementary information.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lesley Castillo, Adelaide IJ Young
Supplementary information
The online version of this article (10.1038/s41388-019-1091-0) contains supplementary material, which is available to authorized users.
References
- 1.Rawla Prashanth, Sunkara Tagore, Gaduputi Vinaya. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World Journal of Oncology. 2019;10(1):10–27. doi: 10.14740/wjon1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raphael Benjamin J., Hruban Ralph H., Aguirre Andrew J., Moffitt Richard A., Yeh Jen Jen, Stewart Chip, Robertson A. Gordon, Cherniack Andrew D., Gupta Manaswi, Getz Gad, Gabriel Stacey B., Meyerson Matthew, Cibulskis Carrie, Fei Suzanne S., Hinoue Toshinori, Shen Hui, Laird Peter W., Ling Shiyun, Lu Yiling, Mills Gordon B., Akbani Rehan, Loher Phillipe, Londin Eric R., Rigoutsos Isidore, Telonis Aristeidis G., Gibb Ewan A., Goldenberg Anna, Mezlini Aziz M., Hoadley Katherine A., Collisson Eric, Lander Eric, Murray Bradley A., Hess Julian, Rosenberg Mara, Bergelson Louis, Zhang Hailei, Cho Juok, Tiao Grace, Kim Jaegil, Livitz Dimitri, Leshchiner Ignaty, Reardon Brendan, Van Allen Eliezer, Kamburov Atanas, Beroukhim Rameen, Saksena Gordon, Schumacher Steven E., Noble Michael S., Heiman David I., Gehlenborg Nils, Kim Jaegil, Lawrence Michael S., Adsay Volkan, Petersen Gloria, Klimstra David, Bardeesy Nabeel, Leiserson Mark D.M., Bowlby Reanne, Kasaian Katayoon, Birol Inanc, Mungall Karen L., Sadeghi Sara, Weinstein John N., Spellman Paul T., Liu Yuexin, Amundadottir Laufey T., Tepper Joel, Singhi Aatur D., Dhir Rajiv, Paul Drwiega, Smyrk Thomas, Zhang Lizhi, Kim Paula, Bowen Jay, Frick Jessica, Gastier-Foster Julie M., Gerken Mark, Lau Kevin, Leraas Kristen M., Lichtenberg Tara M., Ramirez Nilsa C., Renkel Jeremy, Sherman Mark, Wise Lisa, Yena Peggy, Zmuda Erik, Shih Juliann, Ally Adrian, Balasundaram Miruna, Carlsen Rebecca, Chu Andy, Chuah Eric, Clarke Amanda, Dhalla Noreen, Holt Robert A., Jones Steven J.M., Lee Darlene, Ma Yussanne, Marra Marco A., Mayo Michael, Moore Richard A., Mungall Andrew J., Schein Jacqueline E., Sipahimalani Payal, Tam Angela, Thiessen Nina, Tse Kane, Wong Tina, Brooks Denise, Auman J. Todd, Balu Saianand, Bodenheimer Tom, Hayes D. Neil, Hoyle Alan P., Jefferys Stuart R., Jones Corbin D., Meng Shaowu, Mieczkowski Piotr A., Mose Lisle E., Perou Charles M., Perou Amy H., Roach Jeffrey, Shi Yan, Simons Janae V., Skelly Tara, Soloway Matthew G., Tan Donghui, Veluvolu Umadevi, Parker Joel S., Wilkerson Matthew D., Korkut Anil, Senbabaoglu Yasin, Burch Patrick, McWilliams Robert, Chaffee Kari, Oberg Ann, Zhang Wei, Gingras Marie-Claude, Wheeler David A., Xi Liu, Albert Monique, Bartlett John, Sekhon Harman, Stephen Yeager, Howard Zaren, Judy Miller, Breggia Anne, Shroff Rachna T., Chudamani Sudha, Liu Jia, Lolla Laxmi, Naresh Rashi, Pihl Todd, Sun Qiang, Wan Yunhu, Wu Ye, Jennifer Smith, Roggin Kevin, Becker Karl-Friedrich, Behera Madhusmita, Bennett Joseph, Boice Lori, Burks Eric, Carlotti Junior Carlos Gilberto, Chabot John, Pretti da Cunha Tirapelli Daniela, Sebastião dos Santos Jose, Dubina Michael, Eschbacher Jennifer, Huang Mei, Huelsenbeck-Dill Lori, Jenkins Roger, Karpov Alexey, Kemp Rafael, Lyadov Vladimir, Maithel Shishir, Manikhas Georgy, Montgomery Eric, Noushmehr Houtan, Osunkoya Adeboye, Owonikoko Taofeek, Paklina Oxana, Potapova