Abstract
Children at increased genetic risk for type 1 diabetes (T1D) after environmental exposures may develop pancreatic islet autoantibodies (IA) at a very young age. Metabolic profile changes over time may imply responses to exposures and signal development of the first IA. Our present research in The Environmental Determinants of Diabetes in the Young (TEDDY) study aimed to identify metabolome-wide signals preceding the first IA against GAD (GADA-first) or against insulin (IAA-first). We profiled metabolomes by mass spectrometry from children’s plasma at 3-month intervals after birth until appearance of the first IA. A trajectory analysis discovered each first IA preceded by reduced amino acid proline and branched-chain amino acids (BCAAs), respectively. With independent time point analysis following birth, we discovered dehydroascorbic acid (DHAA) contributing to the risk of each first IA, and γ-aminobutyric acid (GABAs) associated with the first autoantibody against insulin (IAA-first). Methionine and alanine, compounds produced in BCAA metabolism and fatty acids, also preceded IA at different time points. Unsaturated triglycerides and phosphatidylethanolamines decreased in abundance before appearance of either autoantibody. Our findings suggest that IAA-first and GADA-first are heralded by different patterns of DHAA, GABA, multiple amino acids, and fatty acids, which may be important to primary prevention of T1D.
Introduction
Primary metabolites and complex lipid concentrations in children’s blood may reflect genetic and environmental exposure variations that contribute to the development of diseases. Research of the prodrome to type 1 diabetes (T1D) has indicated a strong association of the metabolome or lipidome in healthy infants who later developed pancreatic islet autoantibodies (IA) as markers of islet autoimmunity and progressed to T1D (1–3). In a first investigation, the appearance of autoantibodies against insulin (IAA) and GAD (GADA) was preceded by diminished ketoleucine and elevated glutamic acid (4). Subsequently, it was reported that, in children born to a father or a mother with T1D, autoantibody-positive children had higher levels of odd-chain triglycerides (TGs) and polyunsaturated fatty acid–containing phospholipids at first autoantibody appearance than autoantibody-negative children (5). Children who had a first-degree relative with T1D and developed autoantibodies by 2 years of age also had twofold lower levels of methionine compared with those who developed autoantibodies in later childhood or remained autoantibody negative (1,5,6). However, the studies referenced above mostly identified biomarkers for IA and T1D based either on longitudinal blood samples at and after seroconversion (5) or on samples at only one time point prior to seroconversion (4). The study in 3 used longitudinal metabolome in plasma prior to IA only for the children with two or three autoantibodies, without considering the subtypes of first-appearing IA. For prediction of IA for the use of primary prevention therapy, it is critical to analyze the preseroconversion metabolome in longitudinal samples after birth, as well as to compare metabolic patterns preceding IA between different first-appearing autoantibodies.
The Environmental Determinants of Diabetes in the Young (TEDDY) is a multicenter prospective cohort study to identify environmental factors that trigger or protect against the development of islet autoimmunity and T1D. The first end point is the appearance of a first IA such as IAA, GADA, IA-2A, or zinc transporter 8 (ZnT8)A. The second end point is the clinical onset of diabetes (7,8). The end point for IA is the first persistent confirmed autoantibody, defined as one autoantibody confirmed on two or more consecutive samples (9). The first persistent confirmed, i.e., first-appearing autoantibodies, can be one or a combination of GADA, IAA, and IA-2A. IAA as the first-appearing autoantibody (IAA-first) was associated with HLA DR4-DQ8 and a peak of IAA incidence rate at 1–2 years of age, while GADA as the first-appearing IA (GADA-first) tended to occur in children with DR3-DQ2 and displayed a later incidence peak (10). Furthermore, IAA-first was related to the INS gene polymorphism, while GADA-first was related to polymorphisms in the ERBB3, SH2B3, and BACH2 gene polymorphisms (10). Probiotics before 28 days of age reduced the risk for IAA-first but not GADA-first (11). Hence, we aimed to investigate whether longitudinal metabolome profiles of TEDDY subjects show similar patterns associated with the risk of different first-appearing autoantibodies. The current study used mass spectrometry (MS) with gas chromatography time-of-flight (GC-TOF MS) and liquid chromatography quadrupole time of flight (LC-QTOF MS) to analyze nested case-control TEDDY plasma samples at the West Coast Metabolomics Center, University of California, Davis.
Existing metabolomics studies have revealed that the human metabolome may be affected by age, genetics, seasons, and environmental exposures (12–15). In this study, we performed metabolome-wide trajectory and independent time point analyses, respectively, to identify trajectories and precursory biomarkers per time point associated with prospective IA risk without considering seasonality or environmental exposure factors. Recent research in TEDDY identified genetic risks for IA (16,17), which were also incorporated as covariates in our analyses. In addition to identifying predictive trajectories and biomarkers, we performed metabolite set enrichment analysis based on time points at infancy to show metabolites and lipid clusters enriched for the compounds potentially associated with IA risk.
Research Design and Methods
Mass Spectrum Profiling of Longitudinal Plasma Samples
The TEDDY study enrolled 8,676 children based on HLA-DR-DQ haplogenotypes for the risk of T1D in a prospective cohort. Blood sample collection for participants in TEDDY began at the 3 months of age visit and continued at a 3-month interval up to development of IA before 4 years of age (7,18), although most subjects have randomly missing visits. If a subject develops persistent IA, they continue on the 3-month interval schedule up to the age of 15 years; otherwise, they switch to a 6-month interval schedule. The metabolomes of the TEDDY first nested case-control (NCC) cohort (19,20) subjects for IA were profiled from 10,522 plasma samples. Characteristics of the study population and distribution of plasma samples in the TEDDY NCC cohort have previously been described (9).
The primary metabolite assay in TEDDY was profiled by GC-TOF MS on a LECO Pegasus III instrument, annotated by BinBase (21). The complex lipid assay was analyzed by charged-surface hybrid column with electrospray ionization (CSH-ESI) on Agilent ultrahigh-pressure LC-QTOF MS instruments, annotated with LipidBlast (22). Peaks from GC-TOF MS platform were automatically detected and deconvoluted from coeluting peaks by the LECO ChromaTOF (23) software (v3.0). Raw data output from CSH-ESI-QTOF instruments were processed in an untargeted (qualitative) manner by Agilent software (24) MassHunter Qual (v. B.05.00) to find peaks. Peak features are then imported into Mass Profiler Professional for peak alignments to seek which peaks are present in multiple chromatograms. These peaks are then collated and constrained within the MassHunter quantification software (v. B.05.01) on the accurate mass precursor ion level, using the MS information and the LipidBlast library. The TEDDY Data Coordinating Center applied a comprehensive normalization pipeline, systematic error removal using random forest (SERRF) (25), to the raw quantified intensity of primary metabolites and complex lipids and successfully removed laboratory running order effects. The GC-TOF normalized data contained compounds with minimum abundance levels occurring in >10 samples. The recursive minimum abundances were treated as missing and then filled by the values generated from a label-free metabolomics missing value imputation tool, GMSimpute (26).
Time Course Differential Analysis for Trajectories
The metabolome-wide trajectory analysis was performed by time course differential analysis in the Bioconductor package edge (27,28), which was developed for microarray gene expression time course analysis. The abundances of primary metabolites and complex lipids were first transformed to log2 scale and then fitted by a mixed-effects model. Age points were modeled as a random effect, and the mean abundance over time was a fixed effect modeled by cubic spline. IA case versus control was the biological group factor for comparison, and the matched risk pairs were included as an adjustment covariate. Significance of contrast between IA case and control subjects was determined by the P values of likelihood ratio test on full and null models, where null model included age random effect and the adjustment covariate.
Statistical and Machine Learning Methods for Independent Time Point Biomarkers
For each visit before IA seroconversion, we evaluated the interaction between metabolome and genetics risks using a mixed-effects model (29). The known genetic risk factors were HLA haplogenotypes and five single nucleotide polymorphisms (SNPs), rs2476601 (PTPN22), rs2816316 (RGS1), rs2292239 (ERBB3), rs10517086, and rs3184504 (SH2B3), associated with IA risk, reported in previous TEDDY genetic analyses (16,17). The HLA subtyping for TEDDY-eligible subjects has previously been described (8,17), including nine different haplogenotypes, and are converted to a binary factor—DR3/4 compared with others—for this analysis. The mixed-effects modeling was applied to each compound individually. The known compounds with Benjamini-Hochberg–adjusted (30) P values <0.1 were considered to be associated with a genetic risk.
Since none of the metabolites were found to interact with known genetics risks for IA, we first employed the elastic net conditional logistic regression (CLR) model (31) without genetic risk factors to select a subset of candidate compounds contributing to the risk of IA. This machine learning method combines ridge (32) and lasso (33) regression penalties and used the strata of subjects to account for the nested case-control design. The CLR coefficients were estimated by elastic net penalty constraint with the weight for lasso at α = 0.1. These candidate compounds per time point were selected based on a 10-fold cross validation, implemented in R package clogitL1. Secondly, we applied the CLR without elastic net penalty to each candidate compound, using the available profiles of IA case and control subjects per time point. In this step, HLA DR3/4 and the five IA risk SNPs were included as covariates to account for possible false negatives in the association analysis of metabolites with genetic factors, and the compounds with P value <0.05 were selected as the “potential biomarkers” for end point risk. In the third step, for each time point we reapplied one CLR model to the potential biomarkers simultaneously with HLA DR3/4 and the risk SNPs as covariates and then identified significant biomarkers by the threshold of P < 0.05. This analysis was performed sequentially by time points.
