Abstract
We recently derived mouse expanded potential stem cells (EPSCs) from individual blastomeres by inhibiting the critical molecular pathways that predispose their differentiation1. EPSCs had enriched molecular signatures of blastomeres and possessed the developmental potency for all embryonic and extraembryonic cell lineages. Here, we report the derivation of porcine EPSCs, which express key pluripotency genes, are genetically stable, permit genome editing, differentiate to derivatives of the three germ layers in chimeras, and produce primordial germ cell-like cells in vitro. Under similar conditions, human ESCs and iPSCs can be converted, or somatic cells directly reprogrammed, to EPSCs that display the molecular and functional attributes reminiscent of porcine EPSCs. Significantly, trophoblast stem cell-like cells can be generated from both human and porcine EPSCs. Our pathway-inhibition paradigm thus opens a new avenue for generating mammalian pluripotent stem cells, and EPSCs present an unique cellular platform for translational research in biotechnology and regenerative medicine.
Keywords: pluripotent stem cells, totipotency, preimplantation embryos, porcine, chimeras, human, germ cells, iPS cell, trophoblast, placenta, single cell RNA sequencing, histone methylation, DNA methylation, developmental potential
Introduction
Mouse and human embryonic stem cells (ESCs) that derived from preimplantation embryos2–4 self-renew in long term cultures and differentiate to all embryonic cell lineages in vitro and in mouse chimeras. The development of well-defined culture conditions such as 2i/LIF has substantially facilitated derivation and maintenance of mouse ESCs5, and led to intensive efforts for deriving human ESCs akin to mouse ESCs6,7. It has however been challenging to translate the findings in studying mouse and human cells to establishing ESCs from other mammalian species. The domestic pig shares great genetic, anatomical and physiological similarities with humans, and is considered to be an excellent model for human diseases, cell therapies and even as donor for porcine xenografts. To this date, bona fide porcine ESCs have yet to be established.8–15 The published lines usually do not meet with the stringent criteria for pluripotency and are frequently called “ES-like” cells.
We have recently demonstrated that by targeting key molecular pathways that drive lineage differentiation in the mouse preimplantation embryo, expanded potential stem cells (mEPSCs) displaying a broad propensity for extraembryonic and embryonic lineage differentiation were derived1,16. We hypothesized that a similar experimental paradigm of targeting key developmental pathways might be applied for establishing porcine stem cells from preimplantation embryos. However, little is known about the molecular and signalling mechanisms of porcine early preimplantation embryo development, we thus set out to perform a chemical screen of inhibitors that were used for isolating and maintaining mouse mEPSCs, mouse and human ESCs and to delineate the optimal condition for porcine cells. Our results demonstrate that porcine EPSCs could be established, and that significantly, similar culture conditions permit derivation of human EPSCs.
Results and Discussion
Identification of culture conditions for porcine pluripotent stem cells
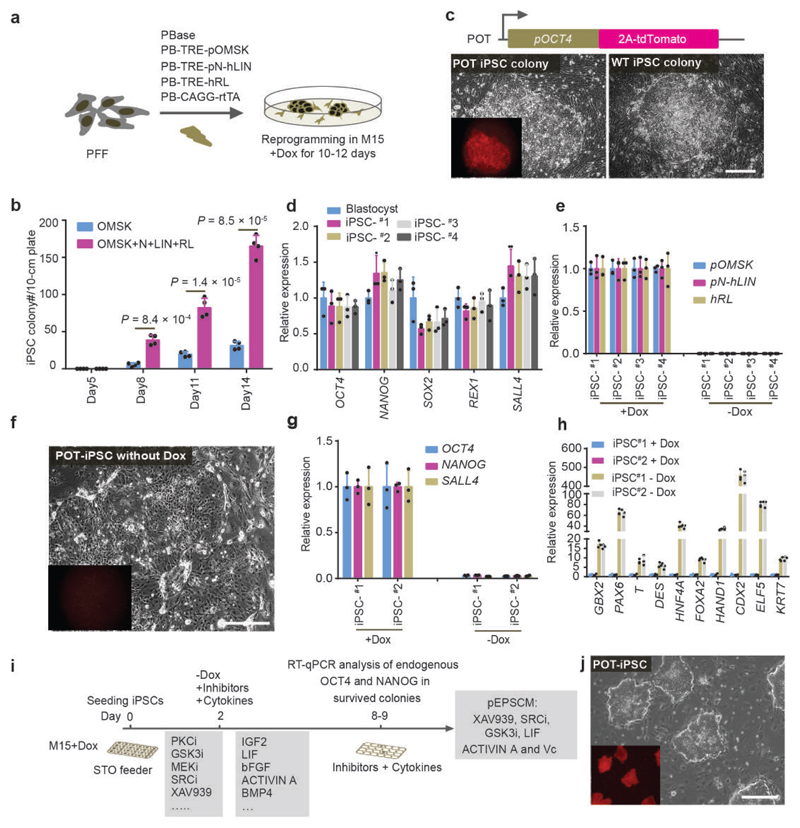
While porcine iPSCs are available, their use for the screen is confounded by the leaky expression of the transgenic reprogramming factors after reprogramming or by low levels of expression of the endogenous pluripotency genes17–20. To overcome this challenge, we generated new porcine iPSCs by expressing Doxycycline (Dox)-inducible eight transcription factors, which substantially improved the efficiency of reprogramming wild-type and transgenic porcine fetal fibroblasts (PFFs), in which a tdTomato cassette had been inserted into the 3’ UTR of the porcine OCT4 (POU5F1) locus (POT PFFs)21, to putative iPSC colonies (Fig. 1a-c). The iPSCs from POT PFFs were OCT4-tdTomato+, (Fig. 1c), and expressed high levels of the endogenous pluripotency factors (Fig. 1d). iPSCs could be passaged as single cells for more than 20 passages in serum-containing medium (M15) plus Dox. Upon Dox removal, the iPSCs differentiated within 4-5 days, concomitant with rapid down-regulation of the exogenous reprogramming factors and endogenous pluripotency genes and with increased expression of both embryonic and extraembryonic cell lineage genes (Fig. 1e-h). These Dox-dependent iPSCs with robust endogenous pluripotency gene expression provided the material for the chemical screen.
Figure 1. Identification of culture conditions for porcine EPSCs.
a. Doxycycline (Dox)-inducible expression of Yamanaka factors OCT4, MYC, SOX2 and KLF4, together with LIN28, NANOG, LRH1 and RARG in porcine PFFs. Stable genomic integration of cDNAs in PFFs was achieved by piggyBac transposition. pOMSK: Porcine OCT4, MYC, SOX2 and KLF4; pN+hLIN: porcine NANOG and human LIN28; hRL: human RARG and LRH1. The reprogrammed colonies were single-cell passaged in the presence of Dox in M15 (15% fetal calf serum). b. Co-expression of LIN28, NANOG, LRH1 and RARG substantially increased the number of reprogrammed colonies from 250,000 PFFs (n = 4 independent experiments). c. Reprogramming of the porcine OCT4-tdTomato knock-in reporter (POT) TAIHU and wide type (WT) German Landrace PFFs to iPSCs. d. The iPSCs lines expressed key pluripotency genes in RT-qPCR analysis. The iPSC lines #1 and #2, and iPSC #3 and #4 were from WT German Landrace and POT PFFs, respectively. e. RT-qPCR analysis of the exogenous reprogramming factors in iPSCs either in the presence of Dox or 5 days after its removal. f. POT iPSCs became Td-tomato negative 5 days after Dox removal. g. RT-qPCR analysis of the expression of endogenous pluripotency genes in iPSCs cultured with or without Dox. h. Expression of lineage genes in porcine iPSCs 5-6 days after DOX removal. Gene expression was measured by RT-qPCR. Relative expression levels are shown with normalization to GAPDH. Experiments were performed 3 times. i. Diagram depicting the screen strategy for identifying culture conditions for porcine pluripotent stem cells using the Dox-dependent iPSC. Small molecule inhibitors and cytokines were selected for various combinations. Cell survival, cell morphology, and expression of endogenous OCT4 and NANOG were employed as the read-outs. j. Images of OCT4-Tdtomato reporter (POT) iPSCs in pEPSCM without Dox. In all RT-qPCR analysis, n=3 independent experiments. All graphs represent the mean ± s.d. P values were computed using a two-tailed t-test. For c, f and j, the experiments were repeated independently three times with similar results. Source data are provided in Supplementary Table 1. Scale bars, 100 μm.
