Abstract
Comparison of epidemiological data on the occurrence of Toxoplasma (T.) gondii tissue cysts in meat is hampered by the lack of standardization and a great variety of methods for molecular detection. Therefore, this study aimed to compare and validate three different polymerase chain reaction (PCR) methods for detection of T. gondii DNA in pork. Analytical performance characteristics of two real time PCRs (qPCRs; Tg-qPCR1, Tg-qPCR2) and one conventional endpoint PCR (cPCR), all targeting the 529 repeated element, were assessed using genomic DNA of three clonal T. gondii types prevailing in Europe and North America. qPCR efficiencies for all three clonal types ranged between 93.8 and 94.4% (Tg-qPCR1) and 94.3–95.6% (Tg-qPCR2). Tg-qPCR1 and Tg-qPCR2 showed an overall PCR performance score of 85% and displayed a similar 95% detection limit of 1.067 and 1.561 genome equivalents per PCR reaction (GE/PCR), respectively. However, T. gondii DNA could be detected at concentrations as low as 0.1 GE/PCR. Reliable quantification is possible over 4 log ranges from 105 to 100 GE/PCR with mean repeatability relative standard deviations of ≤11% and reproducibility relative standard deviations of ≤12.7%. Presumably, both qPCRs are similarly suitable for sensitive and specific detection of T. gondii DNA in pork. In contrast, the cPCR using primer pair TOX5/Tox-8 proved to be highly sensitive with a detection limit of 1.41 GE/PCR, but not suitable for detection of T. gondii DNA in pork as unspecific amplification of porcine DNA was observed resulting in bands with similar size to the desired T. gondii-specific PCR product.
Keywords: Pork, qPCR, 529 RE, Food safety
Highlights
-
•
Comparison of two real time PCRs and one conventional PCR targeting the 529 RE.
-
•
Both real time PCRs show comparable performance and can be considered equivalent.
-
•
qPCRs show 95% detection limits of 1.067 and 1.561 genome equivalents per PCR.
-
•
Reliable quantification is possible over 4 log ranges from 105 to 100 GE/PCR.
-
•
cPCR using primer pair TOX5/Tox-8 showed non-specific amplification of porcine DNA.
1. Introduction
Toxoplasmosis is one of the most common parasitic zoonosis worldwide with approximately 30% of the human population chronically infected (Robert-Gangneux and Darde, 2012). Based on a high disease burden (expressed in Disability Adjusted Life Years), Toxoplasma (T.) gondii globally ranks among the most important foodborne pathogens (Scallan et al., 2015; WHO, 2015). Toxoplasmosis can be acquired horizontally by smear infection or oral ingestion of infectious parasite stages such as consumption of raw or undercooked meat containing highly infectious T. gondii tissue cysts. Water or fresh fruits and vegetables contaminated with T. gondii oocysts represent an additional infection source (Tenter et al., 2000). However, consumption of raw meat is considered as one of the most important risk factors for T. gondii infection in Europe (Kapperud et al., 1996; Cook et al., 2000).
For the assessment of meaningful and representative data on the occurrence of T. gondii tissue cysts in meat, reliable and sensitive methods for molecular detection are essential. However, comparison of epidemiological data is hampered by the great variety of methods for molecular detection of T. gondii described in the literature and lacking standardization. Published molecular techniques include conventional endpoint polymerase chain reaction (PCR), nested PCR, real time PCR (qPCR, either using SYBR green or TaqMan™ probes), and amplified loop-mediated isothermal amplification (LAMP) protocols which are reviewed elsewhere (Liu et al., 2015; Dzib Paredes et al., 2016). Moreover, several target genes have been used to detect T. gondii DNA in different sample types. Some target genes are present in multicopy, such as the B1 gene, the 529 repeated element (RE), the internal transcribed spacer element ITS-1, and 18S rDNA, while other targets represent single-copy genes such as the genes coding for the GRA1, GRA6, SAG1, and SAG2 proteins. In addition, there is also a diversity of published primers and protocols for the same molecular technique using an identical target gene.
This study aimed to compare the analytical and diagnostic performance of one conventional endpoint PCR (cPCR) and two qPCR assays for detection of T. gondii DNA in pork meat samples. The T. gondii (Tg)-qPCR1 (Opsteegh et al., 2010) and Tg-qPCR2 (Talabani et al., 2009) as well as a cPCR (Schares et al., 2008) using the primers TOX5 (Homan et al., 2000) and Tox-8 (Reischl et al., 2003) were chosen, as they are used in different reference laboratories across Europe and described in various studies (Schares et al., 2008; Herrmann et al., 2010; Herrmann, 2012; Gomez-Samblas et al., 2015; Opsteegh et al., 2016; Krücken et al., 2017; Schares et al., 2017; Burrells et al., 2018; Friedrich-Loeffler-Institut, 2018; Schares et al., 2018). Moreover, all three assays target the 529 RE that shows up to 200 to 300 copies per genome and thus generally allows detection of T. gondii DNA with a higher sensitivity compared to other targets genes such as the 35-copy B1 gene (Sterkers et al., 2010). As the number of repeats can vary among different T. gondii strains (Costa and Bretagne, 2012), performance characteristics were assessed using representative strains of the three clonal types prevailing in Europe and North America.
