ABSTRACT
Pro-spermatogonia (SG) serve as the gateway to spermatogenesis. Using single-cell RNA sequencing (RNAseq), we studied the development of ProSG, their SG descendants and testicular somatic cells during the perinatal period in mice. We identified both gene and protein markers for three temporally distinct ProSG cell subsets, including a migratory cell population with a transcriptome distinct from the previously defined T1- and T2-ProSG stages. This intermediate (I)-ProSG subset translocates from the center of seminiferous tubules to the spermatogonial stem cell (SSC) ‘niche’ in its periphery soon after birth. We identified three undifferentiated SG subsets at postnatal day 7, each of which expresses distinct genes, including transcription factor and signaling genes. Two of these subsets have the characteristics of newly emergent SSCs. We also molecularly defined the development of Sertoli, Leydig and peritubular myoid cells during the perinatal period, allowing us to identify candidate signaling pathways acting between somatic and germ cells in a stage-specific manner during the perinatal period. Our study provides a rich resource for those investigating testicular germ and somatic cell developmental during the perinatal period.
KEY WORDS: Single-cell RNA sequencing, Pro-spermatogonia, Gonocyte, Spermatogonial stem cell, Sertoli cells, Peritubular myoid cell, Leydig cells, Testis
Summary: Single-cell RNAseq analysis is used to examine molecular changes in testicular germ and somatic cells during perinatal period transitions, identifying new cell subsets and stage-specific markers.
INTRODUCTION
Pro-spermatogonia (ProSG) – also called gonocytes – are the precursor cells that give rise to spermatogonial stem cells (SSCs). Primary transitional (T1)-ProSG are mitotically quiescent cells that undergo genome-wide shifts in chromatin accessibility, 3D chromatin organization and epigenetic marks (Yamanaka et al., 2019). At postnatal day (P) 1, T1-ProSG begin converting into secondary transitional (T2)-ProSG, which initiate proliferation and are considered the immediate precursor cells that give rise to SSCs (McCarrey, 2013). During the time interval in which this T1-to-T2-ProSG conversion event occurs, these transitioning germ cells migrate from the center of the seminiferous tubule to its periphery (Kluin and de Rooij, 1981; McGuinness and Orth, 1992). This site is where SSCs are thought to first form from T2-ProSG, and it is also where SSCs reside throughout their lifetime (McLean et al., 2003). Thus, the periphery of seminiferous tubules is regarded as the SSC niche (Oatley and Brinster, 2012).
T2-ProSG not only give rise to SSCs but are the transient cell population that generates the ‘first wave of spermatogenesis’, which allows mice to generate sperm as early as ∼5 weeks of age (Bellve et al., 1977). Thus, T2-ProSG are unique cells that have two fates. One fate leads to the rapid production of male gametes and the other fate leads to the production of SSCs and consequent long-term spermatogenesis.
Progress on clarifying the developmental and molecular events that accompany and underlie ProSG development has been hindered, in part, by a dearth of ProSG subset-specific markers. There have been several challenges in identifying ProSG subset-specific markers, including the fact that ProSG subsets exhibit only subtle differences morphologically, they overlap with each other temporally, and they undergo asynchronous changes as they mature in vivo (Culty, 2013; McCarrey, 2013; Vergouwen et al., 1991).
Perinatal testes not only harbor germ cells but also several somatic cell types, including Sertoli cells (SCs), Leydig cells (LCs) and peritubular myoid cells (PTMs). SCs are the only somatic cells in direct contact with germ cells, allowing them to provide many ‘nurse cell functions’ (Griswold, 1998). LCs are located in the connective tissue surrounding the seminiferous tubules, where they produce testosterone, which is crucial for a variety of functions during the fetal, postnatal and adult stages (Griswold and Behringer, 2009). PTMs surround seminiferous tubules, where they provide contractile functions and secrete factors important for spermatogenesis, including GDNF, which is essential for undifferentiated SG to develop in both postnatal and adult mice (Chen et al., 2016). Although the functions of these somatic cell types in spermatogenesis is relatively well established, little is known about their development and function prior to spermatogenesis, particularly at the molecular level (Griswold and Behringer, 2009; Meroni et al., 2019; Nurmio et al., 2012).
Here, we elucidated the nature of the germ and somatic cell types that populate the mouse testes during the perinatal period using single-cell RNA sequencing (scRNAseq) analysis. Recently, several groups, including ours, used scRNAseq analysis to investigate adult spermatogenesis in humans and mice (Guo et al., 2018; Hermann et al., 2018; Sohni et al., 2019; Velte et al., 2019; Wang et al., 2018). Two recent studies used scRNAseq analysis to investigate the steps leading up to spermatogenesis (Law et al., 2019; Liao et al., 2019). Liao et al. identified early postnatal SG subsets, including a CD87+ cell population, from scRNAseq analysis of purified germ cells from P5.5 testes (Liao et al., 2019). Law et al. studied the transcriptome and cellular dynamics that accompany SSC specification through analysis of purified germ cells from multi-lineage reporter mice (Law et al., 2019). Using multiple approaches, including scRNAseq analysis, they defined heterogeneous ProSG and SG populations, along with the associated transcriptomes and putative regulatory networks. Germ-cell transplantation analysis revealed that Id4-eGFP+, but not Id4-eGFP−, E16.5 germ cells were able to rescue spermatogenesis in germ cell-deficient mice, suggesting that Id4-eGFP+ ProSG are fated to become SSCs.
To obtain an unbiased view of the developmental and molecular events occurring during perinatal testicular development, we performed scRNAseq analysis on unfractionated testicular cells from the perinatal period. This analysis, described herein, provides detailed information on both germ and somatic cells during the perinatal period.
RESULTS
Identification of the testicular cell types present during the perinatal period
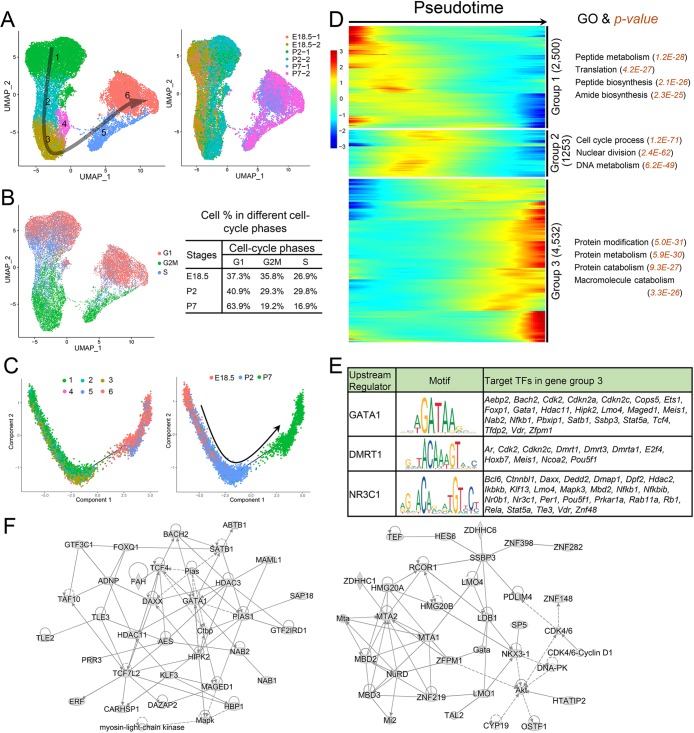
We performed scRNAseq analysis on dissociated cells from freshly isolated whole testes obtained from embryonic day (E) 18.5, P2 and P7 mice. After filtering out poor-quality cells, 50,859 cells remained for subsequent analysis. Using a nonlinear dimensionality-reduction technique – uniform manifold approximation and projection (UMAP) (Becht et al., 2018) – we identified cell clusters corresponding to eight major cell types (Fig. 1A). Some of the gene markers used to define these cell clusters are shown in Fig. 1B. Biological replicates exhibited similar cell distributions (Fig. S1A). Fig. S1B shows standard quality control parameters after filtering. Genes exhibiting enriched expression in each cell subset are shown in Fig. 1C and Table S1. Gene ontology (GO) and ingenuity pathway analyses (IPAs) identified functions enriched for the different cell types (Fig. 1D; Fig. S1C). Many cell clusters exhibited dramatic shifts in gene expression between E18.5 and P7 (Fig. S1A), indicative of dynamic developmental changes during the perinatal period.
Fig. 1.
Identification of the major cell types in E18.5, P2 and P7 mouse testes. (A) UMAP plot of cells from E18.5, P2 and P7 mouse testes. Number of cells in each cluster: germ cells (2898), SCs (23,827), LCs (1910), PTMs (3599), stromal cells (16,704), endothelial cells (1001), macrophage (641) and innate lymphocyte (279). (B) Representative markers for each cell type. (C) Heatmap of differentially expressed genes (DEGs) from the eight major cell types. Top: cell types and cell number; left: the number of DEGs; right: representative enriched genes for each cell type. (D) The most statistically significant gene ontology (GO) terms for each cell type.
Germ cell stages traversing the ProSG-to-SG transition period
We next performed re-clustering analysis on only the germ cells, which revealed the existence of six distinct germ cell clusters (Fig. 2A). Only one cell cluster is present at E18.5 (Fig. 2A). This cell population has a cell-cycle gene expression pattern indicative of non-proliferating cells (Fig. S2A), which is consistent with the evidence that only the proliferative quiescent cell population – called ‘T1-ProSG’ – are present during late embryogenesis (McCarrey, 2013). This T1-ProSG cell cluster expresses genes encoding proteins relevant to DNA methylation (Fig. S2B), consistent with the genome-wide re-establishment of DNA methylation marks at this stage (Tseng et al., 2015). At P2, we identified two cell clusters. One P2 cluster broadly expresses cell-cycle genes (Fig. 2A), corresponding to proliferative T2-ProSG (McCarrey, 2013). The other P2 cluster, which we named ‘intermediate (I)-ProSG’, largely lacks cell-cycle gene expression and has migratory properties (Fig. 2A), as detailed below. At P7, we identified three cell clusters that we named ‘SSC-1’, ‘SSC-2’ and ‘differentiating (Diff)-SG’, based on their expression of known SSC, progenitor and Diff-SG gene markers (Fig. S2C). Genes selectively expressed by these six germ cell clusters are shown in Fig. 2B and Table S1. GO categories significantly enriched in these germ cell clusters are shown in Fig. 2B.
Fig. 2.
