Abstract
Dendrimer nanocarriers are unique hyper-branched polymers with biomolecule-like properties, representing a promising prospect as a nucleic acid delivery system. The design of effective dendrimer-based gene carriers requires considering several parameters, such as carrier morphology, size, molecular weight, surface chemistry, and flexibility/rigidity. In detail, the rational design of the dendrimer surface chemistry has been ascertained to play a crucial role on the efficiency of interaction with nucleic acids. Within this framework, advances in the field of organic chemistry have allowed us to design dendrimers with even small difference in the chemical structure of their surface terminals. In this study, we have selected two different cationic phosphorus dendrimers of generation 3 functionalized, respectively, with pyrrolidinium (DP) and morpholinium (DM) surface groups, which have demonstrated promising potential for short interfering RNA (siRNA) delivery. Despite DP and DM differing only for one atom in their chemical structure, in vitro and in vivo experiments have highlighted several differences between them in terms of siRNA complexation properties. In this context, we have employed coarse-grained molecular dynamics simulation techniques to shed light on the supramolecular characteristics of dendrimer–siRNA complexation, the so-called dendriplex formations. Our data provide important information on self-assembly dynamics driven by surface chemistry and competition mechanisms.
Introduction
RNA-based drugs, including short interfering RNA (siRNA) molecules,1 are particularly promising examples of a modern medicine doctrine approach,2 conceived to minimize side effects. Despite encouraging results shown using siRNA-mediated treatments,3,4 cellular uptake still represents the major issue in the development of effective siRNA therapies. The poor internalization of siRNAs across cell membranes is due to their high molecular weight and negative charge.5 To efficiently achieve intracellular delivery, siRNAs are usually complexed with cationic molecules which generate complexes with a size ranging from tens to a few hundreds of nanometers.6 Several polymeric and lipid nanocarriers have been developed in the literature, including chitosan, cationic lipids, polyethyleneimine, and dendrimers.7−14 In the field of siRNA delivery, cationic phosphorous dendrimers15−18 have proven to be excellent drug carriers for gene-silencing treatments after in vitro and in vivo experiments.4,6,19−21 In a previous study,22 we focused the attention on two different cationic phosphorus dendrimers of generation 3 for siRNA delivery.6 Those dendrimers were functionalized, respectively, with pyrrolidinium (DP) and morpholinium (DM) surface groups.6,22
Several differences between DP and DM have been highlighted in terms of interaction properties with siRNA molecules by both in vitro experiments and molecular modeling. In detail, in vitro experiments indicated DP to have a multivalent character because it efficiently binds more than one siRNA molecule because of its ability to maximize the entropic contribution to the complexation free energy (CFE).22 In addition, DP has also a lower enthalpy contribution to the CFE because of the lower recruitment of its charged terminals, which intrinsically maximizes the ability to complex with more than one siRNA.22 Contrariwise, DM has higher enthalpy contribution to the CFE because it involves a higher number of its charged terminals, which de facto disadvantages the complexation with multiple siRNAs.22 All aforementioned characteristics in fact strongly influence the stoichiometric number of DP and DM, resulting in a different supramolecular binding behavior.
Although atomistic modeling provided a clear picture of how functionalization may drive dendrimer ability to bind siRNA, a complete exploration of dendrimer–siRNA aggregation phenomena is still missing. In particular, to deeply explore the overall dendrimer–siRNA complexation dynamics, molecular systems consisting of more than one siRNA and/or one dendrimer should be simulated. All-atom (AA) molecular dynamics (MD) simulations have limited ability to approach large systems such as supramolecular assemblies and microsecond dynamics of those assemblies. An interesting approach to overcome the abovementioned limitations is the so-called coarse-grained (CG) modeling, which allows to investigate the conformational behavior of supramolecular assemblies and dynamics of microseconds with a reasonable computational effort.23 Within this context, MARTINI CG force field has found a broad range of applications because it combines speed and versatility while maintaining chemical specificity.24−26 MARTINI forcefield has been already applied to model PAMAM27,28 or poly(l-lysine) dendrimers29 and their interaction with the cellular membrane.30−33
In the present study, we carried out CG-MD simulations to provide further crucial information on DP and DM self-assembly phenomena and interaction propensity to siRNA. Our data confirmed how even small changes of the dendrimer surface chemistry may strongly affect the dendrimer interaction mechanism with siRNA, providing a view on how those small chemical changes on the dendrimer surface affect at higher scales the dendriplex superassembly stability. The outcome of this research provides (i) a characterization of DP and DM dendriplexes6,17 and (ii) information to rationally design/optimize the dendrimer surface for tailoring dendrimer–siRNA drug-delivery systems.
Results
In this section, we will show the results regarding the dendrimers self-assembly properties, stoichiometric coefficients, and competition mechanisms.
T1. Dendrimer Self-Aggregation Mechanisms
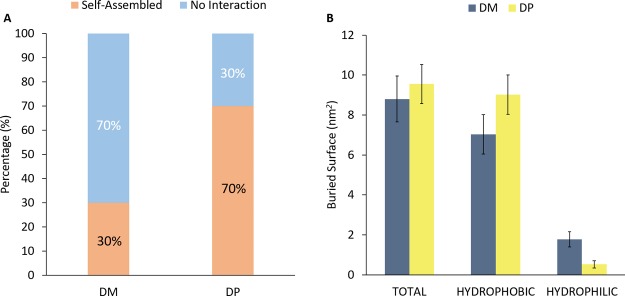
Recent in vitro experiments from our previous work22 highlighted a DP tendency to self-assemble. Here, we have carried out CG simulations to investigate molecular mechanisms describing this behavior (section Methods, category T1). Figure 1A shows the percentage of self-assembled dendrimers. The probability of DP dendrimer to aggregate (70%) was found much higher if compared to DM (30%). Moreover, buried surface (BS) estimation was carried out to evaluate the interacting surface of complexed dendrimers. Figure 1B indicates a quite similar total BS (DP = 9.55 ± 0.98 nm2, DM = 8.80 ± 1.15 nm2) for both DM and DP. As expected, DP–DP complexes were characterized by a slightly higher hydrophobic BS (DP = 9.02 ± 0.97 nm2, DM = 7.03 ± 0.99 nm2) and a lower hydrophilic contact area (DP = 0.53 ± 0.18 nm2, DM = 1.77 ± 0.38 nm2) with respect to DM–DM complexes. The greater tendency shown by DP to interact by hydrophobic contacts is likely driven by apolar terminals on the outer surface. The increased buried hydrophobic area of DP promotes the self-assembly phenomena, in agreement with experimental data.
Figure 1.
Percentage of simulation frames in which dendrimers are found complexed (DP–DP or DM–DM) or free in solution (A). The last 50 ns of all replicas for each dendrimer type were considered as an ensemble trajectory. DP has demonstrated a greater self-aggregation behavior, as it occurs in 70% of the frames; in contrast, DM has exhibited lower attitude to self-aggregate, which occurs in only 30% of the frames. Picture (B) shows the BS between homologous DMs and DPs. In this chart, the hydrophobic variation between the 2DP and 2DM systems is emphasized, which may be the driving force of DP tighter self-assembly.
In addition to the previously BS analysis, dendrimer solvent accessible surface area (SASA) has been also computed in order to clarify the percentage of the dendrimer surface involved in the self-assembly process. Table 1 shows the SASA analysis and the BS/SASA ratio for both the dendrimer type, dividing results in three main categories: total, hydrophobic, and hydrophilic. Accordingly to the Figure 1B bar diagram, the BS/SASA ratio is in perfect correlation with the interaction surface of the dendrimer assembly. Despite DP has lower total SASA, it involves a higher interaction surface, resulting in a higher percentage of the BS/SASA ratio.
