Keywords: bioinformatics, biomarker, inflammation, microRNA, mitochondria, mouse, pathway analysis, proteomics, spinal cord injury; stathmin
Abstract
Our previous study found that microRNA-21a-5p (miR-21a-5p) knockdown could improve the recovery of motor function after spinal cord injury in a mouse model, but the precise molecular mechanism remains poorly understood. In this study, a modified Allen’s weight drop was used to establish a mouse model of spinal cord injury. A proteomics approach was used to understand the role of differential protein expression with miR-21a-5p knockdown, using a mouse model of spinal cord injury without gene knockout as a negative control group. We found that after introducing miR-21a-5p knockdown, proteins that played an essential role in the regulation of inflammatory processes, cell protection against oxidative stress, cell redox homeostasis, and cell maintenance were upregulated compared with the negative control group. Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis identified enriched pathways in both groups, such as the oxidative phosphorylation pathway, which is relevant to Parkinson’s disease, Huntington’s disease, Alzheimer’s disease, and cardiac muscle contraction. We also found that miR-21a-5p could be a potential biomarker for amyotrophic lateral sclerosis, as miR-21a-5p becomes deregulated in this pathway. These results indicate successful detection of some important proteins that play potential roles in spinal cord injury. Elucidating the relationship between these proteins and the recovery of spinal cord injury will provide a reference for future research of spinal cord injury biomarkers. All experimental procedures and protocols were approved by the Experimental Animal Ethics Committee of Shandong University of China on March 5, 2014.
Chinese Library Classification No. R446; R447; R363
Introduction
Spinal cord injury (SCI) involves serious damage to the central nervous system and is dysfunctional in nature (Simon et al., 2019; Yao et al., 2019). The latest research shows that approximately 2.5 million people have varying degrees of SCI, and each year there are more than 130,000 new cases (Thuret et al., 2006). SCI mainly results from traffic trauma, fall injury, work accidents, violence, or sports injuries. To recover from incomplete damage, some sensory neurons can undergo a variety of physical rehabilitative treatments to restore function (Waters et al., 1991; Crozier et al., 1992). SCI prevention, treatment, and rehabilitation of injuries have become a major medical concern. Approximately 20% of patients with spinal fractures have varying degrees of SCI. Currently, treatment of SCI mainly includes drugs, surgery, and immobilization. Treatment methods are chosen according to the severity of the illness, length of the injury, effectiveness of the treatment, and the pathological stage of the specific case. At present, there is no operative treatment to recover lost neurological function after SCI (Cristante et al., 2012; Wei et al., 2019).
MicroRNAs (miRNAs), a class of endogenous small RNAs approximately 20–24 nucleotides in length, have a variety of important regulatory roles in cells. Each miRNA can have multiple target genes, and several miRNAs can regulate the same gene. This complex regulatory network can regulate the expression of multiple genes through a single miRNA, or can finely regulate the expression of a gene through a combination of several miRNAs. It is speculated that miRNAs regulate one-third of human genes. Recent studies have shown that approximately 70% of mammalian miRNAs are located in the regions of transcriptional units (transcription micro RNA; Rodriguez et al., 2004), and most are located within introns (Kim and Nam, 2006). The location of some intronic miRNAs is highly conserved among different species. MiRNAs are not only conserved at gene positions, but also exhibit high sequence homology (Grishok et al., 2001; Lee and Ambros, 2001). MicroRNA-21 (miR-21) not only plays an important role in fibrotic lung diseases, but is also part of an innovative miRNA-based therapeutic method to treat clinically refractory fibrotic diseases, such as idiopathic pulmonary fibrosis. According to our previous research, miR-21 knockdown pointedly upgraded the recovery of motor function after SCI and inhibited neural regeneration, which was suppressed in the miR-21 knockdown group (Wang et al., 2018b). In this study, we used a proteomics approach to understand the status of different proteins in negative control (NC) and miR-21a-5p-knockdown (KD) groups, as well as their interactions and roles after SCI in a mouse model.
Materials and Methods
Animals and SCI models
Eighteen C57BL/6 adult male mice (aged 9–10 weeks) were provided by the Animal Center of Shandong University, China. All mice were maintained in individual cages under controlled conditions of 22–24°C and relative humidity of 40–60%, with a 12-hour light/dark cycle and free access to food and water. All experimental procedures and protocols were approved by the Experimental Animal Ethics Committee of Shandong University of China on March 5, 2014. The experimental procedure followed the United States National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85-23, revised 1996).
Mice were intraperitoneally anesthetized with 10% chloral hydrate (3 mg/kg; Merck, Rahway, NJ, USA). Laminectomy was performed at T8–10 of the thoracic vertebra without any SCI. A moderate collision injury was caused by a modified Allen’s weight drop apparatus (8-g weight at 50 mm, 8 g × 50 mm) knocking on the exposed spinal cord (Liu et al., 2018). The sham group was only subjected to laminectomy without the collision injury.
Treatment of animals
A total of 18 mice were randomly divided into NC and KD groups. In the NC group (n = 9), mice were subjected to SCI and treated with miR-21 NC (RiboBio, Guangzhou, China) at 55 μL/d, 95 nmol/mL (intraspinal injection) for 3 days. In the KD group (n = 9), mice were subjected to SCI and treated with antagomir-21 (intraspinal injection; RiboBio, Guangzhou, China) for 3 days. The markers of successful model establishment included systolic tremor, local edema and congestion, and dural integrity (Liu et al., 2018; Wang et al., 2018). Subsequently, mice were sacrificed, and the injured T8–10 regions of spinal cord were stored in liquid nitrogen.
Protein preparation
Total protein was extracted from spinal cord samples harvested from the site of injury using cell lysate buffer containing 1 mM phenylmethane sulfonyl fluoride and a protein phosphatase inhibitor (Solarbio Life Sciences, Beijing, China). Protein was isolated after washing, crushing, lysis of cells, and finally centrifugation. After protein concentrations were calculated with a NanoDrop spectrometer (Thermo Fisher Scientific, Waltham, MA, USA), protein samples were ready for further processing.
Preparation of samples for liquid chromatography tandem mass spectrometry
For preparation of samples for liquid chromatography tandem mass spectrometry (LC-MS/MS) sample preparation, we performed an in-solution (trypsin) digestion protocol as previously described by Anwar et al. (2019) with minor modification. After trypsin digestion, peptide purification was attained by Ziptip (Millipore, Billerica, MA, USA) as previously described (Zachara et al., 2011).
Data processing and parameters
A high-resolution tandem mass spectroscopy system (NanoLC-Ultra 2D Plus with LTQ Orbitrap Velos Pro, Thermo Fisher Scientific) was used for analysis and data processing of digested peptides. Three biological replicates were prepared for each sample using previously described parameters (Anwar et al., 2019). Raw files from LC-MS were transferred into the built-in Proteome Discoverer software 1.3 (Thermo Fisher Scientific) and searched using the Mascot search engine against mouse proteome sequence databases for data processing. Parameters were adjusted as: (a) in trypsin digestion, with two maximum missed cleavage points permitted; (b) length of the digested peptide: 6–144; (c) precursor mass tolerance of 10 ppm and fragment mass tolerance adjusted to 0.8 Da; (d) in dynamic variation oxidation of methionine and in static modification carbamidomethyl of cysteine were selected; and (e) the false discovery rate rationale was based on q-value < 0.01. The level of peptide confidence for the data filter was adjusted to “high”.
