Abstract
Hundreds of dominant-negative myosin mutations have been identified that lead to hypertrophic cardiomyopathy, and the biomechanical link between mutation and disease is heterogeneous across this patient population. To increase the therapeutic feasibility of treating this diverse genetic population, we investigated the ability of locked nucleic acid (LNA)-modified antisense oligonucleotides (ASOs) to selectively knock down mutant myosin transcripts by targeting single-nucleotide polymorphisms (SNPs) that were found to be common in the myosin heavy chain 7 (MYH7) gene. We identified three SNPs in MYH7 and designed ASO libraries to selectively target either the reference or alternate MYH7 sequence. We identified ASOs that selectively knocked down either the reference or alternate allele at all three SNP regions. We also show allele-selective knockdown in a mouse model that was humanized on one allele. These results suggest that SNP-targeting ASOs are a promising therapeutic modality for treating cardiac pathology.
Keywords: antisense oligonucleotides, myosin, cardiac hypertrophy, cardiomyopathy, nucleic acid, RNA degradation
Introduction
Familial hypertrophic cardiomyopathy (HCM) is a monogenic disease clinically characterized by asymmetrical ventricular hypertrophy, arrhythmias, and progressive heart failure. HCM has a prevalence of 1:500,1 and about 40% of cases are due to autosomal dominant mutations in the MYH7 gene.2 MYH7 encodes the β-myosin heavy chain (β-MHC) protein that acts as a molecular motor to drive active contraction during cardiac systole. More than 300 missense mutations in MYH7 have been linked to HCM pathology, and these mutations are distributed throughout the gene.3,4 There is no common mechanism that links each MYH7 mutation to the HCM phenotype; mutations can affect thick filament formation, sliding velocity, ATPase rate, force, and calcium sensitivity of activation.3,4 Regardless of the exact mutation and its specific effect on actomyosin dynamics, the link between MYH7 mutation and HCM derives from mutant myosin protein that is expressed, stable, and exerts dominant-negative effects.
One classical approach to the treatment of HCM caused by MYH7 mutations is the use of small molecules that counteract the biomechanical effect of the mutation on the actomyosin crossbridge cycle. Because >300 mutations have been identified, a particular small molecule would be efficacious only for treating HCM caused by a single mutation or a subset of mutations that could be proved to alter crossbridge dynamics in the same way. The approach preferred in this study is to not consider the biophysical manifestation of dysfunctional myosin proteins, but to selectively knock down the poison peptide regardless of its downstream effects.
A therapeutic modality that continues to advance is antisense oligonucleotides (ASOs). ASOs can be used to knock down targets of interest by binding to the target RNA and inducing RNA cleavage via RNase H recruitment.5 An ASO targeting apolipoprotein B was recently approved for the treatment of homozygous familial hypercholesterolemia,6 and many others have demonstrated clear clinical benefit in rigorously controlled trials.7 Because there are hundreds of MYH7 mutations linked to HCM, each with relatively low prevalence, it would presently not be practical to develop ASOs that target individual pathogenic mutations. Therefore, we decided to target common single-nucleotide polymorphisms (SNPs) found in the general population. Previous work has shown that SNP-selective knockdown can be achieved with ASOs targeting the huntingtin transcript, both in vitro8 and in vivo.9 We identified three silent SNPs (amino acid sequence is unchanged) in MYH7 that have high heterozygosity across broad demographics and generated ASOs that selectively target either the reference nucleotide or the polymorphism. Designing ASOs to these SNPs enables multiple disease-linked mutations to be targeted with the same antisense compound. Clinically, this approach requires patient haplotyping to determine whether the HCM mutation is on the same allele as the SNP being targeted. Our results show that ASOs targeting human SNPs can distinguish alleles containing single-nucleotide mismatches with both high potency (<100 nM) and high selectivity (>20×). This strategy is therapeutically feasible when a patient harbors the pathogenic MYH7 mutation and the SNP of interest on the same transcript, and this general strategy can be employed for other genetically defined diseases in which SNPs exist within a gene encoding a dominant-negative protein.
Results
SNP Identification
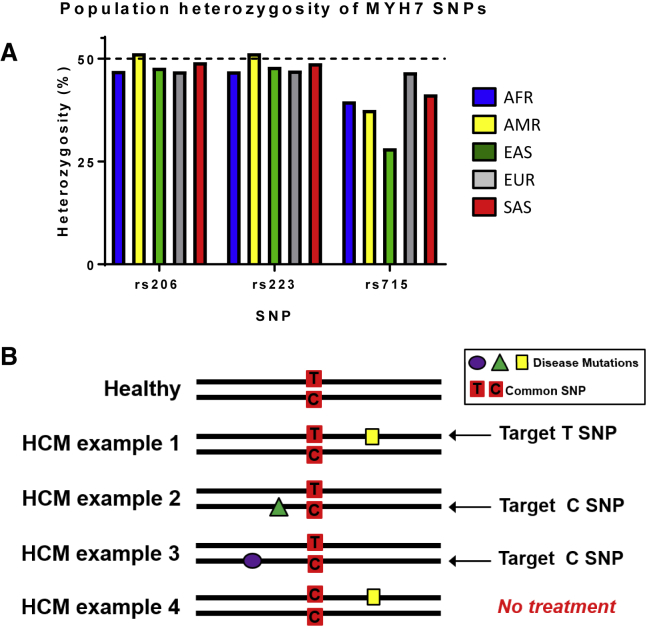
We analyzed the phase 3 1000 Genomes database10 to identify SNPs in the human population that occur with high frequency, i.e., genetic coordinates in MYH7 that contain different nucleotides on each allele (a heterozygous base) in a large fraction of people. We found three SNPs with high heterozygosity: rs2239578 (48%), rs2069540 (48%), and rs7157716 (38%) (Figure 1A). These three common SNPs are found in intron 2, exon 3, and exon 24 of MYH7, respectively, and will be referred to as rs223, rs206, and rs715 throughout this manuscript (Table 1).
