Abstract
Background
The biological and pharmacological effects of BST204, a fermented ginseng extract, have been reported in various disease conditions. However, its molecular action in metabolic disease remains poorly understood. In this study, we identified the antiadipogenic activity of BST204 resulting from its inhibition of the S6 kinase 1 (S6K1) signaling pathway.
Methods
The inhibitory effects of BST204 on S6K1 signaling were investigated by immunoblot, nuclear fractionation, immunoprecipitation analyses. The antiadipogenic effect of BST204 was evaluated by measuring mRNA levels of adipogenic genes and by chromatin immunoprecipitation and quantitative real-time polymerase chain reaction analysis.
Results
Treatment with BST204 inhibited activation and nuclear translocation of S6K1, further decreasing the interaction between S6K1 and histone H2B in 10T1/2 mesenchymal stem cells. Subsequently, phosphorylation of H2B at serine 36 (H2BS36p) by S6K1 was reduced by BST204, inducing an increase in the mRNA expression of Wnt6, Wnt10a, and Wnt10b, which disturbed adipogenic differentiation and promoted myogenic and early osteogenic gene expression. Consistently, BST204 treatment during adipogenic commitment suppressed the expression of adipogenic marker genes and lipid drop formation.
Conclusion
Our results indicate that BST204 blocks adipogenesis of mesenchymal stem cells through the inhibition of S6K1-mediated histone phosphorylation. This study suggests the potential therapeutic strategy using BST204 to combat obesity and musculoskeletal diseases.
Keywords: Adipogenesis, BST204, H2BS36 phosphorylation, S6K1, Wnt genes
1. Introduction
Ribosomal protein S6 kinase 1 (S6K1), a representative downstream substrate of mammalian target of rapamycin (mTOR), induces a number of key catabolic responses, including protein [1], nucleotide [2], [3], and lipid synthesis [4]. In addition to its well-known roles in cellular biology, several studies focused on the physiological role of S6K1. S6K1-deficient mice show resistance to high-fat diet–induced obesity and maintain a lean body shape [5]. This antiobesity phenotype of S6K1-deficient mice originates from enhanced β-oxidation, elevated insulin sensitivity, and impaired adipogenesis [5], [6]. These results present evidence that S6K1 controls energy balance and glucose homeostasis, which are key parts of metabolism, suggesting that S6K1 is a potential target to treat metabolic disorders.
Previously, we further discovered the contributing mechanisms of S6K1 to the development of obesity [7]. During early adipogenesis, activated S6K1 is translocated into the nucleus to interact with histone H2B and phosphorylates H2B at serine 36 (H2BS36), suppressing the expression of Wnt6, Wnt10a, and Wnt10b, which are well-known inhibitors of adipogenesis. This epigenetic role of S6K1 provides a therapeutic target for developing antiobesity medicine.
BST204, a fermented ginseng extract, has recently been investigated because of its multiple therapeutic effects. Its effect on resisting inflammation is associated with inhibition of NO production and COX2 expression, factors responsible for promoting inflammation [8], [9]. Moreover, its effects on inhibiting cancer cell proliferation and invasion were also revealed in our previous work. Treatment with BST204 causes the cell cycle of colon cancer cells to halt at the G1 phase, with concomitantly increased levels of the tumor suppressor genes, p53 and p21 [10]. In a more recent study, BST204 was found to improve cancer-related fatigue. Such phenotype was hypothesized to be related with reduced levels of tumor necrosis factor-α, interleukin-6, aspartate transaminase (AST), alanine aminotransferase (ALT), and creatinine (CRE) [11]. Although these studies have shown the effect of BST204 on cancer cells and macrophages, its influence on cell fate decision in multipotent stem cells has not yet been revealed.
Considering the diverse biological effects of BST204 and previous studies presenting its inhibitory impact on S6K1 signaling [8], [9], we took a closer look at its effect on controlling adipogenesis via the S6K1-mediated epigenetic regulation. Here, we found that BST204 had remarkable inhibitory effects on the activation of S6K1 and S6K1-mediated epigenetic regulation of Wnt genes. Furthermore, treatment with BST204 interrupted adipogenesis of mesenchymal stem cells (MSCs) by reducing the expression level of adipogenic genes, while inducing myogenesis. These findings present the antiadipogenic activity of BST204, which derives from inhibition of S6K1-mediated histone phosphorylation.
2. Materials and methods
2.1. Preparation of the fermented ginseng extract, BST204
BST204 was provided by the Green Cross WellBeing, Co, Ltd. (Seongnam, Korea), and it was manufactured according to a patented technology and earlier study [11]. Briefly, the harvested ginseng was extracted with ethanol repeatedly followed by incubation with an enzyme containing ginsenoside-β-glucosidase. After acid hydrolysis of the residue, the reactant was purified with HP20 resin, followed by washing out first with distilled water and finally with 95% ethanol. The ethanol fractions, containing ginsenoside Rg3 and Rh2, were concentrated and were designated as BST204. As a result of HPLC-UV analysis, the ginsenoside content of BST204 was found to be 10.95% of Rg3 and 7.22% of Rh2. The NMR data and structure of BST204 are shown in the earlier study [12].
2.2. Cell culture and differentiation
C3H10T1/2 (10T1/2) cells, a mouse MSC line, were grown in Dulbecco's Modified Eagle's Medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S). Adipogenic differentiation was performed as previously described [7]. For commitment to preadipocytes, 10T1/2 cells were incubated for 4 d in DMEM with 10% FBS, 1% P/S, and 10 μg/ml of bone morphogenetic protein 4 (BMP4). For terminal differentiation of preadipocytes into adipocytes, the cells were further incubated in DMEM with 10% FBS, 1% P/S, 0.5mM 3-isobuyl-1-methylxanthine, 1μM dexamethasone, and 1 μg/ml of insulin, followed by replacing the medium with DMEM containing 10% FBS, 1% P/S, and 1 μg/ml of insulin every other day.
2.3. Immunoblotting and immunoprecipitation
For immunoblotting, each sample was subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Proteins were transferred to polyvinylidene difluoride membranes using semidry transfer (Bio-Rad, USA). The membranes were incubated overnight with the indicated primary antibodies, followed by incubation with horseradish peroxidase–conjugated secondary antibodies for 1 h (Abcam, UK). The signals were detected using chemiluminescence reagents (iNtRON, Korea). For immunoprecipitation, the cells were lysed with IP lysis buffer [HEPES 40mM (pH 7.4) containing 120mM NaCl, 1mM EDTA, 50mM NaF, 1.5mM Na3VO4, 10mM β-glycerophosphate, 0.3% CHAPSO, and protease inhibitors]. The lysates were centrifuged for 20 min at 13000 rpm and 4°C. Then, the supernatants were incubated with the appropriate antibodies overnight at 4°C, followed by incubation with Anti-Rabbit Ig IP beads (TrueBlot; Rockland Immunochemicals, USA) for 1 h at 4°C. The beads were spun down for 1 min at 2000 rpm and washed three times with IP wash buffer (IP lysis buffer without CHAPSO). The proteins were eluted by boiling for 5 min in Laemmli buffer (Bio-Rad) and subjected to immunoblotting.
