Abstract
TEAD proteins constitute a family of highly conserved transcription factors, characterized by a DNA-binding domain called the TEA domain and a protein-binding domain that permits association with transcriptional co-activators. TEAD proteins are unable to induce transcription on their own. They have to interact with transcriptional cofactors to do so. Once TEADs bind their co-activators, the different complexes formed are known to regulate the expression of genes that are crucial for embryonic development, important for organ formation (heart, muscles), and involved in cell death and proliferation. In the first part of this review we describe what is known of the structure of TEAD proteins. We then focus on two members of the family: TEAD1 and TEAD2. First the different transcriptional cofactors are described. These proteins can be classified in three categories: i), Cofactors regulating chromatin conformation, ii), Cofactors able to bind DNA, and iii), Transcriptional cofactors without DNA binding domain. Finally we discuss the recent findings that identified TEAD1 and 2 and its coactivators involved in cancer progression.
Keywords: TEAD, TEA, Transcription Factor, Cancer, VGLL, YAP
1. Introduction
The TEAD family of transcription factors was first identified through the purification and cloning of the first mammalian TEF factor, TEF1 (TEAD1). TEAD1 was originally shown to regulate the transcription of the early and late promoters of the simian virus 40 (SV40). Specifically TEAD1 can bind to the GT-IIC and Sph enhansons of the SV40 enhancer (Davidson et al., 1988; Xiao et al., 1991). In vertebrates the TEAD family consists of four members, TEAD1 (TEF-1, NTEF-1), TEAD2 (ETF, ETEF-1, TEF-4), TEAD3 (DTEF-1, TEF5, ETFR-1), and TEAD4 (RTEF1, TEF-3, ETFR-2, FR-19) (Table 1).
Table 1.
Characteristics of the TEAD genes. Based on NCBI, CCDS, MGI and Ensembl databases. Values indicated in the table correspond to the principal isoform. Indicated alternative isoforms correspond to the principal isoforms (APPRIS) annoted into the Ensembl database.
| Species | Gene | Aliases | Chromosome | CCDS code | Ensembl transcript principal isoform. | Ensembl transcript ID-Alternative isoforms | Pre-spliced transcript lenght(kb) |
Spliced transcript lenght (nt) | Exons | Coding exons |
protein lenght (aa) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | TEAD1 | TEAD-1, AA, REF1, TEF-1, TCF-13, TCF13, NTEF-1 | 11 | 7810.2 | ENST00000527636.5 | ENST00000526600.1; ENST00000361985.6; ENST00000334310.10; ENST00000598810.5; ENST00000601519.5; ENST00000311227.6; ENST00000593945.5; ENST00000402886.7 | 263.44 | 2544 | 13 | 11 | 426 |
| TEAD2 | ETF, TEF-4, TEF4, TEAD-2 | 19 | 58671.1 | ENST00000377214.8 | 19.84 | 2440 | 11 | 11 | 450 | ||
| TEAD3 | TEAD-3, TEF-5, DTEF-1, TEF5, TEAD5, ETFR-1 | 6 | 47414.1 | ENST00000338863.11 | 23.48 | 2983 | 13 | 12 | 435 | ||
| TEAD4 | RTEF-1, hRTEF-1B, TCF13L1, TEF3, TEF-3, TEFR-1, EFTR-2, RTEF1 | 12 | 31729.1 | ENST00000359864.6 | ENST00000358409.6; ENST00000397122.6 | 81.26 | 1690 | 13 | 11 | 434 | |
| Mouse | Tead1 | 2610024B07Rik, B230114H05Rik, Gtrgeo5, mTEF-1, Tcf13, TEAD-1, TEF-1 | 7 | 52365.1 | ENSMUST00000059768.16 | ENSMUST00000069256.1; ENSMUST00000106638.8; ENSMUST00000084705.11; ENSMUST00000164363.7; ENSMUST00000165036.7 | 227.49 | 9964 | 14 | 13 | 436 |
| Tead2 | ETF, Etdf, TEF4, TEF-4, TEAD-2 | 7 | 71957.1 | ENSMUST00000097216.3 | ENSMUST00000097216.3 | 15.71 | 1233 | 11 | 11 | 410 | |
| Tead3 | TEAD-3, TEF-5, Tcf13r2, DTEF-1, ETFR-1 | 17 | 28577.3 | ENSMUST00000080572.13 | ENSMUST00000114799.7; ENSMUST00000154873.7; ENSMUST00000156862.1 | 19.14 | 2616 | 13 | 12 | 439 | |
| Tead4 | ETFR-2a, Rtef1, TEF-3, Tef3, TEAD-4, ETFR-2, Etfr2, FR-19, Tefr1, Tcf13r1, Tefr, Tefr1a | 6 | 39646.1 | ENSMUST00000006311.12 | ENSMUST00000112157.3; ENSMUST00000130454.7 | 73.67 | 3076 | 12 | 11 | 427 | |
| Rat | Tead1 | TEF-1 | 1 | - | ENSRNOT00000090042.1 | ENSRNOT00000021020.4 | 137.44 | 1607 | 13 | 13 | 436 |
| Tead2 | 1 | - | ENSRNOT00000028086.4 | ENSRNOT00000090838.1 | 15.88 | 2135 | 12 | 11 | 445 | ||
| Tead3 | LOC294299 | 20 | - | ENSRNOT00000000607.6 | 22.70 | 5874 | 13 | 12 | 372 | ||
| Tea4 | 4 | ENSRNOT00000050834.6 | 47.09 | 5253 | 11 | 11 | 361 |
TEAD proteins share a highly conserved DNA binding domain (DBD) called the TEA domain (Andrianopoulos and Timberlake, 1991). The TEA domain (77 amino acids) is also referred to as the ATTS domain because of the 4 proteins first found to harbor this motif (AbaA andTEC1 in yeast, TEF1/TEAD1 in vertebrates and Scalloped in fly)(Campbell et al., 1992). AbaA regulates the development of the asexual spores in Aspergilus nidulans and terminates vegetative growth, (Andrianopoulos and Timberlake, 1994), TEC1 is involved in the activation of the Ty1 retrotransposon in Saccharomyces cerevisiae, (Laloux et al., 1990) and the Drosophila gene scalloped (sd) plays important roles during wing development (Bray, 1999). The comparison of the TEA domain in different species (from C. elegans to human) reveals a remarkable degree of conservation (Anbanandam et al., 2006). The high degree of conservation observed among TEAD proteins indicate the important adaptive role played in different organisms. (Table 2 and 3)
Table 2.
TEAD1 paralogs. Amino acid and nucleotide comparison was extracted from the NCBI’s Homologene database (http://www.ncbi.nlm.nih.gov).
| Gene | Identity to humain TEAD1 (%) | ||
|---|---|---|---|
| Species | NCBI reference sequence | Protein | DNA |
| P. troglodytes | NP_068780.2 | 100.0 | 99.7 |
| M. mulatta | XP_001171551.1 | 98.0 | 97.5 |
| C. lupus | XP_002799676.1 | 99.5 | 94.7 |
| B. taurus | XP_005216143.1 | 100.0 | 95.7 |
| M. musculus | NP_001160056.1 | 99.3 | 91.8 |
| R. norvegicus | NP_001185518.1 | 99.5 | 92.3 |
| G. gallus | NP_001186334.1 | 98.5 | 87.7 |
| X. tropicalis | XP_002943057.2 | 95.6 | 81.3 |
| D. rerio | XP_005172421.1 | 91.7 | 80.2 |
| D. melanogaster | NP_001259581.1 | 69.6 | 63.2 |
| A. gambiae | XP_003437016.1 | 71.5 | 65.1 |
| C. elegans | NP_495186.3 | 45.8 | 51.0 |
Table 3.
TEAD2 paralogs. Amino acid and nucleotide comparison was extracted from the NCBI’s Homologene database (http://www.ncbi.nlm.nih.gov).
| Gene | Identity to humain TEAD2 (%) | ||
|---|---|---|---|
| Species | NCBI reference sequence | Protein | DNA |
| P. troglodytes | XP_512815.4 | 95.3 | 96.3 |
| M. mulatta | XP_001114808.1 | 99.3 | 97.9 |
| C. lupus | XP_001114808.1 | 97.5 | 91.5 |
| B. taurus | XP_005616375.1 | 96.9 | 91.2 |
| M. musculus | NP_001272427.1 | 93.7 | 86.1 |
| R. norvegicus | NP_001100982.1 | 93.9 | 86.7 |
The translation of several TEAD family members may be subject to control as the translation of TEAD1, TEAD3, and TEAD4 are initiated at isoleucine (AUU), leucine (UUG), and isoleucine (AUA) codons, respectively, that lie upstream of the first methionine codon (Jiang et al., 1999; Stewart et al., 1996; Xiao et al., 1991 ). In TEAD2, the methionine (AUG) codon is used for the initiation of translation (Jiang et al., 1999; Yasunami et al., 1995 ).
Several studies characterized the tissue distribution of TEADs (Azakie et al., 2005; Azakie et al., 1996; Jacquemin et al., 1996; Stewart et al., 1996; Xiao et al., 1991; Yasunami et al., 1995; Yasunami et al., 1996; Yockey et al., 1996 ). TEAD1, TEAD3, and TEAD4 are widely expressed in multiple tissues including the skeletal muscle, pancreas, placenta, lung, and heart. In contrast, TEAD2 is selectively expressed in a subset of embryonic tissues including the cerebellum, testis, and distal portions of the forelimb and hindlimb buds as well as the tail bud, but it is essentially absent from adult tissues (Yasunami et al., 1995). TEAD2 expression in the embryo is detectable as early as the 2-cell stage (Kaneko et al., 1997). In this review, we summarize the evolutionary, structural and functional features of the mammalian TEAD1 and TEAD2 proteins and provide some clues on TEAD regulation in both development and cancer.
