Abstract
Grabe et al. celebrate a new mathematical model of the multidrug transporter EmrE, constructed from NMR and stop flow kinetic data.
Have we been projecting “mechanomorphic” ideas onto molecular machines? That is, has our description of these amazing devices been unduly biased by human ideas of machine design? Molecular machines harness the free energy available from different cellular stores (ATP, ion gradients, etc.) to perform essential biological tasks, including synthesizing proteins, propelling the cell through its environment, and pumping molecules across membranes, to name just a few. Our understanding of how machines accomplish these tasks typically resides at the cartoon level: arrows show a single, directed sequence of transitions between the machine’s various states (Fig. 1), much as we might diagram a macroscopic machine, such as a clock. There is no doubt that considerations of total free energy, which must decrease in any process, indicate a tendency for a molecular machine to proceed in a certain direction. But how much else of the typical cartoons drawn in textbooks has been rigorously established? After all, these machines operate in a stochastic thermal environment, potentially visiting states of unknown structure with unknown properties, and possibly performing unknown auxiliary functions. It remains to be seen how the various internal processes are coupled and the extent to which free energy is efficiently transduced. In this vein, a paper by Hussey et al. (from the Henzler-Wildman laboratory) in this issue of the Journal of General Physiology describes a new mathematical model of the EmrE multidrug efflux pump in which few, if any, transitions or states are prohibited. Their analysis shows that different transport regimes can coexist in a single system that is able to self-regulate according to ion and substrate concentrations.
Figure 1.
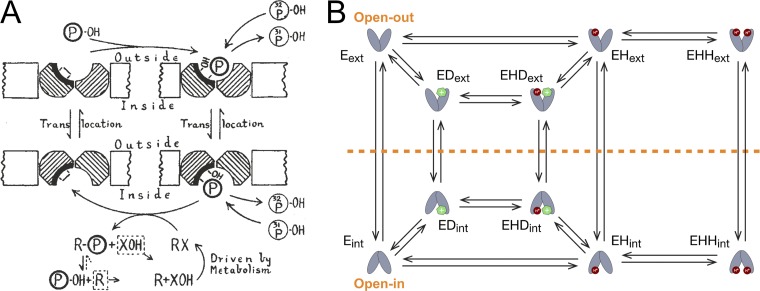
Two different views of transport. (A) Original model of phosphate exchange proposed by Peter Mitchell in the 1950s (Mitchell, 1957). The transporter (dark hash marks) alternates between outward- and inward-facing transitions with phosphate being delivered along the vertical transition on the right and the protein resetting along the left vertical transition. The reaction can only occur in a single file manner along this linear, closed pathway. In this drawing, the transporter creates an internal bond with itself (white rectangle) to satisfy the lack of the bound substrate, but the possibility of an effluxed substrate on the resetting step is mentioned in the paper making the model valid as an antiporter. (B) Free-exchange model of EmrE transport proposed by the Henzler-Wildman laboratory. EmrE is suggested to adopt many more states than the exchanger in A, and these states are more highly connected. Hence, there is not just one linear reaction path through this state space, but instead, many reaction cycles exist with different stoichiometries and varying amounts of “leak.” The red circle is a proton, and the green hexagon is a drug molecule. EmrE is denoted by Eext (external facing) or Eint (internal facing). The symbols E, ED, EHD, EH, and EHH denote EmrE only, drug bound, drug bound with a single proton, and single- and double-bound proton states, respectively.
Concrete examples of machines performing in unexpected ways are well established. The ribosome unbinds many correct transfer RNAs before adding the corresponding amino acid to the growing polypeptide (Blanchard et al., 2004), protein unfolding by the ClpXP protease can reverse under high load (Aubin-Tam et al., 2011), and, on rare occasions, the molecular motor kinesin takes backward steps as it walks along microtubules (Svoboda et al., 1993). In each of these cases, the simple pathway picture breaks down. Nevertheless, the transporter field continues to be dominated by the view that these machines operate along a well-defined, linear cycle, stemming from the seminal alternating-access ideas of Mitchell and Jardetzky (Mitchell, 1957; Jardetzky, 1966). According to their widely accepted schemes, transporter domains rock back and forth between outward-open and inward-open conformations in a single mechanical process, not unlike what we might expect in a human-designed machine. If transporters don’t follow a single pathway, however, the uncoupling that could occur may allow substrates to leak down their concentration gradients, which is what ion channels do. As with the examples discussed already, our ideas tend to be framed in the context of the limited number of structures available, which form the basis for models to explain electrophysiological and biochemical experiments. For instance, the first structure of the sodium-dependent transporter LeuT revealed a “water-tight” occluded state with gates locked to the outside and inside (Yamashita et al., 2005). But there are a small handful of well-studied, classic examples that uphold the notion that transport proteins work with machine precision. For example, years of data revealed how ATP synthase works as a rotary motor (Yoshida et al., 2001) and extensive functional and structural studies showed that LacY works via alternating access (Abramson et al., 2003). Fundamental to the resulting models is a tight coupling of the reaction steps along each cycle. For instance, if two Na+ ions per bound substrate are thought to be present in an x-ray structure then it is often presumed that the stoichiometry is fixed at 2:1, regardless of whether the transporter turnover is fast or slow, or operating close to stall or far from equilibrium.
