Abstract
Objective
The most common kidney cancer, clear cell renal cell carcinoma (ccRCC), is closely associated with obesity. The “clear cell” variant of RCC gets its name from the large lipid droplets that accumulate in the tumor cells. Although renal lipid metabolism is altered in ccRCC, the mechanisms and lipids driving this are not well understood.
Methods
We used shotgun lipidomics in human ccRCC tumors and matched normal adjacent renal tissue. To assess MBOAT7s gene expression across tumor severity, we examined histologically graded human ccRCC samples. We then utilized genome editing in ccRCC cell lines to understand the role of MBOAT7 in ccRCC progression.
Results
We identified a lipid signature for ccRCC that includes an increase in arachidonic acid-enriched phosphatidylinositols (AA-PI). In parallel, we found that ccRCC tumors have increased expression of acyltransferase enzyme membrane bound O-acyltransferase domain containing 7 (MBOAT7) that contributes to AA-PI synthesis. In ccRCC patients, MBOAT7 expression increases with tumor grade, and increased MBOAT7 expression correlates with poor survival. Genetic deletion of MBOAT7 in ccRCC cells decreases proliferation and induces cell cycle arrest, and MBOAT7−/− cells fail to form tumors in vivo. RNAseq of MBOAT7−/− cells identified alterations in cell migration and extracellular matrix organization that were functionally validated in migration assays.
Conclusions
This study highlights the accumulation of AA-PI in ccRCC and demonstrates a novel way to decrease the AA-PI pool in ccRCC by limiting MBOAT7. Our data reveal that metastatic ccRCC is associated with altered AA-PI metabolism and identify MBOAT7 as a novel target in advanced ccRCC.
Keywords: Clear cell renal carcinoma, Lipid, Phosphatidylinositol, Metabolism
1. Introduction
The most lethal and third most common urological cancer in the US is clear cell renal cell carcinoma (ccRCC) [1]. The incident risk of ccRCC is closely associated with obesity [2], and ccRCC tumors are strikingly lipid laden compared to other malignancies [3]. The “clear cell” variant of RCC gets its name from the large intracellular lipid droplets that accumulate in the tumor cells, and in some cases ccRCC cells have a unilocular lipid droplet similar to that seen in adipocytes [4]. Several recent studies demonstrated that altered prostaglandin levels may be involved in ccRCC growth and progression [[5], [6], [7]] and intracellular lipid droplets may play a role in managing oxidative stress in the ccRCC tumor microenvironment [4]. Although renal lipid metabolism is altered in ccRCC, the mechanisms driving this are not well understood.
Phosphatidylinositols (PI) and closely related phosphatidylinositol phosphates (PIPs) are essential for life in eukaryotes [5]. These signaling phospholipids are commonly studied for their importance in cytokinesis, migration, and cell polarity [[6], [7], [8]]. Although many studies have identified key signaling roles for PIPs in cancer progression [[8], [9], [10]], very little is known regarding whether compositional changes in the membrane PI pool, where PIP signaling lipids originate, are involved in ccRCC pathogenesis. Using acyl-CoAs as donors, phospholipids are formed by the de novo Kennedy pathway [12] and diversified by a remodeling pathway known as the Lands' cycle [13]. Although the major enzymes involved in the Kennedy pathway have been well characterized [12], much less is known about the enzymes involved in the Lands' cycle. A unique contributor to the membrane PI pool is membrane bound O-acyltransferase domain containing 7 (MBOAT7), an acyltransferase enzyme that selectively esterifies lysophosphatidylinositol (LPI) lipids to an arachidonyl-CoA to form the arachidonic acid-containing PI (AA-PI) species in the inner leaflet of cell membranes [14,15]. MBOAT7 is a unique contributor to the Lands’ cycle, which is a series of phospholipase-driven deacylation and lysophospholipid acyltransferase-driven reacylation reactions that synergize to alter phospholipid fatty acid compositions, creating membrane asymmetry and diversity. It is important to note that MBOAT7, unlike other lysophospholipid acyltransferses, only diversifies the fatty acid composition of membrane PI species and not phospholipids with other head groups [15,16]. This study demonstrates that increased expression of MBOAT7 in advanced ccRCC is associated with poorer survival and MBOAT7 loss of function diminishes both the proliferative and migratory properties of ccRCC cells. The data support the hypothesis that limiting MBOAT7-driven PI diversification may hold therapeutic promise in ccRCC and potentially other cancers.
2. Methods
2.1. Lipidomic profiling of ccRCC
To understand alterations across a wide range of structurally distinct lipids, we developed a shotgun lipidomics method to measure multiple lipid species as previously described [17]. All of the internal standards were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA). Ten internal standards (12:0 diacylglycerol, 14:1 monoacylglycerol, 17:0 lysophosphatidylcholine, 17:0 phosphatidylcholine, 17:0 phosphatidic acid; 17:0 phosphatidylethanolamine, 17:0 phosphatidylglycerol, 17:0 sphingomyelin, 17:1 lysosphingomyelin, and 17:0 ceramide) were mixed with a final concentration of 100 μM each. Total hepatic lipids were extracted using Bligh and Dyer's method (Bligh and Dyer 1959) with minor modifications. In brief, 50 μL of the 100 μM internal standards were added to tissue homogenates and the lipids were extracted by adding MeOH/CHCl3 (v/v, 2/1) in the presence of dibutylhydroxytoluene (BHT) to limit oxidation. The CHCl3 layer was collected and dried under N2 flow. The dried lipid extract was dissolved in 1 ml of MeOH/CHCl3 (v/v, 2/1) containing 5 mM of ammonium acetate for injection. The solution containing the lipid extract was pumped into a TripleTOF 5600 mass spectrometer (AB Sciex LLC, Framingham, MA, USA) at a flow rate of 40 μL/min for 2 min for each ionization mode. The lipid extracts were analyzed in both positive and negative ion modes for complete lipidome coverage using the TripleTOF 5600 System. Infusion MS/MSALL workflow experiments were conducted consisting of a TOF MS scan from m/z 200–1200 followed by the sequential acquisition of 1001 MS/MS spectra acquired from m/z 200 to 1200 (Simons et al., 2012). The total time required to obtain a comprehensive profile of the lipidome was approximately 10 min per sample. The data were acquired with high resolution (>30,000) and high mass accuracy (∼5 ppm RMS). The data processing using LipidView Software identified 150–300 lipid species covering diverse lipids classes including major glycerophospholipids and sphingolipids. The peak intensities of each identified lipid across all samples were normalized against an internal standard from the same lipid class for the semi-quantitation purpose.
2.2. Targeted quantitation of phosphatidylinositol (PI) and lysophosphatidylinositol (LPI) lipids
A targeted lipidomic assay of LPI and PI lipids was developed using HPLC online electrospray ionization tandem mass spectrometry (LC/ESI/MS/MS). Lipid extracts of the cell lysates were prepared using Folch's extraction and normalized to the total protein.
