SUMMARY
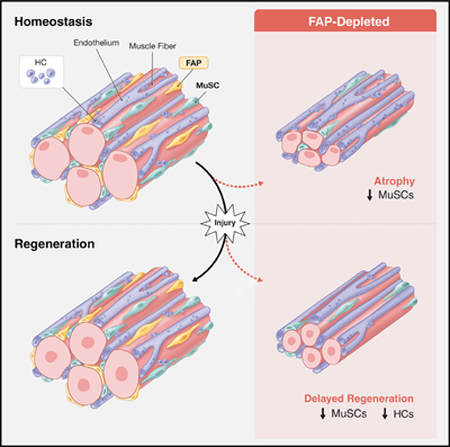
The necessity of mesenchymal stromal cells, called fibroadipogenic progenitors (FAPs), in skeletal muscle regeneration and maintenance remains unestablished. We report the generation of a PDGFRαCreER knockin mouse model that provides a specific means of labeling and targeting FAPs. Depletion of FAPs using Cre-dependent diphtheria toxin expression results in loss of expansion of muscle stem cells (MuSCs) and CD45+ hematopoietic cells after injury and impaired skeletal muscle regeneration. Furthermore, FAP-depleted mice under homeostatic conditions exhibit muscle atrophy and loss of MuSCs, revealing that FAPs are required for the maintenance of both skeletal muscle and the MuSC pool. We also report that local tamoxifen metabolite delivery to target CreER activity in a single muscle, removing potentially confounding systemic effects of ablating PDGFRα+ cells distantly, also causes muscle atrophy. These data establish a critical role of FAPs in skeletal muscle regeneration and maintenance.
In Brief
Wosczyna et al. develop genetic models to target and deplete fibroadipogenic progenitors (FAPs) in skeletal muscle (SkM). Following injury of FAP-depleted SkM, muscle stem cell (MuSC) expansion is impaired, leading to a regenerative deficit. Under homeostatic conditions, FAP-depleted SkM undergoes muscle fiber atrophy, and MuSC numbers decline.
Graphical Abstract
INTRODUCTION
Skeletal muscle is one of the largest organs in the human body and provides a means to generate movement and maintain metabolic homeostasis (Frontera and Ochala, 2015). The remarkable ability of skeletal muscle to regenerate following injury and be maintained over a lifetime is vital to an organism; thus, these processes have been heavily investigated (Tidball, 2011). The majority of these efforts have focused on muscle stem cells (MuSCs; also known as “satellite cells”) because these cells are essential for these processes (Lepper et al., 2011; Murphy et al., 2011; Sambasivan et al., 2011). A population of tissue-resident mesenchymal progenitor cells that lie in the stromal space between myofibers (Joe et al., 2010; Uezumi et al., 2010; Wosczyna et al., 2012) and are generally referred to as fibroadipogenic progenitors (FAPs) have been postulated to be necessary for effective regeneration and maintenance of skeletal muscle because of their ability to support MuSC differentiation in vitro (Heredia et al., 2013; Joe et al., 2010). Further, FAPs have been shown to express immune-modulating cytokines that can cause regulatory T cells to home in on an injured area and to secrete extracellular matrix, all to aid tissue repair (Kuswanto et al., 2016; Lemos et al., 2015). However, the inability to genetically target FAPs in vivo has limited the accurate assessment of their role in muscle regeneration and homeostasis. Here we exploit the highly specific expression of the PDGFRα gene to FAPs in adult skeletal muscle (Tabula Muris Consortium, 2018; Wosczyna et al., 2012; Joe et al., 2010; Uezumi et al., 2010) and generate a knockin mouse model using the endogenous PDGFRα locus to drive a CreER transgene, providing a means to conditionally target FAPs in vivo. We demonstrate that cellular labeling driven by our PDGFRαCreER mouse is specific to the PDGFRα lineage and genetically marks FAPs at a high efficiency. We use this mouse to deplete FAPs and to address the question of FAP necessity in skeletal muscle regenerative and homeostatic processes. We find that FAPs are essential for efficient skeletal muscle regeneration and required for long-term homeostatic maintenance and growth of skeletal muscle.
RESULTS
PDGFRαCreER Knockin Mice Specifically and Efficiently Label FAPs In Vivo
We and others have demonstrated the specific expression of PDGFRα to FAPs in adult skeletal muscle and that the vast majority (~90%) of adult FAPs express this gene (Joe et al., 2010; Uezumi et al., 2010; Wosczyna et al., 2012; Tabula Muris Consortium, 2018). Thus, we created a knockin mouse in which CreER was inserted into the PDGFRα locus using the endogenous start codon to drive the translation of this transgene (Figure S1A). We crossed this mouse with the highly sensitive Cre-dependent GFP reporter R26NG (Yamamoto et al., 2009). Following tamoxifen dosing of PDGFRαCreER;R26NG offspring, we assessed the specificity of the labeling of cells in the PDGFRα lineage in skeletal muscle and the efficiency of labeling of adult FAPs (Figure S1B). The GFP reporter was detected in ~76% of PDGFRα+ cells, and labeling outside of the PDGFRα lineage was not detected (Figure S1C), demonstrating the efficiency and specificity of the model, respectively. Furthermore, ~73% of FAPs were labeled by the GFP reporter (Figures S1D–S1F), consistent with the vast majority of these cells expressing PDGFRα. A small number of cells in other populations express PDGFRα (Figures S1G and S1H; Tabula Muris Consortium, 2018) and, thus, are GFP+ (Figure S1E), but these represented less than 0.7% of the total cells of each fraction. These data confirm that the PDGFRαCreER mouse is a highly specific and effective means to target PDGFRa+ FAPs in vivo.
We next injured skeletal muscle of tamoxifen-dosed mice and determined the cellular response. Consistent with previous reports (Heredia et al., 2013; Lemos et al., 2015), FAP numbers peaked around 3 days post-injury (dpi; Figure S1F). The number of MuSCs and CD45+ hematopoietic cells (HCs) also peaked early, at 3 to 5 dpi, respectively, whereas CD31 + endothelial cells (ECs) increased later in the regenerative response (Figure S1F). By 9 dpi, cell numbers in all populations decreased appreciably, approaching levels in uninjured muscles (Figure S1F).
FAPs Are Necessary for Effective Skeletal Muscle Regeneration
To investigate the role of FAPs in the expansion of MuSCs and HCs in regenerating skeletal muscle, we generated a mouse line, PDGFRαCreER;R26NG/DTA or +/DTA, in which FAPs could be conditionally ablated following tamoxifen treatment and the resulting expression of diphtheria toxin fragment A (Voehringer et al., 2008) in PDGFRα-expressing cells. Following tamoxifen treatment of PDGFRαCreER;R26 NG/DTA or +/DTA mice, hereafter referred to as “ablated mice,” we achieved about 75% depletion of FAPs (Figures 1A and 1B), consistent with our labeling efficiency reported above. Furthermore, other cellular fractions were present in numbers similar to those of genetic controls not receiving tamoxifen (Figure S2A), although there was a minor increase in ECs in ablated mice.
