Abstract
Background
The cellular senescence of primary cultured cells is an irreversible process characterized by growth arrest. Restoration of senescence by ginsenosides has not been explored so far. Rg3(S) treatment markedly decreased senescence-associated β-galactosidase activity and intracellular reactive oxygen species levels in senescent human dermal fibroblasts (HDFs). However, the underlying mechanism of this effect of Rg3(S) on the senescent HDFs remains unknown.
Methods
We performed a label-free quantitative proteomics to identify the altered proteins in Rg3(S)-treated senescent HDFs. Upregulated proteins induced by Rg3(S) were validated by real-time polymerase chain reaction and immunoblot analyses.
Results
Finally, 157 human proteins were identified, and variable peroxiredoxin (PRDX) isotypes were highly implicated by network analyses. Among them, the mitochondrial PRDX3 was transcriptionally and translationally increased in response to Rg3(S) treatment in senescent HDFs in a time-dependent manner.
Conclusion
Our proteomic approach provides insights into the partial reversing effect of Rg3 on senescent HDFs through induction of antioxidant enzymes, particularly PRDX3.
Keywords: Ginsenoside Rg3(S), Human dermal fibroblast, Label-free quantitative proteomics, Restoration, Senescence
1. Introduction
Cellular senescence is one of the hallmarks of aging, which is defined as the process of becoming old [1]. During senescence, cells begin to lose their ability to grow and undergo cell division because of the accumulative damage from reactive oxygen species (ROS) that are generated in mitochondria. In response to ROS over a particular threshold, cells pause their cell cycles, which initiates signaling through replicative senescence pathways, such as p16/Rb and p53/p21 [2]. There might be multiple factors that regulate aging; however, the cellular senescence observed in human dermal fibroblasts (HDFs) can be studied to determine possible causes associated with free radical generation stemming from mitochondrial dysfunction. These free radicals exert age-associated aggravation of cellular damage; however, the age-related senescence in cells and tissues can be prevented by intracellular antioxidant systems, such as superoxide dismutase and peroxiredoxins (PRDXs) [3], [4], [5]. To mitigate ROS-mediated oxidative damage involved in aging, resveratrol, a polyphenol natural product, was introduced and has been extensively applied to improve health by enhancing cellular redox capability [6].
Likewise, ginseng saponins have been suggested to protect against oxidative damage related to aging in senescent mice, tissues, and human cultured cells [7], [8], [9]. Ginseng has been cultivated in North America and eastern Asia—mostly in Korea, northeastern China, Bhutan, and eastern Siberia. Ginseng is a species of perennial plants which belong to the Panax genus in the family, Araliaceae. Panax ginseng (Asian ginseng) and Panax quinquefolius (American ginseng) ginsenosides were determined by liquid chromatography–coupled mass spectrometry (MS) for assessing morphology-based variations [10]. Thus, ginseng is known to be an adaptogenic herb. When American ginseng roots are steamed or heated, the ginsenoside profiles are altered, and the content of ginsenosides, Rh2, Rg2, and Rg3(S/R), is increased. In particular, Rg3, which exhibits strong anticancer activity, was significantly increased [11]. Recently, Rg3-enriched red ginseng extract has been shown to have antiplatelet activity, thus resulting in the alleviation of aging-associated diseases [12]. Rg3 is an effective medicinal compound and can be found as two stereoisomers at C-20: 20(S)-Rg3 and 20(R)-Rg3. Furthermore, a diglucosyl moiety at the C-3 hydroxyl of Rg3 can be degraded into monoglucosyl Rh2 and aglyconic protopanaxadiol (PPD) by glycoside hydrolases expressed in human intestinal bacteria [13].
It was reported that Rg3 prevents oxidative stress–induced astrocytic senescence and inhibits senescence of prostate stromal cells by modulating inflammatory cytokines [14], [15]. In addition to the positive effects on normal cells, Rg3 also promotes senescence and apoptosis of gallbladder cancer cells by activating the p53 pathway [16]. In particular, the Rg3(S) and Rg3(R) stereoisomers exerted different antiphotoaging effects on UV-B–irradiated keratinocytes [17]. Although many positive effects of Rg3 have been reported, the preventative effects of Rg3(S) on HDF cell senescence has not been investigated yet. Herein, senescence-associated β-galactosidase (SA-β-gal) activities and expression levels of senescence marker proteins were compared between HDFs treated with 20(S)-stereotypic Rg3, Rh2, aglyconic PPD, and 20(R)-stereotypic Rg3. In addition, we performed MS-based proteomics experiments using a label-free shotgun approach to identify proteins whose levels are significantly affected by treatment with Rg3(S) in senescent HDFs. The functional ginsenoside proteomics experiments provided several candidate proteins that might be involved in the protective mechanisms associated with Rg3(S) treatment of senescent HDFs. These data also suggest that Rg3(S) could be a potential therapeutic for treating ROS-induced aging.
2. Materials and methods
2.1. Cell culture and chemicals
Primary HDFs were purchased from the American Type Culture Collection (ATCC) (Manassas, VA, USA). Culturing of HDFs was conducted according to the ATCC protocols. Cells were maintained at 37°C in a humidified atmosphere with 5% CO2. The ginsenosides Rg3(S), Rg3(R), Rh2(S), and PPD(S) for treating cells were purchased from the Ambo Institute (Seoul, Korea). HDF cells were grown to 70-80% confluency, washed three times with phosphate-buffered saline (PBS), and treated with 10 μM ginsenosides for 24 h.
2.2. Cell proliferation assays
HDF cells treated with different concentrations of ginsenoside for 24 h were used to measure cell proliferation with a Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer's instructions as previously performed [18]. To examine the toxicities of the ginsenosides, young (passage #6) and senescent (passage #17-18) HDF cells were starved in media without fetal bovine serum (FBS) for 4 h before ginsenoside treatment. The concentrations of Rg3(R), Rh2(S), and PPD(S) were fixed at 20 μM, and Rg3(S) was used in the range of 5-160 μM dissolved in culture medium containing 0.1% dimethyl sulfoxide (DMSO), whereas no ginsenoside with the same medium was considered as the control.
2.3. SA-β-gal assays
SA-β-gal activity assay was conducted as previously described [19]. Senescence Cells Histochemical Staining Kit (Sigma-Aldrich, St. Louis, MO, USA) was used to assess senescence in HDFs following the manufacturer's instructions. Test plates were washed twice with PBS, counterstained with Mayers Hematoxylin staining solution (Sigma-Aldrich, St, Louis, MO, USA) for 5 min at room temperature, and washed twice with PBS. HDFs were imaged by phase contrast on a Leica microscope (Leica Microsystems, Rijswijk, the Netherlands) and recorded at a 100× magnification using a digital color camera. Five hundred randomly selected fibroblasts per sample plate were photographed, and the blue-stained fibroblasts were counted and expressed as a percentage of SA-β-gal–stained cells.
