Abstract

Ponatinib is a multikinase inhibitor that is used to treat chronic myeloid leukemia patients harboring mutated ABL1(T315I) kinase. Due to the potent inhibition of FLT3, RET, and fibroblast growth factor receptors (FGFRs), it is also being evaluated against acute myeloid leukemia (AML), biliary, and lung cancers. The multikinase inhibition profile of ponatinib may also account for its toxicity, thus analogs with improved kinase selectivity or different kinase inhibition profiles could be better tolerated. The introduction of nitrogen into drug compounds can enhance efficacy and drug properties (a concept called “necessary nitrogen”). Here, we introduce additional nitrogen into the benzamide moiety of ponatinib to arrive at nicotinamide analogs. A nicotinamide analogue of ponatinib, HSN748, retains activity against FLT3, ABL1, RET, and PDGFRα/β but loses activity against c-Src and P38α. MNK1 and 2 are key kinases that phosphorylate eIF4E to regulate the protein translation complex. MNK also modulates mTORC1 signaling and contributes to rapamycin resistance. Inhibitors of MNK1 and 2 are being evaluated for anticancer therapy. Ponatinib is not a potent inhibitor of MNK1 or 2, but the nicotinamide analogs are potent inhibitors of MNKs. This illustrates a powerful demonstration of the necessary nitrogen concept to alter both the potency and selectivity of drugs.
Introduction
Ponatinib, developed by Ariad Pharmaceuticals as a multikinase inhibitor, was approved by the Food and Drug Administration (FDA) in 2012.1 It targets many of the various cancer-driver kinases. These include kinases such as ABL1, FLT3, FGFR1-4, and RET. Due to its impressive kinase inhibition profile, it has been shown to potently inhibit various cancers, including chronic myeloid leukemia (CML), acute myeloid leukemia (AML), various fibroblast growth factor receptor (FGFR)- and RET-driven cancers (such as nonsmall cell lung cancer2 and thyroid cancer3). Currently, ponatinib is the only FDA-approved drug for imatinib-resistant CML that harbors the T315I mutation.4 It is also undergoing various clinical trials for AML, lung, and other cancers (NCT02428543; ponatinib for FLT3-ITD acute myelogenous leukemia (PONATINIB-AML),5 NCT02265341; advanced biliary cancer with FGFR2 fusions,6 NCT01813734; ponatinib in advanced NSCLC with RET translocations7).
Despite these impressive arrays of cancer types that ponatinib is currently being evaluated against, the drug is relatively toxic and is associated with cardiovascular adverse events.8 Patients taking ponatinib have also shown side effects of hypertension, platelet dysfunction, and peripheral arterial occlusive disease.9 Other more serious side effects such as myocardial infarction, stroke, and liver failure have occurred in patients taking ponatinib.10 The unfavorable toxicity profile associated with ponatinib could be due to the simultaneous inhibition of cardiovascular-related kinases.11 Herein, we disclose that a nicotinamide analogue of ponatinib (HSN748), whereby the benzamide moiety in ponatinib is replaced with a nicotinamide analog, shows a different kinase inhibition profile to ponatinib. Additionally, the nicotinamide analogue of ponatinib is a better inhibitor of AML cell lines harboring secondary mutations, such as FLT3-ITD, D835Y and FLT3-ITD, F691L, which appear upon prolonged treatment with other FLT3 inhibitors and lead to drug resistance.12
Results and Discussion
Necessary Nitrogen, a High-Level Medicinal Chemistry Design Strategy
The substitution of a −CH group in a hit compound with a N atom in aromatic and heteroaromatic ring systems is a small modification but has potentially large effects on pharmacological profiles. This is due to large changes in molecular and physicochemical properties and intra- and intermolecular interactions. The methyl group scanning is also a high-level medicinal chemistry design strategy, and this has been extensively reviewed.13 Whereas the −CH to −Me or −Me to −CH switch is not accompanied by a big desolvation penalty, a −CH to N switch is accompanied by a large desolvation penalty.14 Despite this penalty, the strategic placement of nitrogen into compounds can lead to dramatic improvement in both potency and drug properties and this has been extensively documented (Figure 1).15−17 A ring nitrogen can form new and stabilizing hydrogen bonding interactions with protein residues, backbone or even form network interactions with water molecules that interact with the protein’s residues or backbone. For a few illustrative examples, Vanotti et al. revealed that a strategic replacement of a phenyl group with a 4-pyridyl group in a cell division cycle 7 (Cdc7) kinase inhibitor improved biochemical activity by >500-fold (compounds 1 to 2, Figure 1). This large effect was attributed to “necessary nitrogen” in the 4-pyridyl substitution making a key hydrogen bonding interaction with the protein backbone (Figure 1a).15
Figure 1.
Examples of the necessary nitrogen concept. Introduction of nitrogen into initial hits led to remarkable activity enhancement as seen in (A) cell division cycle 7 (Cdc7) kinase inhibitors; (B) 5-HT3AR inhibitors; and (C) PLK1 inhibitors.
Esch et al. performed a N-scan on the isoquinoline 5-HT3AR inhibitor 3 (Figure 1b), with pKi = 6.7, and arrived at the quinazoline 4 (Figure 1b) with pKi = 10 against 5-HT3AR (4300-fold enhancement).16 The newly installed N atom was not predicted to engage in any favorable electrostatic interactions for the ligand docking pose in a homology model of the 5-HT3AR binding site (acetylcholine binding protein (AChBP)) yet compound 4 is greater than 2 orders of magnitude more potent than 3. Recently, Remillard et al. showed that the replacement of CH in compound 5 to N in compound 6 increased PLK1 binding (Figure 1c).17
These examples and others prompted us to perform a “nitrogen scan” on ponatinib as well as remove the methyl group on the benzamide core of ponatinib and test the analogs (Figure 2) for inhibition against various kinases and cancers.
Figure 2.
(A) Nitrogen and methyl scan of ponatinib and (B) other ponatinib analogs.
Synthesis of Ponatinib Analogs
HSN748 and other analogs (Figure 2) were synthesized in two linear steps from 5-ethynylnicotinic acid (Figure 3).18−22 By introducing a nitrogen into the benzamide ring (to make a nicotinamide), the calculated log P changed from 4.29 (ponatinib) to 3.62 (HSL420). Removing the methyl group on HSL420 reduced the c log P even further to 3.23 (HSN748).23 The ranking of log P values can be derived from retention times of compounds on C18 columns.24 In concordance with the calculated log P values, the high-performance liquid chromatography (HPLC) retention times of the analogs showed corresponding trends (Table 1).
Figure 3.
Representative synthesis of analogs (HSN748). Conditions: (i) HATU, N,N-diisopropylethylamine (DIPEA), dimethylformamide (DMF), 45 °C, overnight. Yield = 85%; (ii) Pd(PPh3)2Cl2, PPh3, CuI, DMF, triethylamine (TEA), 45 °C, overnight. Yield = 53%. See the Supporting Information for details about other analogs.
