Abstract
Background
Osteogenic differentiation of periodontal ligament stem cells (PDLSCs) is associated with periodontitis. It has been reported that long noncoding RNA X-inactive specific transcript (lncRNA XIST) is upregulated and microRNA-214-3p (miR-214-3p) is downregulated in PDLSCs after osteogenic induction. However, whether XIST is involved in osteogenic differentiation of PDLSCs via miR-214-3p has not been reported.
Material/Methods
The protein expressions of osteogenic markers alkaline phosphatase (ALP), osteocalcin (OCN), and runt-related transcription factor 2 (RUNX2) were examined by Western blot. The levels of miR-214-3p and XIST were determined by qRT-PCR. The relationship between miR-214-3p and XIST was evaluated by luciferase reporter, RNA immunoprecipitation, and RNA pulldown assays.
Results
We found that XIST was increased and miR-214-3p was decreased in PDLSCs after osteogenic stimulation. Silencing of XIST decreased the protein expressions of ALP, OCN, and RUNX2, and also decreased ALP activity. Higher miR-214-3p levels also inhibited osteogenic differentiation of PDLSCs. XIST interacted with miR-214-3p and depletion of miR-214-3p mitigated XIST absence-mediated suppression of osteogenic differentiation.
Conclusions
XIST participates in osteogenic differentiation of PDLSCs by sponging miR-214-3p.
MeSH Keywords: MicroRNAs, Osteosarcoma, Periodontitis
Background
Periodontitis, a chronic inflammatory disease, is caused by destruction of tissues that surround and support the tooth [1,2]. A previous study reported that periodontitis inhibits the differentiation potential of periodontal ligament stem cells (PDLSCs), and treatment using PDLSCs restores the damaged or diseased periodontal tissues [3]. Therefore, better understanding of the molecular mechanisms underlying PDLSCs differentiation could help develop novel medical therapies for periodontitis.
It has been reported that long noncoding RNAs (lncRNAs) participate in biological processes such as stem cell differentiation [4]. Many lncRNAs are differentially expressed during the osteogenic differentiation of PDLSCs [5–7]. For example, anti-differentiation noncoding RNA (ANCR) downregulation inhibits osteogenic differentiation of PDLSCs [8], and lnc Taurine upregulated gene 1 (TUG1) facilitates the expressions of the osteogenic-related gene markers alkaline phosphatase (ALP) level, osteocalcin (OCN), and runt-related transcription factor 2 (RUNX2) in PDLSCs [9]. lncRNA X-inactive specific transcript (XIST) is markedly elevated in osteoblastic-differentiated PDLSCs compared with that of non-differentiated cells [10], suggesting the involvement of XIST in osteoblastic differentiation of PDLSCs. However, little is known about the function of XIST and its potential molecular mechanisms in this process.
It is well-established that lncRNAs play important roles in various diseases by sponging miRNAs [11]. Aberrant expression of miRNAs is also reported to be involved in the development of chronic periodontitis [1]. Li et al. reported that silencing of miR-17 inhibits osteoblastic differentiation of PDLSCs [12]. Another study found that the level of miR-21 is decreased in osteogenically differentiated PDLSCs, and upregulation of miR-21 clearly inhibits osteogenesis of PDLSCs [13]. Zhen et al. reported that depletion of miR-31 promotes the osteogenic differentiation ability of PDLSCs in a high-glucose environment by reducing RUNX2, OSX, and OCN expression levels [14]. These findings indicate the involvement of miRNAs in differentiation of PDLSCs. Recently, miR-214 was reported to be drastically downregulated during the osteoblastic differentiation of PDLSCs [15], and they further demonstrated that overexpression of miR-214 limits differentiation of PDLSCs via negatively regulating mutations in the β-catenin gene. Although many researchers have studied the role of miR-214 in differentiation of PDLSCs, whether lnc RNA XIST is responsible for the osteoblastic differentiation of PDLSCs via interacting with miR-214 has not been explored.
In this study, we assessed the effect of XIST on osteoblastic differentiation of PDLSCs and explored the possible role of miR-214 in this process.
