Abstract
Rationale:
Lower natriuretic peptide (NP) levels may contribute to the development of cardiometabolic diseases. Blacks have lower NP levels than middle-aged and older white adults. A high-carbohydrate challenge causes an upregulation of a negative ANP regulator microRNA-425 (miR-425) which reduces atrial-NP (ANP) levels in whites.
Objectives:
We designed a prospective trial to study racial differences in 1) NP levels among young adults, 2) NP response to a high-carbohydrate challenge, and 3) explore underlying mechanisms for race-based differences.
Methods and Results:
Healthy self-identified blacks and whites received three days of study diet followed by a high-carbohydrate challenge. Gene expression from whole blood RNA was assessed in the trial participants. Additionally, atrial and ventricular tissue samples from the Myocardial Applied Genomics Network (MAGNet) repository were examined for NP system gene expression. Among 72 healthy participants, we found that B-type-NP, N-terminal-pro-B-type-NP (NTproBNP), and mid-regional-pro-ANP (MRproANP) levels were 30%, 47%, and 18% lower in blacks compared with whites (p≤0.01), respectively. The decrease in MRproANP levels in response to a high-carbohydrate challenge differed by race [blacks 23% (95%CI: 19% to 27%) vs. whites 34% (95% CI: 31% to 38%); Pinteraction<0.001], with no change in NTproBNP levels. We did not observe any racial differences in expression of genes encoding for NPs (NPPA/NPPB) or NP signaling (NPR1) in atrial and ventricular tissues. NP processing (corin), clearance (NPR3) and regulation (miR-425) genes were ~3.5, ~2.5, and ~2 fold higher in blacks than whites in atrial tissues, respectively. We also found a 2-and 8-fold higher whole blood RNA expression of gene encoding for Neprilysin (MME) and miR-425 among blacks than whites.
Conclusions:
Racial differences in NP levels are evident in young healthy adults suggesting a state of NP deficiency exists in blacks. Impaired NP processing and clearance may contribute to race-based NP differences. Higher miR-425 levels in blacks motivate additional studies to understand differences in NP downregulation after physiological perturbations.
Clinical Trial Registration:
.
Subject Terms: Biomarkers, Clinical Studies, Mechanism, Physiology, Race and Ethnicity
Keywords: Natriuretic peptide, race, glucose, gene expression, cardiomyocytes
Graphical Abstract
INTRODUCTION
The heart is the primary source of natriuretic peptides (NPs)1 that circulate in two main isoforms; atrial NP (ANP) and B-type NP (BNP).1, 2 Prior studies have revealed substantial heritability of NP levels,3 and suggested a role for common genetic variants4, 5 in elucidating causes for variation in circulating NP levels. Large-scale human genetic studies have put forth the concept of “NP deficiency” (low NP levels) as a biological contributor to the development of cardiometabolic diseases such as hypertension and diabetes.5-7 We and others have observed that blacks have lower resting NP levels as compared with whites in middle-aged and older individuals.8-11 We have also established that the predictive ability of NP as a biomarker may be diminished in blacks with obesity or kidney dysfunction.12 Although these pivotal studies establishing black race as an NP deficient state were conducted in individuals free of prevalent cardiovascular and renal disease, NP levels were examined in older individuals (56 to 64 years),8-11 in whom “subclinical” cardiometabolic disease may exist.
Black individuals are at higher risk for cardiometabolic disease than whites.13 Important factors underlying this excess risk are higher rates of obesity, insulin resistance, and diabetes in blacks as compared to whites.14, 15 Population-based studies have suggested an inverse relationship between clinical components of cardiometabolic disease (i.e., obesity, high blood glucose, and dyslipidemia) and NP levels.16-20 A relative “NP deficiency” in blacks could contribute to an impaired ability to maintain energy homeostasis, lipolysis, and insulin sensitivity, possibly serving as a unifying mechanism underlying racial disparities in cardiometabolic disease.
We have previously shown that in young, healthy whites, a high-carbohydrate challenge causes an acute reduction in circulating ANP levels.21 However, there are no data on how the NP system responds to a carbohydrate challenge in blacks. In an effort to translate insights from human genetic studies into a clinical setting, and to evaluate and implement individualized approaches for the assessment of NPs,22 we conducted a clinical trial to investigate: 1) whether circulating NPs are lower in young healthy blacks as compared with young healthy whites; 2) the change in NPs in response to a high-carbohydrate challenge in blacks and whether there is a difference in the magnitude of change in blacks versus whites; and 3) the mechanisms of race-related NP difference and race-specific NP response to a high-carbohydrate challenge.
METHODS
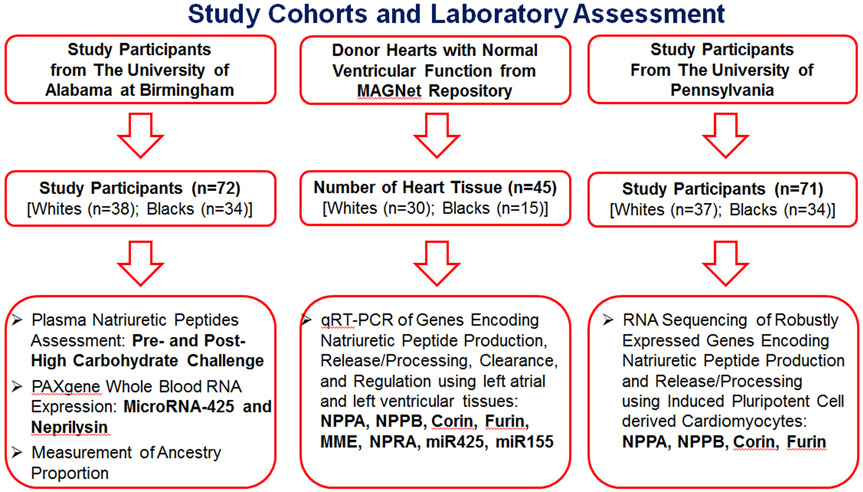
A detailed description of the study populations, trial protocol, and laboratory assessment are provided in Online Method I and Figure 1. Moreover, the data that support the findings of this study will be available from the corresponding author upon reasonable request.
Figure 1.
Study Cohorts and Laboratory Assessment
Study design, setting, and location.
Our study was a single-center, prospective, physiological clinical trial approved by the institutional review board at the University of Alabama at Birmingham (UAB). Participants were recruited from the university campus and surrounding areas. All participants signed informed consent. Atrial and ventricular tissues from organ donor hearts with normal ventricular function in the Myocardial Applied Genomics Network (MAGNet) repository were assessed for the genetic assessment.23 Moreover, induced pluripotent cell-derived cardiomyocytes (iPSC-CMs) from the participants recruited for Phenotyping Lipid traits in iPS derived Hepatocytes Study (PhLiPS study)24 at the University of Pennsylvania (UPenn) were also utilized for the genetic assessment.
Study outcomes and measurements.
The primary outcome of our study was baseline differences in circulating NPs in young healthy blacks and whites. The secondary outcome was the change in NP levels in response to a high-carbohydrate challenge between blacks and whites. We further performed gene expression profiling from whole blood RNA in the trial participants as well as in the atrial and ventricular tissues from the MAGNet repository. Total RNA sequencing (RNA-Seq) of genes associated with NPs were done in iPSC-CMs from healthy white and black adults of PhLiPS study.24
Laboratory measurements.
