Abstract
Alternative splicing of RNAs generates isoform diversity, resulting in different proteins that are necessary for maintaining cellular function and identity. The discovery of alternative splicing has been revolutionized by next-generation transcriptomic sequencing mainly using bulk RNA-sequencing, which has unravelled RNA splicing and mis-splicing of normal cells under steady-state and stress conditions. Single-cell RNA-sequencing studies have focused on gene-level expression analysis and revealed gene expression signatures distinguishable between different cellular types. Single-cell alternative splicing is an emerging area of research with the promise to reveal transcriptomic dynamics invisible to bulk- and gene-level analysis. In this review, we will discuss the technological advances for single-cell alternative splicing analysis, computational strategies for isoform detection and quantitation in single cells, and current applications of single-cell alternative splicing analysis and its potential future contributions to personalized medicine.
Keywords: Single-cell transcriptome analysis, Alternative splicing, Isoform, Percent spliced-in, Next-generation sequencing
1. Introduction
Next-generation sequencing technologies enable high-throughput and genome-wide profiling of the genomic and transcriptomic landscape in various human cell types, during development and differentiation processes, and under physiological and pathological states [1], [2], [3], [4], [5]. Advances in methodologies for DNA and RNA isolation and amplification techniques including the application of microfluidic technologies can now accommodate a small amount of starting DNA and RNA material, enabling the analysis of formalin-fixed paraffin-embedded or archival specimens, rare cell types such as oocytes and cells from early embryos, and more recently from single cells [6], [7], [8]. Up to now, single-cell transcriptomic studies have primarily focused on gene-level expression, where individual gene expression represents the aggregation of isoforms originating from the same gene, to uncover heterogeneity of cells with distinct gene expression signatures and functional states [9], [10], [11], [12], [13], and in response to intrinsic or external signals [14], [15], [16].
The alternative splicing process generates mRNA molecules of different exon composition, and of different length, from the same genetic locus. Alternative splicing is therefore a major driver of protein diversity and represents an additional layer of complexity underlying gene expression profiles [17]. Alternative splicing events include exon-skipping, mutually exclusive exons, intron retention, alternative 5′ and 3′ splice sites, alternative transcription start and end sites, and differential 3′ untranslated region (UTR) usage [18], [19], [20], [21]. A notable example is Bcl-x where an alternative 3′ splice site yields two different isoforms with opposing function; the long isoform Bcl-xL has anti-apoptotic activity, whereas the short isoform Bcl-xs mediates programmed cell death [22]. Bulk RNA-sequencing provides insight into the role of RNA splicing and mis-splicing in tissue and organ development [23], [24] including inherited diseases [25], and in cancer [26], [27]. Nevertheless, bulk RNA-sequencing may not delineate the heterogeneity that exist within a population of cells with similar phenotype, such as rare subpopulations of cells with distinct biological niche and alternative splicing profile [28], [29], [30]. However, the methodology used for bulk RNA-sequencing cannot be immediately applied to single-cell RNA-sequencing due to challenges inherent to RNA-sequencing at the single-cell resolution. These challenges include uneven capturing of the transcript coverage, low molecular capture rate, low cDNA conversion efficiency, limitation in starting materials, and variability of the cell size (amount of RNA molecules inside a cell) that inevitably result in low coverage and high technical noise [31], [32], [33].
In this review, we will discuss technological advances in methodologies for single-cell alternative splicing analysis, with a particular focus on the current computational and statistical approaches used for detection and quantification of alternative splicing (Table 1). We highlight the ways these different approaches complement each other and summarize the current and potential future applications of alternative splicing analysis in single cells.
Table 1.
Summary of computational approaches for detection and quantification of alternative splicing events in single cells.
| Computational method | Software/Statistical method | Read aligner | No. of cells | Cell type(s) | Sample origin | Library preparation | Sequencing platform | Isoform variant analysed | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Developed for bulk short-read RNA-sequencing | MISO | Bowtie | 12 | LNCaP, PC3, T24 | Cell line | Smart-Seq | Genome Analyzer IIx, 150 bp PE | CE | [56] |
| MISO | Bowtie | 18 | BMDCs | Mouse | Smart-Seq | HiSeq 2000, 100 bp PE | CE | [14] | |
| MISO | STAR | 34 | ESCs | Human | Smart-Seq | HiSeq 2000, 100 bp SE | CE | [73] (Data from [74]) | |
| MISO | Bowtie | 18 | BMDCs | Mouse | Smart-Seq | HiSeq 2000, 100 bp PE | CE | [78] (Data from [14]) | |
| VAST-TOOLS | TopHat | 66 | Spermatogenic cells | Mouse | Smart-Seq | HiSeq 4000, 150 bp PE | CE, RI, A5SS, A3SS | [74] | |
| bam2ssj | Bowtie | 10 | GM12878 | Cell line | Smart-Seq | HiSeq 2000, 100 bp SE | SJ | [72] | |
| IPSA | TopHat | 40 | HeLa S3 | Cell line | MIRALCS | HiSeq 2000, 150 bp PE, 50 bp SE | SJ | [61] | |
| Developed for single-cell short-read RNA-sequencing | Custom pipeline | ABI whole transcriptome software tool | 1 | Blastomere | Mouse | Modified single-cell cDNA amplification for microarray | SOLiD sequencer, 50 bp PE | SJ | [54] |
| Custom pipeline | ABI whole transcriptome software tool | 33 | ESCs, ICM, Epiblast, ICM outgrowth cells1 | Mouse | Modified single-cell cDNA amplification for microarray | SOLiD sequencer, 50 bp PE | SJ | [53] | |
| Custom pipeline | Bowtie | 85 | ESCs, embryonic fibroblast | Mouse | STRT | Genome Analyzer IIx, 150 bp PE | SJ | [55] | |
| SingleSplice | GSNAP/GMAP | 182 | ESCs | Mouse | Smart-Seq | HiSeq 2000, 100 bp, PE | Isoform switching | [32] (Data from [70]) | |
| ISOP | Bowtie | 384 96 305 96 |
MDA-MB-231 HTC116 Primary myoblasts Primary glioma |
Cell line Cell line Human Human |
Smart-Seq Smart-Seq Smart-Seq Smart-Seq |
HiSeq 2000, 100 bp PE HiSeq 2000, 150 bp PE HiSeq 2500, 100 bp PE HiSeq 2500, 100 bp PE |
Isoform switching | [69] (Additional data from [71], [110], [111] | |
| Logistic regression | Bowtie STAR STAR |
182 1529 31,831 |
Primary myoblasts ESCs T cells |
Human Human Human |
Smart-Seq Smart-Seq 10x Genomics |
HiSeq 2500, 100 bp PE HiSeq 2000, 100 bp SE NextSeq 500, 75 bp PE |
Isoform switching | [68] (Data from [71], [112], [113] | |
| BRIE | HISAT | 40 | ESCs | Mouse | Smart-Seq | HiSeq 2500, bp, SE | CE | [31] (Data from [114]) | |
| BRIE | STAR | 93 93 |
iPSCs Endoderm |
Human Human |
scM&T-seq | HiSeq 2500 | CE | [86] | |
| BRIE | STAR | 2208 | Oligodendrocytes | Mouse | Smart-Seq | HiSeq X Ten, 50 bp, SE | CE | [29] | |
| BRIE | STAR | 242 | Epithelial breast cancer cells | Human | Smart-Seq | HiSeq 2500, 100 bp PE |
CE | [84] (Data from [115]) | |
| BRIE | Bowtie | 82 | Macrophages | Mouse | Smart-Seq | NextSeq 500, PE | CE | [83] | |
| Expedition | STAR | 63 73 70 |
iPSCs NPCs MNs |
Human | Smart-Seq | HiSeq 2000, 100 bp PE | CE, MXE | [43] | |
| Developed for single-cell long-read RNA-sequencing | Custom pipeline | GMAP | 2 4 |
VLMCs Oligodendrocytes |
Mouse | STRT | PacBio SMRT | CE, A5SS, A3SS, TSS, TTS | [62] |
| Custom pipeline | STARlong | 6627 | Cerebellar cells | Mouse | Smart-Seq | PacBio SMRT | SJ, CE, TSS, TES | [28] | |
| Mandalorion | STAR | 7 | B1a cells | Mouse | Smart-Seq | ONT MinION, 2D | CE, RI, A5SS, A3SS, TSS, TES | [65] | |
| Mandalorion | STAR | 12 | OHCs | Mouse | Smart-Seq | ONT MinION, 1D | CE | [67] | |
| Mandalorion | Minimap2 | 96 | B cells | Human | R2C2 | ONT MinION, 1D | CE, RI, TSS, TES | [66] | |
| IgBLAST, BLASTN | Minimap2 | 6027 | Lymph node cells | Human | Droplet-based (10x Genomics) | ONT MinION, 1D | CE, RI, A5SS, A3SS, TSS, TES | [30] | |
A3SS: Alternative 3′ splice site; A5SS: Alternative 5′ splice site; ABI: Applied Biosystems; BMDCs: Bone-marrow-derived dendritic cells; CE: Cassette exon; ESCs: Embryonic stem cells; ICM: Inner cell mass; IPSA: Integrative Pipeline for Splicing Analyses; iPSCs: Induced pluripotent stem cells; ISOP: ISOform-Patterns; MIRALCS: Microwell full-length mRNA amplification and library construction system; MISO: Mixture of Isoforms; MN: Motor neurons; MXE: Mutually exclusive exons; NPC: Neural progenitor cells; OHC: Outer hair cells; ONT: Oxford Nanopore Technology; PacBio SMRT: Pacific Biosciences Single Molecule Real Time; PE: Paired-end; R2C2: Rolling Circle Amplification to Concatemeric Consensus; RI: Retained-intron; scM&T-seq: Single-cell methylation and transcriptome sequencing; SE: Single-end; SJ: Splice junction; STRT: Single-cell tagged reverse transcription; TES: Transcription end site; TSS: Transcription start site; VLMCs: Vascular and leptomeningeal cells.