2. The structure of TEAD family
2.1. Functional domains of TEAD Factors
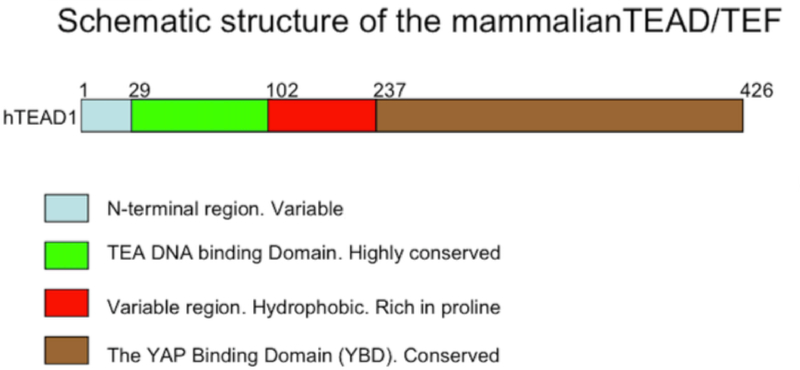
The TEAD proteins are closely related not only in the TEA domain but also in the C-terminal domain called the YAP (Yes/src associated protein) binding domain (YBD), whereas the N-terminal region preceding the TEA domain and the proline-rich region following the TEA domain are more variable. The most conserved region is the TEA domain. The human TEAD factors are more than 99% identical in the DBD (Yoshida, 2008). The C-terminal domain that contains the YAP binding domain (YBD) is also highly conserved (figure 1) (Chen et al.).
Figure 1.
The overall structure of the mammalian TEAD factors is schematized. The domains are defined both by their conservation among the family members and by their amino acid compositions. The numbers represent the amino acid coordinates of each domain in the prototype member TEAD1. The corresponding amino acid coordinates in the TEAD2, TEAD3, and TEAD4 factors are similar.
2.2. The TEA/ATTS DNA binding domain
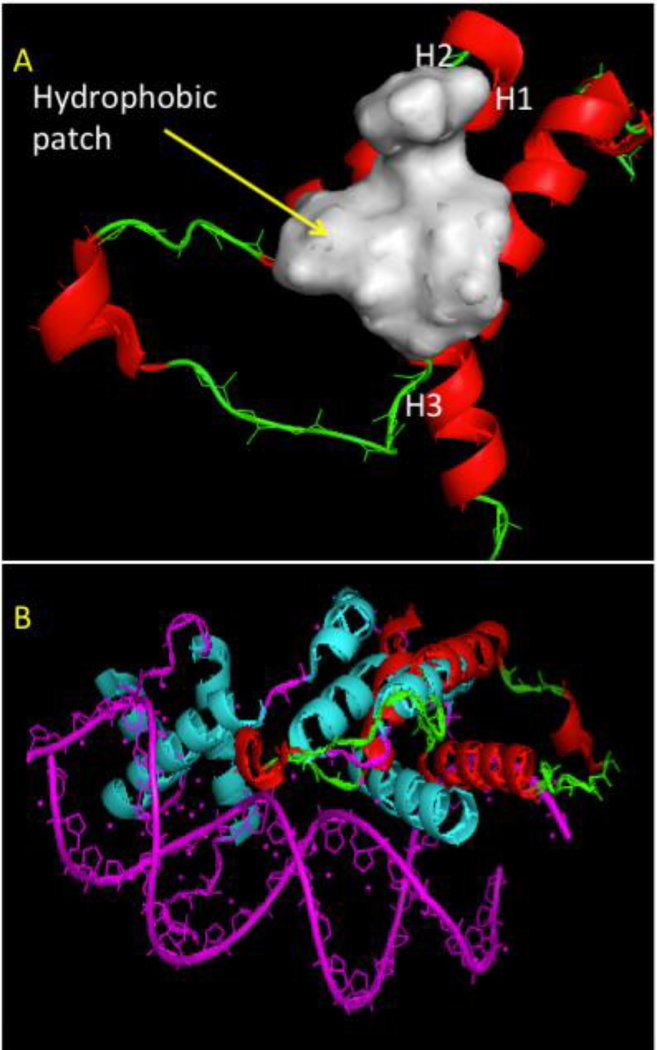
The three dimensional (3D) structure of the TEA domain has been elucidated. This domain is composed of a three-helix bundle with a homeodomain fold (Anbanandam et al., 2006). Structure-function correlations have shown that the L1 loop is essential for cooperative binding of TEAD molecules to tandem duplications of MCAT sites (Anbanandam et al., 2006).
The TEA domain has a folded globular structure made of three α-helices, H1, H2, and H3. H1 and H2 are nearly anti-parallel and pack on either side of the H3. This domain consists of 28 hydrophobic residues with only 12 residues with few hydrophobic contacts composing its core. Subsequently, the TEA domain was predicted to have a low thermodynamic stability. Indeed, the TEA domain unfolds irreversibly with a mid-point of urea denaturation of 2.5M (Anbanandam et al., 2006). The H1-H2 contact creates a hydrophobic patch that consists of 5 amino acids, I23, Y24, L46, Y50, L53 (figure 2A). This surface is likely to be crucial in protein-protein interactions.
Figure 2.
The 3D structure of the TEA domain. A. Hydrophobic surface patch of TEA domain. (B) Superimposition of TEA domain (PDB ID 2HZD) with a structurally homologous protein (MatA1, PDB ID 1AKH, Cyan) bound to DNA (Magenta). TEA: green (Loops) and red (Helices)
The TEA domain binds DNA with high affinity (Anbanandam et al., 2006). The consensus DNA sequence bound by the isolated TEA domain is N[A/T/G]G[AT/C]ATNT and differs from the MCAT sequence. This suggests that other domains of the full-length TEAD proteins participate in binding specificity, perhaps by inducing conformational changes in the TEA domain. Indeed, Jiang et al. have shown that alternative splicing of TEAD1 mRNA in regions immediately after the TEA domain altered its DNA binding properties (Jiang et al., 2000).
The DNA recognition surface is located in the H3 helix and contains three important serines (Anbanandam et al., 2006). This is in agreement with biochemical data showing that the phosphorylation of Ser-102 by protein kinase A (Gupta et al., 2000) or of Ser-91 by protein kinase C diminishes DNA binding activities (Jiang et al., 2001).
2.2.1. Structural conservation of TEA domain
Using DALI, a web server for conservation mapping in 3D (Holm and Sander, 1993, 1995, 1999), we found that the TEA (PDB entry: 2hzd) domain is structurally homologous to about 329 PDB entries from bacteria to mammals ranked by z-score of the statistical significance of the homology (supplemental table 1). It’s important to note that the biological significance of these homologies needs to be examined from case to case. The percentage of structurally homologous amino acids (SHAA) ranges from 3.7 to 95 percent of the hits. Gene ontology analysis using DAVID (Huang et al. 2009), showed that the hit proteins cover a large panel of protein functions and cellular localization. Interestingly, the transcription factors are enriched in homeodomain structures (supplemental table 1). Comparison of TEA domain to the DNA bound homeodomain protein MatA1 showed that they share 90% of structurally homologous amino acids (supplemental table 1) and bind DNA in a similar way (Figure 2B) (Anbanandam et al. 2006). Altogether, this indicates that TEA is a homeodomain structure further confirming the two-decade-old hypothesis of Davidson et al. (Davidson et al., 1988; Xiao et al., 1991); more details in (Anbanandam et al., 2006).
2.2.2. Functional dissection of the TEA domain
The TEA domain was formally identified as a DBD by functional dissection of the human TEAD1 factor (Hwang et al., 1993). The consensus DNA binding site of the TEAD family is 5’-CATTCCA/T-3’ and is called the MCAT element (Cooper and Ordahl, 1985; Farrance et al., 1992 ). TEAD family members bind the double-stranded form of the MCAT element, but not the single-stranded one (Carlini et al., 2002). In contrast to many transcription factors that bind cooperatively to palindromic sites, several members of the TEAD family have been shown to bind cooperatively to tandem repeats, but non-cooperatively to spaced or inverted repeats (Davidson et al., 1988; Jacquemin et al., 1996). The ability to bind cooperatively to tandem, but not spaced, repeats is an intrinsic property of the TEA domain. This cooperativity is required for efficient binding to regulatory elements, such as the SphI+II enhansons of the SV40enhancer or the DF-3 element enhansons in the human Chorionic Somatomammotropin-B (hCS-B) gene enhancer, which are composed of tandemly repeated low-affinity binding sites (Davidson et al., 1988; Jacquemin et al., 1996).
In addition to DNA binding, the TEA domain is also a target of the SV40 large T antigen (TAg) oncoprotein, which interacts with this domain and may modulate the DNA-binding and/or transcriptional properties of TEAD1 (Berger et al., 1996). Interestingly, a single amino acid change in TAg (S189N), which disrupts interaction with the TEA domain, also results in defective transformation function. This suggests that the interactions between TAg and TEA domain factors are involved in cellular transformation and gives interesting cues regarding the regulation of TEAD factors activity.
2.3. The YAP binding domain (YBD)
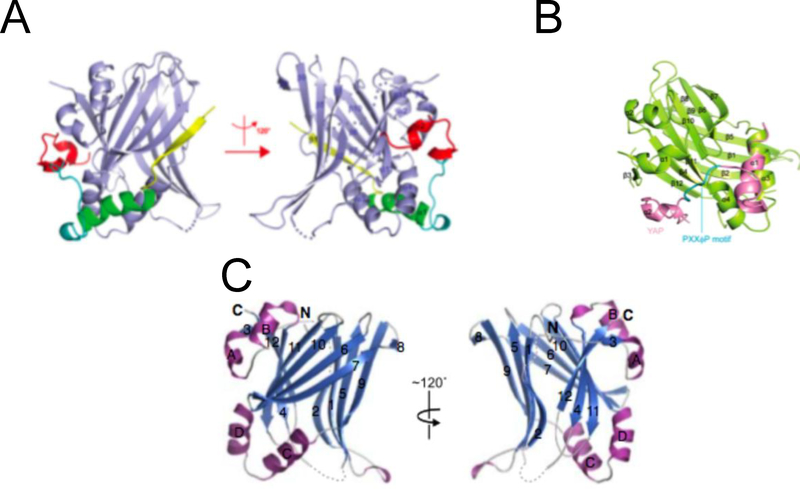
Another important highly conserved domain in TEAD is the C-terminal region. This part of the protein has been shown to mediate interaction with the transcriptional coactivator YAP and was historically called YAP binding domain (YBD). The YBD 3D structure from human TEAD1 and 2 and mouse TEAD4 has been described (Anderson et al.; Li et al.; Tian et al., ). The three structures are strikingly similar and reflect the conservation of the YBD through evolution. The YBD adopts an immunoglobulin-like structure and is composed of 12 β strands and four α helices (Figure 3).