Figure 1. FAPs Are Required for Efficient Skeletal Muscle Regeneration.
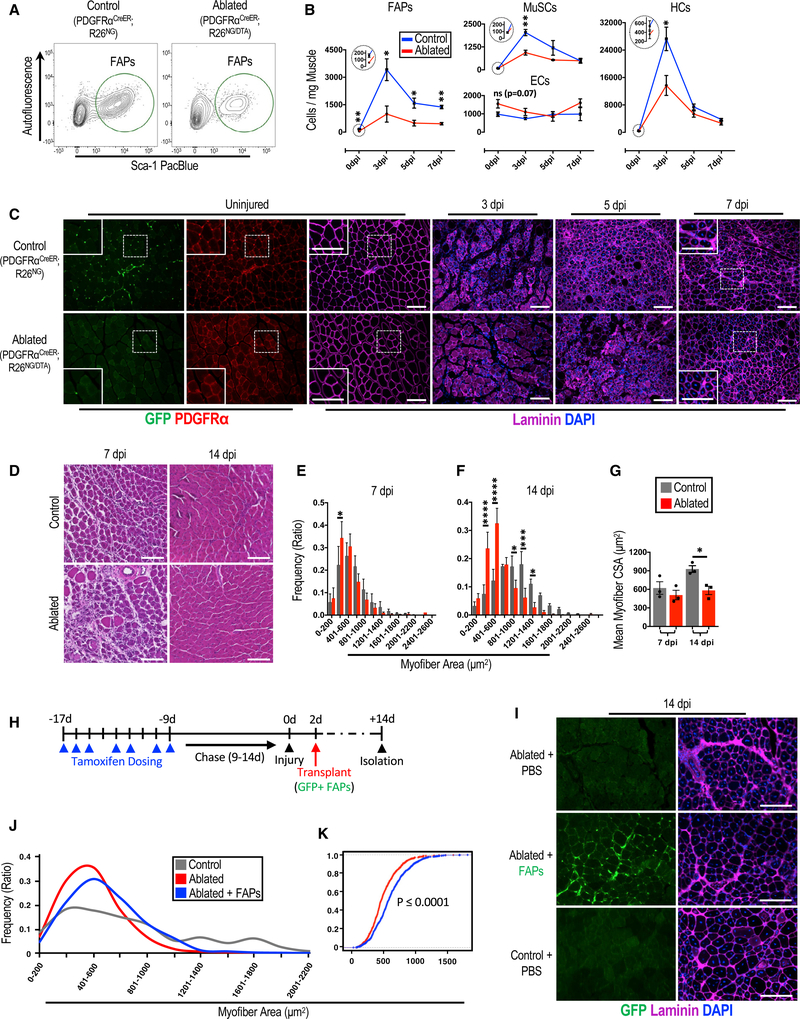
(A-C) The effects of depletion of FAPs on skeletal muscle.
(A) FACS plots showing the efficiency of ablating FAPs.
(B) A 7-day time course examining the influence of FAP ablation on the noted cellular fractions following skeletal muscle injury in ablated (PDGFRαCreER/+; R26NG/DTA or +/DTA) and control mice (n = 5–6 [0 dpi], 4–5 [3 dpi], and 3 [5 and 7 dpi]). The large circles are magnified views of areas in the small circles at 0 dpi for each respective graph.
(C) Immunofluorescence images of TA muscle cross sections from ablated and control mice corresponding to the time points in (B). Note the decrease in PDGFRα+ cells (punctate interstitial staining) in uninjured tissue from ablated mice and impaired myofiber necrosis and regeneration in the ablated tissue at 3 dpi and 7 dpi, respectively.
(D-G) Histological analyses of skeletal muscle regeneration from ablated and control mice.
(D) H&E staining of TA muscle cross sections from ablated and control mice at 7 and 14 dpi.
(E-G) Myofiber cross-sectional area (CSA) plotted as discrete sizes (E, 7 dpi; and F, 14 dpi) and overall means (G) (n = 3).
(H-K) Transplantation of FAPs into skeletal muscle of FAP-ablated mice to rescue regeneration impairment.
(H) Experimental design of the transplantation experiments.
(I) Immunofluorescent images showing GFP+ FAPs transplanted into the TA muscle.
(J) Quantification of myofiber CSAs from each experimental group, shown as a line histogram (n = 3).
(K) An empirical cumulative distribution function graph of all myofiber CSAs from the noted groups, with significance determined by a KS test (n = 681 [ablated] and 857 [ablated + FAPs]).
Scale bars, 100 μm. Error bars represent ± SEM. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
We next injured the lower hindlimb skeletal muscle of control and ablated mice and examined the regenerative response over the subsequent 14 days. FAP numbers in ablated mice remained depleted over the course of the examination, and, interestingly, MuSCs and HCs showed reduced expansion, never reaching the magnitude in cell number as those of their control counterparts (Figure 1B). EC expansion was unaffected over the course of the experiment when comparing cell numbers from ablated and control mice (Figure 1B). This in vivo evidence demonstrates that FAPs are required for normal MuSC and HC expansion in skeletal muscle following injury. Further, examination of cross-sections from tibialis anterior (TA) muscles at 3 dpi revealed impaired necrosis in ablated muscles compared with control muscles. At 5 dpi, the ablated muscle presented necrotic tissue similar to control muscle at 3 dpi, signifying a 2-day delay in this process (Figure 1C). Regeneration was evident at 7 dpi, with both cohorts having centrally nucleated fibers, but myogenic impairment in the form of smaller regenerating fibers was noted in ablated mice (Figures 1C and 1D). This impairment was quantitatively confirmed by comparing the cross-sectional areas (CSAs) of small myofibers from ablated and control mice at 7 dpi (Figure 1E). Furthermore, at 14 dpi, ablated mice demonstrated a significant regenerative deficit, observed as a decrease in both the mean and discrete myofiber CSAs compared with those of control mice (Figures 1F and 1G).
To determine whether the regenerative impairment is specific to the loss of PDGFRα+ cells in skeletal muscle or a consequence of removing PDGFRα+ cells throughout the entire mouse, we performed isotopic transplantations of ablated and control TA muscles into host mice where PDGFRα+ cells remain. TA muscles isolated from FAP-ablated or control mice were transplanted into surgically defined anterior compartments (TA and extensor digitalis longus muscles removed) of the lower limbs of immunocompromised mice (Figure S2B). This assay removes the possible systemic influence of ablating the PDGFRα lineage while supporting a single round of degeneration and regeneration of the transplanted muscle (Carlson and Faulkner, 1989; Grounds and Partridge, 1983; Lepper et al., 2011; Zhang et al., 2014). When comparing TAs from ablated and control mice 14 days following transplantation, a drastic decrease in TA CSAs was observed in those from ablated mice compared with those from controls (Figures S2C and S2D). These data demonstrate the necessity of skeletal muscle-resident PDGFR0α+ cells in the process of muscle regeneration.
