ABSTRACT
Nascent myotubes undergo a dramatic morphological transformation during myogenesis, in which the myotubes elongate over several cell diameters and are directed to the correct muscle attachment sites. Although this process of myotube guidance is essential to pattern the musculoskeletal system, the mechanisms that control myotube guidance remain poorly understood. Using transcriptomics, we found that components of the Fibroblast Growth Factor (FGF) signaling pathway were enriched in nascent myotubes in Drosophila embryos. Null mutations in the FGF receptor heartless (htl), or its ligands, caused significant myotube guidance defects. The FGF ligand Pyramus is expressed broadly in the ectoderm, and ectopic Pyramus expression disrupted muscle patterning. Mechanistically, Htl regulates the activity of Rho/Rac GTPases in nascent myotubes and effects changes in the actin cytoskeleton. FGF signals are thus essential regulators of myotube guidance that act through cytoskeletal regulatory proteins to pattern the musculoskeletal system.
KEY WORDS: FGF signaling, Myogenesis, Organogenesis, Myotube guidance, Rho GTPase, Actin cytoskeleton, Heartless, Drosophila embryogenesis
Summary: This study has identified a novel function for the FGF pathway as a key regulator of myotube guidance during myogenesis, along with a unique experimental framework to further investigate the mechanisms involved.
INTRODUCTION
In mature skeletal muscle, myofibers are perfectly aligned with the skeleton so that muscle contractions can produce coordinated movements. During development, myotubes are directed to specific muscle attachment sites on tendons through the process of myotube guidance, and then mature into correctly aligned myofibers. Compared with our understanding of myoblast cell fate specification, migration and fusion, relatively little is known about the molecular pathways that direct myotube guidance (Maartens and Brown, 2015).
After migrating to sites of myogenesis, myoblasts polarize and mature into nascent myotubes. Polarized nascent myotubes will extend two leading edges in opposite directions, and each leading edge navigates the extracellular environment to identify a muscle attachment site. Through this process of myotube guidance, a single myofiber will be attached to two tendons at the end of myogenesis. The intracellular pathways that reorganize the cytoskeleton during myotube guidance have been characterized in some detail. For example, the RNA-binding protein Hoi-polloi regulates the actin cytoskeleton by modulating Tropomyosin expression (Williams et al., 2015), and the Rho GTPase activating protein Tumbleweed, in combination with the kinesin Pavarotti, reorganizes the microtubule cytoskeleton (Guerin and Kramer, 2009). Although dynamic changes to both the actin and microtubule cytoskeletons are essential for myotube guidance, the extrinsic inputs that regulate cytoskeletal dynamics to guide myotube leading edges toward the correct muscle attachment sites remain incompletely understood.
During Drosophila embryogenesis, Slit-Robo signaling acts as both a chemoattractant to initiate myotube elongation and as a repulsive cue to prevent myoblasts from accumulating at the ventral midline (Kramer et al., 2001). A second signaling pathway that regulates myotube guidance is directed by the orphan transmembrane receptor Kon-tiki (Kon). Kon functions through the intracellular adaptor protein Grip and, although the precise molecular function of Grip during myogenesis is still unclear, it may act as a scaffolding protein to cluster active Kon complexes to the myotube membrane (Schnorrer et al., 2007). Alternatively, Grip may activate intracellular signaling pathways involving small GTPases. Although the vertebrate orthologs of the Slit-Robo and Kon-Grip signaling axes have not been characterized in the context of myogenesis, Wnt11 is required to organize and orient myotubes in the trunk myotome (Gros et al., 2009). In fact, Wnt11 is the only known signaling ligand to direct myotube morphogenesis in vertebrates.
Thirty individual myotubes are specified in each segment of the Drosophila embryo, and each myotube acquires a highly stereotyped morphology (Bate, 1990). Disrupting the Slit-Robo or Kon-Grip signaling pathways affects only a subset of muscles in each segment (Kramer et al., 2001; Schnorrer et al., 2007), which suggests that additional extrinsic inputs are required to direct myotube guidance. To identify the putative signal transduction pathways that regulate myotube guidance, we profiled the transcriptome of nascent embryonic myotubes, and found that transcripts encoding components of the Fibroblast Growth Factor (FGF) pathway were enriched in this cell population. Null mutations in the FGF receptor heartless (htl), or in the FGF ligands pyramus (pyr) and thisbe (ths), caused dramatic myotube guidance defects. htl mutant myotubes that expressed Htl showed largely normal muscle morphology, which argues the role of Htl is cell autonomous. Mechanistically, a loss-of-function allele of the Rho/Rac guanine nucleotide exchange factor pebble (pbl) and a dominant-negative form of Rac1 suppressed the htl myotube guidance phenotype. Rho/Rac GTPases are well-known regulators of the actin cytoskeleton, and Htl is required to restrict Rac activity and in turn F-actin levels in nascent myotubes. This study has identified FGF signaling as an essential component of the myotube guidance pathway that regulates Rac activity to regulate cytoskeletal changes during muscle morphogenesis.
RESULTS
FGF signaling components are enriched in nascent myotubes
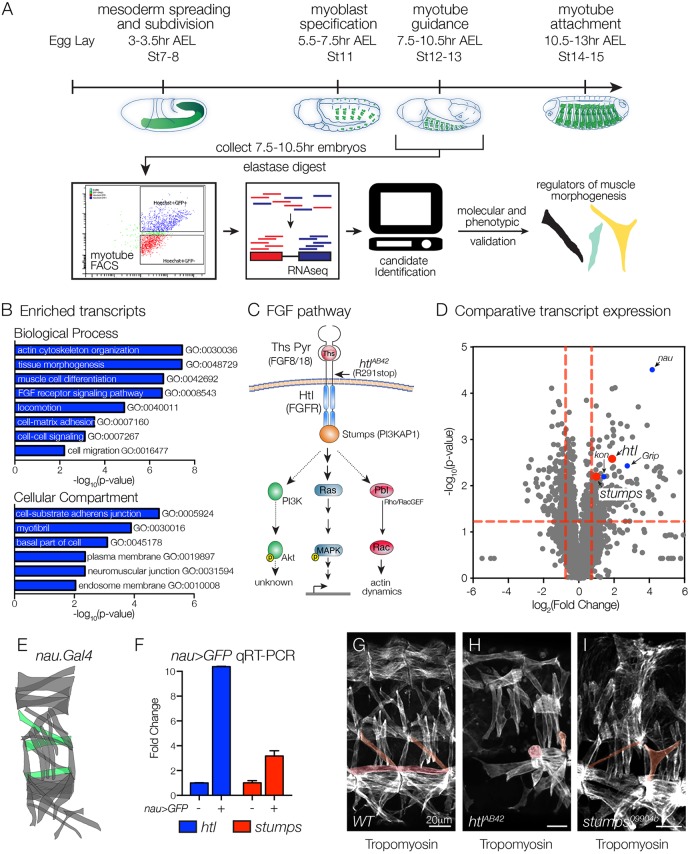
To uncover signal transduction pathways that direct myotube guidance, we devised a fluorescence activated cell sorting and RNA deep sequencing (FACS-seq) strategy to profile the transcriptome of nascent myotubes (Fig. 1A). rp298.GAL4 is broadly expressed in nascent myotubes, and we collected rp298>GFP embryos at 7.5-10.5 h after egg lay (AEL; stage 12-13), sorted GFP+ myotubes and GFP– control cells, and isolated RNA for deep sequencing. This analysis identified 238 transcripts that were significantly enriched in nascent myotubes (Table S1), and the enriched transcripts clustered with a number of Gene Ontology (GO) terms associated with muscle development and function including muscle cell differentiation (GO:0042692), myofibril (GO:0030016) and neuromuscular junction (GO:0031594; Fig. 1B, Table S1).
Fig. 1.
FGF signaling components are enriched in nascent myotubes. (A) Experimental design. Nascent myotubes that expressed rp298>eGFP were FACS sorted and deep sequenced (FACS-seq). Candidate genes were then tested in vivo to identify regulators of myotube guidance. (B) GO analysis of transcripts enriched in GFP+ myotubes compared with GFP–cells. (C) Schematic of the FGF signaling pathway. Vertebrate orthologs are in parentheses. Indirect or putative interactions are shown with dotted lines. (D) Volcano plot of transcripts identified in nascent myotubes. Each data point represents the average values for a single transcript from three biological replicates. Red dashed lines indicate P=0.05. Points above the horizontal line and to the right of the vertical lines are significantly enriched with P<0.05. Red dots indicate FGF signaling components, blue dots indicate known regulators of myogenesis and myotube guidance. (E,F) Molecular validation of FACS-seq results. E shows a diagram of the body wall muscles in a single embryonic segment. nau.Gal4 expressing muscles are shown in green. Nascent myotubes that expressed nau>eGFP were FACS sorted and transcript expression was assayed by qRT-PCR (F). htl and stumps transcripts were significantly enriched in GFP+ myotubes compared with GFP– cells. (G-I) Stage 16 embryos labeled for Tropomyosin to visualize the body wall musculature. htlAB42 (H) and stumps09904b (I) embryos showed multiple body wall muscle defects compared with WT (G). Rounded myotubes and myotubes with attachment site defects are pseudocolored. Embryos are oriented with anterior to the left and dorsal to the top.