Standard solutions: The standards used in this assay were purchased from Avanti Polar Lipids (LPI-16:0, LPI-18:0, LPI-18:1, LPI-20:4, and PI-38:4). The internal standards used for the analyses were LPI-17:1 and PI-34:1-d31, which were also purchased from Avanti Polar Lipids. Standard LPI and PI species at concentrations of 0, 5, 20, 100, 500, and 2000 ng/ml were prepared in 90% methanol containing 2 internal standards at a concentration of 500 ng/ml. A volume of 5 μl was injected into a triple quadrupole mass spectrometer (Shimadzu LCMS-8050) to generate internal standard calibration curves.
HPLC parameters: A silica column (2.1 × 50 mm, Luna Silica, 5 μm, Phenomenex) was used to separate the PI and LPI species. The mobile phases were A (water containing 10 mM ammonium acetate) and B (acetonitrile containing 10 mM ammonium acetate). Mobile phase B at 95% was used from 0 to 2 min at a flow rate of 0.3 ml/min. A linear gradient from 95% B to 50% B from 2 to 8 min was maintained at 50% B from 8 to 16 min, and a linear gradient from 50% B to 95% B from 16 to 16.1 min was maintained at 95% B from 16.1 to 24 min.
Mass spectrometer parameters: The HPLC eluent was directly injected into the Shimadzu LCMS-8050 and the analytes were ionized in the ESI negative mode. The analytes were quantified using selected reaction monitoring (SRM). The SRM transitions (m/z) were 571 → 255 for LPI-16:0, 599 → 283 for LPI-18:0, 597 → 281 for LPI-18:1, 619 → 303 for LPI-20:4, 885 → 241 for PI-38:4, and 583 → 267 for internal standard LPI-17:1 and 866 → 281 for internal standard PI-34:1-d31.
Data analysis: Software LabSolutions LCMS was used to obtain the peak area for both the internal standards and the LPI and PI species. The internal standard calibration curves were used to calculate the concentration of the LPI and PI species in the samples. All of the plasma LPI and PI species were normalized to the PI-34:1-d31 internal standard, all of the tissue LPI species were normalized to the 17-1 LPI internal standard, and all of the tissue PI species were normalized to the PI-34:1-d31 internal standard.
2.3. RNA isolation and quantitative real time-PCR
The total RNA was isolated using the RNeasy or TRIzol isolation methods following the manufacturers’ recommendations (Qiagen and Thermo Fisher). The RNA concentrations were quantified using Nanodrop 2000. The mRNA expression levels were calculated based on the ΔΔ-CT method. qPCR was conducted using an Applied Biosystems 7500 Real-Time PCR System. The primers used for qPCR are available on request.
2.4. Cell lines and cell culture conditions
Caki-1 and 786-O cell lines were obtained from American Type Culture Collection (ATCC) and confirmed to be mycoplasma free. The cells were maintained in McCoy's 5A Medium with 10% FBS and 1% Pen/Strep. The Caki-1 and 786-O expansions were propagated no longer than 18 consecutive passages.
2.5. Guide RNA design and MBOAT7−/− generation
The design of the MBOAT7 sgRNA was optimized using CRISPOR open source software [18]. Synthetic sgRNA were designed to target all of the transcript variants of MBOAT7. Prior to nucleofection, sgMBOAT7 at a working concentration of 30 uM and recombinant Cas9 protein were incubated together at a 6:1 ratio (sgMBOAT7:Cas9 protein) as recommended by the manufacturer (Synthego). Parental Caki-1 and 786-O cells were then nucleofected and single cell isolated for clonal expansion. After growing and passaging cells from a single cell suspension, multiple independent clonal populations were screened for knockout populations. The chromatographs from the clonal populations were screened for editing using a previously described method [19]. The populations were then validated using targeted mass spectrometry for the LPI and PI lipids, mRNA abundance by qPCR, and Western blotting.
2.6. Cell viability assay
At day zero, 2,000 cells were seeded in 96-well plates. The relative cell number was determined using Cell Titer Glo (Promega, Madison, WI, USA), according to the manufacturer's instructions. In this assay, we used a 1:1 ratio of buffer solution to growth media as the endpoint and measured the luminescence after 1 h of incubation. The time points were 48 h, 96 h, and 120 h.
2.7. Cell counting assay
The Caki-1 cells were seeded into a 24-well plate at densities of approximately 20,000 cells/well in 10% FBS containing media. The cells were lifted and counted every day for 5 days. In brief, media was aspirated from the respective cell wells, then trypsinized for 5 min followed by neutralization with serum containing media. The cells were then pelleted at 500×g (rcf) for 5 min. The supernatants were aspirated and then the cells were counted using an automated cell counter (Z series, Beckman Coulter). The data were represented at n = 4 for each cell line for each day was used. Statistical analyses were conducted using one-way ANOVA with Tukey's post-test. Statistical significance was set at p < 0.05.
2.8. Cell cycle analysis
The cell cycle analysis was conducted using a previously described method from BioLegend (Cat# 640914). In brief, we plated 35 mm plates (n = 4 per group) seeded at 200,000 cells for 2 days. The cells were then isolated, washed, and fixed in 70% methanol. The cells were then stained at room temperature in the dark in propidium iodide (Cat# 421301) and Annexin V for 15 min. The cells were then analyzed using LSRFortessa (BD Biosciences). FlowJo software (Tree Star, Inc.) was used to determine the propidium iodide staining after excluding PI+ Annexin V+ apoptotic cells.
2.9. Migration assay
In vitro scratching was performed as previously described [20]. Briefly, prior to seeding the cells, 6-well plates were coated with an extracellular matrix. Approximately 200,000 cells were plated into each well in the 6-well plate. The wells grew to 90–100% confluency. After reaching confluency, the plates were scratched and imaged for 16 h every 10 min. Three separate experiments were conducted with 2 replicates per experiment. The experiments were quantified using the scratch area of the first and last images. The percent change in area was quantified using ImageJ software by using the following formula:
2.10. In vivo ccRCC xenograft studies
MBOAT7+/+ and MBOAT7−/− Caki-1 cell lines were injected into the subcutaneous flank of NSG mice (Jackson Laboratory) at 2.5 million cells per mouse in PBS (n = 10 per group). Once tumors were palpable, digital caliper measurements were used to follow the tumor growth over time. All of the mice studies were approved by the Institutional Animal Care and Use Committee of the Cleveland Clinic.
2.11. Western blotting
Cell tissue lysates were generated using a modified RIPA buffer and western blotting was performed following previously described methods [21].
