Abstract
Background
The lepidopteran Asiatic corn borer (ACB), Ostriniafurnacalis (Guenee), has caused huge economic losses throughout the Asian-Western Pacific region. Usually, chemical pesticides are used for the control, but excessive use of pesticides has caused great harm. Therefore, the inartificial ecotypic pesticides to ACB are extremely essential. In our previous study, we found that panaxadiol saponins (PDS) can effectively reduce the harm of ACB by causing antifeedant activity. Therefore, it is necessary to reveal the biological molecular changes in ACB and the functionary mechanism of PDS.
Methods
We analyzed the global transcription of ACB with different PDS concentration treatment (5 mg/mL, 10 mg/mL, and 25 mg/mL) by high-throughput sequencing and de novo transcriptome assembly method.
Results
PDS treatment could cause the changes of many gene expressions which regulate its signal pathways. The genes in peroxisome proliferator–activated receptor (PPAR) signaling pathway were significantly downregulated, and then, the downstream fatty acid degradation pathway had also been greatly affected.
Conclusion
Through this experiment, we hypothesized that the occurrence of antifeedant action of ACB is because the PDS brought about the downregulation of FATP and FABP, the key regulators in the PPAR, and the downregulation of FATP and FABP exerts further effects on the expression of SCD-1, ACBP, LPL, SCP-X, and ACO, which leads to the disorder of PPAR signaling pathway and the fatty acid degradation pathway. Not only that, PDS treatment leads to enzyme activity decrease by inhibiting the expression of genes associated with catalytic activity, such as cytochrome P450 and other similar genes.
Keywords: Antifeedant, Asiatic corn borer, Fatty acid, Panaxadiol saponins, Peroxisome proliferator–activated receptor
1. Introduction
The lepidopteran Asiatic corn borer (ACB), Ostrinia furnacalis (Guenee), is one of the most widespread and common pests throughout China and other Asian regions and causes great losses to corn, sorghum, millet, and cotton [1]. The annual maize yield losses are estimated to be 6–9 million tons [2]; therefore, it is urgent to carry out the comprehensive management for the ACB. In addition, ACB acts as a devastating pest species for the following reasons: First, the adaptation to many host crops; second, the high fecundity; and third, the high capacity to evolve resistance to Bt [3]. Spraying insecticide is widely used to manage ACB and other agricultural insect pests. But insecticide abuse may cause great harm to the environment, ecological equilibrium, and human health [4]. In Pakistan, because of the abuse of pesticides, the biological diversity is losing, and poisoning the local food chains, more importantly, the local human health have been greatly affected, such as cancer, low fertility, and destruction of the human body's immune system [5]. Chemical insecticides could pollute the tissues of virtually every life form on the earth, the air, the lakes, and the oceans, and then affect organisms that live in polluted environments [6]. According to the US National Academy of Sciences, the bald eagle population in the United States declined primarily because of exposure to dichlorodiphenyltrichloroethane (DDT) and its metabolites [7]. Certain environmental chemicals, including pesticides termed as endocrine disruptors, are known to elicit their adverse effects by mimicking or antagonizing natural hormones in the body, and it has been postulated that their long-term, low-dose exposure is increasingly linked to human health effects such as immune suppression, hormone disruption, diminished intelligence, reproductive abnormalities, and cancer [8], [9]. Therefore, a scientific, biosafe, and environment friendly pesticide to ACB control strategy is extremely essential.
Botanical insecticides are considered to be an effective alternative to synthetic chemical pesticides for pest management because botanicals are assumed to pose little threat to the environment and human health [10]. As a traditional Chinese medicine, ginseng has been widely used in China and other Asian countries, which acts as one of the worldwide botanical medicines [11], [12], [13]. According to our previous research, the extract of ginseng stem and leaf (total ginsenoside) showed inhibitory effect on insect feeding [14]. Therefore, ginseng extract can be used as a candidate plant source insectifuge. Botanical antiinsect agents are a sort of compounds which can inhibit insects but do not directly kill them. Of all the metabolites in plants, there are only a few categories that can cause insect antifeedant action, such as quassins, limonoids, sesquiterpene lactones, and heterogeneous flavonoids [15]. Some of these agents show relative antifeedant activity only in a certain period, but they do affect insect host selection. According to previous studies, botanical antifeedants can be rapidly degraded after application; therefore, it is a kind of environment friendly insecticide and causes little environmental impact [16]. Some research also suggests that the plant extracts showed antifeedant effect as a consequence of the combined effects of several constituents with no individual compound making a major contribution [17]. During the process of coordinated evolution with insects, many secondary metabolites, such as terpenes, alkaloids, and phenols, have been formed in plants [18]. These secondary metabolites can induce insect antifeedant behavior by altering the metabolism of insect hormones, affecting the nervous system, affecting the digestive system, and affecting the respiratory system [19], [20]. Glutathione S-transferase (GST) and acetylcholinesterase (AChE) play important roles in insect metabolism and resistance to study, but the molecular mechanism is still unknown [21].
The ginsenoside can effectively inhibit population and development of O. furnacalis, particularly the third instar. Higher concentration of ginsenosides had significantly longer developmental time [13]. But the main components of ginsenoside can be divided into panaxadiol saponins (PDS) and ginseng three alcohol panaxtrol saponins (PTS) [22], [23]. PDS can produce great physiological and biochemical changes to insects, which showed specific physiological function such as calcium channel blockade [24], anti-free radical actions [25], and regulation of lipid metabolism [26]. We also studied the effect of PDS and PTS on the antifeeding activity of third instar of ACB; the result indicated that the antifeeding activity of PDS was obviously higher than that of PTS. As a result, PDS is used in many medicines, especially in the treatment of cardiovascular diseases and immune responses [27], [28]. However, there is no extensive research in agricultural applications.
In this study, to identify the antifeedant activity regulatory genes, high-throughput sequencing technology was applied to explore the potential antifeedant activity regulated genes. The differentially expressed genes (DEGs) were screened and validated by quantitative polymerase chain reaction (qPCR) to ensure the accuracy of the experiment. Through the analytical result of the transcriptome, we tried to reveal the molecular mechanism of antifeedant activity and put forward some theoretical guidance. To our knowledge, this research was the first reported in application of extract of ginseng stem and leaf in ACB control, and the result of this study might provide a certain theory basis to future research and practical application.
2. Materials and methods
2.1. PDS extraction and antifeedant experiment on ACB
The stems and leaves of Panax ginseng were obtained from Fusong County, Jilin Province, China in 2016, and were stored at room temperature. To prepare the saponins extracts, a total of 1.0 kg of dry powder of P. ginseng stems and leaves was extracted with hot water for 2 h at 100°C. Extracts were added to distilled water to prepare a solution of 100 g/L. Then, the solution was filtered by D101 resin column with distilled water, 35% ethanol and 80% ethanol successively. 80% ethanol eluate was collected and concentrated in vacuo to dryness to obtain the PDS. The contents of ginsenosides of PDS were determined with HPLC, and five kinds of ginsenosides were found, including ginsenosides Rb1, Rc, Rb2, Rb3, and Rd, whose contents were 4.68%, 11.03%, 12.06%, 1.66%, and 53.16%, respectively, for a total of 82.79%. The individual ginsenoside was shown in Table 1. The appropriate amount of PDS was weighed and dissolved in water to establish solutions at different concentrations (2.5 mg/mL, 5.0 mg/mL, 10.0 mg/mL, 25 mg/mL, and 50 mg/mL). Finally, the artificial diets with PDS were stored at 4°C in a refrigerator. The normal artificial diet without adding any ginsenosides was used as the control.