Material and Methods
Cell culture and treatment
This study was approved by the Medical Ethical Committee of the Second Affiliated Hospital, College of Medicine, Zhejiang University. PDLSCs were isolated and cultured as previously described [16]. Briefly, human periodontal ligament tissues were obtained from 6 teenagers (age 13–16 years) without oral or systematic diseases at the Second Affiliated Hospital, College of Medicine, Zhejiang University. The teenagers provided signed informed consent. The PDLSCs were induced for osteogenic differentiation in vitro using osteo-inductive medium (90% Dulbecco’s modified Eagle’s medium, 10% fetal bovine serum, 100 nmol/L dexamethasone, 10 mmol/L β-glycerophosphate, and 50 ug/mL vitamin C; Sigma, St. Louis, MO, USA) [9]. We also assessed the protein expressions of ALP, RUNX2, and OCN.
Plasmid construction
miR-214-3p binding sites of XIST were amplified by PCR and inserted into pMIR-REPORT™ (Thermo Fisher Scientific, Waltham, MA, USA) to construct the XIST wild-type reporter vector (XIST-wt). Small interfering RNA for XIST (si-XIST) or negative control (si-NC), pcDNA vector (vector), and XIST overexpression plasmid (XIST), miR-214-3p mimic (miR-214-3p), miR-214-3p inhibitor (anti-miR-214-3p), or their negative controls were all obtained from Shanghai Genechem (Shanghai, China).
Luciferase reporter assay
XIST-wt or XIST-mut plasmids and miR-214-3p mimic or miR-NC mimic (NC) were co-transfected into cells using lipofectamine 3000 (Thermo Fisher Scientific). Luciferase activity was evaluated using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA).
RNA immunoprecipitation (RIP) and RNA pulldown assays
Cell extracts were incubated with RIP buffer containing magnetic beads conjugated with human anti-Ago2 antibody (Abcam, Cambridge, MA, USA) or negative control. RNA pulldown assay was conducted using Dynabeads® M-280 Streptavidin (Thermo Fisher Scientific). Probes for miR-214-3p of miR-NC were biotinylated (GENEWIZ, Suzhou, China) and then transfected into cells. Then, immunoprecipitated RNAs were isolated. The purified RNAs were subjected to qRT-PCR analysis.
qRT-PCR assay
Total RNAs from cultured cells was extracted using Trizol (Thermo Fisher Scientific). The value of OD260/OD280 was in the range of 1.8~2.1. Moreover, capillary electrophoresis was performed with an Agilent Bioanalyzer and evaluated by RIN (RNA Integrity Number) software scores, with 10 being the best RNA integrity and 0 being the worst. The RIN of RNA in this study was in the range of 7~9. After that, RNA was then reverse transcribed using a First-Strand RT-PCR kit (Invitrogen, Carlsbad, CA, USA). qPCR was performed with LightCycler FastStart DNA MasterPLUS SYBR Green I mix (Roche, Mannheim, Germany) on the ABI-7500 platform (Biosystems, Foster City, CA, USA). In addition, we assessed the melting curves, and a single peak indicated that the primer was specific. U6 and GAPDH were used as internal controls. The expressions of XIST and miR-214-3p were determined using 2−ΔΔCt method.
XIST-F: 5′-CTCCAGATAGCTGGCTAACC-3′,
XIST-R: 5′-AGCTCCTCGGACAGCTGCTAA-3′;
GAPDH-F: 5′-AGAAGGCTGGGGCTCATTTG-3′,
GAPDH-R: 5′-AGGGGCCATCCACAGTCTTC-3′;
miR-214-3p-F, 5′-AGCCACATCGCTCAGACA-3′,
miR-214-3p-R, 5′-CAGACGAGGCTCCGTGGT-3′;
U6-F: 5′-CTCGCTTCGGCAGCACA-3′,
U6-R: 5′-AACGCTTCACGAATTTGCGT-3′.
Alkaline phosphatase activity assay
PDLSCs were lysed after 7 and 14 days of culture in osteogenic medium, and the ALP activity was determined using an ALP assay kit (Abcam) according to the manufacturer’s instructions. Absorbance was evaluated spectrophotometrically at 405 nm.