Table 1. Calculated log P Values and HPLC Retention Times.
| calculated log Pa | HPLC retention time (min)b | |
|---|---|---|
| HSN748 | 3.23 | 10.86 |
| HSL331 | 3.98 | 11.56 |
| HSL338 | 3.32 | 11.63 |
| HSL382 | 3.22 | 12.33 |
| HSL385 | 3.13 | 12.70 |
| HSL407 | 3.14 | 12.73 |
| HSL412 | 3.24 | 12.96 |
| HSL381 | 3.82 | 13.31 |
| HSL420 | 3.62 | 13.46 |
| ponatinib | 4.29 | 13.90 |
Using SwissADME for calculations.
HPLC details: Agilent Eclipse instrument; C18 column (3 μm, 4.6 × 100 mm2); method: 0 → 15 min, 50% B → 100% B (A: 0.1% NH4OH in H2O, B: MeOH), 25 °C. The compound concentration is 100 μM in MeOH.
Anticancer Activities of Nicotinamide Analogs and Ponatinib
With compounds in hand, we proceeded to evaluate the effects of compounds on CML cell lines as well as FLT3, ABL1, and c-Src kinases, which are known to be targets of ponatinib.25 Although ponatinib is not an effective inhibitor of MAPK-interacting kinases (MNK1 and 2), we also evaluated the inhibition of the compounds on MNK1 and MNK2. An unrelated project, using alkynyl nicotinamide compounds, had taught us that alkynyl nicotinamide-containing compounds were MNK2 inhibitors (unpublished work), prompting us to also evaluate the inhibition of ponatinib analogs against MNK enzymes. The mechanistic target of rapamycin (mTOR) is an important drug target as mTOR integrates many stimuli and coordinates the adaptive response of many cellular processes.26 Rapamycin is an inhibitor of mTOR. MNK contributes to rapamycin resistance by sustaining mTORC1 activity upon rapamycin treatment in cancer cells.26 MNKs modulate mTORC1 but not mTORC2 (which includes Rictor, rapamycin-insensitive companion of mTOR) signaling.27,28 Thus, concurrent inhibition of MNK1 and/or MNK2 and any of cancer-driver kinases, such as FLT3, ABL1, RET, BRAF, c-Kit, PDGFRα, and PDGFRβ, could lead to more sustained inhibition of cancer growth. MNK1 and 2 modulate the function of eIF4E (a key player in translational control), which is elevated in human cancers. MNK1 and 2 phosphorylate a conserved serine (Ser209) of eIF4E to modulate function. The inhibition of both MNK1 and 2 has been shown to lead to growth inhibition in cancers.26
With ponatinib as the standard of care for imatinib-resistant CML harboring ABL1(T315I), we were initially interested in how our compounds compared in the imatinib-resistant CML line, KCL22-IR. The compounds were active against the BCR-ABL1 cell lines, K562 and KCL22, with varying activities against KCL22-IR. Removing the methyl and substituting CH with N on the benzamide ring (ponatinib to HSN748) did not affect anticancer properties against the tested CML cell lines. In addition, reversing the amide bond in HSN748 (HSL338) did not result in a change in anticancer activity. Modification of the imidazo[1,2-b]pyridazine moiety resulted in attenuation of CML growth inhibition. Ponatinib, HSN748, HSL338, and HSL420 all share the imidazo[1,2-b]pyridazine ring system, and they all inhibited the CML cell lines K562, KCL22, and KCL22-IR with similar efficacies (Table 2). However, when the imidazo[1,2-b]pyridazine was changed into imidazo[1,2-a]pyridine (HSL381) or imidazo[1,2-a]pyrazine (HSL385), there was a decrease in potency against the CML cell lines with the biggest difference seen in KCL22-IR. The imidazo[1,2-a]pyrimidine-containing compound (HSL382) was inactive against all of the three tested cell lines. Interestingly, when the ring system was changed to imidazol[1,2-a]pyrazine (HSL407), activities against K562 and KCL22 were maintained, but a decrease in activity against KCL22-IR was seen.
Table 2. IC50 of Library against CML Cell Lines.
| IC50 (nM)a |
|||
|---|---|---|---|
| K562 | KCL22 | KCL22-IR | |
| HSN748 | 0.80 ± 0.003 | 1.32 ± 0.05 | 0.23 ± 0.013 |
| HSL420 | 0.06 ± 0.002 | 0.06 ± 0.002 | 0.46 ± 0.01 |
| HSL381 | 0.57 ± 0.003 | 0.89 ± 0.005 | 4.24 ± 0.05 |
| HSL382 | NDb | >100 nM | >100 nM |
| HSL385 | 0.94 ± 0.002 | 2.62 ± 0.06 | 17.2 ± 0.86 |
| HSL338 | 0.45 ± 0.002 | 1.40 ± 0.02 | 4.24 ± 0.05 |
| HSL331 | 0.26 ± 0.002 | 0.63 ± 0.002 | 2.86 ± 0.04 |
| HSL412 | 28.13 ± 0.60 | 28.06 ± 0.37 | >100 nM |
| HSL407 | 1.83 ± 0.02 | 2.93 ± 0.04 | 49.7 ± 2.5 |
| ponatinib | 0.60 ± 0.002 | 0.14 ± 0.003 | 0.66 ± 0.005 |
Experiments done in triplicate.
Not determined.
Compounds Exhibited Potent Inhibition against Key Kinases
Similar trends that were present against the cell lines were also seen when the compounds were profiled against several kinases. Ponatinib had less than 40% inhibition against FLT3(ITD) at 4 nM. However, several of the nicotinamide compounds had inhibition at over 80% such as HSN748 (86%), HSL381 (91%), and HSL338 (84%) at the same concentration. There was also an increase in FLT3(D835Y) inhibition in compounds containing the nicotinamide. Ponatinib had a 39% inhibition against FLT3(D835Y). When the nitrogen was added to make a nicotinamide ring (HSL420), there was no significant change in inhibition. When the methyl was then removed (HSN748), FLT3(D835Y) inhibition increased to 87%. This shows that even small modifications to a kinase inhibitor can result in drastic changes in kinase profiling. Inhibition of c-Src saw the opposite effect. At 111 nM, ponatinib had a 98% inhibition against c-Src. The methyl nicotinamide analogue (HSL420) inhibited c-Src at 32%, whereas HSN748 had less than 10% inhibition. For ABL1(T315I) inhibition, it appears that a benzamide to nicotinamide switch had no effect on inhibition. At 123 nM, both HSL420 and ponatinib had similar inhibition against ABL1 (T315I) at 83 and 90%, respectively. Analogs that contained the imidazo[1,2-b]pyridazine (ponatinib, HSN748, HSL338, and HSL420) system were generally more potent than analogs that did not against ABL1(T315I). Interestingly, when the imidazo[1,2-b]pyridazine was substituted with imidazo[1,2-a]pyridine (HSL381), potency against ABL1(T315I) was maintained at 73%. The compounds had differential activities against MNKs. At 123 nM, ponatinib inhibited MNK2 at 32%, whereas HSN420 and HSN748 inhibited MNK2 at 51 and 85%, respectively. MNK2 inhibition could tolerate minimal changes to the imidazo[1,2-a]pyridine as seen in HSL381 (imidazo[1,2-a]pyridine, 84% inhibition) and HSL385 (imidazo[1,2-a]pyrazine, 72% inhibition).
