Abstract
Ginseng is popularly known to be the king of ancient medicines and is used widely in most of the traditional medicinal compositions due to its various pharmaceutical properties. Numerous studies are being focused on this plant's curative effects to discover their potential health benefits in most human diseases, including cancer— the most life-threatening disease worldwide. Modern pharmacological research has focused mainly on ginsenosides, the major bioactive compounds of ginseng, because of their multiple therapeutic applications. Various issues on ginseng plant development, physiological processes, and agricultural issues have also been studied widely through state-of-the-art, high-throughput sequencing technologies. Since the beginning of the 21st century, the number of publications on ginseng has rapidly increased, with a recent count of more than 6,000 articles and reviews focusing notably on ginseng. Owing to the implementation of various technologies and continuous efforts, the ginseng plant genomes have been decoded effectively in recent years. Therefore, this review focuses mainly on the cellular biomolecular sequences in ginseng plants from the perspective of the central molecular dogma, with an emphasis on genomes, transcriptomes, and proteomes, together with a few other related studies.
Keywords: EST, genome, next-generation sequencing, Panax species, transcriptome
1. Introduction
Decoding the genetics of medicinal plants is significant in understanding their phytochemical constituents with the trace of characterized enzymes in their genome, and the knowledge acquired from each plant is greatly benefiting the pharmaceutical industries in standardizing the development of natural drugs. So far, the knowledge obtained from the history of traditional medicine has taught the importance of medicinal plants and how they are useful in protecting the health of humans from various disorders [1]. Additionally, the current science is continuously acknowledging the benefits of those derived knowledge and impelling the researchers toward evidence-based science from a pseudoscience through empirical search. By these prominently emerging empirical searches, the term “phytochemical genomics” has originated, a discipline that systematically integrates multiple “omics” studies including genomics, transcriptomics, proteomics, and metabolomics [2], [3]. This systematic integration helps researchers to discern the biosynthesis mechanisms of plant-specific phytochemicals. For example, the “gene-to-metabolite” concepts have been successfully applied in Arabidopsis thaliana to characterize its flavonoids and also in other plants to characterize their secondary metabolites [4], [5]. This field of study is becoming extensively acceptable as a proof-of-concept to annotate the array of novel phytochemicals and their biosynthesis mechanisms through derived and multitested hypotheses [5]. The “gene-to-metabolite” concept is more familiar among model plants, as they consist of enormous data sets in comparison to nonmodel plants, such as crops and medicinal plants. To produce similar data structure in nonmodel plants, a theoretical framework that was used to generate a testable hypothesis in model plants was implemented in nonmodel plants using high-throughput technologies—namely, next-generation sequencing technologies and LC/GC-MS/MS–based metabolomics. Advantageously, those high-throughput sequencing technologies are made more accessible to the general plant communities with an affordable cost for implementing the “gene-to-metabolite” concept in nonmodel plant researches [6], [7], [8], [9], [10]. As a result of the overall evolutions and advancements, decoding the complete genome to metabolome for many nonmodel plants were achieved successfully.
The plant ginseng is a nonmodel plant, widely known as an adaptogen belonging to the genus Panax in the family Araliaceae, consisting of 15 species and seven subspecies. The word Panax is derived from the Greek word “panacea,” meaning a cure for all. Moreover, the human-shaped root of Panax ginseng is striking in its identified medicinal properties, and in addition, the plant has a unique correlative factor to the human genome in that its genome size is homologous. Among the 15 species of the genus Panax, only four have been studied widely: P. ginseng (Korean ginseng), Panax notoginseng (Chinese ginseng), Panax quinquefolius (American ginseng), and Panax japonicus (Japanese ginseng). The existing research has focused on the biologically active ingredients of ginseng, including phytochemicals [11], [12] and proteins/peptides [13] for their pharmacological properties [14]. More particularly, the saponin called “Ginsenoside” is the most valuable chemical component of the ginseng plant, having numerous therapeutic benefits on various ailments [14]. Moreover, the ginsenosides exist in various isoforms, and, thus, it has been used desperately for treating multiple conditions, such as cancer, obesity, cardiovascular diseases, neurodegenerative diseases, and diabetes [14], [15]. The content variations of ginsenoside isoforms along with the reported evidence of traditional medicines in the literature have also added different prices in the market upon multiple factors, such as plant type (wild plants), cultivation methods (field grown/mountain grown), and age of the roots [16], [17]. The lifecycle of ginseng and morphology of different Panax species roots are given in Fig. 1, Fig. 2. Various sequencing efforts were conducted to decode the genetics of Panax species and to understand the plant physiology and the biosynthesis of ginsenosides. However, all the sequencing efforts were conducted randomly to elucidate the different physiological process, for which an overall review is not given in the literature so far. A complete evaluation is necessary to select a good quality sample and to design a better structure of the study for future sequencing projects. Therefore, through this review, the studies in the literature till date are organized based on the sequenced biomolecules of Panax species by emphasizing the central molecular dogma, i.e., DNA (genomics), RNA (transcriptomics), and proteins (proteomics) using multiple sequencing technologies.
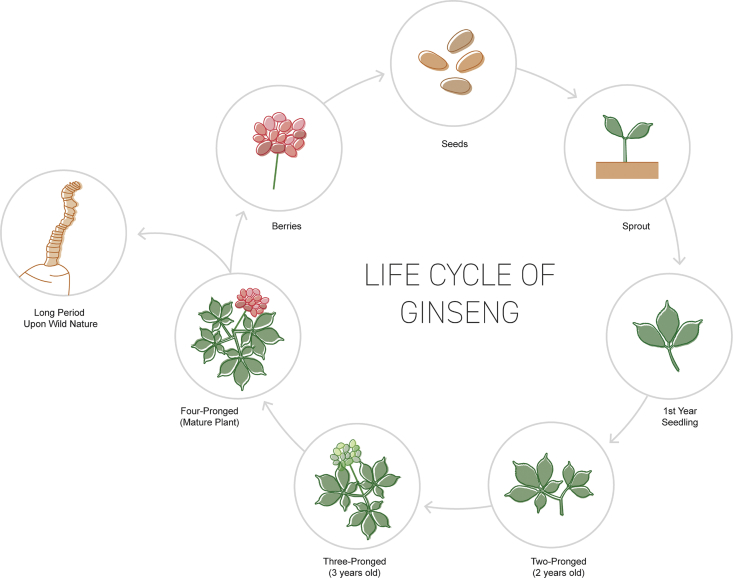
Fig. 1.
Illustration of the P. ginseng developmental life cycle. During the developmental life cycle, the leaf blades gives a clear identification of the plant's age. In the 4th year, the plant reaches the reproductive stage. Berries are collected and long-processed to prepare the seeds. The ginseng roots can live for more than 100 years in the wild; age can be determined by counting the rhizome scars attached to roots.
Fig. 2.
Illustration of different Panax species root types. Because the structure of P. ginseng resembles the shape of a human, it is described as a “man-shaped root.” In traditional medicine, this gave ancient practitioners the confidence to elucidate the yin-yang properties of ginseng according to human-centric philosophies.
2. Genome
Genome sequencing provides a complete overview of the structural organization of functional elements in a given genome. These structural elements carry the knowledge of the evolutionary history of an organism. In the digital era of the early 21st century, nearly complete genetic characterizations of any given species can be determined based on genome sequencing. Significantly, the quality of a reference genome informs the functional factors of various problems that exist in a given species [18]. Concurrently, sequencing technologies, physical mapping, and corresponding computational methods are primarily developed to identify and rapidly construct high-quality genomes [18]. These high-quality, nonfragmented, and chromosomal scales for constructed genomes provide precise knowledge of given species in the context of comparative, functional, and evolutionary aspects. Taking all the above perspectives into consideration, this review is conducted for the genome of the Panax species (Table 1).
Table 1.
Genome sequences of Panax species
| Species | P. ginseng | P. ginseng | P. notoginseng | P. notoginseng | P. notoginseng |
|---|---|---|---|---|---|
| Ploidy | Tetra | Tetra | Di | Di | Di |
| Assembled genome size | 3.5 Gb | 3.12 Gb | 2.39 Gb | 1.85 Gb | 2.36 Gb |
| Genome size estimated (Flow Cytometry/k-mer) | 3.5 | 3.3-3.6 | 2.31 | 2.0 - 2.1 | NA |
| Cultivar/line/cultivation | IR826 | Chunpoong | Green House | Mountain | NA |
| Plant age | 4 yrs (leaves) | 4 yrs (leaves) | NA (leaves) | NA (leaves) | NA |
| Sequencing method(s) | Illumina | Illumina, | Illumina | Illumina | Illumina, |
| Pacbio | Pacbio | ||||
| Raw data size | 391.46 | 1,111 | 1837.6 | 385.28 | 269.07 |
| Scaffolds | 83,074 | 9,845 | 122,131 | 76,517 | 179,913 |
| Total bases | 3,414,349,854 | 2,984,993,682 | 2,394,283,436 | 1,849,578,873 | 2,359,971,642 |
| N50 | 108,708 | 567,017 | 96,155 | 157,811 | 72,374 |
| Gaps | 12.15% | 8.11% | 18.15% | 17.14% | NA |
| Genes/proteins | 42,006 | 59,352 | 36,790 | 34,369 | 35,451 |
| Repeat content in genome | 60% | 79.50% | 75.94% | 61.31% | 51.03% |
| Long terminal repeats | 83.50% | 49% | 66.72% | 95.10% | 90.31 |
| Journal | Giga | Plant Biotechnology | Molecular Plant | Molecular Plant | Revix |
| Science | |||||
| Year | 2017 | 2018 | 2017 | 2017 | 2018 |
2.1. Panax ginseng
P. ginseng was first estimated to be a tetraploidy nuclear genome consisting of 48 chromosomes (2n = 4x = 48) via flow cytometry in 1963. Whereas, other Panax species consist of only 24 chromosomes (2n = 2x = 24), with the exception of P. ginseng and P. quinquefolius. It is assumed that the tetraploidization of these species may have occurred due to natural cross-pollination of two diploidy Panax species [19], [20]. Eventually, following 40 years of research, the genome size was estimated to be 3.12 gigabits (Gb) in 2004 [21]. In the same year, genome sequencing was initiated for P. ginseng through construction of the bacterial artificial chromosome (BAC) library [21]. Duplication and evolutionary events occurring in P. ginseng resulted in a new species—namely, P. quinquefolius—which consists of an increased nuclear genome size of 4.91 gigabytes (GB) [19], [22], [23]. Subsequently, another BAC library was constructed in 2010 to develop a molecular marker for authenticating the Panax species through identification of sequentially tagged sites. In 2017, a minimal number of BAC clones were sequenced with a single-molecule real-time (SMRT) sequencing method [24]. As a result of these continuous efforts, the genome for P. ginseng (chunpoong) in its entirety, having a size of 2.98 GB, was published in 2018, together with a complete assessment of its migration, evolution, and duplication events [25].
