Abstract
Consumption of mothers’ milk is associated with reduced incidence and severity of enteric infections, leading to reduced morbidity in breastfed infants. Fucosylated and sialylated human milk oligosaccharides (HMO) are important for both direct antimicrobial action — likely via a decoy effect — and indirect antimicrobial action through commensal growth enhancement. Bovine milk oligosaccharides (BMO) are a potential source of HMO-mimics as BMO resemble HMO; however, they have simpler and less fucosylated structures. BMO isolated at large scales from bovine whey permeate were modified by the addition of fucose and/or sialic acid to generate HMO-like glycans using high-yield and cost-effective one-pot multienzyme approaches. Quadrupole time-of-flight LC/MS analysis revealed that 22 oligosaccharides were synthesized and 9 had identical composition to known HMO. Preliminary anti-adherence activity assays indicated that fucosylated BMO decreased the uptake of enterohemorrhagic Escherichia coli O157:H7 by human intestinal epithelial Caco-2 cells more effectively than native BMO.
1. Introduction
Human milk is the ideal nourishment for newborns. A highly abundant component of human milk is human milk oligosaccharides (HMO) (Ballard & Morrow, 2013; Kunz, Kuntz, & Rudloff, 2014). HMO are composed of 3 to 22 monosaccharide units for each glycan, including D-glucose (Glc), d-galactose (Gal), N-acetyl-d-glucosamine (GlcNAc), l-fucose (Fuc), and N-acetylneuraminic acid (Neu5Ac) joined through a variety of linkages (German, Freeman, Lebrilla, & Mills, 2008). More than 150 HMO structures have been identified (Chen, 2015; Ninonuevo et al., 2006). HMO usually have a lactose (Galβ1—4Glc) core at the reducing end that sometimes can be elongated with repeating units of N-acetyllactosamine (Galβ1—4GlcNAc) with or without a cap of Galβ1—3GlcNAc. The lactose core can also be decorated by the addition of Neu5Ac in α2—3 and/or α2—6 linkages, and/or Fuc in α1—2, α1—3, and/or α1—4 linkages (Newburg, Ruiz-Palacios, & Morrow, 2005). Because of their unique structural complexity, HMO are indigestible by infants but are consumed by microbiota in the gut. HMO act as selective prebiotics for bacteria that possess the glycosidases required for their degradation and likely also function as decoys for pathogenic bacteria, decreasing the rates of infection (Kunz, Rudloff, Baier, Klein, & Strobel, 2000).
However, for a multitude of reasons, many infants are fed formula as a partial or complete replacement for human milk. Despite their important functions, no formula contains the complete match of HMO structures yet. Formulas are often supplemented with oligosaccharides (OS) derived from plants or produced by bioproduction processes, such as fructooligosaccharides (FOS) and galactooligosaccharides (GOS). Plant-derived OS can have prebiotic effects on the intestinal microbiota (Vandenplas, Greef, & Veereman, 2014), but they lack some of the other beneficial effects provided by HMO, such as direct protection against pathogenic bacteria. This difference can be attributed to the simplicity of the chemical structures of the plant-derived OS used, compared with that of HMO. FOS are derived from inulin, a polysaccharide composed of repeating units of fructose linked by β1—2 bond with a terminal Glc (Tungland, 2003). GOS are usually synthesized from lactose by the addition of repeating units of Ga1 (Albayrak & Yang, 2002). Recently, a few small HMO, including 2’-fucosyllactose (2’FL) and lacto-N-neotetraose (LNnT) have been added to infant formula (Vandenplas et al., 2018). Increasing the diversity and structural complexity of OS in infant formula could potentially lead to improved health outcomes in infants.
Increasing interest in the biological roles of HMO has triggered many efforts to develop synthetic pathways for their production. However, the presence of multiple monosaccharides with a multitude of glycosidic linkages in HMO (Priem, Gilbert, Wakarchuk, Heyraud, & Samain, 2002; Sears & Wong, 2001) makes synthesis complex. Approaches used to synthesize milk OS include chemical, enzymatic, chemoenzymatic and fermentation strategies (Chen, 2015; Nilsson, 1988; Priem et al., 2002; Sears & Wong, 2001).
Significant advances in biocatalytic approaches to produce OS have taken place in the last few decades. Strategies using either glycosidases or glycosyltransferases have been developed. Tremendous advances in glycobiology (Nilsson, 1988; Palcic, 1999; Yu & Chen, 2016) have provided an increasing library of glycosyltransferases and strategies for in-situ generation of sugar nucleotides to allow the synthesis of a diverse array of carbohydrates at large scale with decreased cost. Enzymatic approaches have been successfully developed to synthesize α2—6-linked disialyllacto-N-tetraose (DSLNT) and disialyllacto-N-neotetraose analogs (Yu et al., 2014). These OS have been shown in an animal model to be protective against necrotizing enterocolitis (NEC), a devastating disease affecting an increasing number of premature infants (Yu et al., 2014, 2017).
Although seven HMO including 2’FL, 3-fucosyllactose (3’FL) difucosyllactose (DFL), LNnT, lacto-N-tetraose (LNT), 6’-sialyllactose (6’SL), and 3’-sialyllactose (3’SL) have been successfully synthesized (Inbiose, 2018), they are far from representing the diversity of the OS-structures found in human milk. The development of additional practical and economically feasible approaches to synthesize bioactive OS is useful to enable a better understanding of their functions at the molecular level and to further explore their biological functions, with the goal of bringing these new ingredients to the marketplace.
An alternative strategy to obtain a wide array of OS, to complement the handful structures presently obtainable via synthetic approaches, is to use large-scale membrane filtration to separate them from dairy streams. Bovine milk contains OS termed bovine milk oligosaccharides (BMO). To date, more than 40 OS have been identified and characterized in bovine milk and colostrum (Barile, Meyrand, Lebrilla, & German, 2011; Barile et al., 2009; Sundekilde et al., 2012; Tao, DePeters, German, Grimm, & Lebrilla, 2009; Urashima, Saito, Nakamura, & Messer, 2001). BMO have some structural and compositional resemblance to HMO with some differences (Aldredge et al., 2013; Barile et al., 2010). In addition to the same five monosaccharides that make up HMO (Ga1, Glc, GlcNAc, Fuc, and Neu5Ac), BMO can also contain N-acetyl-d-galactosamine (Ga1NAc) and N-glycolylneuraminic acid (Neu5Gc) (Urashima et al., 2001). The main differences between HMO and BMO are the types of predominant oligosaccharides, and their concentrations in mature milk and colostrum. Human milk has a higher proportion of fucosylated (80%) OS compared with bovine milk (1—5%). On the other hand, sialylated OS are highly abundant in bovine milk, representing about 70%, compared with 10—20% in human milk (Bode, 2012). The concentration of HMO can reach 10—20 g L−1 in colostrum and 5—12 g L−1 in mature milk, whereas the concentration of BMO is much lower, with close to 1 g L−1 in colostrum and trace amounts in mature milk (Coppa et al., 1999; Nakamura et al., 2003; Tao et al., 2009). Although BMO are much less concentrated than HMO in non-processed milk, they are available in large quantities and high concentration in whey permeate, the by-product of whey protein manufacture. Because whey permeate is considered an environmental pollutant and cannot be readily discarded in wastewater, companies have to pay for disposal of whey permeate. In 2017, 242,138 tons of whey protein concentrate were produced in the United States from whey (NASS & USDA, 2018) leaving behind the whey permeate available for BMO recovery.
We report here a new strategy for synthesizing a HMO-like product from BMO, using an enzymatic system to add fucose and sialic acid, converting a waste product of whey protein manufacture to highly valuable ingredients that can be used to improve infant formula functionality.