Day 3 Oct4+Sox2+Nanog+, day 5 Oct4−−Sox2+Nanog+, day 5 Oct4−−Sox2−−Nanog−− outgrowth cells.
2. Technological advances in single-cell alternative splicing
2.1. scRT-PCR and smFISH
The earliest studies used reverse transcription polymerase chain reaction (RT-PCR) and single-cell fluorescence in situ hybridization (smFISH) for detection and quantification of alternative splicing events in single cells [34], [35], [36], [37], [38], [39], [40]. Single cell RT-PCR (scRT-PCR) protocols for investigating alternative splicing events were initially developed for characterizing short isoforms of length <1 kb. This allowed the analysis of exon-level alternative splicing events including exon-skipping [34], [35], [36], [37], [39], [40], mutually exclusive exons [38], and alternative 5′ and 3′ splice sites [34]. On the other hand, long-range single-cell PCR can be used to amplify longer fragments of more than 10 kb [35], [41], [42]. Alternatively, exon-exon junctions can be detected in lieu of sequencing entire exons [43]. The latter is feasible for detecting intron-retaining events, which typically consist of introns spanning several kilobases [34], [38].
smFISH followed by microscopic analysis is a powerful method for in situ single-molecule imaging of RNA splice variants in single cells. smFISH enables counting of single RNA molecules by probing each molecule with multiple short labelled oligonucleotide probes. Usually 30–50 hybridization probes of ~20 nt with different sequences are used for each RNA sequence [44], [45], [46]. In addition to single-molecule quantification of isoforms, smFISH provides temporal and spatial information of the RNA molecules [44], [45], [47]. However, the use of multiple oligonucleotide probes is constrained to target long sequences (>1 kb) and isoforms that vary sufficiently in their sequences [46], [47], [48]. A modified version of smFISH which performs padlock-probe-mediated rolling circle amplification (RNA) prior to imaging of RNA molecules can distinguish isoforms at single-base resolution and quantify isoforms at single-molecule level [49], [50].
Both scRT-PCR and smFISH approaches for alternative splicing analysis in single cells require prior knowledge of RNA sequences and are generally low-throughput and time-consuming. For these reasons, these approaches preclude the discovery of novel alternative splicing events and limit the analysis to a small number of alterative splicing events. Nevertheless, these methods remain useful to validate alternative splicing events detected from next-generation sequencing platforms.
2.2. Short-read RNA-sequencing
Early single-cell cDNA amplification protocols used 3′-end poly(A)-tailing for high-density oligonucleotide microarray analysis which yielded average PCR product lengths of ~0.85 kb [51], [52]. While comprehensive single-cell gene expression profiling was first made practical by using the microarray platform, the analysis was restricted to only gene-level expression analysis of known genes. Subsequent protocols leveraged on next-generation sequencing platforms following single-cell cDNA amplification for high-throughput and cost-efficient characterization of known and novel alternative splicing events in addition to gene expression profiling [53], [54], [55], [56], [57].
A single-cell RNA-sequencing method was introduced to improve cDNA amplification for microarray experiments [53], [54]. The method increased the reverse transcription step during first-strand cDNA amplification and the extension time for PCR, and adding an amine at the 5′ end of PCR primers that enabled the generation of amplified cDNAs of length up to 3 kb from the 3′ end of a transcript [53], [54]. On the other hand, a method for sequencing of mRNA from the 5′ end enabled generation of amplified cDNA of length up to 2 kb from the 5′ end of a transcript [55], [58]. These 3′ and 5′ end-bias methods produced relative short cDNA length and short sequencing reads. Therefore, alternative splicing analysis was restricted to only the identification of the exon-exon junctions and subsequent quantification of junction counts (reads mapping to the exon-exon junctions) [53], [54], [55].
Smart-Seq ameliorates 3′ bias by using SMART template-switching technology following poly(A)-tailing to generate amplified cDNA of length up to 10 kb. The double-stranded cDNA is then simultaneously fragmented and captured (tagged) with synthetic oligonucleotides at both ends to enable barcode adaptors to be appended for multiplexing and downstream sequencing. [56], [59]. Smart-Seq2 further improved read coverage distribution at both 3′ and 5′ ends of transcripts by systematically evaluating a large number of variations in experimental conditions [57], [60]. The main improvements made to Smart-Seq protocol that contributed to increased cDNA yield and length in Smart-Seq2 were the inclusion of a locked nucleic acid (LNA) guanylate in place of a single guanylate at 3′ end of the template switching oligo (TSO), addition of methyl group betaine together with higher MgCl2 concentrations, and incorporating deoxyribonucleotide triphosphates (dNTPs) before RNA denaturation step instead of in the reverse transcription master mix. While early implementation of Smart-Seq and Smart-Seq2 protocols involved only a few cells (<10) [56], integration with microfluidic or microwell platforms increased automation and multiplexing capability up to several hundred of cells [32], [43], [61], [62]. Notably, the microfluidic-based Fluidigm C1 platform enables fully automated cell lysis, reverse transcription, and amplification of full-length RNA molecules in micro-to-nanolitre reaction volume [63], [64].
The improvement of read coverage across full-length transcripts using next-generation short-read sequencing demonstrated, for the first time, characterization of transcriptome-wide exon-level alternative splicing events at the single-cell level, primarily exon-skipping and mutually exclusive events [43], [56], [61]. A notable exception is random displacement amplification sequencing (RamDA-seq) method, the first full-length total RNA-sequencing method for single cells [59]. This method can achieve near uniform full-length coverage of transcripts up to >20 kb for poly(A) and non-poly(A) tail-containing RNAs. The method used a whole-transcriptome amplification that amplifies cDNAs directly from RNA templates and not-so-random primers (NSRs) designed to avoid synthesizing cDNA from rRNAs. NSRs capture both poly(A) and non-poly(A) tail-containing RNAs by multiple priming but do not bind to rRNA sequences as they lack the 6-mers typically present in random hexamer primers that matches the rRNA sequences. Using this approach, the long non-poly(A) isoform of the long non-coding RNA Neat1-001 (>20 kb) and its short poly(A) isoform Neat1-002 were shown to be differentially expressed in mouse embryonic stem cells (mESCs) collected at different time points. Furthermore, recursive splicing, a multistep process of intron removal using cryptic splice sites within long introns, was also detected in pre-mRNAs [59].
2.3. Long-read RNA-sequencing
Relatively high sequencing depth of the massively parallel short-read RNA-sequencing enables the precise identification of splice sites. However, the identification of alternative splicing event was limited at exon-level due to the difficulty of transcript assembly from short sequencing reads [65]. In addition, short-read RNA-sequencing does not take the advantage of the full-length cDNA generated during library preparation, in particular for the protocols utilizing template switching approaches such as the Smart-Seq protocol [56], [57], [60]. Long-read RNA-sequencing can leverage on the full-length cDNA generated from library preparation by directly sequencing these transcripts without prior fragmentation.
The Oxford Nanopore Technologies (ONT) MinION sequencer is a portable device that is based on single-molecule sequencing technology that generates sequencing reads of up to 6 kb [65]. ONT MinION successfully detected 82% of splice sites present in the Spike-in RNA Variant Control Mixes (SIRV) genome annotation [65]. To mitigate the sequencing error rate of ONT MinION, circular consensus principle was introduced using Rolling Circle Amplification to Concatemeric Consensus (R2C2) method to obtain full-length consensus reads [66]. In the R2C2 method, amplified cDNA molecules generated from full-length library preparation protocols such as Smart-Seq2 are first circularized, then amplified, and finally debranched prior to sequencing. Reads generated from the original sequence after circularization are known as subreads and are collapsed computationally to yield a consensus read. R2C2 improved detection of splice sites present in SIRV genome annotation up to 91%. Whilst mismatch errors decreased with the increasing number of subreads, indels were systematically present at homopolymer regions. The high number of sequencing reads generated by ONT MinION relative to PacBio enables quantitation of genes and isoforms and consequently allows for gene expression and isoform profiling in single cells. Although the incorporation of unique molecular identifiers (UMIs) for accurate quantification of isoforms is not recommended due to high sequencing error rate of ONT MinION, gene expression levels generally correlate well when benchmarked against short-read RNA-sequencing gene expression [65], [66].
