Abstract
Background
Epidermal growth factor-containing fibulin-like extracellular matrix protein 2 (EFEMP2), also known as fibulin-4, MBP1 and UPH1, is an extracellular matrix protein associated with a variety of tumors. The purpose of this study was to investigate the prognostic value and the function of EFEMP2 in lung cancer.
Methods
The mRNA and protein expression of EFEMP2 in lung normal and cancer tissues, lung cancer cell lines (A549, H460, H1299 and H1650) and normal epithelial cell line BEAS-2B were evaluated by immunohistochemistry, RT-qPCR and Western blotting. The Public databases (Oncomine and Kaplan-Meier plotter) were used to investigate the prognostic value of EFEMP2 in lung cancer. RNA interference (RNAi) and overexpression transfection were performed to detect the effects of EFEMP2 up- or down-regulation on lung normal and cancer cell proliferation, invasion and metastasis in vitro and in vivo.
Results
EFEMP2 was lowly expressed in lung cancer tissues and cells, and its low expression was associated with malignant phenotype and poor prognosis of lung cancer. The same conclusion had been drawn from the Public databases. EFEMP2 overexpression significantly inhibited the invasion of lung cancer cells, hampered the process of EMT, and decreased the expression and activity of MMP2 and MMP9, while EFEMP2 knockdown remarkably enhanced the invasion of lung cancer cells, promoted EMT, and increased the expression and activity of MMP2 and MMP9.
Conclusion
The low expression of EFEMP2 was detected in lung cancer and was positively correlated with the poor prognosis of patients. EFEMP2 was a tumor suppressor gene that inhibited the progress of lung cancer, which suggested a new research objective for the future studies.
Keywords: EFEMP2, lung cancer, invasion, metastasis, EMT, MMPs
Introduction
Lung cancer occurs in the bronchial mucosal epithelium, and the incidence of lung cancer has been significantly increased in the last 50 years. In the industrial developed countries of Europe and America and some big industrial cities in China, the incidence of lung cancer has ranked first among male malignant tumors, and the incidence of lung cancer in women has also increased rapidly, accounting for the second or third place of women common malignant tumors. There are about 1.8 million new cases of lung cancer worldwide each year, with more than 1.6 million deaths. Lung cancer has become a major disease endangering life and health.1 The early diagnosis of lung cancer is difficult. About 85% of the patients are in the advanced stage of diagnosis, and the effect of surgical treatment is not ideal. The 5-year survival rate is only 16%, which is in sharp contrast to the high survival rate of 90%, 65% and nearly 100% for breast cancer, colon cancer and prostate cancer, respectively.2 Surgical treatment has been recognized as the first choice for the treatment of lung cancer, but most of the patients missed the opportunity of radical surgery, and the invasion, metastasis and poor prognosis of advanced tumors are still difficult to solve. Gene therapy and targeted therapy have been paid more and more attention as a new generation model of tumor therapy. Molecular targeted drugs can specifically select carcinogenic sites and kill tumor cells without affecting the normal tissue cells around them. Therefore, molecular targeted therapy is also known as “biological missiles”. Compared with traditional chemotherapeutic drugs, molecular targeted drugs have the advantages of strong specificity, obvious curative effect and less side effects. However, no specific markers have been found in the treatment of lung cancer, and there is a lack of understanding of the molecular mechanism of invasion and metastasis.3 therefore, clarifying the mechanism of lung cancer invasion and metastasis and finding effective targeting drugs to inhibit invasion and metastasis is essential to fully improve the prognosis of patients.
Epidermal growth factor-containing fibulin-like extracellular matrix protein 2 (EFEMP2, also known as fibulin-4, MBP1 and UPH1), is a member of fibulin glycoprotein family.4–6 Fibulins are a family of seven extracellular matrix proteins characterized by a tandem array of calcium-binding (cb) epidermal growth factor (EGF)-like modules and a fibulin-type C-terminal domain.7–9 EFEMP2 is a 443 amino acid secretory protein and contains of a N-terminal modified cbEGF module, five consecutive cbEGF modules and the FC domain.4 EFEMP2 is often found in tissues and organs that are rich in elastic fibers, including the blood vessels, heart valves, lung and skin.7,10 This gene is necessary for elastic fiber formation and connective tissue development, and plays an important role in stabilizing the extracellular matrix structure.11,12 Defects of EFEMP2 in patients have been linked to many diseases, such as autosomal recessive cutis laxa syndrome, aneurysms and tortuosity of the large arteries, developmental emphysema, diaphragmatic and inguinal hernia, joint laxity and cancer.13–15 The role of EFEMP2 in tumorigenesis has not yet been fully elucidated, both carcinogenic and anti-carcinogenic effects have been reported. High expression of EFEMP2 was associated with tumor progress and poor prognosis in cervical carcinoma,16 ovarian carcinoma,17 glioblastoma18 and osteosarcoma.19 However, in human endometrial carcinoma, EFEMP2 demonstrated inhibitory effects in tumor invasion and metastasis.20 These ostensibly contradictory findings in different cancers may reflect “context-specific” roles of EFEMP2. Little is known about the direct molecular function of EFEMP2 in cancer progressions. Till now, the function of EFEMP2 in lung cancer has not been explored, so the purpose of this study is to detect the expression of EFEMP2 in lung cancer and its relationship with the progress of lung cancer.
Materials and Methods
Tumor Tissues Samples
With the written informed consent of the patients, a total of 415 lung tissue samples (68 normal lung tissue and 347 non-small cell lung cancer) were collected from Shandong Provincial Hospital between 2005 and 2017. Non-small cell lung cancer (NSCLC) patients were diagnosed according to the 8th edition of the tumor-node-metastasis (TNM) classification of lung cancer recommended by the International Association for the Study of Lung Cancer (IASLC) staging committee. None of them had received radiotherapy or chemotherapy before surgery. This study was approved by the Institutional Medical Ethics Committee of Shandong Provincial Hospital Affiliated to Shandong University and all methods were carried out in accordance with the relevant guidelines and regulations.
Cell Lines
A549, H460, H1299, H1650 (human lung cancer cell lines), and BEAS-2B (normal lung epithelial cell line) were obtained from the Cell Resource Center of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. All cells were cultured in Gibco™ Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F-12, SIGMA, RNBG3415, UK) media supplemented with 10% Gibco™ fetal bovine serum (FBS, Australia origin) and 1% Gibco® Antibiotic ((100 µg/mL streptomycin, 100 U/mL penicillin, Millipore, 61207–1, USA), at 37 °C in a 5% CO2 incubator. Cells were routinely tested every 3 months for Mycoplasma with QuickTest Mycoplasma Detection Kit (Biotool, Houston, TX, USA), and cultured for no longer than 15 passages.
Immunohistochemistry (IHC) and Immunocytochemistry (ICC)
For IHC of the samples, tissue blocks were fixed by formalin, embedded in paraffin and then cut into 5-μm thick sections. After xylene dewaxing, gradient ethanol rehydration and high-pressure antigen retrieval, tissue sections were stained with the selected streptavidin–biotin–peroxidase (SP) staining method. For ICC of the samples, cells in logarithmic growth were harvested and seeded into cell culture dishes covered with coverslips. After 24 h of adhesion, the cell coverslips were fixed with 95% ethanol for 30 min and then stained by the same SP staining method as IHC. According to the instructions of the SPlink Detection Kit (ZSGB-BIO, SP-9001, China), the following operations were carried out: All tissue sections and cell coverslips were treated with rabbit monoclonal antibody against human EFEMP2 (1:100 dilution, ab125073, Abcam) at 4°C overnight, and then incubated with biotin-conjugated anti-rabbit secondary antibody for 30 min at 37 °C. Finally, the enzyme substrate 3′,3-diaminobenzidine tetrahydrochloride (DAB reagent kit, ZSGB-BIO, ZLI-9017, China) were used for color development. The presence of brown particles in cell cytoplasm or stroma was confirmed to be positive for EFEMP2. Normal serum was used instead of original antibody as negative control.
Evaluation of IHC and ICC Staining
To evaluate the staining results of IHC and ICC, a semi-quantitative scoring system was used in this study.21 The staining score was calculated according to the staining strength and the proportion of positive stained cells in tissue sections or cell coverslips. The staining strength was scored from 0 to 3: no staining scored 0, weak staining scored 1, medium staining scored 2, and strong staining scored 3. The proportion of positive stained cells was scored from 0 to 4: 0% positive stained cells scored 0, 1%–25% positive stained cells scored 1, 26%–50% positive stained cells scored 2, 51%–75% positive stained cells scored 3, and 76%–100% positive stained cells scored 4. The sum of staining strength and positive cell proportion score was the total staining score (0 to 7). To facilitate statistical analysis, the total staining score was transformed into sum indexes (–), (+), (++), and (+++), corresponding to score 0, (1–3), (4–5), and (6–7), respectively. The sum indexes (–) and (+) were considered low expression and (++) and (+++) were considered high expression. Three pathologists graded each section and coverslip independently; in the case of differences in scores, an agreement was reached through the discussion.
Analysis of the Oncomine and Kaplan-Meier Plotter Public Databases
In this study, Oncomine array datasets (www.oncomine.org) were utilized to compare the expression of EFEMP2 between clinical lung cancer specimens and normal controls. The detailed information of standardization technology and statistical calculation is provided on Oncomine platform. The prognostic value of EFEMP2 in patients with lung cancer was investigated in Kaplan-Meier Plotter public databases (http://kmplot.com/analysis/). The Kaplan-Meier Plotter estimates the survival function from life-time data and visualize the association between the biomarker and the survival, in which the patients are divided into groups based on the parameters. Log rank test is used to compare the survival curves of two or more groups. Hazard ratios with 95% confidence interval and logrank P value are calculated and displayed.
Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
TRIZol reagent (TaKaRa Biotechnology, China) was used to get total RNA, after quantification and dilution to 500 ng/µL, of which 2 µL was reverse transcribed to complementary DNA (cDNA) by PrimeScriptTM RT reagent Kit (TaKaRa). Then RT-qPCR was performed using TB Green™ Premix Ex Taq™ II kit (TaKaRa) by LightCycler 480 System (Roche Diagnostics). A 20-μL reaction system, including 10 µL TB Green Premix Ex Taq II (2× Tli RNaseH Plus), 0.8 µL forward primer (10 µM), 0.8 µL reverse primer (10 µM), 2 µL DNA template (< 100 ng), and 6.4 µL DNAse/RNAse-free water (Sigma-Aldrich), was added into each well of a 96-well plate, and there were three technical replicas. According to the instructions of the manufacturer, the standard two-step PCR reaction program is run, and the quantitative standard curve is generated. Relative quantitative gene expression levels were analyzed by the convenient 2(-Delta Delta C(T)) method.22 The specific forward or reverse primers were designed and synthesized by Takara Biotechnology Co., Ltd, the sequences of them were given in Table 1.
Table 1.
