Abstract
Background
Animal and in vitro experiments implicate the Wnt pathway in cardiac development, fibrosis, vascular calcification, and atherosclerosis, but research in humans is lacking. We examined peripheral blood Wnt pathway gene expression and arterial stiffness in 369 healthy African ancestry men (mean age, 64 years).
Methods and Results
Gene expression was assessed using a custom Nanostring nCounter gene expression panel (N=43 genes) and normalized to housekeeping genes and background signal. Arterial stiffness was assessed via brachial‐ankle pulse‐wave velocity. Fourteen Wnt genes showed detectable expression and were tested individually as predictors of pulse‐wave velocity using linear regression, adjusting for age, height, weight, blood pressure, medication use, resting heart rate, current smoking, alcohol intake, and sedentary lifestyle. Adenomatous polyposis coli regulator of Wnt signaling pathway (APC), glycogen synthase kinase 3β (GSK3B), and transcription factor 4 (TCF4) were significantly associated with arterial stiffness (P<0.05 for all). When entered into a single model, APC and TCF4 expression remained independently associated with arterial stiffness (P=0.04 and 0.003, respectively), and each explained ≈3% of the variance in pulse‐wave velocity.
Conclusions
The current study establishes a novel association between in vivo expression of the Wnt pathway genes, APC and TCF4, with arterial stiffness in African ancestry men, a population at high risk of hypertensive vascular disease.
Keywords: African ancestry, arterial stiffness, gene expression, Wnt pathway
Subject Categories: Gene Expression & Regulation, Vascular Disease, Epidemiology
Clinical Perspective
What Is New?
This is the first study to test the association between broad Wnt pathway gene expression and subclinical cardiovascular disease measures in a population‐based sample.
We found that expression of 2 Wnt genes (APC and TCF4) in peripheral blood samples showed association with pulse‐wave velocity independent of traditional cardiovascular disease risk factors.
Because pulse‐wave velocity is a marker of several changes in the arterial wall, such as cellular hyperplasia, fibrosis, and calcification, further research is needed to determine in what capacity Wnt signaling may be functioning in arterial stiffness and cardiovascular disease.
What Are the Clinical Implications?
These findings in humans add further evidence to basic research experiments that have explored the role of Wnt signaling in the vasculature.
The Wnt pathway gene expression profile, and in particular expression of APC and TCF4, may have clinical application as biomarkers for identifying individuals at risk of cardiovascular disease and/or progression.
Also, because Wnt signaling has many regulatory roles, any therapeutic targets of the Wnt pathway should also define its potential impact on the cardiovascular system.
Introduction
Experiments in animals and in vitro models implicate the Wnt pathway in cardiac development,1, 2, 3 vascular calcification,4, 5, 6 and atherosclerosis.7, 8, 9 Fibrosis and large, dense calcifications occur in the vessel wall10 to stabilize atherosclerotic plaques and in the medial layer of the artery as an early hallmark of the arteriosclerotic process, manifesting as arterial stiffening.8 Previous studies have implicated Wnt pathway genes and proteins in vascular fibrosis and/or calcification, including Dkk1 (dickopf 1),11 Wnt2,12 Wnt3a,12, 13, 14 Wnt5a,12 Wnt7a,13, 15 and Wnt7b.11, 12, 13, 15 It is not currently feasible to measure these pathological features of the vessel wall tissue in population‐based studies; therefore, noninvasive, subclinical cardiovascular disease (CVD) measures, such as arterial pulse‐wave velocity (PWV), are typically used as proxies for these underlying pathological features.16, 17
Despite the wealth of in vitro data, studies on the Wnt pathway and CVD in humans have remained sparse and considered only a few of the many components of the complex Wnt signaling pathway. For example, the existing studies on the Wnt pathway and arterial stiffness are focused on protein‐based assays of circulating Dkk118, 19, 20 and sclerostin.19, 20, 21, 22, 23 These studies have generally found a significant association with arterial stiffness, but research is still lacking on many facets of the Wnt pathway, particularly in individuals of African ancestry who are at high risk of hypertensive vascular disease.24 In the current study, we comprehensively assessed Wnt pathway gene expression in peripheral blood and arterial stiffness in African Caribbean men who have a high burden of hypertension and arterial stiffening.25
Methods
Tobago Health Study
The data that support the findings of this study are available from the corresponding author on reasonable request. All men included in this analysis were from the THS (Tobago Health Study), a population‐based, prospective cohort study of community‐dwelling men aged ≥40 years, residing on the Caribbean island of Tobago.26 Participants for the THS were recruited without regard to health status, and men were eligible if they were ambulatory, not terminally ill, and without a bilateral hip replacement. Men from Tobago are of African ancestry, with low European admixture (<6%).27 A follow‐up visit for the THS, conducted from 2014 to 2017, included assessment of arterial stiffness using brachial‐ankle PWV. At this visit, men underwent assessment for PWV (N=795), and a random subset of 368 men were selected to have Wnt pathway gene expression measured and form the basis for the current analysis. Standard clinical examinations and interviewer‐administered health history questionnaires were administered at all THS visits. Written informed consent was obtained from each man using forms and procedures approved by the University of Pittsburgh Institutional Review Board, the US Surgeon General's Human Use Review Board, and the Tobago Division of Health and Social Services Institutional Review Board.
Brachial‐Ankle PWV
Arterial stiffness was measured as brachial‐ankle PWV using a noninvasive, waveform analyzer (Colin‐VP1000; Omron, Japan/WaveNexus, TX), as previously described.28 Briefly, after 10 minutes of rest in a supine position, occlusion/monitoring cuffs were placed at standardized locations on lightly clothed arms and bare ankles. Then, ECG electrodes were placed on the arms and ankles, while a phonocardiogram was placed on the left edge of the sternum to audibly detect heart sounds. The cuffs were connected to a plethysmographic sensor and an oscillometric pressure sensor, which recorded volume pressure waveforms and blood pressure, respectively. The volume pressure waveforms were collected at the arm (brachial artery) and ankle (tibial artery) over a total sampling time of 10 seconds, with automatic gain analysis and quality adjustment. Brachial‐ankle PWV was calculated as follows: (distance between arterial sites in cm)/(time between the foot of the waveforms in s). The distance was measured using a height‐based formula17 that accounts for the opposite direction of blood flow by subtracting the distance from the brachial artery to the heart. PWV was measured twice on both the right and left sides, and mean PWV was used in this analysis as the average between the 2 left and 2 right PWV measures. Although brachial‐ankle PWV is considered a mixed measure of arterial stiffness because it takes into account both central and peripheral arteries, it has been shown to be more highly correlated with central PWV29 than peripheral PWV,30 and is also predictive of pulse‐pressure31 and cardiovascular events.28, 32
Other Characteristics
Demographic, health history, and anthropomorphic characteristics were assessed by trained staff using interview and clinical examinations. Body weight was measured to the nearest 0.1 kg on a balance beam scale and standing height was measured to the nearest 0.1 cm using a wall‐mounted stadiometer, both without participants wearing shoes. Body mass index was calculated as body weight in kilograms divided by standing height in meters squared. Blood pressure was measured 3 times in a seated position with 10 minutes of rest in between readings using an automated sphygmomanometer (Omron). For this analysis, we present the mean of the second and third readings as average systolic and diastolic blood pressures (SBP and DBP, respectively). Current use of antihypertension medications was assessed at the time of participant interview. Hypertension was defined as mean SBP ≥140 mm Hg, DBP ≥90 mm Hg, or self‐reported use of antihypertension medication. Current self‐reported use of lipid‐lowering therapies was assessed at the time of participant interview. Smoking status was classified as current or not. Alcohol consumption is limited in this cohort sample and was, therefore, coded as consuming >3 drinks per week (yes/no) to identify individuals with greater than average cohort alcohol intake. We modeled sedentary behavior as usually watching ≥14 hours of TV per week versus not.