2.12. Data sharing/open access
The RNAseq dataset produced in this manuscript is available through GEO Datasets at GSE131881.
2.13. Statistical analysis
All of the figures are shown as mean ± SE. To compare three groups, we utilized one-way ANOVA with Tukey's post hoc test. The Mantel–Cox log-rank test was used to compare the survival differences between two groups of patients. When comparing two groups, we used an unpaired Student's t-test or multiple t-tests. The significance cutoffs for all of the tests were p-value <0.05(*), 0.002 (**), 0.0002 (***), and <0.0001 (****).
3. Results
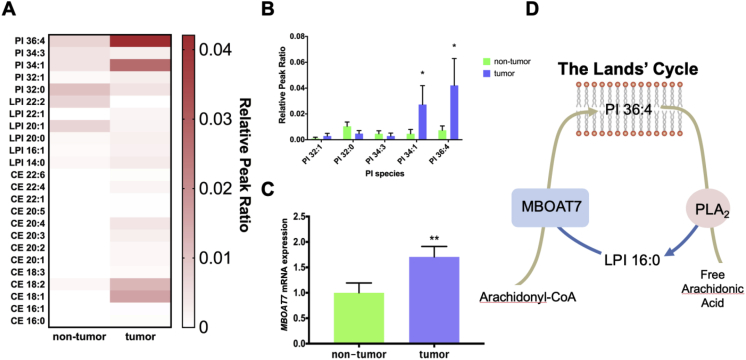
MBOAT7 and its enzymatic product PI 36:4 increased in ccRCC tumors. To study the possible lipid-related drivers of ccRCC, we obtained primary tumor biopsies with matched normal adjacent tissue samples from patients at the Cleveland Clinic and performed untargeted lipidomics (Figure 1A). One of the most significantly increased lipids in the tumors compared to the non-tumors was PI 36:4 (Figure 1B). In parallel, the expression of the LPI acyltransferase enzyme MBOAT7 was elevated (Figure 1C). Previous literature demonstrated that MBOAT7 is the primary LPI acyltransferase enzyme involved in the enrichment of arachidonic acid into PI (AA-PI) [22] within the Lands’ cycle of phospholipid remodeling (Figure 1D). The most striking differences between the groups were the inositol phospholipids; however, other minor changes occurred in the other phospholipid groups (Supplementary Fig. 1).
Figure 1.
Arachidonic acid containing PI (AA-PI) and MBOAT7 are elevated in ccRCC tumors. (A) Heatmap of the most significantly altered lipids in untargeted lipidomic profiling of metastatic ccRCC tumor samples and matched non-tumor samples. (B) Phosphatidylinositol (PI) species in the tumor and non-tumor samples. (C) mRNA levels of MBOAT7, a key acyltransferase determining AA-PI levels. (D) Schematic representation of the Lands' cycle of PI remodeling. Data presented are n = 10 per group, and all of the data are presented as mean ± S.E.M. unless otherwise noted. Student's t-test: *<0.05, **<0.0021, and ***<0.0002.
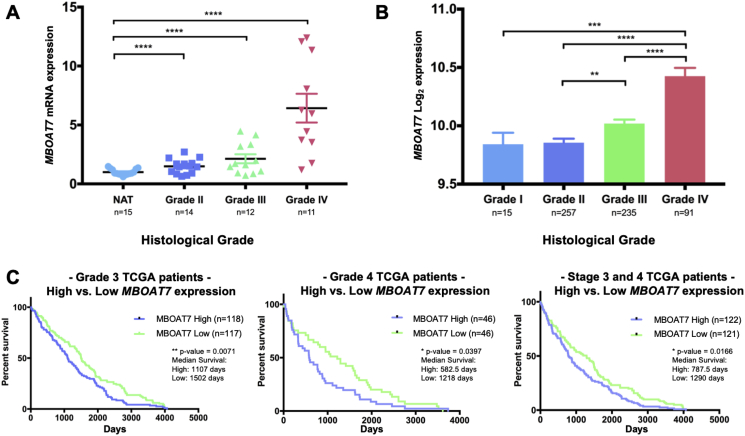
MBOAT7 increased with ccRCC severity and correlated with poor survival. To explore the association of MBOAT7 expression with the histological ccRCC grade, we analyzed nephrectomy samples from the initial surgical resections with age-, BMI-, and sex-matched patients. Interestingly, the MBOAT7 expression increased with the histological ccRCC grade, particularly in Grade 4 cases (Figure 2A). Furthermore, a commonly used ccRCC cell line Caki-1 had an increased expression of MBOAT7 similar to that of the Grade 4 tumor samples (Supplementary Fig. 2). There was no change in phospholipase A2 group IVA (PLA2G4A) expression across the histological tumor grades (Supplementary Fig. 2). We next confirmed the increased MBOAT7 expression with the histological grade in a validation cohort of patient samples. The Cancer Genome Atlas (TCGA) ccRCC cohort (KIRC) demonstrated a similar finding that the MBOAT7 expression increased with the histological severity (Figure 2B). To assess the relationship between MBOAT7 mRNA expression and patient survival, we accessed TCGA datasets and stratified patients into low vs high MBOAT7 mRNA levels. High MBOAT7 expression correlated with a poorer overall survival within the histology matched cohorts or Stage III/IV patients (Figure 2C). The data demonstrate that MBOAT7 and enzymatic substrates or products may play a role in the progression of ccRCC.
Figure 2.
MBOAT7 expression increases with ccRCC severity and correlates with poorer survival. (A) MBOAT7 mRNA expression in ccRCC patients with increasing severity of histological grade in patient tumor biopsies. NAT = normal adjacent tissue. (B) A validation dataset from the cancer genome atlas (TCGA) stratified by pathological staging (I-IV) and pan-cancer normalized. (C) High MBOAT7 expression significantly correlated with a decrease in overall survival in TCGA patients stratified by Grade 3 and Grade 4 histological neoplasms. Similarly, high MBOAT7 expression correlated with a decrease in overall survival with Stage III and IV patients. One-way ANOVA and log-rank test (Mantel–Cox test) ns > 0.1234, *<0.05, **<0.002, and ***<0.0002.