Ponatinib is a More Promiscuous Kinase Inhibitor than HSN748
The initial work with ponatinib analogs indicated that HSN748 and HSL381 had the best-combined ABL1, FLT3, and MNK2 inhibitions. HSN748 was chosen for further studies because it had a lower c log P and HPLC retention time than HSL381 (see Table 1). Therefore, we obtained IC50 for ponatinib and HSN748 against various cancer-associated kinases (Table 3). Interestingly, as already established in the single concentration assay (Figure 4), the inhibition profiles of HSN748 against ABL1 (T315I) and FLT3-ITD were similar to ponatinib, but there were some notable differences with other kinases (Table 3).
Table 3. IC50 of Ponatinib and HSN748 against Several Kinases.
| IC50 (nM)a |
|||
|---|---|---|---|
| kinase | HSN748 | ponatinib | staurosporine |
| ABL1 | 1.1 ± 0.38 | 0.87 ± 0.05 | 379 ± 36.8 |
| ABL1 (T315I) | 11.1 ± 3.83 | 2.5 ± 0.13 | 310 ± 29.5 |
| c-Kit | 96 ± 4.21 | 30 ± 7.5 | 70.31 ± 39.42 |
| c-SRC | >1000 | 4.6 ± 0.57 | 7.06 ± 1.16 |
| FGFR1 | 24.4 ± 3.75 | 6.9 ± 0.29 | 20.1 ± 6.09 |
| FGFR2 | 11.7 ± 0.97 | 6.0 ± 0.61 | 8.54 ± 2.97 |
| FGFR3 | 96.4 ± 3.03 | 25.0 ± 1.68 | 35.4 ± 5.86 |
| FGFR4 | 125.1 ± 0.92 | 46.8 ± 8.64 | 217 ± 63.1 |
| FLT3 (D835Y) | 13.8 ± 1.64 | 176 ± 3.11 | 0.06 ± 0.02 |
| FLT3 (ITD) | 1.5 ± 0.02 | 10.03 ± 0.11 | 1.81 ± 0.34 |
| P38α/MAPK14 | 382.7 ± 17.4 | 86.6 ± 6.31 | b |
| p70S6K/RPS6KB1 | 273.1 ± 8.5 | 200.1 ± 17.5 | 0.80 ± 0.14 |
| PDGFRα | 10.7 ± 5.8 | 3.8 ± 0.29 | 3.88 ± 0.79 |
| PDGFRβ | 10.2 ± 1.02 | 7.01 ± 0.57 | 3.33 ± 0.67 |
| RET | 0.65 ± 0.05 | 0.88 ± 0.07 | 5.54 ± 0.09 |
| MNK1 | 202 ± 13.4 | 3930 ± 707 | 108 ± 2.95 |
| MNK2 | 9.36 ± 0.69 | 268 ± 84.1 | 27.88 ± 11.05 |
IC50 was determined at Reaction Biology (Malvern, PA). [ATP] = 100 μM, experiments done in duplicates.
SB202190 (IC50 = 55.5 ± 47.3) was used as the control for P38a/MAPK14.
Figure 4.
Kinase inhibition against key kinases. Compounds were screened at the following concentrations; FLT3(ITD): at 4 nM, FLT3(D835Y) and c-Src at 111 nM, and ABL1(T315I), MNK1&2 at 123 nM. Percent inhibition is with respect to DMSO control, which is 0% inhibition. Data provided by Reaction Biology Corp.
HSN748 was inactive against c-Src kinase (IC50 > 1 μM), while ponatinib potently inhibited c-Src (IC50 of 4.6 nM), (Table 3). Src has been shown to play various roles in the heart function. For example, Src plays critical roles in maintaining the structure of a myocyte.29 Despite the oncogenic role played by Src in various cancers, its inhibition could also come with the dysregulation of normal cells and platelets.30a It is therefore noteworthy that HSN748 does not inhibit Src, while ponatinib does. c-Kit is an important player in hematopoiesis, and substantial inhibition of c-kit causes myelosuppression.30b The IC50 for ponatinib against c-Kit is 3 times lower than HSN748.
Ponatinib has been shown to inhibit FGFRs, and it is currently undergoing clinical trials for the treatment of biliary cancer with FGFR2 fusion.7 Although many drugs that target FGFRs are currently undergoing clinical trials, FGFRs have important cardiac and liver functions and so their inhibitions could lead to adverse events. Hyperphosphatemia is one major complication that is associated with FGFR inhibition due to interruption to FGF23 signaling.31 Pan-FGFR inhibition has been linked to cardiovascular dysfunction.32 On the other hand, FGFR signaling also promotes AML resistance to drugs.33 Thus, some inhibition of FGFR signaling might enhance the efficacy of AML therapeutics. Ponatinib is slightly more active against FGFR1-4 than HSN748 (Figure 5).
Figure 5.
Dose–response curves of ponatinib and HSN748 against Src and FLT3 (D835Y).
ABL1 and FLT3 are mutated in CML and AML, respectively. Ponatinib and HSN748 have similar activities against ABL1, ABL1 (T315I), and FLT3-ITD. Interestingly, HSN748 has a significantly lower IC50 against FLT3 (D835Y) kinase than ponatinib (compare IC50 of 14 nM for HSN748 versus 173 nM for ponatinib, see Table 1 and Figure 3). Most FLT3 inhibitors used in the clinic show initial efficacy, but within months some patients relapse due to kinase mutation, which reduces the efficacy of treatment.34 The D835Y mutation is one of the most frequent mutations observed in a study using the TKI quizartinib.35 Thus, for drug-resistant AML (due to kinase mutation), HSN748 could be a better treatment option than ponatinib (Table 4).
Table 4. Activities of HSN748 and Ponatinib against FLT3, ABL1, FGFR, and RET-Driven Cancers.