The Sequence of Primer in RT-qPCR
| Primer Name | Specific Sequences |
|---|---|
| EFEMP2 | F:5ʹ-GCTGCTACTGTTGCTCTTGGG-3ʹ R:5ʹ-GGGATGGTCAGACACTCGTTG-3ʹ |
| CDH1 | F:5ʹ-GGATTGCAAATTCCTGCCATTC-3ʹ R:5ʹ-AACGTTGTCCCGGGTGTCA-3ʹ |
| CDH2 | F:5ʹ-CGAATGGATGAAAGACCCATCC-3ʹ R:5ʹ-GCCACTGCCTTCATAGTCAAACACT-3ʹ |
| VIM | F:5ʹ-AACCTGGCCGAGGACATCA-3ʹ R:5ʹ-TCAAGGTCAAGACGTGCCAGA-3ʹ |
| SNAIL | F:5ʹ-GCTCCCTCTTCCTCTCCATACC-3ʹ R: 5ʹ-AAGTCCTGTGGGGCTGATGT-3ʹ |
| SLUG | F: 5ʹ-GAAGCATTTCAACGCCTCCAA-3ʹ R: 5ʹ-GTTGTGGTATGACAGGCATGGAGTA-3ʹ |
| TWIST | F: 5ʹ-CAGCTACGCCTTCTCGGTCT-3ʹ R: 5ʹ-CTGTCCATTTTCTCCTTCTCTGG-3ʹ |
| ACTB | F: 5ʹ-TGGCACCCAGCACAATGAA-3ʹ R: 5ʹ-CTAAGTCATAGTCCGCCTAGAAGCA-3ʹ |
| MMP2 | F: 5ʹ-CTCATCGCAGATGCCTGGAA-3ʹ R: 5ʹ-TTCAGGTAATAGGCACCCTTGAAGA-3ʹ |
| MMP9 | F: 5ʹ-ACGCACGACGTCTTCCAGTA-3ʹ R: 5ʹ-CCACCTGGTTCAACTCACTCC-3ʹ |
Western Blotting
Cells were harvested and lysed by radioimmunoprecipitation assay (RIPA) lysis buffer with phenylmethylsulfonyl fluoride (PMSF) solution (RIPA:PMSF=100:1) on ice for 30 min. Protein samples (40 µg/lane) were separated using 10% sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes. The Western blot membranes were blocked with 5% bovine serum albumin (BSA) for 1 h, and incubated with primary antibodies (EFEMP2, ab125073, Abcam; E-cadherin, N-cadherin, Vimentin, Snail, Slug, #9782, Cell Signaling Technology; Twist, sc-81417 Santa Cruz Biotechnology) at working dilutions (1﹕1000) overnight at 4 °C. The next day, after washing, the membranes were incubated with the corresponding anti-rabbit or mouse secondary antibodies (1﹕2000, Boster Biological Technology) at room temperature for 1 h. Beta-actin (ACTB) and glyceraldehydes-3-phosphate dehydrogenase (GAPDH) are known as a “housekeeping” protein and usually used as a loading control to normalize the levels of target protein. On the following day, the membranes were incubated with the corresponding secondary antibodies (Boster Biology) for 1 h, and developed by the enhanced chemiluminescence (ECL) method (Pierce™ ECL Western Blotting Substrate; Thermo Fisher Scientific, Inc.). Finally, the gray-scale value of the images was measured by Image J (National Institutes of Health, USA).
Lentiviral Transfection for RNA Interference and Overexpression
The lentiviral EFEMP2 cDNA, small hairpin RNA (shRNA) and corresponding negative control vectors were synthesized by Genechem Co., Ltd (Shanghai, China). In order to explore the function of EFEMP2 in lung cancer progress, the expression of EFEMP2 in lung cancer cells H1299 and H1650 were up-regulated by overexpression lentiviral transfection with EFEMP2 cDNA vector, whilst the expression of EFEMP2 in normal lung epithelial cells BEAS-2B were down-regulated by RNAi lentiviral transfection with EFEMP2 shRNA vector. According to the experimental procedures provided by Genechem Co., firstly cells in logarithmic growth were cultured in a 24-well plate for 24 h, then the virus quantity was calculated with an MOI (multiplicity of infection, average virus active units/cell at transfection) value of 100 for overexpression, and 50 for RNAi. To avoid cytotoxicity, the virus mixture was replaced with complete culture medium after 12 h culture. Finally, after 72 h of conventional culture, transfected cells with green fluorescence were observed using the high content imager (Image Xpress Micro Confocal, Molecular Devices). Differential expressions at mRNA and protein levels between the control group and the transfected group, were detected by RT-qPCR and Western blotting to verify the effective transfection.
Cell Growth Curve Assay
Cell growth curve is not only an important index to determine the vitality of cells, but also one of the basic parameters of the biological characteristics of cultured cells. So it can accurately describe the dynamic changes of the number of cells during the whole growth process. The cells with good growth and close to confluence were digested with trypsin and counted after being made into cell suspension with new culture medium. According to the results of the cell count, the cells were inoculated into 24-well plates (1 × 104 cells/well) and cultured in 37 °C and 5% CO2 incubator. From the next day, three wells of cells were taken every 24 h and counted respectively, and their average values were taken for continuous 7 days in a row. According to the results of cell count, the growth curve was drawn with number of living cells as longitudinal coordinate and time as transverse coordinate.
Plate Clone Formation Assay
The cell plate clone formation assay reflects two important traits of cells: population dependence and proliferation ability. In each group, the logarithmic cells were collected and fully blown into single cell suspension to disperse the cells completely. And then the suspension was gradually diluted to ensure that only 500 cells were inoculated into 35mm petri dish and cultured at 37 °C, 5% CO2 and saturated humidity for 2 weeks. When the culture terminated, the clones were visible to the naked eye. Discard the culture medium from the dishes and carefully rinse twice with PBS. The cell clones were fixed by 4% paraformaldehyde for 30 mins and stained by GIMSA staining solution for 10 mins. The number of clones greater than 50 cells was counted under a microscope (low-fold mirror), and the average of three dishes was recorded.
In vitro Cell Invasion and Migration Assay
Transwell chamber, also named as Boyden chamber, consists of two compartments separated by a microporous membrane with a pore size of 8 µm, is commonly used and highly accessible method for examining cell migration and invasion in vitro. The transwell cell migration assay can be used to determine the migration ability of cells toward a chemo-attractant. The transwell cell invasion assay measures both cell chemotaxis and the invasion of cells through ECM (extracellular matrix), which commonly occurred in the process of tumor metastasis.
For cell migration assay, add 150 μL of cell solution (1×106 cells/mL) on top of the filter membrane in the 24-well transwell insert and 600 μL of serum-free conditioned medium of NIH3T3 cells as the chemotactic factor into the bottom of the lower chamber. After culture for 12 h at 37 °C with 5% CO2, pick up the transwell insert from the plate and carefully remove the remaining cells that had not migrated across the membrane. Next, put the transwell insert into 4% paraformaldehyde to fix the migrated cells for 30 min and then stain them by Hematoxylin and Eosin staining kit (Beyotime Biotechnology, China). Finally, under an inverted microscope set at 200×magnification (OLYMPUS, BX63F, Japan), the number of stained cells migrated across the membrane to the other side were enumerated. For cell invasion assay, Matrigel (BD BioCoat), the ECM material, was thawed and liquefied on ice, and then diluted by serum-free medium at a ratio of 1:5, of which 50 μL is added on the top of the membrane in the transwell insert and solidified in a 37 °C incubator for 30 min to form a thin gel layer, simulating the structure and physical properties of ECM. The other steps were the same as the cell migration assay, including the number of cells added on top of the Matrigel coating in the upper chamber and the amount of chemokines added to the lower chamber, as well as fixing, staining and counting methods, but the culture time was 24 h instead of 12 h.
Xenograft Tumor Model in vivo
Female BALB/C/nu/nu mice, 4–6 weeks old and weighed 18–22 g, were purchased from the National Rodent Experimental Animal Seed Center. They were randomly divided into up-regulation group, down-regulation group and corresponding negative control group (n = 5/group), and maintained in a specific pathogen free (SPF) environment with 20–26 °C temperature and 40–60% humidity. Then, 0.3 mL targeted cell suspension (1 × 107 cells) was inoculated subcutaneously on the back and neck of the nude mice. After inoculation, the nude mice were raised in SPF environment for 2months. In the meantime, the professional was responsible for observing and recording tumorigenesis and overall health status of all nude mice. After 8 weeks, all nude mice were sacrificed by carbon dioxide (CO2) euthanasia, and the subcutaneous tumors of each group were measured and dissected. The tumor volume was calculated according to the formula, V (mm3) = long diameter (mm) × short diameter2 (mm2) × 1/2. All mice experiments were approved by the Animal Protection and Use Committee of Shandong Provincial Hospital Affiliated to Shandong University, and performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People’s Republic of China.
Matrix Metalloproteinase (MMP) Zymography Assay
The activities of MMP2 and MMP9 were monitored using the matrix metalloproteinase zymography assay. Zymegraphy assay is a widely used method for detecting protease based on SDS-PAGE electrophoresis and reversed-phase gel staining. SDS-PAGE gel with collagenase substrate was prepared. The sample containing protease was electrolyzed in the gel. After electrophoresis, the gel was incubated with enzyme reaction buffer, and then stained and decolorized. The protease degraded the substrate protein in the gel, thus indicating the position and activity of MMP-2 and MMP-9 at the same time.
Statistical Analysis
Statistical analysis was performed using SPSS Statistics 24.0. Measurement data are expressed as “mean ± SE” and analyzed using two independent sample two-tailed t-test (two groups) and one-way ANOVA (three and more groups, between groups with SNK or Dunnett-t test). However, numeration data were analyzed by χ2 test or Fisher exact test. P < 0.05 (two-sided) was considered statistically significant.
