ABSTRACT
In endolysosomal networks, two hetero-hexameric tethers called HOPS and CORVET are found widely throughout eukaryotes. The unicellular ciliate Tetrahymena thermophila possesses elaborate endolysosomal structures, but curiously both it and related protozoa lack the HOPS tether and several other trafficking proteins, while retaining the related CORVET complex. Here, we show that Tetrahymena encodes multiple paralogs of most CORVET subunits, which assemble into six distinct complexes. Each complex has a unique subunit composition and, significantly, shows unique localization, indicating participation in distinct pathways. One pair of complexes differ by a single subunit (Vps8), but have late endosomal versus recycling endosome locations. While Vps8 subunits are thus prime determinants for targeting and functional specificity, determinants exist on all subunits except Vps11. This unprecedented expansion and diversification of CORVET provides a potent example of tether flexibility, and illustrates how ‘backfilling’ following secondary losses of trafficking genes can provide a mechanism for evolution of new pathways.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: CORVET, Ciliate, Cryomilling, Endosomal trafficking, Evolutionary cell biology, Mass spectrometry, Paralogous expansion, Backfilling
Summary: Analysis in Tetrahymena thermophila reveals remarkable expansion of oligomeric protein complexes that organize the interactions between endosomes, lysosomes and related compartments in ciliates.
INTRODUCTION
Eukaryotic cells contain multiple membrane-bound compartments, with either an endogenous or endosymbiotic origin. Bidirectional transport between compartments is critical for cell function and defects contribute towards disorders including neurodegeneration and cancer (Mellman and Yarden, 2013; Neefjes and van der Kant, 2014). The endolysosomal network refers to a subset of pathways linking endocytic trafficking with degradative and secretory compartments (Huotari and Helenius, 2011). To ensure accurate trafficking, cells deploy intricate mechanisms to ‘license’ interactions between compartments that ultimately allow content mixing via membrane fusion (Kümmel and Ungermann, 2014).
Key determinants for ensuring productive membrane interactions are SNARE proteins, with distinct paralogs present at each compartment (Gerst, 1999). SNARE complex assembly drives formation of a fusion pore, together with complexes called tethers that act upstream of SNAREs (Baker and Hughson, 2016). The homotypic-fusion-and-protein-sorting (HOPS) and class-C-core-vacuole/endosome-tethering (CORVET) complexes are cytoplasmic hetero-hexamers that bridge compartments by binding Rab GTPases at two membranes, and subsequently chaperoning SNARE assembly (Baker and Hughson, 2016; Horazdovsky et al., 1996; Nickerson et al., 2009; Spang, 2016; van der Beek et al., 2019). These reactions have been reconstituted in vitro, but comparably detailed in vivo characterization remains lacking, particularly for CORVET (Ho and Stroupe, 2016; Lobingier and Merz, 2012; Lobingier et al., 2014; Orr et al., 2017; Schwartz et al., 2017). CORVET and HOPS mediate endosome maturation through interaction with Rab5 and Rab7 (Vps21 and Ypt7 in yeast), respectively, promoting fusion of early endosomes (EEs) with multivesicular late endosomes (LEs) and subsequently vacuoles/lysosomes (LLs) (Balderhaar and Ungermann, 2013). HOPS and CORVET share four core subunits: Vps11 (Vps stands for vacuolar protein sorting), Vps16, Vps18 and Vps33 (Nickerson et al., 2009). In addition, each complex contains two specific subunits: Vps3 and Vps8 in CORVET, and Vps39 and Vps41 in HOPS (Peplowska et al., 2007). CORVET may convert into HOPS during endosome maturation by exchanging complex-specific subunits (Ostrowicz et al., 2010; Peplowska et al., 2007), an attractive model since complex-specific subunits bind Rab proteins, and thus are key specificity determinants (Markgraf et al., 2009). Intriguingly, hybrid CORVET–HOPS complexes have been identified in S. cerevisiae (Ostrowicz et al., 2010; Peplowska et al., 2007), but have only been detected under overexpression conditions.
While budding yeast has single genes encoding CORVET and HOPS subunits, the genetic and cell biological landscapes have additional dimensions in metazoa. Two paralogs of VPS33 are present in vertebrates, Drosophila and C. elegans (Gissen et al., 2005); similarly, two VPS16 paralogs are present in mammals and flies. In C. elegans, the two Vps33 paralogs are components of HOPS and CORVET complexes, respectively (Solinger and Spang, 2014), while in mammals and flies the pairs of Vps33 and Vps16 paralogs belong to specific complexes. In mammals, Vps16A and Vps33A belong to HOPS and CORVET, while Vps16B and Vps33B form a distinct complex, the class C homologues in endosome–vesicle interaction (CHEVI) complex (Spang, 2016). CHEVI functions in the biogenesis of α-granules and lamellar bodies, which are mammalian platelet-specific and keratinocyte-specific lysosome-related organelles (LROs) (Bem et al., 2015; Dai et al., 2016; Lo et al., 2005; Rogerson and Gissen, 2018). Similarly, Drosophila Vps16B and Vps33B form a novel complex (Cullinane et al., 2010; Gissen et al., 2004; Hunter et al., 2018; Pulipparacharuvil et al., 2005; Tornieri et al., 2013). Furthermore, HOPS and CORVET subunits may function in stable subcomplexes. A CORVET-independent Vps3–Vps8 subcomplex in HeLa cells functions in recycling β1 integrins (Jonker et al., 2018). In Drosophila, a miniCORVET complex consisting of Vps8, Vps16, Vps18 and Vps33 exists (Lorincz et al., 2016), while mammalian Vps41, but not other HOPS subunits, functions in sorting to secretory granules (Asensio et al., 2013). These studies reveal remarkable flexibility in HOPS and CORVET subunits for a wide range of functions. Nonetheless, virtually all detailed studies have been pursued in a single eukaryotic lineage, the Opisthokonts, which includes both fungi and animals (Lynch et al., 2014). Hence, the full diversity of HOPS/CORVET structure and function remains unexplored. Interestingly, recent studies in Archaeplastida (plants) suggests coupling between CORVET and HOPS may be evolutionarily plastic (Takemoto et al., 2018).
Ciliates are distantly related to both Opisthokonts and Archaeplastids. Together, with dinoflagellates and apicomplexans, ciliates constitute the Alveolate branch of the Stramenopiles–Alveolata–Rhizaria (SAR) supergroup (Adl et al., 2012). The estimated 30,000 morphologically diverse mostly free-swimming species (Adl et al., 2007) contribute to freshwater, marine and terrestrial ecosystems (Gimmler et al., 2016; Warren et al., 2017; Weisse, 2017; Zingel et al., 2019). Ciliates exhibit striking morphological and behavioral complexity for single-celled organisms, and ciliate genomes encode correspondingly large numbers of genes (Hausmann, 1996; Wang et al., 2017). For example, Tetrahymena thermophila expresses roughly the same number of Rab GTPases as in humans, hinting at the diversity of membrane trafficking pathways (Bright et al., 2010; Saito-Nakano et al., 2010), including an elaborate endolysosomal network (Guerrier et al., 2017). Morphological studies suggest at least four pathways for uptake, including clathrin-mediated endocytosis (Elde et al., 2005; Nilsson and Van Deurs, 1983). Phagocytosis begins with phagosome formation at an anterior portal called the oral apparatus, followed by phagolysosome maturation via fusion with multiple classes of endosomes (Jacobs et al., 2006; Nilsson, 1979; Plattner, 2010) that deliver hydrolytic enzymes required to digest the phagolysosome contents. Maturing phagolysosomes are eventually transported to the cell posterior where they release undigested contents via exocytosis (Frankel, 2000). Autophagy-related pathways manifest during conjugation when selected nuclei are eliminated (Akematsu et al., 2014; Davis et al., 1992; Liu and Yao, 2012; Orias et al., 2011). The water-pumping contractile vacuoles are also endolysosomal (Allen, 2000; Plattner, 2015; Bright et al., 2010). Finally, prominent secretory vesicles called mucocysts are LROs (Briguglio et al., 2013; Kaur et al., 2017).
Tetrahymena and related ciliates in the Oligohymenophorea lineage possess an atypical complement of genes encoding endolysosomal tethers and other trafficking genes, indicating a significant bottleneck in ancestors of this lineage that resulted in gene losses (Sparvoli et al., 2018). Most relevant here is that both HOPS-specific subunits were lost, but most other subunits (i.e. the core subunits, as well as the CORVET-specific subunits) were retained and expanded into multiple paralogs (Klinger et al., 2013; Sparvoli et al., 2018). T. thermophila expresses two Vps33, two Vps16, two Vps3, four Vps18 and six Vps8 paralogs. We discovered that the ‘a’ paralog of Vps8 is essential for mucocyst formation (Sparvoli et al., 2018), and our results suggested that a specialized Vps8a-containing tether is used in this pathway. Together with genomic data, these results suggest that expansion of pathway-specific CORVET-related tethers in oligohymenophorean ciliates accompanied elaboration of a complex endolysosomal network. To encapsulate this idea, we suggest the term ‘backfilling’, whereby the retained components expand to occupy a gap in functionality arising from prior gene loss.
Our previous data did not demonstrate that Vps8a belonged to a CORVET complex and if so, whether the six Vps8 paralogs define distinct complexes. Here, we show that Tetrahymena assembles six hexameric CORVET complexes, and that each consists of a unique combination of subunit paralogs. The only subunit for which a single protein contributes to all complexes is Vps11, a key integrator of HOPS/CORVET assembly in other organisms (Plemel et al., 2011). The six Tetrahymena CORVET complexes function at six distinct cellular locations. Vps8 is a key specificity determinant, since complexes differing only in that subunit show differential localization.
RESULTS
Vps8a defines a specific hexameric CORVET complex in Tetrahymena
To ask whether Vps8a associates with subunits forming a classical CORVET complex, we characterized Vps8a-binding proteins. We set out to determine whether Vps8a associated with canonical CORVET subunits by co-expressing FLAG-tagged Vps8a with 6Myc-tagged versions of core subunits. We then precipitated Vps8a and evaluated co-precipitation of the 6Myc-tagged proteins. To avoid non-specific interactions due to overexpression, we tagged all subunits by integrating 3′ epitope tags at the endogenous loci.
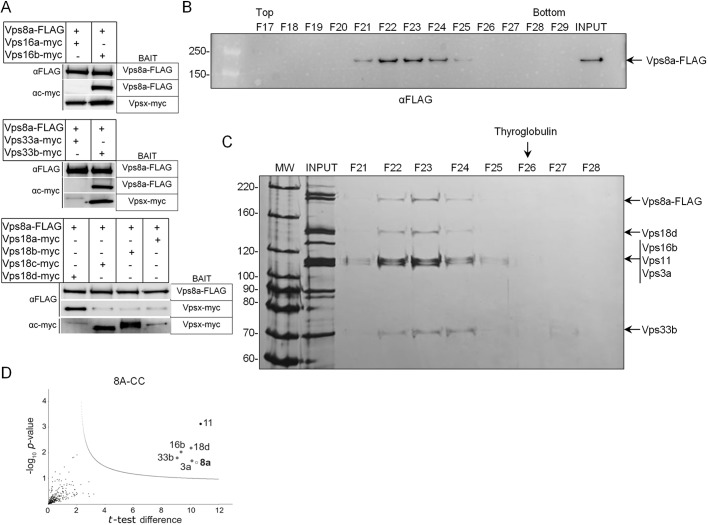
Prior annotation of CORVET subunits encoded by T. thermophila revealed multiple paralogs, including two for VPS16, four for VPS18 and two for VPS33 (Klinger et al., 2013). Single genes were reported for VPS11 and VPS3, although a second VPS3 gene is in fact present (see below). To begin, we asked whether Vps8a associated with Vps16, Vps18 or Vps 33. We tagged all paralogs for each subunit in pairwise combination with Vps8a–FLAG. The results showed that Vps16b, Vps33b and Vps18d, but not the other paralogs of each subunit, could be robustly co-precipitated with Vps8a (Fig. 1A).
Fig. 1.
Vps8a associates with five other subunits in a hexameric CORVET complex. (A) Co-immunoprecipitation of Vps8a–FLAG with Myc-tagged Vps16b, Vps18d and Vps33b subunits. Cells were transformed to endogenously express Vps8a–FLAG in pairwise combination with Myc-tagged Vps16a or Vps16b, Vps33a or Vps33b, and Vps18a, Vps18b, Vps18c or Vps18d. Cell lysates were split and incubated with anti-c-Myc or anti-FLAG beads. SDS-PAGE samples were immunoblotted with anti-c-Myc and anti-FLAG antibodies. (B,C) Sedimentation analysis of 8A-CC. (B) Vps8a–FLAG and associated proteins were immuno-isolated using anti-FLAG beads. Eluted proteins were sedimented in glycerol gradients, and 250 µl fractions were harvested from top to bottom, of which 25 µl aliquots were subjected to SDS-PAGE (4–20% gel). Fractions (F)17–29 (top–bottom) are shown. Vps8a was detected by western blotting with anti-FLAG antibodies. ‘INPUT’ corresponds to 1% of total eluate. F4–F16 (top–bottom) were similarly analyzed (see Fig. S1C). (C) 4% of the total eluate (INPUT) and 35 µl aliquots of gradient fractions (F21 to F28) were subjected to SDS-PAGE (8% gel) and visualized by silver staining. The Vps8a subunit, as well as bands of the expected sizes for five additional CORVET subunits, are identified on the right. Thyroglobulin, sedimented in parallel as a size standard, appeared in F24–F29 (top-bottom), with a peak in F26 (arrow at the top). 8A-CC sediments more slowly than expected for a 727 kDa complex. (D) Mass spectrometric identification of proteins co-isolated with Vps8a. Cryopowders (150 g) from wild-type and Vps8a–FLAG-expressing cells were solubilized and treated as in B, except bound proteins were eluted with LDS sample buffer. The total eluates were prepared for mass spectrometric analysis. On volcano plots, such as the one shown here, proteins falling above the threshold line are considered significant. To generate the plot, the −log10 t-test P-value was plotted versus the t-test difference (difference between means). The cut-off curve is based on the false discovery rate (FDR=0.01) and the artificial factor s0 (s0=1), controlling the relative importance of the t-test P-value and difference between means. The open circle marks the Vps8a subunit used as bait, while the black circle marks the unique Vps11 subunit. The light gray circles indicate specific paralogs of the other four CORVET subunits. Each sample was prepared in duplicate.
Our results suggest that Vps8a belongs to a canonical hexameric complex, but smaller sub-complexes may also exist as described above. To examine this possibility, we immunoisolated the Vps8a-containing CORVET complex by affinity capture using Vps8a–FLAG as bait. To overcome any issues from low expression levels, we adapted methods used in trypanosomes (Obado et al., 2016), yeast (Oeffinger et al., 2007) and mammalian cells (LaCava et al., 2016), where large numbers of cells are rapidly frozen and milled to generate cryopowders. We tested a variety of buffer conditions for solubilizing Vps8a from the cryopowders and final conditions resulted in ∼60% solubilization (data not shown). Vps8a and associated proteins were immunoisolated from cryopowder solutes and eluted with excess FLAG peptide, followed by glycerol gradient centrifugation. Thyroglobulin, yeast alcohol dehydrogenase and bovine serum albumin were sedimented in parallel to provide size standards (Fig. S1A). We identified the fractions containing Vps8a by western blotting (Fig. 1B), and visualized the co-sedimenting proteins in those fractions by silver staining (Fig. 1C). Notably peak fractions 22 and 23 contained prominent bands at the expected sizes for Vps16b, Vps33b and Vps18d, and also for Vps11 and Vps3a, suggesting that a full hexameric complex was present. However, as Vps16b, Vps3a and Vps11 are all between 100–120 kDa, they are difficult to resolve (Fig. 1C). Although the predicted size for the Vps8a-containing CORVET complex (8A-CC) is 727 kDa, and all six subunits are visible by silver staining, the size of the peak glycerol gradient fraction corresponds to ∼550 kDa (Fig. S1B). Importantly, no Vps8a was detected in lower fractions (Fig. S1C), suggesting that this subunit is largely associated with a stable hexameric complex.