Figure 1.
SNP Targeting Strategy
(A) SNP heterozygosity across five genetic super populations. See https://www.internationalgenome.org/category/population/ for details on population descriptions. (B) Developing ASOs for individual HCM mutations is not currently a feasible therapeutic strategy. By targeting SNPs, multiple MYH7 disease-causing mutations can be targeted with a single ASO.
Table 1.
MYH7SNPs
| SNP ID | Sequence (5′–3′) | Position | Allele | Note |
|---|---|---|---|---|
| rs223-T | AGAAAAGCTGAAGCTAGAGTGTTGAAAATCTAGTAAGAC | intron 2 | REF | pre-mRNA |
| rs223-C | AGAAAAGCTGAAGCTAGAGCGTTGAAAATCTAGTAAGAC | intron 2 | ALT | pre-mRNA |
| rs206-C | GCAAAGTCACTGCCGAGACCGAGTATGGCAAGACAGTGA | exon 3 | REF | mRNA |
| rs206-T | GCAAAGTCACTGCCGAGACTGAGTATGGCAAGACAGTGA | exon 3 | ALT | mRNA |
| rs206-C | GCAAAGTCACTGCCGAGACCGAGTATGGCAAGGTGGGTG | exon 3 | REF | pre-mRNA |
| rs206-T | GCAAAGTCACTGCCGAGACTGAGTATGGCAAGGTGGGTG | exon 3 | ALT | pre-mRNA |
| rs715-T | CTGGGCTGGATGAGATCATTGCCAAGCTGACCAAGGAGA | exon 24 | REF | pre-mRNA and mRNA |
| rs715-C | CTGGGCTGGATGAGATCATCGCCAAGCTGACCAAGGAGA | exon 24 | ALT | pre-mRNA and mRNA |
The three SNP regions of interest in human MYH7. The SNP sites are underlined, and the italicized sequence indicates the difference between mRNA and pre-mRNA for the rs206 region (this SNP is near the end of exon 3). LNA gapmers of various length, starting position, and wing design were generated for all eight templates. REF indicates reference allele and ALT indicates alternate allele.
For rs206, the reference nucleotide is cytosine and the SNP is thymine, whereas for rs223 and rs715, the reference is thymine and the SNP is cytosine (Table 1). The designation of a SNP in these cases is somewhat arbitrary because both the reference and alternate allele are common. No other polymorphisms are found within 25 bases upstream or downstream of each SNP. To each of these SNP regions, locked nucleic acid (LNA) gapmer ASOs were designed and synthesized to selectively knock down mRNA containing either cytosine or thymine at the SNP coordinate. This strategy depends on the ability of the ASOs to induce robust degradation of the SNP-matched RNA while minimizing degradation of the SNP-mismatched RNA. This allows for multiple disease-linked mutations to be targeted with the same ASO (Figure 1B). Within each SNP region ASOs were tiled along the transcript, resulting in some ASOs having the position of the SNP in the 5′ end, some in the DNA gap in the middle, and some in the 3′ end. Furthermore, for each position, ASOs from 15 to 20 nt in length were designed, with one to four LNA nucleotides in the 5′ end and two to four in the 3′ end. Varying the SNP position and ASO structure in this manner resulted in 47 ASOs targeting the rs715 SNP (15-C, 32-T), 111 ASOs targeting the rs223 SNP (52-C, 59-T), and 130 ASOs targeting the rs206 SNP (75-C, 55-T) (Table S1).
In Vitro Knockdown
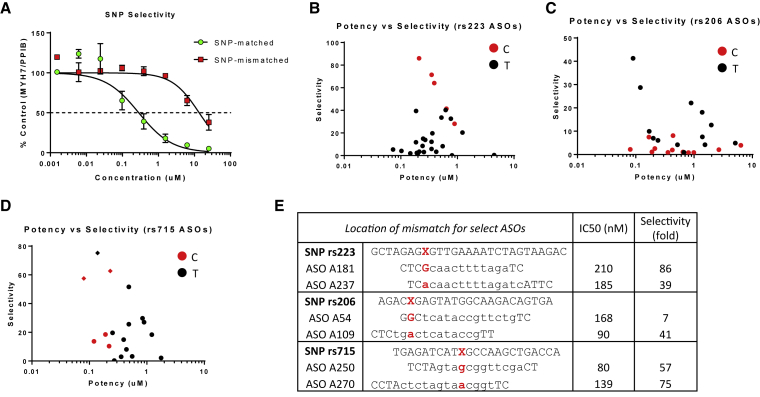
We screened the initial ASO libraries in the QuantiGene 2.0 assay to identify ASOs that exhibit good knockdown of MYH7 RNA. Two human skeletal muscle myoblast cell lines were used; both lines were homozygous at each SNP position, and the lines were perfectly complementary (i.e., one line had C/C at rs206 and T/T at rs223 and rs715, and the other had T/T at rs206 and C/C at rs223 and rs715). ASOs were screened in both cell lines at 5 μM using gymnotic delivery to determine SNP-matched and SNP-mismatched RNA knockdown at a 3-day time point. A non-SNP targeting ASO (S17 in Table S1) was used as a positive control and showed similar activity in both cell lines (88% and 85% knockdown at 5 μM; Figure S1B). This suggests that ASO uptake is similar between the two human myoblast lines. ASOs that showed mild selectivity (>50% knockdown of MYH7 mRNA in the SNP-matched cell line and <25% knockdown in the SNP-mismatched cell line) were selected for follow-up potency determination. Additional ASOs that showed good SNP-matched potency but did not meet the selectivity criteria were also progressed to concentration response curves (CRCs). ASO potency values (IC50) were determined from CRCs using the QuantiGene assay in both the SNP-matched and SNP-mismatched cell lines (Figure 2A). This allows calculation of a selectivity ratio, defined as the ratio of SNP-mismatched potency to SNP-matched potency. In all three SNP regions, ASOs with good potency and selectivity were identified (Figures 2B–2D), highlighting the generalizability of this approach.