2.4. Subcellular fractionation
Cytoplasmic and nuclear extracts were prepared as previously described [13]. In brief, cells were suspended in Buffer A (10mM HEPES containing 1.5mM MgCl2, 10mM KCl, 1mM EDTA, 1mM dithiothreitol (DTT), 0.5 μg/ml of leupeptin, 1mM phenylmethylsulfonyl fluoride (PMSF), 1μM pepstatin A, and 0.05% NP-40), and cytoplasmic extracts were separated by centrifugation at 3000 rpm and 4°C for 10 min. The remaining pellet was resuspended in Buffer B (20mM HEPES containing 1.5mM MgCl2, 420mM KCl, 25% glycerol, 0.2mM EDTA, 1mM DTT, 0.5 μg/ml of leupeptin, 1mM PMSF, and 1μM pepstatin A) and incubated on ice for 30 min. Finally, nuclear extracts were separated by centrifugation at 13000 rpm and 4°C for 20 min.
2.5. Quantitative real-time polymerase chain reaction
Total RNA was extracted from cellular samples using easy-BLUE reagent (iNtRON, Korea). Then 1 μg of total RNA was reverse transcribed into cDNA using a Reverse Transcription Kit (Promega, USA). Quantitative real-time polymerase chain reaction (qPCR) was performed using KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems, USA) and a CFX96 Touch or Chromo4 real-time PCR detector (Bio-Rad). Relative levels of mRNA were normalized to the levels of β-actin mRNA for each reaction. The qPCR primer sequences used are as follows: β-actin forward, 5′-ACGGCCAGGTCATCACTATTG-3′; β-actin reverse, 5′-TGGATGCCACAGGATTCCA-3'; Wnt6 forward, 5′-GCGGAGACGATGTGGACTTC-3′; Wnt6 reverse, 5′-ATGCACGGATATCTCCACGG-3′; Wnt10a forward, 5′-CCACTCCGACCTGGTCTACTTTG-3′; Wnt10a reverse, 5′-TGCTGCTCTTATTGCACAGGC-3′; Wnt10b forward, 5′-GCTGACTGACTCGCCCACCG-3′; Wnt10b reverse, 5′-AAGCACACGGTGTTGGCCGT-3′; PPARγ forward, 5′-GCATGGTGCCTTCGCTGA-3′; PPARγ reverse, 5′-TGGCATCTCTGTGTCAACCATG-3′; Cebpα forward, 5′-CTCCCAGAGGACCAATGAAA-3′; Cebpα reverse, 5′-AAGTCTTAGCCGGAGGAAGC-3′; Adipsin forward, 5′-CATGCTCGGCCCTACATG-3′; Adipsin reverse, 5′-CACAGAGTCGTCATCCGTCAC-3′; Fabp4 forward, 5′-AAGGTGAAGAGCATCATAACCCT-3′; Fabp4 reverse, 5′-TCACGCCTTTCATAACACATTCC-3′; Adipoq forward, 5′-TGTTCCTCTTAATCCTGCCCA-3′; Adipoq reverse, 5′-CCAACCTGCACAAGTTCCCTT-3′; Myf5 forward, 5′-CCAGCCCCACCTCCAACT-3′; Myf5 reverse, 5′-GGGACCAGACAGGGCTGTTA-3′; MyoD forward, 5′-AGTAGAGAAGTGTGCGTGCT-3′; MyoD reverse, 5′-ACGACTTCTATGATGATCCG-3′; Pax7 forward, 5′-GGTGGGGTCTTCATCAATGGTC-3′; Pax7 reverse, 5′-GAACATCCCGGGGTTCTCTCTC-3′; Col1a1 forward, 5′-TCCCAGAACATCACCTATCAC-3′; Col1a1 reverse, 5′-CTGTTGCCTTCGCCTCTGAG-3′; Ocn forward, 5′-AGGGCAATAAGGTAGTGAA-3′; Ocn reverse, 5′-GAGGCTCTGAGAAGCATAAA-3′; Osx forward, 5′-CCCTTCTCAAGCACCAATGG-3′; and Osx reverse, 5′-AAGGGTGGGTAGTCATTTGCATA-3′.
2.6. Chromatin immunoprecipitation and qPCR
Chromatin immunoprecipitation was performed as previously described [14]. In brief, a small portion of the cross-linked, sheared chromatin solution was reserved as the input DNA, and the remainder was subjected to immunoprecipitation overnight at 4°C using the appropriate antibodies. After immunoprecipitation, the recovered chromatin fragments were subjected to qPCR using primer pairs specific for the target gene promoter. The primer sequences are available on request.
2.7. Oil red o staining
Fully differentiated adipocytes were fixed with 10% formalin for 1 h and washed with 60% isopropanol, followed by the incubation with oil red o working solution for 1 h. Then the cells were rinsed with distilled water three times. For the preparation of oil red o stock solution, 300 mg of oil red o powder was dissolved in 100 ml of 99% isopropanol. Thirty milliliter of the stock solution was diluted with 20 ml of distilled water to make oil red o working solution just before use.
2.8. Statistical analysis
Statistical significance was analyzed using Student t test (two tailed) and assessed based on the resulting P-value.