To confirm our interpretation of the glycerol gradient centrifugation and to identify the specific gene products, we immunoisolated Vps8a and associated proteins and analyzed the eluate by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Vps8a-associated proteins prominently include Vps16b, Vps33b, Vps18d, Vps3a and Vps11, with no other paralogs detected (Fig. 1D). Taken together, our results indicate the Vps8a belongs to a hexameric CORVET complex, which we refer to as 8A-CC. However, we note that the non-soluble fraction of Vps8a could potentially participate in a different biochemical complex.
Vps8b–Vps8f associate with distinct subunit combinations and Tetrahymena possesses six distinct CORVET complexes
Vps8a is one of six Vps8 paralogs expressed in Tetrahymena. These paralogs are ancient in origin: the split between even the most-closely related pair (VPS8A and VPS8C) predates the ∼22 million years ago (MYA) divergence between T. thermophila and T. malaccensis (Sparvoli et al., 2018). Moreover, these paralogs have been maintained in multiple species suggesting that they provide important, non-redundant functions. The six VPS8 paralogs in T. thermophila differ in their transcriptional profiles, consistent with functional diversification (Sparvoli et al., 2018). To determine whether each Vps8 paralog belongs to a unique biochemical complex, we expressed each as an endogenously FLAG-tagged fusion. The full-length constructs were confirmed by immunoprecipitation, followed by SDS-PAGE and western blotting (Fig. S2A). We then used cryomilling and immunoisolation as above, followed by SDS-PAGE. In silver-stained polyacrylamide gels of the eluted complexes, we detected multiple bands in the size range expected for CORVET subunits (Fig. S2B), but many bands had distinct migrations from the 8A-CC pulldown bands (Fig. 1C).
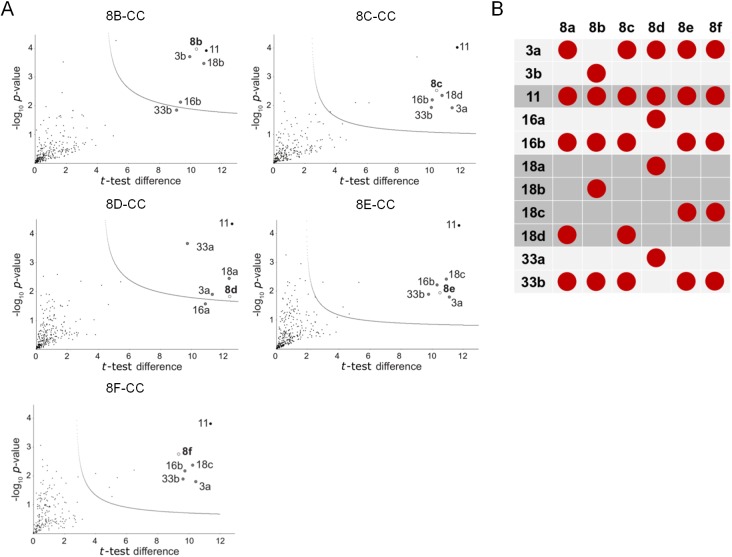
We analyzed each of these immunoisolated mixtures by LC-MS/MS (Fig. 2A). For each Vps8 paralog, the most enriched proteins consisted of the five canonical CORVET subunits, as expected for hexameric complexes. In all cases, a single paralog for each subunit was identified, echoing our findings for 8A-CC. Based on this, the composition of these complexes, which we call 8B-CC, 8C-CC, etc., can be predicted with confidence (Fig. 2B; Table S1). As a cohort, the CORVET complexes share just one gene product, encoded by VPS11. In yeast CORVET and HOPS complexes, Vps11 has a key role in complex assembly (Ostrowicz et al., 2010; Plemel et al., 2011). Furthermore, previous informatics-based analysis suggested that all CORVET complexes contained the identical Vps3 subunit, as only one VPS3 gene was identified (Klinger et al., 2013). However, the LC-MS/MS revealed that 8B-CC contains a distinct and divergent Vps3 paralog.
Fig. 2.
Tetrahymena has six unique CORVET complexes. (A) Volcano plots of mass spectrometry results, identifying the CORVET subunit paralogs associated with FLAG-tagged Vps8b, Vps8c, Vps8d, Vps8e and Vps8f. All samples were prepared as in Fig. 1D, in duplicate. To generate the volcano plot, the −log10 t-test P-value was plotted versus the t-test difference (difference between means). The cut-off curve is based on the false discovery rate (FDR=0.01) and the artificial factor s0 (s0=1), controlling the relative importance of the t-test P-value and difference between means. Significant hits are shown above the threshold line in each plot. (B) Diagram showing the comprehensive composition of CORVET complexes in T. thermophila. Each of the six Vps8 paralogs (top row) is associated with five other subunits, whose identities are indicated in the left column. 8A-CC and 8C-CC share five subunits, as do 8E-CC and 8F-CC. 8B-CC possesses unique Vps8, Vps3 and Vps18 subunit paralogs. 8D-CC possesses unique paralogs of the Vps8, Vps16, Vps18 and Vps33 subunits.
Two pairs of complexes are closely related in subunit composition; 8A-CC and 8C-CC are identical except for the Vps8, with the same relationship existing between 8E-CC and 8F-CC. By contrast 8B-CC and 8D-CC each show an exclusive combination of subunits. Interestingly, the composition of the six CORVET complexes appears largely consistent with evolutionary relationships previously inferred (Sparvoli et al., 2018), in that more closely related Vps8 paralogs can now be seen to belong to complexes that share a larger number of identical subunit paralogs.
The six Tetrahymena Vps8 paralogs have distinct locations
To understand how the six biochemically distinct CORVET tethers contribute to cellular function, we first asked whether they associate with different compartments. We generated cell lines where mNeon was integrated immediately downstream of each VPS8 open reading frame, to express endogenous levels of tagged protein. These integrated constructs were all driven to fixation, to completely replace the wild-type alleles in the somatic macronuclei. Fusion proteins of the expected sizes were detected by immunoprecipitation followed by SDS-PAGE and western blotting, although proteolytic cleavage of some products was also seen, as discussed further below (Fig. S3A). VPS8C and VPS8D are known to be essential genes (Sparvoli et al., 2018). Since the cells relying on mNeon-tagged Vps8c and Vps8d had no detectible growth phenotypes, which was also true for those expressing mNeon-tagged Vps8b, Vps8e and Vps8f, we infer that tagging does not interfere with protein activity. Consistent with this, Vps8a–mNeon is fully functional (Sparvoli et al., 2018).
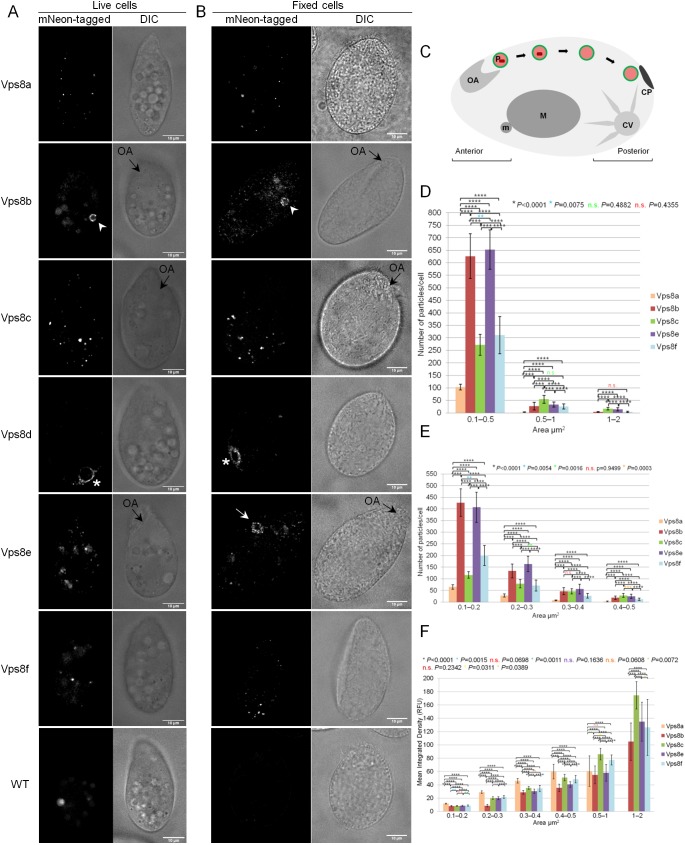
We analyzed the localization of mNeon-tagged Vps8 paralogs under a variety of conditions. First, cells were transferred for 2 h to a medium that reduces the auto-fluorescence in food vacuoles, and then immobilized in agarose dissolved in a Tris buffer (Fig. 3A). In other experiments, cells in standard growth medium were fixed for imaging (Fig. 3B). All six Vps8 paralogs are expressed at low levels (TetraFGD; http://tfgd.ihb.ac.cn) (Xiong et al., 2011a, 2013), so that detecting the mNeon-fusions in whole-cell lysates by western blotting required that they first be concentrated by immunoprecipitation. In our imaging studies, we reproducibly observed stronger fluorescent signals from Vps8a, Vps8c and Vps8d, compared to Vps8b, Vps8e and Vps8f, particularly in live-cell imaging. The weak fluorescence for Vps8e correlates with its apparent partial proteolytic cleavage (Fig. S3A, sixth lane), although we cannot rule out the possibility that proteolytic cleavage occurred during immunoprecipitation, notwithstanding the addition of protease inhibitors during cell lysis. Overall, this microscopy revealed non-identical patterns of fluorescent puncta for five of the six paralogs, and this was consistent with measured differences in the number, size and relative brightness of puncta (Fig. 3D,E,F). With regard to the apparent sizes of the structures associated with the individual paralogs, it is important to note that the differences might reflect variation in the concentration of Vps8 proteins on their target membranes – and hence different intensities of fluorescence – rather than actual size differences between the structures on which they reside.
Fig. 3.
Each Vps8 paralog localizes to distinct cellular compartments. (A) Live-cell imaging of mNeon-tagged Vps8 paralogs revealing their primary distributions. Vps8a localizes to small vesicles. Vps8b localizes to vesicles at the periphery of phagolysosomes (white arrowhead). Vps8c localizes to larger vesicles than those associated with Vps8a (see quantification in D,E), and to tubulovesicular compartments (see Movie 2), both more concentrated in the posterior half of cells (Fig. S3B). Vps8d localizes to the contractile vacuole (asterisk). Vps8e localizes to uniformly small vesicles dispersed in the cytoplasm. Vps8f localizes to few cytoplasmic puncta. Shown for wild-type (WT), Vps8a, Vps8b, Vps8c, Vps8e and Vps8f are single frames from time-lapse videos, with paired differential interference contrast (DIC) images. For Vps8d, a single confocal image was selected from a z-stack. All cells were incubated in S medium for 2 h prior to imaging. For additional details see supplemental videos. (B) Confocal sections of fixed cells expressing mNeon-tagged Vps8 paralogs with paired DIC images. Protein localization in fixed cells showed some differences from that in live cells, as follows. Vps8b–mNeon was associated with phagolysosomes (white arrowhead) but also with dispersed vesicles. Vps8e localized to vesicles at the cell anterior (white arrow), close to the oral apparatus (OA, black arrow in the DIC image). Scale bars: 10 μm. For additional images see Fig. S3D. (C) Tetrahymena cell cartoon. The phagocytic pathway is shown in the upper left to right of the diagram. Food particles (red rectangle) are taken up at the anterior oral apparatus (OA) into food vacuoles/phagosomes (P), from whence they move posteriorly and eventually egest undigested material at the cytoproct (CP). Also in the posterior is a water-pumping compartment, the contractile vacuole (CV). Also shown are the polyploid vegetative macronucleus (M), and diploid germline micronucleus (m). Cell length is 50 μm. (D) Estimation of the number of Vps8-labeled particles with size between 0.1 and 2 μm2, in cells individually expressing mNeon-tagged Vps8a, Vps8b, Vps8c, Vps8e and Vps8f. For all paralogs, most particles are 0.1–0.5 μm2, with fewer particles in the 0.5–1 μm2 and 1–2 μm2 size classes. The analysis was performed on 143, 155, 144, 148 and 143 non-overlapping optical sections for Vps8a, Vps8b, Vps8c, Vps8e and Vps8f, respectively. The reported values represent mean±s.d. number of particles per cell. P-values shown on the figures for each combination of Vps8 paralogs in each size class, were determined by two-tailed t-test. (E) Analysis of the number and size of the Vps8-labeled particles observed in D, limited to those belonging to the 0.1–0.5 μm2 size class. The size distributions of fluorescent puncta for Vps8a, Vps8c and Vps8f were distinct from one another, while those for Vps8b and Vps8e were more similar to one another. (F) Analysis of the brightness of particles in D and E, measuring the integrated density (RFU, relative fluorescence units) for each size class. For each paralog, particle brightness increases with size. Among the smallest size class, Vps8a-labeled particles appear brightest, while Vps8C-labeled particles are brightest in the 0.5–2 μm2 class, which correspond to the larger structures observed in A and B. Vps8b particles are the least bright in all size classes. The analysis was performed as in D.
While fully delineating the localization of these Vps8 paralogs requires additional compartmental markers for Tetrahymena to be developed, we draw some important conclusions. Vps8a–mNeon associates with small heterogenous puncta (Fig. 3A,B, first panels; Movie 1) that correspond to transport vesicles, since they also contain the Sor4 receptor (Sparvoli et al., 2018). The most closely related paralog, Vps8c, also localizes to cytoplasmic puncta, but these are larger and more irregular. In addition, the Vps8c-labeled structures frequently show dynamic tubular extensions (Fig. 3A,B, third panels; see also Movie 2), which are absent or less apparent for the other Vps8 paralog-labeled structures. At least one such tubulovesicular structure was seen in every cell, and they are more frequently located toward the posterior (Fig. S3B). This posterior bias was more obvious in fixed cells, probably because compression of cells under the coverslip reveals more structures in any given focal plane (Fig. 3B, third panel).
