This paper presents the crystal structure of a forced open inward rectifier Kir2.2 channel. Molecular dynamics reveals the details of ion permeation through the open channel.
Abstract
Potassium ion conduction through open potassium channels is essential to control of membrane potentials in all cells. To elucidate the open conformation and hence the mechanism of K+ ion conduction in the classic inward rectifier Kir2.2, we introduced a negative charge (G178D) at the crossing point of the inner helix bundle, the location of ligand-dependent gating. This “forced open” mutation generated channels that were active even in the complete absence of phosphatidylinositol-4,5-bisphosphate (PIP2), an otherwise essential ligand for Kir channel opening. Crystal structures were obtained at a resolution of 3.6 Å without PIP2 bound, or 2.8 Å in complex with PIP2. The latter revealed a slight widening at the helix bundle crossing (HBC) through backbone movement. MD simulations showed that subsequent spontaneous wetting of the pore through the HBC gate region allowed K+ ion movement across the HBC and conduction through the channel. Further simulations reveal atomistic details of the opening process and highlight the role of pore-lining acidic residues in K+ conduction through Kir2 channels.
Introduction
Potassium channels play essential roles in stabilizing membrane potentials and control of numerous physiological phenomena in all cells. Inward rectifier (Kir) potassium channels are a major subfamily of channels that lack the classical four-helix voltage sensing domains of voltage-gated channels, but show voltage dependence of conductance, due to block by intracellular Mg2+ and polyamines at positive membrane potentials, and are controlled by gating in response to regulatory ligand binding (Hibino et al., 2010; Nichols and Lopatin, 1997). Each Kir subfamily exhibits distinct ligand gating; Kir1 and Kir4/5 are controlled by pH, Kir3 by Gβγ proteins, and Kir6 by ADP/ATP and sulfonylurea receptor subunits (Nichols and Lopatin, 1997). Phosphatidylinositol-4,5-bisphosphate (PIP2) binding to a canonical PIP2 binding site (“primary” site) is essential for activation of all Kir channels (D’Avanzo et al., 2010; Hansen et al., 2011; Hilgemann and Ball, 1996). Bulk anionic lipids (PL(-)s) are additional positive allosteric regulators that substantially increase PIP2 sensitivity of the Kir2 subfamily (Cheng et al., 2011; Lee et al., 2013). While several Kir crystal (Hansen et al., 2011; Lee et al., 2016; Tao et al., 2009; Whorton and MacKinnon, 2011, 2013) and recently single particle cryo-EM (Martin et al., 2017) structures have been determined, they do not encompass the full conformational ensemble of functional states of the channel. In particular, the M2 (S6) helix bundle crossing (HBC), located close to the cytoplasm–inner leaflet interface, forms a constriction in all available structures that is too narrow for permeation and therefore indicates a closed state of the channel. Lack of open-state structures significantly limits our understanding of the molecular mechanisms by which PIP2 and other ligands actually open Kir channels, of how pore blockers or other ligands actually influence ion conductance, and the details of conductance itself.
In KcsA and Kir channel crystal structures, residues lining the narrow “bundle-crossing” region where the transmembrane (TM) helices intersect are hydrophobic, and the pore is too narrow for ions or water to be present (Doyle et al., 1998; Hansen et al., 2011; Lee et al., 2016; Nishida et al., 2007; Whorton and MacKinnon, 2011). In voltage-gated Kv channel crystal structures, the channel is typically in an open or activated state (Long et al., 2005, 2007), but channels are predicted to close by formation of a hydrophobic seal formed by the highly conserved Pro-Val-Pro motif (del Camino and Yellen, 2001) at the bundle crossing; hydrophilic, but not hydrophobic, substitutions within this region of Shaker Kv channels (Sukhareva et al., 2003) and of the same region in Kir6 channels (Enkvetchakul et al., 2001) destabilize the closed state. MD simulations over extended timescales (microseconds to milliseconds) indicate that the hydrophobic nature of the Kv inner pore promotes dehydration of the cavity and constriction at the Pro-Val-Pro motif (Jensen et al., 2010), such that de-wetting of the pore halts ion conduction (Jensen et al., 2012). In the so-called twin-pore subfamily of K channels, MD simulations of the TWIK-1 channel also indicate stochastic wetting and de-wetting events associated with two hydrophobic pore-lining leucine residues (Leu146 on TM2 and Leu261 on TM4); mutation of these to polar side chains (asparagine) leads to retention of water in MD simulations and to increased whole-cell currents experimentally (Aryal et al., 2014). Thus, while the potentially obscure functional state of structural conformations determined by x-ray, single particle cryo-EM, or NMR approaches presents a general barrier to progress in molecular-level understanding, MD simulations can provide key functional interpretation of experimentally determined structures and reveal the importance of de-wetting in channel closure at narrow hydrophobic restrictions and the likelihood that specific structures indeed represent open, conductive states (Aryal et al., 2015; Trick et al., 2015).
We recently obtained crystal structures (Lee et al., 2016) of chicken Kir2.2[K62W] mutant protein (KW) in the absence of, or in complex with, PIP2. The K62W mutation endows the equivalent allosteric effect as PL(-) binding, generating physiological sensitivity to activatory PIP2 (Cheng et al., 2011). The Apo-K62W structure (in the absence of PIP2) maintained a high-affinity PIP2 binding site, revealing a “preopen” state stabilized by PL(-) binding. However, even after satisfying both of the recognized lipid requirements for opening, the PIP2-bound K62W structure was still sterically closed at the inner HBC. This suggests that the open state stability is relatively low and perhaps unlikely to be captured in such crystals.
As an alternate approach to capturing the open conformation of eukaryotic Kir channels, we have now introduced an additional mutation (G178D) near the HBC gate that functionally stabilizes open channels, following an approach previously used to obtain an apparently open KirBac3.1 crystal structure (Bavro et al., 2012) and to generate gain-of-function mutants of various other K channels (Brelidze et al., 2003; Nimigean et al., 2003). We then determined crystal structures of these mutant channels with and without bound PIP2. The PIP2-bound Kir2.2[K62W,G178D] (KW/GD) structure was slightly more open (by ∼1.5 Å diameter) at the HBC than the previous PIP2-bound K62W structure (Lee et al., 2016). MD simulations reveal that, when embedded in a lipid membrane, relaxation of this crystal structure, together with side-chain flexibility, results in wetting of the pore through the HBC region. This leads to rapid spontaneous further opening of this region and K+ conductance through the entire channel, revealing novel details of the ion permeation process.
Materials and methods
Cloning, expression, and purification
A single point mutation (G178D) was introduced using QuikChange site-directed mutagenesis (Stratagene Cloning Systems) to both the WT and K62W mutant truncated cKir2.2 channel cDNA and verified by sequencing. Mutant channel proteins were expressed in Pichia pastoris cells as described previously (Tao et al., 2009). Briefly, frozen Pichia cells were broken using a Retsch Model MM400 mixer mill (5 × 3.0 min at 25 cps) and solubilized in lysis buffer (100 mM Decyl β-D-maltopyranoside [DM], 50 mM Tris, pH 7.5, 150 mM KCl, 1 mM EDTA, and 2 mM dithiothreitol [DTT]) at room temperature for 1 h with stirring or rotation. Cell lysate was centrifuged at 30,000 g for 30 min at 10°C. Approximately 750 µl of anti-Flag antibody–conjugated resins per 10 g of cells was added to the supernatant. Binding was performed at 4°C for >1.5 h with gentle rotation. The resin was washed with 20 column volumes of wash buffer (4 mM DM, 50 mM Tris, pH 7.5, 150 mM KCl, 1 mM EDTA, and 2 mM DTT), and protein was released from the resin by PreScission Protease action at 4°C overnight and further purified on a Superdex 200 column (GE Healthcare) equilibrated in SEC buffer (20 mM Tris, pH 7.5, 150 mM KCl, 1 mM EDTA, 4 mM DM, and 20 mM DTT). Peak fractions were combined and concentrated to >5 mg/ml for crystallization experiments and in vitro functional assays.
Electrophysiology
CosM6 cells were transfected with 1–2 µg of pcDNA3.1-Kir2.2-K62W or pcDNA3.1-Kir2.2-K62W-G178D mutants with an addition of 0.4 µg of pcDNA3.1-GFP per 35-mm Petri dish using FuGENE6 (Promega). The cells were used for patching within 1–2 d after transfection. For patch-clamp experiments, symmetrical internal potassium buffer (Kint) was used: 148 mM KCl, 1 mM EGTA, 1 mM K2EDTA, and 10 mM HEPES, pH 7.4. Data were acquired at 15 kHz, low-pass filtered at 5 kHz with Axopatch 1D patch-clamp amplifier, and digitized with Digidata 1320 digitizer (Molecular Devices). Data analysis was performed using the pClamp software suite (Molecular Devices). Pipettes with bubble number (Schnorf et al., 1994) 4.0–5.0 (∼2–4 MOhm in symmetric Kint) were pulled from Kimble Chase 2502 soda lime glass with a Sutter P-86 puller (Sutter Instruments). All measurements were performed on excised inside-out patches at −120 mV membrane potential, or as specified in the text.
Rb+ flux assay
Channel activity of purified recombinant proteins was assessed by 86Rb+ uptake into proteoliposomes. 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) lipids were dissolved (10 mg/ml) in buffer A (450 mM KCl, 10 mM HEPES, 4 mM N-methyl glucamine, and 0.5 mM EGTA, pH 7.4) with 35 mM 3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate. Porcine brain PIP2 was solubilized (2 mg/ml) in 8 mg/ml POPE solution. 1 mg of lipid mixture in 100 µl was incubated at room temperature for 2 h, and 3–10 µg of protein was added and incubated for an additional 20 min. The lipid–protein mixture was passed through partly dehydrated G-50 beads preequilibrated with buffer A to form proteoliposomes. Proteoliposomes were passed through partly dehydrated G-50 beads preequilibrated by buffer B (400 mM sorbitol, 10 mM HEPES, 4 mM NMDG, and 0.5 mM EGTA, pH 7.4) to remove external KCl. For assessing ion selectivity, the internal salt was replaced with Li, Na, Rb, Cs, and NMDG. For blocking experiments, spermine was added only to the external side to a concentration of 10 µM; higher concentrations could not be assessed because of liposome destabilization. 200 µl of buffer B containing 86Rb+ (PerkinElmer) at ∼4.5 nCi was added at time 0. After 10 min, 80-µl samples were collected and passed through cation exchange beads to capture 86Rb+ in the external solution. Uptake was normalized to the maximum uptake in valinomycin, after leak subtraction (uptake into protein-free liposomes). “n” in Fig. 1 represent the number of protein purification, and for each preparation, one to three replica samples were tested for each condition depending on the amount of available proteins. Porcine brain PIP2, POPE, and POPG were purchased from Avanti Polar Lipids.
