Abstract
Analysis of CRISPR-induced mutations at targeted locus can be achieved by polymerase chain reaction amplification followed by parallel massive sequencing. We developed a novel algorithm, named as CRISPRpic, to analyze the sequencing reads for the CRISPR experiments via counting exact-matching and pattern-searching. Compare to the other methods based on sequence alignment, CRISPRpic provides precise mutation calling and ultrafast analysis of the sequencing results. Python script of CRISPRpic is available at https://github.com/compbio/CRISPRpic.
INTRODUCTION
CRISPR is the most widely used technique for genome editing in research and industry (1). After the pioneering development of the Cas9 endonuclease protein encoded by the CRISPR locus in Streptococcus Pyogenes as a toolkit for gene editing in human cells (2–5), various other CRISPR systems in the prokaryotic genome have been characterized (6–9).
For successful genome editing, highly efficient CRISPR-sgRNAs, differing by cell type or target sequence, are required. The standard method for measuring endonuclease-induced mutagenesis efficiency is an enzymatic assay using a mismatch-specific nuclease such as T7E1 endonuclease I (10) or Surveyor nuclease (11). However, this only provides indirect evidence of mutagenesis and can often produce false positive or negative results due to such reasons as poor sensitivity. Thus, additional labor-intensive experiments such as Sanger sequencing are often required to confirm mutant sequences.
The recent development of rapid and inexpensive next generation sequencing technology (NGS) permits convenient and massively parallel measurement of genome editing experiments (3). Tools aligning sequencing reads to the unmodified reference sequence are then used to analyze the data for the desired mutations (12–16). However, sequence alignment requires multiple calculations to identify indels with the highest alignment scores, and frequently produces false calls depending on the sequence context (17,18). We provide a simple, novel, highly accurate and rapid solution to these issues, using the double strand break site generated by programmable nucleases.
Herein, we describe an algorithm for fast and precise analysis of CRISPR-induced mutations via prefixed index counting (CRISPRpic). Use of this algorithm provides multiple improvements for the detection of variants at CRISPR target sites. The algorithm also provides user-friendly customizable input for other applications, i.e. Cpf1 or any other mutation analysis of sequencing reads generated by polymerase chain reaction (PCR)-amplicons. The simple counting algorithm of CRISPRpic allows for sequencing analysis in a low performance computing environment. Taken together, our novel algorithm is a simple and efficient method to analyze the efficiency of CRISPR-induced mutagenesis and increases the feasibility of diverse applications of CRISPR experiments.
MATERIALS AND METHODS
Detailed algorithm of CRISPRpic
Step 1: Build hash tables for exact matching
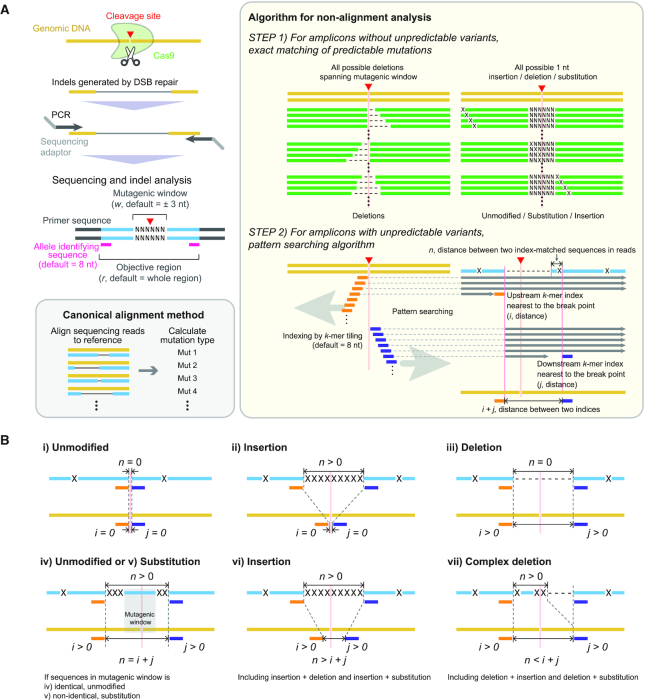
CRISPRpic builds a hash table representing all virtual sequences with all possible mutations encompassing the target site of DSB in the reference sequence, which is the expected amplicon. This target site, or CRISPR mutagenic site, is determined by the window size (w, default = 3) defined by users. In the default case, the mutagenic site encompasses the breakpoint with 3 nt on either side, for a total of 6 nt with a breakpoint in the middle (Figure 1A). We consider only mutations within this mutagenic site as CRISPR-induced mutations. All other mutations, which may be PCR artifacts or sequencing errors, are not considered mutations induced by CRISPR for our purposes.
Figure 1.
Sequencing analysis of CRISPR-induced mutations using CRISPRpic. (A) Schematics representing NGS analysis for CRISPR experiments and CRISPRpic algorithm to analyze them. Two-step algorithm of CRISPRpic or canonical alignment method is shown in yellow or gray box, respectively. (B) Examples of mutations calling by pattern-searching algorithm. Each example of mutation calling is also shown in a logical decision tree in Supplementary Figure S1. Reference, sequencing read and virtual DNA sequence are shown in yellow, blue and green, respectively. Orange and navy indicate k-mer indices.
