Abstract
Acinetobacter baumannii is a highly potent nosocomial pathogen that is associated with increased in-hospital mortality. Here, we investigated the changes in molecular characteristics of carbapenem-resistant A. baumannii (CRAB) isolated from the blood samples of patients admitted to a tertiary hospital in South Korea from January 2009 to July 2015. Whole genome sequencing using the Illumina MiSeq platform and multi-locus sequence typing (MLST) were performed for 98 CRAB clinical isolates. In silico analyses for the prediction of antimicrobial resistance and virulence factor genes were performed. Plasmid sequences, including complete forms, were reconstructed from the sequence reads. Epidemiologic data were collected from the hospital database. MLST using the Oxford scheme revealed 10 sequence types of CRAB, of which ST191 was the dominant type (n = 59). Although blaOXA-23 was shared by most analysed strains, the compositions of antimicrobial resistance determinants differed among sequence types. ST447 and ST451/ST1809 with a few resistance genes were isolated during the later years of the study period. The number of virulence genes increased, while that of ST191 did not change significantly over the investigation period. Intriguingly MLST types, compositions of antimicrobial resistance genes, and virulence genes had no association with clinical outcomes of CRAB bacteraemia. In conclusion, active changes in or accumulations of antimicrobial resistance determinants and virulence genes in CRAB were not observed during the research period. Molecular characteristics of CRAB had no association with clinical outcomes of CRAB bacteraemia.
Introduction
Acinetobacter baumannii is an aerobic gram-negative coccobacillus known for relatively few virulence factors as compared to other gram-negative pathogens [1]. Its ability to acquire various antimicrobial resistance genes makes it a highly successful nosocomial pathogen, which is associated with increased in-hospital mortality [2]. In particular, the prevalence of carbapenem-resistant A. baumannii (CRAB) isolates is on the rise in Asian countries, including South Korea [3,4]. The specific clone, international clone 2 (IC II), is a major clonal group among Korean CRAB isolates [5]. This organism is associated with nosocomial outbreaks and multidrug resistance in association with blaOXA-23-like genes [6]. In a recent study, 75% of A. baumannii clinical isolates were shown to belong to IC II in Korea [7], and IC II was recognised as the second most common cause of bacteraemia from 2012 to 2013 [8].
Considering the threat of the increasing incidence of CRAB infection, prediction of future effects of these isolates is a priority. Whole genome sequencing (WGS) analysis provides unprecedented information about this illusive organism, especially for the characterisation of nosocomial outbreaks [9,10]. WGS provides in-depth epidemiologic knowledge by providing higher resolution in genotyping and suggesting the emergence of new epidemic clones. WGS also helps researchers to understand the various mechanisms underlying the acquisition of antimicrobial resistance genes. Antimicrobial resistance genes were found in alien islands of accessory genomes and were often accompanied by integrases, transposases, or insertion sequences suggestive of their possible acquisition by horizontal gene transfer from other Acinetobacter spp. or bacteria that colonise the same environment [11,12]. Furthermore, although little is known about the virulence factors of this bacterium as compared to the current knowledge about antimicrobial resistance mechanisms [13], the rapid increase in the number of sequenced and annotated genomes has enabled comparative genomic analyses to elucidate clues concerning potential virulence factors.
Previous studies compared the distribution of genes related to virulence and antimicrobial resistance to understand phenotypic differences between isolates [14–16]. However, a comprehensive analysis of the number of virulence or AMR genes in association clinical outcomes has not been performed. This analysis may provide some insight into how this organism is evolving in epidemiologic perspective. Therefore, we attempted a whole genome-association study by sequencing all available isolates from patients with CRAB bacteraemia to characterise the genomic changes over the study period from the epidemiological perspective and investigated the clinical significance of genomic characteristics of CRAB. These investigations of the genetic characteristics, including antimicrobial resistance determinants and virulence factors of CRAB blood isolates, can over time provide insight into how the organism evolves and may suggest clinico-epidemiological implications of genomic characteristics of this organism.
Materials and methods
Ethics statement
This study was approved by the Institutional Review Board (IRB) of Yonsei University Health System Clinical Trial Center (4-2017-0730). Since the study had minimal health risk and the study subjects were anonymised, the Institutional Review Board waived the requirement for written informed consent from the patients.
Collection of bacterial isolates and patient information
Blood isolates from patients with CRAB bacteraemia were collected between January 2009 and July 2015 at intensive care units (ICUs) of Severance Hospital in Seoul, Republic of Korea. CRAB bacteraemia was defined as one or more positive blood cultures for carbapenem-resistant A. baumannii and the presence of clinical features compatible with systemic inflammatory response syndrome. Preliminary species identification and antimicrobial susceptibility tests were conducted with VITEK II system (bioMérieux, Marcy l'Etoile, France). Disc-diffusion testing using antimicrobial discs (Becton Dickinson, Sparks, MD, USA) on cationic-adjusted Mueller–Hinton (MH) agar (Difco Laboratories, Detroit, MI, USA) was subsequently performed as per CLSI guidelines [17]. Minimum inhibitory concentrations (MICs) to three drugs including tigecycline, minocycline and colistin were assessed using VITEK II system. The MICs of imipenem and meropenem were determined with the agar dilution method using MH agar following CLSI guidelines [17]. Additionally, modified Hodge tests and double-disk synergy tests were performed to screen carbapenemase and metallo-β-lactamase activity, respectively.
We collected clinical data, including patients’ age and sex, length of ICU stay, length of hospital stay, pre-existing chronic comorbidities (diabetes, chronic heart failure, chronic liver disease, chronic renal disease, and chronic pulmonary disease), sequential organ failure assessment score, episodes of hospital admission, previous invasive procedures (central line insertion, intubation, continuous renal replacement therapy, and surgery under general anaesthesia), as well as lengths and types of antibiotic treatments. The origin of bacteraemia was determined upon identification of precedent CRAB isolation or evidence of infection before the event of bacteraemia.
Whole genome sequencing (WGS), annotation, prediction of antimicrobial resistance determinants, and identification of virulence genes
The isolates were cultured in Luria Bertani (LB) broth, and genomic DNA was extracted with a PureHelix™ Genomic DNA Prep Kit (cat. no. GCTN100, Nanohelix, Daejeon, Republic of Korea). Paired end sequencing of 98 A. baumannii clinical isolates was carried out using the Illumina MiSeq platform at ChunLab (Seoul, Republic of Korea). Specifically, TruSeq Nano DNA Library Preparation Kit (550 bp library size) and MiSeq Reagent Kit v3 (600 cycles) were used. Using CLC Genomics Workbench v11.0.1 (https://www.qiagenbioinformatics.com/), reads were trimmed (quality limit 0.01, ambiguous limit 1, min length 120 bp, discarding orphan reads) and were de novo assembled. Word and bubble sizes were 64 and 100, respectively. The assembled genome sequences were annotated using NCBI’s Prokaryotic Genome Annotation Pipeline (PGAP) v4.5 [18]. GenBank files produced from PGAP were converted to GFF format, and then subjected to Roary [19] to obtain core genome sequences and their codon-aware alignments, followed by a heuristic method-based automated trimming using trimAl [20]. A phylogenetic tree was constructed using FastTree2 [21] and visualised using iTOL server [22].
To identify antimicrobial resistance genes, the contig sequences were queried against the ResFinder database (https://cge.cbs.dtu.dk/services/data.php; downloaded on June 4, 2018) using Find Resistance v1.01 from the Microbial Genomics Module of CLC Genomics Workbench.
A comprehensive list of virulence factors from A. baumannii was compiled as per the previous studies [23–26]. The list of 182 virulence genes is shown in S1 Table. Distribution of virulence genes across all genomes was obtained using the large-scale blast score ratio (LS-BSR) [27]. A cut-off value of blast score ratio 0.7 was arbitrarily selected to evaluate the existence of each virulence gene.
For comprehensive genomic epidemiological analysis, we used BacWGSTdb (http://bacdb.org/BacWGSTdb) [28] which allows us to find closest isolates that are currently deposited in NCBI GenBank database. Scheme core genome multi-locus sequence typing (cgMLST) was used with a 50 threshold for single nucleotide polymorphism (SNP) and a 50 threshold for MSLT.
Multi-locus sequence typing (MLST) and detection of single nucleotide polymorphism (SNP)
Allelic profiles or STs of each isolate were determined by submitting contig sequences to the PubMLST site (https://pubmlst.org/abaumannii/) according to the Institute Pasteur scheme (MLST-IP) and Oxford Database (MLST-OD).
SNPs were identified by mapping raw sequencing reads to the complete chromosome sequence of A. baumannii strain KBN10P02143 (GenBank CP013924.1), the multidrug strain that completely sequenced for the first time in South Korea, using Snippy (https://github.com/tseemann/snippy).
Analysis of plasmid sequences
Plasmid sequences were constructed either by plasmidSPAdes [29] assembler that uses the read coverage of contigs, or by a custom three-step method consisting of (i) running Plasmid Profiler [30] that first identifies putative plasmid hits from the plasmid database, (ii) mapping of the entire reads to chosen plasmid sequences using SMALT (https://www.sanger.ac.uk/science/tools/smalt-0) and extracting paired reads where at least one end mapped using SAMTOOLS (http://www.htslib.org/), and finally, (iii) assembly of the extracted reads using UniCycler [31]. Assembly statistics and whole-genome assembly results are shown in S2 Table. Complete (also circularised) plasmid sequences were identified from UniCycler assemblies by visualization of assembly graphs using Bandage [32]). Putative protein coding genes were predicted using GeneMarkS [33], and rep genes were identified by hmmsearch (http://hmmer.org/) against Pfam database v31.0 (https://pfam.xfam.org/).
Identification of insertion sequences and transposons
The blaOXA-23 is a major determinant of nosocomial outbreaks of IC II CRAB. The expression of this gene is presumably regulated by several insertion sequences, including ISAba1, which is thought to play the most crucial role in gene expression [34]. ISMapper [35] was used to identify ISAba1 insertion sites on the reference genome of the strain KBN10P02143. Insertion sites of ISAba1 relative to the chromosome of KBN10P02143 are shown in S3 Table. The presence of blaOXA-23-carrying transposons, including Tn2006, Tn2007, Tn2008, and Tn2009, was screened using Primersearch from the EMBOSS package (http://emboss.sourceforge.net/), with primer sets suggested by Chen et al. [36].