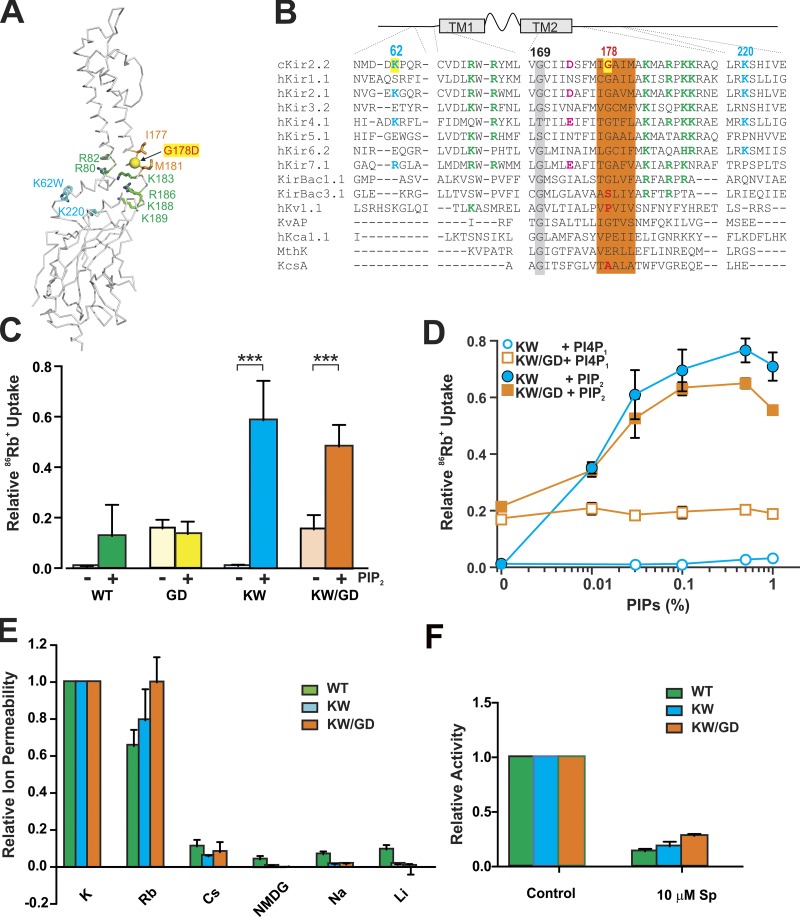
Figure 1.
Force open mutant channel properties in vitro. (A) Backbone trace of Kir2 channel with the side chain of the two residues (I177 and M181) forming the HBC in sticks and the mutated residue G178 in balls. PIP2 binding site residues are shown in sticks, and green and bulk anionic lipid binding site residues are in sticks and cyan. (B) Sequence alignment of selected regions of K+ channels. The residues at the PIP2 binding site are shown in green, and the residues important for secondary anionic lipids in cyan. The residues mutated in this study are highlighted in yellow, and the residues forming the HBC are highlighted in orange. The negatively charged residue in the inner cavity important for rectification, the so-called rectification controller, is shown in pink, and the conserved glycine hinge residues are highlighted in gray. (C) Mutant and control protein activity was assessed in the absence and presence of 0.1% PIP2 in liposomes made with 10% POPG and 90% POPE. Data shows 86Rb+ uptake normalized to the maximum uptake in valinomycin. ***, P < 0.001. (D) Phospholipid specificity was tested for KW and KW/GD proteins. Phosphatidylinositol 4,5-bisphosphate or phosphatidylinositol 4-phosphate was increased from zero to 1% in liposomes formed with 10% POPG/90% POPE. Data show 86Rb+ uptake normalized to the maximum uptake in valinomycin. (E) Ion selectivity was assessed with different ions inside the proteoliposome, at a fixed lipid composition (0.1% PIP2, 10% POPG, and 90% POPE). Data show 86Rb+ uptake normalized to the uptake in K+ gradient. (F) Sensitivity to the pore-blocker spermine was assessed at a fixed lipid composition (0.1% PIP2, 10% POPG, and 90% POPE). Data show 86Rb+ uptake in the absence or presence of 10 µM spermine, normalized to the uptake in the absence of spermine. In C–F, error bars indicate mean ± SEM for n ≥ 3 independent protein preparations with one to three replicates for each preparation (see Materials and methods).
Crystallization and structure determination
PIP2 was solubilized in SEC buffer (5 mM) and added to the concentrated protein sample to 250 µM concentration, ≥30 min before crystallization. Crystals were grown by hanging drop vapor diffusion, by mixing 0.5 µl of protein and 0.5 µl of reservoir solutions. Crystals for the Apo-KW/GD structure were obtained in 180 mM triNaCitrate, 100 mM Tris, pH 7.1, and 27.4% PEG 400 at 20°C; PIP2-KW/GD crystals were obtained in 80 mM triNaCitrate, 100 mM Tris, pH 7.1, and 27.2% PEG 400 at 20°C. Crystals grew at 20°C in 2–3 d, were cryoprotected by 30% (vol/vol) PEG400, and were frozen in liquid nitrogen. X-ray diffraction data (3.6 Å for Apo, 2.81 Å for PIP2− crystals) were collected at the Advanced Photon Source beamlines 24-ID-C and 24-ID-E at wavelength 0.9792 Å under liquid nitrogen stream. Phases were obtained using the program MolRep (Vagin and Teplyakov, 2000) in the CCP4 suite (Winn et al., 2011) through molecular replacement with Apo-K62W mutant crystal structure (PDB ID: 5KUM) as a search model for PIP2-KW/GD crystals, and 5SPC (cKir2.2 WT cocrystallized with pyrophosphatidic acid) for Apo-KW/GD crystals. Iterative model building was performed in COOT (Emsley and Cowtan, 2004), and rounds of refinement were performed with REFMAC (Murshudov et al., 1997). The final Apo-KW/GD model (PDB ID: 6M86) was obtained with R/Rfree of 0.237/0.291 and with 96% of residues in the most favored regions and one in the disallowed region in the Ramachandran plot. The final PIP2-KW/GD model (PDB ID: 6M84) was obtained with R/Rfree of 0.22/0.270 and with 97% of residues in the most favored regions and none in the disallowed region in the Ramachandran plot.
MD simulations
MD simulations and subsequent analyses were performed using the software package Gromacs v5.1.2 (Abraham et al., 2015). As a starting point, the structure was inserted into an equilibrated bilayer membrane consisting of palmitoyloleoylphosphatidylcholine lipids, for which Berger lipid parameters were employed (Berger et al., 1997; Cordomí et al., 2012), surrounded by explicit water molecules (extended simple point charge water model; Berendsen et al., 1987) and 0.2 M potassium chloride (Joung and Cheatham, 2008). Geometry optimization and electrostatic potential calculation of PIP2 (short chain analogue) were performed at the Hartree-Fock/6-31G* level using the program Gaussian09 (Gaussian). The generalized amber force field topologies of the ligand were generated with Antechamber software (Wang et al., 2006) using partial charges from quantum mechanics calculations according to the restrained electrostatic potential approach. The forcefield parameters for PIP2 are included in Dataset S1. Potassium ions were placed in the selectivity filter (SF) at sites S0, S2, and S4, and water molecules were placed at sites S1 and S3 (Åqvist and Luzhkov, 2000). The amber99sb forcefield was used for the protein (Hornak et al., 2006). For the ions, we used corrected monovalent Lennard–Jones parameters for the amber forcefield (Joung and Cheatham, 2008). The cutoff for electrostatic interactions was set to 1.0 nm; long-range electrostatic interactions were treated by the particle-mesh Ewald method (Darden et al., 1993). The cutoff for Lennard–Jones interactions was set to 1.0 nm. The LINCS (linear constraint solver) algorithm was used to constrain bonds (Hess et al., 1997). The simulation temperature was kept constant using a velocity-rescale thermostat (Bussi et al., 2007); protein and lipids as well as the solvent, together with ions, were coupled (τ = 0.1 ps) separately to a temperature bath of 310 K. Similarly, the pressure was kept constant at 1 bar by the Parrinello−Rahman barostat algorithm (coupling constant = 2 ps; Bussi et al., 2007). Steepest descent energy minimization was performed before all simulations. To equilibrate the system, we position restrained all heavy atoms with a force constant (fc) of 1,000 kJ ⋅ mol−1 ⋅ nm−2 and simulated for 20 ns. The Y146 side-chain atoms were restrained using positional restraints throughout the simulations with a fc of 1,000 kJmol−1nm−2. An electric field along the channel pore (40 mV ⋅ nm−1) was applied. With a box length of ∼14.6 nm, this amounts to a transmembrane potential of ∼580 mV. In silico mutation of G178D → G (G178D(G)) was accomplished using Swiss PDB Viewer (Guex and Peitsch, 1997).
Analysis of MD trajectories
Changes in the relative rotation of the cytoplasmic C-terminal domain (CTD) with respect to the transmembrane domain (TMD) were assessed by calculating the torsion angle between two planes. Calculation of the torsion angle required four different points of measurement: the center of mass of the TMD (point 1), the center of mass of the CTD (point 2), the center of mass of one subunit of the TMD (point 3), and the center of mass of one subunit of the CTD (point 4). The first plane is defined by points 1, 2, and 3; the second plane is defined by points 1, 2, and 4. To calculate the changes over simulation time, the angle between these two planes of the first simulation step was defined as 0 (for similar methods, see Linder et al. [2015]).
Before analyzing water occupancies/solvation, all trajectories were aligned at the SF (sequence TTIGYG). The coordinates of protein and potassium ions were saved every 20 ps. Depending on simulation length, this resulted in 10,000 steps (for 200-ns trajectories) or 50,000 steps (for 1-µs trajectories) per run. Along the membrane normal (pore axis z), the area between the intracellular entrance of the channel and the end of the SF was cut into slices of 0.5-Å thickness. Potassium ions inside the channel pore were counted in each slice. Average numbers of resident potassium ions with an applied electric field were plotted against the membrane normal. Based on these occupancies, which entail multi-ion effects, the PMFEB (effective-biased multi-ion potential of mean force) was determined using the following equation: GPMF(z) = −kBT ln[n(z)] (de Groot and Grubmüller, 2001), where G is free energy, kB is the Boltzman constant, T is temperature in Kelvin, and z is location along the pore axis.
PKa calculations
The calculations were performed using PDB2PQR/APBS webserver (Gosink et al., 2014) for KW (5KUM) and KW/GD (6M84) crystal structures and the open KW/GD and KW/GD(G) MD simulated structures sampled at 1,000 ns. The default options, including PARSE force field, were used for the calculations.
Statistical analysis
Statistical significance was analyzed using unpaired t tests unless otherwise stated. Statistical significance of P < 0.05, P < 0.01, and P < 0.001 is indicated by single, double, and triple asterisks, respectively.
Data availability
The datasets and computer code produced in this study are available upon request, and will be deposited in an appropriate public database.
Online supplemental material
Supplementary data include images of electron density in the inner cavity of Apo-KW (5KUK) crystal diffracting to 2.0 Å. Supplementary videos show K ion conduction through KW/GD and KW/GD(G) channels, as well as gating transitions associated with channel opening, and details of knock-on mechanism of K ion permeation through the SF.
Results
Generation of a locked open Kir2.2 channel
To force Kir2.2 channels into an open conformation, we followed the approach used to obtain an apparently open KirBac3.1 structure (Bavro et al., 2012), introducing a charge mutation near the HBC gate in each subunit. By electrostatic repulsion, such introduced charges may provide an energetic push against one another to stabilize the open state. We introduced G178D (Fig. 1, A and B) on both the truncated WT chicken Kir2.2 (Tao et al., 2009) and the KW mutant background (Lee et al., 2016). Purified channel proteins were incorporated into liposomes and characterized functionally by 86Rb+ flux assays. As shown previously (Lee et al., 2016), both WT Kir2.2 and KW channels were inactive in 9:1 POPE:POPG and required 0.1% PIP2 for activity. In contrast, both single-mutant GD and double-mutant KW/GD channels were active in the complete absence of PIP2 (Fig. 1 C), although KW/GD showed further stimulation of activity when PIP2 was present. For unknown reasons, the protein yield of the single GD mutant protein was consistently very low. We speculate that this is due to instability of the tetramer as a result of the additional four negative charges that are concentrated at a very constricted region. We focused further analysis on the KW/GD double-mutant protein, which was expressed at somewhat higher levels, for the following experiments.