Table 1.
Contents of individual ginsenoside from the stems and leaves of PDS.
| No. | Ginsenosides | Content (%) |
|---|---|---|
| 1 | Rb1 | 4.68 ± 0.07 |
| 2 | Rc | 11.03 ± 0.29 |
| 3 | Rb2 | 12.06 ± 0.16 |
| 4 | Rb3 | 1.66 ± 0.18 |
| 5 | Rd | 53.16 ± 0.21 |
| Total | 82.79 ± 1.25 |
Leaf disk test was used to test antifeedant rate of ACB [29]. Freshly laid eggs (<4 h) were collected and placed into petri dishes (diameter × height: 9 cm × 2 cm), and a wet cotton ball was also placed inside each petri dish to maintain the humidity. Newly hatched larvae were collected and separately reared in petri dishes under the incubator chambers (PRS-20; Ningbo Saifu Experimental Instrument Co., Ltd., Zhejiang, China) at 27 ± 1°C, 70 ± 5% relative humidity (RH), and a photoperiod of 14:10, L:D. Ten newly molted third-instar larvae were placed into petri dishes. After starvation for 4 h, six excised corn leaf discs (Φ = 15 mm), soaked in solutions with different concentrations of PDS for 10 s (control discs received distilled water only), were supplied to all larvae. The area of feeding on the leaves was measured every 24 h, and the corresponding nonselective antifeeding activity was calculated. Each test was repeated 10 times.
2.2. ACB development inhibition experiment and RNA-seq sample preparation
Diet incorporation method was used to test development inhibition of ACB [30].
Newly molted third-instar larvae with the same size were selected and weighed and then placed into 24 wells culture boxes. After starvation for 4 h, ACB was fed with artificial diets having different PDS concentrations (0 mg/mL, 2.5 mg/mL, 5.0 mg/mL, 10.0 mg/mL, 25 mg/mL, and 50 mg/mL). Artificial diets were replaced at regular intervals. After 72 h, the larvae were weighed again; then, inhibition rate of weight was calculated. Each test was repeated 10 times. When the development inhibition experiment was completed, all the ACB were stored in liquid nitrogen (−196°C condition) for the following experiments.
2.3. RNA isolation and library preparation for illumina deep sequencing
Three micrograms of purified RNA for every sample was used to construct a sequence library. RNA isolation and purity were performed by trizol (Invitrogen, CA, USA) and 1% agarose gels, respectively, following the Sultan's method [31]. The RNA concentration and purity were measured by Qubit® RNA Assay Kit in QubitFlurometer (CA, USA). After the RNA preparation, a total of not less than 3 μg RNA was used as the input for the preparation of RNA sequencing libraries for each sample. NEBNext RNA Library Prep Kit (New England Biolabs, MA, USA) was used for generating the sequence libraries for Illumina sequence. To distinguish different sequence libraries, the unique indices were used in each library to attribute demultiplexed reads. Briefly, mRNA with poly A was selected by magnetic beads from the total RNA. Purified mRNA was fragmented using bivalent cation under high temperature in 5× NEBNext First Strand Synthesis Reaction mix. Universal primer and M-MuLV Reverse Transcriptase (RNase H) were used to synthesize the first strand, and then, with DNA polymerase I and RNase H, the second strand was synthesized. Proceeding to the next step, the ends of cDNA were modified into blunt ends by exonuclease and polymerase. Subsequently, the blunted DNA was adenylated at the 3′ ends, and the sequence adapter was ligated to the DNA to facilitate hybridization. The sequencing libraries were purified with AMPure XP system (Beckman Coulter, Inc., Beverly, USA), and the fragments of 150–200 base pairs were enriched. Then, adding 3 μl of Uracil-specific excision reagent (USER) Enzyme into the cDNA solution, the reactions were incubated at 37°C for 15 min and then for 5 min at 95°C. PCR was performed with thermostable DNA polymerase. Finally, the PCR products were purified, and the library quality was assessed on the Agilent Bioanalyzer 2100 system (Agilent Technologies, Palo Alto, CA, USA). The clustering of sequence libraries was performed on a cBot Cluster Generation System. After cluster generation, the enriched cDNA was sequenced using an Illumina Hiseq 4000 platform (Illumina, San Diego, CA, USA).
2.4. Bioinformatic analysis
2.4.1. Sequence data quality control
Raw data (raw reads) of fastq format were first conducted for quality control, processed through an in-house perl script. Quality of the sequence reads was verified with FastQC (version 0.10.0) software (Cock et al, 2010). In this step, the data were filtered according to the following indicators: reads containing more than 10% ploy-N should be removed; the reads containing more than 50% low-quality bases (Q ≤ 20) should be removed; reads containing sequence adapters should be removed. After the data quality control process, clean data were obtained and used for the subsequent data analysis.
2.4.2. De novo transcriptome assembly
Trinity [32]was used to assemble a reference transcriptome of ACB with the clean reads of three libraries. The read1 files from all of the three libraries were pooled into a single file, and similarly, the read2 files were merged into one read2.fq file. Transcriptome assembly was performed using the pooled read1.fq and read2.fq files using the Trinity algorithm with the min_kmer_cov set 2, and all the other parameters were set to their default values.
2.4.3. Transcriptome annotation
The annotation of assembled transcriptome was performed via six databases: NCBI nonredundant protein sequences (NR), National Coalition Building Institute (NCBI) nonredundant nucleotide sequences (NT), Swiss-Prot, Protein family (Pfam), Clusters of Orthologous Groups (COGs) of proteins, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Ontology (GO). Swiss-Prot annotation was performed with an E-value threshold of 1.0E-5 and for COG/KOG. E-value threshold of 1.0E-3 was set to predict the function of genes. Blast2GO software program [33] was used to perform GO annotations defined. The pathway assignments were determined based on the KEGG database using BLASTX with an E-value threshold of 1.0E-5.
2.4.4. Differential expression, gene ontology, and KEGG pathway enrichment analysis
For sequence data in each sample, edgeR software package was used to statistically analyze the read counts with adjusted through one scaling normalized factor. Gene expression levels for each sample were counted using RSEM [34].To get the global gene expression pattern in different conditions, we performed pairwise comparisons using the DEseq (2010) R package for screening DEGs [35]. The resulting p values were adjusted using the q values [36]. Differences were considered significant if q value < 0.005. To compare the unigene expression levels, each unigene was further normalized by fragments per kilobase of exon model per million mapped reads [37]. In addition, the DEGs were also used to deduce expression patterns using self-organizing map clustering analysis based on the log2 (ratios) value (fragments per kilobase of exon model per million mapped reads analysis) [38].GO enrichment analysis of the DEGs was performed using the GOseq R package [39]. KEGG is a database that can exhibit the high-level functions and biological system processes including cell, organism, and ecosystem, which contains a large number of biological process information in molecular level including large-scale molecular datasets generated by high-throughput sequencing [40]. KOBAS software was used to test the significance of the KEGG pathway's enrichment in the DEGs [41].
2.5. Real-time reverse-transcription PCR confirmation of illumina sequencing data
Quantitative reverse-transcription PCR (RT-qPCR) was executed on an Applied Biosystems 7500 system (Applied Biosystems, CA, USA) using three replications according to previous reports [42], [43]. All the PCR reactions were carried out in a total volume of 20 μL with the RsActin gene as the internal control [44]. The relative gene expression levels were calculated using the 2−ΔΔCT method.