Statistical analysis
Baseline characteristics were compared using Mann–Whitney U test or independent samples t-test for continuous variables, and by χ2 test or Fisher’s exact test for categorical variables. Continuous variables were expressed as means or medians (interquartile ranges), while categorical variables were expressed as numbers with percentages for the description of baseline characteristics. Statistical analyses were performed using SPSS version 21.0 (IBM Corp., Armonk, NY, USA).
Data availability
Genome sequences were submitted to GenBank under BioProject no. PRJNA448358 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA448358). The complete list of strain names and accession numbers are shown in S2 Table. Complete sequences of five representative plasmids (pABAY04001_1A, pABAY09008_1B, pABAY10001_1C, pABAY14012_4D, and pABAY15001_6E) are available from GenBank under the accession numbers MK386680.1- MK386684.1. All 115 complete plasmid sequences and their rep genes are available at https://doi.org/10.6084/m9.figshare.11467023.v1 (figshare).
Results
Overview of A. baumannii isolates
A total of 109 CRAB blood isolates were used in this study. Of these, three samples with possible contamination and eight samples with insufficient clinical data were excluded. The remaining 98 samples were consequently included in the analysis. The isolates exhibited a high rate of resistance for almost all the antibiotics tested: 92.7% were resistant to gentamycin (89/96), 88.8% to amikacin (87/98), 100% to imipenem (98/98), 98.8% to ciprofloxacin (84/85), 97.8% to levofloxacin (96/98), 98.9% to piperacillin-tazobactam (92/93), 98.9% to ceftazidime (96/97), 96.9% to cefepime (95/98), 4.3% to colistin (4/92), and 11.1% to tetracycline (1/9) (S4 Table).
MLST analysis depending on isolation years
An MLST using Pasteur scheme identified 93 strains (94.9%) that were classified into ST2, indicating that most of the strains belonged to IC II. MLST using Oxford scheme was applied to assign 10 STs (ST858, ST369, ST208/ST1806, ST784, ST451/ST1809, ST357, ST368, ST447, ST552, and ST191). ST191 was the most abundant type (59 strains). Strains ABAY12003 and ABAY13017, single variants of ST191 at gyrB and rpoD, respectively, were not assigned any STs. As a result, we found that 91 strains (92.9%), all belonging to IC II, had two incidences of gdhB alleles, 3 and 189, which made distinctions between ST208, ST1806, ST451, and ST1809 ambiguous. Although all ST191 strains had two gdhB alleles, only the ABAY15007 strain harbouring gpi-94 gpi-107 was equivocally assigned to ST191 and ST784.
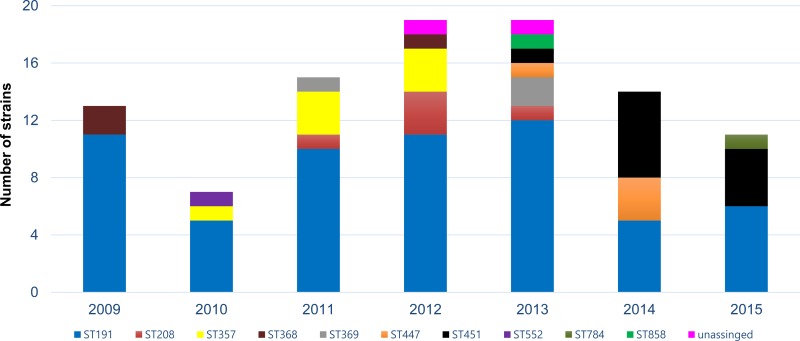
ST191 was evenly isolated throughout the research period. ST368 (n = 3), ST552 (n = 1), ST369 (n = 3), ST357 (n = 7), and ST208/ST1806 (n = 5) were mostly isolated from 2009 to 2012, while ST447 (n = 4), ST451/ST1809 (n = 11), ST858 (n = 1), ST784 (n = 1), and ST191/ST784 (n = 1) were mostly reported from 2013 to 2015 (Fig 1 and Table 1).
Fig 1. Distribution of carbapenem resistance A. baumannii isolates according to sequence types and year of isolation.
The isolates were typed by multi-locus sequence typing (MLST) using Oxford scheme and colored individually according to sequence type. Bar graph represents the individual and cumulative numbers of strains isolated each year.
Table 1. Sequence types of carbapenem resistant A. baumannii (CRAB) and epidemiologic information.
| Group (N) | Phylogenetic clade | Pasteur ST (N) | Oxford ST (N) | Isolated location (N) | Isolated year (N) | Origin of bacteremia |
|---|---|---|---|---|---|---|
| IC II | Clade 1 | ST2 (93) | ST191 (59) | MICU A (15) | 2009 (11) | Respiratory tract (46) |
| MICU B (24) | 2010 (5) | Gastrointestinal tract (3) | ||||
| MICU C (7) | 2011 (10) | Musculoskeletal (1) | ||||
| MICU D (5) | 2012 (11) | Catheter (6) | ||||
| NCU B (1) | 2013 (12) | Other (2) | ||||
| SICU (2) | 2014 (5) | |||||
| CCU A (1) | 2015 (5) | |||||
| CCU B (2) | ||||||
| Clade 2 | ST368 (3) | MICU B (2) | 2009 (2) | Respiratory tract (3) | ||
| CCU A (1) | 2012 (1) | |||||
| Clade 2 | ST357 (7) | MICUA(5) | 2010 (1) | Respiratory tract (7) | ||
| MICUB(1) | 2011 (3) | |||||
| MICU D (1) | 2012 (3) | |||||
| Clade 3 | ST208/ST1806 (5) | MICU B (2) | 2011 (1) | Other (1) | ||
| MICU C (1) | 2012 (3) | |||||
| MICU D (1) | 2013 (1) | |||||
| Clade 3 | ST369 (3) | MICU A (1) | 2011 (1) | Respiratory tract (2) | ||
| MICU D (2) | 2013 (2) | Other (1) | ||||
| Clade 2 | ST858 (1) | MICU C (1) | 2013 (1) | Respiratory tract (1) | ||
| Clade 4 | ST451/ST1809 (11) | MICU A (6) | 2013 (1) | Respiratory tract (9) | ||
| MICU B (4) | 2014 (6) | Urinary tract(1) | ||||
| CCU A (1) | 2015 (4) | Gastrointestinal tract (1) | ||||
| Clade 1 | ST784 (1) | MICU C (1) | 2015 (1) | Musculoskeletal (1) | ||
| Clade 1 | ST191/ST784 (1) | MICU B (1) | 2015 (1) | Respiratory tract (1) | ||
| Non-IC II | NA | ST193 (1) | ST552 (1) | CCU A (1) | 2010 (1) | Respiratory tract (1) |
| NA | ST10 (4) | ST447 (4) | MICU A (2) | 2013 (1) | Respiratory tract (4) | |
| MICU B (2) | 2014 (3) | |||||
| NA | NA | NA | ABAY12003 | MICU A (1) | 2012 (1) | Respiratory tract (1) |
| ABAY13017 | MICU A (1) | 2013 (1) | Catheter (1) |
Data are expressed as number (N). Abbreviation: ST, sequence type; IC II, international clone II; MLST, Multi-locus sequence typing; MICU, medical intensive care unit; NCU, neurologic intensive care unit; SICU, surgical intensive care unit; CCU, cardiac intensive care unit; NA, not applicable.
Core genome-based phylogeny and SNP-based inference of recombination
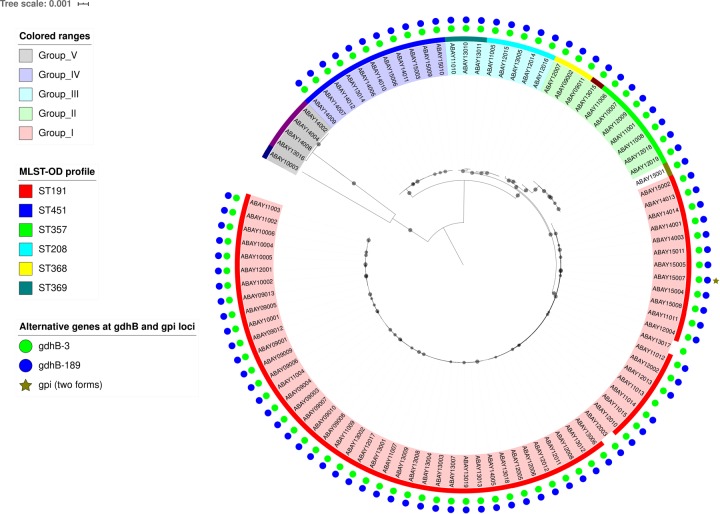
A core genome alignment-based phylogenetic tree was constructed to delineate the in-depth relationships between analysed strains (Fig 2). The analysis resulted in the classification of 98 analysed strains into four major clades. SNP identification relied on read mapping on the reference genome sequence of strain KBN10P02143. Pairwise SNP distances among all strains ranged from 39 to 83,304 bp (5,368 bp median) (S5 Table). Relationships between SNP-based phylogeny and STs were searched. Clade 1 was mostly composed of ST191. The majority of strains classified as clade 2 (ST368, ST357, and ST858) were isolated in the former part of our study period, while those of clade 3 (ST208/ST1806 and ST369) were isolated in the latter part. However, chronological correlation among clades was not identified overall.
Fig 2. Core-gene based phylogenetic tree of 98 A. baumannii strains.
Approximately maximum-likelihood tree was constructed from 2,520 core gene sequences (2,347,373 bp) using FastTree 2. Gray circles on branches represent local support values (0.9–1.0) based on Shimodaira-Hasegawa test.