2. Materials & methods
2.1. Reference material
Genomic DNA of strains representative of the three clonal T. gondii types (strains RH (type 1), ME-49 (type 2), and NED (type 3)) served as reference material and was extracted from ~109 tachyzoites cultivated in MARC-145 cell monolayers. Tachyzoite pellets were incubated in 1 ml lysis buffer (100 mM sodium chloride (NaCl), 25 mM ethylene diamine tetraacetic acid, 0.5% sodium dodecyl sulphate, 10 mM Tris pH 8, 0.1 mg/ml proteinase K) for 12 h at 55 °C. DNA was extracted several times with 1 vol phenol-chloroform-isoamylalcohol (in a 25:24:1 ratio) until the supernatant was without visible contaminants and collected in a fresh 1.5 ml reaction tube. 4 M NaCl was added to yield a final 0.2 M NaCl concentration. DNA was precipitated with 2 vol absolute ethanol, washed with 1 ml 70% (v/v) ethanol, dried, resuspended in 100 μl of molecular grade water and stored at 4 °C. DNA extraction of tachyzoites of Neospora caninum and Besnoitia besnoiti was performed analogously. For Hammondia hammondi and Hammondia heydorni, DNA extraction was performed from oocysts using the NucleoSpin® Soil Kit (MACHEREY-NAGEL GmbH, Germany) according to the manufacturer's instructions. The quality and concentration of DNA extracts were determined using the NanoDrop 1000 Spectrophotometer (Peqlab, Erlangen, Germany). Based on the published genome size of 65.67 Mb of T. gondii strain ME-49 (GenBank Assembly ID GCA_000006565.2.) and a weight of 650 Da for 1 bp, one haploid genome of T. gondii was considered equivalent to 70.88 fg.
2.2. 529 RE quantitative polymerase chain reaction (qPCR)
Tg-qPCR1 and Tg-qPCR2 were carried out as described previously with some modifications (Talabani et al., 2009; Opsteegh et al., 2010). Reactions were performed on a 7500 real-time PCR system (Applied Biosystems, ABI) in a total volume of 25 μl using 10 μl template DNA and 12.5 μl of 2× TaqMan™ universal PCR mastermix (ABI). Oligonucleotides were synthesized by Metabion International AG (Martinsried, Germany) or TIB MOLBIOL Syntheselabor GmbH (Berlin, Germany) and are used at final concentrations listed in Table 1.
Table 1.
Oligonucleotide primers and TaqMan™ probes.
| Assay | Primer name | Gene target | Sequence (5′ - 3′)a | Cycling conditions | Final concentration in PCR (μM) | Reference |
|---|---|---|---|---|---|---|
| cPCR |
Tox-8 | T. gondii 529 RE | CCCAGCTGCGTCTGTCGGGAT | 94 °C - 1 min 35×: 94 °C - 1 min 60 °C - 1 min 72 °C - 1 min 72 °C - 10 min |
0.5 | (Reischl et al., 2003, Schares et al., 2008, Herrmann, 2012) |
| TOX5 | T. gondii 529 RE | CGCTGCAGACACAGTGCATCTGGATT | 0.5 | (Homan et al., 2000, Schares et al., 2008, Herrmann, 2012) | ||
| Tg-qPCR1 |
Tox-9F | T. gondii 529 RE | AGGAGAGATATCAGGACTGTAG | 50 °C - 2 min 95 °C - 10 min 45×: 95 °C -15 s 58 °C - 20 s 72 °C - 30 s |
0.7 | (Reischl et al., 2003, Opsteegh et al., 2010) |
| Tox-11R | T. gondii 529 RE | GCGTCGTCTCGTCTAGATCG | 0.7 | (Reischl et al., 2003, Opsteegh et al., 2010) | ||
| Tox-TP1 | T. gondii 529 RE | FAM™-CCGGCTTGGCTGCTTTTCCT-BHQ-1 | 0.1 | (Opsteegh et al., 2010) | ||
| CIAC-probe | Yersinia pestis caf1 | JOE™-AGCGTACCAACAAGTAATTCTGTATCGATG-BHQ-1 | 0.2 | (Opsteegh et al., 2010) | ||
| Tg-qPCR2 | T. gondii forward | T. gondii 529 RE | TGG TTG GGA AGC GAC GAG AG | 50 °C - 2 min 95 °C - 10 min 55×: 95 °C - 15 s 60 °C - 15 s 72 °C - 30 s |
0.8 | (Talabani et al., 2009) |
| T. gondii reverse | T. gondii 529 RE | CAT CAC CAC GAG GAA AGC GTC | 0.8 | (Talabani et al., 2009) | ||
| T. gondii LNA probe | T. gondii 529 RE | FAM™-AG [+A]GA [+C]AC [+C]GG [+A]ATGCG [+A]T-BHQ-1 | 0.2 | (Talabani et al., 2009) | ||
| pUC 18-F | pUC18/19 | TGT CGT GCC AGC TGC ATT A | 0.075 | (Mäde et al., 2008, Anonymous, 2013, Frentzel et al., 2018) | ||
| pUC 18-R | pUC18/19 | GAG CGA GGA AGC GGA AGA G | 0.075 | (Mäde et al., 2008, Anonymous, 2013, Frentzel et al., 2018) | ||
| Tm-pUC18 | pUC18/19 | JOE™-AAT CGG CCA ACG CGC GG-BHQ-1 | 0.1 | (Mäde et al., 2008, Anonymous, 2013, Frentzel et al., 2018) | ||
TaqMan™ probes are labelled with reporter dyes (5′-end) and quenchers (3′-end): FAM™, 6-carboxyfluorescein; JOE™, 5-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein; BHQ, Black Hole Quencher; LNA probe, locked nucleic acid-substituted TaqMan™ probe.
