Brain injury, especially that caused by stroke, is a leading cause of morbidity and mortality worldwide. Numerous studies on the neuroprotective effects of neural stem cells (NSCs) after brain injury have shown that crosstalk among neural cells, including neuron–glial, glial–glial, NSC–neuronal and NSC–glial communication, plays a major role in post-injury repair. Therefore, a better understanding of the factors involved in the interactions between NSCs and other neural cells should advance the development of therapeutic strategies for brain injury. In this article, we discuss the role of exosomes in the crosstalk between NSCs and microglia, focusing on key exosomal proteins and microRNAs in brain injury progression and repair.
Exosomes in the central nervous system: Exosomes, vesicles that originate from the endocytic pathway, have a diameter of 30–150 nm and participate in inflammatory responses and inflammation-related neuroprotection after brain injury, and their functions are dependent on their origin and contents. Exosomes are involved in innate immune cell crosstalk, microglial training, and the regulation of neuroinflammation. From the extracellular milieu, exosomes may contact target cells by receptor-mediated adhesion to the cellular plasma membrane, followed by endocytic uptake and internalization, or direct fusion of the exosome membrane with the target cell membrane, ultimately releasing their contents into the recipient cell. While numerous studies have focused on astrocyte and neuron-derived exosomes (especially for astrocyte-derived exosomes, given glia are the most abundant cell type in brain, they have received sufficient attention), studies on microglial or NSC-derived exosomes are lacking.
The role of NSC-microglial crosstalk after brain injury: Microglia play a major role in immune responses by functioning as antigen presenting cells. Furthermore, they regulate synaptic pruning and neurogenesis under both physiological and brain injury-related pathophysiological conditions. Microvesicles derived from microglia can stimulate neuronal activity and participate in the propagation of inflammatory signals. To date, numerous studies have shown that microglia and NSCs regulate each other through various signaling pathways.
Impact of microglia-derived exosomes (MDEs) on NSCs: There are a number of protein/RNA markers of MDEs. Microglia and astrocytes release exosomes enriched in inflammatory factors or microRNAs (miRNAs) associated with neurotrophic signals. MDE protein markers can be divided into common exosome proteins and MDE-specific proteins. Common exosome markers include Syntaxin 8, Syntaxin 6, Syntaxin 11, Rab 7, Rab 11, Lamp-1, Lamp-2, CD9, CD81 and CD63. In particular, it has been repeatedly suggested that exosomes can be distinguished from other vesicles by ubiquitous expression of the tetraspanins CD9, CD81 and CD63. Additionally, exosomes may possess cell-specific receptors which determine their target cells. MDE-specific markers include CD13, monocarboxylate transporter-1, major histocompatibility complex class II molecules, membrane tumor necrosis factor-α, and interleukin-1β. Some studies have revealed a role of MDE markers in inflammation following traumatic brain injury, including interleukin-1β and miR-155 (Kumar et al., 2017). Recently, a landmark study showed that microglia can release tau via exosome secretion during the progression of tauopathy (Asai et al., 2015). P2X7 receptors, which have been found on MDEs, could mediate cytokine release from these vesicles. Moreover, in a recent study, MDE treatment of a 3D spheroid glioma culture inhibited tumor invasion (Murgoci et al., 2018). Because of the numerous similarities between NSCs and gliomas, it is conceivable that MDEs may participate in inflammatory injury towards NSCs.
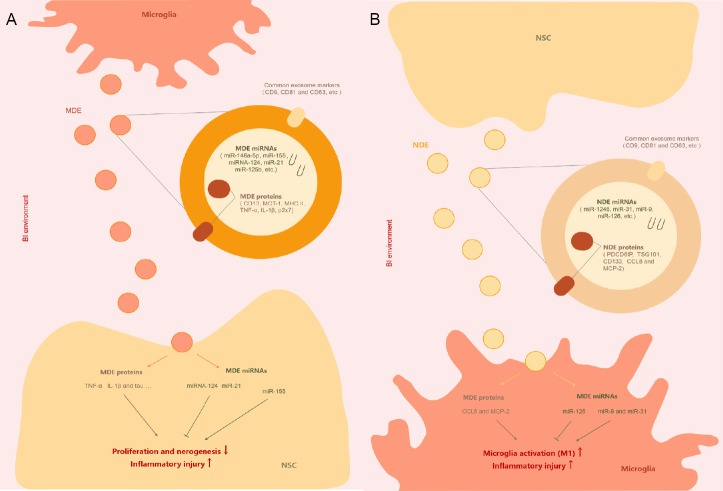
Mature microglial miRNAs are another major MDE cargo that can be released into the extracellular space or into target cells by these vesicles. MDE-specific miRNAs include miR-146a-5p, miR-155, miR-124, miR-21 and miR-125b. In particular, miR-146a-5p, a novel microglial-specific miRNA that is only very weakly expressed in hippocampal neurons, regulates the expression of presynaptic synaptotagmin 1 and postsynaptic neuroligin 1. Therefore, miR-146a-5p may participate in inflammation-induced synaptic changes via microglial–neuronal crosstalk. MiR-155 was recently shown to be involved in microglial activation. After exposure to lipopolysaccharide or interferon-γ, microglial miR-155 expression is significantly increased, possibly inhibiting the suppressor of cytokine signaling 1 pathway and the receptor for activated C kinase 1 pathway, and enhancing M1 polarization (Butovsky et al., 2015). Exosomes from N9 microglia show downregulation of miR-124 at the early stage, but upregulation at the late stage. A recent microarray analysis study showed that miR-124-3p promotes M2 polarization in microglia and inhibits neuronal inflammation in scratch-injured neurons (Huang et al., 2018). MiR-21, a biomarker of various types of microglia and MDEs, has been considered a promising target for therapeutic intervention (Fernandes et al., 2018). MiR-21-3p regulates the EGFR and FGFR signaling pathways in neural development, and may be involved in MDE-mediated neurotoxicity to NSCs in brain injury (Wu et al., 2016). The effects of MDEs on NSCs is summarized in Figure 1A. MDE-derived proteins and miRNAs can both impact NSC proliferation and differentiation in brain injury.
