Abstract
BACKGROUND: Glioblastoma (GBM) is the most malignant primary brain tumor. Relapse occurs regularly, and the clinical behavior seems to be due to a therapy-resistant subpopulation of glioma-initiating cells that belong to the group of cancer stem cells. Aldehyde dehydrogenase (ALDH) has been identified as a marker for this cell population, and we have shown previously that ALDH1A3-positive GBM cells are more resistant against temozolomide (TMZ) treatment. However, it is still unclear how ALDH expression mediates chemoresistance. MATERIALS AND METHODS: ALDH1A3 expression was analyzed in 112 specimens from primary and secondary surgical resections of 56 patients with GBM (WHO grade IV). All patients received combined adjuvant radiochemotherapy. For experimental analysis, CRISPR-Cas9–induced knockout cells from three established GBM cell lines (LN229, U87MG, T98G) and two glioma stem-like cell lines were investigated after TMZ treatment. RESULTS: ALDH1A3 knockout cells were more sensitive to TMZ, and oxidative stress seemed to be the molecular process where ALDH1A3 exerts its role in resistance against TMZ. Oxidative stress led to lipid peroxidation, yielding active aldehydes that were detoxified by ALDH enzymatic activity. During the metabolic process, autophagy was induced leading to downregulation of the enzyme, but ALDH1A3 is upregulated to even higher expression levels after finishing the TMZ therapy in vitro. Recurrent GBMs show significantly higher ALDH1A3 expression than the respective samples from the primary tumor, and patients suffering from GBM with high ALDH1A3 expression showed a shorter median survival time (12 months vs 21 months, P < .05). CONCLUSION: Oxidative stress is an important and clinically relevant component of TMZ-induced therapeutic effects. Cytotoxicity seems to be mediated by aldehydes resulting from lipid peroxidation, and ALDH1A3 is able to reduce the number of toxic aldehydes. Therefore, we present a molecular explanation of the role of ALDH1A3 in therapeutic resistance of human GBM cells.
Introduction
Glioblastoma multiforme (GBM) is the most malignant brain tumor [1]. The standard therapy consists of surgical resection followed by combined radiochemotherapy using the alkylating agent temozolomide (TMZ) [2,3]. Despite advances in multimodal treatment options, the overall prognosis of patients with GBM remains poor, with a median survival of approximately 12-15 months [4]. The unfavorable prognosis is primarily due to regular tumor relapse, and ca. 90% of malignant gliomas regrow at the site of the original tumor location [5]. Strong intrinsic resistance against adjuvant therapy facilitates tumor cell survival, which is the main reason for regular tumor recurrence [6]. A variety of mechanisms of drug resistance have been suggested in GBM [7].
Aldehyde dehydrogenases (ALDHs) are a group of enzymes responsible for metabolizing endogenous and exogenous aldehydes to carboxylic acids. ALDHs have been characterized as markers of cancer stem cells and indicators of worse prognosis in multiple types of cancers [8] [9] [10]. However, it is unclear how ALDH expression mediates chemoresistance since ALDHs are not directly connected to DNA repair pathways.
Here we show that oxidative stress is a relevant component of TMZ-induced therapeutic effects and that ALDH1A3 is involved in the reaction to reactive oxygen species (ROS). ROS can react with the polyunsaturated fatty acids of lipid membranes and induce lipid peroxidation (LPO), which is the major process of oxidative degradation of lipids [11]. Cytotoxicity seems to be mediated by aldehydes resulting from LPO, ultimately leading to cell death, and ALDH1A3 is able to reduce the extent of toxic aldehydes. Therefore, we present a molecular explanation for the role of ALDH1A3 in therapy resistance of human GBM cells. These results are corroborated by clinical data showing increased ALDH1A3 levels in GBM operations from recurrent tumors.
Materials and Methods
Cell Culture, Reagents, and Antibodies
Human glioblastoma cell lines LN229, U87MG, and T98G and glioblastoma stem-like cells X01 and T84 were cultured as previously described [12]. TMZ, N-acetylcysteine (NAC), and bafilomycin were obtained from Sigma (Munich, Germany). 4-Hydroxynonenal (HNE) was from Cayman Chemical (Hamburg, Germany). Antibodies against HNE were obtained from Abcam (England, UK); ALDH1A3 (mouse) and LC3 from Novus Biologicals (Abingdon, UK); ADLH1A3 (rabbit) and p62 from Thermo Fisher Scientific (Munich, Germany); and phospho-H2A.X (Ser139), vinculin, and HRP-conjugated secondary antibody from Cell Signaling Technology (Frankfurt, Germany).
CRISPR-Cas9 Knockout
LN229, U87MG, and T98G cells, respectively, were transfected with CRISPR-Cas9 plasmid pSpCas9(BB)-2A-GFP (PX458) (Addgene plasmid #48138) with ALDH1A3 single-guided RNA as previously described [12].
Cell Viability Assay and Cell Cycle Analysis
LN229, U87MG, and T98G cells, respectively, were seeded in 96-well plates and treated with different concentrations of TMZ (100-500 μM) either alone or with 2 mM NAC for 5 consecutive days. The cell viability was measured by MTT assay (Sigma, Munich, Germany) as previously described [12]. For cell cycle analysis, cells were fixed in cold 70% ethanol for 30 minutes at 4°C and subsequently treated with RNase. Finally, the cells were stained with PI solution (50 μg/ml) for 5-10 minutes at room temperature and analyzed by flow cytometry (FACS Calibur instruments, BD Biosciences, Heidelberg, Germany).