In the screen, over 400 combinations of 20 small molecule inhibitors and cytokines were tested for their ability to maintain Dox-independent porcine iPSCs in the undifferentiated state (Fig. 1i; Supplementary Table 1). A departure was noted from previous reports that naïve mouse ESC medium 2i/LIF5 was able to maintain putative porcine iPSCs22–24: Porcine iPSCs were rapidly lost with 1.0 μM Mek1 inhibitor PD-0325901, irrespective of whether Dox was present or not (Extended Data Fig. 1a-g), indicating that porcine pluripotent stem cells differ from mouse ESCs in the requirement of Mek-ERK signalling5,25. Inhibition of p38 and PKC was also non-conducive for porcine iPSCs (Extended Data Fig. 1f and 1h). Therefore mouse or human naïve ESC conditions5–7 cannot be directly extrapolated to porcine cells. The inhibitors for Mek1/2, p38 and PKC were therefore excluded from the screen. Several conditions were identified that met the screen criteria (Extended Data Fig. 1g), including a minimal requisite condition (#517, porcine EPSC medium: pEPSCM) comprising inhibitors for GSK3 (CHIR99021), SRC (WH-4-023) and Tankyrases (XAV939) (the last two were inhibitors important for mouse EPSCs1), and supplements: Vitamin C (Vc), ACTIVIN A and LIF (Fig. 1i. and Extended Data Fig. 1g and Supplementary Table 1). Under these conditions, the Dox-independent iPSCs (pEPSCiPS) remained undifferentiated in 30 passages, expressed endogenous pluripotency factors at levels comparable to the porcine blastocyst and showed no leaky expression of the exogenous reprogramming factors (Fig. 1j, and Extended Data Fig. 1i-j).
We next repeated the reprogramming experiment by directly culturing the primary colonies in pEPSCM (Extended Data Fig. 2a), and generated 11 stable pEPSCiPS lines from 16 primary colonies (70% efficiency) with six of them having no detectable expression of any of the eight exogenous reprogramming factors but high levels of endogenous pluripotency genes (Extended Data Fig. 2b).
Establishment of porcine EPSCs from preimplantation embryos
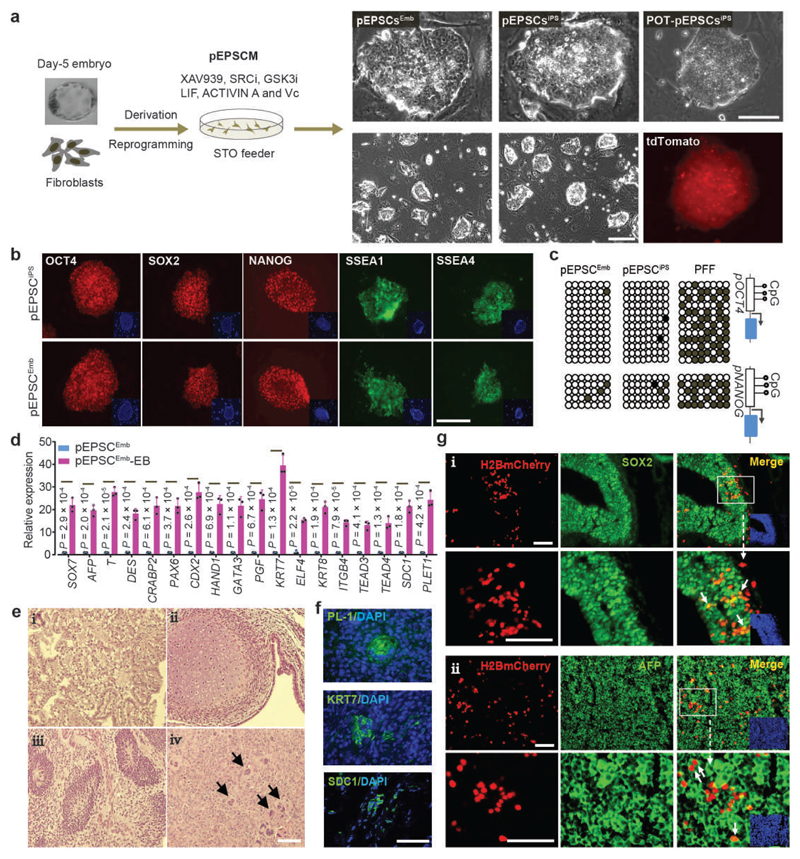
The pEPSCM condition was subsequently employed to derive stem cell lines from porcine preimplantation embryos. A total of 26 lines (pEPSCsEmb, 14 male and 12 female) were established from 76 early blastocysts (5.0 dpc), and 12 cell lines (pEPSCspar) from 252 parthenogenetic blastocysts (Fig. 2a, Supplementary Table 2 and Extended Data Fig. 2c). Similar to pEPSCsiPS, pEPSCsEmb had high nuclear/cytoplasmic ratios, and formed compact colonies with smooth colony edges (Fig. 2a, Extended Data Fig. 2d). pEPSCsEmb were passaged every 3-4 days at 1:8 ratio as single cells, could be maintained for >40 passages on STO feeders without overt differentiation and were genetically stable (Extended Data Fig. 2e). Subcloning efficiency was about 10% at low cell density (2,000 cells per well in a 6-well plate), but routine passaging was performed at high cell density.
Figure 2. Derivation of porcine EPSCs.
a. Left: Schematic diagram of establishment of the pig (Sus Scrofa) EPSCEmb lines from German Landrace day-5 in vivo derived blastocysts on STO feeder cells in pEPSCM, and of pEPSCiPS lines by reprogramming German Landrace PFFs and China TAIHU OCT4-Tdtomato knock-in reporter (POT) PFFs. Right panels: images of established EPSC lines, and a fluorescence image of Td-tomato expression in POT-pEPSCiPS. Three EPSCEmb lines (Male: K3 and K5; Female K1) and two pEPSCiPS lines (#10, #11) were extensively tested in this study. These EPSC lines behaved similarly in gene expression and differentiation. b. Immunostaining detection of pluripotency factors and markers, SSEA-1 and SSEA-4, in pEPSCEmb and pEPSCiPS. c. Bisulphite sequencing analysis of CpG sites in the OCT4 and NANOG promoter regions in PFFs, pEPSCiPS and pEPSCEmb. d. Gene expression in embryoid bodies (EBs, day 7) of pEPSCsEmb. Expression of genes of embryonic and extra-embryonic cell lineages were assessed by RT-qPCR. Relative expression levels were normalized against GAPDH. n=3 independent experiments. Data are mean ± s.d. P values were calculated using a two-tailed t-test. Statistical source data are provided in Supplementary table 10. e. Tissue composition of pEPSCEmb teratoma sections (H&E staining): Examples of glandular epithelium derived from endoderm (i), cartilage derived from mesoderm (ii), immature neural tissue derived from ectoderm, which forms neuroepithelial structures (iii), and large multinucleated cells reminiscent of trophoblasts (arrows in iv). f. PL-1, KRT7 and SDC1 positive cells in pEPSCEmb teratoma sections as revealed by immunostaining. g. Detection of pEPSC descendants in the brain (H2BmCherry+SOX2+) and the liver (H2BmCherry+AFP+) in chimera #16. H2B-mCherry and SOX2 are nuclear localised whereas AFP is a cytoplasmic protein. Boxed areas are shown in higher magnification. Arrows indicate representative cells that were donor cell descendants (mCherry+). DAPI stained nuclei. Additional chimera analyses are presented in Extended Data Fig. 3e-3f. For a-b and e-g, the experiments were repeated independently three times with similar results. Scale bars, 100 μm.
Pluripotency genes were expressed in pEPSCsEmb and pEPSCsiPS at levels comparable to the blastocysts (Fig. 2b and Extended Data Fig. 2b), but were drastically reduced or lost when pEPSCs were cultured in other porcine ESC media previously reported 9–15 (Extended Data Fig. 2f-g). pEPSCs showed extensive DNA demethylation at the OCT4 and NANOG promoter regions (Fig. 2c) and had OCT4 distal enhancer activity (Extended Data Fig. 2h). pEPSCs were amenable for Crispr/Cas9-mediated insertion of an H2B-mCherry expression cassette into the ROSA26 locus (Extended Data Fig. 2i and 2j). In vitro, pEPSCs differentiated to tissues expressing genes representative of the three germ layers and, uniquely, trophoblast genes (Fig. 2d, Extended Data Fig. 2k). In immunocompromised mice, pEPSCsEmb formed mature teratomas with derivatives of the three germ layers, and contained placental lactogen-1 (PL-1)-, KRT7- and SDC1-positive trophoblast-like cells (Fig. 2e-2f). Following incorporation of the pEPSCs into preimplantation embryos and after 48 hours of culture, pEPSCs (marked by H2B-mCherry) had colonized both the trophectoderm and inner cell mass of blastocysts (Extended Data Fig. 3a). Following transfer of the chimeric blastocysts to synchronized recipient sows, a total of 45 conceptuses were harvested from 3 litters at days 26-28 of gestation (Supplementary Table 3, Extended Data Fig. 3b). Flow cytometry analysis of dissociated cells from embryonic and extraembryonic tissues of the chimeras detected mCherry+ cells in 7 conceptuses (Extended Data Fig. 3c, Supplementary Table 4 and Table 5): mCherry+ cells in both the placenta and embryonic tissues in 2 chimeras (#8 and #16); only in embryonic tissues in 3 chimeras (#4, #21 and #34); and exclusively in the placenta of 2 chimeras (#3 and #6). Genomic DNA PCR assays detected mCherry DNA only in those seven mCherry+ chimeras, but not in any other conceptuses (Extended Data Fig. 3d, Supplementary Table 4 and 5). Despite the low contribution of the donor mCherry+ cells, their descendants were found in multiple embryonic tissues and organs that were identified by tissue lineage markers (Fig. 2g and Extended Data Fig. 3e-f). Therefore, pEPSCsEmb and pEPSCsiPS, like mEPSCs, possess an expanded developmental potential for both the embryonic cell lineages and extra-embryonic trophoblast lineages.