Identification of perinatal germ cell clusters. (A) UMAP plot of germ cells from E18.5, P2 and P7 mouse testes. Left, germ cell subsets; right, sample source. Number of cells in each cluster: T1-ProSG (138), I-ProSG (103), T2-ProSG (168), SSC-1 (634), SSC-2 (419) and Diff-SG (440). (B) Heatmap of DEGs from the germ cell subsets. Top: cell types; left: number of DEGs; right: representative enriched biological processes and genes. (C) CDKN2A is a pan germ cell marker during the perinatal stage. Top: the UMAP plot of Cdkn2a expression in all testicular cell types and in germ cells. Bottom: IF analysis of P2 testis section co-stained with antibodies against CDKN2A and TRA98. Scale bar: 20 µm. (D) DNMT3L is a T1-ProSG marker. Left: UMAP and Violin plots of Dnmt3l expression. Right: IF analysis of P0-P5 testes sections co-stained with antibodies against DNMT3L and KI-67. Scale bar: 20 µm. (E) UMAP plots showing expression of candidate gene markers for both I- and T2-ProSG. (F) IF analysis of P2 and P3 testes sections stained with the antibodies shown. The sections were co-stained with an antibody against KI-67. Scale bar: 20 µm. Right: quantification of the candidate markers-positive cells that co-stain or not with KI-67 and are in the indicated compartment at P2. n, number of cells counted. (G) Model of germ cell development during the perinatal period, see text for details.
Inspection of the germ cell-enriched genes (Fig. 1C) suggested that Cdkn2a, which encodes a cyclin-dependent kinase inhibitor, is a pan-ProSG marker (Fig. 2C). Immunofluorescence (IF) analysis showed that its protein product, CDKN2A, is also a pan-ProSG marker, based on its co-expression with the postnatal/adult germ cell marker TRA98 (Tanaka et al., 1998) (Fig. 2C). CDKN2A is expressed on the cell surface (Zhang et al., 1998), raising the possibility that it could be used to purify ProSG. In support, fluorescence-activated cell sorting (FACS)-purified CDKN2A+ cells (∼3.7% of total P2 testicular cells) were enriched for expression of ProSG genes, and largely lacked expression of testicular somatic cells (Fig. S2D).
We found that the Dnmt3l gene is preferentially expressed in T1-ProSG (Fig. 2D), leading to the possibility that its encoded protein specifically labels this ProSG stage. In support, it was previously shown that DNMTL is expressed in ProSG (Sakai et al., 2004), but its specificity for this stage was not determined. Using an antiserum against DNMT3L, we found it strongly stained germ cells at P0 and had dramatically reduced staining at P2 (Fig. 2D), consistent with the expression pattern of T1-ProSG (Culty, 2013; McCarrey, 2013). Co-staining with an antibody against the proliferation marker MKI67 (KI-67) (Gerdes et al., 1983) showed that the vast majority of DNMT3L+ cells are non-proliferative (Fig. 2D), another key characteristic of T1-ProSG (McCarrey, 2013).
Identification of distinct ProSG subsets engaging in proliferation and migration
As described above, we identified a previously unrecognized ProSG subset – I-ProSG – present in the perinatal testis (Fig. 2A). Monocle trajectory analysis showed that I-ProSG are developmentally less advanced than T2-ProSG, placing them at an intermediate position between T1-ProSG and T2-ProSG (Fig. S2E). Many genes involved in cell migration and actin organization exhibit enriched expression in I-ProSG (Fig. S2A,F).
Two genes preferentially expressed in I-ProSG are Elmo1 and Palld (Fig. 2E), which both encode proteins involved in cell migration and cytoskeleton organization (Pogue-Geile et al., 2006; Raftopoulou and Hall, 2004). IF analysis showed that the corresponding proteins, ELMO1 and PALLD, mainly mark germ cells located at the center of seminiferous tubules and are predominantly KI-67 negative (Fig. 2F), consistent with our evidence that I-ProSG are largely non-proliferative (Fig. S2A). The few ELMO1+ and PALLD+ cells at the seminiferous tubule periphery are mostly KI-67+ (Fig. 2F), indicating that I-ProSG undergo proliferative expansion once these cells migrate to the periphery. The ELMO1 and PALLD immunosignal is much dimmer at P3 than at P2 (Fig. 2F), suggesting that these proteins transiently mark migrating ProSG and shut off soon after I-ProSG migrate to the seminiferous tubule periphery.
We found that Etv4 preferentially labels T2-ProSG (Fig. 2E). Its corresponding protein, the transcription factor ETV4, marks T2-ProSG, based on its expression in proliferative (KI-67+) cells at P2 (Fig. 2F). In contrast with I-ProSG (ELMO1+ and PALLD+ cells), which became rare by P3, ETV4+ cells remained abundant at P3 (Fig. 2F), consistent with pseudotime analysis that indicated that T2-ProSG follow I-ProSG developmentally (Fig. S2E). Furthermore, most ETV4+ cells are in the periphery of seminiferous tubules at P2, indicating that most ETV4+ cells have already undergone migration, whereas most I-ProSG (ELMO1+ and PALLD+ cells) are in the center of seminiferous tubules at P2 (Fig. 2F).
Together, these data support a model in which T1-ProSG convert into migratory I-ProSG that translocate from the center of seminiferous tubules to their periphery, whereupon the I-ProSG become proliferative T2-ProSG, which have two fates: (1) conversion into Diff-SG, which undergo the first round of spermatogenesis, and (2) conversion into SSCs for long-term spermatogenesis (Fig. 2G). We note that because ProSG migration to the seminiferous tubule periphery is asynchronous (Kluin and de Rooij, 1981; McGuinness and Orth, 1992), it is not possible, using current methods, to test whether I-ProSG are exclusively migratory.
To validate our cell cluster analysis, we took advantage of two recently published studies that performed scRNAseq analysis on perinatal germ cells (Law et al., 2019; Liao et al., 2019).
Liao et al. examined FACS-purified Oct4-GFP+/KIT− male germ cells from P5.5 mice (Liao et al., 2019). We bioinformatically combined their scRNAseq dataset with ours and found that their P5.5 germ cell cluster overlapped with our P7 cell cluster (Fig. S3A), thus validating our clustering analysis of the P7 time point.
Law et al. examined Tomato fluorescent protein-positive cells from tdTomatoflox_STOP_flox; Blimp-Cre mice (which label primordial germ cells and their descendants) purified by FACS from E16.5 to P6 mice (Law et al., 2019). We combined their scRNAseq datasets with ours (both had similar read depths), but we did not observe a cluster pattern with a coherent temporal sequence, even when using the same sequence alignment tool (data not shown). We then re-analyzed the Law et al. dataset using the same alignment, filtering, normalization and clustering parameters used for our dataset, which revealed nine clusters (Fig. S3B). Analysis of the genes preferentially expressed from each of these clusters (Table S1) revealed that C1 and C3 correspond best to the T1-ProSG cluster we defined; C3 and C4 correspond to I-ProSG; and C4 and C5 correspond to T2-ProSG (Fig. S3C). Consistent with C1 and C3 being the least advanced, most cells in these clusters have a cell-cycle gene expression pattern indicative of their being mitotically quiescent (Fig. S3D), a characteristic of T1-ProSG (McCarrey, 2013). The T1-, I- and T2-ProSG markers we identified preferentially labeled the corresponding Law et al. cell clusters (Fig. S3E). Conversely, the well-established stage-specific germ cell markers used by Law et al. exhibited the expected expression pattern in the cell subsets identified by our study: SSC-associated markers are mainly expressed in the ProSG and SSC-1 clusters, whereas progenitor and differentiation markers are mainly expressed in the SSC-2 and Diff-SG clusters, respectively (Fig. S3F).
Characterization of perinatal undifferentiated SG
As described above, we identified three cell clusters at P7 that we named ‘SSC-1’, ‘SSC-2’ and ‘Diff-SG’ (Fig. 2A). SSC-1 and SSC-2 mainly comprised undifferentiated SG, based on expression of known undifferentiated SG markers (Fig. 2B; Fig. S2C). SSC-1 expressed higher levels of many SSC marker genes (e.g. Gfra1 and Id4) than SSC-2 (Fig. S2C), suggesting that SSC-1 are more primitive than SSC-2. In contrast, SSC-2 express higher levels of progenitor marker genes (e.g. Neurog3, Nanos3 and Rarg) than SSC-1 (Fig. S2C). Although we did not perform a functional assay to ascertain the function of cells in the SSC-1 and -2 clusters, their expression pattern suggests that SSC-1 and SSC-2 are enriched for SSCs and progenitors, respectively.
Re-clustering of the P7 germ cells separated the SSC-1 subset into two cell clusters that we named SSC-s1 and SSC-s2 (Fig. 3A), which differentially express several genes (Fig. 3B; Table S1). SSC-s1 expresses the recently identified SSC markers, Eomes and Cd87 (also known as Plaur) (Fayomi and Orwig, 2018; Liao et al., 2019) (Fig. S4A; Table S1), raising the possibility that SSC-s1 is the more primitive of these two cell clusters, a possibility confirmed by Monocle pseudotime analysis (Fig. 3C,D). Genes involving ‘integrin-mediated signaling’ and ‘vessel development’ are enriched in SSC-s1 (Fig. S4B,C), suggesting these processes may play roles in SSC development.
Fig. 3.
Undifferentiated SG clusters during the SSC establishment period. (A) UMAP plot of SG from P7 mouse testis. Number of cells in each cluster: SSC-s1 (271), SSC-s2 (328), SSC-2 (479) and Diff-SG (418). (B) UMAP plots of DEGs in the indicated SG cell clusters. (C,D) Pseudotime trajectory analysis of the indicated cell clusters. (E) Heatmap of DEGs in different SG subsets, following the trajectory timeline shown in D. Top: pseudotime direction; left: the gene groups (and the number of genes in each); right: representative enriched genes and the most statistically significant GO terms for each cluster. (F) The expression pattern of marker genes along the pseudotime axis. (G) IF analysis of P7 testes sections from Id4-GFP mice stained with the LHX1 antibody. Scale bar: 20 µm. Right: quantification of double-stained cells. n, number of cells counted. (H) FACS plot of P7 mouse Id4-eGFP testicular cells stained with CD82 antibody. The percentage of positive cells are indicated. Left: only stained with secondary antibody (Ctrl).