To test whether this regenerative deficit is specific to the depletion of FAPs, we attempted to restore myofiber CSAs of ablated mice to those of controls by replacing the ablated intramuscular FAPs through cellular transplantation (Figure 1H). The TA muscles of ablated and control mice were injured, and GFP+ FAPs were transplanted 2 days later. Fourteen days following injury, these muscles were isolated, and the distributions of myofiber CSAs were obtained from TA cross-sections (Figure 1I). The myofiber CSA histogram from ablated mice receiving FAPs demonstrated a significant (Kolmogorov-Smirnov [KS] test, p = 5.6 × 10−16) shift away from that of ablated mice receiving only PBS and toward the histogram of control mice (Figures 1J and 1K). This increase in myofiber CSAs in ablated mice receiving donor FAPs demonstrates partial rescue and supports the hypothesis that FAPs are required for efficient skeletal muscle regeneration. Collectively, the regeneration assays, isotopic transplants, and FAP transplantation data are direct evidence of the necessity of FAPs for normal skeletal muscle regeneration.
A previous study that used a TCF4-CreER strain to deplete cells in the body reported impairment of muscle regeneration as a result (Murphy et al., 2011). Because TCF4 has been reported to be a marker for embryonic fibroblasts (Mathew et al., 2011), the authors proposed that the transgenic mouse also targeted adult fibroblasts and, potentially, FAPs. This has led to continued speculation that TCF4 is a marker for FAPs and, thus, that the TCF4-CreER targeted cell depletion data addressed the necessity of FAPs in muscle regeneration (McCullagh and Perlingeiro, 2015; Sambasivan and Tajbakhsh, 2015). To directly address this issue of TCF4 specificity, we analyzed the single-cell transcriptomes from the Tabula Muris Consortium database of all mononuclear cells in skeletal muscle (Figures S1G and S1H; Tabula Muris Consortium, 2018). These data demonstrate that TCF4 expression is detected in MuSCs, FAPs, smooth muscle cells and/or fibroblasts, immune cells, and tenocyte-like cells of skeletal muscle (Figure S1H). This also held true at the level of protein because we detected TCF4 in sorted single cells of all fractionated populations isolated from skeletal muscle using immunocytochemistry and single-cell western blots (Figures S1I–S1L). Collectively, these data demonstrate that TCF4 is not a specific marker for FAPs but, instead, is expressed across multiple cell populations in adult skeletal muscle, precluding its use to target any individual population in adult mice. Therefore, our current study using PDGFRα to target FAPs effectively establishes the necessity of FAPs for effective muscle regeneration.
FAPs Are Essential for Long-Term Homeostatic Maintenance and Normal Growth of Skeletal Muscle
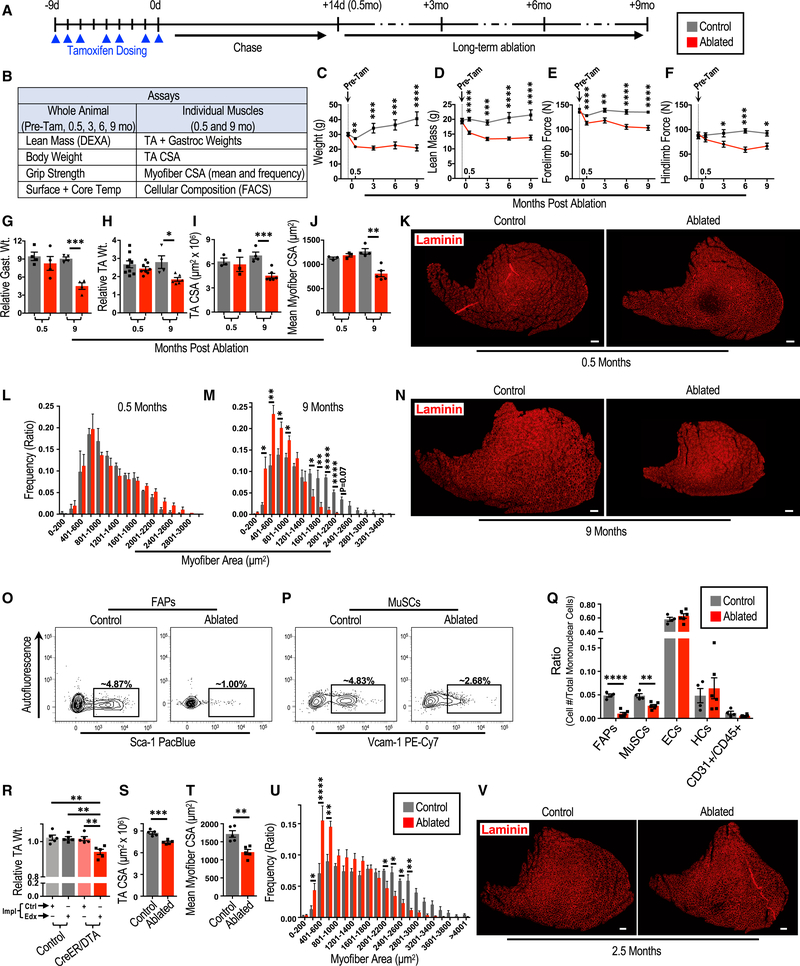
We next examined the consequence of depleting FAPs on the structure and function of skeletal muscle under homeostatic conditions to determine the necessity of FAPs in maintaining skeletal muscle (Figure 2A). We employed multiple assays to assess the integrity and function of skeletal muscle following FAP ablation (Figure 2B). Importantly, all control and experimental mice received tamoxifen to control for its non-specific side effects. Within 2 weeks of FAP depletion, we observed a decrease in weight, a decrease in lean mass, and a decrease in grip strength in ablated mice compared with their control counterparts (Figures 2C–2F). However, when examining individual muscles at the same time point, we detected no significant differences in individual muscle weights, whole muscle CSAs, or myofiber CSAs between ablated and control mice (Figures 2G–2L). Although total body weight and grip strength are multifactorial parameters and can be influenced by non-muscle factors, the decrease in lean body mass suggests that FAPs are required for muscle maintenance and function in the short term.
Figure 2. FAPs Are Necessary for Long-Term Maintenance and Normal Growth of Skeletal Muscle.
(A) Experimental design of the long-term FAP depletion experiments.
(B) Assays used to test the effect of FAP depletion on the whole animal and the noted skeletal muscles.
(C-F) Assessments of animal and muscle parameters (C, weight; D, lean mass; E, forelimb force; and F, hindlimb force) overtime before and after FAP depletion (n = 20–26 [pre-(Tam)], 17–24 [0.5 months], 6–9 [3 months], 5–9 [6 months], and 5–8 [9 months]).
(G-N) Assessment of muscle parameters 0.5 and 9 months after FAP depletion. Wt., weight; Gast., gastrocnemius. n = 4 (G), n = 4–9 (H), n = 3–6 (I and J), and n = 3–5 (L and M).
(K and N) Representative immunofluorescence images (K, 0.5 months; and N, 9 months) of TA cross-sections analyzed in bar graphs of (I)-(N).
(O-Q) The consequence of FAP ablation on cellular fractions of skeletal muscle.