All blood specimens were obtained in EDTA, PAXgene, or gold top tubes, and immediately processed and frozen at −80°C until assayed. Additionally, buffy coats were extracted and stored at −80°C till the end of the study, at which time DNA was isolated for ancestry determination. Serum glucose was measured using a Sirrus Stanbio (Boerne, TX) analyzer using glucose oxidase reagent (minimum detectable limit = 2 mg/dL, intra-assay CV% <2, inter-assay CV% <3). Insulin was measured using a TOSOH 900 AIA (S. San Francisco, CA, minimum detectable limit = 0.5 uU/mL, intra-assay and inter-assay CV% <4). N-terminal pro ANP (NTproANP) is more stable in plasma and has a longer half-life i.e., 60-120 minutes as compared with mature ANP (2-5 minutes).25 Moreover, NTproANP is produced in 1:1 ratio to ANP.1, 2 We and others have measured NTproANP in various previous studies.21, 26-29 However, NTproANP measurements, performed using available immunoassays, have a relatively high intra-plate CV. Therefore, an intermediate portion of NTproANP i.e., mid-regional-proANP (MRproANP) was measured using immunoluminometric sandwich assays (BRAHMS KRYPTOR compact plus, Hennigsdorf, Germany, minimal detectable limit = 2.1 pmol/L, inter-assay CV% <2 and intra-assay CV% ≤6.5).30 Plasma N-terminal-proBNP (NTproBNP) concentration was measured using an electrochemiluminescence immunoassay (Elecsys proBNP, Roche, Indianapolis, IN, minimum detectable limit = 5 pg/mL, inter- and intra-assay CV% <2).21 Plasma BNP was measured by a two-site immunoenzymatic assay (Alere Triage BNP test, minimum detectable limit = 1.0 pg/mL, inter- and intra-assay CV% <10).31 African Americans are an admixed population, therefore ancestry proportion among self-identified blacks was performed. The description for the assessment of African American ancestry is outlined in the Online Method II. Gene expression levels were assessed from whole blood for the study participants at UAB as well as from atrial and ventricular tissues for subjects in the MAGNet repository. A detail description of RNA isolation and mRNA levels assessment are depicted in the Online Method III.
qRT-PCR using atrial and ventricular tissues from the MAGNet repository.
The atrial and ventricular tissues from organ donor hearts with normal ventricular function were obtained from the MAGNet repository.23 A total of 45 (15 from blacks, 30 from whites) left atrial (LA) and left ventricular (LV) samples were assessed (using qRT-PCR) to examine the genes encoding NP production (i.e., NPPA and NPPB), NP processing (i.e., corin and furin), NP signaling receptor (NPR1), and NP clearance [i.e., membrane metallo-endopeptidase (MME) and NP receptor 3 (NPR3)]. The expression of microRNA-425 (miR-425), and microRNA-155 (miR-155) were also assessed in these tissues. Plasma BNP and MRproANP levels were measured in stored plasma samples from a sub-group (8 from blacks, 19 from whites) of these tissue samples from the MAGNet repository.
RNA sequencing using induced pluripotent cell-derived cardiomyocytes in the UPenn cohort.
A multi-ethnic cohort of healthy individuals were recruited (n=91) and mononuclear cells isolated from their peripheral blood were used to generate induced pluripotent stem cells.24 We differentiated the various cell lines into iPSC-CMs using a feeder-free two-dimensional differentiation protocol based on a published procedure that is standard in the field.32 The iPSC-CM colonies were examined for troponin positivity as a measure of quality using flow cytometry. The details of the differentiation protocol and RNA-seq assessment are described in Online Method IV.
Statistical analysis.
Our study was powered to detect the change in ANP levels in response to a high-carbohydrate challenge among black individuals. Based on a prior investigation among whites,21 a high-carbohydrate challenge can reduce circulating plasma ANP levels up to 27%. With an estimated standard deviation of 0.2 (a high estimate) and utilizing prespecified sample size of 30 black individuals,21 our study was powered to detect a change as small as 11% in ANP levels after a high-carbohydrate challenge, with β=0.8 and α=0.05. We expected similar effect size for change in ANP levels after a high-carbohydrate challenge among whites.
All analyses were conducted using STATA, version 14.2 MP (StataCorp LP), and LIMMA R package. Baseline characteristics of participants were compared between blacks and whites using Student’s t-test (normal distribution) or Mann-Whitney test (non-normal distribution) for continuous variables and Pearson χ2 test for categorical variables. The normality of continuous variables was assessed using histogram and Q-Q plots (visual assessment) as well as Shapiro Wilk test (statistical assessment). Plasma NP levels at baseline and after a high-carbohydrate challenge were found to have a non-normal distribution and were log-transformed for the analyses. Linear regression models were used to assess the racial differences in plasma NP levels. Choosing white race as a reference, a relative percentage difference in plasma NP levels with 95% confidence interval (CI) in blacks was calculated using the following formula: (eβ – 1) x 100 (where β is the beta coefficient from linear regression). As the assessment of different NP fragments are not independent to each other, we did not use the statistical correction of multiple testing in the aforementioned models. Linear mixed-effect models excluding missing data (<1%) from 72 individuals were used to assess the effect of high-carbohydrate challenge overtime on plasma NP levels. In these models, we assessed for repeated measurements of NP levels with fixed effects of race, time, and their multiplicative interaction term (race*time), accounting for the correlation among repeated measures in the same person. Participants were treated as random effects in these models. The percentage difference with 95% CI in plasma NP levels after 8 hours in overall as well as by race was calculated: [(eβ – 1) x 100 multiplied by 8 hours (where β is the beta coefficient from linear regression)]. The models mentioned above were further assessed after adjusting for their baseline demographics [i.e., age, sex, body mass index (BMI), insulin].
Gene expression from PAXgene whole blood tubes and atrial/ventricular tissues.
For relative mRNA levels of genes related to NPs, we performed 2-sample t-test with Welch correction for unequal variance. Furthermore, mRNA levels of NPPA, NPPB, corin, furin, NPR1, NPR3, MME, miR-425, and miR-155 levels were log transformed to assess the relative difference between white and black individuals in a multivariable linear regression model (including age, sex, and BMI). A two-sided p<0.05 was considered statistically significant.
RNA sequencing from iPSC-CMs.
RNA sequencing was performed on the Hiseq2500 using 100-bp paired-end reads, and ~30 million read pairs were generated per biological sample. Fastq files were aligned against a human reference (hg19/hGRC37) using the STAR aligner (v2.6). Duplicate reads were removed using MarkDuplicates from Picard tools, and per gene read counts for Ensembl (v75) gene annotations were computed. Expression levels of NPPA, NPPB, corin, and furin in counts per million (CPM) were normalized and transformed using the VOOM procedure in the LIMMA R package. PCA analysis revealed sources of unwanted variation associated with technical sources of variation such as batch and RIN score. Rather than directly adjusting for them, the surrogate variable analysis was used to account for sources of latent (hidden) variation. Using the svaseq function in the R SVA package, 11 significant surrogate variables were determined for the iPSC-CMs samples. Differential gene expression between races was performed with the LIMMA R package using the following linear model, this model adjusts for any sex, age, and surrogate variables:
Y = β0 + β1×race + β2×sex + β3×Age + β4:N-×SVA_1:SVA_N
(where Y is log2 transformed gene expression, the race is either black or white individuals plus adjustments for sex, age, and 11 surrogate variables).
RESULTS
We screened 94 subjects from March 2017 to October 2018 at UAB to examine baseline plasma NP levels and the effects of a high-carbohydrate challenge on the same. The study flow-chart is shown in Online Figure I. A total of n=13 subjects were ineligible after screening (Online Method I). Seven participants picked up meals for the second visit but were unable to attend the protocol, three subjects were found ineligible after the screening (two had high blood glucose levels, and one had anemia), two participants were unable to complete the protocol due to difficulties with the blood draw, and one subject was tested positive for pregnancy on the protocol visit. Our final study sample included 72 individuals, 34 self-reported blacks, and 38 self-reported whites.