Olga, Ramalingam Suresh, Rathmell W. Kimryn, Rieger-Christ Kimberly, Saller Charles, Setdikova Galiya, Shabunin Alexey, Sica Gabriel, Su Tao, Sullivan Travis, Swanson Pat, Tarvin Katherine, Tavobilov Michael, Thorne Leigh B., Urbanski Stefan, Voronina Olga, Wang Timothy, Crain Daniel, Curley Erin, Gardner Johanna, Mallery David, Morris Scott, Paulauskis Joseph, Penny Robert, Shelton Candace, Shelton Troy, Janssen Klaus-Peter, Bathe Oliver, Bahary Nathan, Slotta-Huspenina Julia, Johns Amber, Hibshoosh Hanina, Hwang Rosa F., Sepulveda Antonia, Radenbaugh Amie, Baylin Stephen B., Berrios Mario, Bootwalla Moiz S., Holbrook Andrea, Lai Phillip H., Maglinte Dennis T., Mahurkar Swapna, Triche Timothy J., Van Den Berg David J., Weisenberger Daniel J., Chin Lynda, Kucherlapati Raju, Kucherlapati Melanie, Pantazi Angeliki, Park Peter, Saksena Gordon, Voet Doug, Lin Pei, Frazer Scott, Defreitas Timothy, Meier Sam, Chin Lynda, Kwon Sun Young, Kim Yong Hoon, Park Sang-Jae, Han Sung-Sik, Kim Seong Hoon, Kim Hark, Furth Emma, Tempero Margaret, Sander Chris, Biankin Andrew, Chang David, Bailey Peter, Gill Anthony, Kench James, Grimmond Sean, Johns Amber, Cancer Genome Initiative (APGI Australian Pancreatic, Postier Russell, Zuna Rosemary, Sicotte Hugues, Demchok John A., Ferguson Martin L., Hutter Carolyn M., Mills Shaw Kenna R., Sheth Margi, Sofia Heidi J., Tarnuzzer Roy, Wang Zhining, Yang Liming, Zhang Jiashan (Julia), Felau Ina, Zenklusen Jean C. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32(2):185-203.e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones S., Hruban R. H., Kamiyama M., Borges M., Zhang X., Parsons D. W., Lin J. C.-H., Palmisano E., Brune K., Jaffee E. M., Iacobuzio-Donahue C. A., Maitra A., Parmigiani G., Kern S. E, Velculescu V. E., Kinzler K. W., Vogelstein B., Eshleman J. R., Goggins M., Klein A. P. Exomic Sequencing Identifies PALB2 as a Pancreatic Cancer Susceptibility Gene. Science. 2009;324(5924):217–217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey Peter, Chang David K., Nones Katia, Johns Amber L., Patch Ann-Marie, Gingras Marie-Claude, Miller David K., Christ Angelika N., Bruxner Tim J. C., Quinn Michael C., Nourse Craig, Murtaugh L. Charles, Harliwong Ivon, Idrisoglu Senel, Manning Suzanne, Nourbakhsh Ehsan, Wani Shivangi, Fink Lynn, Holmes Oliver, Chin Venessa, Anderson Matthew J., Kazakoff Stephen, Leonard Conrad, Newell Felicity, Waddell Nick, Wood Scott, Xu Qinying, Wilson Peter J., Cloonan Nicole, Kassahn Karin S., Taylor Darrin, Quek Kelly, Robertson Alan, Pantano Lorena, Mincarelli Laura, Sanchez Luis N., Evers Lisa, Wu Jianmin, Pinese Mark, Cowley Mark J., Jones Marc D., Colvin Emily K., Nagrial Adnan M., Humphrey Emily S., Chantrill Lorraine A., Mawson Amanda, Humphris Jeremy, Chou Angela, Pajic Marina, Scarlett Christopher J., Pinho Andreia V., Giry-Laterriere Marc, Rooman Ilse, Samra Jaswinder S., Kench James G., Lovell Jessica A., Merrett Neil D., Toon Christopher W., Epari Krishna, Nguyen Nam Q., Barbour Andrew, Zeps Nikolajs, Moran-Jones Kim, Jamieson Nigel B., Graham Janet S., Duthie Fraser, Oien Karin, Hair Jane, Grützmann Robert, Maitra Anirban, Iacobuzio-Donahue Christine A., Wolfgang Christopher L., Morgan Richard A., Lawlor Rita T., Corbo Vincenzo, Bassi Claudio, Rusev Borislav, Capelli Paola, Salvia Roberto, Tortora Giampaolo, Mukhopadhyay Debabrata, Petersen Gloria M., Munzy Donna M., Fisher William E., Karim Saadia A., Eshleman James R., Hruban Ralph H., Pilarsky Christian, Morton Jennifer P., Sansom Owen J., Scarpa Aldo, Musgrove Elizabeth A., Bailey Ulla-Maja Hagbo, Hofmann Oliver, Sutherland Robert L., Wheeler David A., Gill Anthony J., Gibbs Richard A., Pearson John V., Waddell Nicola, Biankin Andrew V., Grimmond Sean M. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 5.Johns AL, McKay SH, Humphris JL, Pinese M, Chantrill LA, Mead RS, et al. Lost in translation: returning germline genetic results in genome-scale cancer research. Genome Med. 2017;9:41 10.1186/s13073-017-0430-4. [DOI] [PMC free article] [PubMed]
- 6.Donahue Timothy R., Tran Linh M., Hill Reginald, Li Yunfeng, Kovochich Anne, Calvopina Joseph H., Patel Sanjeet G., Wu Nanping, Hindoyan Antreas, Farrell James J., Li Xinmin, Dawson David W., Wu Hong. Integrative Survival-Based Molecular Profiling of Human Pancreatic Cancer. Clinical Cancer Research. 2012;18(5):1352–1363. doi: 10.1158/1078-0432.CCR-11-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutz Manfred P., Eßer I.B.Silke, Flossmann-Kast Berenike B.M., Vogelmann Roger, Lührs Hardi, Friess Helmut, Büchler Markus W., Adler Guido. Overexpression and Activation of the Tyrosine Kinase Src in Human Pancreatic Carcinoma. Biochemical and Biophysical Research Communications. 1998;243(2):503–508. doi: 10.1006/bbrc.1997.8043. [DOI] [PubMed] [Google Scholar]
- 8.Frame Margaret C. Src in cancer: deregulation and consequences for cell behaviour. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2002;1602(2):114–130. doi: 10.1016/S0304-419X(02)00040-9. [DOI] [PubMed] [Google Scholar]
- 9.Duxbury M. S. Inhibition of Src Tyrosine Kinase Impairs Inherent and Acquired Gemcitabine Resistance in Human Pancreatic Adenocarcinoma Cells. Clinical Cancer Research. 2004;10(7):2307–2318. doi: 10.1158/1078-0432.CCR-1183-3. [DOI] [PubMed] [Google Scholar]
- 10.Morton Jennifer P., Karim Saadia A., Graham Kathryn, Timpson Paul, Jamieson Nigel, Athineos Dimitris, Doyle Brendan, McKay Colin, Heung Man–Yeung, Oien Karin A., Frame Margaret C., Evans T.R. Jeffry, Sansom Owen J., Brunton Valerie G. Dasatinib Inhibits the Development of Metastases in a Mouse Model of Pancreatic Ductal Adenocarcinoma. Gastroenterology. 2010;139(1):292–303. doi: 10.1053/j.gastro.2010.03.034. [DOI] [PubMed] [Google Scholar]
- 11.Chee Cheng Ean, Krishnamurthi Smitha, Nock Charles J., Meropol Neal J., Gibbons Joseph, Fu PingFu, Bokar Joseph, Teston Lois, O'Brien Timothy, Gudena Vinay, Reese Amy, Bergman Mark, Saltzman Joel, Wright John J., Dowlati Afshin, Brell Joanna. Phase II Study of Dasatinib (BMS‐354825) in Patients With Metastatic Adenocarcinoma of the Pancreas. The Oncologist. 2013;18(10):1091–1092. doi: 10.1634/theoncologist.2013-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans T.R.J., Van Cutsem E., Moore M.J., Bazin I.S., Rosemurgy A., Bodoky G., Deplanque G., Harrison M., Melichar B., Pezet D., Elekes A., Rock E., Lin C., Strauss L., O’Dwyer P.J. Phase 2 placebo-controlled, double-blind trial of dasatinib added to gemcitabine for patients with locally-advanced pancreatic cancer. Annals of Oncology. 2017;28(2):354–361. doi: 10.1093/annonc/mdw607. [DOI] [PubMed] [Google Scholar]