One interesting cluster from the GO analysis associated with the term FGF receptor signaling pathway (GO:0008543; Fig. 1B), and included seven transcripts, which encode the FGF receptor Htl, two enzymes that direct the synthesis of FGF co-receptors (heparin sulfate proteoglycans), the Htl intracellular effector proteins Stumps and Rau, a matrix metalloproteinase that regulates FGF signaling, and the modulator of FGF signaling Sprouty (Fig. 1C, Table S1). To further analyze our FACS-seq results, we generated a scatter plot that compared the magnitude of statistical significance in expression with the magnitude of fold change for transcripts in the sorted and control cell populations (Fig. 1D). kon and Grip encode signal transducing proteins known to regulate myotube guidance (Schnorrer et al., 2007); htl and stumps showed similar values on our scatter plot as kon and Grip (Fig. 1D).
The most significantly enriched transcript from our FACS-seq experiment encodes the transcription factor Nautilus (Nau; Fig. 1D); this muscle identity gene is expressed in a subset of nascent myotubes including longitudinal lateral 1 (LL1), ventral lateral 1 (VL1) and ventral oblique 5 (VO5) (Fig. 1E). To validate our large-scale transcriptome profiling study, we used nau.Gal4 to collect stage 12-13 nau>GFP embryos, and sorted GFP+ myotubes and GFP– control cells. Using quantitative real-time PCR (qPCR), we confirmed that htl and stumps were significantly enriched in Nau-expressing myotubes (Fig. 1F). Overall, our FACS-seq experiment highlighted a putative role for the FGF signaling pathway in regulating myotube morphogenesis.
Htl regulates myotube morphogenesis
Founder cells are a specialized population of myoblasts that individually give rise to a single body wall muscle, and Htl is a known regulator of founder cell fate specification (Carmena et al., 1998). We found that htl transcripts were enriched in nascent myotubes several hours after founder cell specification, which suggested that after FGF signals specify a subset of founder cells, subsequent FGF signals direct myotube morphogenesis and perhaps myotube guidance. In support of this hypothesis, embryos homozygous for the amorphic allele htlAB42 had a number of rounded myotubes (Fig. 1G-I), a phenotype previously associated with defects in myotube elongation (Johnson et al., 2013). Embryos homozygous for a strong loss-of-function allele of stumps also showed defects in muscle morphogenesis (Fig. 1I).
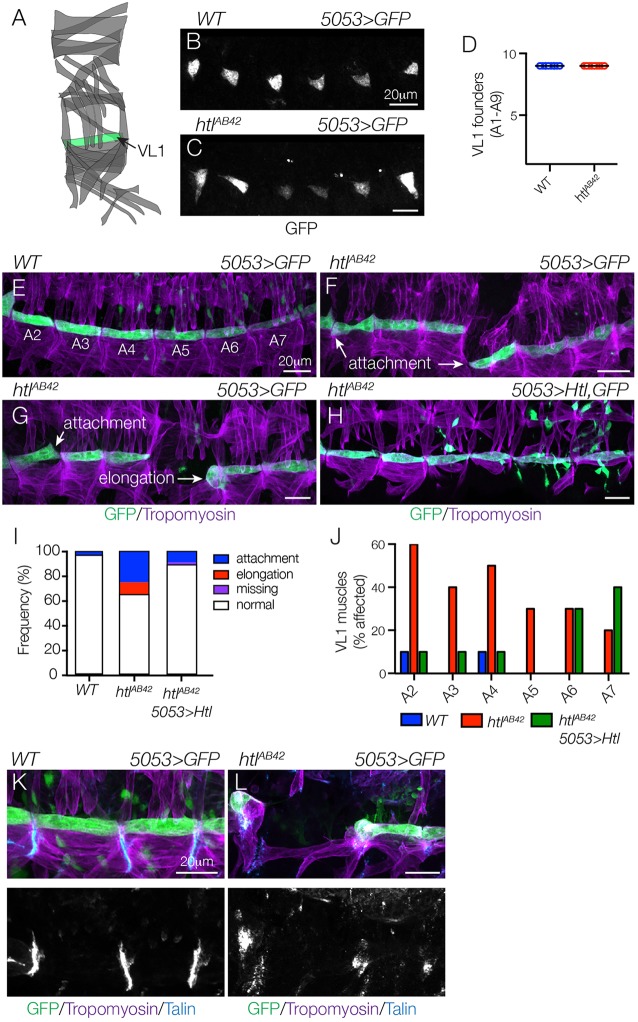
To experimentally distinguish a role for Htl during myotube morphogenesis from its known role in directing founder cell fate specification, we used transgenic lines that are expressed in discrete founder cell populations and subsequently in nascent myotubes. 5053.Gal4 is expressed in VL1 founders (Fig. 2A,B), and htlAB42 embryos showed an equivalent number of 5053>GFP+ VL1 founder cells as wild-type (WT) embryos (Fig. 2B-D). Although we did observe some variability in the 5053>GFP expression levels in htlAB42 embryos, the VL1 founder cell fate was correctly specified. Consistent with the hypothesis that FGF signals direct myotube morphogenesis, htlAB42 5053>GFP+ VL1 muscles showed myotube elongation and muscle attachment site selection defects (Fig. 2E-G), and these defects could be suppressed by expressing Htl in htlAB42 VL1 muscles (Fig. 2H,I). Abdominal segments 2-7 (A2-A7) show the most reproducible muscle pattern in WT embryos (Bate, 1990), so we calculated the frequency of htlAB42 VL1 morphology defects individually in these segments. Myogenic defects were not confined to any subset of segments in htlAB42 embryos, although A2-A4 showed a slightly higher frequency of myogenic phenotypes than A5-A7 (Fig. 2J). htlAB42 VL1 muscles that expressed Htl in A6-A7 did not show an appreciable improvement in muscle morphology, which is likely due to weaker 5053.gal4 expression in segments A6-A7 compared with A2-A5 (Fig. 2E). However, these studies suggest that Htl acts cell autonomously to direct VL1 myotube morphogenesis and not VL1 founder cell fate.
Fig. 2.
Htl acts cell autonomously to direct myotube guidance. (A) Diagram of the body wall muscles in a single embryonic segment (A2-A7), with 5053.Gal4-expressing VL1 muscle shown in green. (B-D) Stage 12 5053>eGFP embryos labeled for GFP to visualize VL1 nascent myotubes. The number of nascent myotubes specified in WT (B) and htlAB42 (C) embryos was equivalent (D). (E-H) Stage 16 5053>eGFP embryos labeled for GFP (green) and Tropomyosin (violet). WT VL1 muscles showed a largely invariant morphology (E). htlAB42 VL1 muscles often attached to the wrong muscle attachment site or failed to elongate (F,G). htlAB42 VL1 muscles that expressed Htl showed normal morphology (H). Mononucleate cells are visceral mesoderm cells that express 5053.Gal4. (I) Histogram of muscle phenotypes in Stage 16 embryos (n=60 per genotype). (J) Histogram of muscle phenotypes in Stage 16 embryos by embryonic segment (n=60 per genotype). (K,L) WT (K) and htlAB42 (L) stage 16 5053>eGFP embryos labeled for Talin (blue), GFP (green) and Tropomyosin (violet). htlAB42 VL1 muscles that failed to elongate will form myotendinous junctions. Embryos are oriented with anterior to the left and dorsal to the top.
Htl regulates myotube guidance
Although the htlAB42 VL1 phenotypes we observed were consistent with a role for Htl in regulating myotube guidance, an alternative explanation is that Htl directs myotendinous junction (MTJ) formation and the htl elongation phenotypes reflect dissociation of VL1 muscles away from tendons after muscle contractions initiate. To distinguish between these possibilities, we first labeled embryos for the MTJ component Talin, which is expressed from mature tendons after myotube attachment (Fig. 2K). Although htlAB42 embryos showed reduced Talin expression compared with WT embryos, we also observed Talin in close proximity to rounded htlAB42 VL1 muscles (Fig. 2L), suggesting that tendon cell fate specification and maturation are largely normal in htlAB42 embryos.
We live imaged longitudinal oblique 1 (LO1) muscle morphogenesis and found that WT myotubes extend a primary leading edge from the posterior of the segment toward the anterior, and that the primary leading edge elongates across a majority of the segment, orthogonal to the anterior-posterior axis (Movie 1). A secondary leading edge also extends a short distance toward the posterior, and each leading edge will ultimately reach a muscle attachment site. htlAB42 LO1 myotubes formed primary and secondary leading edges but, in the case of rounded myotubes, the primary leading edge extended ventrally and failed to reach a muscle attachment site (Movie 1). These studies demonstrate that Htl is required for myotube leading edges to accurately navigate toward muscle attachment sites before MTJ formation, and further argue that Htl directs myotube guidance.
Htl regulates myotube guidance in multiple cell populations
To understand whether Htl acts broadly in the mesoderm to direct myotube guidance, we used slou.GAL4 to track the morphogenesis of five additional muscles in htlAB42 embryos (Fig. 3A-D). The number of slou>GFP+ cells was largely comparable between WT and htlAB42 embryos at the onset of myotube elongation, except that the ventral transverse 1 (VT1) founder cells were not specified (Fig. 3B-D). However, at the end of myogenesis, the dorsal transverse 1 (DT1) and LO1 muscles in htlAB42 embryos showed elongation and attachment site defects (Fig. 3E,F,H).
