Abstract
Haematological malignancies encompass all variations of leukaemia at both the chronic and acute level, together with the specific cell type induced into tumourigenesis. Current diagnostic protocols for leukaemic conditions rely heavily on cytomorphology and other histological examinations from bone marrow aspirates, with the latter being a highly invasive surgical procedure for the patient.
The discovery of microRNAs as one of the key gene regulatory networks in the past two decades has enabled researchers to investigate the possibility of exploiting the identification of dysregulated expression profiles for specific microRNAs present in the leukaemic patient's bloodstream as novel liquid biopsy diagnostic tools. This review article serves to consolidate recent global research efforts aiming to achieve such scopes.
Keywords: microRNA, miRNA, Liquid biopsy, Leukaemia, CML, AML, ALL, CLL
1. Introduction
Haematological malignancies encompass all variations of leukaemia at both the chronic and acute level, together with the specific cell type induced into tumourigenesis. Current diagnostic protocols rely heavily on cytomorphology and other histological examinations from bone marrow aspirates, with the latter being a highly invasive surgical procedure for the patient. Consequently, the need for novel and less/non-invasive diagnostic methodologies is ever present. One highly effective and novel diagnostic method for achieving such a scope is the use of reliable genomic/transcriptomic/noncoding RNA (ncRNA) biomarkers circulating within the bloodstream. In this manner, a standard blood sample can be collected from the leukaemic patient and blood serum/plasma levels for such biomarkers can be rapidly analysed for specific expression profile dysregulations. Such liquid biopsies can be adopted easily within the oncology clinic and also in a cost – effective manner, as long as the specific blood plasma/serum biomarker/s for the specific disease condition is properly identified and experimentally validated in order to ascertain their clinical valorisation.
The use of functional genomics for enabling clinicians to discern multiple tumour characteristics and clinical severity within oncology patients has gradually become established as an essential diagnostic tool across the past 30 years [1]. Consequently, the need for reliable biomarkers for the diagnosis of a specific tumour is ever present. This need includes biomarkers for the suspected tumour's unique phenotypic characteristics, such as metastatic potential and chemoresistance properties, and is considered as pivotal information for the clinical oncologist in order to orchestrate the best theranostic avenues possible for the individual cancer patient. The development of RT-qPCR and microarray technologies has also opened the gates for a more thorough scrutiny of major molecular players in order to identify unique and reproducible dysregulated expression profiles for individual tumour models [2].
MicroRNAs (miRNAs) form key molecular components of the cell and have the valuable ability to implement gene regulation. Consequently, any dysregulation of the miRnome expression profile within the cell can lead to dire consequences and eventual clinical manifestations of disease, depending on which miRNAs are affected. Such miRNA dysregulated expression profiles extend to the level of induction of oncogenesis. Following the initial scientific evidence of miRNA influences in cancer by Calin and colleagues in 2002, this body of evidence has grown exponentially and have rendered miRNAs as highly attractive and novel biomarkers for a vast spectrum of human disease, including cancer [3].
Furthermore, miRNAs are adept biomarkers as they tend to be resilient in varying physical conditions, including wide ranges of temperature and pH, and can withstand degradation within formalin-fixed, paraffin-embedded tissues [4,5]. The stability conferred to miRNAs essentially highlights their potential use as biomarkers in a spectrum of body fluids including serum, plasma, breast milk, saliva and urine [4,[6], [7], [8], [9]]. This renders miRNA diagnostic potential particularly attractive due to the reduced/non-invasiveness of most required sample collection protocols.
The circulation of miRNAs in body fluids occurs either through direct secretion or through protective micro-structures that are secreted into the specific body fluid [10]. Studies revealed that micro-vesicles or exosomes are the preferred source of transport for circulatory miRNAs [11,12]. Micro-vesicles and exosomes are released from endocytic processes in healthy/damaged cells and are an effective means of communication within the body, with approximately 120 miRNAs transported within such micro-vesicles [13]. Furthermore, apoptotic pressure can lead to an exacerbation of exosomal secretion from within tumour cells [[14], [15], [16]]. Direct release of miRNAs is mainly due to damaged cells or cell apoptosis and is not a major player in the generation of circulating miRNAs [10].
2. MiRNAs and cancer
The first indication that miRNAs are linked to cancer was in 2002 [3,17], with reports that the fragile sites and susceptible loci account for 50% of miRNA locations [18]. Since then it has been firmly established within the scientific community that miRNAs are highly influential in cancer pathology [19].