In the abovementioned study, the size of the P. ginseng genome was reestimated to be 3.3-3.6 GB using flow cytometry and k-mer analysis. Similarly, another draft genome for P. ginseng (IR826) was reported in 2017, with an estimated size of 3.5 Gb [26]. By comparing this to a previous iteration of genome sequencing, the size was similar, but the genome assembly was relatively more fragmented. The structural elements of the draft from cultivar IR826 include 42,006 genes with 60% repeat regions, and the selection from cultivar Chunpoong consists of 59,352 genes with 79.52% repeat regions—findings that were predicted from the abovementioned genomic studies. As a result of these outcomes, in average 70% of the P. ginseng species genomes consists of repeats, in comparison to other plants in the angiosperm clade and, more specifically, among medicinal herbs. Thus, the Panax species is well-known for its various pharmacological values, especially the triterpenoid saponins (i.e., ginsenosides), which are only present in the Panax species. The biosynthesis mechanism for ginsenosides is yet to be fully understood. Existing genomic studies have profiled putative ginsenoside pathway genes, attempting to annotate these genes more precisely to uncover the biosynthesis pathways completely. Through these high-throughput sequencing efforts, ∼1502 GB of DNA sequences have been elucidated for P. ginseng to date.
2.2. Panax notoginseng
The P. notoginseng is a diploid genome belonging to the Araliaceae family, consisting of (2n = 2x = 24) chromosomes, half the number in P. ginseng [19], [20]. Genome sequencing for P. notoginseng was recently initiated in the high-throughput sequencing era, with three draft genomes released from 2017 to 2018 [27], [28], [29]. The genome size for P. notoginseng is estimated to be between 2.2 and 3.1 GB by flow cytometry and small compared with the draft genome of P. ginseng, and it makes sense because P. notoginseng is a diploid genome. Nevertheless, the draft genome was constructed for P. notoginseng using more than 100X coverage (790X, 192X, 104X) in short sequences; owing to the repeat complexity, the genome assemblies were highly fragmented. Through these studies, the draft genome for P. notoginseng was constructed in various sizes including 2.39 Gb [29], 1.85 Gb [27], and 2.36 Gb [28]. The drafts of P. notoginseng genomes were also dominated by 75% repeats in one of the three assemblies, which is similar to that of the P. ginseng genome. More specifically, the third draft genome utilizes nearly 13 GB (six-fold) of long-read sequences to improve assembly for enhancing structural annotation. One previous study explains how the diploid genome diverged from the tetraploid genomes even before the occurrence of genome duplication. This hypothesis is reversed in genome studies of P. ginseng [25]. Thus, this theory must be evaluated more precisely in future research. Finally, because of these genomic studies, approximately ∼2492 GB of DNA sequences have been produced for P. notoginseng.
2.3. Organelle genomes
Organelle genomes (any number of organized or specialized structures within a living cell; pertinent organelles in the study of ginseng are chloroplasts and mitochondria) are much smaller than nuclear genomes, consisting of evolutionary and plant physiological information. Evolutionary relationships in a given species are assessed using the chloroplast genome. Most genome-sequencing projects start with these organelle genomes to evaluate preliminary knowledge about the plant and its biological nature. Moreover, organelle genomes contribute greatly to the identification of molecular markers to authenticate the cultivars of P. ginseng [30]. For instance, the single nucleotide polymorphism (SNP) in mitochondrial genes (nad7, cox2) is an authentication marker for the cultivar P. ginseng (chunpoong), differentiating it from other cultivars [30]. A complete chloroplast genome sequence for the Panax species, namely P. ginseng, was initiated in 2004 to elucidate evolutionary relationships in the context of other vascular plants [31]. In this way, evolutionary relationships among other wild ginseng plants were derived [32]. Organelle sequencing has facilitated discovery of nine other Panax cultivars from chloroplast genomes. In addition, six authentication SNP and insertion-deletion polymorphism (indel) markers have been proposed for the P. ginseng cultivars [33]. In 2016 and 2017, complete chloroplast genomes were released for the North American P. quinquefolius [34] and the Panax vietnamensis, respectively [35]. These efforts in genome sequencing have led to the development of DNA barcodes for ginseng from chloroplast genomes to authenticate certain ginseng products on the market. Because the existing universal barcode was not highly reliable for ginseng, a mini-barcode system was eventually proposed for Panax family species using chloroplast genomes [36]. As a benefit of all these sequencing projects, a group of chloroplast genes (i.e., trnC-rps16, trnS-trnG, and trnE-trnM) has emerged as significant in the development of Panax-based molecular markers, as proposed from detailed phylogenomic assessment [37]. Furthermore, genome duplication and diversity in the Panax species were confirmed via chloroplast genomes [38]. Processes through which the Panax species is handling a specific selective pressure have also been assessed with organelle genomes, consequently showing that the mitochondrial genome is more evolved than the chloroplast genome [39]. To date, 36 chloroplast genomes have been submitted to the open-access GenBank sequence database (Table 2).
Table 2.
Organelle genomes of Panax species
| S. No | GenBank ID | Bases | Year | Species | Cultivar |
|---|---|---|---|---|---|
| 1 | AY582139 | 156318 | 2005 | P. ginseng | NA |
| 2 | NC_006290 | 156318 | 2010 | P. ginseng | NA |
| 3 | KF431956 | 156355 | 2015 | P. ginseng | NA |
| 4 | KM088019 | 156248 | 2015 | P. ginseng | Chunpoong |
| 5 | KM067386 | 156356 | 2015 | P. ginseng | Cheongsun |
| 6 | KM067387 | 156355 | 2015 | P. ginseng | Gopoong |
| 7 | KM067388 | 156356 | 2015 | P. ginseng | Gumpoong |
| 8 | KM067389 | 156355 | 2015 | P. ginseng | Jakyung |
| 9 | KM067393 | 156425 | 2015 | P. ginseng | Sunhyang |
| 10 | KM067390 | 156355 | 2015 | P. ginseng | Sunone |
| 11 | KM067391 | 156355 | 2015 | P. ginseng | Sunpoong |
| 12 | KM067392 | 156355 | 2015 | P. ginseng | Sunun |
| 13 | KM067394 | 156241 | 2015 | P. ginseng | Hwangsook |
| 14 | KM088020 | 156355 | 2015 | P. ginseng | Yunpoong |
| 15 | KC686331 | 156354 | 2015 | P. ginseng | Damaya |
| 16 | KC686332 | 156354 | 2015 | P. ginseng | Ermaya |
| 17 | KC686333 | 156354 | 2015 | P. ginseng | Gaolishen |
| 18 | KP036469 | 156188 | 2015 | P. japonicus | NA |
| 19 | NC_028703 | 156188 | 2015 | P. japonicus | NA |
| 20 | KX247146 | 156063 | 2016 | P. japonicus | Bipinnatifidus |
| 21 | MF377620 | 156248 | 2017 | P. japonicus | Bipinnatifidus |
| 22 | KJ566590 | 156387 | 2015 | P. notoginseng | NA |
| 23 | KP036468 | 156466 | 2015 | P. notoginseng | NA |
| 24 | KR021381 | 156387 | 2015 | P. notoginseng | NA |
| 25 | KT001509 | 156324 | 2015 | P. notoginseng | NA |
| 26 | NC_026447 | 156387 | 2015 | P. notoginseng | NA |
| 27 | KM088018 | 156088 | 2015 | P. quinquefolius | NA |
| 28 | KT028714 | 156359 | 2015 | P. quinquefolius | NA |
| 29 | NC_027456 | 156088 | 2015 | P. quinquefolius | NA |
| 30 | KX247147 | 156064 | 2016 | P. stipuleanatus | NA |
| 31 | KY379906 | 156069 | 2017 | P. stipuleanatus | NA |
| 32 | MF377622 | 156090 | 2017 | P. stipuleanatus | NA |
| 32 | MF377622 | 156090 | 2017 | P. stipuleanatus | NA |
| 33 | NC_030598 | 156064 | 2017 | P. stipuleanatus | NA |
| 34 | MF100782 | 156157 | 2018 | P. trifolius | NA |
| 35 | NC_037994 | 156157 | 2018 | P. trifolius | NA |
| 36 | KP036470 | 155993 | 2015 | P. vietnamensis | NA |
| 37 | NC_028704 | 155993 | 2015 | P. vietnamensis | NA |
| 38 | KP036471 | 155992 | 2015 | P. vietnamensis | NA |
| 39 | KU059178 | 155993 | 2016 | P. vietnamensis | NA |
| 40 | MF377621 | 156022 | 2017 | P. vietnamensis | NA |
3. Transcriptome
A transcriptome is a complete representation of all the messenger RNA molecules expressed in the genome of an organism. The fundamental goal of transcriptomics is to understand the protein/gene functions of a given organism—mainly to determine which group of genes is responsible for organism uniqueness. At present, RNA sequencing (RNA-Seq) is the state-of-the-art method in the field of transcriptomics to understand how genes are expressed in an organism. Since completion of the first genome for the plant Arabidopsis thaliana in the year 2000, the genomes of most plants are not sequenced because of their polyploidy and repeat complexities. Nevertheless, RNA-Seq technology has facilitated a way to obtain basic knowledge about genes/proteins for all nonmodel plants on earth. Furthermore, long-read sequencing technologies have aided in uncovering genome-wide, full-length transcriptomes, together with their isoforms, for any nonmodel plant. In parallel, the development of computational methods to analyze and integrate multiple technologies has helped the plant research community to surpass objectives with transcriptome data. In medicinal plants, RNA-Seq plays a vital role in understanding the biosynthesis of unique metabolites. Likewise, most RNA-Seq projects in the genus Panax have been implemented primarily to elucidate ginsenoside biosynthesis in different organs and stages. Of course, RNA-Seq is not limited to this specific aim of investigating how ginsenosides are biosynthesized but are widely used in numerous applications in the Panax species according to other relevant research.