2.4. Measurement of ROS
Intracellular ROS levels were measured using the dye 2′,7′-dichlorodihydrofluorescein diacetate (Abcam, Cambridge, MA, USA) as described elsewhere [20]. HDFs were seeded on a 96-well plate and incubated for 24 h. The cells were starved for 4 h and then treated with a ginsenoside that was diluted in PBS at 37°C for 4 h. Cells were then treated with 200 μM hydrogen peroxide (H2O2) at 37°C for 2 h, and the dye was added to a final concentration of 20 μM. ROS levels in each sample were measured in triplicate. The absorbed and emitted oxidation products were measured at 485 (excitation) and 538 nm (emission), respectively, on a microplate reader (SpectraMax M4 Microplate Reader; Molecular Devices, Sunnyvale, CA, USA).
2.5. Immunoblot analyses
Total protein was isolated using radioimmunoprecipitation assay buffer consisting of 1% (v/v) NP40, 0.1% (w/v) sodium dodecyl sulfate (SDS), 100 μg/mL phenylmethylsulfonyl fluoride, 0.5% (w/v) sodium deoxycholate, protease inhibitors, and phosphatase inhibitors in PBS. Protein concentrations were determined using a BCA Protein Assay Kit (Thermo Fisher Scientific, USA) Proteins were then separated on 10% SDS-polyacrylamide gels and transferred to polyvinylidene fluoride (PVDF) membranes. Membranes were incubated in 5% skim milk with tris-buffered saline Tween (TBST) at room temperature for 1 h and incubated overnight with primary antibodies. The following proteins were probed with the primary antibodies: p16, p21, p53, phospho-p53, caveolin-1, Hsp60, PRDX1-3, and β-actin (Santa Cruz Biotechnology, Dallas, TX, USA). Membranes were washed with TBST and incubated with horseradish peroxidase–conjugated antibody for 1 h. Protein signals were detected using an ECL Detection Kit (BIOFACT, Daejeon, Korea). The signal intensities for each protein band were quantified using the densitometry program, Image J. v1.48. Statistical analyses were performed using Student t test. A p values less than 0.05 were considered statistically significant.
2.6. Tube-gel digestion
A polyacrylamide in-gel digestion of tube gel pieces was performed with slight modification as previously described [21]. In brief, 100 μg of freeze-dried protein was completely solubilized with 6 M urea and denatured at 90°C for 20 min. Proteins were incubated with 10 mM EDTA, 4% SDS, 0.2 M ammonium bicarbonate, and 6 M urea at 37°C for 30 min with intermittent vortexing. The extract was reduced with 200 mM tris (2-carboxyethyl) phosphine hydrochloride and alkylated with 55 mM iodoacetamide at room temperature in the dark for 30 min. The subsequent polymerization, tryptic digestion, and extraction of peptides were same conducted as previously described [21].
2.7. Nano-ultra performance liquid chromatography-MS/MS
The overall procedure was followed by using the previous method with slight modification. Whole protein extracts from HDFs that were treated with Rg3(S) for 2, 6, or 12 h were digested as previously described [22]. The peptides were separated using a nanoACQUITY UPLC Chromatography System (Waters Co., Milford, MA; 75 μm × 250 mm, 1.7 μm particle size, BEH300 C18 RP analytical column, and a 180 μm × 20 mm Symmetry C18 RP 5 μm precolumn). The column temperature was maintained at 30°C. Peptides (5 μL) were injected into the column and eluted with a gradient of mobile phases A and B. Mobile phase A contained 3% acetonitrile and 0.1% formic acid in water, and mobile phase B was composed of 97% acetonitrile and 0.1% formic acid in water. The gradient conditions were as follows: 3% B for 25 min, 3-40% B for 95 min, 40-70% B for 20 min at a flow rate of 300 nL/min and 80% B for 10 min as a rinsing step. For the external calibration, [Glu1]-fibrinopeptide (100 fmol/μL) was delivered as the lock mass compound from the auxiliary pump of the UPLC system at 1 μL/min to the reference sprayer of the NanoLockSpray source in the Synapt‘ HDMSE mass spectrometer (Waters Co., Milford, MA).
2.8. Quantitative real-time polymerase chain reaction
Total RNA was extracted from cultured cells with an RNeasy kit (Qiagen, Redwood City, CA, USA). For cDNA synthesis, replicate, quantitative real-time polymerase chain reaction (PCR) was performed with a QuantiTect SYBR Green PCR Kit (Qiagen, Redwood City, CA, USA). All primers were purchased from Bioneer (Daejeon, Korea). The PCR reactions and cycling protocol were performed following the standard protocol provided by Qiagen. Data were collected and analyzed with the 2−ΔΔCt method for quantification to determine relative mRNA expression levels. Oligonucleotide sequences of the primer sets used in this study are listed in Table S1.
2.9. Bioinformatics analyses
Data-independent analysis mode (LC-MSE) data were processed using PLGS v2.31 (Waters Co., Milford, MA, USA). Peptides were searched against the UniProt database to determine the identities of the proteins (http://www.uniprot.org). The precursor mass tolerance and the fragment tolerance were set to 100 ppm and 0.2 Da, respectively. One missed cleavage with trypsin was allowed during protein identification. Cysteine carbamidomethylation (+57 Da) and methionine oxidation (+16 Da) were chosen as fixed and variable modifications, respectively. The false discovery rates of the peptides identified in PLGS were within 5%. The time-aligned LC-MSE precursor ion masses and retention times were within ±10 ppm and ±30 s, respectively. Proteins were considered identified if at least two peptides were assigned to them. The exact mass retention time signature table was constructed using Expression v2 in PLGS to generate the peptide and protein information, including peptide masses and retention times. MS/MS spectra from the data-independent analysis mode were converted into raw files and pkl-files using PLGS software. The combined .pkl files were interpreted using PLGS v2.31 and MASCOT v2.2. For protein network analyses, Ingenuity Pathway Analysis (Ingenuity Systems Inc., Redwood City, CA, USA) was used to predict the functions and pathways associated with the significantly identified proteins. Relative protein expression levels were described as the fold change at a given time point between untreated cells and those treated with Rg3(S).