PacBio single-molecule real-time sequencing (SMRT) based on the properties of zero-mode waveguides can generate reads of up to 5 kb. Due to its lower sequencing error rates compared to ONT MinION, it can measure splice sites at 5′ and 3′ ends with an accuracy of ±1 bp in External RNA Controls Consortium (ERCC) spike-in [62]. In contrast to ONT MinION which uses 20 bp bins to assess the accuracy of identifying splice sites at 5′ and 3′ ends in SIRV, a control RNA [65]. The high base calling accuracy rate of PacBio enabled it to integrate UMIs to quantify isoforms at the single-molecule level. Counting of isoforms at the single-molecule level revealed a large number of transcripts to constitute singletons, i.e. transcripts supported by a single UMI [28], [62]. The high rate of singletons in PacBio, in part, reflects the low coverage obtained from this platform and therefore preclude differential gene and isoform analysis across individual cells. Hence, the isoform analysis using PacBio has been restricted to the characterization of a limited number of genes [28], [62]. Moreover, multiple deeply sequenced replicates are required for more precise quantitation [28]. Nevertheless, the accurate identification of splice sites has led to the discovery of isoforms not previously captured by publicly available gene annotation databases. In one study, 43%, 71%, and 94% of isoforms detected in mouse cerebellum have at least one splice site not annotated in GENCODE, RefSeq, and UCSC, respectively. These unannotated splice sites represent novel exon-exon junctions linking previously reported splice sites. The parallel analysis of short- and long-read RNA-sequencing enabled validation of these novel splice sites due to the high accuracy and coverage conferred by short-read sequencing [28], [65]. Therefore, full-length isoform information, especially from rare cell types [67], can be a valuable resource to reduce missing gene annotation in publicly available gene annotation databases.
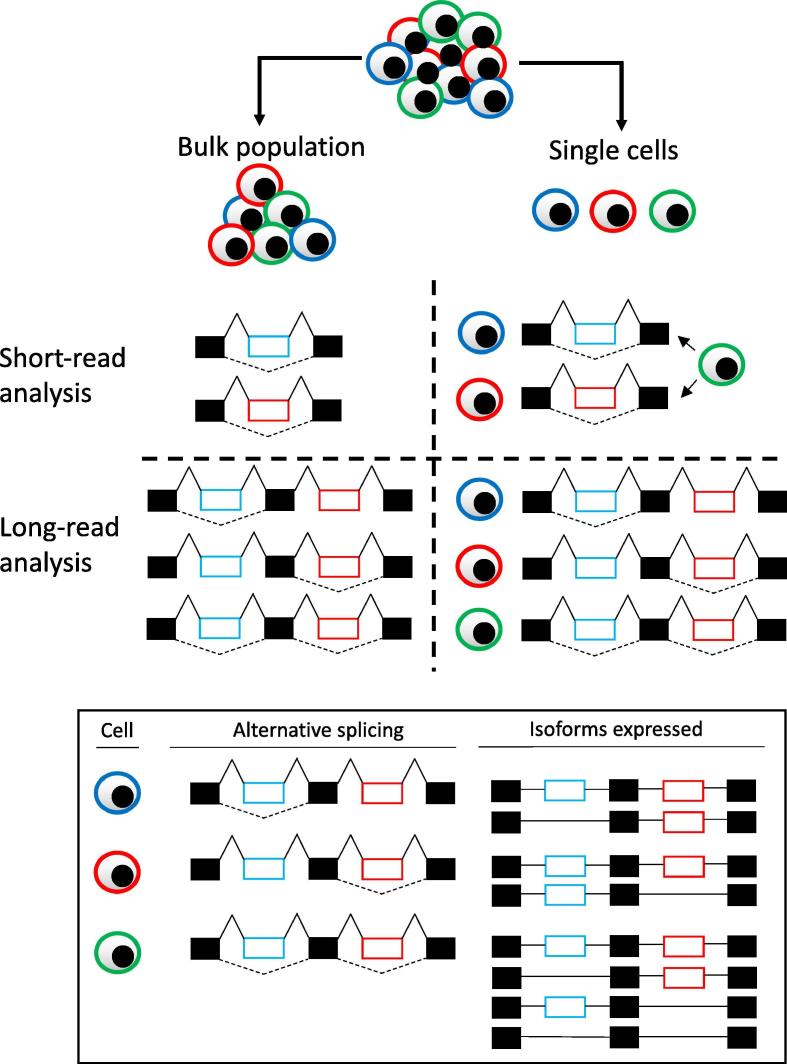
Full-length isoform sequencing enables the characterization of all aspects of isoforms including exon-skipping, alternative 3′ and 5′ splice sites, intron retention and alternative transcription start and end sites, and complex isoform consisting of coordination of multiple exon-level events on the same isoform [28], [30], [62], [65], [66], [67]. It is noteworthy that complex isoforms involving multiple exons may be missed by short-read RNA-sequencing assembly while long-read RNA-sequencing assembly may be able to deconvolute these individual isoforms. This is of particular importance if multiple isoforms of the same gene co-occur in the same cell type (Fig. 1). One such example is the expression of Alzheimer’s disease-associated Bin1 gene in neurons where a recent study identified six alternatively spliced exons on this gene and delineated a range of isoforms containing different combinations of these exons using long-read RNA-sequencing [28]. It is noteworthy that while long-read sequencing has been successful in identifying novel isoforms, in particular complex isoforms, its application on isoform characterization has been restricted to highly-expressed genes, such as cell lineage-specific genes. For example, cell type-specific receptor isoforms were characterized and reported in B cells notwithstanding when poly(A) tail-containing RNA molecules were captured and sequenced in experiments performed by Byrne et al. [65]. As only highly-expressed isoforms are feasible to be analysed with current long-read sequencing technologies, it may be useful to first enriched for isoforms of interest during library preparation prior to sequencing. Indeed, in one such study, T and B cell receptor transcripts were first enriched using hybridization capture approach prior to sequencing on ONT MinION. This enabled extensive characterization of T and B cell receptor isoforms in the immune cell repertoire harvested from the lymph node of a breast cancer patient [30].
Fig. 1.
Unique insights gained through single-cell alternative splicing analysis using short- and long-read RNA-sequencing. For simplicity, two isoforms with one alternative spliced exon (blue and red cells) and one complex isoform with two alternative spliced exons (green cells) illustrated here. Top-left panel: Bulk short-read RNA-sequencing is unable to delineate cell of origin for alternative splicing event. Top-right panel: Single-cell short-read RNA-sequencing is able to delineate cell of origin for each alternative splicing event. With the exception of cells with coordinated alternative splicing event (green) where it will be inferred that there are two isolated alternative splicing events. Bottom-left panel: Bulk long-read RNA-sequencing is able to distinguish isolated and coordinated alternative splicing events but is unable to assign the events to the cell of origin. Bottom-right panel: Single-cell long-read RNA-sequencing is able to distinguish isolated and coordinated alternative splicing events as well as assign the events to the cell of origin. Solid black box represents constitutive exons. Blue and red boxes represent alternatively spliced exons. Solid lines connecting two exons represent no alternative splicing events (no exon-skipping). Dotted lines connecting two exons represent alternative splicing events (exon-skipping). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Computational and statistical approaches for single-cell alternative splicing analysis
3.1. Isoform switching-based gene-level analysis
Most single-cell RNA-sequencing studies quantify gene counts obtained by aggregating or summing isoform-level expressions [43]. Isoform switching event or differential isoform usage is the difference in the isoform ratio of the same gene between groups of cells, and it may not necessarily be reflected by overall changes in gene expression [32]. For example, a gene X consists of isoforms Y and Z and has gene counts of 100 in both cell group 1 and 2. In group 1, isoforms Y and Z constitute 20 and 80 counts, whereas in group 2, isoforms Y and Z constitute 80 and 20 counts, respectively. In this scenario, there would be no detectable change in the gene-expression level, however, there is a clear difference in the isoform usage between these groups of cells. The relative change in individual isoform between difference cell groups is referred to as effect size (direction of change) [68]. Therefore, isoforms with effect sizes in the same direction would indicate no isoform switching, whereas isoforms having effect sizes in the opposite directions would indicate isoform switching. Hence, where one isoform is downregulated and the other is upregulated in one cell group relative to the other, the isoforms are indicated to have effect sizes in the opposite directions and the gene is detected to have undergone isoform switching. Several approaches were developed to perform isoform switching analysis, and by extension, identify overall changes in gene abundances in single cell experiments. These approaches take into account the limitations inherent to single cell experiments, such as high technical noise [32], [69] and 3′ bias transcripts [32], or by taking advantage of the large number of cells in single-cell experiments [68].
SingleSplice identifies genes whose differential isoform usage exceed technical noise in single cells [32]. Technical noise is the random fluctuations of isoform ratios due to reasons other than that of biological sources. Technical noise is caused by a range of factors, such as amplification of cDNA from small amount of starting materials [33]. SingleSplice also overcomes 3′ bias transcript inherent in single-cell experiments by circumventing the need to identify full-length transcripts using the concept of alternative splicing module (ASM) path. First, read alignments are used to build a directed, acyclic splice graph and each path through the graph represents a transcript. Therefore, any change in ratio of these ASM paths would indicate possible isoform switching events. To address high technical noise inherent in single-cell experiments, SingleSplice first builds a distribution of expected variance in coverage due to technical noise for each ASM path (fitted noise distribution). Next, it predicts the expected change in the ratio of these ASM paths by sampling repeatedly from the previous fitted noise distributions. Finally, a gene is considered to have undergone an isoform switching event if the observed change in the ratio of ASM paths exceeds that of the expected change in the ratio of ASM paths due to technical noise alone. Applying SingleSplice to a population of mouse embryonic stem cells [70], cell cycle-associated genes were found to have isoform switching events, and these single cells clustered according to their respective cell cycle stages using these differentially spliced genes [32]. ISOform-Patterns (ISOP) similarly identifies isoform switching events occurring beyond the high technical noise present in single cell experiments [69]. ISOP compares the expression profile of pairs of isoforms and classifies them into one of three main categories: single isoform preference (one isoform is preferentially expressed over the other isoform), bimodal isoform preference (unimodal expression in one of the isoform and a bimodal distribution in the other isoform), and mutually exclusive (either one or the other isoform is expressed, but not both). Application of ISOP on metformin-treated and untreated breast cancer cell line identified a subset of genes with isoform switching events that were missed from differential gene expression analysis [69].
