Abstract
The WRKY proteins are a superfamily of transcription factor that regulate diverse developmental and physiological processes in plants. Completion of the whole-genome sequencing of Aquilaria sinensis allowed us to perform a genome-wide investigation for WRKY proteins. Here, we predicted 70 WRKY genes from the A. sinensis genome and undertaken a comprehensive bioinformatic analysis. Due to their diverse structural features, the 70 AsWRKY genes are classified into three main groups (group I–III), with five subgroups (IIa–IIe) in group II, except two belong to none of them. Distinct expression profiles of AsWRKYs with RNA sequencing data revealed their diverse expression patterns among different tissues and in the process of whole-tree-inducing agarwood formation. Based on the expression characteristics, we predict some AsWRKYs are pseudogenes, and some may be involved in the biosynthesis of agarwood sesquiterpenes as activators or repressors. Among the tested genes treated with MeJA and H2O2, most of them are induced by H2O2, but downregulated by MeJA, implying the complexity of their involvement in signal transduction regulation. Our results not only provide a basic platform for functional identification of WRKYs in A. sinensis but important clues for further analysis their regulation role in agarwood formation.
Subject terms: Plant stress responses, Plant sciences
Introduction
The WRKYs is one of the largest superfamily of transcription factors in higher plants1, which are characterized by their unique WRKYGQK motif at the N-end and the metal chelating zinc finger motif (CX4–5CX22–23HXH or CX7Cx23HXC) at the C-end2,3. They are divided into three subfamilies (Group I, Group II and Group III) according to the number of WRKY domains and category of zinc-finger2. Group I contains two WRKY domains and one C2H2 zinc finger structure; group II contains one WRKY domain and one C2H2 zinc finger structure; group III contains one WRKY domain and one C2HC zinc finger structure. Group II can be divided into five subgroups (IIa-IIe) according to the amino acid sequence. Studies have demonstrated that Group III only exists in higher plants, and most of them are related to plant response to biological stress, while Group I exists not only in higher plants, but also in ferns and some eukaryotic cells that can’t photosynthesize, such as myxomycetes and single-cell protozoa1,4. This suggests that WRKY transcription factor may originate from eukaryotic cells and Group I is the original form.
As transcription factor, WRKYs function by binding to specific sequences on the promoter of target genes. W-box, with consensus sequence (C/T)TGAC(T/C), is the specific recognition site of WRKYs. TGAC is its core conserved sequence, which is directly related to the specificity of WRKY transcription factor and its target downstream. One mutation will significantly reduce the binding activity and even completely disappeared2,5,6. Studies demonstrated that all WRKY proteins could bind to W-box except for SPF17.
Numerous studies have demonstrated that the WRKY transcription factors play important roles in plant response to both biotic and abiotic stress3,8–13, especially in the control of plant disease and pathogen, it works as a core factor involved in plant defense response12. Many studies show that WRKY (e.g. AtWRKY22, AtWRKY29, AtWRKY33) is downstream of MAPK cascade8,14–20. It was found that, using pathogen infection or spraying SA on Arabidopsis, expression of 49 WRKY transcriptional factors out of the 74 were changed8, demonstrating the WRKY transcription factors are involved in many physiological process in plants. At present, 8 WRKY genes (AtWRKY18/38/53/54/58/59/66/70) have been confirmed directly downstream of NPR121.
The WRKY transcription factors also play important roles in secondary metabolite biosynthesis including terpenes, flavonoids and alkaloids22–25. GaWRKY1 in G. arboretum participates in the biosynthesis of gossypol sesquiterpenes by regulating the activity of (+)-δ-Cadinene synthase (CAD1), and MeJA promoted this process22; AaWRKY1, a transcription factor isolated from glandular secretory trichomes in A. annua, binds the W-box of promoter and actives expression of Amorpha-4,11-diene synthase (ADS), a sesquiterpene cyclase that catalyzes the conversion of farnesyl diphosphate into amorpha-4,11-diene in the biosynthesis of the antimalarial artemisin23. Similarly, MeJA strongly induces the expression of AaWRKY1 and ADS in trichomes23. OsWRKY13 can induce the expression of CHS gene, which is related to flavonoid biosynthesis24,25. CrWRKY1 transcription factor regulates indole alkaloid biosynthesis by binding to the TDC gene W-box26; CjWRKY1, the first WRKY transcription factor identified in the biosynthetic pathway of alkaloids, regulates biosynthesis of Isoquinoline Alkaloid (IQA) in Coptis jiponica27. With the development of genome sequencing, medicinal plants have attracted much attention. Herein, an increasing number of WRKY transcription factors will be identified how they function in the regulation of secondary metabolism.
Aquilaria sinensis (Lour.) Gilg is the species of the genus Aquilaria of Thymelaeaceae. Agarwood, the resin portion that is widely used in traditional medicines, perfumes and incense across Middle East, Japan, India, China, and some Southeast Asian countries, is the product of its defensive response to external injuries28–32. We dedicated to the mechanism and technology of agarwood formation, and have completed genome sequencing of A. sinensis. To systematically elucidate the regulatory role of WRKY transcription factors in A. sinensis, in this study, a genome-wide analysis of AsWRKYs was performed. Totally, 70 WRKY transcription factor genes were predicted from A. sinensis genome, and 7 AsWRKY genes that likely regulate agarwood biosynthesis were identified based on their tissue expression patterns and the results of agarwit-treatment and qPCR analyses. Our results provide a foundation for understanding the molecular basis and regulatory mechanisms of WRKY transcription factors in A. sinensis.