Derivation of PGC-like cells (PGCLCs) from pEPSCsEmb
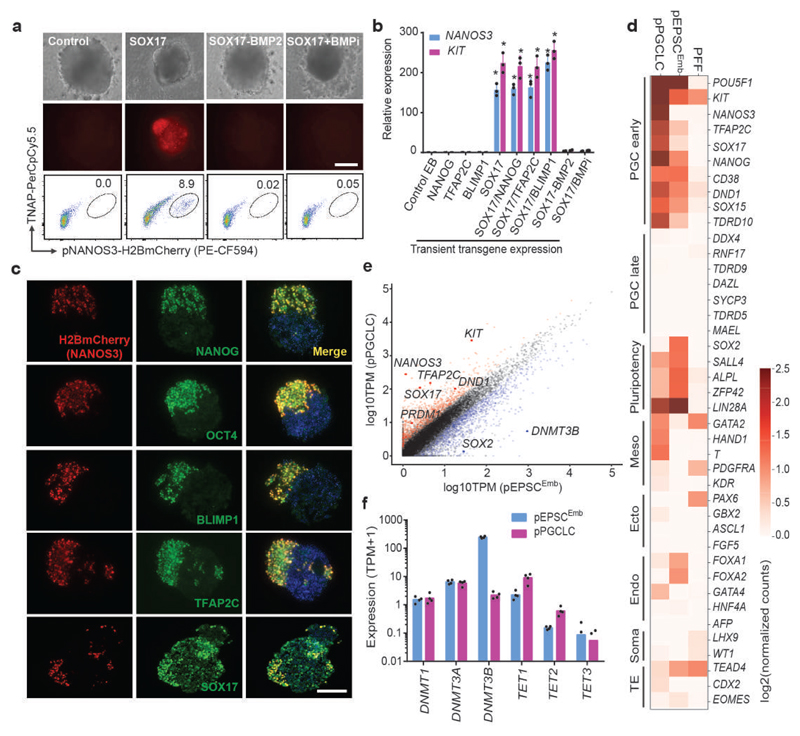
We next asked if pEPSCs had the potential to produce PGCLCs in vitro, similar to mouse and human pluripotent stem cells26–28. In early-primitive streak (PS)-stage porcine embryos (E11.5–E12), the first cluster of porcine PGCs can be detected as SOX17+ cells in the posterior end of the nascent primitive streak, and these cells later co-express OCT4, NANOG, BLIMP1 and TFAP2C28. NANOS3 is an evolutionarily conserved PGC-specific factor29,30 and human NANOS3 reporter ESCs have been used for studying the derivation of PGCLCs27,28. We generated and used a NANOS3-H2B-mCherry pEPSCEmb reporter line to facilitate identification of putative PGCLCs (Extended Data Fig. 4a). After expressing the SOX17 transgene transiently for 12 hours, the reporter cells were allowed to form embryoid bodies (EBs) (Extended Data Fig. 4b), where cell clusters co-expressing NANOS3 (mCherry+) and tissue-nonspecific alkaline phosphatase (TNAP, a PGC marker) were detected within 3-4 days (Fig. 3a).
Figure 3. In vitro generation of PGC-like cells from pEPSCsEmb.
a. Induction of pPGCLC by transiently expressing SOX17 in NANOS3-H2BmCherry reporter pEPSCs. The presence of H2BmCherry+TNAP+ cells in embryoid bodies (EBs) was analysed by FACS. The experiments were repeated independently three times with similar results. b. RT-qPCR analysis of PGC genes in day 3 EBs following pPGCLC induction. Relative expression levels werenormalized against GAPDH. n=3 independent experiments. Data are mean ± s.d. P values were calculated using a two-tailed t-test. Statistical source data are provided in Supplementary table 10. c. Immunofluorescence analysis of PGC factors in the sections of EBs at day 3-4 following pPGCLC induction. The H2BmCherry+ cells co-expressed NANOG, OCT4, BLIMP1, TFAP2C and SOX17. DAPI stained nuclei. Experiments were performed three times. d. RNAseq analysis (Heat map) of sorted H2BmCherry+ of pPGCLC induction shows expression of genes associated with PGCs, pluripotency or somatic lineages (mesoderm, endoderm, and gonadal somatic cells). e. Pair-wise gene expression comparison between pEPSCsEmb and pPGCLCs. Key up-regulated (red) and down-regulated (blue) genes are highlighted. f. Bar plot shows expression of genes related to DNA methylation in pPGCLCs and the parental pEPSCsEmb. Data were from RNAseq of sorted H2BmCherry+ of pPGCLC induction. Scale bars, 100 μm.
The derivation of putative porcine PGCLCs was BMP2/4 dependent (Fig. 3a). Interestingly, different from the reported derivation of human PGCLCs28, expressing NANOG, BLIMP1 or TFAP2C transgenes, either individually or in combination, had no effect on the preponderance of NANOS3+ cells (Extended Data Fig. 4c), whereas co-expression of SOX17 with BLIMP1 appeared to increase NANOS3+ cells (Extended Data Fig. 4c and 4d).
The putative PGCLCs within the EBs expressed PGC-specific genes (Fig. 3b-c, and Extended Data Fig. 4e). Specific RNA-seq analysis of NANOS3+ cells revealed expression of early PGC genes27 (OCT4, NANOG, LIN28A, TFAP2C, CD38, DND1, NANOS3, ITGB3, SOX15 and KIT), and reduced SOX2 expression (Fig. 3d-e). Similar to PGCLC derivation from human ESCs27, DNMT3B was down-regulated in porcine mCherry+/NANOS3+ cells whereas TET1/2 were up-regulated, relative to the parental pEPSCsEmb (Fig. 3e-f).
Establishment of human EPSCs under conditions similar to porcine EPSCs
The finding that inhibition of SRC and Tankyrases is sufficient to convert mouse ESCs to mEPSCs1 and that the same two inhibitors are required for the derivation of pEPSCs raises the possibility that similar in vitro culture conditions may be developed for additional mammalian species. To explore this possibility, we cultured four established human ES cell (hESC) lines (H1, H9, Man1/M1, and Man10/M10 cells)4,31,32 in pEPSCM and passaged them three times. The cells displayed diverse morphologies and heterogeneous expression of OCT4 (Extended Data Fig. 5a).