Oncology patients typically exhibit dysregulated expression levels of cancer-associated miRNAs, with individual members of the miRNome being upregulated or downregulated in expression, consequently bearing unique miRNA expression profiles pertaining to their condition [20]. Furthermore, such dysregulated miRNAs can have oncogenic or tumour suppressive functional roles [[21], [22], [23]].
MiRNA expression profiles are a highly effective diagnostic technique, as demonstrated in a blind study on 22 different tumour types that allowed for tumour classification with an accuracy of 90% or higher when statistically analysed [24]. Furthermore, since circulating miRNAs can be detected in body fluids, this further establishes their role as ideal biomarkers for tumour diagnostics and progress monitoring.
In tandem with the establishment of this novel field of research, a host of platforms for analysis and sequencing were developed to accommodate such scientific endeavours, such as microarray, quantitative polymerase chain reaction and next-generation sequencing technologies [[25], [26], [27]].
3. MiRNA biomarker expression profiles for leukaemic condition diagnostics
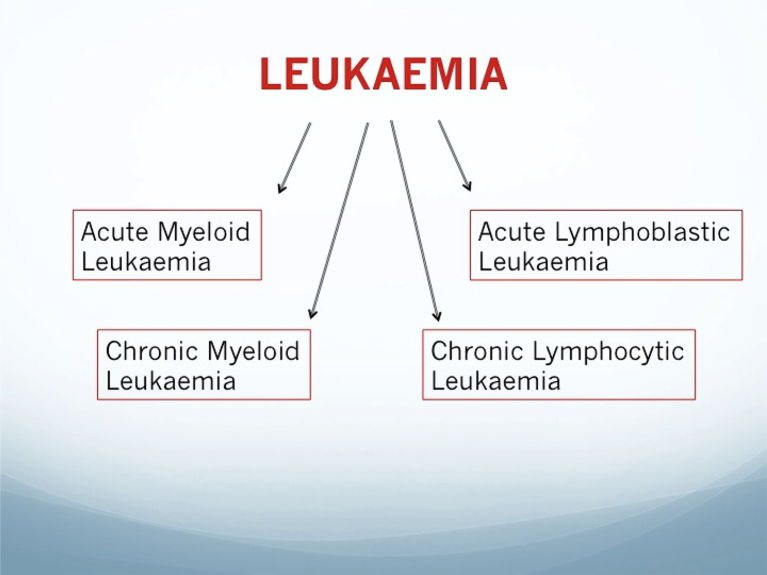
Leukaemic conditions are generally classified according to the originating dysplastic cellular population (myeloid or lymphocytic) and the severity/duration of the condition (acute or chronic, See Fig. 1).
Fig. 1.
Graphical outline of clinically prevalent leukaemic conditions.
The following sections describe briefly each leukaemic manifestation and the compilation of latest evidence pertaining to potential miRNA expression profiles that proving the biggest potential as biomarkers for the specific leukaemic condition.
3.1. Acute myeloid leukaemia (AML)
AML originates in the bone marrow through clonal transformation of myeloid precursors [28]. Studies demonstrated that AML has a network of circulating miRNA biomarkers that can be identified either in serum or plasma, as listed in Table 2 below:
Table 2.
Currently identified circulating/bone marrow miRNA expression profiles for ALL.
| Identified miRNA | Expression | Sample | Reference |
|---|---|---|---|
| miR-92a | Downregulated | Plasma | [29] |
|
miR-638 |
Stable |
Plasma |
|
| miR-128a | Upregulated | Bone marrow | [39] |
| miR-128b | Upregulated | Bone marrow | |
| Let-7b | Downregulated | Bone marrow | |
|
miR-223 |
Downregulated |
Bone marrow |
|
| miR-511 | Upregulated | Plasma | [40] |
| miR-222 | Upregulated | Plasma | |
| miR-34a | Upregulated | Plasma | |
| miR-199a-3p | Downregulated | Plasma | |
| miR-223 | Downregulated | Plasma | |
| miR-221 | Downregulated | Plasma | |
| miR-26a | Downregulated | Plasma | |
| miR-652-3p | Downregulated | Serum | [41] |
| miR-125b-1 | Upregulated | [42] | |
| miR-203 | Downregulated |
Initial studies reporting the potential association of dysregulated miRNA expression profiles with AML manifestation include the investigation carried out by Tanaka and colleagues in 2009 [29]. In this particular study, miRNA microarray analysis from healthy controls and AML patient samples identified a significant down-regulation of miR-92a in the AML patient cohort [29]. Furthermore, miR-638 was found to be stable-expressed in both patient cohorts, allowing the investigators to conclude that the blood plasma expression ratio of miR-92a/miR-638 could be utilised a novel plasma-based biomarker for AML [29].