3. Results
3.1. Antifeedant activity of PDS on third ACB
After PDS treatment, ACB larvae showed strong antifeedant activity. The antifeedant rate increased with the increasing PDS concentration. Significant differences were observed in each treatment group with different PDS concentrations, except for the 2.5 mg/mL and 5 mg/mL group. However, the antifeeding effects of each treatment group were similar on 24 h, 48 h, and 72 h (Table 2).The ACB larval weight data showed that with the increase of concentration, the inhibition rate of weight (72 h) was increased from 5.10% to 87.41%, and significant differences were observed in each treatment group, except for the 2.5 and control check (CK) group (Table 3). On the basis of antifeedant and weight growth inhibition data, 5 mg/mL, 10 mg/mL, 25 mg/mL, and 50 mg/mL were feasible for RNA-seq analysis. But in 50 mg/mL group, the weight of larva was too small for RNA extraction. In summary, 5 mg/mL (C5), 10 mg/mL (C10), and 25 mg/mL (C25) were selected as the final RNA-seq experimental concentration.
Table 2.
Antifeedant activity of PDS.
| Concentration (mg/ml) | 24h |
48h |
72h |
|||
|---|---|---|---|---|---|---|
| Eating area(mm2) | Anti-feeding rate(%) | Eating area(mm2) | Anti-feeding rate(%) | Eating area(mm2) | Anti-feeding rate(%) | |
| 2.5 | 361.60±15.01 | 29.53d | 510.00±33.78 | 44.35d | 399.80±15.91 | 33.98d |
| 5 | 345.24±20.07 | 32.76d | 475.33±33.97 | 47.97d | 391.84±25.47 | 35.46d |
| 10 | 314.67±28.36 | 38.82c | 425.25±24.85 | 53.57c | 343.63±4.73 | 43.35c |
| 25 | 272.15±19.63 | 46.83b | 318.67±26.12 | 65.32b | 312.67±19.50 | 48.43b |
| 50 | 223.27±9.85 | 56.47a | 234.52±22.07 | 74.38a | 275.33±26.02 | 51.93a |
| CK | 513.67±16.92 | – | 916.33±58.59 | – | 602.67±15.28 | – |
Note: The data with different capital letters in same column show extremely significant difference (p≤0.01); the data with different little letters in same column show significant difference (p≤0.05).
Table 3.
Effect of PDS on body weight.
| Concentration (mg/ml) | Initial weight (mg) | Weight after 72h (mg) | Inhibition rate of weight(%) |
|---|---|---|---|
| 2.5 | 32.74±2.32a | 79.92±5.50a | 5.10 |
| 5 | 30.54±2.51a | 70.11±6.47b | 20.41 |
| 10 | 29.42±2.79a | 54.34±5.78c | 49.88 |
| 25 | 30.14±4.84a | 45.39±4.02d | 69.34 |
| 50 | 32.40±32.97a | 38.66±2.78e | 87.41 |
| CK | 33.10±2.27a | 82.82±7.84a | – |
Note: The data with different capital letters in same column show extremely significant difference (p≤0.01); the data with different little letters in same column show significant difference (p≤0.05).
3.2. Transcriptome sequencing and de novo sequence assembly
To obtain the Ostrinia furnacalis transcriptome expression profile with the treatment of different concentration extracts of ginseng (total ginsenoside), 12 Ostrinia furnacalis samples in four groups (three biological repeats in each group): CK group, C5, C10, and C25 extracts treatment were sequenced. In total, 87.71 Giga Illumina sequence data were generated, and each sample was not less than 6.51 Giga (Table 4). Illumina sequencing data from ACB were deposited in the NCBI sequence read archive (SRA) database under accession number SRP093447. In this study, Trinity was used to assemble the 584,748,982 sequenced reads. These reads were assembled into about 153,740 transcripts and 104,575 unigenes with the mean length of 990 and 727 nt, and N50 length was 2096 and 1466 nt for transcripts and unigenes, respectively (Table 5). Correlation of gene expression pattern between biological repeated samples is an important index to test the reliability of the experiment; general correlation coefficient is used to check the consistency of all the biological repeats. The results reflected that the mean value of correlation coefficient was 0.771, 0.789, 0.667, and 0.623 in the CK, C5, C10, and C25, respectively (Fig. 1). This showed that with the increase of the extracts of ginseng concentration, there was a slight decrease in the reproducibility of repeated samples.
Table 4.
Assemble length distribution.
| Transcripts | Unigenes | |
|---|---|---|
| Number | 153740 | 104575 |
| Min Length | 201 nt | 201 nt |
| Max Length | 29016 nt | 29016 nt |
| Mean Length | 428nt | 336 nt |
| N50 | 2096 nt | 1466 nt |
| Total Nucleotides | 152138941 | 75990654 |
Table 5.
Illumina sequence data summary.
| Sample | Raw Reads | Clean reads | Clean bases | Error(%) | Q20(%) | Q30(%) | GC(%) |
|---|---|---|---|---|---|---|---|
| CK_1 | 47969924 | 46727642 | 7.01G | 0.02 | 97.18 | 92.74 | 48.71 |
| CK_2 | 48210238 | 46983082 | 7.05G | 0.02 | 97.26 | 92.88 | 47.39 |
| CK_3 | 47621326 | 46338836 | 6.95G | 0.02 | 97.22 | 92.86 | 48.81 |
| C10_1 | 51912874 | 50507622 | 7.58G | 0.02 | 97.3 | 93.04 | 48.05 |
| C10_2 | 54575068 | 53123724 | 7.97G | 0.02 | 97.24 | 92.88 | 47.86 |
| C10_3 | 43434834 | 42308338 | 6.35G | 0.02 | 97.16 | 92.72 | 47.98 |
| C25_1 | 53046134 | 51653988 | 7.75G | 0.02 | 97.21 | 92.85 | 47.72 |
| C25_2 | 50670778 | 49335294 | 7.4G | 0.02 | 97.2 | 92.83 | 47.43 |
| C25_3 | 48802216 | 47529824 | 7.13G | 0.02 | 97.22 | 92.87 | 48.21 |
| C5_1 | 46545984 | 45367348 | 6.81G | 0.02 | 97.36 | 93.08 | 48.36 |
| C5_2 | 48159566 | 46831892 | 7.02G | 0.02 | 97.22 | 92.85 | 48.22 |
| C5_3 | 43800040 | 42646128 | 6.4G | 0.02 | 97.31 | 93.03 | 48.56 |
Fig. 1.
Mean value of correlation coefficient of biological repeated samples. The darker color indicates the greater value of the correlation coefficient.
CK, control check.
3.3. Sequence annotation
The number of unigenes annotated in at least one of the NR, NT, Swiss-Prot, KEGG, COG, and GO databases was 31,698. We got 25,119 (24.02%), 15,160 (14.49%), 10,213 (9.76%), 18,382 (17.57%), 19,675 (18.81%), 19,940 (19.06), and 12,377 (11.83%) unigenes which annotated to the NR, NT, KO, Swiss-Prot, Pfam, GO, and KOG database, respectively. The comparisons of unigene annotation in the NT, NR, KOG, GO, and Pfam databases were showed in a Venn diagram (Fig. 2). BLASTx homology searches of all the annotated 31,698 unigenes showed that 25,123 (79.26%) had homologous genes in the nonredundant (NR) protein database. The best match percentage (27.8%) was with Bombyx mori sequences, followed by sequences from Danaus plexippus (15.5%), Plutella xylostella (14.6%), Sus scrofa (5.6%), and Homo sapiens (4.6%) (Fig. 3).