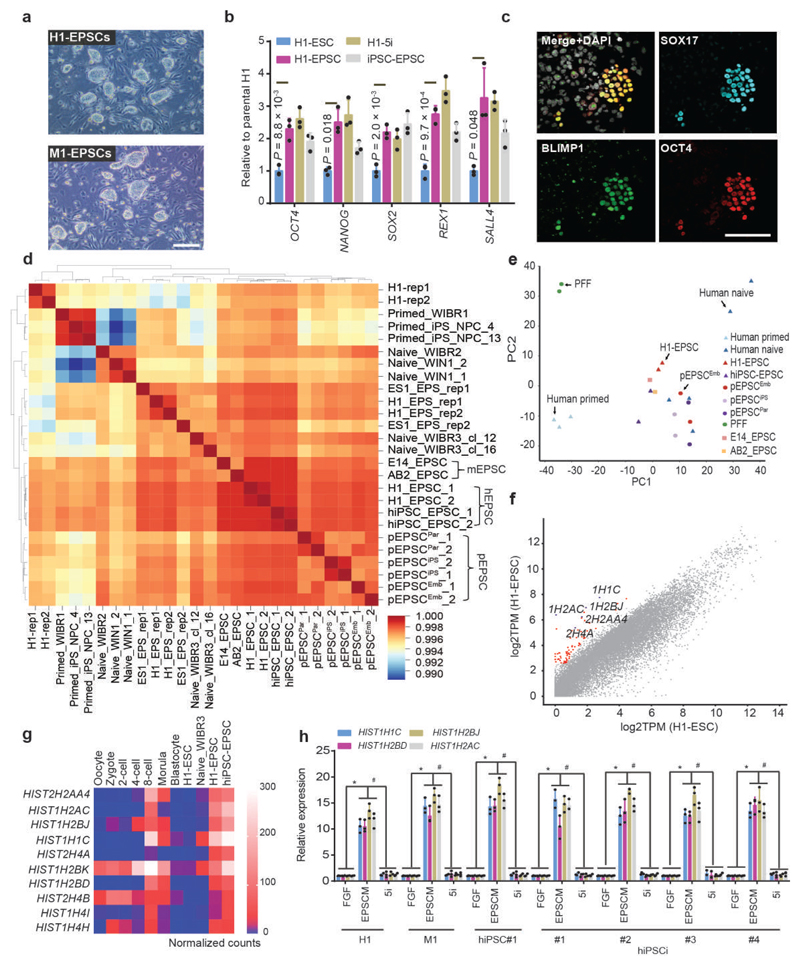
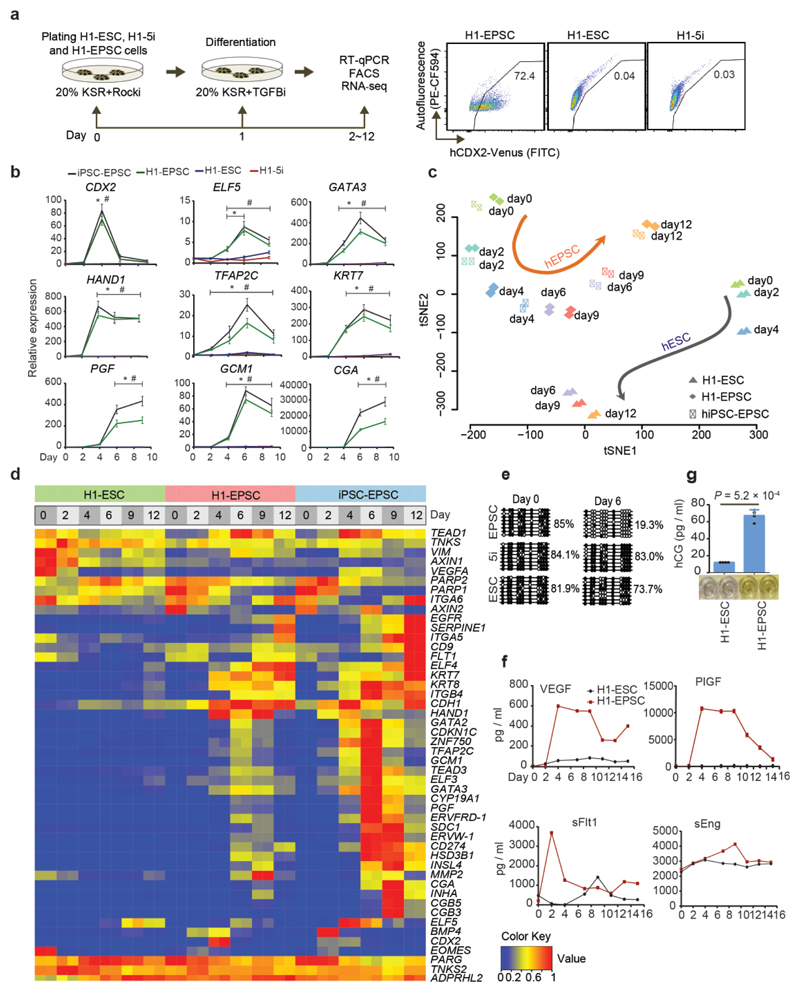
Removing ACTIVIN A (20.0 ng/ml) from pEPSCM led to considerably fewer cell colonies formed from H1 (<1.0%) and M1 (5.0%) ESCs, while none from H9 or M10 (Extended Data Fig. 5a), which reflects the inherent between-line heterogeneity of human ESCs33,34. With further refinement of the culture conditions (for example, replacing WH-4-023 with another SRC inhibitor A419259 in hEPSCM, see Methods), morphologically homogenous and stable cell lines were established from single-cell sub-cloned H1 (H1-EPSCs) and M1 cells (M1-EPSCs) (Fig. 4a). Karyotype analysis of H1 and M1 cells grown in hEPSCM on STO feeders revealed genetic stability (at passage 25 post conversion from the parental hESCs, Extended Data Fig. 5b). When human primary iPSC colonies reprogrammed from fibroblasts were directly cultured in hEPSCM, around 70% of the picked colonies could be established as stable iPSC lines (iPSC-EPSCs) (Extended Data Fig. 5c), which expressed pluripotency markers with no obvious leakiness of the exogenous reprogramming factors in about half of the lines (Fig. 4b and Extended Data Fig. 5d). The H1-EPSCs proliferated more robustly than the H1 ESCs cultured in standard FGF-containing medium (H1-ESC, primed) or under naïve 5i/L/A conditions (H1-naïve ESC)7 (Extended Data Fig. 5e), and were tolerant of single cell passaging with about 10% single cell sub-cloning efficiency in the transient presence of ROCKi. Cell survival at passaging was substantially improved in the presence of 5.0ng/ml ACTIVIN A or by splitting the cells at higher density. Human EPSCs expressed pluripotency genes at higher levels than the H1-ESCs (Fig. 4b) and minimal levels of lineage markers (Extended Data Fig. 5f). Expression of core pluripotency factors and surface markers in human EPSCs was confirmed by immunostaining (Extended Data Fig. 5g). H1-EPSCs differentiated to derivatives of the three germ layers in vitro and in vivo (Extended Data Fig. 5h-i). Moreover, H1-EPSCs were successfully differentiated to PGCLCs using in vitro conditions developed for germ cell competent hESCs or iPSCs27,28 (Fig. 4c and Extended Data Fig. 5j).
Figure 4. Establishment of human EPSCs.
a. Images of the established H1-EPSCs or M1-EPSCs (passage 25). The experiments were repeated independently three times with similar results. b. Expression of pluripotency genes in H1-ESCs, H1-naïve ESCs (5i), H1-EPSCs and iPSC-EPSCs. Data are mean ± s.d. (n = 3). P values were computed for two-tailed t-test. c. EBs of H1-EPSCs to PGCLCs immunostained for SOX17, BLIMP1 and OCT4. Scale bar: 100 μm. d. Hierarchical clustering of gene expression (bulk RNAseq) of EPSCs, and other human pluripotent stem cells. Correlation matrix was clustered using Spearman correlation and complete linkage. Data sets are: pEPSCPar: porcine parthenogenetic EPSCs. E14 and AB2-EPSCs: mouse EPSCs (ref. 1); Human primed ESCs (WIBR1, iPS_NPC_4 and iPS_NPC_13) and naïve ESCs (WIBR2, WIBR3_cl_12, WIBR3_cl_16, WIN1_1 and WIN1_2) (Ref. 7 and 35); Human primed H1 ES cell (H1-rep1 and H1-rep2) and extended pluripotent stem (EPS) cells (H1_EPS_rep1, H1_EPS_rep2, ES1_EPS_rep1 and ES1_EPS_rep2) (ref. 36). e. Principal component analysis (PCA) of bulk RNA-seq data of EPSCs, human primed and naïve ESCs, and PFFs. Human naïve (n=5), human primed (n=3), H1-EPSC (n=2), hiPSC-EPSC (n=2), pEPSCEmb (n=2), pEPSCiPS (n=2), pEPSCPar (n=2), PFF (n=2), E14_EPSC (n=2) and AB2_EPSC (n=1). n= biologically independent experiments. f. Pair-wise comparison of gene expression between H1-ESCs and H1-EPSCs, showing the highly expressed genes (>8 folds) in hEPSCs (total 76, red dots) and representative histone genes (blue dots). g. Heatmap showing expression of selected histone genes in human ESCs, EPSCs and preimplantation embryos. RNAseq data of human ESCs were from ref. 35, whereas embryo cell data were from ref. 37. h. RT-qPCR analysis of four histone 1 cluster genes in seven human ESC or iPSC lines cultured under three conditions. hiPSC lines were from the HIPSC project (http://www.hipsci.org): #1, HPSI1113i-bima_1; #2, HPSI1113i-qolg_3; #3, HPSI1113i-oaaz_2; #4, HPSI1113i-uofv_1. Relative expression levels are shown with normalization to GAPDH. n = 3 independent experiments. Data are mean ± s.d. *P <0.01 compared with the FGF condition cultured cells. #P <0.01 compared with 5i condition cultured cells. Experiments were performed three times. Statistical source data and precise P values are provided in Supplementary table 10.
Our results demonstrate that porcine and human EPSCs could be derived and maintained using the similar set of small molecule inhibitors. Global gene expression profiling revealed that pEPSCs and hEPSCs were clustered together, and were distinct from PFFs or other human pluripotent stem cells1,35,36 (Fig. 4d-e). Both porcine and human EPSCs expressed high levels of key pluripotency genes, low levels of somatic cell lineage genes, PAX6, T, GATA4 and SOX7, or placenta-related genes (PGF, TFAP2C, EGFR, SDC1 and ITGA5) (Extended Data Fig. 6a-d). Consistent with the high levels of global DNA methylation of pEPSCs and hEPSCs (Extended Data Fig. 6e), DNA methyltransferase genes DNMT1 and DNMT3A and DNMT3B were highly expressed, whereas TET1, TET2 and TET3 were expressed at lower levels (Extended Data Fig. 6f-g). Among the highly expressed 76 genes (>8-fold increase) in H1-EPSC in comparison to H1-ESCs, 17 genes encode histone variants with 15 belonging to the histone cluster 1 (Fig. 4f and Supplementary Table 6). nterestingly, these histone genes were expressed at low levels in 5i and primed human ESCs but were highly expressed in human 8-cell and morula stage embryos (Fig. 4g). The significantly higher expression of these histone genes was confirmed in additional hEPSC lines (Fig. 4h). The biological significance of this observation remains to be investigated.