WT Kir2 channels are specifically activated by PIP2, while other naturally occurring phosphoinositides tend to show inhibitory effects (D’Avanzo et al., 2010). Sensitivity to PIP2 was similar for KW and KW/GD channels, with apparent kD of ∼0.03 (wt/wt) %, which corresponds to ∼0.02 mol% in each case. Neither channel was sensitive to phosphatidylinositol 4-phosphate at up to 1 (wt/wt) %, corresponding to ∼0.7 mol%, indicating that phosphoinositide specificity was not altered by the forced open G178D mutation (Fig. 1 D). In these 86Rb flux assays, ion selectivity and spermine block (Lopatin et al., 1994) were also indistinguishable between WT, KW, and KW/GD, with or without PIP2 (Fig. 1, E and F), indicating that, whether separately or in combination, the G178D mutation does not markedly alter sensitivity to regulatory ligands, other than increasing the relative stability of the channel open state.
Crystal structures of Apo-KW/GD and PIP2-KW/GD
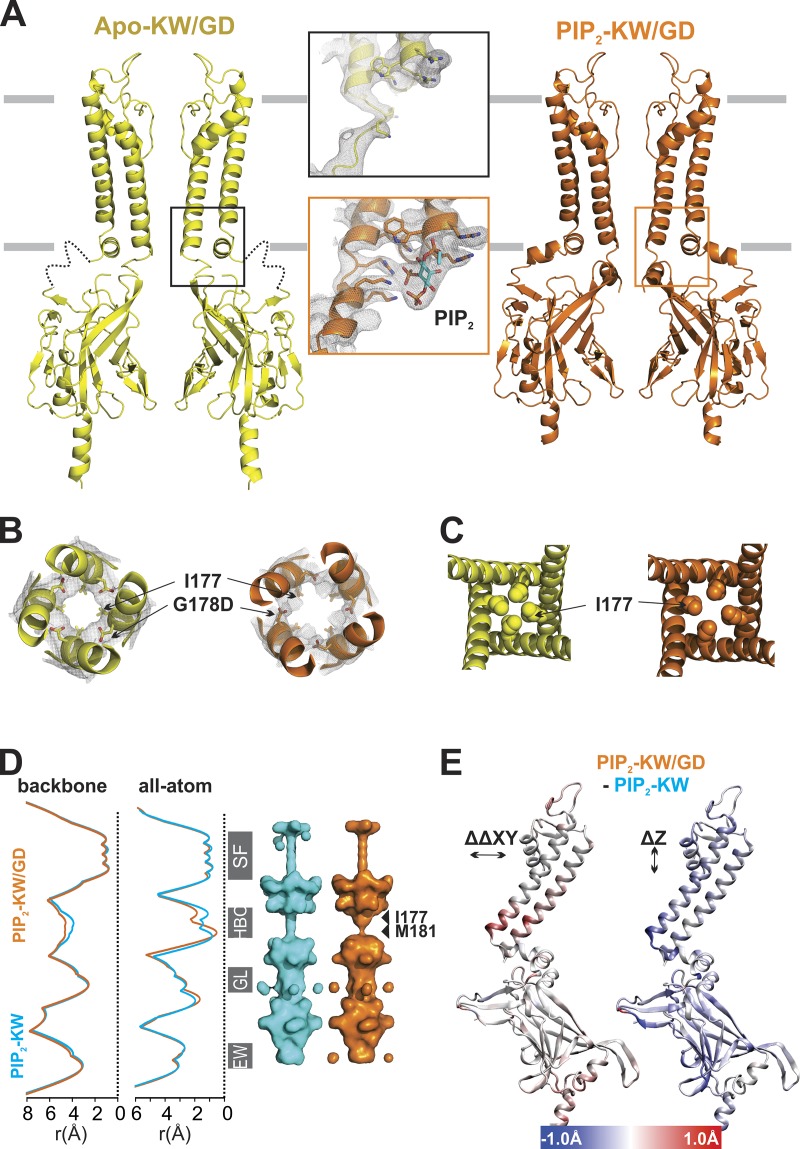
KW/GD mutant crystals were obtained in both the absence (Apo-KW/GD) and presence (PIP2-KW/GD) of di-oleoyl PIP2 (18:1). Crystal growth conditions were similar, although high concentrations (>130 mM) of triNaCitrate were required for the growth of Apo crystals, while lower triNaCitrate concentrations (<100 mM) facilitated PIP2-KG/WD crystal formation. X-ray diffraction data were collected at 3.6 Å resolution, and a final structural model was obtained with R value of 23.7% and Rfree value of 29.1% for Apo-KW/GD. PIP2-KW/GD crystals diffracted to 2.8 Å, and the structural model was refined with R value of 22% and Rfree value of 27% (Fig. 2, A–C, see Materials and methods, and details of crystals and models in Table 1). In both proteins, the G178D side chains face the inner helix of the neighboring subunit (Fig. 2 B), rather than pointing directly into the pore axis. However, an obvious difference between Apo-KW/GD and PIP2-KW/GD structures is that the CTD is disengaged from the TMD in the Apo-KW/GD structure (“loose” conformation), with an unstructured and unoccupied PIP2 binding site, similar to the Apo-WT structure (PDB ID: 3JYC; Tao et al., 2009), whereas the CTD is tightly engaged with the TMD in the PIP2-KW/GD structure (“compact” conformation), with strong PIP2 density in the fully formed binding site (Fig. 2 A, inset).
Figure 2.
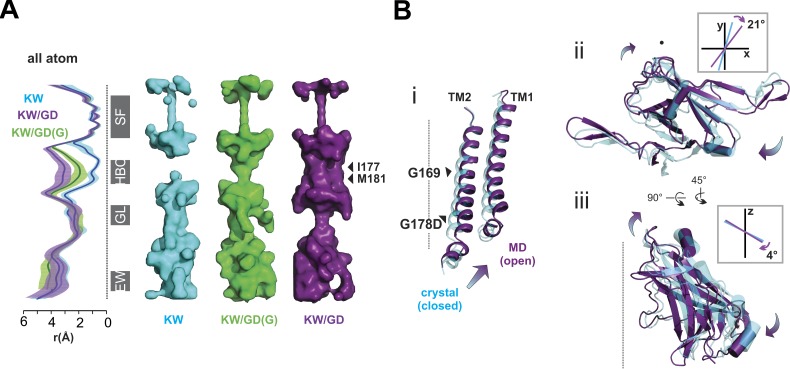
Crystal structures of forced open mutant channels. (A) Left: Apo-KW/GD (yellow) and PIP2-KW/GD (orange) crystal structures are shown in ribbon diagram. For clarity, only two subunits are shown for the side view. The local conformations of the PIP2 binding site region are shown in the inset, with 2fo-fc electron density contoured at 1 σ. (B) Bottom-up views (in the plane of the membrane) shown in ribbon diagrams with electron densities contoured at 1 σ, with mutant G178D and HBC forming I177 residues drawn explicitly in sticks. (C) Top-down view of each structure in the plane of the membrane shows the HBC region with I177 side chains depicted in balls. (D) Pore-radius profiles of PIP2-KW (cyan) and PIP2-KW/GD (orange) crystal structures computed by HOLE2 are shown with backbone atoms (left) or all atoms including side chain (right). Pore-lining surfaces of the PIP2-KW (cyan) and PIP2-KW/GD (orange) crystal structures are shown with the major constrictions designated: SF; HBC (I177, M181); GL, G-loop; and EW (vertically aligned to the ribbon diagrams in C). (E) Diagonal Cα distance changes (ΔΔCα = PIP2-KW/GD[ΔCα] − PIP2-KW[ΔCα]) in the lateral dimension (the xy plane; left) and vertical movement along the z axis of Cα atoms (right) mapped on a single Kir2 subunit. The color bar shows the ranges of distance change.
Table 1. Crystallographic data and refinement statistics.
| Item | Apo-KWGD (6M86) | PIP2-bound KWGD (6M84) |
|---|---|---|
| Data collection | ||
| Space group | I4 | I4 |
| Cell dimensions | ||
| a, b, c (Å) | 82.852, 82.852, 189.109 | 82.878, 82.787, 182.976 |
| α = β = γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50 − 3.6 (3.73 − 3.60) | 91.95 − 2.80 (2.90 − 2.80) |
| Unique reflections (n) | 7,432 | 15,066 |
| Rmerge* | 0.079 (0.742) | 0.059 (0.947) |
| I/σI | 273.6/9.7 (5.9/2.1) = 29.1 (2.8) | 168.4/5.6 (4.3/2.0) = 30.1 (2.2) |
| Completeness (%) | 99.9 (100.0) | 99.9 (99.9) |
| Redundancy | 3.4 (3.2) | 3.4 (3.3) |
| Refinement | ||
| Resolution (Å) | 50 − 3.6 | 50 − 2.80 |
| Reflections (n) | 7,040 | 14,242 |
| Rwork/Rfree | 0.237/0.291 | 0.222/0.270 |
| Atoms | ||
| Protein | 2,578 | 2,620 |
| Ligand/ion | 0/5 | 52/6 |
| Water | 5 | 19 |
| B-factors | ||
| Protein | 187.0 | 98.5 |
| Ligand/ion | 0/151.3 | 164.3/101.5 |
| Water | 138.704 | 81.7 |
| Root mean square deviation | ||
| Bond length (Å) | 0.004 | 0.005 |
| Bond angles (°) | 0.851 | 1.027 |
Rmerge = SUM(|I − <I>|)/SUM(I).
As in previous Kir2.2 structures, the disengaged Apo-KW/GD structure is closed and nonconducting, due to a steric constriction throughout the HBC region (Fig. 2 C). The PIP2-KW/GD structure is subtly, but noticeably, different from all other structures, including the counterpart PIP2-bound KW crystal structure (PIP2-KW, 5KUM; Lee et al., 2016) at the HBC location. Structural changes due to the GD mutation are localized near the HBC, resulting in a minor (∼1 Å) lateral expansion at the bottom of the TM1 and TM2 helices (Fig. 2, B and C). Pore dimensions analyzed by HOLE2 (Smart et al., 1996) and HOLLOW (Ho and Gruswitz, 2008) show a widening at residue 177 due to this M2 helix backbone displacement (Fig. 2 D). The pore is still narrowed immediately below this by M181 sidechain projections, but flexibility in the M181 sidechain could lead to rapid widening of this region. Since hydration, or wetting, of hydrophobic residues that form the gate is a prerequisite for ion channel opening (Aryal et al., 2015; Klesse et al., 2019; Trick et al., 2015), the slight expansion at the backbone (Fig. 2, D and E) might then be sufficient to allow water permeation at the HBC and thereby render the channel open. A recently developed program, CHAP (Klesse et al., 2019), predicts conducting state based on both physical dimensions and hydrophobicity of pore lining residues. CHAP suggests that the current crystal structure is likely to be closed, due to the two hydrophobic layers (hydrophobicity 0.336 for I177 and 0.183 for M181) and small pore radii (0.222 nm at I177 and 0.088 nm at M181). The G178D mutation introduces hydrophobicity of −1, the lower limit, between these two layers of hydrophobic residues, and may significantly affect the wetting process. To assess this, we performed all-atom MD simulations of the PIP2-KW/GD crystal structure (see Materials and methods). In these simulations, K+ ions were initially placed in the S0, S2, and S4 positions of the SF, with intervening water molecules, and one K+ ion was placed in the inner cavity. Unlike MD simulations of previous Kir2.2 crystal structures (Lee et al., 2016), which stayed closed unless external force was applied to pull open the HBC gate (Li et al., 2015), the TM helices and M181 sidechains spontaneously moved outwards, resulting in HBC gate expansion of ∼5 Å (Fig. 3 A) and HBC wetting in all PIP2-KW/GD simulations. This in turn led to permeation of potassium ions through the HBC gate as well as the SF, at rates in a similar order to those measured experimentally, as described in detail below.