3. Results
3.1. BST204 inhibits the activation and nuclear translocation of S6K1
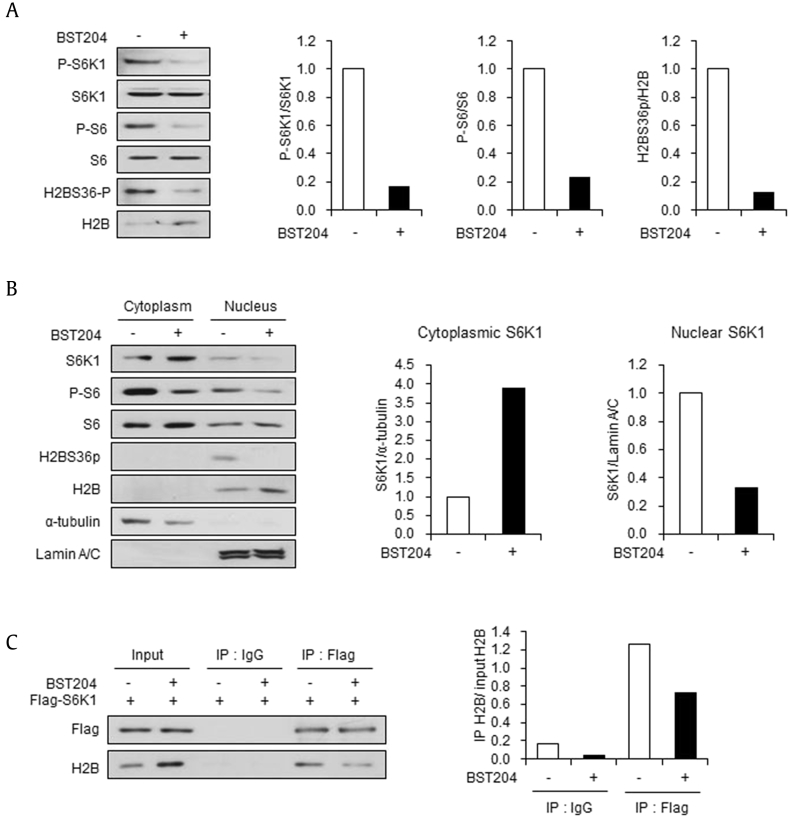
Previously, it has been reported that treatment with 50 μg/ml of BST204 inhibits lipopolysaccharide (LPS)-induced activation of S6K1 in macrophages [8], [9]. Therefore, we examined the effects of BST204 on S6K1 signaling in 10T1/2 MSCs. On treatment with BST204, phosphorylation of S6K1 at threonine 389 (T389), a marker of mTOR-dependent activation, was abolished (Fig. 1A). Phosphorylation of ribosomal protein S6, a representative downstream substrate of S6K1, was also diminished by BST204 treatment (Fig. 1A). Given that nuclear translocation of S6K1 is dependent on mTOR-mediated activation [15], [16], we next examined whether BST204 could regulate the subcellular localization of S6K1. The subcellular fractionation data showed that the level of cytoplasmic S6K1 increased on BST204 treatment, while nuclear S6K1 decreased (Fig. 1B). Additionally, phosphorylation of S6 was reduced in both subcellular locations (Fig. 1B). These data indicate that BST204 blocks mTOR-dependent activation of S6K1 and further nuclear import of activated S6K1 from the cytoplasm.
Fig. 1.
BST204 inhibits activation and nuclear translocation of S6K1. (A) Immunoblot analysis of 10T1/2 cells treated with or without BST204 (50 μg/ml) for 24 h. (B) Immunoblot analysis of cytoplasmic and nuclear extracts from 10T1/2 cells treated with or without BST204 (50 μg/ml) for 24 h. (C) Immunoblot analysis of IgG or Flag immunoprecipitates (IP) and whole cell lysates (Input) from Flag-S6K1–expressing 10T1/2 cells treated with or without BST204 (50 μg/ml) for 24 h.
3.2. BST204 inhibits phosphorylation of H2BS36 by S6K1
In our earlier study, we discovered that nuclear S6K1 directly interacts with H2B and phosphorylates H2B at serine 36 [7]. To determine the effects of BST204 on the interaction between S6K1 and H2B, we performed immunoprecipitation assay with a Flag antibody in 10T1/2 cells expressing Flag-S6K1. Consistent with results showing the inhibitory effects of BST204 on the nuclear translocation of S6K1 (Fig. 1B), BST204 treatment disabled S6K1 from binding to H2B (Fig. 1C). Moreover, we observed that phosphorylated H2BS36 (H2BS36p) was ablated in both the whole cell lysates (Fig. 1A) and nuclear fractions (Fig. 1B) of BST204-treated cells. Thus, these results indicate that BST204 represses S6K1-mediated phosphorylation of H2BS36 by inhibiting the interaction between nuclear S6K1 and H2B.
3.3. BST204 inhibits the suppression of Wnt genes caused by S6K1-mediated H2BS36 phosphorylation
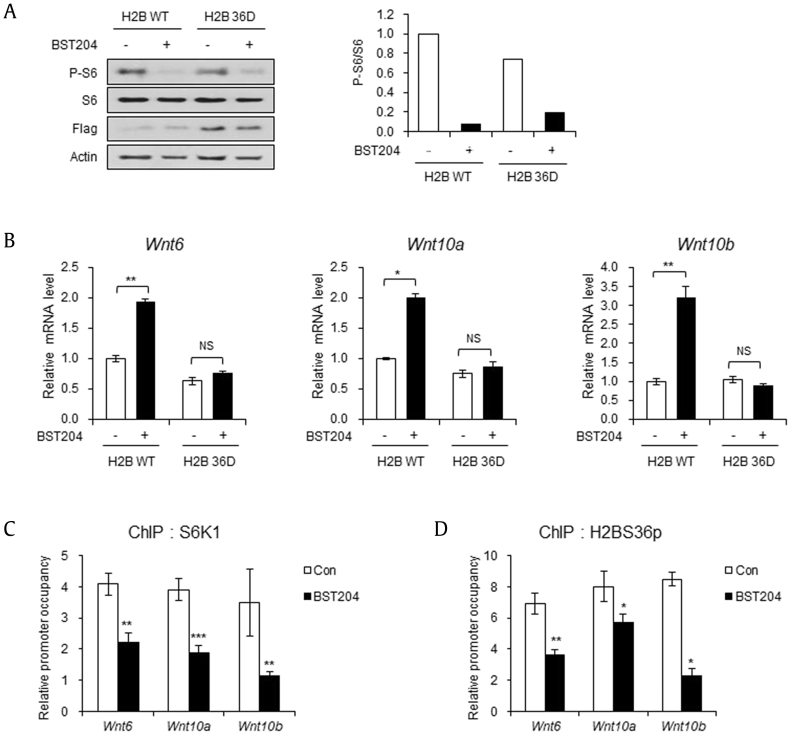
Among the Wnt ligands, Wnt6, Wnt10a, and Wnt10b are well known blockers of adipogenic commitment [17]. Previously, we discovered that S6K1 negatively regulates the expression of Wnt genes through phosphorylation of H2BS36, facilitating adipogenic commitment [7]. Thus, we investigated whether BST204 inhibits the regulation of Wnt genes by S6K1-mediated H2BS36 phosphorylation using the phosphomimetic mutant H2BS36D that contains a single-site mutation of serine 36 to aspartate. Phosphorylation of S6 was also ablated by BST204 in both 10T1/2 cells expressing wild-type H2B and the phosphomimetic mutant of H2B (H2BS36D) (Fig. 2A). However, the mRNA levels of Wnt genes were increased only in wild-type H2B–expressing cells, whereas the expression of H2B 36D did not alter the mRNA level of Wnt genes (Fig. 2B). These data show that restricted H2BS36 phosphorylation was responsible for the increase of Wnt gene expression on BST204 treatment.
Fig. 2.