Results
Identification of WRKY transcription factors in A. sinensis
The WRKYs are one of the largest families of plant transcription factors. The number of WRKY is different in different species. Among the lower plants, there are fewer WRKYs ranging from a few to dozens, for example, single in the unicellular green alga1 and 35 in Spike moss Selaginella moellendorffii33, while the higher plants ranging from dozens to hundreds, for example, 74 in Arabidopsis1 and almost 200 in soybean34. With the completion of genome sequencing, the number of WRKY transcription factors in many species has been revealed. Here, a total of 70 genes in the A. sinensis genome were identified as WRKY transcription factors that encoded 70 proteins. We named these members based on their order in the scaffold. Among these proteins, AsWRKY54 was identified to be the smallest protein with 115 amino acids (aa), whereas the largest one was AsWRKY66 (1505 aa). The molecular weights of the proteins also varied according to protein size ranging from 12.7 kDa (AsWRKY54) to 166.7 kDa (AsWRKY66).
Gene structure, conserved motifs and chromosomal location of AsWRKYs
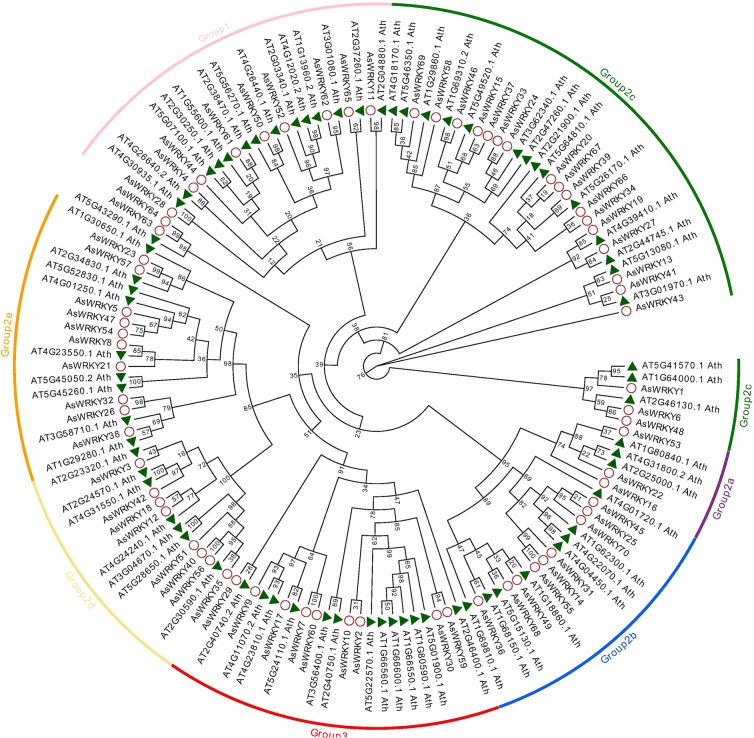
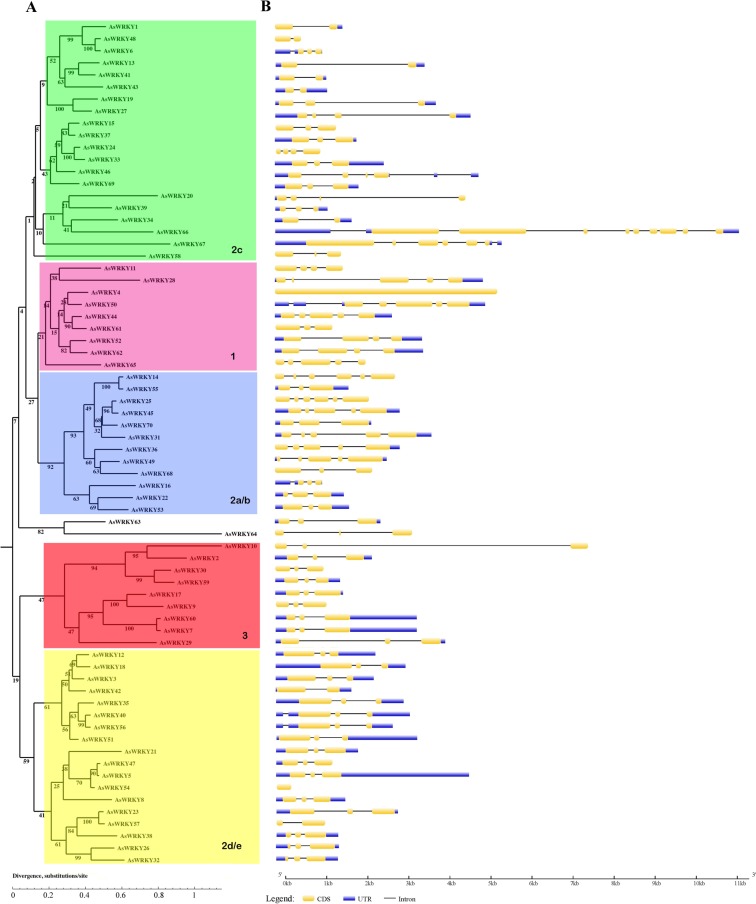
WRKYs are classified into three main groups typically according to their numbers of WRKY domains and zinc finger-like structures35. To look into the structural diversity of AsWRKYs, we first constructed a phylogenetic tree based on the full-length AsWRKY polypeptide sequences, and they were categorized into seven subfamilies as shown by Eulgem et al., 2000 (Fig. 1). Sequences from Arabidopsis WRKY members were included in our analysis as references. Of them, nine belong to group I, which have two WRKY domains (WRKYGQK) and two zinc-finger structure (CX4CX22HX1H, CX4CX23HX1H); nine belong to group III which have single WRKY domain (WRKYGQK) and one zinc-finger structure (CX7CX23HX1C); the other members are belong to groups II, and they have single WRKY domain. Though most of the sequences harbored the well conserved WRKYGQK motif and zinc finger motifs, variants were present in three of the groups II sequences, AsWRKY20, -67, -68, they each has 1 mismatched amino acid within the WRKY motif. The group II is sub-divided into 5 subgroup a-e. Specifically, AsWRKY63, AsWRKY64 were not assigned to any of the subgroup, which is consistent with the phylogenetic tree built on the basis of full-length AsWRKY genes using the neighbor-joining method in MEGA 4.0 (Fig. 2A). They are in the branch with one of the Arabidopsis At5G43290.1, which does not belong to any group in Arabidopsis, too2. This phenomenon is not special that exists in other plants. For example, among the 81 WRKYs in Solanum lycopersicum, 15 belong to group I, 52 belong to group II, 11 belongs to group III, and 3 belong to none of group36. The different WRKY subgroups from A. sinensis, G. raimondii, Arabidopsis and S. lycopersicum are described in Table 1.