Table 1. Dendrimers SASA and the Respective Ratio between BS and SASA Expressed in Percentage.
| SASA total (nm2) | SASA hydrophobic (nm2) | SASA hydrophilic (nm2) | BS/SASA total (%) | BS/SASA hydrophobic (%) | BS/SASA hydrophilic (%) | |
|---|---|---|---|---|---|---|
| DM | 81.26 ± 3.13 | 52.76 ± 3.23 | 28.50 ± 1.45 | 10.83 | 13.32 | 6.21 |
| DP | 73.57 ± 2.83 | 60.75 ± 3.04 | 12.82 ± 0.80 | 12.98 | 14.85 | 4.14 |
T2. siRNA–Dendrimer Binding Stoichiometry
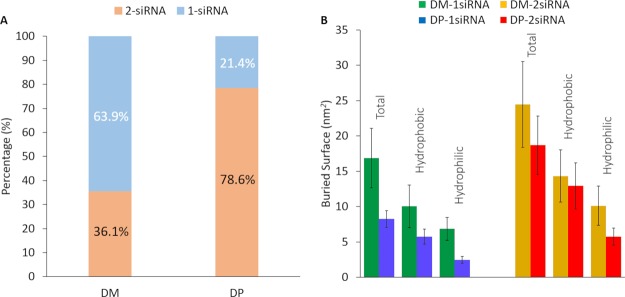
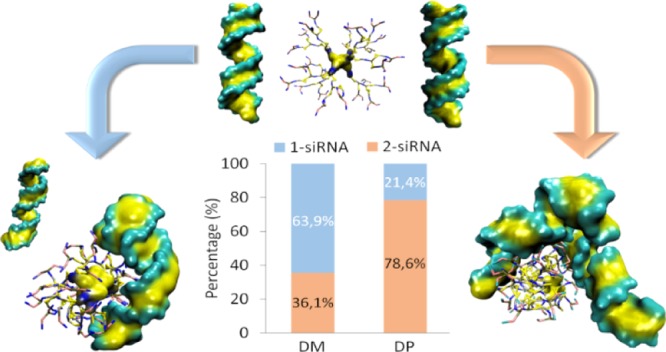
In vitro experiments and AA simulations suggested DP–siRNA stoichiometry higher than 2, whereas DM–siRNA stoichiometry lower than 1, which definitively crown the DP dendrimer as the most efficient nanocarrier. In this work, an extensive CG-MD investigation was carried out to deeply elucidate molecular reasons behind this feature (set up described in section Methods, category T2). Overall, 15 replicas of 500 ns were carried out for each system, considering the last 50 ns for all the following analyses. Figure 2A shows the probability of each dendrimer to be in contact with 1 and 2 siRNA, respectively. Results indicated that DP was able to maximize the probability to bind 2 siRNA molecules (78.6% of the MD frames—Supporting Movie 1), if compared with DM (36.1% of the MD frames—Supporting Movie 2). Figure 2B shows the BS, that is, the interaction surface, between dendrimer and siRNA in stable dendriplex assemblies. In these CG-MD simulations, dendriplex can be composed by 1 dendrimer and 1 siRNA or 1 dendrimer and 2 siRNA. The BS was always larger in case of DM–siRNA dendriplex (DM–1 siRNA = 16.89 ± 4.20 nm2, DM–2 siRNA = 24.46 ± 6.11 nm2; DP–1 siRNA = 8.25 ± 1.18 nm2, DP–2 siRNA = 18.69 ± 4.15 nm2). It is worth noting that BS is 2.27 higher in case of DP binding 2 siRNA (BSDP–2 siRNA/BSDP–1 siRNA > 2), whereas the BS is only 1.45 higher for DM binding 2 siRNA (BSDM–2 siRNA/BSDM–1 siRNA < 1.5). This evidence is again in agreement with a higher stoichiometry of DP in binding siRNA, as suggested in the recent literature.22
Figure 2.
(A) Complexation occurrences bar diagram for both types of dendrimer with siRNA. DP can interact with 2 siRNAs in 78.6% of the frames, instead DM may complex with 2 siRNA in the 35.7% of the frames. (B) Dendrimer–siRNAs BS analysis computed for DM–1 siRNA (green), DM–2 siRNA (orange), DP–1 siRNA (blue), DP–2 siRNA (red).
To complete the picture, terminal-siRNA distance analysis has been performed in order to extract fruitful information on the interaction behavior of the different DM and DP characterizing beads. In greater detail, Table 2 shows the number of Q0, N0, and C1 dendrimer terminal beads present within three main distance intervals from the siRNAs: “primary interaction (d ≤ 0.6 nm)” indicates a strong stabilization range where terminal beads are primary involved in the dendrimer–siRNA complexation. The “Secondary interaction (0.6 nm < d ≤ 1.1 nm)” range indicates an interval where the terminal beads are still involved in interaction with siRNA by van der Waals and Coulomb forces. “Free terminals (d > 1.1 nm)” indicate that no interaction is occurring between beads and siRNA beyond this range. Remarkably, in both cases, where 1 or 2 siRNA are complexed, DM involves a higher number of Q0 and N0 terminal beads for complexation purposes, whereas DP utilizes a lower number of Q0 and C1 terminal beads. Interestingly, the N0 bead of the DM terminals is employed for primary stabilization tasks in a better way (DM–1 siRNA N0/Q0 = 0.73, DM–2 siRNA N0/Q0 = 0.72), rather than the C1 bead of the DP terminals (DP–1 siRNA C1/Q0 = 0.54, DP–2 siRNA C1/Q0 = 0.47).
Table 2. Number of Q0, N0, and C1 Dendrimer Terminal Beads Present within Three Main Ranges from siRNAs.
| Bead | primary interaction d ≤ 0.6 nm | secondary interaction 0.6 nm < d ≤ 1.1 nm | free terminals d > 1.1 nm | |
|---|---|---|---|---|
| DM–1 siRNA | Q0 | 14.6 | 6.1 | 27.3 |
| N0 | 10.6 | 10.3 | 27.1 | |
| DP–1 siRNA | Q0 | 8.4 | 4.4 | 35.2 |
| C1 | 4.5 | 9.0 | 34.5 | |
| DM–2 siRNA | Q0 | 10.8 | 5.7 | 31.4 |
| N0 | 7.8 | 8.7 | 31.5 | |
| DP–2 siRNA | Q0 | 10.0 | 4.5 | 33.5 |
| C1 | 4.7 | 10.6 | 32.7 |
T3. Dendrimer Competition Mechanisms
In this section, we have investigated possible competition phenomena which may affect the ability of DM and DP to complex in dendriplex assemblies (set up described in section Methods, category T3). To address this point, 15 replicas of 500 ns were carried out for each system, considering the last 50 ns for all the following analyses.