BST204 inhibits Wnt gene suppression by S6K1-mediated H2BS36 phosphorylation. (A) Immunoblot analysis of 10T1/2 cells transfected with wild-type (WT) or phosphomimetic (36D) H2B vector and incubated with or without BST204 (50 μg/ml, 24 h). (B) The mRNA levels of the Wnt6, Wnt10a, and Wnt10b genes in 10T1/2 cells transfected with WT or 36D H2B vector and incubated with or without BST204 (50 μg/ml, 24 h). (C) 10T1/2 cells were treated with or without BST204 (50 μg/ml) for 24 h. ChIP assay was performed with an S6K1 antibody followed by real-time PCR with primers for the promoter regions of the Wnt6, Wnt10a, and Wnt10b genes. (D) 10T1/2 cells were treated with or without BST204 (50 μg/ml) for 24 h. ChIP assay was performed with a phosphorylated H2BS36 antibody followed by real-time PCR with primers for the promoter regions of the Wnt6, Wnt10a, and Wnt10b genes. Data represent mean ± SEM for n = 3. *P < 0.05, **P < 0.01, ***P < 0.001.
ChIP, chromatin immunoprecipitation; SEM, standard error of the mean.
Earlier, we discovered that S6K1 directly binds to the promoter regions of the Wnt6, Wnt10a, and Wnt10b genes, suppressing transcription of these genes through H2BS36 phosphorylation [7]. Expectedly, chromatin immunoprecipitation analysis showed that the recruitment of S6K1 to the promoter regions of Wnt6, Wnt10a, and Wnt10b genes was abrogated on BST204 treatment (Fig. 2C). In parallel, we observed that BST204 treatment significantly decreased the enrichment of H2BS36p at the Wnt gene promoters (Fig. 2D). These results support that the elevated expression of Wnt genes by BST204 is due to the inhibition of S6K1-mediated H2B36p at promoter regions of the genes.
3.4. BST204 inhibits adipogenesis of MSCs
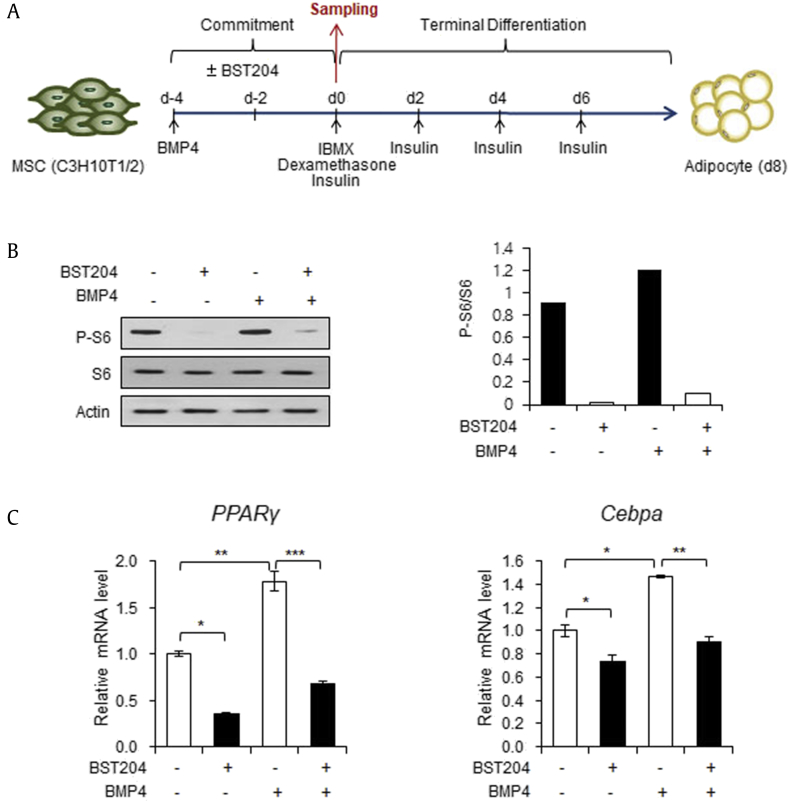
Earlier studies have described that activation of Wnt signaling in MSCs suppresses commitment to the adipocytic lineage [18], [19]. Thus, we next examined the effects of BST204 on adipogenic commitment by inducing differentiation from 10T1/2 MSCs into adipocytes through two distinguishable stages (Fig. 3A): (i) commitment to adipogenic progenitors, which is induced by BMP4 treatment for 4 d and (ii) terminal differentiation into fully differentiated adipocytes, which is induced by an adipogenic medium [20], [21]. First, we confirmed the inhibitory effect of BST204 on S6K1 signaling after treatment of 10T1/2 cells with BMP4 and BST204 for 4 d. BST204 treatment inhibited S6 phosphorylation both in the presence and absence of BMP4 (Fig. 3B). A previous study showed that S6K1-mediated reduction of Wnt gene expression during early adipogenesis drives upregulation of the adipogenic transcription factors, PPARγ and Cebpα [7]. Consistently, treatment of 10T1/2 cells undergoing adipogenic commitment process with BST204 blocked expression of PPARγ and Cebpα (Fig. 3C).
Fig. 3.
BST204 inhibits the adipogenic commitment from mesenchymal stem cells. (A) Schematic representation of 10T1/2 cell differentiation into adipocytes. The protein and mRNA samples used in Fig. 3 were extracted from 10T1/2 cells at d0. (B) Immunoblot analysis of 10T1/2 cells treated with or without BST204 (50 μg/ml) for 4 days in the presence or absence of BMP4, which is inducer of adipogenic commitment. (C) The mRNA levels of the PPARγ and Cebpα genes in 10T1/2 cells treated with or without BST204 (50 μg/ml) for 4 days in the presence or absence of BMP4. Data represent mean ± SEM for n = 3. *P < 0.05, **P < 0.01, ***P < 0.001.
MSC, mesenchymal stem cell; SEM, standard error of the mean.