Baseline characteristics of participants are shown in Online Table I. The mean age of participants was 27 years. We studied the similarly equal proportion of men and women in both groups, with a mean BMI of ~27 kg/m2 (Online Table I). The mean systolic BP was 115 mmHg (IQR, 108-124 mmHg) in blacks and 112 mmHg (105-118 mmHg) in whites (Online Table I). A total of 24 black individuals provided consent to isolate DNA for ancestry proportion analyses. The proportion of African ancestry among self-identified blacks in our study was ~90%.
Racial differences in natriuretic peptide levels among young, healthy adults.
The median BNP and NTproBNP levels in blacks were 17 pg/mL (IQR: 13-23 pg/mL) and 14 pg/mL (IQR: 14-23 pg/mL), while in whites were 22 pg/mL (IQR: 18-33 pg/mL) and 33 pg/mL (IQR: 18-53 pg/mL), respectively (Online Table I). In the unadjusted analysis, log plasma BNP and NTproBNP levels were 28% (95% CI: 11% to 42%, p=0.002) and 51% (95% CI: 34% to 66%, p<0.001) lower in black participants compared with whites, respectively (Table 1). After adjusting for age, sex, BMI, and insulin, the differences in plasma BNP and NTproBNP levels in blacks compared with whites remained similar (30% lower for BNP and 47% lower for NTproBNP) (Table 1). The median MRproANP levels were significantly lower in blacks as compared with whites 43 pmol/L (IQR: 33-55 pmol/L) vs. 52 pmol/L (IQR: 45-65 pmol/L), p=0.02 (Online Table I). Log plasma MRproANP levels were significantly lower in both unadjusted (18%; 95% CI: 3% to 31%, p=0.02) as well as multivariable-adjusted model (18%; 95% CI: 2% to 31%, p=0.03) (Table 1).
Table 1.
Racial differences in baseline plasma natriuretic peptides levels in young healthy blacks and whites.
| Whites | Blacks β (95% CI) |
% Difference |
p-value | |
|---|---|---|---|---|
| Plasma NTproBNP | ||||
| Unadjusted | Reference | −0.72 (−1.09 to −0.35) | −51% | <0.001 |
| Age, sex, BMI, Insulin | Reference | −0.63 (−0.98 to −0.28) | −47% | 0.001 |
| Plasma BNP | ||||
| Unadjusted | Reference | −0.33 (−0.54 to −0.12) | −28% | 0.002 |
| Age, sex, BMI, Insulin | Reference | −0.35 (−0.57 to −0.13) | −30% | 0.002 |
| Plasma MRproANP | ||||
| Unadjusted | Reference | −0.20 (−0.37 to −0.03) | −18% | 0.02 |
| Age, sex, BMI, Insulin | Reference | −0.20 (−0.37 to −0.02) | −18% | 0.03 |
Multivariable linear regression models using natural log-transformed NP as the dependent variable and race as the independent variable was used to assess statistical difference by race. White race was chosen as reference. Values shown were beta coefficient (95% confidence interval) which were on the log NP scale. Percentage lower than whites was the estimated % difference in NP levels in blacks versus whites which was calculated by using the formula (eβ−1)*100 assuming all other variables in the model remained constant.
Racial differences in natriuretic peptide response to a high-carbohydrate challenge.
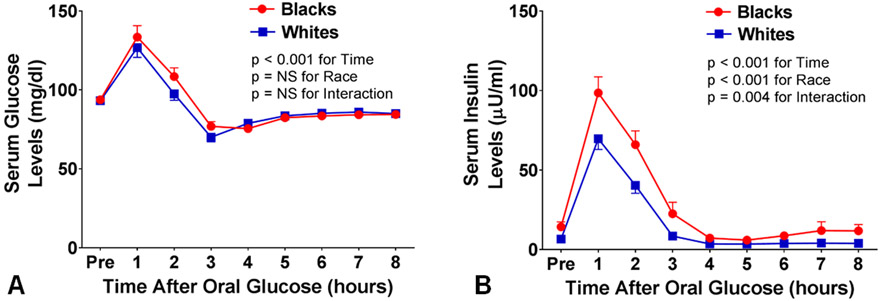
Fasting serum glucose levels were similar between the groups (93.7 ± 2.0 mg/dL in blacks versus 93.1 ± 1.0 mg/dL in whites, p=0.81) (Online Table I). After a high-carbohydrate challenge, serum glucose levels increased and peaked at 1 hour with levels of 129.9 ± 7.3 mg/dL in blacks and 126.8 ± 6.2 mg/dL in whites (Figure 2, Panel A). Fasting serum insulin levels were higher in blacks compared with whites (14.3 ± 3.2 versus 6.6 ± 0.6 μU/mL, p=0.01) (Online Table I). Black individuals had a greater insulin resistance as compared with whites (HOMA-IR: 3.6 ± 1.0 in blacks versus 1.4 ± 0.1 in whites, p=0.03). Serum insulin levels peaked at 1 hour and were 98.5 ± 10.1 μU/mL in blacks compared to 69.7 ± 6.7 μU/mL in whites (p=0.004 for interaction) (Figure 2, Panel B). Black individuals showed hyperinsulinemia to glucose load suggesting reduced insulin sensitivity.
Figure 2.
Serum glucose (Panel A) and insulin (Panel B) levels at baseline and in response to a high-carbohydrate challenge in black and white Individuals. Data are presented as mean ± SEM. 2-way analysis of variance was used to generate p-values for serum glucose and serum insulin by race and time, and to assess for interaction with race and time.
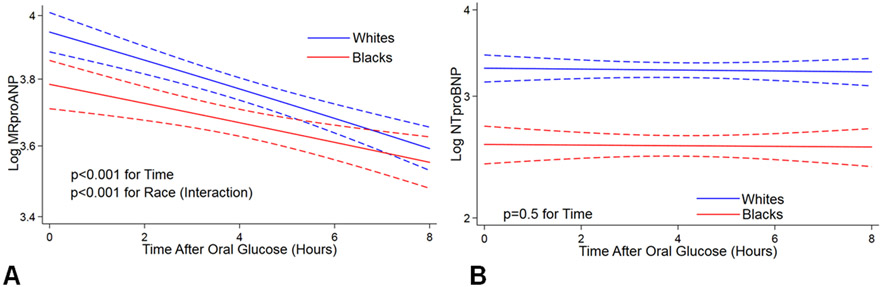
After a high-carbohydrate challenge, there was a significant decrease in plasma MRproANP levels in all participants in both unadjusted and multivariable-adjusted models (i.e., age, sex, BMI, and insulin). In the multivariable-adjusted model, we observed a 29% (47 pmol/L to 33 pmol/L, p<0.001) decrease in the circulating MRproANP levels 8-hours after a high-carbohydrate challenge. The reduction in plasma MRproANP levels after a high-carbohydrate challenge differed significantly by race (p<0.001, in a multivariable-adjusted model). Therefore, we further examined the reduction in plasma MRproANP levels stratified by race. We observed a 34% (52 pmol/L to 34 pmol/L, p<0.001) and 23% (43 pmol/L to 33 pmol/L, p<0.001) reduction in plasma MRproANP levels in whites and blacks, respectively (Figure 3, Panel A). Plasma NTproBNP levels did not change after a high-carbohydrate challenge over time in both blacks and whites (p=0.5 for time) (Figure 3, Panel B).
Figure 3.
Differences in plasma MRproANP (Panel A) and NTproBNP (Panel B) in response to a high-carbohydrate challenge by race. Linear mixed models using natural log-transformed MRproANP and NTproBNP as the dependent variable and Time as the independent variable were used. Values are predicted log MRproANP (Panel A) and log NTproBNP (Panel B) with 95% confidence interval.
Linear mixed models adjusted for age, sex, body mass index, and serum insulin levels were used to assess p-value by race.