In a similar study, conducted by Wang and colleagues in 2011, peripheral blood mononuclear cells (PBMCs) miRNA microarray analysis in healthy and AML patient cohorts revealed a correlation between miR-29a and miR-142-3p down-regulation and AML [30]. Furthermore, this correlation was confirmed through quantitative real-time PCR (RT-qPCR) analysis for the two miRNAs in 52 diagnosed AML patients and 100 normal controls [30].
Later studies, such as the investigation carried out by Fayyad-Kazan and colleagues in 2013, identified additional dysregulated miRNA expression profiles associated with AML [31]. In this case, RT-qPCR-based miRNA microarrays were employed to analyse blood plasma samples from 20 newly diagnosed AML patients and 20 AML remission patients (deemed to be the control cohort) [31]. Following the microarray results, revealing a list of six dysregulated miRNAs in the newly diagnosed AML cohort (see Table 1), further RT-qPCR analysis was employed as a validation method, with receiver-operator characteristic curve (ROC) analysis to confirm more robustly the significantly dysregulated miRNA elements [31]. Such statistically robust analyses resulted in the conclusion that two specific miRNAs present in blood plasma, namely miR-150 and miR-342, are highly dysregulated in AML patients, and with the added observation that AML remission patient cohort expression levels for these two miRNAs were similar to that of healthy control individuals [31].
Table 1.
Currently identified circulating miRNA expression profiles for AML.
| Identified miRNA | Expression | Sample | Reference |
|---|---|---|---|
| miR-92a | Downregulated | Plasma | [29] |
|
miR-638 |
Stable |
Plasma |
|
| miR-29a | Downregulated | Plasma | [30] |
|
miR-142-3p |
Downregulated |
Plasma |
|
| miR-150 | Downregulated | Plasma | [31] |
| miR-342 | Downregulated | Plasma | |
| Let-7d | Downregulated | Plasma | |
| miR-339 | Downregulated | Plasma | |
| miR-523 | Upregulated | Plasma | |
|
Let-7b |
Upregulated |
Plasma |
|
| miR-10a-5p | Upregulated | Serum | [32] |
| miR-93-5p | Upregulated | Serum | |
| miR-129-5p | Upregulated | Serum | |
| miR-155-5p | Upregulated | Serum | |
| miR-181b-5p | Upregulated | Serum | |
|
miR-320d |
Upregulated |
Serum |
|
| miR-125b | Upregulated | Serum | [33] |
| miR-192 | Downregulated | Serum | [34] |
| miR-217 | Downregulated | Serum | [35] |
| miR-223 | Downregulated | Serum | [36] |
Another study, performed in 2013 by Zhi and colleagues, serum samples from 20 AML patients and 20 healthy controls were collected and utilised for Solexa sequencing analysis, with eventual identified dysregulated miRNAs validated by RT-qPCR analysis [32]. Apart from the validated miRNA expression profile (see Table 1) associated with AML, the investigators also revealed that miR-181b-5p can be utilised as a reliable serum miRNA biomarker for overall survival within AML patient cohorts [32].
Interestingly, a later study focusing on serum exosomal miRNA expression profiles for AML identified an up-regulation of miR-125b to be highly associated with increased AML patient relapse and elevated risk of AML-induced mortality, leading miR-125b to be proposed by the investigators as a novel poor-prognosis biomarker for intermediate-risk AML [33].
A separate study carried out in 2018 by Tian and colleagues, tested the hypothesis that down-regulated miR-192 expression (a known oncomiR) could be exploited as a prognostic biomarker for paediatric AML [34]. The study involved serum sample collection from 97 paediatric AML patients and 50 healthy controls, followed by specific RT-qPCR analysis for miR-192 [34]. The results of this study confirmed that miR-192 serum expression level was severely down-regulated in the paediatric AML patient cohort and can consequently be employed as a highly reliable prognostic biomarker in pediatric AML cases [34].
In a parallel study hypothesising the prognostic value of miR-217 in AML, serum samples from 89 AML patients and 60 healthy controls were collected and consequently analyses for miR-217 expression using RT-qPCR [35]. In this particular study, miR-217 was found to be severely down-regulated in the AML patient cohort, with corresponding Kaplan-Meier analyses revealing that miR-217 down-regulated expression was correlated to reduced overall/disease-free survival in the AML patient cohort, rendering it a poor prognosis serum-based miRNA biomarker [35].
