Figure 3.
CISAL Forms DNA: RNA Triplex Structure with the BRCA1 Promoter
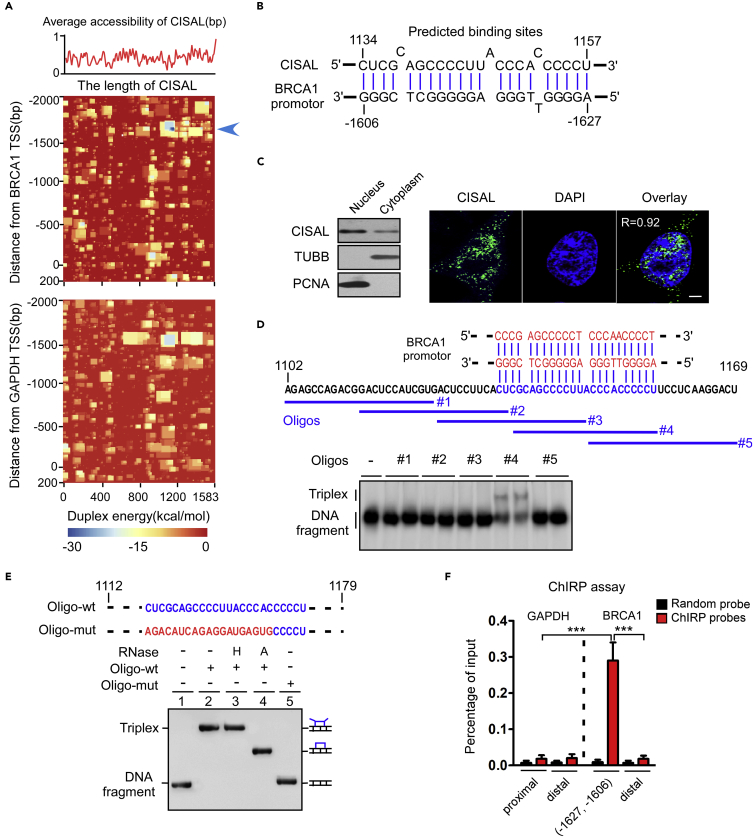
(A) Binding potential between CISAL and BRCA1 or GAPDH promoter regions using IntaRNA. The red curve shows the average probability of single-stranded RNA. Heatmap represents the base-pairing energy for an RNA/RNA duplex model for seed-based regions along the CISAL transcript and 2,000 bp upstream and 200 bp downstream of the TSS of BRCA1 (upper) and GAPDH (lower).
(B) Representation of the predicted interaction of CISAL sequence and the promoter DNA region of BRCA1 at the lowest free energy loci.
(C) Northern blotting (left panel) and FISH (right panel) revealing that CISAL was located in both the nucleus and cytoplasm but predominantly in the nucleus. Scale bar, 3 μm.
(D) Oligo #4 forms a triplex structure with the BRCA1 DNA promoter. The indicated RNA oligos (to #5) were incubated with a biotin-labeled DNA promoter fragment (−1655, −1586) and the formation of DNA: RNA triplexes were monitored by EMSA. The tiled 20 nt RNA oligos (blue) are displayed above.
(E) DNA:RNA triplexes are resistant to RNase H digestion. Triplexes (lanes 2–4), formed by incubating a biotin-labeled BRCA1 DNA promoter fragment (−1685, −1566) with 68-nt wild RNA oligos (1112, 1179) (Oligo-wt), were treated with 30 U of RNase H (H) or RNase A (A) and analyzed by EMSA. Lane 5 shows DNA fragments incubated with the mutant RNA oligos (Oligo-mut).
(F) ChIRP analysis of CISAL in the regulatory regions of BRCA1(−1627, −1606) but not GAPDH. The cross-linked CAL-27 cell lysates were incubated with biotinylated DNA probes against CISAL, and the binding complexes were recovered using streptavidin-conjugated magnetic beads. qPCR was performed to detect enrichment of the specific regulatory regions associated with CISAL.
***p<0.001, one-way ANOVA followed by Dunnett's tests for multiple comparisons. Data are represented as mean ±SEM.