Figure 1.
The impact of MDEs on NSCs (A) and the impact of NDEs on microglia (B) in the brain injury (BI) environment.
CCL8: Chemokine (C-C motif) ligand 8; IL-1β: interleukin-1β; MDEs: microglia-derived exosomes; MCP-2: monocyte chemoattractant protein-2; MCT-1: monocarboxylate transporter-1; MHC: major histocompatibility complex; miRNA: microRNA; NDEs: NSC-derived exosomes; NSCs: neural stem cells; TNF-α: tumor necrosis factor-α.
Impact of NSC-derived exosomes (NDEs) on microglia: Exosomes produced by stem cells are involved in multiple processes. Exosomes can exert beneficial effects during inflammatory injury to enhance tissue repair. The therapeutic effects of NDEs have been investigated in experimental necrotizing enterocolitis and neurodegenerative disorders. They promote neuroprotection in murine and porcine models of stroke and control the process of aging in the hypothalamus. Therefore, NDEs are highly likely to mediate signals from NSCs to microglia. A recent study reported that NDEs selectively target microglia and function as a microglial morphogen (Morton et al., 2018). Additionally, NDEs activate a microglial transcriptional network, resulting in an intercellular feedback loop. NDEs have been clearly shown to modulate microglial activation during brain injury.
NDEs can be characterized by their protein and miRNA markers. In addition to common exosome markers (such as CD63, CD9 and TSG101), NDEs specifically contain PDCD6IP (Alix). Notably, the primary cilium of the neuroepi-thelial stem cell also contains the NSC-specific marker CD133. Furthermore, proinflammatory cytokine signaling in NSCs stimulates the interferon-γ/Stat1 pathway, which results in the export of specific components (such as monocyte chemoattractant protein-2 and chemokine ligand 8) by NDEs. In microglia, chemokine ligand 8 and monocyte chemoattractant protein-2 are targets of miR-146a during human immunodeficiency virus-1 infection, and they play a role in microglial activation.
A recent next generation sequencing analysis study identified a unique set of miRNAs in NDEs, and among these, miR-1246 was the most abundant, followed by miR-4488 and miR-4508 (Stevanato et al., 2016). The overwhelming majority of these miRNAs had no clear relationship to microglia, although several may impact microglia via other targets. The second top-secreted miRNA, miR-31, is enriched in CD11c+ microglia and is associated with microglial activation. Another enriched miRNA, miR-9, can also promote microglial activation by targeting monocyte chemotactic protein-1-induced protein-1. In addition, NDEs enriched in the miR-17-92 cluster enhance neuroplasticity and functional recovery after stroke (Xin et al., 2017). A recent study using an oxygen-glucose deprivation model reported a set of differential NDE-derived miRNAs, including miR-203a, miR-126 and miR-3195 (Zhang et al., 2018). These differential miRNAs may help in the development of novel strategies for the diagnosis and treatment of brain injury. In particular, miR-126-containing exosomes are known to inhibit microglial activation and the expression of inflammatory factors in vivo and in vitro. Thus, NDEs may have potential in microglial modulation, and it might be advantageous to investigate exosomes from other types of stem cells in the future. Indeed, exosomes derived from adipose-derived stem cells significantly attenuate cerebral infarction by suppressing autophagy and promoting M2 microglial/macrophage polarization. During brain injury, NDEs secreted by NSCs may exert various complex (bidirectional) effects on microglia (Figure 1B).