ROS Assay
ROS production was measured using the OxiSelect Intracellular ROS Assay Kit (Cell Biolabs, Heidelberg, Germany). LN229, U87MG ,and T98G cells, respectively, were seeded on black 96-well plates and treated with TMZ (100-500 μM) either alone or with 2 mM NAC for 4 consecutive days. Subsequently, cells were treated with DCFH-DA/media solution at 37°C for 30 minutes followed again by TMZ (100-500 μM) either alone or with NAC treatment for 30 minutes. The fluorescence was read with a fluorometric plate reader at 480 nm/530 nm.
Lipid Peroxidation Detection
Lipid peroxidation was investigated by measuring malondialdehyde (MDA) levels using MDA Assay Kit (Sigma, Munich, Germany). Briefly, cells were homogenized on ice in 300 μl of the MDA Lysis Buffer containing 3 μl BHT. A total of 200 μl supernatant from each sample was mixed with 600 μl TBA solution and incubated at 95°C for 60 minutes; 200 μl from each reaction mixture was taken for colorimetric detection (532 nm).
Western Blot Analysis, Aldefluor Assay, and Proximity Ligation Assay (PLA)
Western blot analysis, Aldefluor assay (Stem Cell Technologies, Cologne, Germany), and PLA using Duolink In Situ Red Starter Kit Mouse/Rabbit (Sigma, Munich, Germany) were performed as previously described [12].
Immunofluorescence
Glioblastoma cell lines LN229, U87MG, and T98G, respectively, grown on glass coverslips were treated with 300 μM TMZ for 5 consecutive days and then fixed 30 minutes after last TMZ treatment or 1 hour after 30-μM HNE treatment. After washing, cells were blocked in blocking buffer (PBS, 1% BSA, 0.1%Triton X-100, 0.01%Tween, 0.02% sodium azide, 2.5% donkey serum) for 30 minutes at room temperature. Subsequently, cells were incubated with primary antibodies (anti-HNE 1:500 and anti-p62 1:500) overnight at 4°C followed by second antibody incubation (1:500). Finally, cells were stained with Hoechst dye (dilute 1:10,000 in PBS) and mounted with Aqua Poly/Mount (PolySciences), ready for fluorescence imaging (Zeiss HBO 100 Upright Fluorescence Microscope, Munich, Germany).
Immunohistochemistry of GBM Tissue Samples
A total of 112 specimens from 56 patients with GBM (IDH wild-type, WHO grade IV) were collected with patients’ consent (according to the TUM medical faculty guidelines for tissue preservation) at the Klinikum Rechts der Isar, Munich, Germany. All patients underwent primary and secondary surgical resection, and standard adjuvant combined chemoradiotherapy with TMZ after primary surgery. Immunohistochemistry was performed as previously described [13]. Briefly, tissue sections (5 μm) were stained with ALDH1A3 antibody (1:600) and assigned expression scores using a modified immunoreactivity score system [14]: negative (0), ≤10% (1), 11%-25% (2), 26%-50%, and ≥51% (4).
Statistical Analysis
Three independent experiments of each assay were conducted to validate results. t tests were used for statistical analysis for comparison of unpaired groups and normally distributed data. P values <.05 were regarded as statistically significant.
Results
ALDH1A3 Knockout Glioblastoma Cell Lines Are More Sensitive to TMZ, But the Difference Can Be Eliminated by Scavenging Oxidative Stress Products
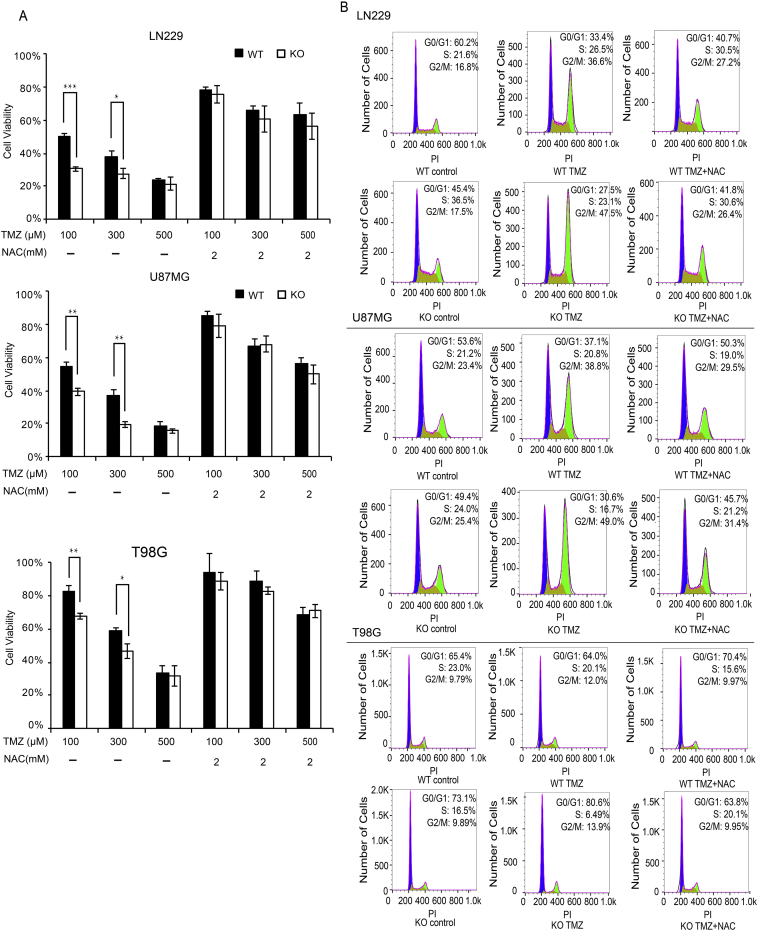
After treatment with different dosages of TMZ (100 μM-500 μM) either alone or with NAC for 5 consecutive days, the survival rates of ALDH1A3 WT and KO cells were measured by MTT assays (Figure 1A). ALDH1A3 KO cells were more sensitive to TMZ compared with WT cells, and this difference was significant especially at dosages ≤300 μM. The elimination of oxidative stress products by NAC led to dramatic improvement of the cell survival rates and diminished the difference between WT and KO cells in both LN229 and U87MG cells. T98G cells were more resistant against TMZ compared with the two other cell lines. These results were corroborated by cell cycle analysis (Figure 1B). G2/M arrest after TMZ treatment was more pronounced in ALDH1A3 KO cells compared with WT cells in all three cell lines, and the differences were diminished by the addition of NAC. All results indicate that the cytotoxicity of TMZ may partly be due to oxidative stress and that ALDH1A3 plays an important role in the response to oxidative stress–induced cytotoxicity. Using an antibody directed against the phosphorylated form of histone H2A.X as an indicator of DNA damage, we observed no significant differences in DNA damage between WT and KO cells after treatment with increasing dosages (100 μM-500 μM) of TMZ (data not shown).