3. Results and discussion
3.1. Rg3(S) restores cellular senescence of HDFs
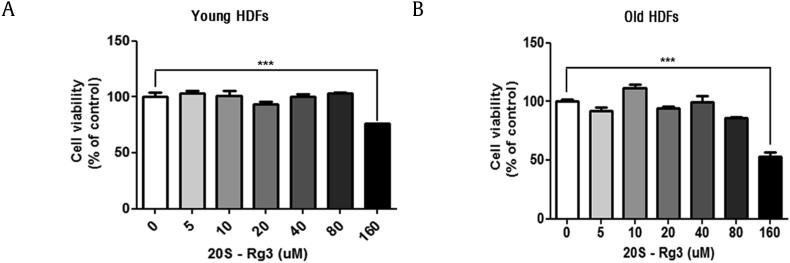
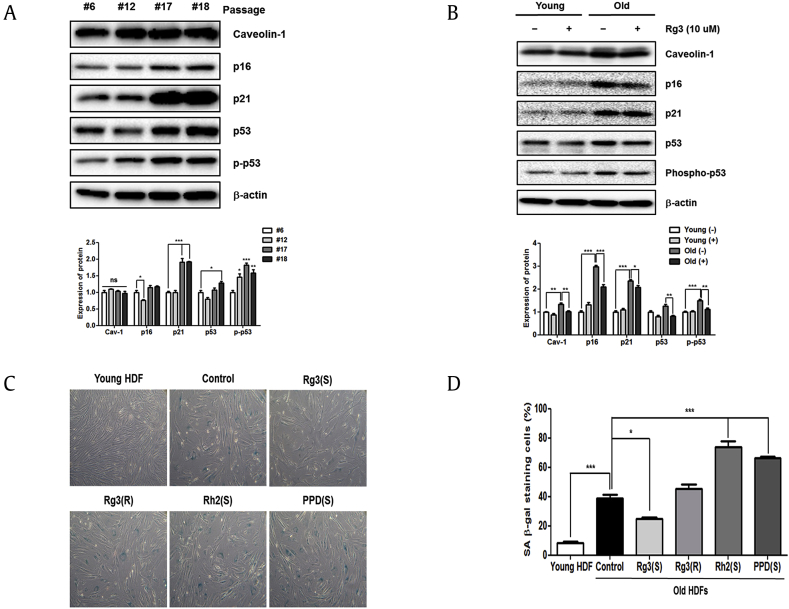
To determine the optimal concentration of Rg3(S) for treating HDFs, we first examined cell viability rates of young and old cells by MTT assays. Cell cytotoxicity of Rg3(S) was examined on passage #6 (young) and #17 (old) HDFs using 5-160 μM Rg3(S) for 24 h. In both young and old HDFs, no cytotoxicity was observed for concentrations up to 80 μM. However, the high dose of 160 μM Rg3(S) decreased the cell viabilities to 25% and 44% in young and old HDFs, respectively (Fig. 1). Contrary to the treatment of 160 μM Rg3(S), the treatment of 10 μM Rg3(S) in old HDFs showed the highest cell viability. Thus, the concentration of Rg3(S) was fixed at 10 μM Rg3(S). Next, to observe the possible replicative senescence, HDF cells derived from ATCC were continuously cultured by changing the culture medium from passage #6 to #18. In general, p16/RB and p53/p21 are considered the classical replicative senescence pathways that are activated in response to external stressors [2]. In addition, senescent cells can undergo caveolin-1–mediated cell cycle arrest through a p53/p21-dependent pathway [23]. The immunoblot analyses revealed increased levels of p16, p21, and p53 in passage #17 and #18 cells (Fig. 2A). Quantification of the relative intensities indicated that the expression level of p21 in passage #17 HDFs was approximately twofold higher than the levels observed in passage #6 HDFs at p < 0.001 (Fig. 2A). In addition, the expression levels of p53, and p-p53 in passage #18 cells were increased 1.3- (p < 0.05) and 1.7-fold (p < 0.01), respectively, compared with the levels observed for passage #6 cells. To clarify whether Rg3(S) affects HDF cell senescence, the levels of senescence markers were compared between untreated cells and those treated with Rg3(S) for both young and old HDFs. The levels of all the senescence proteins, such as caveolin-1, p16, and p53/p21, were decreased at least at p < 0.01 in Rg3(S)-pretreated old HDFs, whereas the overall expression levels of senescence proteins were not significantly altered in Rg3(S)-pretreated young HDFs (Fig. 2B). Quantitation of the immunoblots revealed significant decreases in p16, p21, and p-p53 expression levels in old HDFs treated with Rg3(S). This result suggests that Rg3(S) partially restores senescence through both p53/p21- and p16-dependent pathways, similar to previous studies showing inhibition of cellular senescence in vitro and in vivo by Rg1 [24], [25].
Fig. 1.
Cell cytotoxicity of Rg3(S) in HDFs. No cytotoxicity was observed in (A) young and (B) old HDFs treated with up to 80 μM of Rg3(S). Partial cytotoxicity was observed at 160 μM of Rg3(S). HDFs were pretreated with Rg3(S) at the given concentration for 24 h at 37°C. Rg3(S) was diluted in DMSO (0.1%, v/v), and DMSO alone was used as a control. Cell viability was measured following the CCK-8 protocol.
HDFs, human dermal fibroblasts.
Fig. 2.
Immunoblotting of senescence proteins and SA-β-gal staining of Rg3(S)-treated senescent HDFs. (A) Immunoblot images of senescence marker expression in successively passaged HDFs. (B) Rg3(S)-treated young (passage #6) and old (passage #17) HDFs. HDFs were pretreated with 10 μM Rg3(S) for 24 h at 37°C. Rg3(S) was diluted in DMSO (0.1%, v/v), and DMSO alone was used as a control. β-Actin was used as a loading control. (C) SA-β-gal staining of senescent HDFs pretreated with Rg3(S). (D) Quantification of SA-β-gal staining of Rg3(S)-treated old HDFs. Young (passage #6) and old (passage #17) HDFs were incubated with 10 μM of the PPD-type ginsenosides, Rg3(S), Rg3(R), Rh2(S) and PPD(S), for 24 h. A representative image of SA-β-gal stained cells is shown. The percentages of SA-β-gal–positive cells were calculated by SABIA (ebiogen, Seoul, Korea) using optical microscopy images. Data represent the mean ± SD of three independent experiments. ∗∗Statistical significant of p < 0.01.
HDFs, human dermal fibroblasts; PPD, protopanaxadiol; SA-β-gal, senescence-associated β-galactosidase; SD, standard deviation.