Differences in patient characteristics among STs
Patients were hospitalised in different types of ICUs, and the locations of each isolated ST are shown in Table 1. Most strains were identified in MICU, and no notable relationship between isolated locations and STs was reported. The mean age of the enrolled patients was 60.62 ± 14.63 years, and male patients predominated (70.4%, n = 69). Chronic obstructive pulmonary disease was the most commonly found underlying morbidity (93.9%, n = 92), and chemotherapy was the most common cause of immunosuppression (39.8%, n = 39). Ventilator care was performed in 81.6% of patients (n = 80). The most common source of bacteraemia was the respiratory tract (79.6%, n = 78). The median number of hospitalisation days was 30 [19–64.2], and patients used antibiotics for a median treatment length of 30.5 [16–59.3] days. The all-cause 28-day mortality was 80.6% (n = 79). No significant differences were observed in clinical characteristics among STs except for the following. Patients with strains typed as ST447 Patients with strains typed as ST447 spent less time in the hospital (17 [17–41] days versus 30 [19–64.2] days, p < 0.05) and there were fewer males among patients with strains typed as ST191 (40 [63.5%] versus 29 [82.9%], p < 0.05) (Table 2).
Table 2. Baseline characteristics of patients with carbapenem resistant A.baumannii bacteremia, grouped by sequence types.
| Chracteristics | Grouped by sequence type | ||||||
|---|---|---|---|---|---|---|---|
| ST191 | ST447 | Total | |||||
| ST191(N = 59) | Non ST191(N = 39) | p-value | ST447(n = 4) | Non ST447(n = 94) | p-value | ||
| Age, years | 59.27±14.70 | 63.13±14.39 | 0.21 | 77.0±10.74 | 60.09±14.41 | 0.05 | 60.62±14.63 |
| Gender, male | 37(62.7) | 32(84.2) | 0.02 | 3(75.0) | 66(71.0) | 0.67 | 69(70.4) |
| Comorbidities | |||||||
| DM | 43(72.9) | 32(82.1) | 0.23 | 2(50.0) | 73(77.7) | 0.24 | 75(67.5) |
| COPD | 55(93.2) | 37(94.9) | 0.56 | 4(100.0) | 88(93.6) | 0.77 | 92(93.9) |
| CHF | 49(83.1) | 34(87.2) | 0.42 | 4(100.0) | 79(84.0) | 0.51 | 83(84.7) |
| HTN | 36(61.0) | 26(66.7) | 0.40 | 2(50.0) | 60(63.8) | 0.47 | 62(63.3) |
| CRF | 42(71.2) | 32(82.1) | 0.18 | 3(75.0) | 71(75.5) | 0.69 | 74(75.5) |
| Chronic liver disease | 52(88.1) | 37(94.8) | 0.24 | 4(100.0) | 85(90.4) | 0.60 | 89(90.8) |
| Malignancy | 35(59.3) | 22(56.4) | 0.56 | 2(50.0) | 55(58.5) | 0.67 | 57(58.2) |
| Immunosuppressed status | |||||||
| Transplantation | 7(11.9) | 8(21/1) | 0.18 | 0(0.0) | 15(16.1) | 0.50 | 15(15.3) |
| Steroid usea | 20(33.9) | 10(26.3) | 0.29 | 0(0.0) | 30(32.3) | 0.22 | 30(30.6) |
| Chemotherapy | 23(39.0) | 16(42.1) | 0.46 | 2(50.0) | 37(39.8) | 0.53 | 39(39.8) |
| APACHE II | 21.32±8.16 | 21.26±7.18 | 0.97 | 23.25±3.50 | 21.22±7.881 | 0.35 | 21.17±7.81 |
| Admission episode(≥ once) | 42(71.19) | 25(64.10) | 0.27 | 3(75.0) | 64(68.1) | 0.62 | 67(68.4) |
| Source of infection | |||||||
| Respiratory tract | 46(82.1) | 32(88.9) | 0.29 | 4(100.0) | 74(78.7) | 0.35 | 78(79.6) |
| Other | 13(22.0) | 7(17.9) | 0(0.0) | 20(21.3) | |||
| Time to bacteremia | 21(12–41) | 19(12–41) | 0.38 | 12(8.2–37) | 20.5(12–41) | 0.21 | 20.5(12–41) |
| Invasive procedure | |||||||
| Ventilator | 47(79.7) | 33(86.8) | 0.27 | 4(100.0) | 76(81.7) | 0.47 | 80(81.6) |
| CRRTx | 18(30.5) | 11(28.9) | 0.53 | 2(50.0) | 27(29.0) | 0.35 | 29(29.6) |
| C-line insertion | 52(88.1) | 35(92.1) | 0.40 | 4(100.0) | 83(89.2) | 0.64 | 87(88.8) |
| Operation | 21(35.6) | 8(21.1) | 0.09 | 0(0.0) | 29(31.2) | 0.24 | 29(29.6) |
| Hospital stay (days) | 32(20–57) | 27(17–71) | 0.77 | 17(17–41) | 30(19–64.2) | 0.04 | 30(19–64.2) |
| ICU stay (days) | 22(11–38) | 19(8–52) | 0.34 | 17(15.5–33.5) | 21(9.8–40.5) | 0.34 | 21(9.8–40.5) |
| Duration of antibiotics | 31(17–57) | 25(16–69) | 0.84 | 16.5(16–41) | 30.5(16–59.3) | 0.07 | 30.5(16–59.3) |
| Duration of susceptible antibiotics | 0(0–9) | 8(0–17) | 0.36 | 4.5(0.3–10.8) | 2(0–11.25) | 0.76 | 2(0–11) |
| Susceptible antibiotics useb | 4(6.8) | 7(18.4) | 0.08 | 1(25) | 10(10.8) | 0.39 | 11(11.2) |
| All cause 28 day mortality | 52(88.1) | 27(71.1) | 0.16 | 4(100.0) | 75(80.6) | 0.81 | 79(80.6) |
Data of sequence types only with statistical significance are shown. Data are expressed as the mean ± SD / median (Q1-Q3) or N (%). Abbreviation: DM, diabetes mellitus; COPD, chronic obstructive pulmonary disease; CHF, congestive heart failure; HTN, hypertension; CRF, chronic renal failure; APACHE, acute physiology and chronic health evaluation; CRRTx., continuous renal replacement therapy.
*signifies having statistical significance with p-value<0.05 when compared to groups of the rest.
a Steroid use defined by use of ≥20mg prednisolone for ≥2weeks.
b We consider antibiotics susceptible when phenotypically susceptible antibiotics were used for more than half of time after culture recovered.
Analysis using BacWGSTdb based on MLST strategy revealed that 48 (48/98, 49.0%) strains had no closely related strains. The other strains had close relationships with clinical isolates identified in South Korea. The only strain that was linked to isolates outside of South Korea was ABAY09002 (closely related to GeneBank JRQX01, isolated in Thailand in January, 2010). The rates of strains that showed no relatedness with previously identified strains were as follows: ST191, 40.0% (23/59); ST368, 66.6% (2/3); ST552, 100%(1/1); ST357, 71/43%(5/7); ST208/ST1806, 100%(5/5); ST369, 100%(3/3); ST451/ST1809, 9.1%(1/11); ST8958, 100%(1/1); ST784, 100%(1/1); ST191/ST784, 100%(1/1); ST447,25.0%(1/4); Not applicable (NA), 100%(2/2).
Differences in antimicrobial resistance determinants according to STs and year of isolation
Among 29 AMR genes investigated, an average of 16.64 genes were possessed by each strain. The gene blaOXA-23 was identified in all strains except for ABAY11010 (ST369) and ABAY12016 (ST208/ST1806). Among class D carbapenem-hydrolysing class D β-lactamase genes, blaOXA-82 and blaOXA-66 were found in ABAY11010 and ABAY12016, respectively.
The composition of antimicrobial resistance determinants within each ST was relatively constant, especially in ST191. As we compared the first half of the research period with the latter half, only strA (0%, n = 0 versus 18.2%, n = 4; p = 0.02) showed increased prevalence. Furthermore, we observed a decrease in the prevalence of aac(3)-Ia (9.1%, n = 2 versus 91.9%, n = 34; p < 0.01), armA (86.4%, n = 19 versus 100%, n = 37; p = 0.04), and blaTEM-1D (18.2%, n = 4 versus 97.3%, n = 36; p < 0.01). The comparison of sources of bacteraemia showed that aadA1b was more common in isolates from the respiratory tract than in isolates from any other sources (51.9%, n = 14 versus 0%, n = 0; p < 0.01) (Table 3).
Table 3. Composition of antimicrobial resistant genes of all sequence types combined and sequence type 191 according to isolation date or source of bacteremia.