Gene expression in atrial and ventricular tissues from the MAGNet repository.
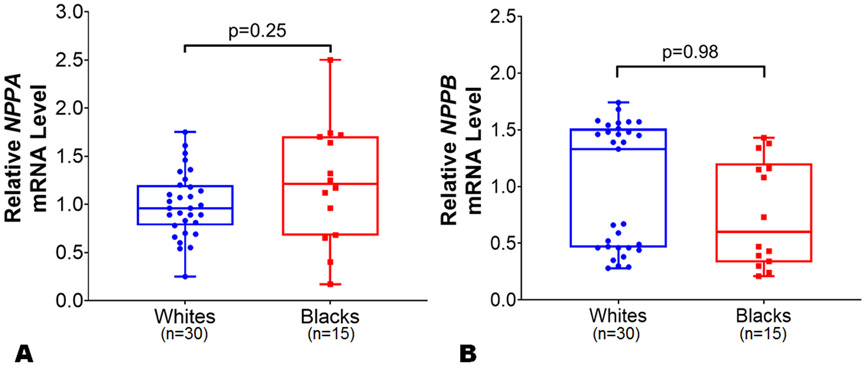
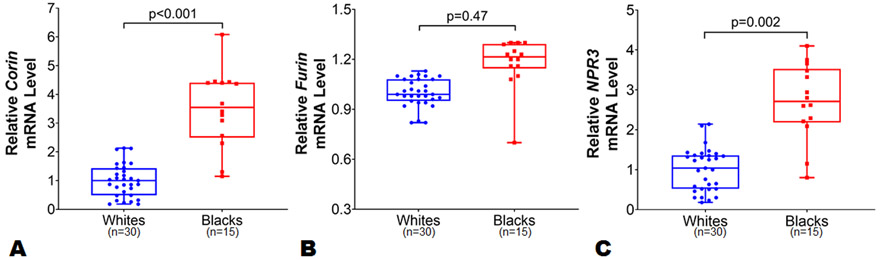
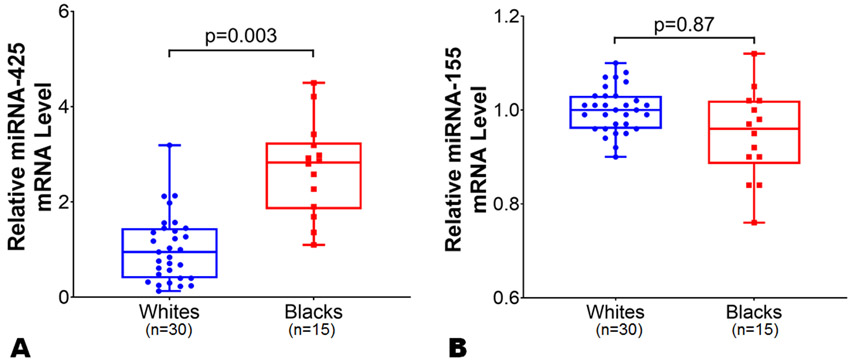
To assess potential mechanisms for racial differences in NP levels, the expression data of genes encoding NPs (i.e., NPPA and NPPB), NP processing (i.e., corin and furin), NP signaling receptor (NPR1), and NP clearance (i.e., MME and NPR3) in LA and LV tissue samples were compared in blacks vs. whites. The mean age of the study individuals from MAGNet repository was 57 years. The proportion of African ancestry among black individuals (n=15) was ~77%. We observed similar NPPA (p=0.25) and NPPB (p=0.98) expression levels by race in the LA (Figure 4, Panels A-B) in an unadjusted as well as multivariable-adjusted analyses. The relative expression levels of corin, NPR3, miR-425, and miR-155 were higher in the atrium than in the ventricle tissue samples which may reflect the physiological role these genes play in NP (particularly ANP) processing, clearance, and regulation. In multivariable model after adjusting for age, sex, and BMI, we observed a ~3.5-fold higher corin expression levels in LA among blacks as compared to whites (Figure 5, Panel A; p=0.003). The expression levels of furin were similar in blacks compared to whites in the LA (Figure 5, Panel B; p=0.47). The expression levels of NP signaling receptor i.e. NPR1 were similar between blacks and whites in the LA (Online Figure II; p=0.15). The expression levels of NPR3 were ~2.5 fold higher in LA tissues among blacks as compared with whites (Figure 5, Panel C; p=0.002). However, the expression levels of NPPA (p=0.17), NPPB (p=0.60), corin (p=0.09), furin (p=0.95), NPR1 (p=0.79), and NPR3 (0.95) were similar between blacks and whites in LV tissues from the MAGNet repository. Heart is not the primary site for MME expression (Online Figure III),33 and no differences were observed in the expression levels of MME in LA and LV tissues across both races. The expression of miR-425 was ~2-fold higher in LA tissue in blacks (p=0.003) as compared with whites (Figure 6, Panel A), while similar in LV tissue between blacks and whites. However, miR-155 expression in LA (Figure 6, Panel B) and LV tissue was similar between blacks and whites (p=0.87 for both).
Figure 4.
Differences in NPPA and NPPB expression levels between black and white individuals in left atrial tissue. The NPPA (Panel A) and NPPB (Panel B) expression levels were normalized to the housekeeping control gene 18s. The NPPA and NPPB expression levels in blacks (n=15) were relative to white (n=30) individuals from atrial tissues from organ donor hearts with normal ventricular function in the Myocardial Applied Genomics Network (MAGNet) repository. Data are presented as mean ± SEM. 2-sample t-test with Welch correction for unequal variance was used to assess the p-value by race.
Figure 5.
Differences corin, furin and NPR3 expression levels between black and white individuals in left atrial tissue. The corin (Panel A), furin (Panel B) and NPR3 (Panel C) expression levels were normalized to the housekeeping control gene 18s. The corin, furin and NPR3 expression levels in blacks (n=15) were relative to white (n=30) individuals from atrial tissues from organ donor hearts with normal ventricular function in the Myocardial Applied Genomics Network (MAGNet) repository. Data are presented as mean ± SEM. Data are presented as mean ± SEM. 2-sample t-test with Welch correction for unequal variance was used to assess the p-value by race.
Figure 6.
Differences in microRNA-425 and microRNA-155 expression levels between black and white individuals in left atrial tissue. The microRNA-425 (Panel A) and microRNA-155 (Panel B) expression levels were normalized to the housekeeping control gene U6. The microRNA-425 and microRNA-155 expression levels in blacks (n=15) were relative to white (n=30) individuals from atrial tissues from organ donor hearts with normal ventricular function in the Myocardial Applied Genomics Network (MAGNet) repository. Data are presented as mean ± SEM. Data are presented as mean ± SEM. 2-sample t-test with Welch correction for unequal variance was used to assess the p-value by race.
Additionally, a total of 27 plasma samples (blacks=8, whites=19) which were available for assessment were examined for BNP and MRproANP levels. We found that BNP and MRproANP levels were 70% (95% CI: 50% to 94%) and 21% (95% CI: 8% to 34%) lower in blacks as compared with whites, respectively.
Gene expression in induced pluripotent stem cells derived cardiomyocytes in the UPenn cohort.