We examined the possible roles of Rg3(S) and other Rg3-derived ginsenosides—Rg3(R) (Rg3(S) enantiomer), Rh2(S) (monoglucosyl form of PPD(S)), and PPD(S) (aglycone form of Rg3(S))—in the senescent HDFs by SA-β-gal staining. Rg3(S) exhibited the lowest level of SA-β-gal staining among the tested compounds (Fig. 2C and D). In particular, quantification of SA-β-gal–stained cells revealed that young and old HDFs were 8% and 40% positive for β-galactosidase, respectively. However, cell staining of old HDFs treated with Rg3(S) was significantly decreased to 25%, at p < 0.05 which is partway between the phenotypes observed for the young and old senescent cells. However, 75% of old HDFs treated with Rh2(S) were stained, which was a 1.9-fold increase compared with control old HDFs. Thus, the structure–function relationship of PPD-type ginsenoside restoration against senescence suggests that the diglucosyl moiety at the C-3 hydroxyl and the stereoisomer (S) type at C-20 of Rg3 play key roles in restoring partially the senescence process of HDFs. Stereospecific effects of Rg3 were previously reported; Rg3(R) was suggested to be a stronger antioxidant and illicit a stronger immune response [26]. In contrast to Rg3(R), the 20(S) form of Rg3 has a greater solubility, is much less toxic, and has antidiabetic activity [27], [28]. These results are in good agreement with the observation that 20(S)-Rg3, not 20(R)-Rg3, decreased UV-B–induced intracellular ROS levels in keratinocytes [17].
3.2. Specific decrease of ROS production in HDFs by Rg3(S)
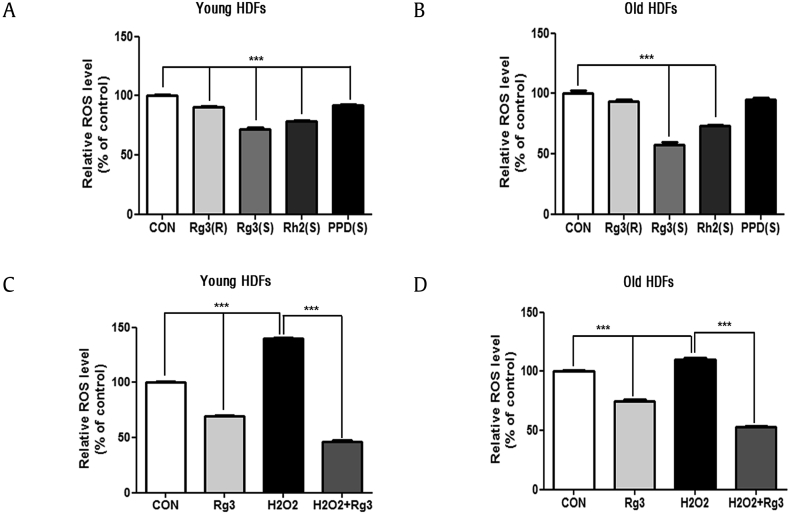
As ROS can induce oxidative stress and promote the cellular senescence, we evaluated whether Rg3(S) affects the production of ROS in HDFs. ROS levels in HDFs were measured by dichlorofluorescein to compare the effects of the PPD-type ginsenosides. Rg3(S) significantly decreased intact ROS levels of young and old HDFs by up to 45% and 55% at p < 0.001, respectively, compared with the control HDFs (Fig. 3A and B). Rh2(S) decreased ROS levels to approximately 30% in both young and old HDFs. However, neither Rg3(R) nor PPD(S) affected ROS levels. Thus, Rg3(S) was the most effective ROS suppressor. These results corroborated our SA-β-gal staining results (Fig. 2C and D). Next, we examined whether Rg3(S) could attenuate H2O2-induced ROS production externally by measuring the scavenging activity of Rg3(S) in young and old HDFs cotreated with H2O2 and Rg3(S). H2O2 significantly increased the ROS signal to 40% in young HDFs, whereas the signal was slightly increased to 10% in old HDFs (Fig. 3C and D). Thus, young HDFs appears to be more sensitive to H2O2 than old HDFs. Interestingly, co-treatment of Rg3(S) and H2O2 decreased ROS signal dramatically to 60% and 50% in young and old cells, respectively, compared with singly H2O2-treated cells. These data suggest that Rg3(S) lowers ROS levels and could be a possible therapeutic for treating oxidative stress-induced senescence. These results closely agree with a previous report suggesting that the ginsenosides, Rh2 and Rg3, inhibit cell proliferation and induce cell apoptosis by increasing mitochondrial ROS levels in human leukemia Jurkat cells [29]. Thus, the decreased SA-β-gal staining and ROS levels by Rg3(S) are consistent with the conclusion that Rg3(S) attenuates H2O2-induced ROS production in HDFs in a stereo-specific manner. Recently, the attenuation of H2O2-mediated oxidative stress by treatment with Rg3 was reported for HepG2 human hepatocarcinoma cells and in an in vitro sepsis model [30], [31].
Fig. 3.
Native and H2O2-induced ROS production in human dermal fibroblasts (HDFs) treated with the indicated PPD-type ginsenosides. (A) Young (passage #6) and (B) old (passage #17) HDFs were pre-incubated with 10 μM of the PPD-type ginsenosides, Rg3(S), Rg3(R), Rh2(S) and PPD(S), for 24 h. (C) Young (passage #6) and (D) old (passage #17) HDFs were incubated with 10 μM of Rg3(S) for 12 h. Cells were stained with dichlorofluorescein diacetate, fixed, and immediately analyzed using a multianalytic validation system (SpectraMax M4). The data represent the mean ± SD of three independent experiments. ∗∗Statistical significant of p < 0.01.
H2O2, hydrogen peroxide; PPD, protopanaxadiol; ROS, reactive oxygen species; SD, standard deviation.
3.3. Proteomic alteration of old HDFs by Rg3(S)
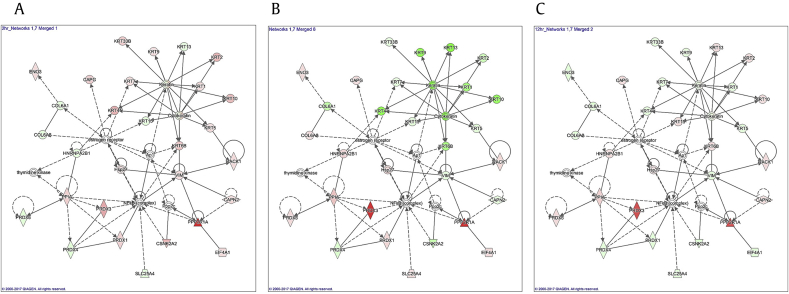
To investigate protein expression levels during an Rg3(S)-treatment time course, senescent HDFs were analyzed by nano-UPLC-QTOF tandem MS using data-independent MSE technology as previously described [32]. Of the original 1327 peptides and 599 proteins observed in the first identification step, 157 human proteins were identified from assignment of two more peptides among the triplicate experiments. The average ratios of 2-h, 6-h, and 12-h treatment of Rg3(S) versus the untreated group were 1.1-, 2.3-, and 1.8-fold, respectively. Thus, the 6-h treatment of Rg3(S) in senescent HDFs revealed the most dramatic differences in the expression of individual proteins. A list of the identified whole proteins is presented in Table S1. Significantly different levels of proteins at p-value < 0.05 were identified: 78 (2 h vs. 0 h), 100 (6 h vs. 0 h), and 60 (12 h vs. 0 h). After 6 h of Rg3(S) treatment, 40 significantly upregulated and 16 significantly downregulated proteins with more than a 1.5-fold difference in level were identified. The whole proteins that were different at each time point were subjected to protein network analyses using Ingenuity Pathway Analysis. The main subnetwork 1 containing PRDX3 belonged to subnetwork 7 was displayed in a time-course pattern of Rg3(S) treatment (Fig. 4). As shown in the protein networks, keratin cluster proteins (Krt-1, -2, -4, -5, -6B, -9, -10, -13, -15, -33B) within subnetwork 1 appeared to fluctuate in expression but were mostly downregulated at 6 h of Rg3(S) treatment. These findings are in good agreement with a previous study suggesting that the ginsenoside, F2, downregulates keratins, such as Krt2, Krt10, Krt16, in human gastric carcinoma cells [33]. It was suggested that de novo synthesis of Rg3(S)-responsive regulatory proteins was triggered and caused the downregulation of structural proteins, such as keratins, in senescent HDFs.