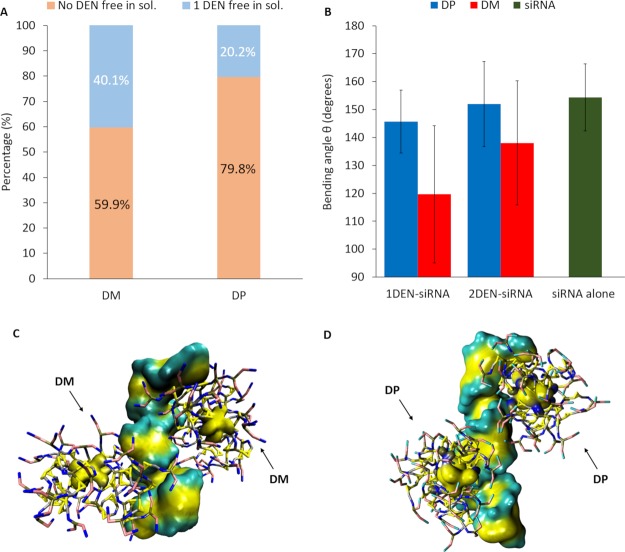
Figure 3A depicts the probability of the DP and DM dendrimers to be found in dendriplex or free in solution. Interestingly, DP dendrimers are found free in solution only with probability 0.2 among all replicas, whereas DM can be found free in solution with a higher probability up to 0.4. This implies that siRNA–DM complexes are in general constituted by 1 siRNA and 1 dendrimer.
Figure 3.
Picture (A) shows the DM and DP aggregation percentage bar diagram with siRNA and the percentage of 1 DM and DP free in solution. DP dendrimers are found free in solution only with probability 20% among all replicas, whereas DM can be found free in solution with a higher probability up to 40%. Picture (B) shows the bending angle “θ” bar diagram of 1DEN–siRNA, 2DEN–siRNA e siRNA alone. Pictures (C,D) show, respectively, 2DMs and 2DPs in close complexation with 1 siRNA.
It is worth noting that the higher probability to find DP dendrimers aggregated in dendriplexes is also driven by DP self-aggregation properties (already highlighted by in vitro experiments22 and shown in Figure 1). This unique DP property is crucial for promoting dendriplex stabilization and growth also through dendrimer–dendrimer contacts. In this sense, the DP self-aggregation tendency should be interpreted as a complexation promoting feature and not a competition mechanism. To complete the picture, a contact analysis has been performed in order to evaluate if the first contact occurs between the two dendrimers or between the siRNA and the dendrimers. The distance below which the molecules are considered as stably complexed is 0.6 nm. In detail, aggregation occurs first between the two dendrimers in 2 replicas out of total 15 (15.4%) for 2DP–siRNA systems, whereas it occurs in 1 replica out of total 15 (7.2%) for two DM–siRNA systems.
It is worth mentioning that DM–siRNA complexation is stabilized by a siRNA deformation while wrapping around dendrimer, as also shown by AA simulation of the previous literature.22 The abovementioned DM–siRNA complexation feature is also detectable in present CG simulations (Figure 3, Supporting Information S.6). Figure 3B quantifies the siRNA bending angle “θ” in three cases: 1DEN–siRNA dendriplex, 2DEN–siRNA dendriplex and siRNA alone. Interestingly, the results show that the siRNA bending angle, both when it interacts with one and two DPs, is close to the conformation assumed by siRNA when alone in water. In contrary, siRNA structure deformation reached lower bending angles both in the case of complexing with one and two DMs. However, siRNA is able to wrap around only one between the two bound DM (Figure 3C, Supporting Information S.6). This aspect suggests that only one DM will be stabilized in the complexation, whereas the other will be more likely able to detach. In this sense, we highlight a competition mechanism in DM–siRNA complexation, which is not present in DP–siRNA complexation (Figure 3D, Supporting Information S.6). All these evidences suggest some competition phenomena among dendrimers when an excess of dendrimers are present in the solutions, as usual in experimental studies. In detail, considering that the dendrimer–siRNA molar ratio is always higher than 1,22 the competition mechanisms can alter certain chemical parameters, including the stoichiometric coefficients.
Discussion
Dendrimers are polymeric hyperbranched nanocarriers characterized by the globular shape with high monodispersity and high degree of versatility, which have outstanding features for nanomedicine.34−44 Within this context, polycationic dendrimers have proven to be an excellent drug delivery carrier for gene therapy strategies.45,46 In this paper, we have employed CG MD to investigate complexation dynamics of siRNA with two different type of polycationic phosphorous dendrimers, namely, the pyrrolidinium and morpholinium dendrimers. MD has already shown to be a powerful tool to investigate nanoscale phenomena driving macromolecules properties from the molecular to supramolecular scale.44,47−51 Here, our in silico study aims at exploring molecular features driving supramolecular complexation in terms of assemble mechanisms and competition phenomena. In this framework, dendrimers or dendrons self-assembled supramolecular nanostructures reduce the difficulty in overcoming the plasmatic membrane, resulting in an increased cellular uptake of siRNAs.52−54 Particular importance must be given to the dendriplex size.6,17 More precisely, the size of dendriplex plays a key role in avoiding problems such as little effectiveness or excessive toxicity on the transfected cells.55 In this study, we have selected two different cationic phosphorus dendrimers of generation 3, functionalized, respectively, with pyrrolidinium (DP) and morpholinium (DM) surface groups, which demonstrated a promising potential for siRNA delivery.6 Our data have demonstrated that DP has an increased capacity to self-assembly rather than DM. Such behavior is related with the different dendrimers’ chemical nature of the terminal beads, which for DM is polar and for DP is completely apolar. Because systems are immersed in an aqueous solvent, the behavior of nonpolar particles to aggregate is increased, and probably it results in our demonstrated tighter self-assembly showed by DP. Further studies on the size and the polydispersity index of the DP aggregates could be indicated to improve the control and the prediction of this sensitive characteristic, with the aim of avoiding adverse events. Another important feature to investigate in order to modulate the dendriplex size and conformation is the dendrimer complexation behavior with siRNA molecules.56 In this context, cationic dendrimer–siRNA aggregates formation are mainly driven by electrostatic interactions.57 Therefore, tuning up the peripheral positive charge density and the terminal chemical structure of dendrimers may strongly influence the binding attitude with siRNAs.58 Recent study has highlighted how even small difference in dendrimer chemical composition can affect the enthalpy and entropic contribution of DP and DM, drastically changing their stoichiometric values.22 It has been already highlighted how the dendrimer multivalence behavior is intrinsically connected to the enthalpy contribution and the entropic penalty in the binding free energy.59−61 In our research, we have demonstrated that DP has an increased capacity to complex with 2 siRNAs, while DM has increased attitude to complex with 1 siRNA. Such a different behavior may indicate that DP can reorganize in a more efficient way its branches in order to bind 2 siRNAs. On the other hand, DM may suffer from higher conformational change when it complexes with 1 siRNA. This assumption is supported by the BS analysis (Figure 2B), which confirms that DM has a greater interaction surface both when it is complexed with 1 or 2 siRNAs. The higher BS values exhibited by DM are due to its capability to flex the siRNA structure, a behavior which is almost totally absent for the DP.22 Because DP has decreased the BS surface and employs a lower number of terminals for complexation purposes, it has an increased efficiency in binding siRNA rather than DM.6,22 In addition, the lower number of terminals employed by DP can be also useful in detachment of siRNAs once inside the cell, which can lead to an amplified bioactivity. On the other hand, we have shown that DP presents self-assembly mechanisms even in the presence of siRNA double-strained filaments. Considering that in vitro experiments are executed with an excess of dendrimer buffer,6,22 we can suggest that DP concentration within the dendriplex will be probably higher in comparison of DM concentration within the dendriplex. According to the recent literature, positively charged nanoparticles and aggregates have much greater propensity to translocate through cell membranes than negatively or neutral charged ones.62−64 In addition, the superficial positive charged molecules have proved to be also strongly correlated to cellular uptake and cytotoxicity.65−67 The higher presence of DPs into the dendriplex nanoparticles can result in a higher capability of masking the siRNA negative charge, leading in an increased cellular internalization. On the other hand, tuning properly the dendrimer hydrophobic/hydrophilic affinity with membranes68,69 can also aid the efficient translocation of dendriplexes inside the cell.62 In this framework, DP increased apolar character rather than DM, may also play a crucial role in the cellular uptake process, because of the increased hydrophobic affinity with the plasmatic membrane. Taken all together, it is reasonable to assume that DP increased binding multivalence and self-assembly attitude, with no significant competition phenomena, lead to higher dendriplex stabilization and neutralization which ultimately can improve the cell transfection.