To define the molecular events involved in perinatal SG development, we identified genes statistically enriched in cells along the trajectory pseudotime axis. This analysis identified four distinct patterns of gene expression dynamics (Fig. 3E). Group-1 genes are expressed in the most primitive SG and thus are candidates to be involved in SSC formation. Significantly enriched GO categories for group-1 genes include ‘system development’ and ‘signaling transduction’ (Fig. 3E). Group-2 and -3 genes are mainly expressed in progenitors; enriched GO categories corresponding to these two gene expression pattern groups include ‘metabolism’, ‘biosynthesis’ and ‘cell surface receptor signaling’. Group-4 genes are mainly expressed in Diff-SG; enriched GO categories include ‘metabolism’ and ‘cell cycle process’, which is consistent with the evidence that differentiating SG undergo proliferative expansion (Suzuki et al., 2012).
To identify novel SSC markers, we screened the genes in group 1 for those with known functions in stem cells. This screen identified Lhx1 and Cd82 as fulfilling this criterion (Fig. 3B,F). To check their specificity further, we leveraged a published scRNAseq dataset from P6 germ cells purified based on the Id4-eGfp signal (Hermann et al., 2018). This revealed that both Lhx1 and Cd82 are mainly expressed in Id4-eGFPbright cells (Fig. S4D,E), which are widely regarded as the most specific mouse SSC marker currently known (Helsel et al., 2017). To determine the specificity of the proteins encoded by these two genes, we examined LHX1 and CD82 protein expression relative to Id4-eGFP. IF analysis showed that virtually all LHX1+ cells were also Id4-eGFP+ (Fig. 3G), implying that LHX is highly selective for SSCs. Interestingly, only ∼69% of Id4-eGFP+ cells were LHX1+ (Fig. 3G), raising the possibility that LHX1 marks a subset of SSCs. Using two different antisera against CD82, we found that CD82 also labels Id4-eGFP+ cells, but because these antisera also stained the SC cytoplasm (Fig. S4F), we were not able to quantify the percentage of positive germ cells using IF analysis. As an alternative approach, we performed FACS analysis with one of these antisera and found that CD82 marked a highly select group of testicular cells (∼0.48% of the total), mainly Id4-eGFPbright cells (Fig. 3H). Together, these results suggest that both LHX1 and CD82 are SSC marker proteins.
Identification of stage-specific signaling pathways and transcription factors (TFs) during perinatal germ cell development
To identify signaling pathways that are potentially involved in perinatal germ cell development, we screened for signaling component genes exhibiting stage-specific expression in the seven germ clusters we identified. This revealed many signaling pathways predicted to be selectively active in specific stages (Fig. S5A; Table S1).
To define TFs that may be involved in regulating genes in these cell subsets, we took advantage of IPA, which uses known consensus DNA-binding sites to define TF targets (Krämer et al., 2014). Fig. S5B provides a list of the TF genes predicted to target genes preferentially expressed in each subset. Their putative target genes in each cell subset are listed in Table S1. Most of these TFs are expressed in the cell subsets in which they are predicted to act (Fig. S5C). Some of these TFs are predicted to act in several cell subsets [e.g. MYC and TP53 (TRP53)], but most display selectivity, including the pluripotency factor, POU5F1, which is predicted to act in I-ProSG; and the transcriptional repressor, REST, which is predicted to act in SSC-2. Some of the TFs are directly involved in signaling pathways (e.g. SMAD4 is the core factor in the TGFβ signaling pathway).
As another approach to define key TFs important during perinatal germ cell development, we identified TFs exhibiting enriched expression in each cell subset we defined (Table S1). Some of these TF genes, including Sall4, Bcl6b, Dmrtc2, Lhx1, T and Dmrt1, were previously shown to have roles in SSCs (Chan et al., 2017; Gegenschatz-Schmid et al., 2017; Oatley et al., 2006; Zhang et al., 2016).
Comparison of SSC-dependent and -independent waves of spermatogenesis
To determine whether there are differences in SG between the first (SSC-independent) and subsequent (SSC-dependent) waves of spermatogenesis, we combined our P7 testes scRNAseq dataset with published testes scRNAseq datasets from older (postnatal and adult) mice (Grive et al., 2019). The SG subsets were annotated as ‘SSC-1’, ‘SSC-2’ and ‘Diff-SG’, based on the gene markers we used to define cell clusters in our P7 scRNAseq datasets. Pearson correlation analysis showed that the SSC-1 subset from these different ages (P6 to adult) are highly related (Fig. S5D). The same was the case for the SSC-2 subset (Fig. S5D). This suggests that SSCs and progenitors exhibit few differences in gene expression as they transition from the neonatal period to adult spermatogenesis. In contrast, the Diff-SG subset exhibited detectable differences in gene expression between the first wave (before P30) and later stages (P30 and adult; Fig. S5D), indicative of marked changes in the molecular events that occur during the first and subsequent waves of SG differentiation. Table S1 lists the unique gene signatures for each SG subset at each stage.
We also analyzed spermatocytes (SPCs) for age-dependent differences in gene expression. Pearson correlation analysis showed that the expression pattern of SPCs when they first emerge (P14) (Bellve et al., 1977) is significantly different from SPCs derived from older mice, including P18 mice (Fig. S5E). This suggests that early SPCs generated during the first round of spermatogenesis differ from those generated later. Consistent with first-round SPCs exhibiting a unique pattern of expression, GO analysis revealed that ‘translation’ and ‘RNA splicing’ functions are enriched in P14 SPCs, whereas adult SPCs are enriched for ‘gametogenesis’-related functions (Fig. S5F).
Somatic cell heterogeneity and development during the perinatal period
To analyze the development of somatic cells during the perinatal period, we independently re-clustered the three known major somatic cell types from our scRNAseq datasets.
SCs
Re-clustering of the SCs from the E18.5, P2 and P7 testicular datasets generated six SC clusters (Fig. 4A), suggesting that SCs undergo discrete step-wise shifts in gene expression as they develop during the perinatal period. E18.5 and P2 SCs are in common clusters, whereas P7 SCs are in distinct clusters (Fig. 4A), suggesting that SCs undergo a dramatic change between P2 and P7. Consistent with this, we identified many more uniquely expressed genes at P7 (257) than at E18.5 and P2 (81 and 17, respectively) (q<0.01; Table S2). Consistent with the known gradual decline in SC proliferation during early postnatal development (Vergouwen et al., 1991), we observed a progressive decline in the percentage of proliferating SCs between E18.5 and P7 (as defined by the expression of cell-cycle genes; Fig. 4B).
Fig. 4.
Identification of SC clusters during the perinatal period. (A) UMAP plot of SCs from E18.5, P2 and P7 mouse testes. Left: SC clusters; right: sample source. Arrow indicates the inferred developmental direction from the analysis shown in C. Number of cells in each cluster: 1 (8693), 2 (2358), 3 (3273), 4 (566), 5 (1647) and 6 (5242). (B) Left: UMAP plot inferring the cell-cycle phase based on expression of a large set of G2/M- and S-phase genes (Kowalczyk et al., 2015). Right: percentages of SCs in different cell-cycle phases from different stages. (C) Monocle pseudotime trajectory analysis of the SC clusters. Left: SC clusters; right: segregated by perinatal age. (D) Heatmap of DEGs from different SC subsets following the trajectory timeline shown in C. Top: pseudotime directions; right: the number of DEGs and the representative biological processes and P-values. (E) Left: upstream regulator (TF) genes exhibiting enriched expression in group 3 (shown in D) that are predicted (by the IPA program) to target group 3 TF genes. Middle: target sequences predicted by the ENCODE database. Right: TF genes predicted to be regulated by the indicated upstream regulator. Znf48 is also known as Zfp553. (F) Gene networks inferred from transcription factors enriched in the gene cluster 3 shown in D.
Pseudotime trajectory analysis revealed a linear trajectory of SCs from E18.5 to P2 to P7 (Fig. 4C). Three patterns of gene expression dynamics were identified along the pseudotime trajectory, each with statistically enriched GO functions (Fig. 4D) and signaling pathways (Table S2). These three gene groups tended to correspond to the developmental age of the SCs, with group-1, -2 and -3 genes being enriched in most primitive, intermediate and later stage of SCs, respectively. We identified 148, 98 and 281 TF genes expressed in groups 1, 2 and 3, respectively (Table S2). Several group-3 TF genes, including Gata1, Dmrt1 and Nr3c1, are predicted to target other TF genes in group 3 (Fig. 4E), suggesting the existence of TF cascades in developing SCs. Indeed, IPA predicted that group-3 TFs form complex regulatory networks (Fig. 4F).
LCs
Re-clustering of LCs generated three distinct cell clusters, with each cluster largely representing a different time point (Fig. S6A). Genes involved in ‘apoptosis’ are enriched in the P2 LC cluster (Fig. S6B), consistent with the timing of fetal LC loss (Griswold and Behringer, 2009). Pseudotime trajectory analysis showed a linear trajectory of LCs from E18.5 to P2 to P7 (Fig. S6C). Three patterns of gene expression dynamics were identified along the pseudotime trajectory, each with statistically enriched GO functions (Fig. S6D) and signaling pathways (Table S2). We identified 52, 67 and 35 TF genes expressed in groups 1, 2 and 3, respectively (Table S2). Fig. S6E shows a gene network inferred by Ingenuity analysis to be formed by group-2 TFs in LCs. Key nodes in this inferred network are AKT (AKT1) and FOXO1, raising the possibility that AKT signaling and the FOXO1 TF have key roles in the fetal-to-adult LC transition (Griswold and Behringer, 2009).