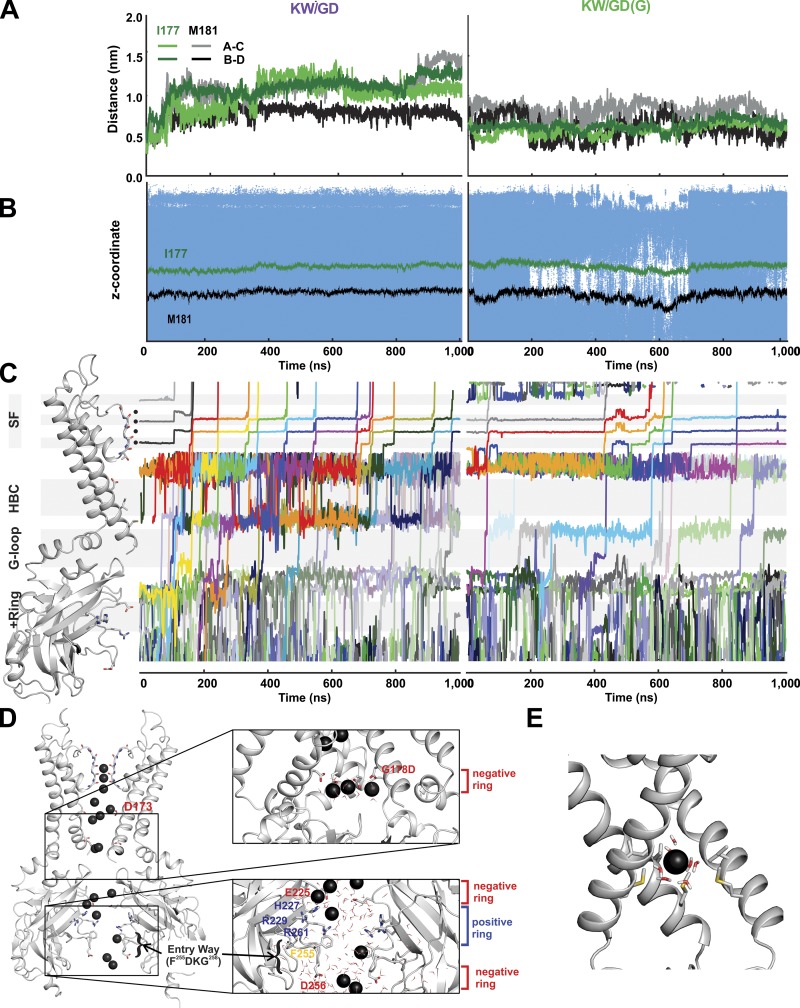
Figure 3.
Ion flux through open conducting structures. (A) The evolution of minimal distances of residues I177 and M181 between two opposing subunits over the course of 1-µs simulation in KW/GD (left) and KW/GD(G) (right). (B) Location of water molecules (represented as small blue dots) within the pore near the HBC gate over the course of 1-µs simulation in KW/GD (left) and KW/GD(G) (right). The HBC gate location is indicated by the I177 (green) and M181 (black) Cα positions. (C) Time trajectories of individual K+ ions (varying colors) are shown from the EW of the pore to beyond the S0 position in the SF (vertical positions match the ribbon diagram on the left). (D) A snapshot of K+ ion distribution along the open pore during PIP2-KW/GD MD simulation. Two opposing subunits are shown in ribbons, and K+ ions and water molecules in the pore are shown in black balls and sticks, respectively. Residues of interest are shown in sticks and labeled. Zoom-in views show the rings of acidic (negative) and basic (positive) residues along the pore. (E) Snapshot of a solvated ion passing the HBC gate. TM2 helices are represented as gray cartoon; the subunit in the front is omitted for clarity. Residues I177 and M181 as well as water molecules are shown in sticks. The K+ ion at the HBC is represented as a black sphere.
K+ ion conduction through the open channel
To gain insight to the permeation mechanism through open Kir2 channels, we performed repeated simulations (for details, see Table 2) of the PIP2-KW/GD structure. Channel opening resulted from spontaneous expansion at the HBC (Fig. 3 A, left) and consequently uninterrupted water accessibility through the entire HBC region (Fig. 3 B, left). In all (six of six) simulations, the HBC opened within 50 ns and, following the first 50 ns of simulation, the shortest diagonal distances at the HBC measured on average 8.14 ± 2.5 Å at the I177 site and 10.7 ± 2.9 Å at M181. Fig. 3 C (left) illustrates the progressive location of individual ions through the pore during one long (1,000-ns) simulation. During such simulations, K+ ion distributions within the channel reached essentially steady state, allowing assessment of the mean location of K+ ions in the open, conducting channel. During permeation, K+ ions were highly localized near the negative charges that underlie weak rectification (E225; on average 3.4, maximum 5) within the CTD, as well as near the rectification controller negative charges (D173; on average 4.7, maximum 6) in the inner cavity. They were also localized near the introduced D178 (on average 3.2, maximum 5) at the HBC gate (Fig. 3 C, left). Over the entire 2.8 μS of simulation, the averaged K+ ion distributions reveal preferred locations (Fig. 4 A, left), implying apparent energy barriers at the G-loop, the HBC, and the SF (Fig. 4 A, right). The most substantial apparent energy barriers are in the SF (4.3 kcal/mol) and are likely to provide the major rate-limiting steps for K+ conduction, although in absolute terms, these are still only slightly higher than what would be expected for diffusion-limited ion flux (Bernèche and Roux, 2001) and hence will not preclude high K+ conductance through the channel.
Table 2. Overview of performed simulations.
| Simulation setup | Number of runs | Simulation time/run (ns) | Sum of simulation time (ns) | Number of ions passed through the SF | Unitary conductance (pS) |
|---|---|---|---|---|---|
| KWGD restrained HBC gate | 1 | 500 | 500 | 0 | 0 |
| KWGD(G) start | 1 | 100 | 100 | 0 | 0 |
| KWGD | 4 | 200 | 2,800 | 6,4,1,0 | 8.43, 5.62, 1.41, 0 |
| 2 | 1,000 | 14, 6 | 3.94, 1.69 | ||
| KWGD(G) after 50 ns | 10 | 200 | 3,000 | 4, 1, 1, 0, 0, 0, 0, 0, 0, 0 | 5.62, 1.41, 1.41, 0, 0, 0, 0, 0, 0, 0 |
| 1 | 1,000 | 6 | 1.69 |
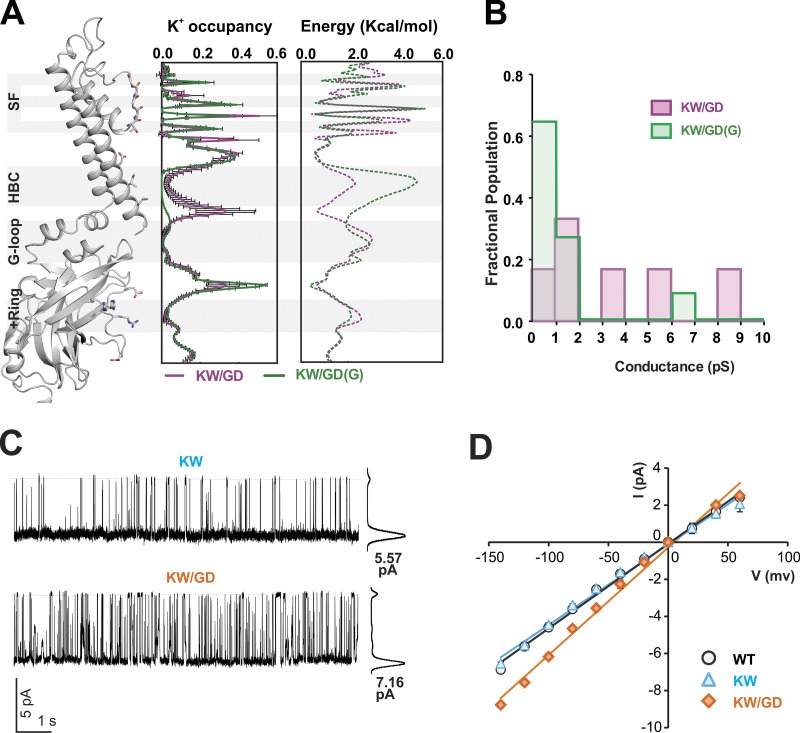
Figure 4.
Ion flux through KW/GD(G) open conducting structures. (A) Left: Average (±SD) ion occupancy along the pore was computed over the resampled simulations after excluding 500, 400, 200, and 0 ns from the concatenated 2.8- and 3-µs simulations of KW/GD and KW/GD(G), respectively, from which the average apparent free energy profile for potassium ions along the pore axis was obtained (right) via an inverse Boltzmann transformation of the K+ ion density profile obtained under the influence of applied electric field. (B) Estimated conductance from individual KW/GD and KW/GD(G) simulations. (C) Single-channel currents recorded from CosM6 cells at −120 mV and 150 mM symmetrical K+ condition using inside-out patch configuration at pH 7.5. This figure is representative of six single-channel recordings of WT Kir2.2 and KW/GD channels (combined duration of >3 min for each). (D) Average single-channel current–voltage relationships (of four or more patches excised from different cells in each case); error bars (where visible) represent mean ± SEM of values obtained from individual records.
These simulations illustrate elementary steps underlying outward movement of K+ ions through the channel (Fig. 3, C and D). Ions enter the cytoplasmic pore from the intracellular side by free diffusion, with no preferred path, until they reach a narrow region, which we term the entry way (EW; Fig. 2 D), formed by the short helices (F255DKG258, Fig. 3 D), which obliges the ions to pass in single file to reach the ring of negative charges at E225. In between these two regions, positively charged amino acids (H227, R229, and R261) form a positive charge ring (+Ring, Fig. 3 D) that prohibits more than one positively charged ion at a time from passing through. Multiple ions are coordinated by the E225 residues, but ions then transition in single file to the region between the G-loop gate and the HBC gate, where up to five ions are again coordinated by the G178D residues. The ions pass the HBC in a fully solvated state (Fig. 3 E), but in single file, to reach the inner cavity (for a full conduction event, see Video 1). As discussed above, major energy barriers are present in the SF, in particular between the S3 and S2 positions (Fig. 4 A), leading to residence times for K+ ions in S3 of 8–131 ns. Nevertheless, in the two longest (1,000-ns) simulations, 14 and 6 ions, respectively, passed completely through the channel, while in the shorter (200-ns) simulations, 6, 4, 1, and 0 ions passed through, corresponding to conductances ≤8 pS (Fig. 4 B; average 3.5 ± 1.7 pS). Although lower, this is a similar order of magnitude to experimentally measured open Kir2.2 channel conductance (∼35 pS in symmetric 140 mM [K+]; Takahashi et al., 1994; Fig. 4, C and D).
Video 1.
K+ ion permeation through an open PIP2-KW/GD channel. KW/GD protein is shown in cartoon with the nearest subunit omitted for clarity. The residues forming the HBC are explicitly shown in sticks (I177, M181). Water molecules are shown in sticks, and K+ ions are represented as gray spheres, with one K+ ion highlighted in yellow illustrating complete K+ ion permeation event starting from the bottom entrance to the extracellular side of the protein.