Western blot assay
Cells were lysed using RIPA buffer (Thermo Fisher Scientific) and proteins were evaluated by BCA assay kit (Solarbio, Beijing, China) and then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Next, proteins were transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). After that, membranes were incubated with primary antibodies (ALP, RUNX2, OCN, GAPDH; Abcam). All protein bands were visualized with an enhanced chemiluminescence (ECL) kit (Solarbio).
Statistical analysis
The experiments in this study were repeated 3 times (n=3). All statistical analyses were performed using SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA). Data are presented as mean±standard deviation. The differences were measured using the t test or one-way ANOVA. Differences were considered to be significant at P<0.05.
Results
XIST was elevated while miR-214-3p was decreased in PDLSCs after osteogenic induction
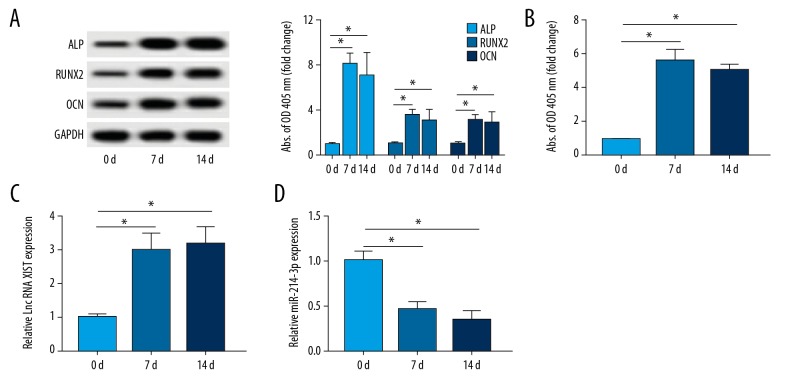
Firstly, we evaluated the expressions of XIST and miR-214-3p in response to the osteogenesis of PDLSCs. As demonstrated in Figure 1A, the osteogenic differentiation-related genes ALP, RUNX2, and OCN were detected. A significant increase in protein expressions of ALP, RUNX2, and OCN was observed after osteoblast induction at 7 or 14 days. ALP activity was also increased at 7 or 14 days compared that of 0 days (Figure 1B). These data suggest that the osteoblastic phenotype of PDLSCs was established. Moreover, the results of qRT-PCR suggested that the level of XIST was upregulated at 7 or 14 days in PDLSCs cultured in osteo-inductive medium compared with that at 0 days (Figure 1C). By contrast, miR-214-3p was decreased in osteoblastic phenotype (Figure 1D). These results suggested that XIST and miR-214-3p are involved in the development of the osteoblastic phenotype of PDLSCs.
Figure 1.
XIST was increased and miR-214-3p was decreased in PDLSCs after induction of osteogenesis. (A) The expressions of osteogenic-related proteins ALP, RUNX2, and OCN in PDLSCs after 7 days and 14 days osteogenic induction were determined by Western blot. (B) The activity of ALP was determined. (C, D) The abundances of lnc RNA XIST and miR-214-3p in osteogenically induced PDLSCs were determined by qRT-PCR. * P<0.05.
XIST downregulation inhibited osteogenic differentiation of PDLSCs
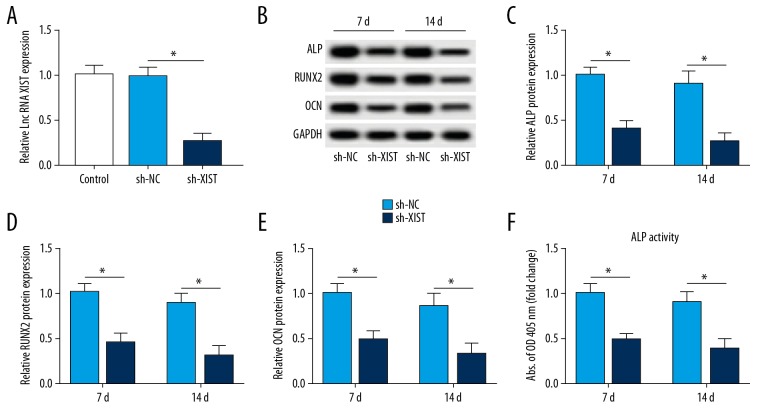
To explore XIST function in PDLSC osteogenesis, we interfered with the expression of XIST using sh-XIST. The transfection efficiency revealed that the introduction of sh-XIST markedly induced the inhibition of XIST compared with that of the sh-NC group (Figure 2A). As compared with the sh-NC group, the expression of ALP was reduced by sh-XIST transfection (Figure 2B, 2C). The same change was also observed in RUNX2 and OCN of the sh-XIST group compared with the sh-NC group (Figure 2B, 2D, 2E). ALP activity was also lower in the sh-XIST group than in the sh-NC group (Figure 2F). These findings suggested that the depletion of XIST suppressed the differentiation of PDLSCs in osteogenesis.