| IC50 (nM) |
||
|---|---|---|
| cell line | HSN748 | ponatinib |
| MV-4-11 | 0.07 | 0.09 |
| MOLM13-res | 1.73 | 24.1 |
| MOLM14 | 0.25 | 0.45 |
| MOLM14-D835Y | 0.69 | 52.6 |
| MOLM14-F691L | 0.18 | 6.8 |
| K562 | 0.8 | 0.6 |
| KCL22 | 1.32 | 0.14 |
| KCL22-IR | 0.23 | 0.66 |
| LC2/Ad | 41 | 35.1 |
| H520 | 838.6 | 128.2 |
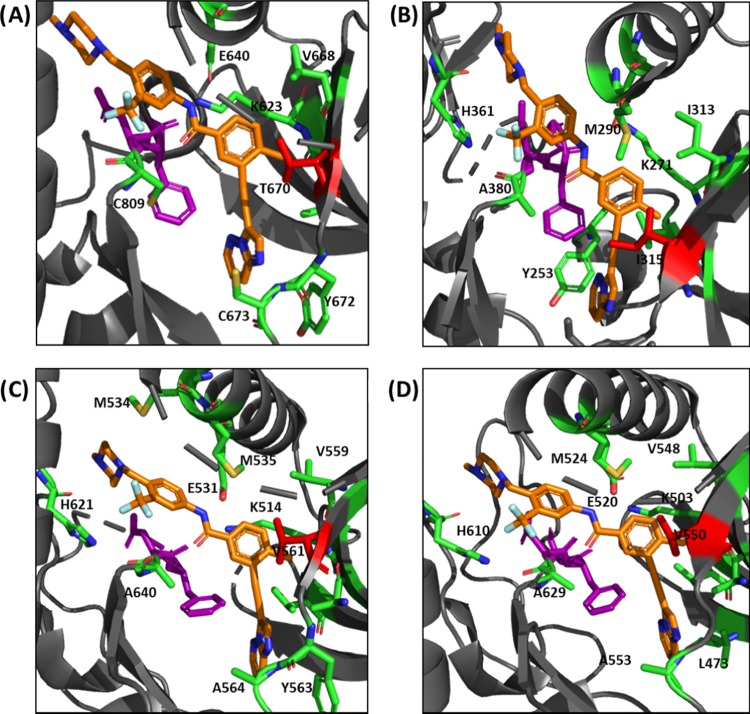
It is interesting that a −CH to −N switch in the benzamide of ponatinib can lead to such dramatic kinase selectivities and potencies. Analyses of the crystal structures of ponatinib bound to several kinases (Figure 6) did not reveal an obvious hydrogen bonding interaction between the introduced nitrogen and the active site residue, which would enhance binding. We however do not discount a role for an uncharacterized active site water molecule to mediate such interactions. In a beautiful work by Boxer et al., it was shown that the cyano moiety in bosutinib is engaged in the water-mediated hydrogen network.36 Future work, which solves the crystal structures of target kinases bound to HSN748, will help clarify how the nicotinamide nitrogen enhances binding.
Figure 6.
Active sites of various kinases with bound ponatinib (orange). Gatekeeper residues of each kinase are shown in red, DGF-motifs are shown in purple, amino acid residues within 4 Å of ponatinib are shown in green. (A) KIT, PDBID: 4V0I; (B) ABL1 with the T315I mutation, PDBID: 3IK3; (C) FGFR1, PDBID: 4V01; and (D) FGFR4, PDBID: 4UXQ.
We proceeded to test whether the degree of inhibition of FLT3, ABL1, RET, and FGFR-driven cancers by ponatinib and HSN748 mirrored the order of kinase inhibition. The IC50 for growth inhibition by both compounds against MV-4-11 (FLT3), K562 (ABL1), and LC2/ad (RET) was similar. HSN748 was better at inhibiting quizartinib-resistant AML (MOLM14-D835Y cell line) than ponatinib (compare IC50 of 0.69 nM for HSN748 and 52.6 nM for ponatinib). For the gilteritinib-resistant AML cell line, Molm14 (ITD, F691L), HSN748 was also more potent than ponatinib (IC50 of 0.18 nM for HSN748 and 6.8 nM for ponatinib).
Conclusions
We have shown that the replacement of the benzamide moiety of ponatinib with nicotinamide results in enhanced activity against drug-resistant AML, while reducing the inhibition of off-target kinases, such as Src, Kit, and FGFRs. HSN748 is a lead compound that warrants further consideration as an antileukemia compound, with a potentially less toxic profile than ponatinib. We are aware that idiosyncratic toxicities in real patient populations cannot be predicted based on in vitro kinase selectivity alone. Nonetheless, the fact that some level of kinase selectivity has been achieved is in the right direction toward a safer ponatinib analog. Ongoing work will conduct further preclinical toxicity experiments on this new ponatinib analog, with an eye toward clinical translation.
Experimental Section
Solvents and starting materials were purchased from commercial sources and used without further purifications.
General Cell Culture Procedure
The human cell lines were cultured using RPMI-1640 (Gibco), supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals), 1% glutaMAX, and 1% penicillin/streptomycin.
Cells were seeded with 2 × 103 cells in each well of a 96-well plate and incubated for 24 h. Cells were then treated for 72 h with serial dilutions of the desired compound. After the 72 h, cells were treated with the CellTiter-Blue Cell Viability Assay. The cells were then incubated for 3 h, and absorbance at 570 nm was measured using a microplate reader. IC50 values were then determined with GraphPad Prism.
General Kinase Assay Procedure (Provided by Reaction Biology)
The reaction buffer was composed of the following:
20 mM N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid (Hepes; pH 7.5), 10 mM MgCl2, 1 mM ethylene glycol tetraacetic acid (EGTA), 0.02% Brij35, 0.02 mg/mL bovine serum albumin (BSA), 0.1 mM Na3VO4, 2 mM dithiothreitol (DTT), and 1% dimethyl sulfoxide (DMSO).
The reaction procedure was as follows:
To the kinase and corresponding substrate (see the Supporting Information, Table S1 for details) in the reaction buffer were added compounds (100% DMSO stock solution) via acoustic technology (Echo500; nanoliter range). The mixture was incubated at room temperature for 20 min, after which 33P-ATP was added to the mixture (final ATP concentration of 100 μM) to start the reaction. The reaction mixture was incubated at room temperature for 2 h, and the degree of substrate phosphorylation was measured via the P81 filter-binding method.
Amide Coupling
5-Ethynyl-N-(4-((4-methylpiperazin-1-yl)methyl)-3-(trifluoromethyl)phenyl)nicotinamide, 1
A solution of 5-ethynylnicotinic acid (250 mg, 1.7 mmol, 1 equiv) and HATU (710 mg, 1.87 mmol, 1.1 equiv) in DMF (5 mL) was brought to 0 °C, and DIPEA (0.83 mL, 3 equiv) and 4-((4-methylpiperazin-1-yl)methyl)-3-(trifluoromethyl)aniline (464 mg, 1.7 mmol, 1 equiv) were added. The reaction was then moved to 50 °C and stirred for 12 h. After 12 h, the reaction was diluted with ethyl acetate (50 mL) and washed with water (3 × 20 mL) and brine solution (20 mL). Combined organic layers were dried over anhydrous sodium sulfate, filtered, and concentrated in vacuo. The pure product was then obtained via column chromatography. Yield = 85%. 1H nuclear magnetic resonance (NMR; 500 MHz, DMSO-d6) δ 10.70 (s, 1H), 9.07 (d, J = 2.2 Hz, 1H), 8.86 (d, J = 2.0 Hz, 1H), 8.41 (t, J = 2.1 Hz, 1H), 8.17 (d, J = 2.2 Hz, 1H), 8.00 (dd, J = 8.5, 2.2 Hz, 1H), 7.71 (d, J = 8.5 Hz, 1H), 4.58 (s, 1H), 3.55 (s, 2H), 2.36 (s, 8H), 2.15 (s, 3H); 13C NMR (126 MHz, DMSO-d6) δ 163.8, 154.9, 148.9, 138.4, 138.2, 133.0, 131.8, 130.3, 123.9, 118.9, 117.7, 85.7, 80.2, 57.9, 55.2, 53.1, 46.1; high-resolution mass spectrometry (HRMS; ESI+): calcd for C21H22F3N4O (M + H+) 403.1740, found 403.1740.