Fig. 3.
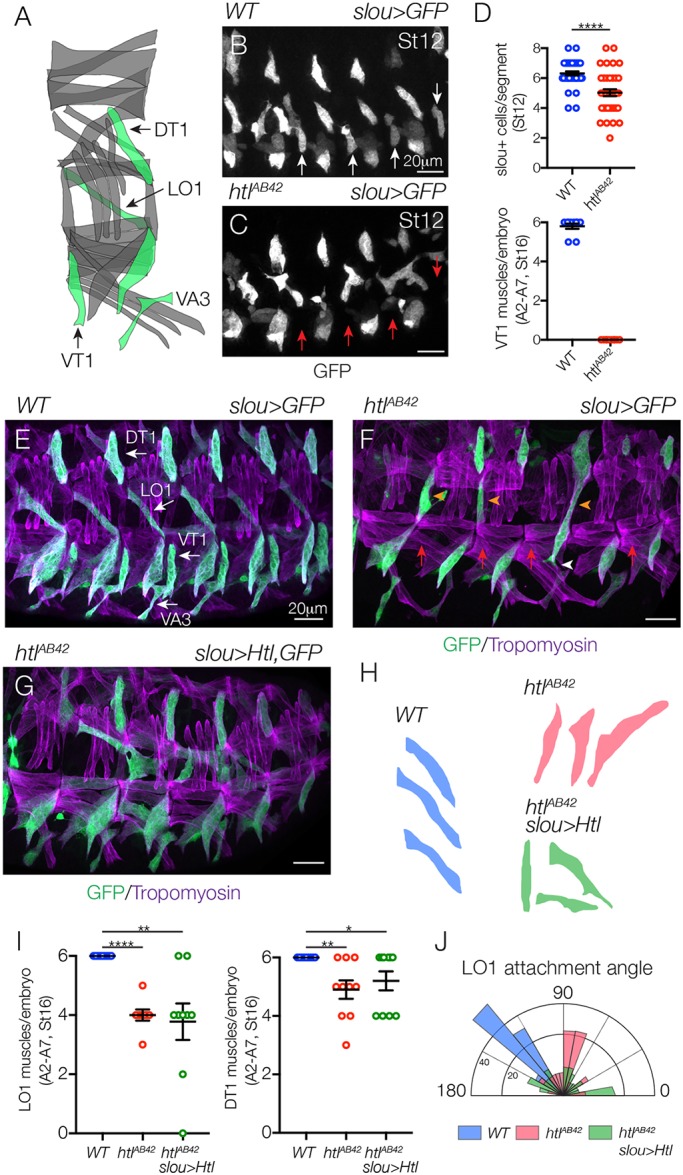
Htl acts cell autonomously and non-cell autonomously in a subset of myotubes. (A) Diagram of body wall muscles that expressed slou.Gal4 (green). (B,C) St12 slou>eGFP embryos labeled for GFP. One nascent myotube per segment was absent in htlAB42 embryos (C, red arrows) compared with WT. (D) Muscle quantification (A2-A7). htlAB42 embryos failed to specify VT1 founder cells. (E-G) Stage 16 slou>eGFP embryos labeled for GFP (green) and Tropomyosin (violet). WT LO1, DT1, VA3 and VT1 muscles (E, white arrows) showed a largely invariant morphology. htlAB42 LO1 muscles (F) acquired a lateral morphology (orange arrowheads) and overextended ventrally (white arrowhead). htlAB42 DT1 muscles also showed defective morphology and VT1 muscles were absent in htlAB42 embryos (red arrows). htlAB42 LO1 and DT1 muscles that expressed Htl showed improved, but not WT, morphology (G). (H) Representative traces of LO1 muscles. (I) Number of LO1 (left) and DT1 (right) muscles in stage 16 embryos. (J) Radial density plot of LO1 muscle attachment angles in stage 16 embryos. Micrographs were rotated to typical anterior-posterior/dorsal-ventral orientation, and LO1 myotube attachment angles were measured using ImageJ software. Myotube attachment angles were binned in 10° increments; each slice represents the percent of myotubes in a given bin (n≥35 muscles per genotype). *P<0.05, **P<0.01, ****P<0.0001 (unpaired, two-tailed Student's t-test). Data are mean±s.e.m. Embryos are oriented with anterior to the left and dorsal to the top.
Live imaging of LO1 muscle morphogenesis showed that WT LO1 myotubes first extend a primary leading edge dorsally and perpendicular to the anterior-posterior axis. The primary leading edge then makes a dramatic turn toward the anterior and extends parallel to the anterior-posterior axis (Movie 2). This circuitous path underlies the final oblique morphology of the LO1 muscle. The secondary leading edge extends ventrally only a short distance and each leading edge will arrive at a muscle attachment site. htlAB42 LO1 myotubes showed three distinct phenotypes. In most cases, the primary LO1 leading edge failed to turn to the anterior, which resulted in an LO1 muscle with longitudinal morphology (Movie 2). htlAB42 LO1 primary leading edges also extended ventrally instead of dorsally (Movie 3), and htlAB42 LO1 myotubes even failed to specify a primary leading edge and both ends of the myotube extended incorrectly (Movie 4). These live imaging studies further support a role for Htl in directing LO1 myotube guidance. More striking, however, was the observation that htlAB42 LO1 leading edges often arrived at the incorrect muscle attachment site, and then moved to a different position in the embryo (Movies 3 and 4). This observation suggests that myotubes have the ability to distinguish among muscle attachment sites and that leading edge proximity to an attachment site alone is not sufficient to dictate attachment site choice.
To understand whether Htl acts cell autonomously, we expressed Htl in htlAB42 LO1 myotubes with slou.Gal4, and this restricted expression partially restored htlAB42 LO1 muscles to WT morphology (Fig. 3G-I). To accurately quantify LO1 muscle morphogenesis, we calculated an attachment angle for individual myotubes (Fig. 3J). WT LO1 muscles had an oblique attachment angle that ranged between 120-150°, whereas a large proportion of htlAB42 LO1 muscles (45.6%) had a lateral attachment angle between 70-90° (Fig. 3J). Only 22.2% of htlAB42 LO1 muscles that expressed Htl had an attachment angle between 70-90° (Fig. 3J), suggesting that the role of Htl during LO1 myotube guidance is partially cell autonomous.
We next considered the possibility that myotube guidance is an interdependent process and that loss of Htl activity in one myotube population might affect the morphogenesis of other myotube populations. To test this possibility, we broadly expressed Htl in all correctly specified founder cells of htlAB42 embryos with rp298.Gal4. Broad Htl expression largely restored muscle morphology in htlAB42 embryos, but LO1 myotubes did not acquire the stereotypically oblique morphology seen in WT embryos (Fig. S1A,B). It appeared to be possible that slou.Gal4 and rp298.Gal4 did not restore Htl expression to WT levels in htlAB42 LO1 myotubes; however, htl mRNA expression was not consistently different among the dorsal, medial and ventral myoblasts at the onset of myotube morphogenesis (Fig. S1C,D). Our rescue and expression experiments indicate that the mechanisms directing LO1 myotube guidance are partially distinct from those directing VL1 myotube guidance, which could depend on a number of additional factors such as cell-cell interactions within the mesoderm (see Discussion). Nonetheless, our studies with slou.Gal4 argue that Htl regulates myotube guidance in multiple cell populations.
Pyr and Ths regulate myotube guidance
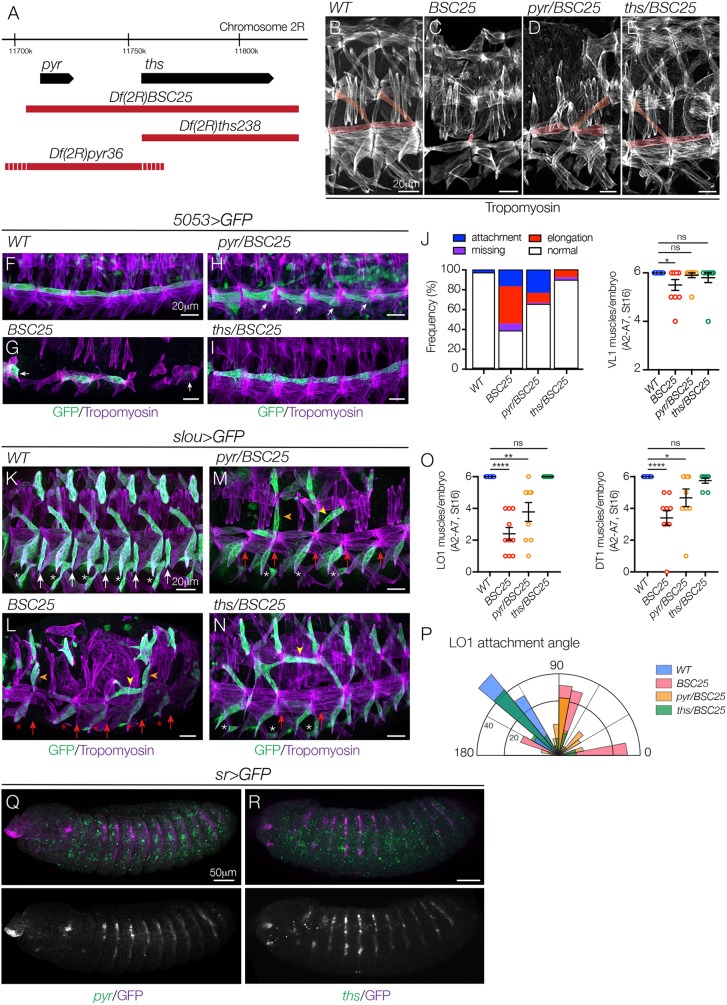
The Htl ligands are encoded by two closely linked genes pyr and ths, and both loci are uncovered by Df(2R)BSC25 (Fig. 4A). Df(2R)BSC25 embryos showed extensive myogenic defects (Fig. 4B,C), as did embryos transheterozygous for Df(2R)BSC25 and smaller deficiencies that specifically delete pyr [Df(2R)pyr36] or ths [Df(2R)ths238] (Fig. 4D,E). We used 5053.Gal4 and slou.Gal4 to further characterize the role of pyr and ths during myogenesis, and found that both ligands are required for VL1 (Fig. 4F-J) and LO1 (Fig. 4K-P) elongation and attachment site selection, although pyr/Df(2R)BSC25 embryos showed much stronger phenotypes than ths/Df(2R)BSC25 embryos (Fig. 4J,P). Consistent with these loss-of-function studies, embryos that expressed Pyr broadly in the ectoderm (under the control of 69B.Gal4) or in the posterior of each segment (under the control of en.Gal4) showed extensive defects in muscle morphogenesis, whereas embryos that expressed Ths in the ectoderm showed largely normal muscle patterning (Fig. S2A-E,I).