Figure 3.
The 3D structure of YBD from (A) Li et al, 2010. (B) Chen et al, 2010. The YBD was crystallized together with the TEAD binding domain of YAP (Yes/Src associated protein kinase). (C) Tian et al, 2010.
The β strands form two β sheets that pack against each other to form a β sandwich with one β sheet composed of strands β1, β2, β5,β8, β9 and the other consisting of β3, β4, β6, β7, β10, β11, β12. The α helices form two helix-turn-helix motifs where each one connects two β strands.
2.4. The Proline-rich domain
Although the Proline-rich region is not conserved at the primary sequence level, all TEAD family members share a Proline-rich domain (16–25% of conservation). The Proline-rich domain of rat TEAD1 is only 20% identical with that of rat TEAD4 (Mahoney et al., 2005). The Proline-rich domain is also required for full interaction with the transcriptional coactivators YAP and TAZ (Vassilev et al., 2001). Therefore, the Proline-rich domain probably accounts for the differential interaction of the TEAD proteins with YAP/TAZ.
2.5. The N-terminal region
The N-terminal region of TEAD proteins is amongst the less conserved in the TEAD family. The TEAD1 N-terminal region has a net negative charge and contains a high concentration of Serines, which are potential phosphorylation sites, and is also required for TEAD1 interaction with the transcription factor MAX (Gupta et al., 2000). Furthermore, the full transcriptional activation by TEAD1 requires its N-terminal region to synergize with the Proline rich-region and the YBD, probably by forming a functional transactivation surface (Hwang et al., 1993).
3. Cofactors of TEAD1/2 proteins
The observation that TEAD proteins cannot by themselves induce target genes transcription led to the demonstration that they require cofactors to do so (Xiao et al., 1991). Xiao et al found that expression of a TEAD1 reporter was repressed in HeLa cells both when TEAD1 or a TEAD protein lacking the TEA domain were overexpressed, suggesting that the overexpressed proteins titrated a cofactor required for transcriptional activity.
Thereafter many cofactors that interact with TEAD1 and TEAD2 to regulate gene expression were identified. They can be classified as:
cofactors regulating chromatin conformation,
other DNA-binding transcription factors,
cofactors that cannot bind to DNA but provide transcriptional activity
3.1. Cofactors regulating chromatin conformation
3.1.1. SRC
The p160 family of steroid receptor coactivators (SRCs) consists of three evolutionary conserved coregulators of transcription: SRC-1 (NCOA-1, RIP160), SRC-2 (NCOA-2, TIF-2, GRIP-1), and SRC-3 (NCOA-3, AIB-1, TRAM-1, pCIP). These proteins are critical components of transcriptional complexes of many nuclear receptors and are able to potentiate the activity of transcription factors. Furthermore, the SRC family plays a role of pleiotropic “master regulators” of steroid hormone receptor, including estrogen receptor (ER) and androgen receptor (AR), and are necessary for cell proliferation, survival and metabolism.
SRC can bind to histone acetyltransferases and methyltransferases through a highly conserved domain, and unwind chromatin to allow access and rapid formation of hierarchical protein assemblies which form the transcription preinitiation complex.
SRCs belongs to another class of transcriptional regulators that harbor a basic helix-loop-helix-Per-Arnt-Sim (bHLH-PAS) domain. This domain mediates diverse functions such as DNA binding, homodimerization and heterodimerization of proteins, signal sensing and transduction.
In a yeast two-hybrid screen using the bHLH-PAS domain of SRC1 as bait and GST pull-down assays, Belindia and Parker revealed that TEAD1 and 2 interact with SRC1. Functional assays in HeLa cells demonstrated that all members of the SRC family could be coactivator of TEAD proteins. Nevertheless, the repression observed when a TEAD factor is overexpressed is not abolished by SRC1 co-transfection, indicating that other limiting cofactors are needed (Belandia and Parker, 2000).
3.1.2. PARP
PARP proteins (Poly-ADP ribose polymerase) are nuclear proteins that can associate with the chromatin and are able to create posttranslational modifications by inducing the synthesis of a poly (ADP-ribose) chain when activated. PARPs are implicated in multiple processes including DNA repair, replication regulation, recombination control, control of cell death and differentiation and regulation of transcription.
PARPs can bind to specific DNA sequences next to the 5’ of the MCAT site of a set of genes expressed in muscles. Moreover it has been shown that inhibition of PARPs creates a strong downregulation of the transcriptional activity of MCAT sequences in muscle cell lines (Butler and Ordahl, 1999). Furthermore, in vitro studies showed that TEAD1 interacts with PARPs to regulate smooth muscle α-actin expression. Thus, PARPs and TEADs form a complex that could positively affect the expression of genes in muscle. Other experiments showed that PARP can ADP-ribosylate TEAD proteins and make the chromatin context favorable to transcription through histone modification.
3.2. Cofactors able to bind DNA
3.2.1. SRF
Serum response factor (SRF) is a member of the MADS (MCM1/Agamous/Deficiens/SRF) box transcription factor family. This protein regulates the activity of many immediate-early genes, for example c-fos, and thereby participates in cell cycle regulation, apoptosis, cell growth, and cell differentiation. SRF is a weak transcriptional activator and needs to physically interact with other tissue-specific coactivators to regulate gene expression.
SRF regulates expression of smooth muscle-specific genes during embryonic development and later on. Smooth muscle is a good example of SRF interaction with cofactors. Indeed, in smooth muscle, SRF binds to bHLH proteins from the MyoD family and other cofactors such as myocardin to regulate gene expression (Gupta et al., 2001).
Moreover, in this tissue, SRF can regulate the smooth muscle α-actin expression, and it is interesting to note that SRF promoter harbor an SRE sequence type (SRF responsive element) near the MCAT domain. Thereby at this genomic location, SRF and TEAD proteins have been shown to interact through their DNA binding domain, respectively the MADS domain and the TEA domain. In vitro studies have demonstrated that this interaction leads to the activation of the smooth muscle α-actin promoter in embryonic vascular smooth muscle cells and myofibroblasts (Maclellan et al., 1994).
However, the interaction of TEAD1/2 and SRF is not required for the promoter activity in adult differentiated vascular smooth muscle cells. Interestingly in adult differentiated vascular smooth muscle cells TEAD1 competes with myocardin for binding to SRF, resulting in a disruption of myocardin and SRF interactions and thereby attenuating expression of smooth muscle-specific genes. In these cells TEAD1 interacts with and titrates its cofactors, thus acting as an inhibitor to induce differentiation.
3.2.2. MEF2
The myocyte enhancer factor 2 (MEF2) is a transcription factor that is able to regulate cell differentiation, proliferation, morphogenesis, survival and apoptosis of a wide range of cell types. MEF2 belongs to a conserved family of proteins. Saccharomyces cerevisiae, Drosophila and Caenorhabditis elegans possess a single Mef2 gene, whereas vertebrates have four, Mef2a, b, c and d. This transcription factor was first discovered in drosophila cardiomyocytes. In vertebrates, MEF2 is also expressed in lymphocytes, neural crest, smooth muscle, endothelium and bone, and different teams claim that MEF2 proteins are ubiquitous (Arnold et al., 2007; Black et al., 1997; Edmondson et al., 1994; Martin et al., 1993; McDermott et al., 1993; Pollock and Treisman, 1991; Yu et al., 1992). MEF2 harbors a MADS box domain in N-terminus similar to the one present in SRF. Structurally, a domain called MEF2 is present in the protein next to the MADS domain. Together these two domains enable protein dimerization, DNA binding and interaction with cofactors. TEAD proteins and MEF2 interact physically (Maeda et al., 2002). The first studies conducted in Drosophila showed that the homologue of MEF2 interacts with Scalloped (TEAD1 homologue) to activate transcription of genes expressed in flight muscles (Bernard et al., 2009; Deng et al., 2009). Interestingly, in mammals the mechanism is different and more complex. Indeed in CV-1 cells (African monkey kidney cells), the binding of MEF2 in the DNA induces and potentiates TEAD1 and TEAD2 recruitment at MCAT sequences that are adjacent to MEF2 binding sites. However, co-transfection of MEF2 and TEAD1 in C2C12 myoblasts results in a repression of the MLC2v (Myosin Light Chain 2 v) and βMHC (β-myosin heavy chain) promoters, which MEF2 is otherwise capable of inducing on its own. These results demonstrate a dual action of the complex that seems to be specific to the promoters of different genes. Finally in vivo and in vitro assays demonstrate that the binding sites for MEF2 and TEAD proteins in the myocardin enhancer are required for complete enhancer function during cardiac and smooth muscle development in vivo.
3.2.3. MAX
MAX is a phosphoprotein characterized by a helix-loop-helix domain and a leucine zipper domain (bHLH-LZ). This protein was identified thanks to its interaction with MYC, another bHLH-LZ protein (Blackwood and Eisenman, 1991).
In vitro and in vivo experiments (GST pull-down and immunoprecipitation) demonstrated the binding of MAX to TEAD1. Once this complex is formed, these two proteins can regulate the alpha-myosin heavy chain (α-MHC) gene expression (Gupta et al., 1997). Furthermore, transfection of both proteins allows a 3 to 4 times higher α-MHC enhancer activity compared with that obtained when transfecting each protein alone. These results demonstrate a synergistic effect of the formation of the TEAD1-MAX complex on the activation of the α-MHC enhancer (Gupta et al., 1997).
Finally, in vitro experiments examining the role of the β-myosin heavy chain (βMyHC) distal muscle CAT (MCAT) element in muscle fiber type-specific expression and mechanical overload (MOV) responsiveness revealed the existence of a TEAD1, PARP, and Max complex. This multiprotein complex has been suggested to maintain basal expression level of endogenous βMyHC gene in adult skeletal muscle.