Single cell RNA-seq (scRNAseq) reveals substantially homogenous EPSC cultures
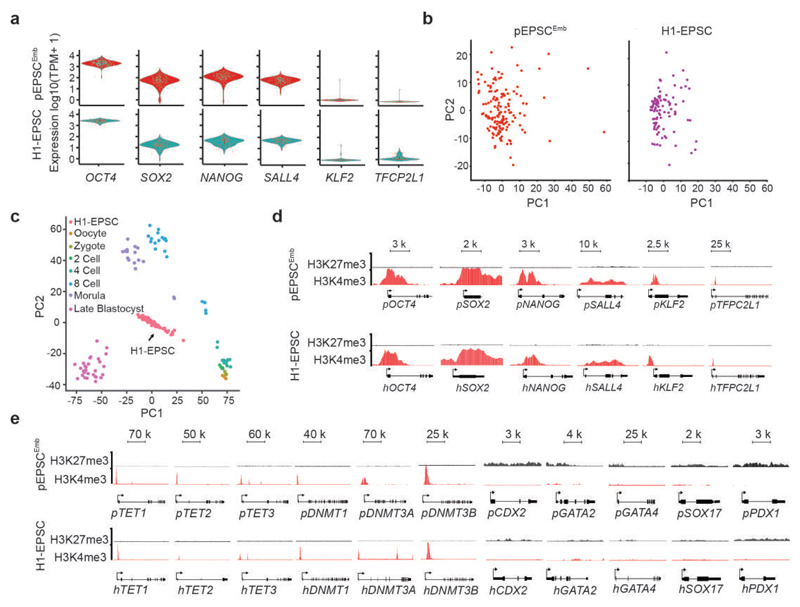
EPSCs expressed uniform levels of the core pluripotency factors (Fig. 5a) and were generally homogenous cells in culture in the context of single-cell transcriptome (Fig. 5b). Mouse EPSCs had enriched transcriptomic features of 4-cell to 8-cell blastomeres1. scRNAseq analysis suggested that hEPSCs were transcriptionally, as well as the histone gene expression profiles, more similar to human 8-cell to morula stage embryos37,38 than other stages of preimplantation embryos (Fig. 5c, and Extended Data Figure 6h; Fig. 4g-hand Extended Data Fig. 6i). Interestingly, single-cell transcriptome also revealed low expression of naïve pluripotency factors such as KLF2 in EPSCs (Fig. 5a and Extended Data Fig. 6a-b), which is also not highly expressed in human early preimplantation embryos39. Although KLF2, TET1, TET2 and TET3 were weakly expressed in both pEPSCs and hEPSCs (Extended Data Fig. 6a-b and 6f-g), their promoter regions were characterized by active H3K4m3 histone marks (Fig. 5d-e). In contrast to pluripotency genes, in both porcine and human EPSCs, the cell lineage gene loci (e.g. CDX2, GATA2, GATA4, SOX7 and PDX1) had high H3K27me3 and low H3K4me3 marks, respectively (Fig. 5e).
Figure 5. Molecular features of porcine and human EPSC.
a. Violin plots show the distribution and the probability density the scRNAseq expression of pluripotency genes in pEPSCsEmb (top panel, n=150) and human H1-EPSCs (lower panel, n=96). n represents the number of cells in each plot. b. PCA of global gene expression pattern (by scRNAseq) of pEPSCsEmb (left panel, n=150) and H1-EPSCs (right panel, n=96). n represents the number of cells in each plot. c. PCA and comparison of gene expression assessed by scRNAseq of human H1-EPSCs and human preimplantation embryos (ref. 39). H1-EPSCs (n=96), oocyte (n=3), zygote (n=3), 2 cell (n=6), 4 cell (n=12), 8 cell (n=20), morulae (n=16), Late blastocyst (n=30). d. ChIP-seq analysis of H3K27me3 and H3K4me3 marks at pluripotency gene loci in pEPSCsEmb and human H1-EPSCs. e. Histone modifications (H3K4me3 and H3K27me3) at the loci for genes encoding enzymes involved in DNA methylation and demethylation and for cell lineage genes. For d and e, experiments were performed three times with similar results.
Human and porcine EPSCs have similar signalling requirements
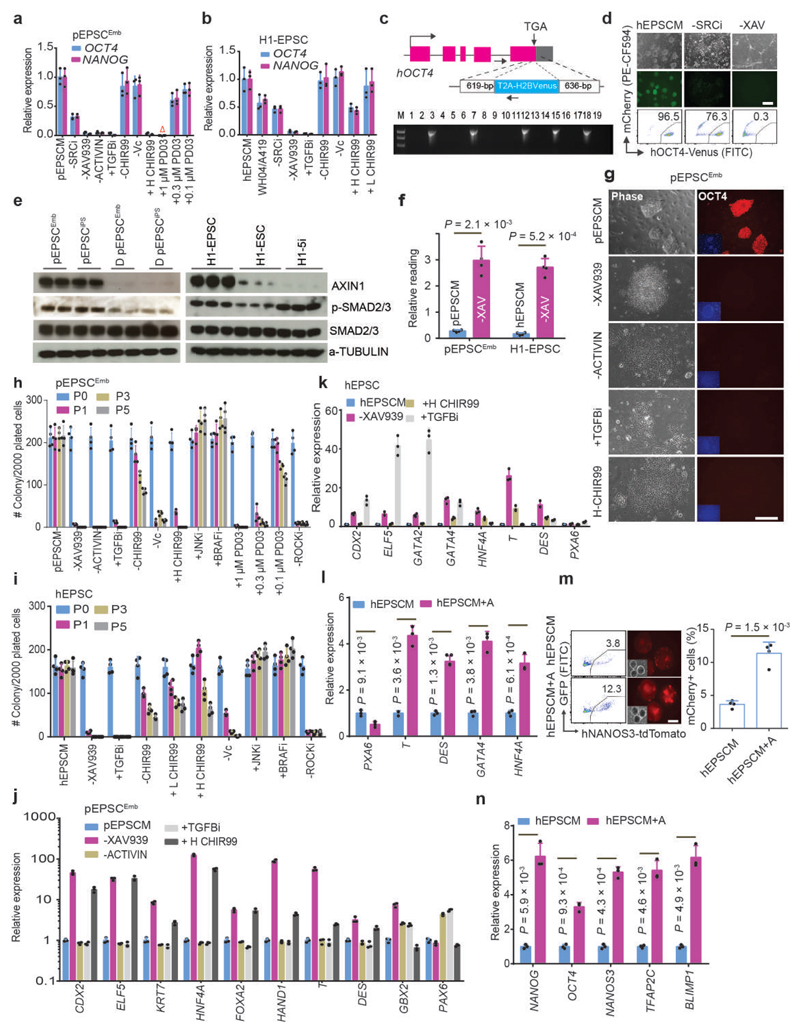
To identify signalling requirements in EPSCs, we removed individual components from the culture medium. Removal of the SRC inhibitor WH-4-023 or A419259 reduced expression of pluripotency factors in both EPSCs (Fig. 6a-d). Notably, in hEPSCs, using the SRC inhibitor WH-4-023 instead of A419259 led to lower pluripotency gene expression (Fig. 6b). Similar to mEPSCs, XAV939 enhanced AXIN1 protein content (Fig. 6e), and reduced canonical WNT activities in both EPSCs (Fig. 6f). Withdrawal of XAV939 caused collapse of pluripotency and differentiation of these EPSCs (Fig. 6a-b, 6d, and 6g-k). SMAD2/3 were phosphorylated in EPSCs (Fig. 6e). Either removing ACTIVIN A from pEPSCM or adding the TGFβ inhibitor SB431542 resulted in massive cell loss and down-regulation of pluripotency factors in pEPSCs (Fig. 6a, 6g, 6h and 6j). hEPSCs did not require exogenous TGFB in culture but inhibiting TGFβ induced rapid cell differentiation with preferential expression of trophoblast genes CDX2, ELF5 and GATA2 (Fig. 6b, 6i and 6k). At a relatively low concentration of ACTIVIN A (5.0ng/ml), hEPSCs showed a stronger propensity for embryonic mesendoderm lineage differentiation (Fig. 6l) and generated more NANOS3-tdTomato+ PGCLCs (Fig. 6m-n). Removing CHIR99021 and Vitamin C from EPSCM did not affect pluripotency gene expression but reduced the number of colonies formed from single cells (Fig. 6a-b and 6h-i), whereas a high CHIR99021 concentration (3.0μM) induced differentiation of both EPSCs (Fig. 6a, 6h and 6j), similar to that in human or rat naïve cells6,40. JNK and BRAF inhibition might improve culture efficiency but was not essential (Fig. 6h-i). Mouse naïve ESCs is cultured 1.0μM Mek1/2 inhibitor PD0325901 (Ref. 5). We noticed that even 0.1 μM PD0325901 decreased pEPSC survival as measured by colony formation in serial passaging (Fig. 6h).