Figure 1.
Cell survival and cell cycle analysis after TMZ treatment
(A) Quantitative results of MTT assays in LN229, U87MG, and T98G cells, respectively. ALDH1A3 depletion led to a significant decrease of cell survival at 100 μM and 300 μM TMZ. The addition of NAC neutralized the effect and eliminated the difference between ALDH1A3 WT and KO cells in both LN229 and U87MG cells (100 μM TMZ, approx.50% WT vs 30% KO to 80% in both WT and KO in LN229 cells, approx.55% WT vs 40% KO to 80% in both WT and KO in U87MG cells. 300 μM TMZ, approx.40% WT vs 30% KO to 60% in both WT and KO in LN229 cells, approx. 35% vs 20% to 70% in both WT and KO in U87MG cells). In T98G, the addition of NAC improved the overall survival rate to approx. 80%-90% at TMZ dosages ≤300 μM and approx. 70% at 500 μM. Untreated cells served as control for 100% viability. Three independent experiments were conducted for each treatment group; each bar indicates the mean ± SD. *, P < .05; **, P < .01;***, P < .001.
(B) Cell cycle analysis of LN229, U87MG, and T98G cells, respectively. Treatment with 300 μM TMZ led to significant G2/M arrest, and the effect was more pronounced in KO cells. The addition of NAC reduced the cell cycle arrest and diminished the difference between ALDH1A3 WT and KO cells.
WT, wild type; KO, knockout.
Higher Levels of HNE-Modified Proteins and MDA Are Detected in ALDH1A3 Knockout Cells Compared with WT Cells After TMZ Treatment
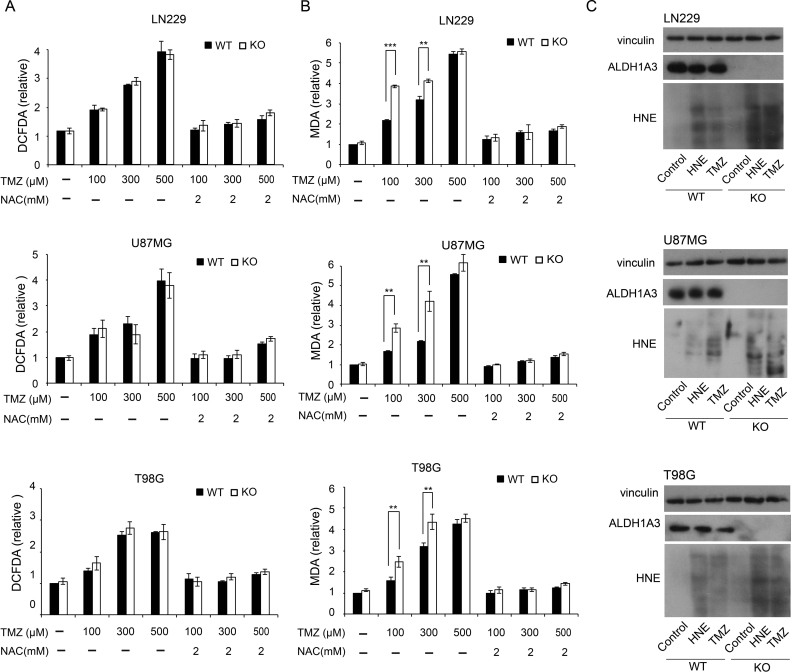
The amount of free radical molecules detected by in vitro ROS/RNS assay increased under TMZ treatment in a dose-dependent manner, but no significant differences between ALDH1A3 WT and KO cells were visible (Figure 2A), indicating that ALDH1A3 mainly acts downstream of ROS production. The addition of NAC led to significant reduction to control levels without TMZ treatment.
Figure 2.
Oxidative stress and lipid peroxidation after TMZ treatment
To investigate ROS production in ALDH1A3 WT and KO cells, in vitro ROS/RNS assay was conducted based on the oxidation of DCFH to highly fluorescent 2′,7′-dichlorodihydrofluorescein. The OxiSelect TBARS assay based on MDA-TBA adducts was used to investigate lipid peroxidation levels in cells. Quantitative results of ROS/RNS assay (A) and TBARS assay (B). TMZ treatment led to significant increase of ROS production (A) and MDA content (B) in all three cell lines. No difference of ROS was observed between ALDH1A3 WT and KO cells, but the difference of MDA content was significant in all three cell lines. The addition of NAC dramatically counteracted the effect of TMZ and eliminated the difference between ALDH1A3 WT and KO cells. Each bar indicates the mean ± SD. *, P < .05; **, P < .01;***, P < .001.
