Abstract
Salmonella typhimurium is a pathogenic gram-negative bacterium, which is found primarily in the intestinal lumen. It often causes diarrhea in infants and young children and leads to food poisoning. Drug resistance of Salmonella typhimurium presented serious complications in clinical patients. In this study, we investigated the antibiotic susceptibility of Salmonella typhimurium standard strain L forms to the third and forth generation cephalosporins, in order to control and eliminate Salmonella typhimurium L forms in infection treatment. Salmonella typhimurium L forms were induced by β-lactam antibiotic cefazolin in the culture medium of bacterial L forms. The antibiotic susceptibility of Salmonella typhimurium L forms was analyzed by K-B drug susceptibility testing. The change trend of drug susceptibility and resistance of Salmonella typhimurium L forms was obtained in accordance with USA clinical and laboratory standards institute (CLSI) evaluation data and statistical analysis. Drug resistance of Salmonella typhimurium L forms showed little increasing trend compared with their parent bacteria. The L form inhibition zone was smaller than in the parent bacteria. However, the drug susceptibility of L forms of Salmonella typhimurium to the third and forth generation cephalosporins remained sensitive.The antibiotic susceptibility of Salmonella typhimurium L forms to the third and forth generation cephalosporins remains sensitive, and the combined use of multi-antibiotics is a convenient and effective method to reduce Salmonella typhimurium L forms occurrence.
Subject terms: Antibiotics, Bacterial structural biology
Introduction
Because of the immature immunity of infants and young children, bacterial acquired antibiotic resistance and poor sanitation, Salmonella typhimurium (S. typhimurium) has become the main pathogen in nosocomial and foodborne infection1–3. Salmonella typhimurium possesses endotoxin, enterotoxin and extracellular enzymes as well as other pathogenic factors, so that Salmonella typhimurium has strong pathogenicity4,5. After the use of antibiotics, it was easy to make Salmonella typhimurium form into cell wall-defective bacteria named bacterial L forms6–8.
BacteriaL L forms still have the ability to cause disease, such as leading to the delay of chronic infection, and decreased susceptibility to antibacterial drugs which act on cell walls9,10. In these experiments, we studied the resistance of Salmonella typhimurium standard strain L forms to advanced cephalosporins and judged the susceptibility degree of Salmonella typhimurium L forms to the third and fourth generation cephalosporins, in order to guide the rational use of clinical antibacterial drugs. The study will provide the basic theory for controlling Salmonella typhimurium infection.
Materials and Methods
Bacterial strain
Salmonella typhimurium standard strain CMCC50115 was purchased from the National Institute for the Control of Pharmaceutical and Biological Products.
Reagents and instruments
Gram staining solution was purchased from Nanjing Jiancheng Bioengineering Institute. Antibiotic Drug sensitive tablets were purchased from OXOID company, UK, including third-generation cephalosporins, such as cefotaxime (CTX), ceftriaxone (CRO), cefoperazone (CFP), ceftazidime (CAZ), and fourth generation cephalosporin cefepim (FEP). Mueller-Hinton (MH) agar was used for determination of bacterial susceptibility to antibiotics by using agar paper diffusion method. 100 ml MH agar medium was prepared and made with 1.7 g agar, 0.2 g beef extract powder, 0.15 g soluble starch, 1.75 g casein hydrolysate, and afterwards regulating pH 7.4, and autoclaving at 121 °C for 15 min. The MH agar medium was inverted into the plate for antibiotic drug susceptibility testing. Soft agar plates for bacterial L forms culture were prepared by using improved MH agar plates with 1% agar and 2% NaCl. A mounted digital microscope (Nikon, Japan) was used for observing bacterial L forms and taking photos of micrographs.
Salmonella typhimurium L form Induction
Salmonella typhimurium was induced into stable L-forms by cefazolin drug discs with 30 µg/tablet. Gram stain was used to observe the microscopic morphology of Salmonella typhimurium and its L forms.