(O and P) Representative FACS plots of (O) MuSCs and (P) FAPs 9 months after FAP ablation (quantified in Q).
(Q) Quantification of mononuclear cell composition of skeletal muscle 9 months after FAP ablation (n = 4–6).
(R-U) Assessment of muscle parameters (R, TA weight; S, TACSA; T, mean my ofiber CSA; and U, my ofiber CSA distribution)following localized FAP depletion in TA muscles. Impl, implant; Ctrl, control-PCL implants; Edx, endoxifen-PCL implants; CreER;DTA, PDGFRαCreER;R26 NG/DTA or +/DTA mice. n = 5 (R-T) and n = 7 (U). (V) Representative immunofluorescent images of TA muscle cross-sections analyzed in (R)-(U).
Ablated and control notations in (S)-(V) refer to TA muscles targeted with Edx-PCL from CreER;DTA mice and control mice, respectively. Scale bars, 200 μm. Error bars represent ± SEM. *p ≤ 0.05, **p ≤ 0.01,***p ≤ 0.001, ****p ≤ 0.0001.
Because skeletal muscle under homeostatic conditions experiences minimal cellular turnover in the short term (Chakkalakal et al., 2012; Fry et al., 2015), we tested whether mice depleted for FAPs would begin to exhibit a phenotype in the long term at the level of individual muscles. We ablated FAPs and then aged these and control mice for 9 months. We examined total body weight, lean body mass, functional grip strength, and skin and core body temperatures as a surrogate for metabolic rate (Figures 2C–2F; Figure S2E). Ablated mice had significantly less lean mass and generated less force at 3, 6, and 9 months compared with control mice (Figures 2D–2F). Skin and core temperatures were similar between ablated and control cohorts throughout the time course, with only a minor difference in core temperature noted within 2 weeks following FAP depletion (Figure S2E). In contrast to assessments done 2 weeks following depletion, the weights of individual muscles, whole muscle CSAs, and myofiber CSAs were significantly decreased in ablated mice compared with control mice 9 months following tamoxifen treatment (Figures 2G–2J, 2M, and 2N), all signs of skeletal muscle atrophy. Further, muscles of ablated mice failed to grow like those of their control counterparts during these 9 months (Figures 2D and 2I–2N; compare 0.5 with 9 months). Together, these data demonstrate that FAPs are necessary for long-term maintenance and normal growth of skeletal muscle.
Because we demonstrated the need for FAPs in the cellular expansion of MuSCs and HCs in regeneration, we decided to investigate the consequence of long-term FAP depletion under homeostatic conditions on the cellular constituents of skeletal muscle (Figures 2O–2Q). Importantly, the depletion of FAPs was maintained over the 9-month experimental time course (Figures 2O and 2Q). EC and HC numbers were not significantly affected compared with control counterparts (Figure 2Q). However, examination of the MuSC compartment revealed an approximately 45% reduction in MuSCs in ablated mice over these 9 months compared with control mice (Figures 2P and 2Q). These data demonstrate that FAPs are required for long-term maintenance of the MuSC pool.
Depleting FAPs Locally Results in Atrophy of the Targeted Muscle under Homeostatic Conditions
Because PDGFRα is not only a marker for FAPs in skeletal muscle but also a marker for similar stromal cells in other tissues (Tabula Muris Consortium, 2018), we depleted these cells systemically in our abovementioned ablation experiments (Figure 2A). Thus, systemic consequences of whole-body mesenchymal stromal cell depletion could possibly influence skeletal muscle over the course of our 9-month experiments intended to investigate the necessity of FAPs in skeletal muscle maintenance. To address this issue and to remove the potential for a systemic influence originating from depletion of these cells in other organs, we developed a focal tamoxifen metabolite delivery system to specifically target a single muscle. We implanted a biocompatible polymer (polycaprolactone [PCL]) impregnated with endoxifen (Edx), a tamoxifen metabolite, in the subcutaneous space adjacent to the TA muscle of PDGFRαCreER;R26NG mice to limit effective concentrations of Edx to the region adjacent to the polymer. We then assessed recombination efficiency among mononuclear cells isolated from the targeted TA muscle and other tissues throughout the body (Figure S2F). We demonstrate a recombination efficiency of ~50% in the targeted TA muscle but no appreciable recombination in distant tissues, including the bone marrow (Figures S2G and S2H), indicating that Edx does not reach sufficient levels systemically to induce recombination. These data establish this procedure as an effective means to limit CreER-dependent recombination to a local area.
We next implemented this technique in PDGFRαCreER; R26+/dta (CreER;DTA) and control mice to test whether depletion of FAPs specifically in a TA muscle would result in atrophy of that muscle, similar to our whole-body depletion experiments. We assessed TA muscles targeted with our control or Edx-PCL biomaterial up to 3 months following implantation (Figure S2F). We detected a significant decrease in the weights of TA muscles from CreER;DTA mice targeted with Edx-PCL implants compared with contralateral TA muscles targeted with control implants and compared with TA muscles from control mice targeted with Edx-PCL or control implants (Figure 2R). Importantly, there was no difference in weight between TAs from CreER;DTA mice that received control implants and TA muscles from control mice that received Edx-PCL or control implants (Figure 2R), suggesting the absence of systemic effects from local depletion of FAPs in the targeted TA muscle. We next examined cross-sections of Edx-PCL-targeted TA muscles from CreER;DTA and control mice and found that there was an approximately 50% decrease in PDGFRα+ cells, confirming depletion of FAPs (Figure S2I). We further revealed that ablated TA muscles had smaller CSAs and smaller myofiber CSAs than control TA muscles (Figure 2R–V). These data indicate that local depletion of FAPs results in atrophy of the targeted skeletal muscle, further supporting the conclusion that FAPs are necessary for the maintenance of skeletal muscle.
DISCUSSION
Using our newly generated PDGFRαCreER mouse, we depleted FAPs in vivo and determined the necessity of these cells in skeletal muscle regeneration, homeostatic maintenance, and normal growth. We demonstrate that these cells are required in the early stages of skeletal muscle regeneration, supporting MuSC and HC expansion in the area of injury. We also demonstrate that FAPs are required for long-term homeostatic maintenance of adult skeletal muscle as well as homeostatic maintenance of the MuSC pool. Our findings establish FAPs as beneficial components of these processes, and they are of particular interest, given that FAPs are also thought to be a key source of detrimental fatty and fibrotic infiltration in skeletal muscle observed in muscular dystrophies and the normal aging process (Tedesco et al., 2017). It will be especially interesting to determine the beneficial influence of FAPs on MuSCs and HCs in the regenerative milieu and on MuSCs and myofibers in long-term maintenance and growth of skeletal muscle; therapeutically targeting these mechanisms may provide a means to aid skeletal muscle regeneration or help maintain skeletal muscle over the normal course of aging.