Statistical analysis
DAVID 6.8 (https://david.ncifcrf.gov/), DAVID 6.7 (https://david-d.ncifcrf.gov/), and KOBAS 3.0 (http://kobas.cbi.pku.edu.cn/) online databases were used for KEGG pathway and protein clustering analyses. Statistical analysis was performed using GraphPad Prism Version 6.01 software (GraphPad, San Diego, CA, USA). Data are expressed as the mean ± SD and Log2 Fold Change (Log2FC). Gene Ontology (GO) of all proteins was observed via online protein ontology database. Student’s t-test was used for statistical significance. A P-value < 0.05 was considered statistically significant.
Results
Up- and downregulated proteins in NC and KD groups
Proteomics evaluation revealed the presence of 267 common proteins in NC and KD groups, as well as 65 and 127 unique proteins in NC and KD groups, respectively (Figure 1A).
Figure 1.

Overall proteome and GO slim between KD and NC groups.
(A) Venn diagram of total proteins in NC and KD groups: Only common proteins were selected from both biological repeats of NC and KD groups, and then further compared with each other using a Venn diagram. A total of 267 common proteins were present in both NC and KD groups, while 127 proteins were unique to the KD group and 65 proteins were unique to the NC group. (B) GO slim of KD and NC groups: the top six gene ontologies of all three categories are presented. miR-21a-5p was involved functions of all three categories (molecular function, cellular component, and biological processes). GO: Gene Ontology; KD: knockdown; NC: negative control.
The ontologies of all proteins from KD and NC groups (394 and 332, respectively) were evaluated via online protein ontology database. Within GO slim, three domains were identified: molecular function, biological process and cellular component, and molecular function. Within molecular function, most proteins were involved in the binding of ions, proteins, and drugs. Within cellular components, several proteins were involved in various components, such as cell parts, cytoplasm, and protein coating complex. For biological process, we found proteins involved in developmental process, multicellular organismal process, and cell communication (Figure 1B).
Among all proteins from KD and NC groups (394 and 332, respectively), we identified significantly differentially expressed proteins on the bases of coverage. Our results revealed that different proteins were significantly upregulated in the KD group, such as protein S100-A8, acyl-CoA-binding proteins, peroxiredoxin-1, vimentin, phosphoglycerate kinase 1, and translationally controlled tumor protein (Table 1 and Figure 2).
Table 1.
Up-regulated proteins in the knockdown and negative control groups
| Protein ID | Description | Gene ID | Coverage (mean value) | Difference (fold change) | P-value | |
|---|---|---|---|---|---|---|
| Knockdown group | Negative control group | |||||
| P27005 | Protein S100-A8 | S100a8 | 87.64 | 41.02 | 2.14 | 0.0617 |
| D3Z563 | Acyl-CoA-binding protein | Dbi | 77.78 | 0 | N.A | N.A |
| P35700 | Peroxiredoxin-1 | Prdx1 | 73.62 | 42.46 | 1.73 | 0.0326 |
| P20152 | Vimentin | Vim | 71.78 | 57.83 | 1.24 | 0.2971 |
| P09411 | Phosphoglycerate kinase 1 | Pgk1 | 71.58 | 68.71 | 1.04 | 0.2025 |
| P09528 | Ferritin heavy chain | Fth1 | 67.31 | 50.83 | 1.32 | 0.1714 |
| P63028 | Translationally-controlled tumor protein | Tpt1 | 67.74 | 56.11 | 1.21 | 0.0946 |
| Q91VW3 | SH3BGR glutaredoxin | Sh3bgrl3 | 56.99 | 0 | N.A | N.A |
| Q9QUH0 | Glutaredoxin-1 | Glrx | 53.27 | 39.72 | 1.34 | 0.0473 |
Figure 2.
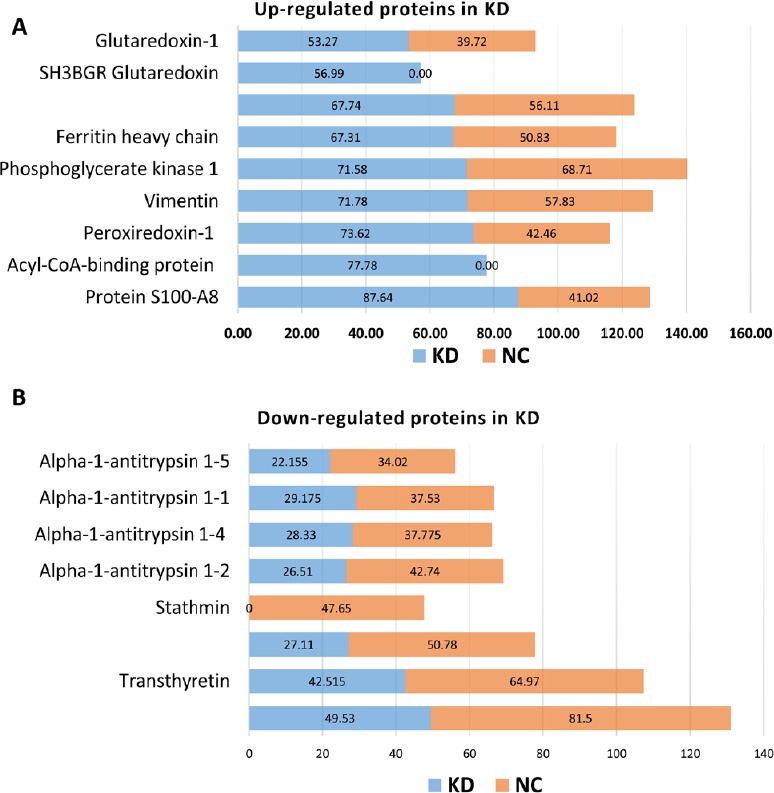
Up- and downregulated proteins in the KD group.
(A) Several proteins, such as S100-A8, acyl-CoA-binding proteins, peroxiredoxin-1, and vimentin, were upregulated in the KD group compared with the NC group. S100-A8 plays a prominent role in the regulation of inflammatory processes, immune response, and antioxidant activity. We found that protein levels of both S100-A8 and S100-A9 were upregulated two-fold in the KD group compared with the NC group. (B) Alpha-1-antitrypsin 1-1, 1-2, 1-4, and 1-5, along with stathmin and neurofilaments (medium and heavy), were downregulated in the KD group compared with the NC group. KD: Knockdown; NC: negative control.