Conclusions
In the present study, we adopted the MARTINI force-filed to shed light on the different supramolecular behavior of two cationic phosphorous dendrimers, namely, DM and DP, in complexing siRNA. We have shown how small surface modification might lead in significant changing on the dendrimer–siRNA complexation dynamics. The results indicate that DP is significantly more efficient in binding 2 siRNAs, while DM has increased attitude to complex with 1 siRNA. In this framework, we also highlighted a competition mechanism in DM–siRNA interaction, which is not present in DP–siRNA interaction. The outcome of this research provides fruitful insight in order to deeply understand the mechanisms driving the supramolecular dendriplex formation. In conclusion, this multiscale computational work paves the way for a future investigation of the dendriplex structures formed by the large-scale interaction of dendrimers and nucleic acids.
Methods
Dendrimers CG Models
Dendrimers CG models, in terms of coordinates and topology, were generated starting from AA trajectories of DM and DP by grouping AA coordinates and mapping them in CG beads as made in several previous works.24−27,29,51
AA-MD set up and results concerning conformational stability are reported in our previous work and summarized in Supporting Information S.1. The AA simulations of single dendrimer in water will be used here as reference for validation of CG dendrimer models.
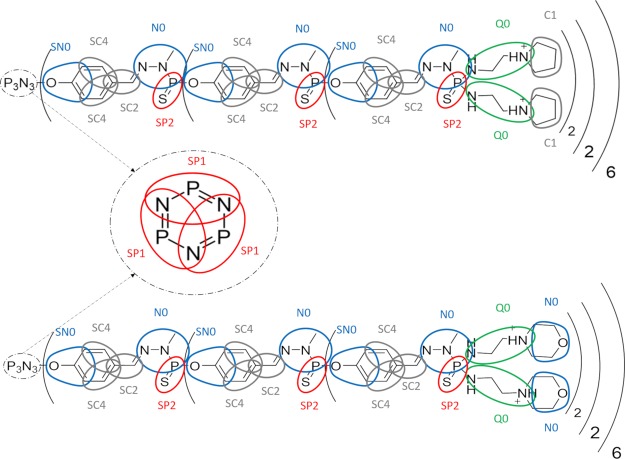
CG-AA maps for DM and DP dendrimers are shown in Figure 4. More details about the AA group division and bead identification is reported in Supporting Information S.2. Nonbonded parameters assigned to each CG bead have been taken from the MARTINI forcefield.24 AA trajectories and AA-CG maps were used as input for PyCGTOOL70 in order to generate the dendrimer CG-bonded terms topology.71,72
Figure 4.
DP (top picture) and DM (bottom picture) maps applied to the AA structure. Different bead polarity is highlighted with different color: apolar beads are indicated in gray, nonpolar beads in blue, polar beads in red, and charged beads in green.
Dendrimer CG Model Validation
As mentioned above, the dendrimer CG models were validated by comparing the dendrimer AA and CG dendrimer conformational dynamics. In a greater detail, AA-MD and CG-MD simulations have been performed on the same molecular system (dendrimer in water) for both DP and DM.
The tuning of bonded parameters was done using the iterative modified Boltzmann inversion (ImBI) technique (Supporting Information S.3), to derive potentials of the bonded terms in order to match the topological parameters of reference atomistic models. Structural conformation of both CG dendrimer models was evaluated in comparison with AA trajectories, by measuring mean and standard deviation of the radius of gyration and root mean square fluctuations.
The ImBI requires several steps of CG dynamics and a topology refinement on the basis of a comparison with system properties obtained at the CG level and the same properties obtained by AA-MD (target trajectory).48 Concerning the CG level, each dendrimer (DP and DM) was positioned in the center of a dodecahedron box filled with normal water beads, using 0.21 nm as van der Waals distance, and ions (Na+ and Cl–) at a physiological concentration (0.15 M). To prevent unwanted CG water freezing, 20% of normal P4 water beads were replaced with special-type BP4 antifreeze water beads.24
Then, each simulation step of the ImBI was performed as follows. The system (dendrimer in water) was energy-minimized by 2000 steps of the steepest descent energy minimization algorithm. A 100 ps position-restrained MD was performed at 320 K using the v-rescale73 thermostat in the NVT ensemble. Then, a 5 ns position-restrained MD was performed at 320 K and 1 atm using Berendsen barostat in the NPT ensemble.74 Finally, a 100 ns MD without position restraints was performed in the NPT ensemble coupling the system by the Parrinello–Rahman75 barostat and the v-rescale73 thermostat. Atom velocities were randomly initialized following a Maxwell–Boltzmann distribution. Long-ranged electrostatic interactions were calculated at every step with the reaction-field method, using a relative dielectric constant value of 15, with a cut-off of 1.1 nm.76 A cut-off of 1.1 nm was also applied to Lennard–Jones interactions.76 The LINCS77 algorithm approach allowed an integration time step of 10 fs.
Validation analysis was performed taking into account the last 20 ns of production run, following two main strategies: (A) suit the bonded parameters as similar as possible to the atomistic models and (B) try to optimize the conformational features of the dendrimers as close as possible to the atomistic simulations.70,78,79
The detailed validation procedure that has been adopted can be found in the Supporting Information S.4.
Generation of siRNA CG Model
The parametrization followed to obtain the siRNA CG model is based on the MARTINI DNA force-field extension80 adapted in order to achieve a correct implementation of the RNA properties.78 The siRNA CG model was created with a soft elastic network which has allowed the building of correct double-stranded siRNA’s structure.78 In detail, the soft model has been found in good agreement with the experimental persistence length and helical parameters for dsRNA molecules.78 The maximum recommended time-step in order to maintain the simulations stability is 10 fs.78
CG MD of Dendrimer–Dendrimer and Dendrimer–siRNA Complexation
CG MD developed in this work can be divided into three main categories (T1, T2, and T3). For each category, a different molecular system has been considered.
T1. Investigation of Dendrimer Self-Aggregation Mechanisms
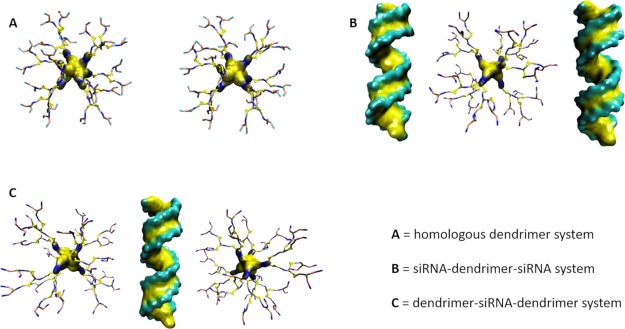
Two homologues, for example, 2DM molecules (or 2DP molecules), have been positioned at a minimum distance of 3 nm from each other (Figure 5A). The simulation test aims at investigating DP and DM self-aggregation tendency.
Figure 5.
Representation of the three simulated systems (T1, T2, T3) for each dendrimer type; (A) two homologues dendrimers in water (T1), (B) siRNA–dendrimer–siRNA in water (T2), (C) dendrimer–siRNA–dendrimer in water (T3). Water and ions are not shown in the picture.