In 2019, another study on 131 AML patients investigated the serum expression profile of miR-223 using RT-qPCR [36]. The results of this study concluded that down-regulated serum expression of miR-223 is linked with AML [36].
3.2. Acute lymphoblastic leukaemia (ALL)
This leukaemic condition arises from within lymphocyte precursors and presents itself with multiple genomic aberrations, including the Philadelphia (Ph) chromosome [37]. Other aberrations at the genomic and transcriptomic levels linked to paediatric ALL include IKZF1, ARID5B, CEBPE, and CDKN2A [38]. A number of miRNAs were identified within recent scientific literature as biomarkers in ALL, as summarised in Table 2 below.
The study conducted by Tanaka and colleagues (described above in section 3.1) also confirmed the clinical value of the expression ratio of miR-92a/miR-638 for potential employment as a biomarker for ALL [29].
The comprehensive study carried out by Mi and colleagues screened 154 leaukaemic bone marrow samples, followed by whole genome miRNA expression profiling analyses and RT-qPCR validation analysis on all identified miRNA candidates demonstrating extreme dysregulated expression [39]. The results of this study concluded that miR-128a, miR-128b, let-7b and miR-223 are all potential miRNA biomarkers in AML/ALL, with the down-regulated expression of let-7b and miR-223 in ALL (on comparison to AML bone marrow samples) allows for leukaemic subtype identification [39].
The separate miRNA expression profiling investigation conducted by Luna-Aguirre and colleagues in 2015 determined a total of 77 miRNAs to be dysregulated in ALL from 39 ALL plasma samples, with seven miRNAs (see Table 2) being highly significant [40]. Furthermore, miR-511 was confirmed as the most reliable plasma miRNA biomarker for ALL from this study [40].
The recent study carried out by Jiang and colleagues in 2018 employed low density miRNA arrays for screening paediatric ALL plasma samples, followed by RT-qPCR validation [41]. The results of this particular study revealed that miR-652-3p expression was negatively correlated to paediatric ALL progression in this patient cohort, suggesting that miR-652-3p can be possible utilised as a potential biomarker and future drug target in paediatric ALL [41].
Another study in 2018, carried out by Swellam and colleagues, focused on the expression profile of miR-125b-1 and miR-203 in paediatric ALL cases [42]. The investigators that using such an expression profile (see Table 2), from both investigated miRNAs, as biomarkers in paediatric ALL was highly sensitive in the patient cohort examined in this study [42].
3.3. Chronic myeloid leukaemia (CML)
This specific leukaemic condition is also afflicted with the Philadelphia gene (Ph), derived from the translocated Abelson gene (ABL) at chromosome 9 and the Breakpoint Cluster Region (BCR) gene on chromosome 22 [43]. This creates a BCR-ABL complex that is responsible for the actual CML clinical condition [44]. The disease is depicted by an increase in the number of white blood cells originating from the myeloid blood stem cells and occurs typically in middle-aged adults. Varying miRNA biomarker expression profiles were identified in recent scientific literature, both at diagnosis (Table 3) and also in a number of patients who responded to treatment [45].
Table 3.
Currently identified circulating/bone marrow miRNA expression profiles for CML.
| Identified miRNA | Expression | Sample | Reference |
|---|---|---|---|
| miR-96 | Upregulated | Bone marrow/Cell lines | [46] |
| miR-10a | Downregulated | Bone marrow/Cell lines | |
| miR-150 | Downregulated | Bone marrow/Cell lines | |
|
miR-151 |
Downregulated |
Bone marrow/Cell lines |
|
| miR-16-1 | Upregulated | Peripheral blood | [47] |
| miR-15a | Upregulated | Peripheral blood | |
| miR-101 | Upregulated | Peripheral blood | |
| miR-568 | Upregulated | Peripheral blood | |
| miR-155 | Downregulated | Peripheral blood | |
| miR-106 | Downregulated | Peripheral blood |
The study carried out by Agirre and colleagues on CML cell line models and bone marrow samples from CML patients led to the identification of four potential miRNA biomarkers for CML [46]. The most significant revelation was that miR-10a down-regulation was associated with CML cellular proliferation and that one of the main target genes of miR-10a was upstream stimulatory factor 2 (USF2), which is a cellular growth inducer in CML [46].