Protein S100-A8, an S100 family member that acts as a calcium-binding protein with antioxidant activity (Choi et al., 2014), was previously reported to play a role in tumor-stromal interactions and arthritis as a pro-inflammatory factor (Kane et al., 2003; Basso et al., 2014). Migration inhibitory factor-related proteins 8 (MRP8) and 14 (MRP14), two binding proteins highly present in neutrophils that influence macrophages (Odink et al., 1987; Pechkovsky et al., 2000; Ryckman et al., 2003), are used as biomarkers to detect autoimmune diseases and inflammation in bacterial infections (Frosch et al., 2000; Pechkovsky et al., 2000; Ryckman et al., 2003). These proteins, which belong to the family of damage-associated molecular pattern molecules, play a role in protection against oxidative tissue damage. During inflammation, excessive oxidative modifications of S100-A8 and S100-A9 eliminate their chemotactic characteristics; this restrains leukocyte conscription and turns off inflammation (Lim et al., 2011).
Peroxiredoxins (PRDXs) are immune modulators that belong to a ubiquitous family of antioxidant enzymes (Pylvas et al., 2010). In prokaryotes and eukaryotes, PRDXs are present in cytosol, whereby they have a significant role in various signaling pathways, as well as influence over many other biological activities of cells, such as immune response, differentiation, proliferation, apoptosis, oxidation-sensitive protein protection, redox signaling, and regulation of cellular H2O2 (Kang et al., 1998; Seo et al., 2000; Rhee et al., 2005; Cong et al., 2018). PRDX1 also plays a role in the recovery of SCI. Upregulation of PRDX1, which is directly associated with fibroblast growth factor 1, regulates the level of reactive oxygen species and autophagy during SCI. In addition, PRDX1 reportedly has a main role in astrocyte and microglia proliferation after SCI (Huang et al., 2015; Li et al., 2018). Our proteomics results suggested that knockdown of miR-21a-5p increases PRDX expression, which is helpful in the recovery of SCI. Vimentin is a class-III intermediate and intracellular filament protein with important roles in various cellular processes, such as migration and adhesion (Eckes et al., 1998; Tsuruta and Jones, 2003; Ivaska et al., 2007). Extracellular vimentin plays a role in SCI recovery by increasing axonal growth activity and motor function. Indeed, through several signaling factors in neurons, vimentin works like a neurotrophic factor or adhesive factor (Shigyo and Tohda, 2016). We also confirmed through western blot assay that knockdown of miR-21a-5p upregulated the expression level of vimentin compared with NC. In contrast, stathmin was downregulated after miR-21a-5p knockdown (data not shown).
Protease inhibitors represent a highly significant mechanism for the regulation of proteolytic activity. Serpins, a vast family of proteins with a conserved structure, have been found across different domains, including animals, viruses, plants, bacteria, and archaea (Silverman et al., 2001; Roberts et al., 2004). Vertebrate serpins are part of essential biological processes such as inflammation, blood coagulation, angiogenesis, and tumor inhibition (van Gent et al., 2003). In the KD group, alpha-1-antitrypsin was downregulated. A detailed description of these proteins is listed in Table 2.
Table 2.
Down-regulated proteins in the knockdown and negative control groups
| Protein ID | Description | Gene ID | Coverage (mean value) | Difference (fold change) | P-value | |
|---|---|---|---|---|---|---|
| Knockdown group | Negative control group | |||||
| P07309 | Transthyretin | Ttr | 42.52 | 64.97 | 1.53 | 0.3469 |
| P22599 | Alpha-1-antitrypsin 1-2 | Serpina1b | 26.51 | 42.74 | 1.61 | 0.9686 |
| P07758 | Alpha-1-antitrypsin 1-1 | Serpina1a | 29.18 | 37.53 | 1.29 | 0.0678 |
| Q00897 | Alpha-1-antitrypsin 1-4 | Serpina1d | 28.33 | 37.78 | 1.33 | 0.0637 |
| Q00898 | Alpha-1-antitrypsin 1-5 | Serpina1e | 22.16 | 34.02 | 1.54 | 0.254 |
| A0A0R4J036 | Neurofilament 3, medium | Nefm | 49.53 | 81.5 | 1.65 | 0.0112 |
| P19246 | Neurofilament heavy polypeptide | Nefh | 27.11 | 50.78 | 1.87 | 0.0317 |
| P54227 | Stathmin | Stmn1 | 0 | 47.65 | N.A | N.A |
Neurofilaments are typically comprised of three intermediary filament proteins: light (NefL), medium (NefM), and heavy (NefH), which play a role in neuronal caliber. In addition, NefH has a vital role in mature axons that is not sub-served by the two smaller neurofilaments proteins (Laser-Azogui et al., 2015). After knockdown of miR-21a-5p, NefH and NefM were downregulated.
Data consist of unique proteins that appear only in the NC and KD groups, along with their sub-cellular localizations and biological functions
We analyzed data from both groups and identified unique proteins in each group of samples (Figure 3). From the first group of MS/MS data, we found 467 proteins in NC-1 and 548 proteins in KD-1. A total of 327 common proteins were identified between samples, as well as 140 unique proteins in NC-1 and 221 unique proteins in KD-1 (Figure 3). From the second group of MS/MS data, 594 and 587 proteins were found from NC-2 and KD-2, respectively, with 419 common proteins between these samples, and 175 and 168 unique proteins in NC-2 and KD-2, respectively (Figure 3).
Figure 3.
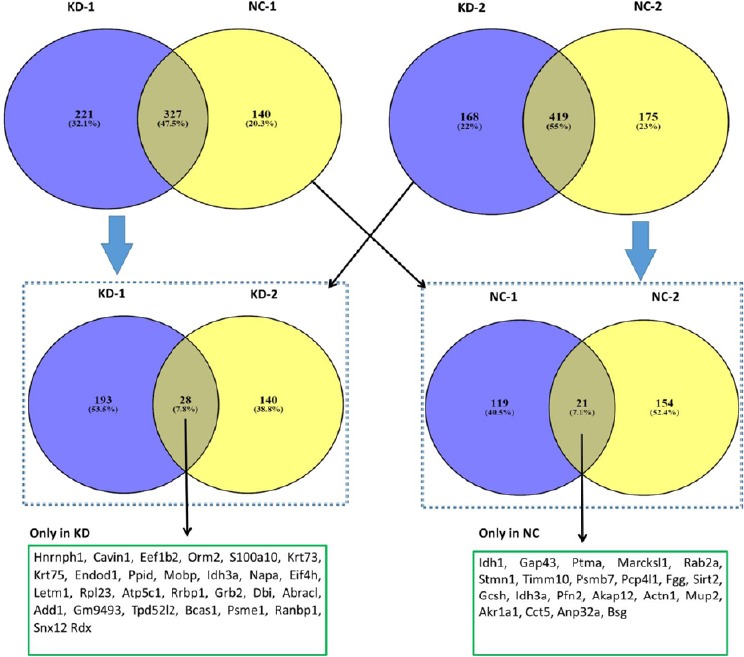
Identified proteins down- or upregulate in a mouse spinal cord injury model with knockdown of miR-21a-5p.
Venn diagrams of enriched proteins identified in both NC and KD groups. There were 221 and 140 unique proteins identified in KD-1 and NC-1, respectively; whereas, 168 and 175 proteins were identified in KD-2 and NC-2, respectively. We identified 28 proteins in both KD-1 and KD-2 (KD only), as well as 21 proteins in both NC-1 and NC-2 (only NC). A list of these proteins is presented within the boxes (only KD and only NC) as their gene names. KD: Knockdown; NC: negative control.