T2. Investigation of siRNA–Dendrimer Binding Stoichiometry
A system consisting of 2 siRNA and 1 dendrimer (DM or DP) has been built by positioning each molecule at a minimum distance of 2 nm from each other. The simulation test aims at investigating molecular reasons behind a different stoichiometry shown by DP and DM in binding siRNA (Figure 5B).
T3. Investigation of Dendrimer Competition Mechanisms in Binding siRNA
A system consisting of 2 dendrimers and 1 siRNA has been built by positioning each molecule at a minimum distance of 2 nm from each other. The simulation test aims at investigating if some kind of competition mechanism may affect the dendrimer ability to bind siRNA (Figure 5C).
All CG simulations have been carried out in CG modeled explicit water and ions as described below. In all the cases, the molecular system was positioned in the center of a dodecahedron box with a minimum periodic images distance no smaller than 2.0 nm and solvated with nonpolarizable water beads. After that Na–Cl ions were added at a concentration of 0.15 M, such as human extracellular ions concentration, forming systems with a total amount of about 45,000 interacting beads. MARTINI v_2.1-dna forcefield24 was adopted for CG simulations. To avoid freezing of the solvent beads, 20% of normal water beads (P4) was replaced with heavy water beads (BP4).24 Long-ranged electrostatic interactions were calculated at every step choosing the reaction-field method, using a relative dielectric constant value of 15, with a cut-off of 1.1 nm.76 A cut-off of 1.1 nm was also applied to vdW interactions.76
Each system was energy-minimized by 2000 steps of steepest descent energy minimization algorithm. A 100 ps position-restrained MD was performed at 320 K using v-rescale73 thermostat in the NVT ensemble. Then was performed a 5 ns position-restrained MD at 320 K and 1 atm using Berendsen74 barostat in the NPT ensemble, giving the time to equilibrate the system density. Atom velocities were randomly initialized following a Maxwell–Boltzmann distribution. Finally, 10 MD replicas for category T1 and 15 MD replicas for both T2 and T3 were performed for 500 ns each, without position restrains, in the NPT ensemble using Parrinello–Rahman75 barostat. Each replica was characterized by different atom velocities at the beginning of the MD simulation. All the performed analyses were done considering the last 50 ns of each replica.
GROMACS 2018.381,82 package was used for all MD simulations. The visual MD (VMD)83 package was used for the visual inspection of the simulated systems and for systems snapshot rendering.
Acknowledgments
Authors gratefully acknowledge the financial support from the Swiss National Foundation through the grant IZCNZ0-174842 (“Computational modeling to characterize and optimize dendrimer-based nanocarriers for siRNA delivery in Chronicle Obstructive Pulmonary Disease”). The authors would also like to acknowledge the financial support provided by COST-European Cooperation in Science and Technology, to the COST Action MP1404: Simulation and pharmaceutical technologies for advanced patient-tailored inhaled medicines (SimInhale). This work was also supported by the Swiss National Supercomputing Centre (CSCS).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b03908.
DP maximizes the probability to bind 2 siRNA molecules (AVI)
DP maximizes the probability to bind 2 siRNA molecules if compared with DM (AVI)
AA MD of DM and DP; features of CG maps; modified Boltzmann inversion; topological parameter validation; siRNA bending angle; and dendrimer competition phenomena (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Kim D. H.; Rossi J. J. Strategies for Silencing Human Disease Using RNA Interference. Nat. Rev. Genet. 2007, 8, 173–184. 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- Kaczmarek J. C.; Kowalski P. S.; Anderson D. G. Advances in the Delivery of RNA Therapeutics: From Concept to Clinical Reality. Genome Med. 2017, 9, 60. 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koli U.; Krishnan R. A.; Pofali P.; Jain R.; Dandekar P. SiRNA-Based Therapies for Pulmonary Diseases. J. Biomed. Nanotechnol. 2014, 10, 1953–1997. 10.1166/jbn.2014.1928. [DOI] [PubMed] [Google Scholar]
- Jain A.; Mahira S.; Majoral J. P.; Bryszewska M.; Khan W.; Ionov M. Dendrimer Mediated Targeting of SiRNA against Polo-like Kinase for the Treatment of Triple Negative Breast Cancer. J. Biomed. Mater. Res., Part A 2019, 107, 1933–1944. 10.1002/jbm.a.36701. [DOI] [PubMed] [Google Scholar]
- Aagaard L.; Rossi J. J. RNAi Therapeutics: Principles, Prospects and Challenges. Adv. Drug Delivery Rev. 2007, 59, 75–86. 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr A.; Tsapis N.; Andreana I.; Chamarat A.; Foged C.; Delomenie C.; Noiray M.; El Brahmi N.; Majoral J.-P.; Mignani S.; Fattal E. Anti-Inflammatory Effect of Anti-TNF-α SiRNA Cationic Phosphorus Dendrimer Nanocomplexes Administered Intranasally in a Murine Acute Lung Injury Model. Biomacromolecules 2017, 18, 2379–2388. 10.1021/acs.biomac.7b00572. [DOI] [PubMed] [Google Scholar]
- Iranpur Mobarakeh V.; Modarressi M. H.; Rahimi P.; Bolhassani A.; Arefian E.; Atyabi F.; Vahabpour R. Optimization of Chitosan Nanoparticles as an Anti-HIV SiRNA Delivery Vehicle. Int. J. Biol. Macromol. 2019, 129, 305–315. 10.1016/j.ijbiomac.2019.02.036. [DOI] [PubMed] [Google Scholar]
- Harvey R. D.; Bourgeois S.; Pietzonka P.; Desire L.; Fattal E. Microencapsulation of Nanoparticulate Complexes of DNA with Cationic Lipids and Polymers in Pectin Beads for Targeted Gene Delivery. Nanobiotechnology 2005, 1, 071–082. 10.1385/nbt:1:1:071. [DOI] [Google Scholar]
- Xue H.; Guo P.; Wen W.-C.; Wong H. Lipid-Based Nanocarriers for RNA Delivery. Curr. Pharm. Des. 2015, 21, 3140–3147. 10.2174/1381612821666150531164540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattal E.; Barratt G. Nanotechnologies and Controlled Release Systems for the Delivery of Antisense Oligonucleotides and Small Interfering RNA. Br. J. Pharmacol. 2009, 157, 179–194. 10.1111/j.1476-5381.2009.00148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa G.; La Rotonda M. I. Nano and Microtechnologies for the Delivery of Oligonucleotides with Gene Silencing Properties. Molecules 2009, 14, 2801–2823. 10.3390/molecules14082801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruxel F.; Cojean S.; Bochot A.; Teixeira H.; Bories C.; Loiseau P.-M.; Fattal E. Cationic Nanoemulsion as a Delivery System for Oligonucleotides Targeting Malarial Topoisomerase II. Int. J. Pharm. 2011, 416, 402–409. 10.1016/j.ijpharm.2011.01.048. [DOI] [PubMed] [Google Scholar]