In this analysis, we did not select the biomarkers from compound-wise multiple hypothesis testing because the limited number of preseroconversion metabolic profiles at a later age may result in reduced testing power and no detected biomarkers for a uniform adjusted P value threshold across time points. In addition, it is hard to select the optimal uniform threshold of adjusted P values at different time points, while varying thresholds across time points are also prone to bias. Furthermore, conditional logistic regression modeling of multiple potential biomarkers may lead to false negatives in lipidomics biomarkers, due to the strong correlation among complex lipids. Therefore, we also performed the metabolite set enrichment analysis as follows.
Metabolite Set Enrichment Analysis
The metabolites and lipids were further investigated for the association with IA risk by metabolite set enrichment analysis using ChemRICH (34). We first used the InChiKeys of primary metabolites and complex lipids to identify the corresponding PubChem CID and SMILES ID via the Chemical Translation Service (35) and the PubChem Identifier Exchanger tool. Next, for each compound per infant age visit (3, 6, and 9 months), we tested the association against the risk of IA via the CLR model with HLA genotypes and risk SNPs as covariates. We used the smallest P value among these three visits per compound and the corresponding odds ratio as the input for ChemRICH, since the compound-wise P values at one single visit may result in few enriched clusters.
Data and Resource Availability
The data sets generated and analyzed during the current study will be made available in the National Institute of Diabetes and Digestive and Kidney Diseases Central Repository at https://www.niddkrepository.org/studies/teddy.
Results
Longitudinal Metabolome in TEDDY Nested Case-Control Design
The NCC cohort in TEDDY consists of 2124 subjects grouped into multiple sets. Each set or “pair” contains a case subject (either IA or T1D positive) and control subjects matched by clinical sites, T1D family history (first-degree relative), and sex (9). The case:control ratio within each matched pair in the TEDDY metabolomics study is 1:3 for both IA and T1D. In the current study, we included subjects in the NCC cohort only for IA (414 case and 1,234 matched control subjects), ignoring their progression to T1D.
The details about enrolling eligible TEDDY subjects, the first NCC cohort for IA and T1D, and the collection of longitudinal plasma samples are provided in the flowchart in Supplementary Fig. 1. Blood samples at visits after seroconversion were not included in the present analysis. The number of IA case subjects having plasma samples up to end point within 3 years (36 months) after birth is shown in Table 1. The numbers in boldface type in Table 1 represent case subjects with plasma samples available at seroconversion, which may be less than the total number of case subjects per end point age. A complete summary for the metabolome profiles of all IA case subjects in the TEDDY NCC cohort is provided in Supplementary Table 1. The total number of IA case subjects dropped to ≤38 if preseroconversion profiles at or after the 39-month visit were analyzed. The 36-month visit was the latest age point after birth in which at least 50 IA case subjects had concurrent plasma samples available and their first-appearing autoantibody had not been positively confirmed. Therefore, for maintaining statistical power, most of our downstream analyses included preseroconversion visits up to 3 years of age.
Table 1.
Number of available plasma samples by 3 years (36 months) of age for TEDDY subjects developing IA
| IA case subjects’ end point age (visit month) | Total number of samples | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3M | 6M | 9M | 12M | 15M | 18M | 21M | 24M | 27M | 30M | 33M | 36M | >36M | ||
| Time point (visit month) | ||||||||||||||
| 3M | 14 | 14 | 37 | 28 | 33 | 26 | 24 | 23 | 26 | 13 | 7 | 12 | 49 | 306 |
| 6M | 14 | 44 | 29 | 30 | 23 | 27 | 19 | 25 | 16 | 13 | 12 | 51 | 303 | |
| 9M | 48 | 26 | 32 | 26 | 29 | 21 | 29 | 16 | 14 | 14 | 55 | 310 | ||
| 12M | 29 | 28 | 29 | 27 | 22 | 27 | 15 | 14 | 17 | 48 | 256 | |||
| 15M | 34 | 23 | 28 | 20 | 28 | 15 | 13 | 17 | 53 | 231 | ||||
| 18M | 31 | 33 | 19 | 31 | 15 | 14 | 16 | 55 | 214 | |||||
| 21M | 33 | 18 | 25 | 15 | 16 | 12 | 54 | 173 | ||||||
| 24M | 26 | 26 | 13 | 17 | 14 | 46 | 142 | |||||||
| 27M | 31 | 15 | 14 | 13 | 50 | 123 | ||||||||
| 30M | 19 | 16 | 13 | 48 | 96 | |||||||||
| 33M | 18 | 13 | 54 | 85 | ||||||||||
| 36M | 16 | 51 | 67 | |||||||||||
| Total number of case subjects | 15 | 18 | 52 | 35 | 38 | 35 | 38 | 27 | 32 | 19 | 19 | 17 | 69 | 2,306 |
The numbers in boldface type on the table diagonal indicate case subjects with plasma samples available at seroconversion. Plasma samples after IA seroconversion (below the table diagonal) were not included in the current analysis. M, months.
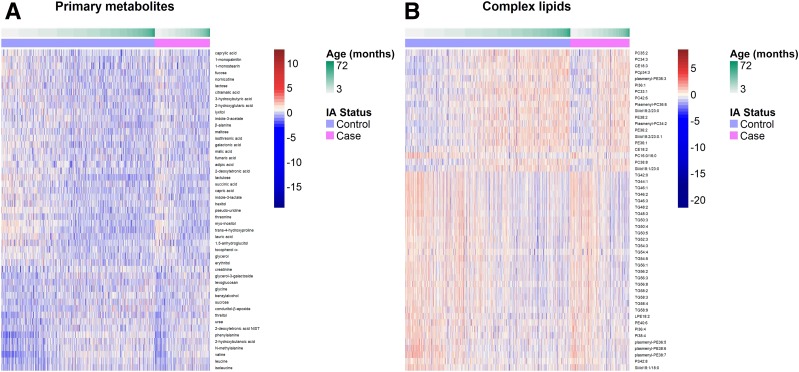
The longitudinal metabolomes for matched control subjects in the TEDDY NCC design were also profiled via mass spectrometry platforms, the available visits for which may not be identical to those of case subjects. In total, the metabolome-wide profiles for all subjects in the TEDDY NCC cohort revealed 144 known primary metabolites and 213 known complex lipids. Downstream statistical analyses need to be performed on metabolites and lipids, respectively, because of different instruments used in profiling. Furthermore, the longitudinal abundance of certain metabolites or lipids up to IA seroconversion in TEDDY subjects displayed visible age patterns (Fig. 1), such as glycine, valine, leucine, isoleucine, 2‐hydroxybutyric acid in Fig. 1A, TGs, phosphatidylethanolamines (PEs), phosphatidylcholines (PCs), and plasmenyl-PEs in Fig. 1B. For better visualization, we plotted the top metabolites and lipids in Fig. 1, selected by the smallest P values of the Spearman correlation test for compound abundance in relation to the 3-month age. This age effect on the TEDDY children’s metabolome may be confounding with underlying case to control contrast over time. Hence, we needed to identify the compounds with differentiated trajectories between case and control subjects in the presence of a common age effect.
Figure 1.
Age effect on the longitudinal metabolome of TEDDY subjects in the nested case-control cohort for islet autoimmunity.
Differentiated Preseroconversion Trajectories
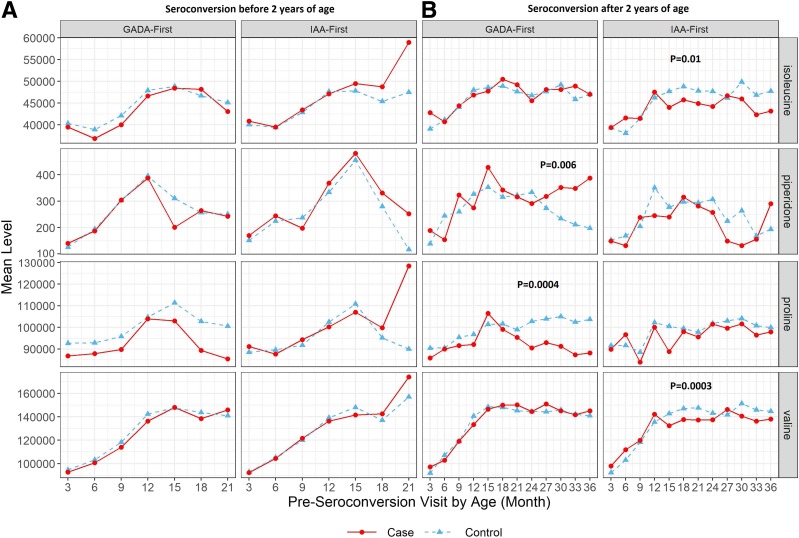
We compared the trajectories of known compounds between IA case and control subjects via a metabolome-wide trajectory differential analysis based on matched pairs for GADA-first and IAA-first, respectively. The case subjects in each autoantibody subset had seroconversion at various ages ranging from 6 months to 72 months. The preseroconversion visits used in this trajectory analysis were at no later than 36 months of age. A trajectory might be “falsely” identified as different if the fold change across visits after 36 months of age was biased due to limited IA cases (e.g., <15 in IAA-first). In the first step, the significance of case compared with control over time per compound was evaluated by the likelihood ratio test P value, listed in Supplementary Tables 2 and 3. Second, we reapplied the analysis to matched pairs with end point age before and after 2 years of age, individually, confirming that valine (P = 0.0003) and isoleucine (P = 0.01) were still different in subjects who experienced IAA-first after 2 years of age, while piperidone (P = 0.006) and proline (P = 0.0004) remained different for GADA-first after 2 years of age. On the other hand, none of the metabolic trajectories were found different with statistical significance (P < 0.01) before 2 years of age for either autoantibody.