In order to identify false-negative results due to PCR inhibition, internal amplification controls (IAC) were included in both qPCRs. For Tg-qPCR1, a PCR product (189 bp) based on the Yersinia pestis caf1 gene was generated as previously described (Opsteegh et al., 2010) and used as a competitive IAC. However, the CIAC was used at a lower final concentration of 0.008 fg (40 copies) per qPCR reaction to ensure stable CIAC amplification with Cq values of 33–35 and to reduce inhibition of T. gondii DNA amplification to a minimum. In Tg-qPCR2, pUC18 DNA (Thermo Fisher Scientific) was included as a non-competitive IAC at a final concentration of 1.45 fg (500 copies) per qPCR reaction to enable stable amplification with Cq values of 31–33. Cycling conditions for both qPCR assays are listed in Table 1. In each run, a non-template control was included and genomic DNA of T. gondii strain RH corresponding to 100 GE/reaction served as positive control.
2.3. Analysis of qPCR runs
The 7500 Fast Systems Software (ABI) was used to calculate the quantification cycle (Cq). After adjusting the threshold line for each target and each run independently in the middle of the exponential phase, the mean threshold ΔRn value over all runs was calculated and evaluated as the optimal threshold value for the corresponding target gene. For standardization during the validation process, the threshold lines were set manually to the established ΔRn values of 0.7 (529 RE, Tg-qPCR1), 0.08 (529 RE, Tg-qPCR2), 0.03 (CIAC, Tg-qPCR1), and 0.03 (pUC18, Tg-qPCR2), respectively.
All samples which showed exponential amplification with a Cq value <40 were scored positive for T. gondii DNA. Samples without exponential amplification or a Cq value ≥40 were scored negative, if amplification of the internal amplification control (IAC) could be observed.
2.4. Analytical specificity and sensitivity of qPCR assays
The specificity of the assays was determined using 1 ng of DNA of related parasitic species belonging to the subfamily of Toxoplasmatinae including Neospora caninum, Besnoitia besnoiti, Hammondia hammondi, and Hammondia heydorni.
The analytical sensitivity of both qPCR assays was determined by serially diluting genomic T. gondii DNA of strains representative of the three clonal types to obtain 105, 104, 103, 102, 10, 1, 0.75, 0.5, 0.25, and 0.1 T. gondii genome equivalents (GE) per PCR reaction. The dilution was prepared in water containing 20 ng/μl porcine DNA resulting in a background of 200 ng matrix DNA per PCR reaction when using 10 μl samples as template DNA (BVL, 2016). Dilution series were freshly prepared before each run and examined with both qPCRs on the same day. Serial dilutions were analyzed in triplicates in four qPCR runs resulting in 12 replicate measurements for each dilution point.
The 95% detection limit was estimated by probit analysis (IBM SPSS Statistics 21). A PCR performance score was determined by dividing the number of positive replicates by the total number of replicates (Robert-Gangneux et al., 2017).
2.5. Precision, PCR efficiency & limit of quantification (LOQ) of qPCR assays
Standard curves were generated based on tenfold dilution series from 105 GE to 10 GE by plotting the mean Cq-value in one run against the logarithm of the template concentration as the GE number per PCR reaction. The slope (s) of the corresponding linear regression line was used to calculate the PCR efficiency E with the formula E = 10–1/s − 1 (Bustin et al., 2009). PCR efficiencies were calculated for each run and clonal type DNA (triplicates).
To evaluate the precision, measured Cq values of each replicate were converted into the actual template concentration using the corresponding standard curves. Based on these back calculated concentrations the coefficient of variation (CV(%) = 100× SD/mean) was calculated for each dilution point (Bustin et al., 2009; Kralik and Ricchi, 2017). To evaluate intra-assay variance, the relative repeatability standard deviation (Rsdr) was determined as the CV of each dilution point in each run.
To evaluate the inter-assay variance, the relative reproducibility standard deviation (RsdR) was determined as the CV of each dilution point over different runs (Bustin et al., 2009; Broeders et al., 2014). The limit of quantification (LoQ) was defined as the lowest concentration at which replicates over four runs show a mean Rsdr ± SD ≤25% on back calculated GE numbers (Kralik and Ricchi, 2017).
2.6. Performance of the Tg-qPCR1 and Tg-qPCR2 assays on pork meat samples
To assess the performance of both qPCR assays using field samples, 38 pork meat products from conventionally raised pigs (35 meat cutlets, 2 minced meat, 1 sausage) were purchased at retail markets and subjected to DNA extraction and examination by both qPCR assays. To generate T. gondii-positive samples, 25 mg portions of 12 negatively tested samples were spiked with a low (100 GE/25 mg sample, n = 4), medium (1000 GE/25 mg sample, n = 4), and high (10,000 GE/25 mg sample, n = 4) amount of T. gondii DNA (strain ME-49) before DNA extraction.
DNA was extracted from 25 mg meat using the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer's instructions. DNA was eluted with 100 μl of molecular grade water and stored at 4 °C. In each experiment a negative extraction control using 25 μl molecular grade water was included to exclude possible cross-contaminations.
2.7. 529 RE conventional endpoint PCR analysis
The cPCR was performed similar to a previously described cPCR (Schares et al., 2008; Herrmann, 2012; Friedrich-Loeffler-Institut, 2018). Amplifications were carried out on a 2720 Thermal Cycler (ABI) in a total volume of 25 μl with 1× DreamTaq buffer (2 mM MgCl2), 0.25 mM of each deoxynucleoside triphosphate, 0.5 μM of each of the primers TOX5 and Tox-8 (Table 1), 20 μg/ml bovine serum albumin (New England Biolabs), 1 U DreamTaq DNA Polymerase (Thermo Fisher Scientific) and 10 μl template DNA. 10 μl PCR product were separated by electrophoresis on 1.2% agarose and visualized using GelRed® in a final concentration of 0.008% (v/v) (Biotium).