Fig. 4.
Protein networks of Rg3(S)-treated senescent human dermal fibroblasts (HDFs). The directly interacting networks of proteins from old HDFs treated with 10 μM of Rg3(S) for (A) 2 h, (B) 6 h, and (C) 12 h were determined by Ingenuity Pathway Analysis. At each time point, networks 1 (main subnetwork) and 7 (peroxyredoxin-3–containing subnetwork) were merged to determine the molecular functional group. Red and green symbols indicate the proteins that are upregulated and downregulated, respectively, after Rg3(S) treatment.
The relative abundancies of Rg3(S)-induced core proteins with significantly different expressions are listed in Table 1. PPP2R1A (serine/threonine protein phosphatase 2A) levels were the most affected by Rg3(S), which was increased by 2.7-, 26.3-, and 11.9-fold at 2, 6, and 12 h, respectively. PPP2R1A interacts with VIM, CAPN2, and EIF4A1 and is also suggested to connect with the core of the NF-κB complex. Upregulation of PPP2R1A by Rg3(S) was consistent with the previous observation that protein phosphatase A2 inactivation promotes the accumulation of NF-κB–dependent oxidative stress and upregulation of inflammatory processes during aging [34]. We focused on the marginal network proteins containing PRDX isotypes and PPIA (peptidyl-prolyl cis-trans isomerase A) because the mitochondrial peroxiredoxin, PRDX3, was dramatically increased by 26.3-fold and PPIA was increased by 2.3-fold after 6 h of treatment with Rg3(S). Interestingly, our finding is quite consistent with a previous report suggesting that downregulation of PRDX3 induces cellular senescence of human trophoblasts [35]. Thus, it is notable that Rg3(S) can induce expression of ROS-scavenging antioxidant enzymes in mitochondria to prevent the progression of senescence in old HDFs.
Table 1.
Relative abundance of core proteins with the significantly differential protein expression in Rg3(S)-pretreated senescent HDFs
| Protein name | Symbol | UnitProt | Peptide count | Ratioa |
p Valueb |
||||
|---|---|---|---|---|---|---|---|---|---|
| 2 h/0 h | 6 h/0 h | 12 h/0 h | 2 h/0 h | 6 h/0 h | 12 h/0 h | ||||
| Serine/threonine protein phosphatase 2A | PPP2R1A | P30153 | 2 | +5.73 | +26.25 | +11.9 | 0.15 | 0.00 | 0.06 |
| Peroxiredoxin-3, mitochondrial | PRDX3 | P30048 | 3 | +1.88 | +22.52 | +11.59 | 0.59 | 0.02 | 0.10 |
| Peptidyl-prolyl cis-trans isomerase A | PPIA | P62937 | 5 | +1.41 | +2.26 | +1.83 | 0.09 | 0.00 | 0.01 |
| Heat shock protein β-1 (Hsp27) | HSPB1 | P04792 | 4 | +1.02 | +1.85 | +1.55 | 0.72 | 0.00 | 0.00 |
| Heterogeneous nuclear ribonucleoproteins A2/B1 | HNRNPA2B1 | P22626 | 6 | −1.10 | +1.82 | +1.45 | 0.61 | 0.00 | 0.38 |
| Peroxiredoxin-1 | PRDX1 | Q06830 | 6 | +1.38 | +1.71 | −1.06 | 0.12 | 0.01 | 0.88 |
| Receptor of activated protein C kinase 1 | RACK1 | P63244 | 6 | +1.13 | +1.69 | +1.26 | 0.05 | 0.01 | 0.18 |
| Peroxiredoxin-6 | PRDX6 | P30041 | 7 | −1.16 | +1.63 | +1.33 | 0.05 | 0.03 | 0.11 |
| Casein kinase II subunit α′ | CSNK2A2 | P19784 | 5 | +1.73 | −1.69 | −1.75 | 0.00 | 0.00 | 0.04 |
| Collagen α-1(VI) chain | COL6A1 | P12109 | 11 | −2.33 | −1.75 | −1.72 | 0.00 | 0.00 | 0.01 |
| Keratin type II, cytoskeletal 6B | KRT6B | P48669 | 10 | +1.55 | −2.27 | +1.04 | 0.01 | 0.00 | 0.40 |
| Keratin type II, cytoskeletal 1 | KRT1 | P04264 | 33 | +1.20 | −2.38 | +1.04 | 0.01 | 0.00 | 0.40 |
| Keratin type I, cytoskeletal 10 | KRT10 | P13645 | 29 | +1.64 | −2.94 | +1.05 | 0.00 | 0.00 | 0.57 |
| Keratin type II, cytoskeletal 4 | KRT4 | P19013 | 6 | +1.10 | −3.33 | −1.37 | 0.30 | 0.00 | 0.00 |
| Keratin type I, cytoskeletal 9 | KRT9 | P35527 | 19 | +1.26 | −3.33 | −1.33 | 0.02 | 0.00 | 0.30 |
| Keratin type I, cytoskeletal 13 | KRT13 | P13646 | 5 | −1.03 | −3.70 | +1.03 | 0.90 | 0.00 | 0.93 |
Ratio measured by MSE methods. Sign (+) and (−) represent the fold change of upregulation and downregulation, respectively.
P-value calculated from individual peak intensities from all tryptic peptides detected per protein.
3.4. Validation of aging-related proteins in HDF cells treated with Rg3(S)
From the label-free, comparative proteomic analyses, PRDXs, such as PRDX1, PRDX3, PRDX4, and PRDX6, were identified throughout 2–12 h treatment of Rg3(S) in senescent HDFs. The expression level of mitochondrial PRDX3 was dramatically increased by 22.5-fold after 6 h of treatment with Rg3(S), whereas that of the other PRDXs only fluctuated or were slightly downregulated (Table S1). As the protein network in Fig. 4 shows, cytosolic PRDX1 was a highly abundant antioxidant enzyme, which can interact with the secretory form of PRDX4 [36]. PRDX3 is located in the mitochondrial matrix where it regulates H2O2 levels and subsequent signaling processes [37]. PRDXs are ROS-scavenging antioxidant enzymes that are considered to protect against aging. However, few proteomic analyses of the effect of mitochondrial-type PRDX3 on aging in HDFs have been studied.