Vps8b–mNeon fluorescence appears in both live and fixed cells, as a tightly spaced array of puncta at the periphery of large circular structures. Their size and shape are sufficient to mark them as probable food vacuoles (Fig. 3A,B, second row; Movie 3), since there are no other similarly sized organelles in Tetrahymena. In addition, a substantial fraction of Vps8b localizes to isolated small cytoplasmic puncta visible in fixed cells, and, to a lesser extent, in favorable focal planes of live cells (Fig. S3C). Vps8d–mNeon fluorescence was strikingly concentrated at the contractile vacuole, a tubulovesicular organelle localized toward the cell posterior that functions in osmoregulation (Fig. 3A,B, forth row; Movie 4). Vps8e–mNeon fluorescence in live cells appears in numerous small puncta throughout the cell cytoplasm (Fig. 3A, fifth row; Movie 5). However, in some fixed cells, puncta were concentrated around a single circular structure near the cell anterior, close to the oral apparatus where food vacuoles are formed (Fig. 3B, fifth row). Finally, Vps8f–mNeon puncta, although almost undetectable in live cells (Fig. 3A, sixth row; Movie 6), are clearly visible in fixed cells (Fig. 3B, sixth row). These puncta are highly heterogeneous, and show no obvious pattern. Additional images of fixed cells showing the localization of the mNeon-tagged Vps8 paralogs are presented in Fig. S3D. For images of live cells, the increased contrast that was used to enhance those paralogs with weak signals also enhanced the background fluorescence. This included auto-fluorescence within food vacuoles, also seen in cells not expressing any fluorescent proteins (Fig. 3A,B, seventh row).
Differences observed in the appearance of Vps8b, Vps8e and Vps8f in live versus fixed cells might be related to the fact that fixed cells were obtained from growing cultures, while optimal live-cell imaging was achieved using partially starved cultures. Tetrahymena undergo rapid physiological changes when shifted from growth medium to non-nutrient medium, and these may be particularly relevant for CORVET complexes, which are associated with feeding via phagosome formation and maturation (Sparvoli et al., 2018).
Vps8b and Vps8e localize to food vacuoles at two different stages
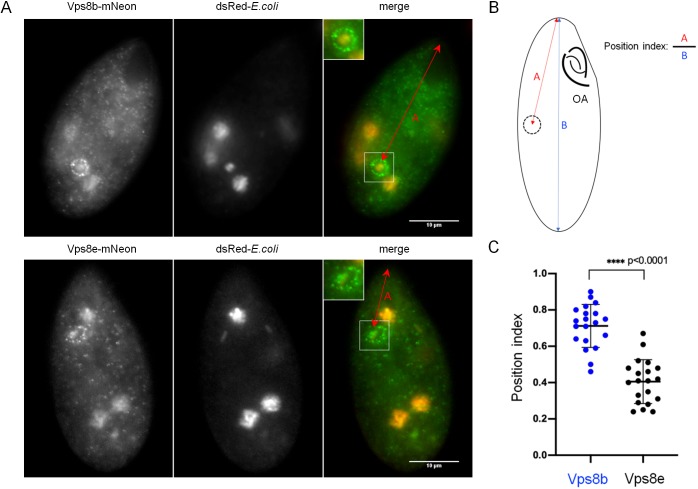
Tetrahymena are avid bacterivores and rapidly concentrate bacteria via ciliary beating at the anterior-positioned oral apparatus. From the base of the anterior oral apparatus bacteria are taken up by phagocytosis into newly formed phagosomes called food vacuoles. The food vacuoles then mature as they move posteriorly, and as their contents are digested (Nilsson, 1979). Based on their fluorescence patterns, a fraction of Vps8b–mNeon and Vps8e–mNeon puncta appeared to localize to food vacuoles. To confirm this, we labeled food vacuoles by incubating Tetrahymena with dsRed-expressing E. coli. Note that not all food vacuoles will be labeled under these conditions, since phagocytosis can also occur without bacterial ingestion. Both Vps8b–mNeon and Vps8e–mNeon were concentrated in ∼60% of the cells around clearly labeled food vacuoles (Fig. 4A), while in the remaining cells, the fluorescent puncta were dispersed. To ask whether Vps8b and Vps8e were associated with food vacuoles at different stages of maturation, we took advantage of the fact that vacuoles form in the cell anterior, and move posteriorly as they mature. For each cell in which we detected a food vacuole associated with Vps8b or Vps8e, we measured the distance of that vacuole from that anterior end of the cell as well as the cell length, and calculated an index to report the relative vacuole position (Fig. 4B). Notably, Vps8e-labeled phagosomes were positioned closer to the anterior end, while those labeled by Vps8b were positioned more posteriorly (Fig. 4C). Representative images of cells used for the quantification are shown in Fig. S4A,B. These results strongly suggest a role for 8E-CC and 8B-CC at successive stages in the formation and maturation of food vacuoles. Our analysis was limited to fixed cells, because the semi-starvation conditions that favor live-cell imaging inhibit food vacuole formation. We found that Vps8b-associated food vacuoles, but not those associated with Vps8e, could still be found in cells starved for several hours, consistent with the idea that Vps8b associates with old food vacuoles (data not shown).
Fig. 4.
Biased posterior versus anterior localization of Vps8b versus Vps8e to bacteria-containing phagosomes. (A) Cells expressing Vps8b–mNeon or Vps8e–mNeon were fed with E.coli expressing dsRed and fixed. The patterns of Vps8b and Vps8e are similar to those in live cells (Fig. 3). Red arrows indicate distance used for analysis as in B,C. Scale bars: 10 μm. (B,C) Calculating a position index for Vps8b- and Vps8e-labeled phagosomes. The position index corresponds to the distance of a vacuole to the tip of the cell divided by the cell length. The oral apparatus (OA) defines the anterior of the cell. The plot for Vps8b- and Vps8e-labeled phagosomes reveals clear posterior versus anterior biases, respectively (two-tailed t-test). Individual data points and the mean±s.d. is shown.
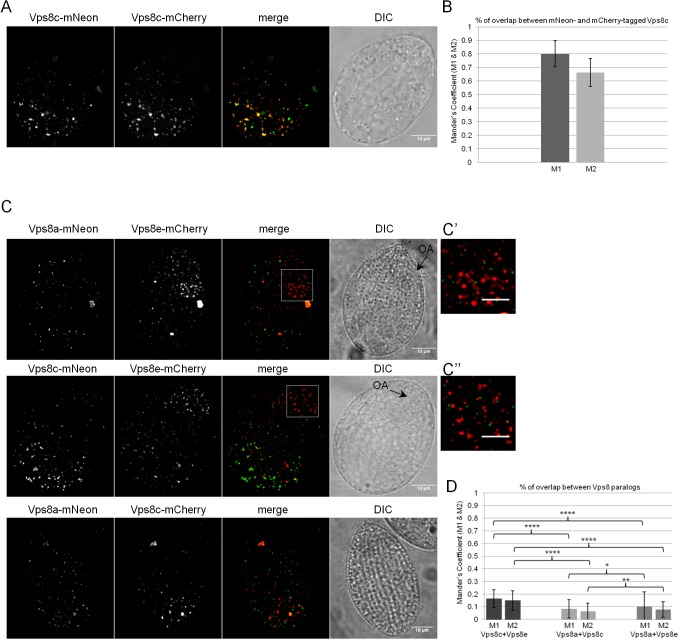
The evolutionarily related Vps8a and Vps8c, and by inference 8A-CC and 8C-CC, show very limited overlap
HOPS and CORVET in other lineages share four subunits, but they differ in their two Rab-binding subunits and therefore are targeted to different compartments (Solinger and Spang, 2013). Hybrid CORVET–HOPS complexes that differ from either HOPS or CORVET at only a single Rab-binding subunit have been detected in S. cerevisiae, but these complexes are of unknown significance (Peplowska et al., 2007). In considering the cohort of CORVET complexes in Tetrahymena, one striking inference is that the evolutionary replacement of the Vps8 subunit is sufficient to provide novel function, even when all other subunits remained identical. That is, although 8A-CC and 8C-CC are identical except Vps8, they localize to two seemingly different structures. The same is true for the 8E-CC and 8F-CC pair.
To understand this further, we focused on the most recently diverged paralogs, Vps8a and Vps8c, and by inference 8A-CC and 8C-CC. The above results showed that Vps8a appeared to localize primarily to different structures than Vps8c, but we could not rule out the possibility for a significant overlap in localization. To examine this possibility more rigorously, we generated cell lines in which complementary pairs of fluorescently tagged CORVET subunits were simultaneously expressed. As a positive control, we created cells simultaneously expressing Vps8c–mNeon and Vps8c–mCherry. In such cells, there was the expected extensive overlap between the red and green signals (Fig. 5A,B). In our negative controls, we found the expected limited overlap between co-expressed Vps8e–mCherry, which was concentrated near the cell anterior (see magnified insets in Fig. 5C′,C″; Fig. S5A), and both Vps8a–mNeon and Vps8c–mNeon (Fig. 5C, upper and middle panels, respectively, and Fig. 5D). Importantly, cells simultaneously expressing Vps8a–mNeon and Vps8c–mCherry also showed very limited overlap between their signals (Fig. 5C, lower panel, and Fig. 5D). The expression of full-length fusions in each cell line was confirmed by SDS-PAGE and western blotting (Fig. S5B,C). This result indicates that the steady-state localization of these CORVET complexes is primarily determined by the Vps8 subunit, rather than any other subunit.
Fig. 5.
8A-CC and 8C-CC localize to non-overlapping compartments. (A) Cells expressing Vps8c–mNeon and Vps8c–mCherry at the VPS8C and VPS8A loci, respectively. Shown is a maximum-intensity projection of a confocal z-stack of a fixed cell. Paired DIC images are confocal cross sections. (B) Mean±s.d. values of the colocalization coefficients M1 and M2 for Vps8c–mNeon and Vps8c–mCherry were calculated with 261 non-overlapping images using the Fiji-JACoP plugin. The two fusion proteins largely overlap. (C) Cells co-expressing the following combinations of tagged proteins: Vps8e–mCherry with Vps8a-mNeon (top); Vps8e–mCherry with Vps8c–mNeon (middle); Vps8a–mNeon with Vps8c–mCherry (bottom). Shown are maximum intensity projections of confocal z-stacks of fixed cells. The boxed regions in top and middle panels (magnified in C′ and C″) highlight Vps8e-related vesicles close to the oral apparatus (black arrows labeled OA in the paired DIC images). (D) Mean±s.d. values of the colocalization coefficients M1 and M2 for Vps8a with Vps8e, Vps8c with Vps8e, and Vps8a with Vps8c were calculated with 246, 337 and 399 non-overlapping images/sample as in B. P-values for M1 and M2 coefficients are shown. ****P<0.0001; *P=0.0113; **P=0.0048 (two-tailed t-test). The paralogs localize to largely non-overlapping cellular structures. Scale bars: 10 μm (main images); 5 μm (C′,C″).
Vps8a–mNeon undergoes limited proteolytic cleavage in cells, as detected by western blotting (Fig. S3A, second lane). We were concerned that the distribution of intact Vps8a–mNeon might not reflect the full distribution of Vps8a in cells. We took advantage of our previous finding showing that Vps8a tagged with GFP is functional and does not undergo proteolytic cleavage (Sparvoli et al., 2018; Fig. S5D). We therefore compared the number of fluorescent puncta in Vps8a–GFP versus Vps8a–mNeon cells (Fig. S5E). Since there was no significant difference (Fig. S5F), we conclude that Vps8a–mNeon provides a suitable reporter for the localization of the endogenous protein, and that Vps8a and Vps8c are chiefly localized to non-overlapping structures.
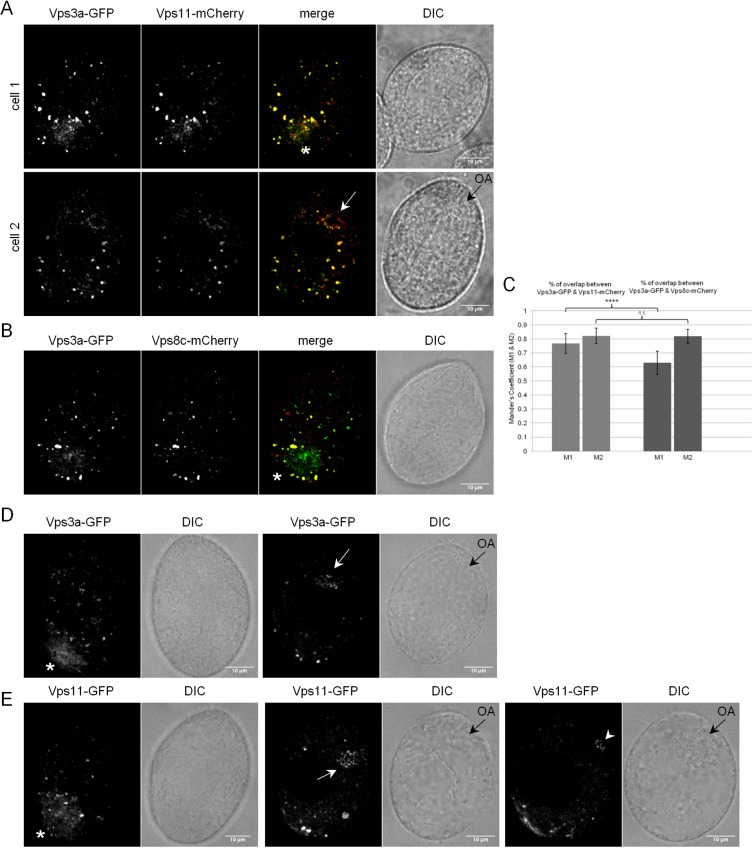
To obtain additional evidence that these Vps8 proteins belong to full CORVET complexes, we tested the inference that the shared subunits Vps11 and Vps3a are present in diverse compartments labeled by the set of Vps8 paralogs. In cells that co-express Vps3a–GFP with Vps11–mCherry, at their endogenous loci, the proteins are highly colocalized (Fig. 6A,C). Additionally, as expected Vps3a colocalizes extensively with Vps8c (Fig. 6B,C). Most importantly, cells individually expressing either GFP-tagged Vps3a or Vps11 showed labeling at a diverse set of structures, including cytoplasmic vesicles of different sizes, phagolysosome-related structures similar to those labeled by Vps8b (only for Vps11–GFP) and Vps8e, and the contractile vacuole (Fig. 6D,E).
Fig. 6.
Vps3a and Vps11 localize to a wider range of compartments than individual Vps8 subunits. (A) Cells co-expressing Vps3a–GFP and Vps11–mCherry. The two subunits largely colocalize at heterogeneous structures, including vesicles (white arrow, cell 2) close to the oral apparatus resembling those associated with Vps8e (black arrow, cell 2) and at the contractile vacuole (white asterisk, cell 1). (B) Cells co-expressing Vps3a–GFP and Vps8c–mCherry. The two subunits significantly overlap on heterogeneous vesicles. In addition, Vps3a–GFP alone is visible at structures, including the contractile vacuole (asterisk). (C) Mean±s.d. percentages of overlap (Mander's coefficients M1 and M2) for Vps3a with Vps11 and Vps3a with Vps8c were derived from 325 and 321 non-overlapping images using the Fiji-JACoP plugin, respectively. Vps3a extensively overlaps with both Vps11 (left columns) and Vps8c (right columns). P-values for M1 and M2 coefficients are shown. ****P<0.0001; n.s., P=0.5135 (two-tailed t-test). (D,E) Confocal sections of cells expressing Vps3a–GFP or Vps11–GFP. The overall distributions of Vps3a and Vps11 resembles that of the combination of individual Vps8 paralogs, and include heterogeneous cytoplasmic vesicles, the contractile vacuole (asterisk), and rings of vesicles near the oral apparatus (black arrows in the paired DIC images) like those labeled by Vps8e (white arrow). Vps11 but not Vps3a is also found at rings of vesicles around food vacuoles (arrowhead), like those labeled by Vps8b. The images showing contractile vacuoles are surface sections, while all others are cell mid-sections. Scale bars: 10 μm.