Fig. 2.
Unigenes annotation in different database. Different colors represent different databases or intersection of different databases.
GO, Gene Ontology; NR, NCBI nonredundant protein sequences; NT, NCBI nonredundant nucleotide sequences; Pfam, Protein family.
Fig. 3.
The percentage of annotated unigenes when matched with different species. Different colors represent different species.
3.4. DEGs analysis of Ostriniafurnacalis with different extracts concentration treatments
To better investigate the biological mechanism of antifeedants in ACB, it is critical to identify the DEGs among different extracts concentration treatments. A high number of unigenes were affected with different extracts concentration treatments. Of all the genes, only the genes that adjusted p value not more than 0.001 were considered significant. The DEGs were compared under the CK, C5, C10, and C25 group. There were 377 DEGs between group C5 and CK; 1,119 DEGs between group C10 and CK; whereas in the comparisons between C25 and CK, 1,906 DEGs were differently expressed (Fig. 4). In this study, DEGs with higher expression in PDS treatment compared with CK were represented as “upregulated” genes, whereas those with lower expression levels in PDS treatment were “downregulated” genes. The expression levels of 162, 342, and 693 genes were upregulated in the C5, C10, and C25, respectively, whereas the other 215, 777, and 1,213 genes showed downregulated. Overall, the amount of downregulated genes was more than the upregulated genes. The hierarchical cluster analysis showed that expression patterns were more similar in C5 and CK than those in C25 and C10 (Fig. 5). But there was still a big difference in C5 and CK.
Fig. 4.
Statistical result of differently expressed genes. (A) The common and specific DEGs in different comparison (C5 versus CK, C10 versus CK, and C25 versus CK). (B) The upregulated and downregulated genes in the comparison of C5 and CK. (C) The upregulated and downregulated genes in the comparison of C10 and CK. (D) The upregulated and downregulated genes in the comparison of C25 and CK.
CK, control check; DEG, differently expressed gene.
Fig. 5.
Cluster analysis of differentially expressed genes. Red indicates high expression, and blue indicates low expression. Color from red to blue, said log10 (FPKM+1) from large to small.
CK, control check; FPKM, fragments per kilobase of exon model per million mapped reads.
3.5. Functional distribution of DEGs
To investigate the biological event that DEGs mainly involved during bolting development, a functional categorization was carried out with the GO analysis (Fig. 6).
Fig. 6.
GO functional categorization. X-axis represents the GO terms; Y-axis represents the number of DEGs. BP, MF, and CC represent biological process, molecular function, and cellular component, respectively.
CK, control check; DEG, differently expressed gene; GO, Gene Ontology.
The clustering results, GO enrichment of downregulated DEGs, and the total DEGs were obtained in each comparison. The GO enrichment results (C5 vs. CK, C10 vs. CK, C25 vs. CK, C10 vs. C5, C25 vs. C5, and C25 vs. C10) were shown in supplementary 1 to 6. In the GO clustering results, GO terms “catalytic activity”, “oxidoreductase activity”, “hydrolase activity”, “iron ion binding”, “fatty acid metabolic process”, “lipid metabolic process,” and “NADP metabolic process” were significantly enriched in comparison of extracts treatment and control, especially in the high concentration treatment (C10 and C25, q ≤ 0.001). In the GO terms aforementioned, the top enriched term was catalytic activity, which belongs to the molecular function category. The result showed that there were greater differences in the result of enriched GO terms in both the comparisons, C10 versus CK and C25 versus CK, than the comparison between C5 and CK, not just in the number of DEGs. In other words, with the increase in the concentration of extracts, the difference between treatment and control group was becoming wider. In catalytic activity GO term, 84 significantly downregulated genes were found (q < 0.05), including the key enzymes in drug metabolism cytochrome P450.
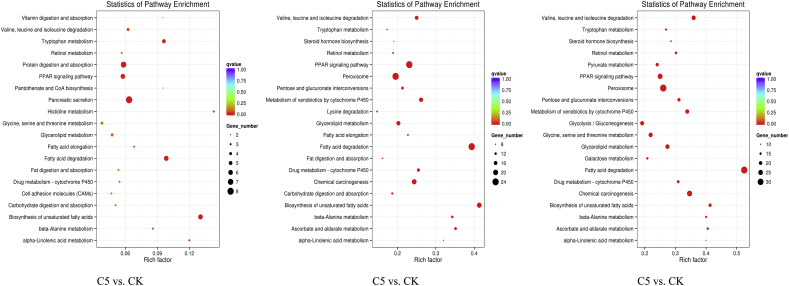
The antifeedants reaction of Ostrinia furnacalis is a coreaction result of many inner-cell metabolic pathways. KEGG pathway analysis was carried out to judge the DEGs function and pathways enrichment. Fatty acid degradation, biosynthesis of unsaturated fatty acids, and peroxisome proliferator–activated receptor (PPAR) signaling pathways were the critical signaling pathways in the top-10 enriched pathways (q ≤ 0.05) in extracts treatment, compared with CK(Fig. 7). The KEGG pathway enrichment of DEGs in extracts treatment was shown in supplementary 7 to 12. The KEGG pathway of PPAR signaling pathway showed statistical significance (q ≤ 0.01) when the extracts treatment was compared with the control group, whereas the comparisons with that of the C10 versus C5 and C25 versus C10 did not show any significant enrichment results in PPAR signaling pathway (q ≥ 0.05). The results showed that 6, 24, and 26 differently expressed unigenes were detected in C5, C10, and C25, respectively, compared with CK. According to the annotation results, the genes FATP and FABP (key genes in PPAR signaling pathway) both showed downregulated in extracts treatment. Meanwhile, the genes SCD, ACO, SCP, and so forth in the pathway also showed differential expressions. In addition, the glutathione metabolism pathway was also significantly enriched. Interestingly, in the glutathione metabolism pathway, all the DEGs were downregulated after PDS treatment, which included the key genes GST, CARP, and GPX.
Fig. 7.
KEGG functional categorization. The vertical axis represents the name of the pathway; the horizontal axis represents pathway corresponding rich factor. The size of q value was represented by the color of the point; the smaller the q value, the closer to the color of red.
CK, control check; PPAR, peroxisome proliferator–activated receptor.