- Dzmitruk V.; Apartsin E.; Ihnatsyeu-Kachan A.; Abashkin V.; Shcharbin D.; Bryszewska M. Dendrimers Show Promise for SiRNA and MicroRNA Therapeutics. Pharmaceutics 2018, 10, 126. 10.3390/pharmaceutics10030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso G.; Deriu M. A.; Patrulea V.; Borchard G.; Möller M.; Danani A. Free Energy Landscape of SiRNA-Polycation Complexation: Elucidating the Effect of Molecular Geometry, Polymer Flexibility, and Charge Neutralization. PLoS One 2017, 12, e0186816 10.1371/journal.pone.0186816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loup C.; Zanta M.-A.; Caminade A.-M.; Majoral J.-P.; Meunier B. Preparation of Water-Soluble Cationic Phosphorus-Containing Dendrimers as DNA Transfecting Agents. Chem.—Eur. J. 1999, 5, 3644–3650. . [DOI] [Google Scholar]
- Caminade A.-M.; Majoral J.-P. Water-Soluble Phosphorus-Containing Dendrimers. Prog. Polym. Sci. 2005, 30, 491–505. 10.1016/j.progpolymsci.2005.01.001. [DOI] [Google Scholar]
- Ferenc M.; Pedziwiatr-Werbicka E.; Nowak K.; Klajnert B.; Majoral J.-P.; Bryszewska M. Phosphorus Dendrimers as Carriers of SiRNA-Characterisation of Dendriplexes. Molecules 2013, 18, 4451–4466. 10.3390/molecules18044451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminade A.-M.; Zibarov A.; Cueto Diaz E.; Hameau A.; Klausen M.; Moineau-Chane Ching K.; Majoral J.-P.; Verlhac J.-B.; Mongin O.; Blanchard-Desce M. Fluorescent Phosphorus Dendrimers Excited by Two Photons: Synthesis, Two-Photon Absorption Properties and Biological Uses. Beilstein J. Org. Chem. 2019, 15, 2287–2303. 10.3762/bjoc.15.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briz V.; Serramia M. J.; Madrid R.; Hameau A.; Caminade A.-M.; Majoral J. P.; Munoz-Fernandez M. A. Validation of a Generation 4 Phosphorus-Containing Polycationic Dendrimer for Gene Delivery Against HIV-1. Curr. Med. Chem. 2012, 19, 5044–5051. 10.2174/0929867311209025044. [DOI] [PubMed] [Google Scholar]
- Shcharbin D.; Dzmitruk V.; Shakhbazau A.; Goncharova N.; Seviaryn I.; Kosmacheva S.; Potapnev M.; Pedziwiatr-Werbicka E.; Bryszewska M.; Talabaev M.; Chernov A.; Kulchitsky V.; Caminade A.-M.; Majoral J.-P. Fourth Generation Phosphorus-Containing Dendrimers: Prospective Drug and Gene Delivery Carrier. Pharmaceutics 2011, 3, 458–473. 10.3390/pharmaceutics3030458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminade A.-M.; Turrin C.-O.; Majoral J.-P. Biological Properties of Phosphorus Dendrimers. New J. Chem. 2010, 34, 1512–1524. 10.1039/c0nj00116c. [DOI] [Google Scholar]
- Deriu M. A.; Tsapis N.; Noiray M.; Grasso G.; El Brahmi N.; Mignani S.; Majoral J.-P.; Fattal E.; Danani A. Elucidating the Role of Surface Chemistry on Cationic Phosphorus Dendrimer-SiRNA Complexation. Nanoscale 2018, 10, 10952–10962. 10.1039/c8nr01928b. [DOI] [PubMed] [Google Scholar]
- Ingólfsson H. I.; Lopez C. A.; Uusitalo J. J.; de Jong D. H.; Gopal S. M.; Periole X.; Marrink S. J. The Power of Coarse Graining in Biomolecular Simulations. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2014, 4, 225–248. 10.1002/wcms.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink S. J.; Risselada H. J.; Yefimov S.; Tieleman D. P.; de Vries A. H. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B 2007, 111, 7812–7824. 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- Marrink S. J.; Tieleman D. P. Perspective on the Martini Model. Chem. Soc. Rev. 2013, 42, 6801–6822. 10.1039/c3cs60093a. [DOI] [PubMed] [Google Scholar]
- Riniker S.; Allison J. R.; Van Gunsteren W. F. On Developing Coarse-Grained Models for Biomolecular Simulation: A Review. Phys. Chem. Chem. Phys. 2012, 14, 12423–12430. 10.1039/c2cp40934h. [DOI] [PubMed] [Google Scholar]
- Zhong T.; Ai P.; Zhou J. Structures and Properties of PAMAM Dendrimer: A Multi-Scale Simulation Study. Fluid Phase Equilib. 2011, 302, 43–47. 10.1016/j.fluid.2010.09.037. [DOI] [Google Scholar]
- Tian W.-d.; Ma Y.-q. Coarse-Grained Molecular Simulation of Interacting Dendrimers. Soft Matter 2011, 7, 500–505. 10.1039/C0SM00696C. [DOI] [Google Scholar]
- Rahimi A.; Amjad-Iranagh S.; Modarress H. Molecular Dynamics Simulation of Coarse-Grained Poly(L-Lysine) Dendrimers. J. Mol. Model. 2016, 22, 1–10. 10.1007/s00894-016-2925-0. [DOI] [PubMed] [Google Scholar]
- Lee H.; Larson R. G. Molecular Dynamics Simulations of PAMAM Dendrimer-Induced Pore Formation in DPPC Bilayers with a Coarse-Grained Model. J. Phys. Chem. B 2006, 110, 18204–18211. 10.1021/jp0630830. [DOI] [PubMed] [Google Scholar]
- Wang Y.-L.; Lu Z.-Y.; Laaksonen A. Specific Binding Structures of Dendrimers on Lipid Bilayer Membranes. Phys. Chem. Chem. Phys. 2012, 14, 8348–8359. 10.1039/c2cp40700k. [DOI] [PubMed] [Google Scholar]
- Tian W.-d.; Ma Y.-q. Insights into the Endosomal Escape Mechanism via Investigation of Dendrimer-Membrane Interactions. Soft Matter 2012, 8, 6378–6384. 10.1039/c2sm25538c. [DOI] [Google Scholar]
- Lee H.; Larson R. G. Coarse-Grained Molecular Dynamics Studies of the Concentration and Size Dependence of Fifth- and Seventh-Generation PAMAM Dendrimers on Pore Formation in DMPC Bilayer. J. Phys. Chem. B 2008, 112, 7778–7784. 10.1021/jp802606y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignani S.; Rodrigues J.; Roy R.; Shi X.; Ceña V.; El Kazzouli S.; Majoral J.-P. Exploration of Biomedical Dendrimer Space Based on In-Vitro Physicochemical Parameters: Key Factor Analysis (Part 1). Drug Discovery Today 2019, 24, 1176–1183. 10.1016/j.drudis.2019.02.014. [DOI] [PubMed] [Google Scholar]