- 13.Renouf Daniel J., Moore Malcolm J., Hedley David, Gill Sharlene, Jonker Derek, Chen Eric, Walde David, Goel Rakesh, Southwood Bernadette, Gauthier Isabelle, Walsh Wendy, McIntosh Lynn, Seymour Lesley. A phase I/II study of the Src inhibitor saracatinib (AZD0530) in combination with gemcitabine in advanced pancreatic cancer. Investigational New Drugs. 2010;30(2):779–786. doi: 10.1007/s10637-010-9611-3. [DOI] [PubMed] [Google Scholar]
- 14.Cardin Dana B., Goff Laura W., Chan Emily, Whisenant Jennifer G., Dan Ayers G., Takebe Naoko, Arlinghaus Lori R., Yankeelov Thomas E., Berlin Jordan, Merchant Nipun. Dual Src and EGFR inhibition in combination with gemcitabine in advanced pancreatic cancer: phase I results. Investigational New Drugs. 2017;36(3):442–450. doi: 10.1007/s10637-017-0519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young AI, Law AM, Castillo L, Chong S, Cullen HD, Koehler M, et al. MCL-1 inhibition provides a new way to suppress breast cancer metastasis and increase sensitivity to dasatinib. Breast Cancer Res. 2016;18:125. 10.1186/s13058-016-0781-6. [DOI] [PMC free article] [PubMed]
- 16.Strasser Andreas, Cory Suzanne, Adams Jerry M. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. The EMBO Journal. 2011;30(18):3667–3683. doi: 10.1038/emboj.2011.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abulwerdi F., Liao C., Liu M., Azmi A. S., Aboukameel A., Mady A. S. A., Gulappa T., Cierpicki T., Owens S., Zhang T., Sun D., Stuckey J. A., Mohammad R. M., Nikolovska-Coleska Z. A Novel Small-Molecule Inhibitor of Mcl-1 Blocks Pancreatic Cancer Growth In Vitro and In Vivo. Molecular Cancer Therapeutics. 2013;13(3):565–575. doi: 10.1158/1535-7163.MCT-12-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chowdry RP, Sica GL, Kim S, Chen Z, Goodman A, Alexis D, et al. Phosphorylated Bcl-2 and Mcl-1 as prognostic markers in small cell lung cancer. Oncotarget. 2016. 10.18632/oncotarget.7485.
- 19.Goodwin C M, Rossanese O W, Olejniczak E T, Fesik S W. Myeloid cell leukemia-1 is an important apoptotic survival factor in triple-negative breast cancer. Cell Death & Differentiation. 2015;22(12):2098–2106. doi: 10.1038/cdd.2015.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leverson J D, Zhang H, Chen J, Tahir S K, Phillips D C, Xue J, Nimmer P, Jin S, Smith M, Xiao Y, Kovar P, Tanaka A, Bruncko M, Sheppard G S, Wang L, Gierke S, Kategaya L, Anderson D J, Wong C, Eastham-Anderson J, Ludlam M J C, Sampath D, Fairbrother W J, Wertz I, Rosenberg S H, Tse C, Elmore S W, Souers A J. Potent and selective small-molecule MCL-1 inhibitors demonstrate on-target cancer cell killing activity as single agents and in combination with ABT-263 (navitoclax) Cell Death & Disease. 2015;6(1):e1590–e1590. doi: 10.1038/cddis.2014.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinn Bridget A, Dash Rupesh, Azab Belal, Sarkar Siddik, Das Swadesh K, Kumar Sachin, Oyesanya Regina A, Dasgupta Santanu, Dent Paul, Grant Steven, Rahmani Mohamed, Curiel David T, Dmitriev Igor, Hedvat Michael, Wei Jun, Wu Bainan, Stebbins John L, Reed John C, Pellecchia Maurizio, Sarkar Devanand, Fisher Paul B. Targeting Mcl-1 for the therapy of cancer. Expert Opinion on Investigational Drugs. 2011;20(10):1397–1411. doi: 10.1517/13543784.2011.609167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bamburg James R. Proteins of the ADF/Cofilin Family: Essential Regulators of Actin Dynamics. Annual Review of Cell and Developmental Biology. 1999;15(1):185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 23.Wang J.T., Song L.Z., Li L.L., Zhang W., Chai X.J., An L., Chen S.L., Frotscher M., Zhao S.T. Src controls neuronal migration by regulating the activity of FAK and cofilin. Neuroscience. 2015;292:90–100. doi: 10.1016/j.neuroscience.2015.02.025. [DOI] [PubMed] [Google Scholar]