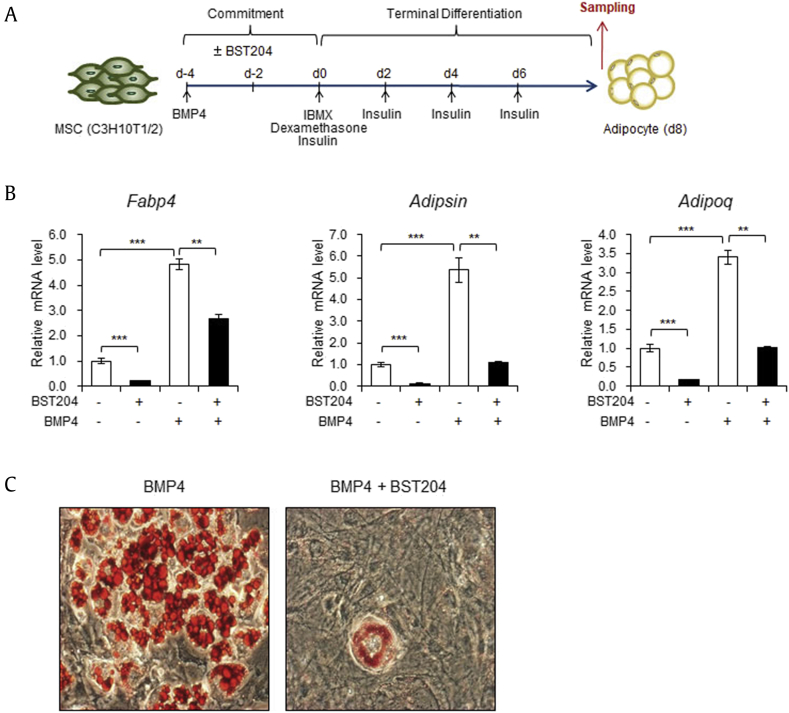
Next, we measured the expression levels of fully differentiated adipocytic marker genes at the end point of adipogenesis from 10T1/2 cells that were incubated with or without BST204 during commitment (Fig. 4A). In line with the decrease in adipogenic transcription factors at the progenitor stage seen with BST204 treatment (Fig. 3C), the mRNA levels of the adipocytic marker genes, Fabp4, Adipsin, and Adipoq, were also markedly reduced when cells were incubated with BST204 during adipogenic commitment (Fig. 4B). Interestingly, even in the cells not exposed to BMP4 during the commitment, the adipogenic gene expressions were decreased by BST204 (Fig. 3, Fig. 4B), indicating that BST204 also affects the signaling in spontaneously differentiating cells without BMP4. Furthermore, when the cells were exposed to BST204 during commitment, lipid droplets in adipocytes were much less accumulated than in those cells incubated with only BMP4, as visualized by oil red o staining (Fig. 4C). Collectively, these results demonstrate that BST204 treatment during adipogenic commitment disturbs differentiation of MSCs into adipocytes.
Fig. 4.
Exposure to BST204 during adipogenic commitment disturbs adipogenesis of mesenchymal stem cells. (A) Schematic representation of 10T1/2 cell differentiation into adipocytes. The protein and mRNA samples used in Fig. 4 were extracted after terminal differentiation of 10T1/2 cells. (B) The mRNA levels of the Fabp4, Adipsin, and Adipoq genes in fully differentiated adipocytes that were treated with or without BST204 (50 μg/ml) during adipogenic commitment in the presence or absence of BMP4. (C) Microscopic image of fully differentiated adipocytes that were treated with or without BST204 (50 μg/ml) during adipogenic commitment. Data represent mean ± SEM for n = 3. *P < 0.05, **P < 0.01, ***P < 0.001.
MSC, mesenchymal stem cell; SEM, standard error of the mean.
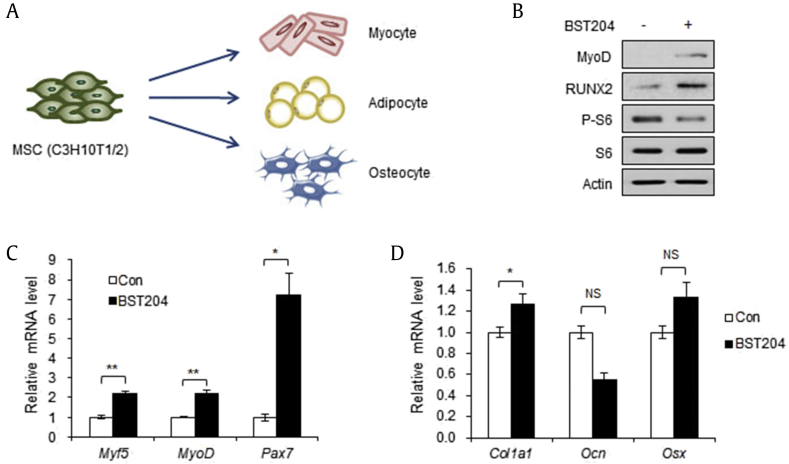
3.5. BST204 promotes myogenesis and osteogenesis of MSCs
MSCs have capacity to differentiate into a variety of cell types, including myocytes, osteocytes, chondrocytes, and adipocytes (Fig. 5A). As Wnt signaling contributes to osteogenesis or myogenesis while blocking adipogenesis [18], [19], we next assessed the capability of BST204 to control cell fate of MSCs by measuring the expression levels of myogenic and osteogenic lineage marker genes. When 10T1/2 cells were treated with BST204 for 24 h, the protein levels of myogenic marker (MyoD) and osteogenic marker (RUNX2) were enhanced, as examined by the immunoblotting assay (Fig. 5B). The mRNA levels of myogenic genes, Myf5, MyoD, and Pax7, were also significantly increased in response to BST204 treatment (Fig. 5C). However, the mRNA levels of osteogenic genes were partially increased on BST204 treatment (Fig. 5D). Col1a1 that is expressed at the early stage of osteogenesis increased only, whereas the expression of Ocn and Osx, relatively late-stage marker genes, did not change (Fig. 5D). These data suggest that BST204 has an impact on the cell fate determination of multipotent cells, promoting differentiation into other cell lineages than adipocytes.
Fig. 5.
BST204 promotes myogenic gene expression in mesenchymal stem cells. (A) MSCs have capacity to differentiate into diverse cell types. (B) Immunoblot analysis of 10T1/2 cells treated with or without BST204 (50 μg/ml, 24 h). (C) The mRNA levels of the Myf5, MyoD, and Pax7 genes in 10T1/2 cells treated with or without BST204 (50 μg/ml, 24 h). (D) The mRNA levels of the Col1a1, Ocn, and Osx genes in 10T1/2 cells treated with or without BST204 (50 μg/ml, 24 h). Data represent mean ± SEM for n = 3. *P < 0.05, **P < 0.01, ***P < 0.001.
MSC, mesenchymal stem cell; SEM, standard error of the mean.
4. Discussion
Ginsenosides, the major pharmacological component in the roots of ginseng, are divided into two structural groups: panaxadiols (Rb1, Rb2, Rc, Rd, Rg3, Rh2, and Rh3) and panaxatriols (Re, Rf, Rg1, Rg2, and Rh1) [22]. On physical or chemical processes, such as heating and enzymatic treatment, the contents and properties of ginseng extract can be altered. For instance, steaming ginseng at high degrees increases the contents of Rg3, improving the anticancer effects of the extract [23], [24]. Fermentation with β-glucosidase also enhances the content of minor ginsenosides including Rg3, Rh2, F2, and compound K, which are more pharmacologically active than the major ginsenosides, Rb1, Rb2, Rd, Re, and Rg1 [25], [26], [27]. In the present study, we examined the effects of a fermented ginseng extract, BST204, which contains abundant Rh2 and Rg3, on adipogenic lineage determination. Although several studies have reported the anti-inflammatory and antiproliferative activity of BST204 [8], [9], [10], [11], its effects on adipocytes and metabolism have not yet been clearly defined. Here, we demonstrated that treatment of MSCs with BST204 during adipogenic commitment led to a remarkable decline in the expression of adipogenic transcription factors, PPARγ and Cebpα (Fig. 3). Exposure to BST204 in the commitment stage also impaired the terminal differentiation toward adipocytes, as demonstrated by reduced expression of adipogenic genes and oil drop deposits (Fig. 4).