Logistic regression approach takes the advantage of the large number of cells in single-cell RNA-sequencing experiments to detect genes with differential isoform usage [68]. The large sample size of single-cell experiments can be leveraged to accurately fit the logistic regression model. Individual isoform abundance is first quantified for each cell from different populations and the linear combination of isoform quantifications (differences in abundance between cell groups with direction of change taken into account) determines the overall effect size. Positive effect size indicates higher transcript expression relative to the reference group. Conversely, negative effect size indicates lower transcript expression relative to the reference group. Hence, genes consisting of isoforms with effect size in opposite direction would indicate isoform switching events, whereas the simple sum of isoform abundances without taking into account direction of change may lead one to conclude there were no differences in the isoform usage. Application of logistic regression to myogenic precursors and differentiating myeloblasts identified genes involved in myogenesis to have differential isoform usage [68], [71].
Both SingleSplice and logistic regression may identify differential isoform usage for two or more isoforms, whereas ISOP infers isoform-switching events from only a pair of isoforms. ISOP requires full-length isoform expressions to infer isoform-switching events, which may not always be attainable from single-cell RNA-sequencing. It is also noteworthy that ISOP and SingleSplice require the use of ERCC spike-in to establish levels of technical noise while logistic regression is the only approach, among the three methods, to demonstrate its utility on RNA-sequencing data generated from both full-length library protocol (e.g. Smart-Seq) and 3′-bias library protocol (e.g. Chromium platform from 10x genomics) [32], [68], [69]. Taken together, identifying genes with isoform switching events can identify hidden subpopulation of cells in a seemingly homogenous population of cells and differentially regulated genes between different groups of cells that would otherwise been missed using only the gene-level expression information.
3.2. Computational methods originally developed for bulk RNA-sequencing
Computational methods applied to early single-cell studies for detection and quantification of exon inclusion rates were originally developed for and benchmarked against short reads from bulk RNA-sequencing data. The exon inclusion rate can be divided into exon- and intron-centric. The inclusion rate of an alternative exon is often presented as the percent spliced-in (PSI or Ψ) index and takes any value between 0 and 1 [14], [56], [61], [72], [73], [74]. The inclusion rate of an exon or Ψ index represents the relative expression of alternatively spliced isoforms (the inclusion isoform) and it in turns reflect the proportion of reads supporting alternative splicing over total reads [75], [76]. Therefore, a Ψ of 0.5 would reflect half of all reads supporting the inclusion isoform, whereas a Ψ of 1 would reflect all reads supporting the inclusion isoform. On the other hand, a Ψ of 0 would reflect the absence of any reads supporting the inclusion isoform.
Mixture-of-isoforms (MISO) model is a probabilistic framework that considers reads aligned to bodies of the alternative exon and its immediate flanking constitutive exons, as well as reads aligned to the junctions between the alternative exon and immediate flanking constitutive exons. In paired-end RNA sequencing, MISO further considers information about length distribution to improve the estimates of Ψ. MISO provides Bayesian confidence intervals in addition to point estimates of Ψ to reflect the uncertainty in the Ψ estimation for each exon [77]. Early single-cell studies used MISO for estimating exon inclusion rate and restricted their analysis to only known or annotated splicing events [14], [56]. RNA-sequencing of full-length cDNA from a panel of cancer cell lines and subsequent detection and quantification of exon inclusion levels using MISO demonstrated heterogeneity in alternative splicing events different between cell types. Furthermore, the improvement of read coverage across the entire length of isoforms with an overall increased in read coverage of RNA-sequencing of full-length cDNA increased the number of detected alternative splicing events by twofold compared to RNA-sequencing with 3′-end bias [53], [56]. On the other hand, RNA-sequencing of full-length cDNA of individual cells derived from a single population of mouse bone-marrow-derived dendritic cells (BMDCs) showed variability in exon inclusion rate. Notably, these individual cells demonstrated bimodal expression pattern of alternative exons, i.e. either inclusion or exclusion of the alternative exon [14]. Collectively, early alternative splicing analysis in single cells have demonstrated the power of single cell RNA-sequencing to unravel heterogeneity in alternative splicing events between different cell types and across cells originating from the same population. The latter would have not been possible with the alternative splicing analysis at the bulk level.
Weighted-log-likelihood expectation maximization method on isoform quantification (WemIQ) is another exon-centric approach for Ψ estimation in bulk RNA-sequencing experiments [78]. WemIQ corrects for bias in RNA-sequencing by assigning different weights to reads from different genomic region according to degree of sequencing bias. The bias parameter makes no assumption about the bias source and format. This is in contrast to some methods that assume a constant bias factor for each relative position of genes or only correct for sequence-specific bias caused by random hexamer priming [79], [80]. Applying WemIQ to the aforementioned mouse BMDC dataset [14] revealed larger variation among exon bias within a gene in single-cell RNA-sequencing compared to bulk RNA-sequencing. Correction of heterogenous bias pattern through WemIQ decreased cell-to-cell expression variability, suggesting that differences in expression profile between single cells may be attributed to both technical and biological factors.
Intron-centric approach for alternative splice site detection, unlike exon-centric approach, ignores reads aligning to exon bodies. Instead, only splice junction reads are considered when computing Ψ. In intron-centric approach, Ψ is split into two indices: Ψ5 and Ψ3, and the calculation of these indices is based on conditional probability. Ψ5 is the number of reads supporting the splicing event from the 5′ (donor)-splice site to the immediate 3′ (acceptor)-splice site relative to the combined number of reads supporting splicing from 5′-splice site to any 3′-splice sites. Similarly, Ψ3 is the number of reads supporting the splicing event from the 3′-splice site to the immediate 5′-splice site relative to the combined number of reads supporting splicing from 3′-splice site to any 5′-splice sites. [61], [81]. Intron-centric estimation of Ψ was applied to single cells from a lymphoblastoid cell line to detect novel splice junctions in single cells [72]. Using conservative approach where at least one of the 5′- or 3′-splice sites has already been annotated, 35% of novel junctions were observed to connect two annotated exons, whereas 60% of novel junctions connected an annotated exon to an unannotated exon. This was also the first study to include spike-in standards for splice site detection, and “novel junctions” were similarly observed in these controls, suggesting quality control measures should be performed when detecting novel alternative splicing events in single cells.
Intron-centric approach for inferring alternative splicing events relies on reads spanning beyond the alternative spliced exon and its immediate flanking exons. Hence, intron-centric approach assumes uniform coverage across the isoform length, which is not always achievable in single-cell RNA-sequencing. Both exon-centric and intron-centric approach infer alternative splicing events from sequencing reads alone and therefore does not address the limitations of low and uneven coverage prevalent in single-cell RNA-sequencing [77], [78], [81]. Although early single-cell studies that utilised computational tools originally developed for bulk RNA-sequencing largely restricted alternative splicing analysis to known or annotated events, most of these studies did not perform independent validation of these alternative splicing events, such as using smFISH or scRT-PCR. Nevertheless, these studies represent a proof-of-concept for alternative splicing detection and quantification in single cells.
3.3. Prediction-based approaches for single-cell RNA-sequencing
The most pronounced computational challenges in single-cell RNA-sequencing are low coverage, high dropout rate and increased technical noise compared to bulk RNA-sequencing [32]. These challenges are not incorporated into the probabilistic methods based on mixture modelling such as MISO, which only considers aligned-reads to form the likelihood of the Bayesian model to estimate Ψ [77], [82]. Therefore, mixture modelling based on aligned-reads alone does not accurately predict Ψ at low coverage. To overcome this problem, Bayesian Regression for Isoform Estimation (BRIE) extends this mixture model approach, such as that of MISO, to not only consider aligned-reads but also to incorporate a Bayesian regression module to automatically learn an informative prior distribution directly from the data [31]. BRIE integrates an informative prior distribution derived from sequence features together with likelihood derived from aligned-reads for Ψ estimation. These features include seven-hundred and thirty-five splicing regulatory features predictive of exon-skipping events derived from a training dataset comprising of >20,000 and >9,000 high-quality exon-skipping events from GENCODE human and mouse gene annotation, respectively. As a consequence, the informative prior distribution enables BRIE to estimate Ψ more accurately at low coverage. On the other hand, at high coverage, the probabilistic model based on aligned-reads dominates in Ψ estimation, whereas Bayes’ theorem is used to trade off imputation and quantification at the region of intermediate coverage. Unsurprisingly, Ψ values estimated with an informative prior distribution demonstrated superior correlation with Ψ values computed from bulk RNA-sequencing reads compared to Ψ values estimated with an uninformative prior distribution [31]. In addition to characterizing exclusion and inclusion rates across identical cells, BRIE is also able to identify events that are differentially spliced between identical cells by comparing all possible pairs of cells [29], [31], [83], [84]. Nevertheless, it is computationally costly and unfeasible to compare Ψ values between all possible pairs of cells, particularly in experiments involving large number of single cells [31], [85].