Figure 6. The requirement of individual components in EPSCM.
a-b. Gene expression in pEPSCsEmb (a) and H1-EPSCs (b) analysed by RT-qPCR. “-SRCi, -XAV939, -ACTIVIN, -Vc, -CHIR99”: removing individually; “+TGFBi, +H-CHIR99, +PD03”: adding SB431542, 3.0 μM CHIR99021, or MEK1/2 inhibitor PD0325901, respectively. “WH04/A419”: replacing A419259 with another SRC inhibitor, “WH-4-23. +L-CHIR99”: 0.2 μM in hEPSCM. Porcine and human EPSC media contain 0.2 μM and 1.0 μM CHIR99021, respectively. Red triangle: no cell survived. c. Targeting the H2B-Venus cassette to the OCT4 last coding exon in H1-EPSCs with the stop codon TGA being deleted. Five of 19 colonies genotyped were correctly targeted. d. The effects of removing WH-4-023 (-SRCi) or XAV939 (-XAV) for 7 days assessed by Venus+ reporter by fluorescence microscopy and flow cytometry. e. Western blot analysis of AXIN1 and phosphorylation of SMAD2/3 in EPSCs. EPSCs had higher levels of AXIN1 and pSMAD2/3 (for TGFβ signalling) than the differentiated (D) EPSCEmb or primed H1-ESCs. f. TOPflash analysis for canonical Wnt signalling activity in EPSCs. Removing XAV939 (pEPSCM-X and hEPSCM-X) for 5 days increased TOPflash activity. g. Bright-field and immunofluorescence images showing pEPSCsEmb cultured with the indicated changes in medium component. Cells were stained for OCT4 and DAPI. h-i. Quantitation of AP+ colonies formed from 2,000 pEPSCsEmb (h) or H1-EPSCs (i) cultured on STO feeders with different medium components. The colonies were scored for 5 consecutive passages. -ROCKi: passaging EPSCs without the ROCK inhibitor Y27632. j-k. RT-qPCR analysis of the expression of lineage genes in pEPSCsEmb (j) or hEPSCs (k) following removal of XAV939 or ACTIVIN A, inhibition of TGFβ signalling by SB431542, or treatment with 3.0 μM CHIR99021. l. The effects of supplementing 5.0 ng/ml ACTIVIN A on gene expression in EBs generated from H1-EPSCs. m-n. Effects of 5.0ng/ml ACTIVIN A on PGCLC (Tdtomato+) production from the NANOS3-Tdtomato reporter EPSCs assessed by FACS (m) and RT-qPCR (n). Relative expression levels were normalized to GAPDH. All graphs represent the mean ± s.d. For a-b, j-l and n, n = 3 independent experiments. For f, h-i and m, n = 4 independent experiments. For d-e and g, the experiments were repeated independently three times with similar results. P values were computed by two-tailed t-test. Statistical source data are presented in Supplementary table 10. Scale bars: 100 μm.
hEPSCs have potent potential to trophoblasts
We further investigated differentiation of hEPSCs to trophoblasts by generating the CDX2-Venus reporter line (Extended Data Fig. 7a). Inhibiting TGFβ by SB431542 resulted in ~70% of the reporter cells being CDX2-Venus+ (Fig. 7a), whereas essentially no CDX2-Venus+ cells were detected if the reporter cells were previously cultured in FGF or under the 5i naïve ESC conditions. Consistently, trophoblast gene levels were rapidly increased in differentiated H1-EPSCs and iPSC-EPSCs but not in H1-ESCs or H1-5i naïve cells (Fig. 7b). Addition of BMP4, which promotes differentiation of human ESCs to putative trophoblasts41, induced a much higher level of expression of trophoblast genes in EPSCs than in H1-ESCs or H1-5i naïve ESCs (Extended Data Fig. 7b). Inhibiting FGF and TGFβ signalling while in parallel activating BMP4 was reported to effectively induce trophoblast differentiation of human ESCs42,43. Under these conditions, expression of trophoblast genes, especially the late trophoblast genes GCM1, CGA and CGB, was much higher in H1-EPSCs than in H1-ESCs, whereas naïve 5i hESCs displayed no sign of trophoblast differentiation (Extended Data Fig. 7c). Global gene expression analysis demonstrated that under TGFβ signalling inhibition H1-EPSCs and iPSC-EPSCs followed a differentiation trajectory distinct from that of H1-ESCs (Fig. 7c), and that in cells differentiated from EPSCs, but not from H1-ESCs, genes associated with trophoblast development or function were highly expressed including: (1) BMP4 (Day 2-4); (2) Syncytin-1 (ERVW-1) and Syncytin-2 (ERVFRD-1) that promote cytotrophoblast fusion into syncytiotrophoblast; (3) p57 (encoded by CDKN1C) 44,45; (4) CD274 (encoding PD-L1 or B7-H1); and (5) EGFR46 (Fig. 7d).
Figure 7. Trophoblast differentiation potential of hEPSCs.
a. Left panel: differentiation of hEPSCs to trophoblasts under TGFβ inhibition. Right panel: flow cytometry analysis of trophoblast differentiation of CDX2-H2B-Venus reporter EPSCs, collected 4 days after TGFβ inhibition. The CDX2-H2B-Venus reporter EPSCs were also cultured in conventional FGF-containing hESCs medium or 5i-naïve medium and differentiated under TGFβ inhibition and examined by flow cytometry. The experiments were repeated independently three times with similar results. b. The dynamic changes in the expression of trophoblast genes during hEPSC differentiation (sampled at 2-day intervals for 12days) were assayed by RT-qPCR. Relative expression levels were normalized against GAPDH. n = 3 independent experiments. Data are mean ± s.d. *P <0.01 compared with H1-ESC cells. #P <0.01 compared with H1-5i cells. The precise P values are presented in Supplementary table 10. c. tSNE analysis of RNA-seq data of the differentiating human ESCs (n = 2) and iPSC-EPSCs (n = 4) treated with TGFβ inhibitor SB431542. RNAs were sampled from cells at Day 0-12 of differentiation. The of H1-EPSCs and hiPSC-EPSCs showed different trajectory of differentiation from H1-ESCs. d. Heatmap shows changes in the expression of trophoblast-specific genes in differentiating H1-ESCs (green), H1-EPSCs (red) and iPSC-EPSCs (blue) collected at several time points of culture for RNAseq analysis. e. DNA demethylation at the promoter region of the ELF5 locus in differentiating H1-EPSCs and other cell types following 6 days of SB431542 treatment. Cells from H1-ESCs, H1-naïve ESCs (5i) showed no discernible DNA demethylation at the ELF5 promoter. f. Secreted hormones from trophoblasts derived from H1-EPSCs induced by TGFβ inhibition (SB431542). VEGF, PLGF, sFlt-1and sEng were measured in the conditioned media for culturing the differentiating EPSCs or ESCs for 16 days following a 48-h SB431542 treatment. g. hCG produced by trophoblasts from SB431542-treated EPSCs or ESCs at day 10 of differentiation, measured by ELISA. n = 4 independent experiments. Data are mean ± s.d. P values were calculated using a two-tailed t-test. Statistical source data are presented in Supplementary table 10.
We next performed Pearson correlation coefficient analysis of the transcriptome of cells differentiated under TGFβ inhibition with published reference data of primary human trophoblasts (PHTs) and human placenta tissues43, which again revealed the similarity between cells differentiated from hEPSCs and PHTs and the placenta (Extended Data Fig. 7d). The differentiated cells from H1-EPSCs expressed human trophoblast specific miRNAs (C19MC miRNAs: hsa-miR-525-3p, hsa-miR-526b-3p, hsa-miR-517-5p, and hsa-miR-517b-3p)47 (Extended Data Fig. 7e-f), displayed DNA demethylation at the ELF5 locus48,49 (Fig. 7e), and produced abundant amounts of placental hormones (Fig. 7f-g).
One key mechanism for derivation and maintenance of EPSCs of mouse, porcine and human is blocking poly(ADP-ribosyl)ation activities of PARP family members TNKS1/2 using small molecule inhibitors such as XAV93950,51. In human cells, poly(ADP-ribose) in proteins is removed by poly(ADP-ribose) glycohydrolase (PARG) and ADP-ribosylhydrolase 3 (ARH3)52. Genetic inactivation of Parp1/2 and TNKS1/2 in the mouse caused trophoblast phenotypes53, whereas inactivating Parg led to loss of functional trophectoderm and trophoblast stem cell (TSCs)54. In hEPSCs, PARG-deficiency did not appear to cause noticeable changes in EPSCs but adversely affected trophoblast differentiation (Extended Data Fig. 7g-j), which may indicate an evolutionally conserved mechanism for EPSCs and trophoblast development between mouse and human.