The trajectories differentiating IA case from control subjects with P value <0.01 in both steps were plotted (Fig. 2) based on end point age, displaying consistently higher or lower mean abundance level across multiple age points before the onset of each autoantibody. According to the mean abundance and P values (Fig. 2), during 12–24 months of age constantly reduced levels of isoleucine and valine were potential signals for the onset of IAA-first after 2 years of age, while a similar and stable pattern in proline was found in subjects having GADA-first onset after 2 years of age. GADA-first case subjects with the end point before 2 years of age also displayed reduced levels of proline at preseroconversion visits (Fig. 2A), although statistical power in this subset analysis was limited due to low incidence rate of early-age GADA-first (10). An increased level of piperidone throughout 2–3 years of age for the later onset of GADA-first was also observed (Fig. 2B). The higher abundance in piperidone may be a result of dietary patterns, since piperidone is derived from piperidine, which is mainly present in peppers, barley, and flavoring agents (36). These top-differentiated preseroconversion trajectories for IAA-first or GADA-first compared with control subjects might be associated with genetic risk factors, and the association between independent time point levels of these metabolites and future risk of IA is still unknown. Therefore, we further performed an independent time point analysis on subsets of metabolites or lipids and multiple genetic risks simultaneously to identify IA-risk metabolic biomarkers.
Figure 2.
Metabolites with preseroconversion trajectories identified as top differentiated in the two-step time course analysis. Mean abundance of metabolites per age point is plotted for matched pairs who had preseroconversion measurement available and experienced seroconversion before 2 years of age (A) or after 2 years of age (B). P values <0.01 in the second step of time course analysis are presented on the plots.
Precursory Biomarkers for IA at Independent Time Points
We selected independent time points by two scenarios to investigate whether the association between IA risk and compounds is age dependent or related to time to seroconversion, i.e., the preseroconversion visits up to 3 years of age or the visits within 1 year prior to seroconversion. Age at each time point or visit for matched case and control subjects was identical in this analysis. The identified precursory metabolic biomarkers for each first-appearing autoantibody were summarized in Tables 2 and 3 along with P values and odds ratios. Higher levels of dehydroascorbic acid (DHAA) (oxidized vitamin C) after birth (3 months of age) significantly increased the prospective risk of either GADA-first (P = 0.028) or IAA-first (P = 0.0048) (with odds ratio 1.34 [Table 2]). Furthermore, γ-aminobutyric acid (GABA) after birth was found to be associated with future risk of IAA-first only (P = 0.0113). Moreover, our analysis found the risk of IAA-first or GADA-first to be negatively associated with amino acid alanine (Tables 2 and 3) and higher risk of GADA-first to be associated with reduced level of amino acid methionine at 6 months of age (Table 2). We also identified fatty acids at different time points as IA precursory biomarkers for GADA-first onset, i.e., lauric acid and linoleic acid and palmitoleic acid (Tables 2 and 3), which might be a result of daily diet exposures. Intermediate compounds in a branched-chain amino acid (BCAA) metabolic pathway, i.e., α-ketoisocaproic acid and α-ketoisovaleric acid (Table 3), were precursors for GADA-first and IAA-first, respectively.
Table 2.
GADA-first or IAA-first precursory biomarkers per age point regardless of time to seroconversion
| Time after birth | Metabolite | Autoantibody | Odds ratio | Lower 95% | Upper 95% | P value |
|---|---|---|---|---|---|---|
| 3 months | Lactulose | GADA-first | 0.7899 | 0.6556 | 0.9516 | 0.0131 |
| DHAA | GADA-first | 1.3351 | 1.0306 | 1.7294 | 0.0286 | |
| Ethanolamine | GADA-first | 1.223 | 1.0183 | 1.4689 | 0.0313 | |
| Methanolphosphate | GADA-first | 0.674 | 0.461 | 0.9854 | 0.0418 | |
| Lauric acid | IAA-first | 0.6002 | 0.4439 | 0.8116 | 0.0009 | |
| Diglycerol | IAA-first | 0.5564 | 0.3900 | 0.7937 | 0.0012 | |
| DHAA | IAA-first | 1.3360 | 1.0922 | 1.6341 | 0.0048 | |
| γ-aminobutyric acid | IAA-first | 1.3399 | 1.0684 | 1.6804 | 0.0113 | |
| Uric acid | IAA-first | 1.4010 | 1.0013 | 1.9600 | 0.0491 | |
| 6 months | Methionine | GADA-first | 0.5441 | 0.3695 | 0.8011 | 0.0020 |
| ε caprolactam | GADA-first | 0.5790 | 0.3951 | 0.8485 | 0.0051 | |
| Diglycerol | GADA-first | 1.6853 | 1.1112 | 2.5561 | 0.0140 | |
| 5-methoxytryptamine | IAA-first | 0.7152 | 0.5813 | 0.88 | 0.0015 | |
| Lactamide | IAA-first | 1.2384 | 1.0309 | 1.4877 | 0.0223 | |
| Itaconic acid | IAA-first | 1.4184 | 1.0385 | 1.9372 | 0.0280 | |
| 9 months | 2,3-dihydroxybutanoic acid NIST | GADA-first | 0.4850 | 0.3064 | 0.7678 | 0.0020 |
| 2-hydroxyglutaric acid | GADA-first | 1.5644 | 1.1141 | 2.1966 | 0.0098 | |
| Xylitol | GADA-first | 0.6113 | 0.3835 | 0.9745 | 0.0386 | |
| Pelargonic acid | IAA-first | 0.4489 | 0.2596 | 0.7762 | 0.0042 | |
| 5-methoxytryptamine | IAA-first | 1.2375 | 1.0523 | 1.4553 | 0.0100 | |
| 1-monoolein | IAA-first | 1.2602 | 1.0564 | 1.5033 | 0.0102 | |
| Threonic acid | IAA-first | 1.7957 | 1.1424 | 2.8225 | 0.0112 | |
| Uridine | IAA-first | 1.5648 | 1.0794 | 2.2685 | 0.0181 | |
| 12 months | Xylose | GADA-first | 0.7681 | 0.6143 | 0.9603 | 0.0206 |
| Lactic acid | IAA-first | 1.5210 | 1.1226 | 2.0607 | 0.0068 | |
| Cystine | IAA-first | 0.5912 | 0.3860 | 0.9057 | 0.0157 | |
| Alanine | IAA-first | 0.6456 | 0.4371 | 0.9536 | 0.0279 | |
| 15 months | Arachidonic acid | GADA-first | 1.4607 | 1.0818 | 1.9724 | 0.0134 |
| Levoglucosan | IAA-first | 1.3530 | 1.0599 | 1.7271 | 0.0152 | |
| 18 months | Lactose | GADA-first | 0.5541 | 0.3336 | 0.9204 | 0.0226 |
| Benzoic acid | IAA-first | 0.4786 | 0.2550 | 0.8983 | 0.0218 | |
| 21 months | Hippuric acid | GADA-first | 0.7381 | 0.5896 | 0.9239 | 0.0080 |
| Ethanolamine | GADA-first | 0.8050 | 0.6689 | 0.9689 | 0.0218 | |
| 24 months | Ribose | GADA-first | 1.8469 | 1.1923 | 2.8608 | 0.006 |
| ε-caprolactam | IAA-first | 2.8974 | 1.3804 | 6.0818 | 0.0049 | |
| 3-hydroxybutyric acid | IAA-first | 0.6948 | 0.5206 | 0.9272 | 0.0134 | |
| Uracil | IAA-first | 2.1945 | 1.0151 | 4.7443 | 0.0457 | |
| 27 months | trans-4-hydroxyproline | GADA-first | 0.3086 | 0.1175 | 0.8107 | 0.0170 |
| Pyruvic acid | GADA-first | 0.7914 | 0.6332 | 0.9891 | 0.0398 | |
| 30 months | Proline | GADA-first | 0.0638 | 0.0144 | 0.2822 | 0.0003 |
| Piperidone | GADA-first | 2.0026 | 1.3209 | 3.0362 | 0.0011 | |
| Lauric acid | GADA-first | 0.2793 | 0.1140 | 0.6844 | 0.0053 | |
| Linoleic acid | GADA-first | 0.4816 | 0.2579 | 0.8993 | 0.0218 | |
| 33 months | Octadecanol | GADA-first | 4.9297 | 1.2056 | 20.1565 | 0.0264 |
NIST, National Institute of Standards and Technology.