PTMs
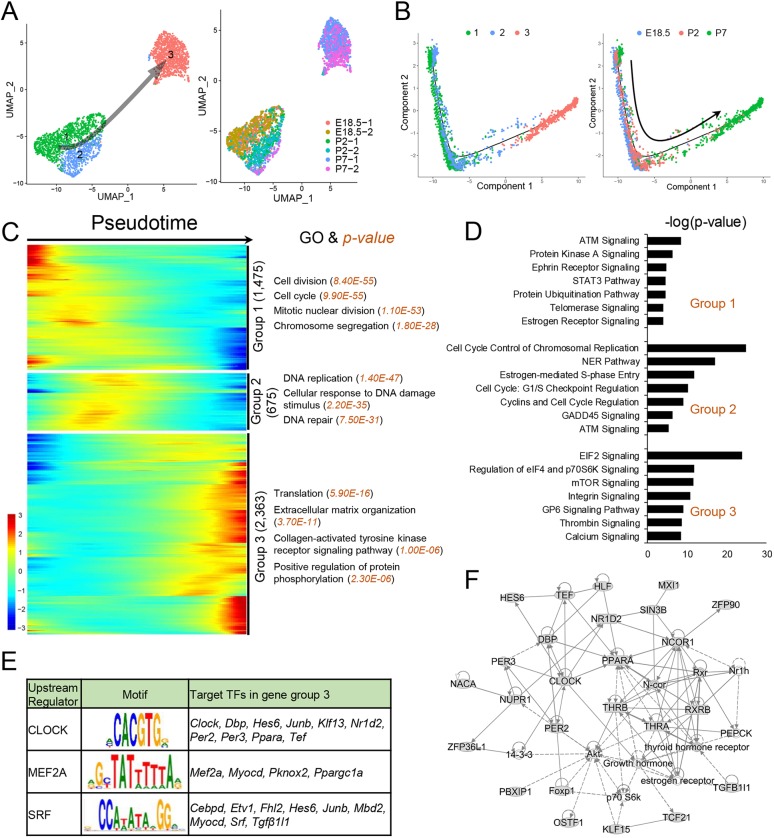
Re-clustering of PTMs generated three distinct cell clusters, with each cluster representing a different time point (Fig. 5A). Enriched genes from E18.5 and P2 partially overlapped, indicative of only a modest shift in gene expression between these time points, whereas enriched genes in the P7 cluster were largely unique, indicative of a relatively dramatic shift in PTM gene expression between P2 and P7 (Table S2). Pseudotime analysis showed a linear trajectory of myoid cells from E18.5 to P7 (Fig. 5B). Three patterns of gene expression dynamics were identified along the pseudotime trajectory, each with statistically enriched GO functions (Fig. 5C) and signaling pathways (Fig. 5D). We identified 131, 53 and 123 TF genes exhibiting enriched expression in groups 1, 2 and 3, respectively (Table S2). Several group-3 TF genes, including Clock, Mef2a and Srf, are predicted to target other TF genes in group 3 (Fig. 5E), suggesting the existence of TF cascades in developing PTMs. Indeed, group-3 TFs are predicted to form complex gene networks comprising many factors, including thyroid hormone receptor and estrogen receptor (Fig. 5F). This raises the possibility that hormone signaling is involved in PTM development during the perinatal period.
Fig. 5.
Identification of PTM clusters during the perinatal period. (A) UMAP plot of PTMs from E18.5, P2 and P7 mouse testes. Left: PTM clusters; right: developmental stage information. Arrow indicates the inferred developmental direction from the analysis shown in C. Number of cells in each cluster: 1 (1068), 2 (532) and 3 (1227). (B) Pseudotime trajectory analysis of the PTM clusters. Left: PTM clusters; right: segregated by perinatal age. (C) Heatmap of DEGs from different PTM subsets following the trajectory timeline shown in B. Top: pseudotime direction; right: the number of DEGs and the representative biological processes and P-values. (D) The top enriched signaling pathways of DEGs as defined in C. (E) Left: upstream regulator (TF) genes exhibiting enriched expression in group 3 (shown in C) that are predicted (by the IPA program) to target group 3 TF genes. Middle: target sequences predicted by the ENCODE database. Right: TF genes predicted to be regulated by the indicated upstream regulator. (F) Gene network inferred from transcription factors enriched in the gene cluster 3 shown in C.
Evidence for a dramatic burst in Hippo signaling during the ProSG-to-SG transition
Our analysis, described above, indicates that both germ cells and somatic cells undergo dynamic gene expression changes during the perinatal period. To define signaling pathways temporally regulated during this period, we used IPA to identify signaling pathways predicted to be active in germ cells, SC, LCs and PTMs at E18.5, P2 and P7 (Fig. 6A). The most statistically significant signaling pathway identified in germ cells was the Hippo pathway. In support of the possibility that the Hippo pathway is active in germ cells during the perinatal period, we found that several Hippo signaling component genes are expressed in germ cells during the perinatal period, with a peak at P2 (Fig. 6B). To test directly whether there is active Hippo signaling in perinatal germ cells, we performed IF analysis of YAP (YAP1), which is regarded as the best marker of Hippo signaling, as this transcriptional co-activator translocates into the nucleus of cells to transcriptionally activate Hippo pathway target genes (Meng et al., 2016). IF analysis showed that YAP is nuclear in perinatal germ cells, indicative of active Hippo signaling. Intriguingly, its nuclear localization is temporally regulated during perinatal development (Fig. 6C; Fig. S7). Between P0 and P2, there is a dramatic switch in the pattern of YAP localization from virtually no nuclear YAP+ germ cells at P0 to nearly 100% nuclear YAP+ germ cells at P2. The percentage of cells with a bright YAP signal increased between P2 and P3, followed by a decrease by P5. By P7, only ∼10% of germ cells expressed nuclear YAP. Together, these data suggest that there is a transient burst in Hippo signaling in germ cells between ∼P2 and ∼P5.
Fig. 6.
Molecular events during perinatal somatic and germ cell development. (A) Top enriched signaling pathways in germ and somatic cells at different stages. (B) Dot plot showing expression of genes related to the Hippo signaling pathway in germ cells. Color intensity indicates average expression level; circle size indicates the percentage of cells expressing the gene. (C) Percentage of TRA98+ cells that have YAP+ nuclei in the P0-P7 testis sections shown in Fig. S7. The signals were divided into bright and dim, based on signal intensity calculated using ImageJ (with 50% as the cut-off). Right: number of cells counted. (D) Dot plot showing expression of receptor-ligand signaling in the indicated cell types. (E) Violin plots showing expression of GDNF signaling receptors (in germ cells) and ligand (in SCs) at different stages. (F) Violin plots showing expression of a BMP signaling receptor (in germ cells) and ligand (in LCs).
Ligand-receptor signaling pairs expressed during the ProSG-to-SG transition
One possible role of somatic cells during the perinatal period is to participate in signaling events, just as they do in the adult testis (Takase and Nusse, 2016; Yang et al., 2013b). To investigate this possibility, we screened for ligand/receptor gene pairs expressed in germ and somatic cell populations during the perinatal period. This revealed the expression of genes involved in a number of signaling pathways, including GDNF, Hedgehog, CXC chemokine, Notch, FGF, TGFβ, WNT, PDGF, LIF and IGF (Fig. 6D; Fig. S8). Perinatal germ cells express genes encoding receptors for the GDNF, TGFβ and WNT signaling pathways (Gfra1/Ret, Bmpr1a and Fzd3, respectively), indicating that they likely respond to signals from other cells through these receptors. Perinatal germ cells also appear to secrete signaling factors, as they express genes encoding ligands for the Notch, TGFβ and PDGF signaling pathways (Dlk1, Tgfb1 and Pdgfb, respectively). Likewise, SCs express genes encoding both ligands and receptors, including ligands for the GDNF, Hedgehog, TGFβ, PDGF and LIF signaling pathways (Gdnf, Dhh, Tgfb1, Pdgfb and Ctf1, respectively), and receptors for Notch, TGFβ, WNT, and IGF signaling pathways (Notch2, Bmpr1a/Tgfbr1/Acvr1, Fzd3 and Sh2b2, respectively). LCs and PTMs also express genes encoding ligands and receptors for signaling pathways, as detailed in Fig. 6D and Fig. S8.
To ascertain when these intercellular pathways are likely to function during perinatal development, we examined the expression pattern of the ligand/receptor pairs. This revealed temporal specificity for several of these intercellular cell-cell signaling pathways. For example, Gdnf, which encodes the GDNF signaling ligand, is expressed in E18.5 SCs, whereas Gfra1 and Ret, which encode the GDNF signaling receptors, do not initiate expression until after birth (Fig. 6E), suggesting that GDNF signaling is not active in germ cells until after birth (GDNF may have non-germ-cell roles before birth). As another example, Bmp2 and Bmpr1a, which encode a BMP signaling ligand and a BMP receptor, respectively, both initiate expression between E18.5 and P2 (Fig. 6F). Because the co-expression of the ligand and receptor is induced in different cell types, this suggests coordinated gene regulation to mediate BMP signaling between LCs and ProSG in a temporally specific manner.
DISCUSSION
In this study, we used scRNAseq to molecularly characterize testicular germ and somatic cells during the perinatal period. We charted the progress of SSC precursor cells – ProSG – as they differentiate, migrate, undergo proliferative expansion, and convert into SSCs (Fig. 7). We also defined SG subsets present during the perinatal period, and defined stage-specific markers for both ProSG and SG subsets. We also characterized the development of somatic cells during the perinatal stage. Finally, we identified TFs and signaling pathways that are likely to function in and between germ and somatic cell subsets during the perinatal period. Together, our study provides a useful resource for understanding perinatal testicular development.
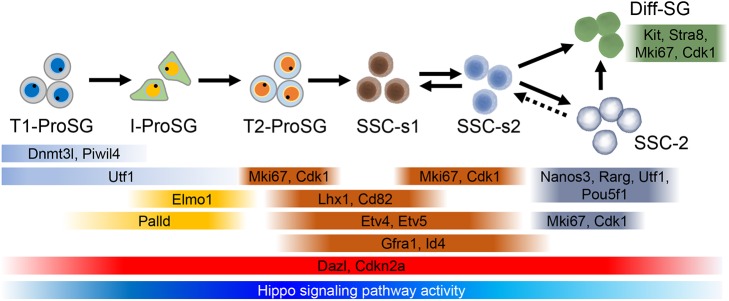
Fig. 7.
Model of ProSG-to-SG development. Black arrows indicate the direction of development. The expression pattern of marker genes (both previously known and those discovered in this study) are indicated. Note that the cell clusters with the ‘SSC’ designation contain undifferentiated SG that express SSC marker genes but for which SSC functional activity remains to be tested.
Identification of ProSG subsets and specific markers
Our scRNAseq analysis identified three distinct stages of ProSG development during the perinatal period. Two of these stages – T1-ProSG and T2-ProSG – were previously known (McCarrey, 2013), but no markers that specifically label these stages had been previously defined. We filled this gap by identifying both gene and protein markers for T1- and T2-ProSG, as well as a pan-ProSG marker. The T1-ProSG protein marker we identified is DNMT3L, a DNA methylation-promoting protein that restores DNA methylation marks erased during the primordial germ cell stage of germ-cell development (Hajkova, 2011) and is crucial for spermatogenesis (Bourc'his and Bestor, 2004; La Salle et al., 2007; Vlachogiannis et al., 2015; Webster et al., 2005). We identified the transcription factor ETV4 as a marker of T2-ProSG. T2-ProSG also express the Etv4 gene paralog Etv5 (Table S1), which is known to promote the proliferation of germline stem cells in vitro (Wu et al., 2011) and early postnatal SG in vivo (Tyagi et al., 2009), suggesting that this transcription factor stimulates SSC self-renewal. By analogy, Etv4 and Etv5 may participate in the proliferative expansion of T2-ProSG and SSC genesis.