General Sonogashira Procedure
A solution of bromo compound (1 equiv), Pd(PPh3)2Cl2 (10 mol %), CuI (5 mol %), and triphenylphosphine (10 mg) in triethylamine was deoxygenated using argon gas. A deoxygenated solution of 5-ethynyl-N-(4-((4-methylpiperazin-1-yl)methyl)-3-(trifluoromethyl)phenyl)nicotinamide (1.2 equiv) in DMF was added slowly over 10 min to the solution. The reaction temperature was increased to 55 °C and stirred for 12 h. The reaction was then diluted with ethyl acetate. The organic layer was washed with water, saturated NH4Cl, and brine. Combined organic layers were dried with anhydrous sodium sulfate, filtered, and concentrated in vacuo. The pure product was obtained by flash column chromatography.
5-(Imidazo[1,2-b]pyridazin-3-ylethynyl)-N-(4-((4-methylpiperazin-1-yl)methyl)-3-(trifluoromethyl)phenyl)nicotinamide, HSN748
Reaction was performed using the standard Sonagashira coupling procedure. Yield = 53%. 1H NMR (500 MHz, DMSO-d6) δ 10.77 (s, 1H), 9.10 (d, J = 2.2 Hz, 1H), 8.97 (d, J = 2.0 Hz, 1H), 8.72 (d, J = 2.9 Hz, 1H), 8.55 (s, 1H), 8.27 (d, J = 8.6 Hz, 2H), 8.19 (s, 1H), 8.03 (d, J = 8.4 Hz, 1H), 7.72 (d, J = 8.5 Hz, 1H), 7.41 (dd, J = 9.2, 4.5 Hz, 1H), 3.56 (s, 2H), 2.38 (s, 8H), 2.16 (s, 3H). 13C NMR (126 MHz, DMSO-d6) δ 163.7, 154.0, 149.0, 145.6, 140.3, 139.4, 138.3, 137.6, 133.0, 131.8, 130.3, 126.7, 124.0, 119.9, 119.0, 117.7, 111.6, 94.9, 81.1, 57.9, 55.1, 53.1, 46.1; HRMS (ESI+): calcd for C27H25F3N7O (M + H+) 520.2067, found 520.2066.
5-(Imidazo[1,2-b]pyridazin-3-ylethynyl)-6-methyl-N-(4-((4-methylpiperazin-1-yl)methyl)-3-(trifluoromethyl)phenyl)nicotinamide, HSL420
Reaction was performed using the standard Sonagashira coupling procedure. Pd(PPh3)2Cl2 (10 mol %) was used as the catalyst. Yield = 38%. 1H NMR (500 MHz, DMSO-d6) δ 10.69 (s, 1H), 8.99 (d, J = 2.2 Hz, 1H), 8.73 (dd, J = 4.3, 2.2 Hz, 1H), 8.49 (d, J = 2.3 Hz, 1H), 8.28–8.24 (m, 2H), 8.19 (d, J = 2.5 Hz, 1H), 8.04 (d, J = 8.4 Hz, 1H), 7.70 (d, J = 8.5 Hz, 1H), 7.40 (ddd, J = 9.1, 4.5, 1.9 Hz, 1H), 3.56 (s, 2H), 2.80 (s, 3H), 2.41 (s, 8H), 2.21 (s, 3H). 13C NMR (126 MHz, DMSO-d6) δ 163.8, 162.7, 148.4, 145.6, 140.3, 139.1, 138.4, 137.7, 132.8, 131.8, 127.8, 126.7, 124.0, 119.8, 117.8, 111.8, 95.2, 83.7, 57.8, 55.0, 52.8, 45.8, 24.0. HRMS (ESI+): calcd for C28H27F3N7O (M + H+) 534.2224, found 534.2219.
5-(Imidazo[1,2-a]pyridin-3-ylethynyl)-N-(4-((4-methylpiperazin-1-yl)methyl)-3-(trifluoromethyl)phenyl)nicotinamide, HSL381
Reaction was performed using the standard Sonagashira coupling procedure. Pd(PPh3)2Cl2 (10 mol %) was used as the catalyst. Yield = 24%. 1H NMR (500 MHz, DMSO-d6) δ 10.81 (s, 1H), 9.11–9.02 (m, 2H), 8.80 (d, J = 6.7 Hz, 1H), 8.62 (d, J = 2.2 Hz, 1H), 8.21 (d, J = 2.2 Hz, 1H), 8.09 (s, 1H), 8.05 (dd, J = 8.5, 2.2 Hz, 1H), 7.73 (t, J = 9.4 Hz, 2H), 7.46 (dd, J = 9.0, 6.7 Hz, 1H), 7.17 (t, J = 6.7 Hz, 1H), 3.58 (s, 2H), 2.48 (s, J = 1.7 Hz, 8H), 2.25 (s, 3H). 13C NMR (126 MHz, DMSO-d6) δ 164.0, 153.9, 148.4, 139.5, 138.3, 137.4, 132.8, 131.8, 130.3, 128.1, 127.8, 127.5, 126.7, 125.8, 124.0, 123.6, 119.4, 118.0, 117.7, 114.5, 96.0, 81.4, 57.7, 54.8, 52.5, 45.5; HRMS (ESI+): calcd for C28H26F3N6O (M + H+) 519.2115, found 519.2111.
5-(Imidazo[1,2-a]pyrimidin-3-ylethynyl)-N-(4-((4-methylpiperazin-1-yl)methyl)-3-(trifluoromethyl)phenyl)nicotinamide, HSL382
Reaction was performed using the standard Sonagashira coupling procedure. Pd(PPh3)2Cl2 (10 mol %) was used as the catalyst. Yield = 29%. 1H NMR (500 MHz, DMSO-d6) δ 10.92 (s, 1H), 9.28 (dd, J = 6.8, 2.0 Hz, 1H), 9.12 (d, J = 2.2 Hz, 1H), 9.06 (d, J = 2.0 Hz, 1H), 8.71 (dt, J = 4.2, 2.4 Hz, 2H), 8.24 (d, J = 2.2 Hz, 2H), 8.08 (dd, J = 8.5, 2.2 Hz, 1H), 7.72 (d, J = 8.5 Hz, 1H), 7.30 (dd, J = 6.7, 4.2 Hz, 1H), 3.57 (s, 2H), 2.42 (s, 8H), 2.22 (s, 3H). 13C NMR (126 MHz, DMSO-d6) δ 163.9, 153.9, 152.6, 149.0, 148.7, 140.4, 138.4, 137.8, 135.3, 132.8, 131.8, 130.2, 128.0, 124.1, 123.6, 119.1, 117.8, 110.8, 106.4, 95.7, 80.6, 57.8, 54.9, 52.7, 45.7. HRMS (ESI+): calcd for C27H25F3N7O (M + H+) 520.2067, found 520.2063.