Figure 2.
Evaluation of SNP-Selective ASOs from Initial Library in Skeletal Muscle Myoblast Cell Lines
(A) Example of concentration-response curves for ASO A181 from the rs223-C sub-library showing high SNP selectivity (mean ± SD). Selectivity is defined as the IC50 in SNP-mismatched cells divided by the IC50 in SNP-matched cells. (B–D) Potency and selectivity evaluated at day 10 are plotted for (B) rs223, (C) rs206, and (D) rs715 ASOs from the initial library. ASOs targeting the C and T allele of a given SNP are shown as red and black dots, respectively. In the rs715 ASO plot, the three diamond symbols indicate the ASOs selected for redesigns. (E) Examples of ASOs targeting each SNP region that have good in vitro potency and selectivity. The SNP is shown in red, with X = T or C. For the ASO sequences, LNA is shown as uppercase letters and DNA is lowercase. For each SNP region, a C-targeting and T-targeting ASO is shown. Notice that the mismatch can be in either the DNA “gap” or the LNA wings of the ASO. The full list of ASOs can be found in the Supplemental Information.
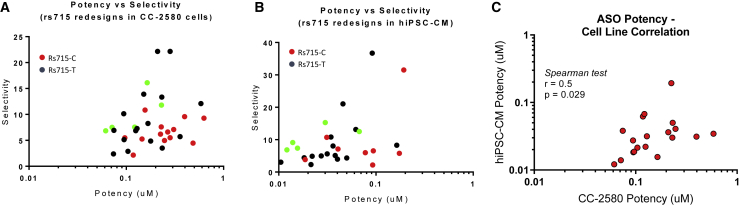
Because allele selectivity was shown at all three SNP regions, we decided to focus on ASOs targeting the rs715 SNP region because of sequence homology between human, dog, and cynomolgus monkey. We also developed a droplet digital PCR (ddPCR) assay that enabled us to measure allele-specific mRNA knockdown in cells that are heterozygous at the rs715 SNP position (T on one allele, C on the other). This assay used multiplexed PCRs to simultaneously measure allele-specific potency in SNP-heterozygous cell lines. We generated 450 LNA gapmer redesigns based on three ASOs from the initial rs715 library that exhibited good potency and selectivity (two ASOs, A249 and A250, targeting the rs715-C SNP and one, A270, targeting rs715-T). The redesigns are shown in Table S1. Transcript start site was maintained, but ASO lengths varied from 17 to 19 nt. Furthermore, the number and position of LNA modifications within each ASO were varied, with LNA and DNA interspersed. All ASOs had between 4 and 15 consecutive DNAs to allow for RNase H binding and cleavage, with the majority of ASOs containing between 5 and 7 consecutive DNAs. These ASOs were tested at 500 nM in human myoblasts that are heterozygous at the rs715 SNP position (CC-2580 cells; Lonza), with mRNA levels determined 6 days after compound addition (Figures S2A–S2C). From these single-point data, a subset of ASOs was selected for follow-up potency determinations in two rs715 SNP-heterozygous cell lines: human myoblasts (CC-2580) and human induced pluripotent stem cell (hiPSC)-derived cardiomyocytes (iCell2; CDI). Potent and selective ASOs were identified that target either the rs715-C or rs715-T SNP (Figure 3).
Figure 3.
Evaluation of SNP-Selective ASOs from Redesign Library Targeting the rs715 SNP
(A and B) Potency and selectivity evaluated in (A) human myoblast CC-2580 cells and (B) hiPSC-derived cardiomyocytes. ASOs targeting the C and T allele of a given SNP are shown as red and black dots, respectively. The five green dots indicate the C-targeting ASOs selected for evaluation in mice. (C) Correlation between potencies in CC-2580 cells and iPSC-derived cardiomyocyte (iPSC-CM) cells (Spearman rank order test: r = 0.5, p = 0.029).
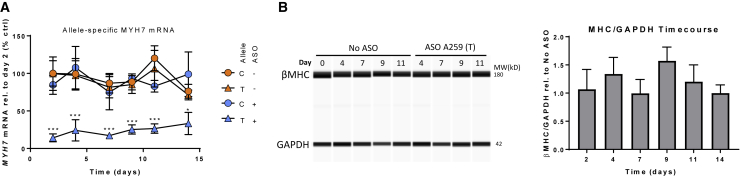
To determine whether allele compensation occurs during allele-selective knockdown, we performed a time-course study in iPSC-derived cardiomyocytes. These cells are heterozygous at the rs715 SNP position and were treated with 250 nM ASO A259 (see Table S1 for sequence), a potent and selective ASO targeting the rs715-T SNP. ASO A259 does not knock down the SNP-mismatched allele but does give strong knockdown of the SNP-matched allele (Figure 4A). This experiment shows in vitro allele-selective mRNA knockdown at up to 2 weeks following ASO addition. These experiments also included replicate cell plates for quantifying the effect of ASO addition on MYH7 protein (β-MHC). Protein lysates at all time points were probed with an antibody that recognizes β-MHC, but not α-MHC (iCell2 cells also contain α-MHC, which is encoded by the MYH6 gene). Reduction in β-MHC was not seen at any time point (Figure 4B), which suggests compensation at the level of translation. However, it must be noted that myosin turnover is quite slow half-life (t1/2 ~15 days in vivo11); measurement of β-MHC levels at extended time points would provide a more thorough understanding of myosin dynamics in iPSC-derived cardiomyocytes.
Figure 4.