First, all possible deletions encompassing the mutagenic window are generated. Some deletions, occurring at different positions, may appear identical when short common sequence motifs (i.e. micro-homology regions) are present in the reference sequence. In this case, we treat them as single deletion events with multiple alignment positions. Second, the hash table includes single nucleotide substitutions, deletions and insertions at all reference sequence positions. Only substitutions, deletions and insertions within the defined CRISPR mutagenic window are counted as mutations while all other outside changes are designated ‘unmodified’.
Step 2: Build two sets of k-mer indices for pattern searching
Some amplicons with unknown variants not induced by the endonuclease cannot be matched exactly to any of the sequences in the hash table. We employ pattern searching for these unmatched amplicons using a k-mer ‘index’ (default length of k is 8). The program generates a set of k-mer indices from the middle breakpoint and tiles upstream in 1 nt increments (Figure 1A). The index order represents the distance from the breakpoint. For example, the first index among the upstream set is located to the right of the breakpoint while the second index refers to the subsequent nucleotide. Next, we examine the unique representation of each k-mer index among all k-mer indices. If a k-mer appears more than once, we skip pattern searching for this k-mer index given the possibility of a false positive.
If the outside index originated from inside the window by skipping, classification is challenging. We keep all indices within the mutagenic window to facilitate the analysis. Thus, we increase the length of the k-mer by 1 nt until all indices within the mutagenic window are unique. For instance, all three k-mer indices must be unique among all indices in the upstream set when the size of mutagenic window is 3. We build the downstream set of k-mer indices in the same way.
Step 3: Identify and select amplicons
The input data for CRISPRpic are single reads in FASTQ formats originating from PCR amplicons. The paired-end reads can be converted to single-end reads by a program called FLASH. We process only reads meeting the following criteria: they contain either one of two adaptor sequences; both the first and last 8 nt of the reference sequence are present after removing adaptor sequences. Based on this processing, we determine the individual amplicon reads and their fractions of the total dataset.
Step 4: Classify amplicon read sequences
First, we examine if all distinctive amplicons are identical to any of the virtual mutant sequences and the reference sequence in the hash table. If they match, they will be classified accordingly in the hash table. Second, all reads not identical to one of the sequences in hash table will be classified by pattern searching. The classification is determined by the following five variables in relation to the given mutagenic window size:
i index, ordinal number of upstream k-mer index, which was first found in the amplicon
i shift-count, number of upstream k-mer skipped
j index, ordinal number of downstream k-mer index, which was first found in the amplicon
j shift-count, number of downstream k-mer skipped
n, the length of remaining sequence between two k-mers in the amplicon
The ordinal number of each index indicates the distance from the breakpoint. For example, the ordinal number of the first index is zero, representing the right breakpoint in the reference sequence. CRISPRpic initiates a search from the upstream k-mer in order until a k-mer is found in the amplicon. As mentioned above, we skip the non-unique k-mers in the amplicon. The number of skipped k-mers are designated as the i-shift count. CRISPRpic repeats the same procedure for downstream k-mers. When no k-mers are found in either the upstream or downstream indices for the amplicon, we designate the read as ‘NA’. After finding upstream and downstream k-mers in the amplicon, CRISPRpic examines the sequences between the two identified k-mer in the amplicons.
After identifying the five parameters above, we eliminate those amplicon sequences having [i index > w and i index – i shift-count < w] or [j index > w and j index – j shift-count]. This step is taken because mutation identification is a challenge in these cases. Otherwise, we classify reads by rules depicted in the logical flow chart (see below) using the following three numbers: i ( = i index – i shift-count), j ( = j index – j shift-count) and n.
Step 5: Classify variants
We describe the parameters used to identify a variant in Figure 1B and Supplementary Figure S1. Each case is considered as follows:
-
Classify reads as reference ‘wild-type’ and not CRISPR-modified when i, j and n equal zero.
Described simply, this is the case when the first k-mer in both upstream and downstream are found and there is nothing between these two k-mers in the amplicon. This also means that the 16 nt centered at the breakpoint (i.e. the targeted mutagenic site) are identical to the reference sequence. These sequences are not matched to any of the possible mutant sequences due to unknown variants existing somewhere outside the mutagenic window of the amplicon.
-
Classify reads as insertions when i and j equal zero, but n is larger than zero.
This happens when some sequences are inserted at the breakpoint. In this case, the first upstream and downstream k-mers are found, but additional sequences between the two indices (n > 0) exist in the amplicon due to insertion. CRISPRpic does not separately classify them as insertions with a deletion or insertions with a substitution when n > 0, as it is unclear whether they are generated by a different event or accidentally identical to the reference sequence.
-
Classify reads as a deletion when n equals zero, but i or j is not equal to zero.
This indicates that some sequences were deleted at the breakpoint. For example, the second upstream and fourth downstream k-mers will be found when 1 and 3 nt are deleted upstream and downstream of the breakpoint, respectively. However, no remaining sequence between the two k-mers will exist on the amplicon. At last, there are complicated cases where m > 0 and n > 0, where m = i + j. Their classification is determined by the rules in the logic flowchart.
