Abstract
Porphyromonas gulae is a major periodontal pathogen in dogs, which can be transmitted to their owners. A major virulence factor of P. gulae consists of a 41-kDa filamentous appendage (FimA) on the cell surface, which is classified into three genotypes: A, B, and C. Thus far, inhibition of periodontal disease in dogs remains difficult. The present study assessed the inhibitory effects of a combination of clindamycin and interferon alpha (IFN-α) formulation against P. gulae and periodontal disease. Growth of P. gulae was significantly inhibited by clindamycin; this inhibition had a greater effect on type C P. gulae than on type A and B isolates. In contrast, the IFN-α formulation inhibited the expression of IL-1β and COX-2 elicited by type A and B isolates, but not that elicited by type C isolates. Furthermore, periodontal recovery was promoted by the administration of both clindamycin and IFN-α formulation to dogs undergoing periodontal treatment; moreover, this combined treatment reduced the number of FimA genotypes in oral specimens from treated dogs. These results suggest that a combination of clindamycin and IFN-α formulation inhibit P. gulae virulence and thus may be effective for the prevention of periodontal disease induced by P. gulae.
Subject terms: Bacteriology, Drug development
Introduction
Periodontal disease is a common infection in dogs1, which is characterised by chronic inflammation of periodontal tissue2. Periodontal disease affects 44% to 64% of all dogs; this proportion increases to 85% in dogs older than 4 years of age3,4. Periodontal disease is caused by the formation of biofilm due to the growth of bacteria in the gingival sulcus1. This biofilm elicits an abnormal host immune responses, which is followed by destruction of periodontal tissues such as periodontal ligament and alveolar bone, eventually leading to tooth loss1,5.
Among periodontal pathogens, Porphyromonas gulae is the bacterial species most often associated with periodontal disease in dogs6. FimA (41-kDa fimbriae) is expressed on the cell surface by strains of P. gulae bacteria obtained from dogs7,8. FimA proteins are divided into three genotypes (A, B, and C), based on differences in their putative amino acid sequences8. Among these fimA genotypes, type C P. gulae is considered to be the most virulent, as it is predominant in the oral cavities of dogs with severe periodontitis8. Interestingly, type C P. gulae is also prevalent in the oral cavities of dogs with mitral regurgitation9, which implies that P. gulae may be associated with some types of systemic disease.
Some periodontal pathogens can disrupt the host innate immune response, which results in the exacerbation of periodontal disease10. Notably, overexpression of interleukin-1β (IL-1β), cyclooxygenase-2 (COX-2), interleukin-8 (IL-8), and transforming growth factor-β1 (TGF-β1) from gingival cells is closely related to periodontal tissue injury11–13. Thus, methods to control the inflammatory response and to eliminate periodontal bacteria are considered to be important for the inhibition of periodontal disease.
Interferon-α (IFN-α), classified as a type I interferon, is generally secreted to combat infection14. IFN-α has also been produced as a pharmaceutical agent which is used for treatment of autoimmune and infectious diseases in humans15. In addition, a canine IFN-α formulation (InterBerryα®; Hokusan Co. Ltd., Higashihiroshima, Japan) has been commercially available as a pharmaceutical agent for periodontal treatment in animals since 2014. The administration of canine IFN-α formulation to the oral cavity of dogs has been reported to improve gingivitis symptoms and reduce the number of bacteria in the Porphyromonas genus16. However, there have been few studies focused on the role of IFN-α in treatment of periodontal disease in dogs.
The recommended approach for prevention and treatment of periodontal disease involves maintenance of oral hygiene by the owner and professional periodontal treatment by veterinarians1,5. Antibiotics are generally prescribed in combination with periodontal treatment, such as scaling and root planing, with the aim of reducing the number of pathogenic bacteria17. In addition to the use of antibiotics, periodontal recovery relies on the control of inflammatory responses within infected periodontal tissue18. Therefore, methods capable of suppressing the host inflammatory immune system should be developed for use in periodontal treatment, in addition to combined application of antibiotic medication and professional periodontal treatment. However, the efficacy of anti-inflammatory treatment of periodontal tissue infected with P. gulae has not yet been investigated.
The present study analysed the inhibitory effect of clindamycin, an antibiotic frequently used in periodontal treatment of dogs, on the growth of P. gulae strains according to their fimA genotypes. In addition, the study investigated whether an IFN-α formulation could inhibit the overexpression of inflammatory responses from gingival epithelial cells induced by P. gulae infection. Finally, the study analysed the effects of combined treatment with clindamycin and IFN-α formulation on the periodontal condition and on the levels of P. gulae with each fimA genotype that were present within oral specimens from dogs.
Results
Inhibitory effects of clindamycin on growth of P. gulae strains
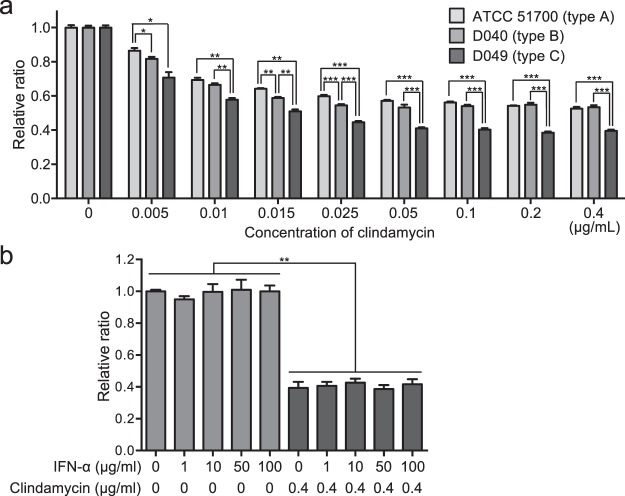
Clindamycin has been used for the treatment of periodontal disease in dogs19. Therefore, we analysed whether clindamycin was effective for P. gulae with each fimA genotype. Growth of all P. gulae strains tested (fimA genotypes A, B, and C) was significantly reduced in the presence of more than 0.005% of clindamycin compared with growth without clindamycin (P < 0.05) (Fig. 1a). Clindamycin inhibited the bacterial growth of the P. gulae strains in a dose-dependent manner. The inhibitory effect on P. gulae D049 (type C) was significantly greater than on P. gulae ATCC 51700 (type A) and P. gulae D040 (type B) in the presence of each concentration of clindamycin (P < 0.05). The inhibitory effect on P. gulae D040 (type B) was significantly greater than on P. gulae ATCC 51700 (type A) in the presence of 0.005%, 0.015%, and 0.025% of clindamycin. When the bacterial growth of P. gulae was analysed in the presence of IFN-α formulation, each concentration of the IFN-α formulation showed no effect on the growth of P. gulae (Fig. 1b). In addition, the effect of clindamycin on the growth of P. gulae was not inhibited by use of the IFN-α formulation.
Figure 1.
Bacterial growth of P. gulae strains. (a) Bacterial growth of P. gulae strains in the presence of various concentrations of clindamycin. Bacterial growth without clindamycin was defined as the baseline. (b) Bacterial growth of P. gulae strain D049 in the presence of IFN-α formulation and clindamycin. Bacterial growth without IFN-α formulation and clindamycin was defined as the baseline. Data are shown as the mean ± SD of three independent experiments. There were significant differences as determined by using analysis of variance with Bonferroni correction (*P < 0.05, **P < 0.01, ***P < 0.001).