Subsequently, we compared unique proteins in NC-1 and NC-2 samples, as well as KD-1 and KD-2 samples. We found two common proteins between NC-1 and NC-2 unique proteins, and 28 common proteins between KD-1 and KD-2. These results suggested that 21 proteins are only present in the NC group (Table 3). Similarly, 28 proteins were only present in the KD group (Table 4).
Table 3.
List of unique proteins present only in the negative control group
| Protein AC | Protein name | Gene name | Pfam name | Product size (bp) |
|---|---|---|---|---|
| O88844 | Isocitrate dehydrogenase [NADP] cytoplasmic | Idh1 | Isocitrate/isopropylmalate dehydrogenase | 414 |
| P06837 | Neuromodulin | Gap43; Basp2 | IQ calmodulin-binding motif; Neuromodulin; Gap junction protein N-terminal region | 227 |
| P26350 | Prothymosin alpha | Ptma | Prothymosin/parathymosin family | 111 |
| P28667 | MARCKS-related protein | Marcksl1; Mlp, Mrp | MARCKS family | 200 |
| P53994 | Ras-related protein Rab-2A | Rab2a; Rab2 | Ras family | 212 |
| P54227 | Stathmin | Stmn1; Lag, Lap18, Pr22 | Stathmin family | 149 |
| P62073 | Mitochondrial import inner membrane translocase subunit Tim10 | Timm10; Tim10 | Tim10/DDP family zinc finger | 90 |
| P70195 | Proteasome subunit beta type-7 precursor | Psmb7; Mmc14 | Proteasome subunit; Proteasome beta subunits C terminal | 277 |
| Q6W8Q3 | Purkinje cell protein 4-like protein 1 | Pcp4l1 | 68 | |
| Q8VCM7 | Fibrinogen gamma chain precursor | Fgg | Fibrinogen beta and gamma chains, C-terminal globular domain; Fibrinogen alpha/beta chain family | 436 |
| Q8VDQ8-2 | Isoform 2 of NAD-dependent protein deacetylase sirtuin-2 | Sirt2 | ||
| Q91WK5 | Glycine cleavage system H protein, mitochondrial precursor | Gcsh | Glycine cleavage H-protein | 170 |
| Q9D6R2 | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial precursor | Idh3a | Isocitrate/isopropylmalate dehydrogenase | 366 |
| Q9WTQ5-2 | Isoform 2 of A-kinase anchor protein 12 | Akap12 | ||
| Q9JJV2 | Profilin-2 | Pfn2 | Profilin | 140 |
| A1BN54 | Alpha actinin 1a | Actn1; Actn1a | Calponin homology; Spectrin repeat; | 887 |
| A2AKN9 | Major urinary protein 2 | Mup2; Mup4 | Lipocalin / cytosolic fatty-acid binding protein family | 180 |
| B1AXW3 | Aldo-keto reductase family 1 member A1 | Akr1a1 | Aldo/keto reductase family | 203 |
| E0CZA1 | T-complex protein 1 subunit epsilon | Cct5 | TCP-1/cpn60 chaperonin family | 199 |
| F6UFG6 | Acidic leucine-rich nuclear phosphoprotein 32 family member A | Anp32a | Leucine-rich repeat | 138 |
| J3QP71 | Basigin | Bsg | 197 |
Table 4.
List of unique proteins present only in the knockdown group
| Protein AC | Protein name | Gene name | Pfam name | Product size (bp) |
|---|---|---|---|---|
| O35737 | Heterogeneous nuclear ribonucleoprotein H | Hnrnph1; Hnrph | RNA recognition motif | 449 |
| O54724 | Caveolae-associated protein 1 | Cavin1; Ptrf | PTRF/SDPR family | 392 |
| O70251 | Elongation factor 1-beta | Eef1b; Eef1b2 | EF-1 guanine nucleotide exchange domain | 225 |
| P07361 | Alpha-1-acid glycoprotein 2 precursor | Orm2; Agp-2, Orm-2 | Lipocalin / cytosolic fatty-acid binding protein family | 207 |
| P08207 | Protein S100-A10 | S100a10; Cal1l | S-100/ICaBP type calcium binding domain | 97 |
| Q6NXH9 | Keratin, type II cytoskeletal 73 | Krt73; Kb36 | Intermediate filament protein | 539 |
| Q8BGZ7 | Keratin, type II cytoskeletal 75 | Krt75; Kb18 | Intermediate filament protein; Keratin type II head | 551 |
| Q9D2P8-4 | Isoform 4 of Myelin-associated oligodendrocyte basic protein | Mobp | ||
| Q8C522 | Endonuclease domain-containing 1 protein precursor | Endod1 | 501 | |
| Q9CR16 | Peptidyl-prolyl cis-trans isomerase D | Ppid | Cyclophilin type peptidyl-prolyl cis-trans isomerase/CLD | 370 |
| Q9D6R2-2 | Isoform 2 of Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial | Idh3a | ||
| Q9DB05 | Alpha-soluble NSF attachment protein | Napa; Snapa | Soluble NSF attachment protein, SNAP | 295 |
| Q9Z2I0 | Mitochondrial proton/calcium exchanger protein precursor | Letm1 | LETM1-like protein | 738 |
| A2A6F8 | 60S ribosomal protein L23 | Rpl23 | 60 | |
| A2AKV0 | ATP synthase | Atp5c1 | ATP synthase | 101 |
| A2AVJ7 | Ribosome-binding protein 1 | Rrbp1 | Ribosome receptor lysine/proline rich region | 1464 |
| B1AT92 | Growth factor receptor-bound protein 2 | Grb2 | SH2 domain; SH3 domain | 203 |
| D3Z563 | Acyl-CoA-binding protein | Dbi | Acyl CoA binding protein | 63 |
| E9QMV2 | Costars family protein ABRACL | Abracl | Costars | 81 |
| F6RDR0 | Alpha-adducin (Fragment) | Add1 | 160 | |
| F6SVV1 | 40S ribosomal protein S7 | Gm9493 | Ribosomal protein S7e | 192 |
| F6VQ81 | Tumor protein D54 (Fragment) | Tpd52l2 | Tumour protein D52 family | 161 |
| F7BNZ5 | Breast carcinoma-amplified sequence 1 homolog | Bcas1 | 379 | |
| G3UWN9 | Proteasome activator complex subunit 1 (Fragment) | Psme1 | Proteasome activator pa28 alpha subunit; Proteasome activator pa28 beta subunit | 173 |
| Q9WUK2-2 | Isoform short of eukaryotic translation initiation factor 4H | Eif4h | ||
| H7BX22 | Ran-specific GTPase-activating protein | Ranbp1 | RanBP1 domain | 153 |
| Q6ZWQ5 | Sorting nexin 12 | Snx12 | PX domain | 162 |
| Q7TSG6 | Radixin | Rdx | FERM central domain | 389 |
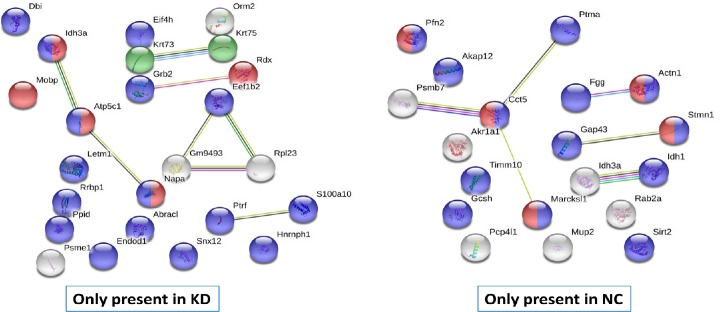
Interactions between unique proteins that appeared only in KD and NC groups were next examined using the STRING database (v11). We found that protein-protein interactions between proteins that only appeared in the KD group showed a strong interaction for myelin sheath and acetylation (Figure 4). For proteins that only appeared in the NC group, the protein-protein interaction showed a strong interaction with cytoskeletal binding proteins. This interaction network also showed that miR-21a-5p knockdown induced unique protein expression compared with the NC group. As mentioned above, these unique proteins have strong interactions for myelin sheath, indicating that miR-21a-5p–KD may be helpful to recover myelin in various scleroses.