In this issue, Hussey and coworkers present a compelling mathematical analysis of the EmrE multidrug efflux pump that explicitly addresses the functional consequences of this transporter’s ability to adopt “off pathway” conformations. Their model is constructed from precise NMR and stop flow kinetic experiments performed in the Henzler-Wildman laboratory (Morrison et al., 2015; Robinson et al., 2017) and others (Adam et al., 2007; Gayen et al., 2016), which have provided unprecedented insight into the detailed mechanism of this transporter. Traditional membrane transport studies, by contrast, are rather imprecise from a structural point of view. If the transporter is electrogenic, patch clamp electrophysiology coupled with radioactive uptake assays can sometimes be used to determine the current-voltage properties of the transporter, revealing kinetic behavior, stoichiometries, and regulatory elements (Loo et al., 2006). However, for transporters that fail to express in oocytes, such as bacterial transporters, radioactive uptake assays in proteoliposomes are the primary tool, and only in cases when enough protein can be expressed. These assays have been used to determine the stoichiometry of transporter (Fitzgerald et al., 2017), but precise timescale information is not preserved, and the orientation of the protein in the membrane and the states it adopts remain unknown. The Henzler-Wildman group has successfully exploited the relatively small size of EmrE, and their ability to express it in large quantities sufficient for NMR experiments, to tease apart different conformational states, the rates between these states, and how these rates depend on environmental conditions.
The Henzler-Wildman model, termed the “free-exchange model” (Fig. 1 B), allows for standard exchange of ion (H+ in this case) and substrate, as well as cotransport. It can be thought of as a more weakly coupled version of a standard transport model, in which few if any transitions or states are prohibited (Zuckerman, 2019; Hill, 2005); for example, inward–outward alternation is permitted in any binding state. Thus, leak or “slippage” pathways, in which ions or molecules pass through the transporter down their gradients uncoupled to any other process, are possible in this model. As the authors’ analysis shows, different transport regimes can then coexist in a single system and are essentially “self-regulated” according to ion and substrate conditions, rather than being controlled externally by, for example, a kinase or endogenous lipid binding. A single set of intrinsic transporter rate constants can cause the efflux of some drugs and import of others. Thus, for some drugs, the transporter acts as an antiporter, while for others, it switches mode to be a symporter. Further, with only moderate biasing of key rate constants the model can behave as a highly coupled transporter with ideal stoichiometry, explaining how certain experimental conditions may make it appear that the system has a fixed stoichiometry, while other conditions alter this view.
The idea of “slippage,” in which the targeted process (e.g., substrate transport) is not fully coupled to the driving process (e.g., downhill ion flow), has been explored theoretically for some time. Notably, Terrell Hill emphasized such imperfect coupling in his remarkable short book on biochemical cycles (Hill, 2005). In addition to the dissipation of free energy as heat, which must accompany any uni-directional process in the cell, slippage entails additional energy loss. In ion-driven transport, for example, slippage would imply that some ions traverse the membrane down their gradient without accomplishing substrate transport. Just such an event was observed in molecular simulations of the sugar symporter vSGLT, in which the bound sugar molecule was released to the extracellular space from an open inward-facing state, while the ion was released to the cytoplasm down its concentration gradient (Adelman et al., 2016). There is clear experimental evidence for the phenomenon of slippage. Notably, the oxidative phosphorylation process can be regulated or mutated to shift the balance between ATP synthesis and heat production (Wallace, 2005), and single-molecule transport studies have revealed previously unappreciated H+ leak states in the AHA2 H+ pump (Veshaguri et al., 2016). A transporter that switches between states with perfect ion-substrate coupling and states with poor coupling will exhibit time-averaged ion-substrate stoichiometries that are not integers. But while noninteger experimental stoichiometries are found in almost every published biophysical study of transporters, the values are often rounded to the nearest whole number. We suggest that these discrepancies, in some cases, may reveal more complex or imperfectly coupled transport. For instance, some systems are known to exhibit varying stoichiometry under different conditions, such as the V-ATPase at different pH values (Kettner et al., 2003) and systems recently reviewed by the Poolman laboratory (Henderson et al., 2019).