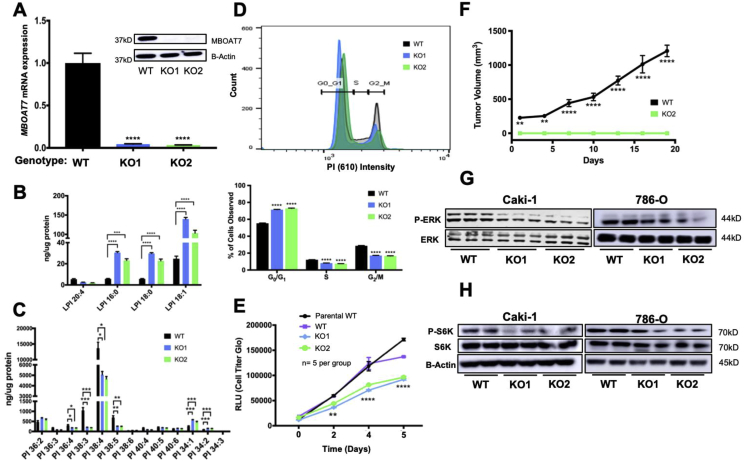
Generation and validation of MBOAT7 loss of function in ccRCC cell lines. To assess the role of MBOAT7 in ccRCC, we utilized CRISPR/Cas9 to knockout MBOAT7 in the Caki-1 and 786-O ccRCC cell lines. Following clonal isolation, we confirmed knockout using sequencing across exon 6 (Supplementary Fig. 2) as well as quantification of the mRNA and protein abundance (Figure 3A). We then validated this loss of enzymatic function with targeted mass spectrometry of MBOAT7 LPI substrates and PI product species in targeted cells. MBOAT7 acts on LPI 16:0 to generate a series of AA-PI species including PI 36:4. Thus, because of MBOAT7deletion, the levels of LPI substrates were significantly increased in the MBOAT7−/− clones (Figure 3B) and the set of AA-PI (36:4, 38:4, 38:5) products were significantly attenuated (Figure 3C). MBOAT7 deficiency led to a 4-fold increase in the total LPI species and an approximately 50% reduction in the total PI (Supplementary Fig. 3). The functional changes resulting from MBOAT7 knockout were finally validated by providing exogenous 3H-aracidonic acid to MBOAT7 null cells. Both MBOAT7−/− clones exhibited a ∼90% reduction in AA incorporation into PI species (Supplementary Fig. 4).
Figure 3.
Genetic deletion of MBOAT7 in ccRCC results in reduced arachidonic acid-PI, cell cycle, proliferation, and growth factor signaling. (A) Generation of Caki-1 MBOAT7 knockout (KO) cell lines KO1 and KO2. (B) Targeted mass spectrometry of Caki-1 WT, KO1, and KO2 cell lines showed increased LPI substrate in MBOAT7 KO1 and KO2. (C) Targeted PI mass spectrometry of Caki-1 WT, KO1, and KO2 showed significantly decreased arachidonic acid containing PI (AA-PI). (D) Cell cycle analysis in WT, KO1, and KO2 cell lines (n = 4 per group). (E) Proliferation assay (Cell Titer Glo) of Caki-1 MBOAT7 KO cell lines, WT, and parental Caki-1 plated at 2,000 cells/well at day 0. The results were confirmed by three independent experiments. (F) In vivo xenograft model with subcutaneous injection of MBOAT7 WT vs KO2 tumor growth showed decreased engraftment of MBOAT7 KO in vivo. (G) Western blotting of asynchronous signaling via phospho-ERK1/2 and total-ERK1/2. (H) Western blotting of phospho-S6K and total-S6K signaling in WT, KO1, and KO2 cell lines. All of the results shown are representative of 3–5 biological replicates that were repeated in 2 independent experiments. Multiple and Student's t-test with p-value significance: ns > 0.1234, *<0.05, **<0.002, ***<0.0002, and ****<0.0001.
Loss of MBOAT7 was associated with decreased cell proliferation, cell cycle, and in vivo tumor formation. To assess the phenotypic changes in MBOAT7+/+ and MBOAT7−/−, we conducted cell proliferation, cell cycle, and subcutaneous xenograft growth experiments. When the cell cycle was analyzed, the MBOAT7−/− cells increased in the G0/G1 phase and decreased in the S and G2/M phases (Figure 3D). In parallel, the MBOAT7−/− cells showed a significant reduction in proliferation up to ∼35% over 5 days (Figure 3E). Pharmacological inhibition of MBOAT7 with thimerosal showed striking reductions in the in vitro cell viability (Supplementary Fig. 3). To test whether this had any effect on tumor growth in vivo, we conducted in vivo xenograft experiments. Approximately 8 weeks post-injection, the MBOAT7+/+ cells had reached the endpoint in the tumor growth experiment, but the MBOAT7−/− failed to develop a palpable tumor (Figure 3F). A common growth signaling pathway related to ccRCC cellular proliferation is the mitogen-activated protein kinase (MAPK) cascade [[23], [24], [25]]. In an asynchronous culture, the MBOAT7−/− cells had reduced phospho-ERK1/2 levels (Figure 3G). Upon synchronizing the cells, the Caki-1 MBOAT7−/− cells demonstrated a blunted platelet-derived growth factor stimulated response in phospho-ERK1/2 activation (Supplementary Fig. 3). The MBOAT7−/− cells had reduced P–S6K activation (Figure 3H).
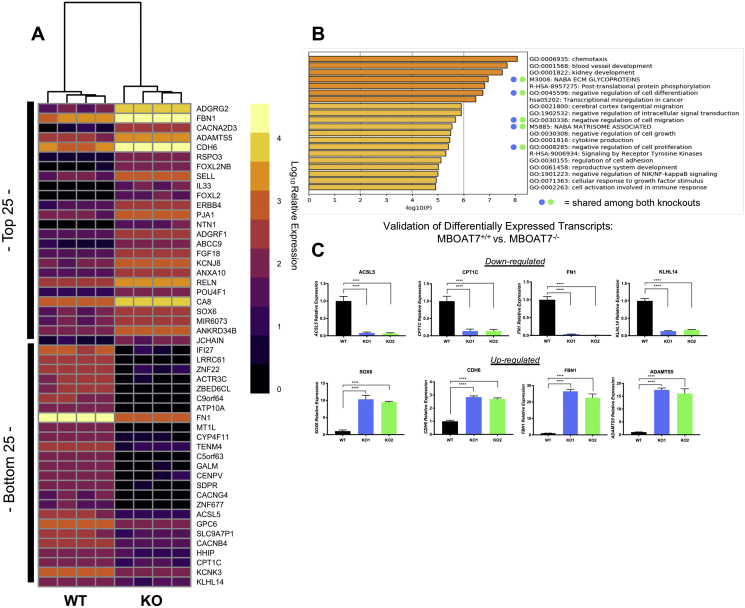
MBOAT7−/− ccRCC cells had differentially regulated motility, proliferation, and matrix organization. We conducted an unbiased RNAseq of the MBOAT7+/+ and MBOAT7−/− Caki-1 cells. The principal component analyses (PCA) of both knockout populations demonstrated that the majority of the variations were defined by the loss of MBOAT7 function (Supplementary Fig. 5). The unbiased analysis demonstrated a group of genes conserved and differentially regulated among both knockout populations compared to wild type (Figure 4A). A pathway analysis of both knockouts revealed a conserved significant enrichment in NABA ECM glycoproteins, negative regulation of cell differentiation, negative regulation of cell migration, and negative regulation of cell proliferation (Figure 4B). To validate the RNAseq dataset, we conducted qRT-PCR on four upregulated (SOX6, CDH6, FBN1, and ADAMTS5) and four downregulated (ACSL5, CPT1C, FN1, and KLHL14) transcripts (Figure 4C). Functionally, to follow up the RNAseq pathway results, we conducted an in vitro migration assay to assess the potential differences in motility.