2. Materials and methods
2.1. Materials
BMO concentrates, produced by a previously published method (Charbonneau et al., 2016), were kindly provided by Hilmar Ingredients (Hilmar, CA, USA). Bovine colostrum whey permeate was provided by La Belle, Inc. (Bellingham, WA, USA) and the BMO were isolated by nanofiltration and diafiltration as previously reported (Cohen, Barile, Liu, & de Moura Bell, 2017; de Moura Bell et al., 2018). Nickel-nitrilotriacetic acid agarose (Ni2+-NTA agarose) was from Qiagen (Valencia, CA, USA). The enzymes for sialylation and fucosylation were over-expressed in an Escherichia coli expression system and purified by Ni2+-NTA affinity columns (Bai et al., 2019; Sugiarto et al., 2012). The enzymes used for donor-monosaccharide synthesis were Neisseria meningitidis CMP-sialic acid synthetase (NmCSS) (Yu, Yu, Karpel, & Chen, 2004), bifunctional Bacteroides fragilis NCTC9343 (ATCC 25285) enzyme that has both l-fucokinase and GDP-fucose pyrophosphorylase activities (BfFKP) (Yi et al., 2009; Yu, Lau, Li, Sugiarto, & Chen, 2012), and Pasteurella multocida inorganic pyrophosphatase (PmPpA) (Lau et al., 2010). P. multocida α2—3-sialyltransferase 1 M144D mutant (PmST1 M144D) (Sugiarto et al., 2012) and Helicobacter pylori NCTC 11639 α1—3-fucosyltransferase (Hp3FT) (Bai et al., 2019; Sun et al., 2007) were used as glycosyltransferases for transferring Neu5Ac and Fuc, respectively. About 100 mg of NmCSS and PmST1 M144D, 10 mg of FKP, 5 mg of Hp3FT, and 500 mg PmPpA were purified from 1-L E. coli expression systems. A variety of solid phase extraction materials were used to purify OS from whey products and from post-enzymatic reactions mixtures. C8 columns (DSC-C8 Discovery, 3 mL tube capacity, 500 mg bed weight; Supelco, Bellefonte, PA, USA) were used to remove peptides; C18 columns (DSC-C18 Discovery, 3 mL tube capacity, 500 mg bed weight; Supelco) were used to remove nucleotides; and nonporous graphitized carbon cartridges (GCC-SPE, 150 mg carbon, 4-mL tube capacity, Alltech, Deerfield, IL, USA) were used to remove salts, lactose and selectively elute OS.
Caco-2 cells (ATCC) were routinely maintained in DMEM (+10% FBS and Pen/Strep) in T75 flasks (Nalgene, Rochester, NY), and seeded in Costar (12-well) TC-Treated Flat bottom plates for bacterial uptake measurements. High-resolution mass spectra (HRMS) used to monitor enzymatic reaction progress were recorded on a Thermo Electron LTQ-Orbitrap Hybrid MS (Thermo Scientific US, Waltham, MA, USA) at the Mass Spectrometry Facility at the University of California, Davis, CA, USA.
2.2. Characterization of BMO
To select an appropriate source of BMO for enzymatic glycosylation, complete glycoprofiling (type and relative abundances) of seven BMO sources was carried out using an Agilent 6520 Quadrupole Time-of-Flight (Q-TOF) LC/MS with a microfluidic nano-electrospray graphitized carbon column chip (Agilent Technologies, Santa Clara, CA) as previously described (Meyrand et al., 2013).
OS from colostrum whey permeate and BMO-concentrates provided by industrial collaborators were further purified as previously described (Barile et al., 2010). Briefly, samples were diluted in nanopure water (18.2 MΩ ionic purity) and centrifuged at 4 °C for 30 min at 4000 × g to remove remaining lipids. Four volumes of 2:1 (v/v) chloroform:methanol were added to the fat-free fraction, and the emulsion was centrifuged at 4000 × g for 30 min at 4 °C. The lower chloroform layer and protein pellet were discarded. The upper layer containing BMO was collected. Two volumes of pure ethanol were added to the BMO-fraction and the remaining protein was precipitated at 4 °C overnight. The samples were centrifuged at 4000 × g for 30 min at 4 °C, and the protein-free supernatant was collected and dried in a vacuum centrifuge at 37 °C. BMO were rehydrated in 1 mL nanopure water. Residual peptides were removed using C8 columns. The cartridges were conditioned with three column volumes (cv) of pure HPLC-grade acetonitrile (ACN) followed by three cv of nanopure water. The carbohydrate-rich solution was loaded onto the cartridge and the peptide-free eluate was collected. The BMO were further purified by GCC-SPE. Prior to use, the GCC-SPE cartridge was activated with 3 cv of 80% ACN, 0.05% trifluoroacetic acid (TFA), and equilibrated with 3 cv nanopure water. The carbohydrate-rich solution was loaded onto the cartridge, and salts were removed by washing with three cv of nanopure water at a flow rate of 1 mL min−1. For BMO-profiling, samples were eluted from the cartridge using 2 cv of 20% ACN in water followed by 2 cv of 40% ACN/0.1% TFA in water. Eluted fractions were dried by vacuum centrifugation at 37 °C and reconstituted in 100 μL of nanopure water. MS analysis was performed in Q-TOF LC/MS with a graphitized carbon column nano-chip as previously described (Meyrand et al., 2013) with some modifications: flow rate for the nanopump was 0.4 μL min −1, and the electro-spray capillary voltage ranged between 1500 and 1900V.
2.3. Oligosaccharide identification
A previously assembled list (Dallas et al., 2014) consisting of 49 BMO was used to create an in-house library to screen all BMO sources for automated matching of masses and retention times. Manual inspection of each OS peak was performed with the “extracted ion chromatogram” feature from Mass Hunter software (Version 6.0, Agilent Technologies, 2014) with a 20 ppm mass error.
2.4. One-pot two-enzyme sialylation of BMO
Enzymatic sialylation of BMO was performed at 37 °C in an one-pot two-enzyme (OP2E) system containing Tris-HCl buffer (100 mm, pH 8.0), BMO (50 mm, an average molecular mass of 600 Daltons was used for calculating the concentration), cytidine triphosphate (1.5 equivalents versus BMO, molar ratio), Neu5Ac (1.25 equivalents versus BMO, molar ratio) with NmCSS, and PmST1 M144D in the presence of MgCl2 (10 mm). To optimize the efficiency of the sialylation reaction, various mass ratios of NmCSS and PmST1 M144D versus BMO and different reaction times (4 h and 24 h) were tested in small-scale reactions. After reactions, the mixtures were analyzed by HRMS to rapidly confirm the success of sialylation before proceeding with further purification for Q-TOF LC/MS system. In small-scale reactions, 1.5 mg (50 mm) of BMO were used as a starting material. The mass ratios of BMO versus NmCSS and PmST1 M144D were 200:1:2 (R1), 200:1:4 (R2), 100:1:1 (R3), and 100:1:2 (R4), respectively. The reactions were incubated for 4 h or 24 h. In large-scale reactions, 6.0 g (50 mm) of BMO and recombinant NmCSS (60.0 mg), and PmST1 M144D (120.0 mg) were used, and the mass ratio of BMO versus NmCSS and PmST1 M144D was 100:1:2. The reaction was incubated in an incubator shaker for 24 h at 37 °C with agitation at 100 rpm. The reaction mixture without enzymes was used as a control.
2.5. One-pot three-enzyme fucosylation of BMO
Enzymatic fucosylation of BMO was performed in Tris-HCl buffer (pH 7.5, 100 mm) in an one-pot three-enzyme (OP3E) system containing BMO (6.0 g, 50 mm), ATP (1.5 equivalents versus BMO, molar ratio), GTP (1.5 equivalents versus BMO, molar ratio), Fuc (1.5 equivalents versus BMO, molar ratio), BfFKP (25 mg), PmPpA (25 mg), Hp3FT (25 mg), and MgCl2 (10 mm). The reaction was incubated in an incubator shaker at 37 °C for 24 h with agitation at 100 rpm. The reaction mixture without enzymes was used as a control.