- Barrett T.; Ravizzini G.; Choyke P.; Kobayashi H. Dendrimers in Medical Nanotechnology: Application of Dendrimer Molecules in Bioimaging and Cancer Treatment. IEEE Eng. Med. Biol. Mag. 2009, 28, 12–22. 10.1109/MEMB.2008.931012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriu M. A.; Popescu L. M.; Ottaviani M. F.; Danani A.; Piticescu R. M. Iron Oxide/PAMAM Nanostructured Hybrids: Combined Computational and Experimental Studies. J. Mater. Sci. 2016, 51, 1996–2007. 10.1007/s10853-015-9509-8. [DOI] [Google Scholar]
- Tomalia D. A.; Reyna L. A.; Svenson S. Dendrimers as Multi-Purpose Nanodevices for Oncology Drug Delivery and Diagnostic Imaging. Biochem. Soc. Trans. 2007, 35, 61–67. 10.1042/bst0350061. [DOI] [PubMed] [Google Scholar]
- Bullen H. A.; Hemmer R.; Haskamp A.; Cason C.; Wall S.; Spaulding R.; Rossow B.; Hester M.; Caroway M.; Haik K. L. Evaluation of Biotinylated PAMAM Dendrimer Toxicity in Models of the Blood Brain Barrier: A Biophysical and Cellular Approach. J. Biomater. Nanobiotechnol. 2011, 02, 485–493. 10.4236/jbnb.2011.225059. [DOI] [Google Scholar]
- Fruchon S.; Poupot M.; Martinet L.; Turrin C.-O.; Majoral J.-P.; Fournié J.-J.; Caminade A.-M.; Poupot R. Anti-Inflammatory and Immunosuppressive Activation of Human Monocytes by a Bioactive Dendrimer. J. Leukocyte Biol. 2009, 85, 553–562. 10.1189/jlb.0608371. [DOI] [PubMed] [Google Scholar]
- Blattes E.; Vercellone A.; Eutamene H.; Turrin C.-O.; Theodorou V.; Majoral J.-P.; Caminade A.-M.; Prandi J.; Nigou J.; Puzo G. Mannodendrimers Prevent Acute Lung Inflammation by Inhibiting Neutrophil Recruitment. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 8795–8800. 10.1073/pnas.1221708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledall J.; Fruchon S.; Garzoni M.; Pavan G. M.; Caminade A.-M.; Turrin C.-O.; Blanzat M.; Poupot R. Interaction Studies Reveal Specific Recognition of an Anti-Inflammatory Polyphosphorhydrazone Dendrimer by Human Monocytes. Nanoscale 2015, 7, 17672–17684. 10.1039/c5nr03884g. [DOI] [PubMed] [Google Scholar]
- Tian F.; Lin X.; Valle R. P.; Zuo Y. Y.; Gu N. Poly(Amidoamine) Dendrimer as a Respiratory Nanocarrier: Insights from Experiments and Molecular Dynamics Simulations. Langmuir 2019, 35, 5364–5371. 10.1021/acs.langmuir.9b00434. [DOI] [PubMed] [Google Scholar]
- Caminade A.-M. Phosphorus Dendrimers for Nanomedicine. Chem. Commun. 2017, 53, 9830–9838. 10.1039/C7CC04949H. [DOI] [PubMed] [Google Scholar]
- Janaszewska A.; Klajnert-Maculewicz B.; Marcinkowska M.; Duchnowicz P.; Appelhans D.; Grasso G.; Deriu M. A.; Danani A.; Cangiotti M.; Ottaviani M. F. Multivalent Interacting Glycodendrimer to Prevent Amyloid-Peptide Fibril Formation Induced by Cu(II): A Multidisciplinary Approach. Nano Res. 2018, 11, 1204–1226. 10.1007/s12274-017-1734-9. [DOI] [Google Scholar]
- Zheng M.; Pavan G. M.; Neeb M.; Schaper A. K.; Danani A.; Klebe G.; Merkel O. M.; Kissel T. Targeting the Blind Spot of Polycationic Nanocarrier-Based SiRNA Delivery. ACS Nano 2012, 6, 9447–9454. 10.1021/nn301966r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan G. M.; Posocco P.; Tagliabue A.; Maly M.; Malek A.; Danani A.; Ragg E.; Catapano C. V.; Pricl S. PAMAM Dendrimers for SiRNA Delivery: Computational and Experimental Insights. Chemistry 2010, 16, 7781–7795. 10.1002/chem.200903258. [DOI] [PubMed] [Google Scholar]
- Bidone T. C.; Kim T.; Deriu M. A.; Morbiducci U.; Kamm R. D. Multiscale Impact of Nucleotides and Cations on the Conformational Equilibrium, Elasticity and Rheology of Actin Filaments and Crosslinked Networks. Biomech. Model. Mechanobiol. 2015, 14, 1143–1155. 10.1007/s10237-015-0660-6. [DOI] [PubMed] [Google Scholar]
- Deriu M. A.; Shkurti A.; Paciello G.; Bidone T. C.; Morbiducci U.; Ficarra E.; Audenino A.; Acquaviva A. Multiscale Modeling of Cellular Actin Filaments: From Atomistic Molecular to Coarse-Grained Dynamics. Proteins 2012, 80, 1598–1609. 10.1002/prot.24053. [DOI] [PubMed] [Google Scholar]
- Havelka D.; Deriu M. A.; Cifra M.; Kučera O. Deformation Pattern in Vibrating Microtubule: Structural Mechanics Study Based on an Atomistic Approach. Sci. Rep. 2017, 7, 4227. 10.1038/s41598-017-04272-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soncini M.; Vesentini S.; Ruffoni D.; Orsi M.; Deriu M. A.; Redaelli A. Mechanical Response and Conformational Changes of Alpha-Actinin Domains during Unfolding: A Molecular Dynamics Study. Biomech. Model. Mechanobiol. 2007, 6, 399–407. 10.1007/s10237-006-0060-z. [DOI] [PubMed] [Google Scholar]
- Deriu M. A.; Cangiotti M.; Grasso G.; Licandro G.; Lavasanifar A.; Tuszynski J. A.; Ottaviani M. F.; Danani A. Self-Assembled Ligands Targeting TLR7: A Molecular Level Investigation. Langmuir 2017, 33, 14460–14471. 10.1021/acs.langmuir.7b03168. [DOI] [PubMed] [Google Scholar]
- Isaacman S.; Buckley M.; Wang X.; Wang E. Y.; Liebes L.; Canary J. W. Targeted Amplification of Delivery to Cell Surface Receptors by Dendrimer Self-Assembly. Bioorg. Med. Chem. Lett. 2014, 24, 1290–1293. 10.1016/j.bmcl.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez-Miranda V.; Araya-Durán I.; Camarada M. B.; Comer J.; Valencia-Gallegos J. A.; González-Nilo F. D. Self-Assembly of Amphiphilic Dendrimers: The Role of Generation and Alkyl Chain Length in SiRNA Interaction. Sci. Rep. 2016, 6, 29436. 10.1038/srep29436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.; Posocco P.; Liu X.; Cheng Q.; Laurini E.; Zhou J.; Liu C.; Wang Y.; Tang J.; Col V. D.; Yu T.; Giorgio S.; Fermeglia M.; Qu F.; Liang Z.; Rossi J. J.; Liu M.; Rocchi P.; Pricl S.; Peng L. Mastering Dendrimer Self-Assembly for Efficient SiRNA Delivery: From Conceptual Design to In Vivo Efficient Gene Silencing. Small 2016, 12, 3667–3676. 10.1002/smll.201503866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.; Shao N.; Wang Y.; Cheng Y. Clustering Small Dendrimers into Nanoaggregates for Efficient DNA and SiRNA Delivery with Minimal Toxicity. Adv. Healthcare Mater. 2016, 5, 584–592. 10.1002/adhm.201500679. [DOI] [PubMed] [Google Scholar]
- Shakya A.; Dougherty C. A.; Xue Y.; Al-Hashimi H. M.; Banaszak Holl M. M. Rapid Exchange Between Free and Bound States in RNA–Dendrimer Polyplexes: Implications on the Mechanism of Delivery and Release. Biomacromolecules 2016, 17, 154–164. 10.1021/acs.biomac.5b01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L. B.; Mortensen K.; Pavan G. M.; Kasimova M. R.; Jensen D. K.; Gadzhyeva V.; Nielsen H. M.; Foged C. Molecular Characterization of the Interaction between SiRNA and PAMAM G7 Dendrimers by SAXS, ITC, and Molecular Dynamics Simulations. Biomacromolecules 2010, 11, 3571–3577. 10.1021/bm101033g. [DOI] [PubMed] [Google Scholar]