Preparation of Salmonella typhimurium and L form bacterial liquid
The bacteria in the logarithmic growth phase were obtained by conventional bacterial culture. 108 CFU/ml bacterial liquid was prepared with sterile physiological saline to a 0.5 McFarland standard concentration. Salmonella typhimurium L forms were subcultured under cefazolin induction, to obtain fresh cultures. 108 CFU/ml L form bacterial liquid was prepared with sterile 3% NaCl hypertonic saline, and counts were measured by McFarland turbidimetry.
Drug susceptibility test
According to the national clinical test operating protocol for drug susceptibility testing in vitro, the first preparation of test broth was made after culturing at 37 °C for 6 hours. The bacterial turbidity degree was regulated to 0.5 McFarland units. The tested bacteria were scribbled and uniform coating on MH agar and agar disc diffusion method was used for drug susceptibility testing. The five kinds of cephalosporin susceptibility paper disc were evenly affixed to the center and surrounding of the plate aseptically. After the conventional bacterial culture, the diameters of the inhibition zones were measured. Referring to the national antibacterial drug susceptibility test criteria, drug susceptibility was determined specifically.
Statistical analysis
The experiment was repeated three times, and continuous variables are expressed as mean ± standard deviation. Between the two groups the data difference was analyzed by the way of using paired t test. A P value of < 0.05 was considered statistically significant.
Results
Gram staining results
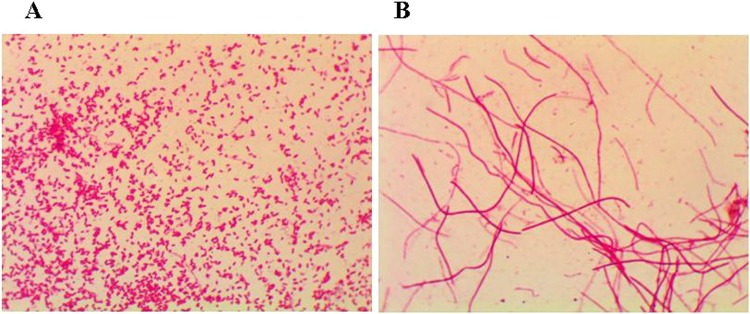
As shown in Fig. 1, the Salmonella typhimurium strain was negative Gram stain and medium size bacillus, and Salmonella typhimurium L forms were negative Gram stain and filamentous bacillus. Salmonella typhimurium L forms were often coiled into groups.
Figure 1.
Gram staining morphological observation of Salmonella typhimurium and its L forms (Gram stain, magnification: 1000×). (A) Salmonella typhimurium is negative Gram stain and medium size bacillus. (B) Salmonella typhimurium L forms are negative Gram stain and filamentous bacillus.
Susceptibility test results
Salmonella typhimurium L forms colonies appeared around drug sensitive paper. The Salmonella typhimurium L forms colonies were tiny and gray, (shown in Fig. 2B). According to the criteria of bacterial drug susceptibility, Salmonella typhimurium standard strain L forms susceptibility to the third-generation cephalosporins, such as cefotaxime (CTX), ceftriaxone (CRO), cefoperazone (CFP) and ceftazidime (CAZ) remained sensitive. Salmonella typhimurium standard strain L forms’ susceptibility to the fourth generation cephalosporin cefepime (FEP) also was maintained at the level of susceptibility. Although the Salmonella typhimurium L forms’ inhibition zones were smaller than the original bacteria, there is no significant difference from the comparison of the Salmonella typhimurium original bacteria and L forms. Results are shown in Fig. 3.
Figure 2.
Drug susceptibility test cultures of Salmonella typhimurium and its L forms. (A) Salmonella typhimurium drug susceptibility test culture. (B) Salmonella typhimurium L forms drug susceptibility test culture.
Figure 3.

Drug susceptibility test results of Salmonella typhimurium and its L forms in connection with different cephalosporins Although the Salmonella typhimurium L forms inhibition zones were smaller than the original bacteria, there is no significant difference from the comparison of the Salmonella typhimurium original bacteria and L forms.
Observation of salmonella typhimurium L form colonies
Salmonella typhimurium L form colonies can appear around drug sensitive paper, showed in Fig. 4A. In the L form colonies, Salmonella typhimurium L forms Gram stain showed negative Gram stain and filamentous bacillus, showed in Fig. 4B.