2.6. Sequential OPME fucosylation and sialylation of BMO
For modifying BMO by both fucosylation and sialylation, sialylation was carried out after fucosylation. To achieve this, upon the completion of fucosylation, the reaction mixture was added to an equal volume of pre-chilled ethanol and the resulting mixture was incubated at −20 °C for 2 h to precipitate the enzymes. The mixture was centrifuged at 4000 × g for 30 min at 4 °C, and the supernatant was collected and subjected to rotary evaporation to remove ethanol. One-half of the resulting mixture was used for sialylation using the large-scale sialylation conditions described in section 2.4. The reaction mixture without enzymes was used as a control.
2.7. Partial purification of enzymatically modified BMO using activated charcoal chromatography
All reactions were quenched by adding an equal volume of ice-cold ethanol, and the mixtures were kept at −20 °C for at least 2 h to precipitate the enzymes. The mixtures were centrifuged at 4000 × g for 30 min at 4 °C. The supernatants were collected, subjected to rotary evaporation to remove solvent, and dissolved in water (0.5—2 mL). After centrifugation, the supernatant was loaded onto an activated-charcoal column (4 cm × 50 cm, 10—40 mesh, Acros Organics, NJ, US) (before packing the column, the charcoal was soaked in 95% ethanol for 4 h) pre-equilibrated with 10 cv of water. The column was washed with 5 cv of water, 3 cv of 10% ethanol, followed by 3 cv of 50% ethanol. The fractions were analyzed by HRMS and those containing the desired products were collected, lyophilized to dryness for further purification, or used for functional assays of inhibition of Caco-2 cell uptake of E. coli by enzymatically-modified BMO.
2.8. C18 column chromatography purification of partially-purified sialylated BMO
The lyophilized powder of partially-purified sialylated BMO was dissolved in water (0.5—2 mL) and purified using a C18 column (43 g) on a CombiFlash® Rf 200i system (Teledyne ISCO, Lincoln, NE, USA). The column was eluted at a flow rate of 40 mL min−1 with water for 6 min, then a gradient of 0—100% acetonitrile in water containing 0.05% formic acid over 12 min, followed by 100% acetonitrile for 2 min. The fractions containing BMO were collected and lyophilized to dryness.
2.9. Identifying novel sialylated and/or fucosylated oligosaccharides
After enzymatic glycosylation, the control (no enzymes) and modified BMO samples were purified. Modified BMO were extracted as follows: samples were centrifuged at 4 °C for 30 min at 4000 × g to recover the supernatant, two volumes of ethanol were added to the BMO fraction, and the remaining proteins were precipitated at 4 °C overnight. The samples were centrifuged at 4 °C for 30 min at 4000 × g, and the enzyme-free supernatant was collected and dried in a vacuum centrifuge. Samples were rehydrated in nanopure water. Residual nucleotides were removed using C18 columns. The cartridges were conditioned with three cv of pure HPLC-grade ACN followed by three cv of nanopure water. The carbohydrate-rich solution was loaded onto the cartridge, and the nucleotide-free eluate was collected. The monosaccharides were removed from the product OS by GCC-SPE. Prior to use, the GCC-SPE cartridge was activated with 3 cv of 80% ACN, 0.05% TFA and equilibrated with 3 cv of nanopure water. The carbohydrate-rich solution was loaded onto the cartridge, and salts and monosaccharides were removed by washing with six cv of nanopure water at a flow rate of 1 mL min−1 (Meyrand et al., 2013). The samples were each eluted with 2 cv of 20% ACN in water followed by 2 cv of 40% ACN/0.1% TFA in water to collect the BMO. Eluted fractions were dried by vacuum centrifugation at 37 °C, resuspended in water, and characterized by a Q-TOF LC/MS with a graphitized carbon column chip, as described in section 2.2.
To detect differences between samples before and after enzymatic modification, the peak area of each known OS that could accept an enzymatic monosaccharide addition was measured before and after the reaction. Due to instrument variations, peak areas were standardized by the base peak chromatogram of each corresponding sample.
2.10. Bacterial uptake assay
The percentage of E. coli uptake of by Caco-2 cells was compared after incubation in the presence of the glycosylated and the control unmodified BMO samples. As a negative control, Glc alone was used. Intestinal epithelial cell line Caco-2 cells were cultured in DMEM supplemented with 10% FBS and Pen/Strep. For assays, confluent cells were washed 3× with PBS, trypsinized, and seeded in 12-well plates at a seeding density of 1E5 cells well−1. Three days post-seeding, confluent cells were washed with DMEM to remove serum and antibiotics. DMEM or BMO prepared from a filter-sterilized 200 mg mL−1 BMO-stock solution in DMEM at a final concentration of 15 mg mL−1 were added to the individual wells. After 16 h of conditioning, the cells were infected with freshly grown enterohemorrhagic E. coli O157:H7 (EHEC) for 2 h at a Mechanism of Injury of 50—100. The unbound cells were washed with PBS buffer, and the Caco-2 cells were subsequently treated with Gentamicin (100 μg mL−1) for 2 h to remove surface-associated bacteria. The cells were washed 2× with PBS buffer prior to lysis with PBS/0.5% Triton to recover internalized bacteria. The resulting lysates were serially diluted, and cfu mL−1 were quantitated by growth on trypticase soy agar plates after overnight incubation at 37 °C.
3. Results and discussion
3.1. Selection of a sample of BMO for glycosylation
The glycoprofiling of seven BMO samples from different sources was carried out to identify and characterize glycans according to their mass and retention time on Q-TOF LC/MS. A representative chromatogram for one of these samples is shown in Supplementary material Fig. S1. The number of sites in BMO that can be potentially modified by enzymatic α1—3-fucosylation and/or α2—3-sialylation (Table 1) was determined by analyzing the structure and composition of oligosaccharides in each sample.
Table 1.
Fucosyl and sialyl linkages that can be added using glycosyltransferases in one-pot multienzyme systems in this study.
| Monosaccharide to be added | Obtainable linkages |
|---|---|
| Fuc | Fucα1—3Glc |
| Fucα1—3GlcNAc | |
| Neu5Ac | Neu5Acα2—3Gal |
The criterion for the selection of the best-suited sample for enzymatic glycosylation was based on the absence of lactose, the total number of fucosylation and/or sialylation sites on BMO, and the relative abundance of these modifiable BMO, as well as the absence of Neu5Gc, a sialic acid form that is biosynthesized by animals but not by humans (Bardor, Nguyen, Diaz, & Varki, 2005). As shown in Table 2, among seven samples tested, sample ID 5 with the absence of lactose, high total BMO, and high potential fucosylation and sialylation sites was the best candidate for enzymatic fucosylation and sialylation to produce HMO mimics.
Table 2.
Characterization of seven samples of BMO by Q-TOF LC/MS.
| ID | Product description | Lactose presence | BMO |
Potential fucosylation sites | Potential sialylation sites | |||
|---|---|---|---|---|---|---|---|---|
| Fucosylated | Sialylated | Neutral non-fucosylated | Total | |||||
| 1 | Colostrum | Yes | 2 | 8 | 28 | 38 | 67 | 61 |
| 2 | Ultra filtered colostrum | Yes | 0 | 6 | 31 | 37 | 58 | 62 |
| 3 | Product prototype neutral OS dominant | No | 0 | 5 | 8 | 13 | 17 | 16 |
| 4 | Product prototype acid OS dominant batch 1 | No | 0 | 5 | 25 | 30 | 48 | 45 |
| 5 | Product prototype acid OS dominant batch 2 | No | 1 | 7 | 27 | 35 | 49 | 47 |
| 6 | Product prototype (non-dominant fraction) | Yes | 0 | 5 | 14 | 19 | 30 | 24 |
| 7 | Lactose free product prototype (non-dominant fraction) | No | 0 | 6 | 18 | 24 | 39 | 33 |
Thirty-five BMO were identified in sample ID 5, with only seven sialylated, one fucosylated, and the rest being neutral non-fucosylated OS. The BMO identified in ID5 are presented in Table 3. The most abundant OS were those with 3 hexoses (Hex); 2 Hex, and 1 N-acetylhexosamine (HexNAc); 2 Hex and 1 Neu5Ac; and 3 Hex and 1 Neu5Ac. The largest OS had up to six monosaccharide units, providing opportunities to produce larger structures resembling those present in HMO populations (Wu, Grimm, German, & Lebrilla, 2011; Wu, Tao, German, Grimm, & Lebrilla, 2010).