(C) Western blots using antibodies against ALDH1A3 and HNE-modified proteins. Vinculin served as loading control. The cells were treated with 100 μM TMZ for 5 days. The proteins were isolated 30 minutes after last treatment; 30-μM HNE treatment for 1 hour served as positive control. More HNE-modified proteins could be observed in ALDH1A3KO cells than WT cells.
Using OxiSelect TBARS assay, TMZ induced an increase of the cytotoxic aldehyde MDA, and MDA levels were significantly higher in ALDH1A3 KO cells compared with WT cells. This result shows that ALDH1A3 is effective in detoxifying MDA. The effect abrogates at 500 μM TMZ due to ALDH1A3 downregulation by autophagy. NAC co-treatment dramatically reduced the production of active aldehyde MDA to control levels without TMZ treatment in both WT and KO cells.
HNE is a major product of lipid peroxidation and confers cytotoxic effects of oxidative stress. By Western blot analysis using an anti-HNE antibody, we investigated the presence of HNE-modified proteins after TMZ treatment. In line with OxiSelect TBARS assay, the amount of HNE-modified proteins in ALDH1A3 KO cells was higher than in WT cells, further verifying the detoxifying role of ALDH1A3 (Figure 2C).
Overall, our results suggest that ALDH1A3 might play a major role by detoxifying toxic aldehydes of oxidative stress and lipid peroxidation.
TMZ-Induced Oxidative Stress Leads to Autophagy and ALDH1A3 Degradation
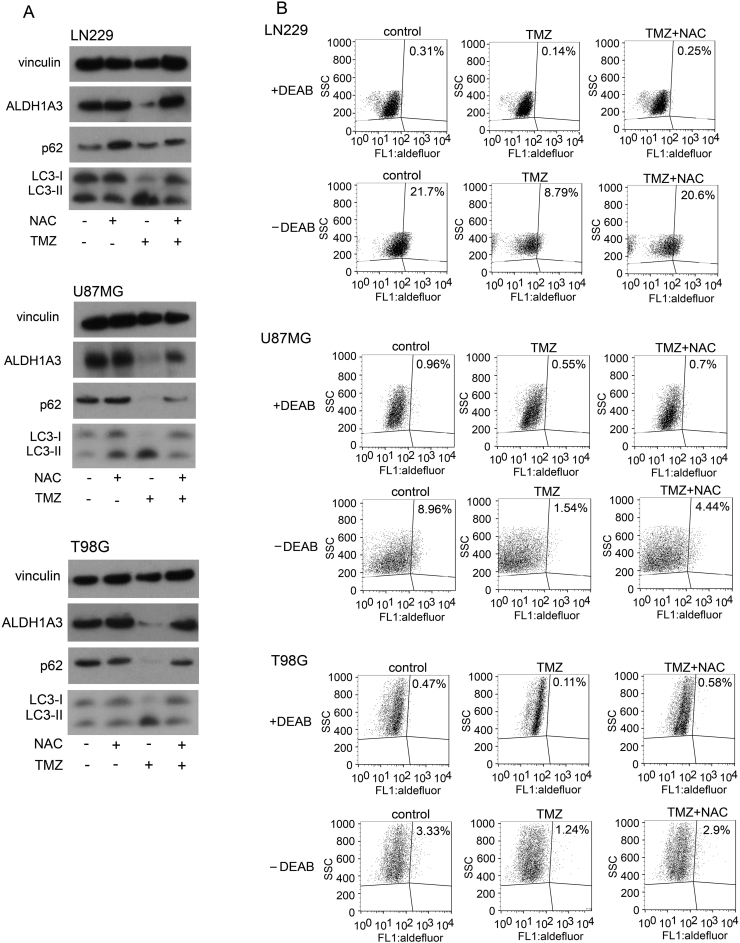
We found a significant reduction of ALDH1A3 protein and an enhancement of autophagy after TMZ treatment that could be blocked by NAC treatment (Figure 3A). Most importantly, ALDH1A3 levels were also significantly recovered by the oxidative stress scavenger. In line with this observation, aldefluor assay showed decreased ALDH enzyme activity with TMZ treatment alone but also recovered after NAC treatment (Figure 3B). The oxidative stress scavenger NAC restored ALDH activity and diminished the difference between WT and KO cells in every cell line. These results show that TMZ induces autophagy and that oxidative stress is most likely the reason for autophagy and ALDH1A3 degradation.
Figure 3.
ALDH1A3 and autophagy after TMZ treatment.
(A) Western blots after treatment with 500 μM TMZ or 2 mM NAC or in combination for 5 days. Downregulation of ALDH1A3, upregulation of LC3-II, and downregulation of p62 were observed in all three cell lines after TMZ treatment but could be restored by co-treatment with NAC.
(B) Aldefluor assays after treatment with 500 μM TMZ either alone or with 2 mM NAC for 5 consecutive days. ALDH activity decreased after TMZ treatment but could be restored by NAC co-treatment. ALDH activity decreased from 21.7% to 8.79% with TMZ treatment alone and recovered to 20.6% with TMZ/NAC co-treatment in LN229 cells. ALDH activity decreased from 8.96% to 1.54% with TMZ treatment alone and recovered to 4.44% with TMZ/NAC co-treatment in U87MG cells. ALDH activity decreased from 3.33% to 1.24% with TMZ treatment alone and recovered to 2.9% with TMZ/NAC co-treatment in T98G cells. Diethylaminobenzaldehyde served as negative control.