The RNA-seq data in human iPSC-CMs comprised of 37 white (49% women) and 34 black (68% women) individuals were assessed. The median age of white and black individuals was 27 years (IQR: 24-30) and 32 years (IQR: 27-41), respectively. Among black individuals, the proportion of African ancestry was ~72%. We found that more than 80% of iPSC-CMs showed troponin positivity by flow cytometry. Furthermore, PCA assessment, before and after the adjustment, outlined in the Online Figure IV. We observed similar NPPA (p=0.66) and NPPB (p=0.67) expression levels by race in the iPSC-CMs in an unadjusted as well as multivariable-adjusted analysis (Online Figure V, Panel A and B). Additionally, we did not observe any difference in the NP processing genes i.e. corin (p=0.73) and furin (p=0.79) as expression levels were similar between white and black individuals in iPSC-CMs (Online Figure V, Panel C and D). We observed that the transcript levels of genes encoding for NP clearance (i.e., MME and NPR1) were low (10 times lower than NPPA, ref gene) in iPSC-CMs in both races, which is consistent with gene-tissue expression database.33-35
Whole Blood mRNA expression in the clinical trial participants.
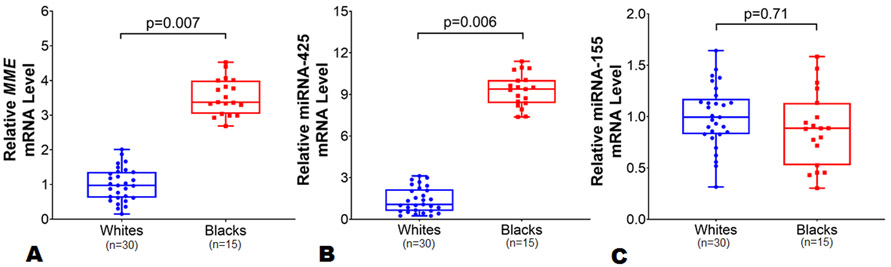
Whole blood samples collected in a PAXgene RNA tube from 49 study participants at UAB who provided consent for the genetic study were analyzed. The baseline characteristics of these 49 study participants are shown in Online Table II. Gene expression profiling performed for all of the aforementioned genes revealed robust expression (CTs <27 indicative of abundant target nucleic acid) only for MME, miR-425 and miR-155 genes, indicating that whole blood mRNA is a good source to study transcript levels of these two genes. Also, the expression of MME (Online Figure III), miR-425 and miR-155 in whole blood is well recognized.4, 36, 37 Of note, MME (known as Neprilysin) is responsible for NP clearance, and we have previously shown that miR-425 is a negative regulator of ANP. Among these 49 study participants (20 black and 29 white individuals), we observed a 3-fold higher MME mRNA levels among black individuals compared with whites (p=0.007) (Figure 7, panel A). Similarly, in multivariable adjusted analysis including age, sex, and BMI, we observed 2-fold higher MME mRNA levels in blacks compared with whites (p=0.009).
Figure 7.
Baseline differences in MME, microRNA-425, and microRNA-155 expression levels in whole blood by race. The MME (Panel A) expression levels was normalized to the housekeeping control gene 18s, while miR-425 (Panel B) and miR-155 (Panel C) expression levels were normalized to the single nucleotide U6. The MME, miR-425, and miR-155 expression levels in blacks (n=20) were relative to white (n=29) individuals. Data are presented as mean ± SEM. Data are presented as mean ± SEM. 2-sample t-test with Welch correction for unequal variance was used to assess the p-value by race.
Next, we investigated whether the race-specific response in MRproANP levels to a high-carbohydrate challenge was related to differential miR-425 and miR-155 expression by race. Among a sub-group (n=49) of clinical trial participants, black individuals had 9-fold higher miR-425 expression levels as compared with whites at baseline (p=0.006) (Figure 7, panel B). In the multivariable-adjusted model, we observed ~8 fold higher miR-425 expression levels in blacks as compared with whites (p<0.001). However, we did not observe any difference in miR-155 expression levels by race (p=0.71) (Figure 7, panel C).
DISCUSSION
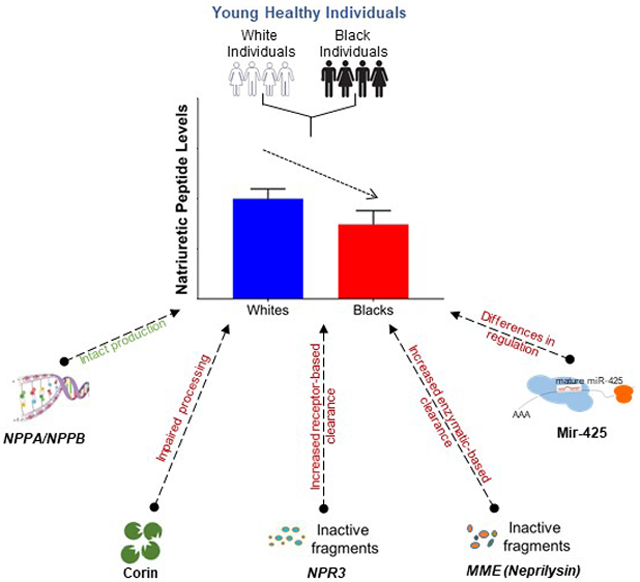
In this clinical trial, we found that baseline levels of both N-terminal and mature BNP were significantly lower in blacks compared with whites in “young” healthy adults. We also observed that the circulating plasma MRproANP levels were significantly lower in blacks compared with whites. Additionally, we found that a high-carbohydrate challenge acutely decreases the circulating plasma MRproANP levels in both healthy blacks and whites with no change in NTproBNP levels. However, the decrease in circulating plasma MRproANP levels after a high-carbohydrate challenge was ~50% greater in magnitude in whites compared with blacks. We observed that the expression levels of genes encoding for NPs (NPPA/NPPB) were similar between blacks and whites in atrial and ventricular tissues as well as in iPSC-CMs. However, the expression of genes affecting NP processing (corin), clearance (NPR3/MME) and negative-regulation (miR-425) were higher among blacks as compared with whites in atrial tissues from the MAGNet repository. Additionally, levels of MME (Neprilysin; NP clearance enzyme) mRNA and miR-425 (a negative regulator of ANP) in whole blood were 2- and 8-fold higher in blacks compared with whites. Taken together, these findings suggest that black individuals harbor an NP-deficient state that may be attributable to the negative regulation of NP transcript, impaired NP processing, and enhanced clearance of circulating NPs.
In previous work done by us and others, it has been shown that the black race is associated with lower circulating NP levels as compared with white individuals.8-11 However, the mean age of participants in the population-based cohorts in which this hypothesis was examined was in the fifth to sixth decade of life8-11 when individuals typically accumulate subclinical cardiovascular and kidney disease. Our physiologic study is the first to provide support to the hypothesis that black race is an “NP deficient” state by demonstrating lower NP levels from a young age. Both NPs i.e., NTproBNP/BNP and MRproANP were found to be similarly lower in blacks compared to whites. Interestingly, pro- and mature NPs are known to have different clearance mechanisms,38 thus one may assume that impaired synthesis/production likely accounts for the observed lower NP levels in blacks. However, we did not observe any differences in the qRT-PCR from atrial and ventricular tissues as well as RNA-seq expression levels of NPPA/NPPB (NP encoding genes) in iPSC-CMs by race. In fact, the expression levels of NPR3 (clearance receptor) and blood expression levels of Neprilysin (an enzyme responsible for cleavage of NPs)37 were higher in black individuals compared with whites. These findings put forward the notion that increased clearance rather than differences in production may be a cause of “NP deficiency” in blacks. Additionally, the expression levels of corin were higher in blacks as compared with whites. However, a previous study did not detect any increase in corin enzymatic activity despite higher corin mRNA and protein expression.39 We speculate that impaired NP processing may also contribute to lower NP levels among blacks. Nonetheless, further work is needed to understand the mechanisms behind regulation of NP processing including corin activity.40, 41 In summary, increased clearance and impaired processing in NPs are possible reason behind differences in circulating NP levels by race. Additionally, our data suggest the differences in the regulators of NP system such as miR-425 may play a role in how the NP system responds to physiological perturbations such as a high-carbohydrate challenge, which needs to be investigated further in future studies.