Fig. 4.
The FGF ligands Pyr and Ths direct myotube guidance. (A) Genomic organization of pyr and ths. Sequences deleted by Df(2R)BSC25, Df(2R)pyr36 and Df(2R)ths238 are shown in red (dashes reflect breakpoint uncertainty). Df(2R)pyr36 does not disrupt ths transcript expression (Kadam et al., 2009). (B-E) Stage 16 embryos labeled for Tropomyosin. Df(2R)BSC25 (C) and Df(2R)pyr36 embryos (D) showed severe body wall muscle defects, including missing and disorganized muscles, compared with WT (B). Df(2R)ths238 embryos (E) showed mild myogenic defects. VL1 and LO1 muscles are pseudocolored. (F-I) Stage 16 5053>eGFP embryos labeled for GFP (green) and Tropomyosin (violet). VL1 muscles in Df(2R)BSC25 (G) and Df(2R)pyr36 embryos (H) showed elongation and attachment site defects, compared with WT (F). VL1 muscles in Df(2R)ths238 embryos were largely normal (I). White arrows show incorrect attachment sites. (J) Quantification of VL1 muscle phenotypes in stage 16 embryos (n≥54 muscles per genotype). Left shows a histogram of muscle phenotypes. Right shows the number of VL1 muscles. (K-N) Stage 16 slou>eGFP embryos labeled for GFP (green) and Tropomyosin (violet). LO1 muscles in Df(2R)BSC25 (L) and Df(2R)pyr36 embryos (M) acquired lateral (orange arrowheads) and transverse (yellow arrowheads) morphologies similar to htlAB42 LO1 muscles, compared with WT (K). LO1 muscles in Df(2R)ths238 embryos also showed morphological defects (N). White asterisks, VA3 muscles; red asterisks, missing VA3 muscles; white arrows, VT1 muscles; red arrows, missing VT1 muscles. (O) Quantification of LO1 (left) and DT1 (right) muscles. (P) Radial density plot of LO1 muscle attachment angles in stage 16 embryos (n≥35 muscles per genotype). (Q,R) Stage 12 sr>eGFP embryos labeled for pyr (Q) or ths (R) mRNA (green) and GFP (violet). pyr is expressed in the three ectodermal domains per segment and is excluded from Sr+ tendon cells (Q). ths expression in the ectoderm is modest but detectable immediately anterior to tendon cells (R). ns, not significant. *P<0.05, **P<0.01, ****P<0.0001 (unpaired, two-tailed Student's t-test). Data are mean±s.e.m. See also Fig. S2. Embryos are oriented with anterior to the left and dorsal to the top.
Stripe (Sr)-expressing tendon cells define the anterior segment border, and we found that pyr is expressed in the ectoderm posterior to the segment border in three distinct domains along the dorsal-ventral axis (Fig. 4Q). ths expression is less organized, although ths can be detected in the most posterior ectoderm cells of some segments (Fig. 4R). Primary myotube leading edges originate in the posterior of the segment and extend across the segment to make muscle attachments at the anterior segment border (Movies 1 and 2). Myotube leading edges could therefore be traveling along the pyr expression domain.
Unknown chemotactic cues from tendon cells are thought to guide myotube leading edges to muscle attachment sites (Maartens and Brown, 2015; Schnorrer et al., 2007), and Pyr can be chemotactic under certain contexts (Kadam et al., 2012). When we misexpressed Pyr in tendon cells (under the control of sr.Gal4) the muscle pattern was largely unchanged, although the dorsal acute 1 and dorsal oblique 1 muscles showed some attachment site defects (Fig. S2G-I). Nonetheless, our results suggest that Pyr and Ths are expressed outside of tendon cells and function to guide myotubes to muscle attachment sites.
We next asked whether FGF signaling could be acting indirectly during myotube guidance by either regulating tendon cell specification or by simply acting as a permissive chemokinetic signal for myotubes to respond to other chemotactic cues. To test these possibilities, we characterized tendon cell fate using sr.Gal4 and found tendon cells are specified correctly in Df(2R)BSC25 embryos (Fig. S3A,B). We also expressed constitutively active (CA)-Htl broadly in the founder cells of Df(2R)BSC25 embryos, but this did not improve the Df(2R)BSC25 myogenic phenotype (Fig. S3C,D). Thus, activating Htl alone is not sufficient to promote myotube guidance in the absence of FGF ligands, suggesting that FGF signals are instructive during myogenesis.
htl genetically interacts with pbl, a guanine nucleotide exchange factor (GEF)
FGF receptors (FGFRs) regulate multiple intracellular signaling pathways to direct cell fate specification and tissue morphogenesis, including the Mitogen-activated protein kinase (MAPK) and Protein kinase B (AKT) cascades, as well as the Rho/Rac family of small GTPases (Muha and Müller, 2013; Ornitz and Itoh, 2015) (Fig. 1C). To identify the mechanism by which Htl directs myotube guidance, we first assayed phospho (p)-MAPK and pAKT levels. A well-characterized antibody has been used to identify pMAPK in Drosophila embryos and, although we detected pMAPK in many of the tissues previously described (Gabay et al., 1997), we did not detect appreciable pMAPK in nascent myotubes (Fig. S4A). A Drosophila-specific pAKT antibody has also been developed (see Materials and Methods), but this antibody does not work well for immunohistochemistry. However, using western blot, we found that pAKT levels were not significantly changed in pyr/ths embryo lysates compared with controls (Fig. S4B). Although these expression studies do not exclude a role for MAPK and AKT signaling during myotube guidance, they do suggest that FGF signaling regulates myotube guidance through a MAPK- and AKT-independent mechanism.
We next checked for genetic interactions between htl and the Rho/Rac GEF pbl (van Impel et al., 2009). Remarkably, the loss-of-function allele pbl3 dominantly suppressed overall muscle morphology defects in htlAB42 embryos (Fig. 5A-D), and specifically suppressed attachment site defects in htlAB42 VL1 muscles (Fig. 5E-H). As pbl suppressed the myotube guidance phenotype in htl embryos, we reasoned that the role of Htl is to restrict Pbl activity in nascent myotubes. To functionally test this possibility, we expressed dominant-negative (DN) and CA-Rac1 in VL1 founder cells with 5053.Gal4. WT VL1 myotubes that expressed DN-Rac1 showed normal morphology (Fig. 5I,M), but VL1 myotubes that expressed CA-Rac1 showed elongation and muscle attachment site defects at a frequency similar to that of htlAB42 VL1 muscles (Fig. 5J,M). In addition, htlAB42 VL1 myotubes that expressed DN-Rac1 showed fewer myotube guidance defects than htlAB42 VL1 muscles (Fig. 5K,M), and htlAB42 VL1 myotubes that expressed CA-Rac1 showed significantly more myotube guidance defects than htlAB42 VL1 muscles (Fig. 5L,M). In fact, htlAB42 VL1 myotubes that expressed CA-Rac1 most often attached to the VL3 muscle attachment sites (Fig. 5L), which we interpret as a qualitative enhancement of the htlAB42 phenotype. Overall, these results are consistent with a role for Htl in restricting Pbl activity to direct myotube guidance.
Fig. 5.