3.6. Expression profile analysis by RT-qPCR
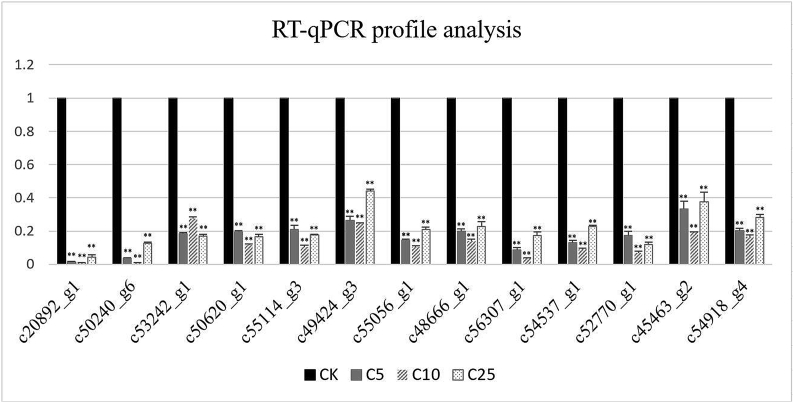
To validate the differential expression patterns of DEGs, the transcriptional level of 13 unigenes was examined by real-time quantitative PCR. The 13 unigenes included the transcript of FATP (c50620_g1 and c55114_g3) and FABP (c49424_g3) in PPAR signaling pathway and cytochrome P450 (c20892_g1, c50240_g6, and c53242_g1) in the catalytic activity GO terms. SCD-1 (c55056_g1, c48666_g1) and ACO (c56307_g1, c54537_g1, and c52770_g1), as the downstream gene of FATP and FABP which can influence the function of fatty acid degradation pathway, were also verified in this study. All the primers designed to the ACB were shown in Table 6. These selected genes were related to fatty acid degradation and PPAR signaling pathway. The results of RT-PCR were consistent with the RNA-Seq result; all the unigenes were significantly downregulated when compared with CK (p < 0.01, Fig. 8).
Table 6.
Primer for qRT-PCR.
| Unigene | Primer sequences (5ʹ-3ʹ) | Annealing temperature (°C) | Product length (bp) |
|---|---|---|---|
| c20892_g1 | F: GTGCCTTTTATCTTCGCCTACT | 57.8 | 142 |
| R: GGAACGCCTCAGAAAGATTG | 57.3 | ||
| c50240_g6 | F: AGATTTCTACTACTTCAGCAGCCG | 59.6 | 116 |
| R: GGTTTTGACGCAACACCTTC | 57.9 | ||
| c53242_g1 | F: TTCGCAGACTTTACAACGGAC | 58.1 | 134 |
| R: CTCAACCCTCTGATGAAGTCGT | 58.3 | ||
| c50620_g1 | F: CAACAAAATGGGAGCAATAGG | 57.7 | 75 |
| R: TCTACTTTCAGAATAGCGATGGG | 58.5 | ||
| c55114_g3 | F: ATGCTGTGGTCTATGGTGTTGA | 57.8 | 234 |
| R: CTTTATTTGGGTCGTAGCCTTC | 58.1 | ||
| c49424_g3 | F: GTGCCAAGGTCAAGTCAGTGT | 57.2 | 129 |
| R: CGGTCATCACAGCCTTCAAC | 58.5 | ||
| c55056_g1 | F: GGTTTCCACAACTACCACCACA | 59.7 | 142 |
| R: CGTCGGAGACCGTTTTCAA | 59.2 | ||
| c48666_g1 | F: TCCTTACGGTATGTTTGGCG | 58.8 | 230 |
| R: TGTCTGCTTTCTGGACTGGG | 58.2 | ||
| c56307_g1 | F: TACGACGCAAAGACTGAGGAA | 58.5 | 158 |
| R: TGAACTATGAAGGGGTGGATG | 57.3 | ||
| c54537_g1 | F: GAAGCAGTATTTCCCCGATTT | 58.0 | 229 |
| R: TTGGACAGAGTACATGCGTTTG | 58.8 | ||
| c52770_g1 | F: TAACGGGTTTCTTGGCTTTG | 58.2 | 248 |
| R: GTTCTGGTCCATCCTTTCTCAC | 57.7 | ||
| c45463_g2 | F: GTTTTGGCGGTGGTGACTT | 58.0 | 128 |
| R: GGCTTCTTGGTTTACATCCCT | 57.8 | ||
| c54918_g4 | F: GACTGATTGACGAGCCACTACTT | 58.0 | 93 |
| R: TCTTCTCCGCCTTGGTCTTA | 57.5 | ||
| Of- actin | F: ATCATCACCAACTGGGACGA | 58.0 | 139 |
Fig. 8.
Expression profile analysis by RT-qPCR. The data represent the mean value of the results of three to nine independent experiments. *p < 0.05. **p < 0.01 compared with untreated control.
CK, control check; RT-qPCR, quantitative reverse-transcription polymerase chain reaction.
4. Discussion
Ginsenosides represent the major bioactive components of ginseng. These triterpenoid saponins have shown to exert numerous beneficial effects on the human body [45], [46], [47], [48]. In the 72 h after PDS treatment, the antifeedant rate increased first and then decreased, and the average antifeedant rate had the highest value in 48 h. This may be related to the tolerance of larvae to drugs. Similar results were also found in the study by Zhang et al [14]. However, the mechanism of PDS bioactive to ACB is not clear. We performed a transcriptome analysis of ACB treated with different concentrations. The reference genome of ACB has not yet been published, which created a great limit to the functional genomics research, but the transcription information of no reference genome insect can be achieved because the de novo RNA-seq assembly technology has matured. Four groups were set in this study, including CK group, C5, C10, and C25 ginseng extract treatments to research the molecular mechanisms of ginseng extract effectiveness.
In this study, 87.71 Giga Illumina sequence data were generated, and for each sample not less than 6.51 Giga, the amount of sequence data was enough for analyzing de novo transcriptome assembly [49]. For ACB, as a nonmodel insect without a reference genome sequence, the assembly by Trinity was better than that of other programs [32]. In the result of transcriptome sequencing and sequence assembly, these reads were assembled into 104,575 unigenes longer than 200 nt, which was close to the research by Liu et al in 2014 [50]. The size distribution indicated that the ratio of unigenes with a length of 200–1000 bp was 83.3%, while the length of 17,399 (13.51%) unigenes was more than 1000 bp. It was significantly larger than that in previous insect transcriptome projects [51], [52].