Nanostring nCounter Analysis of Wnt Pathway Gene Expression
Because there have been no previous studies of Wnt pathway gene expression in humans, Wnt pathway genes (Table S1) were selected for inclusion on a custom gene expression panel based on expression bioinformatics databases, such as the Gene Expression Atlas (http://www.ebi.ac.uk), and required expression in the vasculature and/or atherosclerotic plaques. We also performed a thorough literature review, including review of model organism and in vitro experiment studies, to select Wnt pathway genes that were most likely to be associated with CVD (N=43 selected genes). Gene expression was measured using a custom CodeSet for the selected Wnt genes in the Nanostring nCounter Analysis System (NanoString Technologies, Seattle, WA).33 The NanoString technology uses a digital color‐coded barcode tag with single‐molecule imaging that can detect and count hundreds of unique transcripts per reaction. The correlation between technical replicates using the nCounter system is 0.999, and it has coefficients of variation between 3% and 15%, depending on the number of mRNA transcripts per cell. In addition to the 43 Wnt pathway genes, we included 3 reference or housekeeping genes (Table S1) to control for background signal.
RNA Extraction and Quantification of Gene Expression
Peripheral blood was drawn at the time of the visit into PAXgene Blood RNA collection tubes (PreAnalytiX, Hombrechtikon, Switzerland), according to manufacturer's protocol. Samples were frozen at −80°C and shipped to the University of Pittsburgh overnight on dry ice. Samples were stored at −80°C until extraction and isolation, which was conducted using the PAXgene Blood RNA kit (PreAnalytiX), according to the manufacturer's protocol on the whole peripheral blood samples. No cellular fractionation was performed. All extracted samples were assessed for RNA sample quality and concentration using DropSense96 quantification (Trinean NV/SA, Gentbrugge, Belgium). For the nCounter gene expression analysis, 100 ng of total mRNA was aliquoted at 20 ng/μL for each participant. The data resulting from the nCounter system are expressed as number of molecules (ie, counts), per standardized input of sample. The resulting Wnt gene expression counts were normalized to 3 housekeeping genes (Table S1) after removing the background signal using an automated feature of the nSolver software (NanoString Technologies). Only genes with detectable expression in the peripheral blood were used in the analysis.
Statistical Analysis
Brachial‐ankle PWV and Wnt gene expression counts were assessed for normality and transformed to a normal distribution using a log transformation, as needed. Means and SDs or frequencies were calculated for covariates and gene expression counts in the total sample and stratified by high versus low PWV status, on the basis of splitting the sample at the median PWV (1586 cm/s). Differences in characteristics between high and low PWV status were tested with t tests, Wilcoxon rank‐sum tests, or χ2 tests, as appropriate.
We used multiple linear regression to test the association of individual Wnt gene expression counts (predictors) with PWV (outcome) and reported as the effect on PWV per 1‐SD greater expression. We provide unadjusted, age‐adjusted, and fully adjusted models that adjusted for age, height, weight, SBP, DBP, antihypertensive medication use, resting heart rate, lipid‐lowering medication use, current smoking, alcohol use, and sedentary behavior. All analyses were performed using SAS, version 9.4 (Cary, NC).
To determine if there was underlying structure in the Wnt pathway gene expression profile of these men, we also conducted exploratory factor analysis using maximum likelihood methods with direct varimax rotation using proc factor in SAS. We retained the 3‐factor solution based on model fit statistics and required gene expressions to load at ≥|0.3| to be included in any factor. Similar to the individual Wnt gene expression analysis, we used multiple linear regression to test the association of individual Wnt gene expression factors (predictors) with PWV (outcome) and reported results as the effect on PWV per 1‐SD greater Wnt gene expression factor. We provide unadjusted, age‐adjusted, and fully adjusted models that adjusted for age, height, weight, SBP, DBP, antihypertensive medication use, heart rate, lipid‐lowering medication use, current smoking, alcohol use, and sedentary behavior.
Results
Study Characteristics
Men were aged 63.8 years on average (range, 51–89 years; Table 1) and 65% had hypertension. Mean brachial‐ankle PWV was 1586 cm/s (interquartile range, 1388–1841 cm/s). Men with high PWV based on a median split were older, had higher blood pressures and heart rate, and were more likely to be using lipid‐lowering medications compared with those with low PWV (all P<0.01). However, they also tended to be shorter and weigh less (both P<0.02; Table 1).
Table 1.
Characteristics of the Study Population by Median Arterial Stiffness
| Characteristics | Overall (N=368) | Low PWV (N=183) | High PWV (N=185) | P Valuea |
|---|---|---|---|---|
| Age, y | 63.8±8.5 | 60.4±6.8 | 67.1±8.7 | <0.001 |
| Height, cm | 175.2±7.0 | 176.6±6.7 | 173.8±6.9 | <0.001 |
| Weight, kg | 84.7±15.1 | 86.5±15.6 | 82.8±14.3 | 0.018 |
| BMI, kg/m2 | 27.5±4.4 | 27.7±4.5 | 27.4±4.3 | 0.584 |
| SBP, mm Hg | 142.8±21.7 | 134.4±17.6 | 151.4±22.1 | <0.001 |
| DBP, mm Hg | 79.7±12.0 | 77.2±10.4 | 82.4±13.0 | <0.001 |
| Hypertension, % | 65.3 | 48.1 | 83.1 | <0.001 |
| HR, bpm | 67.5±12.5 | 64.0±11.1 | 71.0±12.8 | <0.001 |
| Current smoking, % | 6.2 | 8.1 | 4.4 | 0.139 |
| Consume >4 alcohol drinks/wk, % | 14.4 | 11.9 | 16.9 | 0.168 |
| Watch >14 h of TV/wk, % | 51.1 | 46.7 | 55.7 | 0.085 |
| Antihypertensive medication use, % | 43.1 | 32.4 | 54.1 | <0.001 |
| Cholesterol‐lowering medication use, % | 15.7 | 10.3 | 20.8 | 0.005 |
| PWV, cm/sa | 1586 (1388–1841) | 1388 (1286–1478) | 1841 (1720–2046) | NA |
Characteristics shown as mean±SD or percentage, as appropriate, and as median (interquartile range) for PWV. BMI indicates body mass index; bpm, beats per minute; DBP, diastolic blood pressure; HR, heart rate; NA, not applicable; PWV, pulse‐wave velocity; SBP, systolic blood pressure.
Unadjusted P values shown from t tests of the difference in participant characteristics by median split PWV.
Wnt Pathway Peripheral Blood Gene Expression
Of the 43 tested Wnt genes, 14 were detected in peripheral blood samples (Table 2; Table S2). Genes with the greatest expression counts included the following: β catenin 1 (CTNNB1; mean count, 1270), transcription factor 7 (TCF7; mean count, 984), lymphoid enhancer binding factor 1 (LEF1; mean count, 910), glycogen synthase kinase 3β (GSK3B; mean count, 853), and axin 1 (AXIN1; mean count, 499). In unadjusted analyses, men with high PWV based on a median split had greater expression of adenomatous polyposis coli regulator of Wnt signaling pathway (APC), AXIN1, cadherin 2 (CDH2), CTNNB1, and GSK3B, and lower expression of axin 2 (AXIN2) and transcription factor 4 (TCF4; all P<0.05; Table 2) compared with men with low PWV.