K+ conduction through open WT KW/GD(G) channels
To examine the effect of the introduced G178D negative charges on pore structure and K+ conduction, we performed additional simulations on channels that were back mutated in silico at residue 178 from Asp to the native Gly (KW/GD(G)), after 50-ns presimulations of the PIP2-KW/GD crystal structure, i.e., after initial wetting of the HBC gate had occurred. Importantly, the pore then maintained a predominantly wetted state, with only transient de-wetting, and channels continued to conduct throughout the full length of the subsequent simulations. In total, 10 replica 200-ns simulations and one 1,000-ns simulation were performed. In all cases, although the minimum distances at the HBC were slightly narrower than in PIP2-KW/GD simulations at the level of residue 177, the PIP2-KW/GD(G) structures remained open and conducting in these simulations (Fig. 3, A–C, right; for a full conduction event, see Video 2). In these PIP2-KW/GD(G) simulations, K+ ions still localized near residues E225 (average 3.5, maximum 5) in the CTD and residues D173 (average 3.7, maximum 5) in the inner cavity, but now ions were much less likely to localize near residue 178 (average ∼1, maximum 2). Otherwise, ion distributions in, and ion fluxes through, the KW/GD(G) channel were quite similar to KW/GD (Figs. 3 C and 4 A). K+ ion distributions averaged over the total 3,000 ns of simulation implied apparent energy barriers that were similar to those in KW/GD (Fig. 4 A, right), except at the HBC gate, where the slightly narrower opening of ∼6.3 Å and concomitant occasional de-wetting (Fig. 3 B, right) resulted in a slightly higher apparent free energy barrier (Fig. 4 A, right), and lower net conductances (Fig. 4 B; average 0.9 ± 0.5 pS).
Video 2.
K+ ion permeation through an open PIP2-KW/GD(G) channel. Similar to Video 1, illustrating complete K+ ion conduction through KW/GD(G) structure.
The structural basis of conducting states
As a consequence of decreased ion occupancy in the pore, the above simulations indicate lower conduction for KW/GD(G) (in which the “forced open” channel is back-mutated to a WT pore without the four acidic Asp residues at the HBC gate) than for KW/GD channels. We experimentally assessed the role of the introduced Asp residues in channel conductance, with single-channel recordings of KW/GD and KW proteins expressed in Cosm6 cells (Fig. 4, C and D). Even though absolute unitary conductance levels predicted above by MD simulations were lower than the experimentally measured values, the relative effect of the introduction of acidic Asp residues at position 178 was strikingly similar: the KW/GD mutant channel conductance was ∼30% higher than that of WT channels (Fig. 4, C and D), quite similar to the ∼40% higher conductance of KW/GD in the simulations.
As representative open-state structures, the last snapshots of 1,000-ns PIP2-KW/GD and PIP2-KW/GD(G) MD simulations are compared with the last snapshot of 100-ns PIP2-KW MD simulations (Fig. 5 A), in which the PIP2-KW channel stayed closed, as reflected in the pore radius (by HOLE2) and pore shape (by HOLLOW). The conformational changes that generate a conducting channel are qualitatively similar in both KW/GD and KW/GD(G): the prohibitive narrowing at residue 181 is removed in both, but the HBC region is considerably wider in the former (Fig. 5 A). TM2 helix bending brings about slight upward and counterclockwise conformational translation (Fig. 5 B i): bending at the highly conserved glycine hinge (residue G169, Fig. 1 B) with average bending values of 14.7° ± 10.1° for PIP2-KW/GD and 10.9° ± 8.0° for PIP2-KW/GD(G) and additional helix bending at G178 location, with average values of 9.5°± 5.4° for PIP2-KW/GD and 6.5° ± 4.3° for PIP2-KW/GD(G), computed by VMD Plugin Bendix (Dahl et al., 2012).
Figure 5.
Kir2 channel bundle crossing gating. (A) PIP2-KW/GD and PIP2-KW/GD(G) MD simulated open structures were compared with the closed PIP2-KW MD simulated structures. Left: Pore-radius profiles of 1-µs PIP2-KW/GD_MD (purple), 1-µs PIP2-KW/GD(G)_MD (green), and 100-ns PIP2-KW_MD (cyan) were computed by HOLE2 with all atoms including side chain, and the averages and standard deviations are shown. Right: The pore-lining surfaces of PIP2-KW/GD_MD at 1 µs (purple), PIP2-KW/GD(G)_MD at 1 µs (green), and PIP2_KW_MD at 100 ns computed by HOLLOW are shown with the major constriction sites designated: SF; HBC (I177, M181); GL, G-loop; and EW. (B) Closed PIP2-KW crystal structure (cyan) and open PIP2-KW/GD_MD (purple) structure are overlaid; arrows indicate predominant conformational changes from closed to open state. (B i) TM1 and TM2 helices are shown in ribbon diagram. (B ii) Top-down view of CTDs in ribbon diagram. The central vectors of the C-linker helices projected on the xy-plane are overlaid to show a twisting motion in the inset. (B iii) Side view of CTD in plane of the membrane normal. The central vectors of the C-linker helices projected on the z-axis are overlaid to show a tilting motion in the inset.
A top-down view of these snapshots (Fig. 5 B ii) reveals CTD motions in conducting channels relative to the crystal structure (highlighted by a morph video based on PIP2-KW crystal structure and KW/GD(G) MD open structure, Video 3). Relative to the crystal structure, these include clockwise whole CTD rotational motions about the central pore axis that converged to 4.7° ± 1.6° for KW/GD and 4.5° ± 2.25° for KW/GD(G) simulations. Also very noticeable was twisting of individual subunit CTDs around an axis within each subunit, with the pivot point at the c-linker (Fig. 5 B ii). A side view of the CTD reveals additional tilting motions that pull the top of the CTD away from the pore axis, while the bottom of the CTD is pulled toward the pore axis (Fig. 5 B iii) in the conducting channel.
Video 3.
Gating transition from a closed to an open state. PIP2-KW crystal structure is taken as closed state, while the last snapshot of 1-µs PIP2-KW/GD(G) MD simulation is taken as open state. The two structures are aligned to maximize overlap of the SF backbone atoms (142–147). The closed state structure is shown in silver, and the structures in transition from the closed to the open state are shown in light green.
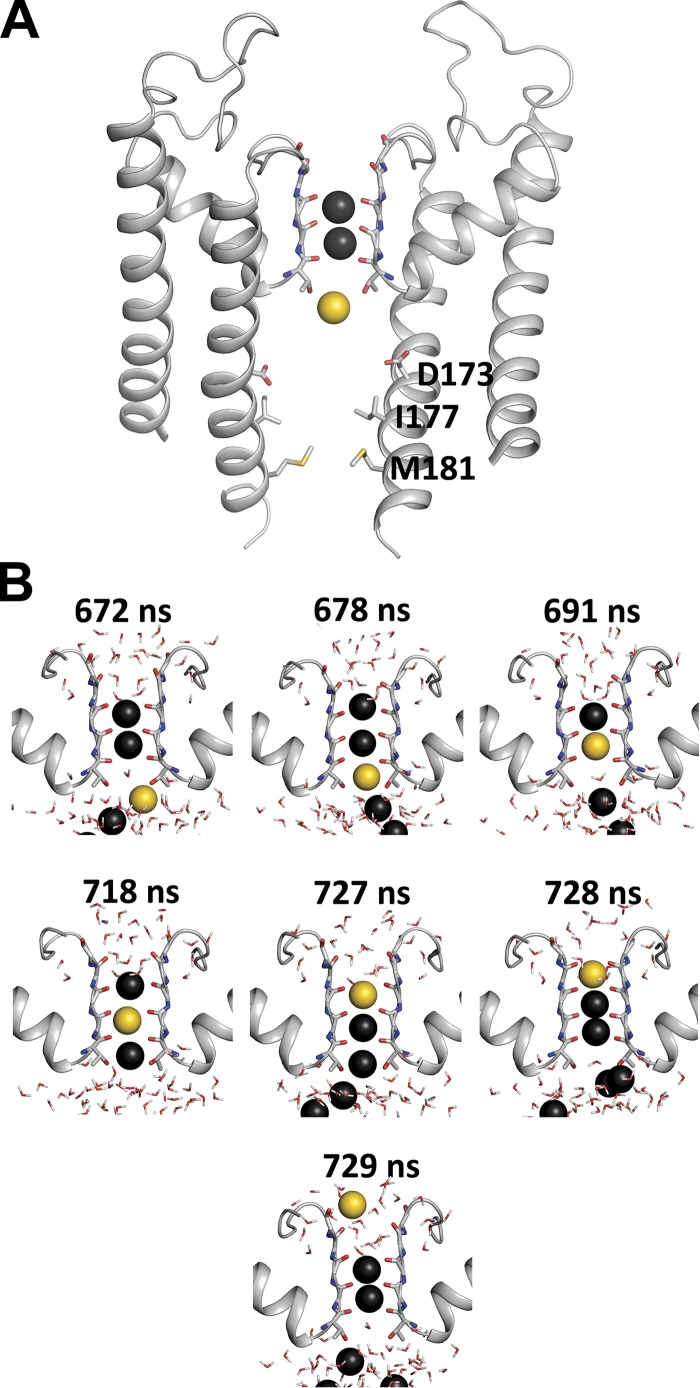
Direct knock-on permeation
The inner cavity contained four to five ions, localized at the level of residues D173 (the rectification controller) and T143, toward the top of the SF (see Figs. 3 C and 4 A) during steady-state KW/GD or KW/GD(G) simulations. Following initiation of each KW/GD simulation, with one K+ ion placed in the inner cavity, transitions from the inner cavity to SF site S4 (formed by the carbonyl oxygen atoms of T143), did not begin until after the four D173 charges were balanced by four K+ ions. Once an ion entered the SF site S4, the ion in site S3 was rapidly pushed via a direct knock-on upwards to S2, leading to a rapid exit of the ion initially located at site S2 to the extracellular side, via S1 and S0 (Fig. 6 and Video 4). Thus, while initial conditions were set up with K+ ions in sites S0, S2, and S4, and water molecules in sites S1 and S3, S2 and S3 were always rapidly and preferably occupied by K+ ions (Fig. 6), leaving sites S0, S1, and S4 filled with water molecules. Importantly, except for the water molecules initially placed in S1 and S3, no additional water entered or permeated the SF throughout the simulations. Thus, our simulations implicate direct knock-on as the underlying mechanism for high conductivity of K+ ions through the channel (Fig. 6 and Video 4), as also seen in other recent simulation studies (Kopec et al., 2018; Köpfer et al., 2014; see Discussion).
Figure 6.
Direct knock-on K ion movement in SF. (A) Pseudo–steady-state conformation of the SF with two resident K+ ions (black) in S2 and S3, immediately before permeation event initiated by entry of inner cavity K+ ion (yellow). For the SF, the backbone of residues IGYG (residues 144 to 147) and the full T143 are shown as sticks, as well as the rectification controller residue D173. For clarity, only two opposing domains are shown. (B) Snapshots of the SF during the ensuing conduction event. Surrounding water represented as sticks. Consecutive simulation snapshots show the complete permeation of the yellow K+ ion, followed by additional K+ ions (black) from the inner cavity, with no intervening water molecules.
Video 4.
K+ ion permeation at the SF. For better visibility, only two opposing subunits of the Kir2.2 are represented as sticks. K+ ions are represented as spheres. Ions that are already within, or that enter, the SF are colored consecutively, from silver via bronze, gold, and brown to black. Ions that are close to the SF but do not enter it within the simulation are colored white. Water is represented as sticks. Although ion binding site S4 is occasionally occupied by water, only K+ ions in this site lead to subsequent ion advancement through the filter.