Conclusions and perspectives: Exosomes are key intercellular communication devices. MDEs and NDEs mediate inflammatory injury, but also exert neuroprotective effects. In general, it seems that the crosstalk is more related to proliferation than differentiation and to inflammatory injury than neuroprotection. Numerous critical neuroprotective and differentiation factors in MDEs and NDEs likely remain to be identified. Furthermore, the mechanisms by which exosomal proteins and miRNAs modulate the activation of microglia and the proliferation/differentiation of NSCs remain to be clarified. Therefore, additional studies are needed for the application of exosomes in brain injury prevention and treatment.
This study was supported by Lanzhou Science and Technology Bureau Project, Lanzhou, Gansu Privince, China, No. 2018-4-68 (to BRH) and Cuiying Scientific and Technological Innovation Program of Lanzhou University Second Hospital, China, No. CY2018-MS08 (to BRH).
Footnotes
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Patel B, Yu J, Song LP; T-Editor: Jia Y
References
- 1.Asai H, Ikezu S, Tsunoda S, Medalla M, Luebke J, Haydar T, Wolozin B, Butovsky O, Kugler S, Ikezu T. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat Neurosci. 2015;18:1584–1593. doi: 10.1038/nn.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butovsky O, Jedrychowski MP, Cialic R, Krasemann S, Murugaiyan G, Fanek Z, Greco DJ, Wu PM, Doykan CE, Kiner O, Lawson RJ, Frosch MP, Pochet N, Fatimy RE, Krichevsky AM, Gygi SP, Lassmann H, Berry J, Cudkowicz ME, Weiner HL. Targeting miR-155 restores abnormal microglia and attenuates disease in SOD1 mice. Ann Neurol. 2015;77:75–99. doi: 10.1002/ana.24304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandes A, Ribeiro AR, Monteiro M, Garcia G, Vaz AR, Brites D. Secretome from SH-SY5Y APPSwe cells trigger time-dependent CHME3 microglia activation phenotypes, ultimately leading to miR-21 exosome shuttling. Biochimie. 2018;155:67–82. doi: 10.1016/j.biochi.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Huang S, Ge X, Yu J, Han Z, Yin Z, Li Y, Chen F, Wang H, Zhang J, Lei P. Increased miR-124-3p in microglial exosomes following traumatic brain injury inhibits neuronal inflammation and contributes to neurite outgrowth via their transfer into neurons. FASEB J. 2018;32:512–528. doi: 10.1096/fj.201700673R. [DOI] [PubMed] [Google Scholar]
- 5.Kumar A, Stoica BA, Loane DJ, Yang M, Abulwerdi G, Khan N, Kumar A, Thom SR, Faden AI. Microglial-derived microparticles mediate neuroinflammation after traumatic brain injury. J Neuroinflammation. 2017;14:47. doi: 10.1186/s12974-017-0819-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morton MC, Neckles VN, Seluzicki CM, Holmberg JC, Feliciano DM. Neonatal subventricular zone neural stem cells release extracellular vesicles that act as a microglial morphogen. Cell Rep. 2018;23:78–89. doi: 10.1016/j.celrep.2018.03.037. [DOI] [PubMed] [Google Scholar]
- 7.Murgoci AN, Cizkova D, Majerova P, Petrovova E, Medvecky L, Fournier I, Salzet M. Brain-cortex microglia-derived exosomes: nanoparticles for glioma therapy. Chemphyschem. 2018;19:1205–1214. doi: 10.1002/cphc.201701198. [DOI] [PubMed] [Google Scholar]
- 8.Stevanato L, Thanabalasundaram L, Vysokov N, Sinden JD. Investigation of content, stoichiometry and transfer of miRNA from human neural stem cell line derived exosomes. PLoS One. 2016;11:e0146353. doi: 10.1371/journal.pone.0146353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu YE, Parikshak NN, Belgard TG, Geschwind DH. Genome-wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder. Nat Neurosci. 2016;19:1463–1476. doi: 10.1038/nn.4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xin H, Katakowski M, Wang F, Qian JY, Liu XS, Ali MM, Buller B, Zhang ZG, Chopp M. MicroRNA cluster miR-17-92 cluster in exosomes enhance neuroplasticity and functional recovery after stroke in rats. Stroke. 2017;48:747–753. doi: 10.1161/STROKEAHA.116.015204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang G, Chen L, Guo X, Wang H, Chen W, Wu G, Gu B, Miao W, Kong J, Jin X, Yi G, You Y, Su X, Gu N. Comparative analysis of microRNA expression profiles of exosomes derived from normal and hypoxic preconditioning human neural stem cells by next generation sequencing. J Biomed Nanotechnol. 2018;14:1075–1089. doi: 10.1166/jbn.2018.2567. [DOI] [PubMed] [Google Scholar]