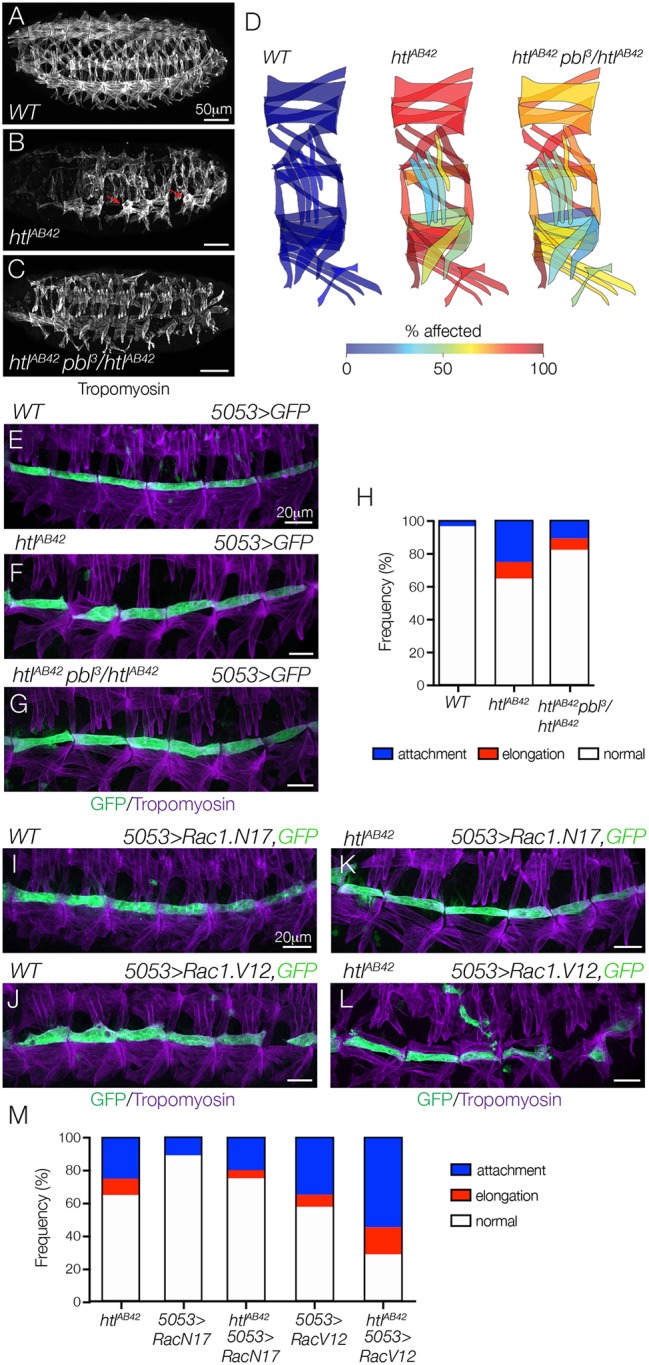
Htl regulates myotube guidance through the Pbl-Rac signaling pathway. (A-C) Stage 16 embryos labeled for Tropomyosin. Body wall muscle defects in htlAB42 embryos (B, red arrows) were suppressed in htlAB42 embryos heterozygous for pbl3 (C), compared with WT (A). (D) Heat map showing the frequency of muscle defects (n=60 segments per genotype) in WT (left) and htlAB42 (middle) embryos, and htlAB42 embryos heterozygous for pbl3 (right). (E-G) Stage 16 5053>eGFP embryos labeled for GFP (green) and Tropomyosin (violet). htlAB42 VL1 myotube guidance defects (F) were suppressed in htlAB42 embryos heterozygous for pbl3 (G), compared with WT (E). (H) Histogram of muscle phenotypes in stage 16 embryos (n≥54 muscles per genotype). (I-L) Stage 16 5053>eGFP embryos labeled for GFP (green) and Tropomyosin (violet). VL1 myotubes that expressed DN Rac1 (Rac1.N17, I) showed normal morphology; VL1 myotubes that expressed CA Rac1 (Rac1.V12, J) showed guidance defects. htlAB42 myotube guidance defects were suppressed in VL1 muscles that expressed Rac1.N17 (K); htlAB42 myotube guidance defects were dramatically enhanced in VL1 muscles that expressed Rac1.V12 (L). (M) Histogram of muscle phenotypes in stage 16 embryos (n≥54 muscles per genotype). See also Fig. S4. Embryos are oriented with anterior to the left and dorsal to the top.
Although LO1 myotubes appeared to develop under different constraints than VL1 myotubes (Fig. 3), we hypothesized that Rho GTPases regulate a common pathway that directs both LO1 and VL1 myotube guidance. Similar to VL1 myotubes, LO1 myotubes that expressed CA-Rac1 showed severe guidance defects, whereas LO1 myotubes that expressed DN-Rac1 showed normal morphology (Fig. S4C-F). These results are consistent with studies of axon morphogenesis in which neurons that expressed CA-Rac1, but not DN-Rac1, showed defects in axon outgrowth (Luo et al., 1994), suggesting that limiting Rac1 activity is a common mechanism essential for cellular outgrowth and guidance.
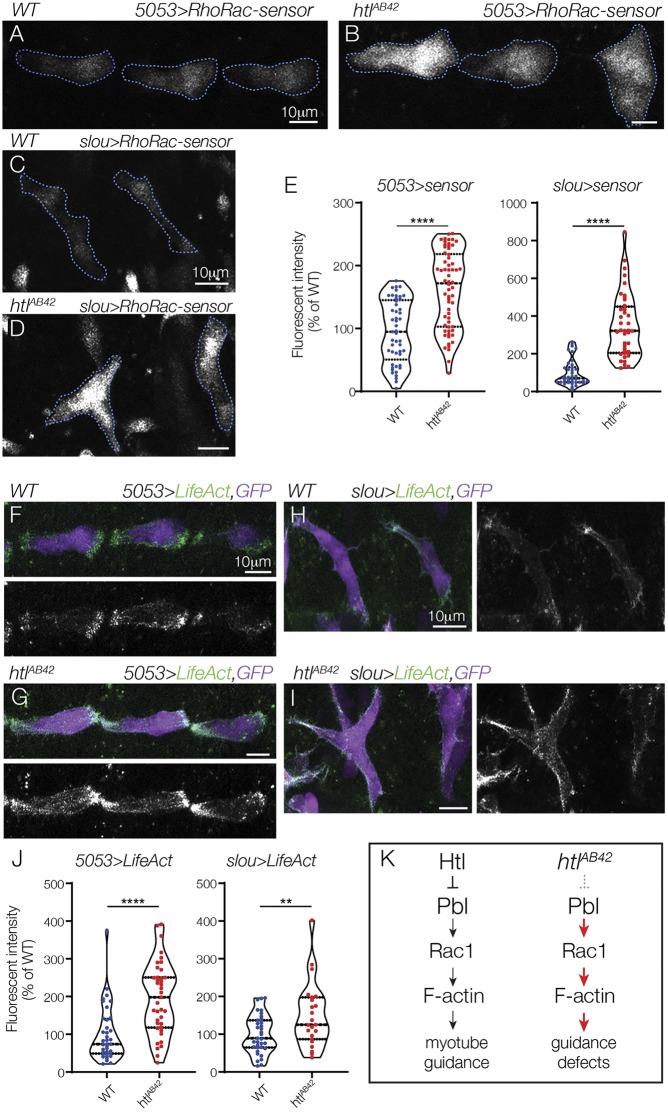
Htl limits Rho/Rac activity and F-actin assembly in nascent myotubes
To directly visualize Rho/Rac activity in vivo, we expressed a Rho/Rac biosensor in VL1 and LO1 myotubes. Fluorescence from the biosensor acts as a readout of Rho/Rac activity (Abreu-Blanco et al., 2014), and htlAB42 VL1 and LO1 myotubes showed a two- to threefold increase in biosensor fluorescence compared with WT controls (Fig. 6A-E). As the Rho/Rac family of small GTPases regulates actin dynamics (Bustelo et al., 2007), we hypothesized that F-actin levels might also be affected in htlAB42 myotubes. In WT VL1 and LO1 myotubes, F-actin accumulated at the leading edges and was largely excluded from the lateral membrane domains (Fig. 6F,H). In htlAB42 VL1 and LO1 myotubes, F-actin accumulated at the leading edges, but also along the lateral membrane domains (Fig. 6G,I). In addition, htlAB42 myotubes showed significantly more internal F-actin than control myotubes (Fig. 6J). From these studies, we propose a model in which Htl restricts Pbl activity, Rho/Rac activation and, ultimately, F-actin assembly, to direct myotube leading edges to correct muscle attachment sites (Fig. 6K).
Fig. 6.
Htl restricts Rho/Rac activity during myotube guidance. (A,B) Live stage 13 5053>RhoRac-sensor::eGFP embryos imaged for GFP. htlAB42 VT1 myotubes (B) showed more GFP fluorescence than WT VT1 myotubes (A). (C,D) Live stage 13 slou>RhoRac-sensor::eGFP embryos imaged for GFP. htlAB42 LO1 myotubes (D) showed more GFP fluorescence than WT LO1 myotubes (C). (E) Violin plots of 5053>RhoRac-sensor (left) and slou>RhoRac-sensor (right) fluorescence. (F,G) Live stage 13 5053>LifeAct::RFP, eGFP embryos imaged for GFP (violet) and RFP (green). F-actin accumulated primarily at the leading edges of WT VT1 myotubes (F), whereas it was not restricted to the leading edges of htlAB42 VT1 myotubes (G). (H,I) Live stage 13 slou>LifeAct::RFP, eGFP embryos imaged for GFP (blue) and RFP (green). WT LO1 myotubes accumulated F-actin at the leading edges (H), whereas F-actin was not restricted to the leading edges of htlAB42 LO1 myotubes, and showed dramatic enrichment at the lateral membrane domains (I). (J) Violin plots of LifeAct fluorescence, as described in E. (K) Proposed mechanism for Htl-mediated myotube guidance. Each data point in violin plots represents normalized fluorescence for a single myotube. The median and interquartile ranges are shown (dotted horizontal lines). **P<0.01, ****P<0.0001 (unpaired, two-tailed Student's t-test). Data are mean±s.e.m. Embryos are oriented with anterior to the left and dorsal to the top.