Figure 4.
Protein-protein network interactions present only in NC or KD groups.
For the KD group, 24 various proteins were inserted into the STRING online database (version #11) to examine protein-protein interactions; 24 nodes and 8 edges were achieved for this interaction, with a protein-protein interaction enrichment P-value of 0.05. Network nodes represent proteins. Colored lines connecting nodes correspond to types of proof used in calculation: green represents neighborhood evidence, bright blue indicates database evidence, red indicates fusion evidence, black represents co-expression evidence, purple represents experimental proof, blue indicates co-occurrence evidence, and yellow represents text-mining evidence. For the KD group, blue spheres represent acetylation, red spheres represent proteins that play a role in the myelin sheath, and green spheres represent keratin type II. In total, 21 unique proteins from the NC group were inserted into the STRING online database to identify protein-protein interactions; 19 nodes and 6 edges of protein-protein were achieved for this interaction, with an enrichment P-value of 0.01. Blue spheres represent protein binding, while red spheres symbolize cytoskeletal protein binding. KD: Knockdown; NC: negative control.
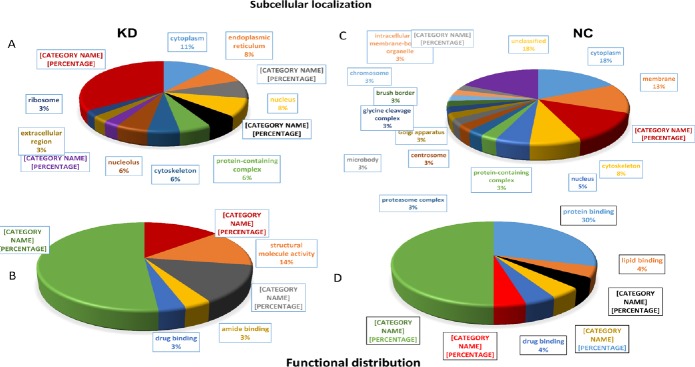
Cellular and molecular functions of proteins are shown in Figure 5. In both NC and KD groups, cytoplasmic proteins were detected. Others were classified into endoplasmic reticulum, mitochondria, and nucleus (Figure 5A and B). Although a similar distribution was observed in both groups, there was a difference in the biological functions of proteins. A few proteins, such as nucleic acid binding and amide binding, were found only in the KD group (Figure 5C), whereas those related to lipid binding, chromatin binding, and transporter activity were only found in the NC group (Figure 5D).
Figure 5.
Subcellular localization and functional distribution of unique proteins present only in KD (A and C) and NC (B and D) groups.
KD: Knockdown; NC: negative control.
In the second part of our analysis, we examined the potential role of miR-21a-5p in different pathways and potential correlation with the NC group. According to analysis via ShinyGO v0.50, DAVID database, and KOBAS 3.0, we found various KEGG pathways in which unique proteins of the KD group played a significant role, such as Huntington’s disease, Parkinson’s disease, carbon metabolism, and oxidative phosphorylation. Proteins highly enriched in the KD group are shown in Figure 6. After knockdown of miR-21a-5p, we found that expression levels of proteins associated with key eurodegenerative diseases such as Huntington’s, Parkinson’s, and Alzheimer’s were deregulated compared with NC samples.
Figure 6.
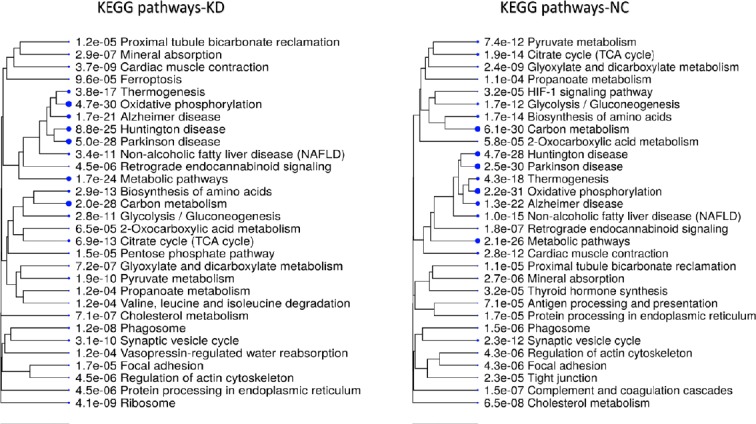
KEGG pathway hierarchical clustering trees.
Pathways such as Huntington’s disease, Parkinson’s disease, oxidative phosphorylation, and carbon metabolism were significantly altered after knockdown of miR-21a-5p in the mouse model of spinal cord injury. KD: Knockdown; NC: negative control.
Amyotrophic lateral sclerosis (ALS) is an untreatable and deadly disease that involves steady loss of motor neurons in different parts of the nervous system, such as the brain stem, cerebral cortex, and spinal cord (Ricci et al., 2018). Previously, Benigni et al. (2016) confirmed significant downregulation of miR-21a-5p in humans with ALS. We also found that in ALS, miR-21a-5p was downregulated (Figure 7). Numerous reasons may underlie the gathering of neurofilaments in neurodegenerative diseases, such as proteolysis, mutations in neurofilaments, dysregulation of neurofilament gene expression, defective axonal transport and irregular posttranslational modifications. Neurofilament accumulation is also possible because of exposure to toxic agents (Perrot and Julien, 2010).
Figure 7.

Amyotrophic lateral sclerosis, pathway ID# mmu05014.
During amyotrophic lateral sclerosis, miR-21a-5p was downregulated at the mitochondrial level, as were all three main neurofilaments (light, medium and heavy), compared with the negative control group. Yellow boxes represent proteins in the knockdown group, while green represents proteins that were not present in our data. MiR-21a-5p can be used as a biomarker to more comprehensively understand and identify the pathogenesis of amyotrophic lateral sclerosis.