Time-Course Study of SNP-Selective Knockdown
(A) mRNA knockdown in hiPSC-derived cardiomyocytes was evaluated at six time points over a 2-week period using allele-specific droplet digital PCR. At day 0, 250 nM rs715-T targeting ASO A259 was added via gymnosis. SNP-matched knockdown is seen for up to 2 weeks, whereas the SNP-mismatched allele does not show knockdown. Data were normalized to the no ASO day 2 time point for each allele. Mean ± SD from three independent experiments is shown. Significance between T alleles (no ASO versus ASO) was determined by two-way ANOVA followed by Sidak’s multiple comparisons test (*p < 0.05, ***p < 0.001). (B) Quantification of β-MHC in iCell2 human iPSC-derived cardiomyocyte (hiPSC-CM) with and without addition of ASO A259. β-MHC is not reduced at any time point, suggesting protein compensation by the SNP-mismatched allele. Bar graph represents three independent experiments (mean ± SD). Protein levels were normalized to the no ASO group at each time point.
In Vivo Knockdown
Lastly, we were interested in determining whether these ASOs could selectively knock down target mRNA in vivo. We generated a genetically engineered mouse model with the human MYH7 sequence inserted at the rs715 SNP region. Because the predominant myosin isoform in mouse heart is fast α-MHC,12 the human rs715-C SNP region was inserted into the mouse Myh6 gene. This was a 57-bp replacement at the rs715 SNP coordinate (i.e., the location of the SNP plus 32 nt upstream and 24 nt downstream; see Figure S3). This genetic modification did not change the predicted amino acid sequence of mouse α-MHC. This mouse line was heterozygous for the humanized MYH7 fragment because it contained wild-type (WT) Myh6 on the other allele. Mouse Myh6 contains a thymine base at the rs715 SNP coordinate, so the rs715-C ASOs are predicted to not target the WT allele. In addition, there is another mismatch six bases upstream of the SNP (guanine in human, adenine in mouse), which gives a 2-bp mismatch between the ASO-targeting sequences and the WT allele (see Figure S3 for details). Five ASOs targeting the rs715-C SNP were tested in vivo in these heterozygous humanized mice.
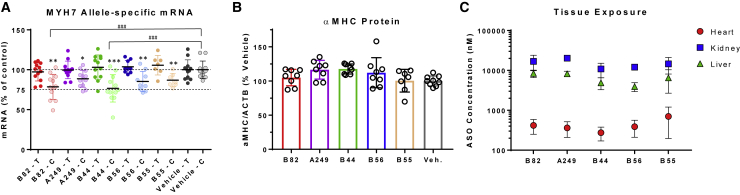
Mice were dosed subcutaneously with 30 mg/kg ASO (or saline) on days 0, 1, and 2, and the animals were sacrificed on day 9. Target mRNA knockdown was determined in left ventricular tissue; in all five ASO-treated groups, a significant reduction in humanized rs715-C mRNA compared with WT Myh6 mRNA was observed (Figure 5A). In addition, mice treated with the ASOs B82 or B44 showed a significant reduction of rs715-C mRNA compared with rs715-C mRNA levels in mice treated with saline. These results demonstrate that allele-selective knockdown can be achieved in cardiac tissue. Protein was quantified using a custom antibody specific for α-MHC (gene product of Myh6). No significant difference in total α-MHC was seen for any ASO-treated group compared with saline-treated controls (Figure 5B). ASO concentrations were determined in left ventricle, kidney, and liver using ASO-specific ELISA assays (Figure 5C). As expected, exposure values were significantly higher in kidney and liver. Two of the five ASOs (B44 and B56) were associated with a significant elevation of liver injury markers (AST, ALT, and alkaline phosphatase) and weight loss in a subset of animals (Figure S4), suggesting that these ASOs were not well tolerated. In contrast, ASOs B82, A249, and B55 showed normal weight gain (except for one animal in the B55 group) and no effect on liver or kidney injury markers, suggesting that ASOs with an improved safety and tolerability profile can be identified under the treatment conditions used in the current study.
Figure 5.
Study of SNP-Selective Knockdown in Mice
(A) Allele-specific mRNA quantitation from mouse left ventricle (LV; 8-week-old males) 1 week following ASO dosing (*p < 0.05, **p < 0.01, ***p < 0.001 comparing C and T allele abundance within a group as determined by t test). All five compounds significantly reduce the C allele compared with the T allele. Two compounds give significant knockdown of the C allele compared with the C allele in the saline group (###p < 0.001 comparing with saline C allele as determined by one-way ANOVA followed by Dunnett’s multiple comparisons test). n = 7–12 per group. (B) Total α-MHC protein quantitation from mouse LV 1 week following ASO dosing. No significant difference was seen relative to the saline group, as determined by one-way ANOVA followed by Dunnett’s multiple comparisons test. An α-MHC-specific antibody was used that showed no reactivity to recombinant β-MHC. (C) ASO concentrations in heart (LV), liver, and kidney. On average, ASO concentration is 37× higher in kidney and 16× higher in liver compared with heart. Mean ± SD shown.
Discussion
ASOs provide a treatment approach with potential to lower mutant protein and address underlying disease pathology in patients harboring pathological missense mutations. In the case of MYH7, hundreds of different mutations are linked to the development of HCM, and targeting individual pathogenic mutations using ASOs is unrealistic. Instead, we designed LNA gapmer ASOs to SNPs found with high heterozygosity across the general population. Due to sequence conservation between human, dog, and cynomolgus monkey, we focused on ASOs targeting the rs715 SNP and generated a library of ASO redesigns based on the best hits from the initial rs715 library. It should be noted that pre-clinical studies in large animals would be of limited value unless the species of interest was heterozygous (T and C) at the rs715 SNP position. For example, evaluation of an rs715-T targeting ASO in animals that are homozygous for rs715-T would result in an overestimation of toxicity because the ASO would knock down MYH7 mRNA that was transcribed from either allele. However, evaluation of an rs715-C ASO in the same animal would enable evaluation of non-MYH7-related toxicity because MYH7 mRNA levels would not be reduced by ASO-mediated RNase H degradation. For pre-clinical studies in a species that had high heterozygosity at the SNP of interest (i.e., a species whose SNP status mimicked the human population), animals would need to be sequenced to confirm SNP zygosity prior to inclusion in the study.