Adipogenesis is finely controlled through a complex network of active and repressive histone marks [28]. Several ginsenosides have been reported to regulate histone modifications via histone deacetylases [29], [30], [31], [32], histone methyltransferase [33], and kinase [34]. However, the epigenetic regulation of adipogenesis by ginsenosides has not been reported. Earlier, we revealed that nuclear S6K1 phosphorylates H2B at serine 36 to suppress the expression of Wnt genes, which are blockers of adipogenesis [7]. In line with our previous findings, we here observed that BST204 treatment inhibited the activation and nuclear translocation of S6K1, which sequentially disturbed the interaction between H2B and S6K1 (Fig. 1). Reduced binding to H2B resulted in hypophosphorylation of H2BS36 both at the global level (Figs. 1A, 1B) and at the local level on the promoter regions of Wnt genes (Fig. 2D). BST204-mediated Wnt gene suppression was attributed to impairment of H2BS36 phosphorylation by S6K1, which was assessed by substituting a phosphomimetic mutant form of H2B (H2BS36D) (Figs. 2A and 2B). Considering its significant influence on S6K1-mediated histone phosphorylation and subsequent gene suppression, our findings suggest BST204 to be a potential epigenetic modulator and provide a novel molecular mechanism of action underlying its pharmacological activity.
Another important issue we present here is that BST204 induces the expression of other cell lineage markers, including myogenic genes and early-stage osteogenic gene (Fig. 5). Several studies have identified ginsenosides that facilitate myogenesis or osteogenesis, including Rg1 [35], [36], [37], [38], Re [39], and Rh2 [40]. Additionally, in this present study, we assessed the effects of BST204 on cell fate conversion of MSCs into adipogenic, myogenic, and osteogenic lineages. In the early stage of cell fate determination, Wnt ligands play a critical role in promoting myogenic or osteogenic differentiation, while disturbing adipogenesis [18], [19]. Our data show that BST204 treatment to MSCs led to increase in the expression of myogenic and early osteogenic genes, accompanied by the suppression of Wnt genes. These data provide better understanding of the way BST204 modulates plasticity of multipotent cells.
To avoid the side effects and rebound weight gain often associated with existing weight loss remedies, there has been much interest in discovering naturally derived dietary compounds that display antiobesity effects. As part of this research, numerous studies have evaluated the impact of ginsenosides, either alone or in combination with other, on diverse factors involved in obesity [41]. Hwang et al. reported that both Rh2 and Rg3 impede adipogenesis from 3T3-L1 preadipocytes by activating the AMP-activated protein kinase (AMPK) signaling pathway and suppressing PPARγ signaling [42], [43]. In contrast, Rh2 promoted adipogenesis by activating glucocorticoid receptors in another study [44]. Despite the inconsistency among some in vitro analyses, a majority of studies in mice and human have indicated that ginseng extract protects against high-fat diet–mediated body weight gain and exerts beneficial effects on diverse physiological indicators associated with metabolic homeostasis [45], [46], [47], [48], [49], [50], [51]. In addition to these previous works, we currently show the inhibitory efficacy of BST204 on de novo adipocyte generation through disruption of the mTOR-S6K1-H2B signaling cascade (Fig. 6). Therefore, our findings suggest that BST204 is a promising therapeutic option for the treatment or prevention of obesity and related musculoskeletal diseases.
Fig. 6.
Molecular model underlying mechanism of action of BST204. During early adipogenesis of mesenchymal stem cells to preadipocytes, S6K1-mediated H2BS36 phosphorylation suppresses the expression of Wnt6, Wnt10a, and Wnt10b, facilitating early adipogenesis. BST204 inhibits S6K1-mediated H2BS36 phosphorylation and suppression of Wnt genes, finally attributing to prevention of adipogenesis.
Conflicts of interest
All authors have no conflicts of interest to declare.
Acknowledgments
The authors thank Jong Sun Kang (Sungkyunkwan University) for the cell line. This work was financially supported through grants from the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (2012R1A5A2A28671860, 2017R1A2B3002186, and 2017R1A6A3A04001986).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.jgr.2018.08.002.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Ma X.M., Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Sahra I., Howell J.J., Asara J.M., Manning B.D. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robitaille A.M., Christen S., Shimobayashi M., Cornu M., Fava L.L., Moes S., Prescianotto-Baschong C., Sauer U., Jenoe P., Hall M.N. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 4.Owen J.L., Zhang Y., Bae S.H., Farooqi M.S., Liang G., Hammer R.E., Goldstein J.L., Brown M.S. Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc Natl Acad Sci USA. 2012;109:16184–16189. doi: 10.1073/pnas.1213343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Um S.H., Frigerio F., Watanabe M., Picard F., Joaquin M., Sticker M., Fumagalli S., Allegrini P.R., Kozma S.C., Auwerx J. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 6.Carnevalli L.S., Masuda K., Frigerio F., Le Bacquer O., Um S.H., Gandin V., Topisirovic I., Sonenberg N., Thomas G., Kozma S.C. S6K1 plays a critical role in early adipocyte differentiation. Dev Cell. 2010;18:763–764. doi: 10.1016/j.devcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yi S.A., Um S.H., Lee J., Yoo J.H., Bang S.Y., Park E.K., Lee M.G., Nam K.H., Jeon Y.J., Park J.W. S6K1 phosphorylation of H2B mediates EZH2 trimethylation of H3: a determinant of early adipogenesis. Mol Cell. 2016;62:443–452. doi: 10.1016/j.molcel.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seo J.Y., Lee J.H., Kim N.W., Her E., Chang S.H., Ko N.Y., Yoo Y.H., Kim J.W., Seo D.W., Han J.W. Effect of a fermented ginseng extract, BST204, on the expression of cyclooxygenase-2 in murine macrophages. Int Immunopharmacol. 2005;5:929–936. doi: 10.1016/j.intimp.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Seo J.Y., Lee J.H., Kim N.W., Kim Y.J., Chang S.H., Ko N.Y., Her E., Yoo Y.H., Kim J.W., Lee B.Y. Inhibitory effects of a fermented ginseng extract, BST204, on the expression of inducible nitric oxide synthase and nitric oxide production in lipopolysaccharide-activated murine macrophages. J Pharm Pharmacol. 2005;57:911–918. doi: 10.1211/0022357056497. [DOI] [PubMed] [Google Scholar]