Figure 4.
RNA sequencing in MBOAT7 deficient ccRCC cells reveals alterations in cell cycle, matrix organization, and cell migration. (A) Heatmap of the top 25 and bottom 25 differentially expressed transcripts from unbiased RNAseq. (B) Pathway analysis revealed regulation of migration, cell proliferation, and extracellular organization from the top 250 significantly upregulated transcripts. (C) qPCR validation of selected genes altered in MBOAT7 null Caki-1 cells. RNAseq and qPCR validation data represent n = 4 per group biological replicates. Student's t-test and p-value: *<0.05, **<0.002, ***<0.0002, and ****<0.0001.
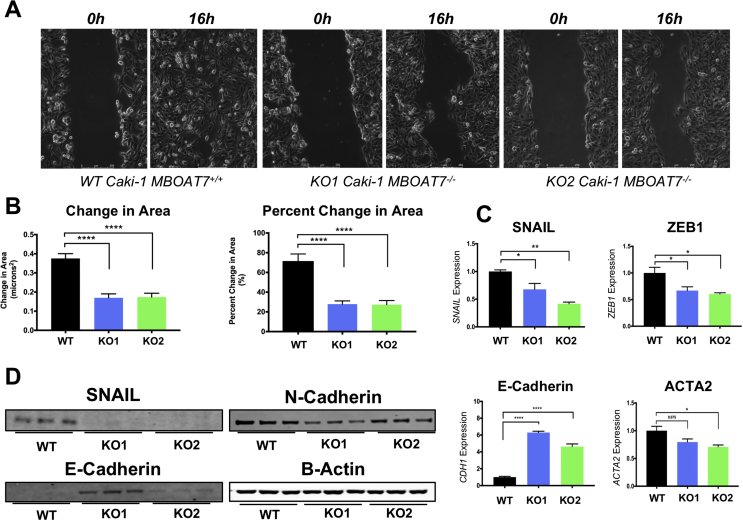
MBOAT7−/− ccRCC cells showed decreased migration and less mesenchymal phenotype. The RNAseq pathway analysis suggested decreased migration and matrix organization. Using an in vitro scratch assay, we found that MBOAT7−/− clones demonstrated a ∼50% reduction in motility (Figure 5B). Videos of the MBOAT7 loss of function also demonstrate a slower and disorganized migratory pattern (Supplementary Movie 1-3). To further explore potential differences in motility using qPCR, we assessed the expression of specific mesenchymal and epithelial genes SNAIL, ZEB1, ACTA2, and CDH1. The MBOAT7−/− had decreased expression of mesenchymal genes SNAIL, ZEB1, and ACTA2, yet increased expression of the epithelial marker E-cadherin (CDH1) (Figure 5C). At the protein level, a similar loss of mesenchymal markers and increased epithelial markers occurred in the MBOAT7−/− cells (Figure 5D). These data suggest that MBOAT7-driven phospholipid remodeling is involved in determining epithelial to mesenchymal transition (EMT).
Figure 5.
MBOAT7 regulates in vitro migration and epithelial/mesenchymal markers. (A) Representative micrographs of MBOAT7+/+ and MBOAT7−/− cells showing in vitro scratch assay (time 0 and time 16 h). See the online supplement for time lapse images. (B) Quantification of change in area and percent change in area in the in vitro scratches. (C) qPCR of MBOAT7+/+ and MBOAT7−/− demonstrated altered expression of epithelial and mesenchymal marker genes. (D) Western blotting showed a decrease in mesenchymal markers and increase in epithelial markers with the loss of MBOAT7. All of the results shown are representative of one experiment that was replicated in at least 2–3 independent experiments (n = 2–5 per group). Student's t-test and p-value: *<0.05, **<0.002, ***<0.0002, and ****<0.0001
Supplementary video related to this article can be found at https://doi.org/10.1016/j.molmet.2020.01.011
The following are the supplementary data related to this article:
4. Discussion
Similar to other solid tumors, ccRCC is primarily driven by the loss of key tumor suppressors such as von Hippel-Lindau (VHL) and the activation of oncogenes [[26], [27], [28]]. Closely associated with these genetic alterations, lipid metabolic processes are dramatically altered in the ccRCC tumor microenvironment [3,4,[29], [30], [31], [32], [33], [34], [35], [36]]. The “clear cell” subtype gets its name from cholesterol ester-rich lipid droplets that accumulate in the tumor cells [35,36]. Although ccRCC can be characterized as a “lipid metabolic disease” [[37], [38], [39]], it remains incompletely understood which lipid metabolic targets are therapeutically tractable. This study identified MBOAT7-driven PI remodeling as a new potential drug target. The key findings from this study are (1) in addition to the accumulation of cholesteryl ester [35,36], metastatic ccRCC is characterized by the accumulation of AA-PI lipids and the elevated expression of AA-PI synthesizing enzyme MBOAT7; (2) MBOAT7 expression is elevated in high grade ccRCC, and high MBOAT7 expression is associated with poor survival; (3) the genetic deletion of MBOAT7 in ccRCC cells results in reduced AA-PI levels and reciprocal increases in substrate LPIs; (4) MBOAT7 null ccRCC cells exhibit reduced proliferation associated with cell cycle arrest and fail to form tumors in vivo; (5) MBOAT7 null ccRCC cells have reduced growth factor-driven MAPK activation; and (6) unbiased RNA sequencing reveals that MBOAT7/AA-PI may play a role in regulating ccRCC migration and epithelial to mesenchymal transition (EMT). Collectively, our findings identify a key new lipid metabolic dependency in ccRCC and show that the inhibition of MBOAT7-driven remodeling of membrane PI lipids may have therapeutic potential in ccRCC patients.