DISCUSSION
This study has uncovered a crucial role for FGF signaling in regulating myotube guidance. We found that the FGF receptor Htl acts cell autonomously in nascent myotubes to direct elongation and muscle attachment site selection. The FGF ligands Pyr and Ths are required for myotube guidance and appear to acts as paracrine signals from the ectoderm to the developing musculature. Mechanistically, Htl acts through the Rho/Rac GEF Pbl to restrict Rho/Rac activity and, in turn, to regulate the actin cytoskeleton. These studies provide novel insights into the mechanisms by which FGF signaling regulates the cytoskeleton to direct cellular guidance and tissue morphogenesis.
Functional role for Htl in myotube guidance
The Htl receptor was originally identified as a regulator of mesoderm spreading in Drosophila (Beiman et al., 1996; Gisselbrecht et al., 1996), and later as a component of the founder cell specification gene regulatory network (Carmena et al., 1998; Michelson et al., 1998). Our study confirms the observations that Htl regulates the specification of a subset of founder cells, in particular the VT1 founders, and identifies a novel function for Htl during myotube guidance that is genetically separable from its role in founder cell specification (Figs 2 and 3).
Perhaps the most surprising result from our studies is that htl myotube leading edge could reach a muscle attachment site and then translocate to alternative attachment sites (Movies 3 and 4). This observation argues that htl myotubes recognized some muscle attachment sites as incorrect and attempted to compensate. If proximity to a muscle attachment site alone was sufficient to initiate a myotube-tendon interaction, then htl myotubes could establish a MTJ with the first muscle attachment site encountered. We speculate that there is a myotendinous code that makes some myotube-tendon interactions permissive and others restrictive. In this model, a single myotube would have multiple permissive attachment sites, which we most convincingly observed in htl CA-Rac1 VL1 myotubes (Fig. 5L), and the role of Htl is to guide a myotube leading edge to a specific, permissive attachment site. The molecules that govern the myotendinous code are unknown, but our prediction is that permissive myotube-attachment site interactions are regulated by heterophilic interactions between cell-surface proteins that are uniquely expressed in subsets of tendons and nascent myotubes.
Htl suppresses Rho/Rac activity during myotube guidance
FGF receptors regulate multiple intracellular pathways. The vertebrate MAPK, AKT and Rho/Rac signaling pathways are activated in response to FGF signals and all three pathways influence cell migration, cell elongation and organ morphogenesis (Bénazéraf et al., 2010; Fera et al., 2004; Harding and Nechiporuk, 2012; Huebner et al., 2016; Jeong et al., 2016; Sato et al., 2011). In Drosophila, the intracellular pathways downstream of FGF receptors have been remarkably understudied compared with most other signal transducing receptors (Muha and Müller, 2013). We began to fill this knowledge gap by showing that the Rho/Rac GEF Pbl is an essential effector of Htl during myotube guidance. Previous genetic epistasis studies established that Pbl acts downstream of Htl during mesoderm spreading (Schumacher et al., 2004), but owing to the complexity of mesoderm-spreading phenotypes it was unclear whether Htl was an activator or repressor of Pbl activity. More recently, Rho and Rac were shown to be direct substrates of Pbl (van Impel et al., 2009), and biosensors were developed to visualize active Rho and Rac GTPases in vivo (Abreu-Blanco et al., 2014). Using these tools and insights, we found that the Pbl-Rho/Rac signaling axis is negatively regulated by Htl in nascent myotubes (Fig. 5A-M and Fig. 6A-E), which in turn reduces overall F-actin levels and localizes F-actin to myotube leading edges (Fig. 6F-K). In addition to these mechanistic insights, we have identified the Rho/Rac biosensor – a novel readout for FGF pathway activity in Drosophila that, to our knowledge, is the only reporter of FGF receptor activity other than pMAPK.
It remains unclear how limiting Rho/Rac activity can promote myotube elongation toward the correct muscle attachment site. One possibility is that active Htl receptor complexes accumulate along the lateral myotube membranes and focus Rho/Rac activity toward the leading edges. However, in WT myotubes Rho/Rac activity was not restricted to the leading edges (Fig. 6A). A more likely explanation is that crosstalk among the Rho family of GTPases, which includes Rho, Rac and Cdc42, localizes the individual family members and affects overall cytoskeletal dynamics. For example, pharmacological inhibition of Rho caused the Cdc42 expression domain to restrict and the Rac expression domain to expand in a Drosophila model of wound healing (Abreu-Blanco et al., 2014). In addition, dynamic but staggered accumulation of Rho and Rac to the leading edges of migratory cells is thought to drive the cytoskeletal changes that underlie the membrane expansion and retraction necessary for migration (Machacek et al., 2009). It appears plausible that Htl could inhibit Rho/Rac activity transiently to generate the waves of leading edge expansion and retraction necessary for myotube guidance, and that the Rho/Rac biosensor is not sensitive to these subtle dynamics. Alternatively, Htl could restrict Rho/Rac activity in a more static fashion to promote restricted Cdc42 accumulation at the myotube leading edges.
FGF and Rho/Rac signaling also regulate cellular elongation outside of myotube guidance. For example, FGF2 promotes mammary epithelial tube elongation in organoid cultures through Rac1- and MAPK-dependent mechanisms. In this system, Rac1 inhibition caused epithelial branches to collapse after FGF2-induced elongation, but Rac1 activation alone was insufficient to induce branch elongation (Huebner et al., 2016). Unlike myotubes, mammary epithelial tubes comprise dozens of cells, and pERK was enriched in cells at the branch tips, which suggests cells in the mammary epithelia have a differential response to exogenous FGF2.
Rac1 also has a well-characterized role in regulating the actin cytoskeleton during axon outgrowth and guidance (Lundquist, 2003; Luo et al., 1994), which more closely approximates myotube elongation and muscle attachment site selection as both systems involve the morphogenesis of a single cell. Consistent with our results, CA-Rac1 inhibits axon outgrowth whereas DN-Rac1 has little effect on neuron morphogenesis (Luo et al., 1994). FGF signaling can induce neurite outgrowth (Baum et al., 2016; Saffell et al., 1997; Williams et al., 1994), and Rac1 has been implicated in regulating neuron morphogenesis downstream of FGF2 (Park et al., 2007). Our study has drawn a number of parallels between myotube guidance and axon guidance and predicts that intracellular regulation of Rho/Rac activity may be a common mechanism by which FGF signals regulate cellular elongation and guidance.
A non-cell autonomous role for Htl
We were surprised that htl LO1 myotube guidance defects could not be rescued in embryos that expressed Htl under the control of slou.Gal4. One explanation is that slou.Gal4 is expressed at high levels in LO1 founder cells. It is possible that slou.Gal4 activated UAS.Htl at levels that are much higher than endogenous htl, which prevented a robust rescue of htl LO1 myotube guidance defects.
Alternatively, Htl could have both cell autonomous and non-cell autonomous functions during LO1 myotube guidance. For example, Htl could be required to induce substrate expression for LO1 myotube guidance. The substrates for myotube guidance have not been characterized in detail, but nascent myotubes are in close proximity to the ectoderm, to fusion competent myoblasts in the somatic mesoderm and to multiple cell types in the visceral mesoderm. Nascent LO1 myotubes are separated from the ectoderm by three lateral transverse myotubes (Fig. 3A), so the substrate for LO1 myotube guidance is likely expressed in mesodermal cells. In the visceral mesoderm, caudal visceral mesoderm (CVM) cells migrate along the trunk visceral mesoderm (TVM), and Htl performs distinct functions in each cell type. In the CVM, Htl transduces chemotactic FGF signals for directed migration, but in the TVM Htl directs integrin expression, which is the putative substrate for CVM migration (Macabenta and Stathopoulos, 2019). FGFs also regulate substrate expression in vertebrates. FGF2-induced substrate expression dramatically enhances axon regrowth across central nervous system injuries in mammals (Anderson et al., 2018). Our studies show the primary LO1 myotube leading edge travels a circuitous route (Movie 2), and one exciting possibility is that Htl-mediated FGF signals direct the expression of substrates that guide the LO1 leading edge to its muscle attachment site.
FGFs and muscle patterning
Ectopic Pyr or Ths expression in the salivary gland is sufficient to misdirect CVM migration in Df(2R)BSC25 embryos (hereafter pyr/ths embryos), which suggests either ligand can act as a long-range chemoattractant (Kadam et al., 2012). In contrast, myotubes were preferentially responsive to Pyr overexpression, which suggests that body wall muscles interpret FGF signals differently than visceral muscles (Fig. S2). As Pyr and Ths expression was not restricted to tendon cells (Fig. 4Q,R), our studies support a model in which broad FGF expression in the ectoderm acts at short range to direct myotube leading edges toward the correct muscle attachment sites. In either event, the mechanisms by which Pyr and Ths direct myotube guidance appear to differ from those that regulate CVM migration.