3.1. Panax ginseng
Transcriptome sequencing for P. ginseng was initiated in the year 2003, using Sanger sequencing through the construction of expressed sequence tags (ESTs) from different plant organs, including roots (i.e., different ages and types), leaves, flower buds, and other parts of plants. Later, extension of the same project took place in which roots were sequenced following treatment with methyl jasmonic acid to assess for differential expression patterns in terpenoid backbone gene biosynthesis [17], [40], [41], [42], [43], [44]. Initially, from the year 2011 to 2014, a 454-pyrosequencing technology was used for tissue-specific and hormone treatments to improve throughput in P. ginseng transcriptomes. While these transcriptome data sets from 454-pyrosequencing technology were similar to data sets obtained with the Sanger method, coverage of the genes was insufficient [45], [46], [47]. Hence, to improve throughput and gene coverage on a cost-effective basis, another next-generation short-read sequencing technology (Illumina) was utilized from 2014 to 2016 to sequence a large-scale transcriptome [48], [49], [50], [51], [52], [53], [54]. Even though the short-read sequencing technology worked for P. ginseng, it was not effective in obtaining full-length transcripts insofar as the genome is covered with ∼70% repeats. Therefore, subsequent researchers have used a long-read single-molecule real-time technique for different tissues to overcome these repeat complexities [55]. To date, 16 transcriptome projects have been performed using de novo analysis from 2003 to 2018 (Table 3). Overall analysis of existing transcriptomic studies shows that they were conducted according to three strategies: (1) to reveal that ginsenoside content varies with age and root cultivation method, (2) to observe tissue-specific and cultivar-specific transcripts/genes, and (3) to study how a plant responds to environmental stress (Table 3).
Table 3.
Descriptions of transcriptome sequences of Panax species
| S. No | Type of sequencing | Species | Cultivars | No. of samples | Samples | Type | Project type | Year | References |
|---|---|---|---|---|---|---|---|---|---|
| 1 | EST-Sanger | Pg | NA | 5 | Rh (4-Y), Sh (in-vitro culture), R (4-Y), Sh (3-weeks-old seedling), S (green-color stage – One month old), | D | DE | 2003 | [42] |
| 2 | EST-Sanger | Pg | NA | 1 | HR-treated with 10 uM MeJA | D | DE | 2005 | [40] |
| 3 | EST-Sanger | Pg | NA | 1 | L (4-Y) | D | DE | 2006 | [41] |
| 4 | EST-Sanger | Pg | NA | 3 | HR, R (14-Y, 4-Y) | D | DE | 2010 | [17] |
| 5 | EST-Sanger | Pg | NA | 2 | Embryonic callus, F. buds | D | DE | 2011 | [44] |
| 6 | 454-Pyro-Seq | Pg | NA | 1 | R (11-Y) | D | DE | 2011 | [45] |
| 7 | 454-Pyro-Seq | Pg | NA | 4 | R, S, L, F (4-Y) | D | DE | 2013 | [47] |
| 8 | 454-Pyro-Seq | Pg | Yanpoong | 5 | MeJA-treated AR (6-Y) (control, 2h, 6h, 12h, and 24h) | C | DE | 2014 | [46] |
| 9 | Illumina | Pg | Chunpoong & Cheongsun | 2 | AR | D | DE | 2014 | [54] |
| 10 | Illumina | Pg | Damaya | 2 | MeJA-treated AR (control, treated) – 4-Y | C | DE | 2015 | [53] |
| 11 | Illumina | Pg | Damaya | 18 | R, S, L (Benzoic acid stress) | C | DE | 2015 | [49] |
| 12 | Illumina | Pg | Damaya | 19 | R (fiber, leg, epiderm, cortex, arm), Rh, S, L (peduncle, blade, pedicel), F (peduncle, pedicel, flesh), S and R (5-Y, 12-Y, 18-Y, and 25-Y) | D | DE | 2015 | [51] |
| 13 | Illumina | Pg | Chunpoong | 4 (3 Rs) | R (1-Y), R–6Y (main body, lateral & rhizome) | D | DE | 2015 | [48] |
| 14 | Illumina | Pg | NA | 1 | R (leaf expansion period) | D | DE | 2016 | [50] |
| 15 | Illumina | Pg | Damaya | 1 | 3-Y R inoculated with Cylindrocarpon destructans (0 to 12 days) | C | DE | 2016 | [52] |
| 16 | PacBio | Pg | Cheonmyeong | 4 | F, L, S and R (4-Y) | D | DE | 2017 | [55] |
| 17 | Illumina | Pg | IR826 | 3 (3 Rs) | Periderm, cortex, and stele | D | G | 2017 | [26] |
| 18 | Illumina | Pg | Chunpoong & Yunpoong | 2 | R (1-Y) (1-3 weeks heat treatment) | D | RG | 2018 | [57] |
| 19 | Illumina & PacBio | Pg | Chunpoong | 58 | Seeds, L, S, F (Stress: salt, cold, drought, MeJA) | D | G | 2018 | [56] |
| 20 | 454-Pyro-Seq | Pn | NA | 1 | R (4-Y) | D | DE | 2011 | [58] |
| 21 | Illumina | Pn | NA | 3 | L, R, F (3-Y plant) | D | DE | 2015 | [59] |
| 22 | Illumina | Pn | NA | 52 | Roots from Seedlings with arsenic treatment | D | DE | 2016 | [60] |
| 23 | Illumina | Pn | NA | 6 | F, L, fruit, S, primary and secondary R | D | G | 2017 | [29] |
| 24 | Illumina | Pn | NA | 5 | R, S, L, F, and Rh | D | G | 2017 | [27] |
| 25 | Illumina | Pn | NA | 8 | L & R(1,2,3-Y), F(2,3-Y) | D | G | 2018 | [28] |
| 26 | EST-Sanger | Pq | NA | 4 | F, L, R, S (4-Y) | D | DE | 2010 | [61] |
| 27 | 454-Pyro-Seq | Pq | NA | 1 | L (4-Y) | D | DE | 2010 | [62] |
| 28 | 454-Pyro-Seq | Pq | NA | 1 | R (4-Y) | C | DE/TR | 2013 | [63] |
| 29 | Illumina | Pq | NA | 1 | Seeds | C | DE/TR | 2015 | [64] |
| 30 | Illumina | Pq | NA | 1 | Roots (5-Y) | C | G | 2016 | [65] |
| 31 | Illumina | Pj | NA | 1 | Rh (6-Y) | D | DE/TR | 2015 | [73] |
| 32 | Illumina | Pj | NA | 5 | F, L, sec R, Rh_Young, Rh_Old (7-Y) | D | TR | 2016 | [72] |
| 33 | Illumina | Pv | NA | 1 | R | D | TR | 2015 | [74] |
| 34 | EST-Sanger | Ps | NA | 3 | L, S, and Rh – 5-Y | D | TR | 2016 | [71] |
| 35 | Illumina | Pz | NA | 4 | R (fiber, main), L, S | D | DE | 2018 | [75] |
Pg, Panax ginseng; Pn, Panax notoginseng; Pq, Panax quinquefolium; Pv, Panax vietnamensis; Pj, Panax japonicus; Ps, Panax sokpayensis; Pz, Panax zingiberensis.
L, leaf; S, stem; Sh, shoot; R, root; Rh, rhizome; E, embryo; F, flower; AR, adventitious roots; HR, hairy roots; MeJA, methyl jasmonic acid; D, discrete; C, continuous; Seq, sequencing; Tech, technology; Y, year(s); DE, de novo; TR, transcriptome; G, genome; RG, reference genome; Rs, replicate.