Results
Expression of EFEMP2 in Normal Lung Tissue and Non-Small Cell Lung Cancer (NSCLC)
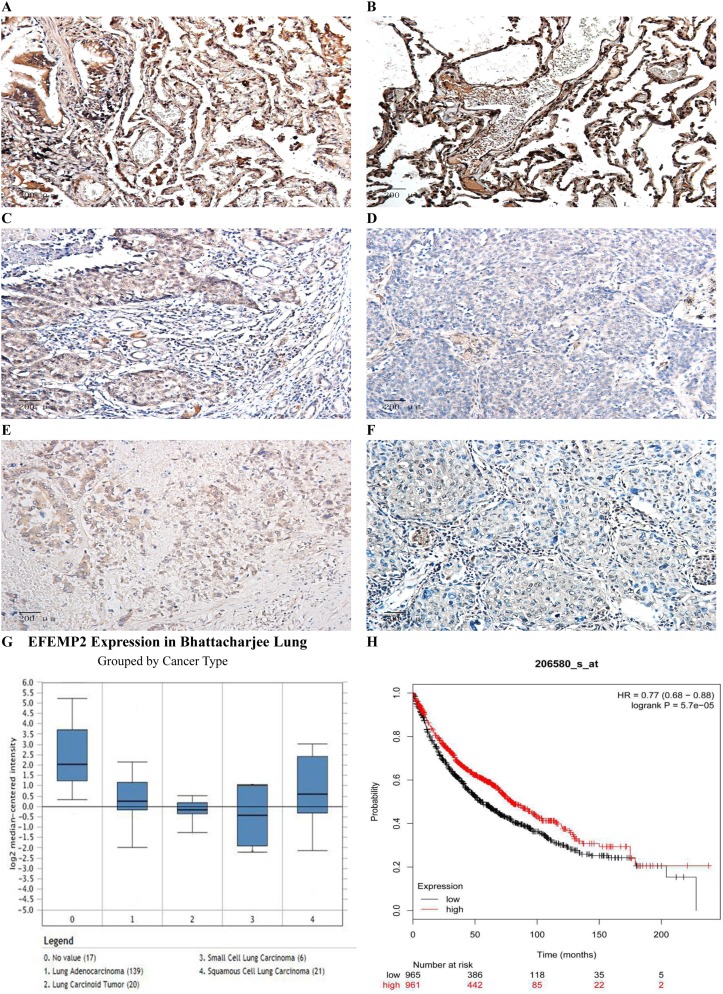
The results of IHC showed that the expression of EFEMP2 in normal lung tissue (Figure 1A and B) was significantly higher than that of NSCLC (Figure 1C–F), and the staining was mainly focused on the cell membrane and cytoplasm. The percentage of EFEMP2 high-expression in normal lung tissue was 80.9% (55/68), which was significantly higher than that of NSCLC (14.4%) (50/347), see Table 2. Compared with poorly differentiated squamous cell carcinoma and adenocarcinoma (Figure 1D and F), EFEMP2 was highly expressed in well-differentiated squamous cell carcinoma and adenocarcinoma (Figure 1C and E).
Figure 1.
Expressions of EFEMP2 in lung normal and cancer tissues and analysis of the public databases.
Notes: EFEMP2 expressions of (A, B) normal lung tissue, (C) well-differentiated squamous cell carcinoma, (D) poorly differentiated squamous cell carcinoma, (E) well-differentiated adenocarcinoma and (F) poorly differentiated adenocarcinoma were measured by IHC. Magnification ×200. (G) In Bhattacharjee Lung Dataset of Oncomine, EFEMP2 mRNA expression in No 0. normal lung was 4.334 times higher than that in No 1. Lung Adenocarcinoma, 6.171 times higher than that in No 2. Lung Carcinoid Tumor, 8.097 times higher than that in No 3. Small Cell Lung Carcinoma, and 3.390 times higher than that in No 4. Squamous Cell Lung Carcinoma (P < 0.05). (H) Analysis of the public Kaplan-Meier plotter database of lung cancer, patients with high EFEMP2 expression (red line) had a much better prognosis than those with low EFEMP2 expression (black line).
Table 2.
Expression of EFEMP2 in Normal Lung Tissue and NSCLC
| Tissue Type | N | EFEMP2 High n (%) | EFEMP2 Low n (%) | χ2 | P |
|---|---|---|---|---|---|
| Normal lung tissue | 68 | 55 (80.9) | 13 (19.1) | 132.931 | 0.000 |
| NSCLC | 347 | 50 (14.4) | 297 (85.6) |
Relationship Between the Expression of EFEMP2 and Clinicopathological Features of NSCLC Patients
The age of the NSCLC patients was from 30 to 86 years, with an average of 60 years old. There was no significant difference in the high expression rate of EFEMP2 between patients aged ≤ 60 years and > 60 years old with NSCLC (P > 0.05). Similarly, there was also no significant difference between female and male patients (P > 0.05). The expression level of EFEMP2 was not correlated with NSCLC histologic types, such as squamous cell carcinoma, adenocarcinoma, and adenosquamous carcinoma (P > 0.05), but related to histological grade of squamous cell carcinoma, pathological subtype of adenocarcinoma, TNM clinical stage and lymph node metastasis (P < 0.05). The expression of EFEMP2 in well-differentiated squamous cell carcinoma (I grade) was significantly higher than that in poorly differentiated squamous cell carcinoma (III grade). EFEMP2 showed significantly higher expression in lepidic adenocarcinomas (well-differentiated) than in solid and micropapillary adenocarcinomas (poorly differentiated) (P < 0.05). The high expression of EFEMP2 was negatively correlated with late clinical stage, low histological grade and positive lymph node metastasis in NSCLC patients, as shown in Table 3.
Table 3.
Relationship Between the Expression of EFEMP2 and Clinicopathological Features of NSCLC Patients
| Clinicopathological Features | N | EFEMP2 High n (%) | EFEMP2 Low n (%) | χ2 | P |
|---|---|---|---|---|---|
| Age | 0.580 | 0.446 | |||
| ≤60 | 156 | 20 (12.8) | 136 (87.2) | ||
| >60 | 191 | 30 (15.7) | 161 (84.3) | ||
| Gender | 0.294 | 0.588 | |||
| Male | 189 | 29 (15.3) | 160 (84.7) | ||
| Female | 158 | 21 (13.3) | 137 (86.7) | ||
| TNM clinical stages | 10.898 | 0.001 | |||
| I and II stage | 168 | 35 (20.8) | 133 (79.2) | ||
| III and IV stage | 179 | 15 (8.4) | 164 (91.6) | ||
| Pathological type | 1.157* | 0.561* | |||
| Squamous cell carcinoma | 126 | 19 (15.1) | 107 (84.9) | 10.790** | 0.005** |
| I grade | 24 | 8 (33.3) | 16 (66.7) | ||
| II grade | 45 | 8 (17.8) | 37 (82.2) | ||
| III grade | 57 | 3 (5.3) | 54 (94.7) | ||
| Adenocarcinoma | 177 | 27 (15.3) | 150 (84.7) | 11.991*** | 0.017*** |
| Lepidic-predominant | 33 | 11 (33.3) | 22 (66.7) | ||
| Acinar-predominant | 38 | 6 (15.8) | 32 (84.2) | ||
| Papillary-predominant | 37 | 5 (13.5) | 32 (86.5) | ||
| Solid-predominant | 35 | 2 (5.7) | 33 (94.3) | ||
| Micropapillary-predominant | 34 | 3 (8.8) | 31 (91.2) | ||
| Adenosquamous carcinoma | 44 | 4 (9.1) | 40 (90.9) | ||
| Lymph node status (N) | 22.858 | 0.000 | |||
| N0 | 143 | 36 (25.2) | 107 (74.8) | ||
| N+ | 204 | 14 (6.9) | 190 (93.1) |
Notes: *There was no significant difference in the high expression rate of EFEMP2 among different NSCLC pathological types, such as squamous cell carcinoma, adenocarcinoma, and adenosquamous carcinoma (P > 0.05). **There was a significant difference among I-III histological grade of squamous cell carcinoma, with the increase of the histological grade, the expression of EFEMP2 was decreased (P < 0.05). ***There was a significant difference among pathological subtype of adenocarcinoma, the expression of EFEMP2 in lepidic adenocarcinomas (well-differentiated) was significantly higher than that in other type of adenocarcinomas (P < 0.05).
Analysis of the Public Databases
Based on the analysis of Oncomine datasets, we found that in Bhattacharjee Lung Dataset, EFEMP2 mRNA expression in normal lung (17) was 4.334 times higher than that in Lung Adenocarcinoma (139), 3.390 times higher than that in Squamous Cell Lung Carcinoma (21), 6.171 times higher than that in Lung Carcinoid Tumor (20), and 8.097 times higher than that in Small Cell Lung Carcinoma (6) (P < 0.05), as shown in Figure 1G and Table 4. There were similar conclusions came from Selamat Lungo, Hou Lung, Landi Lung, Su Lung, Stearman Lung and Okayama Lung Oncomine datasets, which revealed that the mRNA expression of EFEMP2 was significantly reduced in lung carcinomas (Table 4). The relationship between the expression of EFEMP2 and the prognosis of patients with lung cancer was analyzed by public database Kaplan-Meier plotter. The patients were divided into two groups as having higher or lower expression of EFEMP2. Bonferroni multiple testing correction was applied when generating the P value. Survival curve was drawn with survival time as abscissa and survival probability as ordinate. The median survival time (survival probability was 0.5) in over expression group was much longer than that in lower expression group, and logrank P < 0.05 indicated that the good prognostic effect of the over expression EFEMP2 was highly significant. In one word, the prognosis of patients with high expression of EFEMP2 was significantly better than that with low expression of EFEMP2 (HR=0.77 (0.68–0.88), logrank P=5.7e-05). The low expression of EFEMP2 indicated poor prognosis in lung cancer patients (Figure 1H).
Table 4.