Comparison of gene expression among the different treatment groups in the present experiment is helpful for identification of candidate genes underlying response to different concentrations extract of ginseng treatment in ACB. In this study, we detected a total of 2,125 unigenes which were differentially expressed between the comparison of C5 versus CK, C10 versus CK, and C25 versus CK (q.value < 0.005| log2.Fold_change|>1). This result showed that the higher concentration of ginseng extract treatment, the greater number of DEGs, and the main components of ginseng extracts (total ginsenoside) were PDS [53], which indicated that higher concentration of PDS treatment can lead to greater transcription impact on ACB. To further unravel the significantly altered biological processes on PDS stress, the upregulated and downregulated genes were subjected to the GO term enrichment analysis. There were 13, 72, and 117 GO terms that showed significant enrichment in the comparison of C5, C10, and C25 versus CK, respectively (q value < 0.01), but no significantly enriched GO terms were found in the upregulated genes. Of all the enriched GO terms, iron ion binding (GO:0005506) was a common one in the three comparisons, which was not significant in the comparison of C10 versus C5, C25 versus C5, and C25 versus C10. Meanwhile, catalytic activity (GO:0003824) was also significantly enriched GO terms (q value < 0.01). The results indicated that a large number of catalytic activity related genes were downregulated, which may lead to the decreased of catalytic activity. Among the genes that were clustered into enzyme activity, the downregulated expression that was proved may cause enzyme activity to decrease [54]. From the results, it was speculated that ginseng PDS may lead to the alteration of enzyme activation, and reduced form of iron (Fe2+) was necessary for some enzyme catalytic activity [55], [56], which matched with our result of the iron ion binding GO term. In addition, P450 enzyme played a key role in the catalytic activity regulation of enzyme [57], [58]. In this study, PDS showed inhibitory effects on the expression of cytochrome P450, and most of the catalytic activity–enriched unigenes were associated with cytochrome P450. Similarly, a similar phenomenon has been found in previous studies [59], [60]. Therefore, we hypothesized that the inhibitory effect of ginsenoside on the enzyme activity was mainly achieved by downregulating the expression of cytochrome P450. The unigenes associated with cytochrome P450 were also verified by PCR; the result also showed the downregulation was matched with the sequence result. The PPAR was a ligand-dependent nuclear receptor that has been implicated in the modulation of critical aspects of development and homeostasis [61], including adipocyte differentiation, glucose metabolism [62], [63], and macrophage development and function [64]. PPAR had three subtypes (PPARalpha, beta/delta, and gamma) showing different expression patterns in vertebrates [65], [66]. Each of them was encoded in a separate gene and binds fatty acids and eicosanoids. There were a lot of research results showing that ginseng can cause changes in the PPAR pathway, which was one of the key pathways of ginsenoside pharmacological action [67], [68], [69]. The metabolism and synthesis of glutathione play an important role in insect growth and development, and GSTs were an important component of insect detoxifying enzymes [70]. A notable example of the function of glutathione (GSH) in insect intaking plants was the detoxifying effect of detoxifying enzymes on allyl isothiocyanate, and the previous studies have shown that isothiocyanates have insecticidal activity [71]. In this study, glutathione metabolism pathways were significantly affected by the PDS, and the GSTs showed downregulated in the C5, C10, and C25. PDS showed a powerful influence on the PPAR signaling pathway (ko03320). The PPARs have been proved that they can adjust the balance of glutathione system in vivo to improve oxidative stress [72]. Based on our results, PDS initiated changes of the DEGs in PPAR signaling pathway. When the content of PPARs was changed, the glutathione metabolism pathway was further affected, of which the key regulatory gene GST was downregulated. The instability of GSH content leads to the enrichment of isothiocyanates content in insects, which has a great influence on the growth and development, and even death. In addition, PPAR can mediate induction of the fatty acid hydroxylases of the cytochrome P450 4A family by many acidic chemicals [73], which was also confirmed in our results in the inhibition of the cytochrome P450 expression, and then affected the activity of enzymes subsequently. In the present study, the key gene in PPAR signaling pathway FATP and FABP was downregulated with the treatment of PDS and then caused the expression of downstream genes including SCD-1, ACBP, LPL, SCP-X, and ACO, which directly affected the downstream changes in fatty acid metabolism. The KEGG pathway fatty acid degradation (ko00071) was greatly affected (Fig. 9). To a certain extent, fatty acid metabolism was disturbed, due to the weight of ACB reduction. From all the results, it is concluded that PDS caused antifeedant effect by inhibiting the expression of genes in PPAR signaling pathway and then affected the downstream glutathione metabolism and fatty acid degradation pathway. The FATP and FABP were the key regulators in the PPAR, the downregulation of which caused the SCD-1, ACBP, LPL, SCP-X, and ACO expression to decrease. Not only that, PDS treatment led to enzyme activity decrease by inhibiting the expression of genes associated with catalytic activity, such as cytochrome P450 and other similar genes. By this aforementioned biological effect, PDS played a big role in the antifeedants action of ACB.
Fig. 9.
DEGs in PPAR signaling pathway. Genes in green box represent downregulated genes, and genes in red box represent upregulated genes.
DEG, differently expressed genes; PPAR, peroxisome proliferator–activated receptor; CPT, carnitine O-palmitoyltransferase.
Conflicts of interest
All authors have no conflicts of interest to declare.
Acknowledgments
This study was financially supported by Chinese National Agro-scientific Research in the Public Interest (201303111) and International Cooperation Program of Jilin Province Science and Technology Department (20160414008GH).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.jgr.2017.12.002
Contributor Information
Yonghua Xu, Email: xuyonghua777@yeah.net.
Lianxue Zhang, Email: ZLX863@163.com.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Nafus D.M., Schreiner I.H. Review of the biology and control of the Asian corn borer, Ostrinia furnacalis (Lep: Pyralidae) Trop Pest Manage. 1991;37:41–56. [Google Scholar]
- 2.He K., Wang Z., Zhou D., Wen L., Song Y., Yao Z. Evaluation of transgenic Bt corn for resistance to the Asian corn borer (Lepidoptera: Pyralidae) J Econ Entomol. 2003;96:935–940. doi: 10.1093/jee/96.3.935. [DOI] [PubMed] [Google Scholar]
- 3.Zhang T., He K., Wang Z. Transcriptome comparison analysis of Ostrinia furnacalis in four developmental stages. Sci Rep. 2016;6:e35008. doi: 10.1038/srep35008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hernandez M., Margalida A. Pesticide abuse in Europe: effects on the Cinereous vulture (Aegypius monachus) population in Spain. Ecotoxicology. 2008;17:264–272. doi: 10.1007/s10646-008-0193-1. [DOI] [PubMed] [Google Scholar]
- 5.lqbal Z., Zia K., Ahrnad A. Pesticide abuse in Pakistan and associated human health and environmental risks. Pak J Agric Sci. 1997;34:4. [Google Scholar]