5-(Imidazo[1,2-a]pyrazin-3-ylethynyl)-N-(4-((4-methylpiperazin-1-yl)methyl)-3-(trifluoromethyl)phenyl)nicotinamide, HSL385
Reaction was performed using the standard Sonagashira coupling procedure. Pd(PPh3)2Cl2 (10 mol %) was used as the catalyst. Yield = 39%. 1H NMR (500 MHz, DMSO-d6) δ 10.85 (s, 1H), 9.21 (d, J = 1.5 Hz, 1H), 9.11 (dd, J = 18.4, 2.1 Hz, 2H), 8.88 (dd, J = 4.5, 1.5 Hz, 1H), 8.69 (d, J = 2.2 Hz, 1H), 8.28 (s, 1H), 8.21 (d, J = 2.2 Hz, 1H), 8.14 (d, J = 4.5 Hz, 1H), 8.05 (dd, J = 8.4, 2.2 Hz, 1H), 7.72 (d, J = 8.5 Hz, 1H), 3.57 (s, 2H), 2.48–2.28 (m, 8H), 2.23 (s, 3H). 13C NMR (126 MHz, DMSO-d6) δ 163.9, 154.1, 148.9, 143.7, 140.9, 140.4, 138.3, 137.9, 132.9, 131.8, 131.3, 130.3, 127.8, 124.0, 123.6, 119.9, 118.8, 117.8, 108.9, 96.7, 79.8, 57.8, 54.9, 52.7, 45.7. HRMS (ESI+): calcd for C27H25F3N7O (M + H+) 520.2067, found 520.2072.
N-(5-(Imidazo[1,2-b]pyridazin-3-ylethynyl)pyridin-3-yl)-4-((4-methylpiperazin-1-yl)methyl)-3-(trifluoromethyl)benzamide, HSL338
Reaction was performed using the standard Sonagashira coupling procedure. Pd(PPh3)2Cl2 (10 mol %) was used as the catalyst. Yield = 29%. 1H NMR (500 MHz, DMSO-d6) δ 10.76 (s, 1H), 8.97–8.90 (m, 1H), 8.71 (dd, J = 4.4, 1.5 Hz, 1H), 8.57–8.51 (m, 1H), 8.45 (t, J = 2.2 Hz, 1H), 8.28 (d, J = 1.8 Hz, 1H), 8.25 (ddd, J = 8.4, 5.4, 1.7 Hz, 3H), 7.95 (d, J = 8.1 Hz, 1H), 7.40 (dd, J = 9.2, 4.4 Hz, 1H), 3.69 (s, 2H), 2.50–2.37 (m, 8H), 2.26 (s, 3H). 13C NMR (126 MHz, DMSO) δ 165.1, 146.5, 145.5, 142.0, 139.2, 135.9, 133.5, 132.2, 131.3, 129.2, 128.0, 127.7, 126.6, 125.6, 123.5, 119.8, 119.0, 111.8, 95.3, 80.2, 57.8, 54.8, 52.7, 45.6. HRMS (ESI+): calcd for C28H25F3N7O (M + H+) 520.2067, found 520.2067.
3-(Imidazo[1,2-b]pyridazin-3-ylethynyl)-N-(4-((4-methylpiperazin-1-yl)methyl)-3-(trifluoromethyl)phenyl)benzamide, HSL331
Reaction was performed using the standard Sonagashira coupling procedure. Pd(PPh3)2Cl2 (10 mol %) was used as the catalyst. Yield = 36%. 1H NMR (500 MHz, DMSO-d6) δ 10.64 (s, 1H), 8.71 (dd, J = 4.4, 1.6 Hz, 1H), 8.25 (dd, J = 9.2, 1.6 Hz, 1H), 8.23–8.20 (m, 3H), 8.07–8.00 (m, 2H), 7.82 (dt, J = 7.7, 1.4 Hz, 1H), 7.70 (d, J = 8.5 Hz, 1H), 7.64 (t, J = 7.8 Hz, 1H), 7.39 (dd, J = 9.2, 4.4 Hz, 1H), 3.56 (s, 2H), 2.39 (s, 8H), 2.18 (s, 3H). 13C NMR (126 MHz, DMSO-d6) δ 165.2, 145.4, 139.0, 138.6, 135.4, 134.6, 132.6, 131.7, 130.5, 129.8, 129.1, 127.8, 126.6, 125.9, 124.0, 123.7, 122.4, 119.6, 117.8, 97.9, 78.0, 57.8, 55.0, 52.8, 45.8. HRMS (ESI+): calcd for C28H26F3N6O (M + H+) 519.2113, found 519.2114.
N-(4-((4-Methylpiperazin-1-yl)methyl)-3-(trifluoromethyl)phenyl)-5-(pyrazolo[1,5-a]pyrimidin-6-ylethynyl)nicotinamide, HSL412
Reaction was performed using standard the Sonagashira coupling procedure. Pd(PPh3)2Cl2 (10 mol %) was used as the catalyst. Yield = 32%. 1H NMR (500 MHz, DMSO-d6) δ 10.86 (s, 1H), 9.58 (d, J = 2.0 Hz, 1H), 9.13 (d, J = 2.2 Hz, 1H), 8.97 (d, J = 2.0 Hz, 1H), 8.71 (d, J = 2.1 Hz, 1H), 8.55 (t, J = 2.2 Hz, 1H), 8.34 (d, J = 2.3 Hz, 1H), 8.21 (d, J = 2.2 Hz, 1H), 8.05 (dd, J = 8.5, 2.2 Hz, 1H), 7.72 (d, J = 8.5 Hz, 1H), 6.84 (d, J = 2.2 Hz, 1H), 3.58 (s, 2H) 2.50–2.34 (m, 8H), 2.28 (s, 3H). 13C NMR (126 MHz, DMSO-d6) δ 163.8, 154.4, 151.3, 149.0, 147.1, 147.0, 139.2, 138.3, 138.1, 132.8, 131.9, 130.3, 127.8, 125.8, 124.0, 119.0, 117.8, 104.5, 98.0, 89.2, 87.6, 57.7, 54.7, 52.4, 45.3. HRMS (ESI+): calcd for C28H26F3N6O (M + H+) 520.2067, found 520.2063.
5-(Imidazo[1,2-a]pyrazin-5-ylethynyl)-N-(4-((4-methylpiperazin-1-yl)methyl)-3-(trifluoromethyl)phenyl)nicotinamide, HSL407
Reaction was performed using the standard Sonagashira coupling procedure. Pd(PPh3)2Cl2 (10 mol %) was used as the catalyst. Yield = 86%. 1H NMR (500 MHz, DMSO-d6) δ 10.87 (s, 1H), 9.17 (d, J = 2.0 Hz, 3H), 8.76 (q, J = 2.5 Hz, 1H), 8.57 (s, 1H), 8.32 (s, 1H), 8.21 (d, J = 2.2 Hz, 1H), 8.07–8.00 (m, 2H), 7.72 (d, J = 8.5 Hz, 1H), 3.57 (s, 2H), 2.47–2.27 (m, 8H), 2.22 (s, 3H). 13C NMR (126 MHz, DMSO-d6) δ 163.8, 154.8, 149.7, 143.7, 140.0, 138.7, 138.3, 136.8, 134.2, 133.0, 131.8, 130.4, 128.1, 125.8, 124.0, 117.9, 117.7, 115.4, 114.9, 96.8, 82.9, 57.8, 54.9, 52.7, 45.7, 40.5, 40.3, 40.1, 40.0, 39.8, 39.6. 13C NMR (126 MHz, DMSO-d6) δ 163.7, 154.8, 149.7, 143.7, 140.0, 138.7, 138.3, 136.7, 134.2, 132.9, 131.8, 130.4, 128.1, 124.0, 123.6, 117.9, 117.7, 115.4, 114.9, 96.8, 82.9, 57.8, 54.9, 52.7, 45.7. HRMS (ESI+): calcd for C27H25F3N7O (M + H+) 520.2067, found 520.2062.