3.2.4. AP-1
The transcription factor activator protein 1 (AP-1) is a heterodimer composed of different proteins belonging to different families such as c-Fox, c-Jun and ATF. This heterodimeric transcription factor directly bind to the DNA and regulates the transcription of genes involved in a wide range of cellular processes including proliferation, apoptosis and cell differentiation.
Recently using Chip-Seq in different types of cancer cell lines two different studies showed a genomic co-occupancy of AP-1 and TEAD proteins (Zanconato et al., 2015; Liu et al., 2016). Besides in-vitro studies demonstrated a direct interaction between this two transcription factors. Furthermore this co-occupancy was correlated with active enhancers regions and promoters and a cooperation between AP-1 and TEAD proteins on the regulatory elements of several target genes was established (Zanconato et al., 2015; Liu et al., 2016). The clustering of the target genes shared by the two transcription factors demonstrated an enrichment in genes associated with cell adhesion, motility, and migration was identified in the Chip-Seq data, suggesting an involvement for the AP-1/TEAD transcriptional complex in promoting tumorigenesis (Zanconato et al., 2015; Liu et al., 2016).
3.3. Transcriptional cofactors without DNA binding domain
3.3.1. YAP and TAZ
Several studies both in mammals and Drosophila have identified YAP and TAZ as essential coactivators of TEAD proteins. By using affinity chromatography strategy, Vassilev and collaborators identified YAP as an essential cofactor of the four TEAD factors and demonstrated that TEAD transcriptional activity requires YAP (Vassilev et al., 2001). TAZ, a transcriptional coactivator paralogous to YAP was then identified as a co-factor able to interact both in vitro and in vivo with all TEAD proteins and to increase TEAD transcriptional activity (Mahoney et al., 2005).
YAP/TAZ are effectors of the Hippo tumor suppressor pathway and restrict organ growth by keeping in check cell proliferation and promoting apoptosis. Components of this pathway were first discovered in Drosophila but are highly conserved in mammals. The core of this pathway is formed by the two kinases MST1/2 and LATS1/2 and the adaptor proteins WW45 and MOB1. When activated, MST1/2 interacts with WW45 to phosphorylate and activate LATS1/2 and MOB which in turn phosphorylate YAP or TAZ. Phosphorylation of YAP/TAZ generates a 14–3-3 binding site allowing its exclusion from the nucleus through association with 14–3-3 proteins (Yu et al., 2015; Zhao et al., 2010).
The interaction between YAP/TAZ and TEAD implicates the N-terminus domain of YAP and the C-terminus domain of TEAD factors (Vassilev et al., 2001). The crystal structures of YAP/TEAD2 and YAP/TEAD4 have both been determined. YAP and TEAD interact through three highly conserved major interfaces where interface 3 is the most critical (Chen et al., 2010a; Chen et al., 2010b; Tian et al., 2010).
Several studies both in Drosophila and in mammals indicated that TEAD factors play an essential role mediating the influence of the Hippo pathway on tissue growth. In Drosophila, it has been shown that induction of tissue growth by Yorkie (YKI) requires Scalloped (Goulev et al., 2008; Wu et al., 2008; Zhao et al., 2008). Using ChIP-on-chip experiments, Zhao et al. have shown that more than 80% of YAP binding sites overlap with TEAD1 binding sites (Zhao et al., 2008). Furthermore, silencing of TEAD1/3/4 by siRNAs in MCF10A cells inhibits hallmarks of oncogenic transformation induced by YAP overexpression, such as anchorage-independent growth or EMT (Zhao et al., 2008). Similar results were obtained with TAZ by using TEAD-binding deficient mutant TAZ (Zhang et al., 2009). Furthermore, in mice, a dominant negative form of TEAD unable to interact with YAP, or small molecules such as Verteporfin that inhibits TEAD/YAP interaction, suppress hepatocarcinoma and hepatomegaly induced by YAP overexpression (Liu-Chittenden et al., 2012).
3.3.2. Vestigial like proteins
The Vestigial-like (VGLL) proteins belong to a family of conserved proteins. The first member of this family, encoded by the gene vestigial (vg), was identified in Drosophila. It was shown that VG interacts with the product of scalloped (sd) gene, the orthologue of TEAD in drosophila, to form a bipartite transcription factor where SD binds DNA and VG activates transcription (Simmonds et al., 1998; Zider et al., 1998). Since then, four Vg-like (VGLL) genes have been identified in mammals whose products are able to interact with all TEAD proteins (Chen et al.). Interestingly, interactions between TEAD and VGLL proteins are mediated through a short domain (25 amino-acids), named TONDU (TDU) domain, strongly conserved between mammalian and Drosophila VG (Vaudin et al., 1999). The Human VGLL1 can substitute for VG during wing development in Drosophila despite lack of conservation at the amino-acid outside of the TDU domain (Vaudin et al., 1999).
The structures of the YAP/TEAD complex and of the VGLL1/TEAD4 complex have been determined (Pobbati et al., 2012). Interestingly, although VGLL and YAP proteins have different amino acid sequences, they share structural similarities and interact with the same interfaces in TEADs (Pobbati et al., 2012). Namely, YAP interacts with TEADs through three interfaces (Chen et al., 2010b; Li et al., 2010; Tian et al., 2010) and two of these (interfaces 1 and 2) are involved in the interaction with VGLL (Pobbati et al., 2012). In accordance with these results there is a competition between VGLL1 and YAP to bind to TEAD (Pobbati et al., 2012).
The precise function of the interactions between TEADs and VGLLs is still poorly understood. It has been shown that TEAD/VGLL1 complexes promote anchorage-independent cell proliferation in prostate cancer lines suggesting a role in cancer progression (Pobbati et al., 2012). VGLL2 is expressed in skeletal muscles and plays a role in muscle differentiation (Chen et al., 2004a; Gunther et al., 2004). It has been shown that VGLL2 expression increases during muscle differentiation in mice. Importantly, VGLL2 activates TEAD1-dependent muscle promoter upon C2C12 differentiation and enhances MyoD-mediated myogenic in 10T1/2, implicating VGLL2 as a cofactor of TEAD1 (Chen, Maeda, Mullett, & Stewart, 2004) (Gunther et al., 2004).
The mechanism whereby VGLL2 affects TEAD activity is still not fully understood but evidence suggests that VGLL2–2 can alter the DNA binding activities of different TEAD factors (Chen et al., 2004a).
VGLL4 is the only VGLL factor that contains 2 TDU domains and may be involved in TEADl-related gene regulation in the heart (Chen et al., 2004b). TEAD1 is involved in cardiac development and TEAD1 homozygous mutant mice die at embryonic day 11.5 (E11.5) with cardiac abnormalities (Chen et al., 1994). Interestingly, TEAD1 has been implicated in the activation of cardiac muscle fetal genes in response to hypertrophic signals and VGLL4 prevents alphal-adrenergic activation of gene expression in cardiac myocytes (Chen et al., 1994).
Recent data have shown that VGLL4 probably plays an essential role in TEAD transcriptional activity. In Drosophila, TGI, the homolog of VGLL4, confers a repressor activity to SD (Koontz et al., 2013). Importantly, YKI, the transcriptional effector of the Hippo pathway in Drosophila competes with TGI for SD binding and relieves SD/TGI-mediated repression (Koontz et al., 2013). This repressive role is likely conserved in mammals since VGLL4 repressed TEAD/YAP transcriptional activity (Koontz et al., 2013). Furthermore, when overexpressed in mouse liver, YAP induces a hepatomegaly associated with hepatocarcinoma development (Dong et al., 2007). This effect of YAP is linked to its interaction with TEAD and the disruption of this interaction inhibits YAP oncogenic hypertrophy and activity (Liu-Chittenden et al., 2012). By using double transgenic mice allowing overexpression of both YAP and VGLL4, it has been shown that VGLL4 suppresses YAP-induced hepatomegaly and hepatocarcinoma (Koontz et al., 2013). This new role of VGLL4 as a key player in TEAD repressive function suggests that in Drosophila and perhaps in mammals, TEAD/VGLL4 acts as a default transcriptional repressor and its interaction with other cofactors (YAP or VGLL1–3) could switch TEAD from a repressor to an activator state.
4. Normal and pathological function of TEAD1/2
The role of TEAD proteins is important during development and their expression is detected as early as the 2-cell stage in the mouse embryo. (Wang and Latham, 2000). TEADs have different functions and are required for cardiogenesis (Chen et al., 1994), myogenesis (Yoshida, 2008), the development of the neural crest (Milewski et al., 2004), notochord (Sawada et al., 2005) and trophectoderm (Yagi et al., 2007), but also play a role in cancer (Pobbati and Hong, 2013). TEAD genes have a distinct but not mutually exclusive expression patterns. Almost all tissues express at least one TEAD gene, some express all four of them. In this section we will focus on TEAD1/2 functions in normal and pathological conditions in mammals.
4.1. Physiological roles of TEAD1 and TEAD2 in mammal
4.1.1. TEAD1
TEAD1 loss-of-function is embryonic lethal in the mouse, due to severe heart defects (Chen et al., 1994). Histological examination of these embryonic hearts revealed an abnormally thin ventricle wall with a reduced number of trabeculae, demonstrating that TEAD1 is necessary for heart formation. It has been shown that TEAD1 facilitates the expression of cardiac-specific genes, suggesting that TEAD1 is important for myocardium differentiation. However in TEAD1 null mutant embryos only cardiac muscle growth is affected, cardiogenesis is initiated and the expression of several potential TEAD1 targets appears to be normal, indicating that differentiation is not impaired.
Beside cardiac genes, TEAD1 also regulates several genes involved in mouse, as well as porcine, skeletal and smooth muscle formation and function (X. Xu et al., 2008). TEAD1 can bind the consensus MCAT sequence (5’-CATTCCT-3’) (Jiang et al., 2000) in the promoter of smooth muscle gene (α-actin of smooth muscles) (Swartz et al., 1998), skeletal muscles genes (α-actin of skeletal muscles) (Benhaddou et al., 2012; Karns et al., 1995), α and β myosin heavy chain (Rindt et al., 1993), as well as the cardiac muscular genes troponin T and I (Mar and Ordahl, 1988). Interestingly, a recent study showed in a mouse skeletal myoblast cell line (C2C12 cells) a role of TEAD1 in myoblast differentiation (Wang et al., 2013). Moreover, a ChIP-on-chip assay identified new target genes of TEAD1, such as FoxO3a (Qiu, Wang, Liu, Xu, & Liu, 2011) Mrpl21, Ndufa6 and Ccne1. Up-regulation of Mrpl21 (involved in cellular differentiation) and downregulation of Ccne1 (involved in cellular proliferation) are consistent with TEAD1 regulating C2C12 differentiation via a suppression of proliferation.