Identification of a migrating ProSG subset
We identified a largely non-proliferative ProSG cell cluster – I-ProSG – that our evidence indicates undergoes migration from the center to the periphery of seminiferous tubules (Fig. 2G). As further support for this notion, I-ProSG express a large cadre of cell migration and actin organization genes (Fig. S2F). The discovery of the I-ProSG cell subset leads us to propose a refined model for SSC establishment in which the conversion of T1-ProSG to I-ProSG initiates a migratory event that places these germ cells in a niche that triggers their subsequent conversion into SSCs (Fig. 2G). Given that we found that most I-ProSG are in the center of seminiferous tubules, we cannot rule out the possibility that they are predominantly a ‘priming stage’ to prepare ProSG for migration, rather than migratory cells per se. However, we provide evidence that at least some I-ProSG are migratory, as we found some cells with I-ProSG markers that are in the periphery.
In both mice and rats, ProSG undergo migration to the seminiferous tubule periphery at approximately the same time after birth as they re-initiate mitosis (Kluin and de Rooij, 1981; McGuinness and Orth, 1992). It is not clear whether these two events are mechanistically connected. One possibility is that migration-promoting signals are received by T1-ProSG in the center of the seminiferous tubule, which transforms these cells into migratory I-ProSG. Once they migrate to the basement membrane in the periphery of the seminiferous tubule, I-ProSG receive growth signals present in this microenvironment, which drives these cells to convert into proliferative T2-ProSG. The identification of I-ProSG-specific markers will be invaluable to address this and other models. Interestingly, the two protein markers we identified as specific for I-ProSG – ELMO1 and PALLD – are both known to drive cell migration in mammalian cell lines (Goicoechea et al., 2008; Grimsley et al., 2004), which raises the intriguing possibility that ELMO1 and PALLD not only mark I-ProSG, but also promote the migration of these cells in vivo.
I-ProSG not only transiently express genes involved in migration, but other gene categories as well (Table S1). Perhaps most striking is their expression of many genes involved in intracellular signal transduction, including Hspb1, Mast4, Map4k4, Plcg2, Prkci, Pkn2, Srpk2, Stk39, Sgk1, Socs2 and Tulp4. This raises the possibility that I-ProSG activate specific signaling pathways in response to extracellular stimuli, perhaps to guide them in migration. I-ProSG also downregulate several epigenetic modification-related genes, such as Dnmt3l, Dnmt3a, Kdm1b and Piwil4. Perhaps this down-regulatory response results from a checkpoint mechanism that senses when epigenetic marks are fully restored. According to this model, only developing germ cells that pass the checkpoint are allowed to migrate to the SSC niche and form SSCs.
Undifferentiated SG heterogeneity
Several recent scRNAseq studies – focusing on the adult testis – have obtained evidence that undifferentiated SG are not a homogeneous population, but instead a collection of subsets in subtly different states, each with different patterns of gene expression (Green et al., 2018; Grive et al., 2019; Guo et al., 2018; Hermann et al., 2018; Sohni et al., 2019; Tan and Wilkinson, 2019; Wang et al., 2018). How this transcriptome complexity is initially established is not known. Using scRNAseq to analyze germ cells during the SSC establishment period, we identified two cell clusters of undifferentiated SG – SSC-s1 and SSC-s2 – that express distinct patterns of genes, raising the possibility that SSCs are heterogeneous even when first formed.
We found that the SSC-s1 cell cluster is the most naïve, based on Monocle psuedotime trajectory analysis and the expression pattern of known SSC markers (i.e. Eomes and Plaur) and SSC markers identified in this study (e.g. Lhx1 and Cd82). Although the functional significance of this difference in gene expression is not known, one possible explanation is that SSC-s1 and SSC-s2 differ in their endurance as stem cells. Given that SSC-s1 is more primitive than SSC-s2, it may contain cells that tend to retain SSC activity for longer than SSC-s2 cells. Another possibility is that SSC-s1 and SSC-s2 are interconvertible states, in alignment with studies suggesting that mouse undifferentiated SG stages are in equilibrium (Hara et al., 2014).
Among the genes enriched in the SSC-s1 cell cluster are genes involved in integrin-mediated signaling (Fig. S4B), which has roles in virtually all aspects of stem cell niche interactions (Marthiens et al., 2010). Indeed, homing of mouse SSCs to their niche in the seminiferous tubule periphery is known to depend on β1-integrin (Kanatsu-Shinohara et al., 2008). Thus, integrins may function in the initial generation of these naïve cells, their maintenance, and/or their conversion into other SSC states. Genes related to vessel development are also enriched in SSC-s1 cells (Fig. S4C), which raises the possibility that these cells are in close physical proximity to lymphatic endothelial cells and respond to extracellular factors from these cells. In support of this hypothesis, it was recently reported that SSC density is tightly regulated by the supply of FGFs from lymphatic endothelial cells (Kitadate et al., 2019).
Testicular somatic cell development
Our scRNAseq analysis of testicular somatic cells during the perinatal period provides a reference for understanding the development of these cells. In the case of SCs, GO analysis revealed that many genes involved in ‘male gonad development’ and ‘SC differentiation’ are enriched at P7 (Table S2), consistent with the evidence that SCs are differentiating during the early postnatal period (Walker, 2003). Many of the genes exhibiting transiently elevated expression mid-way through the perinatal period (group 2, as defined by pseudotime trajectory analysis) are involved in cell proliferation (Fig. 4D; Table S2), and thus are candidates to drive the proliferation of SCs during early postnatal development (Baker and O'Shaughnessy, 2001). Genes exhibiting elevated expression at a later development stage (group 3) encode proteins involved in ubiquitylation, PI3K/AKT signaling and integrin signaling (Table S2), raising the possibility that these pathways are important for SC maturation. Indeed, several of the TF genes in group 3 (Table S2) have known roles in SC development, e.g. Ar, Dmrt1, Sox8 and Sox9 (Barrionuevo et al., 2009; Raymond et al., 2000; Skinner et al., 1988), or lead to male infertility in gene-knockout experiments, e.g. Nr0b1, Rara, Rb1, Rxrb and Sox3 (Costa et al., 1997; Stickels et al., 2015; Vernet et al., 2008; Weiss et al., 2003; Yang et al., 2013a). Based on the timing of their expression, group-3 TF genes are likely to be expressed in SCs in contact with newly emergent SSCs and differentiating SG, raising the possibility that some group-3 TFs have roles in SSC establishment and the first wave of spermatogenesis, respectively.
LCs are unique in that they exist as two distinct populations – fetal and adult – both of which produce testosterone, but otherwise have distinct characteristics (Baker and O'Shaughnessy, 2001). We found that genes involved in ‘apoptosis’ are enriched in the P2 LC cluster (Fig. S6B), consistent with the timing of fetal LC loss (Griswold and Behringer, 2009). Indeed, TF genes expressed transiently in LCs during the mid-phase of the perinatal period (group-2 TF genes, as defined by pseudotime trajectory analysis; Fig. S6D; Table S2) have known roles in apoptosis, e.g. Mid1, Tnfaip3 and Erf (Collison et al., 2013; Honma et al., 2009; Oikawa and Yamada, 2003). Other group-2 TFs are involved in fetal LC differentiation, e.g. Arx (Miyabayashi et al., 2013), or lead to male infertility in gene-knockout experiments, e.g. Hoxd11, Arntl and Atm (Alvarez et al., 2008; Boulet and Capecchi, 2002; Xu et al., 1996).
Little is known about the development of PTMs (Nurmio et al., 2012). We found that P7 PTMs are enriched for genes involved in actin cytoskeleton organization (Table S2), raising the possibility that one or more of these genes drive the rearrangement of actin filaments that occurs in PTMs postnatally (Maekawa et al., 1996). Among the TF genes transiently upregulated late in PTMs (group 3, as defined by pseudotime analysis; Fig. 5C) is Ar, which is known to be important for PTM development (Welsh et al., 2012). Many cell-cycle, nuclear division, and chromatid segregation genes exhibit enriched expression in E18.5 and P2 PTMs, and thus are candidates to drive the proliferation of PTMs, which occurs around birth in mice (Vergouwen et al., 1991).
Cell signaling during perinatal testicular development
Our scRNAseq analysis allowed us to identify many signaling pathways that are candidates to be active during ProSG development and SSC genesis (Fig. S5A). Some signaling pathways enriched in I-ProSG are known to be involved in germ cell migration, including JAK/Stat (Brown et al., 2006), PDGF (Smith et al., 2005) and CXCR4 (Molyneaux et al., 2003), raising the possibility that these signaling pathways have a role in the migration of I-ProSG from the center to the periphery of the seminiferous tubule. Using a well-established Hippo signaling assay, we found that Hippo signaling is specifically activated in germ cells during the time window when ProSG undergo conversion into differentiating SG and SSCs (between ∼P2 and P5). This raises the possibility that Hippo signaling has roles in one or both of these conversion events. As evidence against this, a recent study showed that conditional knockout of Yap in primordial germ cells did not result in any obvious defects in mouse spermatogenesis (Abou Nader et al., 2019), but this may be because of rescue by the Yap paralog Taz (Meng et al., 2016).
We identified a large number of ligands and their cognate receptors expressed in perinatal somatic and germ cell subsets (Fig. 6D; Fig. S8). Several of these ligand-receptor pairs have been shown to function in spermatogenesis (Mäkelä and Toppari, 2017; Takase and Nusse, 2016; Yang et al., 2013b), but, to our knowledge, none has yet been shown to operate in events that occur in germ or somatic cells during the perinatal period. For example, we found that the gene encoding the growth factor GDNF is expressed in perinatal SCs, and its two receptors, GFRA1 and RET, are expressed in T2-ProSG and SSC-1, raising the possibility that GDNF-mediated signaling is important for the generation and initial expansion of SSCs, thereby expanding its role beyond adult SSC self-renewal (Oatley et al., 2007). We found that the CXC chemokine ligand, Cxcl12, is highly expressed in both stromal cells and PTMs, and its receptor, Cxcr4, is expressed in ProSG, LCs and innate lymphatic cells. This raises the possibility that CXC chemokine signaling may be involved in the establishment of SSC niche, as this pathway is known to promote SSC homing to their niche in the seminiferous tubule periphery in adult mice (Niu et al., 2016). A role for FGF signaling is suggested by our finding that that the FGF receptor gene Fgfr2 is expressed in germ cells, LCs, PTMs and stromal cells, and the FGF ligand genes Fgf2 and Fgf7 are expressed in both stromal cells and PTMs during the perinatal period. This is consistent with the possibility that emerging SSCs are regulated by stromal cells and PTMs, as the supply of FGFs is known to tightly regulate SSC density (Kitadate et al., 2019). Other signaling pathways implicated as being active during perinatal development include the Hedgehog, Notch, TGFβ, WNT, PDGF, LIF and IGF pathways (Fig. S8).