During ALS, three neurofilaments (NefL, NefM, and NefH), which exist as polypeptides at the mitochondria level, are important for the radial development of axons during growth, stabilization of axon caliber, and communication of electrical impulses within axons (Yuan et al., 2012). These proteins were downregulated by miR-21a-5p KD compared with NC, which further confirms the possibility of its use as a disease biomarker. In addition, downregulation of miR-21a-5p is involved in the mitochondrial apoptotic pathway, which yields further insight into the processes responsible for motor neuron degradation (Laser-Azogui et al., 2015).
Discussion
Proteomic approaches can provide new ways to understand the role of miR-21a-5p in various diseases. During this study, we found that many proteins upregulated in the KD group compared with the NC group play a key role in inflammatory regulation, response towards immunity, and antioxidant activity. Between KD and NC groups, there were 394 and 332 significant proteins, respectively. We identified significant proteins in each group on the bases of coverage and validated our results with western blot assays, which also indicated upregulated levels of some proteins, such as vimentin, compared with NC. Meanwhile, stathmin was downregulated in the KD group compared with the NC group. S100-A8 protein plays a role in tumor-stromal interactions and, in arthritis, acts as a pro-inflammatory factor. In humans, the simplest defense mechanism is inflammation. During this process, local and external toxic or dangerous materials are removed to safeguard the body. However, when this system disproportionately responds, for instance during extreme or long-term inflammation, tissue damage can occur. Several studies have confirmed that S100-A8 and S100-A9 proteins have significant importance in the expansion of inflammation and its maintenance (Wang et al., 2018a). In this study, comparative analysis revealed significant upregulation of S100-A8 (about two-fold) in the KD group compared with the NC group. Stathmin downregulation is reportedly necessary for macrophage activation (Patel et al., 2009; Lachkar et al., 2010; Xu and Harrison, 2015; Nouar et al., 2016). The results of this study showed stathmin downregulation with miR-21a-5p knockdown, indicating that macrophage activation contributed to the recovery of SCI.
Myelination is vital for quick and precise impulse transmission within the vertebrate nervous system, and the thickness of myelin is based on the size of the axon fiber (Michailov et al., 2004; Monk et al., 2015). Myelin sheaths are a significantly stretched and altered plasma membrane that enfold around the nerve axon in a helical style (Raine, 1984; Thurnherr et al., 2006; Benninger et al., 2007; Nodari et al., 2007). We found that the protein-protein interaction between unique proteins of the KD group, which were missing from the NC group, were associated with the myelin sheath. Moreover, proteins highly enriched in the KD group play a significant role in pathways such as Huntington’s disease, Parkinson’s disease, carbon metabolism, and oxidative phosphorylation. Neurofilaments, which are important for neuronal caliber, were downregulated in the KD group compared with the NC group.
ALS is a fatal neurological disease as a result of the death of voluntary muscles controlled by the neurons that degenerate (Robberecht and Philips, 2013). Approximately 90% of ALS is diagnosed as periodic (due to some accident, usually traffic- or sports-related), while the remaining 10% is genetically inherited (Renton et al., 2014; Paez-Colasante et al., 2015; Benigni et al., 2016; Morgan and Orrell, 2016). The key role of miR-21a-5p in ALS makes it a useful biomarker for further studies, especially as authenticated biomarkers for ALS are lacking and microRNAs are a modern molecular tool with significant connections to diseases such as ALS (Waller et al., 2017). Recently, the microRNA miR-218 was revealed to be a prospective biomarker for ALS (Hoye et al., 2017). As small non-coding RNA molecules (8–22 nucleotides), miRNAs control the expression of various genes at the post-transcriptional level in a situational manner (Viader et al., 2011; Kye and Goncalves Ido, 2014). Importantly, miRNAs are also convenient biomarkers because they are consistently found within body fluids (Saraiva et al., 2017).
This comparative study between two groups (KD and NC) provides an overall idea about the presence of significant differentially expressed proteins. However, differentially expressed proteins in KD and NC groups detected by MS were only partially verified. Moreover, the molecular mechanism by which miR-21-5p regulates these differentially expressed proteins is unclear; future experiments will be conducted to identify this mechanism. Although further studies are needed, our results support the potential utility of miR-21 as a biomarker for recovery of SCI, ALS, and other neurological diseases.
Footnotes
Conflicts of interest: No financial or other benefits from commercial sources for the work reported on in the manuscript were received by any of the authors which could create a potential conflict of interest or the appearance of a conflict of interest with regard to the work.
Financial support: None.
Institutional review board statement: All experimental procedures and protocols were approved by the Experimental Animal Ethics Committee of Shandong University of China on March 5, 2014.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
C-Editor: Zhao M; S-Editors: Wang J, Li CH; L-Editors: Deusen AV, Robens J, Qiu Y, Song LP; T-Editor: Jia Y
References
- 1.Anwar MN, Li ZF, Gong Y, Singh RP. Omics studies revealed the factors involved in the formation of colony boundary in myxococcus xanthus. Cells. 2019;8:E530. doi: 10.3390/cells8060530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basso D, Bozzato D, Padoan A, Moz S, Zambon CF, Fogar P, Greco E, Scorzeto M, Simonato F, Navaglia F, Fassan M, Pelloso M, Dupont S, Pedrazzoli S, Fassina A, Plebani M. Inflammation and pancreatic cancer: molecular and functional interactions between S100A8, S100A9, NT-S100A8 and TGFbeta1. Cell Commun Signal. 2014;12:20. doi: 10.1186/1478-811X-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benigni M, Ricci C, Jones AR, Giannini F, Al-Chalabi A, Battistini S. Identification of miRNAs as potential biomarkers in cerebrospinal fluid from amyotrophic lateral sclerosis patients. Neuromol Med. 2016;18:551–560. doi: 10.1007/s12017-016-8396-8. [DOI] [PubMed] [Google Scholar]