Gain-of-function experiments showed that a sustained increase of TEAD1 protein in mouse induces an age-dependent heart dysfunction (Tsika et al., 2010). Moreover the anatomical analysis of hearts coming from mice overexpressing TEAD1 revealed a decrease in the left ventricular power output that correlated with increased βMyHC (β myosin heavy chain) protein as well as decreases in p-GSK-3β and nuclear β-catenin. Finally, mice overexpressing TEAD1 do not tolerate stress as they die over a 4-day period after surgical induction of pressure overload. These studies provide the first in vivo evidence that increased TEAD1 can induce defects typical of cardiac muscle repairs (remodeling) consecutive to cardiomyopathy and heart failure.
4.1.2. TEAD2
TEAD2 is the most abundant TEAD protein expressed during the first seven days of development in mouse. TEAD2 RNA has been observed as early as the 2-cell-stage during embryonic development. Specific increase in polysomal abundance of mouse TEAD2 mRNA is coincident with the first appearance of TEADs activity in the early embryo (Wang and Latham, 2000). Later during development TEAD2 regulates neural development through the direct control of Pax3 expression (Kaneko et al., 2007). Indeed, Pax3 is an essential gene involved in the migration and cellular differentiation of neural crest cells. TEAD2 and its coactivator YAP are coexpressed with Pax3 in the dorsal neural tube, and a mutation of the TEAD2 binding site in a Pax3 enhancer abolishes neural formation (Milewski et al., 2004). In addition, TEAD2 also regulates Foxa2 expression during the development of the notochord (Sawada et al., 2005), confirming its role in neural development.
Other data suggest an essential role for TEAD2 in neuron proliferation. Indeed, TEAD2 is expressed in proliferating neuroepithelial fetal cells, and not in post-mitotic neurons (Jacquemin et al., 1996). Accordingly, TEAD2 expression decreases in perinatal brain compared to fetal brain at E12.5 (embryonic day 12.5).
Functional roles of TEAD2 were deduced from gene inactivation studies performed in mice. However there were conflicting reports about the results of these experiments. In one study, defects in neural tube closure were observed (Kaneko et al., 2007), whereas in another TEAD2 null embryos appeared normal (Sawada et al., 2008). Interestingly, this last study uncovered functional redundancy between TEAD1 and TEAD2, suggesting that TEAD genes can compensates each other’s absence or reduction in some embryonic tissues. Although Tead2−/− mutants appeared normal, Teadl−/−; Tead2−/− double mutant embryos showed general growth retardation and severe morphological abnormalities starting at E8.5, indicating that TEAD1 and TEAD2 have redundant functions in development. The Teadl−/−; Tead2−/− embryos displayed defects in mesoderm development, especially in the notochord. Interestingly, double mutant embryos showed also a reduction of cellular proliferation and increased apoptosis, suggesting the implication of TEAD1 and 2 in both processes.
4.2. TEAD1 and TEAD2 roles in cancer
Deregulated cell proliferation and apoptosis are two significant aspects of neoplasia. Given the implication of TEAD1 in both phenomena, its involvement in several types of cancer has been investigated (Table 4).
Table 4.
Summary of TEAD family expression in malignancies. When known cofactors, as well as target genes, are reported.
| Cancer Type | TEAD gene | Expresion in Cancer | Target genes | Cofactors | Reference |
|---|---|---|---|---|---|
| Pancreatic cancer | TEAD1 | Up-regulated | mesothelin | Hucl et al., 2007 | |
| Prostate cancer | TEAD1 | Up-regulated | Knight et al., 2008) | ||
| Kaposi sarcoma | TEAD1 | Up-regulated | www.ebi.ac.uk/gxa | ||
| Basal-like breast cancers | TEAD4 | Up-regulated | Han et al., 2008; | ||
| Breast cancer | TEAD1 | down-regulated | Landin Malt et al., 2012 | ||
| Breast cancer | TEADs | Up-regulated | CTGF/CYR61/Myc | TAZ/YAP/AP1 | Zanconato et al. 2015 Lamar, J.M. 2012 |
| Fallopian tube carcinoma | TEAD4 | Up-regulated | Nowee et al., 2007 | ||
| Germ cell tumors | TEAD4 | Up-regulated | Skotheim et al., 2006 | ||
| Renal cancer | TEAD1 | down-regulated | Landin Malt et al., 2012 | ||
| Bladder cancer | TEAD1 | down-regulated | Landin Malt et al., 2012 | ||
| Endometrial cancer | TEAD1 | Up-regulated | Livin/BIRC1 | ?/YAP1 | Landin Malt et al., 2012 and 2013 |
| Ovarian cancer | TEADs | Up-regulated | YAP1 | Xia et al., 2014 | |
| Cutaneous Melanoma | TEAD1/4 | Up-regulated | YAP1/AP1 | Yuan et al., 2015/ Verfaillie et al., 2015 | |
| Liver cancer | TEAD1 | Up-regulated | AXL receptor kinase | YAP1 | Xu, M.Z. et al. 2011, Liu-Chittenden, Y et al. 2012 |
| Colon cancer | TEAD4 | Up-regulated | vimentin | YAP1 | Liu, Y. et al 2015 |
| Gastric cancer | TEAD1/4 | Up-regulated | VGLL4/YAP | Li et al., 2015 |
4.2.1. Pancreatic cancer
In 2007 Hucl et al. identified a TEAD1 binding site next to the mesothelin (MSLN) gene promoter and demonstrated that TEAD1 regulates MSLN expression. Furthermore MSLN was found to be a nearly ubiquitous, diagnostically and therapeutically useful marker for pancreatic cancer (Hucl et al., 2007), underlining the relevance of studying the role of TEADs in oncogenesis. Interestingly, in this context too TEAD1 did not seem to act on its own. TEAD1 gain and loss-of-function experiments in cultured cell lines both result in the same inhibition effect on MSLN expression, suggesting the existence of a limiting cofactor needed for MSLN regulation. This cofactor is as yet unknown, but could provide another important marker for pancreatic cancer.
Another study in mouse pancreatic cancer cell lines revealed the existence of several TEAD1 isoforms, due to alternative splicing. These truncated TEAD1 isoforms are closely related to the ones found in human pancreatic cancers (Zuzarte et al., 2000) demonstrating the important contribution of TEAD1 as a marker or a putative therapeutic target.
4.2.2. Prostate cancer
The study of different prostate cancers suggested a correlation between high TEAD1 expression and poor clinical outcome (Knight et al., 2008). Knight et al. notably demonstrated a direct link between TEAD1 expression and cellular proliferation in this type of cancer, suggesting a functional role for TEAD1 in tumor growth.
4.2.3. Other cancers
TEADs expression levels have been reported for different types of cancer. A 300-fold increase in TEAD1 levels has been reported in Kaposi sarcoma (www.ebi.ac.uk/gxa). The increased TEAD expression is also observed in basal-like breast cancers, (Han et al., 2008; Richardson et al., 2006), breast cancer (Lamar et al., 2012; Zanconato et al., 2015) fallopian tube carcinoma (Nowee et al., 2007), liver cancer (M. Z. Xu et al., 2011)(Liu-chittenden et al., 2012), colon cancer (Liu et al., 2015), gastric cancer (Li et al., 2015) and germ cell tumors (Skotheim et al., 2006).
In contrast, TEAD1 expression is downregulated in some other breast cancer types and in renal or bladder cancers (www.oncomine.org).
These contradictory observations are consistent with some of our recent findings that reconcile these seemingly opposing roles of TEADs in cancers. We found that both up and downregulation of TEAD1 indirectly cause an increase in the expression of the anti-apoptotic protein Livin (Landin Malt et al., 2012). Livin belongs to a family of highly conserved intracellular proteins, the Inhibitor of Apoptosis Proteins (IAP), and up-regulation of its expression appears to favor cancer progression. Furthermore, we also demonstrate that another members of this family, BIRC1 or NAIP, is positively regulated by TEAD1 through its interaction with YAP (Landin Malt et al., 2013). Together our results demonstrate that depending on the target gene considered and on the cofactor that TEAD1 binds to, both its up regulation and down regulation can lead to cancer formation.
Two recent reports described an interesting involvement of TEAD1/YAP in ovarian and melanoma tumors. In the first study, the authors successfully isolated ovarian cancer stem-cell-like cells from mouse tumor xenografts that were derived from human ovarian cancer cells, and demonstrated that these cells had cancer stem-cell characteristics (Xia et al., 2014). They demonstrated that YAP and TEAD1 promoted self-renewal of ovarian cancer-initiated cell (OCIC). Moreover, they observed that YAP and TEADs were required to maintain the expression of specific genes possibly involved in OCICs’ sternness and chemoresistance. These results suggested that YAP and TEADs were coactivated in cancer stem cells and that their coordinated interactions may have a key role in cancer stem cell pluripotency and self-renewal regulation.
The second study performed on human Cutaneous Melanoma (CM) showed that the survival of CM patients was associated with specific genetic variations in genes belonging to the Hippo pathway, including variants in YAP, TEAD1 and TEAD4 (Yuan et al., 2015). This is the first report suggesting that genetic variants of Hippo pathway genes, particularly YAP, TEAD1 and TEAD4, may independently or jointly modulate survival of CM patients.
However, other cofactors could be involved in TEAD’s role in tumor progression. Using a genome-wide approach on human melanoma tumor biopsies, a recent landmark study showed for the first time that AP-1 and TEAD share many of their targets in invasive type melanoma cells, indicating that these factors may act cooperatively to regulate gene expression. (Verfaillie et al., 2015). Based on these results, the authors suggest that the AP-1/TEAD complex could be considered as a new master regulator of the invasive gene network.