The muscle morphogenesis phenotypes we identified in htlAB42 embryos were present in pyr/ths embryos, but the expressivity of the myogenic phenotypes was more severe in pyr/ths embryos (e.g. Fig. 2I and Fig. 4J). Founder cell specification was also differentially affected in htlAB42 and pyr/ths embryos. For example, VT1 muscles were absent in both htlAB42 and pyr/ths embryos, whereas VA3 muscles were absent only in pyr/ths embryos (Fig. 3F and Fig. 4L). Interestingly, we did not observe any myogenic phenotypes in embryos mutant for a second FGF receptor Breathless (Fig. S5). Our studies argue that Pyr and Ths signal through Htl to regulate myotube guidance and to specify a subset of founder cells, but that Pyr and Ths may also signal through an Htl-independent mechanism to further direct myogenesis, which could include a previously uncharacterized FGF receptor, or combinatorial signaling with other guidance pathways such as Slit or Kon.
In summary, our study has identified a novel function for the FGF pathway during myogenesis, and has established a unique experimental framework to further investigate the discrete molecular mechanisms by which FGF signaling directs cellular guidance and the physical interactions between cells during organogenesis. Although FGF signaling is known to play an important role in promoting myoblast migration out of the somitic mesoderm in vertebrates, a function for FGF signaling during muscle morphogenesis has yet to be defined. Future studies of FGF signaling in both Drosophila and vertebrate systems will be broadly applicable toward understanding how cell shape changes are modulated by extracellular signaling pathways and may uncover much anticipated insights into how vertebrate skeletal muscles acquire spectacular shapes to complement a myriad of body plans.
MATERIALS AND METHODS
Drosophila genetics
The stocks used in this study include htlAB42, stumps09904b, Df(2R)BSC25, pbl3, P{UAS-htl}, P{UAS-htl.λ}, P{Gal4-tey5053A}, P{GMR40D04-GAL4}attP2 (slou.Gal4), P{GMR57C12-GAL4}attP2 (nau.Gal4), P{UAS-Rac1.V12}, P{UAS-Rac1.N17}, P{UAS-Lifeact-RFP}, P{UAS-Pak.RBD-GFP}30, P{UAS-eGFP} (Bloomington Drosophila Stock Center), P{Gal4-kirrerP298} (Nose et al., 1998), and P{UAS-pyr}, P{UAS-ths}, Df(2R)pyr36 and Df(2R)ths238 (Kadam et al., 2009). Cyo, P{Gal4-Twi}, P{2X-UAS.eGFP}, Cyo, P{wg.lacZ}, TM3, P{Gal4-Twi}, P{2X-UAS.eGFP}, and TM3, P{ftz.lacZ} balancers were used to genotype embryos.
FACS-seq
Approximately 200 mg of rp298.GAL4, UAS.eGFP embryos were collected 7-10 h AEL and dissociated as previously described (Bryantsev and Cripps, 2012). The cell suspension was incubated with Hoechst (1 µl/ml, Invitrogen) and sorted on a FACSAria cell sorter. Minimum fluorescent intensity gates were established for GFP and Hoechst by standard methods. GFP+, Hoechst+ cells were collected for the experimental population and GFP–, Hoechst+ cells were collected for the control population. Immediately after sorting, RNA was extracted with the RNeasy Mini kit (Qiagen). cDNA libraries were generated with the TruSeq stranded mRNA sample library kit (Illumina) and sequenced using 50 bp single-reads on the Illumina HiSeq 2000 system. Three biological replicates for each cell population were sequenced in parallel and reads were screened with a custom quality control program and mapped to the Drosophila genome with Genomic Short-Read Nucleotide Alignment Program (GSNAP) using the Cufflinks method.
Bioinformatic and statistical analysis
For RNA-seq, the number of fragments per kilobase of transcript per million mapped reads (FPKM) was calculated using principal component analysis (PCA) and the relative expression for each transcript. Differential open reading frame (ORF) transcription between experimental and control samples was identified by calculating fold changes (FC) in FPKMs and analysis of variance (ANOVA). Transcripts with FPKM values ≥25, FC ≥1.1 and a P-value ≤0.05 in the experimental versus control populations were considered enriched and analyzed with the Database for Annotation, Visualization and Integrated Discovery (DAVID) to cluster transcripts according to biological process (BP) and cellular component (CC). Statistical analysis of embryonic phenotypes was performed using GraphPad Prism 8 software, and significance was determined with the unpaired, one-tailed Student's t-test. Sample sizes are indicated in the figure legends. Data collection and data analysis were routinely performed by different authors to prevent potential bias. All individuals were included in data analysis.
Immunohistochemistry and in situ hybridization
Antibodies used include α-Mef2 (gift from R. Cripps, San Diego State University, CA, USA, 1:2000), α-Tropomyosin (Abcam, MAC141, 1:600), α-dpERK (Millipore Sigma, MAPK-YT, 1:200), α-GFP (Torrey Pines Biolabs, TP-401, 1:600) and α-βgal (Promega, Z3781, 1:100). HRP-conjugated secondary antibodies in conjunction with the TSA system (Molecular Probes, B40922) were used to detect primary antibodies, according to the manufacturer's specifications. Antibody staining was performed as previously described (Johnson et al., 2013).
We then performed in situ hybridization as previously described (Williams et al., 2015), except Dig-labeled probes were detected with HRP-conjugated α-Dig (Roche, 11207733910, 1:2000) in conjunction with the TSA system (Molecular Probes). Probe templates were generated by cloning PCR products from genomic DNA of P{UAS-htl}, P{UAS-pyr} and P{UAS-ths} flies into pBS.KS. Templates were validated by Sanger sequencing.
Imaging and image analysis
All images were generated with an LSM800 confocal microscope. For time-lapse imaging, dechorionated stage 12 embryos were mounted in halocarbon oil and scanned at 6 min intervals. For other live imaging, embryos were dechorionated, mounted in phosphate buffered saline with Tween 20 (PBT) and directly scanned. Control and mutant embryos were prepared and imaged in parallel where possible, and confocal imaging parameters were maintained between genotypes throughout this study. Fluorescence analysis and muscle morphology was analyzed with ImageJ software. Fluorescent values were calculated by dividing the mean myotube fluorescence (gray value) by the background fluorescence; fluorescent intensity was then normalized to the mean WT value.
Quantitative RT-PCR
Embryonic cells were collected by FACS, and RNA was extracted with the RNeasy Mini kit (Qiagen). cDNA was generated using Superscript IV (Life Technologies) and qPCR was performed with SYBR Select Master Mix using an ABI Prism 7000 (Life Technologies). qPCR reactions were run in triplicate and normalized to RpL32 expression.
Western blotting
Embryos were collected at 7-10 h AEL, dechorionated and suspended in lysis buffer [20 mM Tris (pH 7.5), 150 mM NaCl, 1% Triton-X and protease inhibitors]. Cells were lysed with a hand-held homogenizer, and large debris was removed by 10 min centrifugation (12,000 g). Protein quantification of the resulting lysates was performed using Qubit Fluorometric Quantitation (Life Technologies). Western blots were performed with α-mouse-AKT (Cell Signaling Technology, 9272, 1:5000) and α-Drosophila-pAKT (Cell Signaling Technology, 4054, 1:2000) as previously described (Mokalled et al., 2010), and imaged using the ChemiDoc XRS+ system (Bio-Rad).
Supplementary Material
Acknowledgements
We thank Helen McNeill and Mayssa Mokalled for critical reading of the manuscript, and Richard Cripps, Angelike Stathopoulos and the Drosophila community for stocks and reagents. We are also grateful to the reviewers for the insights and suggestions that improved our manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.N.J.; Methodology: A.N.J.; Formal analysis: S.Y., A.W., K.L.J., A.N.J.; Investigation: S.Y., A.W., Y.D., J.M.V., A.N.J.; Resources: A.N.J.; Data curation: S.Y., A.W., A.N.J.; Writing - original draft: A.N.J.; Writing - review & editing: S.Y., A.W., A.N.J.; Visualization: A.N.J.; Supervision: S.Y., A.N.J.; Project administration: A.N.J.; Funding acquisition: A.N.J.
Funding
A.N.J. was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIH R01AR070299), the University of Washington Musculoskeletal Research Center (NIH P30 AR074992) and a Boettcher Foundation Webb-Waring Biomedical Research Award. Deposited in PMC for release after 12 months.