Figure 2.
Effect of XIST depletion on differentiation of PDLSCs after osteogenesis. (A) qRT-PCR analysis of XIST in PDLSCs transfected with sh-XIST or sh-NC. (B–F) The protein levels of osteogenic-related proteins and ALP activity in XIST depletion-induced PDLSCs. * P<0.05.
Overexpression of miR-214-3p suppressed osteogenic differentiation of PDLSCs
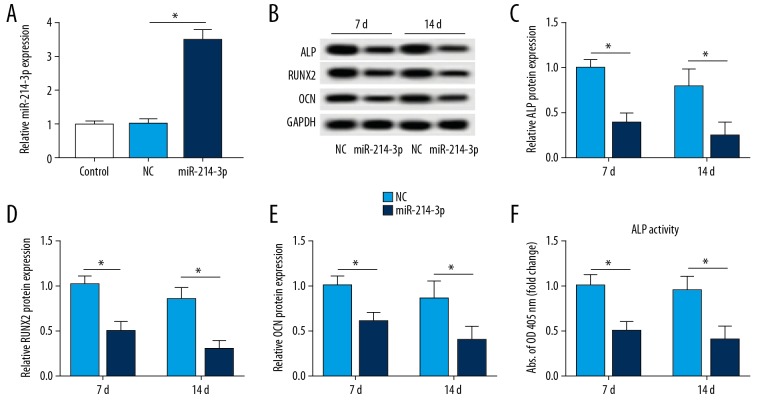
The miR-214-3p level was conspicuously upregulated in cells transfected with miR-214-3p mimic (Figure 3A). Moreover, the results of Western blot analysis disclosed that overexpression of miR-214-3p remarkably decreased the protein level of ALP (Figure 3B, 3C). miR-214-3p upregulation also inhibited the expressions of RUNX2 and OCN compared to NC group (Figure 3B, 3D, 3E). Overexpression of miR-214-3p also decreased ALP activity (Figure 3F). Interestingly, our data suggested that gain of miR-214-3p or XIST ablation has the same effect on PDLSCs differentiation.
Figure 3.
Effect of miR-214-3p upregulation on differentiation of PDLSCs after osteogenesis. (A) The level of miR-214-3p was determined in PDLSCs transfected with miR-214-3p mimic or miR-NC (NC). (B–F) Western blot analysis of ALP, RUNX2, and OCN in miR-214-3p upregulation-mediated PDLSCs and the detection of ALP activity. * P<0.05.