The Differential Analysis of EFEMP2 mRNA Expression in Normal Lung and Lung Carcinoma Tissue in Oncomine Datasets
| Datasets | Lung Carcinoma vs Normal | Fold Change | P |
|---|---|---|---|
| Bhattacharjee Lung | Lung Adenocarcinoma (139) Lung (17) |
−4.334 | 4.28E-5 |
| Squamous Cell Lung Carcinoma (21) Lung (17) |
−3.390 | 0.004 | |
| Lung Carcinoid Tumor (20) Lung (17) |
−6.171 | 3.10E-6 | |
| Small Cell Lung Carcinoma (6) Lung (17) |
−8.097 | 2.60E-4 | |
| Selamat Lung | Lung Adenocarcinoma (58) Lung (58) |
−2.439 | 6.27E-18 |
| Hou Lung | Large Cell Lung Carcinoma (19) Lung (65) |
−2.600 | 2.45E-10 |
| Lung Adenocarcinoma (45) Lung (65) |
−1.431 | 1.06E-4 | |
| Squamous Cell Lung Carcinoma (27) Lung (65) |
−1.510 | 1.47E-4 | |
| Landi Lung | Lung Adenocarcinoma (58) Lung (49) |
−1.416 | 1.45E-13 |
| Su Lung | Lung Adenocarcinoma (27) Lung (30) |
−1.543 | 3.06E-6 |
| Stearman Lung | Lung Adenocarcinoma (20) Lung (19) |
−1.387 | 3.07E-4 |
| Okayama Lung | Lung Adenocarcinoma (226) Lung (20) |
−1.536 | 3.99E-9 |
Expression of EFEMP2 in Normal Lung Epithelial Cells and 4 Lung Cancer Cell Lines
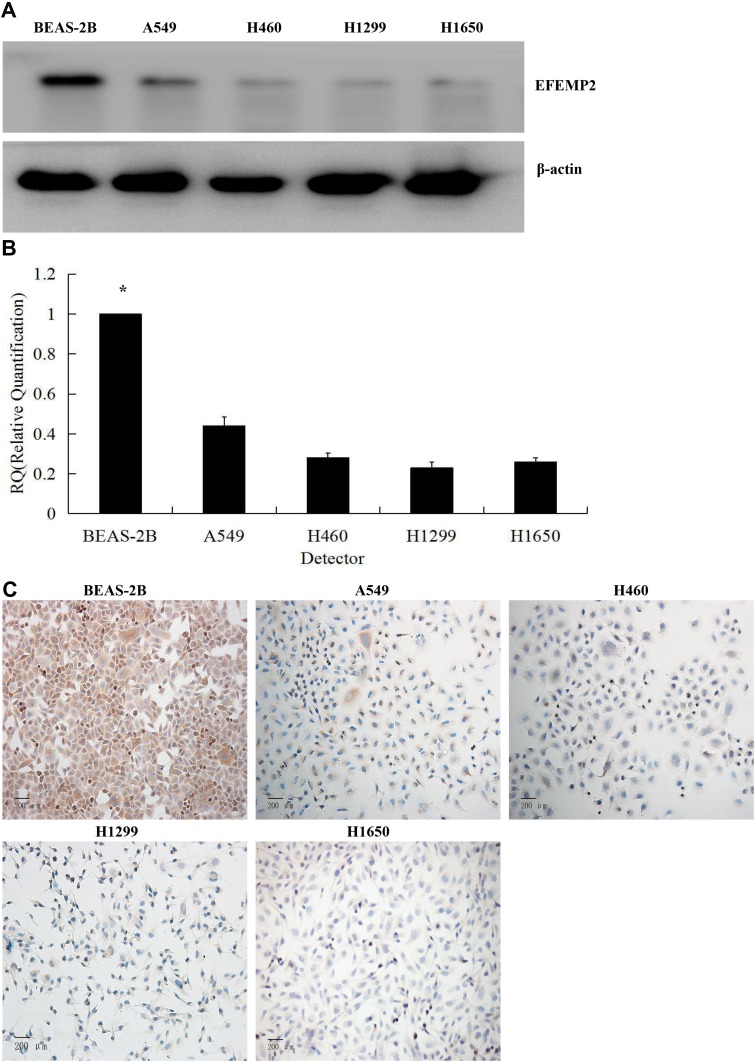
The results of Western blotting (Figure 2A) and RT-qPCR (Figure 2B) showed that the expression of EFEMP2 in 4 lung cancer cell lines (A549, H460, H1299 and H1650) was significantly lower than that in normal lung epithelial cell line BEAS-2B, at both mRNA and protein levels. We could get the same result from ICC (Figure 2C), and the staining was also focused on the cell membrane and cytoplasm, which was consistent with the IHC results of lung cancer tissue. The downward trend of EFEMP2 expression from normal lung to lung cancer in both tissues and cells suggested that the down-regulation of EFEMP2 expression was closely related to the malignant phenotype of lung cancer cells.
Figure 2.
Expression of EFEMP2 in normal lung epithelial cells and 4 lung cancer cell lines.
Notes: The expression of EFEMP2 in normal lung epithelial cell line BEAS-2B and 4 lung cancer cell lines (A549, H460, H1299 and H1650) were measured by (A) Western blotting (cropped blot), (B) RT-qPCR and (C) ICC staining. Magnification ×200. *P < 0.05.
Verification of Lentiviral Transfer Efficiency for EFEMP2 Knockdown and Overexpression
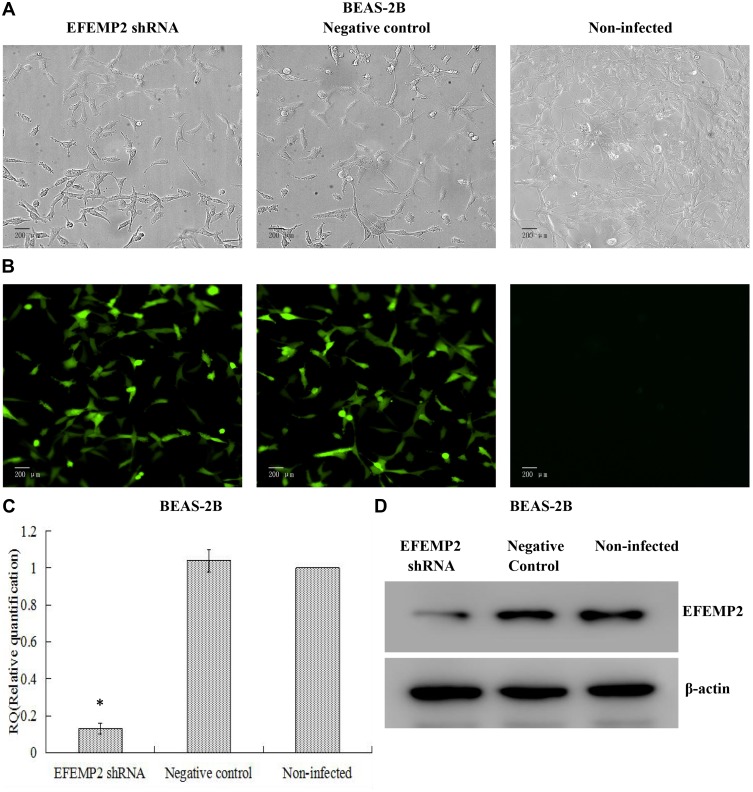
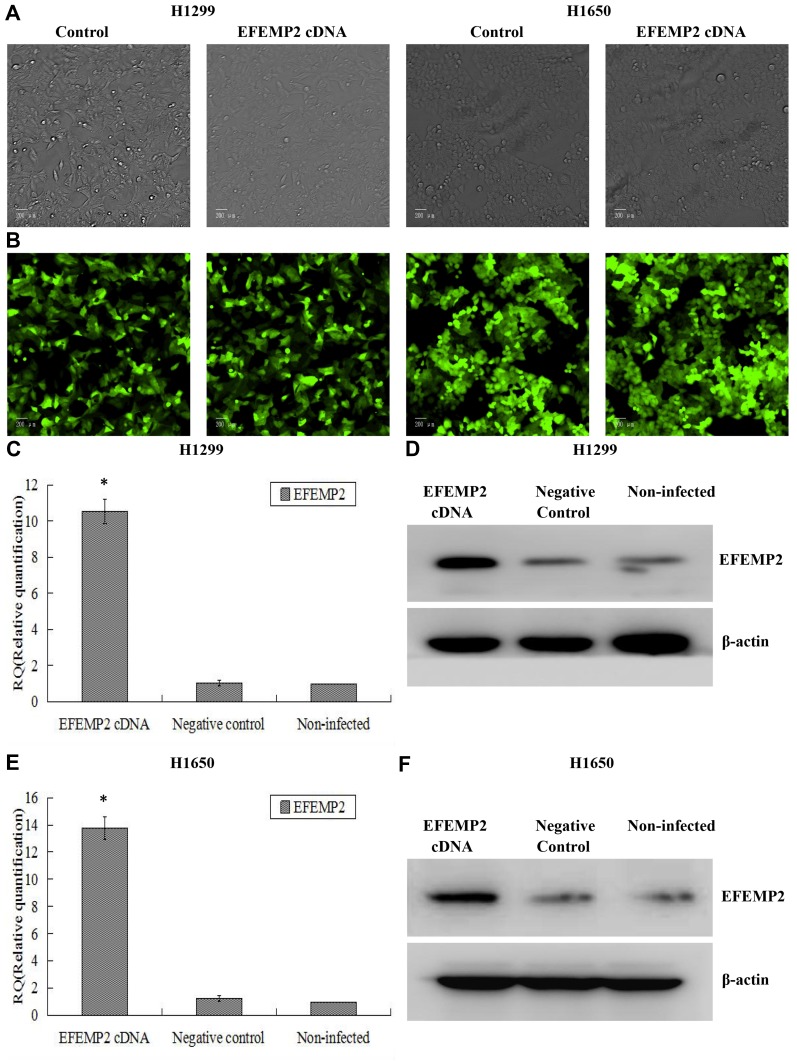
Based on the fact that EFEMP2 was highly expressed in normal lung epithelial cells and lowly expressed in lung cancer cells, we increased EFEMP2 expression in lung cancer cell lines H1299 and H1650 and decreased EFEMP2 expression in normal lung epithelial cell line BEAS-2B, in order to detect the function of EFEMP2 in proliferation, migration, invasion and metastasis of lung cancer cells in vivo and in vitro. For RNAi experiment, after 72 h of normal lung epithelial cell transfection, the rate of GFP positive cells under fluorescence microscope was more than 80%, which meant that the lentiviral transfer efficiency was higher (Figure 3A and B). Next, by RT-qPCR (Figure 3C) and Western blotting (Figure 3D), we confirmed that the expression of EFEMP2 was remarkably down-regulated in BEAS-2B cells transfected with EFEMP2 shRNA, and there was no significant difference between the negative control shRNA transfected cells and the uninfected cells. For overexpression experiment, we also affirmed the successful and effective transfection in the lung cancer cell lines H1299 and H1650, with more than 80% GFP positive cells (Figure 4A and B) and up-regulation of EFEMP2 expression in EFEMP2 cDNA transfected cells at mRNA (Figure 4C and E) and protein (Figure 4D and F) levels.
Figure 3.
Verification of lentiviral transfer efficiency for EFEMP2 knockdown in normal lung epithelial cells.
Notes: (A) Phase contrast images and (B) GFP fluorescence images showed effective lentiviral transfer efficiencies in normal lung epithelial cell line BEAS-2B, with more than 80% GFP positive cells. The expression of EFEMP2 in EFEMP2 shRNA transfected cells, negative control shRNA transfected cells and uninfected cells were measured by (C) RT-qPCR and (D) Western blotting (cropped blot). Magnification ×200. *P < 0.05.
Figure 4.
Verification of lentiviral transfer efficiency for EFEMP2 overexpression in lung cancer cells.
Notes: (A) Phase contrast images and (B) GFP fluorescence images showed effective lentiviral transfer efficiencies in lung cancer cell lines H1299 and H1650, with more than 80% GFP positive cells. The expression of EFEMP2 in EFEMP2 shRNA transfected H1299 cells, negative control shRNA transfected H1299 cells and uninfected H1299 cells were measured by (C) RT-qPCR and (D) Western blotting (cropped blot). The expression of EFEMP2 in EFEMP2 shRNA transfected H1650 cells, negative control shRNA transfected H1650 cells and uninfected H1650 cells were measured by (E) RT-qPCR and (F) Western blotting (cropped blot). Magnification ×200. *P < 0.05.