Vps8a and Vps8c localize to non-equivalent Rab7-labeled compartments
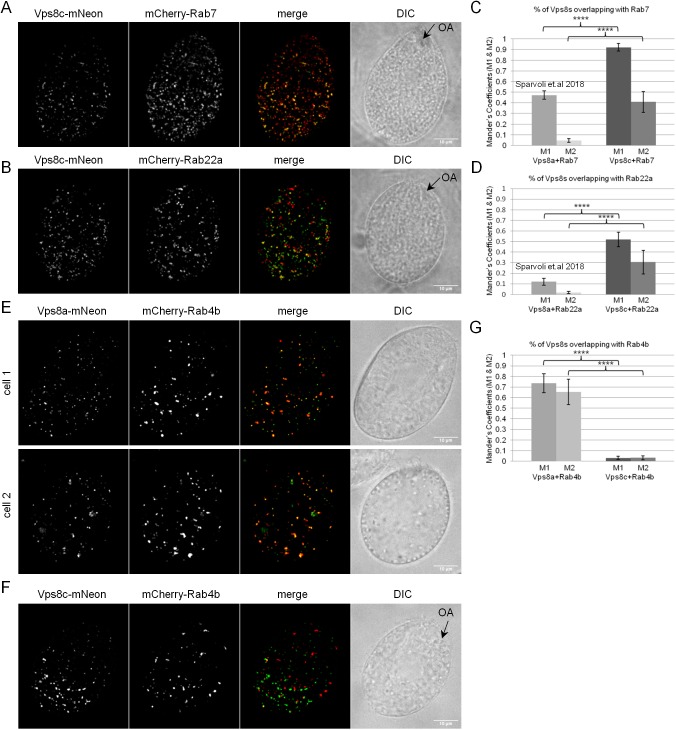
In other organisms, CORVET acts at Rab5-positive compartments, while HOPS functions at Rab7-positive compartments. Rab5 has been lost in the Tetrahymena lineage, but T. thermophila expresses the related Rab22a (Bright et al., 2010). We previously reported the surprising finding that Vps8a shows negligible colocalization with Rab22a but substantial overlap with Rab7 (Sparvoli et al., 2018). To determine whether this was also the case for Vps8c, we expressed N-terminal mCherry-tagged Rab7 (Fig. 7A) and Rab22a (Fig. 7B) in Vps8c–mNeon-expressing cells, and measured the extent of overlap. Vps8c colocalized more strongly with both Rabs than Vps8a (Fig. 7C,D), and in particular overlapped extensively with Rab7 (Fig. 7C). This overlap could also be seen at dynamic tubule-vesicular structures (Fig. S6A,B; Movie 7). Since Vps8a and Vps8c both significantly colocalize with Rab7, but very little with one another, there must be additional localization determinants present.
Fig. 7.
8A-CC and 8C-CC associate with distinct endosomes. (A,B) Cells co-expressing Vps8c–mNeon together with either mCherry–Rab7 (upper panel) or the Rab5 homolog mCherry–Rab22a (lower panel). Rab transgene expression was induced with 1 μg/ml CdCl2 for 2 h in SPP. Shown (including E,F) are maximum intensity projections of z-stacks of fixed cells. The DIC images are confocal cross sections for clarity. (C,D) Percentages of overlap (Mander's coefficients M1 and M2) for Vps8c with Rab7 and Vps8c with Rab22a were calculated using Fiji-JACoP plugin. The mean±s.d. M1 and M2 values for Vps8c were derived from 268 and 232 non-overlapping images for Rab22a and Rab7 samples, respectively. (E,F) Cells co-expressing Vps8a–mNeon (upper panel) or Vps8c–mNeon (lower panel) with mCherry-Rab4b. (G) Mean±s.d. percentages of overlap (Mander's coefficients M1 and M2) for Vps8a with Rab4b, and Vps8c with Rab4b, were calculated with 158 and 139 non-overlapping images/sample, respectively, using the Fiji-JACoP plugin. In contrast to Vps8c–mNeon, Vps8a–mNeon shows extensive colocalization with Rab4b. Scale bars: 10 μm. ****P<0.0001 (two-tailed t-test).
We sought other Rab proteins that could act as determinants in the Vps8a-dependent pathway of mucocyst formation. We previously used a genome-wide approach to identify genes upregulated in periods of stimulated mucocyst formation (Haddad et al., 2002; A.P.T., unpublished). By searching these data for Rabs, we identified a Rab4/Ypt31homolog (Rab4b; Bright et al., 2010), that is upregulated 11-fold under conditions of induced mucocyst formation. Moreover, the transcriptional profile of the RAB4B gene, under a variety of cell culture conditions, is strikingly similar to a variety of established mucocyst-associated genes, suggesting these genes are co-regulated (Fig. S6C). To determine whether Rab4b might contribute towards Vps8a localization, we endogenously tagged Rab4b with mCherry at its N-terminus, and co-expressed in cells with Vps8a–mNeon or Vps8c–mNeon (Fig. 7E and Fig. 7F, respectively). Strikingly, Vps8a, but not Vps8c, showed strong colocalization with Rab4b (Fig. 7G).
Taken together, our results suggest that all six Vps8 paralogs, and by inference their parent CORVET complexes, are individually specialized for distinct trafficking pathways in Tetrahymena. Specialization extends to even the most recently diverged paralogs, namely, 8A-CC and 8C-CC, which can be considered sibling complexes and are largely non-overlapping in their distribution. 8C-CC shows modest overlap with early endosomal Rab22a, and strong colocalization with late endosomal Rab7. 8A-CC, which colocalizes partially with Rab7, has strong colocalization with Rab4b, which is a recycling endosome marker.
DISCUSSION
The loss of the HOPS tethering complex was accompanied in a sub-family of ciliates by expansions in the number of CORVET complex subunits. In T. thermophila, we show here that there are six biochemically distinct CORVET complexes, which we call 8A-CC, 8B-CC, etc., each possessing a different Vps8 paralog. While detailed functional analysis has yet to be achieved, our localization data strongly suggest that the complexes diversified to associate with distinct compartments or subdomains of the same compartment. That is, five of the six endogenously tagged Vps8 paralogs localize to structures that are recognizably different from one another in live cells. Consistent with the idea of functional specialization, there is differential expression of genes expressing subunits that are specific to individual complexes, as seen in whole-genome transcriptional profiling over a range of culture conditions.
The CORVET complexes are expressed at very low levels, but we could nonetheless isolate all six complexes by taking advantage of the cryomilling technique that has been used in other organisms. Determining the subunit composition of all six complexes allowed us to make some observations about the pattern of diversification within CORVET genes. In two pairs of complexes, the only difference is the Vps8 subunit, and for 8A-CC and 8C-CC there is minimal colocalization. CORVET targeting depends upon Rab protein interactions via the two Rab-binding Vps8 and Vps3 subunits (Epp and Ungermann, 2013). In Tetrahymena, at least, the Rab binding by these subunits is not functionally equivalent; specifically since 8A-CC and 8C-CC share Vps3a, they would be expected to colocalize if Vps3 were a primary targeting determinant. Since this is not observed, the Vps8 subunit is likely to be the subunit primarily responsible for the differential steady-state location of most CORVET complexes. This idea is broadly consistent with experiments in yeast HOPS showing that the Rab-binding subunits Vps39 and Vps41 have different binding properties (Lürick et al., 2017). Vps39 and Vps41 are positioned at opposite ends of the extended barbell-like cryo-electron microscopy (cryoEM) structure of yeast HOPS, consistent with independent binding and with the idea that relative Rab affinities could determine steady-state localization (Brocker et al., 2012). A current assumption is that CORVET and HOPS have similar overall structures, given the molecular similarities, but this remains to be demonstrated. The barbell structure was determined from cross-linked complexes using cryoEM, while analysis of non-cross-linked complexes suggests considerable flexibility, with the barbell being just one of several relevant structures (Chou et al., 2016; Kuhlee et al., 2015). Therefore, a second and non-mutually exclusive potential explanation of our results is that alternative conformations for CORVET bias Rab binding in favor of Vps8.
In either model, the non-colocalization of different CORVETs bearing the identical Vps3a subunit could be explained if binding of Vps3 to its cognate Rab is rapidly followed by membrane fusion, that is, bivalent tethering is a short-lived state. Alternatively, non-colocalization could be explained if Vps3a recognizes a Rab present on a variety of membranes, for example, sub-compartments of an organelle. In this scenario, additional contacts might refine the targeting of each CORVET complex to an individual sub-compartment. We observed that although both 8C-CC and 8A-CC individually colocalize with Rab7, as discussed further below, they show minimal colocalization with one another. This suggests that multiple distinct populations of Rab7-positive endosomes are present, similar to distinct subpopulations of early endosomes in mammals (Perini et al., 2014). Consistent with this idea, Tetrahymena Rab7 labels highly mobile cytoplasmic vesicles but also the contractile vacuole and phagosome-associated vesicles (D.S., unpublished data), the last consistent with biochemical data (Jacobs et al., 2006). It is thus possible that Rab7 is present at all CORVET-positive structures.
In yeast and animals, CORVET or HOPS subunits also engage in non-Rab-based interactions with target membranes (Fratti et al., 2004; Ho and Stroupe, 2016; Stroupe et al., 2006). Similarly, combinatorial interactions could explain how different CORVET complexes in Tetrahymena are differentially recruited to Rab7-positive membranes. In this regard, it is notable that several CORVET subunit paralogs in Tetrahymena are larger than their homologs in Opisthokonts, potentially offering novel structures for interactions. Vps8a and Vps8e are 22% and 58% larger, respectively, than their S. cerevisiae ortholog. Size variation for Vps8 is also notable in plants where Arabidopsis Vps8 is 46% larger than in yeast. By contrast Vps11, a subunit with an organizing role in assembly of the HOPS/CORVET core (Ostrowicz et al., 2010; Plemel et al., 2011), has remained nearly invariant in size suggesting significant selective pressure for conservation.
An implication of CORVET composition in Tetrahymena is that most tethers are not acting as homotypic tethers, unlike CORVET in other organisms that have been analyzed. That is, in the five complexes containing Vps3a, the five different Vps8 paralogs are unlikely to all share the Vps3a Rab-binding specificity. As heterotypic tethers, the Tetrahymena complexes may shed light on the nature of hybrid CORVET–HOPS complexes that have been reported in yeast, which have the potential to bind both Rab5 and Rab7 (Peplowska et al., 2007). These hybrids have been hypothesized to represent intermediates in CORVET-to-HOPS switching during endosome maturation, in a mechanism involving step-wise substitution of complex-specific subunits on the shared core, as discussed further below. However, we note that such hybrid CORVET–HOPS tethers have to date only been detected in yeast, and under conditions in which subunit overexpression could potentially result in non-physiological complexes.
The pattern of subunit variation between complexes suggests that the core Vps18 subunit may determine which Vps8 paralog is included. Among Vps8 paralogs, Vps8a and Vps8c are relatively closely related, as are Vps8e and Vps8f, while Vps8b and Vps8d are more divergent from the others (Sparvoli et al., 2018). Based on data here, the cores containing Vps18d form complexes that also contain Vps8a or Vps8c, while cores containing Vps18c instead assemble with Vps8e or Vps8f subunits. The single cores containing Vps18a or Vps18b assemble with the highly unrelated Vps8d and Vps8b subunits, respectively. The idea that Vps18 paralogs determine the inclusion of specific Vps8 paralogs is consistent with genetic, biochemical and structural mapping of subunit interactions (Guo et al., 2013; Hunter et al., 2017; Plemel et al., 2011; Brocker et al., 2012; Chou et al., 2016).
The function of 8A-CC was analyzed in previous work (Sparvoli et al., 2018), while results in this paper provide hints about functions of the other five CORVET complexes. At least two are associated with the pathway of phagocytosis and food vacuole formation. 8E-CC is targeted to vesicles associated with phagosomes, which are likely to be newly forming, based on their anterior position. It may be required for their tethering and fusion, since Δvps8e cells accumulate an excess of small endocytic vesicles (Sparvoli et al., 2018). 8B-CC is associated with vesicles found at the periphery of phagolysosomes at a later stage in the pathway. Interestingly, we observed a notable accumulation of endocytic vesicles around phagosomes in Δvps8b cells (Sparvoli et al., 2018), suggesting the existence of a class of endosomes that require 8B-CC for fusion but not for docking. The function of 8F-CC is not apparent from its localization. However, in preliminary experiments, we saw strongly similar phagocytosis defects in Δvps8e and Δvps8f cells, suggesting that 8F-CC is also required for early steps in phagocytosis.
8D-CC is associated with the contractile vacuole, a Rab11a-positive (and therefore endolysosomal) organelle which is nonetheless not known to be involved in trafficking of endocytic or secreted proteins (Allen, 2000; Bright et al., 2010; Plattner, 2015). Contractile vacuole activity in Tetrahymena is based on repeated cycles of membrane fusion and fission between tubules and a central bladder, and 8D-CC may contribute to that process. Interestingly, contractile vacuole CORVET possesses the largest number of exclusive subunits (four of six) among the Tetrahymena complexes, including a unique paralog of the SNARE-binding Vps33 subunit. The second relatively unique complex, with three exclusive subunits, is 8B-CC. Interestingly, all six subunits defining 8D-CC were strongly detected by mass spectrometry, but the Vps8d subunit was significantly more abundant than the other five based on analysis by silver staining. One possibility is that Vps8d partially exists as a monomer, for which we have preliminary evidence. HOPS and CORVET sub-complexes have been identified in other organisms, as outlined above, and may be highly lineage specific. One important source of ambiguity in characterizing sub-complexes using biochemical approaches is that a substantial fraction of CORVET remains attached to membranes under non-denaturing conditions.
Our understanding of Rab-binding specificities of CORVET in Tetrahymena is limited to 8A-CC and 8C-CC. In yeast and animals, CORVET binds early endosomal Rab5/Vps21 via the Vps3 and Vps8 subunits (Balderhaar et al., 2013; Epp and Ungermann, 2013; Markgraf et al., 2009; Peplowska et al., 2007), while HOPS subunits Vps41 and Vps39 bind late endosomal Rab7/Ypt7 (Plemel et al., 2011; Wurmser et al., 2000). These specificities are maintained in plants (Takemoto et al., 2018) but not Tetrahymena, since the CORVET subunit Vps8a associates with Rab4b, a marker for recycling endosomes, and with Rab7 rather than the Rab5 homolog Rab22a. Interestingly, Vps8 in HeLa cells similarly associates with Rab4, as part of a sub-complex that provides a function distinct from holo-CORVET (Jonker et al., 2018). Moreover, the association of Vps8a with Rab4b-containing endosomal domains, and its function in cargo delivery to mucocysts is consistent with the role of early endosome-derived tubules in cargo delivery to melanosomes (Delevoye et al., 2016, 2009; Dennis et al., 2015), suggesting potential conservation in LRO biogenesis pathways. Although the overall organization of endosomal trafficking in Tetrahymena remains to be analyzed, Rab22a is probably a bona fide early endosomal marker (Bright et al., 2010). We found, relative to Vps8a, that Vps8c colocalizes even more strongly with Rab7, but in addition colocalizes with Rab22a and not Rab4b. In cases of strong overlap, FRET between mNeon and mCherry could reduce green emission.