Although nature may not be able to avoid a certain amount of slippage, evolution is a very effective survival-oriented process; has it therefore learned to exploit slippage? Beyond oxidative phosphorylation–driven heating, another famous example of slippage is the “kinetic proofreading” or “editing” processes that enable significantly enhanced fidelity to a template in transcription, translation, and DNA duplication (Hopfield, 1974; Fersht, 1977). In each of these cases, free energy is seemingly “wasted” in a partial reversal of the process, which ultimately results in improved fidelity to the template. Importantly, partial reversal is a mechanistic requirement for enhanced selectivity, and free energy is traded for information. In yet another example where slippage presents advantages, some transporters are so effective at accumulating substrate that they risk cellular lysis (Postma et al., 1990), thus it has been suggested that slippage may act as a safety valve to limit the osmotic pressure that a transporter can generate (Henderson and Poolman, 2017; Henderson et al., 2019).
Phylogenetic analysis suggests a role for imperfect transporters as evolutionary intermediates. In the early 1990s, Marger and Saier noted homologous families of transmembrane facilitators could be grouped into five clusters based on sequence similarity (Marger and Saier, 1993), including uniporters, symporters, and antiporters that appeared to evolve from one or more common ancestors. Extending this concept, Miller and Accardi realized that the ClC family of membrane proteins, while all adopting the same structural fold, evolved into either Cl−/H+ antiporters or Cl− channels (Accardi and Miller, 2004). Staying within the same symporter cluster, the SGLTs evolved subtypes with different stoichiometries (2:1 for hSGLT1 or 1:1 for hSGLT2) that have different functional properties and expression patterns tuned to specific tissues (Wright et al., 2017). When considering the divergent evolution of all of these transporters, pressing questions arise: Did any intermediate ancestor have variable stoichiometry, or were any capable of both symport and antiport? If so, do extant transporters retain these properties? Henzler-Wildman and coworkers show that EmrE clearly does, and it remains to be seen whether mechanistic heterogeneity in molecular machines provides additional unknown benefits that we have yet to uncover.
Despite the fascinating possibilities that are hinted at in a paradigm embracing slip, including alternative pathways and variable stoichiometries, we want to conclude with a word of caution. Teasing out these phenomena from experiments can be difficult, sometimes leading to conflicting results. For instance, only after careful analysis with the correct substrate and the right experiments were Coady and coworkers able to show an invariant 2:1 stoichiometry for the Na+/monocarboxylate cotransporter SMCT1 (Coady et al., 2007), which had previously been reported to have a variable stoichiometry. Thus, there is no substitute for meticulous experimentation with a critical eye.
Acknowledgments
Merritt C. Maduke served as editor.
This work was supported by grants from the National Institutes of Health R01 GM089740 (to M. Grabe and J.M. Rosenberg) and the National Science Foundation MCB 1715823 (to D.M. Zuckerman).
The authors declare no competing financial interests.
References
- Abramson J., Smirnova I., Kasho V., Verner G., Iwata S., and Kaback H.R.. 2003. The lactose permease of Escherichia coli: overall structure, the sugar-binding site and the alternating access model for transport. FEBS Lett. 555:96–101. 10.1016/S0014-5793(03)01087-1 [DOI] [PubMed] [Google Scholar]
- Accardi A., and Miller C.. 2004. Secondary active transport mediated by a prokaryotic homologue of ClC Cl- channels. Nature. 427:803–807. 10.1038/nature02314 [DOI] [PubMed] [Google Scholar]
- Adam Y., Tayer N., Rotem D., Schreiber G., and Schuldiner S.. 2007. The fast release of sticky protons: kinetics of substrate binding and proton release in a multidrug transporter. Proc. Natl. Acad. Sci. USA. 104:17989–17994. 10.1073/pnas.0704425104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman J.L., Ghezzi C., Bisignano P., Loo D.D., Choe S., Abramson J., Rosenberg J.M., Wright E.M., and Grabe M.. 2016. Stochastic steps in secondary active sugar transport. Proceedings of the National Academy of Sciences of the United States of America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin-Tam M.E., Olivares A.O., Sauer R.T., Baker T.A., and Lang M.J.. 2011. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 145:257–267. 10.1016/j.cell.2011.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard S.C., Gonzalez R.L., Kim H.D., Chu S., and Puglisi J.D.. 2004. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 11:1008–1014. 10.1038/nsmb831 [DOI] [PubMed] [Google Scholar]