We observed that plasma NTproBNP levels remained steady while MRproANP levels were acutely decreased after a high-carbohydrate challenge among black and white individuals, confirming that a high-carbohydrate challenge causes ANP suppression in both races. These data of reduction in MRproANP levels with no effect on NTproBNP were consistent with our previous work.21 This suggests that pathways regulating the two peptides of the NP system (ANP and BNP) may differ upon challenging the NP system with a physiological perturbation.21 Additionally, the magnitude of reduction in the circulating ANP levels (~34%) after a high-carbohydrate challenge in whites noted in the current trial was similar to our previous study (~30%).21
In our prior study, we observed that one of the underlying mechanisms of ANP suppression involved a glucose-induced increase in the expression of a negative regulator of ANP production, miR-425.21 The difference in the magnitude of suppression of ANP levels among white (~34%) compared with black (~23%) individuals raises the possibility that there may be underlying biological differences in how heart responds to a high-carbohydrate challenge. By examining the baseline expression of miR-425 levels by race, we determined that the race-specific ANP response to a high-carbohydrate challenge could be associated with the racial difference in the miR-425 levels. Future studies will be required to investigate whether mechanistic pathways such as the race-based differences in NP regulation by miR-425, assessment of miR-425 levels in different cellular and extracellular compartments as well as glucose-induced expression of miR-425 differ by race.
Previous studies have explored the response of NPs to a high-carbohydrate challenge.42, 43 Goetze examined six healthy individuals and found a similar decrease in proANP levels after a high-carbohydrate challenge.43 Similarly, Asferg et al. observed a reduction in ANP levels after a high-carbohydrate challenge in obese men over the period of 2 hours.42 Both these studies were limited by smaller sample sizes, short duration of follow up, and incomplete profiling of the NP system. To the best of our knowledge, no prior study has examined the ANP response after a high-carbohydrate challenge in young healthy black individuals, and further compared the degree of ANP suppression with whites.
Public health implications in an era of precision medicine.
The awareness of NP deficiency being associated with a higher risk of hypertension5, 6 and diabetes44 came from human genetics. However the impact of NP deficiency in the development of cardiometabolic disease has not been fully realized. Also, it has been known that ANP has a higher affinity for NPR1,45 therefore, one can postulate that lower ANP levels in blacks (as demonstrated in our study by lower MRproANP levels) may have significant functional implications. Reversing NP deficiency in blacks using NP augmenting therapies may mitigate the racial disparities in cardiometabolic disease. There are NP analogs available,46 and treatments such as Neprilysin inhibitors (LCZ696) which augment NP levels have recently been approved for heart failure.47, 48 The precision medicine approach of reversing NP deficiency in blacks prior to the development of cardiometabolic disease will need to be tested.
Our study suggests that the endocrine system (i.e., NP system) of the heart in white individuals may be more vulnerable to dietary habits such as high-carbohydrate intake. Since ANP has well known favorable cardiometabolic effects, prevention strategies for cardiometabolic disease using dietary interventions should be individualized based on racial profile and genetic makeup.
Limitations.
One could conceive that the biggest limitation of our study is the generalizability of our results. Our study protocol required strict adherence to a 3-day standardized diet, and a mandatory fasting the night before the protocol and for 8 hours following consumption of the high-carbohydrate drink. The complexity of our protocol limited the sample size. However, it has provided us an advantage to reduce inherent variation that is present in larger studies where NP levels are measured on a random salt background. Our findings need to be replicated in populations of different ethnicities such as Asians, Hispanics, etc., individuals with cardiovascular disease such as heart failure, and individuals from different age groups such as younger children and adolescents. We did not measure mature ANP among study participants due to the known analytical issues with the measurement.49, 50 Our study had an adequate sample size to study the main effect of a high-carbohydrate challenge on ANP suppression in black and white individuals. However, larger sample size is needed to investigate the differences in ANP suppression in subgroups (such as lean versus obese) of black and white individuals. Also, there is an ongoing debate on whether race is a biological construct. We recognize that recruitment in our clinical trial was based on self-identification of blacks which is an admixed population. To overcome this limitation, we ran ancestry proportions on the self-identified blacks in our trial and found that participants had an average of 90% African ancestry.
Conclusions.
Young healthy black individuals have lower NP levels which may be due to an impaired processing or increased receptor/enzymatic clearance rather than differences in the production. Lower NP levels in black individuals may be an under-recognized biological determinant contributing to health disparities. Dynamic decreases in ANP levels in both blacks and whites with perturbations such as a high-carbohydrate challenge demonstrates a key physiologic link in the cardiometabolic axis, the effects of which need to be further investigated. Our study indicates that whites have lower baseline miR-425 levels and higher MRproANP reduction after a high-carbohydrate diet. Efforts to decrease the burden of cardiometabolic disease should be individualized based on clinical risk factors with race-based personalized prevention strategies.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
Blacks have lower resting natriuretic peptide (NP) levels than middle-aged and older white individuals.
A high-carbohydrate challenge reduces circulating atrial-NP levels in young healthy whites.
What New Information Does This Article Contribute?
Young healthy black individuals have lower NP levels as compared with whites suggesting that black race is a NP deficiency state.
Diminished processing and/or increased receptor/enzymatic clearance may be a reason behind racial differences in NP levels.
White individuals had a higher decrease in atrial-NP levels than blacks after a high-carbohydrate challenge indicating that the NP response to physiological stress maybe regulated differently in blacks and whites.
The study establishes a key pathophysiologic link of the role NPs may play in the cardiometabolic health.
Middle-aged and older black individuals are known to have lower resting NP levels, and a high carbohydrate challenge reduces circulating atrial-NP levels among young healthy whites. This study demonstrates that young black individuals have lower NP levels compared to whites and diminished processing or increased receptor/enzymatic clearance may explain the racial difference in NP levels. This study outlines that white individuals have a greater decline in atrial-NP levels after a high carbohydrate challenge compared to black individuals.
Acknowledgments
SOURCES OF FUNDING
The Research was partly supported by the National Center for Advancing Translational Research of the National Institutes of Health under award number UL1TR001417.Dr. Pankaj Arora is supported by National Institutes of Health Mentored Patient-Oriented Research Award 1K23HL146887-01. Dr. Nirav Patel is supported by National Institutes of Health grant 5T32HL129948-02. Tissue procurement in the MAGNet consortium was supported by a grant from the National Institutes of Health to Drs. Margulies and Cappola R01HL105993.
Nonstandard Abbreviations and Acronyms:
- NP
Natriuretic Peptide
- ANP
Atrial Natriuretic Peptide
- BMI
Body Mass Index
- BNP
B-type Natriuretic Peptide
- CRU
Clinical Research Unit
- iPSC-CM
Induced Pluripotent Cell Derived Cardiomyocytes
- MAGNet
Myocardial Applied Genomics Network
- MME
Membrane Metallo-Endopeptidase
- MRproANP
Mid Regional Pro Atrial Natriuretic Peptide
- NPR1
Natriuretic peptide receptor 1
- NPR3
Natriuretic peptide receptor 3
- NTproBNP
N-terminal Pro B-Type Natriuretic Peptide
- PCR
Polymerase Chain Reaction
Footnotes
DISCLOSURES
Dr. Margulies receives research grant support from Sanofi-Aventis, Merck Sharp and Dohme and GlaxoSmithKline and has consulted for MyoKardia and Luitpold Pharmaceuticals. Drs. Wang and Pankaj Arora are named as coinventors on a patent application relating to the use of miRNAs for the treatment of hypertension and other disorders. None of the authors had any conflicts of interest or financial disclosures to declare.