- 6.Aktar M.W., Sengupta D., Chowdhury A. Impact of pesticides use in agriculture: their benefits and hazards. Interdiscip Toxicol. 2009;2:1–12. doi: 10.2478/v10102-009-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liroff R.A. 2000. Balancing risks of DDT and Malaria in the global POPs treaty. [Google Scholar]
- 8.Brouwer A., Longnecker M.P., Birnbaum L.S., Cogliano J., Kostyniak P., Moore J., Schantz S., Winneke G. Characterization of potential endocrine-related health effects at low-dose levels of exposure to PCBs. Environ Health Perspect. 1999;107:639–649. doi: 10.1289/ehp.99107s4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurley P.M. Mode of carcinogenic action of pesticides inducing thyroid follicular cell tumors in rodents. Environ Health Perspect. 1998;106:437–445. doi: 10.1289/ehp.98106437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isman M.B. Botanical insecticides, deterrents, and repellents in modern agriculture and an increasingly regulated world. Annu Rev Entomol. 2006;51:45–66. doi: 10.1146/annurev.ento.51.110104.151146. [DOI] [PubMed] [Google Scholar]
- 11.Jia L., Zhao Y. Current evaluation of the millennium phytomedicine–ginseng (I): etymology, pharmacognosy, phytochemistry, marketand regulations. Curr Med Chem. 2009;16:2475–2484. doi: 10.2174/092986709788682146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park J.G., Kang W.S., Park K.T., Park D.J., Aravinthan A., Kim J.H., Cho J.Y. Anticancer effect of joboksansam, Korean wild ginseng germinated from bird feces. J Ginseng Res. 2016;40:304–308. doi: 10.1016/j.jgr.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu T., Shen X., Yu H., Sun L., Lin W., Zhang C. Water-soluble ginseng oligosaccharides protect against scopolamine-induced cognitive impairment by functioning as an antineuroinflammatory agent. J Ginseng Res. 2016;40:211–219. doi: 10.1016/j.jgr.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang A.H., Tan S.Q., Zhao Y., Lei F.J., Zhang L.X. Effects of total ginsenosides on the feeding behavior and two enzymes activities of mythimna separata (Walker) larvae. Evid Based Complement Alternat Med. 2015;45:1828. doi: 10.1155/2015/451828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan L., Ren L., Chen F., Feng Y., Luo Y. Antifeedant activity of Ginkgo biloba secondary metabolites against Hyphantria cunea larvae: mechanisms and applications. Plos One. 2016;11:e0155682. doi: 10.1371/journal.pone.0155682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S.Q., Fang Y.L., Zhang Z.N. Studies and applications of botanical insect antifeedants. Entomol Knowl. 2005;5:491–496. [Google Scholar]
- 17.Khater H.F. Prospects of botanical Biopesticides in insect pest management. J ApplPharm Sci. 2012;3:641–656. [Google Scholar]
- 18.Chen Y., Zhong X., Chen L. Chinese herbal medicine resources with Bacteriostatic and insecticidal activities and their application as pesticides. Agric Sci Technol. 2013;14:1307–1314. 18. [Google Scholar]
- 19.Han J.Y., Zhang L.Z., Ji M.S. Research progress of botanical insecticides. Chin Agric Sci Bull. 2011;27:229–233. [Google Scholar]
- 20.Vos R.M., Van Bladeren P.J. Glutathione S-transferases in relation to their role in the biotransformation of xenobiotics. Chem Biol Interact. 1990;75:241–265. doi: 10.1016/0009-2797(90)90069-y. [DOI] [PubMed] [Google Scholar]
- 21.Zhang A.H., Tan S.Q., Zhao Y., Lei F.J., Zhang L.X. Effects of total ginsenosides on the feeding behavior and two enzymes activities of mythimna separata (Walker) larvae. Evidence-Based Complementary Altern Med. 2015:e451828. doi: 10.1155/2015/451828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu K., Zhang Y., Wang Y., Ling P., Xie X., Jiang C., Zhang Z., Lian X.Y. Ginseng Rb fraction protects glia, neurons and cognitive function in a rat model of neurodegeneration. Plos One. 2014;9:e101077. doi: 10.1371/journal.pone.0101077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu YP, Qian XD, Wang WX. Effect of panaxadiol saponin and panaxtrol saponin on proliferation of human bone marrow hemopoietic progenitor cells. Chin J Integr Med2004;24:127–129, 32. [PubMed]
- 24.Zhong G.G., Sun C.W., Li Y.Y., Qi H., Zhao C.Y., Jiang Y., Wang X.M., Yang S.J., Li H. Calcium channel blockade and anti-free-radical actions of panaxadiol saponins Rb1, Rb2, Rb3, Rc, and Rd. Acta Pharmacol Sinica. 1995;16:255–260. [PubMed] [Google Scholar]
- 25.Xie X.S., Liu H.C., Yang M., Zuo C., Deng Y., Fan J.M. Ginsenoside Rb1, a panoxadiol saponin against oxidative damage and renal interstitial fibrosis in rats with unilateral ureteral obstruction. Chin J Integr Med. 2009;15:133–140. doi: 10.1007/s11655-009-0133-9. [DOI] [PubMed] [Google Scholar]
- 26.Wang J., Cui C., Fu L., Xiao Z., Xie N., Liu Y., Yu L., Wang H., Luo B. Genomic expression profiling and bioinformatics analysis on diabetic nephrology with ginsenoside Rg3. Mol Med Rep. 2016;14:1162–1172. doi: 10.3892/mmr.2016.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim T.H., Lee S.M. The effects of ginseng total saponin, panaxadiol and panaxatriol on ischemia/reperfusion injury in isolated rat heart. Food Chem Toxicol. 2010;48:1516–1520. doi: 10.1016/j.fct.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 28.Song X., Bao S., Wu L., Hu S. Ginseng stem-leaf saponins (GSLS) and mineral oil act synergistically to enhance the immune responses to vaccination against foot-and-mouth disease in mice. Vaccine. 2009;27:51–55. doi: 10.1016/j.vaccine.2008.10.030. [DOI] [PubMed] [Google Scholar]
- 29.Müller-Schwarze D. Springer; New York: 2009. Leaf Disk test: Bioassay of Effect of Tannins on insect feeding behavior. [Google Scholar]
- 30.Nathan S.S., Sehoon K. Effects of Melia azedarach L. extract on the teak defoliator Hyblaea puera Cramer(Lepidoptera: hyblaeidae) Crop Prot. 2006;25:287–291. [Google Scholar]
- 31.Zhang Y.W., Li Yan. Cytochrome P450 research review. J Jilin Med Coll. 2005;26:4–6. [Google Scholar]
- 32.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conesa A., Gotz S., Garcia-Gomez J.M., Terol J., Talon M., Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 34.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L., Feng Z., Wang X., Wang X., Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 36.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., Van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamayo P., Slonim D., Mesirov J., Zhu Q., Kitareewan S., Dmitrovsky E., Lander E.S., Golub T.R. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci U S A. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:480–484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao X., Cai T., Olyarchuk J.G., Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 42.Nie S., Xu L., Wang Y., Huang D., Muleke E.M., Sun X., Wang R., Xie Y., Gong Y., Liu L. Identification of bolting-related microRNAs and their targets reveals complex miRNA-mediated flowering-time regulatory networks in radish (Raphanus sativus L.) Sci Rep. 2015;5:e14034. doi: 10.1038/srep14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu L., Wang Y., Liu W., Wang J., Zhu X., Zhang K., Yu R., Wang R., Xie Y., Zhang W. De novo sequencing of root transcriptome reveals complex cadmium-responsive regulatory networks in radish (Raphanus sativus L.) Plant Sci. 2015;236:313–323. doi: 10.1016/j.plantsci.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 44.Xu Y., Zhu X., Gong Y., Xu L., Wang Y., Liu L. Evaluation of reference genes for gene expression studies in radish (Raphanus astivus L.) using quantitative real-time PCR. Biochem Biophys Res Commun. 2012;424:398–403. doi: 10.1016/j.bbrc.2012.06.119. [DOI] [PubMed] [Google Scholar]