- 4.Benninger Y, Thurnherr T, Pereira JA, Krause S, Wu X, Chrostek-Grashoff A, Herzog D, Nave KA, Franklin RJ, Meijer D, Brakebusch C, Suter U, Relvas JB. Essential and distinct roles for cdc42 and rac1 in the regulation of Schwann cell biology during peripheral nervous system development. J Cell Biol. 2007;177:1051–1061. doi: 10.1083/jcb.200610108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi DK, Li ZJ, Chang IK, Yeo MK, Kim JM, Sohn KC, Im M, Seo YJ, Lee JH, Kim CD, Lee Y. Clinicopathological roles of S100A8 and S100A9 in cutaneous squamous cell carcinoma in vivo and in vitro. Arch Dermatol Res. 2014;306:489–496. doi: 10.1007/s00403-014-1453-y. [DOI] [PubMed] [Google Scholar]
- 6.Cong N, Huang W, Yuan JP, Li GZ, Zhai GS, Li BS. Peroxiredoxin1 promotes cell proliferation, migration and invasion of colorectal cancer via p38MAPK signaling. Eur Rev Med Pharmacol Sci. 2018;22:1922–1928. doi: 10.26355/eurrev_201804_14715. [DOI] [PubMed] [Google Scholar]
- 7.Cristante AF, Barros Filho TEPd, Marcon RM, Letaif OB, Rocha IDd. Therapeutic approaches for spinal cord injury. Clinics (Sao Paulo) 2012;67:1219–1224. doi: 10.6061/clinics/2012(10)16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crozier KS, Cheng LL, Graziani V, Zorn G, Herbison G, Ditunno JF., Jr Spinal cord injury: prognosis for ambulation based on quadriceps recovery. Paraplegia. 1992;30:762–767. doi: 10.1038/sc.1992.147. [DOI] [PubMed] [Google Scholar]
- 9.Eckes B, Dogic D, Colucci-Guyon E, Wang N, Maniotis A, Ingber D, Merckling A, Langa F, Aumailley M, Delouvee A, Koteliansky V, Babinet C, Krieg T. Impaired mechanical stability, migration and contractile capacity in vimentin-deficient fibroblasts. J Cell Sci. 1998;111:1897–1907. doi: 10.1242/jcs.111.13.1897. [DOI] [PubMed] [Google Scholar]
- 10.Frosch M, Strey A, Vogl T, Wulffraat NM, Kuis W, Sunderkotter C, Harms E, Sorg C, Roth J. Myeloid-related proteins 8 and 14 are specifically secreted during interaction of phagocytes and activated endothelium and are useful markers for monitoring disease activity in pauciarticular-onset juvenile rheumatoid arthritis. Arthritis Rheum. 2000;43:628–637. doi: 10.1002/1529-0131(200003)43:3<628::AID-ANR20>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 11.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 12.Hoye ML, Koval ED, Wegener AJ. MicroRNA profiling reveals marker of motor neuron disease in ALS models. J Neurosci. 2017;37:5574–5586. doi: 10.1523/JNEUROSCI.3582-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S, Liu X, Zhang J, Bao G, Xu G, Sun Y, Shen Q, Lian M, Huang Y, Cui Z. Expression of peroxiredoxin 1 after traumatic spinal cord injury in rats. Cell Mol Neurobiol. 2015;35:1217–1226. doi: 10.1007/s10571-015-0214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivaska J, Pallari HM, Nevo J, Eriksson JE. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp Cell Res. 2007;313:2050–2062. doi: 10.1016/j.yexcr.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 15.Kane D, Roth J, Frosch M, Vogl T, Bresnihan B, FitzGerald O. Increased perivascular synovial membrane expression of myeloid-related proteins in psoriatic arthritis. Arthritis Rheum. 2003;48:1676–1685. doi: 10.1002/art.10988. [DOI] [PubMed] [Google Scholar]
- 16.Kang SW, Baines IC, Rhee SG. Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J Biol Chem. 1998;273:6303–6311. doi: 10.1074/jbc.273.11.6303. [DOI] [PubMed] [Google Scholar]
- 17.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Kye MJ, Goncalves Ido C. The role of miRNA in motor neuron disease. Front Cell Neurosci. 2014;8:15. doi: 10.3389/fncel.2014.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lachkar S, Lebois M, Steinmetz MO, Guichet A, Lal N, Curmi PA, Sobel A, Ozon S. Drosophila stathmins bind tubulin heterodimers with high and variable stoichiometries. J Biol Chem. 2010;285:11667–11680. doi: 10.1074/jbc.M109.096727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laser-Azogui A, Kornreich M, Malka-Gibor E, Beck R. Neurofilament assembly and function during neuronal development. Curr Opin Cell Biol. 2015;32:92–101. doi: 10.1016/j.ceb.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Wang Q, Cai H, He Z, Wang H, Chen J, Zheng Z, Yin J, Liao Z, Xu H, Xiao J, Gong F. FGF1 improves functional recovery through inducing PRDX1 to regulate autophagy and anti-ROS after spinal cord injury. J Cell Mol Med. 2018;22:2727–2738. doi: 10.1111/jcmm.13566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim SY, Raftery MJ, Geczy CL. Oxidative modifications of DAMPs suppress inflammation: the case for S100A8 and S100A9. Antioxid Redox Signal. 2011;15:2235–2248. doi: 10.1089/ars.2010.3641. [DOI] [PubMed] [Google Scholar]
- 24.Liu R, Wang W, Wang S, Xie W, Li H, Ning B. microRNA-21 regulates astrocytic reaction post-acute phase of spinal cord injury through modulating TGF-β signaling. Aging (Albany NY) 2018;10:1474–1488. doi: 10.18632/aging.101484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michailov GV, Sereda MW, Brinkmann BG, Fischer TM, Haug B, Birchmeier C, Role L, Lai C, Schwab MH, Nave KA. Axonal neuregulin-1 regulates myelin sheath thickness. Science. 2004;304:700–703. doi: 10.1126/science.1095862. [DOI] [PubMed] [Google Scholar]
- 26.Monk KR, Feltri ML, Taveggia C. New insights on Schwann cell development. Glia. 2015;63:1376–1393. doi: 10.1002/glia.22852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan S, Orrell RW. Pathogenesis of amyotrophic lateral sclerosis. Br Med Bull. 2016;119:87–98. doi: 10.1093/bmb/ldw026. [DOI] [PubMed] [Google Scholar]
- 28.Nodari A, Zambroni D, Quattrini A, Court FA, D’Urso A, Recchia A, Tybulewicz VL, Wrabetz L, Feltri ML. Beta1 integrin activates Rac1 in Schwann cells to generate radial lamellae during axonal sorting and myelination. J Cell Biol. 2007;177:1063–1075. doi: 10.1083/jcb.200610014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nouar R, Breuzard G, Bastonero S, Gorokhova S, Barbier P, Devred F, Kovacic H, Peyrot V. Direct evidence for the interaction of stathmin along the length and the plus end of microtubules in cells. FASEB J. 2016;30:3202–3215. doi: 10.1096/fj.201500125R. [DOI] [PubMed] [Google Scholar]
- 30.Odink K, Cerletti N, Bruggen J, Clerc RG, Tarcsay L, Zwadlo G, Gerhards G, Schlegel R, Sorg C. Two calcium-binding proteins in infiltrate macrophages of rheumatoid arthritis. Nature. 1987;330:80–82. doi: 10.1038/330080a0. [DOI] [PubMed] [Google Scholar]
- 31.Paez-Colasante X, Figueroa-Romero C, Sakowski SA, Goutman SA, Feldman EL. Amyotrophic lateral sclerosis: mechanisms and therapeutics in the epigenomic era. Nat Rev Neurol. 2015;11:266–279. doi: 10.1038/nrneurol.2015.57. [DOI] [PubMed] [Google Scholar]