In addition to sequence features, incorporating DNA methylation profiles as an informative prior demonstrated modest improvement in Ψ estimation compared to when either factors was used as informative prior alone [86]. It is noteworthy that sequence features were more informative compared to DNA methylation profile for predicting Ψ. Using DNA methylation profile alone as informative prior distribution confers limited benefits in predicting Ψ, suggesting that sequence features dominate in the Bayesian model. Moreover, DNA methylation profiles are often cell-type specific as shown in single-cell methylation and transcriptomic sequencing (scM&T-seq) of induced pluripotent stem cells (iPSCs) and endoderm cells [86]. Therefore, unlike sequence features, DNA methylation profiles are not universally applicable across different cell types. Taken together, the informative prior information such as that of genomic and epigenetic features can increase accuracy of Ψ prediction when combined with likelihood terms computed from aligned-reads. It would be of particular interest to uncover additional factors that may serve as informative priors. One such potential factor is chromatin accessibility [31], which can be profiled by ATAC (assay for transposase-accessible chromatin) sequencing method. This method currently allows to assess genome-wide chromatin accessibility at the single-cell level [87].
While Ψ estimation using prediction-based approach may mitigate, to some extent, the challenges posed by the absence of data (drop-out genes) or low-confidence data (low coverage) [31], it is noteworthy that presenting Ψ as an estimation does not represent true biological phenomenon [43]. For example, a Ψ value of 0.05 should indicate that 5% of transcripts include the alternative exon while the other 95% skips the alternative exon. As a case in point, Ψ estimation of the Pdgfa exon 6 showed variability in exon inclusion rate (~0–0.8 Ψ) in single cells derived from oligodendrocytes from the spinal cord of mice induced with experimental autoimmune encephalomyelitis (EAE) and that of control mice. However, independent validation of this exon-skipping event using scRT-PCR in bulk spinal cord from EAE mice showed high expression of this exon (~10–60 normalized mRNA expression), whereas bulk spinal cord tissues from control mice showed no expression of this exon (~0 normalized mRNA expression) [29]. In another example, variable Dectin-2 exon 3 inclusion rate was observed in stage 1 and 2 macrophages infected with Candida albicans, where all but one cell was observed to have Ψ values above 0. However, visual-based inspection of coverage distribution in a genome browser revealed the absence of coverage at the exon 3 in ~20% and ~6.5% of stage 1 and 2 infected macrophages, respectively [83]. These suggest that although the use of informative prior comes at the cost of biasing results at low coverage regions, it is nevertheless an effective approach for more accurate Ψ prediction, especially at regions with moderate-to-high coverage [29], [31], [83]. One possible approach to reduce potential false positive detection of alternative splicing events at low coverage regions is to apply stringent filtering criteria to restrict analysis to alternatively spliced exons with coverage above a user-defined threshold and expressed in more than a user-defined percentage or number of cells sequenced [86].
3.4. Read-based approaches for single-cell RNA-sequencing
The Expedition suite creates a custom alternative splicing index from splice junction information to detect and quantify alternative splicing events based on aligned-reads only from short-read RNA-sequencing data [43], [88]. As the Ψ values are computed from aligned-reads only, it is more reflective of the biological exon inclusion rate compared to Ψ values estimated from prediction-based approaches [31], [77]. Expedition suite can be used to more accurately characterize the distribution of exon inclusion rate in the form of “modalities” across a population of single cells. Modalities can be classified into five categories consisting of excluded (Ψ values concentrated mainly around 0), included (Ψ values concentrated mainly around 1), bimodal (Ψ values concentrated around both 0 and 1), middle (Ψ values concentrated around 0.5), and multimodal (uniform distribution of Ψ values from 0 to 1) [43], [86].
Early single-cell studies applied computational approaches for bulk RNA-sequencing to determine Ψ values in small sample sizes of single cells (~10–20 cells) [14], [56], [72]. These studies reported that the majority of cells expressed either one or the other isoform but rarely both, reflective of included and excluded modalities. This suggests that included and excluded modalities may be the most prevalent modalities. Indeed, using Expedition to compute Ψ values and subsequently applying a beta distribution to represent these Ψ values in the large sample size of iPSC cells (>60 cells), it was observed that the included and excluded modalities represented ~50% and 29% of all modalities, whereas bimodal, multimodal, and middle modalities represented ~20%, 1%, and <1% of all modalities, respectively [43]. Prediction-based approach such as BRIE may underestimate splicing heterogeneity at low coverage [31], [77]. For example, alternatively spliced exons with low inclusion rates (Ψ values concentrated mainly around 0) have been shown to have lower variance compared to the alternatively spliced exons with intermediate-to-high inclusion rates [73], [89]. Different thresholds may be applied prior to Ψ estimation using prediction-based approach to circumvent this and enable the characterization of modalities across a population of single cells. These thresholds may include the inclusion of high quality annotation of alternative splicing events and events defined manually by a user-defined cut-off based on the number of reads and cells supporting the events [86]. After applying thresholds prior to Ψ estimation and subsequent characterization of Ψ distribution as a function of mean-variance in iPSC cells, included and excluded modalities were similarly observed to be the most prevalent modalities comprising of ~52% and ~31%, respectively. Furthermore, bimodal, multimodal, and middle modalities represented ~7%, ~8%, and ~2%, respectively, of all modalities. Therefore, combining prediction-based approach for Ψ estimation and carefully selected thresholds prior to Ψ estimation may increase statistical power for the detection of rare modalities, namely multimodal and middle modalities.
Taken together, combining computational approaches developed for single cells and large population of single cells can unravel heterogeneity in exon inclusion rate distribution across single cells that would otherwise be missed when analysing small populations of single cells. This is particularly true for less frequent modalities such as bimodal, multimodal, and middle modalities. It is known that the bimodal and multimodal modalities are critically relevant during cellular differentiation. An analysis of iPSC differentiation into neural progenitor cells (NPCs) and motor neurons (MNs) showed that >99% of alternative splicing events either switched from bimodal or multimodal state, or switched toward a bimodal or multimodal state [43].
Although the utility of read-based approach for computing Ψ values is limited to isoforms expressed at moderate-to-high levels, it may be more precise in reflecting true biological Ψ values compared to Ψ values estimated from prediction-based approach where informative priors are used to infer Ψ at low coverage region [31], [43]. This may be an important consideration when assessing the consistency between computed Ψ values and results from independent validation using experimental or visual-based inspection from computational approaches.
3.5. Visual-based inspection of alternative splicing events
Visual-based inspection of read coverage from RNA-sequencing enables the assessment of bioinformatics methods used to compute gene and isoform expression levels [29], [31], [83], [84]. Genome browsers such as Integrative Genomics Viewer (IGV) and University of California Santa Cruz (UCSC) Genome Browser enable the visualization of read coverage distribution over defined genomic regions [90], [91], [92], [93]. Read coverage distribution is represented as bar graphs where the height of the bar graph is proportional to the number of reads at each genomic coordinate. The extension to this functionality is the display of arcs that represent splice junctions connecting exons. The width of the arc is proportional to the number of reads split across splice junctions. Visual-based presentation that includes both splice junction information in addition to read coverage distribution is called a sashimi plot [77]. IGV and ggsashimi generate sashimi plots from the input Binary Alignment Map (BAM) files using a user-friendly and command-line interface, respectively [90], [91], [94].
The presentation of multi-panel coverage distributions or multiple sashimi plots is not a feasible approach for the large sample sizes (>1000 cells) from single-cell RNA-sequencing experiments. As a consequence, sashimi plots for only a subset of cells are usually displayed and do not capture cell-to-cell variability in alternative splicing events [31], [83], [84]. Furthermore, it is difficult to capture cell-to-cell variability in alternative splicing events even when the coverage distribution of all single cells is displayed in a multi-panel format [43], [83]. One possible approach for visual-based inspection of alternative splicing events across different groups of single cells is to aggregate all single cells by their respective groups [29], [67], [94]. However, merging cells by their respective groups does not take cell-to-cell variation in sequencing depth and group-to-group variation in number of single cells into account. This limits meaningful visual-based inspection of alternative splicing events across different groups of single cells.
One approach to accentuate cell-to-cell heterogeneity in alternative splicing events in single cells is to use a heatmap to display the read coverage distribution for all cells. Millefy displays read coverage distribution across a large number of single cells using heatmap to demonstrate cell-to-cell heterogeneity in gene expression [59]. However, Millefy does not consider split reads or junction reads and hence is not suitable for visualizing Ψ values and corresponding alternative splicing frequencies in single cells. We have developed VALERIE (Visualizing alternative splicing events in single-cell ribonucleic acid (RNA)-sequencing experiments; https://github.com/wenweixiong/VALERIE) to incorporate both split reads and non-split reads to compute Ψ values at each defined nucleotide position, for example at an alternatively spliced exon, and presents these values in a heatmap. Furthermore, an average value, such as mean, is used as the summary statistic for Ψ values for user-defined group of cells. This allows for the overall comparison of alternative splicing rates in different groups of cells. Finally, pair-wise comparison, such as t-test, is performed for Ψ values at each nucleotide position to assess the significant difference in Ψ values between different groups of cells. Therefore, VALERIE can capture within- and between-group cell-to-cell heterogeneity and subsequently allows for meaningful visual-based inspection of alternative splicing events between different groups of cells (Fig. 2).