Cytokine and enzyme expression in gingival epithelial cells infected with P. gulae
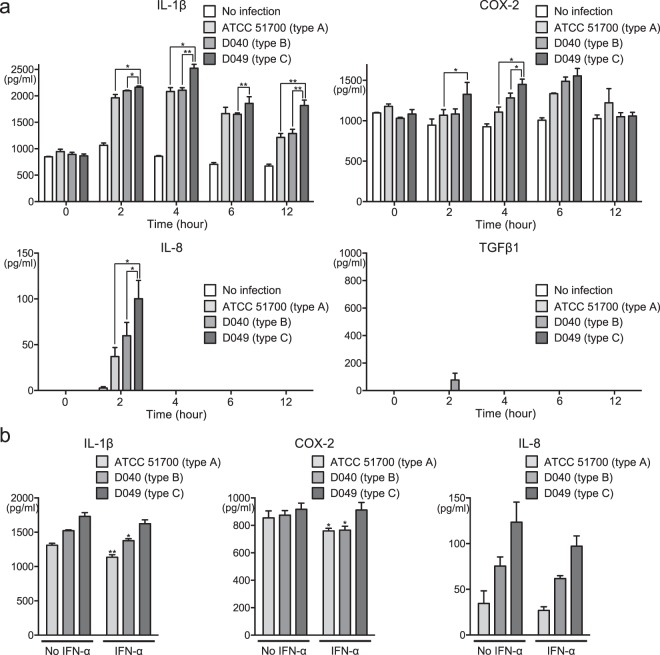
P. gingivalis, a species closely related to P. gulae, induces inflammatory disorders which cause aggravation of periodontal disease20. Gingival epithelial cells play an important role in preventing bacterial invasion deeper into tissue21; however, changes in the host inflammatory response produced by gingival epithelial cells exposed to P. gulae remain unknown. The relative ratios of mRNA expression levels of IL-1β, COX-2, IL-8, and TGF-β1 in Ca9-22 human gingival epithelial cells infected with P. gulae strains were analysed with their respective levels at 0 h after P. gulae infection defined as 1.0. IL-1β, COX-2, and IL-8 levels in the presence of each P. gulae strain were highest at 2 h after infection; these levels were significantly higher than those in uninfected cells (P < 0.001) (Supplementary Fig. 1a). In addition, at 2 h after infection, IL-1β expression induced by P. gulae D049 (type C) infection was significantly higher than that induced by P. gulae ATCC 51700 (type A) or P. gulae D040 (type B) (P < 0.05); a similar trend was observed with respect to COX-2. IL-8 expression tended to be higher in the presence of strain D049 (type C) than in the presence of strains ATCC 51700 (type A) or D040 (type B); however, there were no significant differences among the strains. There was no change in the expression level of TGF-β1 at any time after P. gulae infection. Subsequently, we measured the protein levels of IL-1β, COX-2, IL-8, and TGF-β1 in P. gulae-infected Ca9-22 cells (Fig. 2a). In Ca9-22 cells infected with each strain of bacteria, increased protein levels of IL-1β, COX-2, and IL-8 were observed, whereas protein expression of TGF-β1 was nearly absent, regardless of the presence of the bacteria. Similar to the mRNA analysis, expression levels of IL-1β, COX-2, and IL-8 induced by strain D049 (type C) infection were significantly higher than expression levels induced by strains ATCC 51700 (type A) or D040 (type B).
Figure 2.
Production of cytokines or COX-2 enzyme from Ca9-22 cells infected with P. gulae strains. (a) Amounts of cytokine or enzyme production at multiple time points. Data are shown as the mean ± SD of three independent experiments. There were significant differences in cytokine and enzyme production, as determined by using analysis of variance with Bonferroni correction (*P < 0.05 and **P < 0.01). (b) Amounts of cytokine or enzyme production in the presence of IFN-α formulation. Data are shown as the mean ± SD of three independent experiments. There were significant differences in cytokine and enzyme production, relative to cells that did not receive the IFN-α formulation, upon infection with each P. gulae strain, as determined by using analysis of variance with Bonferroni correction (*P < 0.05).
IFN-α has been reported to be effective in chronic and infectious diseases22, and oral administration of an IFN-α formulation improved gingival inflammation in dogs with periodontal disease16. Therefore, each P. gulae strain and an IFN-α formulation were simultaneously added to respective cultures of Ca9-22 cells, and the expression levels of IL-1β, COX-2, and IL-8 were analysed 2 h after incubation. Before the experiment, the IFN-α formulation, clindamycin, and combined treatment with the IFN-α formulation and clindamycin were confirmed to have no effect on the growth of uninfected Ca9-22 cells (Supplementary Fig. 2a,b). Treatment with the IFN-α formulation significantly reduced the mRNA expression levels and protein production levels of IL-1β and COX-2 in Ca9-22 cells that were infected with P. gulae ATCC 51700 (type A) and P. gulae D040 (type B), respectively (P < 0.05); no changes in IL-8 expression were observed (Supplementary Fig. 1b and Fig. 2b). In addition, such changes in expression levels were not observed in Ca9-22 cells that were infected with P. gulae D049 (type C).
Changes in periodontal conditions before and after clinical treatment
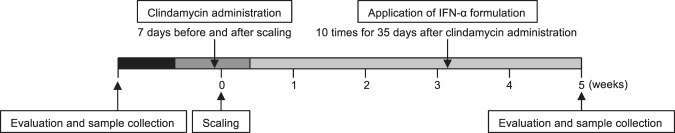
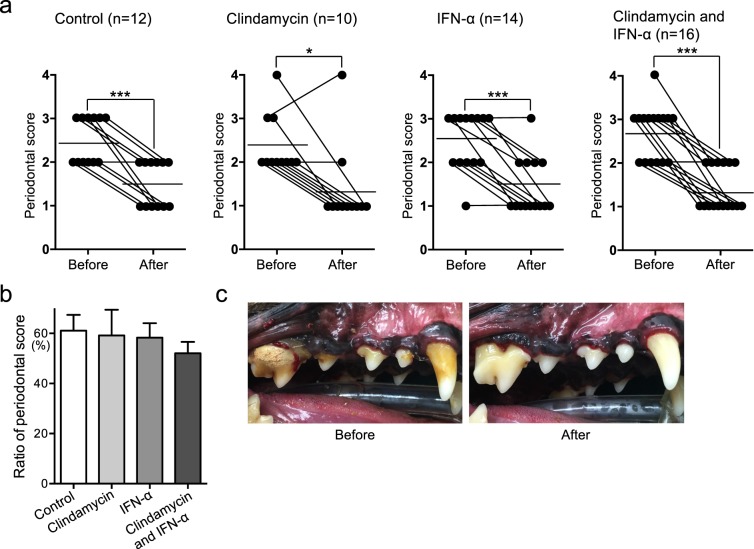
The schedule of treatment is summarized in Fig. 3. Fifty-two dogs were divided into four groups with or without pharmaceutical treatments, as follows: no pharmaceutical treatment (control group; n = 12), clindamycin treatment (clindamycin group; n = 10), IFN-α formulation treatment (IFN-α group; n = 14), and clindamycin and IFN-α formulation combined treatment (combination group; n = 16) (Table 1). In the clindamycin group, clindamycin was ingested for a total of 7 days (4 days before and 3 days after scaling). In the IFN-α group, an IFN-α formulation (InterBerryα®; Hokusan Co. Ltd.) was applied to the gingival margin of all teeth, 10 times total over 35 days. Gingival and periodontal scores of dogs were assessed at the beginning and end of clinical treatment, in accordance with previously described methods23. Periodontal scores in all groups after the treatment were significantly lower than those before treatment (P < 0.05) (Fig. 4a). In addition, the relative ratios were calculated for periodontal scores after periodontal treatment compared with those before periodontal treatment; the combination group had the lowest relative ratio, although there were no significant differences among groups (Fig. 4b). As shown in Fig. 4c, accumulations of dental plaque and dental calculus around teeth were reduced and gingival conditions were improved after periodontal treatment in the combined group.