Table 2.
Distribution of Normalized Wnt Gene Expression Counts and Exploratory Factors by Median Arterial Stiffness
| Wnt Gene | Overall (N=368) | Low PWV (N=185) | High PWV (N=183) | Difference, % | P Valuea |
|---|---|---|---|---|---|
| APC, counts | 157.4±35.6 | 152.5±33.7 | 162.2±36.8 | 6.2 | 0.012 |
| AXIN1, counts | 499.2±93.4 | 487.3±83.8 | 511.2±101.2 | 4.8 | 0.047 |
| AXIN2, counts | 82.7±33.7 | 85.6±31.8 | 79.9±35.5 | −6.9 | 0.012 |
| CDH2, counts | 18.7±6.3 | 18.2±6.8 | 19.2±5.7 | 5.3 | 0.018 |
| CTNNB1, counts | 1269.5±152.8 | 1243.2±146.9 | 1296.1±154.8 | 4.2 | 0.008 |
| FZD1, counts | 26.3±9.0 | 26.0±8.7 | 26.4±9.3 | 1.5 | 0.780 |
| FZD6, counts | 20.2±6.0 | 19.9±5.6 | 20.5±6.3 | 3.0 | 0.299 |
| GSK3B, counts | 852.6±159.0 | 825.4±147.1 | 879.7±166.5 | 6.4 | 0.005 |
| KREMEN1, counts | 119.2±89.2 | 116.3±85.6 | 121.6±92.8 | 4.4 | 0.397 |
| LEF1, counts | 910.2±318.2 | 925.7±302.1 | 895.0±334.5 | −3.4 | 0.102 |
| SFRP2, counts | 19.9±8.8 | 19.6±9.8 | 20.2±7.7 | 3.0 | 0.061 |
| TCF4, counts | 110.5±39.5 | 116.1±43.9 | 104.8±33.6 | −10.2 | 0.009 |
| TCF7, counts | 984.3±318.5 | 994.8±312.5 | 975.1±325.3 | −2.0 | 0.248 |
| WNT16, counts | 20.3±14.0 | 21.1±19.1 | 19.4±5.4 | −8.4 | 0.439 |
Characteristics shown as mean±SD and as the percentage difference in Wnt gene expression counts in high compared with low PWV men, as categorized by a median split. APC indicates adenomatous polyposis coli regulator of Wnt signaling pathway; AXIN, axin; CDH2, cadherin 2; CTNNB1, catenin β 1; FZD, frizzled class receptor; GSK3B, glycogen synthase kinase 3β; KREMEN1, kringle‐containing transmembrane protein 1; LEF1, lymphoid enhancer binding factor 1; PWV, pulse‐wave velocity; SFRP2, secreted frizzled related protein 2; TCF, transcription factor; WNT16, Wnt family member 16.
Unadjusted P values from Wilcoxon nonparametric tests of a difference not equal to 0.
Wnt Pathway Gene Expression and Arterial Stiffness
In age‐adjusted models, men with high PWV had greater expression of APC and lower expression of AXIN2, LEF1, and TCF4 (all P<0.05; Table 3). After additional adjustment for height, weight, SBP, DBP, antihypertensive medication use, heart rate, lipid‐lowering medication use, current smoking, alcohol use, and sedentary behavior, both APC and TCF4 expression remained significantly associated with PWV in opposite directions (P=0.01 for both; APC positive direction, TCF4 negative direction). In addition, greater GSK3B expression was associated with greater PWV (P=0.048). When APC, TCF4, and GSK3B were entered together in the fully adjusted model, only APC and TCF4 remained significantly associated with PWV (Table 4). Specifically, a 1‐SD greater expression of APC was associated with 44.4 cm/s greater PWV (P=0.035), while at the same time, a 1‐SD lower expression of TCF4 was associated with 43.2 cm/s greater PWV (P=0.003). The effects of APC and TCF4 expression were similar in magnitude to a ≈3‐year higher or lower age, respectively.
Table 3.
Association of Individual Wnt Gene Expression Counts With Arterial Stiffness
| Wnt Gene | Unadjusted | Age Adjusted | Fully Adjusteda | |||
|---|---|---|---|---|---|---|
| β, cm/s | P Value | β, cm/s | P Value | β, cm/s | P Value | |
| APC | 68.7 | <0.001 | 41.7 | 0.032 | 42.3 | 0.010 |
| AXIN1 | 35.7 | 0.051 | 6.2 | 0.709 | 10.3 | 0.463 |
| AXIN2 | −51.1 | 0.003 | −35.3 | 0.020 | −8.9 | 0.487 |
| CDH2 | 33.4 | 0.055 | 15.6 | 0.289 | 2.3 | 0.766 |
| CTNNB1 | 46.7 | 0.016 | 10.3 | 0.655 | 18.9 | 0.235 |
| FZD1 | 23.4 | 0.247 | 5.7 | 0.831 | 9.7 | 0.548 |
| FZD6 | 23.4 | 0.233 | 18.1 | 0.308 | 14.4 | 0.344 |
| GSK3B | 63.2 | 0.002 | 29.4 | 0.157 | 33.2 | 0.048 |
| KREMEN1 | 41.8 | 0.037 | 17.5 | 0.397 | 22.6 | 0.179 |
| LEF1 | −63.2 | <0.001 | −34.8 | 0.049 | −11.3 | 0.600 |
| SFRP2 | 18.0 | 0.294 | −0.5 | 0.967 | −1.4 | 0.995 |
| TCF4 | −73.1 | <0.001 | −61.2 | <0.001 | −37.7 | 0.007 |
| TCF7 | −59.8 | 0.002 | −33.3 | 0.070 | −9.7 | 0.729 |
| WNT16 | −1.0 | 0.888 | −1.6 | 0.845 | −8.5 | 0.457 |
Values shown as the β coefficient for pulse‐wave velocity per SD greater Wnt gene expression. APC indicates adenomatous polyposis coli regulator of Wnt signaling pathway; AXIN, axin; CDH2, cadherin 2; CTNNB1, catenin β 1; FZD, frizzled class receptor; GSK3B, glycogen synthase kinase 3β; KREMEN1, kringle‐containing transmembrane protein 1; LEF1, lymphoid enhancer binding factor 1; SFRP2, secreted frizzled related protein 2; TCF, transcription factor; WNT16, Wnt family member 16.
Fully adjusted models include adjustment for age, height, weight, systolic blood pressure, diastolic blood pressure, antihypertensive medication use, heart rate, lipid‐lowering medication use, current smoking, alcohol use, and sedentary behavior.
Table 4.