In a more recent study conducted by Fallah and colleagues, peripheral blood leukocyte samples from 50 CML patients were profiled using stem loop RT-qPCR techniques [47]. The results of this study elucidated a unique miRNA expression profile within the CML patient cohort, consisting of four up-regulated miRNAs and two down-regulated miRNAs (see Table 3) [47].
3.4. Chronic lymphocytic leukaemia (CLL)
In CLL, tumourigenesis occurs within lymphoid blood stem cells, leading to the accumulation of B-lymphocytes in lymph and bone marrow. CLL is the most prevalent leukaemic malignancy, characterised by a spectrum of chromosomal aberrations that include the deletion of the 13q, 11q, 17p and 6q gene or trisomy of the 12q chromosomal segment [48]. A number of miRNAs that affect oncogenes associated with CLL were identified within recent scientific literature. One such study focused on patients with variable ZAP-70 protein levels and mutated/unmutated IgVh gene, elucidating 13 miRNAs as playing a major role on such gene dysregulated expression profiles [49]. In a separate meta-analysis study on previously published scientific literature, miR-155 was identified as the most prevalent oncomiR in B-Cell malignancies and has the potential to be used as a diagnostic and prognostic marker [50]. Additional circulating miRNA biomarkers (Table 4) include the study performed by Moussay and colleagues [51]. In this particular investigation, blood plasma from 41 B-CLL patients were subjected to low density miRNA array profiling, with the consequent result of four miRNAs being highly dysregulated in the B-CLL patient cohort [51].
Table 4.
Currently identified circulating miRNA expression profiles for CLL.
| Identified miRNA | Expression | Sample | Reference |
|---|---|---|---|
| miR-150 | Upregulated | Plasma | [51] |
| miR-150* | Upregulated | Plasma | |
| miR-29a | Upregulated | Plasma | |
|
miR-135a* |
Upregulated |
Plasma |
|
|
miR-192 |
Downregulated |
Peripheral blood |
[52] |
| miR-155 | Upregulated | Peripheral blood | [53] |
| miR-363 | Upregulated | Plasma | [54] |
| miR-16 | Upregulated | Plasma |
Other novel findings include the study conducted by Cui and colleagues focused on the possible influences of miR-155 in CLL [53]. The results of this study, which analysed peripheral blood samples from a total of 265 CLL patients for miR-155 expression level, revealed that there is an up-regulated expression of miR-155 and this can lead to poor prognosis in CLL due to the miR-155 exacerbated influence on B-cell receptor signalling [53].
4. Conclusions and perspectives
Since their discovery, miRNAs have undoubtedly demonstrated to be a valuable diagnostic tool for the clinician, especially within the oncology clinical setting. Furthermore, selected miRNA biomarkers can also serve as potential drug targets for novel miRNA-based therapies for such devastating leukaemic conditions. The newfound possibility of exploiting miRNA biomarkers present in easily accessible body fluids, such as the bloodstream, has opened the doors to novel research niches that include the development of liquid biopsy diagnostics. Notwithstanding, the gauntlet has been thrown down to the global research community in order to identify and validate further potential biomarkers apart from miRNAs, such as long non-coding RNAs and other non-coding RNA families. Such a myriad of biomarker networks will certainly serve to widen not only the knowledge on the molecular interactions of life threatening conditions such as leukaemia, but also to enhance the repertoire of tools available for utilisation by the clinical oncologist. It is the authors’ firm belief that liquid biopsy diagnostics will, in the very near future, become firmly established within the global oncology clinical setting – not just for the conditions described above, but for all disease conditions whereby selected circulating miRNAs have proven to be reliable and precise biomarkers of disease progression.