- 10.Park J.W., Lee J.C., Ann S., Seo D.W., Choi W.S., Yoo Y.H., Park S.H., Choi J.Y., Um S.H., Ahn S.H. A fermented ginseng extract, BST204, inhibits proliferation and motility of human colon cancer cells. Biomol Ther. 2011;19:211–217. [Google Scholar]
- 11.Park H.J., Shim H.S., Kim J.Y., Kim J.Y., Park S.K., Shim I. Ginseng purified dry extract, BST204, improved cancer chemotherapy-related fatigue and toxicity in mice. Evid Based Complement Alternat Med. 2015;2015:197459. doi: 10.1155/2015/197459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang H., Yoo G., Kim H.S., Kim J.Y., Kim S.O., Yoo Y.H., Sung S.H. Implication of the stereoisomers of ginsenoside derivatives in the antiproliferative effect of HSC-T6 cells. J Agric Food Chem. 2012;60:11759–11764. doi: 10.1021/jf303714c. [DOI] [PubMed] [Google Scholar]
- 13.Park J.W., Lee J.C., Ha S.W., Bang S.Y., Park E.K., Yi S.A., Lee M.G., Kim D.S., Nam K.H., Yoo J.H. Requirement of protein l-isoaspartyl O-methyltransferase for transcriptional activation of trefoil factor 1 (TFF1) gene by estrogen receptor alpha. Biochem Biophys Res Commun. 2012;420:223–229. doi: 10.1016/j.bbrc.2012.02.072. [DOI] [PubMed] [Google Scholar]
- 14.Lee J.C., Kang S.U., Jeon Y., Park J.W., You J.S., Ha S.W., Bae N., Lubec G., Kwon S.H., Lee J.S. Protein L-isoaspartyl methyltransferase regulates p53 activity. Nat Commun. 2012;3:927. doi: 10.1038/ncomms1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosner M., Hengstschläger M. Nucleocytoplasmic localization of p70 S6K1, but not of its isoforms p85 and p31, is regulated by TSC2/mTOR. Oncogene. 2011;30:4509–4522. doi: 10.1038/onc.2011.165. [DOI] [PubMed] [Google Scholar]
- 16.Rosner M., Schipany K., Hengstschläger M. p70 S6K1 nuclear localization depends on its mTOR-mediated phosphorylation at T389, but not on its kinase activity towards S6. Amino Acids. 2012;42:2251–2256. doi: 10.1007/s00726-011-0965-4. [DOI] [PubMed] [Google Scholar]
- 17.Wang L., Jin Q., Lee J.E., Su I.H., Ge K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc Natl Acad Sci USA. 2010;107:7317–7322. doi: 10.1073/pnas.1000031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cawthorn W.P., Bree A.J., Yao Y., Du B., Hemati N., Martinez-Santibañez G., MacDougald O.A. Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a β-catenin-dependent mechanism. Bone. 2012;50:477–489. doi: 10.1016/j.bone.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christodoulides C., Lagathu C., Sethi J.K., Vidal-Puig A. Adipogenesis and WNT signalling. Trends Endocrinol Metab. 2009;20:16–24. doi: 10.1016/j.tem.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Q.Q., Lane M.D. Adipogenesis: from stem cell to adipocyte. Annu Rev Biochem. 2012;81:715–736. doi: 10.1146/annurev-biochem-052110-115718. [DOI] [PubMed] [Google Scholar]
- 21.Huang H., Song T.J., Li X., Hu L., He Q., Liu M., Lane M.D., Tang Q.Q. BMP signaling pathway is required for commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc Natl Acad Sci USA. 2009;106:12670–12675. doi: 10.1073/pnas.0906266106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tachikawa E., Kudo K., Harada K., Kashimoto T., Miyate Y., Kakizaki A., Takahashi E. Effects of ginseng saponins on responses induced by various receptor stimuli. Eur J Pharmacol. 1999;369:23–32. doi: 10.1016/s0014-2999(99)00043-6. [DOI] [PubMed] [Google Scholar]
- 23.Kim W.Y., Kim J.M., Han S.B., Lee S.K., Kim N.D., Park M.K., Kim C.K., Park J.H. Steaming of ginseng at high temperature enhances biological activity. J Nat Prod. 2000;63:1702–1704. doi: 10.1021/np990152b. [DOI] [PubMed] [Google Scholar]
- 24.Wang C.Z., Zhang B., Song W.X., Wang A., Ni M., Luo X., Aung H.H., Xie J.T., Tong R., He T.C. Steamed American ginseng berry: ginsenoside analyses and anticancer activities. J Agric Food Chem. 2006;54:9936–9942. doi: 10.1021/jf062467k. [DOI] [PubMed] [Google Scholar]
- 25.Kim J.K., Cui C.H., Yoon M.H., Kim S.C., Im W.T. Bioconversion of major ginsenosides Rg1 to minor ginsenoside F1 using novel recombinant ginsenoside hydrolyzing glycosidase cloned from Sanguibacter keddieii and enzyme characterization. J Biotechnol. 2012;161:294–301. doi: 10.1016/j.jbiotec.2012.06.021. [DOI] [PubMed] [Google Scholar]
- 26.Lee S.J., Kim Y., Kim M.G. Changes in the ginsenoside content during the fermentation process using microbial strains. J Ginseng Res. 2015;39:392–397. doi: 10.1016/j.jgr.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park S.E., Na C.S., Yoo S.A., Seo S.H., Son H.S. Biotransformation of major ginsenosides in ginsenoside model culture by lactic acid bacteria. J Ginseng Res. 2017;41:36–42. doi: 10.1016/j.jgr.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge K. Epigenetic regulation of adipogenesis by histone methylation. Biochim Biophys Acta. 2012;1819:727–732. doi: 10.1016/j.bbagrm.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shan X., Fu Y.S., Aziz F., Wang X.Q., Yan Q., Liu J.W. Ginsenoside Rg3 inhibits melanoma cell proliferation through down-regulation of histone deacetylase 3 (HDAC3) and increase of p53 acetylation. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Q., Li J., Feng Z., Zhao L., Luo L., You Z., Li D., Xia J., Zuo G., Chen D. Effect of ginsenoside Rh2 on the migratory ability of HepG2 liver carcinoma cells: recruiting histone deacetylase and inhibiting activator protein 1 transcription factors. Mol Med Rep. 2014;10:1779–1785. doi: 10.3892/mmr.2014.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z.H., Li J., Xia J., Jiang R., Zuo G.W., Li X.P., Chen Y., Xiong W., Chen D.L. Ginsenoside 20(s)-Rh2 as potent natural histone deacetylase inhibitors suppressing the growth of human leukemia cells. Chem Biol Interact. 2015;242:227–234. doi: 10.1016/j.cbi.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Wan Q., Ma X., Zhang Z.J., Sun T., Xia F., Zhao G., Wu Y.M. Ginsenoside reduces cognitive impairment during chronic cerebral hypoperfusion through brain-derived neurotrophic factor regulated by epigenetic modulation. Mol Neurobiol. 2017;54:2889–2900. doi: 10.1007/s12035-016-9868-4. [DOI] [PubMed] [Google Scholar]