| Genes | ST191 | All | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Source of bacteremia | Isolated year | Isolated year | |||||||
| Respiratory tract(n = 46) | Other(n = 13) | p-value | 2009-2012(n = 37) | 2013-2015(n = 22) | p-value | 2009-2012(n = 54) | 2013-2015(n = 44) | p-value | |
| Aminoglycoside | |||||||||
| aac(3)-Ia | 29(63.0) | 7(53.8) | 0.39 | 34(91.9) | 2(9.1) | <0.01 | 38(70.4) | 3(6.8) | <0.01 |
| aac(6')-Iaf | 0(0.0) | 0(0.0) | 1.00 | 0(0.0) | 0(0.0) | 1.00 | 2(3.7) | 0(0) | 0.30 |
| aac(6')-Ib | 44(95.7) | 13(100) | 0.60 | 36(97.3) | 21(95.5) | 0.61 | 50(92.6) | 29(65.9) | <0.01 |
| aac(6')-Ib3 | 44(95.7) | 13(100) | 0.60 | 37(100) | 20(90.9) | 0.14 | 51(94.4) | 28(63.6) | <0.01 |
| aadA1 | 45(97.8) | 13(100) | 0.78 | 37(100) | 21(95.5) | 0.37 | 51(94.4) | 29(65.9) | <0.01 |
| aadA1b | 14(51.9) | 0(0.0) | <0.01 | 10(27.0) | 4(18.1) | 0.57 | 30(55.6) | 17(68.6) | 0.07 |
| ant(3'')-Ia | 44(95.7) | 13(100) | 0.60 | 37(100) | 20(90.9) | 0.14 | 51(94.4) | 28(63.6) | <0.01 |
| aph(3')-Ia | 0(0.0) | 0(0.0) | 1.00 | 0(0.0) | 0(0.0) | 1.00 | 6(11.1) | 9(20.5) | 0.16 |
| aph(3'')-Ib | 0(0.0) | 0(0.0) | 1.00 | 0(0.0) | 0(0.0) | 1.00 | 14(25.9) | 15(34.1) | 0.26 |
| aph(6)-Id | 0(0.0) | 0(0.0) | 1.00 | 0(0.0) | 0(0.0) | 1.00 | 14(25.9) | 15(34.1) | 0.26 |
| aph(3')-VIb | 0(0.0) | 0(0.0) | 1.00 | 0(0.0) | 0(0.0) | 1.00 | 3(5.6) | 1(2.3) | 0.39 |
| aph(3')-Vij | 1(2.2) | 0(0.0) | 0.78 | 1(2.7) | 0(0.0) | 0.63 | 4(7.4) | 1(2.3) | 0.25 |
| armA | 43(93.5) | 13(100) | 0.47 | 37(100) | 19(86.4) | 0.04 | 51(94.4) | 37(84.1) | 0.09 |
| strA | 4(8.7) | 0(0.0) | 0.36 | 0(0.0) | 4(18.2) | 0.02 | 14(25.9) | 21(47.7) | 0.02 |
| b-lactam | |||||||||
| blaADC-25 | 46(100) | 13(100) | 1.00 | 37(100) | 22(100) | 1.00 | 53(98.1) | 40(90.9) | 0.12 |
| blaOXA-23 | 45(97.8) | 13(100) | 0.78 | 37(100) | 21(95.5) | 0.37 | 53(98.1) | 43(97.7) | 0.69 |
| blaOXA-66 | 46(100) | 13(100) | 1.00 | 37(100) | 22(100) | 1.00 | 52(96.3) | 39(90.7) | 0.24 |
| blaOXA-68 | 0(0.0) | 0(0.0) | 1.00 | 0(0.0) | 0(0.0) | 1.00 | 0(0) | 3(6.8) | 0.09 |
| blaOXA-82 | 0(0.0) | 0(0.0) | 1.00 | 0(0.0) | 0(0.0) | 1.00 | 1(1.9) | 2(4.5) | 0.42 |
| blaOXA-120 | 1(2.2) | 0(0.0) | 0.78 | 1(2.7) | 0(0.0) | 0.67 | 1(1.9) | 0(0) | 0.55 |
| blaPER-1 | 0(0.0) | 0(0.0) | 1.00 | 0(0.0) | 0(0.0) | 1.00 | 3(5.6) | 1(2.3) | 0.39 |
| blaTEM-1D | 33(71.7) | 7(53.8) | 0.19 | 36(97.3) | 4(18.2) | <0.01 | 47(87.0) | 12(27.3) | <0.01 |
| Microlide | |||||||||
| mph(E) | 37(80.4) | 13(100) | 0.08 | 35(94.6) | 15(68.2) | 0.01 | 49(90.7) | 31(70.5) | 0.01 |
| msr(E) | 41(89.1) | 12(92.3) | 0.60 | 34(91.9) | 19(86.4) | 0.40 | 48(88.9) | 37(84.1) | 0.34 |
| Sulphonamide | |||||||||
| sul1 | 45(97.8) | 12(92.3) | 0.40 | 37(100) | 20(90.9) | 0.14 | 51(94.4) | 28(63.6) | <0.01 |
| sul2 | 0(0.0) | 0(0.0) | 1.00 | 0(0.0) | 0(0.0) | 1.00 | 12(22.2) | 17(38.6) | 0.06 |
| Chloramphenicol | |||||||||
| catB8 | 44(95.7) | 12(92.3) | 0.53 | 35(94.6) | 21(95.5) | 0.69 | 49(90.7) | 29(65.9) | <0.01 |
| Quinolone | |||||||||
| aac(6')-Ib-cr | 40(86.9) | 12(92.3) | 0.35 | 45(97.8) | 17(81.0) | 0.12 | 50(92.6) | 29(65.9) | <0.01 |
| Tetracycline | |||||||||
| tet(B) | 0(0.0) | 0(0.0) | 1.00 | 0(0.0) | 0(0.0) | 1.00 | 11(20.4) | 18(40.9) | 0.02 |
Data are expressed as N (%). Values with statistical significance (p-value<0.05) are expressed in boldface.
However, we observed clear distinctions in the composition of AMR genes among ST types. ST447 and ST451/ST1809, which were the dominant STs in the latter part of our study, had the least amount of AMR genes. In these types, aac(6')-Ib, aac(6')-Ib, aadA1, ant(3'')-Ia, and sul1 were less frequently detected, while sul2 and tet(B) were the more frequently observed genes. The ST that contained the most abundant AMR genes was ST208/ST1806. This ST possessed the class A extended-spectrum β-lactamase gene blaPER-1, which is a rare contributor of antimicrobial resistance in A. baumannii, among others (Table 4).
Table 4. Composition of antimicrobial resistance genes according to sequence types.
| Gene | Sequence types | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ST191 (n = 59) | ST357 (n = 7) | ST368 (n = 3) | ST208/ST1806 (n = 5) | ST552 (n = 1) | ST858 (n = 1) | ST369 (n = 3) | ST784 (n = 1) | ST191/ST784 (n = 1) | ST451/ST1809 (n = 11) | ST447 (n = 4) | NA (n = 2) | |
| Aminoglycoside | ||||||||||||
| aac(3)-Ia | 36(61.0) | 0(0) | 3(100) | 0(0.0) | 0(0.0) | 0(0.0) | 1(33.3) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 2(100) |
| aac(6')-Iaf | 0(0.0) | 0(0) | 2(66.7) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) |
| aac(6')-Ib | 57(96.6) | 7(100) | 1(33.3) | 5(100) | 0(0.0) | 1(100) | 3(100) | 1(100) | 1(100) | 0(0.0) | 1(25.0) | 2(100) |
| aac(6')-Ib3 | 57(96.6) | 7(100) | 1(33.3) | 5(100) | 0(0.0) | 1(100) | 3(100) | 1(100) | 1(100) | 0(0.0) | 1(25.0) | 2(100) |
| aadA1 | 58(98.3) | 7(100) | 1(33.3) | 5(100) | 0(0.0) | 1(100) | 3(100) | 1(100) | 1(100) | 0(0.0) | 1(25.0) | 2(100) |
| aadA1b | 29(50.0) | 2(28.6) | 3(100) | 4(80.0) | 0(0.0) | 0(0.0) | 1(33.3) | 1(100) | 1(100) | 0(0.0) | 1(25.0) | 0(0.0) |
| ant(3'')-Ia | 57(96.6) | 7(100) | 1(33.3) | 5(100) | 0(0.0) | 1(100) | 3(100) | 1(100) | 1(100) | 0(0.0) | 1(25.0) | 2(100) |
| aph(3')-Ia | 0(0.0) | 3(42.9) | 0(0.0) | 4(80.0) | 0(0.0) | 1(100) | 0(0.0) | 0(0.0) | 0(0.0) | 7(63.6) | 0(0.0) | 0(0.0) |
| aph(3'')-Ib | 0(0.0) | 7(100) | 2(66.7) | 5(100) | 0(0.0) | 1(100) | 3(100) | 0(0.0) | 0(0.0) | 11(100) | 0(0.0) | 0(0.0) |
| aph(6)-Id | 0(0.0) | 7(100) | 2(66.7) | 5(100) | 0(0.0) | 1(100) | 3(100) | 0(0.0) | 0(0.0) | 11(100) | 0(0.0) | 0(0.0) |
| aph(3')-VIb | 0(0.0) | 0(0) | 0(0.0) | 4(80.0) | 0(0.0) | (0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) |
| aph(3')-Vij | 1(1.7) | 0(0) | 0(0.0) | 4(80.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) |
| armA | 56(94.9) | 7(100) | 1(33.3) | 5(100) | 0(0.0) | 1(100) | 3(100) | 1(100) | 1(100) | 10(90.9) | 1(25.0) | 2(100) |
| strA | 4(6.8) | 7(100) | 2(66.7) | 5(100) | 0(0.0) | 1(100) | 2(66.7) | 0(0.0) | 0(0.0) | 11(100) | 1(25.0) | 1(50.0) |
| b-lactam | ||||||||||||
| blaADC-25 | 59(100) | 7(100) | 3(100) | 5(100) | 0(0.0) | 1(100) | 3(100) | 1(100) | 1(100) | 11(100) | 0(0.0) | 2(100) |
| blaOXA-23 | 58(98.3) | 7(100) | 3(100) | 5(100) | 1(100) | 1(100) | 3(100) | 1(100) | 1(100) | 11(100) | 4(100) | 2(100) |
| blaOXA-66 | 59(100) | 7(100) | 3(100) | 5(100) | 0(0.0) | 1(100) | 1(33.3) | 1(100) | 1(100) | 11(100) | 1(25.0) | 2(100) |
| blaOXA-68 | 0(0.0) | 0(0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 3(75.0) | 0(0.0) |
| blaOXA-82 | 0(0.0) | 0(0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 2(66.7) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) |
| blaOXA-120 | 1(1.7) | 0(0) | 0(0.0) | 0(0.0) | 1(100) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) |
| blaPER-1 | 0(0.0) | 0(0) | 0(0.0) | 4(80.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) | 0(0.0) |
| blaTEM-1D | 40(67.8) | 5(71.4) | 3(100) | 3(60) | 0(0.0) | 0(0.0) | 1(33.3) | 0(0.0) | 0(0.0) | 7(63.6) | 0(0.0) | 1(50.0) |
| Macrolide | ||||||||||||
| mph(E) | 50(84.7) | 7(100) | 1(33.3) | 5(100) | 0(0.0) | 1(100) | 3(100) | 1(100) | 1(100) | 10(90.9) | 0(0.0) | 1(50.0) |
| msr(E) | 53(89.8) | 7(100) | 1(33.3) | 5(100) | 0(0.0) | 1(100) | 3(100) | 1(100) | 1(100) | 10(90.9) | 1(25.0) | 2(100) |
| Sulphonamide | ||||||||||||
| sul1 | 57(96.6) | 7(100) | 1(33.3) | 5(100) | 0(0.0) | 1(100) | 3(100) | 1(100) | 1(100) | 0(0.0) | 1(25.0) | 2(100) |
| sul2 | 0(0.0) | 6(85.7) | 2(66.7) | 5(100) | 0(0.0) | 1(100) | 0(0.0) | 0(0.0) | 0(0.0) | 11(100) | 4(100) | 0(0.0) |
| Chloramphenicol | ||||||||||||
| catB8 | 56(94.9) | 7(100) | 0(0.0) | 5(100) | 0(0.0) | 1(100) | 3(100) | 1(100) | 1(100) | 0(0.0) | 1(25.0) | 2(100) |
| Quinolone | ||||||||||||
| aac(6')-Ib-cr | 52(88.1) | 7(100) | 1(33.3) | 5(100) | 0(0.0) | 1(100) | 3(100) | 1(100) | 1(100) | 0(0.0) | 1(25.0) | 2(100) |
| Tetracycline | ||||||||||||
| tet(B) | 0(0.0) | 7(100) | 2(66.7) | 1(20) | 0(0.0) | 1(100) | 2(66.7) | 0(0.0) | 0(0.0) | 11(100) | 4(100) | 0(0.0) |
Data are expressed as N (%). Values with statistical significance (p-value<0.05) when compared to groups of the rest are expressed in boldface.