Classify complex variants when they do not fall in the aforementioned categories on the first analysis pass.
We compare the expected distance (m, |i|+|j|) of two identified k-mers in the reference sequence with the length of remaining sequence (n). There are three possibilities between m and n; (i) n > m, (ii) n = m and (iii) n < m.
First, amplicons are classified as insertions when n is larger than m. This occurs when the insertion occurs simultaneously with some other event. Second, amplicons are classified as either substitutions or unmodified when n equals m. In this case, we examine if the remaining sequence is identical to the mutagenic site sequence. If they are identical, amplicons are classified as unmodified, otherwise, they are classified as substitutions. Third, amplicons are classified as either complex deletions or unmodified when n is smaller than m. In this case, we examine the deletion's location relative to the breakpoint. If the deletion occurs outside the mutagenic site, they are classified as complex deletions as they have additional variants within n. CRISPRpic does not distinguish deletions with insertion or deletions with substitution from complex deletion as it is uncertain how they were generated. Otherwise, they are classified as unmodified comparing the sequences of n with sequences in mutagenic window.
Calculate the frequency of deletions at all positions in the amplicon
Multiple deletion events at different positions of the reference sequence result in the same sequence in the presence of ‘micro-homology’ sequences at the deletion junction. Micro-homology refers to repeated short sequence motifs, and can produce multiple alignments to the reference sequence. We use the count of the deletion sequence for each position divided by the total number of multiple alignments containing the position. For instance, we observed a deletion sequence 100 times that was derived from five different alignments of deletions. All positions deleted in all five alignments are counted 100 times as well. However, the deletion at the most upstream can occur in only one alignment, so it was counted only 20 times (100/5).
These deletions associated with micro-homology motifs tend to be more prevalent than random deletions due to sequencing artifacts (19). We use the number of redundant indices from each read to calculate the number of micro-homology motifs (number of nucleotides in micro-homology = possible alignment -1).
Simulation with virtual sequencing reads
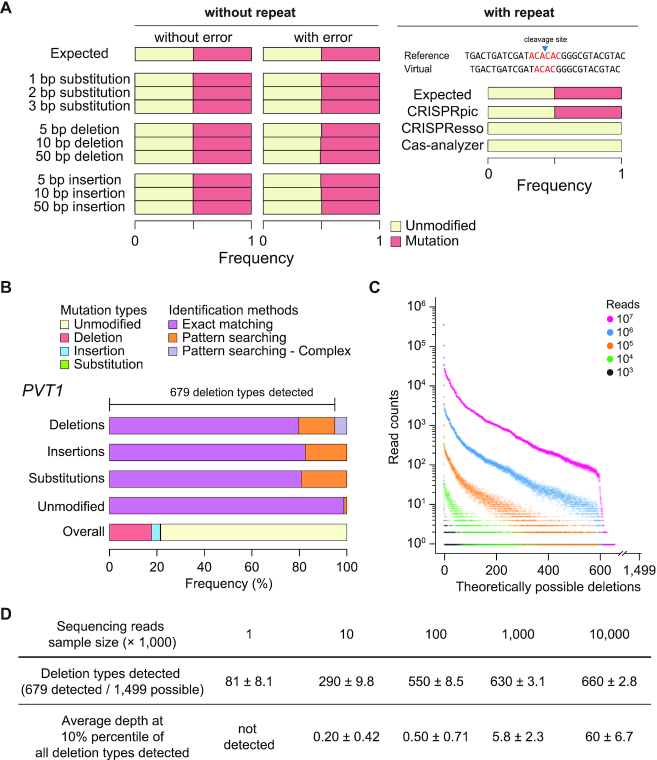
In order to test CRISPRpic, we analyzed virtual sequencing reads consisting of 1000 reads from unmodified DNA and 1000 reads containing substitutions, deletions or insertions of various lengths. We evaluated CRISPRpic using simulation data from CRISPResso. We downloaded Supplementary Data 4 from https://media.nature.com/original/nature-assets/nbt/journal/v34/n7/extref/nbt.3583-S12.zip. In addition, we generated simulation data using a simulator program called ‘ART’ (20), which was also used in CRISPResso, with the following reference amplicon sequence; AATGTCCCCCAATGGGAAGTTCATCTGGCACTGCCCAGATCGATCGTAGCTGTGACTGACTGATCGATACA∧CACGGGCGTACGTACACGTACGTAGCTGAGTAAGAATGGCTTCAAGAGGCTCGGCTGTGGTT. ∧ indicates the location of the DSB while the underlined CGG is a PAM site. We generated mut.fa containing a mutated sequence with AC deletion around DSB. The simulation data with sequencing error was made by ART with the following command: art_illumina -ss MSv1 -i mut.fa -amp -o mut_seq -p -l 100 -f 1000 –sam. The final FASTQ files have 1,000 wild-type sequences and 1000 mutant sequences respectively. This simulation data is also available at Github (https://github.com/compbio/CRISPRpic).