Declaration of competing interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- 1.Goncalves R., Warner W.A., Luo J., Ellis M.J. New concepts in breast cancer genomics and genetics. Breast Cancer Res. 2014;16:460. doi: 10.1186/s13058-014-0460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teutsch S.M., Bradley L.A., Palomaki G.E., Haddow J.E., Piper M., Calonge N., Dotson W.D., Douglas M.P., Berg A.O. EGAPP working group, the evaluation of genomic applications in practice and prevention (EGAPP) initiative: methods of the EGAPP working group. Genet. Med. 2009;11:3–14. doi: 10.1097/GIM.0b013e318184137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., Rassenti L., Kipps T., Negrini M., Bullrich F., Croce C.M. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O'Briant K.C., Allen A., Lin D.W., Urban N., Drescher C.W., Knudsen B.S., Stirewalt D.L., Gentleman R., Vessella R.L., Nelson P.S., Martin D.B., Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xi Y., Nakajima G., Gavin E., Morris C.G., Kudo K., Hayashi K., Ju J. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13:1668–1674. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kosaka N., Izumi H., Sekine K., Ochiya T. microRNA as a new immune-regulatory agent in breast milk. Silence. 2010;1:7. doi: 10.1186/1758-907X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michael A., Bajracharya S.D., Yuen P.S.T., Zhou H., Star R.A., Illei G.G., Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park N.J., Zhou H., Elashoff D., Henson B.S., Kastratovic D.A., Abemayor E., Wong D.T. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin. Canc. Res. 2009;15:5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber J.A., Baxter D.H., Zhang S., Huang D.Y., Huang K.H., Lee M.J., Galas D.J., Wang K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zen K., Zhang C.-Y. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med. Res. Rev. 2012;32:326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 11.Kosaka N., Iguchi H., Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Canc. Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter M.P., Ismail N., Zhang X., Aguda B.D., Lee E.J., Yu L., Xiao T., Schafer J., Lee M.-L.T., Schmittgen T.D., Nana-Sinkam S.P., Jarjoura D., Marsh C.B. Detection of microRNA expression in human peripheral blood microvesicles. PloS One. 2008;3 doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 14.Taylor D.D., Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 15.Lodes M.J., Caraballo M., Suciu D., Munro S., Kumar A., Anderson B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PloS One. 2009;4:e6229. doi: 10.1371/journal.pone.0006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutwein P., Stoeck A., Riedle S., Gast D., Runz S., Condon T.P., Marmé A., Phong M.-C., Linderkamp O., Skorokhod A., Altevogt P. Cleavage of L1 in exosomes and apoptotic membrane vesicles released from ovarian carcinoma cells. Clin. Canc. Res. 2005;11:2492–2501. doi: 10.1158/1078-0432.CCR-04-1688. [DOI] [PubMed] [Google Scholar]
- 17.McManus M.T. MicroRNAs and cancer. Semin. Canc. Biol.. 2003;13:253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 18.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Canc. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 19.Di Leva G., Garofalo M., Croce C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth C., Rack B., Müller V., Janni W., Pantel K., Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12:R90. doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bousquet M., Harris M.H., Zhou B., Lodish H.F. MicroRNA miR-125b causes leukemia. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21558–21563. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia H.-Y., Wang Y.-X., Yan W.-T., Li H.-Y., Tian Y.-Z., Wang S.-M., Zhao H.-L. MicroRNA-125b functions as a tumor suppressor in hepatocellular carcinoma cells. Int. J. Mol. Sci. 2012;13:8762–8774. doi: 10.3390/ijms13078762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J., Zhang K.-Y., Liu S.-M., Sen S. Tumor-associated circulating microRNAs as biomarkers of cancer. Molecules. 