Fully Adjusted Effects of Wnt Pathway Gene Expression and Traditional Cardiovascular Risk Factors on Arterial Stiffness
| Covariate | Unit | β, cm/s | % PWV Difference Explained | P Value |
|---|---|---|---|---|
| Agea | 5 y | 69.2 | 4.2 | <0.001 |
| Heighta | 7.0 cm | −27.1 | −1.6 | 0.062 |
| Weighta | 15.1 kg | −24.2 | −1.5 | 0.213 |
| SBPa | 21.7 mm Hg | 116.8 | 7.1 | <0.001 |
| DBPa | 12.0 mm Hg | 3.9 | 0.2 | 0.710 |
| Antihypertensive medication use | 1 | −10.1 | −0.6 | 0.789 |
| Heart ratea | 12.5 bpm | 89.7 | 5.4 | <0.001 |
| Current smoking | 1 | −61.5 | −3.7 | 0.276 |
| Consume >4 alcohol drinks/wk | 1 | 68.1 | 4.1 | 0.088 |
| Watch >14 hours of TV/wk | 1 | 9.5 | 0.6 | 0.722 |
| Cholesterol‐lowering medication use | 1 | 106.7 | 6.5 | 0.012 |
| APC a | 35.6 count | 44.4 | 2.7 | 0.035 |
| GSK3B a | 159.0 count | 5.5 | 0.3 | 0.892 |
| TCF4 a | 39.5 count | −43.2 | −2.6 | 0.003 |
APC indicates adenomatous polyposis coli regulator of Wnt signaling pathway; DBP, diastolic blood pressure; GSK3B, glycogen synthase kinase 3β; PWV, pulse‐wave velocity; SBP, systolic blood pressure; TCF4, transcription factor 4.
Results shown per 1‐SD greater covariate value, except for age, which was modeled as per a 5‐year greater age.
Exploratory Factor Analysis of Wnt Gene Expression and Arterial Stiffness
Exploratory factor analysis identified 3 factors underlying the Wnt gene expression profile (Table 5). All factors were normalized to have a mean of 0 with an SD of 1 unit. The first factor, which explained 19% of the variance in Wnt gene expression, consisted of GSK3B, CTNNB1, APC, KREMEN1 (kringle‐containing transmembrane protein 1), and AXIN1. The second factor, which explained the most variance in Wnt gene expression at 28%, consisted of TCF4, LEF1, and AXIN2. The third factor, which only explained 2% of the variance in Wnt gene expression, consisted of frizzled class receptor 6 (FZD6), CDH2, SFRP2 (secreted frizzled related protein 2), and Wnt family member 16 (WNT16). Of note, FZD1 and TCF4 did not load onto any factor ≥0.3. These loadings largely reflected the underlying correlation structure between the Wnt genes (Table S2). In fully adjusted analyses of these Wnt gene expression factors and arterial stiffness, factor 1 was significantly associated with greater PWV, such that a 1‐SD greater expression of factor 1 was associated with 34.7‐cm/s greater PWV (P=0.03; Figure). Although neither factor 2 nor factor 3 was significantly associated with PWV in fully adjusted models, higher factor 2 was associated with lower PWV in unadjusted (P=0.002) and age‐adjusted models (P=0.04; Figure).
Table 5.
Exploratory Factor Analysis Loadings
| Wnt Gene | Factor 1 | Factor 2 | Factor 3 |
|---|---|---|---|
| APC | 0.723 | … | … |
| AXIN1 | 0.520 | … | … |
| AXIN2 | … | 0.807 | … |
| CDH2 | … | … | 0.620 |
| CTNNB1 | 0.796 | … | … |
| FZD1 a | … | … | … |
| FZD6 | … | … | 0.621 |
| GSK3B | 0.968 | … | … |
| KREMEN1 | 0.557 | … | … |
| LEF1 | … | 0.920 | … |
| SFRP2 | … | … | 0.461 |
| TCF4 a | … | … | … |
| TCF7 | … | 0.970 | … |
| WNT16 | … | … | 0.343 |
| Variance explained, %b | 19.3 | 28.0 | 1.9 |
APC indicates adenomatous polyposis coli regulator of Wnt signaling pathway; AXIN, axin; CDH2, cadherin 2; CTNNB1, catenin β 1; FZD, frizzled class receptor; GSK3B, glycogen synthase kinase 3β; KREMEN1, kringle‐containing transmembrane protein 1; LEF1, lymphoid enhancer binding factor 1; SFRP2, secreted frizzled related protein 2; TCF, transcription factor; WNT16, Wnt family member 16.
FZD1 and TCF4 did not load onto any factor at ≥0.3.
Weighted variance of Wnt gene expression explained by each factor.
Figure 1.
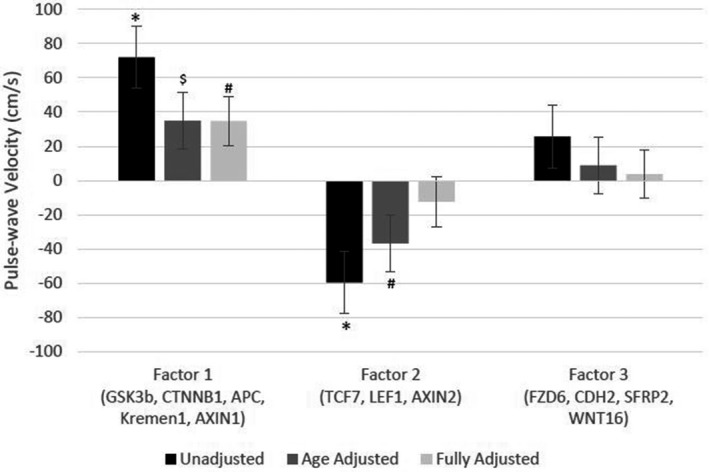
Association of Wnt gene expression factors with arterial stiffness. The graph shows the adjusted β coefficients and SE bars for the difference in pulse‐wave velocity per 1‐SD greater expression of each Wnt pathway gene factor. Unadjusted (black bars), age‐adjusted (dark gray bars), and fully adjusted (light gray bars) models are shown. Fully adjusted models include adjustment for age, height, weight, systolic blood pressure, diastolic blood pressure, antihypertensive medication use, heart rate, lipid‐lowering medication use, current smoking, alcohol use, and sedentary behavior. APC, adenomatous polyposis coli regulator of Wnt signaling pathway; AXIN, axin; CDH2, cadherin 2; CTNNB1, catenin β 1; FZD6, frizzled class receptor 6; GSK3B, glycogen synthase kinase 3β; Kremen1, kringle‐containing transmembrane protein 1; LEF1, lymphoid enhancer binding factor 1; SFRP2, secreted frizzled related protein 2; TCF7, transcription factor 7; WNT16, Wnt family member 16. $ P=0.07, +P<0.05, *P<0.001.
Discussion
This is the first study to comprehensively assess the association of Wnt pathway gene expression and arterial stiffness in humans. We found that of the 43 Wnt mRNAs tested, 14 were detected in peripheral blood from our African ancestry study sample and may suggest that they function in human circulation. Of the detectable mRNAs, 2 (APC and TCF4) showed significant associations with arterial stiffness, as assessed by PWV, even after accounting for traditional CVD risk factors. These relationships were also identified through exploratory factor analyses. These results highlight the biologic complexity underlying the Wnt pathway's effect on the vascular system and the importance of assessing multiple pathway components in determining overall Wnt pathway signaling status.