Figure 1.
Polygenetic tree of AsWRKY proteins and homologous in Arabidopsis.
Figure 2.
Phylogenetic tree and gene structure of A. sinensis WRKY genes. (A) The phylogenetic tree was constructed with MEGA 4.1 software by the neighbor-joining (NJ) method with 1,000 bootstrap replicates. (B) Exon/intron structure of AsWRKY genes: the introns, exons and UTR are represented by black lines, yellow boxes and blue boxes, respectively.
Table 1.
The number of the main three groups and their subgroups, as well as the total number of WRKY genes in A. sinensis, G. raimondii and Arabidopsis.
| Group | subgroup | AsWRKYs | GaWRKYs | AtWRKYs | SiWRKYs |
|---|---|---|---|---|---|
| I | 9 | 20 | 14 | 15 | |
| II | IIa | 3 | 7 | 3 | 5 |
| IIb | 9 | 16 | 8 | 8 | |
| IIc | 20 | 37 | 18 | 16 | |
| IId | 8 | 15 | 7 | 6 | |
| IIe | 10 | 13 | 1 | 17 | |
| III | 9 | 12 | 13 | 11 | |
| none group | 2 | 2 | 3 | ||
| Total | 70 | 120 | 74 |
As gene structure is a typical imprint of evolution within a gene family, we analyzed the WRKY genes in A. sinensis using tools at the GSDS website. A detailed illustration of the gene structures is shown in Fig. 2B. The number of exons ranged from 1 to 10, and more than half of AsWRKYs have 3 exons and 2 introns. Each group showed similar structure, especially group III with group 2d/e (Fig. 2B), showing that almost all these members have tree exons and two introns. Most member in group I and group 2a/b contain three or four introns, while group 2c varies from one to nine. This phenomenon is also found in other plants, such as populus37, soybean38, and setaria39, demonstrating the conservation of the structure in WRKY family. It is also noted a few members, such as AsWRKY66, AsWRKY4 and AsWRKY54, showed quite different protein structures compared with other members. AsWRKY66 has the largest numbers 10 exons and 9 introns. However, AsWRKY4 and AsWRKY54 have only 1 exon without containing any intron, and AsWRKY4 is the longest exon. This loss of introns was considered as the result of intron turnover or due to reverse transcription of the mature mRNA followed by homologous recombination with intron-containing alleles35,37. It has been suggested that the transcription efficiency of genes may be related to the length of introns, and introns are relatively free to evolve faster than exons40. We also found that seven genes, including AsWRKY10, AsWRKY13, AsWRKY19, AsWRKY20, AsWRKY27, AsWRKY28, AsWRKY29, have longer intron than others, especially AsWRKY10 has the longest intron more 6 kb (Fig. 2B), demonstrating mRNA transcripts would span over especially long intron for these genes.
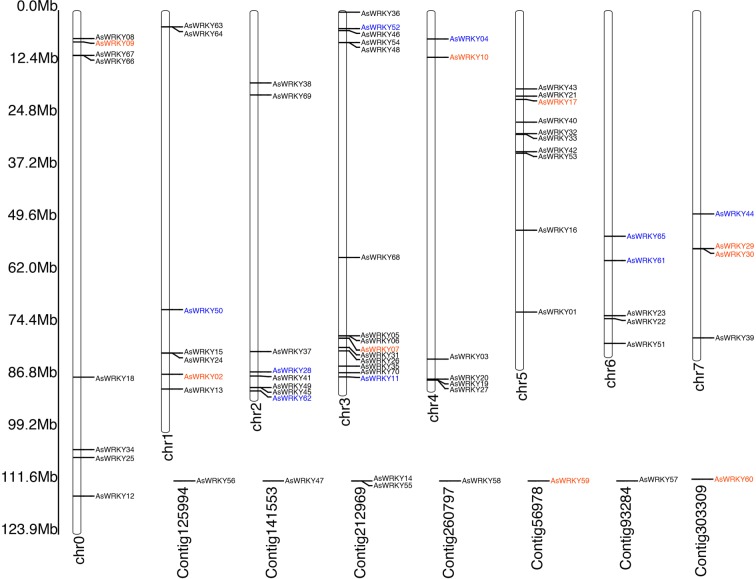
The distribution of the AsWRKY genes on chromosomes was shown in Fig. 3. They were unevenly distributed throughout all eight chromosomes, and the number on each chromosome was not related to its length. Chromosome 3 (Chr 3) possessed the largest number of WRKY genes (14 genes) followed by Chr 5 (10 genes). Due to assembly of genome, 8 genes (AsWRKY14/47/55/56/57/58/59/60) could not be located on the chromosome.
Figure 3.
Distribution of AsWRKY genes on eight A. sinensis chromosomes. The chromosome number is indicated at the bottom of each chromosome. The genes marked in different color indicate the group of the WRKY family (I, blue; II, red; III, black).
