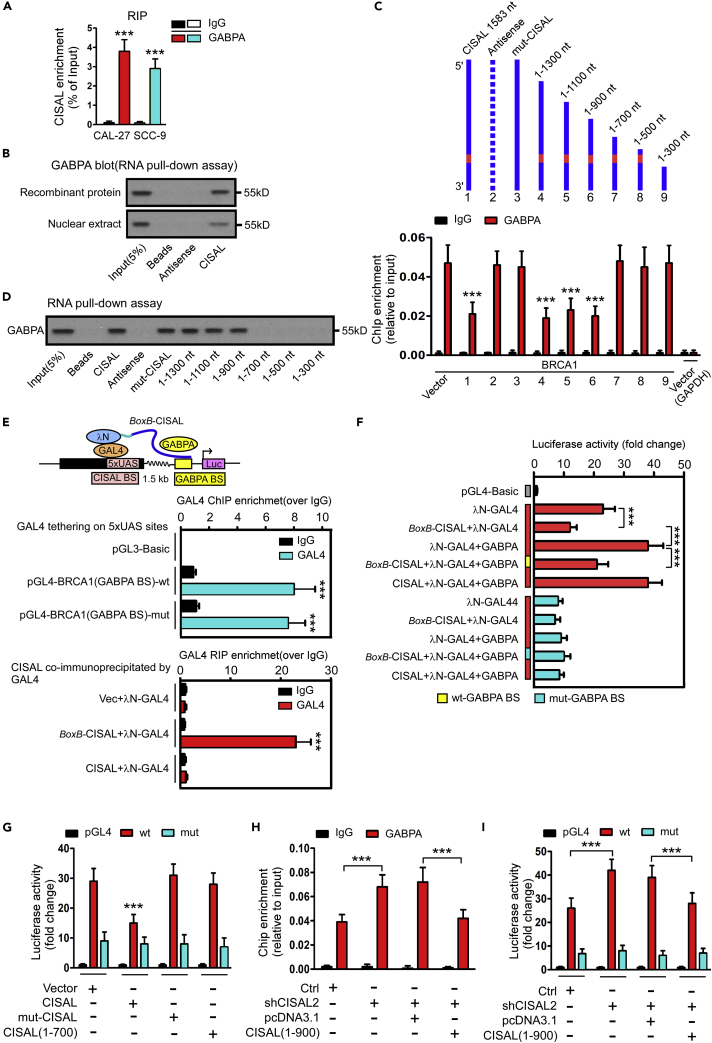
Figure 5.
CISAL Sequesters GABPA away from Regulatory Binding Sites at BRCA1 Promoter
(A) RT-qPCR analysis of CISAL enrichment by GABPA in the RIP assay in CAL-27 and SCC-9 cells. Normal IgG was used as a nonspecific control.
(B) Western blot analysis showing that CISAL associates with GABPA, as indicated by the pull-down assay with in vitro translated GABP or nuclear extracts of CAL-27 cells. Antisense CISAL was used as a negative control RNA in the pull-down assay.
(C and D) Serial deletions of CISAL were used in ChIP-qPCR analysis (C) and RNA pull-down assays (D) to identify valid length of CISAL, required for physical interaction with GABPA and for sequestering GABPA away from the its downstream binding sites at BRCA1 promoter in CAL-27 cells. Immunoprecipitated DNA was measured by real-time PCR with primers to amplify the BRCA1 promoter region, including the GAPDH locus as a negative control.
(E) Schematic diagram of the x-tethering system on CISAL, which is upstream of GABPA binding sites in BRCA1 promoter-linked luciferase (Luc). CISAL-binding sequence was substituted with 5xUAS and a chimeric RNA by fusing CISAL to BoxB viral RNA BoxB-CISAL. N-GAL4 fusion protein tethers BoxB-CISAL to the 5xUAS sites. Middle panel indicate efficiency of GAL4 tethering at 5xUAS region by ChIP-qPCR analysis. Lower panel shows that BoxB-CISAL instead of CISAL co-immunoprecipitated by GAL4.
(F) Luciferase assay shows BoxB-CISAL repressed BRCA1 promoter activity, which was rescued upon enhancement of GABPA expression in CAL-27 cells with wild GABPA-binding sites transfection and using mutant GABPA-binding sites as a negative control.
(G) Site-directed mutagenesis of 1–700 nt of CISAL leads to a loss of the effect on BRCA1 promoter activity in CAL-27 cells.
(H and I) Forced expression of the truncated CISAL (1–900) abolished the increase of GABPA occupancy (H) and BRCA1 transcriptional activity (I) by silencing endogenous CISAL in CAL-27 cells.
***p< 0.001 by 2-tailed Student's t test (A, C, E, and G–I) or 1-way ANOVA followed by Dunnett's tests for multiple comparisons (F). Data are represented as mean ±SEM.