Data availability
Normalized read counts for all mapped transcripts are available from the Dryad Digital Repository at https://doi.org/10.5061/dryad.j0zpc869m (Johnson, 2020). Raw RNA-seq data are no longer available.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.183624.supplemental
References
- Abreu-Blanco M. T., Verboon J. M. and Parkhurst S. M. (2014). Coordination of Rho family GTPase activities to orchestrate cytoskeleton responses during cell wound repair. Curr. Biol. 24, 144-155. 10.1016/j.cub.2013.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. A., O'Shea T. M., Burda J. E., Ao Y., Barlatey S. L., Bernstein A. M., Kim J. H., James N. D., Rogers A., Kato B. et al. (2018). Required growth facilitators propel axon regeneration across complete spinal cord injury. Nature 561, 396-400. 10.1038/s41586-018-0467-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate M. (1990). The embryonic development of larval muscles in Drosophila. Development 110, 791-804. [DOI] [PubMed] [Google Scholar]
- Baum P., Vogt M. A., Gass P., Unsicker K. and von Bohlen und Halbach O. (2016). FGF-2 deficiency causes dysregulation of Arhgef6 and downstream targets in the cerebral cortex accompanied by altered neurite outgrowth and dendritic spine morphology. Int. J. Dev. Neurosci. 50, 55-64. 10.1016/j.ijdevneu.2016.03.002 [DOI] [PubMed] [Google Scholar]
- Beiman M., Shilo B. Z. and Volk T. (1996). Heartless, a Drosophila FGF receptor homolog, is essential for cell migration and establishment of several mesodermal lineages. Genes Dev. 10, 2993-3002. 10.1101/gad.10.23.2993 [DOI] [PubMed] [Google Scholar]
- Bénazéraf B., Francois P., Baker R. E., Denans N., Little C. D. and Pourquié O. (2010). A random cell motility gradient downstream of FGF controls elongation of an amniote embryo. Nature 466, 248-252. 10.1038/nature09151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryantsev A. L. and Cripps R. M. (2012). Purification of cardiac cells from Drosophila embryos. Methods 56, 44-49. 10.1016/j.ymeth.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustelo X. R., Sauzeau V. and Berenjeno I. M. (2007). GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays 29, 356-370. 10.1002/bies.20558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena A., Gisselbrecht S., Harrison J., Jimenez F. and Michelson A. M. (1998). Combinatorial signaling codes for the progressive determination of cell fates in the Drosophila embryonic mesoderm. Genes Dev. 12, 3910-3922. 10.1101/gad.12.24.3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fera E., O'Neil C., Lee W., Li S. and Pickering J. G. (2004). Fibroblast growth factor-2 and remodeled type I collagen control membrane protrusion in human vascular smooth muscle cells: biphasic activation of Rac1. J. Biol. Chem. 279, 35573-35582. 10.1074/jbc.M400711200 [DOI] [PubMed] [Google Scholar]
- Gabay L., Seger R. and Shilo B. Z. (1997). MAP kinase in situ activation atlas during Drosophila embryogenesis. Development 124, 3535-3541. [DOI] [PubMed] [Google Scholar]
- Gisselbrecht S., Skeath J. B., Doe C. Q. and Michelson A. M. (1996). heartless encodes a fibroblast growth factor receptor (DFR1/DFGF-R2) involved in the directional migration of early mesodermal cells in the Drosophila embryo. Genes Dev. 10, 3003-3017. 10.1101/gad.10.23.3003 [DOI] [PubMed] [Google Scholar]
- Gros J., Serralbo O. and Marcelle C. (2009). WNT11 acts as a directional cue to organize the elongation of early muscle fibres. Nature 457, 589-593. 10.1038/nature07564 [DOI] [PubMed] [Google Scholar]
- Guerin C. M. and Kramer S. G. (2009). RacGAP50C directs perinuclear gamma-tubulin localization to organize the uniform microtubule array required for Drosophila myotube extension. Development 136, 1411-1421. 10.1242/dev.031823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding M. J. and Nechiporuk A. V. (2012). Fgfr-Ras-MAPK signaling is required for apical constriction via apical positioning of Rho-associated kinase during mechanosensory organ formation. Development 139, 3130-3135. 10.1242/dev.082271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner R. J., Neumann N. M. and Ewald A. J. (2016). Mammary epithelial tubes elongate through MAPK-dependent coordination of cell migration. Development 143, 983-993. 10.1242/dev.127944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong W., Lee J., Bazer F. W., Song G. and Kim J. (2016). Fibroblast growth factor 4-induced migration of porcine trophectoderm cells is mediated via the AKT cell signaling pathway. Mol. Cell. Endocrinol. 419, 208-216. 10.1016/j.mce.2015.10.020 [DOI] [PubMed] [Google Scholar]
- Johnson A. (2020). Transcript expression in embryonic myoblasts. Dryad Digital Repository. 10.5061/dryad.j0zpc869m [DOI] [Google Scholar]
- Johnson A. N., Mokalled M. H., Valera J. M., Poss K. D. and Olson E. N. (2013). Post-transcriptional regulation of myotube elongation and myogenesis by Hoi Polloi. Development 140, 3645-3656. 10.1242/dev.095596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S., McMahon A., Tzou P. and Stathopoulos A. (2009). FGF ligands in Drosophila have distinct activities required to support cell migration and differentiation. Development 136, 739-747. 10.1242/dev.027904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S., Ghosh S. and Stathopoulos A. (2012). Synchronous and symmetric migration of Drosophila caudal visceral mesoderm cells requires dual input by two FGF ligands. Development 139, 699-708. 10.1242/dev.068791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S. G., Kidd T., Simpson J. H. and Goodman C. S. (2001). Switching repulsion to attraction: changing responses to slit during transition in mesoderm migration. Science 292, 737-740. 10.1126/science.1058766 [DOI] [PubMed] [Google Scholar]
- Lundquist E. A. (2003). Rac proteins and the control of axon development. Curr. Opin. Neurobiol. 13, 384-390. 10.1016/S0959-4388(03)00071-0 [DOI] [PubMed] [Google Scholar]
- Luo L., Liao Y. J., Jan L. Y. and Jan Y. N. (1994). Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8, 1787-1802. 10.1101/gad.8.15.1787 [DOI] [PubMed] [Google Scholar]
- Maartens A. P. and Brown N. H. (2015). The many faces of cell adhesion during Drosophila muscle development. Dev. Biol. 401, 62-74. 10.1016/j.ydbio.2014.12.038 [DOI] [PubMed] [Google Scholar]
- Macabenta F. and Stathopoulos A. (2019). Migrating cells control morphogenesis of substratum serving as track to promote directional movement of the collective. Development 146, dev177295 10.1242/dev.177295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G. L., Hahn K. M. and Danuser G. (2009). Coordination of Rho GTPase activities during cell protrusion. Nature 461, 99-103. 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson A. M., Gisselbrecht S., Zhou Y., Baek K.-H. and Buff E. M. (1998). Dual functions of the heartless fibroblast growth factor receptor in development of the Drosophila embryonic mesoderm. Dev. Genet. 22, 212-229. [DOI] [PubMed] [Google Scholar]
- Mokalled M. H., Johnson A., Kim Y., Oh J. and Olson E. N. (2010). Myocardin-related transcription factors regulate the Cdk5/Pctaire1 kinase cascade to control neurite outgrowth, neuronal migration and brain development. Development 137, 2365-2374. 10.1242/dev.047605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muha V. and Müller H.-A. (2013). Functions and mechanisms of Fibroblast Growth Factor (FGF) signalling in Drosophila melanogaster. Int. J. Mol. Sci. 14, 5920-5937. 10.3390/ijms14035920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nose A., Isshiki T. and Takeichi M. (1998). Regional specification of muscle progenitors in Drosophila: the role of the msh homeobox gene. Development 125, 215-223. [DOI] [PubMed] [Google Scholar]
- Ornitz D. M. and Itoh N. (2015). The fibroblast growth factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 4, 215-266. 10.1002/wdev.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. B., Kim E. J., Yang E. J., Seo S. R. and Chung K. C. (2007). JNK- and Rac1-dependent induction of immediate early gene pip92 suppresses neuronal differentiation. J. Neurochem. 100, 555-566. 10.1111/j.1471-4159.2006.04263.x [DOI] [PubMed] [Google Scholar]
- Saffell J. L., Williams E. J., Mason I. J., Walsh F. S. and Doherty P. (1997). Expression of a dominant negative FGF receptor inhibits axonal growth and FGF receptor phosphorylation stimulated by CAMs. Neuron 18, 231-242. 10.1016/S0896-6273(00)80264-0 [DOI] [PubMed] [Google Scholar]
- Sato A., Scholl A. M., Kuhn E. B., Stadt H. A., Decker J. R., Pegram K., Hutson M. R. and Kirby M. L. (2011). FGF8 signaling is chemotactic for cardiac neural crest cells. Dev. Biol. 354, 18-30. 10.1016/j.ydbio.2011.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer F., Kalchhauser I. and Dickson B. J. (2007). The transmembrane protein Kon-tiki couples to Dgrip to mediate myotube targeting in Drosophila. Dev. Cell 12, 751-766. 10.1016/j.devcel.2007.02.017 [DOI] [PubMed] [Google Scholar]
- Schumacher S., Gryzik T., Tannebaum S. and Müller H.-A. J. (2004). The RhoGEF pebble is required for cell shape changes during cell migration triggered by the Drosophila FGF receptor heartless. Development 131, 2631-2640. 10.1242/dev.01149 [DOI] [PubMed] [Google Scholar]
- van Impel A., Schumacher S., Draga M., Herz H.-M., Großhans J. and Müller H. A. J. (2009). Regulation of the Rac GTPase pathway by the multifunctional Rho GEF Pebble is essential for mesoderm migration in the Drosophila gastrula. Development 136, 813-822. 10.1242/dev.026203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E. J., Furness J., Walsh F. S. and Doherty P. (1994). Activation of the FGF receptor underlies neurite outgrowth stimulated by L1, N-CAM, and N-cadherin. Neuron 13, 583-594. 10.1016/0896-6273(94)90027-2 [DOI] [PubMed] [Google Scholar]
- Williams J., Boin N. G., Valera J. M. and Johnson A. N. (2015). Noncanonical roles for Tropomyosin during myogenesis. Development 142, 3440-3452. 10.1242/dev.117051 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.