As expected, CRISPRpic identified a 50% mutation frequency in the simulation data, which was comparable to frequencies produced by other software packages. Next, we generated virtual sequencing reads containing 2 bp repeats. Mutation calling becomes more challenging when micro-homology sequence or repetitive sequences are present within the cleavage site or breakpoint, because sequence alignment of repetitive sequences is ambiguous, resulting in biased or false mutation calling. In this simulation, CRISPRpic again identified precisely 50% mutation frequency, demonstrating that the alignment-free CRISPRpic algorithm can analyze repetitive sequences appropriately.
Information of computing environment
All analysis was performed with the following specifications, 2.5 GHz AMD Opteron 6380, 512 GB 1600Mhz DDR3, Linux 4.4.0–122 generic #146-Ubuntu SMP. CRISPRpic was also tested on a personal laptop computer with 2.3GHz Intel Core i5, 8 GB 2133 MHz LPDDR3, macOS High Sierra 10.13.4. CRISPRpic is designed to use one core from the CPU to analyze the sequencing reads.
Software version used in this study
CRISPRpic was written with Python 2.7, and can be run by Python 3.7 as well. Paired-end sequencing reads were merged using Flash 1.2.11 prior to running CRISPRpic. In order to compare the analysis results, sequencing reads were also analyzed using various software with the following parameters unless described separately: CRISPRpic with (-w 3 -s 8); CRISPResso and CRISPResso2, 1.0.8 with (-w 6 -a [amplicon sequence] -g [target sequence without PAM] for SpCas9; -w 10 –cleavage_offset 1 -a [amplicon sequence] -g [target sequence without PMA] for AsCpf1); Cas-Analyzer, web version (http://www.rgenome.net/cas-analyzer/) with ([checked in the ‘or use both ends’], [1 for minimum frequency], [3 for WT marker]); CRISPR-GA, web version http://crispr-ga.net/index.php; CRISPR-DAV with all defaults.
Manual inspection for comparison between programs
When manually inspected, the inspector could not determine clearly a specific variant call in many cases. To eliminate this ambiguity, we developed the following rules for assignment: (i) Mutations are only classified as unmodified, insertion, substitution or deletion; and (ii) If the sequencing read is assigned to multiple categories, we prioritize insertions or deletions above wild-type or substitution. As a test, this manual inspection was done in a blinded fashion.
RESULTS
Overview of CRISPRpic algorithm
We developed a computational tool, CRISPRpic, that counts each possible mutation in the sequencing reads without alignments. Identification of Indels by sequence alignment CRISPRpic is based on three unique properties of gene edit experiments: (i) Sequencing reads have fixed ends originating from PCR primer pairs, (ii) CRISPR/Cas9 induces a double strand break (DSB) at a predictable position within the target sequence and (iii) Mutations should encompass the DSB site (Figure 1A). These features enable the prediction of a majority of the possible mutation spectrum and therefore their efficient identification. The default input to the program is the list of amplicon sequences, the guide RNA sequences located within each amplicon, and the type of endonuclease with a defined breakpoint such as CRISPR/Cas9 from different bacterial species: SpCas9 or AsCpf1, etc. Using these parameters, CRISPRpic has the flexibility to analyze genomic alterations produced by several different enzymes covering a variety of DBS positions. Our pipeline implements the following steps (Figure 1A); (i) build hash table and set of k-mers, (ii) identify and select amplicons with their frequencies and (iii) classify amplicons using hash tables or pattern searching of k-mers. Most mutations were as predicted, however, some unknown variants were produced by PCR or sequencing errors, and some sequencing reads were not identical to the prediction. In order to classify reads with unpredicted variants, we designed a pattern searching algorithm using distance of the k-mer indices in the references and sequencing reads (Figure 1A). Altogether, CRISPRpic is designed to follow a logical decision tree using either exact-matching or pattern-searching, allowing for non-ambiguous mutation calling (Figure 1B and Supplementary Figure S1).
Precise mutation calling by CRISPRpic
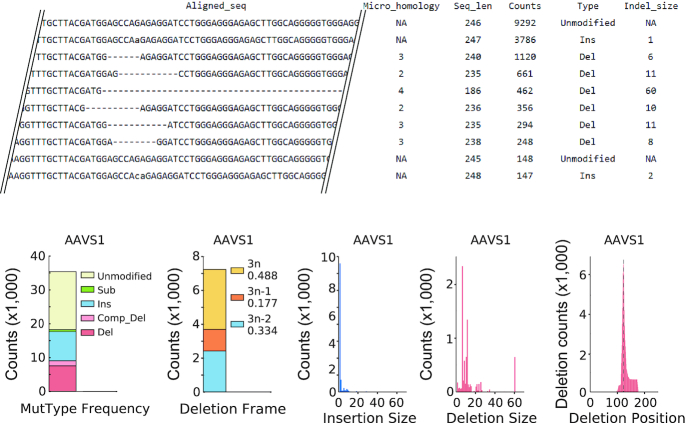
In order to evaluate CRISPRpic's accuracy, we tested CRISPRpic using virtual and real sequencing data. First, we analyzed virtual sequencing reads consisting of 1000 reads from unmodified DNA and 1000 reads containing substitutions, deletions or insertions of various lengths (Figure 2A). As expected, CRISPRpic identified a 50% mutation frequency in the simulation data, comparable to other software packages. Next, we hypothesized that mutation calling may become more challenging when repetitive sequences are present within the mutagenic window, because sequencing alignment of repetitive sequences is ambiguous. We generated virtual sequencing reads containing 2 bp repeats (Figure 2A). In this simulation, CRISPRpic again identified precisely 50% mutation frequency whereas alignment-based software such as CRISPResso and Cas-analyzer had errors. This result demonstrates that the alignment-free CRISPRpic algorithm can analyze repetitive sequences precisely. Indeed, CRISPRpic can easily differentiate the number of micro-homology bases from the number of redundant index sequences, which is an important parameter for predicting the mutation pattern of programmable nucleases and CRISPR applications (21). Therefore, CRISPRpic provides an output table containing information on micro-homology as well as length, counts, mutation calling and mutation length for each sequencing read (Figure 3). CRISPRpic also generates graphical summaries representing counts of mutation types, counts of deletion frame, length of indels and deletion count per nucleotide.