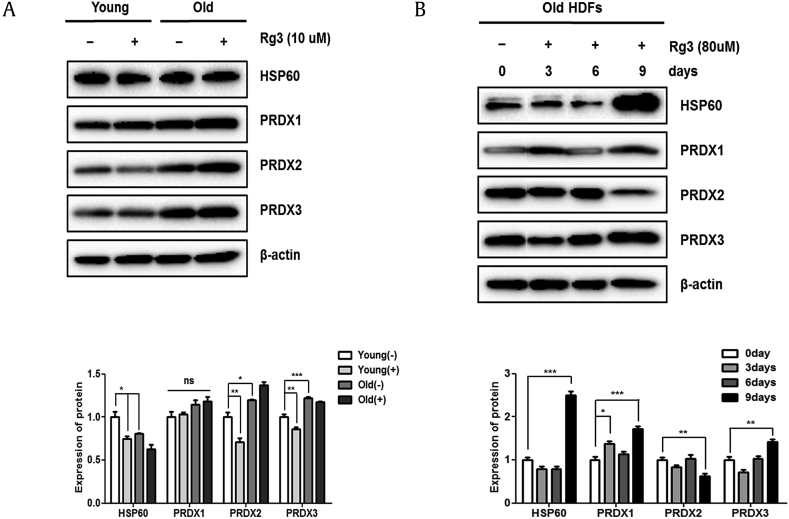
To validate the Rg3(S)-dependent alterations in PRDX isoform and PPIA levels identified by the proteomic analyses, quantitative real-time PCR was performed using a gene-specific primer set. Overall, mRNA expression patterns were similar to those observed in the proteomic analyses, except for PRDX4 (Table S3). PRDX1 and PRDX3 manifested very similar expression patterns, and the changes were statistically significant. These discrepancies between mRNA levels and protein abundances can be attributed to different transcriptional and translational regulations [38]. The abundances of PRDX isoforms were measured by immunoblotting whole cell lysates of senescent HDFs treated with Rg3(S) between 24 h and 9 days. As shown in Fig. 5A, differences in expression levels of PRDXs between young and old HDFs were observed only for PRDX2 and PRDX3. In particular, the protein expression levels of PRDX3 in HDFs treated with 80 μM Rg3(S) for 9 days increased by 40%, as shown in Fig. 5B. The expression levels of PRDX1 also increased by 80% in that the same condition, but the overall abundance of PRDX1 was lower than that of PRDX3. PRDX2—the supplementary form—decreased after 9 days of treatment with 80 μM Rg3(S). Therefore, it seems to be regulated differently by Rg3(S) during the cellular senescence of HDFs. No differences were observed in Hsp60 abundance but were observed between untreated senescent HDFs and those treated with Rg3(S). However, when HDFs were treated with the cell-tolerant concentration of 80 μM Rg3(S) for 9 days, a dramatic increase in Hsp60 level was observed. This result is similar to previous studies that showed mitochondrial Hsp60 increased with age in mollusks and serum Hsp60 increased in aged individuals [39], [40]. It is notable that oxidative stress induced a decrease in mitochondrial Hsp60 levels and a concomitant increase in cytosolic Hsp60 in young HDFs but not in old cells [41]. It remains to be clarified the possible translocation of Hsp60 and the protective mechanism of Rg3(S) in senescent HDFs later. We have also observed that Caenorhabditis elegans that fed on Rg3(S) had longer life spans (data not shown). Similarly, there were several reports that suggested PRDXs are involved in life span extension. Knockout mice with PRDX1 showed shortened life spans, and PRDX2 silencing decreased the life span of C. elegans [42], [43]. In addition, protein levels of mitochondrial PRDX3 were shown to be reduced during aging process [44]. Thus, Rg3(S) appears to induce mitochondrial PRDX expression to prevent further aging in HDFs and partly restores their functions to those observed in younger HDFs. Taken together, PRDX3 could be a key player in Rg3(S)-induced partial restoration of cellular senescence in HDFs. This systematic study of ginsenoside biochemistry combined with functional proteomics will provide valuable insights into understanding senescence with respect to prevention and the possible therapeutic application of ginseng saponins.
Fig. 5.
Immunoblotting of Rg3(S)-induced proteins in human dermal fibroblasts (HDFs). (A) Young (passage #6) and old (passage #17) HDFs were pretreated with 10 μM Rg3(S) for 24 h. (B) Old HDFs were incubated with a high dose of Rg3(S) (80 μM) for 3 to 9 days. At each time point, cells were collected, and the expression levels of Hsp60, PRDX, PRDX2, and PRDX3 were analyzed by immunoblotting. To compare relative expression levels, β-actin was used as loading control. Quantification of protein expression levels was performed using ImageJ. Statistical significance: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
PRDX, peroxiredoxin.
Conflicts of interest
All authors have no conflicts of interest to declare.
Acknowledgements
The present study was supported by the Korea Basic Science Institute research grant (C39712) awarded to J.S. Choi and 2017R1D1A1B03034936. Also, this work was supported by Innopolis Foundation through Technology Commercialization services, funded by Ministry of Science and ICT (2019JBRD0004) to I.S. Jang. This work was partly supported by the Korean Government (MEST; 2015K1A1A2028365, 2015M3A9C 4076321) through the Brain Korea 21 Plus Project, Republic of Korea, which was awarded to H.J. Kwon.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.jgr.2018.07.008.
Contributor Information
Ho Jeong Kwon, Email: kwonhj@yonsei.ac.kr.
Jong-Soon Choi, Email: jschoi@kbsi.re.kr.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
List of identified proteins in Rg3(S)-treated senescent human dermal fibroblasts (HDFs). Here, the ratios of 2 h or 6 h or 12 h vs. 0 h were measured by MSE methods.
DNA sequences of target genes for quantitative RT-PCR.
Comparisons of mRNA and proteome levels in Rg3(S)-treated senescent human dermal fibroblasts (HDFs) for 6 h. Relative expression ratios (6 h/0 h) were calculated for the 10 μM of Rg3(S)-treated group by dividing by the levels observed for the control. Statistical significance: (*) P < 0.05, (**) P < 0.01, ns, not significant.