In yeast and animals, the mechanism of endosome maturation involves CORVET-dependent recruitment of a guanine nucleotide exchange factor (GEF) that activates Rab7, which then recruits HOPS (Nordmann et al., 2010). By this mechanism, Rab5-to-Rab7 conversion during endosome maturation is linked with the engaging of successive tethers. This mechanism also exists in plants, although recent evidence suggests that the HOPS function in some plant pathways does not depend upon this kind of GTPase switch (Takemoto et al., 2018). In Tetrahymena, our results beg the question of whether there is maturation-linked switching between distinct CORVET complexes like 8C-CC and 8A-CC. In both Opisthokonts and Archaeplastids, the Rab-switching complex that links CORVET and HOPS is the Mon1–Ccz1 heterodimer, where Ccz1 possesses GEF activity (Cui et al., 2014; Kiontke et al., 2017; Nordmann et al., 2010). Interestingly, while Tetrahymena has an unambiguous Mon1 homolog, there is no convincing ortholog for Ccz1, similar to what has been reported in C. elegans (Morris et al., 2018).
The expansion and functional diversification of CORVET in ciliates provides a potent example of mechanisms underlying new trafficking pathways. Loss of HOPS in the lineage leading to Tetrahymena may indicate a simplification of pre-existing transport pathways. Such reductions in pathway complexity, relative to that present in the last eukaryotic common ancestor (LECA), are supported by a collapse in diversity of pan-eukaryote Rab and TBC-domain-containing GTPase-activating proteins (Elias et al., 2012; Gabernet-Castello et al., 2013). We hypothesize that expansion of lineage-specific Rab proteins, which account for ∼40% of the repertoire (RabX in Elias et al., 2012), likely reflects subsequent pressures in ciliates that also drove diversification of CORVET-mediated endosomal pathways, but which could only be met by expansions of CORVET subunits. This is fully consistent with the organelle paralogy model proposed previously whereby expansions in trafficking gene families facilitate the emergence of new pathways (Dacks and Field, 2007), and is also reminiscent of the evolution of multiple early endosomal pathways in many lineages, including yeasts and kinetoplastida, where Rab5 paralogs have arisen independently of each other. We suggest that a ‘backfilling’ mode, involving expansion following loss, is a potentially underappreciated aspect of trafficking diversity, especially as a number of key trafficking proteins are frequently, and independently, lost from multiple lineages.
MATERIALS AND METHODS
Cell culture
Tetrahymena thermophila strains used in this work are shown in Table S2. Cells were grown overnight in SPP (2% proteose peptone, 0.1% yeast extract, 0.2% dextrose and 0.003% ferric-EDTA) supplemented with 250 µg/ml penicillin G, 250 µg/ml streptomycin sulfate, and 0.25 μg/ml amphotericin B fungizone, to medium density (1×105–3×105 cells/ml). For biolistic transformation, growing cells were subsequently starved in 10 mM Tris-HCl buffer, pH 7.4, for 18–20 h. Fed and starved cells were both kept at 30°C with agitation at 99 rpm, unless otherwise indicated. For live microscopy, cells were transferred to S medium (0.2% yeast extract and 0.003% ferric-EDTA) for 2 h prior to imaging. Culture densities were measured using a Z1 Coulter Counter (Beckman Coulter Inc.).
Endogenous tagging of the Vps8 paralogs with mNeon fluorescent tags
Two mNeonGreen fluorescent tags were integrated at the C-termini of the macronuclear open-reading frames (ORFs) of VPS8B, VPS8C, VPS8D, VPS8E and VPS8F via homologous recombination using linearized pVPS8B-2mNeon-6myc-Neo4, pVPS8C-2mNeon-6myc-Neo4, pVPS8D-2mNeon-6myc-Neo4, pVPS8E-2mNeon-6myc-Neo4 and pVPS8F-2mNeon-6myc-Neo4 created as follows. The C terminal 745 bp, 763 bp, 763 bp, 745 bp and 468 bp from the VPS8B, VPS8C, VPS8D, VPS8E and VPS8F genomic loci (minus the stop codon), respectively, were amplified by PCR and cloned in digested p2mNeon-6myc-Neo4 vector (Sparvoli et al., 2018) at the SacI/MluI sites by Quick Ligation (New England Biolabs Inc.). Subsequently, the 758 bp, 744 bp and 799 bp 3′ UTRs of VPS8B, VPS8C and VPS8D, respectively, were cloned in the VPS8-specific p2mNeon-6myc-Neo4 vector at the XhoI/ApaI, while the 808 bp, 793 bp 3′ UTRs of VPS8E and VPS8F, were cloned at the EcoRV/XhoI sites. The final constructs were digested with SacI and KpnI prior to biolistic transformation of CU428.1. The primers are listed in Table S3.
Endogenous tagging of the Vps8 paralogs with FLAG epitope
The FLAG-ZZ tag, containing 3xFLAG, followed by the TEV (Tobacco Etch Virus cysteine protease) cleavage site and the IgG binding domain of protein A (ZZ-domain), was integrated at the C-termini of VPS8B, VPS8C, VPS8D, VPS8E and VPS8F macronuclear ORFs by homologous recombination, using pVPS8B-FLAG-ZZ-Neo4, pVPS8C-FLAG-ZZ-Neo4, pVPS8D-FLAG-ZZ-Neo4, pVPS8E-FLAG-ZZ-Neo4 and pVPS8F-FLAG-ZZ-Neo4 created as follows. The 5′ ends and the 3′ UTRs of VPS8B, VPS8C and VPS8D were removed from pVPS8B-2mNeon-6myc-Neo4, pVPS8C-2mNeon-6myc-Neo4, pVPS8D-2mNeon-6myc-Neo4 vectors by using SacI/NheI and XhoI/ApaI, and cloned in the corresponding sites in pVPS11-FLAG-ZZ-Neo4, to replace the 5′ end and the 3′ UTR of VPS11, respectively. The same strategy was used to clone the 5′ ends and the 3′ UTRs of VPS8E and VPS8F at the SacI/MluI and EcoRV/XhoI sites in pVPS8A-FLAG-ZZ-Neo4 (Sparvoli et al., 2018) to replace the 5′ end and the 3′ UTR of VPS8A, respectively. The final constructs were digested with SacI and KpnI prior to biolistic transformation of CU428.1.
Co-expression of c-Myc-tagged Vps16a, Vps16b, Vps33a, Vps33b, Vps18a, Vps18b, Vps18c or Vps18d with Vps8a–FLAG
The 6Myc tag was integrated at the C-termini of VPS16A, VPS16B, VPS18A, VPS18B, VPS18C and VPS18D, and at the N-termini of VPS33A and VPS33B by homologous recombination at the macronuclear ORFs, using pVPS16A-6c-myc-Chx, pVPS16B-6c-myc-Chx, pVPS18A-6c-myc-Chx, pVPS18B-6c-myc-Chx, pVPS18C-6c-myc-Chx, pVPS18D-6c-myc-Chx, p6c-myc-VPS33A-Chx and p6c-myc-VPS33B-Chx, respectively, created as follows. PCR was used to amplify the 5′ ends (730–794 bp minus the stop codon) and the 3′ UTRs (576–794 bp) of VPS16A, VPS16B, VPS18A, VPS18B, VPS18C and VPS18D. The 5′ ends and the 3′ UTR amplicons were cloned in the p6c-myc-Chx vector (Sparvoli et al., 2018) by Quick Ligation (New England Biolabs Inc.) at the SacI/NheI and XhoI/ApaI sites, respectively. VPS33A and VPS33B macronuclear ORFs were PCR-amplified and inserted by In-Fusion cloning (Clontech, Mountain View, CA) in the linearized p6c-myc-Chx vector at the SpeI site. PCR was used to amplify the 670–792 bp of 5′ and 3′ UTRs of VPS33A and VPS33B. The 5′ and 3′ UTRs amplicons were then cloned in the corresponding p6c-myc-Chx vector containing the appropriate VPS33 gene, by Quick Ligation at SacI-XbaI/BamHI and XhoI/ApaI sites, respectively. The final vectors pVPS16A-6c-myc-Chx, pVPS16B-6c-myc-Chx, pVPS18A-6c-myc-Chx, pVPS18B-6c-myc-Chx, pVPS18C-6c-myc-Chx, pVPS18D-6c-myc-Chx were digested with SacI and KpnI, p6c-myc-VPS33A-Chx with SacI and ApaI, p6c-myc-VPS33B-Chx with XbaI and KpnI, prior to biolistic transformation of Vps8a-FLAG-ZZ expressing cells. All primers are listed in Table S3.
Endogenous tagging of Vps8c and Vps8e with mCherry in Vps8a–mNeon- and Vps8c–mNeon-expressing cells
3mCherry2HA tag was integrated at the C-termini of VPS8C and VPS8E macronuclear ORFs by homologous recombination using pVPS8C-3mCherry2HA-Chx and pVPS8E-3mCherry2HA-Chx. To construct the pVPS8C-3mCherry2HA-Chx, the 3FLAG-ZZ-Neo4 fragment in pVPS8C-FLAG-ZZ-Neo4 was replaced with 4491 bp of the 3mCherry2HA-Chx fragment digested with NheI and XhoI from pVPS11-3mCherry2HA-Chx (see below), and cloned at the corresponding sites in pVPS8C-FLAG-ZZ-Neo4 by Quick Ligation (New England Biolabs Inc.) pVPS8E-3mCherry2HA-Chx was obtained by first cloning the 2742 bp of the 3mCherry2HA-3′UTR-BTU1 fragment from pVPS8C-3mCherry2HA-Chx at the NheI and PstI sites in pVPS8E-FLAG-ZZ-Neo4, and then by replacing the Neo4 drug resistance cassette with the PCR-amplified Chx cassette at the PstI and EcoRV sites via Quick Ligation. The final constructs were then linearized with SacI and KpnI, and pVPS8C-3mCherry2HA-Chx was biolistically transfected into Vps8a-mNeon-expressing cells, while linearized pVPS8E-3mCherry2HA-Chx was transfected into Vps8c-mNeon and Vps8a-mNeon expressing cells. The primers are listed in Table S3.
Expression of mCherry-tagged Vps8c at the VPS8A locus in Vps8c–mNeon-expressing cells
A second 2mCherry2HA-tagged copy of VPS8C was introduced in the VPS8A locus in Vps8c–mNeon-expressing cells by homologous recombination using p5′UTR-VPS8A-VPS8C-2mCherry2HA-Chx. To create p5′UTR-VPS8A-VPS8C-2mCherry2HA-Chx, we used p5′UTR-VPS8A-VPS8C-FLAG-ZZ-Chx as the starting vector. Briefly, for the construction of p5′UTR-VPS8A-VPS8C-FLAG-ZZ-Chx, we first replaced the Neo4 drug resistance cassette in pVPS8A-FLAG-ZZ-Neo4 (Sparvoli et al., 2018) with Chx at the SpeI and EcoRV sites, via digestion and Quick Ligation, to generate the pVPS8A-FLAG-ZZ-Chx vector. A 5218-bp-long gene block was PCR amplified, including the VPS8C macronuclear ORF together with the 813-bp-long 5′ UTR of VPS8A. The latter was digested with SacI and HindIII and cloned in pVPS8A-FLAG-ZZ-Chx at the corresponding sites, to integrate the additional copy of the VPS8C gene into the VPS8A genomic locus by homologous recombination. The reverse primer for the 5′ UTR-VPS8A cloning contained a PmeI site upstream the HindIII site; thus, the VPS8C gene ORF was cloned between PmeI and MluI sites of the corresponding vector by Quick Ligation (New England Biolabs Inc.). We then replaced the FLAG-ZZ-3′UTR-BTU1 fragment in p5′UTR-VPS8A-VPS8C-FLAG-ZZ-Chx with 2mCherry2HA-3′UTR-BTU1, obtained by digesting pGRL3-2mCherry2HA-Chx vector with BamHI and XmaI. The fragment was cloned by Quick Ligation into the BamHI/XmaI-linearized p5′UTR-VPS8A-VPS8C-FLAG-ZZ-Chx vector. The final construct p5′UTR-VPS8A-VPS8C-2mCherry2HA-Chx was linearized with SacI and KpnI prior to biolistically transforming Tetrahymena. Primers are listed in Table S3.
Expression of GFP-tagged Vps3a and Vps11
Monomeric enhanced GFP (mEGFP) was integrated at the C-termini of VPS3A and VPS11 macronuclear ORFs via homologous recombination using pVPS3A-mEGFP-Neo4 and pVPS11-mEGFP-Neo4, respectively. PCR was used to amplify the 5′ ends (666–778 bp minus the stop codon) and the 3′ UTRs (785–690 bp) of VPS3A and VPS11. The 5′ end and the 3′ UTR amplicons were cloned in the pmEGFP-Neo4 vector (Briguglio et al., 2013) by Quick Ligation (New England Biolabs Inc.) at the SacI/NheI and XhoI/ApaI sites, respectively. The final vectors pVPS3A-mEGFP-Neo4 and pVPS11-mEGFP-Neo4 were digested with SacI and KpnI prior to biolistic transformation of CU428.1.
Expression of mCherry-tagged Vps11 or Vps8c in cells expressing Vps3a–GFP
Vps11–mCherry and Vps8c–mCherry were integrated at the corresponding endogenous loci in cells expressing Vps3a–GFP by homologous recombination, using pVPS11-3mCherry2HA-Chx and the previously mentioned pVPS8C-3mCherry2HA-Chx vectors. 3mCherry2HA was integrated at the C-terminus of the VPS11 macronuclear ORF via homologous recombination using the pVPS11-3mCherry2HA-Chx vector. The vector was constructed by cloning the PCR-amplified 5′ end (778 bp minus the stop codon) and 3′ UTR (690 bp) of VPS11 into the p3mCherry2HA-Neo4 vector (Sparvoli et al., 2018), at the SacI/NheI and XhoI/ApaI sites, respectively. The resulting pVPS11-3mCherry2HA-Neo4 vector was then digested with PstI and XhoI to replace the Neo4 resistance cassette with the Chx cassette. The final vectors pVPS8C-3mCherry2HA-Chx and pVPS11-3mCherry2HA-Chx were linearized with SacI and KpnI prior to biolistic transformation of Vps3a–GFP-expressing cells.
Co-expression of mCherry–Rab4b with Vps8a–mNeon and Vps8c–mNeon
The 2HA3mCherry tag was integrated at the N-terminus of the RAB4B (TTHERM_01097960) macronuclear ORF via homologous recombination using p2HA3mCherry-RAB4B-Chx. Constructs p6c-myc-RAB4B-Chx and p2HA3mCherry-RAB4B-ncvb were used as templates to construct the final Rab4b vector. First, the 5′UTR of RAB4B was PCR-amplified, digested with SacI and BamHI, and subsequently cloned by Quick Ligation (New England Biolabs Inc.) at the corresponding sites in p6c-myc-RAB4B-Chx, to replace the 5′UTR and thereby introduce a PmeI site upstream of the BamHI site. RAB4B has a PmeI site within the genomic sequence, and this was used to linearize p6c-myc-RAB4B-Chx containing the new 5′UTR, and thus to introduce the 2767 bp 2HA3mCherry-N-terminal RAB4B fragment. The latter was obtained by digesting the p2HA3mCherry-RAB4B-ncvb vector with PmeI. The correct orientation of the fragment was tested using MfeI and SpeI. The final p2HA3mCherry-RAB4B-Chx vector was linearized with SacI and KpnI prior to biolistic transformation of Vps8a–mNeon- and Vps8c–mNeon-expressing cells. The primers are listed in Table S3.