STAR⋆METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Thomas Rando (rando@stanford.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Animals were maintained on a mixed C57BL6/FVB background and received tamoxifen treatment between 3–6 months of age. For the long-term homeostasis studies, all mice received tamoxifen between 3–4 months of age so that at 9 months following ablation, mice were 12–13 months of age. Both males and females were used. The PDGFRαCreER/+ knock-in mice were made in collaboration with GenOway Transgenic Services (Lyons, France) under contract (see Figure S1A). R26NG mice were created by mating r26nzg/nzg mice (Jackson Labs, 012429) with R26Flp/Flp mice (Jackson Labs, 009086) and resulting offspring were bread to establish the R26NG line (Yamamoto et al., 2009). R26dta/dta (009669), NSG (005557), and UBC-GFP (004353) mice were purchased from Jackson Labs. Mice referred to as ablated have the following genotype: PDGFRαCreER/+;R26NG/DTA or +/DTA. Control mice in ablated experiments were littermates that did not carry both CreER and DTA transgenes as to allow for tamoxifen dosing of all mice with the exception of “-TAM” mice in Figure S2A. Animals were housed in the Veterinary Medical Unit at Veterans Affairs Palo Alto Health Care System (VAPAHCS), a specific pathogen free facility with a 12 hour on/off light cycle. Mouse procedures and care were approved by the Institutional Animal Care and Use Committee of the VAPAHCS and were in accordance with the Administrative Panel on Laboratory Animal Care of Stanford University.
Cell Lines
Mouse 3T3 fibroblasts (ATCC, CRL-1658) were grown in DMEM containing 10% FBS and 1X penicillin/streptomycin. Growing cells were allowed to proliferate until ~70% confluency and then split for propagation.
METHOD DETAILS
Tamoxifen administration
Mice received ~0.1 mg/gm of tamoxifen solution (50 mg/ml in 7.5% ethanol/corn oil) by intraperitoneal (IP) injection. Seven injections were administered over nine days, with a day of rest on the fourth and seventh day. A chase period was given for ≥ 9 days before any experimental protocols were commenced to allow for tamoxifen to be fully metabolized and minimize any chance of further Cre dependent recombination. Both control and experimental mice were treated with tamoxifen with the exception of the “-TAM” in Figure S2A that received only vehicle (corn oil). Pre-tamoxifen (“Pre-Tam”) assays were completed in the week prior to the first IP dose of tamoxifen.
Local delivery of tamoxifen metabolite
Endoxifen hydrochloride hydrate (“Edx,” Sigma) was resuspended in methoxypolyethylene glycol 350 (PEG, Sigma) and then blended with molten polycaprolactone (PCL, Sigma) at a 1:3 g/g ratio (polymers described in Winternitz et al., 1996). After insuring that these materials were completely blended, the molten mixture was quickly poured into a plastic mold to achieve a thin (1–2 mm) layer, which was allowed to solidify at room temperature. Implantable bars were then carefully cut and weighed to achieve bars that would contain ~125 μg of Edx. Bars without Edx were prepared using a similar method and were used for control implants. For the implantation of bars, mice were anesthetized and a small incision was made over the TA muscle. Edx/PCL or control bars were inserted into the subcutaneous space with great care as to not compromise the integrity of the muscle. Left limbs received Edx/PCL bars while right contralateral limbs received control bars, and this was done for both experimental and control mice. Incisions were closed by suture and skin glue (3M).
Flow cytometry
Hindlimb muscle was isolated from mice at the noted times in the figures. For samples that were used to determine cells per milligram tissue, only the lower hindlimb muscles were dissected. Isolated muscles were minced with scissors and then enzymatically digested with 760 U/ml Collagenase Type II (Worthington) for 45 minutes in a 37°C shaking water bath. Samples were then washed with 10% horse serum in Ham’s F-10 (wash medium), and further digested in 1000 U/ml Collagenase Type II and 11 U/ml Dispase (GIBCO) in wash medium for 30 minutes in a 37°C shaking water bath. Wash medium was added and then the sample was triturated with a 20-gauge needle, washed, and passed through a 40 μm cell strainer to obtain a single cell suspension.
Adipose tissue depots (interscapular brown, perigonadal visceral, posterior subcutaneous) were carefully dissected, minced with scissors, and then enzymatically digested with 760 U/ml Collagenase Type II and 1U/ml Dispase (GIBCO) in DMEM for 30–45 minutes in a37°C shaking water bath. Samples were then washed with wash medium and passed sequentially through 100 μm and 40 μm cell strainers to obtain single cell suspensions. Bone marrow samples were isolated from femurs and tibias by exposing the intramedullary cavities with scissors, and these cavities were then flushed directly into 35 U/ml Collagenase Type II and 1 U/ml Dispase in DMEM and digested for 30 minutes in a 37°C shaking water bath.
Cells were stained with fluorescent conjugated antibodies (dilutions: 1:100 (Biolegend) or 1:500 (eBiosicence); see Antibodies in Key Resources Table) on a nutating mixer for 30 minutes at 4°C, washed in wash medium and filtered through a 30 mm cell strainer tube (Falcon). Cells were analyzed or sorted on a BD Biosciences LSR II flow cytometer equipped with a High Throughput Sampler (HTS) system, and with 405 nm, 488 nm, 532 nm, and 633 nm lasers, a BD Biosciences FACSAria II with 405 nm, 488 nm, 633 nm lasers or a FACS Aria III with 355 nm, 405 nm, 488 nm, 532 nm, and 633 nm lasers. Strict gating schemes were determined and applied based on no antibody or fluorescence minus one controls. Sorted cells were collected in wash medium and reanalyzed for purity. Representative FACS plots can be found in Figures S2J and 2K.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Donkey anti-goat secondary antibody, alexa fluor 1488 | Invitrogen | Cat#A-11055; RRID:AB_142672 |
| Donkey anti-goat secondary antibody, alexa fluor 555 | Invitrogen | Cat#A-21432; RRID:AB_141788 |
| Donkey anti-goat secondary antibody, alexa fluor 647 | Invitrogen | Cat#A-21447; RRID:AB_141844 |
| Donkey anti-rat secondary antibody, alexa fluor 488 | Invitrogen | Cat#A-21208; RRID:AB_141709 |
| Donkey anti-rat secondary antibody, DyLight 550 | Invitrogen | Cat#SA5-10027 RRID:AB_2556607 |