2014;19:1912–1938. doi: 10.3390/molecules19021912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenfeld N., Aharonov R., Meiri E., Rosenwald S., Spector Y., Zepeniuk M., Benjamin H., Shabes N., Tabak S., Levy A., Lebanony D., Goren Y., Silberschein E., Targan N., Ben-Ari A., Gilad S., Sion-Vardy N., Tobar A., Feinmesser M., Kharenko O., Nativ O., Nass D., Perelman M., Yosepovich A., Shalmon B., Polak-Charcon S., Fridman E., Avniel A., Bentwich I., Bentwich Z., Cohen D., Chajut A., Barshack I. MicroRNAs accurately identify cancer tissue origin. Nat. Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 25.Liu C.-G., Calin G.A., Meloon B., Gamliel N., Sevignani C., Ferracin M., Dumitru C.D., Shimizu M., Zupo S., Dono M., Alder H., Bullrich F., Negrini M., Croce C.M. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson G., Nour A.A., Nolan T., Huggett J., Bustin S. Minimum information necessary for quantitative real-time PCR experiments. Methods Mol. Biol. 2014;1160:5–17. doi: 10.1007/978-1-4939-0733-5_2. [DOI] [PubMed] [Google Scholar]
- 27.Gao L., Jiang F. MicroRNA (miRNA) profiling. Methods Mol. Biol. 2016;1381:151–161. doi: 10.1007/978-1-4939-3204-7_8. [DOI] [PubMed] [Google Scholar]
- 28.Nunes A. de L., Paes C. de A., Murao M., Viana M.B., De Oliveira B.M. Cytogenetic abnormalities, WHO classification, and evolution of children and adolescents with acute myeloid leukemia. Hematol. Transfus. Cell Ther. 2019 doi: 10.1016/j.htct.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka M., Oikawa K., Takanashi M., Kudo M., Ohyashiki J., Ohyashiki K., Kuroda M. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PloS One. 2009;4 doi: 10.1371/journal.pone.0005532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F., Wang X.-S., Yang G.-H., Zhai P.-F., Xiao Z., Xia L.-Y., Chen L.-R., Wang Y., Wang X.-Z., Bi L.-X., Liu N., Yu Y., Gao D., Huang B.-T., Wang J., Zhou D.-B., Gong J.-N., Zhao H.-L., Bi X.-H., Yu J., Zhang J.-W. miR-29a and miR-142-3p downregulation and diagnostic implication in human acute myeloid leukemia. Mol. Biol. Rep. 2012;39:2713–2722. doi: 10.1007/s11033-011-1026-5. [DOI] [PubMed] [Google Scholar]
- 31.Fayyad-Kazan H., Bitar N., Najar M., Lewalle P., Fayyad-Kazan M., Badran R., Hamade E., Daher A., Hussein N., ElDirani R., Berri F., Vanhamme L., Burny A., Martiat P., Rouas R., Badran B. Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. J. Transl. Med. 2013;11:31. doi: 10.1186/1479-5876-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhi F., Cao X., Xie X., Wang B., Dong W., Gu W., Ling Y., Wang R., Yang Y., Liu Y. Identification of circulating microRNAs as potential biomarkers for detecting acute myeloid leukemia. PloS One. 2013;8 doi: 10.1371/journal.pone.0056718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang L., Deng T., Wang D., Xiao Y. Elevated serum exosomal miR-125b level as a potential marker for poor prognosis in intermediate-risk acute myeloid leukemia. Acta Haematol. 2018;140:183–192. doi: 10.1159/000491584. [DOI] [PubMed] [Google Scholar]
- 34.Tian C., Zhang L., Li X., Zhang Y., Li J., Chen L. Low miR-192 expression predicts poor prognosis in pediatric acute myeloid leukemia. Canc. Biomarkers. 2018;22:209–215. doi: 10.3233/CBM-170657. [DOI] [PubMed] [Google Scholar]
- 35.Yan J., Wu G., Chen J., Xiong L., Chen G., Li P. Downregulated miR-217 expression predicts a poor outcome in acute myeloid leukemia. Canc. Biomarkers. 2018;22:73–78. doi: 10.3233/CBM-170936. [DOI] [PubMed] [Google Scholar]
- 36.Yu G., Yin Z., He H., Zheng Z., Chai Y., Xuan L., Lin R., Wang Q., Li J., Xu D. Low serum miR-223 expression predicts poor outcome in patients with acute myeloid leukemia. J. Clin. Lab. Anal. 2019 doi: 10.1002/jcla.23096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papadantonakis N., Advani A.S. Recent advances and novel treatment paradigms in acute lymphocytic leukemia. Ther. Adv. Hematol. 2016;7:252–269. doi: 10.1177/2040620716652289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inaba H., Greaves M., Mullighan C.G. Acute lymphoblastic leukaemia. Lancet. 2013;381:1943–1955. doi: 10.1016/S0140-6736(12)62187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mi S., Lu J., Sun M., Li Z., Zhang H., Neilly M.B., Wang Y., Qian Z., Jin J., Zhang Y., Bohlander S.K., Le Beau M.M., Larson R.A., Golub T.R., Rowley J.D., Chen J. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luna-Aguirre C.M., de la Luz Martinez-Fierro M., Mar-Aguilar F., Garza-Veloz I., Treviño-Alvarado V., Rojas-Martinez A., Jaime-Perez J.C., Malagon-Santiago G.I., Gutierrez-Aguirre C.H., Gonzalez-Llano O., Salazar-Riojas R., Hidalgo-Miranda A., Martinez-Rodriguez H.G., Gomez-Almaguer D., Ortiz-Lopez R. Circulating microRNA expression profile in B-cell acute lymphoblastic leukemia. Canc. Biomarkers. 2015;15:299–310. doi: 10.3233/CBM-150465. [DOI] [PubMed] [Google Scholar]