As a result, all existing transcriptome data sets are formulated with one of two bioinformatics analysis strategies—discrete or continuous. Notably, only two studies were continuously simulated to understand environmental stress mechanisms in P. ginseng—specifically, how P. ginseng root reacts to benzoic acid in the context of autotoxicity [49] and how it responds to root-rot diseases induced by Cylindrocarpon destructants [52]. In contrast, all other transcriptomic studies have been based on discrete models. The major concept of almost all the previous research is elucidation of ginsenoside biosynthesis enzymes and their expression patterns in different tissues and age-groups of roots. Most projects have focused highly on the de novo model, and continued focus on this perspective will depend on interpretations of final results. At this point, our focus on one specific species may be minimized with the help of two different versions (chunpoong and yunpoong) of reference draft genomes. In these two draft versions, the chunpoong draft is optimized more precisely via co-expression networks. Overall, this study has contributed the highest number of publicly available transcriptome libraries with organized and user-friendly databases [56]. Reaching the end of the P. ginseng de novo transcriptome era, the next dimension will be the reference transcriptome, involving the mining of novel genes and functional annotations for predicted genes. In this emerging discipline, the first study uses the chunpoong draft to predict heat-stress responsive genes in the two cultivars (chunpoong and yunpoong). The resulting hypotheses are that CAB, FAD, and WRKY genes will be valid candidates for further characterization [57]. Other advantages of using this reference genome are that it will help researchers to organize outcomes in a genome-centric manner and in detailed characterization of various problems associated with P. ginseng.
3.2. Panax notoginseng
Initiation of the P. notoginseng transcriptome occurred in 2011, using 454 pyrosequencing, to identify genes responsible for ginsenoside biosynthesis along with the simple sequence repeat (SSR) markers from their roots [58]. To date, three transcriptome studies have been reported along with the abovementioned Illumina study (i.e., research conducted using an Illumina sequencer to study alkaloid and ginsenoside biosyntheses in different organs of plants) [59]. Another study on the environmental toxicity of plants was conducted to determine how ginseng regulates the response of ginsenosides and flavonoids to arsenic-based heavy metal stress [60]. In addition, organ-specific transcriptomes for P. notoginseng are generated in three different genomic studies [27], [28], [29]. Unlike P. ginseng, research in this species focuses highly on different secondary metabolites such as flavonoids and alkaloids. Thus, this aspect of research includes fewer studies in genome-wide expression analysis.
3.3. Panax quinquefolius
Transcriptome research for the P. quinquefolius species was initiated in 2010 through the construction of ESTs. Subsequently, different organs of the species were sequenced with Sanger sequencing technology [61]. Concurrently, in 2010, a root transcriptome was sequenced with 454 pyrosequencing to study ginsenoside biosynthesis in detail through elicitation with methyl jasmonate (MeJA) [62]. Sequentially, three other studies were formulated to understand the different molecular mechanisms involved in P. quinquefolius including (1) ginsenoside biosynthesis in various developmental stages [63], (2) seed dormancy mechanisms [64], and (3) expression of ginsenoside biosynthesis transcripts following MeJA treatments at different points in time [65]. Additionally, another study has assessed a SNP common to two Panax species (i.e., P. ginseng and P. quinquefolius). Finally, high-throughput transcriptomic studies for this species were generated to develop continuous data sets rather than discrete samples. These continuous data sets are an additional milestone and are vital in constructing co-expression networks for P. quinquefolius, which may help to characterize the additional genes responsible for ginsenoside biosynthesis. Unlike many other Panax species, however, P. quinquefolius does not have a reference genome. Regardless, the current transcriptome data set may aid researchers in conducting a systematic bioinformatics analysis to unpack multiple hypotheses on ginsenoside biosynthesis.
3.4. MicroRNAs
MicroRNAs (miRNAs) are noncoding RNAs (21-24 bases) possessing multiple regulatory roles, particularly in posttranscriptional modifications. They are involved in the process of 3′-prime, 2′-O-methylation in plants, whereas in animals, the analogous function occurs in the 5′-prime. Additionally, molecular signatures left from Dicer and Argonaute proteins machines are considered to be core features in the establishment of computational methods of miRNA predictions in plants and animals specifically. In Panax species, these computational methods have been used along with deep-sequencing technologies for profiling miRNAs. Initially, in the years 2012 and 2013, miRNAs were profiled from plant tissues of P. ginseng aged 4-5 years [66], [67]. That study resulted in very few miRNAs (i.e., 73 conserved and 28 nonconserved [67] and 69 conserved miRNAs [66]) in comparison to other plants, which we now know is because the predictions therein were based on the transcriptome for which it was conducted rather than the genome. Furthermore, younger tissues are more highly recommended for profiling miRNAs compared with mature tissues because organogenesis occurs more widely in immature tissues than in mature tissues. Another study also reports few miRNAs with variability across age groups of roots (1-3 years of age) in P. ginseng [68]. In P. notoginseng, the function of an individual miRNA (i.e., miR156) was identified as a critical regulator for different sizes of roots [69]. Also reported the essential regulatory role of miR171 in pentatricopeptide repeat proteins, which have a necessary function in the completion of degradome sequencing [70]. Finally, the data set generated in the work of Mathiyalagan et al. [66] has been utilized with the P. ginseng genome to predict/annotate miRNAs [25].
3.5. Other Panax species
According to the National Center for Biotechnology Information Taxonomy classifications, the genus Panax has 15 species and six subspecies. Among them, only seven species have a transcriptome for understanding ginsenoside biosynthesis. With addition to above three, the four other species are Panax sokpayensis, P. vietnamensis, Panax zingiberensis, and P. japonicus. Newly reported from Sikkim, near the Indian Himalayan valley, the species P. sokpayensis consists of minimally expressed sequence tags generated as a result of suppression subtractive hybridization and Sanger sequencing methods. Likewise, Sanger methods were used for assessing differentially expressed transcripts in the rhizomes and leaves of P. sokpayensis [71], which was later added as an essential data set to the Panax family. P. japonicus is common in Japan, where it has been used as an alternative to P. ginseng for more than 1000 years. In comparison to other Panax species, P. japonicus is abundant in oleanane-type saponins [72]. Recently, transcriptome data from five different tissues of P. japonicus were generated using Illumina, and the transcripts responsible for ginsenoside biosynthesis—namely, cytochrome and glycosyltransferase—were identified for this species [72]. Similarly, in China, another species-specific set of transcriptomes in rhizomes was sequenced [73]. P. vietnamensis, also known as Vietnamese ginseng, is rich in ocotillol-type saponins and, more specifically, in majonoside R2. The transcriptome for the root of this species was generated using Illumina, and it was further assessed for ginsenoside biosynthesis pathways [74]. P. zingiberensis is an endangered Panax species native to South China. This species is highly abundant in oleanane and dammarane-type ginsenosides. Transcriptomes from these plant tissues have been recently generated to study ginsenoside biosynthesis pathways [75]. In fact, most of the transcriptome projects for the species mentioned here were conducted to more specifically reveal and understand the genes/transcripts responsible for biosynthesis in the novel and species-specific saponins in comparison to other research factors.
4. Proteome
The development of multiple “omics” platforms has advanced the annotation of genomes at different levels of the molecular central dogma. These technological advancements have resulted with a knowledge on multiple genomes and transcriptomes for the Panax species. In contrast, only a few studies have been conducted to elucidate proteomes for various applications. The proteome was initially constructed to discern P. ginseng from P. quinquefolius using two-dimensional electrophoresis (2-DE). That research uses samples that were collected from different tissues and cultured cells of the two relevant Panax species [76]. Later, comprehensive proteome/peptide fingerprinting was conducted for P. ginseng from hairy root cultures via 2-DE. Subsequently, those 2-DE spots were sequenced using matrix-assisted laser desorption/ionization, electrospray ionization quadrupole, and time-of-flight mass spectrometry. Finally, tissue-specific proteins were identified by comparing peptides from those sequences with ESTs for functional annotations [77]. Similarly, in other proteomic studies, high-light responsive proteins were identified from P. ginseng to elucidate protective mechanisms because ginseng is a shade-grown plant [78]. Likewise, growth-responsive proteins from P. ginseng roots aged 1-5 years were elucidated [79], and proteomes related to cultivated and wild-type roots were identified using 2D and iTRAQ systems, respectively [80], [81]. These are the primary proteome elucidation projects in Panax species for elucidating physiological processes and authentication markers. The limited number of existing studies helps us understand the challenges faced in building the proteome library. Additionally, problems associated with library construction have been illuminated, including sampling, high-throughput proteome sequencing, annotation, differential expression, and integration with genomics/transcriptomics (proteogenomics) [82]. Some existing research has helped in the elucidation of proteomes, which are responsible for secondary and species-specific secondary metabolites biosynthesis [83]. However, the data sets have not been deposited in the public repository properly; hence, the research outcomes on Panax species proteomes are unorganized and nonreusable.