Discussion
The mechanism of Kir channel opening
K channels gate in response to channel-specific stimuli, including water-soluble or membrane-soluble ligands and/or membrane potentials (Grandi et al., 2017). Rather than interacting directly with the pore domain, these stimuli typically interact with or transform regulatory domains, which then effect opening or closing of the pore. In Kir channels, gating is controlled by diverse ligands and regulators (Na+, Gβγ, ethanol, ATP, sulfonylurea receptor subunits) in addition to the conserved primary agonist, PIP2 (Hibino et al., 2010). In each case, these ligands bind to the cytoplasmic CTD to control opening and closure of the channel through physical coupling to the gate, a steric restriction located at the bottom of the transmembrane pore domain. Multiple high-resolution structures of different eukaryotic Kir channel family members have been obtained, but always in a closed or nonconducting state (Hansen et al., 2011; Lee et al., 2016; Tao et al., 2009; Whorton and MacKinnon, 2011, 2013). In an effort to overcome this limitation, we here generated a mutant Kir2.2 channel (KW/GD) that is essentially locked open functionally and obtained high-resolution crystal structures. These structures show a minor expansion at the HBC gate compared with previous Kir2.2 crystal structures, but when placed in a membrane and freed from crystallographic contacts in silico, the structures undergo rapid spontaneous expansion at the HBC, enabling water and K+ ion flux through the HBC gate as well as through the SF.
Ligand-dependent movements of the Kir CTD with respect to the TMD have been reported in crystallographic, FRET, and simulation studies (Whorton and MacKinnon, 2011, 2013; Bavro et al., 2012; Wang et al., 2012, 2016). Consistent with these studies, Kir channel crystal structures determined in complex with different combinations of ligands suggest that two major conformational changes occur concurrent with channel opening: CTD rotation/twisting and CTD tethering to the TMD. In our MD simulations, even though the CTD rotations fluctuate in both directions, we also observe net gating-dependent clockwise rotational motions of the CTD relative to the TMD in comparison to crystal structures.
More interestingly, we observed rigid body twisting and tilting of individual subunit CTDs around their own axes with the pivot at the C-linker of each subunit (Fig. 5 B). Such rigid body motions are in accordance with changes in FRET between CTDs during KirBac1.1 opening and closing (Wang et al., 2012, 2016), which require subunit CTD motions relative to one another and would not be registered by whole CTD rotational motions about the pore axis, since the latter involve no diagonal distance changes.
The second obvious gating-associated conformational change is tethering of the CTD to the TMD to form a compact structure (Hansen et al., 2011; Lee et al., 2016; Tao et al., 2009). The first PIP2-bound Kir2.2 structure (PDB ID: 3SPI) suggested that PIP2 may be responsible for causing this vertical motion of the CTD by inducing formation of the PIP2 binding site from the unstructured Apo-structure (PDB ID: 3JYC). However, subsequent GIRK2 crystal structures, which adopted a compact conformation even in the absence of PIP2 (PDB ID: 3SYO; Whorton and MacKinnon, 2011), suggest that other factors could lead to generation of the compact CTD conformation. Our recent crystal structures show that Kir2.2 also adopts the compact conformation in the absence of PIP2, if the requirement for additional tethering of the CTD to the membrane at a second, low-affinity, PL(-) site (Cheng et al., 2011; Lee et al., 2013) is met (Lee et al., 2016). Thus the primary effect of PIP2 binding, rather than being to induce the compact conformation, may be to stabilize the compact conformation, thereby facilitating additional opening movements.
The number of ions in the pore
While all K channels exhibit specificity for K+ ions over Na+ ions, each K channel subfamily is unique in single-channel properties (Naranjo et al., 2016), with conductance of the single channel being dependent on rate-limiting structures within the permeation path. Multiple studies of various K channel subtypes have converged to the same conclusion that negative charges lining the pore below the SF as well as steric dimensions (width and length) of the hydrophobic regions of the pore act as major determinants of single-channel conductance, by varying K+ ion accessibility to the SF (Brelidze et al., 2003; Díaz-Franulic et al., 2015; Naranjo et al., 2016; Nimigean et al., 2003). The Kir channel pore is uniquely long (∼7 nm), due to the cytoplasmic extension (∼4 nm) that is provided by the intracellular CTD (Naranjo et al., 2016) and also lined by several charged residues (three acidic and three basic residues per subunit). Point mutations that reduce the negative charges in the CTD decrease K+ conductance, while reduction in positive charges in the CTD increase K+ conductance (Fujiwara and Kubo, 2006), implying that local K+ concentrations below the SF are critical. Experimentally, we show here that introduction of additional negative charges at the residue 178 positions results in higher single-channel conductance for the KW/GD mutant, compared with the KW and WT counterparts (Fig. 4, C and D). In addition to showing how the D178 negative charges cause conformational changes that generate the open conducting state (see above), MD simulations of KW/GD versus the back-mutated KW/GD(G) indicate that the D178 negative charges lower the barrier for conductance through the HBC gate region (1.9 vs. 3.5 kcal ⋅ mol−1), increasing K+ ion concentrations in the inner cavity (compare Figs. 3 C, left and right) which then increase unitary conductance by increasing the rate of ion movement into and through the SF.
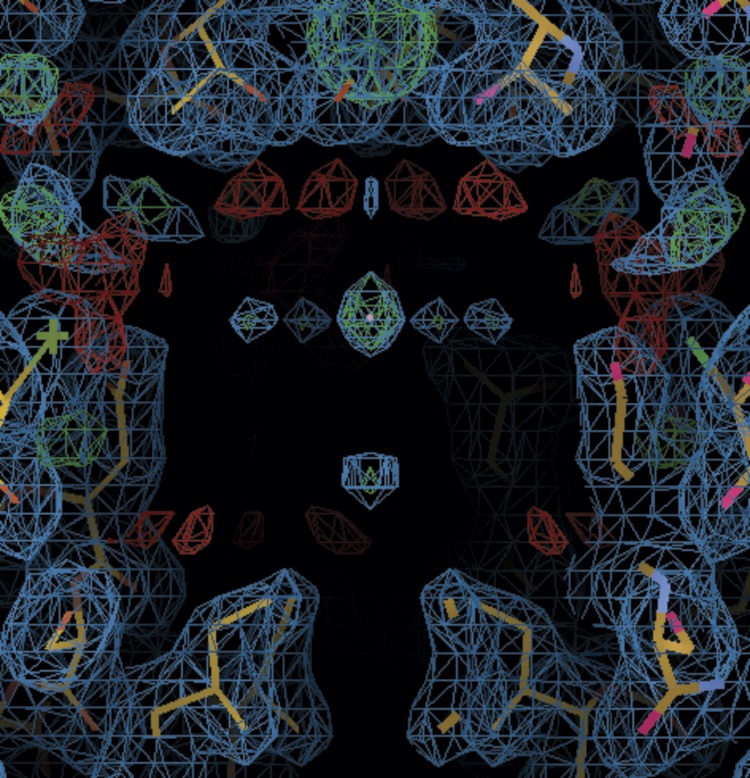
It is important to note that the above interpretation, i.e., that multiple ions can be simultaneously present at a given depth within the pore, is at least superficially inconsistent with interpretations of crystallographic studies. Multiple high-resolution crystal structures indicate that K+ ions are vertically aligned along the central pore axis, with essentially no more than one ion present simultaneously at any location (Lee et al., 2016; Xu et al., 2009), including within the inner cavity (Tao et al., 2009), although a crystal structure of Kir2.2[K62W] at 2.0 Å resolution (PDB ID: 5KUM; Lee et al., 2016) indicates at most two K+ ions in the inner cavity (Fig. S1). By contrast, the KW/GD MD simulations predict K+ ion occupancy in the open conducting Kir2.2 inner cavity as high as 4.7, with 3.5 K+ ions continuously present near E225 in the cytoplasmic cavity. It is conceivable that acidic residues within the channel pore may be protonated due to close apposition (Bradley et al., 2012), and predicted pKa values by PDB2PQR/APBS webserver (Gosink et al., 2014) are indeed close to pH 7.0 for D173 and pH 6.0 for E225 in the closed KW and KW/GD crystal structures (Table 3). These values are much higher than the pKa values of these residues in solution (∼pH 3.7). Thus it is possible that, at least in closed channels, not all acidic residues are charged. Countering this notion, cKir2.2 structures can clearly hold at least two to three positive charges in the inner cavity and cytoplasmic cavity based on heavy atom (Sr2+, Eu3+) anomalous diffraction (Tao et al., 2009), although this could be explained by a decrease in apparent pKa values in the presence of strong positive charges, i.e., the protonation state of the acidic residues may be sensitive to the presence of ion charges such that the inner cavity could accommodate different ion charges without undergoing any conformational changes.
Figure S1.
Electron density in the inner cavity of Apo-KW (5KUK) crystal diffracting to 2.0 Å. 2fo-fc map in cyan is contoured at 1 σ, while fo-fc maps (green or red) are contoured at 3 σ. Gray circles indicate putative K+ ion densities.
Table 3. Predicted pKa values of ionizable residues along the pore axis by APBS.
| Residue | PIP2-KW (xtal:closed) | PIP2-KW/GD (xtal:closed) | PIP2-KW/GD (MD:open) | PIP2-KW/GD(G) (MD:open) |
|---|---|---|---|---|
| D173 | 7.04 | 7.12 | 6.50 | 5.44 |
| 7.32 | 7.21 | 5.83 | 5.90 | |
| 7.04 | 7.12 | 6.50 | 6.77 | |
| 7.32 | 7.21 | 5.88 | 5.87 | |
| G178D | 9.92 | 5.99 | ||
| 9.39 | 5.77 | |||
| 9.92 | 6.38 | |||
| 9.39 | 5.43 | |||
| E225 | 5.72 | 3.32 | 9.35 | 8.81 |
| 6.09 | 5.89 | 7.21 | 6.24 | |
| 5.72 | 3.32 | 5.09 | 2.54 | |
| 6.09 | 5.89 | 7.75 | 6.85 | |
| E300 | 2.11 | 3.54 | 5.92 | 3.45 |
| 2.11 | 5.69 | 0.67 | 6.50 | |
| 2.11 | 3.54 | 3.92 | 2.64 | |
| 2.11 | 5.69 | 3.09 | 6.32 | |
| D256 | 3.95 | 4.00 | 4.30 | 4.38 |
| 4.01 | 3.87 | 3.98 | 4.01 | |
| 3.95 | 4.00 | 4.16 | 4.17 | |
| 3.92 | 3.87 | 2.69 | 2.82 |
However, additional experimental findings support assignation of full negative charge to these acidic residues in the MD simulations. First, the Kir6.2 channel inner cavity with four introduced acidic charges at the rectification controller (N160D) can enclose spermine4+ (Phillips and Nichols, 2003), suggesting that four positive charges can cohabitate with and be balanced by four negatively charged aspartate residues. Second, the voltage dependence of spermine block of Kir6.2 is directly dependent on the number of negative charges in the inner cavity, whether changed by the number of mutant N160D residues or by adding positive charge via MTSEA (2-aminoethyl methanethiosulfonate hydrobromide) or MTSET (2-(trimethylammonium)ethyl methanethiosulfonate, bromide) modification (Kurata et al., 2010). The only straightforward interpretation of such results is that four negative charges in the inner cavity are required for steep spermine-induced rectification and that, at least in the open channel, the rectification controller residues are indeed charged.