3. Results
3.1. Analytical validation of qPCR assays
3.1.1. Specificity
Neither the Tg-qPCR1 nor Tg-qPCR2 showed cross-reaction with DNA of related non-target species, confirming the analytical specificity of both assays.
3.1.2. Sensitivity & limit of detection (LoD)
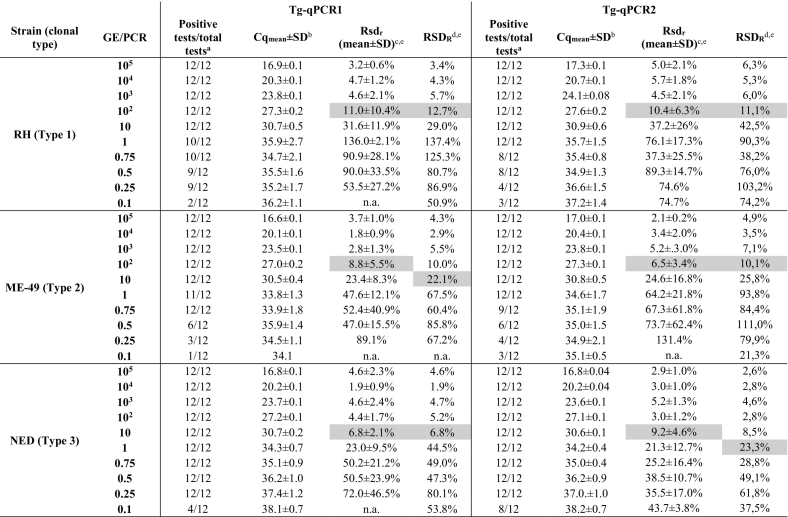
In both qPCR assays, T. gondii DNA could be detected at concentrations as low as 0.1 GE/PCR with mean Cq values between 34.1 and 38.1 (Tg-qPCR1) and 35.1–38.2 (Tg-qPCR2) for all three clonal type strains (Table 2). Considering the results of all three types, both qPCRs showed an identical PCR performance score of 85% (Table 3) and a similar 95% detection limit of 1.067 GE (95% CI: 0,773-1789) and 1.561 GE (95% CI: 0,998-3506) for Tg-qPCR1 and Tg-qPCR2, respectively. The probit regression model fitted the data adequately (Tg-qPCR1: Pearson χ2 = 3637, p = 0.89; Tg-qPCR2: Pearson χ2 = 6.454, p = 0.597) (Table 3). However, Tg-qPCR2 showed a slightly higher sensitivity, as it was able to detect a higher percentage of replicates (39%, 14/36) at low concentrations of 0.1 GE/PCR compared to Tg-qPCR1, which was positive in only 19% (7/36) of the replicates (Table 2, Table 3). Interestingly, DNA of T. gondii NED (type 3) could be detected with higher sensitivity compared to the other two clonal type strains (Table 2, Table 3). As all strains showed nearly identical Cq values at higher DNA concentrations, this phenomenon cannot be explained by differences in the DNA concentration or by different numbers of the 529 RE repeats.
Table 2.
Analytical sensitivity and precision of Tg-qPCR1 and Tg-qPCR2 for each of the three clonal types of Toxoplasma gondii prevailing in Europe and North-America.
GE/PCR, number of genome equivalents per PCR reaction; Cq, quantification cycle; SD, standard deviation; n.a., not applicable. a 12 replicates were obtained by testing triplicates in four runs. b Mean Cq values of 12 replicates and standard deviations are shown. c Rsdr, relative repeatability standard deviation calculated for each run and averaged over four runs. d RsdR, relative reproducibility standard deviations calculated over four runs. e The lowest concentrations with acceptable Rsdr and RsdR values ≤25% are highlighted in grey.
Table 3.
Performance characteristics of Tg-qPCR1 and Tg-qPCR2 for each of the three clonal types of Toxoplasma gondii prevailing in Europe and North-America.
| Clonal type |
Type 1 |
Type 2 |
Type 3 |
All types |
||||
|---|---|---|---|---|---|---|---|---|
| Assay | Tg-qPCR1 | Tg-qPCR2 | Tg-qPCR1 | Tg-qPCR2 | Tg-qPCR1 | Tg-qPCR2 | Tg-qPCR1 | Tg-qPCR2 |
| P 0.1 GE | 17% | 25% | 8% | 25% | 33% | 67% | 19% | 39% |
| LOD95% (95%CI) | 1.669 GE (0.87–10.71) | 1.943 GE (1.03–10.53) | 1.178 GE (0.80–2.68) | 2.002 GE (1.06–10.70) | 0.351 GE (0.23–1.29) | 0.273 GE (0.17–8.40) | 1.067 GE (0.77–1.79) | 1.561 GE (1.00–3.51) |
| E% (mean ± SD) | 94.4 ± 1.2 | 95.6 ± 1.8 | 94.1 ± 1.5 | 95.1 ± 2.9 | 93.8 ± 1.0 | 94.3 ± 0.5 | 94.1 ± 1.2 | 95.0 ± 1.9 |
| R2(mean) | 0,9999 | 0,9998 | 0,9999 | 0,9997 | 0,9993 | 0,9999 | 0,9997 | 0,9998 |
| LOQ | 100 GE | 100 GE | 100 GE | 100 GE | 10 GE | 10 GE | 100 GE | 100 GE |
| Range with RsdR ≤25% | 105–102 GE | 105–102 GE | 105–10 GE | 105–102 GE | 105–10 GE | 105-1GE | 105–102 GE | 105–102 GE |
| PCR performance score | 83% | 79% | 78% | 78% | 93% | 97% | 85% | 85% |
GE, genome equivalents per PCR reaction; P 0.1 GE, detection probability of 0.1 GE/PCR; LOD95%, 95% detection limit as the lowest amount of template DNA which was detected in 95% of the replicates; 95%CI, 95% confidence interval; E%, mean of PCR efficiencies of 4 runs using triplicates; SD, standard deviation; R2(mean), averaged R2 value over 4 runs, standard curves were generated over the linear dynamic range from 10 to 105 GE/reaction; LOQ, limit of quantification; Rsdr, relative repeatability standard deviation; RsdR, relative reproducibility standard deviation; PCR performance score, number of positive replicates/number of total replicates considering all dilution points.