Table 3.
GADA-first or IAA-first precursory biomarkers per time point within 1 year before seroconversion
| Months to IA | Metabolite | Autoantibody | Odds ratio | Lower 95% | Upper 95% | P value |
|---|---|---|---|---|---|---|
| 3 months | Palmitoleic acid | GADA-first | 1.2282 | 1.0374 | 1.4541 | 0.017 |
| Salicylaldehyde | GADA-first | 0.7085 | 0.5268 | 0.9529 | 0.0227 | |
| Xylose | GADA-first | 0.7646 | 0.5905 | 0.9902 | 0.0419 | |
| Lactamide | IAA-first | 1.3493 | 1.1034 | 1.6501 | 0.0035 | |
| Indole-3-lactate | IAA-first | 1.4403 | 1.0667 | 1.9448 | 0.0172 | |
| Xylose | IAA-first | 1.1904 | 1.0095 | 1.4037 | 0.0382 | |
| 6 months | N-methylalanine | GADA-first | 0.4313 | 0.2363 | 0.7874 | 0.0062 |
| Salicylaldehyde | GADA-first | 1.3796 | 1.0315 | 1.845 | 0.0301 | |
| Tagatose | IAA-first | 0.844 | 0.7253 | 0.982 | 0.0282 | |
| Tocopherol α | IAA-first | 2.1471 | 1.2686 | 3.6338 | 0.0044 | |
| 9 months | Salicylaldehyde | GADA-first | 1.3981 | 1.048 | 1.8653 | 0.0227 |
| Phosphate | GADA-first | 2.1096 | 1.1297 | 3.9393 | 0.0191 | |
| 2-ketoisovaleric acid | IAA-first | 1.3308 | 1.0596 | 1.6714 | 0.014 | |
| 12 months | Hexitol | GADA-first | 0.5098 | 0.332 | 0.7829 | 0.0021 |
| Alanine | GADA-first | 0.2094 | 0.0736 | 0.5958 | 0.0034 | |
| N-acetylmannosamine | GADA-first | 0.4875 | 0.2908 | 0.8173 | 0.0064 | |
| Phosphoethanolamine | GADA-first | 1.7059 | 1.1561 | 2.5171 | 0.0071 | |
| Xylose | GADA-first | 1.4224 | 1.0944 | 1.8488 | 0.0084 | |
| 2-ketoisocaproic acid | GADA-first | 0.4103 | 0.2098 | 0.8025 | 0.0093 | |
| 1-monoolein | GADA-first | 1.3622 | 1.069 | 1.736 | 0.0125 | |
| 1,2-dihydroxycylohexane NIST | IAA-first | 1.379 | 1.0262 | 1.8532 | 0.0331 | |
| Levoglucosan | IAA-first | 1.2465 | 1.0139 | 1.5323 | 0.0365 |
NIST, National Institute of Standards and Technology.
Few precursory biomarkers were found in lipids compared with primary metabolites, as strong correlation among lipids alleviated the significance of lipid biomarkers fitted simultaneously in a conditional logistic regression model. In addition, primary metabolite biomarkers were rarely detected after 18 months of age for IAA-first, while GADA-first metabolite biomarkers were present up to 33 months. This contrast of age points between GADA-first and IAA-first metabolic biomarkers is consistent with the mean onset ages for these autoantibodies, i.e., 33 months for GADA-first and 18.3 months for IAA-first. Reduced sample size at later age points may also lead to the absence of metabolic biomarkers. To verify this possibility, we further visualized the contrast between the metabolite profiles of IA case and control subjects prior to seroconversion at 3-, 12-, 24-, and 36-month visits, individually, using partial least squares regression with discriminant analysis (37), presented by principal components plots in Supplementary Fig. 2. The contrast between case and control subjects was greater at 2–3 years of age compared with earlier visits. Therefore, it was the reduced statistical power of limited preseroconversion profiles at 2–3 years of age that resulted in few biomarkers detected for future risk of IAA-first.
Primary Metabolites and Complex Lipid Sets Enriched for Prospective IA Risk
The third result based on longitudinal metabolome profiles is primary metabolite or complex lipid clusters enriched for compounds potentially associated with IA-risk. The input and output information of ChemRICH is listed in Supplementary Tables 4 and 5. The top enriched compound clusters were selected by a threshold of adjusted P value <0.05 (Table 4). The results showed an overlap and some difference in the enriched compound clusters between GADA-first and IAA-first. Specifically, unsaturated TGs, unsaturated PCs, PEs were enriched for both autoantibodies, while sugar alcohols and butyrates were enriched only for IAA-first and plasmalogens, sphingomyelins, unsaturated diglycerides were enriched only for GADA-first. For either first-appearing autoantibody, unsaturated TGs and PEs decreased prior to future seroconversion, while the butyrate cluster (containing GABA) was positively associated with the risk of IAA-first, confirming a similar result from the independent time point analysis.
Table 4.
Top compound clusters enriched for IA-risk primary metabolites or complex lipids at infant age
| Compound cluster name | Cluster size | P values | Adjusted P values | Key compound | Altered metabolites | Increased | Decreased |
|---|---|---|---|---|---|---|---|
| GADA-first | |||||||
| Unsaturated TGs | 53 | 4E-14 | 1E-12 | TG62:1 | 5 | 0 | 5 |
| Unsaturated PCs | 40 | 5E-07 | 9E-06 | Plasmenyl-PC38:5 | 6 | 1 | 5 |
| Plasmalogens | 8 | 4E-05 | 0.0005 | Plasmenyl-PE36:6 | 2 | 0 | 2 |
| Sphingomyelins | 14 | 0.0005 | 0.005 | SM(d18:1/18:1) | 2 | 1 | 1 |
| PEs | 13 | 0.002 | 0.012 | PE38:2 | 3 | 0 | 3 |
| Unsaturated diglycerides | 7 | 0.002 | 0.012 | DG38:5 | 2 | 0 | 2 |
| IAA-first | |||||||
| Unsaturated TGs | 53 | 5E-14 | 2E-12 | TG52:4 | 11 | 0 | 11 |
| Sugar alcohols | 11 | 1E-05 | 0.0002 | Glycerol | 3 | 1 | 2 |
| Unsaturated phosphatidylcholines | 40 | 9E-05 | 0.0011 | PC32:1 | 8 | 3 | 5 |
| Butyrates | 4 | 0.0013 | 0.0091 | γ-aminobutyric acid | 3 | 2 | 1 |
| PEs | 13 | 0.0082 | 0.056 | LPE18:2 | 4 | 1 | 3 |
Discussion
The TEDDY study offers a robust analysis of the metabolome in 417 infants who developed IA, with primarily either IAA only (49%) or GADA only (33%) as the first appearing autoantibodies. The subjects who experienced both IAA and GADA (14%) as the first appearing autoantibodies were not considered in the current analysis. Our aim in the current study was to discover longitudinal metabolic patterns preceding different first appearing IA in the presence of the well-known age effect on metabolic profiles (3,5).