Effects of EFEMP2 Up- or Down-Regulation on Lung Normal and Cancer Cell Migration and Invasive Activities in vitro
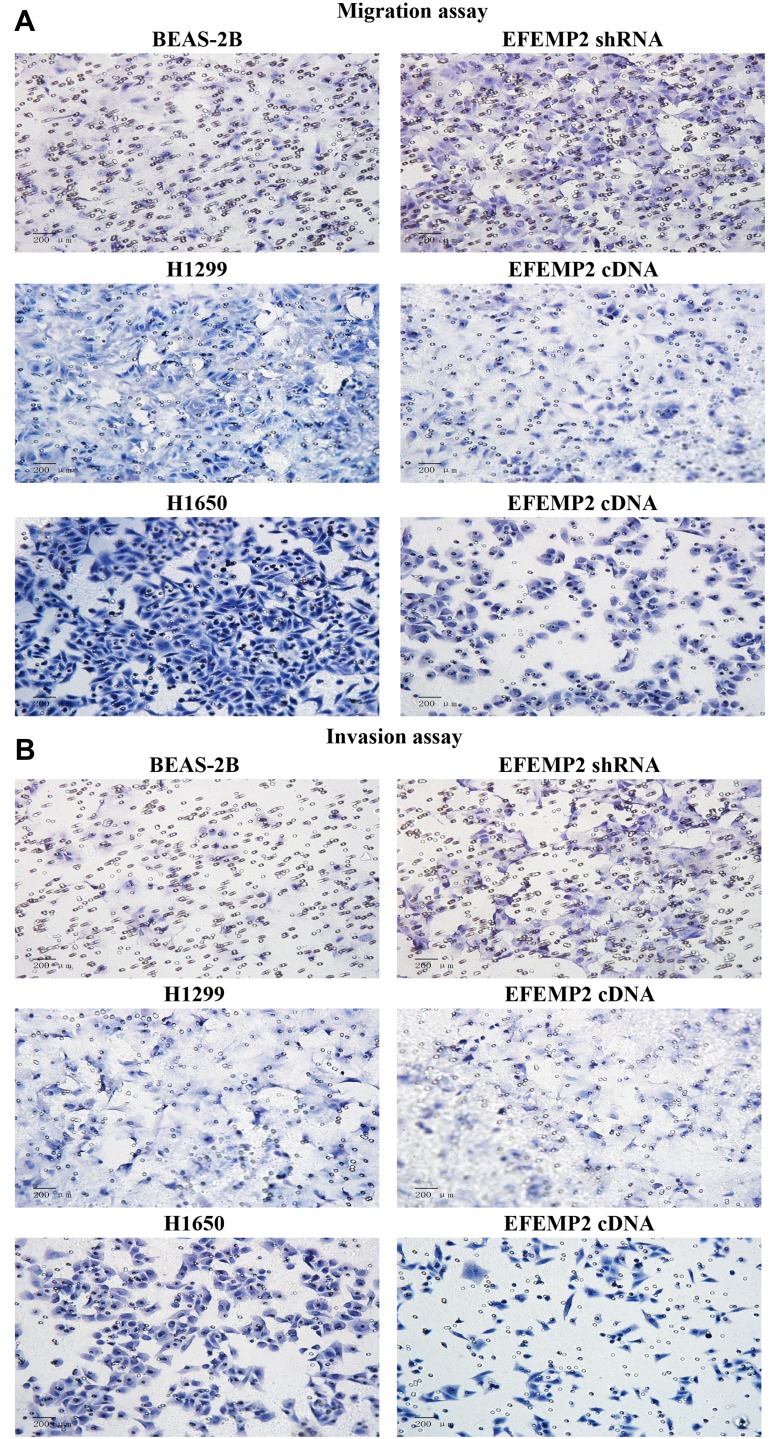
We evaluated the effects of EFEMP2 up- or down-regulation on lung normal and cancer cell migration and invasion activity by cell migration and invasion assay using Transwell chambers. By comparing the average number of cells migrated through the PVPF membrane or invaded through the Matrigel among different groups, cell migration and invasion activity could be determined. In the cell migration assay (Figure 5A), when EFEMP2 expression was down-regulated, more cells migrated through the PVPF membrane in the EFEMP2 shRNA transfected group than in the control group, in contrast, when EFEMP2 expression was up-regulated, fewer cells migrated through the PVPF membrane in the EFEMP2 cDNA transfected group than in the control group. Thus, EFEMP2 down-regulation increased the migration capacity of normal lung epithelial cells, whereas EFEMP2 up-regulation inhibited the migration capacity of lung cancer cells. Meanwhile, in the cell invasion assay (Figure 5B), when EFEMP2 expression was down-regulated, the average number of EFEMP2 shRNA-transfected cells that invaded through the Matrigel were considerably higher than that of the negative control cells, on the contrary, when EFEMP2 expression was up-regulated, the average number of EFEMP2 cDNA transfected cells that invaded through the Matrigel were significantly less than that of the negative control cells. Therefore, EFEMP2 down-regulation increased the invasive capacity of normal lung epithelial cells, while EFEMP2 up-regulation inhibited the invasive capacity of lung cancer cells. In conclusion, EFEMP2 down-regulation promoted the migration and invasion ability of normal lung cells, while EFEMP2 up-regulation inhibited the migration and invasion ability of lung cancer cells.
Figure 5.
Effects of EFEMP2 up- or down-regulation on lung normal and cancer cell migration and invasive activities in vitro.
Notes: (A) Images of cell migration assay performed in control shRNA or EFEMP2 shRNA transfected BEAS-2B cells, and control cDNA or EFEMP2 cDNA transfected H1299 and H1650 cells, by Boyden chambers without Matrigel. (B) Images of cell invasion assay performed in control shRNA or EFEMP2 shRNA transfected BEAS-2B cells, and control cDNA or EFEMP2 cDNA transfected H1299 and H1650 cells, by Boyden chambers coated with Matrigel. When EFEMP2 expression was down-regulated, more cells migrated through the PVPF membrane or invaded through the Matrigel in the EFEMP2 shRNA transfected group than in the control group, in contrast, when EFEMP2 expression was up-regulated, fewer cells migrated through the PVPF membrane or invaded through the Matrigel in the EFEMP2 cDNA transfected group than in the control group. In summary, EFEMP2 down-regulation promoted invasion and migration abilities of normal lung BEAS-2B cells, otherwise EFEMP2 up-regulation inhibited invasion and migration abilities of lung cancer H1299 and H1650 cells. Magnification ×200.
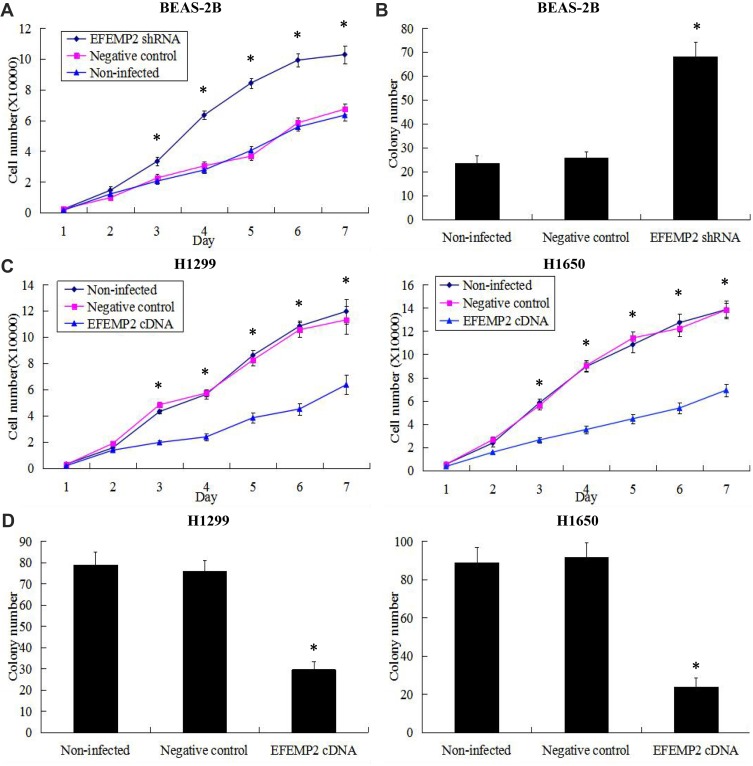
Effects of EFEMP2 Up- or Down-Regulation on Lung Normal and Cancer Cell Proliferation and Clonogenicity
The growth curves reflected the ability of cell growth and proliferation, and the plate clone formation assay reflected cell population dependence and clone formation potential. These two experiments were carried out to detect the effects of EFEMP2 up- or down-regulation on lung normal and cancer cell proliferation and clonogenicity. After RNAi experiment, growth curves showed that EFEMP2 shRNA-transfected cells grew faster and had a better proliferative ability than the negative control cells (Figure 6A). At the same time, the results of the plate clone formation assay showed that the average number of colonies formed by EFEMP2 shRNA-transfected cells was considerably higher than that formed by negative control cells (Figure 6B). In summary, EFEMP2 down-regulation distinctly promoted the proliferation and clonogenicity of normal lung epithelial cells. After overexpression experiment, compared with the negative control cells, EFEMP2 cDNA-transfected cells grew slowly (Figure 6C) and formed fewer clones (Figure 6D), which meant that EFEMP2 overexpression significantly inhibited the proliferation and clonogenicity of lung cancer cells.
Figure 6.
Effects of EFEMP2 up- or down-regulation on lung normal and cancer cell proliferation and clonogenicity.
Notes: Cell proliferation ability was evaluated by growth curve, and cell clonogenicity ability was evaluated by plate clone formation assay. (A) EFEMP2 down-regulation distinctly promoted cell proliferative abilities of normal lung epithelial cell line BEAS-2B. (B) The colony numbers formed by EFEMP2 shRNA transfected BEAS-2B cells were much more than that formed by the negative control cells. There was no significant difference between the negative control shRNA transfected cells and the uninfected cells. (C) EFEMP2 up-regulation significantly inhibited cell proliferative abilities of both lung cancer cell lines H1299 and H1650. (D) The colony numbers were markedly down-regulated in EFEMP2 cDNA transfected H1299 and H1650 cells, compared to negative control groups. There was no significant difference between the negative control cDNA transfected cells and the uninfected cells. EFEMP2 knockdown evidently promoted normal lung cell proliferation and clonogenicity, meanwhile, EFEMP2 overexpression distinctly inhibited lung cancer cell proliferation and clonogenicity. *P < 0.05.