| Donkey anti-rabbit secondary antibody, alexa fluor 555 | Invitrogen | Cat#A-31572; RRID:AB_162543 |
| Donkey anti-mouse secondary antibody, alexa fluor 647 | Invitrogen | Cat#A-31571; RRID:AB_162542 |
| APC anti-mouse CD31 monoclonal antibody | BioLegend | Clone#: MEC13.3; Cat#102510; RRID:AB_312917 |
| FITC anti-mouse CD31 monoclonal antibody | BioLegend | Clone#: MEC 13.3; Cat#102506, RRID:AB_312913 |
| CD31 monoclonal antibody, PE-Cyanine7 | eBioscience, Thermo Fisher Scientific | Clone#: 390; Cat#25-0311-81; RRID:AB_2734962 |
| APC anti-mouse CD45 monoclonal antibody | BioLegend | Clone#: 30-F11; Cat#103112; RRID:AB_312977 |
| FITC anti-mouse CD45 monoclonal antibody | BioLegend | Clone#: 30-F11; Cat#103108; RRID:AB_312973 |
| Pacific Blue anti-mouse Sca-1 monoclonal antibody | BioLegend | Clone#: D7 Cat#108120; RRID:AB_493273 |
| PE/Cy7 anti-mouse VCAM-1 monoclonal antibody | BioLegend | Clone#: 429 (MVCAM.A); Cat#105719; RRID:AB_2214047 |
| APC rat IgG2a, Isotype control monoclonal antibody | BioLegend | Clone#: RTK2758; Cat#400511 |
| FITC rat IgG2a, Isotype control monoclonal antibody | BioLegend | Clone#: RTK2758; Cat#400505; RRID:AB_2736919 |
| Pacific Blue rat IgG2a, Isotype control monoclonal antibody | BioLegend | Clone#: RTK2758; Cat#400527 |
| Mouse PDGF R alpha polyclonal antibody | R&D Systems | Cat#AF1062; RRID:AB_2236897 |
| Anti-Laminin 2 alpha monoclonal antibody | Abcam | Clone#: 4H8-2 Cat#ab11576, RRID:AB_298180 |
| Anti-Laminin polyclonal antibody | Sigma-Aldrich | Cat#L9393, RRID:AB_477163 |
| TCF4 polyclonal antibody | Novus | Cat#NBP1-88633 RRID:AB_11021504 |
| GAPDH monoclonal antibody | Novus | Cione:1D4 Cat#NB300-221; RRID:AB_10077627 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma | Cat#T5648-5G |
| Endoxifen Hydrochloride Hydrate | Sigma | Cat#E8284-25MG |
| Polycaprolactone | Sigma-Aldrich | Cat#440752-250G |
| Methoxypolyethylene glycol 350 | Sigma-Aldrich | Cat#M6768-250G |
| BaCl2 | Sigma-Aldrich | Cat#B-0750 |
| Critical Commercial Assays | ||
| Standard Single-Cell West Kit | Protein Simple | K600 |
| Small Single-Cell West Kit | Protein Simple | K500 |
| Deposited Data | ||
| Tabula Muris Consortium Single Celi Droplet RNA Sequencing | Tabula Muris Consortium, 2018 | https://figshare.com/articies/Singie-ceii_RNA-seq_data_from_microfluidic_emuision_v2_/5968960 |
| Experimental Models: Cell Lines | ||
| Mouse 3T3 Fibroblasts | ATCC | CRL-1658 |
| Experimental Models: Organisms/Strains | ||
| PDGFRαCreER/+ | This paper | NA |
| R26NG | See derivation in methods; Yamamoto et al., 2009 | NA |
| R26NZG | The Jackson Laboratory; Yamamoto et al., 2009 | Jax stock # 012429 |
| R26Flp | The Jackson Laboratory; Farley et al., 2000 | Jax stock # 009086 |
| R26DTA | The Jackson Laboratory; Voehringer et al., 2008 | Jax stock # 009669 |
| NSG | The Jackson Laboratory; Shultz et al., 2005 | Jax stock # 005557 |
| UBC-GFP | The Jackson Laboratory; Schaefer et al., 2001 | Jax stock # 004353 |
| Oligonucleotides | ||
| Primer sequences | See Table S1 | N/A |
| SMARTpool: ON-TARGETplus Mouse Tcf4 siRNA | Dharmacon | #L-043160-02 |
| SMARTpool: ON-TARGETplus Non-targeting siRNA | Dharmacon | #D-001810-10 |
| Software and Algorithms | ||
| R software package Seurat | Butler et al., 2018 | https://satijalab.org/seurat/ |
| BD FACSDiva 8.0 | BD Biosciences | N/A |
| BZ-X Viewer version 01.03.00.05 | Keyence Corporation | N/A |
| BZ-X Analyzer version 1.3.0.3 | Keyence Corporation | N/A |
| Improvision Volocity 6.3.0 | Perkin Elmer | N/A |
| QDR 12 | Hologic, Inc | N/A |
| Scout 2.0.5 | ProteinSimple | https://www.proteinsimple.com/scout/downloads/ |
| GIMP 2.8 | N/A | https://www.gimp.org |
| ImageJ | N/A | https://imagej.nih.gov/ij/ |
| Prism 7 | GraphPad Software, Inc | N/A |
Muscle injury and tissue processing
Mice were anesthetized with isoflurane and 90 μL of 1.2% barium chloride (BaCl2) solution was distributed over ~5 injections with an insulin syringe (BD) into the lower hindlimb. The injection was followed by multiple piercings with the same needle to aid in distribution of the BaCl2. Injured and uninjured muscles were isolated at the times noted in the figures. All muscle weights were normalized to tibia lengths. Tibialis anterior (TA) muscles were isolated and fixed in 0.5% paraformaldehyde in PBS for 5–7 days at 4°C, washed with PBS, and incubated in 30% sucrose solution prior to freezing in Tissue Tek OCT compound (Electron Microscopy Sciences) for sectioning. Cross sections were obtained on a Leica cryostat. Samples were initially trimmed ~1500–2000 μm from the posterior end and then 12 μm sections were collected every 350 μm.
Immunofluorescence
Cryosections were washed and rehydrated in PBS, permeabilized with 0.1% Triton X-100, blocked with blocking buffer (10% donkey serum and 1% BSA in PBS) containing 0.1% Tween-20, and incubated overnight with primary antibodies (dilutions: PDGFRa −1:100; Laminin - 1:250 (AbCam), 1:500 (Sigma), 1:100 -TCF4; see Antibodies in Key Resources Table) in blocking buffer at 4°C. Samples were then washed with PBS and stained with secondary Alexa Fluor conjugated antibodies (all at a dilution of 1:500, see Antibodies in Key Resources Table) in blocking buffer at room temperature, washed with PBS, stained with DAPI, and coverslipped with FluoroGel mounting media (Electron Microscopy Sciences). Sorted cells were plated in wells treated with extracellular matrix (ECM, Sigma) and cultured in growth medium (20% fetal bovine serum, 10% horse serum, 1X penicillin/streptomycin, 1 ng/ml fibroblast growth factor, DMEM) overnight to allow for adherence to the ECM coated dish surface. Cells were then fixed with 4% PFA in PBS and stained using the same methods as with cryosections.
TA and myofiber cross sectional areas
Total TA cross sectional areas were measured with the BZ-Analyzer software by carefully tracing the TA section as defined by laminin immunofluorescence staining and then using the area measurement tool. For myofiber CSAs, Volocity Quantification Software (PerkinElmer) was used to measure individual laminin-stained myofiber cross-sectional areas. Myofibers exhibiting strong laminin staining were measured with the magic wand ROI tool (5%−7% voxel tolerance) while myofibers with weaker laminin staining were traced with the freehand ROI tool. A range of 150–300 representative myofibers were measured per sample from three separate sections each spaced ~1 mm apart and centered at the mid-belly of the TA.