This direction of effect recapitulates the known Wnt pathway biological features in that some genes lead to upregulation of Wnt pathway target genes, whereas others downregulate it. For example, most of the Wnt pathway genes with positive associations between their expression and PWV, such as those that clustered together in factor 1 (APC, AXIN1, and GSK3B), form a protein complex with β‐catenin (CTNNB1; also in factor 1) in the absence of Wnt signaling to prevent the transcription of Wnt target genes.34 In contrast, most of the Wnt pathway genes with negative associations between their expression and PWV, such as those that clustered together in factor 2 (LEF1, TCF7, and AXIN2) and TCF4, generally code for proteins that, in the presence of Wnt signaling, cluster together and allow transcription factors to translocate to the nucleus to upregulate Wnt pathway target genes.34 Axin‐2 is a slightly different story because its protein form functions to downregulate Wnt signaling,35 but the gene AXIN2 is also a target of Wnt signaling36; therefore, it makes sense that its gene expression tracks closely with LEF1 and TCF7. Given the cross‐sectional nature of our data, we cannot say for certain whether this Wnt expression pattern caused the arterial stiffness, thereby suggesting that Wnt signaling protects the vessels from stiffening, or whether these data may depict a negative feedback loop that downregulated Wnt signaling, causing arteries to stiffen. Last, Wnt signals via both canonical and noncanonical pathways.37 Most of the research referenced herein is about the canonical Wnt signaling cascade; however, when canonical signaling is reduced, noncanonical signaling is concomitantly increased and may, in fact, be a significant driver in CVD, as well.38, 39 Nonetheless, the results from this study are the first to suggest that Wnt pathway signaling is associated with human arterial stiffness.
Previous studies of the Wnt pathway and arterial stiffness generally studied 1 or 2 circulating Wnt inhibitor proteins, such as SOST (sclerostin) or Dkk1. SOST and Dkk1 reduce Wnt pathway signaling.34 However, they represent only a single aspect of the complex Wnt signaling pathway. Most,19, 20, 21, 23 but not all,18, 22 of these studies found circulating SOST to be positively associated with greater arterial stiffness, which is in alignment with findings from the current study. However, previous results for Dkk1 were generally null,19, 20, 23 although one showed an inverse relationship.18 In contrast, the current study used a pathway‐wide approach to obtain a broader view of Wnt pathway gene expression and its potential association with arterial stiffness.
We found that 2 Wnt genes were significantly associated with arterial stiffness: APC and TCF4. Previous research on APC and arterial stiffness is limited, although some studies have implicated APC in atherosclerotic fibrous cap stability,40 cardiac repair41 and development,42 hypertrophic cardiomyopathy,43 and cardiomyocyte proliferation.44 Because the APC protein is largely intracellular, it has not previously been assessed in population‐based studies and this is the first report of its in vivo expression and potential association with human vascular disease measures.
On the other hand, TCF4, which is also known as TCF7L2, has a genetic variant (rs7903146) that was identified in 2007 as a strong risk factor for diabetes mellitus and impaired fasting glucose.45 This genetic association has been replicated in many studies and has also been shown to be associated with diabetic coronary artery atherosclerosis46, 47 and ankle‐brachial index.48 To our knowledge, TCF4 genetic variants have not been studied in relationship to arterial stiffness. The action of TCF4 in coronary artery disease appears to be through vascular smooth muscle cell plasticity49, 50 and proliferation,51 which could also impact arterial wall stiffness. In sensitivity analyses, we found that the addition of diabetes mellitus to our full model slightly attenuated (≈7%) the reported associations between TCF4 expression and arterial stiffness, although the magnitude of the relationship was similar and still significant (P=0.004; data not shown). Like APC, the TCF4 protein is largely intracellular and has not previously been assessed in population‐based studies, so this report represents a novel finding that extends the known associations of TCF4 and vascular disease to arterial stiffness.
Factor analysis identified 2 main factors that explained most of the variation in Wnt gene expression patterns. Factor 1 appears to consist mostly of genes responsible for Wnt signal transduction, whereas factor 2 appears to consist mostly of genes responsible for transcriptional regulation of Wnt signaling target genes. In addition, factor analysis identified a third factor that only accounted for a small percentage of variation in Wnt gene expression, and appears to consist mostly of genes involved in cell‐surface Wnt signaling. Although factors 1 and 2 were significantly associated with arterial stiffness in minimally adjusted models, only factor 1 was significant in models adjusted for CVD risk factors. Interestingly, the Wnt gene with strongest individual association between its expression and arterial stiffness, TCF4, did not load onto any factor at ≥0.3 and it had fairly low levels of correlation to other Wnt genes (maximum, 0.26 with AXIN2; Table S2). This may suggest that the strong association of TCF4 with arterial stiffness may be operating through additional mechanisms other than canonical Wnt signaling.
As with any gene expression study, the results are reflective only of the tissue and population sample they are derived from: in this case, peripheral blood from African ancestry men. We could not perform blood cell sorting or counting for determining the cellular composition of the blood samples, so our results reflect expression profiles of a mixture of circulating cells. However, as our outcome of interest is arterial stiffness, peripheral blood is a reasonable tissue source52, 53 given the noninvasive collection method and its constant contact with the vessel wall.54 We found a large number (29 of 43) of the tested Wnt pathway genes did not have detectable expression, so we cannot comment on their potential association with vascular disease. We also cannot comment on Wnt pathway genes that were not included on this custom array panel, which was designed to include only genes with previous evidence for involvement in CVD processes and/or expression in the vascular wall. It is possible that some of these genes may be expressed and function only in the vessel wall, rather than the circulation.12, 13, 14, 15 It is also possible that expression profiles may differ by ethnicity, exposures, or physiologic conditions, such as inflammation, which we were unable to control. However, as individuals of African ancestry are at particularly high risk of vascular disease with poor hypertension control, including these THS men,25 this represents an important population segment for human molecular research. There may be a biologic difference in Wnt expression profiles by sex, as previously reported,55, 56 and future studies will be needed to compare the current results in women. Last, it is unclear what is driving the differences in Wnt gene expression (whether that be genetic or epigenetic variation, or a combination of both). Additional studies should also be performed to assess the impact of Wnt gene expression on other forms of subclinical and clinical disease and outcomes.
We found the expression of 2 circulating Wnt pathway genes, APC and TCF4, was strongly associated with arterial stiffness, even after adjustment for traditional CVD risk factors. Greater expression of APC, a gene involved in downregulating Wnt pathway target gene expression, was associated with greater arterial stiffness; whereas greater expression of TCF4, a gene involved in upregulating Wnt pathway target gene expression, was associated with lower arterial stiffness. These human‐derived expression data suggest that Wnt pathway signaling is involved in vascular stiffness and may represent a novel risk factor and pharmacologic target for improving arterial health.
Sources of Funding
This work was supported by grant K01‐HL125658 (principal investigator: Dr Kuipers) from the National Heart, Lung, and Blood Institute; grant R01‐DK097084 (principal investigator: Dr Miljkovic) from the National Institute of Diabetes and Digestive and Kidney Diseases; and grant R01‐AR049747 (principal investigator: Dr Zmuda) from the National Institute of Arthritis and Musculoskeletal and Skin Diseases.
Disclosures
None.
Supporting information
Table S1. List of All Genes Tested on the Nanostring nCounter Gene Expression Panel
Table S2. Spearman Correlation Coefficients Between Expressed Wnt Genes
Acknowledgments
The authors would like to thank all supporting staff from the THS (Tobago Health Study) Office and University of Pittsburgh Ultrasound Research Laboratory.
(J Am Heart Assoc. 2020;9:e014170 DOI: 10.1161/JAHA.119.014170.)