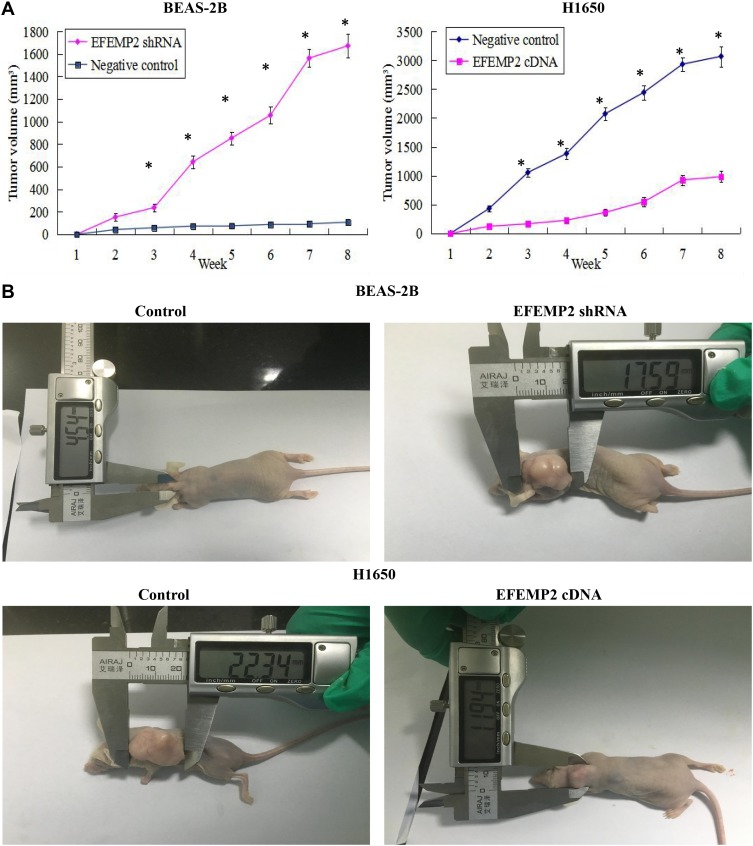
Effects of EFEMP2 Up- or Down-Regulation on Tumor Growth in the Nude Mice
The model of xenotransplantation was set up in the nude mice to further evaluate the effects of EFEMP2 up- or down-regulation on tumor growth in vivo. Five nude mice in each group were inoculated subcutaneously with EFEMP2 shRNA-transfected BEAS-2B cells, EFEMP2 cDNA-transfected H1650 cells and their corresponding negative control cells, respectively. After that all mice were raised in the SPF environment for 8 weeks. The tumors formed by EFEMP2 shRNA-transfected BEAS-2B cells grew faster than that formed by the negative control cells, whereas the EFENP2 cDNA transfected group showed a clear delay in both tumor size and growth rate in vivo (Figure 7A). the subcutaneous tumors formed by EFEMP2 shRNA-transfected BEAS-2B cells were larger than that formed by the negative control cells, in contrast, the average subcutaneous tumor size in the EFENP2 cDNA transfected group was significantly smaller than that in the negative control group (Figure 7B). Collectively, EFEMP2 knockdown could promote tumor formation and growth in the nude mice, whereas EFEMP2 up-regulation could inhibit tumor growth in vivo.
Figure 7.
Effects of EFEMP2 up- or down-regulation on tumor growth in the nude mice.
Notes: (A) The growth tumors profile of control or EFEMP2 shRNA transfected BEAS-2B cells and control or EFEMP2 cDNA transfected H1650 cells were observed continuously for 8 weeks. The tumors formed by EFEMP2 shRNA-transfected BEAS-2B cells grew faster than that formed by the negative control cells, whereas the EFENP2 cDNA transfected group showed a clear delay in both tumor size and growth rate in vivo. (B) Images of xenotransplantation tumor after subcutaneous inoculation of control or EFEMP2 shRNA transfected BEAS-2B cells and control or EFEMP2 cDNA transfected H1650 cells. The subcutaneous tumors formed by EFEMP2 shRNA-transfected BEAS-2B cells were larger than that formed by the negative control cells, in contrast, the average subcutaneous tumor size in the EFENP2 cDNA transfected group was significantly smaller than that in the negative control group. EFEMP2 knockdown could promote tumor formation and growth in the nude mice, whereas EFEMP2 up-regulation could inhibit tumor growth in vivo. *P<0.05.
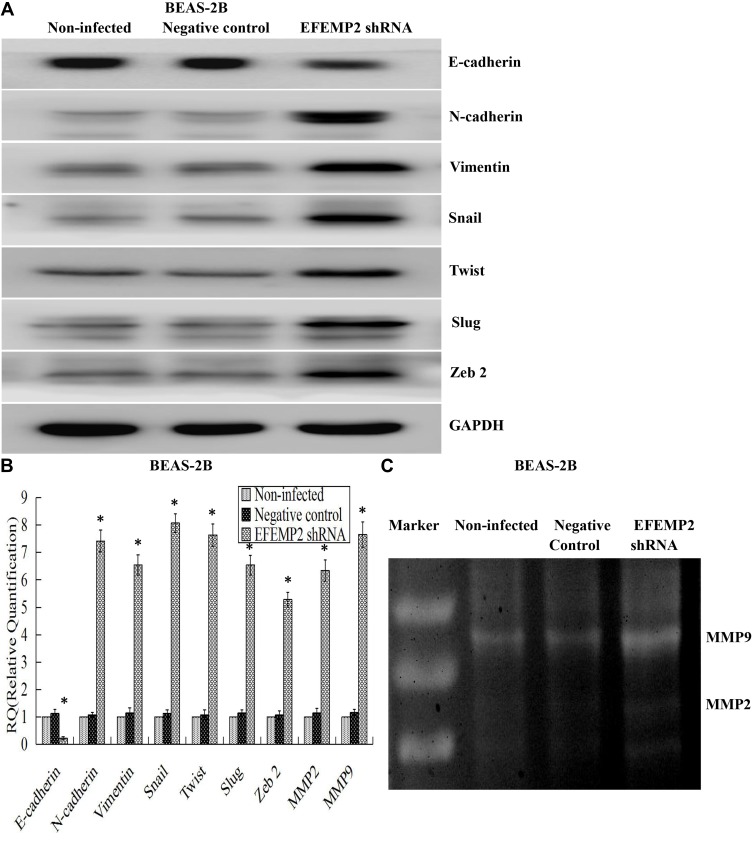
Effects of EFEMP2 Up- or Down-Regulation on the EMT-Related Genes and Matrix Metalloproteinases (MMPs)
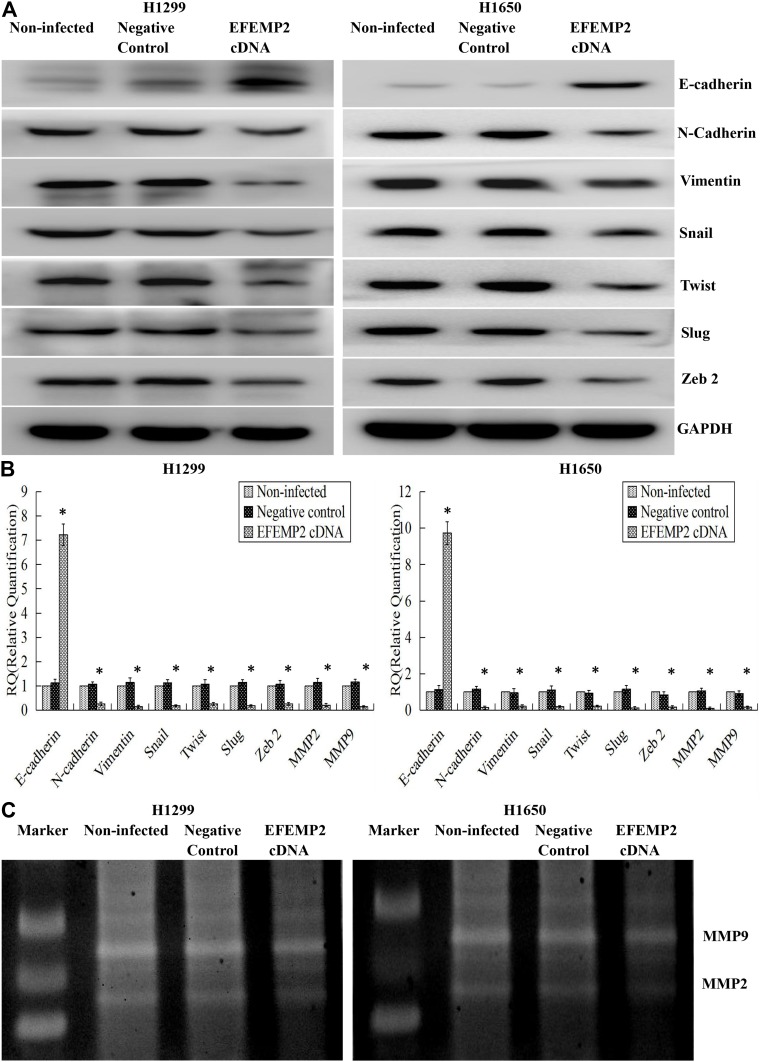
EMT is a special biological process, in which epithelial cells would lose cell polarity and epithelial phenotype and transform into mesenchymal cells with the phenotype of high migration and invasion and the ability of degrading the extracellular matrix (ECM). EMT plays an important role in embryonic development, chronic inflammation, tissue reconstruction, fibrotic diseases and cancer metastasis.23,24 From our present experiments, we found that EFEMP2 up- or down-regulation could affect the proliferation and invasion capacity of lung cancer cells in vitro and in vivo; therefore, we doubted whether EFEMP2 up- or down-regulation would affect the EMT related genes and MMPs. By Western blotting (Figure 8A), we found that at the protein level, in the EFEMP2 shRNA-transfected BEAS-2B cells, EFEMP2 down-regulation significantly decreased the expression of the epithelial hallmark E-cadherin, and increased the expression of the mesenchymal hallmarks N-cadherin and Vimentin and transcription factors Snail, Slug and Twist; and at the mRNA level, we got the same conclusion through RT-qPCR (Figure 8B). MMPs can almost degrade various protein components in ECM, destroy the histological barrier of tumor cell invasion, and play a key role in tumor invasion and metastasis. By RT-qPCR and MMP zymography assay, we found that the mRNA expression of MMP2 and MMP9 was significantly increased in EFEMP2 shRNA-transfected BEAS-2B cells (Figure 8B), and their activities were significantly higher in the EFEMP2 shRNA-transfected group than in the negative control group (Figure 8C). In summary, EFEMP2 knockdown could promote EMT and enhance the expression and activity of MMP2 and MMP9. Conversely, in EFEMP2 cDNA-transfected lung cancer cells, EFEMP2 up-regulation remarkably increased the expression of E-cadherin and decreased the expression of N-cadherin, Vimentin, Snail, Slug and Twist, at protein (Figure 9A) and mRNA levels (Figure 9B). In addition, compared with the negative control cells, EFEMP2 cDNA-transfected lung cancer cells had weaker mRNA expression of MMP2 and MMP9 (Figure 9B), and their activities were significantly inhibited by EFEMP2 up-regulation (Figure 9C). Therefore, EFEMP2 overexpression could hamper the process of EMT and weaken the expression and activity of MMP2 and MMP9 in lung cancer.
Figure 8.
Effects of EFEMP2 down-regulation on the EMT-related genes and MMPs.