Tissue expression analysis of AsWRKYs
For the character that WRKY are widely involved in the developmental and physiological process of plant, expression profile of these AsWRKYs were analyzed from transcriptome data in 7 tissues including agarwood, root, branch, stem, old leaves, temder leaves, bud and flower. As shown in Fig. 4, the expression heatmap in different tissues are clustered. It is found that most of the AsWRKYs has a low expression level in temder leaves, some of them highly expressed in branch, stem and root, some expressed highly in bud and flower, some expressed highly only in old leaves, fully demonstrating the functional diversity of WRKYs. It is note that, 6 WRKYs (AsWRKY7, -9, -10, -54, -63, -64) were not expressed in none of the tissues, 18 were not expressed in agarwood, and 5 (AsWRKY13, -38, -49, -58, -69) were specifically highly expressed in agarwood, which is presumed closely related to the formation of agarwood. Interestingly, we also found that all of the AsWRKYs in group I expressed highly in all of the 7 tissues without obvious different expression level, which may indicate their fundamental roles in different cell-types in A. sinensis. All of group III has very low expression level in agarwood, branch, stem and root, even not expressed in agarwood. AsWRKYs expressed highly in agarwood belong to group II, correctly most of them (AsWRKY13, -34, -37, -58, -66, -69) belong to 2c, except to AsWRKY49 belongs to 2b and AsWRKY38 belongs to 2e. These results demonstrated that group III may not be involved in the agarwood formation, and the group II plays important regulatory role. It has been noted that gene expression patterns can provide important clues for gene function, these findings above may facilitate to identify the function of AsWRKYs and lead to discover their roles in plant growth and development, and in the process of agarwood formation as the biological functions of almost all of the AsWRKYs remain to be elucidated.
Figure 4.
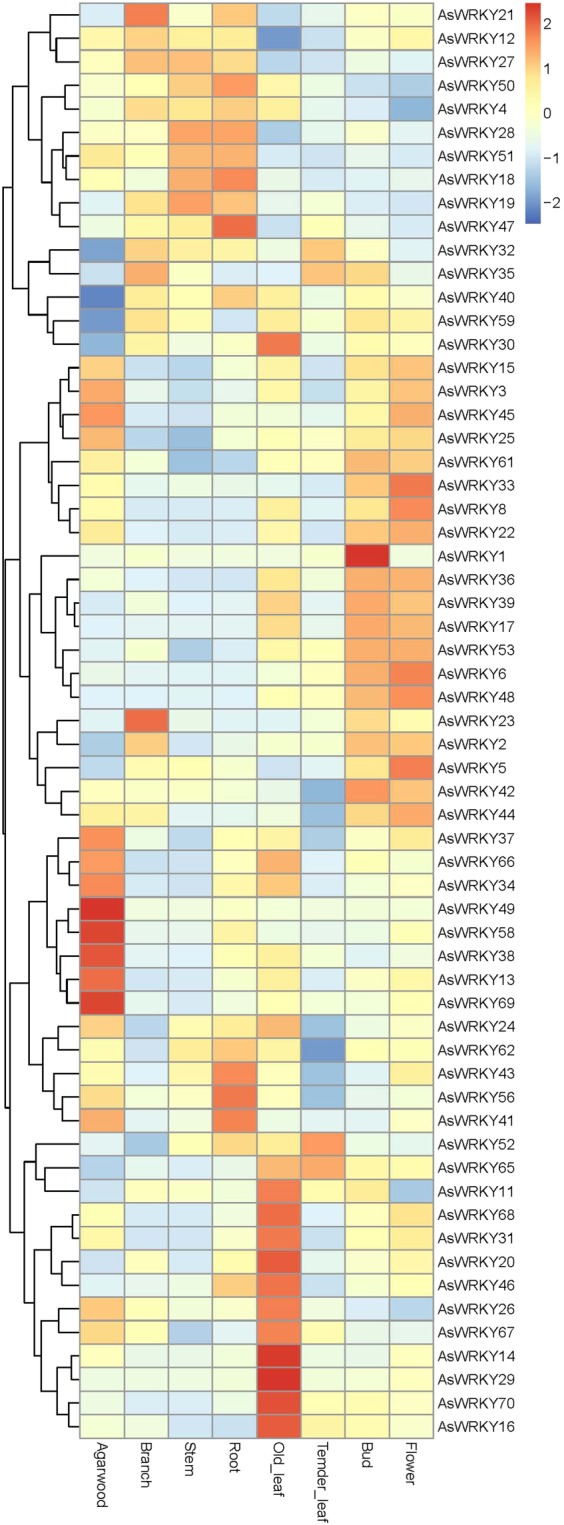
Heat map of the AsWRKY genes expression profiles in different tissues. All gene expression levels were transformed to scores ranging from −2 to 2 and were colored blue, white, or red to represent low, moderate, or high expression levels, respectively.
Expression profiles of AsWRKYs in the process of whole-tree-inducing agarwood formation
It has been demonstrated that WRKY TFs were involved in the activation of plant defense systems in response to stimuli3. Wound-induced agawood formation is the production of defense response in A. sinensis. To investigate whether AsWRKYs are involved in agarwood formation process, we analyzed the transcriptome data which performed using the whole-tree inducing materials, including different treatment-time and different layers. From the data and heat map (Fig. 5), it was found that there was a clear cluster, showing that some WRKYs highly expressed in all of the four layers (healthy layer—H, agarwood layer—AT and A, transition layer—T and decomposed layer–D), some WRKYs were highly expressed in agarwood layers, which we were concerned about. Of these different expression genes, some induced at the early stage (before 24 H), and some were induced later, especially only expressed in A21M, such as AsWRKY49, AsWRKY68 and AsWRKY69. Combined with the tissue expression patterns, it was predicted that AsWRKY13, AsWRKY25, AsWRKY34, AsWRKY38, AsWRKY49, AsWRKY58, AsWRKY69 are likely important positive regulators in agarwood formation. Moreover, we also found some WRKYs (such as AsWRKY21, AsWRKY23) maybe the negative regulators in agarwood formation, for their expression were down-regulated after wound treatment. Moreover, some WRKYs could not be detected in all of layers, they are AsWRKY7, AsWRKY9, AsWRKY10, AsWRKY54, AsWRKY63, and AsWRKY64. Combined with the tissue expression profile, the six genes were not be detected, and their RPKM values are zero. Herein, they are speculated to be pseudogenes.