Abbreviation: ST, sequence type; NA, not applicable
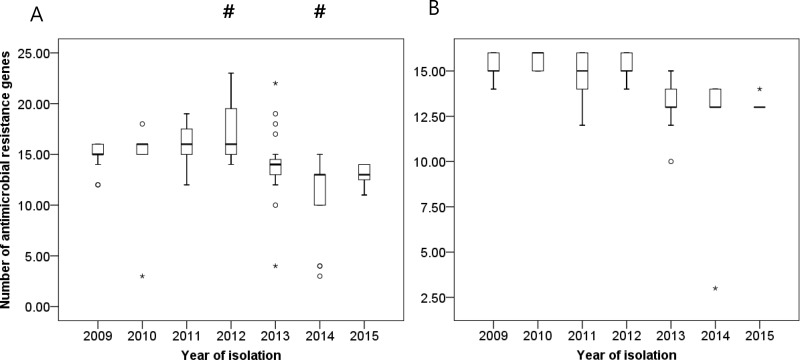
The comparison of the total numbers of AMR genes based on the year of isolation showed fluctuating results. The strains isolated in 2012 showed the most abundant AMR genes, while those found in 2014 exhibited the least amount of these genes. In the latter part of the study period, aac(3)-Ia, aac(6')-Ib, aac(6')-Ib3, aadA1, ant(3'')-Ia,blaTEM-1D, mph(E), sul1, and aac(6')-Ib-cr were significantly decreased, while strA and tet(B) were increased. The change was not significant upon separation and chronological comparison of only ST191 (Fig 3A and 3B).
Fig 3. Changes in numbers of antimicrobial resistance genes according to year of isolation.
Bar graph represents the medians, percentiles (25th and 75th) and 95% confidence intervals: (A) when all sequence types were included and (B) when only sequence type 191 was included. Circle (○) and asterisk (*) symbols indicate extreme values. Pound (#) indicates a statistically significant result (p<0.05) as compared to the rest of the year.
Differences in virulence genes according to STs and year of isolation
From the literature survey, we compiled a list of 182 potential virulence genes [37]. Of all genes identified, 107 genes were possessed by all of the isolated strains. The most differences among STs were found in the ACICU_00075-ACICU_00087 (immune evasion), bap, csuA, casuA/B, csuB, csuC, csuD, csuE, pgaD (biofilm formation), and vgrG1, vgrG2, and vgrG4 (Type IV protein secretion system) genes (S6 Table). We separated ST191 and searched for epidemiologic differences to find that only carO, related to porin, was more frequently detected in the samples originating from the respiratory tract. No statistically significant chronological change was observed (Table 5). The ST that possessed the most abundant virulence genes was ST208/ST1809, which had more ACICU_00075-ACICU_00087, pgaD, blaPER-1, and hcp genes compared to the others. The second most common type was ST451/ST1809, which possessed more ACICU_00075-ACICU_00080, ACICU_00086, ACICU_00087, pgaD, pclC1, and hcp. ST552 had the fewest virulence genes and was devoid of blaOXA-24, ABUW_1966, ACICU_00074-ACICU_00087, fhaC, pgaD, blaPER-1, omp33-36, plcC1, and hcp. The ST that had the second fewest virulence genes was ST447 (S6 Table).
Table 5. Composition of virulence genes of all sequence types combined and sequence type 191 according to isolation date or source of bacteremia.
| Genes | ST191 | ALL | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Source of bacteremia | Isolated year | Isolated year | |||||||
| Respiratory tract(n = 46) | Other(n = 13) | p-value | 2009-2012(n = 37) | 2013-2015(n = 22) | p-value | 2009-2012(n = 37) | 2009-2012(n = 37) | p-value | |
| Outer membrane vesicle | |||||||||
| A1S_0009 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| A1S_0116 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 50(92.6) | 39(88.6) | 0.37 |
| A1S_1180 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| A1S_1321 | 45(97.8) | 13(100) | 0.78 | 36(97.3) | 22(100) | 0.63 | 53(98.1) | 44(100) | 0.55 |
| A1S_1386 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| A1S_1510 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 53(98.1) | 44(100) | 0.55 |
| A1S_1921 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| A1S_2470 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| A1S_2525 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| A1S_3143 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| A1S_3175 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| A1S_3411 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ACV72173.1 | 43(93.5) | 13(100) | 0.47 | 34(91.9) | 22(100) | 0.24 | 46(85.2) | 43(97.7) | 0.03 |
| ACV72174.1 | 43(93.5) | 13(100) | 0.47 | 34(91.9) | 22(100) | 0.24 | 47(87.0) | 43(97.7) | 0.06 |
| ADB23465.1 | 43(93.5) | 13(100) | 0.47 | 34(91.9) | 21(95.5) | 0.52 | 47(87.0) | 43(97.7) | 0.06 |
| ADB23466.1 | 39(84.8) | 12(92.3) | 0.43 | 31(83.8) | 20(90.9) | 0.36 | 40( | 39( | |
| ADB23467.1 | 43(93.5) | 12(92.3) | 0.64 | 34(91.9) | 21(95.5) | 0.52 | 47(87.0) | 42(95.5) | 0.14 |
| ADB23468.1 | 43(93.5) | 13(100) | 0.47 | 34(91.9) | 22(100) | 0.24 | 47(87.0) | 43(97.7) | 0.06 |
| ADB23470.1 | 43(93.5) | 13(100) | 0.47 | 34(91.9) | 22(100) | 0.24 | 47(87.0) | 43(97.7) | 0.06 |
| ADB23471.1 | 40(87.0) | 13(100) | 0.21 | 33(89.2) | 20(90.9) | 0.60 | 45(83.3) | 41(93.2) | 0.126 |
| ADB23472.1 | 37(80.4) | 12(92.3) | 0.29 | 28(75.7) | 21(95.5) | 0.07 | 41(75.9) | 40(90.9) | 0.04 |
| ADB23473.1 | 42(91.3) | 13(100) | 0.36 | 33(89.2) | 22(100) | 0.15 | 46(85.2) | 43(97.7) | 0.03 |
| GADB23474.1 | 43(93.5) | 13(100) | 0.47 | 34(91.9) | 22(100) | 0.24 | 46(85.2) | 43(97.7) | 0.03 |
| GADB23475.1 | 43(93.5) | 13(100) | 0.47 | 34(91.9) | 22(100) | 0.24 | 47(87.0) | 43(97.7) | 0.06 |
| blaOXA-24 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 0(0) | 0(0) | 1 |
| Antibiotic resistance | |||||||||
| ABUW_1156 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 53(98.1) | 40(90.9) | 0.12 |
| ABUW_1499 | 43(93.5) | 13(100) | 0.47 | 36(97.3) | 20(90.9) | 0.31 | 45(83.3) | 38(86.4) | 0.45 |
| ABUW_1520 | 41(89.2) | 13(100) | 0.27 | 35(94.6) | 19(86.4) | 0.26 | 45(83.3) | 26(59.1) | <0.01 |
| ABUW_1645 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ABUW_1672 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ABUW_1673 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 53(98.1) | 44(100) | 0.55 |
| ABUW_1692 | 45(97.8) | 13(100) | 0.78 | 36(97.3) | 22(100) | 0.63 | 53(98.1) | 44(100) | 0.55 |
| ABUW_1755 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ABUW_1768 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ABUW_1849 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ABUW_1851 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ABUW_1966 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 0(0) | 0(0) | 1 |
| ABUW_2074 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ABUW_2550 | 45(97.8) | 13(100) | 0.78 | 36(97.3) | 22(100) | 0.63 | 53(98.1) | 44(100) | 0.55 |
| Aromatic hydrocarbon metabolism | |||||||||
| ABUW_2090 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ABUW_2123 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ABUW_2236 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ABUW_2349 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ABUW_2370 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ABUW_2374 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| benP1 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| paaI1 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| paaY | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| pcaC | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| pcaD1 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| pcaU | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| Transcriptional regulation | |||||||||
| ABUW_2520 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ABUW_2544 | 45(97.8) | 13(100) | 0.78 | 36(97.3) | 22(100) | 0.63 | 52(96.3) | 44(100) | 0.30 |
| Immune evasion | |||||||||
| ACICU_00074 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ACICU_00075 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 4(7.4) | 12(27.3) | <0.01 |
| ACICU_00076 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 4(7.4) | 12(27.3) | <0.01 |
| ACICU_00077 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 4(7.4) | 12(27.3) | <0.01 |
| ACICU_00078 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 4(7.4) | 12(27.3) | <0.01 |
| ACICU_00079 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 4(7.4) | 12(27.3) | <0.01 |
| ACICU_00080 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 4(7.4) | 12(27.3) | <0.01 |
| ACICU_00081 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 4(7.4) | 1(2.3) | 0.25 |
| ACICU_00082 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 4(7.4) | 1(2.3) | 0.25 |
| ACICU_00083 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 4(7.4) | 1(2.3) | 0.25 |
| ACICU_00084 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 4(7.4) | 1(2.3) | 0.25 |
| ACICU_00085 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 4(7.4) | 1(2.3) | 0.25 |
| ACICU_00086 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 7(13.0) | 15(34.1) | 0.01 |
| ACICU_00087 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 7(13.0) | 15(34.1) | 0.01 |
| ACICU_00088 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ACICU_00089 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ACICU_00091 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ACICU_00092 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| lpsB | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| lptE | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| lpxA | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| lpxB | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| lpxC | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| lpxD | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| lpxL | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| lpxM | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| eps | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 43(79.6) | 42(95.5) | 0.02 |
| pgi | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ptk | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ptp | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| Regulation | |||||||||
| abaI | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 50(92.6) | 39(88.6) | 0.37 |
| abaR | 43(93.5) | 11(84.6) | 0.30 | 36(97.3) | 18(81.8) | 0.06 | 49(90.7) | 35(79.5) | 0.10 |
| bfmR | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| bfmS | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| Killing of host cells | |||||||||
| abeD | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| envZ | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| fhaB | 45(97.8) | 13(100) | 0.78 | 36(97.3) | 22(100) | 0.63 | 52(96.3) | 37(84.1) | 0.04 |
| fhaC | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 1(1.9) | 0(0) | 0.55 |
| Biofilm formation | |||||||||