Figure 3.
Schedule of periodontal treatment of dogs in the present study, using clindamycin and IFN-α formulation.
Table 1.
Dog breeds included in the present study.
| Prescription | Breeds | Age | Sex | Weight | fimA genotype | |
|---|---|---|---|---|---|---|
| pre | post | |||||
| Control (n = 12) | Cavalier King Charles Spaniel | 10Y6M | Female | 10.1 | B | A |
| Miniature Dachshund | 9Y11M | Male | 6 | (—) | A | |
| Miniature Dachshund | 9Y4M | Female | 5.3 | A | A | |
| Miniature Dachshund | 12Y6M | Female | 5.5 | A | A | |
| Yorkshire Terrier | 13Y8M | Female (spay) | 2.7 | A | A | |
| Corgi | 8Y0M | Male | 12.3 | B, C | B, C | |
| Chihuahua | 10Y3M | Male (castration) | 2.8 | (—) | A | |
| Toy Poodle | 8Y0M | Male (castration) | 3.4 | A | A | |
| Mix | 1Y5M | Male | 3 | A | A | |
| Miniature Dachshund | 14Y0M | Female (spay) | 4.6 | A | B, C | |
| Toy Poodle | 3Y7M | Male | 3.2 | A, B, C | C | |
| Miniature Dachshund | 13Y6M | Male (castration) | 6.9 | B | B | |
| Clindamycin (n = 10) | Toy Poodle | 13Y5M | Male | 5.4 | A, C | A |
| English Cocker Spaniel | 11Y7M | Female (spay) | 10.8 | A, C | C | |
| Toy Manchester | 9Y0M | Male (castration) | 4.3 | C | (—) | |
| Mix | 13Y0M | Female | 5.5 | C | B, C | |
| Italian Greyhound | 10Y2M | Female (spay) | 5.1 | A, B, C | A | |
| Yorkshire Terrier | 9Y8M | Male (castration) | 1.2 | A, C | (—) | |
| Shiba | 7Y10M | Female (spay) | 7.5 | A, C | A | |
| Pomeranian | 14Y3M | Male (castration) | 4.1 | B | B | |
| Papillon | 13Y0M | Female (spay) | 3.3 | Untypeable | B | |
| Toy Poodle | 12Y1M | Male | 8.3 | A, C | A, C | |
| IFN-α (n = 14) | Miniature Dachshund | 10Y6M | Female (spay) | 5.9 | A | (—) |
| Shetland Sheepdog | 10Y7M | Male | 11.3 | C | (—) | |
| Miniature Dachshund | 11Y11M | Male | 5.1 | A | A | |
| Miniature Dachshund | 9Y10M | Male | 4.3 | A | A | |
| Mix | 12Y0M | Female | 4.1 | A | (—) | |
| Miniature Dachshund | 7Y1M | Female (spay) | 5.7 | A | (—) | |
| Cavalier King Charles Spaniel | 5Y2M | Male (castration) | 9.8 | A | A | |
| Toy Poodle | 9Y7M | Female | 5.5 | A, B, C | C | |
| American Cocker Spaniel | 12Y1M | Female | 7.5 | A, C | A | |
| Miniature Dachshund | 15Y10M | Female (spay) | 2.9 | A, B, C | B, C | |
| Miniature Dachshund | 15Y4M | Female (spay) | 4.1 | A, B, C | A, B, C | |
| Miniature Schnauzer | 4Y8M | Male (castration) | 5.8 | C | C | |
| Toy Poodle | 9Y10M | Female (spay) | 3.8 | A | A | |
| Chihuahua | 9Y10M | Male | 2.7 | A | A | |
| Clindamycin and IFN-α (n = 16) | Japanese Terrier | 11Y8M | Female | 13.4 | B | (—) |
| Yorkshire Terrier | 11Y9M | Male (castration) | 2.8 | A, C | A | |
| Miniature Dachshund | 13Y9M | Male (castration) | 4.3 | A, B, C | B | |
| Toy Poodle | 8Y8M | Male (castration) | 2.8 | B | (—) | |
| Petit Basset Griffon Vendeen | 7Y11M | Male | 12.6 | C | B, C | |
| Pomeranian | 11Y5M | Male | 4.4 | B, C | (—) | |
| Bearded Collie | 11Y11M | Female | 16.7 | A, C | A | |
| Toy Poodle | 6Y9M | Male | 3 | B, C | (—) | |
| Toy Poodle | 7Y5M | Male (castration) | 6.7 | A, B, C | A | |
| Toy Poodle | 7Y11M | Male (castration) | 5.4 | C | (—) | |
| Toy Poodle | 13Y0M | Male (castration) | 4.6 | B | B | |
| Toy Poodle | 15Y2M | Female (spay) | 4.9 | C | Untypeable | |
| Miniature Poodle | 10Y0M | Female | 6.4 | B | (—) | |
| Pug | 7Y0M | Female | 8.7 | A, B, C | A, C | |
| Toy Poodle | 10Y10M | Male (castration) | 3.4 | Untypeable | Untypeable | |
| Miniature Dachshund | 13Y11M | Female (spay) | 6.6 | A | A | |
Figure 4.
Periodontal conditions before and after clinical treatment. (a) Periodontal scores before and after treatment. There were significant differences as determined by using analysis of variance with Bonferroni correction (*P < 0.05, ***P < 0.001). (b) Ratio of periodontal scores after treatment relative to those before treatment. (c) Representative images of periodontal condition before and after treatment with clindamycin and IFN-α formulation.