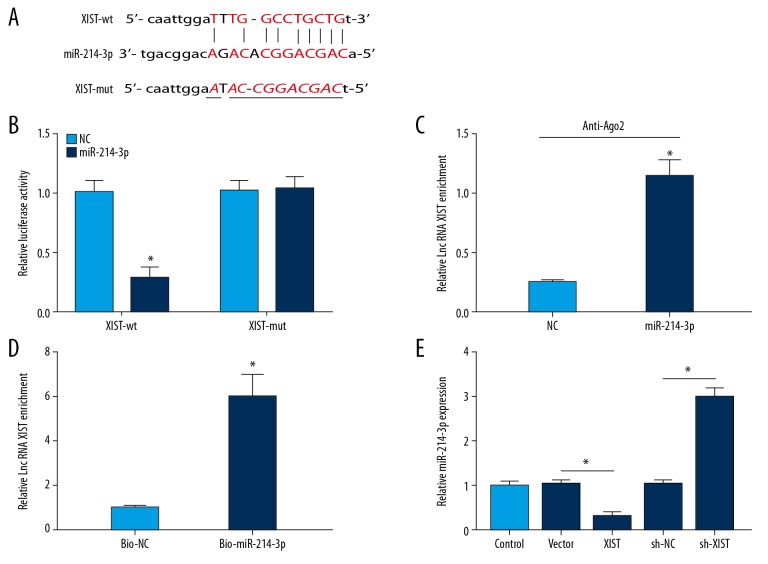
XIST negatively regulated the expression of miR-214-3p
The above results suggest that miR-214-3p is involved in the function of XIST. Subsequently, the binding sites between miR-214-3p and XIST were predicted using starBase 2.0 (Figure 4A), indicating that miR-214-3p is a potential target of XIST. To test our hypothesis, we performed luciferase reporter assay to assess the relationship between them. As expected, the luciferase activity was significantly decreased by the combination miR-214-3p mimic and XIST-wt plasmid. However, no evident change was observed in the XIST-mut group (Figure 4B). RIP assay indicated that XIST was notably enriched in the Ago2 pellet of the miR-214-3p group in contrast to the NC group (Figure 4C). Then, cells were treated with bio-miR-214-3p or bio-NC, and then subjected to RNA pulldown assay. XIST were captured by M-280 streptavidin magnetic beads and biotin-labeled miRNAs. As demonstrated in Figure 4D, XIST was pulled down in groups transfected with bio-miR-214-3p compared to those transfected with bio-NC. Additionally, we examined the effect of XIST on the expression of miR-214-3p. qRT-PCR data showed that delivery of XIST overexpression plasmid significantly reduced the expression of miR-214-3p, whereas loss of XIST enhanced the expression of miR-214-3p (Figure 4E). Hence, these results demonstrated that miR-214-3p is a target of XIST and XIST can affect osteogenic differentiation by sponging miR-214-3p.
Figure 4.
XIST interacts with miR-214-3p. (A, B) The binding sites between XIST and miR-214-3p were predicated, and luciferase activity was used to confirm the relationship between them. (C) The enrichment of XIST in cells after Ago2 or IgG RIP assay. (D) The enrichment of XIST in the samples pulled down by biotinylated miR-214-3p. (E) The effect of XIST overexpression or knockdown on miR-214-3p expression. * P<0.05.
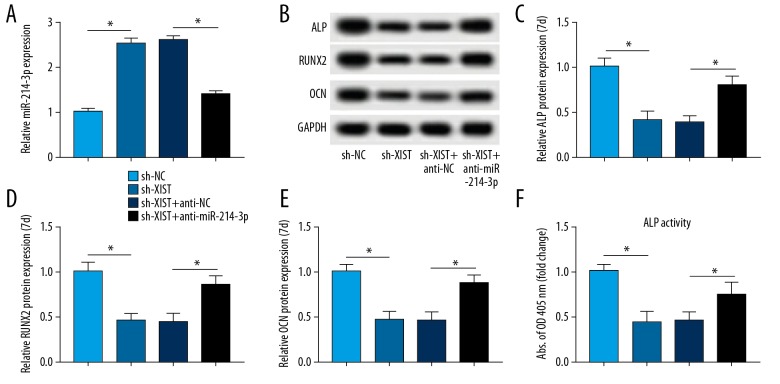
Loss of miR-214-3p rescues XIST depletion-mediated function on the differentiation of PDLSCs in osteogenesis
A rescue-of-function experiment was performed to evaluate whether XIST is involved in differentiation of PDLSCs in osteogenesis by regulating miR-214-3p. The anti-miR-214-3p and sh-XIST were co-transfected into cells, and the results of qRT-PCR suggested that the depletion of XIST promoted the expression of miR-214-3p, which was abated by treatment with anti-miR-214-3p (Figure 5A). ShRNA-mediated knockdown of XIST repressed the expression of osteogenic differentiation markers. However, silencing of miR-214-3p reduced the XIST descend-induced effect (Figure 5B–5F). In conclusion, our data suggest that XIST positively regulated osteogenic differentiation by suppressing miR-214-3p.
Figure 5.