Figure 2.
Pilot analysis using CRISPRpic for indel identification. (A) Simulation analysis using CRISPRpic. Simulation results using virtual sequencing reads with or without repetitive sequences around the cleavage site. AC repeats are shown in red. (B) Mutation analysis of sequencing reads from the PVT1 locus targeted by SpCas9. Bar plots showing proportions of mutation callings by exact matching or pattern searching. (C) Distribution of all theoretically possible deletions at the PVT1 locus by sample size of sequencing reads (n = 10, technical replicates). (D) Summary for coverage and sequencing depth of detection of deletions.
Figure 3.
Example of output files of CRISPRpic.
Next, we tested CRISPRpic to analyze 20 million reads of the human PVT1 locus amplified from human cells treated with CRISPR/SpCas9 (22) (Figure 2B). CRISPRpic successfully classified 94.8% or 5.2% of the reads by exact-matching or pattern-matching method, respectively. Only 0.003% of reads were not appropriately classified. From this sequencing data, we sampled the sequencing reads ranged from the 103 to the 107 reads. As CRISPRpic made all theoretically possible deletions in the hash table, we analyzed the frequencies of the deletion alleles. Theoretically, 1499 types of deletions can be generated in this amplicon, while 679 deletion types were detected (Figure 2C). The frequency of each deletion allele varied by sample size and saturated at a sample size of 107 reads. This analysis suggests that mutant allele-based studies require more than 107 reads (Figure 2D).
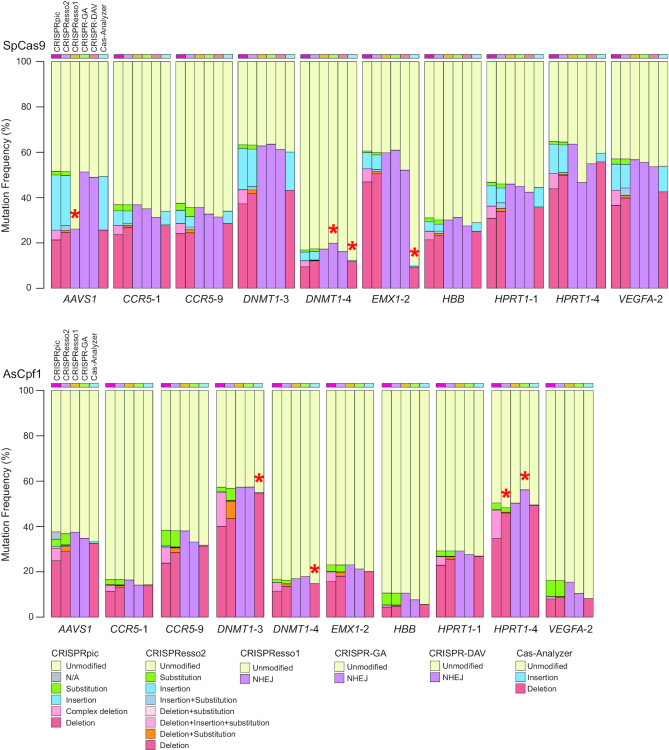
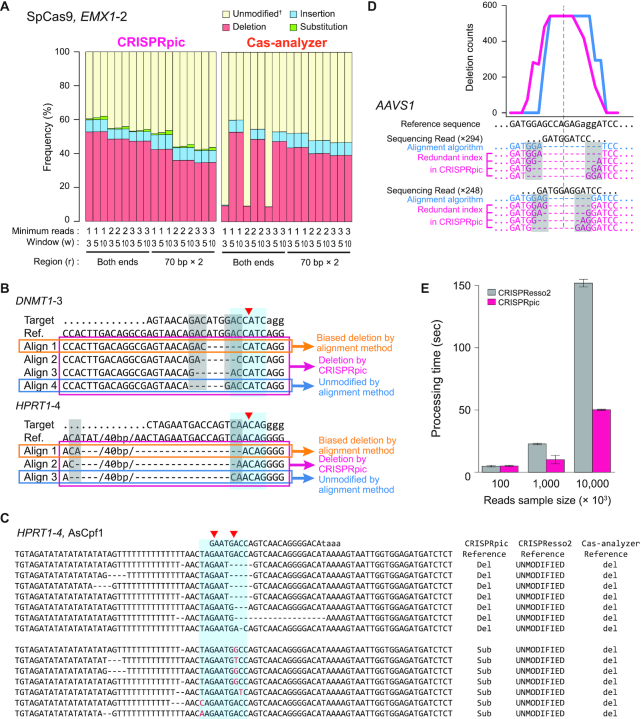
We further analyzed sequencing reads from 10 different loci in the human genome targeted by two different types of CRISPR (23). The actual mutation frequency is unknown for the sequencing reads generated from genomic DNA, so we compared the mutation frequency analyzed by the following six programs: CRISPRpic, CRISPResso2, CRISPResso, CRISPR-GA, CRISPR-DAV and Cas-Analyzer (Figure 4). CRISPRpic successfully assigned the vast majority of the sequencing reads (>99.99% of the total reads from 20 targeted loci) to a single prefixed classification. In this analysis, other programs showed outlying indel frequencies compared to average frequencies for at least one locus: AAVS1(SpCas9) for CRISPResso, HPRT1-4 (AsCpf1) for CRISPResso2, DNMT1-4 (SpCas9) and HPRT1-4 (AsCpf1) for CRISPR-GA, DNMT1-4 (SpCas9), EMX1-2 (SpCas9), DNMT1-3 (AsCpf1) and DNMT1-4 (AsCpf1) for Cas-Analyzer.
Figure 4.
Comparison of analysis by different programs for CRISPR-induced mutations. Mutation frequencies analyzed by CRISPRpic in addition to other programs for 10 different target loci treated by SpCas9 (upper) or AsCpf1 (lower). Red asterisks indicate outliers located 1.5 times outside the interquartile range above the upper quartile and below the lower quartile.