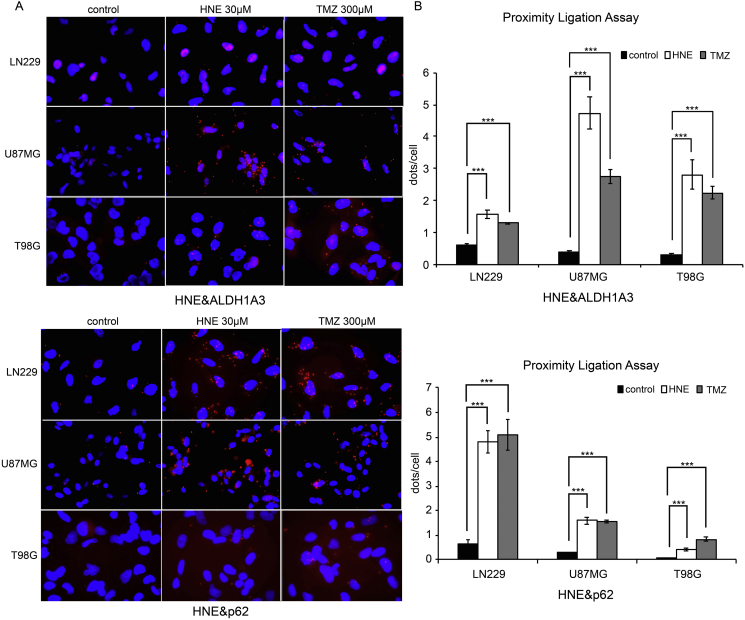
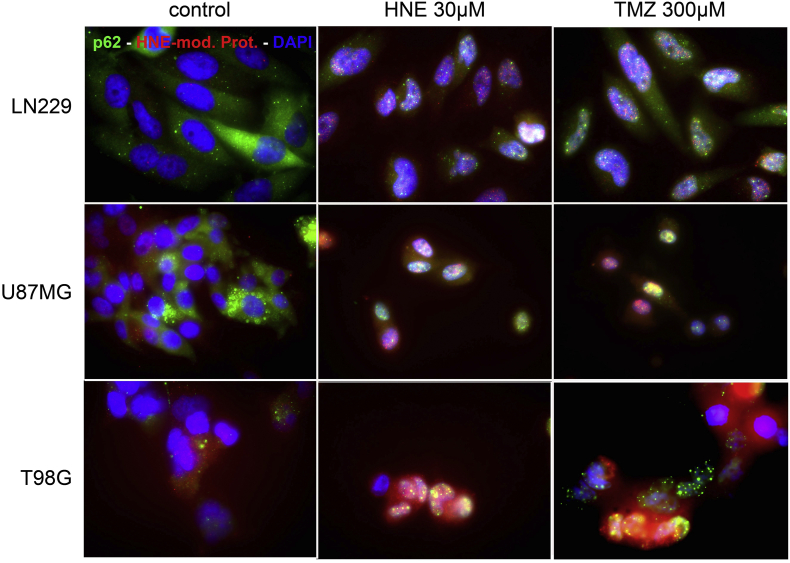
We hypothesized that the end products of lipid peroxidation could be the direct cause of autophagy since autophagy is directly involved in the clearance of defective proteins. By PLA, we observed a proximity between HNE-modified proteins and both p62 and ALDH1A3 (Figure 4). In addition, co-immunofluorescence of p62 and HNE confirmed the colocalization (Figure 5). These results indicate that HNE-modified proteins are processed to autophagy and final degradation. ALDH1A3 is downregulated in the same process either due to aldehyde binding during enzyme function or by direct HNE modification.
Figure 4.
Detection of the proximity of HNE-modified proteins and ALDH1A3 and p62.
(A) PLAs of cells treated with 300 μM TMZ for 5 consecutive days; 30-μM HNE treatment for 1 hour served as positive control. The red dots demonstrate the proximity of HNE-modified proteins and ALDH1A3 and p62, respectively. More red dots were observed in TMZ- and HNE-treated groups, indicating that HNE-modified proteins and ALDH1A3 and p62, respectively, might have direct interactions (microscopic magnification 40×). Remarks: The red dots appear in the nuclei because of overlay effect (cytoplasm surrounds the nucleus).
(B) Quantitative results of PLA. Three independent experiments were conducted. Each bar indicates the mean ± SD. *, P < .05; **, P < .01;***, P < .001.
Figure 5.
Detection of the colocation of HNE-modified proteins and p62.
Double immunofluorescence of cells treated with 300 μM TMZ for 5 consecutive days; 30-μM HNE treatment for 1 hour served as positive control. Alexa Fluor 488 dye represents p62; Alexa Fluor 594 dye represents HNE-modified protein. HNE or TMZ treatment led to colocation of HNE-modified proteins and p62.