Acknowledgments
The authors thank Prof. Neil Shah for providing drug-resistant AML cell lines MOLM14 (ITD, D835Y) and MOLM14 (ITD, F691L). The authors also thank Prof. Robert Kirken (University of Texas at El Paso) for donating KCL22 and KCL22-IR.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b03223.
1H and 13C NMR spectra; HPLC traces of compounds; reaction biology conditions (PDF)
Author Present Address
∥ J. Michael Bishop Institute of Cancer Research, 9-1 Keyuan Boulevard South, GaoXin District, Chengdu, China (N.N.).
Author Contributions
H.O.S. designed the project. E.L., E.F.Y.C., and N.N. performed experiments. H.O.S. and E.L. wrote the manuscript. All authors reviewed the manuscript.
The authors thank Purdue University, Purdue University Center for Cancer Research, and Elks Foundation for financial support. NMR and MS data were acquired by the NMR and MS facilities supported by NIH P30 CA023168.
The authors declare the following competing financial interest(s): H.O.S. is a co-founder of KinaRx LLC, a start-up company interested in developing therapies for malignant neoplastic diseases.
Supplementary Material
References
- U.S. Food & Drug Administration . FDA Approves Pronatinib. https://www.fda.gov/drugs/informationondrugs/approveddrugs/ucm381452.htm.
- Ren M.; Hong M.; Lui G.; Wang H.; Patel V.; Biddinger P.; Silva J.; Cowell J.; Hao Z. Noel FGFR inhibitor ponatinib suppresses growth of non-small cell lung cancer cells overexpressing FGFR1. Oncol. Rep. 2013, 29, 2181–2190. 10.3892/or.2013.2386. [DOI] [PubMed] [Google Scholar]
- De Falco V.; Buonocore P.; Muthu M.; Torregrossa L.; Basolo F.; Billaud M.; Gozgit J. M.; Carlomagno F.; Santoro M. Ponatinib (AP24534) is a novel potent inhibitor of oncogenic RET mutants associated with thyroid cancer. J. Clin. Endocrinol. Metab. 2013, 98, E811–E819. 10.1210/jc.2012-2672. [DOI] [PubMed] [Google Scholar]
- Gibbons D. L.; Pricl S.; Kantargian H.; Cortes J.; Quintas-Cardama A. The rise and fall of gatekeep mutations? The BCR-ABL T315I paradigm. Cancer 2012, 118, 293–299. 10.1002/cncr.26225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH U.S. National Library of Medicine . NCT02428543, Ponatinib for FLT3-ITD Acute Myelogenous Leukemia (PONATINIB-AML). https://clinicaltrials.gov/ct2/show/NCT02428543.
- NIH U.S. National Library of Medicine . NCT02265341, Ponatinib Hydrochloride in Treating Patients with Advanced Biliary Cancer with fgfr2 Fusions. https://clinicaltrials.gov/ct2/show/NCT02265341.
- NIH U.S. National Library of Medicine . NCT01813734, Ponatinib in Advanced NSCLC w/RET Translocation. https://clinicaltrials.gov/ct2/show/NCT01813734.
- Aghel N.; Delgado D. H.; Lipton J. H. Cardiovascular toxicities of BCR-ABL tyrosine kinase inhibitors in chronic myeloid leukemia: preventive strategies and cardiovascular surveillance. Vasc. Health Risk Manage. 2017, 13, 293–303. 10.2147/VHRM.S108874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasvolsky O.; Leader A.; Iakobishvili Z.; Wasserstrum Y.; Kornowski R.; Raanani P. Tyrosine kinase inhibitor associated vascular toxicity in chronic myeloid leukemia. Cardio-Oncology 2015, 1, 5 10.1186/s40959-015-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banavath H. N.; Sharma O. P.; Kumar M. S.; Baskaran R. Identification of novel tyrosine kinase inhibitors for drug resistant T315I mutant BCR-ABL: a virtual screening and molecular dynamics simulations study. Sci. Rep. 2014, 4, 6948 10.1038/srep06948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moslehi J. J.; Meininger M. Tyrosine kinase inhibitor-associated cardiovascular toxicity in chronic myeloid leukemia. J. Clin. Oncol. 2015, 35, 4210–4218. 10.1200/JCO.2015.62.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. D.; Zimmerman E. I.; Wang Y. D.; Orwick S.; Zatechka D. S.; Buaboonnam J.; Neale G. A.; Olsen S. R.; Enemark E. J.; Shurtleff S.; Rubnitz J. E.; Mullighan C. G.; Inaba H. Emergence of polyclonal FLT3 tyrosine kinase domain mutations during sequential therapy with sorafenib and sunitinib in FLT3-ITD-positive acute myeloid leukemia. Clin. Cancer Res. 2013, 19, 5758–5768. 10.1158/1078-0432.CCR-13-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro E. J.; Kummerle A. E.; Fraga C. A. M. The methylation effect in medicinal chemistry. Chem. Rev. 2011, 111, 5215–5246. 10.1021/cr200060g. [DOI] [PubMed] [Google Scholar]
- Pennington L. D.; Moustakas D. T. The necessary nitrogen atom: a versatile high-impact design element for multiparameter optimization. J. Med. Chem. 2017, 60, 3552–3579. 10.1021/acs.jmedchem.6b01807. [DOI] [PubMed] [Google Scholar]
- Vanotti E.; Amici R.; Bargiotti A.; Berthelsen J.; Bosotti R.; Ciavolella A.; Cirla A.; Cristiani C.; D’Alessio R.; Forte B.; Isacchi A.; Martina K.; Menichincheri M.; Molinari A.; Montagnoli A.; Orsini P.; Pillan A.; Roletto F.; Scolaro A.; Tibolla M.; Valsasina B.; Varasi M.; Volpi D.; Santocanale C. Cdc7 kinase inhibitors: Pyrrolopyridinones as potential antitumor agents. 1. Synthesis and structure-activity relationships. J. Med. Chem. 2008, 51, 487–501. 10.1021/jm700956r. [DOI] [PubMed] [Google Scholar]