4.2.4. Cancer therapy
The 3D structures of TEAD-YAP complexes have recently been studied. Three papers have been published in the same year reporting the structural characterization of TEAD alone or TEAD-YAP complexes showing the importance of these factors for the scientific community. The available 3D structures are the human TEAD2 YBD (Tian et al., 2010), mouse TEAD4/YAP complex (Chen et al., 2010), and human TEAD1/YAP2 complex (Li et al., 2010). Furthermore, these studies reported the impact of introducing mutations at the interface allowing the TEAD-YAP interaction and demonstrated its crucial role in mediating the Hippo pathway. This inhibition is expected to have less side effects than classical drug design approaches that focus on inhibition of kinases and membrane receptors. In fact, inhibition/activation of the Hippo pathway at upper levels of the kinase cascade will have an effect on the TEAD-YAP complex but will undoubtedly affect other components of the pathway, leading to deregulation of other pathways and thus resulting in side effects. Conversely, specific regulation of the Hippo pathway at its lowest level, which is the TEAD-YAP complexes, is expected to affect only this complex but no other components of the pathway. Therefore inhibition of this interaction is very relevant to cancer therapy and may open new therapeutic routes by targeting transcription factors effectors of signaling pathways rather than upstream kinases regulators. Pioneering experiments performed by Liu-Chittenden et al. demonstrated that these approaches were relevant (Liu-Chittenden et al., 2012). By using transgenic mice, they showed that expression of a dominant negative form of TEAD1 deleted for its DNA-binding domain was able to suppress hepatomegaly and hepatocarcinoma induced by YAP overexpression, or the bile duct over-proliferation induced by NF2/Merlin knockout in liver. These results indicated that TEAD/YAP interaction was a possible drug target to inhibit the effect of YAP or Hippo deregulation on cancer progression. By screening a drug library, they identified Verteporfin, a photosensitizer belonging to porphyrin molecule family as a strong inhibitor of TEAD/YAP interaction. Most importantly, they demonstrated that treatment of mice with Verteporfin was able to inhibit hepatomegaly and hepatocarcinoma induced by YAP overexpression or NF2/Merlin knockout (Liu-Chittenden et al., 2012). Other small molecules able to disrupt TEAD/YAP complexes have also been identified recently (Pobbati et al., 2015). Pobbati et al. identified a central pocket in TEAD1 that is a target for small molecules. By screening a drug library, they identified Flufenamates, nonsteroidal anti-inflammatory drugs, as small molecules able to disrupt the interaction between TEAD and YAP and to reduce cell proliferation and migration induced by TEAD/YAP (Pobbati et al., 2015).
Another therapeutic approach developed by Zhou and co-workers has been recently described in (Zhou, Z. et al., 2015). It is based on small stabilized cyclic peptides that mimic YAP-like peptides targeting the interface 3 regions of TEADs, which is involved in the interaction with YAP. The aim is to specifically disrupt the interaction between TEAD and YAP (Pobbati et al., 2015). However, one major challenge, before using these cyclic peptides for therapeutic purpose, is to increase their cellular uptake. A similar approach has been developed by Jiao et al. in which they used peptides mimicking the 2 TDU domains present in the VGLL4 protein (Jiao et al., 2014). It has been shown that VGLL4, a member of the VGLL family, is able to compete with YAP for the binding of TEAD and can act as a tumor suppressor in several human cancer such as pancreatic adenocarcinoma (Mann et al., 2012) or gastric cancer (Jiao et al.,2014). They produced small peptides called Super-TDU containing the tandem TDU domains found in VGLL4 and showed that they were able to suppress tumor growth of human gastric cancer cells injected in nude mice, and to inhibit tumor growth in a murine model of gastric cancer induced by H. Pylori infection (Jiao et al.,2014).
5. Post-translational regulation of the TEAD proteins
Post-translational regulation of TEAD1 and 2 proteins has been little studied. Concerning TEAD2 only bioinformatics evidence allows us to identify a putative ubiquitination site at Lys75 and several phosphorylation sites. However, no data is available to validate these sites and identify the function of these post-translational modifications. For TEAD1 some results are available.
5.1. Protein Kinase A (pKA)
β-adrenergic / cAMP signaling induces the expression of the gene encoding α - MyHC through an EM domain containing an E-Box and an MCAT motif (Gupta et al., 1996). Once the AMPc signaling is on, the pKA become active and phosphorylate TEAD1 at Serine 102 downstream of the TEA domain in a non-conserved region of TEAD proteins.
If TEAD1 Serine 102 is mutated, there is no more transcriptional activation of the αMyHC gene. However, when TEAD1 is phosphorylated by pKA there is a considerable reduction of its affinity for DNA, suggesting that the binding of TEAD1 to the MCAT domain has a repressive effect on α - MyHC promoter activity (Gupta et al., 1996).
5.2. Protein Kinase C pKC
Experiments in the BeWo cell line correlated a decrease in pKC activity with an increase in transcriptional activity mediated by the GTIIC site and TEAD1. In contrast, an increase of pKC decreased this transcriptional activity. In vitro assays revealed that pKC phosphorylates TEAD1 on serine and threonine next to the last alpha loop in the TEA domain. Thus it has been hypothesized that pKC phosphorylates TEAD1 and decreases its binding to the GTIIC enhancer (Jiang et al., 2001).
5.3. Palmitoylation
Recent studies uncovered a new post-transcriptional modification of the TEAD proteins. S-palmitoylation is a much conserved process that exists in all eukaryotic organisms and consists in the attachment of 16-carbon fatty acids onto cysteine residues via a reversible thioester linkage (Chamberlain and Shipston, 2015; Hannoush, 2015). In mammals 23 palmitoyltransferases permit the attachment of palmite residues to a conserved cysteine at the C-terminus of TEAD1, 2, 3 and 4. This posttranslational modification permits proper establishment of tertiary structure of TEAD proteins and ensures their stability. Moreover it has been suggested that the lack of palmitoylation also alters the sub-localization of TEAD proteins in the nuclear membrane (Noland et al., 2016).
6. Conclusions
TEAD1 and TEAD2 are highly conserved members of the TEAD family and play redundant roles during development. The interaction with several transcriptional co-activators allow TEAD protein to regulate a multitude of process including differentiation, apoptosis, proliferation and chromatin modification. These biological process are known to be often deregulated in several types of cancer therefore TEAD proteins may constitute relevant therapeutic targets. Recently the elucidation of the structure of the interaction between TEAD and its cofactors permitted the identification of several molecules and peptides allowing, in mouse, the regression of cancer. These recent studies are mainly focused on the interaction between TEAD and it transcriptional co activator YAP due to the implication of the Hippo pathway, a tumor suppressor pathway that regulates YAP shuttling to the nucleus. Therefore there is still questions that remains unanswered. Very little is known about the proper transcriptional and post-translational regulation of TEAD1 and 2. Finally the study of the interaction with other transcriptional co-activators could lead to the finding of new drugs regulating the activity of TEAD proteins
Together, all these results and future studies will open new avenues of research in cancer therapy.
Supplementary Material
Supplemental Table 1. Structural homologs of TEA domain.
Highlights.
TEAD proteins constitute a family of highly conserved transcription factors.
TEAD1/2 have a crucial role during embryonic development
TEAD1/2 regulate organ formation, cell death and proliferation.
TEAD1/2 are deregulated in several types of cancer.
TEAD1/2 could constitute good candidates for therapeutic targets.
Acknowledgments
This review and the corresponding Gene Wiki article are written as part of the Gene Wiki Review series--a series resulting from a collaboration between the journal GENE and the Gene Wiki Initiative. The Gene Wiki Initiative is supported by National Institutes of Health (GM089820). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE. The authors would like to thank the “Groupement des Entreprises Françaises dans la Lutte contre le Cancer » (G.E.F.L.U.C.), subvention 2015. We thank Jerome Collignon for help with the English.