We inspected the reads classified differently by other programs. First, we found that alignment-based programs showed false mutation calling based on the parameters at a particular locus (Figure 5A). Second, most erroneous cases of mutation calling by alignment methods occurred when the micro-homology sequences were present at the border of deleted sequences. When DSBs are repaired in living cells, micro-homology-mediated deletions occur more frequently than they would randomly (19). CRISPRpic algorithm can correctively classify mutations harboring micro-homology sequences. Sequence-aligning algorithms randomly choose only one alignment whereas multiple alignments are possible due to the micro-homology sequences (Figure 5B). CRISPRpic works by reproducing the CRISPR-induced mutagenesis in silico, enabling unbiased mutation calling when the micro-homology sequences are present at the border of the mutagenic window. Furthermore, CRISPRpic is designed following a logical decision tree, thus always providing a pre-designed mutation classification (Supplementary Figure S1). Third, we also found that some erroneous mutation calling occurred for sequencing reads harboring sequence variants not encompassing the mutagenic window. CRISPRpic also successfully excludes such variants caused by PCR or sequencing errors rather than CRISPR-induced editing (Figure 5C). At last, micro-homology sequences result in an inaccurate distribution of overall deletion pattern by aligning algorithms (Figure 5D). CRISPRpic presents possible deletions in contrast to alignment-based methods which show a positional bias. Taken together, CRISPRpic showed more precise analysis of mutation frequency independent of sequence context.
Figure 5.
Non-alignment algorithm of CRISPRpic provides precise analysis of CRISPR-induced mutations (A) Comparison of mutation frequencies for EMX1-2 locus by different parameters of CRISPRpic or Cas-Analyzer. †Cas-analyzer does not distinguish substitutions from unmodified alleles. (B) Example of classification of deletions harboring micro-homology sequences. Blue or gray box indicates mutagenic window (±3 bp from breakpoint, red triangle) or micro-homology sequences, respectively. (C) Example of erroneous mutation calling by alignment method. (D) Unbiased analysis of deletion pattern by CRISPRpic. Two sequencing reads from the AAVS1 locus were extracted from actual output files of CRISPRpic or CRISPResso as an example. Micro-homology sequences are marked in the gray box. Pink or blue line indicates deletion distribution for each nucleotide position analyzed by CRISPRpic or CRISPResso, respectively. Dashed line indicates the breakpoint. (E) Bar plots showing processing time for analysis of the PVT1 locus using CRISPRpic or CRISPResso2.
Ultrafast analysis of mutations by CRISPRpic
In addition to precise mutation calling, CRISPRpic required less analysis time compared to alignment-based methods (Figure 5E). We noted that, except for CRISPResso2, other programs took several days to analyze millions of reads. Because CRISPRpic is designed for simple counting and k-mer searching rather than multiple calculations, it therefore does not require high computing performance. Therefore, CRISPRpic could perform analysis of 20 million reads in only one minute on a personal laptop computer. To our best knowledge, CRISPRpic is the fastest algorithm for analyzing mutations in amplicon sequencing.