Derivation of TSC-like cells from human and porcine EPSCs
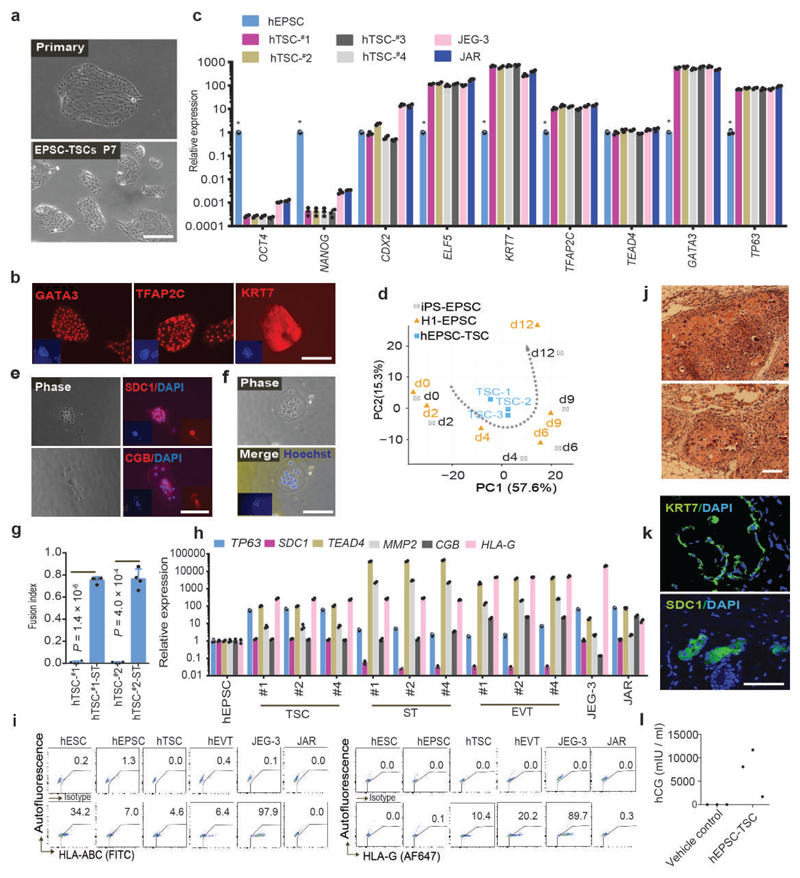
When hEPSCs (ESC-converted-EPSCs and iPSC-EPSCs) were cultured in human TSC conditions46 with low cell density (2,000 cells/3.5cm dish), colonies with TSC morphology formed after 7-9 days (Fig. 8a). These colonies were picked and expanded into stable cell lines under TSC conditions with up to 30% efficiency in line establishment. These hEPSC-derived TSC-like cells were referred in this study as hTSCs. In contrast, no lines were established from human H1 or M1 ESCs under the hTSC condition, whether they were originally cultured under primed or naïve ESCs conditions. The hTSCs expressed trophoblast transcription regulators: GATA3 and TFAP2C but had down-regulated pluripotency genes (Fig. 8b-c), and showed enriched transcriptomic features of day-4 differentiated human EPSCs under TGFβ inhibition (Fig. 8d). Following the published protocols46, we were able to differentiate hTSCs to multi-nucleated syncytiotrophoblasts (ST) and HLA-G+ extravillous trophoblasts (EVT) (Fig. 8e-i). Once injected into immunocompromised mice, hTSCs formed lesions with cells positively stained for SDC1 and KRT7 (Fig. 8j-k). Additionally, high levels of hCG were detected in blood of the mice with hTSC-lesions but not in control mice injected with vehicle only (Fig.8l). Although neither EPSCs expressed high levels of placenta development-related genes (Extended Data Fig. 6c-d), both displayed enriched H3K4me3 at these loci (Extended Data Fig. 8a), clearly underpinning EPSCs’ trophoblast potency. Under hTSC conditions, stable TSC-like lines could also be derived from pEPSCsEmb (pTSCs). Extended Data Fig. 8b). pTSCs were similar to hTSCs in gene expression profiles and the propensity of lesion formation in immunocompromised mice (Extended Data Fig. 8c-f). When introduced into porcine preimplantation embryos, descendants of pTSCs were found in the trophectoderm and expressed GATA3 and CDX2 (Extended Data Fig. 8g). Our results therefore provide compelling evidence that human and porcine EPSCs possessed expanded potential that encompasses the trophoblast lineage.
Figure 8. Derivation of trophoblast stem cell-like cells from hEPSCs.
a. Phase-contrast images of primary TSC colonies formed from individual hEPSCs (left) and of TSCs at passage 7 (right). b. Expression of trophoblast markers GATA3, TFAP2C and KRT7 in EPSC-TSCs detected by immunostaining. Nuclei were stained with DAPI. Similar results were obtained with four independent EPSC-TSC lines. c. RT-qPCR analysis of pluripotency and TSC genes in four EPSC-derived TSC lines and their parental hEPSCs. JEG3 and JAR are trophoblast cell lines. n = 3 independent experiments. Data are mean ± s.d.. *p < 0.01 compared to TSCs. The precise P values are presented in Supplementary table 10. d. PCA of gene expression of hTSCs (n = 3) and of cells differentiated from human EPSCs (n = 2) under TGFβ inhibition at several time points. hTSCs showed enriched transcriptomic features of day-4 differentiated EPSCs. e. Immunostaining of SDC1 and CGB in hTSC-dervied syncytiotrophoblasts. DAPI stained the nucleus. f. Phase-contrast and Hoechst staining images of multinucleated hTSC-derived syncytiotrophoblasts. g. The fusion index of forming syncytiotrophoblasts from hTSCs calculated as the number of nuclei in syncytia/total number of nuclei. n = 4 independent experiments. Data are mean ± s.d. h. RT-qPCR analysis of trophoblast-specific genes in syncytiotrophoblast (ST) and extravillous trophoblast (EVT) derived from three hTSC lines. Expression levels were normalized against GAPDH. n = 3 independent experiments. Data are mean ± s.d. i. Flow cytometry detection of HLA-ABC and HLA-G in hESCs, hEPSCs, hTSCs, and hTSC-derived EVT cells (protocol of ref. 46). The choriocarcinoma cells JEG-3 and JAR represent the extravillous and villous trophoblast cells, respectively. JEG-3 cells expressed HLA-G, HLA-C and HLA-E, but not JAR cells. j. H&E staining of lesions formed by hTSCs engrafted subcutaneously in NOD-SCID mice. k. Confocal images of immunostaining for SDC1- or KRT7-positive cells in hTSC-derived lesions. DAPI stained the nucleus. l. Serum hCG levels in six NOD-SCID mice 7 days after hTSC engraftment (n = 3) or injection of vehicle only (n = 3). For a-b, e-f and h-l, the experiments were repeated independently three times with similar results. Statistical source data are presented in Supplementary table 10. Scale bars: 100 μm.
In conclusion, EPSCs of mouse, pig and human can now be established under similar in vitro culture conditions. These stem cells share common molecular features and possess expanded potency for both embryonic and extraembryonic cell lineages that are generally not seen in the conventional embryo-derived or induced pluripotent stem cells. EPSCs therefore represents a unique state of cellular potency. The successful generation of EPSCs produces new tools for investigation of embryonic development and opens new avenues for translational research in biotechnology, agriculture, and genomics and regenerative medicine.
Supplementary Material
Acknowledgements
We thank colleagues of the Wellcome Trust Sanger Institute core facilities for generous support (James Bussell, Yvette Hooks, N. Smerdon, B. L. Ng and J. Graham), and Professor Ashley Moffett for critical comments. We acknowledge the following funding and supports: The Wellcome Trust (grant numbers 098051 and 206194) to the Sanger Institute, and The University of Hong Kong internal funding (P. Liu); Wellcome Trust Clinical PhD Fellowship for Academic Clinicians (D.J.R.); PhD fellowship (Portuguese Foundation for Science and Technology, FCT (SFRH/BD/84964/2012) (L.A.), Marie Sklodowska-Curie Individual Fellowship (M.A.E.-M.); BBSRC (grant BB/K010867/1), Wellcome Trust (grant 095645/Z/11/Z), EU EpiGeneSys, and BLUEPRINT (W.R.); Chongqing Agriculture Development Grant (17407 for L.P.G., Z.H.L., and Y.H.; REBIRTH project No. 9.1, Hannover Medical School (MHH) (for H.N.); Shuguang Planning of Shanghai Municipal Education Commission (16SG14) and The National Key Research and Development Program (2017YFA0104500) (L. Lu); China Postdoctoral Science Foundation (grant 2017M622795) (D.C.); Shenzhen Municipal Government of China (DRC-SZ [2016] 884) (Z.S.); NHMRC of Australia (Senior Principal Research Fellowship, Grant 1110751 (P.P.L.T.); GRF of Hong Kong (17119117 and 17107915) and National Natural Science Foundation (81671579 for L. Lu; 31471398 for W.S.B.Y.; and U1804281 for Y. Zhang). The authors thank Dr. Bjoern Petersen for performing the surgical embryo transfers and staff of the experimental pig facility in Friedrich-Loeffler-Institut Mariensee for competent and enduring assistance.