The mechanism of K+ ion conduction
Mechanisms of ion translocation through the SF of various voltage gated K+ channels, as well as through KcsA, have been interrogated previously using MD simulations (Åqvist and Luzhkov, 2000; Bernèche and Roux, 2001; Furini and Domene, 2009; Köpfer et al., 2014). In general, following the original KcsA crystal structure, which indicated alternating water and K+ ions within the sequential SF binding sites, most studies have assumed that conduction involves concerted movement of ions and water through the filter, requiring both electric repulsion between K+ ions and diffusional movement of water molecules driven by osmotic pressure (Åqvist and Luzhkov, 2000; Bernèche and Roux, 2001). However, it has also been suggested that less coordinated transitions may be involved (Åqvist and Luzhkov, 2000; Furini and Domene, 2009), and more recent studies have suggested that, rather than alternating ions and water being obligatory, K+ ions can occupy adjacent sites in the SF, with conductance then being via a direct ion–ion knock-on mechanism (Kopec et al., 2018; Köpfer et al., 2014; Langan et al., 2018). In our simulations, K+ ions unambiguously permeated the open Kir2.2 channel via a direct knock-on mechanism, in which an ion entering the S4 site pushed the ion ahead to site S3 and then S2, leading to exit of the outermost ion on the extracellular side without accompanying water permeation. Although we applied an unphysiologically high voltage (580 mV), previous simulations (Köpfer et al., 2014), performed with voltages ranging from 90 to 560 mV, revealed no voltage dependence of the conductance mechanism. Our recent simulations of Kir3.2 channel conductance (Bernsteiner et al., 2019) also showed strict knock-on conductance, with proportional reduction in conduction at lower voltages, using the same approach.
This conclusion clearly contradicts the long-recognized reality that water molecules traverse K+ channels in addition to the permeant ion (Alcayaga et al., 1989). Intriguingly, however, the coupling ratio between water and K+ is typically lower in more K+-selective channels (Kopec et al., 2018) and declines as [K+] increases (Ando et al., 2005), consistent with water movement being reduced as K+ occupancy increases. Our recent single-molecule FRET studies indicate that the canonical K+-selective conformation of the KirBac1.1 SF is stabilized by K+ ions specifically, and that significant SF dilation, resulting in a nonselective conformation, occurs at lower [K+] (Wang et al., 2019). It is unknown how generalizable such findings are, but they lead us to speculate that the apparent coupling of water and K+ ion flux in K channels might be a result of the SF cycling between two distinct conformations, one in which water-independent K+ permeation occurs through the canonical SF conformation via a direct K+-ion knock-on mechanism, and one in which water permeates through the dilated SF conformation.
While a direct knock-on mechanism is a clear finding here, one important caveat is that the Tyr side chain in the SF loop has to be constrained to maintain a conductive SF conformation otherwise, as noted by others, flipping of peptide bonds can result in a nonconducting distortion (Domene et al., 2004; Haider et al., 2007). Multiple experimental studies indicate that subtle conformational changes or mutations at this residue have significant effects on permeation. For instance, conservative mutation from Tyr (Y) to Phe (Y145F) in KcsA, as well as to other residues (Liu et al., 2001), affects apparent channel open times, while in KATP (Kir6.2) channels, conservative mutation of the equivalent Phe at residue 133 to Tyr results in nonfunctional channels (Proks et al., 2001). Cryo-EM structures of WT and inactivation-deficient S631A mutant hERG (Kv11.1) channels (Schönherr and Heinemann, 1996; Wang and MacKinnon, 2017) also suggest that subtle conformational changes of the aromatic side chain position of the equivalent F627 residue could underlie channel inactivation. We thus speculate that subtly unaccounted for energetics might bias MD simulations into such pore-inactivated states when the Tyr side chain is unconstrained. We currently have no experimental data to support this, but speculate that perhaps such a state might be the “low K” state identified in single-molecule FRET studies (Wang et al., 2019).
In conclusion, despite constitutive activity of recombinant Kir2.2[K62W, G178D] (KW/GD), KW/GD crystal structures remained in a nonconducting state. However, in subsequent MD simulations (in which crystallographic constraints are removed), KW/GD channel pores spontaneously widened at the HBC gate, resulting in wetting at the HBC region, high K+ concentrations in the inner cavity, and K+ ion permeation through the SF via strict ion-ion knock-on, at rates comparable to measured ion conductance. When channels were back-mutated to the WT Gly at residue 178 after wetting had occurred, KW/GD(G) channels remained in a predominantly open and conducting state, but with intermittent de-wetted phases and reduced overall conductance, qualitatively paralleling lower experimentally measured single-channel conductance in WT versus KW/GD channels.
Supplementary Material
Acknowledgments
Merritt C. Maduke served as editor.
We thank the staff at Advanced Photon Source beamlines 23-ID-C/E, especially K. Rajashankar, K. Perry, and N. Sukumar, for assistance at the synchrotron. The computational results presented have been achieved using the Vienna Scientific Cluster and XSEDE. We thank Peter Zangerl for computational advice and scripting.
This work used NE-CAT beamlines (National Institutes of Health, National Institute of General Medical Sciences grant GM103403), a Pilatus detector (National Institutes of Health grant RR029205) and an Eiger detector (National Institutes of Health grant OD021527) at Advanced Photon Source (U.S. Department of Energy grant DE-AC02-06CH11357). This work was supported by National Institutes of Health grant HL140024 (to C.G. Nichols), and Austrian Science Fund grants W1232 (to E-M. Zangerl-Plessl, H. Bernsteiner, and A. Stary-Weinzinger) and I-2101-B26 (to A. Stary-Weinzinger).
The authors declare no competing financial interests.
Author contributions: E-M. Zangerl-Plessl, S-J. Lee, A. Stary-Weinzinger, and C.G. Nichols conceived the project. S-J. Lee and G. Maksaev carried out experiments. E-M. Zangerl-Plessl and H. Bernsteiner carried out molecular modeling and analyses. F. Ren and P. Yuan collected the diffraction data, which was analyzed by S-J. Lee. E-M. Zangerl-Plessl, S-J. Lee, A. Stary-Weinzinger, and C.G. Nichols wrote the paper, which was edited by H. Bernsteiner, G. Maksaev, F. Ren, and P. Yuan.
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., and Lindahl E.. 2015. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1–2:19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Alcayaga C., Cecchi X., Alvarez O., and Latorre R.. 1989. Streaming potential measurements in Ca2+-activated K+ channels from skeletal and smooth muscle. Coupling of ion and water fluxes. Biophys. J. 55:367–371. 10.1016/S0006-3495(89)82814-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando H., Kuno M., Shimizu H., Muramatsu I., and Oiki S.. 2005. Coupled K+-water flux through the HERG potassium channel measured by an osmotic pulse method. J. Gen. Physiol. 126:529–538. 10.1085/jgp.200509377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åqvist J., and Luzhkov V.. 2000. Ion permeation mechanism of the potassium channel. Nature. 404:881–884. 10.1038/35009114 [DOI] [PubMed] [Google Scholar]
- Aryal P., Abd-Wahab F., Bucci G., Sansom M.S.P., and Tucker S.J.. 2014. A hydrophobic barrier deep within the inner pore of the TWIK-1 K2P potassium channel. Nat. Commun. 5:4377 10.1038/ncomms5377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryal P., Sansom M.S., and Tucker S.J.. 2015. Hydrophobic gating in ion channels. J. Mol. Biol. 427:121–130. 10.1016/j.jmb.2014.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavro V.N., De Zorzi R., Schmidt M.R., Muniz J.R., Zubcevic L., Sansom M.S., Vénien-Bryan C., and Tucker S.J.. 2012. Structure of a KirBac potassium channel with an open bundle crossing indicates a mechanism of channel gating. Nat. Struct. Mol. Biol. 19:158–163. 10.1038/nsmb.2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen H.J.C., Grigera J.R., and Straatsma T.P.. 1987. The missing term in effective pair potentials. J. Phys. Chem. 91:6269–6271. 10.1021/j100308a038 [DOI] [Google Scholar]
- Berger O., Edholm O., and Jähnig F.. 1997. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 72:2002–2013. 10.1016/S0006-3495(97)78845-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernèche S., and Roux B.. 2001. Energetics of ion conduction through the K+ channel. Nature. 414:73–77. 10.1038/35102067 [DOI] [PubMed] [Google Scholar]
- Bernsteiner H., Zangerl-Plessl E.M., Chen X., and Stary-Weinzinger A.. 2019. Conduction through a narrow inward-rectifier K+ channel pore. J. Gen. Physiol. 151:1231–1246. 10.1085/jgp.201912359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley J., O’Meara F., Farrell D., and Nielsen J.E.. 2012. Highly perturbed pKa values in the unfolded state of hen egg white lysozyme. Biophys. J. 102:1636–1645. 10.1016/j.bpj.2012.02.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelidze T.I., Niu X., and Magleby K.L.. 2003. A ring of eight conserved negatively charged amino acids doubles the conductance of BK channels and prevents inward rectification. Proc. Natl. Acad. Sci. USA. 100:9017–9022. 10.1073/pnas.1532257100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G., Donadio D., and Parrinello M.. 2007. Canonical sampling through velocity rescaling. J. Chem. Phys. 126:014101 10.1063/1.2408420 [DOI] [PubMed] [Google Scholar]
- Cheng W.W.L., D’Avanzo N., Doyle D.A., and Nichols C.G.. 2011. Dual-mode phospholipid regulation of human inward rectifying potassium channels. Biophys. J. 100:620–628. 10.1016/j.bpj.2010.12.3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordomí A., Caltabiano G., and Pardo L.. 2012. Membrane Protein Simulations Using AMBER Force Field and Berger Lipid Parameters. J. Chem. Theory Comput. 8:948–958. 10.1021/ct200491c [DOI] [PubMed] [Google Scholar]
- D’Avanzo N., Cheng W.W., Doyle D.A., and Nichols C.G.. 2010. Direct and specific activation of human inward rectifier K+ channels by membrane phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 285:37129–37132. 10.1074/jbc.C110.186692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl A.C., Chavent M., and Sansom M.S.. 2012. Bendix: intuitive helix geometry analysis and abstraction. Bioinformatics. 28:2193–2194. 10.1093/bioinformatics/bts357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T., York D., and Pedersen L.. 1993. Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- de Groot B.L., and Grubmüller H.. 2001. Water permeation across biological membranes: mechanism and dynamics of aquaporin-1 and GlpF. Science. 294:2353–2357. 10.1126/science.1062459 [DOI] [PubMed] [Google Scholar]
- del Camino D., and Yellen G.. 2001. Tight steric closure at the intracellular activation gate of a voltage-gated K(+) channel. Neuron. 32:649–656. 10.1016/S0896-6273(01)00487-1 [DOI] [PubMed] [Google Scholar]
- Díaz-Franulic I., Sepúlveda R.V., Navarro-Quezada N., González-Nilo F., and Naranjo D.. 2015. Pore dimensions and the role of occupancy in unitary conductance of Shaker K channels. J. Gen. Physiol. 146:133–146. 10.1085/jgp.201411353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domene C., Grottesi A., and Sansom M.S.. 2004. Filter flexibility and distortion in a bacterial inward rectifier K+ channel: simulation studies of KirBac1.1. Biophys. J. 87:256–267. 10.1529/biophysj.104.039917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., and MacKinnon R.. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. 10.1126/science.280.5360.69 [DOI] [PubMed] [Google Scholar]