MATERIALS AND METHODS
Animals
This study was carried out in strict accordance with the Guidelines of the Institutional Animal Care and Use Committee (IACUC) at the University of California, San Diego. The protocol was approved by the IACUC at the University of California, San Diego (permit number: S09160). All C57BL/6 mice (female: 7-10 weeks; male: 8-16 weeks) were housed under a 12 h light:12 h dark cycle and provided with food and water ad libitum.
Testis sample preparation
Each sample analyzed by scRNAseq consisted of testicular cells pooled from two pups. Single testicular cells were isolated using a two-step enzymatic digestion protocol described previously (Sohni et al., 2019; Valli et al., 2014), with minor modifications. In brief, the tunica albugineas of fresh mouse testes were removed and the testes mechanically disrupted and enzymatically digested with 1 mg ml−1 collagenase type IV (Worthington Biochemical Corporation) in Hank's Balanced Salt Solution (HBSS; Gibco) at 37°C. The tubules were gravity sedimented and washed with HBSS and digested in 0.25% Trypsin-EDTA (Thermo Fisher) and Deoxyribonuclease I (Worthington Biochemical Corporation). The suspension was triturated vigorously ten times, incubated at 37°C for 5 min, followed by repeat trituration and incubation. The digestion was stopped by adding the same volume of αMEM+10% fetal bovine serum (FBS) medium and the cells were sequentially size-filtered through 70 μm and 40 μm strainers (Thermo Fisher) and pelleted by centrifugation at 600 g for 3 min. The dead cells were removed using the ClioCell Dead Cell Removal kit (Amsbio) following the manufacturer's instructions.
10x Genomics library preparation
Single testicular cells were washed once in PBS and re-suspended in 0.3% FBS in PBS for loading on the 10x Chromium chip. Cell capturing, and library preparation was carried as per kit instructions [Chromium Single Cell Kit (v2 chemistry)]. In brief, 10,000 cells were targeted for capture per sample; after cDNA synthesis, 12-14 cycles were used for library amplification. The resultant libraries were size selected, pooled and sequenced using 2×100 paired-end sequencing protocol on an Illumina HiSeq 4000 instrument. The libraries initially underwent shallow sequencing to access quality and to adjust subsequent sequencing depth based on the capture rate and unique molecular indices (UMI) detected. All sequencing was performed at the Institute of Genomic Medicine at UCSD.
De-multiplexed raw sequencing reads were processed and mapped to the mouse genome (mm10) using Cell Ranger software (v3.0.2) with default parameters. Quality control metrics from these data are listed in Table S3. We filtered raw count matrices by excluding cells expressing fewer than 200 detectably expressed genes and genes expressed in fewer than three cells and each library was tagged with a library batch ID and combined across independent experiments using the Seurat package (Butler et al., 2018) in R. To check the quality of the single-cell data and to remove multiplets, we performed Seurat-based filtering of cells based on three criteria: number of detected features (nFeature_RNA) per cell, number of UMIs expressed per cell (nCount_RNA) and mitochondrial content, using the following threshold parameters: nFeature_RNA (200 to 6000), and percentage of mitochondrial genes expressed (<0.2%). For the detailed analysis of each cell type, we applied more stringent filtering parameters (i.e. mitochondrial content percent cut-off was set to 0.15). In addition, we used a known lineage marker profile to ensure that the same barcode was not assigned to two cells of different lineages (multiplets), as well as to rule out the possibility of captured cell-free droplets. Gene expression values were log normalized and regressed by the mitochondrial expression (‘percent.mt’) and cell cycle (‘S.Score’ and ‘G2M.Score’) using the ‘SCTransform’ function. Batch correction was performed using the ‘JackStraw’ functions in the Seurat package.
To identify cell clusters, we employed UMAP (Becht et al., 2018). The number of the cells in each cluster are summarized in Table S4. The ‘FindMarkers’ function (a Wilcoxon rank sum test) was used to determine differential gene expression between clusters (set at minimum expression in 25% of cells). The ‘CellCycleScoring’ function was used to infer cell cycle phase, as this program determines relative expression of a large set of G2/M- and S-phase genes (Kowalczyk et al., 2015). The ‘DoHeatmap’ function was used to generate an expression heatmap for given cells and features. Pearson correlation analysis of cell clusters was performed using the ‘CellScatter’ function. GO analysis (DAVID v6.8) was done using top differentially (positively) expressed genes with a P-adjusted cut-off of 0.01. Lists of differentially expressed genes were analyzed by IPA to identify biological pathways that are significantly over-represented among the genes in each list.
Cell trajectory analysis
Single-cell pseudotime trajectories were constructed with the Monocle 2 package (v2.10.1) (Qiu et al., 2017) according to the provided documentation (http://cole-trapnell-lab.github.io/monocle-release/). UMI counts were modeled as a negative binomial distribution. Genes defining the trajectory are those with high dispersion across cells (mean_expression≥0.01; dispersion_empirical≥1). The discriminative dimensionality reduction with trees (DDRTree) method was used to reduce data to two dimensions. Differentially expressed genes were identified and used for dynamic trajectory analysis [false discovery rate (FDR)<0.05], which ordered cells in pseudotime. The ‘plot_pseudotime_heatmap’ function was used to generate heatmaps.
Immunofluorescence analysis
Sections were deparaffinized twice in xylene, followed by serial dilutions of ethanol. Unmasking was performed with IHC-TekTM epitope retrieval solution using a steamer (IHC World) for 40 min. Blocking was performed by incubating with 5% serum (from the species in which the secondary antibody was raised) for 1 h at room temperature. The sections were then incubated overnight with the primary antibody (Table S5) at 4°C and incubated with secondary antisera (see Table S5 for a list) for 1 h at room temperature. The nuclei were counterstained with DAPI, a coverslip was placed over the sections with mounting medium, and the images were viewed using a Leica DMI4000 B fluorescence microscope.
FACS and qRT-PCR analysis
After dissecting single testicular cells, cells were re-suspended in staining buffer (PBS+3% FBS) and stained with the CDKN2A antibody for 20 min on ice, washed with staining buffer, incubated with secondary antibodies for 20 min on ice, re-suspended in staining buffer and sorted by FACS. For FACS, gating was set based on size (forward scatter, FSC) to remove small debris and doublets. We then used unstained and secondary antibody only (primary omitted)-stained cells as negative controls for gating unstained and false positive-stained cells, respectively.
cDNAs were generated using the Iscript reverse transcriptase (RT) kit, according to the manufacturer's protocol (Bio-Rad). qRT-PCR was performed using an iCycler real-time PCR machine according to the manufacturer's protocol (Bio-Rad). The production of the amplicon was measured by SYBR green fluorescence and the threshold cycle (Ct) values were calculated. Ct values obtained were normalized to Ct values for the ribosomal protein Rpl19 gene. Three independent biological replicates were performed.
Statistical methods
The details of the statistical method used for identifying the differential gene expression and pseudotime trajectory analysis are provided in the detailed methods above. Quantification of the immunostainings was performed by counting the positively stained cells in different fields of view. The number of cells counted is indicated on the respective figure or its figure legend.
Supplementary Material
Acknowledgements
We thank the UCSD Institute for Genomic Medicine for technical support and the San Diego Supercomputer Center for providing data analysis resources. We also thank the Developmental Studies Hybridoma Bank (created by the NICHD and maintained at The University of Iowa, Department of Biology) for the LHX1 antibody generated by Dr David R. Soll.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: K.T.; Methodology: K.T., H.-W.S.; Software: K.T.; Validation: K.T.; Formal analysis: K.T.; Investigation: K.T.; Resources: K.T., H.-W.S.; Data curation: K.T.; Writing - original draft: K.T.; Writing - review & editing: K.T., M.F.W.; Supervision: M.F.W.; Project administration: M.F.W.; Funding acquisition: M.F.W.
Funding
This work was supported by the National Institutes of Health (R01 GM119128 to M.F.W.) and the Lalor Foundation (K.T.). Deposited in PMC for release after 12 months.