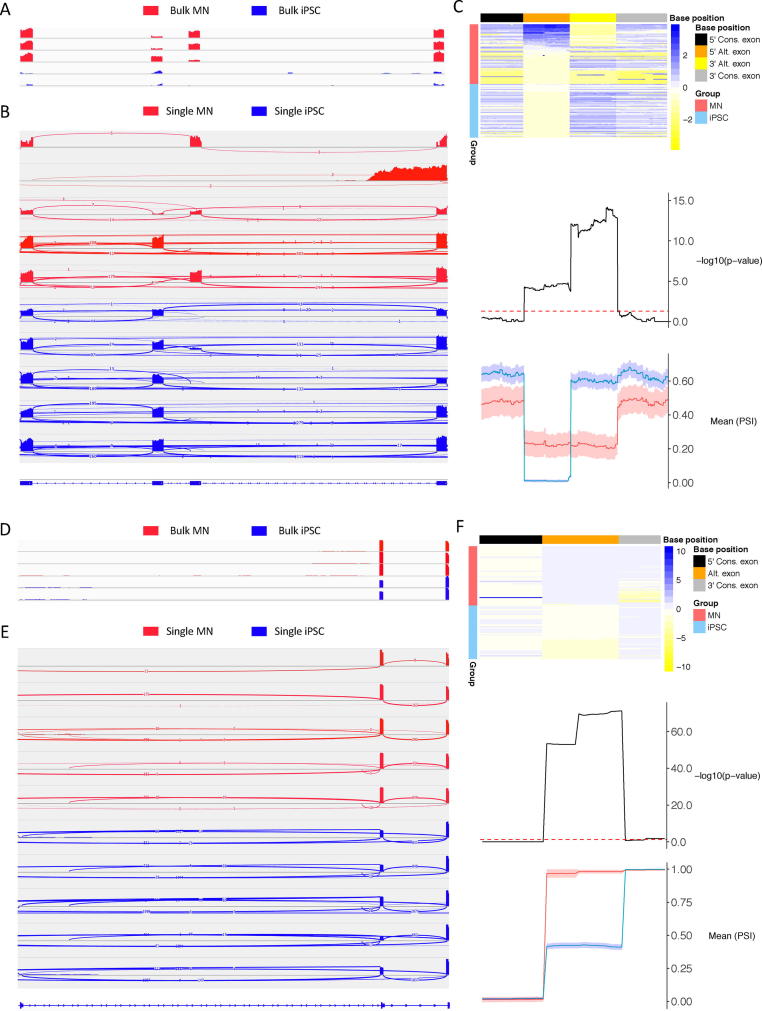
Fig. 2.
The comparison of different visualization methods of alternative splicing events in 69 motor neurons (MNs) and 63 induced-pluripotent stem cells (iPSCs) from Song et al. using short-read RNA-sequencing data [43]. (A-C) Mutually exclusive exon 9 and 10 of PKM gene. Alternative splicing event validated using smFISH previously. iPSCs almost exclusively express exon 10 while MNs predominantly express exon 9. (A) IGV display of coverage distribution of mutually exclusive exon 9 and 10 together with flanking constitutive exons from 3 MN (red) and 2 iPSC (blue) bulk samples show inconsistency in relative coverage for exon 9 and 10 across MN samples. Specifically, MN sample 1 and 2 show higher exon 9 coverage whereas MN sample 3 show higher exon 10 coverage. (B) IGV display of coverage distribution of mutually exclusive exon 9 and 10 together with flanking constitutive exons from 5 MN (red) and 5 iPSC (blue) representative cells show inconsistency in relative coverage for exon 9 and 10 across MN cells. Specifically, MN sample 1, 3, and 5 show higher exon 9 expression, MN sample 4 show higher exon 10 expression, whereas MN sample 2 had no detectable coverage across both exon 9 and 10. (C) VALERIE display of PSI values for all MN (red) and iPSC (blue) cells in heatmap annotated with mutually exclusive exon 9 (orange) and 10 (yellow) and flanking constitutive exons (black and grey). MN exon 9 with higher PSI compared to iPSC where MN exon 10 with lower PSI compared to iPSC. Differences in PSI for mutually exclusive exon 9 and 10 between MN and iPSC cell groups are statistically significant. On the other hand, there is no statistical difference in PSI values of both constitutive exons between MN and iPSC cell groups. (D-F) Exon 6 skipping of RPS24 gene. Alternative splicing event validated using sc-qPCR. MNs express higher levels of exon 6 compared to iPSCs. (D) IGV display of coverage distribution of alternative spliced exon 6 together with flanking constitutive exons from 3 MN (red) and 2 iPSC (blue) bulk samples show consistently higher coverage of exon 6 in MN compared to iPSC. (E) IGV display of coverage distribution of alternative spliced exon 6 together with flanking constitutive exons from 5 MN (red) and 5 iPSC (blue) representative cells show consistently higher coverage of exon 6 in MN compared to iPSC. (F) VALERIE display of PSI values for all MN (red) and iPSC (blue) cells in heatmap annotated with alternatively spliced exon 6 (orange) and flanking constitutive exons (black and grey). MN exon 6 with higher PSI compared to iPSC. Differences in PSI for alternatively spliced exon 6 between MN and iPSC cell groups are statistically significant. On the other hand, there is no statistical difference in PSI values for both constitutive exons between MN and iPSC cell groups. VALERIE standardizes the display of base position in 5′-to-3′ direction, focuses on informative exonic regions by excluding long intronic sequences with no splicing events, and displays PSI values rather than coverage information. Two-sided t-test used as statistical test for comparing PSI values at each genomic coordinate. IGV: Integrative Genome Browser. VALERIE: Visualizing alternative splicing events in single-cell ribonucleic acid (RNA)-sequencing experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. Full-length isoform analysis in single cells
Most of single-cell alternative splicing analysis have been conducted on short-read RNA-sequencing data. This is reflected by the paucity of computational tools available to analyse alternative splicing events from long-read RNA-sequencing data. Mandalorion is a software package for isoform detection and quantification from Nanopore sequencing reads and it was first developed for analysing 2D sequencing data [65]. In 2D sequencing, both strands of the molecule are ligated with a hairpin adapter and each strand is sequenced sequentially [95]. However, Mandalorion no longer supports 2D sequencing reads analysis. The recent version of Mandalorion supports analysis of sequencing reads generated from the more accurate R2C2 approach [66] and an experimental version of Mandalorion supports 1D sequencing reads analysis [67]. In contrast to 2D sequencing, 1D sequencing involves sequencing one strand of the molecule and thus generates lower quality sequencing data compared to 2D sequencing, albeit at higher sequencing efficiency [95]. To date, there is no published software available for isoform detection and quantification from PacBio sequencing reads. Instead, custom scripts were developed by individual studies [28], [62]. The lack of a unified framework for isoform detection and quantification from long-read RNA-sequencing in single cells reflects the infancy of this area of research and presents an opportunity to develop robust and potentially novel computational tools for single-cell alternative splicing analysis using long-read RNA-sequencing data.
4. Motivation for the alternative splicing analysis in single cells
Comprehensive studies for alternative splicing analysis in single cells have involved characterizing alternative splicing events during neuronal differentiation, across different subtypes of neuronal cells, and in immune cells [28], [43], [62], [65], [66], [86]. The initial focus on alternative splicing in individual neurons was motivated by preceding decades of studies that demonstrated alternative exons play critical roles in multiple aspects of neuronal development including neuronal migration, axon guidance, and synapse formation [23]. Characterization of alternative splicing events in immune cells demonstrated alternative splicing as a source for diversity in T and B cell-specific surface receptors [30], [65], [66]. On the other hand, several single-cell studies firstly focused on the overall gene expression analysis and then performed the alternative splicing analysis as another layer of information to investigate the cellular heterogeneity driven by distinct alternative splicing events [29], [56], [72], [74], [83].
Cancer biology is one potential area of research that can be further advanced by single-cell alternative splicing analysis. RNA mis-splicing may arise from novel splice sites created by somatic mutations (cis-acting) and mutations specific to genes involved in the splicing machinery (trans-acting) [25]. Aberrantly spliced genes may introduce premature stop codons and consequently degradation by the nonsense-mediated decay pathway, which yield short peptides that are ultimately presented on the cell surface through MHC-I pathway (on the condition the peptide is compatible with the individual patient’s human leukocyte antigen (HLA) type) [20], [96]. Comprehensive alternative splicing analysis across multiple tumour types from The Cancer Genome Atlas have showed both cis- and trans-associated aberrant alternative splicing events to correlate with neoepitopes presentation and immune signatures [97], [98], [99]. Moreover, several aberrantly spliced cancer driver genes were found to be more prevalent in splicing factor-mutated samples [97], thus providing additional actionable candidates for targeted therapy.