- 41.Jiang Q., Lu X., Huang P., Gao C., Zhao X., Xing T., Li G., Bao S., Zheng H. Expression of miR-652-3p and effect on apoptosis and drug sensitivity in pediatric acute lymphoblastic leukemia. BioMed Res. Int. 2018;2018 doi: 10.1155/2018/5724686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swellam M., Hashim M., Mahmoud M.S., Ramadan A., Hassan N.M. Aberrant expression of some circulating miRNAs in Childhood acute lymphoblastic leukemia. Biochem. Genet. 2018;56:283–294. doi: 10.1007/s10528-018-9844-y. [DOI] [PubMed] [Google Scholar]
- 43.Aliano S., Cirmena G., Fugazza G., Bruzzone R., Palermo C., Sessarego M. Standard and variant Philadelphia translocation in a CML patient with different sensitivity to imatinib therapy. Leuk. Res. Rep. 2013;2:75–78. doi: 10.1016/j.lrr.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mineo M., Garfield S.H., Taverna S., Flugy A., De Leo G., Alessandro R., Kohn E.C. Exosomes released by K562 chronic myeloid leukemia cells promote angiogenesis in a Src-dependent fashion. Angiogenesis. 2012;15:33–45. doi: 10.1007/s10456-011-9241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Stefano C., Mirone G., Perna S., Marfe G. The roles of microRNAs in the pathogenesis and drug resistance of chronic myelogenous leukemia (Review) Oncol. Rep. 2016;35:614–624. doi: 10.3892/or.2015.4456. [DOI] [PubMed] [Google Scholar]
- 46.Agirre X., Jiménez-Velasco A., San José-Enériz E., Garate L., Bandrés E., Cordeu L., Aparicio O., Saez B., Navarro G., Vilas-Zornoza A., Pérez-Roger I., García-Foncillas J., Torres A., Heiniger A., Calasanz M.J., Fortes P., Román-Gómez J., Prósper F. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol. Canc. Res. 2008;6:1830–1840. doi: 10.1158/1541-7786.MCR-08-0167. [DOI] [PubMed] [Google Scholar]
- 47.Fallah P., Amirizadeh N., Poopak B., Toogeh G., Arefian E., Kohram F., Hosseini Rad S.M.A., Kohram M., Teimori Naghadeh H., Soleimani M. Expression pattern of key microRNAs in patients with newly diagnosed chronic myeloid leukemia in chronic phase. Int. J. Lab. Hematol. 2015;37:560–568. doi: 10.1111/ijlh.12351. [DOI] [PubMed] [Google Scholar]
- 48.Döhner H., Stilgenbauer S., Benner A., Leupolt E., Kröber A., Bullinger L., Döhner K., Bentz M., Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl. J. Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 49.Calin G.A., Ferracin M., Cimmino A., Di Leva G., Shimizu M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M., Iuliano R., Palumbo T., Pichiorri F., Roldo C., Garzon R., Sevignani C., Rassenti L., Alder H., Volinia S., Liu C., Kipps T.J., Negrini M., Croce C.M. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 50.Due H., Svendsen P., Bødker J.S., Schmitz A., Bøgsted M., Johnsen H.E., El-Galaly T.C., Roug A.S., Dybkær K. miR-155 as a biomarker in B-cell malignancies. BioMed Res. Int. 2016;2016:9513037. doi: 10.1155/2016/9513037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moussay E., Wang K., Cho J.-H., van Moer K., Pierson S., Paggetti J., Nazarov P.V., Palissot V., Hood L.E., Berchem G., Galas D.J. MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 2011;108:6573–6578. doi: 10.1073/pnas.1019557108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fathullahzadeh S., Mirzaei H., Honardoost M.A., Sahebkar A., Salehi M. Circulating microRNA-192 as a diagnostic biomarker in human chronic lymphocytic leukemia. Canc. Gene Ther. 2016;23:327–332. doi: 10.1038/cgt.2016.34. [DOI] [PubMed] [Google Scholar]
- 53.Cui B., Chen L., Zhang S., Mraz M., Fecteau J.-F., Yu J., Ghia E.M., Zhang L., Bao L., Rassenti L.Z., Messer K., Calin G.A., Croce C.M., Kipps T.J. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood. 2014;124:546–554. doi: 10.1182/blood-2014-03-559690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alharthi A., Beck D., Howard D.R., Hillmen P., Oates M., Pettitt A., Wagner S.D. An increased fraction of circulating miR-363 and miR-16 is particle bound in patients with chronic lymphocytic leukaemia as compared to normal subjects. BMC Res. Notes. 2018;11:280. doi: 10.1186/s13104-018-3391-9. [DOI] [PMC free article] [PubMed] [Google Scholar]