- 33.Li Q., Li B., Dong C., Wang Y., Li Q. 20(S)-Ginsenoside Rh2 suppresses proliferation and migration of hepatocellular carcinoma cells by targeting EZH2 to regulate CDKN2A-2B gene cluster transcription. Eur J Pharmacol. 2017;815:173–180. doi: 10.1016/j.ejphar.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 34.Yang J., Yuan D., Xing T., Su H., Zhang S., Wen J., Bai Q., Dang D. Ginsenoside Rh2 inhibiting HCT116 colon cancer cell proliferation through blocking PDZ-binding kinase/T-LAK cell-originated protein kinase. J Ginseng Res. 2016;40:400–408. doi: 10.1016/j.jgr.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Go G.Y., Lee S.J., Jo A., Lee J., Seo D.W., Kang J.S., Kim S.K., Kim S.N., Kim Y.K., Bae G.U. Ginsenoside Rg1 from Panax ginseng enhances myoblast differentiation and myotube growth. J Ginseng Res. 2017;41:608–614. doi: 10.1016/j.jgr.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu Y., Zhou J., Wang Q., Fan W., Yin G. Ginsenoside Rg1 promotes osteogenic differentiation of rBMSCs and healing of rat tibial fractures through regulation of GR-dependent BMP-2/SMAD signaling. Sci Rep. 2016;6:25282. doi: 10.1038/srep25282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin L.H., Cheng W.X., Qin Z.S., Sun K.M., Zhong M., Wang J.K., Gao W.Y., Yu Z.H. Effects of ginsenoside Rg-1 on the proliferation and osteogenic differentiation of human periodontal ligament stem cells. Chin J Integr Med. 2015;21:676–681. doi: 10.1007/s11655-014-1856-9. [DOI] [PubMed] [Google Scholar]
- 38.Wang P., Wei X., Zhang F., Yang K., Qu C., Luo H., He L. Ginsenoside Rg1 of Panax ginseng stimulates the proliferation, odontogenic/osteogenic differentiation and gene expression profiles of human dental pulp stem cells. Phytomedicine. 2014;21:177–183. doi: 10.1016/j.phymed.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 39.Kim H.M., Kim D.H., Han H.J., Park C.M., Ganipisetti S.R., Valan Arasu M., Kim Y.O., Park C.G., Kim B.Y., Soung N.K. Ginsenoside Re promotes osteoblast differentiation in mouse osteoblast precursor MC3T3-E1 cells and a Zebrafish model. Molecules. 2016;22 doi: 10.3390/molecules22010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D.Y., Jung M.S., Park Y.G., Yuan H.D., Quan H.Y., Chung S.H. Ginsenoside Rh2(S) induces the differentiation and mineralization of osteoblastic MC3T3-E1 cells through activation of PKD and p38 MAPK pathways. BMB Rep. 2011;44:659–664. doi: 10.5483/BMBRep.2011.44.10.659. [DOI] [PubMed] [Google Scholar]
- 41.Li Z., Ji G.E. Ginseng and obesity. J Ginseng Res. 2018;42:1–8. doi: 10.1016/j.jgr.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang J.T., Kim S.H., Lee M.S., Kim S.H., Yang H.J., Kim M.J., Kim H.S., Ha J., Kim M.S., Kwon D.Y. Anti-obesity effects of ginsenoside Rh2 are associated with the activation of AMPK signaling pathway in 3T3-L1 adipocyte. Biochem Biophys Res Commun. 2007;364:1002–1008. doi: 10.1016/j.bbrc.2007.10.125. [DOI] [PubMed] [Google Scholar]
- 43.Hwang J.T., Lee M.S., Kim H.J., Sung M.J., Kim H.Y., Kim M.S., Kwon D.Y. Antiobesity effect of ginsenoside Rg3 involves the AMPK and PPAR-gamma signal pathways. Phytother Res. 2009;23:262–266. doi: 10.1002/ptr.2606. [DOI] [PubMed] [Google Scholar]
- 44.Niu C.S., Yeh C.H., Yeh M.F., Cheng J.T. Increase of adipogenesis by ginsenoside (Rh2) in 3T3-L1 cell via an activation of glucocorticoid receptor. Horm Metab Res. 2009;41:271–276. doi: 10.1055/s-0028-1103277. [DOI] [PubMed] [Google Scholar]
- 45.Lee Y.S., Cha B.Y., Yamaguchi K., Choi S.S., Yonezawa T., Teruya T., Nagai K., Woo J.T. Effects of Korean white ginseng extracts on obesity in high-fat diet-induced obese mice. Cytotechnology. 2010;62:367–376. doi: 10.1007/s10616-010-9288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee H., Park D., Yoon M. Korean red ginseng (Panax ginseng) prevents obesity by inhibiting angiogenesis in high fat diet-induced obese C57BL/6J mice. Food Chem Toxicol. 2013;53:402–408. doi: 10.1016/j.fct.2012.11.052. [DOI] [PubMed] [Google Scholar]
- 47.Yuan H.D., Quan H.Y., Jung M.S., Kim S.J., Huang B., Kim D.Y., Chung S.H. Anti-diabetic effect of pectinase-processed ginseng radix (GINST) in high fat diet-fed ICR mice. J Ginseng Res. 2011;35:308–314. doi: 10.5142/jgr.2011.35.3.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song Y.B., An Y.R., Kim S.J., Park H.W., Jung J.W., Kyung J.S., Hwang S.Y., Kim Y.S. Lipid metabolic effect of Korean red ginseng extract in mice fed on a high-fat diet. J Sci Food Agric. 2012;92:388–396. doi: 10.1002/jsfa.4589. [DOI] [PubMed] [Google Scholar]
- 49.Kim C.M., Yi S.J., Cho I.J., Ku S.K. Red-koji fermented red ginseng ameliorates high fat diet-induced metabolic disorders in mice. Nutrients. 2013;5:4316–4332. doi: 10.3390/nu5114316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwon D.H., Bose S., Song M.Y., Lee M.J., Lim C.Y., Kwon B.S., Kim H.J. Efficacy of Korean red ginseng by single nucleotide polymorphism in obese women: randomized, double-blind, placebo-controlled trial. J Ginseng Res. 2012;36:176–189. doi: 10.5142/jgr.2012.36.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song M.Y., Kim B.S., Kim H. Influence of Panax ginseng on obesity and gut microbiota in obese middle-aged Korean women. J Ginseng Res. 2014;38:106–115. doi: 10.1016/j.jgr.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.