- 32.Patel PC, Fisher KH, Yang EC, Deane CM, Harrison RE. Proteomic analysis of microtubule-associated proteins during macrophage activation. Mol Cell Proteomics. 2009;8:2500–2514. doi: 10.1074/mcp.M900190-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pechkovsky DV, Zalutskaya OM, Ivanov GI, Misuno NI. Calprotectin (MRP8/14 protein complex) release during mycobacterial infection in vitro and in vivo. FEMS Immunol Med Microbiol. 2000;29:27–33. doi: 10.1111/j.1574-695X.2000.tb01501.x. [DOI] [PubMed] [Google Scholar]
- 34.Perrot R, Julien JP. maldistribution of neurofilaments, disease pathogenesis, and amyotrophic lateral sclerosis. Neurology. 2010;5:30–34. [Google Scholar]
- 35.Pylvas M, Puistola U, Kauppila S, Soini Y, Karihtala P. Oxidative stress-induced antioxidant enzyme expression is an early phenomenon in ovarian carcinogenesis. Eur J Cancer. 2010;46:1661–1667. doi: 10.1016/j.ejca.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Raine CS. Morphology of Myelin and Myelination. In: Morell P, editor. Myelin. Boston, MA: Springer US; 1984. pp. 1–50. [Google Scholar]
- 37.Renton AE, Chio A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhee SG, Kang SW, Jeong W, Chang TS, Yang KS, Woo HA. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr Opin Cell Biol. 2005;17:183–189. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Ricci C, Marzocchi C, Battistini S. MicroRNAs as biomarkers in amyotrophic lateral sclerosis. Cells. 2018;7:E219. doi: 10.3390/cells7110219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robberecht W, Philips T. The changing scene of amyotrophic lateral sclerosis. Nat Rev Neurosci. 2013;14:248–264. doi: 10.1038/nrn3430. [DOI] [PubMed] [Google Scholar]
- 41.Roberts TH, Hejgaard J, Saunders NF, Cavicchioli R, Curmi PM. Serpins in unicellular eukarya, archaea, and bacteria: sequence analysis and evolution. J Mol Evol. 2004;59:437–447. doi: 10.1007/s00239-004-2635-6. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryckman C, Vandal K, Rouleau P, Talbot M, Tessier PA. Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J Immunol. 2003;170:3233–3242. doi: 10.4049/jimmunol.170.6.3233. [DOI] [PubMed] [Google Scholar]
- 44.Saraiva C, Esteves M, Bernardino L. MicroRNA: basic concepts and implications for regeneration and repair of neurodegenerative diseases. Biochem Pharmacol. 2017;141:118–131. doi: 10.1016/j.bcp.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 45.Seo MS, Kang SW, Kim K, Baines IC, Lee TH, Rhee SG. Identification of a new type of mammalian peroxiredoxin that forms an intramolecular disulfide as a reaction intermediate. J Biol Chem. 2000;275:20346–20354. doi: 10.1074/jbc.M001943200. [DOI] [PubMed] [Google Scholar]
- 46.Shigyo M, Tohda C. Extracellular vimentin is a novel axonal growth facilitator for functional recovery in spinal cord-injured mice. Sci Rep. 2016;6:28293. doi: 10.1038/srep28293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O’Donnell E, Salvesen GS, Travis J, Whisstock JC. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 48.Simon F, Floros N, Ibing W, Schelzig H, Knapsis A. Neurotherapeutic potential of erythropoietin after ischemic injury of the central nervous system. Neural Regen Res. 2019;14:1309–1312. doi: 10.4103/1673-5374.253507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thuret S, Moon LD, Gage FH. Therapeutic interventions after spinal cord injury. Nat Rev Neurosci. 2006;7:628–643. doi: 10.1038/nrn1955. [DOI] [PubMed] [Google Scholar]
- 50.Thurnherr T, Benninger Y, Wu X, Chrostek A, Krause SM, Nave KA, Franklin RJ, Brakebusch C, Suter U, Relvas JB. Cdc42 and Rac1 signaling are both required for and act synergistically in the correct formation of myelin sheaths in the CNS. J Neurosci. 2006;26:10110–10119. doi: 10.1523/JNEUROSCI.2158-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsuruta D, Jones JC. The vimentin cytoskeleton regulates focal contact size and adhesion of endothelial cells subjected to shear stress. J Cell Sci. 2003;116:4977–4984. doi: 10.1242/jcs.00823. [DOI] [PubMed] [Google Scholar]
- 52.van Gent D, Sharp P, Morgan K, Kalsheker N. Serpins: structure, function and molecular evolution. Int J Biochem Cell Biol. 2003;35:1536–1547. doi: 10.1016/s1357-2725(03)00134-1. [DOI] [PubMed] [Google Scholar]
- 53.Viader A, Chang LW, Fahrner T, Nagarajan R, Milbrandt J. MicroRNAs modulate Schwann cell response to nerve injury by reinforcing transcriptional silencing of dedifferentiation-related genes. J Neurosci. 2011;31:17358–17369. doi: 10.1523/JNEUROSCI.3931-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waller R, Goodall EF, Milo M, Cooper-Knock J, Da Costa M, Hobson E, Kazoka M, Wollff H, Heath PR, Shaw PJ, Kirby J. Serum miRNAs miR-206, 143-3p and 374b-5p as potential biomarkers for amyotrophic lateral sclerosis (ALS) Neurobiol Aging. 2017;55:123–131. doi: 10.1016/j.neurobiolaging.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang S, Song R, Wang Z, Jing Z, Wang S, Ma J. S100A8/A9 in Inflammation. Front Immunol. 2018a;9:1298–1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang W, Liu R, Su Y, Li H, Xie W, Ning B. MicroRNA-21-5p mediates TGF-β-regulated fibrogenic activation of spinal fibroblasts and the formation of fibrotic scars after spinal cord injury. Int J Biol Sci. 2018b;14:178–188. doi: 10.7150/ijbs.24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waters RL, Adkins RH, Yakura JS. Definition of complete spinal cord injury. Paraplegia. 1991;29:573–581. doi: 10.1038/sc.1991.85. [DOI] [PubMed] [Google Scholar]
- 58.Wei WB, Zou ZL, Zhou BB, Li BL, Qin JY, Feng ZF, Li JN, Ye LY, Wu RM. Selection of injury segments in a rat model of spinal cord injury: network meta-analysis. Zhongguo Zuzhi Gongcheng Yanjiu. 2019;23:650–656. [Google Scholar]
- 59.Xu K, Harrison RE. Down-regulation of stathmin is required for the phenotypic changes and classical activation of macrophages. J Biol Chem. 2015;290:19245–19260. doi: 10.1074/jbc.M115.639625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao X, Zhang Y, Hao J, Duan HQ, Zhao CX, Sun C, Li B, Fan BY, Wang X, Li WX, Fu XH, Hu Y, Liu C, Kong XH, Feng SQ. Deferoxamine promotes recovery of traumatic spinal cord injury by inhibiting ferroptosis. Neural Regen Res. 2019;14:532–541. doi: 10.4103/1673-5374.245480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuan A, Rao MV, Veeranna, Nixon RA. Neurofilaments at a glance. J Cell Sci. 2012;125:3257. doi: 10.1242/jcs.104729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zachara NE, Vosseller K, Hart GW. Detection and analysis of proteins modified by O-linked N-acetylglucosamine. Curr Protoc Mol Biol Chapter 17:Unit 17.6. 2011 doi: 10.1002/0471142727.mb1706s95. [DOI] [PMC free article] [PubMed] [Google Scholar]