References
- 1.Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salama R., Sadaie M., Hoare M., Narita M. Cellular senescence and its effector programs. Genes Dev. 2014;28:99–114. doi: 10.1101/gad.235184.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe K., Shibuya S., Koyama H., Ozawa Y., Toda T., Yokote K., Shimizu T. Sod1 loss induces intrinsic superoxide accumulation leading to p53-mediated growth arrest and apoptosis. Int J Mol Sci. 2013;14:10998–11010. doi: 10.3390/ijms140610998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neumann C.A., Krause D.S., Carman C.V., Das S., Dubey D.P., Abraham J.L., Bronson R.T., Fujiwara Y., Orkin S.H., Van Etten R.A. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defense and tumour suppression. Nature. 2003;424:561–565. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- 5.Han Y.H., Kim H.S., Kim J.M., Kim S.K., Yu D.Y., Moon E.Y. Inhibitory role of peroxiredoxin II (Prdx II) on cellular senescence. FEBS Lett. 2005;579:4897–4902. doi: 10.1016/j.febslet.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 6.Pandey K.B., Rizvi S.I. Resveratrol up-regulates the erythrocyte plasma membrane redox system and mitigates oxidation-induced alterations in erythrocytes during aging in humans. Rejuvenation Res. 2013;16:232–240. doi: 10.1089/rej.2013.1419. [DOI] [PubMed] [Google Scholar]
- 7.Yokozawa T., Satoh A., Cho E.J. Ginsenoside-Rd attenuates oxidative damage related to aging in senescence-accelerated mice. J Pharm Pharmacol. 2004;56:107–113. doi: 10.1211/0022357022449. [DOI] [PubMed] [Google Scholar]
- 8.Zhao H.F., Li Q., Li Y. Long-term ginsenoside administration prevents memory loss in aged female C57BL/6J mice by modulating the redox status and up-regulating the plasticity-related proteins in hippocampus. Neuroscience. 2011;183:189–202. doi: 10.1016/j.neuroscience.2011.03.048. [DOI] [PubMed] [Google Scholar]
- 9.Oh S.J., Oh Y., Rhu I.W., Kim K., Lim C.J. Protective properties of ginsenoside Rb3 against UV-B radiation-induced oxidative stress in HaCaT keratinocytes. Biosci Biotechnol Biochem. 2015;80:95–103. doi: 10.1080/09168451.2015.1075862. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y., Zhao Z., Chen H., Brand E., Yi T., Qin M., Liang Z. Determination of ginsenosides in Asian and American ginsengs by liquid chromatography-quadrupole/time-of-flight MS: assessing variations based on morphological characteristics. J Ginseng Res. 2017;41:10–22. doi: 10.1016/j.jgr.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C.Z., Aung H.H., Zhang B., Sun S., Li X.L., He H., Xie J.T., He T.C., Du W., Yuan C.S. Chemopreventive effects of heat-processed Panax quinquefolius root on human breast cancer cells. Anticancer Res. 2008;28:2545–2551. [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong D., Irfan M., Kim S.D., Kim S., Oh J.H., Park C.K., Kim H.K., Rhee M.H. Ginsenoside Rg3-enriched Red Ginseng Extract Inhibits Platelet Activation and in vivo thrombus formation. J Ginseng Res. 2017;41:548–555. doi: 10.1016/j.jgr.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H., Li Z., Zhou Z., Yang H., Zhong Z., Lou C. Antidepressant-like effects of ginsenosides: a comparison of ginsenoside Rb3 and its four deglycosylated derivatives, Rg3, Rh2, compound K, and 20-protopanaxadiol in mice models of despair. Pharmacol Biochem Behav. 2016;140:17–26. doi: 10.1016/j.pbb.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 14.Hou J., Kim S., Sung C., Choi C. Ginsenoside Rg3 prevents oxidative stress-induced astrocytic senescence and ameliorates senescence paracrine effects on glioblastoma. Molecules. 2017;22:E1516. doi: 10.3390/molecules22091516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng Y., Zhang R., Kong L., Shen Y., Xu D., Zheng F., Liu J., Wu Q., Jia B., Zhang J. Ginsenoside Rg3 inhibits the senescence of prostate stromal cells through down-regulation of interleukin 8 expression. Oncotarget. 2017;8:64779–64792. doi: 10.18632/oncotarget.17616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang F., Li M., Wu X., Hu Y., Cao Y., Wang X., Xiang S., Li H., Jiang L., Tan Z. 20(S)-ginsenoside Rg3 promotes senescence and apoptosis in gallbladder cancer cells via the p53 pathway. Drug Des Devel Ther. 2015;9:3969–3987. doi: 10.2147/DDDT.S84527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim C.J., Choi W.Y., Jung H.J. Stereoselective skin anti-photoaging propertied of ginsenoside Rg3 in UV-B-irradiated keratinocytes. Biol Pharm Bull. 2014;37:1583–1590. doi: 10.1248/bpb.b14-00167. [DOI] [PubMed] [Google Scholar]
- 18.Sohn E.J., Kim J.M., Kang S.H., Kwon J., An H.J., Sung J.S., Cho K.A., Jang I.S., Choi J.S. Restoring effects of natural anti-oxidant quercetin on cellular senescent human dermal fibroblasts. Am J Chin Med. 2018;46:853–873. doi: 10.1142/S0192415X18500453. [DOI] [PubMed] [Google Scholar]
- 19.Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O. A biomarker that identified senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eruslanov E., Kusmartsev S. Identification of ROS using oxidized DCFDA and flow-cytometry. Meth Mol Biol. 2010;594:57–72. doi: 10.1007/978-1-60761-411-1_4. [DOI] [PubMed] [Google Scholar]
- 21.Yu H., Wakim B., Li M., Halligan B., Tint G.S., Patel S.B. Quantifying rat proteins in neonatal mouse brain by ‘tube-gel’ protein digestion label-free shotgun proteomics. Proteome Sci. 2007;5:17. doi: 10.1186/1477-5956-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon J., Park S.H., Park C., Kwon S.O., Choi J.S. Analysis of membrane proteome by data-dependent LC-MS/MS combined with data-independent LC-MSE technique. J Anal Sci Tech. 2010;1:78–85. [Google Scholar]
- 23.Galbiati F., Volonte D., Liu J., Capozza F., Frank P.G., Zhu I., Pestell R.G., Lisanti M.P. Caveolin-1 expression negatively regulates cell cycle progression by inducing G0/G1 arrest via a p53/p21WAF1/Cip1-dependent mechanism. Mol Biol Cell. 2001;12:2229–2244. doi: 10.1091/mbc.12.8.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S.G., Yan M.Z., Zhang D., Ye M., Deng J.J. Effects of ginsenoside Rg1 on the senescence of vascular smooth muscle cells. Genet Mol Res. 2016;15:15038409. doi: 10.4238/gmr.15038409. [DOI] [PubMed] [Google Scholar]