Co-expression of mCherry–Rab7 or mCherry–Rab22a with Vps8c–mNeon
mCherry–Rab7 and mCherry–Rab22a were integrated at the metallothionein (MTT1) genomic locus in cells expressing Vps8c–mNeon by homologous recombination, using the previously described 2HA-3mCherry-RAB7-ncvb and 2HA-3mCherry-RAB22a-ncvb vectors (Sparvoli et al., 2018), and their expression induced with CdCl2. The constructs were linearized with SfiI prior to biolistic transformation.
Biolistic transformation
Tetrahymena transformants were generated and selected after biolistic transformation as previously described (Kaur et al., 2017; Sparvoli et al., 2018). Transformants were serially transferred six times per week in increasing concentrations of drug and decreasing concentrations of CdCl2 (up to 2 mg/ml of paromomycin and 0.1 μg/ml CdCl2; up to 18–21 μg/ml of cycloheximide and 1 μg/ml CdCl2; and up to 90 μg/ml of blasticidin and 0.1 μg/ml CdCl2) for at least 5 weeks before further testing. Successful integration and replacement of all endogenous alleles at each genomic locus was tested by RT-PCR as previously described (Sparvoli et al., 2018). At least three independent transformants were tested for each line.
Co-immunoprecipitations
Vps8a–FLAG-ZZ was co-immunoprecipitated with Vps16b–Myc, Myc–Vps33b or Vps18d–Myc, from detergent-extracted lysates of cells co-expressing Vps8a–FLAG-ZZ with either Vps16a–Myc, Vps16b–Myc, Myc–Vps33a, Myc–Vps33b, Vps18a–Myc, Vps18b–Myc, Vps18c–Myc or Vps18d–Myc, using anti-FLAG beads (EZ view Red Anti-FLAG M2 Affinity Gel, Sigma) and anti-c-Myc beads (Pierce anti-c-Myc Agarose, Thermo Fisher Scientific), respectively, as previously described (Sparvoli et al., 2018). In brief, 300–500 ml cultures were grown overnight to 2×105–3×105 cells/ml. The cells were washed once with 10 mM Tris-HCl, pH 7.4, pelleted and resuspended in cold lysis buffer (20 mM Tris-HCl pH 7.4, 50 mM NaCl, 1 mM MgCl2, 1 mM DTT, 1 mM EGTA, 0.2% NP-40, 10% glycerol and 4% BSA), supplemented with protease inhibitor cocktail tablets (Roche), and gently mixed for 45 min on ice. Lysates were cleared by centrifugation at 35,000 rpm (142,000 g; Beckman Instruments type 45 Ti rotor) for 1.5 h at 4°C, split in two 50 ml-falcon tubes, and separately mixed with 75 μl anti-FLAG and 400 μl anti-c-Myc beads, and pre-incubated with cold lysis buffer for 2 h at 4°C, respectively. The beads were then washed five times with 20 mM Tris-HCl pH 7.4, 1 mM EDTA, 500 mM NaCl, 0.1% NP-40, 1 mM DTT, 10% glycerol, and resuspended in 80 μl of 100°C 2× lithium dodecyl sulfate (LDS) sample buffer containing 40 mM DTT.
Immunoprecipitations from cryomilled cell powders
Tetrahymena were grown overnight to 2×105–3×105 cells/ml, washed once with 10 mM Tris-HCl pH 7.4 and re-pelleted (1000 g for 5 min). Supernatants were rapidly aspirated to leave a dense cell slurry. The slurries were transferred, drop-wise, into liquid nitrogen and milled to powders using a Cryogenic Grinder 6875 Freezer Mill. The cryopowders were resuspended in buffer B4 (20 mM Hepes pH 7.4, 250 mM NaCitrate, 1 mM MgCl2, 0.1% CHAPS and 1 mM DTT), supplemented with protease inhibitor cocktail tablets (Roche), gently mixed for 1 h at 4°C, and then on ice until no solid matter was visible. Lysates were cleared by centrifugation at 35,000 rpm (142,000 g; Beckman Instruments type 45 Ti rotor) for 1.5 h at 4°C, and mixed with anti-FLAG beads (EZ view Red Anti-FLAG M2 Affinity Gel, Sigma), pre-washed with cold lysis buffer, for 2 h at 4°C. The beads were then washed five times with 20 mM Tris-HCl pH 7.4, 1 mM EDTA, 500 mM NaCl, 0.1% NP-40, 1 mM DTT, 10% glycerol supplemented with protease inhibitor cocktail tablets (Roche). Samples destined for silver staining were washed one additional time with elution buffer (20 mM Hepes pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 0.1% CHAPS, 1 mM DTT, 5% glycerol and protease inhibitor tablets). Washed beads were then resuspended in elution buffer, or in 100°C LDS sample buffer containing 40 mM DTT, depending on the purpose of the experiment. For isolation of CORVET complexes for subsequent mass spectrometry, Tetrahymena from an overnight culture were inoculated in 10 l SPP, and grown to 2×105–3×105 cells/ml for 24–26 h at 30°C with agitation at 75 rpm, and powders prepared as described above. A total of 100–150 g of powder and 400 μl of anti-FLAG beads were used for each cell line. Proteins were eluted from the beads with 60 μl 100°C LDS sample buffer containing 40 mM DTT, prior to SDS-PAGE and Coomassie staining. To isolate 8A-CC for subsequent analysis by glycerol gradient centrifugation, we used 30 g of cryopowder per experiment and 75 μl anti-FLAG beads. The complex was eluted by mixing the beads with 250 μl of 450 ng/μl 3×FLAG peptides in elution buffer for 2 h at 4°C. The elution step was repeated with an additional 250 μl of 450 ng/μl 3×FLAG peptides, for 1 h at 4°C. Roughly 450 μl of these eluates were applied to glycerol gradients. For the isolation of CORVET complexes containing Vps8b, Vps8c, Vps8d, Vps8e and Vps8f for SDS-PAGE and silver staining, we used 2 g of cryopowder per sample and 50 μl anti-FLAG beads. Complexes were eluted by mixing the beads with 150 μl of 450 ng/μl 3×FLAG peptides in elution buffer, for 2 h at 4°C. 100°C LDS sample buffer containing 40 mM DTT was added to 150 μl eluates prior to SDS-PAGE.
Immunoprecipitations from cell pellets
This protocol was used to immunoprecipitate FLAG- and mNeon-tagged Vps8 paralogs, Vps8a–GFP, Vps8c–2mCherry, Vps8c–3mCherry and Vps8e–3mCherry, prior to visualization by western blotting. Wild-type cells were processed in parallel as control. Cell cultures (50–100 ml) were grown overnight to 3×105 cells/ml, except for the 3mCherry-tagged Vps8c and Vps8e, which were grown to ∼7×105 cells/ml. Cells were washed once in 10 mM Tris-HCl pH 7.4, pelleted, resuspended in buffer B4 (20 mM Hepes pH 7.4, 250 mM NaCitrate, 1 mM MgCl2, 0.1% CHAPS and 1 mM DTT) supplemented with protease inhibitor cocktail tablets (Roche), and gently mixed for 1 h on ice. The lysates were cleared by centrifugation at 55,000 rpm (164,000 g; Beckman Instruments TLA-100.4 rotor) for 1.5 h at 4°C, and mixed with 20 μl antibody-conjugated beads, pre-washed with cold buffer B4, for 2 h at 4°C. Reagents used were: anti-FLAG beads (EZ view Red Anti-FLAG M2 Affinity Gel, Sigma), anti-c-Myc beads (Pierce anti-c-Myc magnetic beads, Thermo Fisher Scientific), and anti-HA beads (EZ view Red Anti-HA Affinity Gel, Sigma-Aldrich), for FLAG-, mNeon- and mCherry-tagged fusion proteins, respectively. To immunoprecipitate Vps8a–GFP, we used 120 μl anti-GFP beads (GFP-nAb agarose, Allele Biotechnology). Beads were washed five times with 20 mM Tris-HCl pH 7.4, 1 mM EDTA, 500 mM NaCl, 0.1% NP-40, 1 mM DTT and 10% glycerol, and resuspended in 50 μl of 100°C LDS sample buffer containing 40 mM DTT.
Glycerol gradient centrifugation
Continuous 11-ml glycerol gradients were made by layering 10%, 20%, 30% 40% (v/v) glycerol solutions in 14×89 mm tubes (Beckman Instruments). The glycerol solutions were prepared in 20 mM HEPES pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 0.1% CHAPS and 1 mM DTT. The tubes were gently laid on their side for 1.5 h, and then stood upright for overnight at 4°C. Protein samples were overlaid on the gradients and the tubes centrifuged for 18 h at 37,000 rpm (235,000 g) using a SW 41 Ti rotor (Beckman Instruments), at 4°C. Then 250-μl fractions were harvested and analyzed by western blotting and silver staining in the case of the 8A-CC sample, and by Coomassie Blue staining for the fractions containing standards sedimented in parallel (100 μg/ml of each thyroglobulin, yeast alcohol dehydrogenase and BSA were mixed and applied to the gradient). The size of the 8A-CC complex was estimated by plotting the molecular mass of the standards as a function of the corresponding peak fractions.
Western blotting
Protein samples were analyzed by western blotting as previously described (Sparvoli et al., 2018). Mouse mAb anti-GFP (BioLegend), mouse mAb anti-c-Myc (9E10, Sigma-Aldrich), rabbit anti-FLAG (Sigma-Aldrich), and mouse mAb anti-HA (HA.11, BioLegend) antibodies, were diluted 1:5000, 1:5000, 1:2000 and 1:2000, respectively, in blocking solution. Proteins were visualized with either anti-rabbit IgG (whole molecule) conjugated to horseradish peroxidase (Sigma) or ECL horseradish peroxidase-linked anti-mouse-IgG (NA931) (GE Healthcare Life Sciences) secondary antibody diluted 1:20,000, and SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific).
Silver staining
Proteins samples were loaded on 8% Tris-Glycine gels (Invitrogen) and stained with Pierce Silver Stain for Mass Spectrometry (Thermo Fisher Scientific), according to the manufacturer's instructions.
Mass spectrometry
Protein samples were loaded on a 4–20% gel for SDS-PAGE, allowed to migrate for ∼1 cm into the gel, and briefly stained with Coomassie Blue R-250 solution (0.1% w/v Coomassie, 10% acetic acid and 50% methanol). A single 1 cm gel slice per lane was excised from the Coomassie-stained gel, destained, and then subjected to tryptic digest and reductive alkylation. LC-MS/MS was performed by the Proteomic Facility at the University of Dundee. The samples were subjected to LC-MS/MS on a Ultimate3000 nano rapid separation LC system (Dionex) coupled to a LTQ Velos mass spectrometer (Thermo Fisher Scientific). Mass spectra were processed using the intensity-based label-free quantification (LFQ) method of MaxQuant version 1.6.1.0 (Cox et al., 2014; Cox and Mann, 2008) searching the T. thermophila annotated protein database from ciliate.org (Eisen et al., 2006; Stover et al., 2006). The minimum peptide length was set at six amino acids, isoleucine and leucine were considered indistinguishable, and false discovery rates (FDR) of 0.01 were calculated at the levels of peptides, proteins and modification sites based on the number of hits against the reversed sequence database. If the identified peptide sequence set of one protein contained the peptide set of another protein, these two proteins were assigned to the same protein group. Perseus (Tyanova et al., 2016) was used to calculate P-values applying t-test-based statistics and to draw statistical plots. Proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaíno et al., 2016) with the dataset identifier PXD014895.
Feeding with DsRed-bacteria
E. coli expressing DsRed-express2 (Strack et al., 2008) (a gift from Benjamin S. Glick, The University of Chicago, Chicago, IL) were grown in 15 ml LB broth, supplemented with ampicillin, and induced overnight at 37°C with 1 mM IPTG. Tetrahymena expressing Vps8b–mNeon or Vps8e–mNeon were grown in 20 ml SPP overnight to 4×105 cells/ml, and were washed and starved in 10 mM Tris buffer for 3 h. The starved Tetrahymena were incubated with 3% dsRed-expressing E. coli for 5 min at room temperature and then quickly washed by using 10 mM Tris buffer through low speed centrifugation (500 g for 1 min) to remove remaining E. coli prior to cell fixation
Fixed cell imaging
Cells (3×105) endogenously expressing Vps11–GFP, Vps3a–GFP alone or with Vps11–mCherry, or Vps8c–mCherry or mNeon-tagged Vps8 paralogs alone or in combination with mCherry-tagged Vps8c, Vps8e and Rab4b, were washed once with 10 mM Tris-HCl pH 7.4, and fixed with ice-cold 4% paraformaldehyde in 1× PBS for 30 min. Cells fed with dsRed-E.coli were collected by centrifugation, washed once with SPP and fixed. For the simultaneous localization of Vps8c–mNeon with either mCherry–Rab22a or mCherry–Rab7, the expression of the Rab GTPases was induced by incubating the cells with 1 μg/ml CdCl2 for 2 h in SPP, prior to fixation. The visualization of fusion proteins was not enhanced with immunolabeling. Cells were mounted with Trolox (1:1000) to inhibit bleaching and imaged at room temperature on a Marianas Yokogawa-type spinning disc inverted confocal microscope, with an 100× oil objective with NA=1.45, equipped with two photometrics Evolve back-thinned EM-CCD cameras, using Slidebook6 software (Zeiss, Intelligent Imaging Innovations, Denver, CO). Tetrahymena cells expressing Vps8b–mNeon and Vps8e–mNeon and used to quantify the position of Vps8b- and Vps8e-related structures, were imaged with a Carl Zeiss Microscope stand Axio Observer 7 system, with an 100× oil objective with NA=1.46, equipped with a Camera Axiocam 702 mono. z-stack images and z-projection images were denoised, adjusted in brightness and contrast, and colored with the program Fiji (Schindelin et al., 2012).
Live-cell imaging
Tetrahymena expressing the mNeon-tagged Vps8 paralogs, or co-expressing Vps8c–mNeon and mCherry–Rab7, were grown overnight to 1×105–2×105 cells/ml and transferred to S medium for 2 h prior to imaging. The expression of Rab7 was induced by adding 1 μg/ml CdCl2 to the S medium. Cells were immobilized in thin 3% low-melting-point agarose gel pads, as described previously (Kaur et al., 2017), and imaged within 15 min. z-stack images (12 stacks along the z-axis at 0.5 μm intervals) and time-lapse videos (30 frames at 1.24 s/interval for mNeon-tagged Vps8 paralogs shown in the main figure, and 300 frames at 0.17 s/interval for the additional full videos; 200 frames at 0.17 s/interval for Vps8c–mNeon with mCherry-Rab7) were collected at room temperature with a Marianas Yokogawa-type spinning disc inverted confocal microscope as above. Images and movies were denoised, and adjusted in brightness/contrast with the program Fiji (Schindelin et al., 2012). Images shown are single slices/frames for clarity. Videos of Vps8c-mNeon/mCherry-Rab7-expressing cells were created by simultaneously recording in two fluorescent channels, which were subsequently merged in a single multicolor movie using the program Fiji.