Silencing of miR-214-3p abates the effect of XIST downregulation on PDLSCs. (A) The level of miR-214-3p in cells co-transfected with sh-XIST and anti-miR-214-3p. (B–F) Western blot analysis of osteogenic-related proteins in PDLSCs of each group and ALP activity. * P<0.05.
Discussion
Stem cell-based tissue engineering provides novel therapeutic strategies for periodontitis. Several kinds of stem cells, including PDLSCs, have been isolated from human adult teeth and used for tooth and periodontal regeneration [17,18]. ALP is highly expressed during early osteogenic differentiation [19]. Moreover, RUNX2 and OCN have been reported to be specific osteoblast markers during differentiation [16]. In this study, we found that the level of XIST was upregulated in PDLSCs after osteogenic induction, suggesting that XIST participates in the osteogenic differentiation of PDLSCs. lncRNA XIST is involved in the development of many diseases [20,21]. A previous study disclosed that depletion of XIST curbs cell proliferation and enhances radio-sensitivity of nasopharyngeal carcinoma cells by upregulating miR-29c [22]. XIST is also reported to be associated with stemness. Chen et al. revealed that XIST overexpression suppresses osteogenic differentiation of bone marrow mesenchymal stem cells (BMSCs) [10]. However, the role of XIST during the differentiation of PDLSCs in osteogenesis remains largely unknown. We found that the protein expressions of osteoblast markers ALP, OCN, and RUNX2, as well as ALP activity, were decreased after inhibition of XIST. Therefore, we hypothesized that a high level of XIST promotes osteogenic differentiation.
miR-214-3p is linked to progression of many cancers. For instance, strong overexpression of HOXA11-AS enhances hepatocellular carcinoma proliferation and invasion and induces epithelial-mesenchymal transition through downregulating miR-214-3p expression [23]. A prior study showed that lnc MT1JP represses cell proliferation, cycle, and invasion through sponging miR-214-3p in bladder cancer [24]. More importantly, many investigations also indicated that miR-214-3p also is involved in cell differentiation. Du et al. proved that silencing of miR-214 enhances proliferation and differentiation in human hair follicle stem cells [25]. In another study, dysregulation of miR-214-3p expression during myoblast-to-myocyte differentiation was also observed [26]. These findings show that miR-214-3p participates in cell differentiation. Other studies have assessed the role of miR-214-3p in osteogenic differentiation. Liu et al. found that miR-214-3p expression is markedly downregulated during the osteogenic differentiation of hFOB 1.19 cells, and the ectopic expression of miR-214 overexpression inhibits the level of ALP. miR-214 can suppress osteogenesis through inhibiting the expression of baculoviral IAP repeat-containing 7 [27]. Wang et al. reported that miR-214 is decreased during the osteogenic induction of BMSCs. lncRNA Kcnq1ot1 can enhance osteogenic differentiation of BMSCs by serving as a competitive RNA to modulate expression of bone morphogenetic protein-2 via interacting with miR-214 [28]. In addition, miR-214 also has a suppressor role in osteogenic differentiation of PDLSCs [16]. In our study, we found that the level of miR-214-3p was also downregulated in PDLSCs after osteogenic induction. Consistent with prior reports, our data revealed that upregulation of miR-214-3p decreased osteoblast markers, suggesting that the underexpression of miR-214-3p after osteogenic induction is associated with osteogenic differentiation.
In addition, the lncRNA-miRNA regulatory network has a significant role in osteogenic differentiation of PDLSCs [11]. Although a report has shown that miR-214-3p directly binds XIST in epithelial ovarian cancer cells [29], the relationship between XIST and miR-214-3p in PDLSCs has not been reported, and the direct evidence of whether XIST interacts with miR-214-3p in PDLSCs osteogenic differentiation is still lacking. Subsequently, we demonstrated that miR-214-3p contains binding sites with XIST. The qRT-PCR analysis further indicated that XIST negatively regulates miR-214-3p expression. These results suggest that miR-214-3p acts as a sponge of XIST to participate in the XIST-mediated osteogenic differentiation. The rescue-of-function experiments further showed that the absence of miR-214-3p reduced the XIST depletion-mediated inhibition effect of PDLSCs on osteogenic markers ALP, OCN, and RUNX2, suggesting that XIST is involved in osteogenesis of PDLSCs by sponging miR-214-3p.