Figure 5.
Heat map of the AsWRKY genes expression profiles in different layers and different agar-wit treated time. H- the healthy layer; D- the dead layer after Agar-Wit treatment, A- the agarwood layer; T- the transition layer; B- the blocked layer. All gene expression levels were transformed to scores ranging from −2 to 2 and were colored blue, white, or red to represent low, moderate, or high expression levels, respectively.
Investigation of AsWRKYs expressions in response to MeJA and H2O2 treatment exogenously by qRT-PCR
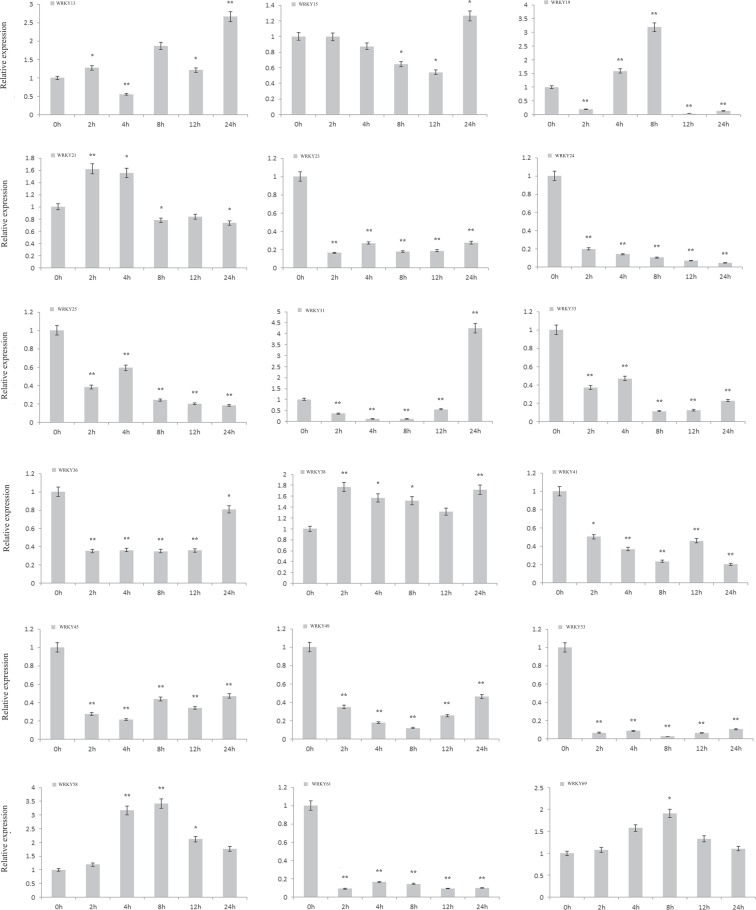
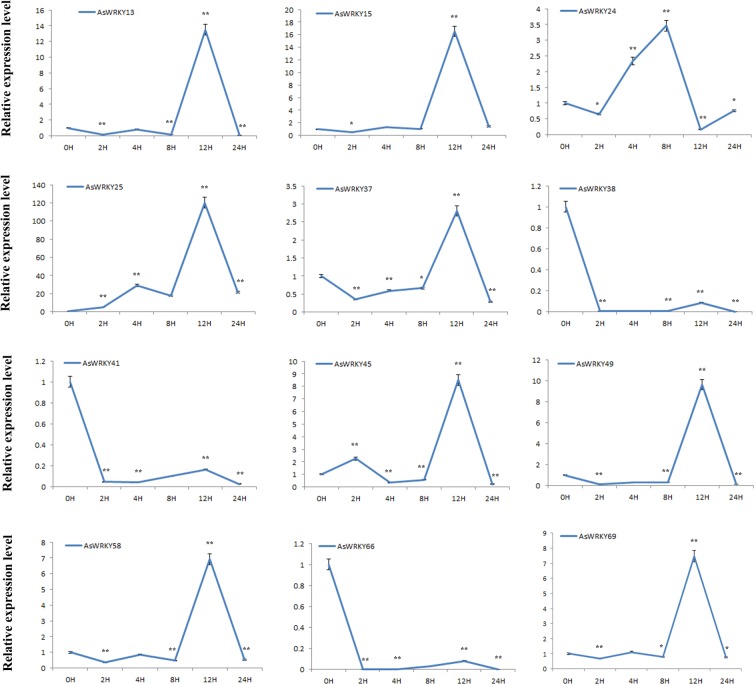
It has been confirmed that MeJA and H2O2 are important signal molecules involved in regulating agarwood formation31,41. To test the above expression analysis, we further detected the expression level of some genes responding to MeJA and H2O2 treatment by real-time PCR. They showed different characters in response to the two treatments. For MeJA treatment (Fig. 6), 6 were up-regulated and 9 down-regulated among the detected genes. Though the induction-fold was not obvious, the decrease rapidly and significantly. AsWRKY23, AsWRKY24, AsWRKY53, AsWRKY61 declined 10 times rapidly within 2 hours and maintained stable until 24 h. AsWRKY21 increased slightly and then decreased steadily, which may be a stress response to the external stimuli, and its regulation may be at the translational level. Expression of AsWRKY15 and AsWRKY38 were hardly affected, demonstrating they might not participate in the wound-induced agarwood formation. AsWRKY38 and AsWRKY69 are induced by MeJA and no decline within the 24 h, implying they are likely positive regulators. For H2O2 treatment by contrast (Fig. 7), most of detected WRKYs are induced by H2O2, except AsWRKY38, AsWRKY41, AsWRKY66, which dropped so sharply to their lowest level that barely detectable in two hours. Of the nine induced genes, AsWRKY25 was found to be the strongest one with showing more 100 times, other eight peaked at 12 h, and then declined rapidly to the lowest level. In general, all the detected genes response more strongly to H2O2 than MeJA, which may consistent with our previous research result that hydrogen peroxide burst triggers accumulation of jasmonates in wound-induced agarwood formation41. These basic analyses provide important clues for further research.