The closely related signaling network of phosphoinositide 3-kinase (PI3K), protein kinase B (AKT), and mammalian target of rapamycin (mTOR) controls many aspects of cancer initiation and progression. Given the central role that the PI3K-AKT-mTOR pathway plays in cancer cell biology, many developmental and approved cancer drugs target key nodes within the pathway [[40], [41], [42]]. Although many drugs that currently target the PI3K-AKT-mTOR pathway are designed for kinase inhibition, signaling through the PI3K-AKT-mTOR pathway is sustained by the metabolism of key lipids such as PI [9,[43], [44], [45], [46]]. The PI pool in the inner leaflet of the plasma membrane is quantitatively small compared to other phospholipids but serves as a key reservoir for growth factor and hormone stimulated generation of phosphatidylinositol phosphate (PIP) lipids [[47], [48], [49]]. Key PI-derived lipids such as phosphatidylinositol 4-phosphate (PI4P), phosphatidylinositol 4,5-bisphosphate (PI-4,5-P2), and inositol 1,4,5-triphosphate (IP3) play critical roles in normal hormones, growth factor signaling, and aberrant signaling processes that drive malignancy [47,48,50,51], and as a result, many anti-cancer drugs aim to alter levels of these critical signaling lipids. However, very little attention has been paid to factors that regulate the abundance or composition of the membrane PI pool where PIPs originate. In this study, we identified a lipid ccRCC signature that is characterized by specific alterations in arachidonic acid-containing membrane PI species (AA-PI), which is likely to have downstream regulatory roles in PIP-dependent signaling. Supporting this hypothesis, it was recently demonstrated that MBOAT7 knockout mice have a selective reduction in arachidonic acid-containing PIP species [15]. The decrease in the PI pool may have a broader impact on cell function than PIP3 formation, as this may also impact vesicular trafficking. The decreased response to growth factor stimulation may be due to decreased PtdIns(4)P in the endosomal compartment, which is important for growth factor receptor sorting [52,53]. Currently, it is incompletely understood how the acyl chain composition of PIPs can impact downstream kinase activation, and our studies suggest that this is an area of research worth exploring across many diverse cancers.
Given the striking lipid accumulation seen in ccRCC, it remains entirely possible that abnormal lipid metabolism in the tumor microenvironment is mechanistically involved in tumor initiation and/or progression. However, the key lipid metabolic targets that are therapeutically tractable must be identified. Prior lipidomic studies identified a striking accumulation of cholesterol esters in the cytosolic lipid droplets of ccRCC patients [3,36]. However, it is unlikely that this inert storage form of lipid plays a causal role in ccRCC pathogenesis and instead it is likely a result of altered cholesterol handling in transformed cells. A recent study suggested that the esterification of fatty acids into triglycerides may provide a “buffer” for the toxic effects of saturated ceramides and acylcarnitines [33]. However, in contrast to the considerable accumulation of cholesterol ester, human ccRCC tumors are not particularly rich in stored triglycerides, demonstrating that this “buffering” capacity is likely limited. There is also evidence that hypoxia-inducible factor (HIF) driven transcriptional repression of carnitine palmitoyltransferase (CPT1A) plays a role in ccRCC tumorigenesis, but re-expressing a silenced gene in cancer cells is challenging. Furthermore, HIF2α hyperactivity in ccRCC cells can promote the expression of the lipid droplet-associated protein perilipin 2 (PLIN2), which facilitates lipid storage and indirectly blunts endoplasmic reticulum (ER) stress pathways [4]. PLIN2 has been identified as a potential urinary biomarker of advance ccRCC [54,55], providing additional rationale for developing therapeutic strategies to intervene in this HIF2α-PLIN2 pathway. Collectively, these recently described lipid metabolic pathways altered in ccRCC provide potential targets for drug discovery. This study expands this list to include MBOAT7 as a potential lipid metabolic target that is therapeutically tractable using several platforms. A semi-selective small molecule MBOAT7 inhibitor (thimerosal) [14] has shown some promise as a potential cancer therapeutic [[56], [57], [58]]. However, thimerosal likely has many enzymatic targets, and additional structure-activity-relationship (SAR) drug discovery efforts are needed to find more selective inhibitors. Collectively, our work suggests that selective MBOAT7 inhibitors may hold promise to blunt the progression of metastatic ccRCC, and these findings may be broadly related to other cancer types in which the PI3K-AKT-mTOR axis is hyperactive.
Author contributions
C.K.A.N., J.D.S., J.D.L., B.I.R., and J.M.B. planned the project, designed the experiments, and wrote the manuscript. C.K.A.N., R.Z., D.J.S., C.A.T., V.V., and D.B. conducted the experiments. C.K.A.N., R.Z., D.J.S., V.V., and D.B. analyzed the data. J.D.L., B.I.R., and J.M.B. provided financial support. All of the authors were involved in editing the final manuscript.
Acknowledgments
The authors thank the patients at Taussig Cancer Center at the Cleveland Clinic who donated their tumor samples to this study and the Lathia Laboratory for constructive directions on this research project. This study was supported in part by grants from the National Institutes of Health R01 HL122283 (J.M.B.), P50 AA024333 (J.M.B.), and P01 HL147823 (J.M.B.). The development of the lipid mass spectrometry methods reported in this study was supported by generous pilot grants from the Clinical and Translational Science Collaborative of Cleveland (4UL1TR000439), the National Center for Advancing Translational Sciences of NIH, the NIH Roadmap for Medical Research, the Case Comprehensive Cancer Center (P30 CA043703), the VeloSano Foundation, and a Cleveland Clinic Research Center of Excellence Award. D.B. was supported by a Case Comprehensive Cancer Center training grant (T32CA059366).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.01.011.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Cohen H.T., McGovern F.J. Renal-cell carcinoma. New England Journal of Medicine. 2005;353(23):2477–2490. doi: 10.1056/NEJMra043172. [DOI] [PubMed] [Google Scholar]
- 2.Lauby-Secretan B., Scoccianti C., Loomis D., Grosse Y., Bianchini F., Straif K. Body fatness and cancer — viewpoint of the IARC working group. New England Journal of Medicine. 2016;375(8):794–798. doi: 10.1056/NEJMsr1606602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzalez R., Goldberg M.E. Origin of intracellular cholesterol in renal-cell carcinoma. Lancet (London, England) 1977;1(8017):912. doi: 10.1016/s0140-6736(77)91245-4. [DOI] [PubMed] [Google Scholar]