P. gulae detection before and after clinical treatment
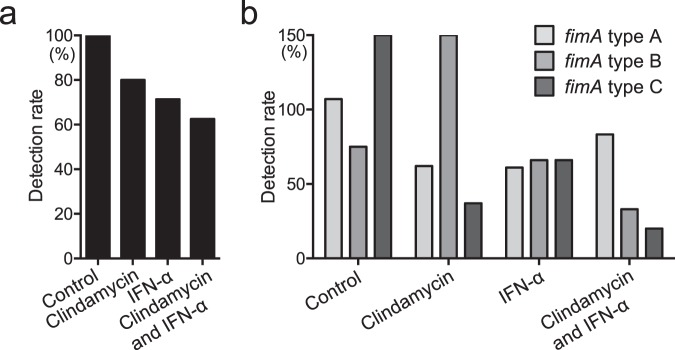
Oral swab specimens were collected from the gingival margin of the maxillary right or left canine and fourth maxillary premolar at the beginning and end of clinical treatment, as described previously24. Distributions of P. gulae and fimA genotype classification in the oral specimens were determined using polymerase chain reaction (PCR)-based methods, as previously described8,25. With the exception of two dogs in the control group, all dogs were positive for P. gulae before periodontal treatment; all P. gulae-positive dogs in the control group were positive for P. gulae after treatment (Table 1, Fig. 5a). Two of 10 dogs (20%) in the clindamycin group and four of 14 dogs (28.6%) in the IFN-α group were negative for P. gulae after treatment. The combination group had the lowest rate of P. gulae detection after treatment: P. gulae was absent from the oral cavity in six of 16 dogs (37.5%). Next, we determined the relative ratio of the detection of each fimA type after treatment, compared with that before treatment. In the control group, only the relative ratio of fimA type B was reduced after treatment (Fig. 5b). In contrast, the relative ratios of fimA types A and C were reduced after treatment in the clindamycin group; the relative ratios of all fimA genotypes were reduced after treatment in the IFN-α and combination groups. Notably, the relative ratios of fimA type C were dramatically reduced after treatment, which were present in 37.5% and 20% of dogs in the clindamycin and combination groups, respectively. When the detection rates of fimA genotypes in each group were compared with those in the control group, detection rates of all fimA genotypes in each group (with the exception of fimA type B in the clindamycin group) were lower than those in the control group.
Figure 5.
Rates of P. gulae detection before and after clinical treatment. (a) Rates of P. gulae detection after treatment, relative to those before treatment. (b) Rates of detection of fimA genotypes after treatment, relative to those before treatment.
Discussion
Periodontal disease is an inflammatory disease caused by bacterial infection5. Bacterial plaque remains at the gingival sulcus between the teeth and gingiva, resulting in gingivitis where the margin of the gingiva becomes red and swollen5. The presence of chronic gingivitis results in the formation of irreversible deep periodontal pockets at the gingival sulcus5. Consequently, alveolar bone resorption and tooth mobility occur, followed by loss of teeth in some instances26. Periodontal disease is present in more than 80% of mature dogs and is one of the most prevalent diseases in dogs26. P. gulae is a major periodontopathic bacteria that has been strongly associated with the deterioration of periodontal disease in dogs7. However, there have been no studies of the potential for preventing periodontal disease in dogs by reducing the pathogenicity of P. gulae. In the present study, we examined the inhibition of P. gulae and periodontal disease by using treatment with clindamycin and IFN-α formulation.
Antibacterial agents are often prescribed for a period of 1 week in combination with periodontal treatment; this approach is useful for reduction of periodontal bacteria17. Among antibiotics prescribed during scaling and root planing treatment of dogs, the lincomycin-derived antibacterial agent clindamycin is widely used for prevention of infection27. In the present study, 0.005–0.4 μg/ml clindamycin was used to assess the inhibitory effect of antibiotic treatment on growth of P. gulae. These low concentrations of clindamycin could inhibit the growth of all fimA types of P. gulae, among which the inhibitory effect most prominently affected the type C P. gulae strain. In the clinical portion of this study, type C P. gulae was also reduced in dogs that received clindamycin treatment. In our previous study, type C P. gulae was frequently detected in dogs with periodontitis, at rates significantly higher than those in healthy dogs8; therefore, clindamycin may be especially effective for dogs with severe periodontitis.
The Ca9-22 cell line was used as an in vitro counterpart of gingival epithelial cells28. Bacterial components or chemical regents have previously been reported to induce several inflammatory responses, such as IL-1β, COX-2, and IL-8, in Ca9-22 cells29,30. In addition, the mRNA expression levels of TGF-β1 in several cell lines were reportedly upregulated by exposure to virus protease, although the level of TGF-β1 mRNA in Ca9-22 was unchanged31. Thus, we examined the effects of P. gulae infection on IL-1β, IL-8, COX-2, and TGF-β1.
IL-1β has rapidly emerged as a key player in the regulation of inflammatory processes, which is capable of augmenting IL-8 production32,33. In addition, IL-1β exhibits an important role in the modulation of other inflammatory cytokines in human gingival epithelial cells infected with P. gingivalis34. COX-2 is the key enzyme involved in prostaglandin synthesis, and is expressed in inflammatory cells35; it is also known as a potent stimulator of bone resorption and is associated with periodontal attachment loss36. IL-8 is a key player in inflammatory conditions with a potent neutrophil recruiting and activating capacity32; it is induced in gingival epithelial cells upon exposure to several types of periodontopathic bacteria37. TGF-β1 has been shown to play a critical role in anti-inflammatory signaling38, which occurs in wound healing and periodontal regeneration11. Our results indicated that P. gulae infection induced overexpression of IL-1β, IL-8, and COX-2, potentially leading to inflammation involved in deterioration of periodontal disease.
Several oral pathogens, such as Fusobacterium nucleatum, Streptococcus sanguinis, and Aggregatibacter actinomycetemcomitans, have been reported to induce increased mRNA expression levels of IL-1β, IL-8, and tumour necrosis factor-α in Ca9-22 cells29,37. Stimulation of immortalised human gingival epithelial cells with vesicles from P. gingivalis bacteria led to upregulation of COX-2 and IL-8 mRNA36. Additionally, stimulation with P. gulae has been shown to induce increased secretion of IL-1β39. In the present study, we found that secretion and expression levels of IL-1β, IL-8, and tumour necrosis factor-α were increased upon infection with P. gulae. These findings suggest that P. gulae infection induces mRNA expression of a wide variety of inflammatory-related proteins in Ca9-22 cells. TGF-β1 mRNA and protein expression were reportedly upregulated by infection with bacteria such as Staphylococcus aureus, Helicobacter pylori, and group A Streptococcus38,40,41. In contrast, infection of human gingival fibroblast cells with P. gingivalis resulted in an increase in mRNA expression levels of TGF-β1, but did not influence the corresponding protein expression levels42. Following P. gulae infection in the present study, TGF-β1 mRNA expression levels were increased, while corresponding protein expression levels were not affected; this suggested that the effects of P. gulae on TGF-β1 expression were similar to those of P. gingivalis. Moreover, these findings indicate that the protein expression levels of TGF-β1 may not always reflect its mRNA expression levels.
Pharmaceutical agents containing IFN-α are widely used as antiviral and anticancer agents, because IFN-α can inhibit the growth of viruses and cancer cells43. In addition to these effects, IFN-α can regulate immune and inflammatory processes44. Therefore, we examined whether a canine IFN-α formulation could suppress inflammatory responses in gingival epithelial cells that had been infected with P. gulae. We found that the IFN-α formulation could reduce the expression of IL-1β and COX-2, which were upregulated by infection with fimA types A and B P. gulae strains. Furthermore, we found that clindamycin was more effective than the IFN-α formulation against the fimA type C P. gulae strain; thus, combined use of an IFN-α formulation and clindamycin may be effective for P. gulae strains, regardless of fimA genotype. In future studies, we plan to analyse effects of the combined use of an IFN-α formulation and clindamycin on the expression of other cytokines and enzymes, as well as on the survival of various periodontopathogenic bacteria.
In the present study, we examined whether an IFN-α formulation is useful for the suppression of inflammatory responses induced by P. gulae infection and whether it may be effective for treatment of periodontal disease in dogs. We applied the IFN-α formulation after removal of dental plaque and dental calculus to allow the IFN-α formulation to penetrate into the periodontal tissues. In addition, we continued the application of the IFN-α formulation for 35 days to suppress chronic inflammation, because most periodontal diseases are chronic diseases45. Although the IFN-α formulation was applied to all dogs for the same duration in the present study, the duration and frequency at which the IFN-α formulation is applied should be determined on the basis of periodontal disease severity in future studies.