| adeF | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| adeG | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| bap | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 53(98.1) | 40(90.9) | 0.12 |
| csuA | 44(95.7) | 13(100) | 0.61 | 37(100) | 20(90.9) | 0.14 | 47(87.0) | 36(81.8) | 0.33 |
| csuA/B | 44(95.7) | 13(100) | 0.61 | 37(100) | 20(90.9) | 0.14 | 47(87.0) | 35(79.5) | 0.23 |
| csuB | 44(95.7) | 13(100) | 0.61 | 37(100) | 20(90.9) | 0.14 | 47(87.0) | 35(79.5) | 0.23 |
| csuC | 44(95.7) | 13(100) | 0.61 | 37(100) | 20(90.9) | 0.14 | 47(87.0) | 36(81.8) | 0.33 |
| csuD | 44(95.7) | 13(100) | 0.61 | 37(100) | 20(90.9) | 0.14 | 47(87.0) | 36(81.8) | 0.33 |
| csuE | 44(95.7) | 13(100) | 0.61 | 36(97.3) | 20(90.9) | 0.31 | 46(85.2) | 36(81.8) | 0.43 |
| pgaA | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 53(98.1) | 44(100) | 0.55 |
| pgaB | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| pgaC | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 40(90.9) | 0.04 |
| pgaD | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 15(27.8) | 19(43.2) | 0.14 |
| adeH | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| Antibiotic resisance | |||||||||
| adeI | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| adeJ | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| adeK | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| blaPER-1 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 3(5.6) | 1(2.3) | 0.39 |
| Transcriptional regulation | |||||||||
| alkR | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| gigA | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| gigB | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| gigC | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| soxR | 45(97.8) | 13(100) | 0.78 | 36(97.3) | 22(100) | 0.63 | 53(98.1) | 44(100) | 0.55 |
| Iron uptake | |||||||||
| barA | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| barB | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| basA | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| basB | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| basC | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| basD | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| basF | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| basG | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| basH | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| basI | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| basJ | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| bauA | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 40(90.9) | 0.04 |
| bauB | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| bauC | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| bauD | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| bauE | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| bauF | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| nfuA | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| Porin | |||||||||
| carO | 46(100) | 11(84.6) | 0.046 | 37(100) | 20(90.9) | 0.14 | 54(100) | 40(90.9) | 0.04 |
| omp22 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| omp33-36 | 0(0.0) | 0(0.0) | 1 | 37(100) | 22(100) | 1 | 0 | 0 | |
| ompR | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| orpD | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| Serun resistance, invasion | |||||||||
| cipA | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| cobA | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| pbpG | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| surA1 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| tuf | 46(10) | 13(100) | 1 | 36(97.3) | 22(100) | 0.63 | 53(98.1) | 44(100) | 0.55 |
| typA | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | |||
| Enzyme | |||||||||
| plcC1 | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 8(14.8) | 14(31.8) | 0.05 |
| plcC2 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| plcD | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| pldA | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| Cysteine metabolism | |||||||||
| cysD | 46(10) | 13(100) | 1 | 54(100) | 44(100) | 1 | |||
| cysE | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| cysH | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| cysI | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| cysN | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| cysQ | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| sulP | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | |||
| Siderophore biosynthesis | |||||||||
| entA | 46(100) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 42(95.5) | 0.20 |
| Neutrophil influx | |||||||||
| gacS | 46(10) | 13(100) | 1 | 54(100) | 44(100) | 1 | |||
| paaA | 45(97.8) | 13(100) | 0.78 | 36(97.3) | 22(100) | 0.63 | 53(98.1) | 44(100) | 0.55 |
| Type II protein secretion system | |||||||||
| gspD | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| Type VI protein secretion system | |||||||||
| vgrG1 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 39(72.2) | 27(61.4) | 0.29 |
| vgrG2 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 52(96.3) | 33(75.0) | <0.01 |
| vgrG3 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 53(98.1) | 44(100) | 0.55 |
| vgrG4 | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 40(74.1) | 27(61.4) | 0.20 |
| hcp | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 12(22.2) | 15(34.1) | 0.14 |
| Type V protein secretion system | |||||||||
| ata | 41(89.1) | 12(92.3) | 0.60 | 31(83.8) | 22(100) | 0.08 | 42(77.8) | 39(88.6) | 0.13 |
| Stress response genes | |||||||||
| kef | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| kefF | 45(97.8) | 13(100) | 0.78 | 36(97.3) | 22(100) | 0.63 | 54(100) | 44(100) | 1 |
| mscS | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| ostB | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| recA | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| resP | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| trkH | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| upsA | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| uspA | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 0(0) | 0(0) | 1 |
| uvrD | 0(0.0) | 0(0.0) | 1 | 0(0.0) | 0(0.0) | 1 | 0(0) | 0(0) | 1 |
| Manganese acquisition system | |||||||||
| mumC | 45(97.8) | 13(100) | 0.78 | 36(97.3) | 22(100) | 0.63 | 52(96.3) | 44(100) | 0.30 |
| mumT | 45(97.8) | 13(100) | 0.78 | 36(97.3) | 22(100) | 0.63 | 52(96.3) | 44(100) | 0.30 |
| Adherence | |||||||||
| ompA | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| Micronutrient acquisition | |||||||||
| znuA | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| znuB | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| znuC | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
| zur | 46(10) | 13(100) | 1 | 37(100) | 22(100) | 1 | 54(100) | 44(100) | 1 |
Data are expressed as N (%). Values with statistical significance (p-value<0.05) are expressed in boldface.
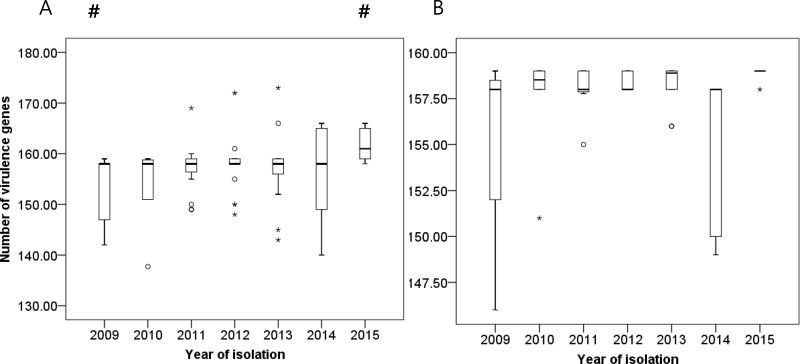
The number of virulence genes was compared according to the year of isolation. The number increased as research period elapsed. The strains isolated in the year 2009 carried fewer virulence genes, while those isolated in 2015 exhibited more virulence genes compared to the rest. We singled out ST191 and observed statistically insignificant results (Fig 4A and 4B).
Fig 4. Changes in numbers of virulence genes according to year of isolation.
Bar graph represents medians, percentiles (25th and 75th), and 95% confidence intervals: (A) when all sequence types were included and (B) when only sequence type 191 was included. Circle (○) and asterisk (*) symbols indicate extreme values. Pound (#) indicates a statistically significant result (p<0.05) as compared to the rest of the year.
Difference in ISAba1 and carbapenemase-encoding transposons on CRAB
ISAba1 belongs to the IS4 family and has been detected upstream of the ampC, blaOXA-23, blaOXA-27, and sul2 antibiotic resistance genes in Acinetobacter species [34]. Insertion sites of ISAba1 were different for each strain, and no common insertion sites were conserved throughout all strains. Insertion sites of ISAba1 that were shared by more than 70% of strains were designated as IS*1, IS*2, IS*3, IS*4, IS*5, IS*6, and IS*7. The compositions of IS*1 through IS*7 differed between STs (S7 Table). ST447 contained fewer insertion sequences compared to the rest, while ST191 carried comparatively more insertion sequences; these incidences remained constant throughout the study period for each insertion sequence (S1 Fig). We could detect common blaOXA-23-ΔATPase modules from all isolates with the exception of ABAY11010 (ST369) and ABAY12016 (ST208/ST1806), which implied that Tn2006, Tn2008, or Tn2009 was widespread among these strains. However, although antimicrobial resistance (AMR) genes were predicted using three sets of contig sequences, which include assemblies obtained by CLC Genomics Workbench (whole-genome assembly), by plasmid SPAdes, and by plasmid profiler-based process, these cannot determine the exact location of AMR genes. ABAY11010 and ABAY12016 originated in the respiratory tract, and had larger number of virulence and AMR genes than average (virulence genes, 160 and 161 copies each; AMR genes, 16 and 23 copies each). Both strains were isolated at different places (MICU D and MICU C each) and times (2011 and 2012 each).
Resistance islands
AbaR1 is a commonly reported sequence from strains isolated in South Korea [38,39]. We searched contig sequences using the 87.7-kb AbaR1 sequence from the strain AYE (CU459141.1) [40] as a query, and found 70 strains to carry a very short AbaR-like sequence. These were only 10,641 bp long after including the disrupted ATPase gene (comM’) at both ends. AbaR-like islands were similar to the 14.5-kb long Tn6021 that is devoid of the multiple antibiotic resistance region found in ATCC 17978 (GenBank CP00521.1). When read mapping on the complete AbaR-like sequence extracted from ABAY09001 (GenBank QHGS01000008.1; 56,153–66,793 bp) was applied, all strains except ABAY13016, ABAY14002, ABAY14004, and ABAY14008 (all categorised as ST447) possessed AbaR-like islands.