5. Population studies
The selection of economical traits in seedlings requires genome markers to facilitate effective breeding schemes by considering various factors such as growth, germination, biomass, secondary metabolites, pathogen resistance, and environmental stress resistance. Our contemporary era of advancement in sequencing technologies such as genotyping by sequencing (GBS) has facilitated multiple opportunities to elucidate genome reliable genetic SNP markers. Along with GBS, the genomic and transcriptome-wide SNP assessment studies have also enhanced the molecular marker identification in Panax species. Initially, 16 nuclear single copy genes were selected from the model plant Arabidopsis thaliana, which was used to derive SNP molecular markers to identify the diversity that exists among the two Panax species i.e., P. ginseng and P. quinquefolium [84]. Later, the SNP markers elucidated from the previous studies were extended with organelle genomes to enhance the quality of the SNP marker and then it used to derive the diversity patterns that exist in cultivated and wild-grown ginseng [33], [85]. As a result research has revealed that the cultivated ginseng has a higher genetic shift in comparison to wild-grown ginseng because of adaptation to light and other selective pressures in cultivation fields and other controlled cultivation environments. This extended data set and the available genome for P. ginseng have facilitated the ways to assess genome-wide variation among the Panax species. Also, the existing research has helped to derive more optimized diversity patterns in the cultivated ginseng in Northern and Southern China [86]. Knowledge of these patterns has resulted in the identification of SNP markers from 97 genes associated with ginseng from Northern China and five genes in ginseng from parts of Southern China. Finally, in P. notoginseng, de novo-based genotype markers were elucidated from the GBS sequencing to identify the province-specific plants [87]. However, still more research focus is needed to optimize the SNP markers for different Panax species to enhance the cultivations.
6. Multifamily genes
Panax-specific secondary metabolites are ginsenosides that belong to triterpenoids derived from the common precursor 2,3-oxidosqualene. There are more than 150 types of ginsenosides, which are further classified into three groups of protopanaxadiol, protopanaxatriol, and oleanane [11]. However, the mevalonate pathway is a precursor for deriving different type of ginsenosides, and the extended putative ginsenoside consists of two major multifamily genes, namely cytochromes and glycosyltransferases, which are essential and remains uncharacterized. These multifamily enzymes are essential to synthesize different types of ginsenosides, which are mainly classified according to the types of carbohydrates present in the functional groups. Almost all sequencing projects—from ESTs to genome projects—aim to reveal the biosynthesis mechanisms of ginsenosides in Panax species. At first, these studies attempted to group the cytochromes and glycosyltransferase from minimal EST data [88], [89]. At this point, from the P. ginseng genome, 383 cytochromes and 226 UDP-glucosyltransferase (UGTs) have been identified; from the P. notoginseng genome, 127 UGTs and 145 cytochromes have been identified [25], [27]. Among these, only 34 genes have been thoroughly characterized to construct ginsenoside biosynthesis pathways [90]. In addition, the transcription factors were estimated to be a regulator of ginsenoside biosynthesis enzymes, which have been assessed in greater detail regarding WRKY proteins [91], nucleotide binding site–encoding gene families [92], and basic-helix-loop-helix transcription factors [93]. Additionally, other than ginsenoside biosynthesis, the receptor-like kinase–encoding gene family has also been identified as one of the most vital gene families for understanding the biotic and abiotic stresses in Panax species [94].
7. Concluding remarks
The overall research efforts conducted to decode the genetics of Panax species till date has facilitated a way to obtain a collective knowledge of the sequenced samples as well as the structure of available data sets at different stages of the central molecular dogma (graphical abstract). Although, these efforts have given the multiple draft version of genomes for only two of all the Panax species [25], [26], [27], [28], [29], still need more improvements in order to obtain the draft versions for the other Panax species. Also, the available draft versions can be enhanced further for achieving the chromosomal level assemblies. As a consequence, those improvements will be more helpful in enhancing the genome-assisted breeding and to yield high productivity by cultivating ginseng in different environments and more extensive geographical regions [87]. More focus and effort toward the development of traits for the nonshaded disease resistance and to obtain enriched secondary metabolites in their earlier stage are in need to reduce the current cultivation pitfalls [57]. Moreover, as explained in graphical abstract, the multiple sequencing techniques were effectively utilized to overcome the throughputs and in the reduction of fragmented assemblies; however, still the complexities such as ploidy, genome size, and high-repeat region coverage of the genome exists in these plants, making it hard to obtain chromosome level assemblies for the Panax species. In the future, the researchers are expected to utilize the contemporary multiple long-read sequencing, scaffolding, and physical mapping technologies along with productive computational algorithms and collaborative efforts to bring out the high-quality reference genome [6], [18], [95].
In addition, while considering transcriptome, the generated data sets were highly supportive in identifying and understanding the genes in Panax species. Two different assembly strategies such as the reference-based and de novo assemblies were utilized mainly for generating the data sets. However, while coming to the functional annotations and expression quantifications, the computational models have played a significant role in the identification of causative genes in the genome from the specified conditions. Further, the researchers should consider the instructions that were proposed using the computational/mathematical models to interpret/predict the candidate transcripts/genes for a given model. For instance, while constructing co-expression networks, the user could consider the sample size, heterogeneity of the selected libraries, and the sequence qualities, which may positively influence the results and accuracy of the prediction model [9]. Besides, in transcriptome differential expressions quantification, the methods were mostly developed for two classes of the data models, i.e., paired (i.e., control vs. case) and continuous models (time serious). Among all the transcriptome data sets, only a few studies have generated data sets for the continuous models [46], [49], [52], where others have majorly generated data sets for the paired models (Table 1). Another crucial difficulty is the limitations in using computational research outcomes directly for further experiments due to the lack of statistical significance, which is eventually reducing the reproducibility via experiments on account of ignoring the sample replicates because most of the quantification methods are optimized only with statistical models [9], [96]. By considering the above proposals, the researchers are requested to give more focus toward sample selection and model of the study by giving importance to the plant's physiological process. The physiological process, such as different developmental stages and also by combining their different stresses should be considered to understand the core-stress responsible genes, which will further promote the biosynthesis of secondary metabolites in the Panax species [97].
Furthermore, most of the sequencing projects in Panax species are only valuing ginsenosides, whereas, other secondary metabolites and bioactive proteins/peptides are overlooked from confirming the superstitious beliefs obtained through the traditional medicinal practices for ages. Hence, concentrating on other biologically active compounds having therapeutic values will further improve the overall value and quality of the ginseng plant on the pharmaceutical market. For example, gintonin—a protein from Panax ginseng is reported to have a potential in activating the G protein-coupled receptors in human disease, which is one of the most vital targets for modern medicinal drugs [13]. Although being one of the important therapeutic molecules of Panax species, the complete biosynthesis pathway of ginsenosides remains uncharacterized [90]. So, to completely characterize its pathway, more sequencing efforts should be taken by implementing the systematic bioinformatics analysis to identify the cytochromes and glycosyl-transferases from the Panax genome or the microbiota associated with these plant species [98], [99].
Eventually, the ginsenoside biosynthesis pathway can also be implemented in the microbial system, such as yeast to produce different type of ginsenosides in a massive quantity using the raw materials from industries as implemented for the opioids in Papaver sominefera [100]. One of the major concerns for taking ginseng as a medicinal drug is their completely unproven myths on its toxic effects. Currently, the protein data for Panax have limitations, where those produced from a few studies are not able to be used for further studies. Hence, in future, the field of proteomics is expected to emerge with optimized high-throughput sequencing techniques to produce large-scale data sets than the earlier ones, and it has to be considered for improving the annotation of Panax genome [10]. Therefore, the researchers in medicinal plant community are highly interested in elucidating the therapeutic molecules more precisely through sequencing technologies for better treatment because the global demand for herbal medicinal products such as ginseng has increased significantly in the recent years.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
This research was supported by a grant from the Korea Institute of Planning & Evaluation for Technology in Food, Agriculture, Forestry, & Fisheries (KIPET NO: 317007-3), Republic of Korea. The authors thank Jeein Kim for support in designing the image of the ginseng life cycle.