There are some limitations to this study. We only explored the roles of XIST and miR-214-3p in vitro, and the functions of XIST and miR-214-3p in vivo needs to be investigated in future. The underlying signaling pathways in XIST/miR-214-3p axis-mediated osteogenic differentiation in PDLSCs also warrant further study.
Conclusions
XIST promotes osteogenic differentiation of PDLSCs by sponging miR-214-3p. This study is the first to show that the XIST/miR-214-3p axis is a novel pathway involved in regulating PDLSCs osteogenic differentiation, providing novel insights into differentiation of PDLSCs in tissue regeneration applications.
Footnotes
Conflicts of interest
None.
Source of support: Departmental sources
References
- 1.Venugopal P, Koshy T, Lavu V, et al. Differential expression of microRNAs let-7a, miR-125b, miR-100, and miR-21 and interaction with NF-κB pathway genes in periodontitis pathogenesis. J Cell Physiol. 2018;233:5877–84. doi: 10.1002/jcp.26391. [DOI] [PubMed] [Google Scholar]
- 2.Panduwawala CP, Zhan X, Dissanayaka WL, et al. In vivo periodontal tissue regeneration by periodontal ligament stem cells and endothelial cells in three-dimensional cell sheet constructs. J Periodontal Res. 2017;52:408–18. doi: 10.1111/jre.12405. [DOI] [PubMed] [Google Scholar]
- 3.Zheng W, Wang S, Wang J, et al. Periodontitis promotes the proliferation and suppresses the differentiation potential of human periodontal ligament stem cells. Int J Mol Med. 2015;36:915–22. doi: 10.3892/ijmm.2015.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou J, Long H, Zhou C, et al. Long noncoding RNA Braveheart promotes cardiogenic differentiation of mesenchymal stem cells in vitro. Stem Cell Res Ther. 2017;8:4. doi: 10.1186/s13287-016-0454-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng Y, Li X, Huang Y, et al. Time series clustering of mRNA and lncRNA expression during osteogenic differentiation of periodontal ligament stem cells. Peer J. 2018;6:e5214. doi: 10.7717/peerj.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu X, Li M, Jin Y, et al. Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genet. 2017;18:100. doi: 10.1186/s12863-017-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Q, Chen L, Cui S, et al. Expression and regulation of long noncoding RNAs during the osteogenic differentiation of periodontal ligament stem cells in the inflammatory microenvironment. Sci Rep. 2017;7:13991. doi: 10.1038/s41598-017-14451-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia Q, Jiang W, Ni L. Down-regulated non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of periodontal ligament stem cells. Arch Oral Biol. 2015;60:234–41. doi: 10.1016/j.archoralbio.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 9.He Q, Yang S, Gu X, et al. Long noncoding RNA TUG1 facilitates osteogenic differentiation of periodontal ligament stem cells via interacting with Lin28A. Cell Death Dis. 2018;9:455. doi: 10.1038/s41419-018-0484-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, Yang L, Ge D, et al. Long non-coding RNA XIST promotes osteoporosis through inhibiting bone marrow mesenchymal stem cell differentiation. Exp Ther Med. 2019;17:803–11. doi: 10.3892/etm.2018.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Wu F, Song Y, et al. Long noncoding RNA related to periodontitis interacts with miR-182 to upregulate osteogenic differentiation in periodontal mesenchymal stem cells of periodontitis patients. Cell Death Dis. 2016;7:e2327. doi: 10.1038/cddis.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, Liu W, Wang H, et al. Mutual inhibition between HDAC9 and miR-17 regulates osteogenesis of human periodontal ligament stem cells in inflammatory conditions. Cell Death Dis. 2018;9:480. doi: 10.1038/s41419-018-0480-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei F, Yang S, Guo Q, et al. MicroRNA-21 regulates osteogenic differentiation of periodontal ligament stem cells by targeting Smad5. Sci Rep. 2017;7:16608. doi: 10.1038/s41598-017-16720-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhen L, Jiang X, Chen Y, et al. MiR-31 is involved in the high glucose-suppressed osteogenic differentiation of human periodontal ligament stem cells by targeting Satb2. Am J Transl Res. 2017;9:2384–93. [PMC free article] [PubMed] [Google Scholar]