Table 3.
Unique BMO in sample ID5 and new oligosaccharides (OS) formed by enzymatic glycosylation of BMO. a
| Starting material |
Newly formed OS |
||
|---|---|---|---|
| Mass (H+) m/z | Composition | Mass (H+) m/z | Composition |
| 505.1764 | 30000 | ||
| 546.2029 | 21000 | ||
| 634.2189 | 20010 | ||
| 651.2342 | 30100 | ||
| 667.2291 | 40000 | ||
| 675.2455 | 11010 | ||
| 708.2558 | 31000 | ||
| 749.2823 | 22000 | ||
| 763.2615 | 10020 | ||
| 780.2768 | 20110 | ||
| 796.2718 | 30010 | ||
| 829.2821 | 50000 | ||
| 837.2983 | 21010 | ||
| 854.3136 | 31100 | ||
| 870.3087 | 41000 | ||
| 895.3402 | 22100 | ||
| 911.3352 | 32000 | ||
| 942.3296 | 30110 | ||
| 958.3245 | 40010 | ||
| 999.3511 | 31010 | ||
| 1016.3664 | 41100 | ||
| 1073.3879 | 42000 | ||
| 1087.3671 | 30020 | ||
| 1104.3824 | 40110 | ||
| 1114.4145 | 33000 | ||
| 1114.4396 | 11500 | ||
| 1128.3937 | 21020 | ||
| 1216.4097 | 20030 | ||
| 1290.4465 | 31020 | ||
| 1307.4618 | 41110 | ||
| 1331.4731 | 22020 | ||
| 1363.488 | 20320 | ||
| 1365.5037 | 42200 | ||
| 1452.4993 | 41020 | ||
| 1510.5412 | 42110 | ||
| 1898.6515 | 80210 | ||
| 2077.7575 | 32420 | ||
| 2078.7528 | 54110 | ||
ID5 is defined in Table 2; numbers shown for each mass species represent the numbers of hexose, N-acetylhexosamine, fucose, N-acetyl neuraminic acid, and N-glycolylneuraminic acid, respectively, in the structure.
3.2. Enzymatic modification of BMO
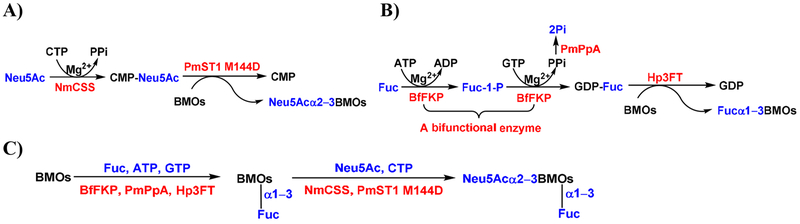
An efficient one-pot multienzyme (OPME) α2—3-sialylation system (Lin, Yuan, Li, & Lin, 2006; Yu et al., 2005) was used to sialylate the selected BMO sample (ID 5). In this system (Fig. 1), inexpensive and commercially available Neu5Ac and cytidine 5’-triphosphate were used by a recombinant NmCSS (Yu et al., 2004) to form cytidine 5’-monophosphate (CMP)-Neu5Ac, the sugar nucleotide donor for sialyltransferases. PmST1 M144D, a mutant of P. multocida α2—3-sialyltransferase (PmST1) (Yu et al., 2005) with decreased donor hydrolysis and sialidase activity (Sugiarto et al., 2012) was used to produce the desired α2—3-linked sialosides resembling those in human milk. Small-scale reactions were carried out by varying the ratios of BMO and the enzymes (NmCSS and PmST1 M144D) used, and the reactions were monitored by HRMS (Supplementary material Table S1 and Fig. S2). Among several conditions tested, the reaction carried out at 37 °C for 24 h with the ratio of BMO:NmCSS:PmST1_M144D = 100:1:2 (by weight) was optimum. Therefore, these conditions were chosen for large-scale sialylation.
Fig. 1.
OPME fucosylation and/or sialylation of BMO. A. One-pot two-enzyme (OP2E) sialylation of BMO; B. One-pot three-enzyme (OP3E) fucosylation of BMO, C. Sequential OPME fucosylation and sialylation of BMO.
Fucosylation of BMO sample ID 5 was carried out using a OP3E α1—3-fucosylation system (Fig. 1B) containing a bifunctional BfFKP (Yi et al., 2009), PmPpA (Lau et al., 2010), and Hp3FT (Bai et al., 2019; Sugiarto et al., 2011, 2012). The bifunctional BfFKP was responsible for the formation of guanosine-5’-diphosphate fucose (GDP-Fuc), the sugar nucleotide donor substrate for fucosyltransferases. PmPpA helped break down the by-product pyrophosphate formed shift the reaction towards the GDP-Fuc formation direction. Hp3FT is a commonly used bacterial fucosyltransferase for the enzymatic and chemoenzymatic synthesis of α1—3-linked fucosides (Bai et al., 2019; Yu et al., 2012). Hp3FT preferentially adds a Fuc residue to the GlcNAc in Galβ1-4GlcNAcβOR (LacNAcβOR) to form Lewisx. Hp3FT can also add a Fuc to the Glc in lactose, although the efficiency was much lower (Bai et al., 2019; Yu et al., 2012). The optimal reaction conditions identified in our study were Tris-HCl buffer (pH 7.5, 100 mm) at 37 °C for 24 h with 25 mg of each enzyme for fucosylating 6 g of BMO (ID 5).
The fucosylated BMO were further α2—3-sialylated using the one-pot two-enzyme system similar to that described above as PmST1 M144D mutant was able to using fucosylated compounds as Lex as acceptor substrates (Sugiarto et al., 2012). The sequential OPME fucosylation and sialylation strategy (Fig. 3C) was used to produce fucosylated and sialylated BMO.
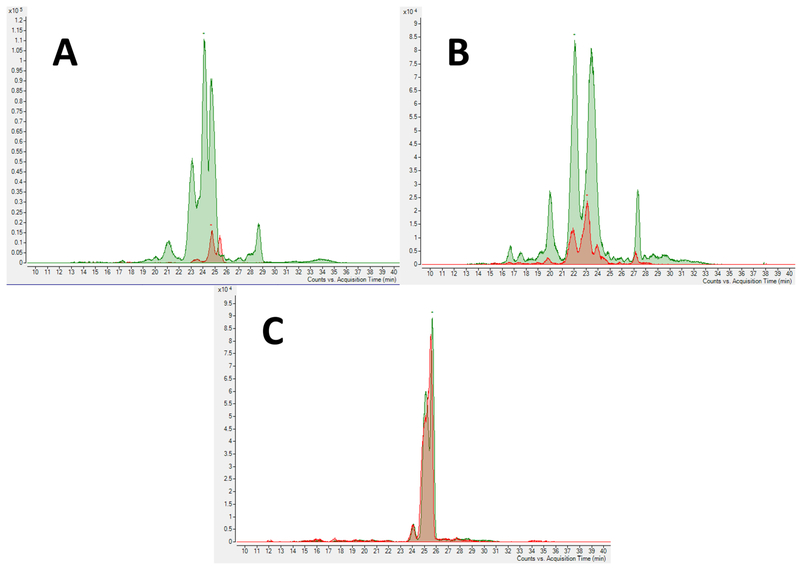
Fig. 3.
Extracted ion chromatogram of 3 hexose 1 sialic acid (796.2718 m/z) before (Red) and after (Green) (A) α2—3-sialylation, (B) after both α1—3-fucosylation and α2—3-sialylation, and (C) after only α1—3-fucosylation.