Data availability
The scRNAseq data generated in this study has been deposited at NCBI's Gene Expression Omnibus database under the accession number GSE130593.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.183251.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.183251.reviewer-comments.pdf
References
- Abou Nader N., Levasseur A., Zhang X., Boerboom D., Nagano M. C. and Boyer A. (2019). Yes-associated protein expression in germ cells is dispensable for spermatogenesis in mice. Genesis 57, e23330 10.1002/dvg.23330 [DOI] [PubMed] [Google Scholar]
- Alvarez J. D., Hansen A., Ord T., Bebas P., Chappell P. E., Giebultowicz J. M., Williams C., Moss S. and Sehgal A. (2008). The circadian clock protein BMAL1 is necessary for fertility and proper testosterone production in mice. J. Biol. Rhythms 23, 26-36. 10.1177/0748730407311254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P. J. and O'Shaughnessy P. J. (2001). Role of gonadotrophins in regulating numbers of Leydig and Sertoli cells during fetal and postnatal development in mice. Reproduction 122, 227-234. 10.1530/rep.0.1220227 [DOI] [PubMed] [Google Scholar]
- Barrionuevo F., Georg I., Scherthan H., Lécureuil C., Guillou F., Wegner M. and Scherer G. (2009). Testis cord differentiation after the sex determination stage is independent of Sox9 but fails in the combined absence of Sox9 and Sox8. Dev. Biol. 327, 301-312. 10.1016/j.ydbio.2008.12.011 [DOI] [PubMed] [Google Scholar]
- Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I. W. H., Ng L. G., Ginhoux F. and Newell E. W. (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38-44. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Bellve A. R., Cavicchia J. C., Millette C. F., O'Brien D. A., Bhatnagar Y. M. and Dym M. (1977). Spermatogenic cells of the prepuberal mouse: isolation and morphological characterization. J. Cell Biol. 74, 68-85. 10.1083/jcb.74.1.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet A. M. and Capecchi M. R. (2002). Duplication of the Hoxd11 gene causes alterations in the axial and appendicular skeleton of the mouse. Dev. Biol. 249, 96-107. 10.1006/dbio.2002.0755 [DOI] [PubMed] [Google Scholar]
- Bourc'his D. and Bestor T. H. (2004). Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96-99. 10.1038/nature02886 [DOI] [PubMed] [Google Scholar]
- Brown S., Zeidler M. P. and Hombría J. E. C.-G. (2006). JAK/STAT signalling in Drosophila controls cell motility during germ cell migration. Dev. Dyn. 235, 958-966. 10.1002/dvdy.20709 [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E. and Satija R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411-420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.-L., La H. M., Legrand J. M. D., Mäkelä J.-A., Eichenlaub M., De Seram M., Ramialison M. and Hobbs R. M. (2017). Germline stem cell activity is sustained by SALL4-dependent silencing of distinct tumor suppressor genes. Stem Cell Rep. 9, 956-971. 10.1016/j.stemcr.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-Y., Willis W. D. and Eddy E. M. (2016). Targeting the Gdnf Gene in peritubular myoid cells disrupts undifferentiated spermatogonial cell development. Proc. Natl. Acad. Sci. USA 113, 1829-1834. 10.1073/pnas.1517994113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collison A., Hatchwell L., Verrills N., Wark P. A. B., de Siqueira A. P., Tooze M., Carpenter H., Don A. S., Morris J. C., Zimmermann N. et al. (2013). The E3 ubiquitin ligase midline 1 promotes allergen and rhinovirus-induced asthma by inhibiting protein phosphatase 2A activity. Nat. Med. 19, 232-237. 10.1038/nm.3049 [DOI] [PubMed] [Google Scholar]
- Costa S. L., Boekelheide K., Vanderhyden B. C., Seth R. and McBurney M. W. (1997). Male infertility caused by epididymal dysfunction in transgenic mice expressing a dominant negative mutation of retinoic acid receptor α1. Biol. Reprod. 56, 985-990. 10.1095/biolreprod56.4.985 [DOI] [PubMed] [Google Scholar]
- Culty M. (2013). Gonocytes, from the fifties to the present: is there a reason to change the name?. Biol. Reprod. 89, 46 10.1095/biolreprod.113.110544 [DOI] [PubMed] [Google Scholar]
- Fayomi A. P. and Orwig K. E. (2018). Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Res. 29, 207-214. 10.1016/j.scr.2018.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegenschatz-Schmid K., Verkauskas G., Demougin P., Bilius V., Dasevicius D., Stadler M. B. and Hadziselimovic F. (2017). DMRTC2, PAX7, BRACHYURY/T and TERT are implicated in male germ cell development following curative hormone treatment for cryptorchidism-induced infertility. Genes (Basel) 8, e267 10.3390/genes8100267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J., Schwab U., Lemke H. and Stein H. (1983). Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int. J. Cancer 31, 13-20. 10.1002/ijc.2910310104 [DOI] [PubMed] [Google Scholar]
- Goicoechea S. M., Arneman D. and Otey C. A. (2008). The role of palladin in actin organization and cell motility. Eur. J. Cell Biol. 87, 517-525. 10.1016/j.ejcb.2008.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. D., Ma Q., Manske G. L., Shami A. N., Zheng X., Marini S., Moritz L., Sultan C., Gurczynski S. J., Moore B. B. et al. (2018). A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-seq. Dev. Cell 46, 651-667.e610. 10.1016/j.devcel.2018.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsley C. M., Kinchen J. M., Tosello-Trampont A.-C., Brugnera E., Haney L. B., Lu M., Chen Q., Klingele D., Hengartner M. O. and Ravichandran K. S. (2004). Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. J. Biol. Chem. 279, 6087-6097. 10.1074/jbc.M307087200 [DOI] [PubMed] [Google Scholar]
- Griswold M. D. (1998). The central role of Sertoli cells in spermatogenesis. Semin. Cell Dev. Biol. 9, 411-416. 10.1006/scdb.1998.0203 [DOI] [PubMed] [Google Scholar]
- Griswold S. L. and Behringer R. R. (2009). Fetal Leydig cell origin and development. Sex. Dev. 3, 1-15. 10.1159/000200077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grive K. J., Hu Y., Shu E., Grimson A., Elemento O., Grenier J. K. and Cohen P. E. (2019). Dynamic transcriptome profiles within spermatogonial and spermatocyte populations during postnatal testis maturation revealed by single-cell sequencing. PLoS Genet. 15, e1007810 10.1371/journal.pgen.1007810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Grow E. J., Mlcochova H., Maher G. J., Lindskog C., Nie X., Guo Y., Takei Y., Yun J., Cai L. et al. (2018). The adult human testis transcriptional cell atlas. Cell Res. 28, 1141-1157. 10.1038/s41422-018-0099-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P. (2011). Epigenetic reprogramming in the germline: towards the ground state of the epigenome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 2266-2273. 10.1098/rstb.2011.0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Nakagawa T., Enomoto H., Suzuki M., Yamamoto M., Simons B. D. and Yoshida S. (2014). Mouse spermatogenic stem cells continually interconvert between equipotent singly isolated and syncytial states. Cell Stem Cell 14, 658-672. 10.1016/j.stem.2014.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsel A. R., Yang Q.-E., Oatley M. J., Lord T., Sablitzky F. and Oatley J. M. (2017). ID4 levels dictate the stem cell state in mouse spermatogonia. Development 144, 624-634. 10.1242/dev.146928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann B. P., Cheng K., Singh A., Roa-De La Cruz L., Mutoji K. N., Chen I.-C., Gildersleeve H., Lehle J. D., Mayo M., Westernströer B. et al. (2018). The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 25, 1650-1667.e1658. 10.1016/j.celrep.2018.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma K., Tsuzuki S., Nakagawa M., Tagawa H., Nakamura S., Morishima Y. and Seto M. (2009). TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood 114, 2467-2475. 10.1182/blood-2008-12-194852 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Takehashi M., Takashima S., Lee J., Morimoto H., Chuma S., Raducanu A., Nakatsuji N., Fässler R. and Shinohara T. (2008). Homing of mouse spermatogonial stem cells to germline niche depends on beta1-integrin. Cell Stem Cell 3, 533-542. 10.1016/j.stem.2008.08.002 [DOI] [PubMed] [Google Scholar]
- Kitadate Y., Jörg D. J., Tokue M., Maruyama A., Ichikawa R., Tsuchiya S., Segi-Nishida E., Nakagawa T., Uchida A., Kimura-Yoshida C. et al. (2019). Competition for mitogens regulates spermatogenic stem cell homeostasis in an open niche. Cell Stem Cell 24, 79-92.e76. 10.1016/j.stem.2018.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluin P. M. and de Rooij D. G. (1981). A comparison between the morphology and cell kinetics of gonocytes and adult type undifferentiated spermatogonia in the mouse. Int. J. Androl. 4, 475-493. 10.1111/j.1365-2605.1981.tb00732.x [DOI] [PubMed] [Google Scholar]
- Kowalczyk M. S., Tirosh I., Heckl D., Rao T. N., Dixit A., Haas B. J., Schneider R. K., Wagers A. J., Ebert B. L. and Regev A. (2015). Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25, 1860-1872. 10.1101/gr.192237.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Green J., Pollard J. Jr and Tugendreich S. (2014). Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523-530. 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Salle S., Oakes C. C., Neaga O. R., Bourc'his D., Bestor T. H. and Trasler J. M. (2007). Loss of spermatogonia and wide-spread DNA methylation defects in newborn male mice deficient in DNMT3L. BMC Dev. Biol. 7, 104 10.1186/1471-213X-7-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law N. C., Oatley M. J. and Oatley J. M. (2019). Developmental kinetics and transcriptome dynamics of stem cell specification in the spermatogenic lineage. Nat. Commun. 10, 2787 10.1038/s41467-019-10596-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J., Ng S. H., Luk A. C., Suen H. C., Qian Y., Lee A. W. T., Tu J., Fung J. C. L., Tang N. L. S., Feng B. et al. (2019). Revealing cellular and molecular transitions in neonatal germ cell differentiation using single cell RNA sequencing. Development 146, dev174953 10.1242/dev.174953 [DOI] [PubMed] [Google Scholar]
- Maekawa M., Kamimura K. and Nagano T. (1996). Peritubular myoid cells in the testis: their structure and function. Arch. Histol. Cytol. 59, 1-13. 10.1679/aohc.59.1 [DOI] [PubMed] [Google Scholar]
- Mäkelä J. and Toppari J. (2017). Spermatogenesis. In Endocrinology of the Testis and Male Reproduction. Endocrinology (ed. Simoni M. and Huhtaniemi I.). pp. 417-455. Cham: Springer. [Google Scholar]
- Marthiens V., Kazanis I., Moss L., Long K. and Ffrench-Constant C. (2010). Adhesion molecules in the stem cell niche--more than just staying in shape? J. Cell Sci. 123, 1613-1622. 10.1242/jcs.054312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarrey J. R. (2013). Toward a more precise and informative nomenclature describing fetal and neonatal male germ cells in rodents. Biol. Reprod. 89, 47 10.1095/biolreprod.113.110502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness M. P. and Orth J. M. (1992). Reinitiation of gonocyte mitosis and movement of gonocytes to the basement membrane in testes of newborn rats in vivo and in vitro. Anat. Rec. 233, 527-537. 10.1002/ar.1092330406 [DOI] [PubMed] [Google Scholar]