Figure 6.
Expression profile of AsWRKYs in MeJA treated calli. A. sinensis calli were transferred to MS containing 100 μM MeJA and sample at appointed times (0 h, 0.5 h, 2 h, 4 h, 6 h, 12 h, 24 h). Expression levels of all detected genes were assayed using real-time PCR analysis and AsGADPH as the internal control. Each value is the mean ± SE of 3 independent biological replicates. Asterisks indicate significant differences between each treatment point and controls (0-h time point) (*P < 0.05, **P < 0.01, Student’s t-tests).
Figure 7.
Expression profile of AsWRKYs in H2O2 treated calli. A. sinensis calli were transferred to MS containing 100 μM H2O2 and sample at appointed times (0 h, 0.5 h, 2 h, 4 h, 6 h, 12 h, 24 h). Expression levels of all detected genes were assayed using real-time PCR analysis and AsGADPH as the internal control. Each value is the mean ± SE of 3 independent biological replicates. Asterisks indicate significant differences between each treatment point and controls (0-h time point) (*P < 0.05, **P < 0.01, Student’s t-tests).
Discussion
WRKY proteins have been detected in various organisms10. Here, we systematically identified 70 WRKY members from whole genome sequences of A. sinensis for the first time. Sequence alignment and phylogenetic analysis classified them into three major groups (I, II, III) based on the WRKY domain and conserved zinc finger-like motif. Of them, group I and group III contains 9 members, respectively, and 50 belong to group II (Fig. 1) that is the largest group. However, in Arabidopsis and poplar, group I houses the largest number of WRKYs; while in rice, group III is the largest. The largest group in A. sinensis is group II, implying that this group may experience more gene duplications during the evolutionary course. At present, WRKYs found in the genomes of Chlamydomonas monocytogenes, non-photosynthetic eukaryotic myxomycetes, unicellular protoplasts, lower plant bryophytes and ferns belong to group I, demonstrating the group I is likely the original protein expression form and may originate from eukaryotic cells before undifferentiated plant kingdom 1.5–2 billion years ago1. Furthermore, by referring to the domain feature and the phylogenic tree (Fig. 2A), it showed three interesting observations: (i) all group I members have two WRKY domain and two zinc finger structure; (ii) AsWRKY20 possess only WRKY domain and lack zinc finger structure, and AsWRKY54 possess only zinc finger structure and lack WRKY domain; (iii) AsWRKY63 and AsWRKY64 belong to none of the group. All these results implied the AsWRKY genes may have experienced WRKY domain loss during the evolution.
In higher plants, the group III WRKY gene accounts for 20% of the family members, but it does not exist in some lower plants such as bryophytes. Studies have showed that almost all of group III WRKYs in Arabidopsis are related to biotic stress response8, which indicates that the group III WRKYs is relatively latest in the evolutionary history of terrestrial plants. However, in A. sinensis, all of group III has very low expression level in agarwood, branch, stem and root, even not expressed in agarwood (Fig. 4), and three of them (AsWRKY7, -9, -10) are considered as pseudogenes, implying the group III members may have little to do with the agarwood formation, which is inconsistent with the Arabidopsis.
Tissue expression analysis showed that group I expressed highly in all of the 7 tissues without obvious different expression level, all of group III has very low expression level in agarwood, branch, stem and root, even not expressed in agarwood. Members highly expressed specifically in agarwood belong to group II. Combined with their expression in the different layers of the agarwit treated, expression of members in group III did not change much during the whole process of agarwood formation, and only AsWRKY61 in group I had highest expression in AT layer. So, AsWRKYs belonging to group II are predicted important regulators that act as activator or repressors of sesquiterpene biosynthesis. The results are consistent with their evolutionary classification that group I are the original proteins participating in various processes of growth and development, while group II has undergone great changes in the course of evolution. Anyhow, these results demonstrating that although WRKYs have conservative domains, their functions and regulations are very complex, and even the differences among members are very large.
There is a general consensus that agarwood can not be formed in healthy A. sinensis tree, only be induced after injury. In this study, we explored the possible role of WRKY in the formation of agarwood through tissue-specific expression and expression profiles in the process of agar-wit treated agarwood formation. From the tissue-expression heat map, it can be seen that AsWRKY13/38/49/58/69 specific expressed in agarwood (Fig. 4), implying they may play a regulatory role in the formation of agarwood. WRKY proteins act as an activator or repressor regulating expression of downstream target genes in many different biological processes14,42–44. Some can function as activator in one pathway but as repressor in another43. The heat map in different layers and different agar-wit treated time showed that AsWRKY13/25/34/38/49/58/69 may positive regulators, while AsWRKY21/23 may function as negative regulators (Fig. 5). Of course, these information needs further verification. Our research group has previously confirmed that MeJA and H2O2 are important signal molecules involved in regulating agarwood formation31,41. Herein, we treated callus with MeJA and H2O2 and detected 15 genes expression of concern (Figs. 6, 7). It was showed that, among the 15 AsWRKYs, 9 are induced by H2O2, 3 induced by MeJA, and some genes showed an opposite response to MeJA and H2O2 treatment. Our previous study showed that hydrogen peroxide burst triggers accumulation of jasmonates in wound-induced agarwood formation41, the WRKY repressors and activators presented in the work will help test the model and contribute to the dissection of the transcriptional complex involved in MeJA and H2O2 signaling.
In summary, the WRKY family in A. sinensis was systematically identified, including their classification, chromosome distribution and their expression profiles in different tissues and response to different external stimulus, using comprehensive computational approaches and real-time PCR analysis. Our results could help to select appropriate candidate genes for further characterization of their functions in A. sinensis.