3.3. Partial purification of enzymatically modified BMO
HMO are a mixture of more than 150 OS that can be recognized by numerous pathogenic bacteria (Coppa et al., 2006). Considering the specificity of glycan recognition of pathogens, the oral supplementation with OS mixtures, such as the reaction products described above, could allow broad-spectrum protection in the gut. BMO enzymatically glycosylated by OPME reactions were purified using an activated charcoal column with or without an additional C18 column to remove free monosaccharides (e.g., Neu5Ac and/or Fuc) and sugar nucleotides. The samples before and after column purifications were analyzed by HRMS using a Thermo Electron LTQ-Orbitrap Hybrid Mass Spectrometer. For OPME fucosylation reactions, as shown in Supplementary material Fig. S3, Fuc-1-P, ADP, AMP, GMP, and GDP-Fuc were completely removed by one-step charcoal purification. For sialylation, the activated charcoal column removed some CMP and all free Neu5Ac (Supplementary material Fig. S4). Applying the samples to a C18 column using a Combiflash® system improved the purity of the product by removing the remaining CMP.
3.4. Identification of novel OS structures
Upon enzymatic modification, the control and modified BMO samples were extracted by C18 and GCC-SPE-columns, and characterized by a Q-TOF LC/MS with a graphitized carbon column nano-chip to identify novel structures.
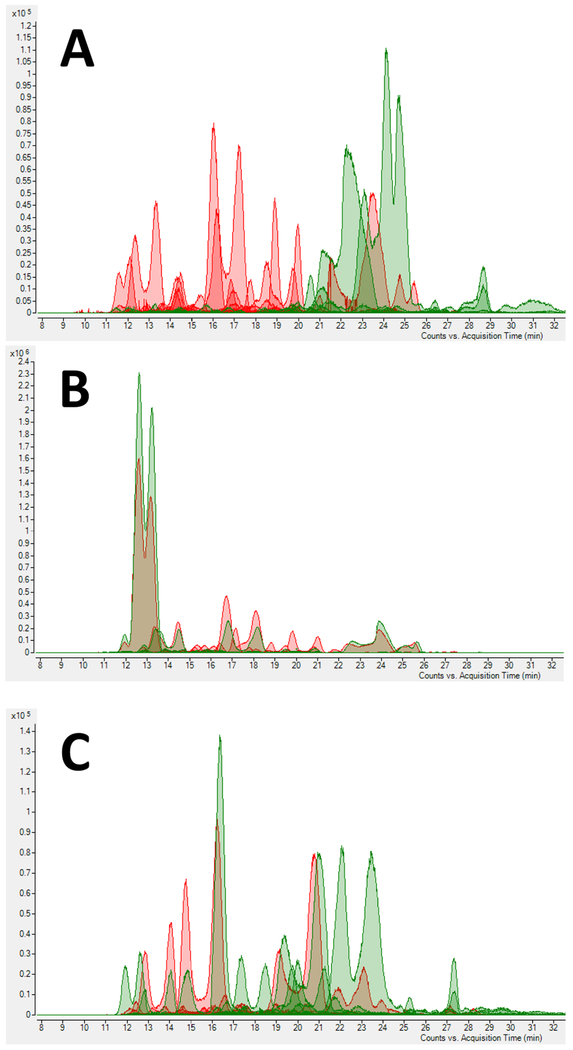
The extracted ion chromatograms of all BMO naturally present in samples before and after enzymatic addition of Neu5Ac (A), Fuc (B), and both (C) are shown in Fig. 2. The disappearance of some peaks corresponded to their use as suitable acceptor substrates by the glycosyltransferases used. Also, increased intensity was observed for some peaks, indicating the production of an additional amount of the compounds from other precursors by the OPME glycosylation reaction.
Fig. 2.
Extracted ion chromatograms of BMO before (Red) and after (Green) (A) α2—3-sialylation, (B) α1—3-fucosylation, and (C) with both α1—3-fucosylation and α2—3-sialylation.
As a result of the successful enzymatic modifications of BMO, 22 OS not reported previously in bovine milk (Aldredge et al., 2013) were identified. The compositions of these OS are shown in Table 3. We were able to increase the maximum size of the OS present in the original source of BMO from DP 6 to DP 11. HMO can reach to DP 12 (Wu et al., 2011, 2010). Thus, increasing the degree of polymerization of BMO represents a shift toward the HMO structural profile. Nine of these 22 new OS (3 Hex 1 Fuc; 2 Hex 1 Fuc 1 Neu5Ac; 4 Hex 1 Neu5Ac; 4 Hex 1 GlcNAc1 Fuc; 3 Hex 1 GlcNAc2 Neu5Ac; 4 Hex 1 GlcNAc1 Fuc 1 Neu5Ac; 2 Hex 2 GlcNAc1 Neu5Ac; 4 Hex 2 GlcNAc2 Fuc, and 4 Hex 2 GlcNAc1 Fuc 1 Neu5Ac) matched known HMO compositions, thus rendering the resulting OS mixture more similar to HMO than the original mixture of BMO from dairy streams. To our knowledge, this is the first time that HMO-like OS mixtures containing several larger di-fucosylated species have been created.
The HMO DSLNT (3 Hex, 1 GlcNAc and 2 Neu5Ac), containing an α2—3- and an α2—6-linked Neu5Ac residues, was reported to lower the rate of NEC in animal models (Yu et al., 2014, 2017). NEC is the most common deadly disease in preterm and low-weight infants (Neu, 1996), with a mortality rate of 15—30% (Berman & Moss, 2011). Although we did not characterize the individual linkagesOS with the same composition as DSLNT were identified in the sialylated BMO. Further studies would be needed to test whether the OS formed have NEC-preventive activity.
In addition to the new OS, OS present in the original BMO sample were formed. The most noticeable increase was for 3 Hex and 1 Neu5Ac after α2—3-sialylation (Fig. 3A) or after both α1—3-fucosylation and α2—3-sialylation (Fig. 3B) reactions. There was no increase in this OS when only fucosylation was performed (Fig. 3C). These were expected outcomes, as this specific isomer of 3 Hex (determined by molecular weight and retention time matching) is a suitable acceptor substrate for PmST1 M144D (Sugiarto et al., 2012) but not a good acceptor for Hp3FT (Bai et al., 2019).
The ability to produce a BMO mixture containing more sialylated glycans than initially present in the BMO source is highly valuable to broaden the spectrum of indirect anti-pathogen action via the decoy mechanism attributed to the complexity of the HMO. Neu5Ac-containing HMO were shown to decrease the attachment, thus infection, of several pathogens, including N. meningitidis, E. coli, Vibrio cholerae, Streptococcus pneumoniae, and Salmonella fyris, among others (Barthelson, Mobasseri, Zopf, & Simon, 1998; Coppa et al., 2006; Hakkarainen et al., 2005; Martín-Sosa, Martín, & Hueso, 2002). Upon enzymatic modification, the sialylated portion of OS in the BMO sample increased from 29.4% (by number of structures) to as high as 51.7%, which, if applied to a formula supplement, may improve the protective efficiency of OS against pathogens.
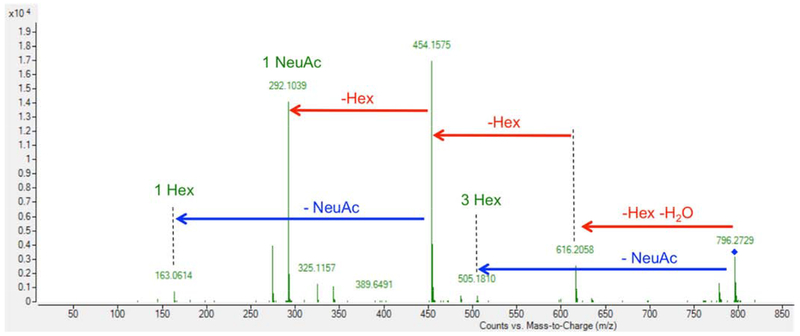
In addition to finding that accurate mass and retention time matched previously assembled milk OS libraries, tandem MS/MS confirmed the compositions of all enzyme-modified BMO. As shown in Fig. 4, the tandem MS/MS for sialylgalactosyllactose, the breakdown of 3 Hex and 1 Neu5Ac to each individual component, confirmed the composition of this OS.