- 4.Qiu B., Ackerman D., Sanchez D.J., Li B., Ochocki J.D., Grazioli A. HIF2α-Dependent lipid storage promotes endoplasmic reticulum homeostasis in clear-cell renal cell carcinoma. Cancer Discovery. 2015;5(6):652–667. doi: 10.1158/2159-8290.CD-14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shewan A., Eastburn D.J., Mostov K. Phosphoinositides in cell architecture. Cold Spring Harbor Perspectives in Biology. 2011;3(8) doi: 10.1101/cshperspect.a004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janetopoulos C., Borleis J., Vazquez F., Iijima M., Devreotes P. Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Developmental Cell. 2005;8(4):467–477. doi: 10.1016/j.devcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 7.Janetopoulos C., Devreotes P. Phosphoinositide signaling plays a key role in cytokinesis. The Journal of Cell Biology. 2006;174(4):485–490. doi: 10.1083/jcb.200603156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hehnly H., Doxsey S. Polarity sets the stage for cytokinesis. Molecular Biology of the Cell. 2012;23(1):7–11. doi: 10.1091/mbc.E11-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitman M., Kaplan D.R., Schaffhausen B., Cantley L., Roberts T.M. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315(6016):239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 10.Fleischman L.F., Chahwala S.B., Cantley L. ras-transformed cells: altered levels of phosphatidylinositol-4,5-bisphosphate and catabolites. Science (New York, N.Y.) 1986;231(4736):407–410. doi: 10.1126/science.3001936. [DOI] [PubMed] [Google Scholar]
- 12.Kennedy E.P., Weiss S.B. The function of cytidine coenzymes in the biosynthesis of phospholipides. Journal of Biological Chemistry. 1956;222(1):193–214. [PubMed] [Google Scholar]
- 13.Lands W.E. Metabolism of glycerolipides; a comparison of lecithin and triglyceride synthesis. Journal of Biological Chemistry. 1958;231(2):883–888. [PubMed] [Google Scholar]
- 14.Gijón M.A., Riekhof W.R., Zarini S., Murphy R.C., Voelker D.R. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. Journal of Biological Chemistry. 2008;283(44):30235–30245. doi: 10.1074/jbc.M806194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson K.E., Kielkowska A., Durrant T.N., Juvin V., Clark J., Stephens L.R. Lysophosphatidylinositol-Acyltransferase-1 (LPIAT1) is required to maintain physiological levels of PtdIns and PtdInsP2 in the mouse. PloS One. 2013;8(3) doi: 10.1371/journal.pone.0058425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee H.-C., Inoue T., Sasaki J., Kubo T., Matsuda S., Nakasaki Y. LPIAT1 regulates arachidonic acid content in phosphatidylinositol and is required for cortical lamination in mice. Molecular Biology of the Cell. 2012;23(24):4689–4700. doi: 10.1091/mbc.E12-09-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gromovsky A.D., Schugar R.C., Brown A.L., Helsley R.N., Burrows A.C., Ferguson D. Δ-5 fatty acid desaturase FADS1 impacts metabolic disease by balancing proinflammatory and proresolving lipid mediators. Arteriosclerosis, Thrombosis, and Vascular Biology. 2018;38(1):218–231. doi: 10.1161/ATVBAHA.117.309660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.-B. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biology. 2016;17(1):148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dehairs J., Talebi A., Cherifi Y., Swinnen J.V. CRISP-ID: decoding CRISPR mediated indels by Sanger sequencing. Scientific Reports. 2016;6 doi: 10.1038/srep28973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang C.-C., Park A.Y., Guan J.-L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nature Protocols. 2007;2(2):329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 21.Brown J.M., Boysen M.S., Chung S., Fabiyi O., Morrison R.F., Mandrup S. Conjugated linoleic acid induces human adipocyte delipidation. Journal of Biological Chemistry. 2004;279(25):26735–26747. doi: 10.1074/jbc.M401766200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shindou H., Shimizu T. Acyl-CoA:Lysophospholipid acyltransferases. Journal of Biological Chemistry. 2009;284(1):1–5. doi: 10.1074/jbc.R800046200. [DOI] [PubMed] [Google Scholar]
- 23.Huang D., Ding Y., Luo W.-M., Bender S., Qian C.-N., Kort E. Inhibition of MAPK kinase signaling pathways suppressed renal cell carcinoma growth and angiogenesis in vivo. Cancer Research. 2008;68(1):81–88. doi: 10.1158/0008-5472.CAN-07-5311. [DOI] [PubMed] [Google Scholar]
- 24.Sang N., Stiehl D.P., Bohensky J., Leshchinsky I., Srinivas V., Caro J. MAPK signaling up-regulates the activity of hypoxia-inducible factors by its effects on p300. Journal of Biological Chemistry. 2003;278(16):14013–14019. doi: 10.1074/jbc.M209702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katz M., Amit I., Yarden Y. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochimica et Biophysica Acta. 2007;1773(8):1161–1176. doi: 10.1016/j.bbamcr.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey S.T., Smith A.M., Kardos J., Wobker S.E., Wilson H.L., Krishnan B. MYC activation cooperates with Vhl and Ink4a/Arf loss to induce clear cell renal cell carcinoma. Nature Communications. 2017;8:15770. doi: 10.1038/ncomms15770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao X., Tan J., Lim K.J., Koh J., Ooi W.F., Li Z. VHL deficiency drives enhancer activation of oncogenes in clear cell renal cell carcinoma. Cancer Discovery. 2017;7(11):1284–1305. doi: 10.1158/2159-8290.CD-17-0375. [DOI] [PubMed] [Google Scholar]
- 28.Sato Y., Yoshizato T., Shiraishi Y., Maekawa S., Okuno Y., Kamura T. Integrated molecular analysis of clear-cell renal cell carcinoma. Nature Genetics. 2013;45(8):860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 29.Fu L., Wang G., Shevchuk M.M., Nanus D.M., Gudas L.J. Generation of a mouse model of Von Hippel-Lindau kidney disease leading to renal cancers by expression of a constitutively active mutant of HIF1α. Cancer Research. 2011;71(21):6848–6856. doi: 10.1158/0008-5472.CAN-11-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du W., Zhang L., Brett-Morris A., Aguila B., Kerner J., Hoppel C.L. HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. Nature Communications. 2017;8(1):1769. doi: 10.1038/s41467-017-01965-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu G.-H., Lou N., Shi H.-C., Xu Y.-C., Ruan H.-L., Xiao W. Up-regulation of SR-BI promotes progression and serves as a prognostic biomarker in clear cell renal cell carcinoma. BMC Cancer. 2018;18(1):88. doi: 10.1186/s12885-017-3761-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez D.J., Steger D.J., Skuli N., Bansal A., Simon M.C. PPARγ is dispensable for clear cell renal cell carcinoma progression. Molecular Metabolism. 2018;14:139–149. doi: 10.1016/j.molmet.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ackerman D., Tumanov S., Qiu B., Michalopoulou E., Spata M., Azzam A. Triglycerides promote lipid homeostasis during hypoxic stress by balancing fatty acid saturation. Cell Reports. 2018;24(10):2596–2605. doi: 10.1016/j.celrep.2018.08.015. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Syafruddin S.E., Rodrigues P., Vojtasova E., Patel S.A., Zaini M.N., Burge J. A KLF6-driven transcriptional network links lipid homeostasis and tumour growth in renal carcinoma. Nature Communications. 2019;10(1):1152. doi: 10.1038/s41467-019-09116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gebhard R.L., Clayman R.V., Prigge W.F., Figenshau R., Staley N.A., Reesey C. Abnormal cholesterol metabolism in renal clear cell carcinoma. Journal of Lipid Research. 1987;28(10):1177–1184. [PubMed] [Google Scholar]