Colocalization analysis
To estimate the extent of colocalization, we calculated the Mander's coefficients M1 and M2 with the Fiji-JACoP plugin, as previously described (Sparvoli et al., 2018); 158, 139, 268 and 232 non-overlapping images/sample were analyzed to determine the correlations between Vps8a and Rab4b, Vps8c and Rab4b, Vps8c and Rab22a, and Vps8c andRab7, respectively; 399, 246 and 337 non-overlapping images/sample were used to calculate the coefficients M1 and M2 for Vps8a with Vps8c, Vps8a with Vps8e, and Vps8c with Vps8e, respectively. To determine the correlations between Vps8c–mNeon and Vps8c–mCherry, 261 non-overlapping images were analyzed to calculate the coefficients M1 and M2; 325 and 321 non-overlapping images/sample were used to calculate the overlap of Vps3a–GFP with either Vps11–mCherry or Vps8c–mCherry, respectively. Mander's coefficients M1 and M2 are reported as mean values.
Particle analysis
To estimate the number of fluorescent puncta in Vps8a–mNeon- and Vps8a–GFP-expressing cells, we used the Fiji tool ‘SpotCounter’, setting the tolerance noise to 1200–1500, and box size to 3. The plugin counts spots by detecting local maxima, which are accepted when the maximum is higher than a user-defined number (tolerance noise) over the average of the four corners of the box. The number of particles was calculated using maximum intensity z-projections, generated from z-stacks of 15 cells/sample, and it is reported as a mean value.
The estimation of the number, the size and the integrated density of particles for Vps8a, Vps8b, Vps8c, Vps8e and Vps8f, were obtained by using the Fiji tool “Analyze Particles” (http://imagej.nih.gov/ij/). The analysis was performed by using 143, 155, 144, 148 and 143 non-overlapping images for Vps8a, Vps8b, Vps8c, Vps8e and Vps8f, respectively, which were corrected for noise and analyzed by setting the threshold to the estimated background value, and then converted into ‘Mask’. The calculation was restricted to different area-based size ranges, selected on the base of the overall population size in Vps8-expressing cells, including particles between 0.1 and 2 μm2.
Transcription profiles
Gene expression profiles were downloaded from the Tetrahymena Functional Genomics Database (TFGD, http://tfgd.ihb.ac.cn/) (Miao et al., 2009; Xiong et al., 2011b). For plotting the graphs, each profile was normalized by setting the maximum expression level of the gene to 1.
Supplementary Material
Acknowledgements
At the University of Chicago, we thank Klaus Nielsen and Jon Staley for help with cryomilling, Matthew Sullivan and Alex Ruthenburg for help with FPLC, Aarthi Kuppannan for help with cell culture, and Vytas Bindokas for help at the Integrated Light Microscopy Core Facility. We thank Naomi Stover (Bradley University) for gene annotation, and Lev Tsypin (Caltech) for stimulating and entertaining discussion. We thank the FingerPrints Proteomic Facility at the University of Dundee for excellent technical support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: D.S., M.Z., M.C.F., A.P.T.; Methodology: D.S., M.Z., C.-Y.C.; Investigation: D.S., M.Z., C.-Y.C.; Data curation: M.Z.; Writing - original draft: D.S., A.P.T.; Writing - review & editing: M.Z., M.C.F., A.P.T.; Supervision: M.C.F., A.P.T.; Project administration: A.P.T.; Funding acquisition: M.C.F., A.P.T.
Funding
Work in APT's laboratory was supported by the National Institutes of Health (NIH) (GM105783) and in MCF's laboratory by a Wellcome Trust Investigator Award (204697/Z/16/Z). Deposited in PMC for release after 6 months.
Data availability
Mass spectrometry data on the CORVET pulldowns have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014895.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.238659.supplemental
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.238659.reviewer-comments.pdf
References
- Adl S. M., Leander B. S., Simpson A. G. B., Archibald J. M., Anderson O. R., Bass D., Bowser S. S., Brugerolle G., Farmer M. A., Karpov S. et al. (2007). Diversity, nomenclature, and taxonomy of protists. Syst. Biol. 56, 684-689. 10.1080/10635150701494127 [DOI] [PubMed] [Google Scholar]
- Adl S. M., Simpson A. G. B., Lane C. E., Lukeš J., Bass D., Bowser S. S., Brown M. W., Burki F., Dunthorn M., Hampl V. et al. (2012). The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59, 429-493. 10.1111/j.1550-7408.2012.00644.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akematsu T., Fukuda Y., Attiq R. and Pearlman R. E. (2014). Role of class III phosphatidylinositol 3-kinase during programmed nuclear death of Tetrahymena thermophila. Autophagy 10, 209-225. 10.4161/auto.26929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R. D. (2000). The contractile vacuole and its membrane dynamics. BioEssays 22, 1035-1042. [DOI] [PubMed] [Google Scholar]
- Asensio C. S., Sirkis D. W., Maas J. W. Jr, Egami K., To T.-L., Brodsky F. M., Shu X., Cheng Y. and Edwards R. H. (2013). Self-assembly of VPS41 promotes sorting required for biogenesis of the regulated secretory pathway. Dev. Cell 27, 425-437. 10.1016/j.devcel.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. W. and Hughson F. M. (2016). Chaperoning SNARE assembly and disassembly. Nat. Rev. Mol. Cell Biol. 17, 465-479. 10.1038/nrm.2016.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar H. J. and Ungermann C. (2013). CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J. Cell Sci. 126, 1307-1316. 10.1242/jcs.107805 [DOI] [PubMed] [Google Scholar]
- Balderhaar H. J., Lachmann J., Yavavli E., Brocker C., Lürick A. and Ungermann C. (2013). The CORVET complex promotes tethering and fusion of Rab5/Vps21-positive membranes. Proc. Natl. Acad. Sci. USA 110, 3823-3828. 10.1073/pnas.1221785110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bem D., Smith H., Banushi B., Burden J. J., White I. J., Hanley J., Jeremiah N., Rieux-Laucat F., Bettels R., Ariceta G. et al. (2015). VPS33B regulates protein sorting into and maturation of alpha-granule progenitor organelles in mouse megakaryocytes. Blood 126, 133-143. 10.1182/blood-2014-12-614677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright L. J., Kambesis N., Nelson S. B., Jeong B. and Turkewitz A. P. (2010). Comprehensive analysis reveals dynamic and evolutionary plasticity of Rab GTPases and membrane traffic in Tetrahymena thermophila. PLoS Genet. 6, e1001155 10.1371/journal.pgen.1001155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briguglio J. S., Kumar S. and Turkewitz A. P. (2013). Lysosomal sorting receptors are essential for secretory granule biogenesis in Tetrahymena. J. Cell Biol. 203, 537-550. 10.1083/jcb.201305086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocker C., Kuhlee A., Gatsogiannis C., Balderhaar H. J., Honscher C., Engelbrecht-Vandre S., Ungermann C. and Raunser S. (2012). Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc. Natl. Acad. Sci. USA 109, 1991-1996. 10.1073/pnas.1117797109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H.-T., Dukovski D., Chambers M. G., Reinisch K. M. and Walz T. (2016). CATCHR, HOPS and CORVET tethering complexes share a similar architecture. Nat. Struct. Mol. Biol. 23, 761-763. 10.1038/nsmb.3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. and Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367-1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N. and Mann M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513-2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Zhao Q., Gao C., Ding Y., Zeng Y., Ueda T., Nakano A. and Jiang L. (2014). Activation of the Rab7 GTPase by the MON1-CCZ1 complex is essential for PVC-to-vacuole trafficking and plant growth in arabidopsis. Plant Cell 26, 2080-2097. 10.1105/tpc.114.123141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinane A. R., Straatman-Iwanowska A., Zaucker A., Wakabayashi Y., Bruce C. K., Luo G., Rahman F., Gürakan F., Utine E., Özkan T. B. et al. (2010). Mutations in VIPAR cause an arthrogryposis, renal dysfunction and cholestasis syndrome phenotype with defects in epithelial polarization. Nat. Genet. 42, 303-312. 10.1038/ng.538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacks J. B. and Field M. C. (2007). Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J. Cell Sci. 120, 2977-2985. 10.1242/jcs.013250 [DOI] [PubMed] [Google Scholar]
- Dai J., Lu Y., Wang C., Chen X., Fan X., Gu H., Wu X., Wang K., Gartner T. K., Zheng J. et al. (2016). Vps33b regulates Vwf-positive vesicular trafficking in megakaryocytes. J. Pathol. 240, 108-119. 10.1002/path.4762 [DOI] [PubMed] [Google Scholar]
- Davis M. C., Ward J. G., Herrick G. and Allis C. D. (1992). Programmed nuclear death: apoptotic-like degradation of specific nuclei in conjugating Tetrahymena. Dev. Biol. 154, 419-432. 10.1016/0012-1606(92)90080-Z [DOI] [PubMed] [Google Scholar]
- Delevoye C., Hurbain I., Tenza D., Sibarita J.-B., Uzan-Gafsou S., Ohno H., Geerts W. J. C., Verkleij A. J., Salamero J., Marks M. S. et al. (2009). AP-1 and KIF13A coordinate endosomal sorting and positioning during melanosome biogenesis. J. Cell Biol. 187, 247-264. 10.1083/jcb.200907122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye C., Heiligenstein X., Ripoll L., Gilles-Marsens F., Dennis M. K., Linares R. A., Derman L., Gokhale A., Morel E., Faundez V. et al. (2016). BLOC-1 brings together the actin and microtubule cytoskeletons to generate recycling endosomes. Curr. Biol. 26, 1-13. 10.1016/j.cub.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis M. K., Mantegazza A. R., Snir O. L., Tenza D., Acosta-Ruiz A., Delevoye C., Zorger R., Sitaram A., de Jesus-Rojas W., Ravichandran K. et al. (2015). BLOC-2 targets recycling endosomal tubules to melanosomes for cargo delivery. J. Cell Biol. 209, 563-577. 10.1083/jcb.201410026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J. A., Coyne R. S., Wu M., Wu D., Thiagarajan M., Wortman J. R., Badger J. H., Ren Q., Amedeo P., Jones K. M. et al. (2006). Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4, e286 10.1371/journal.pbio.0040286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde N. C., Morgan G., Winey M., Sperling L. and Turkewitz A. P. (2005). Elucidation of clathrin-mediated endocytosis in tetrahymena reveals an evolutionarily convergent recruitment of dynamin. PLoS Genet. 1, e52 10.1371/journal.pgen.0010052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias M., Brighouse A., Gabernet-Castello C., Field M. C. and Dacks J. B. (2012). Sculpting the endomembrane system in deep time: high resolution phylogenetics of Rab GTPases. J. Cell Sci. 125, 2500-2508. 10.1242/jcs.101378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp N. and Ungermann C. (2013). The N-terminal domains of Vps3 and Vps8 are critical for localization and function of the CORVET tethering complex on endosomes. PLoS ONE 8, e67307 10.1371/journal.pone.0067307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel J. (2000). Cell biology of Tetrahymena thermophila. Methods Cell Biol. 62, 27-125. 10.1016/S0091-679X(08)61528-9 [DOI] [PubMed] [Google Scholar]
- Fratti R. A., Jun Y., Merz A. J., Margolis N. and Wickner W. (2004). Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J. Cell Biol. 167, 1087-1098. 10.1083/jcb.200409068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabernet-Castello C., O'Reilly A. J., Dacks J. B. and Field M. C. (2013). Evolution of Tre-2/Bub2/Cdc16 (TBC) Rab GTPase-activating proteins. Mol. Biol. Cell 24, 1574-1583. 10.1091/mbc.e12-07-0557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerst J. E. (1999). SNAREs and SNARE regulators in membrane fusion and exocytosis. Cell. Mol. Life Sci. 55, 707-734. 10.1007/s000180050328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimmler A., Korn R., de Vargas C., Audic S. and Stoeck T. (2016). The Tara Oceans voyage reveals global diversity and distribution patterns of marine planktonic ciliates. Sci. Rep. 6, 33555 10.1038/srep33555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissen P., Johnson C. A., Morgan N. V., Stapelbroek J. M., Forshew T., Cooper W. N., McKiernan P. J., Klomp L. W., Morris A. A. M., Wraith J. E. et al. (2004). Mutations in VPS33B, encoding a regulator of SNARE-dependent membrane fusion, cause arthrogryposis-renal dysfunction-cholestasis (ARC) syndrome. Nat. Genet. 36, 400-404. 10.1038/ng1325 [DOI] [PubMed] [Google Scholar]
- Gissen P., Johnson C. A., Gentle D., Hurst L. D., Doherty A. J., O'Kane C. J., Kelly D. A. and Maher E. R. (2005). Comparative evolutionary analysis of VPS33 homologues: genetic and functional insights. Hum. Mol. Genet. 14, 1261-1270. 10.1093/hmg/ddi137 [DOI] [PubMed] [Google Scholar]
- Guerrier S., Plattner H., Richardson E., Dacks J. B. and Turkewitz A. P. (2017). An evolutionary balance: conservation vs innovation in ciliate membrane trafficking. Traffic 18, 18-28. 10.1111/tra.12450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Johnston W., Kovtun O., Mureev S., Bröcker C., Ungermann C. and Alexandrov K. (2013). Subunit organisation of in vitro reconstituted HOPS and CORVET multisubunit membrane tethering complexes. PLoS ONE 8, e81534 10.1371/journal.pone.0081534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad A., Bowman G. R. and Turkewitz A. P. (2002). A new class of cargo protein in Tetrahymena thermophila dense core secretory granules. Eukaryot. Cell 1, 583-593. 10.1128/EC.1.4.583-593.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann K. (1996). Ciliates: Cells as Organisms. Stuttgart: Gustav FischerHausmann, K. [Google Scholar]
- Ho R. and Stroupe C. (2016). The HOPS/class C Vps complex tethers high-curvature membranes via a direct protein-membrane interaction. Traffic 17, 1078-1090. 10.1111/tra.12421 [DOI] [PubMed] [Google Scholar]
- Horazdovsky B. F., Cowles C. R., Mustol P., Holmes M. and Emr S. D. (1996). A novel RING finger protein, Vps8p, functionally interacts with the small GTPase, Vps21p, to facilitate soluble vacuolar protein localization. J. Biol. Chem. 271, 33607-33615. 10.1074/jbc.271.52.33607 [DOI] [PubMed] [Google Scholar]