Fig. 4.
Tandem MS/MS of 3 hexose 1 sialic acid (796.2718 m/z).
HMO are highly fucosylated, reaching 80% fucosylation in some studies (Bode, 2012). Fucosylated milk-OS were associated with anti-pathogenic activity against a variety of microorganisms, such as E. coli, Campylobacter jejuni, V. cholerae, Sal. fyris, and Calicivirus (Coppa et al., 2006; Morrow et al., 2004; Newburg, Pickering, McCluer, & Cleary, 1990; Newburg et al., 2004). The proportion of fucosylated OS in bovine milk is much lower than in human milk (Bode, 2012). In this work, fucosylated OS (by number of structures) increased from 5.9% to 24%, which could lead to an increase in the anti-pathogenic actions of this pool of OS if applied as a supplement to formulas. The composition of the new fucosylated OS formed is presented in Table 4.
Table 4.
Fucosylated oligosaccharides formed by enzymatic fucosylation of BMO. a
| Mass (H+) m/z | OS composition |
|---|---|
| 651.2342 | 30100 |
| 780.2768 | 20110 |
| 895.3402 | 22100 |
| 942.3296 | 30110 |
| 1016.3664 | 41100 |
| 1104.3824 | 40110 |
| 1114.4396 | 11500 |
| 1307.4618 | 41110 |
| 1363.488 | 20320 |
| 1365.5037 | 42200 |
| 1510.5412 | 42110 |
| 1898.6515 | 80210 |
| 2077.7575 | 32420 |
| 2078.7528 | 54110 |
Numbers for OS composition represent the numbers of hexose, N-acetylhexosamine, fucose, N-acetylneuraminic acid, and N-glycolylneuraminic acid, respectively, in the structure.
Several methods have been developed to reproduce the diversity of OS structures naturally found in human milk. However, to date, only a limited number of HMO have been efficiently synthesized in large-scale. Metabolically engineered bacteria, transgenic animals, and chemical and enzymatic synthesis are common methods used in research laboratories. To our knowledge, none of those methods have been able to create the mixture of a diverse pool of OS mimicking HMO as described in this work. HMO and BMO differ in the size of OS structures, with HMO reaching dodecasaccharides and BMO reaching heptasaccharides (Aldredge et al., 2013; Wu et al., 2011, 2010). With the enzymatic glycosylation methods described herein, we produced OS of a size up to undecasaccharides. Using additional OPME fucosylation and sialylation systems, with or without OPME N-acetylglucosaminylation and galactosylation reactions, the size of OS in BMO can be further increased to better mimic HMO. For this reason, we consider the strategy described here a novel and valuable approach towards the objective of achieving a product having structures resembling those of HMO.
3.5. Effects of novel BMO on bacterial uptake
Enterohemorrhagic E. coli is a pathogen that can produce Shiga toxins and cause hemorrhagic colitis and the life-threatening sequelae hemolytic uremic syndrome in humans (Basu et al., 2016; Rojas-Lopez, Monterio, Pizza, Desvaux, & Rosini, 2018). Several serotypes of EHEC are frequently associated with human diseases such as O26:H11, O91:H21, O111:H8, O157:NM, and O157:H7. E. coli O157:H7 is a major foodborne pathogen causing severe disease in humans worldwide. Healthy cattle are a reservoir of E. coli O157:H7. Bovine food products and fresh produce contaminated with bovine waste are the most common sources for disease outbreaks in the United States (Lim, Yoon, & Hovde, 2010).
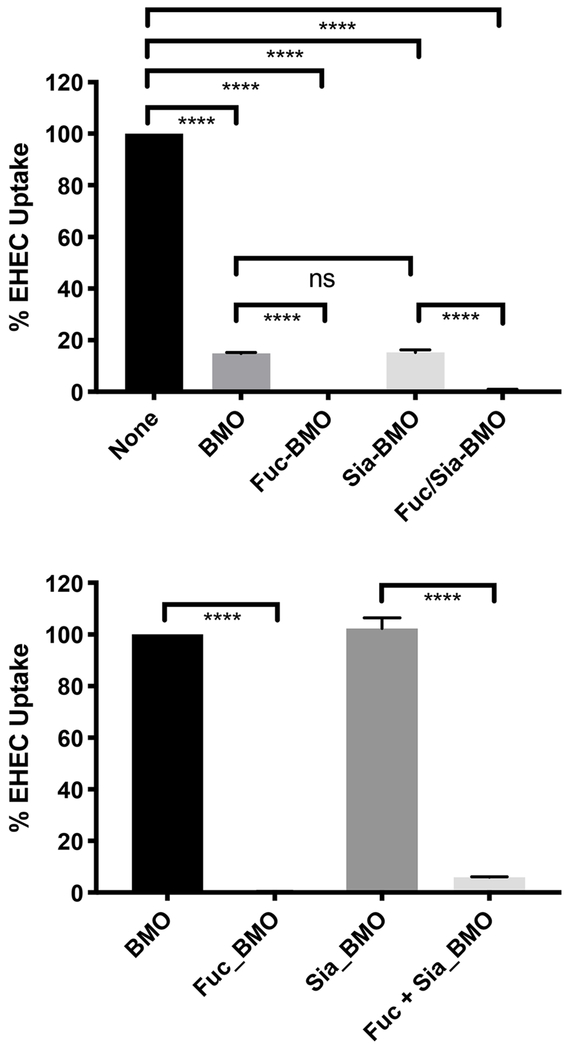
Partially purified oligosaccharides obtained from enzymatic glycosylation reactions were tested in inhibiting the uptake of E. coli O157:H7 by Caco-2 cells. BMO sample (ID5) was used as a control. As shown in Fig. 5, while α2—3-sialylation did not show significant changes, α1—3-fucosylated BMO with or without additional α2—3-sialylation showed improved inhibition effect against the uptake of E. coli O157:H7 by Caco-2 cells, indicating a potential improved anti-infective activity. These data clearly indicate that the presence of BMO pool prevents invasion by pathogenic bacteria like E. coli EHEC. Secondly, it clearly demonstrates (part B), that fucosylation is critical to a dramatic alteration in the uptake of bacteria, since Sia-BMO shows similar activity to BMO, whereas Fuc-BMO is clearly altered. These data add to the biological function of oligosaccharide modification on a pathogenesis outcome. These results are consistent with previous reports that fucosylated OS reduced binding of a variety of pathogenic bacteria to target intestinal cells in vitro (Morrow et al., 2004a,b; Ruiz-Palacios, Cervantes, Ramos, Chavez-Munguia, & Newburg, 2003).
Fig. 5.
Effect of enzymatic addition of sialic acid and/or fucose to BMO on the uptake of E. coli O157:H7 by Caco-2 cells. **** indicate P <0.001 as determined by Graphpad Column analysis (Two tailed t tests) between individual columns; ns, non-significant.
4. Conclusion
Using high-yield, cost-effective one-pot multienzyme sialylation and fucosylation approaches, as well as readily available, inexpensive bovine whey permeates as starting materials, highly-sialylated and/or fucosylated BMO mimicking HMO were produced. Partially-purified fucosylated BMO strongly decreased the adhesion of E. coli O157:H7 to intestinal epithelial cells. The strategy described can be extended to additional enzymatic sialylation, fucosylation, N-acetylglucosaminylation, and galactosylation to generate more complex mixtures of BMO to better mimic the structure and beneficial functions of HMO.
