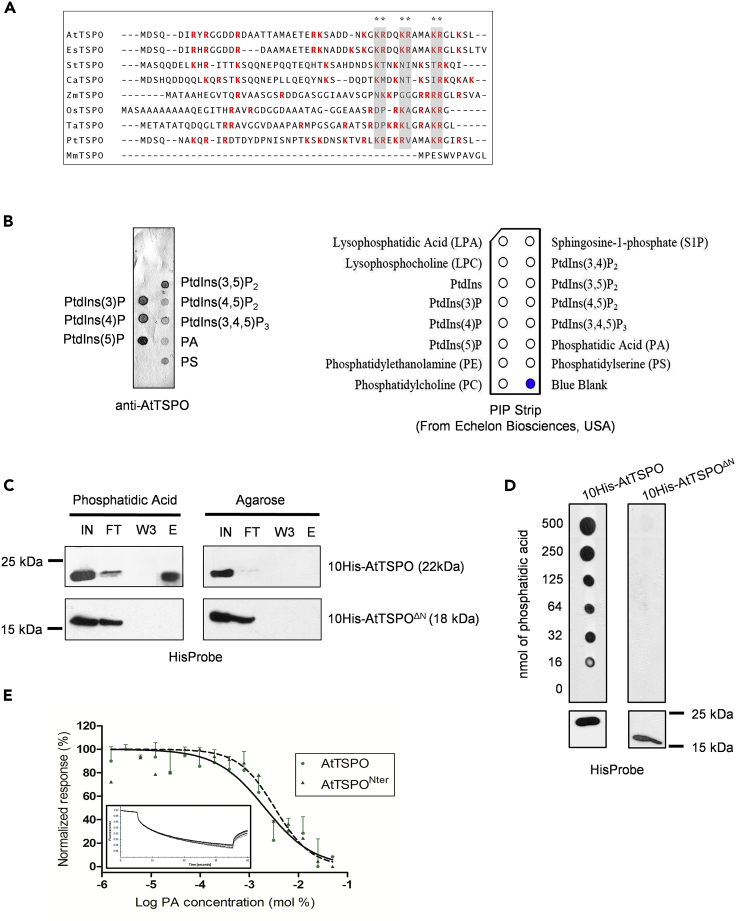
Figure 3.
Purified AtTSPO Binds Defined Anionic Lipids In Vitro and Binding Requires the N-terminus
(A) The N-terminal extension of plant TSPO is positively charged, as shown by ClustalW alignment of the first 50 residues in higher plant (monocot and dicot) TSPO sequences. At, Arabidopsis thaliana; Es, Eutrema salsugineum; St, Solanum tuberosum; Ca, Capsicum annuum; Zm, Zea mays; Os, Oryza sativa; Ta; Triticum aestivum; Pt; Populus trichocarpa. Positively charged amino acids are red (neutral pH) and conserved lysine/arginine residues near the transmembrane domain are indicated by gray rectangles. The mouse (Mus musculus, Mm) TSPO lacks the plant conserved N-terminal extension.
(B) Initial screening of AtTSPO ligands yielded several candidate anionic lipids. AtTSPO purified from yeast was incubated with spotted lipids in PIP-strip overlay assays (right panel) and detected with anti-AtTSPO antibodies (left panel).
(C) The plant-specific N-terminal extension is involved in AtTSPO-anionic lipid interactions in vitro, as shown by lipid-dependent pull-down of AtTSPO. Purified AtTSPO or AtTSPOΔN[IN] were incubated with PA or agarose resin (negative control). Flow-through [FT], wash (last wash W3), and eluted [E] fractions were probed with HisProbe.
(D) Ten histidine-tagged and N-terminally truncated AtTSPOs expressed and solubilized from yeast microsomes were incubated with a gradient of phosphatidic acid concentrations and detected with HisProbe. The bottom panel shows control detection of both proteins.
(E) Thermophoresis analyses of full-length AtTSPO and its N-terminal peptide titrated against different phosphatidic acid concentrations. Protein samples were labeled with the red fluorescent dye NT-647-NHS, and non-linear fitting of labeled full-length protein (continuous line) yielded a Kd of 2.1 ± 0.4 mM, compared with a Kd of 3.3 ± 0.9 mM for the N-terminal peptide (dotted line). The rectangular insert is a representative fluorescence trace. For AtTSPO shown is the mean ± SD of three experiments and for AtTSPOΔN shown is a recorded value from one experiment.