3.1.3. PCR efficiency
Amplification efficiency (E) and the coefficient of determination (R2) for both assays were determined for each run and each strain and derived from standard curves covering the linear dynamic range from 105 to 10 GE/reaction.
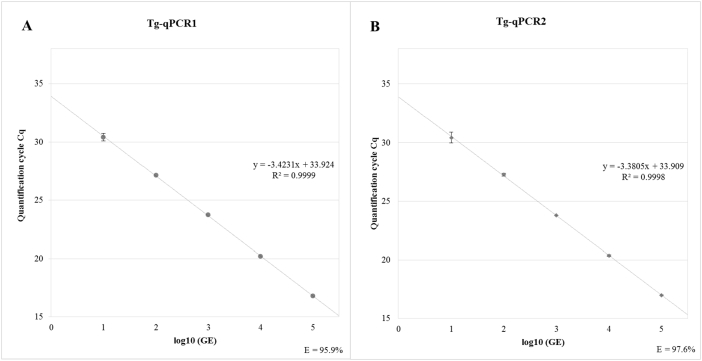
Exemplary standard curves of one run are shown for T. gondii ME-49 (Type 2) in Fig. 1. Amplification of the three clonal type strains in both qPCRs showed comparable PCR efficiencies with average PCR efficiencies ranging between 93.8%–94.4% (Tg-qPCR1) and 94.3–95.6% (Tg-qPCR2) (Table 3). Linear regression revealed average R2 values for all strains and runs over the 5 log range of 0.9997 (Tg-qPCR1) and 0.9998 (Tg-qPCR2) (Table 3).
Fig. 1.
Standard curves of exemplary log-dilution series of genomic DNA of Toxoplasma gondii strain ME-49 diluted in 20 ng/μl porcine DNA (105 GE–10 GE/PCR reaction) obtained in Tg-qPCR1 (A) and Tg-qPCR2 (B). The linear equation of each regression line, the coefficient of determination (R2), and amplification efficiency (E) are displayed in each graph.
3.1.4. Precision & limit of quantification (LOQ)
Considering all clonal type strains, both qPCRs met the required criteria for reliable quantification of Rsdr ≤25% (Broeders et al., 2014; Kralik and Ricchi, 2017) at least over a 4 log range from 105 to 100 GE/reaction (Table 2). The limit of quantification (LOQ) is reached between the log levels 10 and 100 GE/reaction (Table 2, Table 3).
For DNA concentrations ≥100 GE/reaction, both qPCRs show acceptable inter-run reproducibility with RsdR values of ≤12.7% (Tg-qPCR1) and ≤11.1% (Tg-qPCR2) (Table 2, Table 3).
3.2. Performance of the qPCR assays on pork meat samples
Tg-qPCR1 and Tg-qPCR2 showed comparable performance on pork meat samples. All 38 pork meat products were tested negative for T. gondii DNA in both qPCRs, while amplification of the IACs could be observed with mean Cq values of 35.8 (±1.3) and 34.5 (±0,8) for CIAC and pUC18, respectively.
Each of the 12 artificially contaminated pork samples tested positive for T. gondii DNA with comparable Cq values ≤35 and back-calculated GE numbers (Table 4) in both qPCR assays. Generally, the back-calculated GE numbers per PCR in the spiked samples were four to eleven times less than expected.
Table 4.
Performance of Tg-qPCR1 and Tg-qPCR2 on spiked pork meat samples.
| Sample ID | Spiked GE/25 mg meata | Expected GE/PCR | Tg-qPCR1 |
Tg-qPCR2 |
||
|---|---|---|---|---|---|---|
| Cqmean ± SDb | Calculated GE/PCRc | Cqmean ± SDb | Calculated GE/PCRc | |||
| 16-TO-0002 | 104 | 103 | 27.0 ± 0.0 | 97 | 27.7 ± 0.1 | 87 |
| 16-TO-0003 | 104 | 103 | 27.1 ± 0.1 | 94 | 27.6 ± 0.2 | 83 |
| 16-TO-0007 | 104 | 103 | 26.7 ± 0.2 | 118 | 27.1 ± 0.1 | 117 |
| 16-TO-0008 | 104 | 103 | 25.5 ± 0.0 | 269 | 25.8 ± 0.0 | 268 |
| 16-TO-0004 | 103 | 102 | 28.9 ± 0.1 | n.a. | 29.1 ± 0.1 | n.a. |
| 16-TO-0005 | 103 | 102 | 30.5 ± 0.0 | n.a. | 30.7 ± 0.4 | n.a. |
| 16-TO-0006 | 103 | 102 | 29.6 ± 0.2 | n.a. | 30,1 ± 0.1 | n.a. |
| 16-TO-0007 | 103 | 102 | 30.2 ± 0.2 | n.a. | 30.8 ± 0.1 | n.a. |
| 16-TO-0002 | 102 | 101 | 32.8 ± 0.0 | n.a. | 33.3 ± 0.3 | n.a. |
| 16-TO-0003 | 102 | 101 | 35.3 ± 1.7 | n.a. | 34.1 ± 1.2 | n.a. |
| 16-TO-0005 | 102 | 101 | 33.9 ± 0.2 | n.a. | 33.8 ± 2.2 | n.a. |
| 16-TO-0006 | 102 | 101 | 34.1 ± 0.4 | n.a. | 34.1 ± 1.1 | n.a. |
GE/PCR, number of genome equivalents per PCR reaction; Cq, quantification cycle; SD, standard deviation; n.a., not applicable.