- Coady M.J., Wallendorff B., Bourgeois F., Charron F., and Lapointe J.Y.. 2007. Establishing a definitive stoichiometry for the Na+/monocarboxylate cotransporter SMCT1. Biophys. J. 93:2325–2331. 10.1529/biophysj.107.108555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A.R. 1977. Editing mechanisms in protein synthesis. Rejection of valine by the isoleucyl-tRNA synthetase. Biochemistry. 16:1025–1030. 10.1021/bi00624a034 [DOI] [PubMed] [Google Scholar]
- Fitzgerald G.A., Mulligan C., and Mindell J.A.. 2017. A general method for determining secondary active transporter substrate stoichiometry. eLife. 6:6 10.7554/eLife.21016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayen A., Leninger M., and Traaseth N.J.. 2016. Protonation of a glutamate residue modulates the dynamics of the drug transporter EmrE. Nat. Chem. Biol. 12:141–145. 10.1038/nchembio.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., and Poolman B.. 2017. Proton-solute coupling mechanism of the maltose transporter from Saccharomyces cerevisiae. Sci. Rep. 7:14375 10.1038/s41598-017-14438-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R.K., Fendler K., and Poolman B.. 2019. Coupling efficiency of secondary active transporters. Curr. Opin. Biotechnol. 58:62–71. 10.1016/j.copbio.2018.11.005 [DOI] [PubMed] [Google Scholar]
- Hill T.L. 2005. Free energy transduction and biochemical cycle kinetics. Dover Publications, Mineola, NY. [Google Scholar]
- Hopfield J.J. 1974. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. USA. 71:4135–4139. 10.1073/pnas.71.10.4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardetzky O. 1966. Simple allosteric model for membrane pumps. Nature. 211:969–970. 10.1038/211969a0 [DOI] [PubMed] [Google Scholar]
- Kettner C., Bertl A., Obermeyer G., Slayman C., and Bihler H.. 2003. Electrophysiological analysis of the yeast V-type proton pump: variable coupling ratio and proton shunt. Biophys. J. 85:3730–3738. 10.1016/S0006-3495(03)74789-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo D.D.F., Hirayama B.A., Karakossian M.H., Meinild A.-K., and Wright E.M.. 2006. Conformational dynamics of hSGLT1 during Na+/glucose cotransport. J. Gen. Physiol. 128:701–720. 10.1085/jgp.200609643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marger M.D., and Saier M.H. Jr. 1993. A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem. Sci. 18:13–20. 10.1016/0968-0004(93)90081-W [DOI] [PubMed] [Google Scholar]
- Mitchell P. 1957. A general theory of membrane transport from studies of bacteria. Nature. 180:134–136. 10.1038/180134a0 [DOI] [PubMed] [Google Scholar]
- Morrison E.A., Robinson A.E., Liu Y., and Henzler-Wildman K.A.. 2015. Asymmetric protonation of EmrE. J. Gen. Physiol. 146:445–461. 10.1085/jgp.201511404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma E., Verduyn C., Kuiper A., Scheffers W.A., and van Dijken J.P.. 1990. Substrate-accelerated death of Saccharomyces cerevisiae CBS 8066 under maltose stress. Yeast. 6:149–158. 10.1002/yea.320060209 [DOI] [PubMed] [Google Scholar]
- Robinson A.E., Thomas N.E., Morrison E.A., Balthazor B.M., and Henzler-Wildman K.A.. 2017. New free-exchange model of EmrE transport. Proc. Natl. Acad. Sci. USA. 114:E10083–E10091. 10.1073/pnas.1708671114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K., Schmidt C.F., Schnapp B.J., and Block S.M.. 1993. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 365:721–727. 10.1038/365721a0 [DOI] [PubMed] [Google Scholar]
- Veshaguri S., Christensen S.M., Kemmer G.C., Ghale G., Møller M.P., Lohr C., Christensen A.L., Justesen B.H., Jørgensen I.L., Schiller J., et al. 2016. Direct observation of proton pumping by a eukaryotic P-type ATPase. Science. 351:1469–1473. 10.1126/science.aad6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D.C. 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39:359–407. 10.1146/annurev.genet.39.110304.095751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E.M., Ghezzi C., and Loo D.D.F.. 2017. Novel and Unexpected Functions of SGLTs. Physiology (Bethesda). 32:435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Singh S.K., Kawate T., Jin Y., and Gouaux E.. 2005. Crystal structure of a bacterial homologue of Na+/Cl–dependent neurotransmitter transporters. Nature. 437:215–223. 10.1038/nature03978 [DOI] [PubMed] [Google Scholar]
- Yoshida M., Muneyuki E., and Hisabori T.. 2001. ATP synthase–a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2:669–677. 10.1038/35089509 [DOI] [PubMed] [Google Scholar]
- Zuckerman D. 2019. Physical Lens on the Cell Available at: http://www.physicallensonthecell.org/chemical-physics/advanced-cycle-logic (accessed November 15, 2019).