- 36.Clayman R.V., Figenshau R.S., Prigge W.F., Forstrom L., Gebhard R.L. Transport of circulating serum cholesterol by human renal cell carcinoma. The Journal of Urology. 1987;137(6):1262–1265. doi: 10.1016/s0022-5347(17)44478-8. [DOI] [PubMed] [Google Scholar]
- 37.Linehan W.M., Srinivasan R., Schmidt L.S. The genetic basis of kidney cancer: a metabolic disease. Nature Reviews Urology. 2010;7(5):277–285. doi: 10.1038/nrurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez D.J., Simon M.C. Genetic and metabolic hallmarks of clear cell renal cell carcinoma. Biochimica Et Biophysica Acta Reviews on Cancer. 2018;1870(1):23–31. doi: 10.1016/j.bbcan.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu C.-A.A., Klopfer E.I., Ray P.E. Human apolipoprotein L1 (ApoL1) in cancer and chronic kidney disease. FEBS Letters. 2012;586(7):947–955. doi: 10.1016/j.febslet.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esposito A., Viale G., Curigliano G. Safety, tolerability, and management of toxic effects of phosphatidylinositol 3-kinase inhibitor treatment in patients with cancer: a review. JAMA Oncology. 2019 doi: 10.1001/jamaoncol.2019.0034. [DOI] [PubMed] [Google Scholar]
- 41.Fruman D.A., Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nature Reviews Drug Discovery. 2014;13(2):140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu P., Cheng H., Roberts T.M., Zhao J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nature Reviews Drug Discovery. 2009;8(8):627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macara I.G., Marinetti G.V., Balduzzi P.C. Transforming protein of avian sarcoma virus UR2 is associated with phosphatidylinositol kinase activity: possible role in tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(9):2728–2732. doi: 10.1073/pnas.81.9.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaplan D.R., Whitman M., Schaffhausen B., Pallas D.C., White M., Cantley L. Common elements in growth factor stimulation and oncogenic transformation: 85 kd phosphoprotein and phosphatidylinositol kinase activity. Cell. 1987;50(7):1021–1029. doi: 10.1016/0092-8674(87)90168-1. [DOI] [PubMed] [Google Scholar]
- 45.Kaplan D.R., Whitman M., Schaffhausen B., Raptis L., Garcea R.L., Pallas D. Phosphatidylinositol metabolism and polyoma-mediated transformation. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(11):3624–3628. doi: 10.1073/pnas.83.11.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diringer H., Friis R.R. Changes in phosphatidylinositol metabolism correlated to growth state of normal and Rous sarcoma virus-transformed Japanese quail cells. Cancer Research. 1977;37(9):2979–2984. [PubMed] [Google Scholar]
- 47.Carney D.H., Scott D.L., Gordon E.A., LaBelle E.F. Phosphoinositides in mitogenesis: neomycin inhibits thrombin-stimulated phosphoinositide turnover and initiation of cell proliferation. Cell. 1985;42(2):479–488. doi: 10.1016/0092-8674(85)90105-9. [DOI] [PubMed] [Google Scholar]
- 48.Cantley L.C., Whitman M., Chahwala S., Fleischman L., Kaplan D.R., Schaffhausen B.S. Oncogenes and phosphatidylinositol turnover. Annals of the New York Academy of Sciences. 1986;488:481–490. doi: 10.1111/j.1749-6632.1986.tb46580.x. [DOI] [PubMed] [Google Scholar]
- 49.Berridge M.J., Heslop J.P., Irvine R.F., Brown K.D. Inositol trisphosphate formation and calcium mobilization in Swiss 3T3 cells in response to platelet-derived growth factor. Biochemical Journal. 1984;222(1):195–201. doi: 10.1042/bj2220195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitman M.R., Epstein J., Cantley L. Inositol 1,4,5-trisphosphate stimulates phosphorylation of a 62,000-dalton protein in monkey fibroblast and bovine brain cell lysates. Journal of Biological Chemistry. 1984;259(22):13652–13655. [PubMed] [Google Scholar]
- 51.Rittenhouse S.E., Sasson J.P. Mass changes in myoinositol trisphosphate in human platelets stimulated by thrombin. Inhibitory effects of phorbol ester. Journal of Biological Chemistry. 1985;260(15):8657–8660. [PubMed] [Google Scholar]
- 52.Henmi Y., Morikawa Y., Oe N., Ikeda N., Fujita A., Takei K. PtdIns4KIIα generates endosomal PtdIns(4)P and is required for receptor sorting at early endosomes. Molecular Biology of the Cell. 2016;27(6):990–1001. doi: 10.1091/mbc.E15-08-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zoncu R., Perera R.M., Sebastian R., Nakatsu F., Chen H., Balla T. Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(10):3793–3798. doi: 10.1073/pnas.0611733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morrissey J.J., Mellnick V.M., Luo J., Siegel M.J., Figenshau R.S., Bhayani S. Evaluation of urine aquaporin-1 and perilipin-2 concentrations as biomarkers to screen for renal cell carcinoma: a prospective cohort study. JAMA Oncology. 2015;1(2):204–212. doi: 10.1001/jamaoncol.2015.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song Y., Zhong L., Zhou J., Lu M., Xing T., Ma L. Data-independent acquisition-based quantitative proteomic analysis reveals potential biomarkers of kidney cancer. Proteomics. Clinical Applications. 2017;11(11–12) doi: 10.1002/prca.201700066. [DOI] [PubMed] [Google Scholar]
- 56.Chang H.-T., Liu C.-S., Chou C.-T., Hsieh C.-H., Chang C.-H., Chen W.-C. Thimerosal-induced cytosolic Ca2+ elevation and subsequent cell death in human osteosarcoma cells. Pharmacological Research. 2005;52(4):328–333. doi: 10.1016/j.phrs.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 57.Woo K.J., Lee T.-J., Bae J.H., Jang B.-C., Song D.-K., Cho J.-W. Thimerosal induces apoptosis and G2/M phase arrest in human leukemia cells. Molecular Carcinogenesis. 2006;45(9):657–666. doi: 10.1002/mc.20202. [DOI] [PubMed] [Google Scholar]
- 58.Liu S.-I., Huang C.-C., Huang C.-J., Wang B.-W., Chang P.-M., Fang Y.-C. Thimerosal-induced apoptosis in human SCM1 gastric cancer cells: activation of p38 MAP kinase and caspase-3 pathways without involvement of [Ca2+]i elevation. Toxicological Sciences: An Official Journal of the Society of Toxicology. 2007;100(1):109–117. doi: 10.1093/toxsci/kfm205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.