Figure 4.
Salmonella typhimurium L form colonies appeared around drug sensitive paper. (A) Salmonella typhimurium L forms’ drug susceptibility test culture. The Salmonella typhimurium L forms colonies were tiny and gray around drug sensitive paper. (B) Salmonella typhimurium L forms Gram stain showed negative Gram stain and filamentous bacillus.
Discussion
Salmonella typhimurium, commonly known as food poisoning pathogens, has many virulence factors and antibiotic resistance. It has become one of the major pathogens of nosocomial and foodborne infection and food poisoning. And it is more common in food poisoning and sepsis of infants and young children11,12. Salmonella typhimurium L forms often occur in patients after β-lactam antibiotic treatment, resulting in chronic infection. And bacterial L forms were often detected as a pathogen13,14. Therefore, the study on drug susceptibility of Salmonella typhimurium L forms is important to prevent and control Salmonella typhimurium and its L form infection disease.
Bacterial L-form is a cell wall-deficient type of bacteria, and bacterial cell wall peptidoglycan formation is inhibited under various factors in vivo and in vitro. But Bacterial L forms can still grow and reproduce, and they still had pathogenicity. Cephalosporins act on the bacterial cell wall to inhibit the transpeptidase activity, affect the synthesis of the peptidoglycan layer, and exert a bactericidal effect15–17. Cephalosporin antibiotic individuation can induce bacterial variability to L forms and result in prolonged infection in blood and bone marrow. Ceftriaxone is the third generation of cephalosporin antibiotics for the treatment of lower respiratory tract infections, skin and soft tissue infections, sepsis caused by sensitive pathogens. And similar drugs also include ceftazidime, cefotaxime and cefoperazone, as well as cefepime of the fourth generation cephalosporins18,19. This study found that Salmonella typhimurium under cephalosporin treatment can mutate to bacterial L forms, so that the drug inhibition zones of third and fourth generation cephalosporins were reduced. But Salmonella typhimurium L forms remained sensitive to third-generation cephalosporins including cefotaxime, ceftriaxone, cefoperazone and ceftazidime and to the fourth-generation cephalosporin cefepime, in relation to the executive standard of the national antibacterial drug susceptibility test. However, clinical medication may require extended treatment of cephalosporins by means of combination medication with other action route antibiotic, in order to achieve complete pathogen removal, including original bacteria and their bacterial L forms20,21.
Salmonella typhimurium has multi-drug resistance and can naturally resist a variety of antibacterial drugs. The main mechanisms of drug resistance are decreased permeability of the outer membrane, changes in lactamase and penicillin-binding proteins, activation and production of efflux pumps. In recent years, with the increasing number of clinically applied antibacterial drugs, broad-spectrum resistant Salmonella typhimurium bacteria have become more and more widespread22–24. Besides the influence of plasmids, the development of drug resistance depends on the type of chromosomal gene and the gene of the strain. Mutations and environmental choices are closely related. The data show that Salmonella typhimurium L forms still have pathogenicity. Salmonella typhimurium L forms have cell wall defects, so that they are more resistant to cephalosporins acting on cell walls than Salmonella typhimurium original bacteria. Cephalosporins alone can induce bacterial L form production, but combined use of antibiotics that act on ribosomes or nucleic acids can prevent the prolongation of disease caused by the production of L-forms. Therefore, a reasonable combination of antimicrobial agents is an effective method to kill all Salmonella typhimurium and reduce the Salmonella typhimurium L forms’ formation.
According to the antibiotic target site of pathogenic bacteria, the action mechanisms of antibiotics are divided into four categories. They inhibit bacterial cell wall synthesis, or affect cytoplasmic membrane permeability, or inhibit protein synthesis (eg. macrolides, aminoglycosides), or inhibit nucleic acid metabolism (eg. quinolone, sulfonamides). Doctors should be careful in choosing antibiotics, which should follow the principle of combination and interaction of antibiotics, in order to control Salmonella typhimurium infection.