Number of genome equivalents of T. gondii strain ME-49 spiked in 25 mg meat samples.
Mean and SD of duplicate measurements.
GE per PCR were quantified using standard curves from 105 to 10 GE/PCR. Calculation for samples spiked with ≤103GE/25 mg meat was not valid, as Cq values were below the limit of quantification.
3.3. Analytical sensitivity and specificity of the 529 RE conventional endpoint PCR
To determine the analytical sensitivity of the cPCR, dilution series of 1 ng to 1 fg of genomic DNA of T. gondii RH in water were subjected to analysis. In 4/4 experiments, DNA of T. gondii strain RH could be detected with a high sensitivity at concentrations as low as 100 fg DNA, which corresponds to 1.41 GE per PCR reaction.
However, this cPCR using TOX5/Tox-8 as a primer pair was not suitable for the detection of T. gondii in pork meat samples in our hands as this PCR generated unspecific amplicons. When dilutions of genomic T. gondii DNA in porcine DNA, as well as pork meat samples were subjected to PCR analysis, non-specific bands with similar size (472 bp) to the desired T. gondii-specific PCR product (452 bp) were observed. Sanger-sequencing of these non-specific products followed by BLAST analysis revealed cross-amplification of porcine DNA present in different Sus scrofa breeds with this cPCR (data not shown). Attempts to eliminate this non-specific amplification by gradually increasing the annealing temperature from 60 °C of up to 67 °C and/or by reducing the duration of the final extension step from 10 min to 2 min showed no satisfactory results (data not shown). Further optimization experiments have not been conducted.
4. Discussion
In recent literature, the 529 RE has evolved as the common target of choice for molecular detection of T. gondii DNA, as its high copy number of 200–300 enables high sensitivity (Schares et al., 2008; Talabani et al., 2009; Herrmann et al., 2010; Opsteegh et al., 2010; Sterkers et al., 2010; Gomez-Samblas et al., 2015; Opsteegh et al., 2016; Krücken et al., 2017; Schares et al., 2017; Burrells et al., 2018; Friedrich-Loeffler-Institut, 2018; Schares et al., 2018). This study aimed to validate and compare three molecular methods targeting the 529 RE to detect T. gondii DNA in pork meat samples. Tg-qPCR1 was originally described by Reischl et al. (2003) for highly sensitive and specific detection of T. gondii DNA using a pair of FRET hybridization probes on a LightCycler instrument (Reischl et al., 2003). The qPCR was adapted by Opsteegh et al. (2010) using a modified TaqMan™ probe and an altered cycling protocol in addition to the incorporation of an competitive internal amplification control (CIAC) (Opsteegh et al., 2010). In both publications, a high sensitivity with detection limits between 16 and 20 fg DNA was described, which corresponds to 0.23-0-28 GE according to our calculation. In this study, the protocol was further adapted for application on an ABI 7500 system, which could explain the decreased sensitivity observed in our experiments. Although, we were also able to detect T. gondii DNA at low concentrations of up to 0.1 GE/PCR in 19% of the replicates, the 95% detection limit for all three clonal type strains was reached at higher concentrations of 1.067 GE/PCR.
Tg-qPCR2 was originally described by Talabani et al. (2009) to diagnose human ocular toxoplasmosis and has been used to amplify T. gondii DNA in ocular or amniotic fluid (Sterkers et al., 2010; Robert-Gangneux et al., 2017). More recently, Tg-qPCR2 has additionally been used to detect T. gondii tissue cysts in chicken (Opsteegh et al., 2016; Schares et al., 2017; Schares et al., 2018) but has not been validated in detail before. In the original publication, a non-competitive internal amplification control (NCIAC) was included, but not further specified. As the addition of an IAC has been proposed to be mandatory for molecular detection of foodborne pathogens (Hoorfar et al., 2003), we added plasmid pUC18 as target for a NCIAC to Tg-qPCR2, which has already been successfully used in PCRs for detection of Bacillus cereus (Frentzel et al., 2018), Yersinia enterocolitica (Mäde et al., 2008) and Salmonella spp. (Anonymous, 2013).
The direct comparison of Tg-qPCR1 and Tg-qPCR2 in this study revealed that both qPCRs show comparable performance on dilution series of the three clonal type strains of T. gondii in porcine DNA as well as for detection of T. gondii DNA in pork meat samples. The analytical sensitivity of Tg-qPCR2 was slightly higher based on a higher detection probability at low concentrations of 0.1 GE/PCR. A reason for this could be the use of a locked nucleic acid (LNA) probe, which can enhance the fluorescence signal and increase specificity and sensitivity of qPCR assays in comparison to standard TaqMan™ probes (Reynisson et al., 2006). Among others, a further reason for the lower analytical sensitivity of Tg-qPCR1 could be a slight inhibition at low concentrations of target-DNA due to the use of a competitive IAC. Both qPCR assays proved to be highly specific for detection of T. gondii as none of the other tested non-target parasites were detected and no cross-reactivity to DNA extracted from pork meat samples was observed. Additionally, both qPCR assays allow reliable quantification of T. gondii DNA in pork over at least 4 log ranges from 105 to 100 GE/PCR and thus determination of the parasitic load in diverse tissues compared to the cPCR or bioassay.