- Jensen L. B.; Pavan G. M.; Kasimova M. R.; Rutherford S.; Danani A.; Nielsen H. M.; Foged C. Elucidating the Molecular Mechanism of PAMAM-SiRNA Dendriplex Self-Assembly: Effect of Dendrimer Charge Density. Int. J. Pharm. 2011, 416, 410–418. 10.1016/j.ijpharm.2011.03.015. [DOI] [PubMed] [Google Scholar]
- Pavan G. M.; Albertazzi L.; Danani A. Ability to Adapt: Different Generations of PAMAM Dendrimers Show Different Behaviors in Binding SiRNA. J. Phys. Chem. B 2010, 114, 2667–2675. 10.1021/jp100271w. [DOI] [PubMed] [Google Scholar]
- Vasumathi V.; Maiti P. K. Complexation of SiRNA with Dendrimer: A Molecular Modeling Approach. Macromolecules 2010, 43, 8264–8274. 10.1021/ma1012495. [DOI] [Google Scholar]
- Pavan G. M.; Danani A.; Pricl S.; Smith D. K. Modeling the Multivalent Recognition between Dendritic Molecules and DNA: Understanding How Ligand “Sacrifice” and Screening Can Enhance Binding. J. Am. Chem. Soc. 2009, 131, 9686–9694. 10.1021/ja901174k. [DOI] [PubMed] [Google Scholar]
- Fox L. J.; Richardson R. M.; Briscoe W. H. PAMAM Dendrimer—Cell Membrane Interactions. Adv. Colloid Interface Sci. 2018, 257, 1–18. 10.1016/j.cis.2018.06.005. [DOI] [PubMed] [Google Scholar]
- Hong S.; Leroueil P. R.; Janus E. K.; Peters J. L.; Kober M.-M.; Islam M. T.; Orr B. G.; Baker J. R.; Banaszak Holl M. M. Interaction of Polycationic Polymers with Supported Lipid Bilayers and Cells: Nanoscale Hole Formation and Enhanced Membrane Permeability. Bioconjugate Chem. 2006, 17, 728–734. 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
- Gorzkiewicz M.; Deriu M. A.; Studzian M.; Janaszewska A.; Grasso G.; Pułaski Ł.; Appelhans D.; Danani A.; Klajnert-Maculewicz B. Fludarabine-Specific Molecular Interactions with Maltose-Modified Poly (Propyleneimine) Dendrimer Enable Effective Cell Entry of Active Drug Form: Comparison with Clofarabine. Biomacromolecules 2019, 20, 1429–1442. 10.1021/acs.biomac.9b00010. [DOI] [PubMed] [Google Scholar]
- Fröhlich E. The Role of Surface Charge in Cellular Uptake and Cytotoxicity of Medical Nanoparticles. Int. J. Nanomed. 2012, 7, 5577–5591. 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S.; Ershov D.; Gucht J. v. d.; Alink G. M.; Rietjens I. M. C. M.; Zuilhof H.; Marcelis A. T. M. Surface Charge-Specific Cytotoxicity and Cellular Uptake of Tri-Block Copolymer Nanoparticles. Nanotoxicology 2013, 7, 71–84. 10.3109/17435390.2011.633714. [DOI] [PubMed] [Google Scholar]
- Chen L.; Simpson J. D.; Fuchs A. V.; Rolfe B. E.; Thurecht K. J. Effects of Surface Charge of Hyperbranched Polymers on Cytotoxicity, Dynamic Cellular Uptake and Localization, Hemotoxicity, and Pharmacokinetics in Mice. Mol. Pharm. 2017, 14, 4485–4497. 10.1021/acs.molpharmaceut.7b00611. [DOI] [PubMed] [Google Scholar]
- Ionov M.; Gardikis K.; Wróbel D.; Hatziantoniou S.; Mourelatou H.; Majoral J.-P.; Klajnert B.; Bryszewska M.; Demetzos C. Interaction of Cationic Phosphorus Dendrimers (CPD) with Charged and Neutral Lipid Membranes. Colloids Surf., B 2011, 82, 8–12. 10.1016/j.colsurfb.2010.07.046. [DOI] [PubMed] [Google Scholar]
- Klajnert B.; Epand R. M. PAMAM Dendrimers and Model Membranes: Differential Scanning Calorimetry Studies. Int. J. Pharm. 2005, 305, 154–166. 10.1016/j.ijpharm.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Graham J. A.; Essex J. W.; Khalid S. PyCGTOOL: Automated Generation of Coarse-Grained Molecular Dynamics Models from Atomistic Trajectories. J. Chem. Inf. Model. 2017, 57, 650–656. 10.1021/acs.jcim.7b00096. [DOI] [PubMed] [Google Scholar]
- Khalid S.; Piggot T. J.; Samsudin F. Atomistic and Coarse Grain Simulations of the Cell Envelope of Gram-Negative Bacteria: What Have We Learned?. Acc. Chem. Res. 2019, 52, 180–188. 10.1021/acs.accounts.8b00377. [DOI] [PubMed] [Google Scholar]
- Goossens K.; De Winter H. Molecular Dynamics Simulations of Membrane Proteins: An Overview. J. Chem. Inf. Model. 2018, 58, 2193–2202. 10.1021/acs.jcim.8b00639. [DOI] [PubMed] [Google Scholar]
- Bussi G.; Donadio D.; Parrinello M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Berendsen H. J. C.; Postma J. P. M.; Van Gunsteren W. F.; Dinola A.; Haak J. R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. 10.1063/1.448118. [DOI] [Google Scholar]
- Parrinello M.; Rahman A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. 10.1063/1.328693. [DOI] [Google Scholar]
- de Jong D. H.; Baoukina S.; Ingólfsson H. I.; Marrink S. J. Martini Straight: Boosting Performance Using a Shorter Cutoff and GPUs. Comput. Phys. Commun. 2016, 199, 1–7. 10.1016/j.cpc.2015.09.014. [DOI] [Google Scholar]
- Hess B.; Bekker H.; Berendsen H. J. C.; Fraaije J. G. E. M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. . [DOI] [Google Scholar]
- Uusitalo J. J.; Ingólfsson H. I.; Marrink S. J.; Faustino I. Martini Coarse-Grained Force Field: Extension to RNA. Biophys. J. 2017, 113, 246–256. 10.1016/j.bpj.2017.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez C. L.; Petruk A.; Bringas M.; Estrin D. A.; Roitberg A. E.; Marti M. A.; Capece L. Coarse-Grained Simulations of Heme Proteins: Validation and Study of Large Conformational Transitions. J. Chem. Theory Comput. 2016, 12, 3390–3397. 10.1021/acs.jctc.6b00278. [DOI] [PubMed] [Google Scholar]
- Uusitalo J. J.; Ingólfsson H. I.; Akhshi P.; Tieleman D. P.; Marrink S. J. Martini Coarse-Grained Force Field: Extension to DNA. J. Chem. Theory Comput. 2015, 11, 3932–3945. 10.1021/acs.jctc.5b00286. [DOI] [PubMed] [Google Scholar]
- Abraham M. J.; Murtola T.; Schulz R.; Páll S.; Smith J. C.; Hess B.; Lindahl E. Gromacs: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Kutzner C.; Páll S.; Fechner M.; Esztermann A.; Groot B. L.; Grubmüller H. More Bang for Your Buck: Improved Use of GPU Nodes for GROMACS 2018. J. Comput. Chem. 2019, 40, 2418–2431. 10.1002/jcc.26011. [DOI] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: Visual Molecular Dynamics. J. Mol. Graphics 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.