Mutations in splicing factors are enriched in blood neoplasms, in particular myeloid dysplastic and proliferative neoplasms (MDS and MPN), where up to ~50–85% of these patients are found to be carriers of splicing factor mutations of SF3B1, SRSF2, U2AF1, and ZRSR2 [19], [100], [101]. The high variant allele frequency (VAF) of splicing factor mutations and reconstruction of clonal hierarchy from genotypes of bulk samples derived from bone marrow nuclear cells (BMNCs) and genotypes of individual primary human hematopoietic stem and progenitor cells (HSPCs) suggest that mutations in splicing factors are early events arising in the HSPC compartment [19], [100], [102], [103], [104]. Nevertheless, genetic and cellular heterogeneity can exist within the phenotypically identical HSPC compartment [13], [105], [106]. Therefore, single-cell approaches can be used to deconvolute alternative splicing events attributed to splicing factor mutations alone or their interaction with other cancer driver genes mutation, as well as cell- and lineage-specific alternative splicing events.
Taken together, it may be of particular interest to identify aberrant splicing events engendered from different genetic background and in different cellular types from phenotypically homogenous populations, in both solid and blood neoplasm, by leveraging technological advances in parallel single-cell genomic and transcriptomic sequencing to guide development of personalized therapy for cancer patients [103], [107], [108], [109].
Conflict of interest
None.
Acknowledgements
The Clarendon Fund and Oxford-Radcliffe Scholarship in conjunction with WIMM Prize PhD Studentship to W.X.W., Medical Research Council (MRC) Senior Clinical Fellowship and CRUK Senior Cancer Research Fellowship to A.J.M., Medical Research Council (MRC) in conjunction with NIHR Oxford Biomedical Research fellowship to S.T. This work was supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.01.009.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Metzker M.L. Sequencing technologies – the next generation. Nat Rev Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 2.Liu J. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2) doi: 10.1016/j.cell.2018.02.052. 400-416 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International Cancer Genome, C International network of cancer genome projects. Nature. 2010;464(7291):993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Consortium, GT The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wen W.X., Leong C.O. Association of BRCA1- and BRCA2-deficiency with mutation burden, expression of PD-L1/PD-1, immune infiltrates, and T cell-inflamed signature in breast cancer. PLoS One. 2019;14(4) doi: 10.1371/journal.pone.0215381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen-Dumont T. A high-plex PCR approach for massively parallel sequencing. BioTechniques. 2013;55(2):69–74. doi: 10.2144/000114052. [DOI] [PubMed] [Google Scholar]
- 7.McDonough S.J. Use of FFPE-derived DNA in next generation sequencing: DNA extraction methods. PLoS One. 2019;14(4) doi: 10.1371/journal.pone.0211400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nawy T. Single-cell sequencing. Nat Methods. 2014;11(1):18. doi: 10.1038/nmeth.2771. [DOI] [PubMed] [Google Scholar]
- 9.Horning A.M. Single-cell RNA-seq reveals a subpopulation of prostate cancer cells with enhanced cell-cycle-related transcription and attenuated androgen response. Cancer Res. 2018;78(4):853–864. doi: 10.1158/0008-5472.CAN-17-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J. Single-cell gene expression analysis reveals regulators of distinct cell subpopulations among developing human neurons. Genome Res. 2017;27(11):1783–1794. doi: 10.1101/gr.223313.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen Q.H. Single-cell RNA-seq of human induced pluripotent stem cells reveals cellular heterogeneity and cell state transitions between subpopulations. Genome Res. 2018;28(7):1053–1066. doi: 10.1101/gr.223925.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grover A. Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat Commun. 2016;7:11075. doi: 10.1038/ncomms11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drissen R. Distinct myeloid progenitor-differentiation pathways identified through single-cell RNA sequencing. Nat Immunol. 2016;17(6):666–676. doi: 10.1038/ni.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shalek A.K. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498(7453):236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wimmers F. Single-cell analysis reveals that stochasticity and paracrine signaling control interferon-alpha production by plasmacytoid dendritic cells. Nat Commun. 2018;9(1):3317. doi: 10.1038/s41467-018-05784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsson A.J.M. Genomic encoding of transcriptional burst kinetics. Nature. 2019;565(7738):251–254. doi: 10.1038/s41586-018-0836-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manning K.S., Cooper T.A. The roles of RNA processing in translating genotype to phenotype. Nat Rev Mol Cell Biol. 2017;18(2):102–114. doi: 10.1038/nrm.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reyes A., Huber W. Alternative start and termination sites of transcription drive most transcript isoform differences across human tissues. Nucleic Acids Res. 2018;46(2):582–592. doi: 10.1093/nar/gkx1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiozawa Y. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat Commun. 2018;9(1):3649. doi: 10.1038/s41467-018-06063-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smart A.C. Intron retention is a source of neoepitopes in cancer. Nat Biotechnol. 2018;36(11):1056–1058. doi: 10.1038/nbt.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciolli Mattioli C. Alternative 3' UTRs direct localization of functionally diverse protein isoforms in neuronal compartments. Nucleic Acids Res. 2019;47(5):2560–2573. doi: 10.1093/nar/gky1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z. Pro-apoptotic effects of splice-switching oligonucleotides targeting Bcl-x pre-mRNA in human glioma cell lines. Oncol Rep. 2016;35(2):1013–1019. doi: 10.3892/or.2015.4465. [DOI] [PubMed] [Google Scholar]
- 23.Vuong C.K., Black D.L., Zheng S. The neurogenetics of alternative splicing. Nat Rev Neurosci. 2016;17(5):265–281. doi: 10.1038/nrn.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baralle F.E., Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol. 2017;18(7):437–451. doi: 10.1038/nrm.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scotti M.M., Swanson M.S. RNA mis-splicing in disease. Nat Rev Genet. 2016;17(1):19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S.C., Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22(9):976–986. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dvinge H. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16(7):413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta I. Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat Biotechnol. 2018 doi: 10.1038/nbt.4259. [DOI] [PubMed] [Google Scholar]
- 29.Falcao A.M. Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis. Nat Med. 2018;24(12):1837–1844. doi: 10.1038/s41591-018-0236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh M. High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat Commun. 2019;10(1):3120. doi: 10.1038/s41467-019-11049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y., Sanguinetti G. BRIE: transcriptome-wide splicing quantification in single cells. Genome Biol. 2017;18(1):123. doi: 10.1186/s13059-017-1248-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welch J.D., Hu Y., Prins J.F. Robust detection of alternative splicing in a population of single cells. Nucleic Acids Res. 2016;44(8) doi: 10.1093/nar/gkv1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brennecke P. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10(11):1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 34.Kumazaki T. Detection of alternative splicing of fibronectin mRNA in a single cell. J Cell Sci. 1999;112(Pt 10):1449–1453. doi: 10.1242/jcs.112.10.1449. [DOI] [PubMed] [Google Scholar]
- 35.Springer J. Alternative splicing in single cells dissected from complex tissues: separate expression of prepro-tachykinin A mRNA splice variants in sensory neurones. J Neurochem. 2003;85(4):882–888. doi: 10.1046/j.1471-4159.2003.01720.x. [DOI] [PubMed] [Google Scholar]
- 36.Graf E.M. Tissue distribution of a human Ca v 1.2 alpha1 subunit splice variant with a 75 bp insertion. Cell Calcium. 2005;38(1):11–21. doi: 10.1016/j.ceca.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 37.Mechaly I. Molecular diversity of voltage-gated sodium channel alpha subunits expressed in neuronal and non-neuronal excitable cells. Neuroscience. 2005;130(2):389–396. doi: 10.1016/j.neuroscience.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 38.Steinboeck F., Kristufek D. Identification of the cytolinker protein plectin in neuronal cells - expression of a rodless isoform in neurons of the rat superior cervical ganglion. Cell Mol Neurobiol. 2005;25(7):1151–1169. doi: 10.1007/s10571-005-8503-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanumilli S. Alternative splicing generates a smaller assortment of CaV2.1 transcripts in cerebellar Purkinje cells than in the cerebellum. Physiol Genomics. 2006;24(2):86–96. doi: 10.1152/physiolgenomics.00149.2005. [DOI] [PubMed] [Google Scholar]
- 40.Castro M.A. Extracellular isoforms of CD6 generated by alternative splicing regulate targeting of CD6 to the immunological synapse. J Immunol. 2007;178(7):4351–4361. doi: 10.4049/jimmunol.178.7.4351. [DOI] [PubMed] [Google Scholar]
- 41.Zanssen S. Single cell PCR from archival stained bone marrow slides: a method for molecular diagnosis and characterization. J Clin Lab Anal. 2004;18(3):176–181. doi: 10.1002/jcla.20019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rygiel K.A. Triplex real-time PCR–an improved method to detect a wide spectrum of mitochondrial DNA deletions in single cells. Sci Rep. 2015;5:9906. doi: 10.1038/srep09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song Y. Single-cell alternative splicing analysis with expedition reveals splicing dynamics during neuron differentiation. Mol Cell. 2017;67(1) doi: 10.1016/j.molcel.2017.06.003. 148–161 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Femino A.M. Visualization of single RNA transcripts in situ. Science. 1998;280(5363):585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 45.Raj A. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5(10):877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui Y., Liu J., Irudayaraj J. Beyond quantification: in situ analysis of transcriptome and pre-mRNA alternative splicing at the nanoscale. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2017:9(4). doi: 10.1002/wnan.1443. [DOI] [PubMed] [Google Scholar]
- 47.Waks Z., Klein A.M., Silver P.A. Cell-to-cell variability of alternative RNA splicing. Mol Syst Biol. 2011;7:506. doi: 10.1038/msb.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larsson C. In situ detection and genotyping of individual mRNA molecules. Nat Methods. 2010;7(5):395–397. doi: 10.1038/nmeth.1448. [DOI] [PubMed] [Google Scholar]