References
- 1. Cohen ED, Tian Y, Morrisey EE. Wnt signaling: an essential regulator of cardiovascular differentiation, morphogenesis and progenitor self‐renewal. Development. 2008;135:789–798. [DOI] [PubMed] [Google Scholar]
- 2. Tian Y, Cohen ED, Morrisey EE. The importance of Wnt signaling in cardiovascular development. Pediatr Cardiol. 2010;31:342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van de Schans VA, Smits JF, Blankesteijn WM. The Wnt/frizzled pathway in cardiovascular development and disease: friend or foe? Eur J Pharmacol. 2008;585:338–345. [DOI] [PubMed] [Google Scholar]
- 4. Bostrom KI, Rajamannan NM, Towler DA. The regulation of valvular and vascular sclerosis by osteogenic morphogens. Circ Res. 2011;109:564–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shao JS, Aly ZA, Lai CF, Cheng SL, Cai J, Huang E, Behrmann A, Towler DA. Vascular BMP MSX2 Wnt signaling and oxidative stress in arterial calcification. Ann N Y Acad Sci. 2007;1117:40–50. [DOI] [PubMed] [Google Scholar]
- 6. Al‐Aly Z. Arterial calcification: a tumor necrosis factor‐alpha mediated vascular Wnt‐opathy. Transl Res. 2008;151:233–239. [DOI] [PubMed] [Google Scholar]
- 7. Tsaousi A, Mill C, George SJ. The Wnt pathways in vascular disease: lessons from vascular development. Curr Opin Lipidol. 2011;22:350–357. [DOI] [PubMed] [Google Scholar]
- 8. Marinou K, Christodoulides C, Antoniades C, Koutsilieris M. Wnt signaling in cardiovascular physiology. Trends Endocrinol Metab. 2012;23:628–636. [DOI] [PubMed] [Google Scholar]
- 9. Mill C, George SJ. Wnt signalling in smooth muscle cells and its role in cardiovascular disorders. Cardiovasc Res. 2012;95:233–240. [DOI] [PubMed] [Google Scholar]
- 10. Barrett HE, Van der Heiden K, Farrell E, Gijsen FJH, Akyildiz AC. Calcifications in atherosclerotic plaques and impact on plaque biomechanics. J Biomech. 2019;87:1–12. [DOI] [PubMed] [Google Scholar]
- 11. Cheng SL, Shao JS, Behrmann A, Krchma K, Towler DA. Dkk1 and MSX2‐Wnt7b signaling reciprocally regulate the endothelial‐mesenchymal transition in aortic endothelial cells. Arterioscler Thromb Vasc Biol. 2013;33:1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng SL, Behrmann A, Shao JS, Ramachandran B, Krchma K, Arredondo YB, Kovacs A, Mead M, Maxson R, Towler DA. Targeted reduction of vascular MSX1 and msx2 mitigates arteriosclerotic calcification and aortic stiffness in LDLR‐deficient mice fed diabetogenic diets. Diabetes. 2014;63:4326–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shao JS, Sierra OL, Cohen R, Mecham RP, Kovacs A, Wang J, Distelhorst K, Behrmann A, Halstead LR, Towler DA. Vascular calcification and aortic fibrosis: a bifunctional role for osteopontin in diabetic arteriosclerosis. Arterioscler Thromb Vasc Biol. 2011;31:1821–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kirton JP, Crofts NJ, George SJ, Brennan K, Canfield AE. Wnt/beta‐catenin signaling stimulates chondrogenic and inhibits adipogenic differentiation of pericytes: potential relevance to vascular disease? Circ Res. 2007;101:581–589. [DOI] [PubMed] [Google Scholar]
- 15. Cheng SL, Shao JS, Halstead LR, Distelhorst K, Sierra O, Towler DA. Activation of vascular smooth muscle parathyroid hormone receptor inhibits Wnt/beta‐catenin signaling and aortic fibrosis in diabetic arteriosclerosis. Circ Res. 2010;107:271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sutton‐Tyrrell K, Najjar SS, Boudreau RM, Venkitachalam L, Kupelian V, Simonsick EM, Havlik R, Lakatta EG, Spurgeon H, Kritchevsky S, Pahor M, Bauer D, Newman A. Elevated aortic pulse wave velocity, a marker of arterial stiffness, predicts cardiovascular events in well‐functioning older adults. Circulation. 2005;111:3384–3390. [DOI] [PubMed] [Google Scholar]
- 17. Yamashina A, Tomiyama H, Takeda K, Tsuda H, Arai T, Hirose K, Koji Y, Hori S, Yamamoto Y. Validity, reproducibility, and clinical significance of noninvasive brachial‐ankle pulse wave velocity measurement. Hypertens Res. 2002;25:359–364. [DOI] [PubMed] [Google Scholar]
- 18. Thambiah S, Roplekar R, Manghat P, Fogelman I, Fraser WD, Goldsmith D, Hampson G. Circulating sclerostin and dickkopf‐1 (DKK1) in predialysis chronic kidney disease (CKD): relationship with bone density and arterial stiffness. Calcif Tissue Int. 2012;90:473–480. [DOI] [PubMed] [Google Scholar]
- 19. Hampson G, Edwards S, Conroy S, Blake GM, Fogelman I, Frost ML. The relationship between inhibitors of the Wnt signalling pathway (Dickkopf‐1(DKK1) and sclerostin), bone mineral density, vascular calcification and arterial stiffness in post‐menopausal women. Bone. 2013;56:42–47. [DOI] [PubMed] [Google Scholar]
- 20. Yang HY, Wu DA, Chen MC, Hsu BG. Correlation between sclerostin and Dickkopf‐1 with aortic arterial stiffness in patients with type 2 diabetes: a prospective, cross‐sectional study. Diabet Vasc Dis Res. 2019;16:281–288. [DOI] [PubMed] [Google Scholar]
- 21. Chang YC, Hsu BG, Liou HH, Lee CJ, Wang JH. Serum levels of sclerostin as a potential biomarker in central arterial stiffness among hypertensive patients. BMC Cardiovasc Disord. 2018;18:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gaudio A, Fiore V, Rapisarda R, Sidoti MH, Xourafa A, Catalano A, Tringali G, Zanoli L, Signorelli SS, Fiore CE. Sclerostin is a possible candidate marker of arterial stiffness: results from a cohort study in catania. Mol Med Rep. 2017;15:3420–3424. [DOI] [PubMed] [Google Scholar]
- 23. Hsu BG, Liou HH, Lee CJ, Chen YC, Ho GJ, Lee MC. Serum sclerostin as an independent marker of peripheral arterial stiffness in renal transplantation recipients: a cross‐sectional study. Medicine. 2016;95:e3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lackland DT. Racial differences in hypertension: implications for high blood pressure management. Am J Med Sci. 2014;348:135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuipers AL, Miljkovic I, Barinas‐Mitchell E, Cvejkus R, Bunker CH, Wheeler VW, Zmuda JM. Arterial stiffness and hypertension status in Afro‐Caribbean men. J Hypertens. 2019;37:546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hill DD, Cauley JA, Sheu Y, Bunker CH, Patrick AL, Baker CE, Beckles GL, Wheeler VW, Zmuda JM. Correlates of bone mineral density in men of African ancestry: the Tobago bone health study. Osteoporos Int. 2008;19:227–234. [DOI] [PubMed] [Google Scholar]