- Emsley P., and Cowtan K.. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Enkvetchakul D., Loussouarn G., Makhina E., and Nichols C.G.. 2001. ATP interaction with the open state of the K(ATP) channel. Biophys. J. 80:719–728. 10.1016/S0006-3495(01)76051-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y., and Kubo Y.. 2006. Functional roles of charged amino acid residues on the wall of the cytoplasmic pore of Kir2.1. J. Gen. Physiol. 127:401–419. 10.1085/jgp.200509434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furini S., and Domene C.. 2009. Atypical mechanism of conduction in potassium channels. Proc. Natl. Acad. Sci. USA. 106:16074–16077. 10.1073/pnas.0903226106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosink L.J., Hogan E.A., Pulsipher T.C., and Baker N.A.. 2014. Bayesian model aggregation for ensemble-based estimates of protein pKa values. Proteins. 82:354–363. 10.1002/prot.24390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi E., Sanguinetti M.C., Bartos D.C., Bers D.M., Chen-Izu Y., Chiamvimonvat N., Colecraft H.M., Delisle B.P., Heijman J., Navedo M.F., et al. 2017. Potassium channels in the heart: structure, function and regulation. J. Physiol. 595:2209–2228. 10.1113/JP272864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N., and Peitsch M.C.. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 18:2714–2723. 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- Haider S., Khalid S., Tucker S.J., Ashcroft F.M., and Sansom M.S.. 2007. Molecular dynamics simulations of inwardly rectifying (Kir) potassium channels: a comparative study. Biochemistry. 46:3643–3652. 10.1021/bi062210f [DOI] [PubMed] [Google Scholar]
- Hansen S.B., Tao X., and MacKinnon R.. 2011. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature. 477:495–498. 10.1038/nature10370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Bekker H., Berendsen H.J.C., and Fraaije J.G.E.M.. 1997. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [DOI] [Google Scholar]
- Hibino H., Inanobe A., Furutani K., Murakami S., Findlay I., and Kurachi Y.. 2010. Inwardly rectifying potassium channels: their structure, function, and physiological roles. Physiol. Rev. 90:291–366. 10.1152/physrev.00021.2009 [DOI] [PubMed] [Google Scholar]
- Hilgemann D.W., and Ball R.. 1996. Regulation of cardiac Na+,Ca2+ exchange and KATP potassium channels by PIP2. Science. 273:956–959. 10.1126/science.273.5277.956 [DOI] [PubMed] [Google Scholar]
- Ho B.K., and Gruswitz F.. 2008. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8:49 10.1186/1472-6807-8-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., and Simmerling C.. 2006. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 65:712–725. 10.1002/prot.21123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.O., Borhani D.W., Lindorff-Larsen K., Maragakis P., Jogini V., Eastwood M.P., Dror R.O., and Shaw D.E.. 2010. Principles of conduction and hydrophobic gating in K+ channels. Proc. Natl. Acad. Sci. USA. 107:5833–5838. 10.1073/pnas.0911691107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.O., Jogini V., Borhani D.W., Leffler A.E., Dror R.O., and Shaw D.E.. 2012. Mechanism of voltage gating in potassium channels. Science. 336:229–233. 10.1126/science.1216533 [DOI] [PubMed] [Google Scholar]
- Joung I.S., and Cheatham T.E. III. 2008. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 112:9020–9041. 10.1021/jp8001614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klesse G., Rao S., Sansom M.S.P., and Tucker S.J.. 2019. CHAP: A Versatile Tool for the Structural and Functional Annotation of Ion Channel Pores. J. Mol. Biol. 431:3353–3365. 10.1016/j.jmb.2019.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec W., Köpfer D.A., Vickery O.N., Bondarenko A.S., Jansen T.L.C., de Groot B.L., and Zachariae U.. 2018. Direct knock-on of desolvated ions governs strict ion selectivity in K+ channels. Nat. Chem. 10:813–820. 10.1038/s41557-018-0105-9 [DOI] [PubMed] [Google Scholar]
- Köpfer D.A., Song C., Gruene T., Sheldrick G.M., Zachariae U., and de Groot B.L.. 2014. Ion permeation in K+ channels occurs by direct Coulomb knock-on. Science. 346:352–355. 10.1126/science.1254840 [DOI] [PubMed] [Google Scholar]
- Kurata H.T., Zhu E.A., and Nichols C.G.. 2010. Locale and chemistry of spermine binding in the archetypal inward rectifier Kir2.1. J. Gen. Physiol. 135:495–508. 10.1085/jgp.200910253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langan P.S., Vandavasi V.G., Weiss K.L., Afonine P.V., El Omari K., Duman R., Wagner A., and Coates L.. 2018. Anomalous X-ray diffraction studies of ion transport in K+ channels. Nat. Commun. 9:4540 10.1038/s41467-018-06957-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-J., Wang S., Borschel W., Heyman S., Gyore J., and Nichols C.G.. 2013. Secondary anionic phospholipid binding site and gating mechanism in Kir2.1 inward rectifier channels. Nat. Commun. 4:2786 10.1038/ncomms3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.J., Ren F., Zangerl-Plessl E.M., Heyman S., Stary-Weinzinger A., Yuan P., and Nichols C.G.. 2016. Structural basis of control of inward rectifier Kir2 channel gating by bulk anionic phospholipids. J. Gen. Physiol. 148:227–237. 10.1085/jgp.201611616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lü S., Liu Y., Pang C., Chen Y., Zhang S., Yu H., Long M., Zhang H., Logothetis D.E., et al. 2015. Identification of the Conformational transition pathway in PIP2 Opening Kir Channels. Sci. Rep. 5:11289 10.1038/srep11289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder T., Wang S., Zangerl-Plessl E.-M., Nichols C.G., and Stary-Weinzinger A.. 2015. Molecular Dynamics Simulations of KirBac1.1 Mutants Reveal Global Gating Changes of Kir Channels. J. Chem. Inf. Model. 55:814–822. 10.1021/acs.jcim.5b00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.S., Sompornpisut P., and Perozo E.. 2001. Structure of the KcsA channel intracellular gate in the open state. Nat. Struct. Biol. 8:883–887. 10.1038/nsb1001-883 [DOI] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., and Mackinnon R.. 2005. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903. 10.1126/science.1116269 [DOI] [PubMed] [Google Scholar]
- Long S.B., Tao X., Campbell E.B., and MacKinnon R.. 2007. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 450:376–382. 10.1038/nature06265 [DOI] [PubMed] [Google Scholar]
- Lopatin A.N., Makhina E.N., and Nichols C.G.. 1994. Potassium channel block by cytoplasmic polyamines as the mechanism of intrinsic rectification. Nature. 372:366–369. 10.1038/372366a0 [DOI] [PubMed] [Google Scholar]
- Martin G.M., Yoshioka C., Rex E.A., Fay J.F., Xie Q., Whorton M.R., Chen J.Z., and Shyng S.L.. 2017. Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating. eLife. 6:e24149 10.7554/eLife.24149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., and Dodson E.J.. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255. 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- Naranjo D., Moldenhauer H., Pincuntureo M., and Díaz-Franulic I.. 2016. Pore size matters for potassium channel conductance. J. Gen. Physiol. 148:277–291. 10.1085/jgp.201611625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols C.G., and Lopatin A.N.. 1997. Inward rectifier potassium channels. Annu. Rev. Physiol. 59:171–191. 10.1146/annurev.physiol.59.1.171 [DOI] [PubMed] [Google Scholar]
- Nimigean C.M., Chappie J.S., and Miller C.. 2003. Electrostatic tuning of ion conductance in potassium channels. Biochemistry. 42:9263–9268. 10.1021/bi0348720 [DOI] [PubMed] [Google Scholar]
- Nishida M., Cadene M., Chait B.T., and MacKinnon R.. 2007. Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. EMBO J. 26:4005–4015. 10.1038/sj.emboj.7601828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips L.R., and Nichols C.G.. 2003. Ligand-induced closure of inward rectifier Kir6.2 channels traps spermine in the pore. J. Gen. Physiol. 122:795–805. 10.1085/jgp.200308953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proks P., Capener C.E., Jones P., and Ashcroft F.M.. 2001. Mutations within the P-loop of Kir6.2 modulate the intraburst kinetics of the ATP-sensitive potassium channel. J. Gen. Physiol. 118:341–353. 10.1085/jgp.118.4.341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorf M., Potrykus I., and Neuhaus G.. 1994. Microinjection technique: routine system for characterization of microcapillaries by bubble pressure measurement. Exp. Cell Res. 210:260–267. 10.1006/excr.1994.1038 [DOI] [PubMed] [Google Scholar]
- Schönherr R., and Heinemann S.H.. 1996. Molecular determinants for activation and inactivation of HERG, a human inward rectifier potassium channel. J. Physiol. 493:635–642. 10.1113/jphysiol.1996.sp021410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart O.S., Neduvelil J.G., Wang X., Wallace B.A., and Sansom M.S.. 1996. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14:354–360: 376. 10.1016/S0263-7855(97)00009-X [DOI] [PubMed] [Google Scholar]
- Sukhareva M., Hackos D.H., and Swartz K.J.. 2003. Constitutive activation of the Shaker Kv channel. J. Gen. Physiol. 122:541–556. 10.1085/jgp.200308905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Morishige K., Jahangir A., Yamada M., Findlay I., Koyama H., and Kurachi Y.. 1994. Molecular cloning and functional expression of cDNA encoding a second class of inward rectifier potassium channels in the mouse brain. J. Biol. Chem. 269:23274–23279. [PubMed] [Google Scholar]
- Tao X., Avalos J.L., Chen J., and MacKinnon R.. 2009. Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science. 326:1668–1674. 10.1126/science.1180310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trick J.L., Aryal P., Tucker S.J., and Sansom M.S.. 2015. Molecular simulation studies of hydrophobic gating in nanopores and ion channels. Biochem. Soc. Trans. 43:146–150. 10.1042/BST20140256 [DOI] [PubMed] [Google Scholar]
- Vagin A., and Teplyakov A.. 2000. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 56:1622–1624. 10.1107/S0907444900013780 [DOI] [PubMed] [Google Scholar]
- Wang J., Wang W., Kollman P.A., and Case D.A.. 2006. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 25:247–260. 10.1016/j.jmgm.2005.12.005 [DOI] [PubMed] [Google Scholar]
- Wang S., Lee S.-J.L., Maksaev G., Fang X., Zuo C., and Nichols C.G.. 2019. Potassium channel selectivity filter dynamics revealed by single-molecule FRET. Nat. Chem. Biol. 15:377–383. 10.1038/s41589-019-0240-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Lee S.J., Heyman S., Enkvetchakul D., and Nichols C.G.. 2012. Structural rearrangements underlying ligand-gating in Kir channels. Nat. Commun. 3:617 10.1038/ncomms1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Vafabakhsh R., Borschel W.F., Ha T., and Nichols C.G.. 2016. Structural dynamics of potassium-channel gating revealed by single-molecule FRET. Nat. Struct. Mol. Biol. 23:31–36. 10.1038/nsmb.3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., and MacKinnon R.. 2017. Cryo-EM Structure of the Open Human Ether-à-go-go-Related K+ Channel hERG. Cell. 169:422–430.e10. 10.1016/j.cell.2017.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton M.R., and MacKinnon R.. 2011. Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell. 147:199–208. 10.1016/j.cell.2011.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton M.R., and MacKinnon R.. 2013. X-ray structure of the mammalian GIRK2-βγ G-protein complex. Nature. 498:190–197. 10.1038/nature12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242. 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Shin H.G., Szép S., and Lu Z.. 2009. Physical determinants of strong voltage sensitivity of K(+) channel block. Nat. Struct. Mol. Biol. 16:1252–1258. 10.1038/nsmb.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets and computer code produced in this study are available upon request, and will be deposited in an appropriate public database.