The IFN-α formulation can be used in all dogs regardless of age, based on its approval by the Ministry of Agriculture, Forestry and Fisheries. The manufacturer currently recommends that the IFN-α formulation is administered to dogs with a periodontal score of ≤1. However, we have successfully shown that the IFN-α formulation is also effective in dogs with more severe periodontal conditions, which indicates that the IFN-α formulation can be used for treatment of dogs with more severe periodontitis. During the experimental period, no dogs had any problems with gingival or physical conditions due to the administration of the IFN-α formulation. Based on our results, the IFN-α formulation could be used in the treatment of dogs at various ages and with various degrees of periodontal disease.
In the present study, the periodontal score was reduced in all dogs after clinical periodontal treatment. These reductions in periodontal scores were also observed in the control group, which serves as evidence for the importance of mechanical removal of dental plaque and dental calculus by scaling or root planing, consistent with the findings of a previous report46. However, the group that received combined treatment showed the lowest periodontal score among all groups; therefore, combined use of clindamycin and IFN-α formulation may be effective as a supporting method for the improvement of periodontal condition, following mechanical cleaning.
In humans, no IFN-α formulation has been used in periodontal treatment. However, it may be useful to investigate whether combined treatment with antibiotics and an IFN-α formulation can be effective in human periodontal disease, because dog owners sometimes are colonized by P. gulae from their dogs47. In addition, it may be also informative to analyse whether combined treatment with antibiotics and an IFN-α formulation is effective to inhibit P. gingivalis growth, because this bacterial species is detected in both humans and dogs with periodontal disease.
To the best of our knowledge, this is the first study to show that the combination of clindamycin and IFN-α formulation is effective in improving the periodontal condition in dogs. In this study, we showed that combination therapy with clindamycin and IFN-α could improve periodontal conditions and reduce P. gulae in randomly selected dogs. However, in addition to the effects of clindamycin and IFN-α, genetic and environmental factors may have influenced our findings, because only a small number of dogs participated in this study. Therefore, larger clinical studies are needed using more dogs with different backgrounds.
In summary, clindamycin is an effective antibiotic for inhibition of the growth of P. gulae, and the application of an IFN-α formulation can suppress inflammatory responses produced from P. gulae-infected gingival epithelial cells. The combination of clindamycin and IFN-α formulation in the treatment of canine periodontal disease may contribute to improved periodontal conditions in dogs.
Methods
Bacterial strains and cell cultures
P. gulae strains ATCC 51700 (fimA type A), D040 (fimA type B), and D049 (fimA type C) were selected from the stock culture collection in our laboratory7,8,24. Bacterial cells were grown anaerobically at 37 °C for 24 h in trypticase soy broth supplemented with yeast extract (1 mg/ml), haemin (5 μg/ml), and menadione (1 μg/ml), as previously described48; they were then used in the following experiments. Ca9-22 cells (originally isolated from human gingival epithelia) were obtained from the Japanese Collection of Research Bioresources (Tokyo, Japan); these cells were used as an in vitro counterpart of gingival epithelial cells28 because they have been widely used as an in vitro culture model of gingival epithelial cells28,49. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Wako, Osaka, Japan) supplemented with 10% fetal bovine serum at 37 °C in 5% CO2.
Bacterial growth
Bacterial growth was analysed in accordance with previously described methods, with some modifications50–52. Various concentrations of clindamycin were tested for their effects on P. gulae growth: 0.005, 0.01, 0.015, 0.025, 0.05, 0.1, 0.2, and 0.4 μg/ml. In addition, various concentrations of IFN-α formulation were tested: 0, 1, 10, 50, and 100 μg/ml. Clindamycin and IFN-α formulation, separately or in combination, were added to the trypticase soy broth supplemented with yeast extract, haemin, and menadione used for bacterial suspension growth. In addition, overnight cultured P. gulae bacteria were added to the media at a density of 4 × 108 CFU/ml and then cultured at 37 °C for 24 h. Bacterial growth after incubation was measured by determining the optical density at 600 nm using a microplate reader (SH-1000 Lab, Corona Electric, Katsuta, Japan), because the number of P. gulae bacteria in a given suspension has been previously estimated by measurement of the optical density at 600 nm and subsequent extrapolation from a standard curve53. The relative ratio of growth of each P. gulae strain was calculated by comparison with the optical density at 600 nm value of each bacterial broth without clindamycin. All assays were performed in triplicate on three separate occasions (n = 9).
Real-time reverse transcription-polymerase chain reaction (RT-PCR) for quantitative detection of mRNA expression
Ca9-22 cells were incubated in DMEM with 10% fetal bovine serum until confluent. After incubation, the monolayers of Ca9-22 cells were washed three times with serum-free DMEM to remove unattached cells. Additionally, the number of viable cells in each monolayer was determined using trypan blue dye exclusion and cell counting method. IFN-α (100 μg/ml) was preincubated with Ca9-22 cells prior to addition of bacteria. Overnight cultured P. gulae were harvested and washed with sterile phosphate-buffered saline. The bacteria were then diluted to 1 × 108 CFU/ml in DMEM and used to infect Ca9-22 cells. For experiments involving bacterial infection of Ca-22 cells, we used the concept of multiplicity of infection (MOI), which is commonly defined as the ratio of infectious microorganisms to cells in a culture54. Ca9-22 cells were diluted to a density of 1 × 106 and infected with 1 × 108 CFU of respective P. gulae strains at MOI of 100 for 0–12 h. Then, total RNA was extracted from these P. gulae-infected Ca9-22 cells and cDNA was synthesized as described previously55. Briefly, total RNA from Ca9-22 cells was isolated using TRIsure (BIOLINE, Luckenwalde, Germany) and converted into cDNA using an iScript™ cDNA Synthesis kit (Bio-Rad, Hercules, CA, USA) in accordance with the manufacturer’s instructions. cDNAs were amplified using a QuantiFast SYBR Green PCR master mix (Qiagen, Valencia, CA, USA), in accordance with the manufacturer’s instructions. The primers specific for genes encoding IL-1β, COX-2, IL-8, and TGF-β1, which were used in this study, are listed in Table 2. GAPDH was used as a housekeeping control and negative reverse transcription reactions were included in each assay. Expression values for mRNA were quantified by the ΔΔCt method, using GAPDH as the control. All assays were performed in triplicate on three separate occasions (n = 9).
Table 2.