Also, 75 ABAY strains harbor class I integron carrying cat2, aadA1, sul1 and armA. It has been reported that two consecutive copies of integrons are located within a putative resistance island, but read mapping-based analysis (this study) could not reveal how integrons are situated in ABAY chromosomes. The gene organization is similar with class I integron identified from NCGM 237 [41].
Reconstruction of plasmid sequence
Assembly-based methods produced putative plasmidic contigs from all 98 isolates: average total lengths of plasmidic contigs predicted by plasmiSPAdes and plasmid profiler-based unicycler were 129,195.1 bp and 141,235 bp, respectively. Standard deviations of their total lengths were 136,737.5 bp and 71,471.1 bp, respectively. The latter method, which relies on graph-based assembly followed by circularization, identified 115 complete plasmid sequences along with linear and complex type assemblies. We clustered plasmid sequences into groups based on sequence similarity and length, which resulted in five groups ranging from 110,967 bp (group A) to 2,278 bp (group E) (S8 Table). To estimate the presence of all five groups of plasmids across strains, sequencing reads were re-mapped on the representative plasmid sequences (S9 Table). Unexpectedly, we could not find any antimicrobial resistance genes from the five representative plasmid sequences. However, unresolved plasmids in the plasmid profiler-based unicycler assemblies might contain not-yet-identified ones that harbor multiple AMR determinants.
We identified ISAba1, which is known to promote expression of downstream genes, from four complete plasmids in group B. However, no blaOXA-23 was found in their vicinity or elsewhere in the complete plasmid sequences.
Discussion
Since the 1970s, there has been a progressive increase in the antimicrobial resistance for the majority of A. baumannii strains, which were otherwise sensitive to the commonly used antibiotics [42]. By 2007, up to 70% of isolates in certain settings had developed multidrug resistance including resistance to carbapenems, which were once considered as the mainstay against multidrug-resistant A. baumannii infections [43]. In our study, ST191 was the dominant ST during the 7 years of our study and showed stable genomic variations. However, when other STs were combined, a tendency toward increasing virulence genes was observed without additional changes in antimicrobial resistance of CRAB in restricted hospital environments.
ST191 is a predominant strain isolated in South Korea [44] known for expressing blaOXA-23, which is responsible for the high rate of carbapenem resistance. The trait that allows this strain to emerge as a highly successful nosocomial pathogen through frequent modification of genetic components was also manifested in our study. ST191 had higher insertion sequences compared to the other types, indicating the frequent recombination and gene rearrangement events. Also, only 60% of strains involved in our study had closed related strains identified in previous studies. However, no cumulative change in AMR genes and virulence genes was observed over time. The only gene that showed increased prevalence was strA. This gene is related to aminoglycoside resistance and is frequently found in AbaR-like islands along with other genes such as tetA(B), tetR(B), CR2, strB, and orf4b [45]. We observed no simultaneous increase in other AMR genes in this study.
It is worth noting that heterogenous groups of ST were isolated simultaneously. ST208/ST1806 was the sequence type with the most abundant AMR genes; blaPER-1 was found only in this sequence type. Although it is a relatively uncommon gene for A. baumannii, blaPER-1 is frequently reported as being implicated in various virulence mechanisms [46,47]. In our study, this strain was isolated mostly in 2012, and additional spread of this gene was not noted. ST447, which was categorised into international clone 1 (IC I), was isolated in the latter part of our study period. ST447 possessed fewer AMR genes. Since this strain was more frequently isolated from patients with shorter hospital stays, it was introduced from the outside rather than being an indigenous strain. This type was equipped with β-lactam resistance genes without any other resistance genes. As with other strains, ST447 also had blaOXA-23-ΔATPase modules, indicating that it carries Tn2006, Tn2008, or Tn2009. While Tn2006 was mostly transferred by mobile elements, the spread of Tn2008 and Tn2009 was entirely dependent on the clonal dissemination of the bacterial host [44]. In light of ST447 containing all of these transposons, this type might have been under selective antibiotic pressure for a long period of time. It is possible that the previously susceptible strain may have gained genes only for carbapenem resistance during the study period, but the widespread use of carbapenems may have contributed to the emergence of this type. The density of virulence genes was still low, and this type of strain had relatively lower insertion sequences, which are commonly found in other STs. This defies the concern posed by a previous study in South Korea which reported the emergence of a new strain that would provoke the dissemination of more antibacterial resistance [48].
Despite sporadic studies suggesting correlation between the number of virulence genes with organisms’ phenotypic differences [16], the significance of number of virulence genes in association with clinical outcomes and epidemiology has not been investigated. Therefore, the increased number of virulence genes over time shown in our study demands immediate attention. Although previous studies suggested certain associations between antimicrobial resistance and virulence [49], our study demonstrated that linear correlation between the two did not exist. It is worth noting that more than half of the highly significant virulence genes were possessed by all of these carbapenem-resistant strains. A slight increase in the number of virulence genes was reported in 2015, probably due to the emergence of ST451/ST1809. ST451/ST1809 possessed higher number of virulence genes than average (165 [159–166] vs. 158[138–173], p<0.001). A gene specifically related to the functioning of porin (carO) [50] had decreased, which may be attributed to enhanced antimicrobial resistance. However, genes that were previously considered important in forming biofilms and iron utilization (bauA and pgaC) [49,51] decreased in number. The increased genes were ACICU_00075-ACICU_00080, ACICU_00086, and ACICU_00087, which are presumably associated with immune evasion [52]. Although further evaluation is warranted regarding the authentic effects of these putative virulence genes, the gradual accumulation of virulence genes that are transmitted by horizontal transfer emphasises the need for more intensive infection control strategies to prevent re-infections of CRAB.
We failed to observe any significant differences in patient outcomes according to STs. As more than 80% of patients died within 28 days of the first isolation of the organism, it is possible that the patients were in serious condition and hence died before the analysis reached a conclusive stage. However, previous studies also suggested that phenotypical characteristics are not in accordance with genotypical results. It is possible that individual virulence factors may not be important for A. baumannii virulence in human hosts [53], and the same virulence factor may play different roles in different habitats [54]. The expression of virulence-associated genes could be under different regulation in pathogenic and non-pathogenic species [55].
We found no discrepancies between phenotypic and genotypic results in terms of carbapenem resistance. This observation confirmed the results of a previous study, in which WGS accurately identified resistance to β-lactam antibiotics for gram-negative bacterial pathogens [56]. Whether this observation holds true for other types of antimicrobial agents warrants further studies.
The current study had a few limitations. First, only patients who were admitted to ICUs were involved, limiting further epidemiological investigation. Second, the relationships between strains, such as evolutionary linkages, were not determined. Third, detailed comparative genomics to verify mechanisms of acquisition of virulence or AMR genes acquisition were not analysed.
In conclusion, this study confirms the absence of accumulation of antimicrobial resistance determinants while the number of virulence genes increases in CRAB. Genetic transfer between subtypes cannot be ruled out; hence, it is important to be cautious about re-infections.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
Bar graph represents individual and cumulative number of each sequence types according to year of isolation.
(TIF)
Acknowledgments
We would like to thank all of the nursing and laboratory staff as well as the physicians who supported this project. Finally, we give credit to all of the patients who took part in this study. We would also like to thank e-World Editing (https://eworldediting.com/index.php) for providing English language editing service.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Korea HIV/AIDS Cohort Study (Grant No. 2019-ER5101-00), Ministry of Health & Welfare, South Korea (Grant No. HI14C1324), Ministry of Science, ICT & Future Planning, South Korea (Grant No. H-GUARD_2013M3A6B2078952), a Severance Hospital Research fund for Clinical Excellence (SHRC) and the KRIBB initiative program.
References
- 1.Lin MF, Lan CY (2014) Antimicrobial resistance in Acinetobacter baumannii: From bench to bedside. World J Clin Cases 2: 787–814. 10.12998/wjcc.v2.i12.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones CL, Clancy M, Honnold C, Singh S, Snesrud E, et al. (2015) Fatal Outbreak of an Emerging Clone of Extensively Drug-Resistant Acinetobacter baumannii With Enhanced Virulence. Clinical Infectious Diseases 61: 145–154. 10.1093/cid/civ225 [DOI] [PubMed] [Google Scholar]
- 3.Kim DH, Choi J-Y, Kim HW, Kim SH, Chung DR, et al. (2013) Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 in Asia and AbaR-type resistance islands. Antimicrobial agents and chemotherapy 57: 5239–5246. 10.1128/AAC.00633-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung DR, Song J-H, Kim SH, Thamlikitkul V, Huang S-G, et al. (2011) High prevalence of multidrug-resistant nonfermenters in hospital-acquired pneumonia in Asia. American journal of respiratory and critical care medicine 184: 1409–1417. 10.1164/rccm.201102-0349OC [DOI] [PubMed] [Google Scholar]
- 5.Kim YA, Park YS (2018) Epidemiology and treatment of antimicrobialresistant gram-negative bacteria in Korea. Korean J Intern Med 33: 247–255. 10.3904/kjim.2018.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeon H, Kim S, Kim MH, Kim SY, Nam D, et al. (2018) Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates from a Korean hospital that carry blaOXA-23. Infect Genet Evol 58: 232–236. 10.1016/j.meegid.2018.01.003 [DOI] [PubMed] [Google Scholar]
- 7.Kim DH, Jung SI, Kwon KT, Ko KS (2017) Occurrence of Diverse AbGRI1-Type Genomic Islands in Acinetobacter baumannii Global Clone 2 Isolates from South Korea. Antimicrob Agents Chemother 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi JY, Kwak YG, Yoo H, Lee SO, Kim HB, et al. (2016) Trends in the distribution and antimicrobial susceptibility of causative pathogens of device-associated infection in Korean intensive care units from 2006 to 2013: results from the Korean Nosocomial Infections Surveillance System (KONIS). J Hosp Infect 92: 363–371. 10.1016/j.jhin.2015.12.012 [DOI] [PubMed] [Google Scholar]
- 9.Didelot X, Eyre DW, Cule M, Ip CL, Ansari MA, et al. (2012) Microevolutionary analysis of Clostridium difficile genomes to investigate transmission. Genome Biol 13: R118 10.1186/gb-2012-13-12-r118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reuter S, Ellington MJ, Cartwright EJ, Koser CU, Torok ME, et al. (2013) Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA Intern Med 173: 1397–1404. 10.1001/jamainternmed.2013.7734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imperi F, Antunes LC, Blom J, Villa L, Iacono M, et al. (2011) The genomics of Acinetobacter baumannii: insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB Life 63: 1068–1074. 10.1002/iub.531 [DOI] [PubMed] [Google Scholar]
- 12.Antunes LC, Visca P, Towner KJ (2014) Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis 71: 292–301. 10.1111/2049-632X.12125 [DOI] [PubMed] [Google Scholar]
- 13.Cerqueira GM, Peleg AY (2011) Insights into Acinetobacter baumannii pathogenicity. IUBMB Life 63: 1055–1060. 10.1002/iub.533 [DOI] [PubMed] [Google Scholar]
- 14.Imperi F, Antunes LC, Blom J, Villa L, Iacono M, et al. (2011) The genomics of Acinetobacter baumannii: insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB life 63: 1068–1074. 10.1002/iub.531 [DOI] [PubMed] [Google Scholar]
- 15.Thummeepak R, Kongthai P, Leungtongkam U, Sitthisak S (2016) Distribution of virulence genes involved in biofilm formation in multi-drug resistant Acinetobacter baumannii clinical isolates. Int Microbiol 19: 121–129. 10.2436/20.1501.01.270 [DOI] [PubMed] [Google Scholar]
- 16.Wang N, Ozer EA, Mandel MJ, Hauser AR (2014) Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. MBio 5: e01163–01114. 10.1128/mBio.01163-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(2015) Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Disk Susceptibility Tests—Twelfth Edition: Approved Standard M02-A12 CLSI. Wayne, PA, USA.