- 24.Bravo-Cordero Jose Javier, Magalhaes Marco A. O., Eddy Robert J., Hodgson Louis, Condeelis John. Functions of cofilin in cell locomotion and invasion. Nature Reviews Molecular Cell Biology. 2013;14(7):405–415. doi: 10.1038/nrm3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oser Matthew, Condeelis John. The cofilin activity cycle in lamellipodia and invadopodia. Journal of Cellular Biochemistry. 2009;108(6):1252–1262. doi: 10.1002/jcb.22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Opferman Joseph T. Attacking cancer's Achilles heel: antagonism of anti-apoptotic BCL-2 family members. The FEBS Journal. 2015;283(14):2661–2675. doi: 10.1111/febs.13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wheeler Deric L., Iida Mari, Dunn Emily F. The Role of Src in Solid Tumors. The Oncologist. 2009;14(7):667–678. doi: 10.1634/theoncologist.2009-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyamoto Y, Hosotani R, Wada M, Lee JU, Koshiba T, Fujimoto K, et al. Immunohistochemical analysis of Bcl-2, Bax, Bcl-X, and Mcl-1 expression in pancreatic cancers. Oncology. 1999;56:73–82. doi: 10.1159/000011933. [DOI] [PubMed] [Google Scholar]
- 29.Chou Angela, Froio Danielle, Nagrial Adnan M, Parkin Ashleigh, Murphy Kendelle J, Chin Venessa T, Wohl Dalia, Steinmann Angela, Stark Rhys, Drury Alison, Walters Stacey N, Vennin Claire, Burgess Andrew, Pinese Mark, Chantrill Lorraine A, Cowley Mark J, Molloy Timothy J, Waddell Nicola, Johns Amber, Grimmond Sean M, Chang David K, Biankin Andrew V, Sansom Owen J, Morton Jennifer P, Grey Shane T, Cox Thomas R, Turchini John, Samra Jaswinder, Clarke Stephen J, Timpson Paul, Gill Anthony J, Pajic Marina. Tailored first-line and second-line CDK4-targeting treatment combinations in mouse models of pancreatic cancer. Gut. 2017;67(12):2142–2155. doi: 10.1136/gutjnl-2017-315144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotschy András, Szlavik Zoltán, Murray James, Davidson James, Maragno Ana Leticia, Le Toumelin-Braizat Gaëtane, Chanrion Maïa, Kelly Gemma L., Gong Jia-Nan, Moujalled Donia M., Bruno Alain, Csekei Márton, Paczal Attila, Szabo Zoltán B., Sipos Szabolcs, Radics Gábor, Proszenyak Agnes, Balint Balázs, Ondi Levente, Blasko Gábor, Robertson Alan, Surgenor Allan, Dokurno Pawel, Chen Ijen, Matassova Natalia, Smith Julia, Pedder Christopher, Graham Christopher, Studeny Aurélie, Lysiak-Auvity Gaëlle, Girard Anne-Marie, Gravé Fabienne, Segal David, Riffkin Chris D., Pomilio Giovanna, Galbraith Laura C. A., Aubrey Brandon J., Brennan Margs S., Herold Marco J., Chang Catherine, Guasconi Ghislaine, Cauquil Nicolas, Melchiore Fabien, Guigal-Stephan Nolwen, Lockhart Brian, Colland Frédéric, Hickman John A., Roberts Andrew W., Huang David C. S., Wei Andrew H., Strasser Andreas, Lessene Guillaume, Geneste Olivier. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature. 2016;538(7626):477–482. doi: 10.1038/nature19830. [DOI] [PubMed] [Google Scholar]
- 31.Timpson P, McGhee EJ, Erami Z, Nobis M, Quinn JA, Edward M, et al. Organotypic collagen I assay: a malleable platform to assess cell behaviour in a 3-dimensional context. J Vis Exp. 2011;56:e3089. 10.3791/3089. [DOI] [PMC free article] [PubMed]