Supplementary Material
Acknowledgements
This work was supported by National Institute of Health (grants R43 HD083981, R01HD065122, R01AT008759), and USDA:NIFA Hatch project 232719. YL, HY, and XC are co-founders of Glycohub, Inc., a company focused on the development of carbohydrate-based reagents, diagnostics, and therapeutics; this does not affect the objectivity of the work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albayrak N, & Yang S-T (2002). Production of galacto-oligosaccharides from lactose by Aspergillus oryzae β-galactosidase immobilized on cotton cloth. Biotechnology and Bioengineering, 77, 8–19. [DOI] [PubMed] [Google Scholar]
- Aldredge DL, Geronimo MR, Hua S, Nwosu CC, Lebrilla CB, & Barile D (2013). Annotation and structural elucidation of bovine milk oligosaccharides and determination of novel fucosylated structures. Glycobiology, 23, 664–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Wu Z, Sugiarto G, Gadi MR, Yu H, Li Y, et al. (2019). Biochemical characterization of Helicobacter pylori α1—3-fucosyltransferase and its application in the synthesis of fucosylated human milk oligosaccharides. Carbohydrate Research, 480, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard O, & Morrow AL (2013). Human milk composition. Pediatric Clinics of North America, 60, 49–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardor M, Nguyen DH, Diaz S, & Varki A (2005). Mechanism of uptake and incorporation of the non-human sialic acid N-glycolylneuraminic acid into human cells. Journal of Biological Chemistry, 280, 4228–4237. [DOI] [PubMed] [Google Scholar]
- Barile D, Marotta M, Chu C, Mehra R, Grimm R, Lebrilla CB, et al. (2010). Neutral and acidic oligosaccharides in Holstein-Friesian colostrum during the first 3 days of lactation measured by high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Journal of Dairy Science, 93, 3940–9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile D, Meyrand M, Lebrilla CB, & German JB (2011). Examining bioactive components of milk. Sources of complex oligosaccharides (Part 2). Agro Food Industry Hi-Tech, 22, 37–39. [Google Scholar]
- Barile D, Tao N, Lebrilla CB, Coisson J-D, Arlorio M, & German JB (2009). Permeate from cheese whey ultrafiltration is a source of milk oligosaccharides. International Dairy Journal, 19, 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelson R, Mobasseri A, Zopf D, & Simon P (1998). Adherence of Streptococcus pneumoniae to respiratory epithelial cells is inhibited by sialylated oligosaccharides. Infection and Immunity, 66, 1439–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D, Li X-P, Kahn JN, May KL, Kahn PC, & Tumer NE (2016). The A1 subunit of Shiga toxin 2 has higher affinity for ribosomes and higher catalytic activity than the A1 subunit of Shiga toxin 1. Infection and Immunity, 84, 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman L, & Moss RL (2011). Necrotizing enterocolitis: An update. Seminars in Fetal and Neonatal Medicine, 16, 145–150. [DOI] [PubMed] [Google Scholar]
- Bode L (2012). Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology, 22, 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau MR, O’Donnell D, Blanton LV, Totten SM, Davis JCC, Barratt MJ, et al. (2016). Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell, 164, 859–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X (2015). Human milk oligosaccharides (HMOS). In Advances in Carbohydrate Chemistry and Biochemistry, 72, 113–190). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JL, Barile D, Liu Y, & de Moura Bell JMLN (2017). Role of pH in the recovery of bovine milk oligosaccharides from colostrum whey permeate by nanofiltration. International Dairy Journal, 66, 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppa GV, Pierani P, Zampini L, Carloni I, Carlucci A, & Gabrielli O (1999). Oligosaccharides in human milk during different phases of lactation. Acta Paediatrica, 88, 89–94. [DOI] [PubMed] [Google Scholar]
- Coppa GV, Zampini L, Galeazzi T, Facinelli B, Ferrante L, Capretti R, et al. (2006). Human milk oligosaccharides inhibit the adhesion to Caco-2 cells of diarrheal pathogens: Escherichia coli, Vibrio cholerae, and Salmonella fyris. Pediatric Research, 59, 377–382. [DOI] [PubMed] [Google Scholar]
- Dallas DC, Weinborn V, de Moura Bell JMLN, Wang M, Parker EA, Guerrero A, et al. (2014). Comprehensive peptidomic and glycomic evaluation reveals that sweet whey permeate from colostrum is a source of milk protein-derived peptides and oligosaccharides. Food Research International, 63, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moura Bell JMLN, Cohen JL, de Aquino LFMC, Lee H, de Melo VL, Liu Y, et al. , (2018). An integrated bioprocess to recover bovine milk oligosaccharides from colostrum whey permeate. Journal of Food Engineering, 216, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German JB, Freeman SL, Lebrilla CB, & Mills DA (2008). Human milk oligosaccharides: evolution, structures and bioselectivity as substrates for intestinal bacteria In Bier DM, German JB, & Lönnerdal B (Eds.), Personalized nutrition for the diverse needs of infants and children (pp. 205–222). Helsinki, Finland: 62nd Néstle Nutrition Workshop, Pediatric Programme. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkarainen J, Toivanen M, Leinonen A, Frängsmyr L, Strömberg N, Lapinjoki S, et al. (2005). Human and bovine milk oligosaccharides inhibit Neisseria meningitidis pili attachment in vitro. Journal of Nutrition, 135, 2445–2448. [DOI] [PubMed] [Google Scholar]
- Inbiose. (2018). Inbiose is fast tracking 7 HMOs from its portfolio for use in infant formula and health markets. Retrieved September 11, 2019, from https://www.inbiose.com/press-release-7-hmo-portfolio/
- Kunz C, Kuntz S, & Rudloff S (2014). Bioactivity of human milk oligosaccharides In Moreno FJ, & Luz Sanz M (Eds.), Food oligosaccharides: Production, analysis and bioactivity (pp. 1–20). Chichester, West Sussex, UK: J. Wiley & Sons. [Google Scholar]
- Kunz C, Rudloff S, Baier W, Klein N, & Strobel S (2000). Oligosaccharides in human milk: Structural, functional, and metabolic aspects. Annual Review of Nutrition, 20, 699–722. [DOI] [PubMed] [Google Scholar]
- Lau K, Thon V, Yu H, Ding L, Chen Y, Muthana MM, et al. (2010). Highly efficient chemoenzymatic synthesis of beta1-4-linked galactosides with promiscuous bacterial beta1-4-galactosyltransferases. Chemical Communications, 46, 6066–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JY, Yoon J, & Hovde CJ (2010). A brief overview of Escherichia coli O157:H7 and its plasmid O157. Journal of Microbiology and Biotechnology, 20, 5–14. [PMC free article] [PubMed] [Google Scholar]
- Lin S-W, Yuan T-M, Li J-R, & Lin C-H (2006) Carboxyl terminus of Helicobacter pylori α1,3-fucosyltransferase determines the structure and stability. Biochemistry, 45, 8108–8116 [DOI] [PubMed] [Google Scholar]
- Martín-Sosa S, Martín M-J, & Hueso P (2002). The sialylated fraction of milk oligosaccharides Is partially responsible for binding to enterotoxigenic and uropathogenic Escherichia coli human strains. Journal of Nutrition, 132, 3067–3072. [DOI] [PubMed] [Google Scholar]
- Meyrand M, Dallas DC, Caillat H, Bouvier F, Martín P, & Barile D (2013). Comparison of milk oligosaccharides between goats with and without the genetic ability to synthesize αs1-casein. Small Ruminant Research, 113, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow AL, Ruiz-Palacios GM, Altaye M, Jiang X, Guerrero ML, Meinzen-Derr JK, et al. (2004a). Human milk oligosaccharide blood group epitopes and innate immune protection against Campylobacter and calicivirus diarrhea in breastfed infants In Pickering LK, Morrow AL, Ruiz-Palacios GM, & Schanler RJ (Eds.), Protecting infants through human milk (pp. 443–446). New York, NY, USA: Springer; [DOI] [PubMed] [Google Scholar]