Reverse transcription polymerase chain reaction primers used in the present study.
| Specific primer set | Sequence (5′-3′) | Size (bp) | References |
|---|---|---|---|
| GAPDH | GTCTTCACCACCATGGAGAAG | 210 | 58 |
| GTTGTCATGGATGACCTTGGC | |||
| IL-1β | CTTTGAAGCTGATGGCCCTAAA | 101 | 59 |
| AGTGGTGGTCGGAGATTCGT | |||
| COX-2 | CACAGGCTTCCATTGACCAGA | 240 | 36 |
| GTGCTCCAACTTCTACCATGG | |||
| IL-8 | CTTGGCAGCCTTCCTGATTTC | 209 | 36 |
| CCAGACAGAGCTCTCTTCCAT | |||
| TGF-β1 | CACCCGCGTGCTAATGGT | 100 | 60 |
| CTCGGAGCTCTGATGTGTTGAA |
Enzyme-linked immunosorbent assays (ELISAs)
P. gulae strains were used to infect Ca9-22 cells with or without administration of the IFN-α formulation, as described in the above section regarding RT-PCR. ELISAs were performed in accordance with previously described methods, with some modifications56. Ca9-22 cells were stimulated with P. gulae strains in the presence or absence of IFN-α for 24 h. For the quantification of IL-1β, COX-2, IL-8, and TGF-β1 in cell lysate at each time point, sandwich ELISAs were performed using the Human IL-1β ELISA kit (Proteintech Group Inc., Rosemont, IL, USA), Human/mouse total COX-2 DuoSet IC ELISA (R&D Systems Inc., Minneapolis, MN, USA), IL-8 ELISA kit (Proteintech Group Inc.) and TGF-β1 ELISA kit (Proteintech Group Inc.), respectively, in accordance with the manufacturers’ instructions. Absorbance was measured at 450 nm, with correction to 550 nm, using a SH-1000 Lab microplate reader (Corona Electric, Ibaraki, Japan). All assays were performed in triplicate on three separate occasions (n = 9).
Schedule in clinical experiment
The clinical experiment was conducted in full adherence to the Declaration of Helsinki. All study protocols were approved by the Animal Research Committee of Azabu University. Prior to the clinical experiment, all owners were informed of the content of the study and gave written informed consent for approval of their pets’ participation. The schedule for clinical treatment is summarized in Fig. 3. In total, 52 dogs (28 males, 24 females; median age: 10 years [range: 1–15 years]) were enrolled (Table 1); all dogs received periodontal treatment under general anaesthesia. The subjects were divided into four groups with or without pharmaceutical treatments, as follows: no pharmaceutical treatment (control group; n = 12), clindamycin treatment (clindamycin group; n = 10), IFN-α formulation treatment (IFN-α group; n = 14), and combined clindamycin and IFN-α formulation treatment (combination group; n = 16). In the clindamycin group, clindamycin 5 mg/kg was administered via the oral cavity twice per day, beginning 4 days before treatment and ending 3 days after periodontal treatment (a total of 7 days), in accordance with the guidelines of the American Animal Hospital Association57. In the IFN-α group, 2.75 g of IFN-α formulation (InterBerryα®) was applied to the gingival margin of all teeth, 10 times total over 35 days after periodontal treatment. The combination group received both clindamycin and IFN-α formulation treatments, in accordance with the methods described above. Oral swab specimens were collected from the gingival margin of the maxillary right or left canine and fourth premolar using a micro brush (Microapplicator fine, FEED Corporation, Yokohama, Japan), as described previously24, several days before and 5 weeks after periodontal treatment.
Evaluation of periodontal conditions before and after clinical treatment
Periodontal scores were determined by assessment of the gingival margin of the maxillary right or left canine and fourth maxillary premolar, several days before and 5 weeks after periodontal treatment, using a modified version of a previously described method13. For each dog, gingival scores of the maxillary right or left canine and fourth premolar were evaluated visually as follows: (1) no significant findings; (2) mild periodontal disease—gingival swelling, gingival regression, and halitosis; (3) moderate periodontal disease—exposure of root, spontaneous bleeding, and tooth loss; and (4) severe periodontal disease—furcation involvement and fistula formation.
Detection of P. gulae and fimA genotypes in clinical samples
Distributions of P. gulae and fimA genotypes were determined using previously developed PCR-based methods8,25; Table 3 lists the PCR primers used in this study. Bacterial DNA was extracted from each oral specimen using a Gentra Puregene Yeast/Bact. Kit B (Qiagen, Hilden, Germany). PCR analysis was then performed using universal primer sets targeting 16 S rRNA genes to confirm that bacterial DNA was successfully extracted; subsequently, P. gulae and fimA genotypes were determined using respective specific primer sets8,24,25. Amplification reactions were performed using a total volume of 20 µl with 1 µl of template solution and Ex Taq DNA Polymerase (Takara Bio. Inc., Otsu, Japan) with the following cycling parameters: initial denaturation at 95 °C for 4 min; 30 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s; and final extension at 72 °C for 7 min. Amplification reactions were performed in an iCycler thermal cycler (Bio-Rad). The resulting products were separated by electrophoresis on a 1.5% agarose gel-Tris-acetate-EDTA buffer. The gel was stained with 0.5 μg/ml ethidium bromide and photographed under ultraviolet illumination.
Table 3.
Polymerase chain reaction primers used in the present study.
| Specific primer set | Sequence (5′-3′) | Size (bp) | References |
|---|---|---|---|
| Universal primer | |||
| PA | AGA GTT TGA TCC TGG CTC AG | 315 | 61 |
| PD | GTA TTA CCG CGG CTG CTG | ||
| Detection of P. gulae | TTG CTT GGT TGC ATG ATC GG | 314 | 24 |
| GCT TAT TCT TAC GGT ACA TTC ACA | |||
| Specification of fimA type | |||
| Type A fimA | |||
| Pgfim-AF | TGA GAA TAT CAA ATG TGG TGC AGG CTC ACG | 190 | 25 |
| Pgfim-AR | CTT GCC TGC CTT CAA AAC GAT TGC TTT TGG | ||
| Type B fimA | |||
| Pgfim-BF | TAA GAT TGA AGT GAA GAT GAG GGA TTC TTA TGT | 357 | 25 |
| Pgfim-BR | ATT TCC TCA GAA CTC AAA GGA GTA CCA TCA | ||
| Type C fimA | |||
| Pgfim-CF | CGA TTA TGA CCT TGT CGG TAA GAG CTT GGA | 631 | 8 |
| Pgfim-CR | TGT GGC TTC GTT GTC GCA GAA TCC GGC ATG | ||
Statistical analysis
Statistical analyses were conducted using GraphPad Prism 6 (GraphPad Software Inc., La Jolla, CA, USA). Intergroup differences were estimated using analysis of variance with Bonferroni correction. Differences were considered statistically significant at P < 0.05.
Supplementary information
Acknowledgements
This study was supported by the Promotion and Mutual Aid Corporation of Private Schools of Japan, and by a Grant-in-Aid for Matching Fund Subsidy for Private Universities.
Author contributions
R.N., H.I. and H.Y. designed the entire study under the supervision of F.A., M.N. and K.N. H.Y., J.Y. and N.A. collected clinical samples. R.N., H.I., M.S., Y.K., M.M., N.I., S.S., S.Y., S.M. and Y.M. performed the experiments. Data interpretation was conducted by R.N., H.I., H.Y. and K.N. R.N., H.I., H.Y. and K.N. wrote the manuscript, which all authors read and approved.
Data availability
The data are not available for public access because of patient privacy concerns, but are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ryota Nomura, Hiroaki Inaba and Hidemi Yasuda.
Mitsuyuki Shirai and Yukio Kato are deceased.
Supplementary information
is available for this paper at 10.1038/s41598-020-59730-9.