ALDH1A3-Positive Cells Are Enriched in Tumor Relapse In Vivo and After TMZ Treatment In Vitro
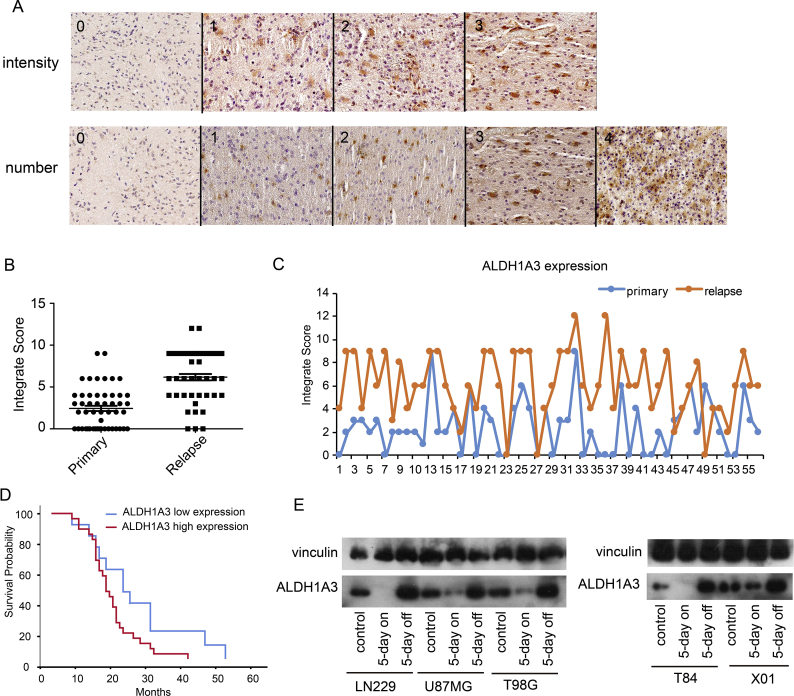
By immunohistochemistry (Figure 6A), specimens of recurrent GBM from 56 GBM patients showed significantly higher ALDH1A3 expression levels than the respective samples from the primary tumor of the same patient (Figure 6, B and C). ALDH1A3-positive cells are more resistant to the TMZ chemotherapy after first surgery, and it is therefore most likely that these cells survive the adjuvant therapy and form the resistant subpopulation responsible for tumor relapse. In 41/56 cases, patients' survival times were available, and we observed that patients suffering from ALDH1A3 high expression tumors have significantly shorter survival times compared with patients with low ALDH1A3 expression tumors based on the analysis of the specimens of the recurrent GBMs (Figure 6D). These findings indicate that high ALDH1A3 expression correlates with worse prognosis.
Figure 6.
ALDH1A3-positive cells formed the resistant subpopulation in the tumor regrowth.
(A) Immunohistochemistry examples for intensity (0 = absent, 1 = weak, 2 = moderate, 3 = strong) and percentage of positive cells (0 = negative, 1 = ≤10%, 2 = 11%-30%, 3 = 31%-50%, 4 = ≥51%). Integrate score = intensity × percentage.
(B) Specimens from recurrent GBM showed higher expression of ALDHA3 than the respective samples of the primary tumors.
(C) The scores of primary tumor and relapse tumor from each patient, respectively.
(D) Patients with ALDH1A3 high expression (integrate score > 4, n = 28) show shorter median survival time than those with ALDH1A3 low expression (integrate score < 5, n = 13) (median survival time for patients with ALDH1A3 high expression: 16 months, those with low expression: 21 months, P < .05).
(E). Western blots of established cell lines (LN229, U87MG, T98G) and glioma stem-like cell lines (T84, X01) after treatment with 500 μM TMZ for 5 consecutive days with and without 5-day recovery time in fresh medium (5 days on = without, 5 days off = with recovery). TMZ treatment diminished ALDH1A3 expression but restored it to higher levels than the control in the recovery groups.
To confirm this hypothesis experimentally, we investigated the ALDH1A3 regulation in established cell lines and in glioma stem-like cell lines (Figure 6E). The population of surviving tumor cells showed both higher ALDH1A3 expression levels after TMZ treatment (500 μM for 5 consecutive days) and recovery for additional 5 days. These results indicate that ALDH1A3 plays an important role in the TMZ chemoresistance phenotype of malignant gliomas.
Discussion
Aldehyde dehydrogenases of the ALDH1 subfamily have previously been identified as markers of cancer stem cells that constitute the most therapy-resistant subpopulation of tumor cells. Moreover, in GBM, ALDH1 is not only a biomarker for this resistant cell population but is directly involved in the molecular mechanisms that lead to TMZ resistance [15]. However, it is still unclear how ALDH expression mediates chemoresistance since ALDHs are not directly connected to DNA repair.
In the present study, we corroborate our previous findings [12] that ALDH1A3 is an important regulator of chemoresistance in glioma cells since ALDH1A3 knockout cells were more sensitive to TMZ treatment compared with wild-type cells. We further show that oxidative stress leads to lipid peroxidation, yielding active aldehydes including MDA and HNE that are detoxified by ALDH enzymatic activity. The reduction of reactive oxygen species by a scavenger blocked the acute cellular effects of TMZ. During the metabolic process, autophagy is induced most likely by the molecular interaction of aldehydes with ALDH1A3 leading to downregulation of the enzyme. Interestingly, ALDH1A3 is restored in the oxidative stress–free time interval after TMZ treatment in vitro, and in vivo recurrent GBMs show significantly higher ALDH1A3 expression than the respective samples from the primary tumor of the same patient. These data indicate that ALDH1A3 plays an important role in the TMZ chemoresistance phenotype of malignant gliomas.
ALDH1A3 depletion in knockout cells rendered the glioblastoma cell lines more sensitive to TMZ. The addition of the oxidative stress scavenger NAC restored the cell viability, diminished G2/M cell cycle arrest, and eliminated the difference between ALDH1A3 WT and KO cells. Therefore, oxidative stress seems to be a critical component of TMZ-mediated cytotoxicity, and ALDH1A3 plays a role in the processes induced by ROS. These results were independent of MGMT status since NAC increased the survival rate in both MGMT-negative LN229 and U87 and in MGMT-positive T98G cells, indicating that oxidative stress effects are independent of the alkylating effect of TMZ. Oxidative stress has not been investigated in detail as therapeutic aspect of chemotherapy, although it is well known that several side effects of chemotherapy are caused by drug-induced oxidative stress. Examples include cardiac toxicity of doxorubicin [16], skeletal myopathy due to long-term administration of azidothymidine [17], and multiorgan toxicity by cis-diamminedichloroplatinum (cisplatin) [18].