References
- 1.Petrovska B.B. Historical review of medicinal plants’ usage. Pharmacognosy Reviews. 2012;6(11):1–5. doi: 10.4103/0973-7847.95849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saito K. Phytochemical genomics—a new trend. Current Opinion in Plant Biology. 2013;16(3):373–380. doi: 10.1016/j.pbi.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Abbai R., Subramaniyam S., Mathiyalagan R., Yang D.C. Functional genomic approaches in plant research. In: Hakeem K.R., Malik A., Vardar-Sukan F., Ozturk M., editors. Plant bioinformatics: decoding the phyta. Springer International Publishing; Cham: 2017. pp. 215–239. [Google Scholar]
- 4.Yonekura-Sakakibara K., Saito K. Functional genomics for plant natural product biosynthesis. Natural Product Reports. 2009;26(11):1466–1487. doi: 10.1039/b817077k. [DOI] [PubMed] [Google Scholar]
- 5.Khater S., Anand S., Mohanty D. In silico methods for linking genes and secondary metabolites: the way forward. Synth Syst Biotechnol. 2016;1(2):80–88. doi: 10.1016/j.synbio.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nature Reviews Genetics. 2016;17:333. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin P., Xu G. Current state-of-the-art of nontargeted metabolomics based on liquid chromatography–mass spectrometry with special emphasis in clinical applications. Journal of Chromatography A. 2014;1374:1–13. doi: 10.1016/j.chroma.2014.11.050. [DOI] [PubMed] [Google Scholar]
- 8.Krumsiek J., Bartel J., Theis F.J. Computational approaches for systems metabolomics. Current Opinion in Biotechnology. 2016;39:198–206. doi: 10.1016/j.copbio.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 9.van Dam S., Võsa U., van der Graaf A., Franke L., de Magalhães J.P. Gene co-expression analysis for functional classification and gene–disease predictions. Briefings in Bioinformatics. 2018;19(4):575–592. doi: 10.1093/bib/bbw139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Restrepo-Pérez L., Joo C., Dekker C. Paving the way to single-molecule protein sequencing. Nature Nanotechnology. 2018;13(9):786–796. doi: 10.1038/s41565-018-0236-6. [DOI] [PubMed] [Google Scholar]
- 11.Qiao Y.-J., Shang J.-H., Wang D., Zhu H.-T., Yang C.-R., Zhang Y.-J. Research of Panax spp. in Kunming Institute of Botany, CAS. Natural Products and Bioprospecting. 2018;8(4):245–263. doi: 10.1007/s13659-018-0176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W-z, Hu Y., Wu W-y, Ye M., Guo D-a. Saponins in the genus Panax L. (Araliaceae): a systematic review of their chemical diversity. Phytochemistry. 2014;106:7–24. doi: 10.1016/j.phytochem.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 13.Im D-s, Nah S-y. Yin and Yang of ginseng pharmacology: ginsenosides vs gintonin. Acta Pharmacologica Sinica. 2013;34:1367. doi: 10.1038/aps.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohanan P., Subramaniyam S., Mathiyalagan R., Yang D.-C. Molecular signaling of ginsenosides Rb1, Rg1, and Rg3 and their mode of actions. Journal of Ginseng Research. 2018;42(2):123–132. doi: 10.1016/j.jgr.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahuja A., Kim J.H., Kim J.-H., Yi Y.-S., Cho J.Y. Functional role of ginseng-derived compounds in cancer. Journal of Ginseng Research. 2018;42(3):248–254. doi: 10.1016/j.jgr.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baeg I.-H., So S.-H. The world ginseng market and the ginseng (Korea) Journal of Ginseng Research. 2013;37(1):1–7. doi: 10.5142/jgr.2013.37.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sathiyamoorthy S., In J.-G., Gayathri S., Kim Y.-J., Yang D.-C. Generation and gene ontology based analysis of expressed sequence tags (EST) from a Panax ginseng C. A. Meyer roots. Molecular Biology Reports. 2010;37(7):3465–3472. doi: 10.1007/s11033-009-9938-z. [DOI] [PubMed] [Google Scholar]
- 18.Dominguez Del Angel V., Hjerde E., Sterck L., Capella-Gutierrez S., Notredame C., Vinnere Pettersson O., Amselem J., Bouri L., Bocs S., Klopp C. Ten steps to get started in genome assembly and annotation. F1000Research. 2018;7 doi: 10.12688/f1000research.13598.1. ELIXIR–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi H.-I.L., Kim N.-H., Lee J., Choi B.S., Kim K.D., Park J.Y., Lee S.C., Yang T.J. Evolutionary relationship of Panax ginseng and P. quinquefolius inferred from sequencing and comparative analysis of expressed sequence tags. Genetic Resources and Crop Evolution. 2013;60(4):1377–1387. [Google Scholar]
- 20.Wen J., Zimmer E.A. Phylogeny and biogeography of Panax L. (the ginseng genus, Araliaceae): inferences from ITS sequences of nuclear ribosomal DNA. Molecular Phylogenetics and Evolution. 1996;6(2):167–177. doi: 10.1006/mpev.1996.0069. [DOI] [PubMed] [Google Scholar]
- 21.Hong C.P., Lee S.J., Park J.Y., Plaha P., Park Y.S., Lee Y.K., Choi J.E., Kim K.Y., Lee J.H., Lee J. Construction of a BAC library of Korean ginseng and initial analysis of BAC-end sequences. Molecular Genetics and Genomics. 2004;271(6):709–716. doi: 10.1007/s00438-004-1021-9. [DOI] [PubMed] [Google Scholar]
- 22.Obae S, West T. Nuclear DNA content and genome size of American Ginseng2012. 4719-4723 p.
- 23.Kim K., Nguyen V.B., Dong J., Wang Y., Park J.Y., Lee S.-C., Yang T.J. Evolution of the Araliaceae family inferred from complete chloroplast genomes and 45S nrDNAs of 10 Panax-related species. Scientific Reports. 2017;7:4917. doi: 10.1038/s41598-017-05218-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang W., Kim N., Lee J., Waminal N.E., Lee S., Jayakodi M., Choi H.I., Park J.Y., Lee J.E., Yang T.J. A glimpse of Panax ginseng genome structure revealed from ten BAC clone sequences obtained by SMRT sequencing platform. Plant Breeding and Biotechnology. 2017;5(1):25–35. [Google Scholar]
- 25.Kim N.-H., Jayakodi M., Lee S.-C., Choi B.-S., Jang W., Lee J., Kim H.H., Waminal N.E., Lakshmanan M., van Nguyen B. Genome and evolution of the shade-requiring medicinal herb Panax ginseng. Plant Biotechnology Journal. 2018;0(0) doi: 10.1111/pbi.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J., Chu Y., Liao B., Xiao S., Yin Q., Bai R., Su H., Dong L., Li X., Qian J. Panax ginseng genome examination for ginsenoside biosynthesis. GigaScience. 2017;6(11):1–15. doi: 10.1093/gigascience/gix093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang D., Li W., Xia E.H., Zhang Q.J., Liu Y., Zhang Y., Tong Y., Zhao Y., Niu Y.C., Xu J.H. The medicinal herb Panax notoginseng genome provides insights into ginsenoside biosynthesis and genome evolution. Mol Plant. 2017;10(6):903–907. doi: 10.1016/j.molp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 28.Fan G., Fu Y., Yang B., Liu M., Zhang H., Liang X., Shi C., Ma K., Wang J., Liu W. bioRxiv; 2018. Sequencing of Panax notoginseng genome reveals genes involved in disease resistance and ginsenoside biosynthesis. [Google Scholar]
- 29.Chen W., Kui L., Zhang G., Zhu S., Zhang J., Wang X., Yang M., Huang H., Liu Y., Wang Y. Whole-genome sequencing and analysis of the Chinese herbal plant Panax notoginseng. Mol Plant. 2017;10(6):899–902. doi: 10.1016/j.molp.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 30.Wang H., Sun H., Kwon W.-S., Jin H., Yang D.-C. Molecular identification of the Korean ginseng cultivar “Chunpoong” using the mitochondrial nad7 intron 4 region. Mitochondrial DNA. 2009;20(2–3):41–45. doi: 10.1080/19401730902856738. [DOI] [PubMed] [Google Scholar]
- 31.Kim K.J., Lee H.L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004;11(4):247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y., Yin J., Guo H., Zhang Y., Xiao W., Sun C., Wu J., Qu X., Yu J., Wang X. The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Frontiers in Plant Science. 2015;5(696) doi: 10.3389/fpls.2014.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim K., Lee S.-C., Lee J., Lee H.O., Joh H.J., Kim N.-H., Park H.-S., Yang T.J. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLOS ONE. 2015;10(6) doi: 10.1371/journal.pone.0117159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Z-j, Li W., Liu Y., Gao L-z. The complete chloroplast genome of North American ginseng, Panax quinquefolius. Mitochondrial DNA Part A. 2016;27(5):3496–3497. doi: 10.3109/19401736.2015.1066365. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen B., Kim K., Kim Y.-C., Lee S.-C., Shin J.E., Lee J., Kim N.H., Jang W., Choi H.I., Yang T.J. The complete chloroplast genome sequence of Panax vietnamensis Ha et Grushv (Araliaceae) Mitochondrial DNA Part A. 2017;28(1):85–86. doi: 10.3109/19401736.2015.1110810. [DOI] [PubMed] [Google Scholar]