Abbreviations
- DBD
DNA Binding Domain
- IAP
Inhibitor of Apoptosis Protein
- LATS
Large Tumour Suppressor
- MST
Mammalian Ste20-like Protein Kinase
- TEA
transcriptional enhancer activator
- TEAD
TEA domain
- VGLL
Vestigial like
- YAP
Yes-Associated Protein
- YBD
YAP Binding Domain
- Yki
Yorkie
Footnotes
The corresponding Gene Wiki entry for this review can be found here:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anbanandam A, Albarado DC, Nguyen CT, Halder G, Gao X, and Veeraraghavan S. 2006. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc Natl Acad Sci U S A. 103:17225–17230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LV, Davison K, Moss JA, Young C, Cullen MJ, Walsh J, Johnson MA, Bashir R, Britton S, Keers S, Argov Z, Mahjneh I, Fougerousse F, Beckmann JS, and Bushby KM. 1999. Dysferlin is a plasma membrane protein and is expressed early in human development. Hum Mol Genet. 8:855–861. [DOI] [PubMed] [Google Scholar]
- Andrianopoulos A, and Timberlake WE. 1991. ATTS, a new and conserved DNA binding domain. Plant Cell. 3:747–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianopoulos A, and Timberlake WE. 1994. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol Cell Biol. 14:2503–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R and Olson EN (2007). MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev. Cell 12,377–389. [DOI] [PubMed] [Google Scholar]
- Azakie A, Lamont L, Fineman JR, and He Y. 2005. Divergent transcriptional enhancer factor-1 regulates the cardiac troponin T promoter. Am J Physiol Cell Physiol. 289:C1522–1534. [DOI] [PubMed] [Google Scholar]
- Azakie A, Larkin SB, Farrance IK, Grenningloh G, and Ordahl CP. 1996. DTEF-1, a novel member of the transcription enhancer factor-1 (TEF-1) multigene family. J Biol Chem. 271:8260–8265. [DOI] [PubMed] [Google Scholar]
- Belandia B, and Parker MG. 2000. Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. The Journal of biological chemistry. 275:30801–30805. [DOI] [PubMed] [Google Scholar]
- Benhaddou A, Keime C, Ye T, Morlon A, Michel I, Jost B, Mengus G, and Davidson I. 2012. Transcription factor TEAD4 regulates expression of myogenin and the unfolded protein response genes during C2C12 cell differentiation. Cell Death Differ. 19:220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger LC, Smith DB, Davidson I, Hwang JJ, Fanning E, and Wildeman AG. 1996. Interaction between T antigen and TEA domain of the factor TEF-1 derepresses simian virus 40 late promoter in vitro: identification of T-antigen domains important for transcription control. J Virol. 70:1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BL, Lu J and Olson EN (1997). The MEF2A 3′ untranslated region functions as a cis-acting translational repressor. Mol. Cell. Biol 17,2756–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood EM, and Eisenman RN. 1991. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 251:1211–1217. [DOI] [PubMed] [Google Scholar]
- Bray S 1999. Drosophila development: Scalloped and Vestigial take wing. Curr Biol. 9:R245–247. [DOI] [PubMed] [Google Scholar]
- Butler AJ, and Ordahl CP. 1999. Poly(ADP-ribose) polymerase binds with transcription enhancer factor 1 to MCAT1 elements to regulate muscle-specific transcription. Molecular and cellular biology. 19:296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Inamdar M, Rodrigues V, Raghavan V, Palazzolo M, and Chovnick A. 1992. The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes Dev. 6:367–379. [DOI] [PubMed] [Google Scholar]
- Carlini LE, Getz MJ, Strauch AR, and Kelm RJ Jr. 2002. Cryptic MCAT enhancer regulation in fibroblasts and smooth muscle cells. Suppression of TEF-1 mediated activation by the single-stranded DNA-binding proteins, Pur alpha, Pur beta, and MSY1. J Biol Chem. 277:8682–8692. [DOI] [PubMed] [Google Scholar]
- Chamberlain LH, and Shipston MJ. 2015. The physiology of protein S-acylation. Physiol Rev. 95:341–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Maeda T, Mullett SJ, and Stewart AF. 2004a. Transcription cofactor Vgl-2 is required for skeletal muscle differentiation. Genesis. 39:273–279. [DOI] [PubMed] [Google Scholar]
- Chen HH, Mullett SJ, and Stewart AF. 2004b. Vgl-4, a novel member of the vestigial-like family of transcription cofactors, regulates alpha1-adrenergic activation of gene expression in cardiac myocytes. The Journal of biological chemistry. 279:30800–30806. [DOI] [PubMed] [Google Scholar]
- Chen L, Chan SW, Zhang X, Walsh M, Lim CJ, Hong W, and Song H. 2010. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 24:290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chan SW, Zhang X, Walsh M, Lim CJ, Hong W, and Song H. 2010a. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 24:290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Loh PG, and Song H. 2010b. Structural and functional insights into the TEAD-YAP complex in the Hippo signaling pathway. Protein Cell. 1:1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Friedrich GA, and Soriano P. 1994. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 8:2293–2301. [DOI] [PubMed] [Google Scholar]
- Cooper TA, and Ordahl CP. 1985. A single cardiac troponin T gene generates embryonic and adult isoforms via developmentally regulated alternate splicing. J Biol Chem. 260:11140–11148. [PubMed] [Google Scholar]
- Davidson I, Xiao JH, Rosales R, Staub A, and Chambon P. 1988. The HeLa cell protein TEF-1 binds specifically and cooperatively to two SV40 enhancer motifs of unrelated sequence. Cell. 54:931–942. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, and Pan D. 2007. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 130:1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Lyons GE, Martin JF and Olson EN (1994). Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis.Development 120,1251–1263 [DOI] [PubMed] [Google Scholar]
- Farrance IK, Mar JH, and Ordahl CP. 1992. M-CAT binding factor is related to the SV40 enhancer binding factor, TEF-1. J Biol Chem. 267:17234–17240. [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, and Zider A. 2008. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 18:435–441. [DOI] [PubMed] [Google Scholar]
- Gunther S, Mielcarek M, Kruger M, and Braun T. 2004. VITO-1 is an essential cofactor of TEF1-dependent muscle-specific gene regulation. Nucleic Acids Res. 32:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta MP, Amin CS, Gupta M, Hay N, and Zak R. 1997. Transcription enhancer factor 1 interacts with a basic helix-loop-helix zipper protein, Max, for positive regulation of cardiac alpha-myosin heavy-chain gene expression. Molecular and cellular biology. 17:3924–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta MP, Davis F, Gupta M, Pogwizd SM, Jayakar D, Bacha E, and Jeevananadam V. 2001. An alternatively spliced isoform of serum response factor is increased human failing hearts: Potential implications in altered myocardial gene expression during overloads. Circulation. 104:197–197.11447086 [Google Scholar]
- Gupta MP, Gupta M, Dizon E, and Zak R. 1996. Sympathetic control of cardiac myosin heavy chain gene expression. Molecular and cellular biochemistry. 157:117–124. [DOI] [PubMed] [Google Scholar]
- Gupta MP, Kogut P, and Gupta M. 2000. Protein kinase-A dependent phosphorylation of transcription enhancer factor-1 represses its DNA-binding activity but enhances its gene activation ability. Nucleic Acids Res. 28:3168–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Jung EM, Cho J, Lee JW, Hwang KT, Yang SJ, Kang JJ, Bae JY, Jeon YK, Park IA, Nicolau M, Jeffrey SS, and Noh DY. 2008. DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes Chromosomes Cancer. 47:490–499. [DOI] [PubMed] [Google Scholar]
- Hannoush RN 2015. Synthetic protein lipidation. Curr Opin Chem Biol. 28:39–46. [DOI] [PubMed] [Google Scholar]
- Hucl T, Brody JR, Gallmeier E, Iacobuzio-Donahue CA, Farrance IK, and Kern SE. 2007. High cancer-specific expression of mesothelin (MSLN) is attributable to an upstream enhancer containing a transcription enhancer factor dependent MCAT motif. Cancer Res. 67:9055–9065. [DOI] [PubMed] [Google Scholar]
- Hwang JJ, Chambon P, and Davidson I. 1993. Characterization of the transcription activation function and the DNA binding domain of transcriptional enhancer factor-1. Embo J. 12:2337–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemin P, Hwang JJ, Martial JA, Dolle P, and Davidson I. 1996. A novel family of developmentally regulated mammalian transcription factors containing the TEA/ATTS DNA binding domain. The Journal of biological chemistry. 271:21775–21785. [DOI] [PubMed] [Google Scholar]
- Jiao S, Wang H, Shi Z, Dong A, Zhang W, Song X, He F, Wang Y, Zhang Z, Wang W, Wang X, Guo T, Li P, Zhao Y, Ji H, Zhang L, Zhou Z.(2014)A Peptide Mimicking VGLL4 Function Acts as a YAP Antagonist Therapy against Gastric Cancer. Cancer Cell 25, 166–180 [DOI] [PubMed] [Google Scholar]
- Jiang SW, Dong M, Trujillo MA, Miller LJ, and Eberhardt NL. 2001. DNA binding of TEA/ATTS domain factors is regulated by protein kinase C phosphorylation in human choriocarcinoma cells. The Journal of biological chemistry. 276:23464–23470. [DOI] [PubMed] [Google Scholar]
- Jiang SW, Trujillo MA, Sakagashira M, Wilke RA, and Eberhardt NL. 2000. Novel human TEF-1 isoforms exhibit altered DNA binding and functional properties. Biochemistry. 39:3505–3513. [DOI] [PubMed] [Google Scholar]
- Jiang SW, Wu K, and Eberhardt NL. 1999. Human placental TEF-5 transactivates the human chorionic somatomammotropin gene enhancer. Mol Endocrinol. 13:879–889. [DOI] [PubMed] [Google Scholar]
- Kaneko KJ, Cullinan EB, Latham KE, and DePamphilis ML. 1997. Transcription factor mTEAD-2 is selectively expressed at the beginning of zygotic gene expression in the mouse. Development. 124:1963–1973. [DOI] [PubMed] [Google Scholar]
- Kaneko KJ, Kohn MJ, Liu C, and DePamphilis ML. 2007. Transcription factor TEAD2 is involved in neural tube closure. Genesis. 45:577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karns LR, Kariya K, and Simpson PC. 1995. M-CAT, CArG, and Sp1 elements are required for alpha 1-adrenergic induction of the skeletal alpha-actin promoter during cardiac myocyte hypertrophy. Transcriptional enhancer factor-1 and protein kinase C as conserved transducers of the fetal program in cardiac growth. The Journal of biological chemistry. 270:410–417. [DOI] [PubMed] [Google Scholar]