- 27. Miljkovic‐Gacic I, Ferrell RE, Patrick AL, Kammerer CM, Bunker CH. Estimates of African, European and native American Ancestry in afro‐Caribbean men on the island of Tobago. Hum Hered. 2005;60:129–133. [DOI] [PubMed] [Google Scholar]
- 28. Venkitachalam L, Mackey RH, Sutton‐Tyrrell K, Patel AS, Boraz MA, Simkin‐Silverman LR, Kuller LH. Elevated pulse wave velocity increases the odds of coronary calcification in overweight postmenopausal women. Am J Hypertens. 2007;20:469–475. [DOI] [PubMed] [Google Scholar]
- 29. Sugawara J, Hayashi K, Yokoi T, Cortez‐Cooper MY, DeVan AE, Anton MA, Tanaka H. Brachial‐ankle pulse wave velocity: an index of central arterial stiffness? J Hum Hypertens. 2005;19:401–406. [DOI] [PubMed] [Google Scholar]
- 30. Tsuchikura S, Shoji T, Kimoto E, Shinohara K, Hatsuda S, Koyama H, Emoto M, Nishizawa Y. Brachial‐ankle pulse wave velocity as an index of central arterial stiffness. J Atheroscler Thromb. 2010;17:658–665. [DOI] [PubMed] [Google Scholar]
- 31. Tomiyama H, Arai T, Koji Y, Yambe M, Motobe K, Zaydun G, Yamamoto Y, Hori S, Yamashina A. The age‐related increase in arterial stiffness is augmented in phases according to the severity of hypertension. Hypertens Res. 2004;27:465–470. [DOI] [PubMed] [Google Scholar]
- 32. Koji Y, Tomiyama H, Yamada J, Yambe M, Motobe K, Shiina K, Yamashina A. Relationship between arterial stiffness and the risk of coronary artery disease in subjects with and without metabolic syndrome. Hypertens Res. 2007;30:243–247. [DOI] [PubMed] [Google Scholar]
- 33. Malkov VA, Serikawa KA, Balantac N, Watters J, Geiss G, Mashadi‐Hossein A, Fare T. Multiplexed measurements of gene signatures in different analytes using the nanostring ncounter assay system. BMC Res Notes. 2009;2:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Badimon L, Borrell‐Pages M. Wnt signaling in the vessel wall. Curr Opin Hematol. 2017;24:230–239. [DOI] [PubMed] [Google Scholar]
- 35. Rennoll SA, Konsavage WM Jr, Yochum GS. Nuclear AXIN2 represses MYC gene expression. Biochem Biophys Res Comm. 2014;443:217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta‐catenin/tcf signaling induces the transcription of AXIN2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grumolato L, Liu G, Mong P, Mudbhary R, Biswas R, Arroyave R, Vijayakumar S, Economides AN, Aaronson SA. Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes Dev. 2010;24:2517–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Albanese I, Yu B, Al‐Kindi H, Barratt B, Ott L, Al‐Refai M, de Varennes B, Shum‐Tim D, Cerruti M, Gourgas O, Rheaume E, Tardif JC, Schwertani A. Role of noncanonical Wnt signaling pathway in human aortic valve calcification. Arterioscler Thromb Vasc Biol. 2017;37:543–552. [DOI] [PubMed] [Google Scholar]
- 39. Cheng SL, Ramachandran B, Behrmann A, Shao JS, Mead M, Smith C, Krchma K, Bello Arredondo Y, Kovacs A, Kapoor K, Brill LM, Perera R, Williams BO, Towler DA. Vascular smooth muscle LRP6 limits arteriosclerotic calcification in diabetic LDLR‐/‐ mice by restraining noncanonical wnt signals. Circ Res. 2015;117:142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eken SM, Jin H, Chernogubova E, Li Y, Simon N, Sun C, Korzunowicz G, Busch A, Backlund A, Osterholm C, Razuvaev A, Renne T, Eckstein HH, Pelisek J, Eriksson P, Gonzalez Diez M, Perisic Matic L, Schellinger IN, Raaz U, Leeper NJ, Hansson GK, Paulsson‐Berne G, Hedin U, Maegdefessel L. Microrna‐210 enhances fibrous cap stability in advanced atherosclerotic lesions. Circ Res. 2017;120:633–644. [DOI] [PubMed] [Google Scholar]
- 41. Arif M, Pandey R, Alam P, Jiang S, Sadayappan S, Paul A, Ahmed RPH. Microrna‐210‐mediated proliferation, survival, and angiogenesis promote cardiac repair post myocardial infarction in rodents. J Mol Med. 2017;95:1369–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rezvani M, Liew CC. Role of the adenomatous polyposis coli gene product in human cardiac development and disease. J Biol Chem. 2000;275:18470–18475. [DOI] [PubMed] [Google Scholar]
- 43. Masuelli L, Bei R, Sacchetti P, Scappaticci I, Francalanci P, Albonici L, Coletti A, Palumbo C, Minieri M, Fiaccavento R, Carotenuto F, Fantini C, Carosella L, Modesti A, Di Nardo P. Beta‐catenin accumulates in intercalated disks of hypertrophic cardiomyopathic hearts. Cardiovasc Res. 2003;60:376–387. [DOI] [PubMed] [Google Scholar]
- 44. Ye B, Hou N, Xiao L, Xu Y, Boyer J, Xu H, Li F. Apc controls asymmetric Wnt/beta‐catenin signaling and cardiomyocyte proliferation gradient in the heart. J Mol Cell Cardiol. 2015;89:287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Raitakari OT, Ronnemaa T, Huupponen R, Viikari L, Fan M, Marniemi J, Hutri‐Kahonen N, Viikari JS, Lehtimaki T. Variation of the transcription factor 7‐like 2 (TCF7L2) gene predicts impaired fasting glucose in healthy young adults: the cardiovascular risk in young Finns study. Diabetes Care. 2007;30:2299–2301. [DOI] [PubMed] [Google Scholar]