In order to safely target MYH7 with this approach, we hypothesized that downregulation of the SNP-matched (poison peptide) allele would be accompanied by upregulation of the SNP-mismatched (WT) allele to maintain normal levels of β-MHC protein. An in vitro time-course study with a potent ASO targeting the T allele showed SNP-matched mRNA knockdown out to 2 weeks without upregulation of the SNP-mismatched mRNA (Figure 4). However, total β-MHC protein is not reduced over this time period, which suggests that upregulation of protein in vitro is occurring at the level of translation on the SNP-mismatched allele. Because myosin turnover (at least in vivo) takes place on the timescale of weeks,11 additional data from longer time points would strengthen this argument.
To assess allele-selective knockdown and protein compensation in vivo, we dosed humanized mice with five ASOs targeting the rs715-C allele. Because Myh6 is the predominant myosin gene expressed in the mouse heart, we humanized one Myh6 allele with the human rs715-C sequence. This resulted in a 2-bp mismatch between the human MYH7 sequence and endogenous Myh6 sequence in the transcript region complementary to the ASOs that were tested in vivo. Our original plan was to humanize one Myh6 allele with the human rs715-T sequence and the other Myh6 allele with the human rs715-C sequence because this would result in a single base pair mismatch between the alleles. Unfortunately, we were unable to generate this fully humanized mouse line (i.e., heterozygous for the human rs715 sequence), although we would not expect any technical challenges in doing so. All five ASOs showed preferential activity toward the SNP-matched (hMYH7) allele without reducing the amount of WT Myh6 transcript. At the protein level, total α-MHC was not significantly different between groups. This could be explained by an increase in translation from either allele (relative to baseline translation rate), although interpretation of these data are limited by the single time point used here (day 9). Because myosin is tightly bound in the thick filaments of the sarcomere, it is thought that myosin turnover is quite slow. Indeed, an isotope enrichment study suggests that the half-life of myosin in rat heart is ~15 days.11 More data are required to determine whether myosin protein levels remain constant following allele-specific mRNA reduction. Maintenance of total myosin protein via compensation through the mismatched (non-targeted) allele is likely necessary for clinical feasibility of this approach. Although the presence of mutant myosin leads to HCM over time, a significant reduction of myosin protein (even if only mutant protein is downregulated) would pose substantial safety concerns and would require thorough drug safety evaluation prior to clinical development. It should be noted that mice hemizygous for Myh6 have ~25% less α-MHC protein compared with WT animals and have altered cardiac function.13 Another challenge is that this approach would require genome phasing to assign the disease mutation to the proper SNP allele. A companion diagnostic would need to be developed to guarantee that the prescribed ASO targets the SNP associated with the poison peptide allele and not the WT allele. The translational feasibility of this approach would be strengthened by evidence of single-base pair selectivity in vivo and evidence of efficacy in pre-clinical models. In addition, the long-term physiological effects of reducing MYH7 with ASOs needs to be evaluated.
For the five ASOs tested in vivo, average exposure was ~400 nM in the heart, and target mRNA knockdown was ~20%. In vitro, these ASOs showed IC50 potencies between 100–200 nM in CC-2580 myoblasts and 10–30 nM in iPSC-derived cardiomyocytes. The fact that in vitro potency does not directly translate to in vivo potency is not surprising and could be because of numerous factors including tissue distribution, cell-type-specific ASO uptake (i.e., cardiomyocytes versus fibroblasts), and subcellular localization. It is also possible that greater target knockdown may occur at longer time points or with higher or more frequent dosing. Enhanced uptake in cardiac tissue might also result in better target knockdown and could be achieved by ASO conjugation with short peptides, fatty acids, or other molecules capable of binding muscle cells.14, 15, 16
In addition to strategies to improve ASO delivery, the discovery of ASOs with increased potency and selectivity in vitro would provide new leads with potential to achieve greater allele-selective target knockdown in vivo. The optimal design and modification chemistry of allele-selective gapmers is not obvious because selectivity is determined by a number of factors, such as the difference in free energy of binding to SNP-matched and -mismatched alleles, SNP-sensitive differences in secondary RNA structure, and the mismatch-sensitive substrate requirements of RNase H.8,17, 18, 19 In the present study, we also did not find a clear consensus for which LNA modification patterns were superior to the others tested. Consequently, we speculate that the selectivity of our preferred LNA gapmers, such as B44 and B82, could be further improved, e.g., by combining several different modification chemistries in the wings,20 as well as in the gap region.21 Additional work will be needed to answer these important questions. Nonetheless, it has been shown that a 28% reduction of mutant myosin transcript via RNA interference inhibits development of the HCM phenotype in mice,22 suggesting that even the modest level of in vivo knockdown shown here may be therapeutically efficacious.
We show that LNA gapmer ASOs can selectively knock down MYH7 mRNA based on the presence of a SNP. By targeting individual SNPs, multiple pathogenic dominant-negative mutations can be targeted with a single antisense therapeutic, provided that the SNP is located on the poison peptide allele. We show proof of concept that peripheral dosing can lead to allele-selective knockdown in cardiac tissue. As genomic sequencing capabilities continue to improve and precision diagnosis of genetically defined diseases becomes more routine, the potential of antisense therapeutics to treat unmet medical diseases will continue to grow.