DISCUSSION
Induction of mutations at targeted locus is one of the most common applications for CRISPR, but their analysis by sequence alignment has multiple issues, including incomplete or erroneous calling of variants and the requirement for high-performance computing. Although larger numbers of researchers use alignment-based programs to analyze CRISPR-induced mutations, a precise and fast method for analysis has not yet been developed. CRISPRpic is based on exact-matching and logical decisions, which provides precise and ultrafast analysis of mutation analysis for CRISPR experiments.
We noted that there were only two cases where CRISPRpic could not classify, known as NA, and only one case where it classified incorrectly. From our analysis of the amplicon sequencing data across 20 loci, we observed only one case as the source of NA; all k-mers in either upstream or downstream were not found in amplicons. This happens when a large deletion left less than k nucleotides at either end or less than k × 2 with some other mutation events. In our analysis of 20 targets, 15 loci did not have any NA and only one locus showed NA greater than 0.001%. CRISPRpic is also designed to classify NA when the i or j index is larger than w but [i index – i shift-count] or [j index – j shift-count] is not less than w. This occurs when sequencing reads have multiple discontinuous variants in and out of the mutagenic window within index length. In this case, it is not clear whether the variants are caused by poor sequencing quality or one mutagenic event leaving long mutations, but coincidently most of them were identical to the reference sequence. This did not occur, however, in the 20 targeted sites we analyzed to develop CRISPRpic.
In our manual inspection, we found 35 sequencing reads out of 8100 input reads (0.43%) counted more than once from the HPRT1-4 locus-treated SpCas9, which was incorrectly identified as a complex deletion although it was a simple deletion. CRISPRpic called a given set of reads as complex deletions while manual examination showed a simple deletion. This classification error occurred because the locus contained AT- or GT-repeat sequences longer than the k-mer on the upstream sequence. In this case, CRISPRpic repeatedly skipping several non-unique k-mers due to AT repeats. This resulted in i not being zero, and subsequent classification as a complex deletion. For cases such as this, we recommend users provide a longer k-mer length which can be simply adjusted as an input command line. However, this only happened to sequencing reads containing additional variants outside the mutagenic window because sequencing reads without such variants are classified correctly first in the exact-matching step.
In summary, CRISPRpic provides fast and precise analysis of CRISPR-induced mutation analysis independent of sequence context, allowing for analyzing the high depth of targeted sequencing data from CRISPR experiments such as high-throughput profiling of CRISPR sgRNA (24), mutagenesis-based functional studies of proteins (25) and regulatory elements in a DNA-centric manner (26). The advent of single cell analysis of CRISPR-mediated perturbations (27–30) further necessitates the need for highly efficient and scalable means of analyzing gene edits. Furthermore, CRISPRpic requires only Python to implement. Thus, CRISPRpic can be easily adapted to other applications such as indel analysis of cancer genome. From the analysis conducted in this study, we suggest that 103 to 104 sequencing reads per target locus are required to accurately evaluate CRISPR efficiency. For allele-based quantitative analysis, we recommend 106 to 107 total reads depending on the mutation frequency to capture all possible deletion types (Figure 2).
Recently, it was reported that CRISR can induce large deletions over several kilobases (31). In order to survey mutations within a long range, longer PCR or sequencing errors can be incorporated, but this may cause false results by alignment followed by wrong mutation calling. CRISPRpic is exceptional for distinguishing variants not induced by CRISPR, making it a standard method for analysis of CRISPR-induced mutations for any type of amplicons.
Taken together, our method facilitates CRISPR-based experiments, provides greater accessibility to novice researchers unfamiliar with the complex nuances of CRISPR modifications in the genome editing field and will increase the analytical throughput of screening for CRISPR-engineered variants across a broad range of projects.
DATA AVAILABILITY
CRISPRpic is implemented in Python 2.7 or 3.4 and the source code are available at Github (https://github.com/compbio/CRISPRpic). All raw sequencing reads used in this study are derived through NCBI Sequencing Read Archive with accession number SRP070794 (23) and Gene Expression Omnibus with accession number GSM2572600 (22).
Supplementary Material
ACKNOWLEDGEMENTS
We thank to Stephanie Greer for her help in testing and depositing CRISPRpic to Github. We also thank to Ahmed Shuaibi, Jiamin Chen, Jing He, Soah Lee, Sue Grimes and Grace Chen for their helps in testing CRISPRpic. We appreciate Ava C. Carter, Semin Lee and Taejoo Hwang helped in preparing this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
National Institutes of Health [R35-CA209919 to H.Y.C., RM1-HG007735 to H.Y.C., P01-HG00205 to H.P.J.]; Ulsan National Institute of Science and Technology [1.190041.01 to S.W.C]. Funding for open access charge: National Institutes of Health [R01-HG006137 to H.P.J].
Conflict of interest statement. The authors filed an intellectual property based on this study. H.Y.C. is a co-founder of Accent Therapeutics, Boundless Bio and an advisor to 10× Genomics, Arsenal Biosciences and Spring Discovery.