Conclusions
The antibiotic susceptibility of Salmonella typhimurium standard strain L forms to the third and forth generation cephalosporins remains sensitive, but its L forms inhibition zones are smaller than the original bacteria. The combined use of multi-antibiotics is a convenient and effective method to reduce Salmonella typhimurium L forms occurrence in order to avoid the occurrence of bacterial L form infection.
Acknowledgements
We would like to thank Anhui Key Laboratory of Infection and Immunity, Bengbu Medical College, China. This work was supported by China grants from the Key Fund Project of Anhui University Natural Science Research (KJ2016A461), and the Scientific Research Innovation Team Project of Anhui Colleges and Universities (2016-40).
Author contributions
L.-H.H., Z.-T. and C.-D.Y. conceived the idea and designed the study protocol. Y.-C.P., C.-Y.F. and Z.-T. contributed with provision of study material. L.-H.H., Y.-C.P. and C.-Y.F. conducted experiments, collected, analysed the data; Z.-T., C.-D.Y. and Z.-J.L. performed statistical analysis, wrote the manuscript. All authors read and approved the final manuscript.
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Cuiping Yang, Huihui Li and Tao Zhang.
Contributor Information
Junli Zuo, Email: zuo-junli@163.com.
Dengyu Chen, Email: chengong131@sina.com.
References
- 1.Tamang MD, et al. Antimicrobial susceptibility and virulence characteristics of Salmonella enterica Typhimurium isolates from healthy and diseased pigs in Korea. J. Food Prot. 2014;77:1481–1486. doi: 10.4315/0362-028X.JFP-14-084. [DOI] [PubMed] [Google Scholar]
- 2.Ranjbar R, Elhaghi P, Shokoohizadeh L. Multilocus Sequence Typing of the Clinical Isolates of Salmonella Enterica Serovar Typhimurium in Tehran Hospitals. Iran J. Med. Sci. 2017;42:443–448. [PMC free article] [PubMed] [Google Scholar]
- 3.Sun J, et al. The Molecular Epidemiological Characteristics and Genetic Diversity of Salmonella Typhimurium in Guangdong, China, 2007–2011. PLoS One. 2014;9:e113145. doi: 10.1371/journal.pone.0113145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lotha R, Sundaramoorthy NS, Shamprasad BR, Nagarajan S, Sivasubramanian A. Plant nutraceuticals (Quercetrin and Afzelin) capped silver nanoparticles exert potent antibiofilm effect against food borne pathogen Salmonella enterica serovar typhimurium and curtail planktonic growth in zebrafish infection model. Microb Pathog. 2018;120:109–118. doi: 10.1016/j.micpath.2018.04.044. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z, et al. Comparative Study on Antibiotic Resistance and DNA Profiles of Salmonella enterica Serovar Typhimurium Isolated from Humans, Retail Foods, and the Environment in Shanghai, China. Foodborne Pathog Dis. 2018;15:481–488. doi: 10.1089/fpd.2017.2414. [DOI] [PubMed] [Google Scholar]
- 6.Wong MH, Yan M, Chan EW, Biao K, Chen S. Emergence of clinical Salmonella enterica serovar Typhimurium isolates with concurrent resistance to ciprofloxacin, ceftriaxone, and azithromycin. Antimicrob Agents Chemother. 2014;58:3752–3756. doi: 10.1128/AAC.02770-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawai Y, Mickiewicz K, Erringto J. Lysozyme Counteracts β-Lactam Antibiotics by Promoting the Emergence of L-Form Bacteria. Cell. 2018;172:1038–1049. doi: 10.1016/j.cell.2018.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf D, Domínguez-Cuevas P, Daniel RA, Mascher T. Cell Envelope Stress Response in Cell Wall-Deficient L-Forms of Bacillus subtilis. Antimicrob Agents Chemother. 2012;56:5907–5915. doi: 10.1128/AAC.00770-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Errington J, Mickiewicz K, Kawai Y, Wu LJ. L-form bacteria, chronic diseases and the origins of life. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2016;371:20150494. doi: 10.1098/rstb.2015.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawai Y, et al. Cell Growth of Wall-Free L-Form Bacteria Is Limited by Oxidative Damage. Curr Biol. 2015;25:1613–1618. doi: 10.1016/j.cub.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song Q, Zhang D, Gao H, Wu J. Salmonella Species’ Persistence and Their High Level of Antimicrobial Resistance in Flooded Man-Made Rivers in China. Microb Drug Resist. 2018;24:1404–1411. doi: 10.1089/mdr.2017.0316. [DOI] [PubMed] [Google Scholar]