References
- 1.Niemiec BA. Periodontal disease. Top. Companion Anim. Med. 2008;23:72–80. doi: 10.1053/j.tcam.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Loesche WJ. Chemotherapy of dental plaque infections. Oral. Sci. Rev. 1976;9:65–107. [PubMed] [Google Scholar]
- 3.Glickman LT, et al. Association between chronic azotemic kidney disease and the severity of periodontal disease in dogs. Prev. Vet. Med. 2011;99:193–200. doi: 10.1016/j.prevetmed.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 4.Wallis C, et al. A longitudinal assessment of changes in bacterial community composition associated with the development of periodontal disease in dogs. Vet. Microbiol. 2015;181:271–282. doi: 10.1016/j.vetmic.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Albuquerque C, et al. Canine periodontitis: the dog as an important model for periodontal studies. Vet. J. 2012;191:299–305. doi: 10.1016/j.tvjl.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 6.Fournier D, et al. Porphyromonas gulae sp. nov., an anaerobic, gram-negative coccobacillus from the gingival sulcus of various animal hosts. Int. J. Syst. Evol. Microbiol. 2001;51:1179–1189. doi: 10.1099/00207713-51-3-1179. [DOI] [PubMed] [Google Scholar]
- 7.Hamada N, et al. Molecular and antigenic similarities of the fimbrial major components between Porphyromonas gulae and P. gingivalis. Vet. Microbiol. 2008;128:108–117. doi: 10.1016/j.vetmic.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 8.Yamasaki Y, et al. Distribution and molecular characterization of Porphyromonas gulae carrying a new fimA genotype. Vet. Microbiol. 2012;161:196–205. doi: 10.1016/j.vetmic.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 9.Shirai M, et al. Distribution of Porphyromonas gulae fimA genotypes in oral specimens from dogs with mitral regurgitation. Res. Vet. Sci. 2015;102:49–52. doi: 10.1016/j.rvsc.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Hajishengallis G, Lambris JD. Complement and dysbiosis in periodontal disease. Immunobiology. 2012;217:1111–1116. doi: 10.1016/j.imbio.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inaba H, Kawai S, Kanayama K, Okahashi N, Amano A. Effect of Enamel matrix derivative on periodontal ligament cells in vitro is diminished by Porphyromonas gingivalis. J. Periodontol. 2004;75:858–865. doi: 10.1902/jop.2004.75.6.858. [DOI] [PubMed] [Google Scholar]
- 12.Phusuntornsakul P, Jitpukdeebodintra S, Pavasant P, Leethanakul C. Vibration enhances PGE2, IL-6, and IL-8 expression in compressed hPDL cells via cyclooxygenase pathway. J. Periodontol. 2018;89:1131–1141. doi: 10.1002/JPER.17-0653. [DOI] [PubMed] [Google Scholar]
- 13.Araújo AA, et al. Gliclazide reduced oxidative stress, inflammation, and bone loss in an experimental periodontal disease model. J. Appl. Oral Sci. 2019;27:e20180211. doi: 10.1590/1678-7757-2018-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boxx GM, Cheng G. The Roles of Type I Interferon in Bacterial Infection. Cell Host Microbe. 2016;19:760–769. doi: 10.1016/j.chom.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benveniste EN, Qin H. Type I interferons as anti-inflammatory mediators. Sci. STKE. 2007;416:pe70. doi: 10.1126/stke.4162007pe70. [DOI] [PubMed] [Google Scholar]
- 16.Ito A, et al. Ability of orally administered IFN-α4 to inhibit naturally occurring gingival inflammation in dogs. J. Vet. Med. Sci. 2010;72:1145–1151. doi: 10.1292/jvms.09-0201. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen D, et al. Effects of treatment with clindamycin hydrochloride on progression of canine periodontal disease after ultrasonic scaling. Vet. Ther. 2000;1:150–158. [PubMed] [Google Scholar]
- 18.Bickel M, Axtelius B, Solioz C, Attström R. Cytokine gene expression in chronic periodontitis. J. Clin. Periodontol. 2001;28:840–847. doi: 10.1034/j.1600-051x.2001.028009840.x. [DOI] [PubMed] [Google Scholar]
- 19.Zetner K, Thiemann G. The antimicrobial effectiveness of clindamycin in diseases of the oral cavity. J. Vet. Dent. 1993;10:6–9. doi: 10.1177/089875649301000204. [DOI] [PubMed] [Google Scholar]
- 20.Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 1998;62:1244–1263. doi: 10.1128/MMBR.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amano A. Disruption of epithelial barrier and impairment of cellular function by Porphyromonas gingivalis. Front. Biosci. 2007;12:3965–3974. doi: 10.2741/2363. [DOI] [PubMed] [Google Scholar]
- 22.Parkin J, Cohen B. An overview of the immune system. Lancet. 2001;357:1777–1789. doi: 10.1016/S0140-6736(00)04904-7. [DOI] [PubMed] [Google Scholar]
- 23.Harvey, C. E. & Emily, P. P. Small Animal Dentistry. 1st ed. Mosby. Year book, St. Loius 413 (1993).
- 24.Kato Y, et al. Molecular detection of human periodontal pathogens in oral swab specimens from dogs in Japan. J. Vet. Dent. 2011;28:84–89. doi: 10.1177/089875641102800204. [DOI] [PubMed] [Google Scholar]
- 25.Nomura R, et al. Diversity of fimbrillin among Porphyromonas gulae clinical isolates from Japanese dogs. J. Vet. Med. Sci. 2012;74:885–891. doi: 10.1292/jvms.11-0564. [DOI] [PubMed] [Google Scholar]
- 26.Fernandes NA, et al. Prevalence of periodontal disease in dogs and owners’ level of awareness - a prospective clinical trial. Rev. Ceres Viçosa. 2012;59:446–451. doi: 10.1590/S0034-737X2012000400003. [DOI] [Google Scholar]
- 27.Sarkiala E, Harvey C. Systemic antimicrobials in the treatment of periodontitis in dogs. Semin. Vet. Med. Surg. 1993;8:197–203. [PubMed] [Google Scholar]
- 28.Evans M, et al. Combined effects of starvation and butyrate on autophagy-dependent gingival epithelial cell death. J. Periodontal Res. 2017;52:522–531. doi: 10.1111/jre.12418. [DOI] [PubMed] [Google Scholar]
- 29.Peyret-Lacombe A, Brunel G, Watts M, Charveron M, Duplan H. TLR2 sensing of F. nucleatum and S. sanguinis distinctly triggered gingival innate response. Cytokine. 2009;46:201–210. doi: 10.1016/j.cyto.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Nagahama Y, et al. Oxidized low-density lipoprotein-induced periodontal inflammation is associated with the up-regulation of cyclooxygenase-2 and microsomal prostaglandin synthase 1 in human gingival epithelial cells. Biochem. Biophys. Res. Commun. 2011;413:566–571. doi: 10.1016/j.bbrc.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Li SW, et al. SARS coronavirus papain-like protease induces Egr-1-dependent up-regulation of TGF-β1 via ROS/p38 MAPK/STAT3 pathway. Sci. Rep. 2016;6:25754. doi: 10.1038/srep25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pease JE, Sabroe I. The role of interleukin-8 and its receptors in inflammatory lung disease: implications for therapy. Am. J. Respir. Med. 2002;1:19–25. doi: 10.1007/BF03257159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabay C, Lamacchia C, Palmer G. IL-1 pathways in inflammation and human diseases. Nat. Rev. Rheumatol. 2010;6:232–241. doi: 10.1038/nrrheum.2010.4. [DOI] [PubMed] [Google Scholar]