Footnotes
Author contributions
X.G. developed the culture conditions for pEPSCs and hEPSCs and performed most of the experiments; MN performed the pig experiments and wrote the paper; DH was critically involved in all porcine experiments; SP provided some pig reprogramming factor genes; XC performed most of the informatics analysis and ST supported XC; DC, SW, JZho, JZhu and ZS analyzed RNAseq data of hEPSCs/ESCs to trophoblasts; ACHC, YLL and WSBY performed teratoma and TSC lesion at the HKU; and MEM and WR performed EPSC DNA methylation analysis; TK and AS helped XG on PGCLC analysis; AH and AA measured hormones in cells differentiated from hEPSCs; LC analysed teratoma sections; YF and FB karyotyped cells; DR, XW, LG, ZL, YH, TN, DW, DP, LLai, GL, DRyan, JY, LA, YY, SGX, YZ, LLu, ZX were involved in refining the culture conditions or intellectual inputs; SJK provided human M1 and M10 ESCs; PPLT provided intellectual inputs and edited the manuscript; HN conceptualized pig experiments, wrote paper, secured funding for the pig part of the experiments; PL conceived the pEPSC culture screen concept, supervised the research and wrote paper.
References
- 1.Yang J, et al. Establishment of mouse expanded potential stem cells. Nature. 2017;550:393–397. doi: 10.1038/nature24052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 3.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 5.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takashima Y, et al. Resetting Transcription Factor Control Circuitry toward Ground-State Pluripotency in Human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Theunissen TW, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell stem cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ezashi T, Yuan Y, Roberts RM. Pluripotent Stem Cells from Domesticated Mammals. Annu Rev Anim Biosci. 2016;4:223–253. doi: 10.1146/annurev-animal-021815-111202. [DOI] [PubMed] [Google Scholar]
- 9.Brevini TA, et al. Culture conditions and signalling networks promoting the establishment of cell lines from parthenogenetic and biparental pig embryos. Stem Cell Rev. 2010;6:484–495. doi: 10.1007/s12015-010-9153-2. [DOI] [PubMed] [Google Scholar]
- 10.Vassiliev I, et al. In vitro and in vivo characterization of putative porcine embryonic stem cells. Cell Reprogram. 2010;12:223–230. doi: 10.1089/cell.2009.0053. [DOI] [PubMed] [Google Scholar]
- 11.Haraguchi S, Kikuchi K, Nakai M, Tokunaga T. Establishment of self-renewing porcine embryonic stem cell-like cells by signal inhibition. J Reprod Dev. 2012;58:707–716. doi: 10.1262/jrd.2012-008. [DOI] [PubMed] [Google Scholar]
- 12.Park JK, et al. Primed pluripotent cell lines derived from various embryonic origins and somatic cells in pig. PLoS One. 2013;8:e52481. doi: 10.1371/journal.pone.0052481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou DR, et al. Derivation of Porcine Embryonic Stem-Like Cells from In Vitro-Produced Blastocyst-Stage Embryos. Sci Rep. 2016;6 doi: 10.1038/srep25838. 25838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue B, et al. Porcine Pluripotent Stem Cells Derived from IVF Embryos Contribute to Chimeric Development In Vivo. PLoS One. 2016;11:e0151737. doi: 10.1371/journal.pone.0151737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Y, Yu T, Cai Y, Wang H. Preserving self-renewal of porcine pluripotent stem cells in serum-free 3i culture condition and independent of LIF and b-FGF cytokines. Cell Death Discov. 2018;4:21. doi: 10.1038/s41420-017-0015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Ryan DJ, Lan G, Zou X, Liu P. In vitro establishment of expanded-potential stem cells from mouse pre-implantation embryos or embryonic stem cells. Nat Protoc. 2019 doi: 10.1038/s41596-018-0096-4. [DOI] [PubMed] [Google Scholar]
- 17.Esteban MA, et al. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J Biol Chem. 2009;284:17634–17640. doi: 10.1074/jbc.M109.008938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ezashi T, et al. Derivation of induced pluripotent stem cells from pig somatic cells. Proc Natl Acad Sci U S A. 2009;106:10993–10998. doi: 10.1073/pnas.0905284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.West FD, et al. Porcine induced pluripotent stem cells produce chimeric offspring. Stem Cells Dev. 2010;19:1211–1220. doi: 10.1089/scd.2009.0458. [DOI] [PubMed] [Google Scholar]
- 20.Petkov S, Glage S, Nowak-Imialek M, Niemann HL. Long-Term Culture of Porcine Induced Pluripotent Stem-Like Cells Under Feeder-Free Conditions in the Presence of Histone Deacetylase Inhibitors. Stem Cells Dev. 2016;25:386–394. doi: 10.1089/scd.2015.0317. [DOI] [PubMed] [Google Scholar]
- 21.Lai S, et al. Generation of Knock-In Pigs Carrying Oct4-tdTomato Reporter through CRISPR/Cas9-Mediated Genome Engineering. PLoS One. 2016;11:e0146562. doi: 10.1371/journal.pone.0146562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, et al. Pluripotent and Metabolic Features of Two Types of Porcine iPSCs Derived from Defined Mouse and Human ES Cell Culture Conditions. PLoS One. 2015;10:e0124562. doi: 10.1371/journal.pone.0124562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Telugu BP, Ezashi T, Roberts RM. Porcine induced pluripotent stem cells analogous to naive and primed embryonic stem cells of the mouse. Int J Dev Biol. 2010;54:1703–1711. doi: 10.1387/ijdb.103200bt. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du X, et al. Barriers for Deriving Transgene-Free Pig iPS Cells with Episomal Vectors. Stem Cells. 2015;33:3228–3238. doi: 10.1002/stem.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, et al. Erk signaling is indispensable for genomic stability and self-renewal of mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2015;112:E5936–E5943. doi: 10.1073/pnas.1516319112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 27.Irie N, et al. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi T, et al. Principles of early human development and germ cell program from conserved model systems. Nature. 2017;546:416–420. doi: 10.1038/nature22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Julaton VT, Reijo Pera RA. NANOS3 function in human germ cell development. Hum Mol Genet. 2011;20:2238–2250. doi: 10.1093/hmg/ddr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gkountela S, et al. The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat Cell Biol. 2013;15:113–122. doi: 10.1038/ncb2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camarasa MV, et al. Derivation of Man-1 and Man-2 research grade human embryonic stem cell lines. In Vitro Cell Dev Biol Anim. 2010;46:386–394. doi: 10.1007/s11626-010-9291-5. [DOI] [PubMed] [Google Scholar]
- 32.Ye J, et al. High quality clinical grade human embryonic stem cell lines derived from fresh discarded embryos. Stem Cell Res Ther. 2017;8:128. doi: 10.1186/s13287-017-0561-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International Stem Cell, I et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 34.Koyanagi-Aoi M, et al. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc Natl Acad Sci U S A. 2013;110:20569–20574. doi: 10.1073/pnas.1319061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Theunissen TW, et al. Molecular Criteria for Defining the Naive Human Pluripotent State. Cell Stem Cell. 2016;19:502–515. doi: 10.1016/j.stem.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y, et al. Derivation of Pluripotent Stem Cells with In Vivo Embryonic and Extraembryonic Potency. Cell. 2017;169:243–257 e225. doi: 10.1016/j.cell.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan L, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 38.Dang Y, et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 2016;17:130. doi: 10.1186/s13059-016-0991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blakeley P, et al. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development. 2015;142:3613. doi: 10.1242/dev.131235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Blair K, Smith A. Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition. Stem cell reports. 2013;1:209–217. doi: 10.1016/j.stemcr.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu RH, et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 42.Amita M, et al. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci U S A. 2013;110:E1212–1221. doi: 10.1073/pnas.1303094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yabe S, et al. Comparison of syncytiotrophoblast generated from human embryonic stem cells and from term placentas. Proc Natl Acad Sci U S A. 2016;113:E2598–2607. doi: 10.1073/pnas.1601630113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chilosi M, et al. Differential expression of p57kip2, a maternally imprinted cdk inhibitor, in normal human placenta and gestational trophoblastic disease. Lab Invest. 1998;78:269–276. [PubMed] [Google Scholar]
- 45.Zhang P, Wong C, DePinho RA, Harper JW, Elledge SJ. Cooperation between the Cdk inhibitors p27(KIP1) and p57(KIP2) in the control of tissue growth and development. Genes Dev. 1998;12:3162–3167. doi: 10.1101/gad.12.20.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okae H, et al. Derivation of Human Trophoblast Stem Cells. Cell Stem Cell. 2018;22:50–63 e56. doi: 10.1016/j.stem.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 47.Lee CQ, et al. What Is Trophoblast? A Combination of Criteria Define Human First-Trimester Trophoblast. Stem Cell Reports. 2016;6:257–272. doi: 10.1016/j.stemcr.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hemberger M, Udayashankar R, Tesar P, Moore H, Burton GJ. ELF5-enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Hum Mol Genet. 2010;19:2456–2467. doi: 10.1093/hmg/ddq128. [DOI] [PubMed] [Google Scholar]
- 49.Ng RK, et al. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol. 2008;10:1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang SM, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 51.Thorsell AG, et al. Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors. J Med Chem. 2017;60:1262–1271. doi: 10.1021/acs.jmedchem.6b00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 53.Hemberger M, et al. Parp1-deficiency induces differentiation of ES cells into trophoblast derivatives. Developmental biology. 2003;257:371–381. doi: 10.1016/s0012-1606(03)00097-6. [DOI] [PubMed] [Google Scholar]
- 54.Koh DW, et al. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci U S A. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.