- McLean D. J., Friel P. J., Johnston D. S. and Griswold M. D. (2003). Characterization of spermatogonial stem cell maturation and differentiation in neonatal mice. Biol. Reprod. 69, 2085-2091. 10.1095/biolreprod.103.017020 [DOI] [PubMed] [Google Scholar]
- Meng Z., Moroishi T. and Guan K.-L. (2016). Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1-17. 10.1101/gad.274027.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroni S. B., Galardo M. N., Rindone G., Gorga A., Riera M. F. and Cigorraga S. B. (2019). Molecular mechanisms and signaling pathways involved in sertoli cell proliferation. Front. Endocrinol. 10, 224 10.3389/fendo.2019.00224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabayashi K., Katoh-Fukui Y., Ogawa H., Baba T., Shima Y., Sugiyama N., Kitamura K. and Morohashi K.-I. (2013). Aristaless related homeobox gene, Arx, is implicated in mouse fetal Leydig cell differentiation possibly through expressing in the progenitor cells. PLoS ONE 8, e68050 10.1371/journal.pone.0068050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux K. A., Zinszner H., Kunwar P. S., Schaible K., Stebler J., Sunshine M. J., O'Brien W., Raz E., Littman D., Wylie C. et al. (2003). The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development 130, 4279-4286. 10.1242/dev.00640 [DOI] [PubMed] [Google Scholar]
- Niu Z., Goodyear S. M., Avarbock M. R. and Brinster R. L. (2016). Chemokine (C-X-C) ligand 12 facilitates trafficking of donor spermatogonial stem cells. Stem Cells Int. 2016, 5796305 10.1155/2016/5796305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurmio M., Kallio J., Adam M., Mayerhofer A., Toppari J. and Jahnukainen K. (2012). Peritubular myoid cells have a role in postnatal testicular growth. Spermatogenesis 2, 79-87. 10.4161/spmg.20067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley J. M. and Brinster R. L. (2012). The germline stem cell niche unit in mammalian testes. Physiol. Rev. 92, 577-595. 10.1152/physrev.00025.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley J. M., Avarbock M. R., Telaranta A. I., Fearon D. T. and Brinster R. L. (2006). Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc. Natl. Acad. Sci. USA 103, 9524-9529. 10.1073/pnas.0603332103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley J. M., Avarbock M. R. and Brinster R. L. (2007). Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J. Biol. Chem. 282, 25842-25851. 10.1074/jbc.M703474200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T. and Yamada T. (2003). Molecular biology of the Ets family of transcription factors. Gene 303, 11-34. 10.1016/S0378-1119(02)01156-3 [DOI] [PubMed] [Google Scholar]
- Pogue-Geile K. L., Chen R., Bronner M. P., Crnogorac-Jurcevic T., Moyes K. W., Dowen S., Otey C. A., Crispin D. A., George R. D., Whitcomb D. C. et al. (2006). Palladin mutation causes familial pancreatic cancer and suggests a new cancer mechanism. PLoS Med. 3, e516 10.1371/journal.pmed.0030516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H. A. and Trapnell C. (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979-982. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftopoulou M. and Hall A. (2004). Cell migration: Rho GTPases lead the way. Dev. Biol. 265, 23-32. 10.1016/j.ydbio.2003.06.003 [DOI] [PubMed] [Google Scholar]
- Raymond C. S., Murphy M. W., O'Sullivan M. G., Bardwell V. J. and Zarkower D. (2000). Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 14, 2587-2595. 10.1101/gad.834100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y., Suetake I., Shinozaki F., Yamashina S. and Tajima S. (2004). Co-expression of de novo DNA methyltransferases Dnmt3a2 and Dnmt3L in gonocytes of mouse embryos. Gene Expr. Patterns 5, 231-237. 10.1016/j.modgep.2004.07.011 [DOI] [PubMed] [Google Scholar]
- Skinner M. K., Fetterolf P. M. and Anthony C. T. (1988). Purification of a paracrine factor, P-Mod-S, produced by testicular peritubular cells that modulates Sertoli cell function. J. Biol. Chem. 263, 2884-2890. [PubMed] [Google Scholar]
- Smith C. A., McClive P. J., Hudson Q. and Sinclair A. H. (2005). Male-specific cell migration into the developing gonad is a conserved process involving PDGF signalling. Dev. Biol. 284, 337-350. 10.1016/j.ydbio.2005.05.030 [DOI] [PubMed] [Google Scholar]
- Sohni A., Tan K., Song H.-W., Burow D., de Rooij D. G., Laurent L., Hsieh T.-C., Rabah R., Hammoud S. S., Vicini E. et al. (2019). The neonatal and adult human testis defined at the single-cell level. Cell Rep. 26, 1501-1517.e1504. 10.1016/j.celrep.2019.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickels R., Clark K., Heider T. N., Mattiske D. M., Renfree M. B. and Pask A. J. (2015). DAX1/NR0B1 was expressed during mammalian gonadal development and gametogenesis before it was recruited to the eutherian X chromosome. Biol. Reprod. 92, 22 10.1095/biolreprod.114.119362 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Ahn H. W., Chu T., Bowden W., Gassei K., Orwig K. and Rajkovic A. (2012). SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev. Biol. 361, 301-312. 10.1016/j.ydbio.2011.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase H. M. and Nusse R. (2016). Paracrine Wnt/beta-catenin signaling mediates proliferation of undifferentiated spermatogonia in the adult mouse testis. Proc. Natl. Acad. Sci. USA 113, E1489-E1497. 10.1073/pnas.1601461113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K. and Wilkinson M. F. (2019). Human Spermatogonial Stem Cells Scrutinized under the Single-Cell Magnifying Glass. Cell Stem Cell 24, 201-203. 10.1016/j.stem.2019.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Pereira L. A. V. D., Nozaki M., Tsuchida J., Sawada K., Mori H. and Nishimune Y. (1998). A germ cell-specific nuclear antigen recognized by a monoclonal antibody raised against mouse testicular germ cells. Int. J. Androl. 20, 361-366. 10.1046/j.1365-2605.1998.00080.x [DOI] [PubMed] [Google Scholar]
- Tseng Y.-T., Liao H.-F., Yu C.-Y., Mo C.-F. and Lin S.-P. (2015). Epigenetic factors in the regulation of prospermatogonia and spermatogonial stem cells. Reproduction 150, R77-R91. 10.1530/REP-14-0679 [DOI] [PubMed] [Google Scholar]
- Tyagi G., Carnes K., Morrow C., Kostereva N. V., Ekman G. C., Meling D. D., Hostetler C., Griswold M., Murphy K. M., Hess R. A. et al. (2009). Loss of Etv5 decreases proliferation and RET levels in neonatal mouse testicular germ cells and causes an abnormal first wave of spermatogenesis. Biol. Reprod. 81, 258-266. 10.1095/biolreprod.108.075200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli H., Sukhwani M., Dovey S. L., Peters K. A., Donohue J., Castro C. A., Chu T., Marshall G. R. and Orwig K. E. (2014). Fluorescence- and magnetic-activated cell sorting strategies to isolate and enrich human spermatogonial stem cells. Fertil. Steril. 102, 566-580.e567. 10.1016/j.fertnstert.2014.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velte E. K., Niedenberger B. A., Serra N. D., Singh A., Roa-DeLaCruz L., Hermann B. P. and Geyer C. B. (2019). Differential RA responsiveness directs formation of functionally distinct spermatogonial populations at the initiation of spermatogenesis in the mouse. Development 146, dev173088 10.1242/dev.173088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergouwen R. P. F. A., Jacobs S. G. P. M., Huiskamp R., Davids J. A. G. and de Rooij D. G. (1991). Proliferative activity of gonocytes, Sertoli cells and interstitial cells during testicular development in mice. J. Reprod. Fertil. 93, 233-243. 10.1530/jrf.0.0930233 [DOI] [PubMed] [Google Scholar]
- Vernet N., Dennefeld C., Klopfenstein M., Ruiz A., Bok D., Ghyselinck N. B. and Mark M. (2008). Retinoid X receptor beta (RXRB) expression in Sertoli cells controls cholesterol homeostasis and spermiation. Reproduction 136, 619-626. 10.1530/REP-08-0235 [DOI] [PubMed] [Google Scholar]
- Vlachogiannis G., Niederhuth C. E., Tuna S., Stathopoulou A., Viiri K., de Rooij D. G., Jenner R. G., Schmitz R. J. and Ooi S. K. T. (2015). The Dnmt3L ADD domain controls cytosine methylation establishment during spermatogenesis. Cell Rep. 11, 990 10.1016/j.celrep.2015.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker W. H. (2003). Molecular mechanisms controlling Sertoli cell proliferation and differentiation. Endocrinology 144, 3719-3721. 10.1210/en.2003-0765 [DOI] [PubMed] [Google Scholar]
- Wang M., Liu X., Chang G., Chen Y., An G., Yan L., Gao S., Xu Y., Cui Y., Dong J. et al. (2018). Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell 23, 599-614.e594. 10.1016/j.stem.2018.08.007 [DOI] [PubMed] [Google Scholar]
- Webster K. E., O'Bryan M. K., Fletcher S., Crewther P. E., Aapola U., Craig J., Harrison D. K., Aung H., Phutikanit N., Lyle R. et al. (2005). Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc. Natl. Acad. Sci. USA 102, 4068-4073. 10.1073/pnas.0500702102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J., Meeks J. J., Hurley L., Raverot G., Frassetto A. and Jameson J. L. (2003). Sox3 is required for gonadal function, but not sex determination, in males and females. Mol. Cell. Biol. 23, 8084-8091. 10.1128/MCB.23.22.8084-8091.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh M., Moffat L., Belling K., de França L. R., Segatelli T. M., Saunders P. T. K., Sharpe R. M. and Smith L. B. (2012). Androgen receptor signalling in peritubular myoid cells is essential for normal differentiation and function of adult Leydig cells. Int. J. Androl. 35, 25-40. 10.1111/j.1365-2605.2011.01150.x [DOI] [PubMed] [Google Scholar]
- Wu X., Goodyear S. M., Tobias J. W., Avarbock M. R. and Brinster R. L. (2011). Spermatogonial stem cell self-renewal requires ETV5-mediated downstream activation of Brachyury in mice. Biol. Reprod. 85, 1114-1123. 10.1095/biolreprod.111.091793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Ashley T., Brainerd E. E., Bronson R. T., Meyn M. S. and Baltimore D. (1996). Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 10, 2411-2422. 10.1101/gad.10.19.2411 [DOI] [PubMed] [Google Scholar]
- Yamanaka S., Nishihara H., Toh H., Eijy Nagai L. A., Hashimoto K., Park S.-J., Shibuya A., Suzuki A. M., Tanaka Y., Nakai K. et al. (2019). Broad heterochromatic domains open in gonocyte development prior to de novo DNA methylation. Dev. Cell 51, 21-34.e25. 10.1016/j.devcel.2019.07.023 [DOI] [PubMed] [Google Scholar]
- Yang Q.-E., Gwost I., Oatley M. J. and Oatley J. M. (2013a). Retinoblastoma protein (RB1) controls fate determination in stem cells and progenitors of the mouse male germline. Biol. Reprod. 89, 113 10.1095/biolreprod.113.113159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.-E., Kim D., Kaucher A., Oatley M. J. and Oatley J. M. (2013b). CXCL12-CXCR4 signaling is required for the maintenance of mouse spermatogonial stem cells. J. Cell Sci. 126, 1009-1020. 10.1242/jcs.119826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Ramsay E. S. and Mock B. A. (1998). Cdkn2a, the cyclin-dependent kinase inhibitor encoding p16INK4a and p19ARF, is a candidate for the plasmacytoma susceptibility locus, Pctr1. Proc. Natl. Acad. Sci. USA 95, 2429-2434. 10.1073/pnas.95.5.2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Oatley J., Bardwell V. J. and Zarkower D. (2016). DMRT1 is required for mouse spermatogonial stem cell maintenance and replenishment. PLoS Genet. 12, e1006293 10.1371/journal.pgen.1006293 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.