In comparison to the bioassay as gold standard for T. gondii detection in meat samples, where several hundred gram of meat are analyzed, molecular methods are generally considered to be less sensitive, as a significantly lower amount of tissue is used for DNA extraction (about 25 mg). However, comparable sensitivity can be achieved when molecular detection is combined with a pepsin or trypsin digestion or magnetic capture to analyze larger sample sizes (Schares et al., 2018).
Taking together, this study showed that both qPCRs, Tg-qPCR1 and Tg-qPCR2 targeting the 529 RE are similarly suited for the detection of T. gondii DNA in pork meat samples. In contrast to the two qPCR assays, the cPCR with primer pair TOX5/Tox-8 was not suitable for detection of T. gondii DNA in pork, as unspecific amplification of porcine DNA was observed. Nevertheless, the described cPCR showed a high sensitivity with a detection limit of 100 fg corresponding to 1.41 GE/PCR. Thus, it could potentially be used for the examination of other food matrices or animal tissues and has e.g. already been applied to detect T. gondii oocysts in fecal samples of cats (Schares et al., 2008; Herrmann et al., 2010). The results underline the importance of validating molecular methods in the presence of specified matrix DNA, before screening of large sample numbers is performed.
Acknowledgments
Acknowledgements
The work was funded by the German Federal Institute for Risk Assessment (BfR 1322-672). We thank Beatrice Schwarz and Sophie Gellert for their excellent technical support.
Conflict of interest statement
The authors declare they have no conflicts of interest.
Contributor Information
Nadja S. Bier, Email: Nadja.bier@bfr.bund.de.
Gereon Schares, Email: Gereon.schares@fli.de.
Annette Johne, Email: Annette.johne@bfr.bund.de.
Annett Martin, Email: Annett.martin@bfr.bund.de.
Karsten Nöckler, Email: Karsten.noeckler@bfr.bund.de.
Anne Mayer-Scholl, Email: Anne.mayer-scholl@bfr.bund.de.
References
- Anonymous . Beuth Verlag GmbH; Köln, Germany: 2013. Microbiology of Food and Animal Feeding Stuffs - Polymerase Chain Reaction (PCR) for the Detection of Food-borne Pathogens - Method for the Detection of Salmonella (DIN 10135:2013-05) [Google Scholar]
- Broeders S., Huber I., Grohmann L., Berben G., Taverniers I., Mazzara M. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol. 2014;37:115–126. [Google Scholar]
- Burrells A., Taroda A., Opsteegh M., Schares G., Benavides J., Dam-Deisz C. Detection and dissemination of Toxoplasma gondii in experimentally infected calves, a single test does not tell the whole story. Parasit. Vectors. 2018;11:45. doi: 10.1186/s13071-018-2632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- BVL . Bundesamt für Verbraucherschutz und Lebensmittelsicherheit; Braunschweig: 2016. Leitlinien zur Einzellabor-Validierung qualitativer real-time PCR Methoden. [Google Scholar]
- Cook A.J., Gilbert R.E., Buffolano W., Zufferey J., Petersen E., Jenum P.A. Sources of Toxoplasma infection in pregnant women: European multicentre case-control study. European Research Network on Congenital Toxoplasmosis. BMJ. 2000;321:142–147. doi: 10.1136/bmj.321.7254.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa J.-M., Bretagne S. Variation of B1 gene and AF146527 repeat element copy numbers according to Toxoplasma gondii strains assessed using real-time quantitative PCR. J. Clin. Microbiol. 2012;50:1452. doi: 10.1128/JCM.06514-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzib Paredes G.F., Ortega-Pacheco A., Rosado-Aguilar J.A., Acosta-Viana K.Y., Guzmán-Marín E., Jiménez-Coello M. Toxoplasma gondii in meat for human consumption – a brief review of the most described strategies for its detection and quantification. In: Makun H., editor. Significance, Prevention and Control of Food Related Diseases. 2016. [Google Scholar]
- Frentzel H., Thanh M.D., Krause G., Appel B., Mader A. Quantification and differentiation of Bacillus cereus group species in spices and herbs by real-time PCR. Food Control. 2018;83:99–108. [Google Scholar]
- Friedrich-Loeffler-Institut . Amtliche Methodensammlung: Meldepflichtige Tierkrankheiten. 2018. Toxoplasma gondii; p. 14. [Google Scholar]
- Gomez-Samblas M., Vilchez S., Racero J.C., Fuentes M.V., Osuna A. Quantification and viability assays of Toxoplasma gondii in commercial “Serrano” ham samples using magnetic capture real-time qPCR and bioassay techniques. Food Microbiol. 2015;46:107–113. doi: 10.1016/j.fm.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Herrmann D.C. Humboldt Universität; Berlin: 2012. Molecular Typing of Toxoplasma gondii Isolates From Cats and Humans in Germany. (Dr. rer. nat Dissertation) [Google Scholar]
- Herrmann D.C., Pantchev N., Vrhovec M.G., Barutzki D., Wilking H., Frohlich A. Atypical Toxoplasma gondii genotypes identified in oocysts shed by cats in Germany. Int. J. Parasitol. 2010;40:285–292. doi: 10.1016/j.ijpara.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Homan W.L., Vercammen M., De Braekeleer J., Verschueren H. Identification of a 200- to 300-fold repetitive 529 bp DNA fragment in Toxoplasma gondii, and its use for diagnostic and quantitative PCR. Int. J. Parasitol. 2000;30:69–75. doi: 10.1016/s0020-7519(99)00170-8. [DOI] [PubMed] [Google Scholar]