Isotopic TA transplants
TAs were isolated from anesthetized tamoxifen treated control or ablated mice and transplanted into the surgically defined anterior compartment of anesthetized NSG mice. The surgically defined anterior compartment was created by opening the overlaying skin from knee to ankle and then removing the TA and extensor digitalis longus muscles of the host just prior to receiving the transplant. The transplant was then fixed in place by suturing the anterior and posterior portions of the TA to the adjacent peroneus longus muscle of the recipient as described (Zhang et al., 2014). The overlaying skin was closed with sutures. Fourteen days following transplantation, the donor TAs were carefully isolated using a dissecting scope (Zeiss) or surgical binocular loupes (Zeiss) to aid in removal of host tissue from the transplanted TA. Due to the delicate nature of the transplanted TAs, host tissue that was strongly adherent was carried through sectioning and edges were defined following imaging.
Transplantation of FAPs
TAs from control or ablated mice were inured with BaCl2 as described above. Two days following injury, a 5 mm incision was made in the skin covering the TA, and 7.5 × 104 GFP+ FAPs isolated by FACS from UBC-GFP mice were suspended in PBS and transplanted into the injured TA using an insulin syringe. The skin was then closed with sutures and skin adhesive (Histoacryl). The TAs of control mice (ablated with no cells and control) received the same volume of PBS but without cells. TAs were isolated 14 dpi (12 days following transplantation) and processed for cryosectioning as described above.
Dual Energy X-Ray Absorptiometry (DEXA)
Lean mass was determined by the Hologic Discovery A DEXA device on the rodent/ rat whole body setting. Mice were anesthetized prior to scanning to ensure accurate body composition readings and a ≤ 2-minute scout-scan (exposure dose is 300 μSv/mouse) was performed. The cephalic region was cropped out of the analyzed region of interest. Bone mineral content (BMC) was removed from total lean mass measurements to yield the replicate values noted in the figures.
Body and individual muscle weights
Mouse body weights were measured with the Ohaus CS Series CS200–001. All weights were determined at approximately the same time of day to reduce feeding and fluid related fluctuations that could alter mass. For individual isolated muscles, relative weights were calculated by normalizing each muscle weight to the tibia length of its respective leg to account for animal size variance.
Body temperatures
Surface temperatures at the central abdomen were measured with an Extech Instruments IR Non-Contact Thermometer (42529). Internal (Core) temperatures were measured by the Extech Instruments Type K Thermometer (421501), with the temperature probe inserted 3 mm into the rectum. All temperatures were determined at approximately the same time of day to reduce circadian cycle related temporal variability.
Grip strength
The BIOSEB grip strength test (BIO-GS3) was used according to BIOSEB instructions for mouse strength assessment. For forelimb measurements, mice were held by the tail and allowed to grab the grid with only their forepaws before being pulled in a consistent parallel motion. For hindlimb measurements, mice were held loosely by the back of the neck and allowed to grab the grid with only their hind paws, before being pulled in a similar motion as used for forelimbs. All strength tests were conducted at a similar time of day in identical environmental conditions. Five strength measurements were taken per mouse with the highest three measurements being averaged for the reported value.
Analysis of single cell RNA-sequencing data from the Tabula Muris Consortium
Microfluidic droplet single cell sequencing (10X Genomics) data for all mononuclear cells of skeletal muscle from the Tabula Muris Consortium (Tabula Muris Consortium, 2018) is available from figshare (https://figshare.com/articles/Single-cell_RNA-seq_data_from_microfluidic_emulsion_v2_/5968960). These data were clustered to define cell populations and analyzed for stromal makers using the R software package Seurat (https://satijalab.org/seurat/).
Single Cell Western Blots
Muscle tissue isolation, processing and cell sorting were completed as describe above. Freshly sorted cells representing all mononuclear cell fractions of skeletal muscle were processed for single cell (SC) western blots in accordance with the manufacturer’s protocol (Milo SC-Western, Protein Simple). Milo scWest chips were rehydrated in 1X Suspension Buffer for ≥ 30 minutes and then single cell suspensions of FACS-isolated cells at a concentration of 1 × 105 cells/ml were loaded on small scWest chips. Cell settling was monitored by brightfield microscopy after ~20 minutes and then gently washed with 1X Suspension Buffer to achieve single-cell occupancies of wells. Cell-loaded chips were then inserted into the Milo System and run with the following parameters: lysis for 10 s, electrophoresis for 75 s and UV capture for 240 s. Antibody probing and washing were performed following the manufacturer’s protocol with antibodies to TCF4 and GAPDH (1:40 dilution, see Antibodies in Key Resources Table). Stained and dried scWest chips were scanned on a GenePix Microarray Scanner and images for each spectral channel were analyzed with Scout Software (Protein Simple). Only lanes positive for denatured GAPDH were used for quantification of 25–60 cells per cell population. To confirm the specificity of the TCF4 antibody in the SC-Western method, cultured 3T3 fibroblasts treated with TCF4 siRNA (see method below) were loaded on standard scWest chips (three replicates per siRNA or control were combined and each used to seed an scWest chip) and processed as stated above. Scout software was used to plot the fluorescence counts around the peak region (Figure S1F) demonstrating a specific knockdown of TCF4 and confirming the specificity of this antibody for TCF4.
Quantitative PCR
Total RNA from transfected 3T3 fibroblasts (ATCC) was isolated with the mirVana Isolation Kit (Invitrogen) in accordance with the manufacturer’s protocol. Isolated RNA was then reverse transcribed with a High-Capacity RNA-to-cDNA Kit (Applied Biosystems) and quantitative PCR was performed using SYBR Green master mix (Roche) and a LightCycler480 II system (Roche). Relative quantification of transcript expression was calculated by the 2-ΔCt method of gene quantification using GAPDH as the reference gene. The following primers were used: TCF4 forward primer - CGAAAAGTTCCTCCGGGTTTG; TCF4 reverse primer - CGTAGCCGGGCT GATTCAT; GAPDH forward primer - TCAAGAAGGTGGTGAAGCAG; GAPDH reverse primer - GTTGAAGTCGCAGGAGACAA.
siRNA knockdown
Mouse 3T3 fibroblasts (ATCC) were transfected with 150 nM of either Smart Pool (Dharmacon) scrambled siRNA or TCF4 siRNA using XtremeGENE transfection reagent (Roche). Cells were harvested from three experimental replicates 72 hours following transfection and analyzed by single cell western analysis and qRT-PCR, both described above.
Microscopy and image processing
Imaging of immunofluorescent samples were conducted on a Zeiss Axio Observer.Z1 inverted microscope equipped with a Hamamatsu ORCA-R2 digital camera and Velocity 6.3.0 imaging software (PerkinElmer), a Zeiss LSM 710 confocal system and Zen acquisition software (Zeiss), or a Keyence BX-X710 all-in-one fluorescence microscope and BZ-X Viewer version 01.03.00.05 and BZ-S Analyzer version 1.3.0.3 software with the sectioning, advanced observation, advanced analysis, measurement, hybrid cell count and macro cell count modules. Images were taken at the lowest allowable exposure to capture fluorescent signal and then processed in GIMP (https://www.gimp.org) or ImageJ (https://imagej.nih.gov/ij/), both open source editing software, for overlays and linear (only) contrast adjustments.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics and graphs
Statistics and graphs were prepared using GraphPad Prism version 7 software. Arithmetic means for each experimental cohort are plotted and error bars represent the standard error of the mean (SEM). The number of replicates (n) for each experiment are reported in the figure legend and refer to biological replicates (exceptions are noted in the figure legends). Statistical significance was assessed using unpaired t tests and significance was reached when the p value was ≤ 0.05. Histograms displaying fitted lines for myofiber spreads were prepared using Microsoft Excel and empirical cumulative distribution functions were prepared using R, with significance determined by a Komolgorov-Smirnov (KS) test.