- 15.Cao F, Zhan J, Chen X, et al. miR-214 promotes periodontal ligament stem cell osteoblastic differentiation by modulating Wnt/betacatenin signaling. Mol Med Rep. 2017;16:9301–8. doi: 10.3892/mmr.2017.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao S, Zhao W, Ou Q, et al. MicroRNA-214 suppresses osteogenic differentiation of human periodontal ligament stem cells by targeting ATF4. Stem Cells Int. 2017;2017 doi: 10.1155/2017/3028647. 3028647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu L, Liu Y, Wang S. Stem cell-based tooth and periodontal regeneration. Oral Dis. 2018;24:696–705. doi: 10.1111/odi.12703. [DOI] [PubMed] [Google Scholar]
- 18.Han J, Menicanin D, Gronthos S, et al. Stem cells, tissue engineering and periodontal regeneration. Aust Dent J. 2014;59(Suppl 1):117–30. doi: 10.1111/adj.12100. [DOI] [PubMed] [Google Scholar]
- 19.Cao FY, Fan JX, Long Y, et al. A smart fluorescence nanoprobe for the detection of cellular alkaline phosphatase activity and early osteogenic differentiation. Nanomedicine. 2016;12:1313–22. doi: 10.1016/j.nano.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Song H, He P, Shao T, et al. Long non-coding RNA XIST functions as an oncogene in human colorectal cancer by targeting miR-132-3p. J BUON. 2017;22:696–703. [PubMed] [Google Scholar]
- 21.Ernst EH, Nielsen J, Ipsen MB, et al. Transcriptome analysis of long non-coding RNAs and genes encoding paraspeckle proteins during human ovarian follicle development. Front Cell Dev Biol. 2018;6:78. doi: 10.3389/fcell.2018.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han Q, Li L, Liang H, et al. Downregulation of lncRNA X inactive specific transcript (XIST) suppresses cell proliferation and enhances radiosensitivity by upregulating mir-29c in nasopharyngeal carcinoma cells. Med Sci Monit. 2017;23:4798–807. doi: 10.12659/MSM.905370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhan M, He K, Xiao J, et al. LncRNA HOXA11-AS promotes hepatocellular carcinoma progression by repressing miR-214-3p. J Cell Mol Med. 2018 doi: 10.1111/jcmm.13633. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu H, Wang S, Zhu H, et al. LncRNA MT1JP functions as a tumor suppressor via regulating miR-214-3p expression in bladder cancer. J Cell Physiol. 2019 doi: 10.1002/jcp.28274. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 25.Du KT, Deng JQ, He XG, et al. MiR-214 regulates the human hair follicle stem cell proliferation and differentiation by targeting EZH2 and Wnt/beta-catenin signaling way in vitro. Tissue Eng Regen Med. 2018;15:341–50. doi: 10.1007/s13770-018-0118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honardoost M, Soleimani M, Arefian E, et al. Expression change of miR-214 and miR-135 during muscle differentiation. Cell J. 2015;17:461–70. doi: 10.22074/cellj.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Li Y, Luo M, et al. MicroRNA-214 inhibits the osteogenic differentiation of human osteoblasts through the direct regulation of baculoviral IAP repeat-containing 7. Exp Cell Res. 2017;351:157–62. doi: 10.1016/j.yexcr.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Wang CG, Liao Z, Xiao H, et al. LncRNA KCNQ1OT1 promoted BMP2 expression to regulate osteogenic differentiation by sponging miRNA-214. Exp Mol Pathol. 2019;107:77–84. doi: 10.1016/j.yexmp.2019.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Wang C, Qi S, Xie C, et al. Upregulation of long non-coding RNA XIST has anticancer effects on epithelial ovarian cancer cells through inverse downregulation of hsa-miR-214-3p. J Gynecol Oncol. 2018;29:e99. doi: 10.3802/jgo.2018.29.e99. [DOI] [PMC free article] [PubMed] [Google Scholar]