Materials and Methods
ASO Synthesis and Purification
LNA-modified gapmers were designed with fully modified phosphorothioate backbones and were synthesized on a MerMade 192X synthesizer (Bioautomation, TX, USA) following standard phosphoramidite protocols. The final 5′-dimethoxytrityl (DMT) group was left on the oligonucleotide. After synthesis, the oligonucleotides were cleaved from the solid support using aqueous ammonia and subsequently deprotected at 65°C for 5 h. The oligonucleotides were purified by solid-phase extraction in TOP DNA cartridges (Agilent, Glostrup, Denmark) using the lipophilic DMT group as a chromatographic retention probe. After eluting impurities, the DMT group was removed by treatment with dichloroacetic acid. As the last step in the purification process, the oligonucleotides were eluted from the cartridge and the eluate was evaporated to dryness. The oligonucleotides were dissolved in phosphate-buffered saline (PBS) and the oligonucleotide concentration in solution determined using Beer-Lambert’s law by calculating the extinction coefficient and measuring UV absorbance. Oligonucleotide identity and purity were determined by reversed-phase ultra-performance liquid chromatography coupled to mass spectrometry (UPLC-MS).
Cell Culture
Human skeletal muscle myoblasts (8220 and NH10-637A in-licensed from Dr. Vincent Mouly23 and CC-2580; Lonza) were seeded in collagen-coated 96-well plates at a density of 15,000 cells/well. Cells were maintained in SKM-M growth media (ZenBio, NC, USA) until confluence, at which point SKM-D differentiation media were used. Cells were cultured for 1 week in differentiation media to allow for myoblast fusion and differentiation into myotubes, with media exchange every other day. One week after switching to differentiation media, ASOs were added to the cells in the absence of transfection reagents (i.e., gymnotic delivery); biological duplicates were used. Cells were lysed at day 3 or 6 for single-point studies and at day 6 or 10 for CRCs. hiPSC-derived cardiomyocytes were purchased from Cellular Dynamics International and cultured according to the manufacturer’s instructions. Cells were seeded in collagen/fibronectin-coated (0.01 mg/mL) 96-well plates at a density of 20,000 cells/well. ASOs dissolved in PBS or water were added 4 days after plating, and media were changed every other day until lysis.
QuantiGene
The QuantiGene 2.0 assay (Affymetrix) was used to quantify RNA abundance of MYH7 (QG probe SA-10161) and the endogenous control (human PPIB probe SA-10003) in each lysate following the manufacturer’s protocol. The QG probes are designed to exonic regions of MYH7 and PPIB. Assay signals were background subtracted and normalized to the endogenous control to correct for cell density and lysis efficiency. MYH7 knockdown is reported relative to no ASO negative control.
RNA Purification and ddPCR
Cells were lysed by removal of media followed by addition of 125 μL PureLink Pro 96 Lysis buffer (12173.001A; Invitrogen) and 125 μL 70% ethanol. RNA was purified according to the manufacturer’s instructions and eluted in a final volume of 50 μL water resulting in an RNA concentration of 10–20 ng/μL. ddPCR was done using Bio-Rad Automatic Droplet Generator (AutoDG) using Automated Droplet Generation Oil for Probes (Bio-Rad) together with the OX200 droplet digital reader. The ddPC Supermix for Probes (No dUTP) (1863024; Bio-Rad) reactions were run according to the manufacturer’s instructions with an annealing temperature of 55.5°C for the human reactions and 55°C for the mouse reactions. The droplets were read in the OX200 droplet digital reader, and the data were analyzed and quantified using the QuantaSoft Analysis Pro Software 1.0.596 (Bio-Rad). The thresholds for defining the different droplet groups in the triplex PCR were set by free hand within the software according to the guidelines. Assays for human SNPs: rs715T (fw_primer 5′-CAGAGGAGATGGCTGG-3′, rev_primer 5′-TGCAGAGCTTTCTTCTCC-3′, probe 5′-CAGCTTGGCAATGATCTC-3′ HEX_IowaBlack); rs715C (fw_primer 5′-CAGAGGAGATGGCTGG-3′, rev_primer 5′-TGCAGAGCTTTCTTCTCC-3′, probe 5′-CAGCTTGGCGATGATCT-3′ FAM_IowaBlack); GAPDH (dHsa CPE5031596, FAM_IowaBlack and dHsa CPE5031597, HEX_IowaBlack) from Bio-Rad. Assays for humanized mouse model were: humanized rs715C myh6 (fw_primer 5′-AAAACCTAACAGAGGAGATG-3′, rev_primer 5′-CTTCTTGCAGAGCTTTCTT-3′, probe 5′-TGAGATCATCGCCAAGC-3′ Hex_IowaBlack); WT myh6 (fw_primer 5′-AAAACCTAACAGAGGAGATG-3′, rev_primer 5′-CTTCTTGCAGAGCTTTCTT-3′, probe 5′-TGAAATCATTGCCAAGCTG-3′ FAM_IowaBlack); and GAPDH (dMmuCPE5195282, FAM_IowaBlack, and dMmuCPE5195283, HEX_IowaBlack) from Bio-Rad.
Statistical Analysis of Concentration-Response Curves
Concentration-response curves of RNA levels after treatment with ASO at eight different concentrations were analyzed by nonlinear least-squares fitting of the two-parameter logistic function using the R software package drc.24 For the two-parameter logistic function, the lower and upper limits are fixed at 0% and 100%, respectively, and the two parameters estimated from each curve are the IC50 value and Hill coefficient. The maximal possible IC50 value was set to the maximal ASO concentration evaluated.