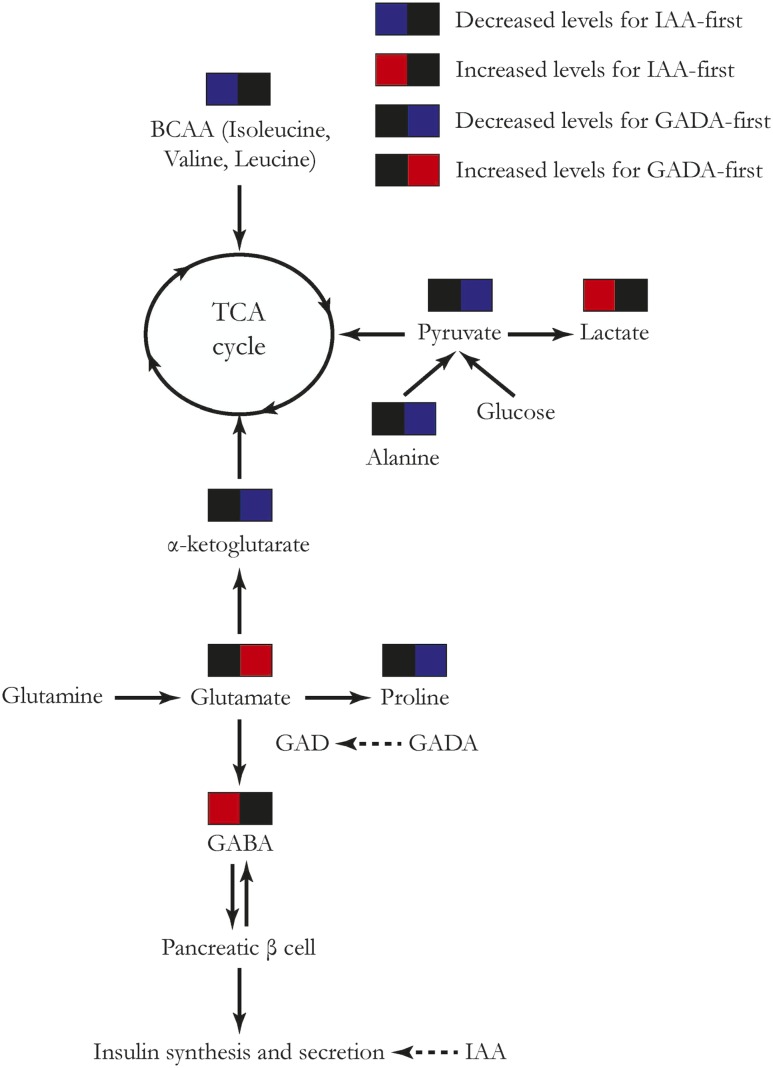
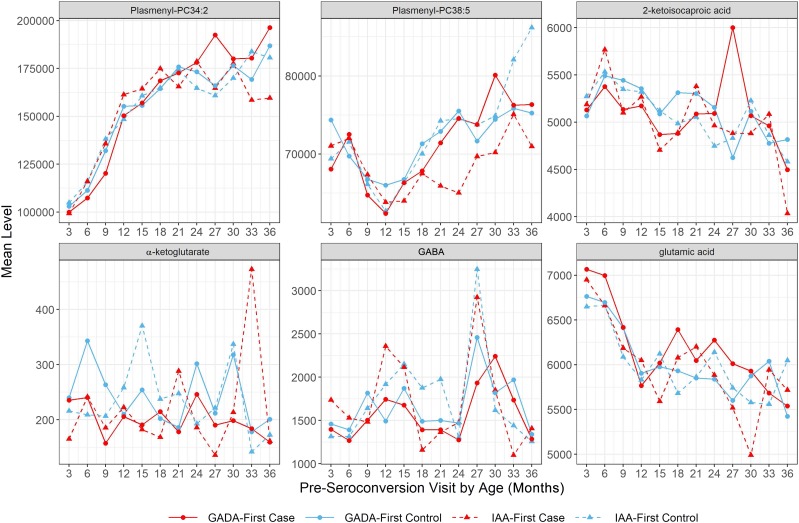
Our data suggest that IAA-first and GADA-first differ in the way that metabolites and lipids precede seroconversion. The significantly lower abundance of isoleucine and valine prior to seroconversion in IAA-first subjects (Fig. 2) is of interest, as isoleucine and valine are BCAAs widely known to potentiate glucose-stimulated insulin secretion (38,39). On the other hand, proline as a nonessential amino acid produced from glutamate cyclization (40) remained at a reduced level across multiple visits before IA (Fig. 2). In addition to the differentiated trajectory, proline was also found to be negatively associated with the risk of GADA-first in independent time point analysis. Proline biosynthesis involves l-glutamate, which interacts with the metabolic pathway of glutamic acid (41), GAD, and GABA (42), as illustrated in Fig. 3. Another critical metabolite derived from glutamic acid, i.e., α-ketoglutarate (42), also displayed lower concentration in GADA-first case compared with control subjects over time before seroconversion (Fig. 4). These results suggest that an enduring decrease in proline and lower α-ketoglutarate level may imply an abnormal glutamic acid metabolism causally related to the underlying change of GAD enzymatic activity in subjects who later developed GADA-first, with relatively higher glutamic acid levels prior to seroconversion (Fig. 4). The elevated level of glutamic acid may also be an indicator for the emergence of GADA-first.
Figure 3.
Metabolic pathways involving trajectory signals and independent time point biomarkers for IA in TEDDY. TCA, tricarboxylic acid.
Figure 4.
Mean abundance of preseroconversion GABA, glutamic acid, glutamine, α-ketoglutarate, leucine, and plasmenyl-PC (ether PC) per age point for case and control subjects in GADA-first and IAA-first groups.
Another major finding in the current study was the association between plasma GABA after birth and the future appearance of IAA-first. This finding was based on both independent time point analysis (Table 2) and enrichment analysis results (Table 4). Why GABA levels would be associated with IAA-first and not GADA-first may be explained by the fact that β-cell GAD65, which produces GABA, may not be affected before GADA onset. The impact of GABA on the pancreatic β-cell function has been thoroughly investigated (43,44). The odds ratio for plasma GABA (Table 2) for IAA-first indicates that higher levels of GABA immediately after birth may be related to β-cell dysfunction, with the appearance of IAA-first possibly related to abnormal insulin synthesis or secretion at early ages. In contrast, we did not find the contribution of GABA to future risk of seroconversion in GADA-first children, providing evidence that GABA has no causal influence on the appearance of GADA-first.
A third major finding of biomarkers for the risk of IA in TEDDY was DHAA after birth. DHAA identified at 3 months of age did not discriminate between GADA-first and IAA-first but showed statistical significance for both autoantibodies. Elevated DHAA or oxidized vitamin C was found to inhibit insulin secretion in mice (45–47), and exposure of isolated mouse pancreatic islets to DHAA or vitamin C reduced the responsiveness of the islets (48) or led to inhibition of insulin secretion from the pancreatic β-cells (47). The results for DHAA in TEDDY samples would seem to be consistent with the existing findings, showing a possible suppressive effect on human pancreatic islet β-cells. Another recent study in TEDDY found immunoassay measurements of plasma vitamin C levels to be associated with lower risk of IAA but not GADA. Further analyses would therefore be required to include immunoassay measurement of DHAA to detail the possible importance of the vitamin C/DHAA ratio and its regulation. Other compounds identified as contributing to IA development in TEDDY were also found to be important metabolic features in existing T1D-related studies, such as amino acids alanine (4) and methionine (5), fatty acids (49), vitamin E (45), sugar alcohols, and unsaturated TGs (5,50). Furthermore, we observed (Table 2) 5-methoxytryptamine at ages 6 months and 9 months contributing to the risk of IAA-first with association altered from positive to negative between 6 and 9 months, prior to and near the age of population-wide IAA-first incidence peak (10). 5-methoxytryptamine is a metabolite of melatonin and serotonin, which have been linked to diabetes and autoimmune disorders in previous studies (51,52).
The current study in TEDDY represents the largest prospective cohort analysis of metabolomes in children at increased genetic risk for T1D and identifies biomarkers for islet autoimmunity (stage I and II) that precedes the clinical onset of diabetes (stage III) (53). Similar observations were found in the Type 1 Diabetes Prediction and Prevention (DIPP) study (4) reporting that changes in GABA, glutamic acid, glutamine, α-ketoglutarate, leucine, and plasmenyl-PCs (ether PCs) were age dependent and could be associated with the onset of GADA and IAA. Based on the TEDDY longitudinal metabolome profiles, we not only confirmed the DIPP findings using average abundance of these compounds across time points but also separated the age and time-to-seroconversion effects (Fig. 4 and Supplementary Fig. 3). The trend of ether PC between 1 and 2 years of age in TEDDY subjects was similar to the change over time before GADA-first onset observed in DIPP, which was the result of overlapping effects of age and time to seroconversion. On the other hand, the decrease in GABA levels within 1 year before seroconversion observed in DIPP was a pattern determined by time to seroconversion instead of age. Furthermore, our results in metabolite enrichment analysis (Table 4) not only agreed with the reduced level of PC, TGs and plasmenyl- (or ether) phospholipids found in individuals who developed T1D (4) but also revealed lower PE and sugar alcohols during infancy associated with future onset of IA.
A limitation to the current study is the quarterly time sampling from 3 months of age and onward. The effect of age on metabolites and complex lipids (4,5) would have been better understood with a more frequent blood sampling, especially in relation to IAA as the first appearing autoantibody. IAA-first has been related to prior infectious episodes both in DIPP (54) and TEDDY (55), and at this early age, statistical analyses will have to take both age-related effects and environmental exposures into account to further delineate the mechanisms that trigger an autoimmune response against insulin.
Current analyses focused on metabolic markers for IA prior to seroconversion and included TEDDY participants who only developed IAA or GADA as the first-appearing autoantibody. It is worthwhile to extend future analyses to participants who experienced multiple autoantibodies either at seroconversion or throughout the follow-up, since the age at development of multiple autoantibodies has been found associated with the risk of progression to T1D (16). Genetics or environmental causes leading to metabolic signals (such as DHAA, GABA, and proline) identified in present analyses were still unknown and should be investigated further jointly with genome-wide SNP data, gut microbiome, and dietary patterns.
Conclusion
These results from metabolome-wide trajectory, independent time point, and enrichment analyses support the notion that the onset of IA as GADA-first or IAA-first in TEDDY children is heralded by distinct metabolic precursors in plasma after birth. The precursory signals for each autoantibody include DHAA; GABA; amino acids proline, alanine, and methionine; and compounds in BCAA metabolism as well as fatty acids. Unsaturated TGs and PEs at infant age were found to be decreased before appearance of either autoantibody. The distinct metabolic patterns for these autoantibodies support the idea that the causes of each type of initial autoimmunity may be different, and may account for the earlier incidence peak of IAA-first compared with that of GADA-first in TEDDY.