- Verheij M. H. P.; Thompson A. J.; van Muijlwijk-Koezen J. E.; Lummis S. C. R.; Leurs R.; de Esch I. J. P. Design, synthesis, and structure–activity relationships of highly potent 5-HT3 receptor ligands. J. Med. Chem. 2012, 55, 8603–8614. 10.1021/jm300801u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remillard D.; Buckley D. L.; Seo H.; Ferguson F. M.; Dhe-Paganon S.; Bradner J. E.; Gray N. S. Dual inhibition of TAF1 and BET bromodomains from the BI-2536 kinase inhibition scaffold. ACS Med. Chem. Lett. 2019, 10, 1443–1449. 10.1021/acsmedchemlett.9b00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.; Zhou J.; Wang C.; Carter-Cooper B.; Yang F.; Larocque E.; Fine J.; Tsuji G.; Chopra G.; Lapidus R. G.; Sintim H. O. Identification of new FLT3 inhibitors that potently inhibit AML cell lines via an azo click-it/staple-it approach. ACS Med. Chem. Lett. 2017, 8, 492–497. 10.1021/acsmedchemlett.6b00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocque E.; Naganna N.; Ma Z.; Opoku-Temeng C.; Carter-Cooper B.; Chopra G.; Lapidus R. G.; Sintim H. O. Aminoisoquinoline benzamides, FLT3 and Src-family kinase inhibitors, potently inhibit proliferation of acute myeloid leukemia cell lines. Future Med. Chem. 2017, 9, 1213–1225. 10.4155/fmc-2017-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocque E. A.; Naganna N.; Opoku-Temeng C.; Lambrecht A. M.; Sintim H. O. Alkynylnicotinamide-based compounds as ABL1 inhibitors with potent activities against drug-resistant CML harboring ABL1(T315I) mutant kinase. ChemMedChem 2018, 13, 1172–1180. 10.1002/cmdc.201700829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganna N.; Opoku-Temeng C.; Choi E. Y.; Larocque E.; Chang E. T.; Carter-Cooper B. A.; Wang M.; Torregrosa-Allen S. A.; Elzey B. D.; Lapidus R. G.; Sintim H. O. Amino alkynylisoquinoline and alkynylnaphthyridine compounds potently inhibit acute myeloid leukemia proliferation in mice. EBioMedicine 2019, 40, 231–239. 10.1016/j.ebiom.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Naganna N.; Sintim H. O. Identification of nicotinamide aminonaphthyridine compounds as potent RET kinase inhibitors and antitumor activities against RET rearranged lung adenocarcinoma. Bioorg. Chem. 2019, 90, 103052 10.1016/j.bioorg.2019.103052. [DOI] [PubMed] [Google Scholar]
- Daina A.; Michielin O.; Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B.; West L. M. Estimating the lipophilicity of natural products using polymer reversed phase HPLC method. J. Liq. Chromatogr. Relat. Technol. 2009, 33, 118–132. 10.1080/10826070903430464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F. H.; Putoczki T. L.; Stylli S. S.; Luwor R. B. Ponatinib: a novel multi-tyrosine kinase inhibition against human malignancies. OncoTargets Ther. 2019, 12, 635–645. 10.2147/OTT.S189391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J.; Lam F.; Proud C.; Wang S. Targeting MNKs for cancer therapy. Oncotarget 2012, 3, 119–131. 10.18632/oncotarget.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. C.; Gromeier M. MNK Controls the mTORC1: Substrate association through regulation of TELO2 binding with mTORC1. Cell Rep. 2017, 18, 1444–1457. 10.1016/j.celrep.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lineham E.; Spencer J.; Morley S. J. Dual abrogation of MNK and mTOR: a novel therapeutic approach for the treatment of aggressive cancer. Future Med. Chem. 2017, 9, 1539–1555. 10.4155/fmc-2017-0062. [DOI] [PubMed] [Google Scholar]
- Destaing O.; Sanjay A.; Itzstein C.; Horne W. C.; Derek T.; De Cammilli P.; Baron R. The tyrosine kinase activity of c-Src regulates actin dynamic and organization of podosomes in osteoclasts. Mol. Biol. Cell 2008, 19, 394–404. 10.1091/mbc.e07-03-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Turro E.; Greene D.; Wijgaerts A.; Thys C.; Lentalgne C.; Bariana T. K.; Westbury S. K.; Kelly A. M.; Selleslag D.; Stephens J. C.; Papadia S.; Simeoni I.; Penkett C. J.; Ashford S.; Attwood A.; Austin S.; Bakchoul T.; Collins P.; Deevi S. V. V.; Favier R.; Kostadima M.; Lambert M. P.; Mathias M.; Millar C. M.; Peerlinck K.; Perry D. J.; Schulman S.; Whitehorn D.; Wittevrongel C.; BRIDGE-BPD Consortium; De Maeyer M.; Rendon A.; Gomez K.; Erber W. N.; Mumford A. D.; Nurgen P.; Stirrups K.; Bradley J. R.; Raymond F. L.; Laffan M. A.; Van Geet C.; Richardson S.; Freson K.; Ouwehand W. H.. A dominant gain-of-function mutation in universal tyrosine kinase SRC causes thrombocytopenia, myelofibrosis, bleeding, and bone pathologies. Sci. Transl. Med. 2016, 8, 328ra30 10.1126/scitranslmed.aad7666. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Galanis A.; Levis M. Inhibition of c-Kit by tyrosine kinase inhibitors. Haematologica 2015, 100, e77–e79. 10.3324/haematol.2014.117028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierro C.; Roden J.; Tabernero J. Fibroblast Growth Factor (FGF) Receptor/FGF inhibitors: novel targets and strategies for optimization of response of solid tumors. Semin. Oncol. 2015, 42, 801–819. 10.1053/j.seminoncol.2015.09.027. [DOI] [PubMed] [Google Scholar]
- Yanochko G. M.; Vitsky A.; Heyen J. R.; Hirakawa B.; Lam J. L.; May J.; Nichols T.; Sace F.; Trajkovic D.; Blasi E. Pan-FGFR inhibition leads to blockade of FGF23 signaling, soft tissue mineralization, and cardiovascular dysfunction. Toxicol. Sci. 2013, 135, 451–464. 10.1093/toxsci/kft161. [DOI] [PubMed] [Google Scholar]
- Karajannis M. A.; Vincent R.; Direnzo R.; Shmelkov S. V.; Zhang F.; Feldman E. J.; Bohlen P.; Zhu Z.; Sun H.; Kussie P.; Rafii S. Activation of the FGFR1β signaling pathway promotes survival, migration and resistance to chemotherapy in acute myeloid leukemia cells. Leukemia 2006, 20, 979–986. 10.1038/sj.leu.2404203. [DOI] [PubMed] [Google Scholar]
- Daver N.; Cortes J.; Ravandi F.; Patel K. P.; Burger J. A.; Konopleva M.; Kantarjain H. Secondary mutations as mediators of resistance to targeted therapy in leukemia. Blood 2015, 125, 3236–3245. 10.1182/blood-2014-10-605808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis M. FLT3 mutations in acute myeloid leukemia: what is the best approach in 2013?. Hematology 2013, 2013, 220–226. 10.1182/asheducation-2013.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson N. M.; Boxer S. G. A conserved water-mediated hydrogen bond network defines bosutinib’s kinase selectivity. Nat. Chem. Biol. 2014, 10, 127–132. 10.1038/nchembio.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.