- Hunter M., Scourfield E. J., Emmott E. and Graham S. C. (2017). VPS18 recruits VPS41 to the human HOPS complex via a RING-RING interaction. Biochem. J. 474, 3615-3626. 10.1042/BCJ20170588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter M. R., Hesketh G. G., Benedyk T. H., Gingras A.-C. and Graham S. C. (2018). Proteomic and Biochemical Comparison of the Cellular Interaction Partners of Human VPS33A and VPS33B. J. Mol. Biol. 430, 2153-2163. 10.1016/j.jmb.2018.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari J. and Helenius A. (2011). Endosome maturation. EMBO J. 30, 3481-3500. 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M. E., DeSouza L. V., Samaranayake H., Pearlman R. E., Siu K. W. M. and Klobutcher L. A. (2006). The Tetrahymena thermophila phagosome proteome. Eukaryot. Cell 5, 1990-2000. 10.1128/EC.00195-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonker C. T. H., Galmes R., Veenendaal T., ten Brink C., van der Welle R. E. N., Liv N., de Rooij J., Peden A. A., van der Sluijs P., Margadant C. et al. (2018). Vps3 and Vps8 control integrin trafficking from early to recycling endosomes and regulate integrin-dependent functions. Nat. Commun. 9, 792 10.1038/s41467-018-03226-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H., Sparvoli D., Osakada H., Iwamoto M., Haraguchi T. and Turkewitz A. P. (2017). An endosomal syntaxin and the AP-3 complex are required for formation and maturation of candidate lysosome-related secretory organelles (mucocysts) in Tetrahymena thermophila. Mol. Biol. Cell 28, 1551-1564. 10.1091/mbc.e17-01-0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke S., Langemeyer L., Kuhlee A., Schuback S., Raunser S., Ungermann C. and Kümmel D. (2017). Architecture and mechanism of the late endosomal Rab7-like Ypt7 guanine nucleotide exchange factor complex Mon1-Ccz1. Nat. Commun. 8, 14034 10.1038/ncomms14034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger C. M., Klute M. J. and Dacks J. B. (2013). Comparative genomic analysis of multi-subunit tethering complexes demonstrates an ancient pan-eukaryotic complement and sculpting in Apicomplexa. PLoS ONE 8, e76278 10.1371/journal.pone.0076278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlee A., Raunser S. and Ungermann C. (2015). Functional homologies in vesicle tethering. FEBS Lett. 589, 2487-2497. 10.1016/j.febslet.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Kümmel D. and Ungermann C. (2014). Principles of membrane tethering and fusion in endosome and lysosome biogenesis. Curr. Opin. Cell Biol. 29, 61-66. 10.1016/j.ceb.2014.04.007 [DOI] [PubMed] [Google Scholar]
- LaCava J., Jiang H. and Rout M. P. (2016). Protein complex affinity capture from cryomilled mammalian cells. J. Vis. Exp. 118, e54518 10.3791/54518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.-L. and Yao M.-C. (2012). Role of ATG8 and autophagy in programmed nuclear degradation in Tetrahymena thermophila. Eukaryot. Cell 11, 494-506. 10.1128/EC.05296-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo B., Li L., Gissen P., Christensen H., McKiernan P. J., Ye C., Abdelhaleem M., Hayes J. A., Williams M. D., Chitayat D. et al. (2005). Requirement of VPS33B, a member of the Sec1/Munc18 protein family, in megakaryocyte and platelet alpha-granule biogenesis. Blood 106, 4159-4166. 10.1182/blood-2005-04-1356 [DOI] [PubMed] [Google Scholar]
- Lobingier B. T. and Merz A. J. (2012). Sec1/Munc18 protein Vps33 binds to SNARE domains and the quaternary SNARE complex. Mol. Biol. Cell 23, 4611-4622. 10.1091/mbc.e12-05-0343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobingier B. T., Nickerson D. P., Lo S.-Y. and Merz A. J. (2014). SM proteins Sly1 and Vps33 co-assemble with Sec17 and SNARE complexes to oppose SNARE disassembly by Sec18. eLife 3, e02272 10.7554/eLife.02272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz P., Lakatos Z., Varga A., Maruzs T., Simon-Vecsei Z., Darula Z., Benko P., Csordas G., Lippai M., Ando I. et al. (2016). MiniCORVET is a Vps8-containing early endosomal tether in Drosophila. eLife 5, e14226 10.7554/eLife.14226.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lürick A., Gao J., Kuhlee A., Yavavli E., Langemeyer L., Perz A., Raunser S. and Ungermann C. (2017). Multivalent Rab interactions determine tether-mediated membrane fusion. Mol. Biol. Cell 28, 322-332. 10.1091/mbc.e16-11-0764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Field M. C., Goodson H. V., Malik H. S., Pereira-Leal J. B., Roos D. S., Turkewitz A. P. and Sazer S. (2014). Evolutionary cell biology: two origins, one objective. Proc. Natl. Acad. Sci. USA 111, 16990-16994. 10.1073/pnas.1415861111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markgraf D. F., Ahnert F., Arlt H., Mari M., Peplowska K., Epp N., Griffith J., Reggiori F. and Ungermann C. (2009). The CORVET subunit Vps8 cooperates with the Rab5 homolog Vps21 to induce clustering of late endosomal compartments. Mol. Biol. Cell 20, 5276-5289. 10.1091/mbc.e09-06-0521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I. and Yarden Y. (2013). Endocytosis and cancer. Cold Spring Harbor Perspect. Biol. 5, a016949 10.1101/cshperspect.a016949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao W., Xiong J., Bowen J., Wang W., Liu Y., Braguinets O., Grigull J., Pearlman R. E., Orias E. and Gorovsky M. A. (2009). Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS ONE 4, e4429 10.1371/journal.pone.0004429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris C., Foster O. K., Handa S., Peloza K., Voss L., Somhegyi H., Jian Y., Vo M. V., Harp M., Rambo F. M. et al. (2018). Function and regulation of the Caenorhabditis elegans Rab32 family member GLO-1 in lysosome-related organelle biogenesis. PLoS Genet. 14, e1007772 10.1371/journal.pgen.1007772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefjes J. and van der Kant R. (2014). Stuck in traffic: an emerging theme in diseases of the nervous system. Trends Neurosci. 37, 66-76. 10.1016/j.tins.2013.11.006 [DOI] [PubMed] [Google Scholar]
- Nickerson D. P., Brett C. L. and Merz A. J. (2009). Vps-C complexes: gatekeepers of endolysosomal traffic. Curr. Opin. Cell Biol. 21, 543-551. 10.1016/j.ceb.2009.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J. R. (1979). Phagotrophy in tetrahymena. In Biochemistry and Physiology of Protozoa, Vol. 2 (ed. Levandowsky M. and Hutner S. H.), pp. 339-379. New York: Academic Press. [Google Scholar]
- Nilsson J. R. and Van Deurs B. (1983). Coated pits and pinocytosis in Tetrahymena. J. Cell Sci. 63, 209-222. [DOI] [PubMed] [Google Scholar]
- Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S. and Ungermann C. (2010). The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20, 1654-1659. 10.1016/j.cub.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Obado S. O., Field M. C., Chait B. T. and Rout M. P. (2016). High-efficiency isolation of nuclear envelope protein complexes from trypanosomes. Methods Mol. Biol. 1411, 67-80. 10.1007/978-1-4939-3530-7_3 [DOI] [PubMed] [Google Scholar]
- Oeffinger M., Wei K. E., Rogers R., DeGrasse J. A., Chait B. T., Aitchison J. D. and Rout M. P. (2007). Comprehensive analysis of diverse ribonucleoprotein complexes. Nat. Methods 4, 951-956. 10.1038/nmeth1101 [DOI] [PubMed] [Google Scholar]
- Orias E., Cervantes M. D. and Hamilton E. P. (2011). Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res. Microbiol. 162, 578-586. 10.1016/j.resmic.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr A., Song H., Rusin S. F., Kettenbach A. N. and Wickner W. (2017). HOPS catalyzes the interdependent assembly of each vacuolar SNARE into a SNARE complex. Mol. Biol. Cell 28, 975-983. 10.1091/mbc.e16-10-0743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowicz C. W., Bröcker C., Ahnert F., Nordmann M., Lachmann J., Peplowska K., Perz A., Auffarth K., Engelbrecht-Vandré S. and Ungermann C. (2010). Defined subunit arrangement and rab interactions are required for functionality of the HOPS tethering complex. Traffic 11, 1334-1346. 10.1111/j.1600-0854.2010.01097.x [DOI] [PubMed] [Google Scholar]
- Peplowska K., Markgraf D. F., Ostrowicz C. W., Bange G. and Ungermann C. (2007). The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev. Cell 12, 739-750. 10.1016/j.devcel.2007.03.006 [DOI] [PubMed] [Google Scholar]
- Perini E. D., Schaefer R., Stöter M., Kalaidzidis Y. and Zerial M. (2014). Mammalian CORVET is required for fusion and conversion of distinct early endosome subpopulations. Traffic 15, 1366-1389. 10.1111/tra.12232 [DOI] [PubMed] [Google Scholar]
- Plattner H. (2010). Membrane trafficking in protozoa SNARE proteins, H+-ATPase, actin, and other key players in ciliates. Int. Rev. Cell Mol. Biol. 280, 79-184. 10.1016/S1937-6448(10)80003-6 [DOI] [PubMed] [Google Scholar]
- Plattner H. (2015). The contractile vacuole complex of protists--new cues to function and biogenesis. Crit. Rev. Microbiol. 41, 218-227. 10.3109/1040841X.2013.821650 [DOI] [PubMed] [Google Scholar]
- Plemel R. L., Lobingier B. T., Brett C. L., Angers C. G., Nickerson D. P., Paulsel A., Sprague D. and Merz A. J. (2011). Subunit organization and Rab interactions of Vps-C protein complexes that control endolysosomal membrane traffic. Mol. Biol. Cell 22, 1353-1363. 10.1091/mbc.e10-03-0260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulipparacharuvil S., Akbar M. A., Ray S., Sevrioukov E. A., Haberman A. S., Rohrer J. and Kramer H. (2005). Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J. Cell Sci. 118, 3663-3673. 10.1242/jcs.02502 [DOI] [PubMed] [Google Scholar]
- Rogerson C. and Gissen P. (2018). VPS33B and VIPAR are essential for epidermal lamellar body biogenesis and function. Biochim. Biophys Acta Mol. Basis Dis. 1864, 1609-1621. 10.1016/j.bbadis.2018.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito-Nakano Y., Nakahara T., Nakano K., Nozaki T. and Numata O. (2010). Marked amplification and diversification of products of ras genes from rat brain, Rab GTPases, in the ciliates Tetrahymena thermophila and Paramecium tetraurelia. J. Eukaryot. Microbiol. 57, 389-399. 10.1111/j.1550-7408.2010.00503.x [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. L., Nickerson D. P., Lobingier B. T., Plemel R. L., Duan M., Angers C. G., Zick M. and Merz A. J. (2017). Sec17 (alpha-SNAP) and an SM-tethering complex regulate the outcome of SNARE zippering in vitro and in vivo. eLife 6, e27396 10.7554/eLife.27396.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger J. A. and Spang A. (2013). Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J. 280, 2743-2757. 10.1111/febs.12151 [DOI] [PubMed] [Google Scholar]
- Solinger J. A. and Spang A. (2014). Loss of the Sec1/Munc18-family proteins VPS-33.2 and VPS-33.1 bypasses a block in endosome maturation in Caenorhabditis elegans. Mol. Biol. Cell 25, 3909-3925. 10.1091/mbc.e13-12-0710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A. (2016). Membrane tethering complexes in the endosomal system. Front. Cell Dev. Biol. 4, 35 10.3389/fcell.2016.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparvoli D., Richardson E., Osakada H., Lan X., Iwamoto M., Bowman G. R., Kontur C., Bourland W. A., Lynn D. H., Pritchard J. K. et al. (2018). Remodeling the specificity of an endosomal CORVET tether underlies formation of regulated secretory vesicles in the ciliate tetrahymena thermophila. Curr. Biol. 28, 697-710.e13. 10.1016/j.cub.2018.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover N. A., Krieger C. J., Binkley G., Dong Q., Fisk D. G., Nash R., Sethuraman A., Weng S. and Cherry J. M. (2006). Tetrahymena genome database (TGD): a new genomic resource for Tetrahymena thermophila research. Nucleic Acids Res. 34, D500-D503. 10.1093/nar/gkj054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack R. L., Strongin D. E., Bhattacharyya D., Tao W., Berman A., Broxmeyer H. E., Keenan R. J. and Glick B. S. (2008). A noncytotoxic DsRed variant for whole-cell labeling. Nat. Methods 5, 955-957. 10.1038/nmeth.1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroupe C., Collins K. M., Fratti R. A. and Wickner W. (2006). Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 25, 1579-1589. 10.1038/sj.emboj.7601051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K., Ebine K., Askani J. C., Krüger F., Gonzalez Z. A., Ito E., Goh T., Schumacher K., Nakano A. and Ueda T. (2018). Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis. Proc. Natl. Acad. Sci. USA 115, E2457-E2466. 10.1073/pnas.1717839115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornieri K., Zlatic S. A., Mullin A. P., Werner E., Harrison R., L'Hernault S. W. and Faundez V. (2013). Vps33b pathogenic mutations preferentially affect VIPAS39/SPE-39-positive endosomes. Hum. Mol. Genet. 22, 5215-5228. 10.1093/hmg/ddt378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M. and Cox J. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731-740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- van der Beek J., Jonker C., van der Welle R., Liv N. and Klumperman J. (2019). CORVET, CHEVI and HOPS - multisubunit tethers of the endo-lysosomal system in health and disease. J. Cell Sci. 132, jcs189134 10.1242/jcs.189134 [DOI] [PubMed] [Google Scholar]
- Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al. (2016). 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 11033 10.1093/nar/gkw880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang Y., Sheng Y., Huang J., Chen X., Al-Rasheid K. A. S. and Gao S. (2017). A comparative study of genome organization and epigenetic mechanisms in model ciliates, with an emphasis on Tetrahymena, Paramecium and Oxytricha. Eur. J. Protistol. 61, 376-387. 10.1016/j.ejop.2017.06.006 [DOI] [PubMed] [Google Scholar]
- Warren A., Patterson D. J., Dunthorn M., Clamp J. C., Achilles-Day U. E. M., Aescht E., Al-Farraj S. A., Al-Quraishy S., Al-Rasheid K., Carr M. et al. (2017). Beyond the “Code”: a guide to the description and documentation of biodiversity in ciliated protists (Alveolata, Ciliophora). J. Eukaryot. Microbiol. 64, 539-554. 10.1111/jeu.12391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisse T. (2017). Functional diversity of aquatic ciliates. Eur. J. Protistol. 61, 331-358. 10.1016/j.ejop.2017.04.001 [DOI] [PubMed] [Google Scholar]
- Wurmser A. E., Sato T. K. and Emr S. D. (2000). New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 151, 551-562. 10.1083/jcb.151.3.551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J., Lu X., Lu Y., Zeng H., Yuan D., Feng L., Chang Y., Bowen J., Gorovsky M., Fu C. et al. (2011a). Tetrahymena gene expression database (TGED): a resource of microarray data and co-expression analyses for Tetrahymena. Sci. China Life Sci. 54, 65-67. 10.1007/s11427-010-4114-1 [DOI] [PubMed] [Google Scholar]
- Xiong J., Yuan D., Fillingham J. S., Garg J., Lu X., Chang Y., Liu Y., Fu C., Pearlman R. E. and Miao W. (2011b). Gene network landscape of the ciliate Tetrahymena thermophila. PLoS ONE 6, e20124 10.1371/journal.pone.0020124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J., Lu Y., Feng J., Yuan D., Tian M., Chang Y., Fu C., Wang G., Zeng H. and Miao W. (2013). Tetrahymena functional genomics database (TetraFGD): an integrated resource for Tetrahymena functional genomics. Database 2013, bat008 10.1093/database/bat008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingel P., Agasild H., Karus K., Buholce L. and Nõges T. (2019). Importance of ciliates as food for fish larvae in a shallow sea bay and a large shallow lake. Eur. J. Protistol. 67, 59-70. 10.1016/j.ejop.2018.10.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.