Supplementary Material
Article Information
Funding. The TEDDY study is funded by the National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Allergy and Infectious Diseases, National Institute of Child Health and Human Development, National Institute of Environmental Health Sciences, Centers for Disease Control and Prevention, and JDRF (grants U01 DK63829, U01 DK63861, U01 DK63821, U01 DK63865, U01 DK63863, U01 DK63836, U01 DK63790, UC4 DK63829, UC4 DK63861, UC4 DK63821, UC4 DK63865, UC4 DK63863, UC4 DK63836, UC4 DK95300, UC4 DK100238, UC4 DK106955, UC4 DK112243, and UC4 DK117483 and contract no. HHSN267200700014C). This work is supported in part by the National Institutes of Health/Clinical and Translational Science Awards to the University of Florida (UL1 TR000064) and the University of Colorado (UL1 TR001082).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. A.L., W.H., M.R., J.-X.S., A.-G.Z., B.A., O.F., and J.P.K conceived and designed the study. Q.L., A.L., and J.P.K. developed an analytical plan and interpreted the results. Q.L. drafted the manuscript. Q.L., H.P., M.D.B., and S.F. conducted analysis programming. Q.L. and S.F. developed statistical software. A.L., M.R., J.-X.S., J.T., and A.-G.Z. facilitated physical data collection from patients. Q.L., H.P., M.D.B., A.L., W.H., M.R., J.-X.S., J.T., A.-G.Z., B.A., O.F., S.F., and J.F.K. reviewed, edited, and approved the final draft of the manuscript. Q.L. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 12th annual RECOMB/ISCB Conference on Regulatory & Systems Genomics, 4–6 November 2019.
Appendix
The members of the TEDDY Study Group are listed below. The numbers listed correspond with the committees as follows: 1Ancillary Studies, 2Diet, 3Genetics, 4Human Subjects/Publicity/Publications, 5Immune Markers, 6Infectious Agents, 7Laboratory Implementation, 8Psychosocial, 9Quality Assurance, 10Steering, 11Study Coordinators, 12Celiac Disease, and 13Clinical Implementation.
Colorado Clinical Center. Marian Rewers, Principal Investigator (PI),1,4,5,6,9,10 Aaron Barbour, Kimberly Bautista,11 Judith Baxter,8,9,11 Daniel Felipe-Morales, Kimberly Driscoll,8 Brigitte I. Frohnert,2,13 Marisa Stahl,12 Patricia Gesualdo,2,6,11,13 Michelle Hoffman,11,12,13 Rachel Karban,11 Edwin Liu,12 Jill Norris,2,3,11 Stesha Peacock, Hanan Shorrosh, Andrea Steck,3,13 Megan Stern, Erica Villegas,2 and Kathleen Waugh6,7,11: Barbara Davis Center for Childhood Diabetes.
Finland Clinical Center. Jorma Toppari, PI,¥^1,4,10,13, Olli G. Simell, Annika Adamsson,^11 Suvi Ahonen,*±§ Mari Åkerlund,*±§ Leena Hakola,* Anne Hekkala,µ† Henna Holappa,µ† Heikki Hyöty,*±6 Anni Ikonen,µ† Jorma Ilonen,¥¶3 Sinikka Jäminki,*± Sanna Jokipuu,^ Leena Karlsson,^ Jukka Kero,¥^ Miia Kähönen,µ†11,13 Mikael Knip,*±5 Minna-Liisa Koivikko,µ† Merja Koskinen,*± Mirva Koreasalo,*±§2 Kalle Kurppa,*±12 Jarita Kytölä,*± Tiina Latva-aho,µ† Katri Lindfors,*12 Maria Lönnrot,*±6 Elina Mäntymäki,^ Markus Mattila,* Maija Miettinen,§2 Katja Multasuo,µ† Teija Mykkänen,µ† Tiina Niininen,±*11 Sari Niinistö,±§2 Mia Nyblom,*± Sami Oikarinen,*± Paula Ollikainen,µ† Zhian Othmani,^ Sirpa Pohjola,µ† Petra Rajala,^ Jenna Rautanen,±§ Anne Riikonen,*±§2 Eija Riski,^ Miia Pekkola,*± Minna Romo,^ Satu Ruohonen,^ Satu Simell,¥12 Maija Sjöberg,^ Aino Stenius,µ†11 Päivi Tossavainen,µ† Mari Vähä-Mäkilä,¥ Sini Vainionpää,^ Eeva Varjonen,^11 Riitta Veijola,µ†13 Irene Viinikangas,µ† and Suvi M. Virtanen*±§2: ¥University of Turku; *Tampere University; µUniversity of Oulu; ^Turku University Hospital; Hospital District of Southwest Finland; ±Tampere University Hospital; †Oulu University Hospital; §National Institute for Health and Welfare, Finland; and ¶University of Kuopio.
Georgia/Florida Clinical Center. Jin-Xiong She, PI,,1,3,4,10 Desmond Schatz,*4,5,7,8 Diane Hopkins,11 Leigh Steed,11,12,13 Jennifer Bryant,11 Katherine Silvis,2 Michael Haller,*13 Melissa Gardiner,11 Richard McIndoe, Ashok Sharma, Stephen W. Anderson,^ Laura Jacobsen,*13 John Marks,*11,13 and P.D. Towe*: Center for Biotechnology and Genomic Medicine, Augusta University; *University of Florida; and ^Pediatric Endocrine Associates, Atlanta.
Germany Clinical Center. Anette G. Ziegler, PI,1,3,4,10 Ezio Bonifacio,*5 Anita Gavrisan, Cigdem Gezginci, Anja Heublein, Verena Hoffmann,2 Sandra Hummel,2 Andrea Keimer,¥2 Annette Knopff,7 Charlotte Koch, Sibylle Koletzko,¶12 Claudia Ramminger,11 Roswith Roth,8 Marlon Scholz, Joanna Stock,8,11,13 Katharina Warncke,13 Lorena Wendel, and Christiane Winkler2,11 from: Forschergruppe Diabetes e.V. and Institute of Diabetes Research, Helmholtz Zentrum München, Forschergruppe Diabetes, and Klinikum rechts der Isar, Technische Universität München; *Center for Regenerative Therapies, TU Dresden; ¶Department of Gastroenterology, Dr. von Hauner Children’s Hospital, Ludwig Maximillians University Munich; and ¥Department of Nutritional Epidemiology, University of Bonn.
Sweden Clinical Center. Åke Lernmark, PI,1,3,4,5,6,8,9,10 Daniel Agardh,6,12 Carin Andrén Aronsson,2,11,12 Maria Ask, Rasmus Bennet, Corrado Cilio,5,6 Helene Engqvist, Emelie Ericson-Hallström, Annika Fors, Lina Fransson, Thomas Gard, Monika Hansen, Hanna Jisser, Fredrik Johansen, Berglind Jonsdottir,11 Silvija Jovic, Helena Elding Larsson,6,13 Marielle Lindström, Markus Lundgren,13 Marlena Maziarz, Maria Månsson-Martinez, Maria Markan, Jessica Melin,11 Zeliha Mestan, Caroline Nilsson, Karin Ottosson, Kobra Rahmati, Anita Ramelius, Falastin Salami, Anette Sjöberg, Birgitta Sjöberg, Malin Svensson, Carina Törn,3 Anne Wallin, Åsa Wimar13, and Sofie Åberg: Lund University.
Washington Clinical Center. William A. Hagopian, PI,1,3,4,5,6,7,10,12,13 Michael Killian,6,7,11,12 Claire Cowen Crouch,11,13 Jennifer Skidmore,2 Masumeh Chavoshi, Rachel Hervey, Rachel Lyons, Arlene Meyer, Denise Mulenga,11 Jared Radtke, Matei Romancik, Davey Schmitt, and Sarah Zink: Pacific Northwest Research Institute.
Pennsylvania Satellite Center. Dorothy Becker, Margaret Franciscus, MaryEllen Dalmagro-Elias Smith,2 Ashi Daftary, Mary Beth Klein, and Chrystal Yates: UPMC Children’s Hospital of Pittsburgh.
Data Coordinating Center. Jeffrey P. Krischer, PI, 1,4,5,9,10 Sarah Austin-Gonzalez, Maryouri Avendano, Sandra Baethke, Rasheedah Brown,11 Brant Burkhardt,5,6 Martha Butterworth,2 Joanna Clasen, David Cuthbertson, Stephen Dankyi, Christopher Eberhard, Steven Fiske,8 Jennifer Garmeson, Veena Gowda, Kathleen Heyman, Belinda Hsiao, Christina Karges, Francisco Perez Laras, Hye-Seung Lee,1,2,3,12 Qian Li,5,12 Shu Liu, Xiang Liu,2,3,8,13 Kristian Lynch,5,6,8 Colleen Maguire, Jamie Malloy, Cristina McCarthy,11 Aubrie Merrell, Hemang Parikh,3 Ryan Quigley, Cassandra Remedios, Chris Shaffer, Laura Smith,8,11 Susan Smith,11 Noah Sulman, Roy Tamura,1,2,11,12,13 Dena Tewey, Michael Toth, Ulla Uusitalo,2 Kendra Vehik,4,5,6,8,13 Ponni Vijayakandipan, Keith Wood, and Jimin Yang2; past staff, Michael Abbondondolo, Lori Ballard, David Hadley, Wendy McLeod, and Steven Meulemans: University of South Florida.