REFERENCES
- 1.ER Levin, DG Gardner and WK Samson. Natriuretic peptides. The New England journal of medicine. 1998;339:321–8. [DOI] [PubMed] [Google Scholar]
- 2.Clerico A, Iervasi G and Mariani G. Pathophysiologic relevance of measuring the plasma levels of cardiac natriuretic peptide hormones in humans. Horm Metab Res. 1999;31:487–98. [DOI] [PubMed] [Google Scholar]
- 3.Wang TJ, Larson MG, Levy D, Benjamin EJ, Corey D, Leip EP and Vasan RS. Heritability and Genetic Linkage of Plasma Natriuretic Peptide Levels. Circulation. 2003;108:13–16. [DOI] [PubMed] [Google Scholar]
- 4.Arora P, Wu C, Khan AM, Bloch DB, Davis-Dusenbery BN, Ghorbani A, Spagnolli E, Martinez A, Ryan A, Tainsh LT, Kim S, Rong J, Huan T, Freedman JE, Levy D, Miller KK, Hata A, del Monte F, Vandenwijngaert S, Swinnen M, Janssens S, Holmes TM, Buys ES, Bloch KD, Newton-Cheh C and Wang TJ. Atrial natriuretic peptide is negatively regulated by microRNA-425. The Journal of Clinical Investigation. 2013;123:3378–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newton-Cheh C, Larson MG, Vasan RS, Levy D, Bloch KD, Surti A, Guiducci C, Kathiresan S, Benjamin EJ, Struck J, Morgenthaler NG, Bergmann A, Blankenberg S, Kee F, Nilsson P, Yin X, Peltonen L, Vartiainen E, Salomaa V, Hirschhorn JN, Melander O and Wang TJ. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat Genet. 2009;41:348–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, van der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL,Wellcome Trust Case Control C, Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di Gregorio A, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Doring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O’Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen AL, McCarthy MI, O’Reilly PF, Peltonen L, Pouta A, de Jong PE, Snieder H, van Gilst WH, Clarke R, Goel A, Hamsten A, Peden JF, Seedorf U, Syvanen AC, Tognoni G, Lakatta EG, Sanna S, Scheet P, Schlessinger D, Scuteri A, Dorr M, Ernst F, Felix SB, Homuth G, Lorbeer R, Reffelmann T, Rettig R, Volker U, Galan P, Gut IG, Hercberg S, Lathrop GM, Zelenika D, Deloukas P, Soranzo N, Williams FM, Zhai G, Salomaa V, Laakso M, Elosua R, Forouhi NG, Volzke H, Uiterwaal CS, van der Schouw YT, Numans ME, Matullo G, Navis G, Berglund G, Bingham SA, Kooner JS, Connell JM, Bandinelli S, Ferrucci L, Watkins H, Spector TD, Tuomilehto J, Altshuler D, Strachan DP, Laan M, Meneton P, Wareham NJ, Uda M, Jarvelin MR, Mooser V, Melander O, Loos RJ, Elliott P, Abecasis GR, Caulfield M and Munroe PB. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang TJ, Larson MG, Levy D, Benjamin EJ, Corey D, Leip EP and Vasan RS. Heritability and genetic linkage of plasma natriuretic peptide levels. Circulation. 2003;108:13–6. [DOI] [PubMed] [Google Scholar]
- 8.Bajaj NS, Gutierrez OM, Arora G, Judd SE, Patel N, Bennett A, Prabhu SD, Howard G, Howard VJ, Cushman M and Arora P. Racial Differences in Plasma Levels of N-Terminal Pro-B-Type Natriuretic Peptide and Outcomes: The Reasons for Geographic and Racial Differences in Stroke (REGARDS) Study. JAMA cardiology. 2018;3:11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta DK, Claggett B, Wells Q, Cheng S, Li M, Maruthur N, Selvin E, Coresh J, Konety S, Butler KR, Mosley T, Boerwinkle E, Hoogeveen R, Ballantyne CM and Solomon SD. Racial differences in circulating natriuretic peptide levels: the atherosclerosis risk in communities study. J Am Heart Assoc. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta DK, Daniels LB, Cheng S, deFilippi CR, Criqui MH, Maisel AS, Lima JA, Bahrami H, Greenland P, Cushman M, Tracy R, Siscovick D, Bertoni AG, Cannone V, Burnett JC, Carr JJ and Wang TJ. Differences in Natriuretic Peptide Levels by Race/Ethnicity (From the Multi-Ethnic Study of Atherosclerosis). The American journal of cardiology. 2017;120:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta DK, de Lemos JA, Ayers CR, Berry JD and Wang TJ. Racial Differences in Natriuretic Peptide Levels: The Dallas Heart Study. JACC Heart Fail. 2015;3:513–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel N, Cushman M, Gutierrez OM, Howard G, Safford MM, Muntner P, Durant RW, Prabhu SD, Arora G, Levitan EB and Arora P. Racial differences in the association of NT-proBNP with risk of incident heart failure in REGARDS. JCI Insight. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Judd SE, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Mackey RH, Magid DJ, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER 3rd, Moy CS, Mussolino ME, Neumar RW, Nichol G, Pandey DK, Paynter NP, Reeves MJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Wong ND, Woo D, Turner MB, American Heart Association Statistics C and Stroke Statistics S. Heart disease and stroke statistics--2014 update: a report from the American Heart Association. Circulation. 2014;129:e28–e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de Ferranti S, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Judd SE, Kissela BM, Lackland DT, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Matchar DB, McGuire DK, Mohler ER 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Willey JZ, Woo D, Yeh RW and Turner MB. Heart disease and stroke statistics--2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–322. [DOI] [PubMed] [Google Scholar]
- 15.Menke A, Casagrande S, Geiss L and Cowie CC. Prevalence of and Trends in Diabetes Among Adults in the United States, 1988-2012. JAMA. 2015;314:1021–9. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez OA, Duprez DA, Bahrami H, Daniels LB, Folsom AR, Lima JA, Maisel A, Peralta CA and Jacobs DR Jr. The associations between metabolic variables and NT-proBNP are blunted at pathological ranges: the Multi-Ethnic Study of Atherosclerosis. Metabolism: clinical and experimental. 2014;63:475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao Y, Shang X, Zhou L, Hu R, Li Y and Ding W. Relationship between N-terminal pro-B-type natriuretic peptide levels and metabolic syndrome. Archives of medical science : AMS. 2011;7:247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang TJ, Larson MG, Keyes MJ, Levy D, Benjamin EJ and Vasan RS. Association of plasma natriuretic peptide levels with metabolic risk factors in ambulatory individuals. Circulation. 2007;115:1345–53. [DOI] [PubMed] [Google Scholar]
- 19.Olsen MH, Hansen TW, Christensen MK, Gustafsson F, Rasmussen S, Wachtell K, Borch-Johnsen K, Ibsen H, Jorgensen T and Hildebrandt P. N-terminal pro brain natriuretic peptide is inversely related to metabolic cardiovascular risk factors and the metabolic syndrome. Hypertension (Dallas, Tex : 1979). 2005;46:660–6. [DOI] [PubMed] [Google Scholar]