Materials and Methods
Plant materials for RNA-Seq
The materials for transcription sequence are seven-year-old A. sinensis stems grown in Hainan branch of IMPLAD (Institute of Medicinal Plant Development). Trees were treated using Whole-tree agarwood-inducing technique (Agar-Wit)45. Samples were collected at the pointed times. The healthy wood was used as control (0 h).
Plant materials for qRT-PCR analysis
The materials for qRT-PCR are A. sinensis suspension. The suspensions have been cultured in dark. 100 μM MeJA or H2O2 was added to the suspension. Samples were taken at the pointed time after treatment. The suspensions without any treatment were used as control. All materials were frozen in liquid N2 and stored at −70 °C until used.
Library preparation, transcriptome sequencing and bioinformatics analysis
Total RNA was isolated from the materials using a TRIzol kit (Invitrogen, USA) and purified to enrich messenger RNA (mRNA). Quantification and qualification RNA was checked to meet the standards for library construction. A total amount of 3 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. The library preparations were sequenced on an Illumina Hiseq platform and 125 bp/150 bp paired-end reads were generated. Raw data (raw reads) were filtered to obtain the clean reads to ensure the quality of the information analysis. Index of the reference genome was built using Bowtie v2.2.3 and paired-end clean reads were aligned to the reference genome using TopHat v2.0.12.
Identification of transcription factor of AsWRKYs and their chromosomal location
Transcription factors are characterized based on iTAK program (http://bioinfo.bti.cornell.edu/tool/itak)46,47. The program first compared species protein sequences against the pfam domain database48 using the HMMER3.1 program (http://hmmer.janelia.org). Meanwhile, GSDS software was used to map the WRKY gene structure. All parameters are default. AsWRKY genes were located on chromosomes according to their positions based on Hic (Chromosome conformation capture) result of A. sinensis genome.
Phylogenetic and sequence feature analysis
WRKY protein blast was performed using the Muscle software, and then build a Neighbour-Joining tree using the TreeBeST of PAMLsoftware (bootstrap = 1000)49. The WRKY genes were classified based on their protein motifs and sequence similarity.
Tissue expression and induced expression analysis
Calculate of the RPKM value of WRKY transcription factors in different tissues and log2(rpkm + 1) as the expression level, after normalization procedure draw the heat map. Similarly, heat maps at different time points in different layers were drawn according to the value of log2(rpkm + 1)/(H-0h -rpkm + 1). H-0h is the control. Here, the data was normalized using the Z-score standardization method (the mean value is 0 and the variance is 1). The expression level of each gene in each tissue (RPKM) was converted to Z fraction: Zi = (Xi-μ)/σ, Zi, Xi, μ and σ represent Z fraction in i tissue, RPKM value and gene expression in i tissue, the mean value and the variance of gene expression.
Expression profiling using qPCR
To analyze gene expression response to treatment of MeJA, and H2O2, exogenously, total RNA was isolated from treated cell suspensions using a Total RNA Rapid Extraction kit RN38-EASYspin Plus (Aidlab). 1-μg aliquot was subjected to first-strand synthesis according to the instructions of PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara). The primers used for real-time PCR are listed in Supplementary Table S1. Analysis was performed essentially according to our previously described procedures31,50.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81573525, 81673549), and the program of CAMS Initiative for Innovative Medicine (CAMS-I2M) (2016-I2M-2-003).
Author contributions
Y.X.: analyzed the data and drafted the manuscript. P.S.: Performed the real-time experiments. X.T.: Prepared the A. sinensis materials. Z.G. and Z.Z.: Participated in the discussion of the result. J.W.: Helped conceive the study, and revised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yan-Hong Xu and Pei-Wen Sun.
Supplementary information
is available for this paper at 10.1038/s41598-020-59597-w.
References
- 1.Ulker B, Somssich IE. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004;7:491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Eulgem T, et al. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 3.Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Bio. 2007;10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 4.Yamasaki K, et al. Structures and evolutionary origins of plant specific transcription factor DNA binding domains. Plant Physio. Biochem. 2008;46:394–401. doi: 10.1016/j.plaphy.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 5.Yu DQ, Chen CH, Chen ZX. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell. 2001;13:1527–1540. doi: 10.1105/TPC.010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciolkowski I, Wanke D, Birkenbihl R, Somssich I. Studies on DNAbinding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008;68:81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 50 upstream regions of genes coding for sporamin and b-amylase from sweet potato. Mol. Gen. Genet. 1994;244:563–571. doi: 10.1007/BF00282746. [DOI] [PubMed] [Google Scholar]
- 8.Dong J, Chen C, Chen Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003;51:21–37. doi: 10.1023/A:1020780022549. [DOI] [PubMed] [Google Scholar]
- 9.Jones JDG, Dang JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 10.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Ren XZ, et al. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010;63:417–429. doi: 10.1111/j.1365-313X.2010.04248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muthamilarasan M, Prasad M. Plant innate immunity: an updated insight into defense mechanism. J. Biosci. 2013;38:433–449. doi: 10.1007/s12038-013-9302-2. [DOI] [PubMed] [Google Scholar]
- 13.Muthamilarasan, M. et al. C2H2-type of zinc finger transcription factors infox tail millet define response to abiotic stresses. Funct. Integr. Genomics 14, 531–543 (2014a). [DOI] [PubMed]
- 14.Asai T, et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 15.Mészáros T, et al. The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J. 2006;48:485–498. doi: 10.1111/j.1365-313X.2006.02888.x. [DOI] [PubMed] [Google Scholar]