Notes: (A) By Western blotting (cropped blot), EMT hallmarks, including E-cadherin, N-cadherin, vimentin, Snail, Slug and Twist, were measured in control or EFEMP2 shRNA transfected BEAS-2B cells. (B) By RT-qPCR, EFEMP2 down-regulation repressed the expression of E-cadherin, and enhanced the expression of N-cadherin, Vimentin, Snail, Slug, Twist, MMP2 and MMP9, at mRNA levels. (C) By MMP zymography assay, the activities of MMP2 and MMP9 were significantly higher in the EFEMP2 shRNA-transfected group than in the negative control group. *P < 0.05.
Figure 9.
Effects of EFEMP2 up-regulation on the EMT-related genes and MMPs.
Notes: (A) By Western blotting (cropped blot), EMT hallmarks, including E-cadherin, N-cadherin, vimentin, Snail, Slug and Twist, were measured in control or EFEMP2 cDNA transfected H1299 and H1650 cells. (B) By RT-qPCR, EFEMP2 up-regulation increased the expression of E-cadherin, and decreased the expression of N-cadherin, Vimentin, Snail, Slug, Twist, MMP2 and MMP9, at mRNA levels. (C) By MMP zymography assay, the activities of MMP2 and MMP9 were significantly inhibited by EFEMP2 up-regulation. EFEMP2 knockdown could promote EMT and enhance the expression and activity of MMP2 and MMP9, while EFEMP2 overexpression could hamper the process of EMT and weaken the expression and activity of MMP2 and MMP9 in lung cancer. *P < 0.05.
Discussion
In our study, we initially detected that the expression of EFEMP2 was down-regulated in lung cancer tissues and cell lines, its low expression was significantly correlated with late clinical stage, low histological grade, positive lymph node metastasis and poor prognosis of NSCLC patients. The effects of EFEMP2 up- or down-regulation on cell proliferation, invasion and metastasis were explored in vitro and in vivo. The results revealed that EFEMP2 played an inhibitory role in the development of lung cancer. EFEMP2 could hamper the process of EMT and reduce the activity of MMPs, and decelerate the proliferation and invasion capacity of lung cancer cells.
The IHC results of lung tissues showed that the high expression rate of EFEMP2 in normal lung tissues was higher than that in lung cancer tissues, and EFEMP2 was highly expressed in well-differentiated carcinoma in contrast with poorly differentiated carcinoma. The high expression of EFEMP2 was negatively associated with poor clinical pathological characteristics and prognosis of lung cancer. We got the same conclusion from the Public database analysis (Oncomine and Kaplan-Meier plotter). On this basis, the expression of EFEMP2 in normal lung epithelial cell line and 4 lung cancer cell lines was further studied by RT-qPCR,Western blotting and ICC. The results showed that the expression of EFEMP2 in 4 lung cancer cell lines was significantly lower than that in normal lung epithelial cell line, which was consistent with the IHC results of lung tissues. So far, the function of EFEMP2 in tumorigenesis had not been fully elucidated, which was complicated and sometimes contradicting, because it displayed both pro- and anti-neoplastic effects depending on different tumor microenvironment. In 1999, EFEMP2 was first identified as a partner of mutant p53 interaction, also called MBP1 (Mutant p53-Binding Protein 1), and was considered as a candidate oncogene.5 Since then, with regard to the role of EFEMP2 in tumorigenesis, there have been some contradictory conclusions in the study of various tumors. Gallagher et al by fluorescence in situ hybridisation analysis, found that EFEMP2 gene was located on chromosome 11q13, which was prone to translocation and rearrangement during tumorigenesis, therefore, it was believed that EFEMP2 played an important role in the development of human cancer. Then they further found that in human colon tumor, the mRNA expression of EFEMP2 in tumors was about 2–7 times higher than that in adjacent normal tissues, which suggested that EFEMP2 gene was associated with human colon tumourigenesis.10 In the same pattern, in colorectal cancer, EFEMP2 was highly expressed even at the early stage, and was identified as a serum biomarker for the early detection of colorectal cancer.25 Increased EFEMP2 expression was also found in human osteosarcoma clinical specimens and cell lines, and EFEMP2 promoted osteosarcoma cell proliferation and invasion and played critical neoplastic roles in tumor development.19,26 In gliomas, EFEMP2 had the carcinogenic potential and its knockdown inhibited the growth and metastasis of cancer cells, high expression of EFEMP2 was related to the poor prognosis of glioblastoma.18,27 EFEMP2 expression was up-regulated in cervical carcinoma and ovarian cancer, and its overexpression was significantly associated with the patient’s adverse clinical and pathological characteristics and poor prognosis.16,17 However, there were some opposite conclusions in the study of other tumors. EFEMP2 was weakly expressed in prostate carcinoma cell lines compared to normal epithelial cells.28 In human endometrial carcinoma, EFEMP2 expression was significantly decreased in cancer tissues and cell lines, which was negatively correlated with malignant phenotype and poor prognosis.20 The loss of EFEMP2 expression was also found in breast cancer and associated with grade histology, which suggested that EFEMP2 could be considered as a potential prognostic biomarker.29 In human bladder cancer, the expression of EFEMP2 was significantly down-regulated in cancer samples and cells, which was negatively correlated with advanced tumor stage, high tumor grade and poor prognosis, and the low expression of EFEMP2 was considered to be a high-risk index of the patient’s survival.30 Our study showed that the role of EFEMP2 in lung cancer was anti-tumor, and EFEMP2 acted as a tumor suppressor gene that inhibited the progress of lung cancer. Whether the high or low expression of EFEMP2 can promote tumorigenesis depends on the type of cancer, and the complexity might originate from the specific microenvironment of each human cancer.31
Epithelial-mesenchymal transition (EMT) refers to the biological process of the epithelial cell being transformed into a cell with a mesenchymal phenotype by a specific procedure. It is mainly characterized by the reduction of the expression of cell adhesion molecules (such as E-cadherin), the transformation of cytokeratin cytoskeleton into the cytoskeleton dominated by vimentin, and the characteristics of the mesenchymal cells in the form. EMT is an important biological process, during which malignant epithelial cells acquired the migration and invasion capacity, so it is very important for the development of many epithelial cancers. There were also contradictory conclusions about the role of EFEMP2 in the process of EMT. In human osteosarcoma, EFEMP2 knockdown could suppress the invasion and metastasis of osteosarcoma cells by inhibiting EMT, meanwhile EFEMP2 up-regulation could enhance the invasion and metastasis of osteosarcoma cells by inducing EMT, which suggested that EFEMP2 promoted the process of EMT in osteosarcoma cells.19 On the contrary, in human bladder cancer and endometrial carcinoma, EFEMP2 played inhibitory roles in the process of EMT. EFEMP2 overexpression could inhibit the proliferation, invasion and metastasis of bladder cancer cells and EMT process, while EFEMP2 down-regulation could promote the proliferation, invasion and metastasis of bladder cancer cells and EMT process.30 In the same pattern, EFEMP2 significantly inhibited the proliferation, invasion and metastasis of endometrial carcinoma cells and EMT, therefore, EFEMP2 had the ability to suppress endometrial cancer progression.20 In our study, by Western blotting and RT-qPCR, we got the same conclusion that EFEMP2 down-regulation significantly decreased the expression of the epithelial hallmark E-cadherin, and increased the expression of the mesenchymal hallmarks N-cadherin and Vimentin and transcription factors Snail, Slug and Twist; reversely, EFEMP2 up-regulation remarkably increased the expression of E-cadherin and decreased the expression of N-cadherin, Vimentin, Snail, Slug and Twist. According to different tumor microenvironments, EFEMP2 played a dual role of inhibition or promotion in the process of EMT. At present, the research on the function of EFEMP2 in tumorigenesis is still in the initial stage, and there are few related literatures. We believe that the focus of future research should be on the upstream and downstream signal pathways of EFEMP2 in cancer progression, as well as the characterization of their intracellular and extracellular partners.
Malignant cells have highly invasive ability to invade adjacent areas and metastasize to distant organs by degrading ECM components. Matrix metalloproteinases, including MMP-2 and MMP-9, play a critical role in local invasion, metastasis and angiogenesis.32 In glioma, the silence of EFEMP2 expression in two glioma cell lines remarkably inhibited cell proliferation, induced cell apoptosis and significantly inhibited the invasive ability of both glioma cells, accompanied by the down-regulation of MMP-2 and MMP-9, which suggested that EFEMP2 could promote the invasion of glioma cells by regulating MMP-2 and MMP-9. Our data indicated that EFEMP2 overexpression notably inhibited the invasion of lung cancer cells and decreased the expression and activity of MMP2 and MMP9, while EFEMP2 knockdown remarkably promoted the invasion of lung normal epithelial cells and enhanced the expression and activity of MMP2 and MMP9. In summary, EFEMP2 could inhibit the invasion of lung cancer cells by weakening the expression and activity of MMP2 and MMP9. It is noteworthy that other fibulin members can also act as modulators of MMPs. EFEMP1 and EFEMP2 have very high homology in structure, and they also have many similar features in the progression of cancer.13,14 As in the case of EFEMP2, EFEMP1 played inhibitory roles in lung cancer cell invasion by hampering the process of EMT and down-regulating cellular MMP-7, MMP-2 and MMP9.33–36 However, in human gliomas, EFEMP1 was significantly up-regulated and promoted tumor cell motility and invasion with elevated expression and activity of matrix metalloproteases, such as MMP-2, MMP-9, ADAMTS-5 and ADAM17.37–39 In conclusion, there are reports for pro-oncogenic and anti-oncogenic roles for EFEMP2, and more in-depth research is needed in the future.
Conclusion
Our study revealed that down-regulated EFEMP2 expression was associated with the malignant phenotype and poor prognosis of lung cancer, which was consistent with the public database analyses. Moreover, EFEMP2 was able to suppress the proliferation, invasion, and metastasis of lung cancer cells in vitro and in vivo by inhibiting EMT and down-regulating MMP-2 and MMP9. The study of the mechanism of EFEMP2 in the invasion and metastasis of lung cancer is of great significance in finding new therapeutic strategies to improve the prognosis of lung cancer patients.
Abbreviations
EFEMP2, Epidermal growth factor-containing fibulin-like extracellular matrix protein 2; EMT, epithelial interstitial transformation; cb, calcium-binding; EGF, epidermal growth factor; NSCLC, Non-small cell lung cancer; TNM, tumor-node-metastasis; IASLC, International Association for the Study of Lung Cancer; DMEM/F-12, Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12; FBS, fetal bovine serum; IHC, immunohistochemistry; ICC, immunocytochemistry; SP staining, streptavidin-biotin-peroxidase staining: DAB, 3′, 3-diaminobenzidine tetrahydrochloride: RT-qPCR¸real time quantitative reverse transcriptase-polymerase chain reaction: cDNA, complementary DNA; RIPA, radioimmunoprecipitation assay; PMSF, phenylmethylsulfonyl fluoride; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel; PVDF, polyvinylidene difluoride: PBS, phosphate buffer saline; BSA, bovine serum albumin; ECL, enhanced chemiluminescence; shRNA, small hairpin RNA; RNAi, RNA interference; MOI, multiplicity of infection; ECM, extracellular matrix; SPF, specific pathogen free; CO2, carbon dioxide; MMP, matrix metalloproteinase.