- 12.Mohamed T, Zhao S, White DG, Parveen S. Molecular characterization of antibiotic resistant Salmonella Typhimurium and Salmonella Kentucky isolated from pre- and post-chill whole broilers carcasses. Food Microbiol. 2014;38:6–15. doi: 10.1016/j.fm.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Wang DN, et al. Salmonella L-forms: formation in human bile in vitro and isolation culture from patients’ gallbladder samples by a non-high osmotic isolation technique. Clin Microbiol Infect. 2015;21(470):e9–16. doi: 10.1016/j.cmi.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Markova ND. L-form bacteria cohabitants in human blood: significance for health and diseases. Discov Med. 2017;23:305–313. [PubMed] [Google Scholar]
- 15.Domingues S, Rosário N, Ben Cheikh H, Da Silva GJ. SAba1 and Tn6168 acquisition by natural transformation leads to third-generation cephalosporins resistance in Acinetobacter baumannii. Infect Genet Evol. 2018;63:13–16. doi: 10.1016/j.meegid.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Ranjbar R, et al. The Study of Genetic Relationship Among Third Generation Cephalosporin-resistant Salmonella enterica Strains by ERIC-PCR. Open Microbiol J. 2013;7:142–145. doi: 10.2174/1874285801307010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campos MJ, et al. Co-occurrence of ACSSuT and cephalosporin resistance phenotypes is mediated by int1-associated elements in nontyphoidal Salmonella enterica from human infections in Spain. Microb Drug Resist. 2013;19:384–391. doi: 10.1089/mdr.2012.0261. [DOI] [PubMed] [Google Scholar]
- 18.Iwamoto M, et al. Ceftriaxone-Resistant Nontyphoidal Salmonella from Humans, Retail Meats, and Food Animals in the United States, 1996–2013. Foodborne Pathog Dis. 2017;14:74–83. doi: 10.1089/fpd.2016.2180. [DOI] [PubMed] [Google Scholar]
- 19.Folster JP, et al. Characterization of Resistance Genes and Plasmids from Outbreaks and Illness Clusters Caused by Salmonella Resistant to Ceftriaxone in the United States, 2011–2012. Microb Drug Resist. 2017;23:188–193. doi: 10.1089/mdr.2016.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anuforom O, Wallace GR, Buckner MM, Piddock LJ. Ciprofloxacin and ceftriaxone alter cytokine responses, but not Toll-like receptors, to Salmonella infection in vitro. J Antimicrob Chemother. 2016;71:1826–1833. doi: 10.1093/jac/dkw092. [DOI] [PubMed] [Google Scholar]
- 21.Zaki NM, Hafez MM. Enhanced Antibacterial Effect of Ceftriaxone Sodium-Loaded Chitosan Nanoparticles Against Intracellular Salmonella typhimurium. AAPS Pharm Sci Tech. 2012;13:411–421. doi: 10.1208/s12249-012-9758-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fàbrega A, Vila J. Salmonella enterica Serovar Typhimurium Skills To Succeed in the Host: Virulence and Regulation. Clin Microbiol Rev. 2013;26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knudsen GM, et al. The role of ClpP, RpoS and CsrA in growth and filament formation of Salmonella enterica serovar Typhimurium at low temperature. BMC Microbiology. 2014;14:208. doi: 10.1186/s12866-014-0208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan D, Mukherjee M, Nayak R, Dutta R, Suar M. Biological and regulatory roles of acid-induced small RNA RyeC in Salmonella Typhimurium. Biochimie. 2018;150:48–56. doi: 10.1016/j.biochi.2018.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.