- 49.Deng R. Highly specific imaging of mRNA in single cells by target RNA-initiated rolling circle amplification. Chem Sci. 2017;8(5):3668–3675. doi: 10.1039/c7sc00292k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren X. SpliceRCA: in Situ Single-Cell Analysis of mRNA Splicing Variants. ACS Cent Sci. 2018;4(6):680–687. doi: 10.1021/acscentsci.8b00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurimoto K. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res. 2006;34(5) doi: 10.1093/nar/gkl050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kurimoto K. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protoc. 2007;2(3):739–752. doi: 10.1038/nprot.2007.79. [DOI] [PubMed] [Google Scholar]
- 53.Tang F. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6(5):468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang F. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 55.Islam S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21(7):1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramskold D. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30(8):777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Picelli S. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10(11):1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 58.Islam S. Highly multiplexed and strand-specific single-cell RNA 5' end sequencing. Nat Protoc. 2012;7(5):813–828. doi: 10.1038/nprot.2012.022. [DOI] [PubMed] [Google Scholar]
- 59.Hayashi T. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nat Commun. 2018;9(1):619. doi: 10.1038/s41467-018-02866-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Picelli S. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9(1):171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 61.Wu L. Full-length single-cell RNA-seq applied to a viral human cancer: applications to HPV expression and splicing analysis in HeLa S3 cells. GigaScience. 2015;4:51. doi: 10.1186/s13742-015-0091-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karlsson K., Linnarsson S. Single-cell mRNA isoform diversity in the mouse brain. BMC Genomics. 2017;18(1):126. doi: 10.1186/s12864-017-3528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durruthy-Durruthy R., Ray M. Using fluidigm C1 to generate single-cell full-length cDNA libraries for mRNA sequencing. Methods Mol Biol. 2018;1706:199–221. doi: 10.1007/978-1-4939-7471-9_11. [DOI] [PubMed] [Google Scholar]
- 64.Sen R. Single-cell RNA sequencing of glioblastoma cells. Methods Mol Biol. 2018;1741:151–170. doi: 10.1007/978-1-4939-7659-1_12. [DOI] [PubMed] [Google Scholar]
- 65.Byrne A. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat Commun. 2017;8:16027. doi: 10.1038/ncomms16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Volden R. Improving nanopore read accuracy with the R2C2 method enables the sequencing of highly multiplexed full-length single-cell cDNA. Proc Natl Acad Sci U S A. 2018;115(39):9726–9731. doi: 10.1073/pnas.1806447115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ranum P.T. Insights into the biology of hearing and deafness revealed by single-cell RNA sequencing. Cell Rep. 2019;26(11) doi: 10.1016/j.celrep.2019.02.053. 3160–3171 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ntranos V. A discriminative learning approach to differential expression analysis for single-cell RNA-seq. Nat Methods. 2019;16(2):163–166. doi: 10.1038/s41592-018-0303-9. [DOI] [PubMed] [Google Scholar]
- 69.Vu T.N. Isoform-level gene expression patterns in single-cell RNA-sequencing data. Bioinformatics. 2018;34(14):2392–2400. doi: 10.1093/bioinformatics/bty100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buettner F. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33(2):155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 71.Trapnell C. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marinov G.K. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res. 2014;24(3):496–510. doi: 10.1101/gr.161034.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Faigenbloom L. Regulation of alternative splicing at the single-cell level. Mol Syst Biol. 2015;11(12):845. doi: 10.15252/msb.20156278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Y. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 2018;28(9):879–896. doi: 10.1038/s41422-018-0074-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brooks A.N. Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res. 2011;21(2):193–202. doi: 10.1101/gr.108662.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Katz Y. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7(12):1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang J., Kuo C.C., Chen L. WemIQ: an accurate and robust isoform quantification method for RNA-seq data. Bioinformatics. 2015;31(6):878–885. doi: 10.1093/bioinformatics/btu757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roberts A. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12(3):R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pervouchine D.D., Knowles D.G., Guigo R. Intron-centric estimation of alternative splicing from RNA-seq data. Bioinformatics. 2013;29(2):273–274. doi: 10.1093/bioinformatics/bts678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Trapnell C. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Munoz J.F. Coordinated host-pathogen transcriptional dynamics revealed using sorted subpopulations and single macrophages infected with Candida albicans. Nat Commun. 2019;10(1):1607. doi: 10.1038/s41467-019-09599-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Manipur I., Granata I., Guarracino M.R. Exploiting single-cell RNA sequencing data to link alternative splicing and cancer heterogeneity: A computational approach. Int J Biochem Cell Biol. 2019;108:51–60. doi: 10.1016/j.biocel.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 85.Arzalluz-Luque A., Conesa A. Single-cell RNAseq for the study of isoforms-how is that possible? Genome Biol. 2018;19(1):110. doi: 10.1186/s13059-018-1496-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Linker S.M. Combined single-cell profiling of expression and DNA methylation reveals splicing regulation and heterogeneity. Genome Biol. 2019;20(1):30. doi: 10.1186/s13059-019-1644-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Buenrostro J.D. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109 doi: 10.1002/0471142727.mb2129s109. 21 29 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dobin A. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan L. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20(9):1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 90.Robinson J.T. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Karolchik D., Hinrichs A.S., Kent W.J. The UCSC genome browser. Curr Protoc Hum Genet. 2011 doi: 10.1002/0471142905.hg1806s71. Chapter 18: p. Unit18 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Haeussler M. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019;47(D1):D853–D858. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Garrido-Martin D. ggsashimi: Sashimi plot revised for browser- and annotation-independent splicing visualization. PLoS Comput Biol. 2018;14(8):e1006360. doi: 10.1371/journal.pcbi.1006360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tyler A.D. Evaluation of Oxford nanopore’s MinION sequencing device for microbial whole genome sequencing applications. Sci Rep. 2018;8(1):10931. doi: 10.1038/s41598-018-29334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schischlik F. Mutational landscape of the transcriptome offers putative targets for immunotherapy of myeloproliferative neoplasms. Blood. 2019;134(2):199–210. doi: 10.1182/blood.2019000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kahles A. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell. 2018;34(2) doi: 10.1016/j.ccell.2018.07.001. 211–224 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Seiler M. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 2018;23(1) doi: 10.1016/j.celrep.2018.01.088. 282–296 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jayasinghe R.G. Systematic analysis of splice-site-creating mutations in cancer. Cell Rep. 2018;23(1) doi: 10.1016/j.celrep.2018.03.052. 270–281 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pellagatti A. Impact of spliceosome mutations on RNA splicing in myelodysplasia: dysregulated genes/pathways and clinical associations. Blood. 2018;132(12):1225–1240. doi: 10.1182/blood-2018-04-843771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yoshida K. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 102.Gerlach M.M. Clonogenic versus morphogenic mutations in myeloid neoplasms: chronologic observations in a U2AF1, TET2, CSF3R and JAK2 'co-mutated’ myeloproliferative neoplasm suggest a hierarchical order of mutations and potential predictive value for kinase inhibitor treatment response. Leuk Lymphoma. 2018;59(8):1994–1997. doi: 10.1080/10428194.2017.1406087. [DOI] [PubMed] [Google Scholar]
- 103.Rodriguez-Meira A. Unravelling intratumoral heterogeneity through high-sensitivity single-cell mutational analysis and parallel RNA sequencing. Mol Cell. 2019;73(6) doi: 10.1016/j.molcel.2019.01.009. 1292–1305 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grinfeld J. Classification and personalized prognosis in myeloproliferative neoplasms. N Engl J Med. 2018;379(15):1416–1430. doi: 10.1056/NEJMoa1716614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Giustacchini A. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat Med. 2017;23(6):692–702. doi: 10.1038/nm.4336. [DOI] [PubMed] [Google Scholar]
- 106.Carrelha J. Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature. 2018;554(7690):106–111. doi: 10.1038/nature25455. [DOI] [PubMed] [Google Scholar]
- 107.Han K.Y. SIDR: simultaneous isolation and parallel sequencing of genomic DNA and total RNA from single cells. Genome Res. 2018;28(1):75–87. doi: 10.1101/gr.223263.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hou Y. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26(3):304–319. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Macaulay I.C. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods. 2015;12(6):519–522. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- 110.Wu A.R. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11(1):41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Muller S. Single-cell sequencing maps gene expression to mutational phylogenies in PDGF- and EGF-driven gliomas. Mol Syst Biol. 2016;12(11):889. doi: 10.15252/msb.20166969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Petropoulos S. Single-cell RNA-Seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. 2016;165(4):1012–1026. doi: 10.1016/j.cell.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zheng G.X. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Scialdone A. Resolving early mesoderm diversification through single-cell expression profiling. Nature. 2016;535(7611):289–293. doi: 10.1038/nature18633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chung W. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun. 2017;8:15081. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.