- 18.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, et al. (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44: 6614–6624. 10.1093/nar/gkw569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. (2015) Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31: 3691–3693. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price MN, Dehal PS, Arkin AP (2010) FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44: W242–245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gebhardt MJ, Gallagher LA, Jacobson RK, Usacheva EA, Peterson LR, et al. (2015) Joint Transcriptional Control of Virulence and Resistance to Antibiotic and Environmental Stress in Acinetobacter baumannii. MBio 6: e01660–01615. 10.1128/mBio.01660-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Zheng D, Liu B, Yang J, Jin Q (2016) VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res 44: D694–697. 10.1093/nar/gkv1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallace L, Daugherty SC, Nagaraj S, Johnson JK, Harris AD, et al. (2016) Use of Comparative Genomics To Characterize the Diversity of Acinetobacter baumannii Surveillance Isolates in a Health Care Institution. Antimicrob Agents Chemother 60: 5933–5941. 10.1128/AAC.00477-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CR, Lee JH, Park M, Park KS, Bae IK, et al. (2017) Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front Cell Infect Microbiol 7: 55 10.3389/fcimb.2017.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahl JW, Caporaso JG, Rasko DA, Keim P (2014) The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. PeerJ 2: e332 10.7717/peerj.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruan Z, Feng Y (2015) BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic acids research 44: D682–D687. 10.1093/nar/gkv1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, et al. (2016) plasmidSPAdes: assembling plasmids from whole genome sequencing data. bioRxiv: 048942. [DOI] [PubMed] [Google Scholar]
- 30.Zetner A, Cabral J, Mataseje L, Knox NC, Mabon P, et al. (2017) Plasmid Profiler: comparative analysis of plasmid content in WGS data. BioRxiv: 121350. [Google Scholar]
- 31.Wick RR, Judd LM, Gorrie CL, Holt KE (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS computational biology 13: e1005595 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wick RR, Schultz MB, Zobel J, Holt KE (2015) Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31: 3350–3352. 10.1093/bioinformatics/btv383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic acids research 29: 2607–2618. 10.1093/nar/29.12.2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, et al. (2006) The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett 258: 72–77. 10.1111/j.1574-6968.2006.00195.x [DOI] [PubMed] [Google Scholar]
- 35.Hawkey J, Hamidian M, Wick RR, Edwards DJ, Billman-Jacobe H, et al. (2015) ISMapper: identifying transposase insertion sites in bacterial genomes from short read sequence data. BMC Genomics 16: 667 10.1186/s12864-015-1860-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y, Gao J, Zhang H, Ying C (2017) Spread of the blaOXA-23-Containing Tn2008 in Carbapenem-Resistant Acinetobacter baumannii Isolates Grouped in CC92 from China. Front Microbiol 8: 163 10.3389/fmicb.2017.00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harding CM, Hennon SW, Feldman MF (2018) Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol 16: 91–102. 10.1038/nrmicro.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee Y, D'souza R, Yong D, Lee K (2016) Prediction of putative resistance islands in a carbapenem-resistant Acinetobacter baumannii global clone 2 clinical isolate. Annals of laboratory medicine 36: 320–324. 10.3343/alm.2016.36.4.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim DH, Ko KS (2015) AbaR-type genomic islands in non-baumannii Acinetobacter species isolates from South Korea. Antimicrobial agents and chemotherapy 59: 5824–5826. 10.1128/AAC.01175-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, et al. (2006) Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2: e7 10.1371/journal.pgen.0020007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng Y, Ruan Z, Shu J, Chen C-L, Chiu C-H (2016) A glimpse into evolution and dissemination of multidrug-resistant Acinetobacter baumannii isolates in East Asia: a comparative genomics study. Scientific reports 6: 24342 10.1038/srep24342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergogne-Berezin E, Towner K (1996) Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clinical microbiology reviews 9: 148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kempf M, Rolain JM (2012) Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: clinical impact and therapeutic options. Int J Antimicrob Agents 39: 105–114. 10.1016/j.ijantimicag.2011.10.004 [DOI] [PubMed] [Google Scholar]
- 44.Yoon EJ, Kim JO, Yang JW, Kim HS, Lee KJ, et al. (2017) The blaOXA-23-associated transposons in the genome of Acinetobacter spp. represent an epidemiological situation of the species encountering carbapenems. J Antimicrob Chemother 72: 2708–2714. 10.1093/jac/dkx205 [DOI] [PubMed] [Google Scholar]
- 45.Kim DH, Ko KS (2015) AbaR-type genomic islands in non-baumannii Acinetobacter species isolates from South Korea. Antimicrob Agents Chemother 59: 5824–5826. 10.1128/AAC.01175-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vahaboglu H, Oztürk R, Aygün G, Coşkunkan F, Yaman A, et al. (1997) Widespread detection of PER-1-type extended-spectrum beta-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrobial agents and chemotherapy 41: 2265–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sechi LA, Karadenizli A, Deriu A, Zanetti S, Kolayli F, et al. (2004) PER-1 type beta-lactamase production in Acinetobacter baumannii is related to cell adhesion. Medical science monitor 10: BR180–BR184. [PubMed] [Google Scholar]
- 48.Selasi GN, Nicholas A, Jeon H, Lee YC, Yoo JR, et al. (2015) Genetic basis of antimicrobial resistance and clonal dynamics of carbapenem-resistant Acinetobacter baumannii sequence type 191 in a Korean hospital. Infect Genet Evol 36: 1–7. 10.1016/j.meegid.2015.09.001 [DOI] [PubMed] [Google Scholar]
- 49.Liu C, Chang Y, Xu Y, Luo Y, Wu L, et al. (2018) Distribution of virulence-associated genes and antimicrobial susceptibility in clinical Acinetobacter baumannii isolates. Oncotarget 9: 21663–21673. 10.18632/oncotarget.24651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernandez-Cuenca F, Smani Y, Gomez-Sanchez MC, Docobo-Perez F, Caballero-Moyano FJ, et al. (2011) Attenuated virulence of a slow-growing pandrug-resistant Acinetobacter baumannii is associated with decreased expression of genes encoding the porins CarO and OprD-like. Int J Antimicrob Agents 38: 548–549. 10.1016/j.ijantimicag.2011.08.002 [DOI] [PubMed] [Google Scholar]
- 51.Hatami Moghadam R, Alvandi A, Akbari N, Jafari P, Abiri R (2018) Assessment of biofilm formation among clinical isolates of Acinetobacter baumannii in burn wounds in the west of Iran. Cell Mol Biol (Noisy-le-grand) 64: 30–34. [PubMed] [Google Scholar]
- 52.Iacono M, Villa L, Fortini D, Bordoni R, Imperi F, et al. (2008) Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob Agents Chemother 52: 2616–2625. 10.1128/AAC.01643-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McConnell MJ, Actis L, Pachon J (2013) Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev 37: 130–155. 10.1111/j.1574-6976.2012.00344.x [DOI] [PubMed] [Google Scholar]
- 54.Antunes LC, Imperi F, Carattoli A, Visca P (2011) Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS One 6: e22674 10.1371/journal.pone.0022674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pallen MJ, Wren BW (2007) Bacterial pathogenomics. Nature 449: 835–842. 10.1038/nature06248 [DOI] [PubMed] [Google Scholar]
- 56.Shelburne SA, Kim J, Munita JM, Sahasrabhojane P, Shields RK, et al. (2017) Whole-Genome Sequencing Accurately Identifies Resistance to Extended-Spectrum beta-Lactams for Major Gram-Negative Bacterial Pathogens. Clin Infect Dis 65: 738–745. 10.1093/cid/cix417 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
Bar graph represents individual and cumulative number of each sequence types according to year of isolation.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.
Genome sequences were submitted to GenBank under BioProject no. PRJNA448358 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA448358). The complete list of strain names and accession numbers are shown in S2 Table. Complete sequences of five representative plasmids (pABAY04001_1A, pABAY09008_1B, pABAY10001_1C, pABAY14012_4D, and pABAY15001_6E) are available from GenBank under the accession numbers MK386680.1- MK386684.1. All 115 complete plasmid sequences and their rep genes are available at https://doi.org/10.6084/m9.figshare.11467023.v1 (figshare).