- 46. Muendlein A, Saely CH, Geller‐Rhomberg S, Sonderegger G, Rein P, Winder T, Beer S, Vonbank A, Drexel H. Single nucleotide polymorphisms of TCF7L2 are linked to diabetic coronary atherosclerosis. PLoS One. 2011;6:e17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sousa AG, Marquezine GF, Lemos PA, Martinez E, Lopes N, Hueb WA, Krieger JE, Pereira AC. tcf7 l2 Polymorphism RS7903146 is associated with coronary artery disease severity and mortality. PLoS One. 2009;4:e7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wassel CL, Lamina C, Nambi V, Coassin S, Mukamal KJ, Ganesh SK, Jacobs DR Jr, Franceschini N, Papanicolaou GJ, Gibson Q, Yanek LR, van der Harst P, Ferguson JF, Crawford DC, Waite LL, Allison MA, Criqui MH, McDermott MM, Mehra R, Cupples LA, Hwang SJ, Redline S, Kaplan RC, Heiss G, Rotter JI, Boerwinkle E, Taylor HA, Eraso LH, Haun M, Li M, Meisinger C, O'Connell JR, Shuldiner AR, Tybjaerg‐Hansen A, Frikke‐Schmidt R, Kollerits B, Rantner B, Dieplinger B, Stadler M, Mueller T, Haltmayer M, Klein‐Weigel P, Summerer M, Wichmann HE, Asselbergs FW, Navis G, Mateo Leach I, Brown‐Gentry K, Goodloe R, Assimes TL, Becker DM, Cooke JP, Absher DM, Olin JW, Mitchell BD, Reilly MP, Mohler ER III, North KE, Reiner AP, Kronenberg F, Murabito JM. Genetic determinants of the ankle‐brachial index: a meta‐analysis of a cardiovascular candidate gene 50k SNP panel in the candidate gene association resource (CARE) consortium. Atherosclerosis. 2012;222:138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Srivastava R, Rolyan H, Xie Y, Li N, Bhat N, Hong L, Esteghamat F, Adeniran A, Geirsson A, Zhang J, Ge G, Nobrega M, Martin KA, Mani A. tcf7 l2 (Transcription factor 7‐like 2) regulation of GATA6 (GATA‐binding protein 6)‐dependent and ‐independent vascular smooth muscle cell plasticity and intimal hyperplasia. Arterioscler Thromb Vasc Biol. 2019;39:250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Srivastava R, Zhang J, Go GW, Narayanan A, Nottoli TP, Mani A. Impaired LRP6‐TCF7L2 activity enhances smooth muscle cell plasticity and causes coronary artery disease. Cell Rep. 2015;13:746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhuang Y, Mao JQ, Yu M, Dong LY, Fan YL, Lv ZQ, Xiao MD, Yuan ZX. Hyperlipidemia induces vascular smooth muscle cell proliferation involving Wnt/beta‐catenin signaling. Cell Biol Int. 2016;40:121–130. [DOI] [PubMed] [Google Scholar]
- 52. Wain LV, Vaez A, Jansen R, Joehanes R, van der Most PJ, Erzurumluoglu AM, O'Reilly PF, Cabrera CP, Warren HR, Rose LM, Verwoert GC, Hottenga JJ, Strawbridge RJ, Esko T, Arking DE, Hwang SJ, Guo X, Kutalik Z, Trompet S, Shrine N, Teumer A, Ried JS, Bis JC, Smith AV, Amin N, Nolte IM, Lyytikainen LP, Mahajan A, Wareham NJ, Hofer E, Joshi PK, Kristiansson K, Traglia M, Havulinna AS, Goel A, Nalls MA, Sober S, Vuckovic D, Luan J, Del Greco MF, Ayers KL, Marrugat J, Ruggiero D, Lopez LM, Niiranen T, Enroth S, Jackson AU, Nelson CP, Huffman JE, Zhang W, Marten J, Gandin I, Harris SE, Zemunik T, Lu Y, Evangelou E, Shah N, de Borst MH, Mangino M, Prins BP, Campbell A, Li‐Gao R, Chauhan G, Oldmeadow C, Abecasis G, Abedi M, Barbieri CM, Barnes MR, Batini C, Beilby J, Blake T, Boehnke M, Bottinger EP, Braund PS, Brown M, Brumat M, Campbell H, Chambers JC, Cocca M, Collins F, Connell J, Cordell HJ, Damman JJ, Davies G, de Geus EJ, de Mutsert R, Deelen J, Demirkale Y, Doney ASF, Dorr M, Farrall M, Ferreira T, Franberg M, Gao H, Giedraitis V, Gieger C, Giulianini F, Gow AJ, Hamsten A, Harris TB, Hofman A, Holliday EG, Hui J, Jarvelin MR, Johansson A, Johnson AD, Jousilahti P, Jula A, Kahonen M, Kathiresan S, Khaw KT, Kolcic I, Koskinen S, Langenberg C, Larson M, Launer LJ, Lehne B, Liewald DCM, Lin L, Lind L, Mach F, Mamasoula C, Menni C, Mifsud B, Milaneschi Y, Morgan A, Morris AD, Morrison AC, Munson PJ, Nandakumar P, Nguyen QT, Nutile T, Oldehinkel AJ, Oostra BA, Org E, Padmanabhan S, Palotie A, Pare G, Pattie A, Penninx B, Poulter N, Pramstaller PP, Raitakari OT, Ren M, Rice K, Ridker PM, Riese H, Ripatti S, Robino A, Rotter JI, Rudan I, Saba Y, Saint Pierre A, Sala CF, Sarin AP, Schmidt R, Scott R, Seelen MA, Shields DC, Siscovick D, Sorice R, Stanton A, Stott DJ, Sundstrom J, Swertz M, Taylor KD, Thom S, Tzoulaki I, Tzourio C, Uitterlinden AG, Volker U, Vollenweider P, Wild S, Willemsen G, Wright AF, Yao J, Theriault S, Conen D, Attia J, Sever P, Debette S, Mook‐Kanamori DO, Zeggini E, Spector TD, van der Harst P, Palmer CNA, Vergnaud AC, Loos RJF, Polasek O, Starr JM, Girotto G, Hayward C, Kooner JS, Lindgren CM, Vitart V, Samani NJ, Tuomilehto J, Gyllensten U, Knekt P, Deary IJ, Ciullo M, Elosua R, Keavney BD, Hicks AA, Scott RA, Gasparini P, Laan M, Liu Y, Watkins H, Hartman CA, Salomaa V, Toniolo D, Perola M, Wilson JF, Schmidt H, Zhao JH, Lehtimaki T, van Duijn CM, Gudnason V, Psaty BM, Peters A, Rettig R, James A, Jukema JW, Strachan DP, Palmas W, Metspalu A, Ingelsson E, Boomsma DI, Franco OH, Bochud M, Newton‐Cheh C, Munroe PB, Elliott P, Chasman DI, Chakravarti A, Knight J, Morris AP, Levy D, Tobin MD, Snieder H, Caulfield MJ, Ehret GB. Novel blood pressure locus and gene discovery using genome‐wide association study and expression data sets from blood and the kidney. Hypertension. 2017;pii: HYPERTENSIONAHA.117.09438. DOI: 10.1161/HYPERTENSIONAHA.117.09438. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marketou ME, Kontaraki JE, Tsakountakis NA, Zacharis EA, Kochiadakis GE, Arfanakis DA, Chlouverakis G, Vardas PE. Arterial stiffness in hypertensives in relation to expression of angiopoietin‐1 and 2 genes in peripheral monocytes. J Hum Hypertens. 2010;24:306–311. [DOI] [PubMed] [Google Scholar]
- 54. Joehanes R, Johnson AD, Barb JJ, Raghavachari N, Liu P, Woodhouse KA, O'Donnell CJ, Munson PJ, Levy D. Gene expression analysis of whole blood, peripheral blood mononuclear cells, and lymphoblastoid cell lines from the Framingham heart study. Physiol Genomics. 2012;44:59–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kouzmenko AP, Takeyama K, Ito S, Furutani T, Sawatsubashi S, Maki A, Suzuki E, Kawasaki Y, Akiyama T, Tabata T, Kato S. Wnt/beta‐catenin and estrogen signaling converge in vivo. J Biol Chem. 2004;279:40255–40258. [DOI] [PubMed] [Google Scholar]
- 56. Gao Y, Huang E, Zhang H, Wang J, Wu N, Chen X, Wang N, Wen S, Nan G, Deng F, Liao Z, Wu D, Zhang B, Zhang J, Haydon RC, Luu HH, Shi LL, He TC. Crosstalk between Wnt/beta‐catenin and estrogen receptor signaling synergistically promotes osteogenic differentiation of mesenchymal progenitor cells. PLoS One. 2013;8:e82436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of All Genes Tested on the Nanostring nCounter Gene Expression Panel
Table S2. Spearman Correlation Coefficients Between Expressed Wnt Genes