- 36.Dong W., Liu H., Xu C., Zuo Y., Chen Z., Zhou S. A chloroplast genomic strategy for designing taxon specific DNA mini-barcodes: a case study on ginsengs. BMC Genetics. 2014;15(1):138. doi: 10.1186/s12863-014-0138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manzanilla V., Kool A., Nguyen Nhat L., Nong Van H., Le Thi Thu H., de Boer H.J. Phylogenomics and barcoding of Panax: toward the identification of ginseng species. BMC Evolutionary Biology. 2018;18(1):44. doi: 10.1186/s12862-018-1160-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi F.-X., Li M.-R., Li Y.-L., Jiang P., Zhang C., Pan Y.-Z., Liu B., Xiao H.-X., Li L.-F. The impacts of polyploidy, geographic and ecological isolations on the diversification of Panax (Araliaceae) BMC Plant Biology. 2015;15(1):297. doi: 10.1186/s12870-015-0669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang P., Shi F.-X., Li M.-R., Liu B., Wen J., Xiao H.-X., Li L.-F. Positive selection driving cytoplasmic genome evolution of the medicinally important ginseng plant genus Panax. Frontiers in Plant Science. 2018;9(359) doi: 10.3389/fpls.2018.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi D.-W., Jung J., Ha Y.I., Park H.-W., In D.S., Chung H.-J., Liu J.R. Analysis of transcripts in methyl jasmonate-treated ginseng hairy roots to identify genes involved in the biosynthesis of ginsenosides and other secondary metabolites. Plant Cell Reports. 2005;23(8):557–566. doi: 10.1007/s00299-004-0845-4. [DOI] [PubMed] [Google Scholar]
- 41.Kim M.K., Lee B.-S., In J.-G., Sun H., Yoon J.-H., Yang D.-C. Comparative analysis of expressed sequence tags (ESTs) of ginseng leaf. Plant Cell Reports. 2006;25(6):599–606. doi: 10.1007/s00299-005-0095-0. [DOI] [PubMed] [Google Scholar]
- 42.Jung J.D., Park H.W., Hahn Y., Hur C.G., In D.S., Chung H.J., Liu J.R., Choi D.W. Discovery of genes for ginsenoside biosynthesis by analysis of ginseng expressed sequence tags. Plant Cell Reports. 2003;22(3):224–230. doi: 10.1007/s00299-003-0678-6. [DOI] [PubMed] [Google Scholar]
- 43.Sathiyamoorthy S., In J.G., Gayathri S., Kim Y.J., Yang D.C. Gene ontology study of methyl jasmonate-treated and non-treated hairy roots of Panax ginseng to identify genes involved in secondary metabolic pathway. Russian Journal of Genetics. 2010;46(7):828–835. [PubMed] [Google Scholar]
- 44.Sathiyamoorthy S., In Jun-Gyo, Lee Byum-Soo, Kwon Woo-Seang, Yang Dong-Uk, Kim Ju-Han, Yang Deok-Chun. Insilico analysis for expressed sequence tags from embryogenic callus and flower buds of Panax ginseng C. A. Meyer. Journal of Ginseng Research. 2011;35(1):21–30. [Google Scholar]
- 45.Chen S., Luo H., Li Y., Sun Y., Wu Q., Niu Y., Song J., Lv A., Zhu Y., Sun C. 454 EST analysis detects genes putatively involved in ginsenoside biosynthesis in Panax ginseng. Plant Cell Reports. 2011;30(9):1593. doi: 10.1007/s00299-011-1070-6. [DOI] [PubMed] [Google Scholar]
- 46.Subramaniyam S., Mathiyalagan R., Natarajan S., Kim Y.-J., Jang M-g, Park J.-H., Yang D.C. Transcript expression profiling for adventitious roots of Panax ginseng Meyer. Gene. 2014;546(1):89–96. doi: 10.1016/j.gene.2014.05.024. [DOI] [PubMed] [Google Scholar]
- 47.Li C., Zhu Y., Guo X., Sun C., Luo H., Song J., Li Y., Wang L., Qian J., Chen S. Transcriptome analysis reveals ginsenosides biosynthetic genes, microRNAs and simple sequence repeats in Panax ginseng C. A. Meyer. BMC Genomics. 2013;14(1):245. doi: 10.1186/1471-2164-14-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jayakodi M., Lee S.-C., Lee Y.S., Park H.-S., Kim N.-H., Jang W., Lee H.O., Joh H.J., Yang T.J. Comprehensive analysis of Panax ginseng root transcriptomes. BMC Plant Biology. 2015;15:138. doi: 10.1186/s12870-015-0527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu B., Long Q., Gao Y., Wang Z., Shao T., Liu Y., Li Y., Ding W. Comprehensive characterization of a time-course transcriptional response induced by autotoxins in Panax ginseng using RNA-Seq. BMC Genomics. 2015;16(1):1010. doi: 10.1186/s12864-015-2151-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu S., Wang S., Liu M., Yang F., Zhang H., Liu S., Wang Q., Zhao Y. De novo sequencing and analysis of the transcriptome of Panax ginseng in the leaf-expansion period. Molecular Medicine Reports. 2016;14(2):1404–1412. doi: 10.3892/mmr.2016.5376. [DOI] [PubMed] [Google Scholar]
- 51.Wang K., Jiang S., Sun C., Lin Y., Yin R., Wang Y., Zhang M. The spatial and temporal transcriptomic landscapes of ginseng, Panax ginseng C. A. Meyer. Scientific Reports. 2015;5:18283. doi: 10.1038/srep18283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao Y., He X., Wu B., Long Q., Shao T., Wang Z., Wei J., Li Y., Ding W. Time-course transcriptome analysis reveals resistance genes of Panax ginseng induced by cylindrocarpon destructans infection using RNA-seq. PLoS One. 2016;11(2) doi: 10.1371/journal.pone.0149408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao H., Nuruzzaman M., Xiu H., Huang J., Wu K., Chen X., Li J., Wang L., Jeong J.H., Park S.J. Transcriptome analysis of methyl jasmonate-elicited Panax ginseng adventitious roots to discover putative ginsenoside biosynthesis and transport genes. International Journal of Molecular Sciences. 2015;16(2):3035. doi: 10.3390/ijms16023035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jayakodi M., Lee S.-C., Park H.-S., Jang W., Lee Y.S., Choi B.-S., Nah G.J., Kim D.S., Natesan S., Sun C. Transcriptome profiling and comparative analysis of Panax ginseng adventitious roots. Journal of Ginseng Research. 2014;38(4):278–288. doi: 10.1016/j.jgr.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jo I.-H., Lee J., Hong C.E., Lee D.J., Bae W., Park S.-G., Ahn Y.J., Kim Y.C., Kim J.U., Lee J.W. Isoform Sequencing provides a more comprehensive view of the Panax ginseng transcriptome. Genes. 2017;8(9):228. doi: 10.3390/genes8090228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jayakodi M., Choi B.-S., Lee S.-C., Kim N.-H., Park J.Y., Jang W., Lakshmanan M., Mohan S.V.G., Lee D.Y., Yang T.J. Ginseng genome database: an open-access platform for genomics of Panax ginseng. BMC Plant Biology. 2018;18(1):62. doi: 10.1186/s12870-018-1282-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jayakodi M., Lee S.-C., Yang T.-J. Comparative transcriptome analysis of heat stress responsiveness between two contrasting ginseng cultivars. Journal of Ginseng Research. 2019;43(4):572–579. doi: 10.1016/j.jgr.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo H., Sun C., Sun Y., Wu Q., Li Y., Song J., Niu Y., Cheng X., Xu H., Li C. Analysis of the transcriptome of Panax notoginseng root uncovers putative triterpene saponin-biosynthetic genes and genetic markers. BMC Genomics. 2011;12(5):S5. doi: 10.1186/1471-2164-12-S5-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu M.-H., Yang B.-R., Cheung W.-F., Yang K.Y., Zhou H.-F., Kwok J.S.-L., Liu G.C., Li X.F., Zhong S., Lee S.M. Transcriptome analysis of leaves, roots and flowers of Panax notoginseng identifies genes involved in ginsenoside and alkaloid biosynthesis. BMC Genomics. 2015;16(1):265. doi: 10.1186/s12864-015-1477-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y., Mi Y., Zhang J., Li Q., Chen L. Illumina-based transcriptomic profiling of Panax notoginseng in response to arsenic stress. Botanical Studies. 2016;57(1):13. doi: 10.1186/s40529-016-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu Q., Song J., Sun Y., Suo F., Li C., Luo H., Liu Y., Li Y., Zhang X., Yao H. Transcript profiles of Panax quinquefolius from flower, leaf and root bring new insights into genes related to ginsenosides biosynthesis and transcriptional regulation. Physiologia Plantarum. 2010;138(2):134–149. doi: 10.1111/j.1399-3054.2009.01309.x. [DOI] [PubMed] [Google Scholar]
- 62.Sun C., Li Y., Wu Q., Luo H., Sun Y., Song J., Lui E.M., Chen S. De novo sequencing and analysis of the American ginseng root transcriptome using a GS FLX Titanium platform to discover putative genes involved in ginsenoside biosynthesis. BMC Genomics. 2010;11(1):262. doi: 10.1186/1471-2164-11-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu D., Austin R.S., Zhou S., Brown D. The root transcriptome for North American ginseng assembled and profiled across seasonal development. BMC Genomics. 2013;14(1):564. doi: 10.1186/1471-2164-14-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qi J., Sun P., Liao D., Sun T., Zhu J., Li X. Transcriptomic analysis of American ginseng seeds during the dormancy release process by RNA-seq. PLoS One. 2015;10(3):e0118558. doi: 10.1371/journal.pone.0118558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J., Li J., Li J., Liu S., Wu X., Li J., Gao W. Transcriptome profiling shows gene regulation patterns in ginsenoside pathway in response to methyl jasmonate in Panax Quinquefolium adventitious root. Scientific Reports. 2016;6:37263. doi: 10.1038/srep37263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mathiyalagan R., Subramaniyam S., Natarajan S., Kim Y.J., Sun M.S., Kim S.Y., Kim Y.J., Yang D.C. Insilico profiling of microRNAs in Korean ginseng (Panax ginseng Meyer) Journal of Ginseng Research. 2013;37(2):227–247. doi: 10.5142/jgr.2013.37.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu B., Wang M., Ma Y., Yuan L., Lu S. High-throughput sequencing and characterization of the small RNA transcriptome reveal features of novel and conserved MicroRNAs in Panax ginseng. PLoS One. 2012;7(9):e44385. doi: 10.1371/journal.pone.0044385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei R., Qiu D., Wilson I.W., Zhao H., Lu S., Miao J., Feng S., Bai L., Wu Q., Tu D. Identification of novel and conserved microRNAs in Panax notoginseng roots by high-throughput sequencing. BMC Genomics. 2015;16(1):835. doi: 10.1186/s12864-015-2010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng Y., Chen K., Xu Z., Liao P., Zhang X., Liu L., Wei K., Liu D., Li Y.F., Sunkar R. Small RNA profiles from Panax notoginseng roots differing in sizes reveal correlation between miR156 abundances and root biomass levels. Scientific Reports. 2017;7(1):9418. doi: 10.1038/s41598-017-09670-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen K., Liu L., Zhang X., Yuan Y., Ren S., Guo J., Wang Q., Liao P., Li S., Cui X. Phased secondary small interfering RNAs in Panax notoginseng. BMC Genomics. 2018;19(Suppl 1):41. doi: 10.1186/s12864-017-4331-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gurung B., Bhardwaj P.K., Talukdar N.C. Subtractive transcriptome analysis of leaf and rhizome reveals differentially expressed transcripts in Panax sokpayensis. Functional & Integrative Genomics. 2016;16(6):619–639. doi: 10.1007/s10142-016-0517-9. [DOI] [PubMed] [Google Scholar]