Digital Western Blot
Lysates from hiPSC-derived cardiomyocytes (iCell2; CDI) treated with ASOs were probed for β-MHC and GAPDH using a digital western blot system (WES, ProteinSimple). A total of 0.5 μg/μL protein was used per lane. A custom antibody specific for β-MHC was generated to allow for discrimination between myosin isoforms (iCell2 cells also contain α-MHC). A commercial antibody was used to measure GAPDH (G9545, 1:10,000; Sigma). Secondary antibody probing and detection via substrate followed standard WES protocols. Lysates from mouse heart were probed with a custom α-MHC-specific antibody and normalized to actin (97373; Abcam).
Mouse Model Generation and In Vivo Study
Because the predominant isoform in mouse heart is Myh6, the human MYH7 sequence (Ensembl: ENSG00000092054) around the rs715-C SNP was inserted into the mouse Myh6 gene (Ensembl: ENSMUSG00000040752) using homologous recombination in C57BL/6J mice (Figure S3). This 57-nt insertion (5′-acagaggagatggctgggctggatgagatcatCgccaagctgaccaaggagaagaaa-3′ replacing 5′-acagaggagatggctgggctggatgaaatcatTgccaagctgaccaaagagaagaaa-3′) is not predicted to affect amino acid sequence (SNP nucleotide shown as uppercase). Mice are homozygous for thymine at the base position that corresponds to the rs715 SNP in humans. Heterozygous mice (human rs715-C)+/− lacking FLP recombinase were used for the in vivo study. All studies were conducted in accordance with the Bristol-Myers Squibb Policy on the Care, Welfare, and Treatment of Laboratory Animals. All protocols were reviewed by the Animal Care and Use Committee at Bristol-Myers Squibb. Eight-week-old male animals were dosed with ASO subcutaneously at 30 mg/kg on days 0, 1, and 2 and were sacrificed on day 9 (n = 7–12 per group). Allele-specific Myh6 mRNA knockdown was measured via ddPCR from RNA isolated from half of the left ventricle. The other half of the left ventricle, in addition to one kidney and a portion of liver, was quick frozen in liquid nitrogen to determine the tissue concentrations of ASO (Oligo ELISA, Exiqon, Denmark). Blood was collected at the time of sacrifice, and serum was isolated. Kidney and liver injury markers (blood urea nitrogen [BUN], creatinine, alkaline phosphatase, aspartate aminotransferase [AST], and alanine transaminase [ALT]) were quantified on a Siemens Advia 1800 automated chemistry analyzer. Statistical significance was determined using one-way ANOVA and Dunnett’s multiple comparisons test.
Author Contributions
Conceptualization, M.L.J., P.H.H., B.R.A., S.C.L., R.E.O., A.C., and L.J.B.; Methodology: M.L.J., P.H.H., B.R.A., S.C.L., and R.A.; Software, P.H.H.; Formal Analysis, M.L.J., P.H.H., B.R.A., S.C.L., and R.A.; Investigation, J.V., B.N., B.R.A., B.K., L.I., Y.C., G.D.-K., and J.N.; Resources, I.M. and S.M.; Writing – Original Draft, M.L.J., P.H.H., S.C.L., and B.R.A.; Writing – Review & Editing, L.J.B.; Supervision, B.R.H., M.H., T.K., R.E.O., J.T., A.C., and L.B.J.
Conflicts of Interest
M.L.J., P.H.H., J.V., B.N., B.R.H., M.H., and T.K. are employees of Roche Pharma Research and Early Development, a part of F. Hoffmann-La Roche Ltd., a company that is developing LNA-modified oligonucleotides for therapeutic purposes. B.R.A., S.C.L., R.E.O., R.A., B.K., J.T., I.M., S.M., L.I., Y.C., J.N., G.D.-K., A.C., and L.J.B. are employees of Bristol-Myers Squibb and may hold company stock or stock options.
Acknowledgments
The authors thank Dong Li, Tong Tang, Yulia Benitex, and Brad Snyder for expert technical assistance.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.01.012.
Supplemental Information
Uppercase indicates LNA; lowercase indicates DNA. ASOs that are referred to directly in the text, figures, or supplement are bolded. The redesign library was based on three rs715 compounds from the first library (A249, A250, and A270). A259 was used for the in vitro time course in hiPSC-CM. The five ASOs that were tested in vivo were A249, B44, B55, B56, and B82. The scanning library listed at the end of the table consists of ASOs targeting sequence regions throughout the human MYH7 gene (i.e. not localized to the three SNP regions) and was used to validate target accessibility. From this scanning library a positive control ASO (S17) was selected for in vitro screens.