REFERENCES
- 1. Kim H., Kim J.S.. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014; 15:321–334. [DOI] [PubMed] [Google Scholar]
- 2. Cho S.W., Kim S., Kim J.M., Kim J.S.. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013; 31:230–232. [DOI] [PubMed] [Google Scholar]
- 3. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al.. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J.. RNA-programmed genome editing in human cells. Elife. 2013; 2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim E., Koo T., Park S.W., Kim D., Kim K., Cho H.Y., Song D.W., Lee K.J., Jung M.H., Kim S. et al.. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017; 8:14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. et al.. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015; 163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu J.J., Orlova N., Oakes B.L., Ma E., Spinner H.B., Baney K.L.M., Chuck J., Tan D., Knott G.J., Harrington L.B. et al.. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature. 2019; 566:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. et al.. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015; 520:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim H.J., Lee H.J., Kim H., Cho S.W., Kim J.S.. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009; 19:1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miller J.C., Holmes M.C., Wang J., Guschin D.Y., Lee Y.L., Rupniewski I., Beausejour C.M., Waite A.J., Wang N.S., Kim K.A. et al.. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007; 25:778–785. [DOI] [PubMed] [Google Scholar]
- 12. Pinello L., Canver M.C., Hoban M.D., Orkin S.H., Kohn D.B., Bauer D.E., Yuan G.C.. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016; 34:695–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Park J., Lim K., Kim J.S., Bae S.. Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics. 2017; 33:286–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guell M., Yang L., Church G.M.. Genome editing assessment using CRISPR Genome Analyzer (CRISPR-GA). Bioinformatics. 2014; 30:2968–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang X., Tilford C., Neuhaus I., Mintier G., Guo Q., Feder J.N., Kirov S.. CRISPR-DAV: CRISPR NGS data analysis and visualization pipeline. Bioinformatics. 2017; 33:3811–3812. [DOI] [PubMed] [Google Scholar]
- 16. Clement K., Rees H., Canver M.C., Gehrke J.M., Farouni R., Hsu J.Y., Cole M.A., Liu D.R., Joung J.K., Bauer D.E. et al.. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 2019; 37:224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lunter G., Rocco A., Mimouni N., Heger A., Caldeira A., Hein J.. Uncertainty in homology inferences: assessing and improving genomic sequence alignment. Genome Res. 2008; 18:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rimmer A., Phan H., Mathieson I., Iqbal Z., Twigg S.R.F., Consortium W.G.S., Wilkie A.O.M., McVean G., Lunter G.. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 2014; 46:912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bae S., Kweon J., Kim H.S., Kim J.S.. Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods. 2014; 11:705–706. [DOI] [PubMed] [Google Scholar]
- 20. Huang W., Li L., Myers J.R., Marth G.T.. ART: a next-generation sequencing read simulator. Bioinformatics. 2012; 28:593–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nakade S., Tsubota T., Sakane Y., Kume S., Sakamoto N., Obara M., Daimon T., Sezutsu H., Yamamoto T., Sakuma T. et al.. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 2014; 5:5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cho S.W., Xu J., Sun R., Mumbach M.R., Carter A.C., Chen Y.G., Yost K.E., Kim J., He J., Nevins S.A. et al.. Promoter of lncRNA Gene PVT1 Is a tumor-suppressor DNA boundary element. Cell. 2018; 173:1398–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim D., Kim J., Hur J.K., Been K.W., Yoon S.H., Kim J.S.. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016; 34:863–868. [DOI] [PubMed] [Google Scholar]
- 24. Kim H.K., Song M., Lee J., Menon A.V., Jung S., Kang Y.M., Choi J.W., Woo E., Koh H.C., Nam J.W. et al.. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat. Methods. 2017; 14:153–159. [DOI] [PubMed] [Google Scholar]
- 25. Findlay G.M., Boyle E.A., Hause R.J., Klein J.C., Shendure J.. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014; 513:120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vierstra J., Reik A., Chang K.H., Stehling-Sun S., Zhou Y., Hinkley S.J., Paschon D.E., Zhang L., Psatha N., Bendana Y.R. et al.. Functional footprinting of regulatory DNA. Nat. Methods. 2015; 12:927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adamson B., Norman T.M., Jost M., Cho M.Y., Nunez J.K., Chen Y., Villalta J.E., Gilbert L.A., Horlbeck M.A., Hein M.Y. et al.. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell. 2016; 167:1867–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dixit A., Parnas O., Li B., Chen J., Fulco C.P., Jerby-Arnon L., Marjanovic N.D., Dionne D., Burks T., Raychowdhury R. et al.. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016; 167:1853–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kalhor R., Mali P., Church G.M.. Rapidly evolving homing CRISPR barcodes. Nat. Methods. 2017; 14:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rubin A.J., Parker K.R., Satpathy A.T., Qi Y., Wu B., Ong A.J., Mumbach M.R., Ji A.L., Kim D.S., Cho S.W. et al.. Coupled single-cell CRISPR screening and epigenomic profiling reveals causal gene regulatory networks. Cell. 2019; 176:361–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kosicki M., Tomberg K., Bradley A.. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018; 36:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CRISPRpic is implemented in Python 2.7 or 3.4 and the source code are available at Github (https://github.com/compbio/CRISPRpic). All raw sequencing reads used in this study are derived through NCBI Sequencing Read Archive with accession number SRP070794 (23) and Gene Expression Omnibus with accession number GSM2572600 (22).