- 34.Eskan MA, et al. Interleukin-1beta modulates proinflammatory cytokine production in human epithelial cells. Infect. Immun. 2008;46:2080–2089. doi: 10.1128/IAI.01428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Font-Nieves M, et al. Induction of COX-2 enzyme and down-regulation of COX-1 expression by lipopolysaccharide (LPS) control prostaglandin E2 production in astrocytes. J. Biol. Chem. 2012;287:6454–6468. doi: 10.1074/jbc.M111.327874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kou Y, et al. Inflammatory responses of gingival epithelial cells stimulated with Porphyromonas gingivalis vesicles are inhibited by hop-associated polyphenols. J. Periodontol. 2008;79:174–180. doi: 10.1902/jop.2008.070364. [DOI] [PubMed] [Google Scholar]
- 37.Shimada T, et al. Differential effects of five Aggregatibacter actinomycetemcomitans strains on gingival epithelial cells. Oral. Microbiol. Immunol. 2008;23:455–458. doi: 10.1111/j.1399-302X.2008.00449.x. [DOI] [PubMed] [Google Scholar]
- 38.Wang B, et al. Induction of TGF-beta1 and TGF-beta1-dependent predominant Th17 differentiation by group A streptococcal infection. Proc. Natl Acad. Sci. USA. 2010;107:5937–5942. doi: 10.1073/pnas.0904831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holden JA, et al. Porphyromonas gulae activates unprimed and gamma interferon-primed macrophages via the pattern recognition receptors Toll-Like Receptor 2 (TLR2), TLR4, and NOD2. Infect. Immun. 2017;85:e00282–17. doi: 10.1128/IAI.00282-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kehrl JH, et al. Transforming growth factor beta is an important immunomodulatory protein for human B lymphocytes. J. Immunol. 1986;137:3855–3860. [PubMed] [Google Scholar]
- 41.Kandulski A, et al. Naturally occurring regulatory T cells (CD4+, CD25high, FOXP3+) in the antrum and cardia are associated with higher H. pylori colonization and increased gene expression of TGF-beta1. Helicobacter. 2008;13:295–303. doi: 10.1111/j.1523-5378.2008.00612.x. [DOI] [PubMed] [Google Scholar]
- 42.Palm E, Demirel I, Bengtsson T, Khalaf H. The role of toll-like and protease-activated receptors in the expression of cytokines by gingival fibroblasts stimulated with the periodontal pathogen Porphyromonas gingivalis. Cytokine. 2015;76:424–432. doi: 10.1016/j.cyto.2015.08.263. [DOI] [PubMed] [Google Scholar]
- 43.Goldstein D, Laszlo J. The role of interferon in cancer therapy: a current perspective. CA Cancer J. Clin. 1988;38:258–277. doi: 10.3322/canjclin.38.5.258. [DOI] [PubMed] [Google Scholar]
- 44.Teijaro JR. Type I interferons in viral control and immune regulation. Curr. Opin. Virol. 2016;16:31–40. doi: 10.1016/j.coviro.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dumitrescu AL. Editorial: Periodontal Disease - A Public Health Problem. Front. Public. Health. 2016;3:278. doi: 10.3389/fpubh.2015.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamath DG, Umesh Nayak S. Detection, removal and prevention of calculus: Literature Review. Saudi Dent. J. 2014;26:7–13. doi: 10.1016/j.sdentj.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamasaki Y, et al. Distribution of periodontopathic bacterial species in dogs and their owners. Arch. Oral Biol. 2012;57:1183–1188. doi: 10.1016/j.archoralbio.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 48.Inaba H, et al. Adhesion and invasion of gingival epithelial cells by Porphyromonas gulae. PLoS One. 2019;14:e0213309. doi: 10.1371/journal.pone.0213309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tada H, et al. Porphyromonas gingivalis induces the production of interleukin-31 by human mast cells, resulting in dysfunction of the gingival epithelial barrier. Cell Microbiol. 2019;21:e12972. doi: 10.1111/cmi.12972. [DOI] [PubMed] [Google Scholar]
- 50.Leke N, Grenier D, Goldner M, Mayrand D. Effects of hydrogen peroxide on growth and selected properties of Porphyromonas gingivalis. FEMS Microbiol. Lett. 1999;174:347–353. doi: 10.1111/j.1574-6968.1999.tb13589.x. [DOI] [PubMed] [Google Scholar]
- 51.Masuda T, Murakami Y, Noguchi T, Yoshimura F. Effects of various growth conditions in a chemostat on expression of virulence factors in Porphyromonas gingivalis. Appl. Env. Microbiol. 2006;72:3458–3467. doi: 10.1128/AEM.72.5.3458-3467.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akcalı A, et al. Exposure of Porphyromonas gingivalis to cortisol increases bacterial growth. Arch. Oral Biol. 2014;59:30–34. doi: 10.1016/j.archoralbio.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 53.Kuboniwa M, et al. Specific antibodies to Porphyromonas gingivalis Lys-gingipain by DNA vaccination inhibit bacterial binding to hemoglobin and protect mice from infection. Infect. Immun. 2001;69:2972–2979. doi: 10.1128/IAI.69.5.2972-2979.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shabram P, Aguilar-Cordova E. Multiplicity of infection/multiplicity of confusion. Mol. Ther. 2000;2:420–421. doi: 10.1006/mthe.2000.0212. [DOI] [PubMed] [Google Scholar]
- 55.Inaba H, et al. Porphyromonas gingivalis promotes invasion of oral squamous cell carcinoma through induction of proMMP9 and its activation. Cell Microbiol. 2014;16:131–145. doi: 10.1111/cmi.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Vries TJ, et al. Periodontal ligament fibroblasts as a cell model to study osteogenesis and osteoclastogenesis in fibrodysplasia ossificans progressiva. Bone. 2018;109:168–177. doi: 10.1016/j.bone.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 57.Inaba H, et al. Identification of hop polyphenolic components which inhibit prostaglandin E2 production by gingival epithelial cells stimulated with periodontal pathogen. Biol. Pharm. Bull. 2008;31:527–530. doi: 10.1248/bpb.31.527. [DOI] [PubMed] [Google Scholar]
- 58.Bellows J, et al. 2019 AAHA dental care guidelines for dogs and cats. J. Am. Anim. Hosp. Assoc. 2019;55:49–69. doi: 10.5326/JAAHA-MS-6933. [DOI] [PubMed] [Google Scholar]
- 59.Kato T, et al. Impaired degradation of matrix collagen in human gingival fibroblasts by the antiepileptic drug phenytoin. J. Periodontol. 2005;76:941–950. doi: 10.1902/jop.2005.76.6.941. [DOI] [PubMed] [Google Scholar]
- 60.Scheres N, Laine ML, de Vries TJ, Everts V, van Winkelhoff AJ. Gingival and periodontal ligament fibroblasts differ in their inflammatory response to viable Porphyromonas gingivalis. J. Periodontal Res. 2010;45:262–270. doi: 10.1111/j.1600-0765.2009.01229.x. [DOI] [PubMed] [Google Scholar]
- 61.Marques da Silva R, et al. Bacterial diversity in aortic aneurysms determined by 16S ribosomal RNA gene analysis. J. Vasc. Surg. 2006;44:1055–1060. doi: 10.1016/j.jvs.2006.07.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are not available for public access because of patient privacy concerns, but are available from the corresponding author on reasonable request.