- 16.Schikora A, et al. N-acyl-homoserine lactone confers resistance toward biotrophic and hemibiotrophic pathogens via altered activation of AtMPK61. Plant Physiol. 2011;157:1407–1418. doi: 10.1104/pp.111.180604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González-Lamothe R, ElOirdi M, Brisson N, Bouarab K. The conjugated auxin indole-3-acetic acid—aspartic acid promotes plant disease development. Plant Cell. 2012;24:762–777. doi: 10.1105/tpc.111.095190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Q, Febres VJ, Jones JB, Moore GA. Responsiveness of different citrus genotypes to the Xanthomonas citri ssp. citri-derived pathogen-associated molecular pattern (PAMP) flg22 correlates with resistance to citrus canker. Mol. Plant Pathol. 2015;16:507–520. doi: 10.1111/mpp.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kloth KJ, et al. AtWRKY22 promotes susceptibility to aphids and modulates salicylic acid and jasmonic acid signaling. J. Exp. Bot. 2016;67(11):3383–3396. doi: 10.1093/jxb/erw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng ZY, et al. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006;48:592–605. doi: 10.1111/j.1365-313X.2006.02901.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang D, Amornsiripanitch N, Dong XN. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. Plos Pathog. 2006;2:123. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu YH, Wang JW, Wang S, Wang JY, Chen XY. Characterization of GaWRKY1, a Cotton Transcription Factor That Regulates the Sesquiterpene Synthase Gene (+)-δ-Cadinene Synthase-A. Plant Physiol. 2004;135(1):507–515. doi: 10.1104/pp.104.038612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma D, et al. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4, 11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 2009;50:2146–2161. doi: 10.1093/pcp/pcp149. [DOI] [PubMed] [Google Scholar]
- 24.Qiu DY, et al. OsWRKY13 mediates rice disease resistance by regulating defense -related genes in salicylate-and jasmonate-dependent signaling. Mol. Plant-Microbe. In. 2007;20(5):492–499. doi: 10.1094/MPMI-20-5-0492. [DOI] [PubMed] [Google Scholar]
- 25.Qiu DY, et al. Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol. Plant. 2008;1(3):538–551. doi: 10.1093/mp/ssn012. [DOI] [PubMed] [Google Scholar]
- 26.Suttipanta N, et al. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011;157:2081–2093. doi: 10.1104/pp.111.181834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schluttenhofer C, Yuan L. Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 2015;167:295–306. doi: 10.1104/pp.114.251769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumeta Y, Ito M. Characterization of δ-guaiene synthases from cultured cells of Aquilaria, responsible for the formation of the sesquiterpenes in agarwood. Plant Physiol. 2010;154(4):1998–2007. doi: 10.1104/pp.110.161828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu PW, et al. Interxylary phloem and xylem rays are the structural foundation of agarwood resin formation in the stems of Aquilaria sinensis. Trees. 2018;33(2):533–542. doi: 10.1007/s00468-018-1799-4. [DOI] [Google Scholar]
- 30.Xu YH, et al. Identification of genes related to agarwood formation: transcriptome analysis of healthy and wounded tissues of Aquilaria sinensis. BMC Genomics. 2013;14:227. doi: 10.1186/1471-2164-14-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu YH, et al. Jasmonic acid is a crucial signal transducer in heat shock induced sesquiterpene formation in Aquilaria sinensis. Sci. Rep. 2016;6:21843. doi: 10.1038/srep21843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu YH, et al. Transcription Factor AsMYC2 Controls the Jasmonate-Responsive Expression of ASS1 Regulating Sesquiterpene Biosynthesis in Aquilaria sinensis (Lour.) Gilg. Plant Cell Physiol. 2017;58(11):1924–1933. doi: 10.1093/pcp/pcx122. [DOI] [PubMed] [Google Scholar]
- 33.Banks JA, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;20:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Chen J, Wang L, Wang S. Genome-Wide Investigation of WRKY Transcription Factors Involved in Terminal Drought Stress Response in Common Bean. Front Plant Sci. 2017;8:380. doi: 10.3389/fpls.2017.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu KL, Guo ZJ, Wang HH, Li J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005;12:9–26. doi: 10.1093/dnares/12.1.9. [DOI] [PubMed] [Google Scholar]
- 36.Huang S, et al. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet Genomics. 2012;287(6):495–513. doi: 10.1007/s00438-012-0696-6. [DOI] [PubMed] [Google Scholar]
- 37.He H, et al. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012;31:1199–1121. doi: 10.1007/s00299-012-1241-0. [DOI] [PubMed] [Google Scholar]
- 38.Yu YC, Wang N, Hu R, Xiang FN. Genome-wide identification of soybean WRKY transcription factors in response to salt stress. Springer Plus. 2016;5:920. doi: 10.1186/s40064-016-2647-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muthamilarasan M, et al. Global analysis of WRKY transcription factor superfamily in Setaria identifies potential candidates involved in abiotic stress signaling. Front plant Sci. 2015;6:910. doi: 10.3389/fpls.2015.00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morello L, Breviario D. Plant spliceosomal introns: not only cut and paste. Current Genomics. 2008;9:227–238. doi: 10.2174/138920208784533629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lv FF, et al. Hydrogen peroxide burst triggers accumulation of jasmonates and salicylic acid inducing sesquiterpene biosynthesis in wounded Aquilaria sinesis. J. Plant Physio. 2019;234–235:167–175. doi: 10.1016/j.jplph.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 42.De Pater S, et al. Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res. 1996;24:4624–4631. doi: 10.1093/nar/24.23.4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE. Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J. 1999;18:4689–4699. doi: 10.1093/emboj/18.17.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C, Chen Z. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physio. 2002;129:706–716. doi: 10.1104/pp.001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei, J. H. et al. Production of agarwood in Aquilaria sinensis trees via transfusion technique. CN101755629B (2010).
- 46.Guo S, et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet. 2013;45(1):51–82. doi: 10.1038/ng.2470. [DOI] [PubMed] [Google Scholar]
- 47.Huang S, et al. Draft genome of the kiwifruit Actinidia chinensis. Nat. Commun. 2013;4(4):2640. doi: 10.1038/ncomms3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Bio Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 50.Robatzek S, Somssich IE. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Gene Dev. 2002;16:1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.