- 72.Rai A., Yamazaki M., Takahashi H., Nakamura M., Kojoma M., Suzuki H., Saito K. RNA-seq Transcriptome analysis of Panax japonicus, and its comparison with other panax species to identify potential genes involved in the saponins biosynthesis. Frontiers in Plant Science. 2016;7(481) doi: 10.3389/fpls.2016.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang S., Wu Y., Jin J., Hu B., Zeng W., Zhu W., Zheng Y., Chen P. De novo characterization of Panax japonicus C. A. Mey transcriptome and genes related to triterpenoid saponin biosynthesis. Biochemical and Biophysical Research Communications. 2015;466(3):450–455. doi: 10.1016/j.bbrc.2015.09.048. [DOI] [PubMed] [Google Scholar]
- 74.Zhang G.-H., Ma C.-H., Zhang J.-J., Chen J.-W., Tang Q.-Y., He M.-H., Xu X.Z., Jiang N.H., Yang S.C. Transcriptome analysis of Panax vietnamensis var. fuscidicus discovers putative ocotillol-type ginsenosides biosynthesis genes and genetic markers. BMC Genomics. 2015;16(1):159. doi: 10.1186/s12864-015-1332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tang Q.-Y., Chen G., Song W.-L., Fan W., Wei K.-H., He S.-M., Zhang G.H., Tang J.R., Li Y., Lin Y. Transcriptome analysis of Panax zingiberensis identifies genes encoding oleanolic acid glucuronosyltransferase involved in the biosynthesis of oleanane-type ginsenosides. Planta. Feb 2019;249(2):393–406. doi: 10.1007/s00425-018-2995-6. [DOI] [PubMed] [Google Scholar]
- 76.Lum J.H.-K., Fung K.-L., Cheung P.-Y., Wong M.-S., Lee C.-H., Kwok F.S.-L., Leung M.C., Hui P.K., Lo S.C. Proteome of Oriental ginseng Panax ginseng C. A. Meyer and the potential to use it as an identification tool. Proteomics. 2002;2(9):1123–1130. doi: 10.1002/1615-9861(200209)2:9<1123::AID-PROT1123>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 77.Kim S.I., Kim J.Y., Kim E.A., Kwon K.-H., Kim K.-W., Cho K., Lee J.H., Nam M.H., Yang D.C., Yoo J.S. Proteome analysis of hairy root from Panax ginseng C. A. Meyer using peptide fingerprinting, internal sequencing and expressed sequence tag data. Proteomics. 2003;3(12):2379–2392. doi: 10.1002/pmic.200300619. [DOI] [PubMed] [Google Scholar]
- 78.Nam M.H., Heo E.J., Kim J.Y., Kim S.I., Kwon K.-H., Seo J.B., Kwon O., Yoo J.S., Park Y.M. Proteome analysis of the responses of Panax ginseng C. A. Meyer leaves to high light: Use of electrospray ionization quadrupole-time of flight mass spectrometry and expressed sequence tag data. Proteomics. 2003;3(12):2351–2367. doi: 10.1002/pmic.200300509. [DOI] [PubMed] [Google Scholar]
- 79.Ma R., Sun L., Chen X., Jiang R., Sun H., Zhao D. Proteomic changes in different growth periods of ginseng roots. Plant Physiology and Biochemistry. 2013;67:20–32. doi: 10.1016/j.plaphy.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 80.Ma R., Sun L., Chen X., Mei B., Chang G., Wang M., Zhao D. Proteomic Analyses Provide Novel Insights into Plant Growth and Ginsenoside Biosynthesis in Forest Cultivated Panax ginseng (F. Ginseng) Frontiers in Plant Science. 2016;7(1) doi: 10.3389/fpls.2016.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun H., Liu F., Sun L., Liu J., Wang M., Chen X., Xu X., Ma R., Feng K., Jiang R. Proteomic analysis of amino acid metabolism differences between wild and cultivated Panax ginseng. Journal of Ginseng Research. 2016;40(2):113–120. doi: 10.1016/j.jgr.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nesvizhskii A.I. Proteogenomics: concepts, applications and computational strategies. Nature Methods. 2014;11:1114. doi: 10.1038/nmeth.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martínez-Esteso M.J., Martínez-Márquez A., Sellés-Marchart S., Morante-Carriel J.A., Bru-Martínez R. The role of proteomics in progressing insights into plant secondary metabolism. Frontiers in Plant Science. 2015;6:504. doi: 10.3389/fpls.2015.00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li M., Wang X., Zhang C., Wang H., Shi F., Xiao H., Li L.-F. A Simple Strategy for Development of Single Nucleotide Polymorphisms from Non-Model Species and Its Application in Panax. International Journal of Molecular Sciences. 2013;14(12):24581. doi: 10.3390/ijms141224581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gordon S.P., Tseng E., Salamov A., Zhang J., Meng X., Zhao Z., Kang D., Underwood J., Grigoriev I.V., Figueroa M. Widespread Polycistronic Transcripts in Fungi Revealed by Single-Molecule mRNA Sequencing. PLOS ONE. 2015;10(7) doi: 10.1371/journal.pone.0132628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li M.-R., Shi F.-X., Li Y.-L., Jiang P., Jiao L., Liu B., Li L.F. Genome-Wide Variation Patterns Uncover the Origin and Selection in Cultivated Ginseng (Panax ginseng Meyer) Genome Biology and Evolution. 2017;9(9):2159–2169. doi: 10.1093/gbe/evx160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pan Y., Wang X., Sun G., Li F., Gong X. Application of RAD sequencing for evaluating the genetic diversity of domesticated Panax notoginseng (Araliaceae) PLoS One. 2016;11(11):e0166419. doi: 10.1371/journal.pone.0166419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Devi B.S.R., Kim Y.-J., Sathiyamoorthy S., Khorolragchaa A., Gayathri S., Parvin S., Yang D.U., Selvi S.K., Lee O.R., Lee S. Classification and characterization of putative cytochrome P450 genes from Panax ginseng C. A. Meyer. Biochemistry (Moscow) 2011;76(12):1347–1359. doi: 10.1134/S000629791112008X. [DOI] [PubMed] [Google Scholar]
- 89.Khorolragchaa A., Kim Y.-J., Rahimi S., Sukweenadhi J., Jang M.-G., Yang D.-C. Grouping and characterization of putative glycosyltransferase genes from Panax ginseng Meyer. Gene. 2014;536(1):186–192. doi: 10.1016/j.gene.2013.07.077. [DOI] [PubMed] [Google Scholar]
- 90.Yang J.-L., Hu Z.-F., Zhang T.-T., Gu A.-D., Gong T., Zhu P. Progress on the studies of the key enzymes of ginsenoside biosynthesis. Molecules. 2018;23(3):589. doi: 10.3390/molecules23030589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nuruzzaman M., Cao H., Xiu H., Luo T., Li J., Chen X., Luo J., San Z., Luo D. Transcriptomics-based identification of WRKY genes and characterization of a salt and hormone-responsive PgWRKY1 gene in Panax ginseng. Acta Biochimica et Biophysica Sinica. 2016;48(2):117–131. doi: 10.1093/abbs/gmv122. [DOI] [PubMed] [Google Scholar]
- 92.Yin R., Zhao M., Wang K., Lin Y., Wang Y., Sun C., Wang Y., Zhang M. Functional differentiation and spatial-temporal co-expression networks of the NBS-encoding gene family in Jilin ginseng, Panax ginseng C.A. Meyer. PLoS One. 2017;12(7) doi: 10.1371/journal.pone.0181596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chu Y., Xiao S., Su H., Liao B., Zhang J., Xu J., Chen S. Genome-wide characterization and analysis of bHLH transcription factors in Panax ginseng. Acta Pharmaceutica Sinica B. 2018;8(4):666–677. doi: 10.1016/j.apsb.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lin Y., Wang K., Li X., Sun C., Yin R., Wang Y., Wang Y., Zhang M. Evolution, functional differentiation, and co-expression of the RLK gene family revealed in Jilin ginseng, Panax ginseng C.A. Meyer. Molecular Genetics and Genomics. 2018;293(4):845–859. doi: 10.1007/s00438-018-1425-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Udall J.A., Dawe R.K. Is it ordered correctly? Validating genome assemblies by optical mapping. The Plant Cell. 2018;30(1):7. doi: 10.1105/tpc.17.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szcześniak M.W., Gaffney D J., Elo L L., Zhang X. A survey of best practices for RNA-seq data analysis. Genome Biology. 2016;17(1):13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gassmann W., Appel H.M., Oliver M.J. The interface between abiotic and biotic stress responses. J Exp Bot. 2016;67(7):2023–2024. doi: 10.1093/jxb/erw110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Quan L.-H., Min J.-W., Sathiyamoorthy S., Yang D.-U., Kim Y.-J., Yang D.-C. Biotransformation of ginsenosides Re and Rg1 into ginsenosides Rg2 and Rh1 by recombinant β-glucosidase. Biotechnology Letters. 2012;34(5):913–917. doi: 10.1007/s10529-012-0849-z. [DOI] [PubMed] [Google Scholar]
- 99.Kim D., Jung M., Ha J.I., Lee Y.M., Lee S.-G., Shin Y., Subramaniyam S., Oh J. Transcriptional profiles of secondary metabolite biosynthesis genes and cytochromes in the leaves of four Papaver species. Data. 2018;3(4) [Google Scholar]
- 100.Galanie S., Thodey K., Trenchard I.J., Filsinger Interrante M., Smolke C.D. Complete biosynthesis of opioids in yeast. Science. 2015;349(6252):1095. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]