Ethics Approval and Consent
This study was approved by the Medical Ethics Committee of Human and Animal Institution of Shandong Provincial Hospital Affiliated to Shandong University. All methods were performed in accordance with the relevant guidelines and regulations. Written informed consent was obtained from the patients involved in the study. All mouse experimental procedures were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People’s Republic of China. The study was carried out in accordance with the principles of the Declaration of Helsinki.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Funding
This work was supported by The Key Research and Development Program of Shandong Province (No. 2017GSF218096, No. 2016GSF201038 and No. 2014GSF118018). Those providing funds had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208 [DOI] [PubMed] [Google Scholar]
- 3.Chaffer CL, Weinberg RA. A perspective on cancer cell. Metastasis. Science. 2011;331(6024):1559–1564. doi: 10.1126/science.1203543 [DOI] [PubMed] [Google Scholar]
- 4.Giltay R, Timpl R, Kostka G. Sequence, recombinant expression and tissue localization of two novel extracellular matrix proteins, fibulin-3 and fibulin-4. Matrix Biol. 1999;18(5):469–480. doi: 10.1016/S0945-053X(99)00038-4 [DOI] [PubMed] [Google Scholar]
- 5.Gallagher WM, Argentini M, Sierra V, Bracco L, Debussche L, Conseiller E. MBP1: a novel mutant p53-specific protein partner with oncogenic properties. Oncogene. 1999;18(24):3608–3616. doi: 10.1038/sj.onc.1202937 [DOI] [PubMed] [Google Scholar]
- 6.Katsanis N, Venable S, Smith JR, Lupski JR. Isolation of a paralog of the Doyne honeycomb retinal dystrophy gene from the multiple retinopathy critical region on 11q13. Hum Genet. 2000;106(1):66–72. doi: 10.1007/s004399900224 [DOI] [PubMed] [Google Scholar]
- 7.de Vega S, Iwamoto T, Yamada Y. Fibulins: multiple roles in matrix structures and tissue functions. Cell Mol Life Sci. 2009;66:1890–1902. doi: 10.1007/s00018-009-8632-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Timpl R, Sasaki T, Kostka G, Chu ML. Fibulins: a versatile family of extracellular matrix proteins. Nat Rev Mol Cell Biol. 2003;4(6):479–489. doi: 10.1038/nrm1130 [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi N, Kostka G, Garbe JH, et al. A comparative analysis of the fibulin protein family. Biochemical characterization, binding interactions, and tissue localization. J Biol Chem. 2007;282(16):11805–11816. [DOI] [PubMed] [Google Scholar]
- 10.Gallagher WM, Greene LM, Ryan MP, et al. Human fibulin-4: analysis of its biosynthetic processing and mRNA expression in normal and tumour tissues. FEBS Lett. 2001;489(1):59–66. doi: 10.1016/S0014-5793(00)02389-9 [DOI] [PubMed] [Google Scholar]
- 11.Horiguchi M, Inoue T, Ohbayashi T, et al. Fibulin-4 conducts proper elastogenesis via interaction with cross-linking enzyme lysyl oxidase. Proc Natl Acad Sci U S A. 2009;106(45):19029–19034. doi: 10.1073/pnas.0908268106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papke CL, Yanagisawa H. Fibulin-4 and fibulin-5 in elastogenesis and beyond: insights from mouse and human studies. Matrix Biol. 2014;37:142–149. doi: 10.1016/j.matbio.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura T. Roles of short fibulins, a family of matricellular proteins, in lung matrix assembly and disease. Matrix Biol. 2018;73:21–33. doi: 10.1016/j.matbio.2018.02.003 [DOI] [PubMed] [Google Scholar]
- 14.Tsuda T. Extracellular interactions between fibulins and Transforming Growth Factor (TGF)-β in physiological and pathological conditions. Int J Mol Sci. 2018;19:9. doi: 10.3390/ijms19092787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher WM, Currid CA, Whelan LC. Fibulins and cancer: friend or foe? Trends Mol Med. 2005;11:336–340. doi: 10.1016/j.molmed.2005.06.001 [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Zhang J, Liu X, Fang R, Zhao Y, Ma D. Overexpression of fibulin-4 is associated with tumor progression and poor prognosis in patients with cervical carcinoma. Oncol Rep. 2014;31(6):2601–2610. doi: 10.3892/or.2014.3139 [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Liu Z, Fang S, et al. Fibulin-4 is associated with tumor progression and a poor prognosis in ovarian carcinomas. BMC Cancer. 2015;15:91. doi: 10.1186/s12885-015-1100-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Chen Q, Chen Z, et al. EFEMP2 is upregulated in gliomas and promotes glioma cell proliferation and invasion. Int J Clin Exp Pathol. 2015;8(9):10385–10393. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang D, Wang S, Chen J, et al. Fibulin-4 promotes osteosarcoma invasion and metastasis by inducing epithelial to mesenchymal transition via the PI3K/Akt/mTOR pathway. Int J Oncol. 2017;50(5):1513–1530. doi: 10.3892/ijo.2017.3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T, Wang M, Fang S, Wang Q, Fang R, Chen J. Fibulin-4 is associated with prognosis of endometrial cancer patients and inhibits cancer cell invasion and metastasis via Wnt/β-catenin signaling pathway. Oncotarget. 2017;8(12):18991–19012. doi: 10.18632/oncotarget.15086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soumaoro LT, Uetake H, Higuchi T, Takagi Y, Enomoto M, Sugihara K. Cyclooxygenase-2 expression: a significant prognostic indicator for patients with colorectal cancer. Clin Cancer Res. 2004;10(24):8465–8471. doi: 10.1158/1078-0432.CCR-04-0653 [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 23.Sun L, Fang J. Epigenetic regulation of epithelial-mesenchymal transition. Cell Mol Life Sci. 2016;73(23):4493–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20(2):69–84. doi: 10.1038/s41580-018-0080-4 [DOI] [PubMed] [Google Scholar]
- 25.Yao L, Lao W, Zhang Y, et al. Identification of EFEMP2 as a serum biomarker for the early detection of colorectal cancer with lectin affinity capture assisted secretome analysis of cultured fresh tissues. J Proteome Res. 2012;11(6):3281–3294. doi: 10.1021/pr300020p [DOI] [PubMed] [Google Scholar]
- 26.Li R, Wang L. Fibulin-4 is a novel Wnt/β-Catenin pathway activator in human osteosarcoma. Biochem Biophys Res Commun. 2016;474(4):730–735. doi: 10.1016/j.bbrc.2016.05.018 [DOI] [PubMed] [Google Scholar]
- 27.Li F, Li Y, Zhang K, et al. FBLN4 as candidate gene associated with long-term and short-term survival with primary glioblastoma. Onco Targets Ther. 2017;10:387–395. doi: 10.2147/OTT.S117165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wlazlinski A, Engers R, Hoffmann MJ, et al. Downregulation of several fibulin genes in prostate cancer. Prostate. 2007;67(16):1770–1780. doi: 10.1002/(ISSN)1097-0045 [DOI] [PubMed] [Google Scholar]
- 29.Motalebzadeh J, Mahjoubi F, Nafissi N, Hashemian M, Taheri M, Hosseinpour Y. FBLN-4 and BCRP genes as two prognostic markers are downregulated in breast cancer tissue. Cancer Biomark. 2017;19(1):51–55. doi: 10.3233/CBM-160335 [DOI] [PubMed] [Google Scholar]
- 30.Zhou Q, Chen S, Lu M, et al. EFEMP2 suppresses epithelial-mesenchymal transition via Wnt/β-catenin signaling pathway in human bladder cancer. Int J Biol Sci. 2019;15(10):2139–2155. doi: 10.7150/ijbs.35541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Obaya AJ, Rua S, Moncada-Pazos A, Cal S. The dual role of fibulins in tumorigenesis. Cancer Lett. 2012;325(2):132–138. doi: 10.1016/j.canlet.2012.06.019 [DOI] [PubMed] [Google Scholar]
- 32.Wyganowska-Świątkowska M, Tarnowski M, Murtagh D, Skrzypczak-Jankun E, Jankun J. Proteolysis is the most fundamental property of malignancy and its inhibition may be used therapeutically (Review). Int J Mol Med. 2019;43(1):15–25. doi: 10.3892/ijmm.2018.3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X, Meng J, Yue W, et al. Fibulin-3 suppresses Wnt/β-catenin signaling and lung cancer invasion. Carcinogenesis. 2014;35(8):1707–1716. doi: 10.1093/carcin/bgu023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu S, Yang Y, Sun YB, Wang HY, Sun CB, Zhang X. Role of fibulin-3 in lung cancer: in vivo and in vitro analyses. Oncol Rep. 2014;31(1):79–86. doi: 10.3892/or.2013.2799 [DOI] [PubMed] [Google Scholar]
- 35.Kim IG, Kim SY, Choi SI, Lee JH, Kim KC, Cho EW. Fibulin-3-mediated inhibition of epithelial-to-mesenchymal transition and self-renewal of ALDH+ lung cancer stem cells through IGF1R signaling. Oncogene. 2014;33(30):3908–3917. doi: 10.1038/onc.2013.373 [DOI] [PubMed] [Google Scholar]
- 36.Kim EJ, Lee SY, Woo MK, et al. Fibulin-3 promoter methylation alters the invasive behavior of non-small cell lung cancer cell lines via MMP-7 and MMP-2 regulation. Int J Oncol. 2012;40(2):402–408. doi: 10.3892/ijo.2011.1191 [DOI] [PubMed] [Google Scholar]
- 37.Nandhu MS, Kwiatkowska A, Bhaskaran V, Hayes J, Hu B, Viapiano MS. Tumor-derived fibulin-3 activates pro-invasive NF-κB signaling in glioblastoma cells and their microenvironment. Oncogene. 2017;36(34):4875–4886. doi: 10.1038/onc.2017.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Hu Y, Liu C, et al. Human fibulin-3 protein variant expresses anti-cancer effects in the malignant glioma extracellular compartment in intracranial xenograft models. Oncotarget. 2017;8(63):106311–106323. doi: 10.18632/oncotarget.22344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu B, Thirtamara-Rajamani KK, Sim H, Viapiano MS. Fibulin-3 is uniquely upregulated in malignant gliomas and promotes tumor cell motility and invasion. Mol Cancer Res. 2009;7(11):1756–1770. doi: 10.1158/1541-7786.MCR-09-0207 [DOI] [PMC free article] [PubMed] [Google Scholar]