References
- 1.Maron B.J., Gardin J.M., Flack J.M., Gidding S.S., Kurosaki T.T., Bild D.E. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 2.Richard P., Charron P., Carrier L., Ledeuil C., Cheav T., Pichereau C., Benaiche A., Isnard R., Dubourg O., Burban M., EUROGENE Heart Failure Project Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 3.Moore J.R., Leinwand L., Warshaw D.M. Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circ. Res. 2012;111:375–385. doi: 10.1161/CIRCRESAHA.110.223842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colegrave M., Peckham M. Structural implications of beta-cardiac myosin heavy chain mutations in human disease. Anat. Rec. (Hoboken) 2014;297:1670–1680. doi: 10.1002/ar.22973. [DOI] [PubMed] [Google Scholar]
- 5.Bennett C.F., Swayze E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 6.Raal F.J., Santos R.D., Blom D.J., Marais A.D., Charng M.J., Cromwell W.C., Lachmann R.H., Gaudet D., Tan J.L., Chasan-Taber S. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;375:998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- 7.Stein C.A., Castanotto D. FDA-Approved Oligonucleotide Therapies in 2017. Mol. Ther. 2017;25:1069–1075. doi: 10.1016/j.ymthe.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skotte N.H., Southwell A.L., Østergaard M.E., Carroll J.B., Warby S.C., Doty C.N., Petoukhov E., Vaid K., Kordasiewicz H., Watt A.T. Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: providing a therapeutic option for all Huntington disease patients. PLoS ONE. 2014;9:e107434. doi: 10.1371/journal.pone.0107434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Southwell A.L., Skotte N.H., Kordasiewicz H.B., Østergaard M.E., Watt A.T., Carroll J.B., Doty C.N., Villanueva E.B., Petoukhov E., Vaid K. In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol. Ther. 2014;22:2093–2106. doi: 10.1038/mt.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papageorgopoulos C., Caldwell K., Schweingrubber H., Neese R.A., Shackleton C.H., Hellerstein M. Measuring synthesis rates of muscle creatine kinase and myosin with stable isotopes and mass spectrometry. Anal. Biochem. 2002;309:1–10. doi: 10.1016/s0003-2697(02)00253-1. [DOI] [PubMed] [Google Scholar]
- 12.Ng W.A., Grupp I.L., Subramaniam A., Robbins J. Cardiac myosin heavy chain mRNA expression and myocardial function in the mouse heart. Circ. Res. 1991;68:1742–1750. doi: 10.1161/01.res.68.6.1742. [DOI] [PubMed] [Google Scholar]
- 13.Jones W.K., Grupp I.L., Doetschman T., Grupp G., Osinska H., Hewett T.E., Boivin G., Gulick J., Ng W.A., Robbins J. Ablation of the murine alpha myosin heavy chain gene leads to dosage effects and functional deficits in the heart. J. Clin. Invest. 1996;98:1906–1917. doi: 10.1172/JCI118992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jirka S.M., Heemskerk H., Tanganyika-de Winter C.L., Muilwijk D., Pang K.H., de Visser P.C., Janson A., Karnaoukh T.G., Vermue R., ’t Hoen P.A. Peptide conjugation of 2′-O-methyl phosphorothioate antisense oligonucleotides enhances cardiac uptake and exon skipping in mdx mice. Nucleic Acid Ther. 2014;24:25–36. doi: 10.1089/nat.2013.0448. [DOI] [PubMed] [Google Scholar]
- 15.Prakash T.P., Mullick A.E., Lee R.G., Yu J., Yeh S.T., Low A., Chappell A.E., Østergaard M.E., Murray S., Gaus H.J. Fatty acid conjugation enhances potency of antisense oligonucleotides in muscle. Nucleic Acids Res. 2019;47:6029–6044. doi: 10.1093/nar/gkz354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Østergaard M.E., Jackson M., Low A., E Chappell A., G Lee R., Peralta R.Q., Yu J., Kinberger G.A., Dan A., Carty R. Conjugation of hydrophobic moieties enhances potency of antisense oligonucleotides in the muscle of rodents and non-human primates. Nucleic Acids Res. 2019;47:6045–6058. doi: 10.1093/nar/gkz360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monia B.P., Johnston J.F., Ecker D.J., Zounes M.A., Lima W.F., Freier S.M. Selective inhibition of mutant Ha-ras mRNA expression by antisense oligonucleotides. J. Biol. Chem. 1992;267:19954–19962. [PubMed] [Google Scholar]
- 18.Lima W.F., Rose J.B., Nichols J.G., Wu H., Migawa M.T., Wyrzykiewicz T.K., Vasquez G., Swayze E.E., Crooke S.T. The positional influence of the helical geometry of the heteroduplex substrate on human RNase H1 catalysis. Mol. Pharmacol. 2007;71:73–82. doi: 10.1124/mol.106.025429. [DOI] [PubMed] [Google Scholar]
- 19.Rukov J.L., Hagedorn P.H., Høy I.B., Feng Y., Lindow M., Vinther J. Dissecting the target specificity of RNase H recruiting oligonucleotides using massively parallel reporter analysis of short RNA motifs. Nucleic Acids Res. 2015;43:8476–8487. doi: 10.1093/nar/gkv759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Østergaard M.E., Southwell A.L., Kordasiewicz H., Watt A.T., Skotte N.H., Doty C.N., Vaid K., Villanueva E.B., Swayze E.E., Bennett C.F. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 2013;41:9634–9650. doi: 10.1093/nar/gkt725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Østergaard M.E., Kumar P., Nichols J., Watt A., Sharma P.K., Nielsen P., Seth P.P. Allele-Selective Inhibition of Mutant Huntingtin with 2-Thio- and C5- Triazolylphenyl-Deoxythymidine-Modified Antisense Oligonucleotides. Nucleic Acid Ther. 2015;25:266–274. doi: 10.1089/nat.2015.0547. [DOI] [PubMed] [Google Scholar]
- 22.Jiang J., Wakimoto H., Seidman J.G., Seidman C.E. Allele-specific silencing of mutant Myh6 transcripts in mice suppresses hypertrophic cardiomyopathy. Science. 2013;342:111–114. doi: 10.1126/science.1236921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mamchaoui K., Trollet C., Bigot A., Negroni E., Chaouch S., Wolff A., Kandalla P.K., Marie S., Di Santo J., St Guily J.L. Immortalized pathological human myoblasts: towards a universal tool for the study of neuromuscular disorders. Skelet. Muscle. 2011;1:34. doi: 10.1186/2044-5040-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritz C., Baty F., Streibig J.C., Gerhard D. Dose-Response Analysis Using R. PLoS ONE. 2015;10:e0146021. doi: 10.1371/journal.pone.0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Uppercase indicates LNA; lowercase indicates DNA. ASOs that are referred to directly in the text, figures, or supplement are bolded. The redesign library was based on three rs715 compounds from the first library (A249, A250, and A270). A259 was used for the in vitro time course in hiPSC-CM. The five ASOs that were tested in vivo were A249, B44, B55, B56, and B82. The scanning library listed at the end of the table consists of ASOs targeting sequence regions throughout the human MYH7 gene (i.e. not localized to the three SNP regions) and was used to validate target accessibility. From this scanning library a positive control ASO (S17) was selected for in vitro screens.