Supplementary Material
Highlights.
A method to target or deplete FAPs locally in skeletal muscle is described
Depletion of FAPs results in a regenerative deficit in skeletal muscle
Depletion of FAPs results in skeletal muscle atrophy
MuSC number declines in muscles in which FAPs have been depleted
ACKNOWLEDGMENTS
We would like to thank current and former members of the Rando laboratory for thoughtful discussions. We also thank the members of the Veterans Affairs/Palo Alto Veterans Institute for Research FACS core for machine maintenance and care. This work was supported by the Glenn Foundation for Aging Research and by grants from the NIH (T32 AG000266 and K99 AG053438 to M.N.W. and P01 AG036695, TR01 AG047820, and R37 (MERIT Award) AG023806 to T.A.R.) and the Department of Veterans Affairs (BLR&D Merit Review to T.A.R.). The graphical abstract was illustrated by Eva Mae Natividad Baucom.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.04.074.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BM, and Faulkner JA (1989). Muscle transplantation between young and old rats: age of host determines recovery. Am. J. Physiol 256, C1262–C1266. [DOI] [PubMed] [Google Scholar]
- Chakkalakal JV, Jones KM, Basson MA, and Brack AS (2012). The aged niche disrupts muscle stem cell quiescence. Nature 490, 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, and Dymecki SM (2000). Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28, 106–110. [PubMed] [Google Scholar]
- Frontera WR, and Ochala J. (2015). Skeletal muscle: a brief review of structure and function. Calcif. Tissue Int 96, 183–195. [DOI] [PubMed] [Google Scholar]
- Fry CS, Lee JD, Mula J, Kirby TJ, Jackson JR, Liu F, Yang L, Mendias CL, Dupont-Versteegden EE, McCarthy JJ, and Peterson CA (2015). Inducible depletion of satellite cells in adult, sedentary mice impairs muscle regenerative capacity without affecting sarcopenia. Nat. Med 21, 76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grounds MD, and Partridge TA (1983). Isoenzyme studies of whole muscle grafts and movement of muscle precursor cells. Cell Tissue Res. 230, 677–688. [DOI] [PubMed] [Google Scholar]
- Heredia JE, Mukundan L, Chen FM, Mueller AA, Deo RC, Locksley RM, Rando TA, and Chawla A. (2013). Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell 153, 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe AWB., Yi L., Natarajan A., Le Grand F., So L., Wang J., Rudnicki MA., and Rossi FMV. (2010). Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol 12, 153–-163.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuswanto W, Burzyn D, Panduro M, Wang KK, Jang YC, Wagers AJ, Benoist C, and Mathis D. (2016). Poor Repair of Skeletal Muscle in Aging Mice Reflects a Defect in Local, Interleukin-33-Dependent Accumulation of Regulatory T Cells. Immunity 44, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos DR, Babaeijandaghi F, Low M, Chang C-K, Lee ST, Fiore D, Zhang R-H, Natarajan A, Nedospasov SA, and Rossi FMV (2015). Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nat. Med 21, 786–794. [DOI] [PubMed] [Google Scholar]
- Lepper C, Partridge TA, and Fan C-M (2011). An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 138, 3639–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew SJ, Hansen JM, Merrell AJ, Murphy MM, Lawson JA, Hutcheson DA, Hansen MS, Angus-Hill M, and Kardon G. (2011). Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development 138, 371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh KJA, and Perlingeiro RCR (2015). Coaxing stem cells for skeletal muscle repair. Adv. Drug Deliv. Rev 84, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, and Kardon G. (2011). Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 138, 3625–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R, and Tajbakhsh S. (2015). Adult Skeletal Muscle Stem Cells. Results Probl. Cell Differ. 56, 191–213. [DOI] [PubMed] [Google Scholar]
- Sambasivan R, Yao R, Kissenpfennig A, Van Wittenberghe L, Paldi A, Gayraud-Morel B, Guenou H, Malissen B, Tajbakhsh S, and Galy A. (2011). Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 138, 3647–3656. [DOI] [PubMed] [Google Scholar]
- Schaefer BC, Schaefer ML, Kappler JW, Marrack P, and Kedl RM (2001). Observation of antigen-dependent CD8+ T-cell/ dendritic cell interactions in vivo. Cell. Immunol 214, 110–122. [DOI] [PubMed] [Google Scholar]
- Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, Kotb M, Gillies SD, King M, Mangada J, et al. (2005). Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J. Immunol 174, 6477–6489. [DOI] [PubMed] [Google Scholar]
- Tabula Muris Consortium. (2018). Single-cell transcriptomic characterization of 20 organs and tissues from individual mice creates a Tabula Muris. Nature 562, 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedesco FS, Moyle LA, and Perdiguero E. (2017). Muscle Interstitial Cells: A Brief Field Guide to Non-satellite Cell Populations in Skeletal Muscle. Methods Mol. Biol 1556, 129–147. [DOI] [PubMed] [Google Scholar]
- Tidball JG (2011). Mechanisms of muscle injury, repair, and regeneration. Compr. Physiol 1, 2029–2062. [DOI] [PubMed] [Google Scholar]
- Uezumi A, Fukada S, Yamamoto N, Takeda S, and Tsuchida K. (2010). Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol 12, 143–152. [DOI] [PubMed] [Google Scholar]
- Voehringer D, Liang H-E, and Locksley RM (2008). Homeostasis and effector function of lymphopenia-induced “memory-like” T cells in constitutively T cell-depleted mice. J. Immunol 180, 4742–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winternitz CI, Jackson JK, Oktaba AM, and Burt HM (1996). Development of a polymeric surgical paste formulation for taxol. Pharm. Res 13, 368–375. [DOI] [PubMed] [Google Scholar]
- Wosczyna MN, Biswas AA, Cogswell CA, and Goldhamer DJ (2012). Multipotent progenitors resident in the skeletal muscle interstitium exhibit robust BMP-dependent osteogenic activity and mediate heterotopic ossification. J. Bone Miner. Res 27, 1004–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Shook NA, Kanisicak O, Yamamoto S, Wosczyna MN, Camp JR, and Goldhamer DJ (2009). A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis 47, 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, King OD, Rahimov F, Jones TI, Ward CW, Kerr JP, Liu N, Emerson CP Jr., Kunkel LM, Partridge TA, and Wagner KR (2014). Human skeletal muscle xenograft as a new preclinical model for muscle disorders. Hum. Mol. Genet 23, 3180–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.