Project Scientist. Beena Akolkar1,3,4,5,6,7,9,10: National Institutes of Diabetes and Digestive and Kidney Diseases.
Other Contributors. Kasia Bourcier5: National Institutes of Allergy and Infectious Diseases. Thomas Briese6: Columbia University. Suzanne Bennett Johnson8,11: Florida State University. Eric Triplett6: University of Florida.
Autoantibody Reference Laboratories. Liping Yu,^5 Dongmei Miao,^ Polly Bingley,*5 Alistair Williams,* Kyla Chandler,* Olivia Ball,* Ilana Kelland,* and Sian Grace*: ^Barbara Davis Center for Childhood Diabetes and *Bristol Medical School, University of Bristol, U.K.
HLA Reference Laboratory. William Hagopian,3 Masumeh Chavoshi, Jared Radtke, and Sarah Zink: Pacific Northwest Research Institute, Seattle, WA (previously Henry Erlich,3 Steven J. Mack, and Anna Lisa Fear: Center for Genetics, Children’s Hospital Oakland Research Institute).
Metabolomics Laboratory. Oliver Fiehn, Bill Wikoff, Brian Defelice, Dmitry Grapov, Tobias Kind, Mine Palazoglu, Luis Valdiviez, Benjamin Wancewicz, Gert Wohlgemuth, and Joyce Wong: West Coast Metabolomics Center.
SNP Laboratory. Stephen S. Rich,3 Wei-Min Chen,3 Suna Onengut-Gumuscu,3 Emily Farber, Rebecca Roche Pickin, Jonathan Davis, Jordan Davis, Dan Gallo, Jessica Bonnie, and Paul Campolieto: Center for Public Health Genomics, University of Virginia.
Footnotes
Members of the TEDDY Study Group are listed in the appendix.
This article contains Supplementary Data online at https://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-0756/-/DC1.
Contributor Information
Collaborators: TEDDY Study Group, Marian Rewers, Aaron Barbour, Kimberly Bautista, Judith Baxter, Daniel Felipe-Morales, Kimberly Driscoll, Brigitte I. Frohnert, Marisa Stahl, Patricia Gesualdo, Michelle Hoffman, Rachel Karban, Edwin Liu, Jill Norris, Stesha Peacock, Hanan Shorrosh, Andrea Steck, Megan Stern, Erica Villegas, Kathleen Waugh, Jorma Toppari, Olli G. Simell, Annika Adamsson, Suvi Ahonen, Mari Åkerlund, Leena Hakola, Anne Hekkala, Henna Holappa, Heikki Hyöty, Anni Ikonen, Jorma Ilonen, Sinikka Jäminki, Sanna Jokipuu, Leena Karlsson, Jukka Kero, Miia Kähönen, Mikael Knip, Minna-Liisa Koivikko, Merja Koskinen, Mirva Koreasalo, Kalle Kurppa, Jarita Kytölä, Tiina Latva-aho, Katri Lindfors, Maria Lönnrot, Elina Mäntymäki, Markus Mattila, Maija Miettinen, Katja Multasuo, Teija Mykkänen, Tiina Niininen, Sari Niinistö, Mia Nyblom, Sami Oikarinen, Paula Ollikainen, Zhian Othmani, Sirpa Pohjola, Petra Rajala, Jenna Rautanen, Anne Riikonen, Eija Riski, Miia Pekkola, Minna Romo, Satu Ruohonen, Satu Simell, Maija Sjöberg, Aino Stenius, Päivi Tossavainen, Mari Vähä-Mäkilä, Sini Vainionpää, Eeva Varjonen, Riitta Veijola, Irene Viinikangas, Suvi M. Virtanen, Jin-Xiong She, Desmond Schatz, Diane Hopkins, Leigh Steed, Jennifer Bryant, Katherine Silvis, Michael Haller, Melissa Gardiner, Richard McIndoe, Ashok Sharma, Stephen W. Anderson, Laura Jacobsen, John Marks, P.D. Towe, Anette G. Ziegler, Ezio Bonifacio, Anita Gavrisan, Cigdem Gezginci, Anja Heublein, Verena Hoffmann, Sandra Hummel, Andrea Keimer, Annette Knopff, Charlotte Koch, Sibylle Koletzko, Claudia Ramminger, Roswith Roth, Marlon Scholz, Joanna Stock, Katharina Warncke, Lorena Wendel, Christiane Winkler, Åke Lernmark, Daniel Agardh, Carin Andrén Aronsson, Maria Ask, Rasmus Bennet, Corrado Cilio, Helene Engqvist, Emelie Ericson-Hallström, Annika Fors, Lina Fransson, Thomas Gard, Monika Hansen, Hanna Jisser, Fredrik Johansen, Berglind Jonsdottir, Silvija Jovic, Helena Elding Larsson, Marielle Lindström, Markus Lundgren, Marlena Maziarz, Maria Månsson-Martinez, Maria Markan, Jessica Melin, Zeliha Mestan, Caroline Nilsson, Karin Ottosson, Kobra Rahmati, Anita Ramelius, Falastin Salami, Anette Sjöberg, Birgitta Sjöberg, Malin Svensson, Carina Törn, Anne Wallin, Åsa Wimar, Sofie Åberg, William A. Hagopian, Michael Killian, Claire Cowen Crouch, Jennifer Skidmore, Masumeh Chavoshi, Rachel Hervey, Rachel Lyons, Arlene Meyer, Denise Mulenga, Jared Radtke, Matei Romancik, Davey Schmitt, Sarah Zink, Dorothy Becker, Margaret Franciscus, MaryEllen Dalmagro-Elias Smith, Ashi Daftary, Mary Beth Klein, Chrystal Yates, Jeffrey P. Krischer, Sarah Austin-Gonzalez, Maryouri Avendano, Sandra Baethke, Rasheedah Brown, Brant Burkhardt, Martha Butterworth, Joanna Clasen, David Cuthbertson, Stephen Dankyi, Christopher Eberhard, Steven Fiske, Jennifer Garmeson, Veena Gowda, Kathleen Heyman, Belinda Hsiao, Christina Karges, Francisco Perez Laras, Hye-Seung Lee, Qian Li, Shu Liu, Xiang Liu, Kristian Lynch, Colleen Maguire, Jamie Malloy, Cristina McCarthy, Aubrie Merrell, Hemang Parikh, Ryan Quigley, Cassandra Remedios, Chris Shaffer, Laura Smith, Susan Smith, Noah Sulman, Roy Tamura, Dena Tewey, Michael Toth, Ulla Uusitalo, Kendra Vehik, Ponni Vijayakandipan, Keith Wood, Jimin Yang, Michael Abbondondolo, Lori Ballard, David Hadley, Wendy McLeod, Steven Meulemans, Beena Akolkar, Kasia Bourcier, Thomas Briese, Suzanne Bennett Johnson, Eric Triplett, Liping Yu Dongmei Miao, Polly Bingley, Alistair Williams, Kyla Chandler, Olivia Ball, Ilana Kelland, Sian Grace, William Hagopian, Masumeh Chavoshi, Jared Radtke, Sarah Zink, Henry Erlich, Steven J. Mack, Anna Lisa Fear, Oliver Fiehn, Bill Wikoff, Brian Defelice, Dmitry Grapov, Tobias Kind, Mine Palazoglu, Luis Valdiviez, Benjamin Wancewicz, Gert Wohlgemuth, Joyce Wong, Stephen S. Rich, Wei-Min Chen, Suna Onengut-Gumuscu, Emily Farber, Rebecca Roche Pickin, Jonathan Davis, Jordan Davis, Dan Gallo, Jessica Bonnie, and Paul Campolieto
References
- 1.Frohnert BI, Rewers MJ. Metabolomics in childhood diabetes. Pediatr Diabetes 2016;17:3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.La Torre D, Seppänen-Laakso T, Larsson HE, et al.; DiPiS Study Group . Decreased cord-blood phospholipids in young age-at-onset type 1 diabetes. Diabetes 2013;62:3951–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jørgenrud B, Stene LC, Tapia G, et al. . Longitudinal plasma metabolic profiles, infant feeding, and islet autoimmunity in the MIDIA study. Pediatr Diabetes 2017;18:111–119 [DOI] [PubMed] [Google Scholar]
- 4.Orešič M, Simell S, Sysi-Aho M, et al. . Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med 2008;205:2975–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pflueger M, Seppänen-Laakso T, Suortti T, et al. . Age- and islet autoimmunity-associated differences in amino acid and lipid metabolites in children at risk for type 1 diabetes. Diabetes 2011;60:2740–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Overgaard AJ, Kaur S, Pociot F. Metabolomic biomarkers in the progression to type 1 diabetes. Curr Diab Rep 2016;16:127. [DOI] [PubMed] [Google Scholar]
- 7.Vehik K, Fiske SW, Logan CA, et al.; TEDDY Study Group . Methods, quality control and specimen management in an international multicentre investigation of type 1 diabetes: TEDDY. Diabetes Metab Res Rev 2013;29:557–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagopian WA, Erlich H, Lernmark A, et al.; TEDDY Study Group . The Environmental Determinants of Diabetes in the Young (TEDDY): genetic criteria and international diabetes risk screening of 421 000 infants. Pediatr Diabetes 2011;12:733–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H-S, Burkhardt BR, McLeod W, et al.; TEDDY Study Group . Biomarker discovery study design for type 1 diabetes in The Environmental Determinants of Diabetes in the Young (TEDDY) study. Diabetes Metab Res Rev 2014;30:424–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krischer JP, Lynch KF, Schatz DA, et al.; TEDDY Study Group . The 6 year incidence of diabetes-associated autoantibodies in genetically at-risk children: the TEDDY study. Diabetologia 2015;58:980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uusitalo U, Liu X, Yang J, et al.; TEDDY Study Group . Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatr 2016;170:20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brewerton TD, Berrettini WH, Nurnberger JI Jr, Linnoila M. Analysis of seasonal fluctuations of CSF monoamine metabolites and neuropeptides in normal controls: findings with 5HIAA and HVA. Psychiatry Res 1988;23:257–265 [DOI] [PubMed] [Google Scholar]