- Knight JF, Shepherd CJ, Rizzo S, Brewer D, Jhavar S, Dodson AR, Cooper CS, Eeles R, Falconer A, Kovacs G, Garrett MD, Norman AR, Shipley J, and Hudson DL. 2008. TEAD1 and c-Cbl are novel prostate basal cell markers that correlate with poor clinical outcome in prostate cancer. Br J Cancer. 99:1849–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz LM, Liu-Chittenden Y, Yin F, Zheng Y, Yu J, Huang B, Chen Q, Wu S, and Pan D. 2013. The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev Cell. 25:388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloux I, Dubois E, Dewerchin M, and Jacobs E. 1990. TEC1, a gene involved in the activation of Ty1 and Ty1-mediated gene expression in Saccharomyces cerevisiae: cloning and molecular analysis. Molecular and cellular biology. 10:3541–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landin Malt A, Cagliero J, Legent K, Silber J, Zider A, and Flagiello D. 2012. Alteration of TEAD1 expression levels confers apoptotic resistance through the transcriptional up-regulation of Livin. PLoS One. 7:e45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landin Malt A, Georges A, Silber J, Zider A, and Flagiello D. 2013. Interaction with the Yes-associated protein (YAP) allows TEAD1 to positively regulate NAIP expression. FEBS Lett. 587:3216–3223. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, Guan KL, and Xu Y. Structural insights into the YAP and TEAD complex. Genes Dev. 24:235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, Guan KL, and Xu Y. 2010. Structural insights into the YAP and TEAD complex. Genes Dev. 24:235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, and Pan D. 2012. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 26:1300–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li H, Rajurkar M, Li Q, Cotton JL, Ou J, Zhu LJ, Goel HL, Mercurio AM, Park JS, Davis RJ, Mao J. 2016. Tead and AP1 coordinate transcription and motility Cell Rep.14(5): 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclellan WR, Lee TC, Schwartz RJ, and Schneider MD. 1994. Transforming Growth-Factor-Beta Response Elements of the Skeletal Alpha-Actin Gene - Combinatorial Action of Serum Response Factor, Yy1, and the Sv40 Enhancer-Binding Protein, Tef-1. Journal of Biological Chemistry. 269:16754–16760. [PubMed] [Google Scholar]
- Maeda T, Chapman DL, and Stewart AF. 2002. Mammalian vestigial-like 2, a cofactor of TEF-1 and MEF2 transcription factors that promotes skeletal muscle differentiation. The Journal of biological chemistry. 277:48889–48898. [DOI] [PubMed] [Google Scholar]
- Mahoney WM Jr., Hong JH, Yaffe MB, and Farrance IK. 2005. The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem J. 388:217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann KM, Ward JM, Yew CC, Kovochich A, Dawson DW, Black MA, Brett BT, Sheetz TE, Dupuy AJ, I. Australian Pancreatic Cancer Genome, Chang DK, Biankin AV, Waddell N, Kassahn KS, Grimmond SM, Rust AG, Adams DJ, Jenkins NA, and Copeland NG. 2012. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America. 109:5934–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar JH, and Ordahl CP. 1988. A conserved CATTCCT motif is required for skeletal muscle-specific activity of the cardiac troponin T gene promoter. Proceedings of the National Academy of Sciences of the United States of America. 85:6404–6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JF, Schwarz JJ and Olson EN (1993). Myocyte enhancer factor (MEF) 2C: a tissue-restricted member of the MEF-2 family of transcription factors. Proc. Natl. Acad. Sci. USA 90,5282–5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott JC, Cardoso MC, Yu YT, Andres V, Leifer D, Krainc D, Lipton SA and Nadal-Ginard B (1993). hMEF2C gene encodes skeletal muscle- and brain-specific transcription factors. Mol. Cell. Biol 13,2564–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewski RC, Chi NC, Li J, Brown C, Lu MM, and Epstein JA. 2004. Identification of minimal enhancer elements sufficient for Pax3 expression in neural crest and implication of Tead2 as a regulator of Pax3. Development. 131:829–837. [DOI] [PubMed] [Google Scholar]
- Noland CL, Gierke S, Schnier PD, Murray J, Sandoval WN, Sagolla M, Dey A, Hannoush RN, Fairbrother WJ, and Cunningham CN. 2016. Palmitoylation of TEAD Transcription Factors Is Required for Their Stability and Function in Hippo Pathway Signaling. Structure. 24:179–186. [DOI] [PubMed] [Google Scholar]
- Nowee ME, Snijders AM, Rockx DA, de Wit RM, Kosma VM, Hamalainen K, Schouten JP, Verheijen RH, van Diest PJ, Albertson DG, and Dorsman JC. 2007. DNA profiling of primary serous ovarian and fallopian tube carcinomas with array comparative genomic hybridization and multiplex ligation-dependent probe amplification. J Pathol. 213:46–55. [DOI] [PubMed] [Google Scholar]
- Pobbati AV, Chan SW, Lee I, Song H, and Hong W. 2012. Structural and functional similarity between the Vgll1-TEAD and the YAP-TEAD complexes. Structure. 20:1135–1140. [DOI] [PubMed] [Google Scholar]
- Pobbati AV, Han X, Hung AW, Weiguang S, Huda N, Chen GY, Kang C, Chia CS, Luo X, Hong W, and Poulsen A. 2015. Targeting the Central Pocket in Human Transcription Factor TEAD as a Potential Cancer Therapeutic Strategy. Structure. 23:2076–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobbati AV, and Hong W. 2013. Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol Ther. 14:390–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R and Treisman R (1991). Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 5,2327–2341. [DOI] [PubMed] [Google Scholar]
- Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, and Ganesan S. 2006. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 9:121–132. [DOI] [PubMed] [Google Scholar]
- Rindt H, Gulick J, Knotts S, Neumann J, and Robbins J. 1993. In vivo analysis of the murine beta-myosin heavy chain gene promoter. The Journal of biological chemistry. 268:5332–5338. [PubMed] [Google Scholar]
- Sawada A, Kiyonari H, Ukita K, Nishioka N, Imuta Y, and Sasaki H. 2008. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Molecular and cellular biology. 28:3177–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada A, Nishizaki Y, Sato H, Yada Y, Nakayama R, Yamamoto S, Nishioka N, Kondoh H, and Sasaki H. 2005. Tead proteins activate the Foxa2 enhancer in the node in cooperation with a second factor. Development. 132:4719–4729. [DOI] [PubMed] [Google Scholar]
- Simmonds AJ, Liu X, Soanes KH, Krause HM, Irvine KD, and Bell JB. 1998. Molecular interactions between Vestigial and Scalloped promote wing formation in Drosophila. Genes Dev. 12:3815–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotheim RI, Autio R, Lind GE, Kraggerud SM, Andrews PW, Monni O, Kallioniemi O, and Lothe RA. 2006. Novel genomic aberrations in testicular germ cell tumors by array-CGH, and associated gene expression changes. Cell Oncol. 28:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AF, Richard CW 3rd, Suzow J, Stephan D, Weremowicz S, Morton CC, and Adra CN. 1996. Cloning of human RTEF-1, a transcriptional enhancer factor-1-related gene preferentially expressed in skeletal muscle: evidence for an ancient multigene family. Genomics. 37:68–76. [DOI] [PubMed] [Google Scholar]
- Swartz EA, Johnson AD, and Owens GK. 1998. Two MCAT elements of the SM alpha-actin promoter function differentially in SM vs. non-SM cells. Am J Physiol. 275:C608–618. [DOI] [PubMed] [Google Scholar]
- Tian W, Yu J, Tomchick DR, Pan D, and Luo X. Structural and functional analysis of the YAP-binding domain of human TEAD2. Proc Natl Acad Sci U S A. 107:7293–7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Yu J, Tomchick DR, Pan D, and Luo X. 2010. Structural and functional analysis of the YAP-binding domain of human TEAD2. Proceedings of the National Academy of Sciences of the United States of America. 107:7293–7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsika RW, Ma L, Kehat I, Schramm C, Simmer G, Morgan B, Fine DM, Hanft LM, McDonald KS, Molkentin JD, Krenz M, Yang S, and Ji J. 2010. TEAD-1 overexpression in the mouse heart promotes an age-dependent heart dysfunction. The Journal of biological chemistry. 285:13721–13735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, and DePamphilis ML. 2001. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 15:1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudin P, Delanoue R, Davidson I, Silber J, and Zider A. 1999. TONDU (TDU), a novel human protein related to the product of vestigial (vg) gene of Drosophila melanogaster interacts with vertebrate TEF factors and substitutes for Vg function in wing formation. Development. 126:4807–4816. [DOI] [PubMed] [Google Scholar]
- Wang Q, and Latham KE. 2000. Translation of maternal messenger ribonucleic acids encoding transcription factors during genome activation in early mouse embryos. Biol Reprod. 62:969–978. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, and Pan D. 2008. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 14:388–398. [DOI] [PubMed] [Google Scholar]
- Xia Y, Zhang YL, Yu C, Chang T, and Fan HY. 2014. YAP/TEAD co-activator regulated pluripotency and chemoresistance in ovarian cancer initiated cells. PLoS One. 9:e109575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao JH, Davidson I, Matthes H, Garnier JM, and Chambon P. 1991. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 65:551–568. [DOI] [PubMed] [Google Scholar]
- Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, and Buonanno A. 2007. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 134:3827–3836. [DOI] [PubMed] [Google Scholar]
- Yasunami M, Suzuki K, Houtani T, Sugimoto T, and Ohkubo H. 1995. Molecular characterization of cDNA encoding a novel protein related to transcriptional enhancer factor-1 from neural precursor cells. J Biol Chem. 270:18649–18654. [DOI] [PubMed] [Google Scholar]
- Yasunami M, Suzuki K, and Ohkubo H. 1996. A novel family of TEA domain-containing transcription factors with distinct spatiotemporal expression patterns. Biochem Biophys Res Commun. 228:365–370. [DOI] [PubMed] [Google Scholar]
- Yockey CE, Smith G, Izumo S, and Shimizu N. 1996. cDNA cloning and characterization of murine transcriptional enhancer factor-1-related protein 1, a transcription factor that binds to the M-CAT motif. J Biol Chem. 271:3727–3736. [DOI] [PubMed] [Google Scholar]
- Yoshida T 2008. MCAT elements and the TEF-1 family of transcription factors in muscle development and disease. Arterioscler Thromb Vasc Biol. 28:8–17. [DOI] [PubMed] [Google Scholar]
- Yu FX, Zhao B, and Guan KL. 2015. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 163:811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Breitbart RE, Smoot LB, Lee Y, Mahdavi V and Nadal-Ginard B (1992). Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 6,1783–1798 [DOI] [PubMed] [Google Scholar]
- Yuan H, Liu H, Liu Z, Zhu D, Amos CI, Fang S, Lee JE, and Wei Q. 2015. Genetic variants in Hippo pathway genes YAP1, TEAD1 and TEAD4 are associated with melanoma-specific survival. Int J Cancer. 137:638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, Xiong Y, Lei QY, and Guan KL. 2009. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. The Journal of biological chemistry. 284:13355–13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, and Guan KL. 2010. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 24:862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, and Guan KL. 2008. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22:1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z et al. 2015. Targeting Hippo pathway by specific interruption of YAP-TEAD interaction using cyclic YAP-like peptides. FASEB J. 29, 724–732. [DOI] [PubMed] [Google Scholar]
- Zider A, Paumard-Rigal S, Frouin I, and Silber J. 1998. The vestigial gene of Drosophila melanogaster is involved in the formation of the peripheral nervous system: Genetic interactions with the scute gene. J Neurogenet. 12:87–99. [DOI] [PubMed] [Google Scholar]
- Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. (2015) Nature Cell Biology 17, 1218–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuzarte PC, Farrance IK, Simpson PC, and Wildeman AG. 2000. Tumor cell splice variants of the transcription factor TEF-1 induced by SV40 T-antigen transformation. Biochim Biophys Acta. 1517:82–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Structural homologs of TEA domain.