- 20.Patel N, Gutierrez OM, Arora G, Howard G, Howard VJ, Judd SE, Prabhu SD, Levitan EB, Cushman M and Arora P. Race-based demographic, anthropometric and clinical correlates of N-terminal-pro B-type natriuretic peptide. International journal of cardiology. 2019;286:145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arora P, Wu C, Hamid T, Arora G, Agha O, Allen K, Tainsh RE, Hu D, Ryan RA, Domian IJ, Buys ES, Bloch DB, Prabhu SD, Bloch KD, Newton-Cheh C and Wang TJ. Acute Metabolic Influences on the Natriuretic Peptide System in Humans. J Am Coll Cardiol. 2016;67:804–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins FS and Varmus H. A new initiative on precision medicine. The New England journal of medicine. 2015;372:793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin H, Dolmatova EV, Morley MP, Lunetta KL, McManus DD, Magnani JW, Margulies KB, Hakonarson H, del Monte F, Benjamin EJ, Cappola TP and Ellinor PT. Gene expression and genetic variation in human atria. Heart Rhythm. 2014;11:266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pashos EE, Park Y, Wang X, Raghavan A, Yang W, Abbey D, Peters DT, Arbelaez J, Hernandez M, Kuperwasser N, Li W, Lian Z, Liu Y, Lv W, Lytle-Gabbin SL, Marchadier DH, Rogov P, Shi J, Slovik KJ, Stylianou IM, Wang L, Yan R, Zhang X, Kathiresan S, Duncan SA, Mikkelsen TS, Morrisey EE, Rader DJ, Brown CD and Musunuru K. Large, Diverse Population Cohorts of hiPSCs and Derived Hepatocyte-like Cells Reveal Functional Genetic Variation at Blood Lipid-Associated Loci. Cell stem cell. 2017;20:558–570.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yandle TG, Richards AM, Nicholls MG, Cuneo R, Espiner EA and Livesey JH. Metabolic clearance rate and plasma half life of alpha-human atrial natriuretic peptide in man. Life sciences. 1986;38:1827–33. [DOI] [PubMed] [Google Scholar]
- 26.Arora P, Reingold J, Baggish A, Guanaga DP, Wu C, Ghorbani A, Song Y, Chen-Tournaux A, Khan AM, Tainsh LT, Buys ES, Williams JS, Heublein DM, Burnett JC, Semigran MJ, Bloch KD, Scherrer-Crosbie M, Newton-Cheh C, Kaplan LM and Wang TJ. Weight loss, saline loading, and the natriuretic peptide system. J Am Heart Assoc. 2015;4:e001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arora P, Wu C, Khan AM, Bloch DB, Davis-Dusenbery BN, Ghorbani A, Spagnolli E, Martinez A, Ryan A, Tainsh LT, Kim S, Rong J, Huan T, Freedman JE, Levy D, Miller KK, Hata A, Del Monte F, Vandenwijngaert S, Swinnen M, Janssens S, Holmes TM, Buys ES, Bloch KD, Newton-Cheh C and Wang TJ. Atrial natriuretic peptide is negatively regulated by microRNA-425. J Clin Invest. 2013;123:3378–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heringlake M, Kox T, Poeling J, Klaus S, Hanke T, Franz N, Eberhardt F, Heinze H, Armbruster FP and Bahlmann L. The effects of physical exercise on plasma levels of relaxin, NTproANP, and NTproBNP in patients with ischemic heart disease. Eur J Med Res. 2009;14:106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galasko G, Collinson PO, Barnes SC, Gaze D, Lahiri A and Senior R. Comparison of the clinical utility of atrial and B type natriuretic peptide measurement for the diagnosis of systolic dysfunction in a low-risk population. J Clin Pathol. 2007;60:570–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melander O, Newton-Cheh C, Almgren P and et al. Novel and conventional biomarkers for prediction of incident cardiovascular events in the community. JAMA. 2009;302:49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra RK, Beatty AL, Jaganath R, Regan M, Wu AH and Whooley MA. B-type natriuretic peptides for the prediction of cardiovascular events in patients with stable coronary heart disease: the Heart and Soul Study. J Am Heart Assoc. 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ and Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1848–E1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gene Expression for MME (ENSG00000196549.6) Gene Page. GTEx Web Site. https://gtexportal.org/home/gene/MME. Accessed on December 12, 2018.
- 34.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gene Expression for NPR3 (ENSG00000113389.11) Gene Page. GTEx Web Site. https://gtexportal.org/home/gene/NPR3. Accessed on December 12, 2018.
- 36.Cutrona G, Leanza N, Ulivi M, Melioli G, Burgio VL, Mazzarello G, Gabutti G, Roncella S and Ferrarini M. Expression of CD10 by human T cells that undergo apoptosis both in vitro and in vivo. Blood. 1999;94:3067–76. [PubMed] [Google Scholar]
- 37.Bayes-Genis A, Barallat J and Richards AM. A Test in Context: Neprilysin: Function, Inhibition, and Biomarker. Journal of the American College of Cardiology. 2016;68:639–653. [DOI] [PubMed] [Google Scholar]
- 38.Potter LR. Natriuretic Peptide Metabolism, Clearance and Degradation. The FEBS journal. 2011;278:1808–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S, Sen S, Young D, Wang W, Moravec CS and Wu Q. Protease corin expression and activity in failing hearts. American journal of physiology Heart and circulatory physiology. 2010;299:H1687–H1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gladysheva IP, Robinson BR, Houng AK, Kovats T and King SM. Corin is co-expressed with pro-ANP and localized on the cardiomyocyte surface in both zymogen and catalytically active forms. Journal of molecular and cellular cardiology. 2008;44:131–42. [DOI] [PubMed] [Google Scholar]
- 41.Liao X, Wang W, Chen S and Wu Q. Role of glycosylation in corin zymogen activation. The Journal of biological chemistry. 2007;282:27728–35. [DOI] [PubMed] [Google Scholar]
- 42.Asferg CL, Nielsen SJ, Andersen UB, Linneberg A, Goetze JP and Jeppesen JL. Serum proatrial natriuretic peptide concentrations during oral glucose-induced acute hyperinsulinemia in lean and obese men. Peptides. 2018. [DOI] [PubMed] [Google Scholar]
- 43.Goetze JP. Plasma proANP decreases after meal intake. Clinical chemistry. 2013;59:1270–1. [DOI] [PubMed] [Google Scholar]
- 44.Pfister R, Sharp S, Luben R, Welsh P, Barroso I, Salomaa V, Meirhaeghe A, Khaw KT, Sattar N, Langenberg C and Wareham NJ. Mendelian randomization study of B-type natriuretic peptide and type 2 diabetes: evidence of causal association from population studies. PLoS Med. 2011;8:e1001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pandey KN. Guanylyl cyclase / atrial natriuretic peptide receptor-A: role in the pathophysiology of cardiovascular regulation. Can J Physiol Pharmacol. 2011;89:557–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dickey DM, Burnett JC Jr., and Potter LR. Novel bifunctional natriuretic peptides as potential therapeutics. The Journal of biological chemistry. 2008;283:35003–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMurray JJ, Packer M, Desai AS, Gong J, Lefkowitz MP, Rizkala AR, Rouleau JL, Shi VC, Solomon SD, Swedberg K, Zile MR, Investigators P-H and Committees. Angiotensin-neprilysin inhibition versus enalapril in heart failure. The New England journal of medicine. 2014;371:993–1004. [DOI] [PubMed] [Google Scholar]
- 48.Solomon SD, Zile M, Pieske B, Voors A, Shah A, Kraigher-Krainer E, Shi V, Bransford T, Takeuchi M, Gong J, Lefkowitz M, Packer M, McMurray JJ and Prospective comparison of AwARBoMOhfwpefI. The angiotensin receptor neprilysin inhibitor LCZ696 in heart failure with preserved ejection fraction: a phase 2 double-blind randomised controlled trial. Lancet (London, England). 2012;380:1387–95. [DOI] [PubMed] [Google Scholar]
- 49.Semenov AG and Katrukha AG. Analytical Issues with Natriuretic Peptides - has this been Overly Simplified? EJIFCC. 2016;27:189–207. [PMC free article] [PubMed] [Google Scholar]
- 50.Morgenthaler NG, Struck J, Thomas B and Bergmann A. Immunoluminometric Assay for the Midregion of Pro-Atrial Natriuretic Peptide in Human Plasma. Clinical chemistry. 2004;50:234–236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.