- Hoorfar J., Cook N., Malorny B., Wagner M., De Medici D., Abdulmawjood A. Making internal amplification control mandatory for diagnostic PCR. J. Clin. Microbiol. 2003;41:5835. doi: 10.1128/JCM.41.12.5835.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapperud G., Jenum P.A., Stray-Pedersen B., Melby K.K., Eskild A., Eng J. Risk factors for Toxoplasma gondii infection in pregnancy. Results of a prospective case-control study in Norway. Am. J. Epidemiol. 1996;144:405–412. doi: 10.1093/oxfordjournals.aje.a008942. [DOI] [PubMed] [Google Scholar]
- Kralik P., Ricchi M. A basic guide to real time PCR in microbial diagnostics: definitions, parameters, and everything. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krücken J., Blümke J., Maaz D., Demeler J., Ramünke S., Antolová D. Small rodents as paratenic or intermediate hosts of carnivore parasites in Berlin, Germany. PLoS One. 2017;12 doi: 10.1371/journal.pone.0172829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Wang Z.D., Huang S.Y., Zhu X.Q. Diagnosis of toxoplasmosis and typing of Toxoplasma gondii. Parasit. Vectors. 2015;8:292. doi: 10.1186/s13071-015-0902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäde D., Reiting R., Strauch E., Ketteritzsch K., Wicke A. A real-time PCR for detection of pathogenic Yersinia enterocolitica in food combined with an universal internal amplification control system. J. Verbr. Lebensm. 2008;3:141–151. [Google Scholar]
- Opsteegh M., Langelaar M., Sprong H., Den Hartog L., De Craeye S., Bokken G. Direct detection and genotyping of Toxoplasma gondii in meat samples using magnetic capture and PCR. Int. J. Food Microbiol. 2010;139:193–201. doi: 10.1016/j.ijfoodmicro.2010.02.027. [DOI] [PubMed] [Google Scholar]
- Opsteegh M., Schares G., Blaga R., Van Der Giessen J. vol. 13. EFSA Supporting Publications; 2016. Experimental Studies on Toxoplasma gondii in the Main Livestock Species (GP/EFSA/BIOHAZ/2013/01) Final Report. (995E-n/a) [Google Scholar]
- Reischl U., Bretagne S., Kruger D., Ernault P., Costa J.M. Comparison of two DNA targets for the diagnosis of Toxoplasmosis by real-time PCR using fluorescence resonance energy transfer hybridization probes. BMC Infect. Dis. 2003;3:7. doi: 10.1186/1471-2334-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynisson E., Josefsen M.H., Krause M., Hoorfar J. Evaluation of probe chemistries and platforms to improve the detection limit of real-time PCR. J. Microbiol. Methods. 2006;66:206–216. doi: 10.1016/j.mimet.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Robert-Gangneux F., Darde M.L. Epidemiology of and diagnostic strategies for toxoplasmosis. Clin. Microbiol. Rev. 2012;25:264–296. doi: 10.1128/CMR.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Gangneux F., Brenier-Pinchart M.-P., Yera H., Belaz S., Varlet-Marie E., Bastien P. Evaluation of toxoplasma ELITe MGB real-time PCR assay for diagnosis of toxoplasmosis. J. Clin. Microbiol. 2017;55:1369–1376. doi: 10.1128/JCM.02379-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R.M., Mahon B.E., Jones T.F., Griffin P.M. An assessment of the human health impact of seven leading foodborne pathogens in the United States using disability adjusted life years. Epidemiol. Infect. 2015;143:2795–2804. doi: 10.1017/S0950268814003185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schares G., Vrhovec M.G., Pantchev N., Herrmann D.C., Conraths F.J. Occurrence of Toxoplasma gondii and Hammondia hammondi oocysts in the faeces of cats from Germany and other European countries. Vet. Parasitol. 2008;152:34–45. doi: 10.1016/j.vetpar.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Schares G., Herrmann D.C., Maksimov P., Matzkeit B., Conraths F.J., Moré G. Chicken line-dependent mortality after experimental infection with three type IIxIII recombinant Toxoplasma gondii clones. Exp. Parasitol. 2017;180:101–111. doi: 10.1016/j.exppara.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Schares G., Koethe M., Bangoura B., Geuthner A.C., Randau F., Ludewig M. Toxoplasma gondii infections in chickens - performance of various antibody detection techniques in serum and meat juice relative to bioassay and DNA detection methods. Int. J. Parasitol. 2018;48:751–762. doi: 10.1016/j.ijpara.2018.03.007. [DOI] [PubMed] [Google Scholar]
- Sterkers Y., Varlet-Marie E., Cassaing S., Brenier-Pinchart M.P., Brun S., Dalle F. Multicentric comparative analytical performance study for molecular detection of low amounts of Toxoplasma gondii from simulated specimens. J. Clin. Microbiol. 2010;48:3216–3222. doi: 10.1128/JCM.02500-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talabani H., Asseraf M., Yera H., Delair E., Ancelle T., Thulliez P. Contributions of immunoblotting, real-time PCR, and the Goldmann-Witmer coefficient to diagnosis of atypical toxoplasmic retinochoroiditis. J. Clin. Microbiol. 2009;47:2131–2135. doi: 10.1128/JCM.00128-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenter A.M., Heckeroth A.R., Weiss L.M. Toxoplasma gondii: from animals to humans. Int. J. Parasitol. 2000;30:1217–1258. doi: 10.1016/s0020-7519(00)00124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2015. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015. (Switzerland) [Google Scholar]