There was no significant difference of ROS production between ALDH1A3 WT and KO cells, indicating that oxidative stress is the upstream factor and ALDH1A3 has no effect on ROS production. Significantly higher MDA levels and higher incidence of HNE-modified proteins in ALDH1A3 KO cells imply that ALDH1A3 mainly contributes by detoxifying bioactive aldehydes from lipid peroxidation. ALDHs are regarded as “aldehyde scavengers” under different biological conditions since they are able to metabolize a wide range of chemically and structurally diverse aldehydes [19,20].
The difference of lipid peroxidation products also diminished between ALDH1A3 WT and KO cells at high concentrations of TMZ treatment. However, we have shown previously that ALDH1A3 decreases in a dose-dependent manner after TMZ treatment by autophagy [12,21]. In the present experiments, ALDH1A3 was almost absent after 500 μM TMZ, and aldehyde levels compensate between ALDH1A3 WT and KO cells at higher TMZ concentrations.
It is not clear how TMZ induces autophagy and how ALDH1A3 could be degraded in this process. We could show that autophagy is caused by TMZ-induced oxidative stress since co-treatment with NAC significantly reduced the level of autophagy. It has been shown by others that aldehydes like HNE are capable of inducing autophagy [22]. The role of toxic aldehydes is under discussion in different pathologies, and it has been suggested in Parkinson's disease that HNE is increased and autophagy is promoted [23,24].
The molecular mechanisms that integrate ALDH1A3 in autophagy have not been investigated in detail. Autophagy is physiologically induced for disposal of damaged molecules or organelles. It is most likely that the formation of aldehyde-modified proteins triggers autophagy after TMZ therapy. ALDH1A3 could be degraded by autophagy as an enzymatic complex bound with an aldehyde or as one of the aldehyde-modified proteins. The association between HNE and ALDH1A3 in autophagy is substantiated by the results of the PLA and co-immunoprecipitation in the present study indicating proximity of both ALDH1A3 and p62 with HNE-modified proteins.
The expression of ALDH1A3 clearly protects tumor cells against TMZ treatment. Here, we show experimentally that ALDH1A3 is restored in the time interval after TMZ treatment, and protein levels were much stronger in TMZ treated cells compared with untreated controls. These effects were also visible in situ. In our cohort of pairs of samples from primary and relapse GBM operations, we found significantly higher ALDH1A3 expression levels in the recurrent tumors, and patients with high ALDH1A3 showed significantly shorter survival times, indicating that ALDH1A3 constitutes the therapy-resistant subpopulation of glioblastoma cells both in vitro and in vivo.
In conclusion, we demonstrate that ALDH1A3 confers chemoresistance of glioblastoma cells against TMZ and that it is mediated by its function of detoxifying activated aldehydes resulting from lipid peroxidation induced by oxidative stress after TMZ treatment. ALDH1A3 protein expression is regulated by autophagy and restored after finishing TMZ treatment. The role of ALDH1A3 seems to be functional not only in vitro but also in vivo since ALDH1A3 is upregulated in recurrent GBM and high expression correlates with shorter survival times.
Acknowledgments
Acknowledgements
We are thankful to Feng Zhang (McGovern Institute for Brain Research at Massachusetts Institute of Technology) for sharing pSpCas9(BB)-2A-GFP (PX458) plasmid. We also thank the DKTK Cell Sorting Facility at the Institute of Clinical Chemistry and Pathobiochemistry, Technical University Munich (Head: Prof. Dr. Jürgen Ruland) for cell sorting.
Conflict of Interest
The authors have declared no conflicts of interest.