- Morrow Ardythe L, Ruiz-Palacios GM, Altaye M, Jiang X, Guerrero ML, Meinzen-Derr JK, et al. (2004b). Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. Journal of Pediatrics, 145, 297–303. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Kawase H, Kimura K, Watanabe Y, Ohtani M, Arai I, et al. (2003). Concentrations of sialyloligosaccharides in bovine colostrum and milk during the prepartum and early lactation. Journal of Dairy Science, 86, 1315–1320. [DOI] [PubMed] [Google Scholar]
- NASS & USDA. (2018). Dairy products (No. ISSN: 1949-0399). Retrieved from https://downloads.usda.library.Cornell.edu/usda-esmis/files/m326m1757/2f75r892j/j9602160s/DairProd-03-01-2018.pdf
- Neu J (1996). Necrotizing enterocolitis: the search for a unifying pathogenic theory leading to prevention. Pediatric Clinics of North America, 43, 409–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburg DS, Pickering LK, McCluer RH, & Cleary TG (1990). Fucosylated oligosaccharides of human milk protect suckling mice from heat-stable enterotoxin of Escherichia coli. Journal of Infectious Diseases, 162, 1075–1080. [DOI] [PubMed] [Google Scholar]
- Newburg DS, Ruiz-Palacios GM, Altaye M, Chaturvedi P, Guerrero ML, Meinzen-Derr JK, et al. (2004). Human milk α1,2-linked fucosylated oligosaccharides decrease risk of diarrhea due to stable toxin of E. coli in breastfed infants In Pickering LK, Morrow AL, Ruiz-Palacios GM, & Schanler RJ (Eds.), Protecting infants through human milk (pp. 457–461). New York, NY, USA: Springer; [DOI] [PubMed] [Google Scholar]
- Newburg DS, Ruiz-Palacios GM, & Morrow AL (2005). Human milk glycans protect infants against enteric pathogens. Annual Review of Nutrition, 25, 37–58. [DOI] [PubMed] [Google Scholar]
- Nilsson KGI (1988). Enzymatic synthesis of oligosaccharides. Trends in Biotechnology, 6, 256–264. [Google Scholar]
- Ninonuevo MR, Park Y, Yin H, Zhang J, Ward RE, Clowers BH, et al. (2006). A strategy for annotating the human milk glycome. Journal of Agricultural and Food Chemistry, 54, 7471–7480. [DOI] [PubMed] [Google Scholar]
- Palcic MM (1999). Biocatalytic synthesis of oligosaccharides. Current Opinion in Biotechnology, 10, 616–624. [DOI] [PubMed] [Google Scholar]
- Priem B, Gilbert M, Wakarchuk WW, Heyraud A, & Samain E (2002). A new fermentation process allows large-scale production of human milk oligosaccharides by metabolically engineered bacteria. Glycobiology, 12, 235–240. [DOI] [PubMed] [Google Scholar]
- Rojas-Lopez M, Monterio R, Pizza M, Desvaux M, & Rosini R (2018). Intestinal pathogenic Escherichia coli: Insights for vaccine development. Frontiers in Microbiology, 9, Article 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, & Newburg DS (2003). Campylobacter jejuni binds intestinal H(O) antigen (Fuc α1, 2Gal β1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. Journal of Biological Chemistry, 278, 14112–14120. [DOI] [PubMed] [Google Scholar]
- Sears P, & Wong CH (2001). Toward automated synthesis of oligosaccharides and glycoproteins. Science, 291, 2344–2350. [DOI] [PubMed] [Google Scholar]
- Sugiarto G, Lau K, Qu J, Li Y, Lim S, Mu S, et al. (2012). A sialyltransferase mutant with decreased donor hydrolysis and reduced sialidase activities for directly sialylating Lewisx. ACS Chemical Biology, 7, 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiarto G, Lau K, Yu H, Vuong S, Thon V, Li Y, et al. (2011). Cloning and characterization of a viral α2–3-sialyltransferase (vST3Ga1-I) for the synthesis of sialyl Lewisx. Glycobiology, 21, 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H-Y, Lin S-W, Ko T-P, Pan J-F, Liu C-L, Lin C-N, et al. (2007). Structure and mechanism of Helicobacter pylori fucosyltransferase. A basis for lipopolysaccharide variation and inhibitor design. Journal of Biological Chemistry, 282, 9973–9982. [DOI] [PubMed] [Google Scholar]
- Sundekilde UK, Barile D, Meyrand M, Poulsen NA, Larsen LB, Lebrilla CB, et al. (2012). Natural variability in bovine milk oligosaccharides from Danish Jersey and Holstein-Friesian breeds. Journal of Agricultural and Food Chemistry, 60, 6188–6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao N, DePeters EJ, German JB, Grimm R, & Lebrilla CB (2009). Variations in bovine milk oligosaccharides during early and middle lactation stages analyzed by high-performance liquid chromatography-chip/mass spectrometry. Journal of Dairy Science, 92, 2991–3001. [DOI] [PubMed] [Google Scholar]
- Tungland BC (2003). Fructooligosaccharides and other fructans: Structures and occurrence, production, regulatory aspects, Food applications, and nutritional health significance In Eggleston G & Cote GL (Eds.), Oligosaccharides in food and agriculture (pp. 135–152). Washington, DC, USA: American Chemical Society [Google Scholar]
- Urashima T, Saito T, Nakamura T, & Messer M (2001). Oligosaccharides of milk and colostrum in non-human mammals. Glycoconjugate Journal, 18, 357–371. [DOI] [PubMed] [Google Scholar]
- Vandenplas Y, Berger B, Carnielli V, Ksiazyk J, Lagström H, Sanchez Luna M, et al. (2018). Human milk oligosaccharides: 2′-fucosyllactose (2′-FL) and lacto-N-neotetraose (LNnT) in infant formula. Nutrients, 10, Article 1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenplas Y, Greef E, & Veereman G (2014). Prebiotics in infant formula. Gut Microbes, 5, 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Grimm R, German JB, & Lebrilla CB (2011). Annotation and structural analysis of sialylated human milk oligosaccharides. Journal of Proteome Research, 10, 856–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tao N, German JB, Grimm R, & Lebrilla CB (2010). Development of an annotated library of neutral human milk oligosaccharides. Journal of Proteome Research, 9, 4138–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Liu X, Li Y, Li J, Xia C, Zhou G, et al. (2009). Remodeling bacterial polysaccharides by metabolic pathway engineering. Proceedings of the National Academy of Sciences of the United States of America, 106, 4207–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, & Chen X (2016). One-pot multienzyme (OPME) systems for chemoenzymatic synthesis of carbohydrates. Organic and Biomolecular Chemistry, 14, 2809–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chokhawala H, Karpel R, Yu H, Wu B, Zhang J, et al. (2005). A multifunctional Pasteurella multocida sialyltransferase: A powerful tool for the synthesis of sialoside libraries. Journal of the American Chemical Society, 127, 17618–17619. [DOI] [PubMed] [Google Scholar]
- Yu H, Lau K, Li Y, Sugiarto G, & Chen X (2012). One-pot multienzyme synthesis of Lewisx and sialyl Lewisx antigens. Current Protocols in Chemical Biology, 4, 233–247). 10.1002/9780470559277.ch110277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Lau K, Thon V, Autran CA, Jantscher-Krenn E, Xue M, et al. (2014). Synthetic disialyl hexasaccharides protect neonatal rats from necrotizing enterocolitis. Angewandte Chemie International Edition, 53, 6687–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Yan X, Autran CA, Li Y, Etzold S, Latasiewicz J, et al. (2017). Enzymatic and chemoenzymatic syntheses of disialyl glycans and their necrotizing enterocolitis preventing effects. Journal of Organic Chemistry, 82, 13152–13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Yu H, Karpel R, & Chen X (2004). Chemoenzymatic synthesis of CMP—sialic acid derivatives by a one-pot two-enzyme system: comparison of substrate flexibility of three microbial CMP-sialic acid synthetases. Bioorganic and Medicinal Chemistry, 12, 6427–6435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.