- 25.Chen C., Mu X.Y., Zhou Y., Shun K., Geng S., Liu J., Wang J.W., Chen J., Li T.Y., Wang Y.P. Ginsenoside Rg1 enhances the resistance of hematopoietic stem/progenitor cells to radiation-induced aging in mice. Acta Pharmacol Sin. 2014;35:143–150. doi: 10.1038/aps.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei X., Su F., Su X., Hu T., Hu S. Stereospecific antioxidant effects of ginsenoside Rg3 on oxidative stress induced by cyclophosphamide in mice. Fitoterapia. 2012;83:636–642. doi: 10.1016/j.fitote.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Park M.W., Ha J., Chung S.H. 20(S)-ginsenoside Rg3 enhances glucose-stimulated insulin secretion and activates AMPK. Biol Pharm Bull. 2008;31:748–751. doi: 10.1248/bpb.31.748. [DOI] [PubMed] [Google Scholar]
- 28.Liu J.P., Lu D., Nicholson R.C., Zao W.J., Li P.Y., Wang F. Toxicity of a novel anti-tumor agent 20(S)-ginsenoside Rg3 via CHOP-mediated DR5 upregulation in human hepatocellular carcinoma cells. Mol Cancer Ther. 2012;12:274–285. doi: 10.1158/1535-7163.MCT-12-0054. [DOI] [PubMed] [Google Scholar]
- 29.Xia T., Wang Y.N., Zhou C.X., Wu L.M., Liu Y., Zeng Q.H., Zhang X.L., Yao J.H., Wang M., Fang J.P. Ginsenoside Rh2 and Rg3 inhibit cell proliferation and induce apoptosis by increasing mitochondrial reactive oxygen species in human leukemia Jurkat cells. Mol Med Rep. 2017;15:3591–3598. doi: 10.3892/mmr.2017.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bak M.J., Jeong W.S., Kim K.B. Detoxifying effect of fermented black ginseng on H2O2-induced oxidative stress in HepG2 cells. Int J Mol Med. 2014;34:1516–1522. doi: 10.3892/ijmm.2014.1972. [DOI] [PubMed] [Google Scholar]
- 31.Xing W., Yang L., Peng Y., Wang Q., Gao M., Yang M., Xiao X. Ginsenoside Rg3 attenuates sepsis-induced injury and mitochondrial dysfunction in liver via AMPL-mediated autophagy flux. Biosci Rep. 2017;39:20170934. doi: 10.1042/BSR20170934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moon Y.J., Kwon J., Yun S.H., Lim H.L., Kim M.S., Kang S.G., Lee J.H., Choi J.S., Kim S.I., Chung Y.H. Proteome analyses of hydrogen-producing hyperthermophilic archaeon Thermococcus onnurineus NA1 in different one-carbon substrate culture conditions. Mol Cell Proteomics. 2012;11:M111. doi: 10.1074/mcp.M111.015420. 015420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao Q., Zhang P.H., Yang J., Xu J.D., Kong M., Shen H., Zhu H., Bai M., Zhou L., Li G.F. iTRAQ-based proteomic analysis of ginsenoside F2 on human gastric carcinoma cells SGC7901. Evidence Based Comp Altern Med. 2016:2635483. doi: 10.1155/2016/2635483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung K.J., Kim D.H., Lee E.K., Song C.W., Yu B.P., Chung H.Y. Oxidative stress induces inactivation of protein phosphatase 2A, promoting proinflammatory NF-κB in aged rat kidney. Free Rad Biol Med. 2013;61:206–217. doi: 10.1016/j.freeradbiomed.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 35.Wu W.B., Menon R., Xu Y.Y., Zhao J.R., Wang Y.L., Liu Y., Zhang H.J. Downregulation of peroxiredoxin-3 by hydrophobic bile acid induces mitochondrial dysfunction and cellular senescence in human trophoblasts. Sci Rep. 2016;6:38946. doi: 10.1038/srep38946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin D.Y., Chae H.Z., Rhee S.G., Jeang K.T. Regulatory role for a novel human thioreoxin peroxidase in NF-B activation. J Biol Chem. 1997;272:30952–30961. doi: 10.1074/jbc.272.49.30952. [DOI] [PubMed] [Google Scholar]
- 37.Rhee S.G., Kang S.W., Jeong W., Chang T.S., Wang K.S., Woo H.A. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr Opin Cell Biol. 2005;17:183–189. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 38.Koussounadis A., Langdon S.P., Um I.H., Harrison D.H., Smith V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci Rep. 2015;5:10775. doi: 10.1038/srep10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivanina A.V., Sokolova I.M., Sukhotin A.A. Oxidative stress and expression of chaperones in aging mollusks. Comp Biochem Physiol B Biochem Mol Biol. 2008;150:53–61. doi: 10.1016/j.cbpb.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Rea I.M., McNerlan S., Pckley A.G. Serum heat shock protein and anti-heat shock protein antibody levels in aging. Exp Gerontol. 2001;36:341–352. doi: 10.1016/s0531-5565(00)00215-1. [DOI] [PubMed] [Google Scholar]
- 41.Lee Y.H., Lee J.C., Moon H.J., Jung Je, Sharma M., Park B.H., Yi H.K., Jhee E.C. Differential effect of oxidative stress on the apoptosis of early and late passage human diploid fibroblasts: implication of heat shock protein 60. Cell Biochem Funct. 2008;26:502–508. doi: 10.1002/cbf.1473. [DOI] [PubMed] [Google Scholar]
- 42.Chen L., Na R., Ran Q. Enhanced defense against mitochondrial hydrogen peroxide attenuates age-associated cognition decline. Neurobiol Aging. 2014;35:2552–2561. doi: 10.1016/j.neurobiolaging.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 43.Olahova M., Veal E.A. A peroxiredoxin, PRDX-2, is required for insulin secretion and insulin/IIS-dependent regulation of stress resistance and longevity. Aging Cell. 2015;14:558–568. doi: 10.1111/acel.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kayashima Y., Yamakawa-Kobayashi K. Involvement of Prx3, a Drosophila ortholog of the thiol-dependent peroxidase PRDX3, in age-dependent oxidative stress resistance. Biomed Res. 2012;33:319–322. doi: 10.2220/biomedres.33.319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of identified proteins in Rg3(S)-treated senescent human dermal fibroblasts (HDFs). Here, the ratios of 2 h or 6 h or 12 h vs. 0 h were measured by MSE methods.
DNA sequences of target genes for quantitative RT-PCR.
Comparisons of mRNA and proteome levels in Rg3(S)-treated senescent human dermal fibroblasts (HDFs) for 6 h. Relative expression ratios (6 h/0 h) were calculated for the 10 μM of Rg3(S)-treated group by dividing by the levels observed for the control. Statistical significance: (*) P < 0.05, (**) P < 0.01, ns, not significant.