- 45.Ahmed T., Raza S.H., Maryam A., Setzer W., Braidy N., Nabavi S.F., de Oliveira M.R., Nabavi S.M. Ginsenoside Rb1 as neuroprotective agent: a review. Brain Res Bull. 2016;125:30–43. doi: 10.1016/j.brainresbull.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 46.Hwang S.H., Lee B.H., Choi S.H., Kim H.J., Won K.J., Lee H.M., Rhim H., Kim H.C., Nah S.Y. Effects of gintonin on the proliferation, migration, and tube formation of human umbilical-vein endothelial cells: involvement of lysophosphatidic-acid receptors and vascular-endothelial-growth-factor signaling. J Ginseng Res. 2016;40:325–333. doi: 10.1016/j.jgr.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee S.E., Park Y.S. Korean Red Ginseng water extract inhibits COX-2 expression by suppressing p38 in acrolein-treated human endothelial cells. J Ginseng Res. 2014;38:34–39. doi: 10.1016/j.jgr.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan H.D., Kim J.T., Kim S.H., Chung S.H. Ginseng and diabetes: the evidences from in vitro, animal and human studies. J Ginseng Res. 2012;36:27–39. doi: 10.5142/jgr.2012.36.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y., Shen D., Zhou F., Wang G., An C. Identification of immunity-related genes in Ostrinia furnacalis against entomopathogenic fungi by RNA-seq analysis. Plos One. 2014;9:e86436. doi: 10.1371/journal.pone.0086436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu S., Wei W., Chu Y., Zhang L., Shen J., An C. De novo transcriptome analysis of wing development-related signaling pathways in Locusta migratoria manilensis and Ostrinia furnacalis (Guenee) Plos One. 2014;9:e106770. doi: 10.1371/journal.pone.0106770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu J.Y., Yang P., Zhang Z., Wu G.X., Yang B. Transcriptomic immune response of Tenebrio molitor pupae to parasitization by Scleroderma guani. Plos One. 2013;8:e54411. doi: 10.1371/journal.pone.0054411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karatolos N., Pauchet Y., Wilkinson P., Chauhan R., Denholm I., Gorman K., Nelson D.R., Bass C., ffrench-Constant R.H., Williamson M.S. Pyrosequencing the transcriptome of the greenhouse whitefly, Trialeurodes vaporariorum reveals multiple transcripts encoding insecticide targets and detoxifying enzymes. BMC Genomics. 2011;12:56. doi: 10.1186/1471-2164-12-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang R.W., Lau K.M., Hon P.M., Mak T.C., Woo K.S., Fung K.P. Chemistry and biological activities of caffeic acid derivatives from Salvia miltiorrhiza. Curr Med Chem. 2005;12:237–246. doi: 10.2174/0929867053363397. [DOI] [PubMed] [Google Scholar]
- 54.Cases S., Smith S.J., Zheng Y.W., Myers H.M., Lear S.R., Sande E., Novak S., Collins C., Welch C.B., Lusis A.J. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc Natl Acad Sci U S A. 1998;95:13018–13023. doi: 10.1073/pnas.95.22.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leung K.W., Wong A.S. Pharmacology of ginsenosides: a literature review. Chin Med. 2010;5:20. doi: 10.1186/1749-8546-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tu L.H., Ma J., Liu H.P., Wang R.R., Luo J. The neuroprotective effects of ginsenosides on calcineurin activity and tau phosphorylation in SY5Y cells. Cell Mol Neurobiol. 2009;29:1257–1264. doi: 10.1007/s10571-009-9421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feyereisen R. Insect P450 enzymes. Annu Rev Entomol. 1999;44:507–533. doi: 10.1146/annurev.ento.44.1.507. [DOI] [PubMed] [Google Scholar]
- 58.Rewitz K.F., Rybczynski R., Warren J.T., Gilbert L.I. The Halloween genes code for cytochrome P450 enzymes mediating synthesis of the insect moulting hormone. Biochem Soc Trans. 2006;34:1256–1260. doi: 10.1042/BST0341256. [DOI] [PubMed] [Google Scholar]
- 59.Henderson G.L., Harkey M.R., Gershwin M.E., Hackman R.M., Stern J.S., Stresser D.M. Effects of ginseng components on c-DNA-expressed cytochrome P450 enzyme catalytic activity. Life Sci. 1999;65:209–214. doi: 10.1016/s0024-3205(99)00407-5. [DOI] [PubMed] [Google Scholar]
- 60.Kim D.S., Kim Y., Jeon J.Y., Kim M.G. Effect of Red Ginseng on cytochrome P450 and P-glycoprotein activities in healthy volunteers. J Ginseng Res. 2016;40:375–381. doi: 10.1016/j.jgr.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang J.T., Welch J.S., Ricote M., Binder C.J., Willson T.M., Kelly C., Witztum J.L., Funk C.D., Conrad D., Glass C.K. Interleukin-4-dependent production of PPAR-gamma ligands in macrophages by 12/15-lipoxygenase. Nature. 1999;400:378–382. doi: 10.1038/22572. [DOI] [PubMed] [Google Scholar]
- 62.Lehmann J.M., Moore L.B., Smith-Oliver T.A., Wilkison W.O., Willson T.M., Kliewer S.A. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 63.Willson T.M., Cobb J.E., Cowan D.J., Wiethe R.W., Correa I.D., Prakash S.R., Beck K.D., Moore L.B., Kliewer S.A., Lehmann J.M. The structure-activity relationship between peroxisome proliferator-activated receptor gamma agonism and the antihyperglycemic activity of thiazolidinediones. J Med Chem. 1996;39:665–668. doi: 10.1021/jm950395a. [DOI] [PubMed] [Google Scholar]
- 64.Ricote M., Li A.C., Willson T.M., Kelly C.J., Glass C.K. The peroxisome proliferator-activated receptor-gamma is a negative regulator of macrophage activation. Nature. 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 65.Mu Q., Fang X., Li X., Zhao D., Mo F., Jiang G., Yu N., Zhang Y., Guo Y., Fu M. Ginsenoside Rb1 promotes browning through regulation of PPARgamma in 3T3-L1 adipocytes. Biochem Biophys Res Commun. 2015;466:530–535. doi: 10.1016/j.bbrc.2015.09.064. [DOI] [PubMed] [Google Scholar]
- 66.Wei S., Li W., Yu Y., Yao F.A.L., Lan X., Guan F., Zhang M., Chen L. Ginsenoside Compound K suppresses the hepatic gluconeogenesis via activating adenosine-5'monophosphate kinase: a study in vitro and in vivo. Life Sci. 2015;139:8–15. doi: 10.1016/j.lfs.2015.07.032. [DOI] [PubMed] [Google Scholar]
- 67.Mollah M.L., Kim G.S., Moon H.K., Chung S.K., Cheon Y.P., Kim J.K., Kim K.S. Antiobesity effects of wild ginseng (Panax ginseng C.A. Meyer) mediated by PPAR-gamma, GLUT4 and LPL in ob/ob mice. Phytother Res. 2009;23:220–225. doi: 10.1002/ptr.2593. [DOI] [PubMed] [Google Scholar]
- 68.Han K.L., Jung M.H., Sohn J.H., Hwang J.K. Ginsenoside 20S-protopanaxatriol (PPT) activates peroxisome proliferator-activated receptor gamma (PPARgamma) in 3T3-L1 adipocytes. Biol Pharm Bull. 2006;29:110–113. doi: 10.1248/bpb.29.110. [DOI] [PubMed] [Google Scholar]
- 69.Banz W.J., Iqbal M.J., Bollaert M., Chickris N., James B., Higginbotham D.A., Peterson R., Murphy L. Ginseng modifies the diabetic phenotype and genes associated with diabetes in the male ZDF rat. Phytomedicine. 2007;14:681–689. doi: 10.1016/j.phymed.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 70.Zou H. South China Normal University; 2013. Study on the function of SlGSTE1 in feeding plants of Spodoptera litura. [Google Scholar]
- 71.Wu H., Zhang G.A., Zeng S., Lin K.C. Extraction of allyl isothiocyanate from horseradish (Armoracia rusticana) and its fumigant insecticidal activity on four stored-product pests of paddy. Pest Manage Sci. 2009;65:1003–1008. doi: 10.1002/ps.1786. [DOI] [PubMed] [Google Scholar]
- 72.Bergamo P., Luongo D., Maurano F., Mazzarella G., Stefanile R., Rossi M. Conjugated linoleic acid enhances glutathione synthesis and attenuates pathological signs in MRL/MpJ-Fas(lpr) mice. J Lipid Res. 2006;47:2382–2391. doi: 10.1194/jlr.M600187-JLR200. [DOI] [PubMed] [Google Scholar]
- 73.Waxman D.J. P450 gene induction by structurally diverse xenochemicals: central role of nuclear receptors CAR, PXR, and PPAR. Arch Biochem Biophys. 1999;369:11–23. doi: 10.1006/abbi.1999.1351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.