References
- 1.van den Bent MJ, Weller M, Wen PY, Kros JM, Aldape K. Chang S. A clinical perspective on the 2016 WHO brain tumor classification and routine molecular diagnostics. Neuro Oncol. 2017;19:614–624. doi: 10.1093/neuonc/now277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., Belanger K., Brandes A.A., Marosi C., Bogdahn U. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Sulman EP, Ismaila N, Chang SM. Radiation therapy for glioblastoma: American Society of Clinical Oncology clinical practice guideline endorsement of the American Society for Radiation Oncology guideline. J Oncol Pract. 2017;13:123–127. doi: 10.1200/JOP.2016.018937. [DOI] [PubMed] [Google Scholar]
- 4.Pearson JRD, Regad T. Targeting cellular pathways in glioblastoma multiforme. Signal Transduct Target Ther. 2017;2 doi: 10.1038/sigtrans.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ringel F., Pape H., Sabel M., Krex D., Bock H.C., Misch M., Weyerbrock A., Westermaier T., Senft C., Schucht P. Clinical benefit from resection of recurrent glioblastomas: results of a multicenter study including 503 patients with recurrent glioblastomas undergoing surgical resection. Neuro Oncol. 2016;18:96–104. doi: 10.1093/neuonc/nov145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osuka S, Van Meir EG. Overcoming therapeutic resistance in glioblastoma: the way forward. J Clin Invest. 2017;127:415–426. doi: 10.1172/JCI89587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagel Z.D., Kitange G.J., Gupta S.K., Joughin B.A., Chaim I.A., Mazzucato P., Lauffenburger D.A., Sarkaria J., Samson L.D. DNA repair capacity in multiple pathways predicts chemoresistance in glioblastoma multiforme. Cancer Res. 2017;77:198–206. doi: 10.1158/0008-5472.CAN-16-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Croker A.K., Rodriguez-Torres M., Xia Y., Pardhan S., Leong H.S., Lewis J.D., Allan A.L. Differential functional roles of ALDH1A1 and ALDH1A3 in mediating metastatic behavior and therapy resistance of human breast cancer cells. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yun X., Zhang K., Wang J., Pangeni R.P., Yang L., Bonner M., Wu J., Wang J., Nardi I.K., Gao M. Targeting USP22 suppresses tumorigenicity and enhances cisplatin sensitivity through ALDH1A3 downregulation in cancer-initiating cells from lung adenocarcinoma. Mol Cancer Res. 2018;16:1161–1171. doi: 10.1158/1541-7786.MCR-18-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park J., Shim J.K., Kang J.H., Choi J., Chang J.H., Kim S.Y., Kang S.G. Regulation of bioenergetics through dual inhibition of aldehyde dehydrogenase and mitochondrial complex I suppresses glioblastoma tumorspheres. Neuro Oncol. 2018;20:954–965. doi: 10.1093/neuonc/nox243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tudek B, Zdzalik-Bielecka D, Tudek A, Kosicki K, Fabisiewicz A, Speina E. Lipid peroxidation in face of DNA damage, DNA repair and other cellular processes. Free Radic Biol Med. 2017;107:77–89. doi: 10.1016/j.freeradbiomed.2016.11.043. [DOI] [PubMed] [Google Scholar]
- 12.Wu W, Schecker J, Wurstle S, Schneider F, Schonfelder M, Schlegel J. Aldehyde dehydrogenase 1A3 (ALDH1A3) is regulated by autophagy in human glioblastoma cells. Cancer Lett. 2018;417:112–123. doi: 10.1016/j.canlet.2017.12.036. [DOI] [PubMed] [Google Scholar]
- 13.Schäfer A., Teufel J., Ringel F., Bettstetter M., Hoepner I., Rasper M., Gempt J., Koeritzer J., Schmidt-Graf F., Meyer B. Aldehyde dehydrogenase 1A1—a new mediator of resistance to temozolomide in glioblastoma. Neuro Oncol. 2012;14:1452–1464. doi: 10.1093/neuonc/nos270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loeffler J.S., Alexander E., 3rd., Hochberg F.H., Wen P.Y., Morris J.H., Schoene W.C., Siddon R.L., Morse R.H., Black P.M. Clinical patterns of failure following stereotactic interstitial irradiation for malignant gliomas. Int J Radiat Oncol Biol Phys. 1990;19:1455–1462. doi: 10.1016/0360-3016(90)90358-q. [DOI] [PubMed] [Google Scholar]
- 15.Xu X., Chai S., Wang P., Zhang C., Yang Y., Yang Y., Wang K. Aldehyde dehydrogenases and cancer stem cells. Cancer Lett. 2015;369:50–57. doi: 10.1016/j.canlet.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 16.Zhang YW, Shi JJ, Li YJ, Wei L. Cardiomyocyte death in doxorubicin-induced cardiotoxicity. Arch Immunol Ther Ex. 2009;57:435–445. doi: 10.1007/s00005-009-0051-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amatore C., Arbault S., Jaouen G., Koh A.C.W., Leong W.K., Top S., Valleron M.A., Woo C.H. Pro-oxidant properties of AZT and other thymidine analogues in macrophages: implication of the azido moiety in oxidative stress. ChemMedChem. 2010;5:296–301. doi: 10.1002/cmdc.200900464. [DOI] [PubMed] [Google Scholar]
- 18.Santos NAG, Catao CS, Martins NM, Curti C, Bianchi MLP, Santos AC. Cisplatin-induced nephrotoxicity is associated with oxidative stress, redox state unbalance, impairment of energetic metabolism and apoptosis in rat kidney mitochondria. Arch Toxicol. 2007;81:495–504. doi: 10.1007/s00204-006-0173-2. [DOI] [PubMed] [Google Scholar]
- 19.Grunblatt E, Riederer P. Aldehyde dehydrogenase (ALDH) in Alzheimer's and Parkinson's disease. J Neural Transm. 2016;123:83–90. doi: 10.1007/s00702-014-1320-1. [DOI] [PubMed] [Google Scholar]
- 20.Pors K, Moreb JS. Aldehyde dehydrogenases in cancer: an opportunity for biomarker and drug development? Drug Discov Today. 2014;19:1953–1963. doi: 10.1016/j.drudis.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Wuerstle S., Schneider F., Ringel F., Gempt J., Lammer F., Delbridge C., Wu W., Schlegel J. Temozolomide induces autophagy in primary and established glioblastoma cells in an EGFR independent manner. Oncol Lett. 2017;14:322–328. doi: 10.3892/ol.2017.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haberzettl P, Hill BG. Oxidized lipids activate autophagy in a JNK-dependent manner by stimulating the endoplasmic reticulum stress response. Redox Biol. 2013;1:56–64. doi: 10.1016/j.redox.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dias V, Junn E, Mouradian MM. The role of oxidative stress in Parkinson's disease. J Park Dis. 2013;3:461–491. doi: 10.3233/JPD-130230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynch-Day MA, Mao K, Wang K, Zhao M, Klionsky DJ. The role of autophagy in Parkinson's disease. Cold Spring Harb Perspect Med. 2012;2:a009357. doi: 10.1101/cshperspect.a009357. [DOI] [PMC free article] [PubMed] [Google Scholar]