- 13.Woitge HW, Knothe A, Witte K, et al. . Circaannual rhythms and interactions of vitamin D metabolites, parathyroid hormone, and biochemical markers of skeletal homeostasis: a prospective study. J Bone Miner Res 2000;15:2443–2450 [DOI] [PubMed] [Google Scholar]
- 14.Bradman A, Quirós-Alcalá L, Castorina R, et al. . Effect of organic diet intervention on pesticide exposures in young children living in low-income urban and agricultural communities. Environ Health Perspect 2015;123:1086–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorburn AN, McKenzie CI, Shen S, et al. . Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun 2015;6:7320. [DOI] [PubMed] [Google Scholar]
- 16.Krischer JP, Lynch KF, Lernmark Å, et al.; TEDDY Study Group . Genetic and environmental interactions modify the risk of diabetes-related autoimmunity by 6 years of age: the TEDDY study. Diabetes Care 2017;40:1194–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krischer JP, Liu X, Lernmark Å, et al.; TEDDY Study Group . The influence of type 1 diabetes genetic susceptibility regions, age, sex, and family history on the progression from multiple autoantibodies to type 1 diabetes: a TEDDY study report. Diabetes 2017;66:3122–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.TEDDY Study Group The Environmental Determinants of Diabetes in the Young (TEDDY) study: study design. Pediatr Diabetes 2007;8:286–298 [DOI] [PubMed] [Google Scholar]
- 19.Stewart CJ, Ajami NJ, O’Brien JL, et al. . Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018;562:583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vatanen T, Franzosa EA, Schwager R, et al. . The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018;562:589–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skogerson K, Wohlgemuth G, Barupal DK, Fiehn O. The volatile compound BinBase mass spectral database. BMC Bioinformatics 2011;12:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kind T, Liu K-H, Lee DY, DeFelice B, Meissen JK, Fiehn O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat Methods 2013;10:755–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnsen LG, Skou PB, Khakimov B, Bro R. Gas chromatography - mass spectrometry data processing made easy. J Chromatogr A 2017;1503:57–64 [DOI] [PubMed] [Google Scholar]
- 24.Croley TR, White KD, Callahan JH, Musser SM. The chromatographic role in high resolution mass spectrometry for non-targeted analysis. J Am Soc Mass Spectrom 2012;23:1569–1578 [DOI] [PubMed] [Google Scholar]
- 25.Fan S, Kind T, Cajka T, et al. . Systematic error removal using random forest for normalizing large-scale untargeted lipidomics data. Anal Chem 2019;91:3590–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Fisher K, Meng W, et al. . GMSimpute: a generalized two-step Lasso approach to impute missing values in label-free mass spectrum analysis. Bioinformatics 2020;36:257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc Natl Acad Sci U S A 2005;102:12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw 2015;67:1–48 [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300 [Google Scholar]
- 31.Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Series B Stat Methodol 2005;67:301–320 [Google Scholar]
- 32.Marquardt DW, Snee RD. Ridge regression in practice. Am Stat 1975;29:3–20 [Google Scholar]
- 33.Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc B 1996;58:267–288 [Google Scholar]
- 34.Barupal DK, Fiehn O. Chemical Similarity Enrichment Analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci Rep 2017;7:14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wohlgemuth G, Haldiya PK, Willighagen E, Kind T, Fiehn O. The Chemical Translation Service--a web-based tool to improve standardization of metabolomic reports. Bioinformatics 2010;26:2647–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorgani L, Mohammadi M, Najafpour GD, Nikzad M. Piperine—the bioactive compound of black pepper: from isolation to nedicinal formulations. Compr Rev Food Sci Food Saf 2017;16:124–140 [DOI] [PubMed] [Google Scholar]
- 37.Bylesjö M, Rantalainen M, Cloarec O, Nicholson JK, Holmes E, Trygg J. OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J Chemom 2006;20:341–351 [Google Scholar]
- 38.Kaneto A, Kosaka K. Effects of leucine and isoleucine infused intrapancreatically on glucagon and insulin secretion. Endocrinology 1972;91:691–695 [DOI] [PubMed] [Google Scholar]
- 39.Yang J, Chi Y, Burkhardt BR, Guan Y, Wolf BA. Leucine metabolism in regulation of insulin secretion from pancreatic beta cells. Nutr Rev 2010;68:270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tapiero H, Mathé G, Couvreur P, Tew KD. II. Glutamine and glutamate. Biomed Pharmacother 2002;56:446–457 [DOI] [PubMed] [Google Scholar]
- 41.Liu W, Hancock CN, Fischer JW, Harman M, Phang JM. Proline biosynthesis augments tumor cell growth and aerobic glycolysis: involvement of pyridine nucleotides. Sci Rep 2015;5:17206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yelamanchi SD, Jayaram S, Thomas JK, et al. . A pathway map of glutamate metabolism. J Cell Commun Signal 2016;10:69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bansal P, Wang S, Liu S, Xiang Y-Y, Lu W-Y, Wang Q. GABA coordinates with insulin in regulating secretory function in pancreatic INS-1 β-cells. PLoS One 2011;6:e26225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ben-Othman N, Vieira A, Courtney M, et al. . Long-term GABA administration induces alpha cell-mediated beta-like cell neogenesis. Cell 2017;168:73–85.e11 [DOI] [PubMed] [Google Scholar]
- 45.Behrens WA, Madere R. Vitamin C and vitamin E status in the spontaneously diabetic BB rat before the onset of diabetes. Metabolism 1991;40:72–76 [DOI] [PubMed] [Google Scholar]
- 46.Senmaru T, Yamazaki M, Okada H, et al. . Pancreatic insulin release in vitamin C-deficient senescence marker protein-30/gluconolactonase knockout mice. J Clin Biochem Nutr 2012;50:114–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergsten P, Moura AS, Atwater I, Levine M. Ascorbic acid and insulin secretion in pancreatic islets. J Biol Chem 1994;269:1041–1045 [PubMed] [Google Scholar]
- 48.Pence LA, Mennear JH. Inhibition effect of dehydroascorbic acid on insulin secretion from mouse pancreatic islets. Toxicol Appl Pharmacol 1979;50:57–65 [DOI] [PubMed] [Google Scholar]
- 49.Peterson LR, Herrero P, McGill J, et al. . Fatty acids and insulin modulate myocardial substrate metabolism in humans with type 1 diabetes. Diabetes 2008;57:32–40 [DOI] [PubMed] [Google Scholar]
- 50.Lamichhane S, Ahonen L, Dyrlund TS, et al. . Dynamics of plasma lipidome in progression to islet autoimmunity and type 1 diabetes - Type 1 Diabetes Prediction and Prevention Study (DIPP). Sci Rep 2018;8:10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hardeland R. Melatonin and inflammation-story of a double-edged blade. J Pineal Res 2018;65:e12525. [DOI] [PubMed] [Google Scholar]
- 52.Lin G-J, Huang S-H, Chen S-J, Wang C-H, Chang D-M, Sytwu H-K. Modulation by melatonin of the pathogenesis of inflammatory autoimmune diseases. Int J Mol Sci 2013;14:11742–11766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Insel RA, Dunne JL, Atkinson MA, et al. . Staging presymptomatic type 1 diabetes: a scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care 2015;38:1964–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sioofy-Khojine A-B, Lehtonen J, Nurminen N, et al. . Coxsackievirus B1 infections are associated with the initiation of insulin-driven autoimmunity that progresses to type 1 diabetes. Diabetologia 2018;61:1193–1202 [DOI] [PubMed] [Google Scholar]
- 55.Lönnrot M, Lynch KF, Elding Larsson H, et al.; TEDDY Study Group . Respiratory infections are temporally associated with initiation of type 1 diabetes autoimmunity: the TEDDY study. Diabetologia 2017;60:1931–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.