- 32.Law AMK, Yin JXM, Castillo L, Young AIJ, Piggin C, Rogers S, et al. Andy’s Algorithms: new automated digital image analysis pipelines for FIJI. Sci Rep. 2017;7:15717. 10.1038/s41598-017-15885-6. [DOI] [PMC free article] [PubMed]
- 33.Waddell Nicola, Pajic Marina, Patch Ann-Marie, Chang David K., Kassahn Karin S., Bailey Peter, Johns Amber L., Miller David, Nones Katia, Quek Kelly, Quinn Michael C. J., Robertson Alan J., Fadlullah Muhammad Z. H., Bruxner Tim J. C., Christ Angelika N., Harliwong Ivon, Idrisoglu Senel, Manning Suzanne, Nourse Craig, Nourbakhsh Ehsan, Wani Shivangi, Wilson Peter J., Markham Emma, Cloonan Nicole, Anderson Matthew J., Fink J. Lynn, Holmes Oliver, Kazakoff Stephen H., Leonard Conrad, Newell Felicity, Poudel Barsha, Song Sarah, Taylor Darrin, Waddell Nick, Wood Scott, Xu Qinying, Wu Jianmin, Pinese Mark, Cowley Mark J., Lee Hong C., Jones Marc D., Nagrial Adnan M., Humphris Jeremy, Chantrill Lorraine A., Chin Venessa, Steinmann Angela M., Mawson Amanda, Humphrey Emily S., Colvin Emily K., Chou Angela, Scarlett Christopher J., Pinho Andreia V., Giry-Laterriere Marc, Rooman Ilse, Samra Jaswinder S., Kench James G., Pettitt Jessica A., Merrett Neil D., Toon Christopher, Epari Krishna, Nguyen Nam Q., Barbour Andrew, Zeps Nikolajs, Jamieson Nigel B., Graham Janet S., Niclou Simone P., Bjerkvig Rolf, Grützmann Robert, Aust Daniela, Hruban Ralph H., Maitra Anirban, Iacobuzio-Donahue Christine A., Wolfgang Christopher L., Morgan Richard A., Lawlor Rita T., Corbo Vincenzo, Bassi Claudio, Falconi Massimo, Zamboni Giuseppe, Tortora Giampaolo, Tempero Margaret A., Gill Anthony J., Eshleman James R., Pilarsky Christian, Scarpa Aldo, Musgrove Elizabeth A., Pearson John V., Biankin Andrew V., Grimmond Sean M. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ADAMS J, CORY S. Bcl-2-regulated apoptosis: mechanism and therapeutic potential. Current Opinion in Immunology. 2007;19(5):488–496. doi: 10.1016/j.coi.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creedon H, Brunton VG. Src kinase inhibitors: promising cancer therapeutics? Crit Rev Oncog. 2012;17:145–59. doi: 10.1615/CritRevOncog.v17.i2.20. [DOI] [PubMed] [Google Scholar]
- 36.Patel Ami, Sabbineni Harika, Clarke Andrea, Somanath Payaningal R. Novel roles of Src in cancer cell epithelial-to-mesenchymal transition, vascular permeability, microinvasion and metastasis. Life Sciences. 2016;157:52–61. doi: 10.1016/j.lfs.2016.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Siyuan, Yu Dihua. Targeting Src family kinases in anti-cancer therapies: turning promise into triumph. Trends in Pharmacological Sciences. 2012;33(3):122–128. doi: 10.1016/j.tips.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Booy E P, Henson E S, Gibson S B. Epidermal growth factor regulates Mcl-1 expression through the MAPK-Elk-1 signalling pathway contributing to cell survival in breast cancer. Oncogene. 2011;30(20):2367–2378. doi: 10.1038/onc.2010.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaguchi Hideki, Wyckoff Jeffrey, Condeelis John. Cell migration in tumors. Current Opinion in Cell Biology. 2005;17(5):559–564. doi: 10.1016/j.ceb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 40.YAMAGUCHI H, PIXLEY F, CONDEELIS J. Invadopodia and podosomes in tumor invasion. European Journal of Cell Biology. 2006;85(3-4):213–218. doi: 10.1016/j.ejcb.2005.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
APGI membership List for Publications 2019