Abstract
Long non-coding small nucleolar RNA host gene 7 (lncRNA SNHG7) is located on chromosome 9q34.3 in length of 984 bp. SNHG7 has been found to play the role of oncogene in varieties of cancers, and its dysregulation has been found to be associated with carcinogenesis and progression. In the present study, we examined the expression of SNHG7 in prostate cancer tissues and in paired adjacent normal prostate tissues, and we further explored the clinical significance and prognostic value of SNHG7 in prostate cancer patients.
A total of 127 prostate cancer tissues were collected from prostate cancer patients who underwent radical prostatectomy between April 2011 and March 2019 at the department of urology, Pudong New Area People's Hospital. Real-time quantitative polymerase chain reaction experiment was performed to detect the relative expressions of SNHG7 in the prostate cancer tissues and normal prostate tissues. The Kaplan–Meier method was used to create survival curves and the log-rank test was used to determine statistical significance. A Cox proportional hazard analysis was used to evaluate the prognostic factors in univariate and multivariate analyses.
Compared with paired adjacent normal prostatic tissues, SNHG7 expression was increased in prostate cancer tissues (P < .001). Increased SNHG7 expression correlated with Gleason score (P = .021), bone metastasis (P = .013), pelvic lymph node metastasis (P = .008), and TNM stage (P = .007). Multivariate Cox regression analyses revealed increased SNHG7 expression was independently associated with a poor prognosis of prostate cancer patients (hazard ratio [HR] = 2.839, 95% confidence interval [CI] = 1.921–8.382, P = .038).
This study showed that lncRNA-SNHG7 was overexpressed in prostate cancer tissues, and it might contributes to the development and progression of prostate cancer. Furthermore, the SNHG7 expression was associated with the prognosis of prostate cancer, suggesting a potential target for the treatment and prognosis of prostate cancer. Nevertheless, the underlying modulatory mechanism by which SNHG7 aggravates prostate cancer progression need to be further studied.
Keywords: expression, lncRNA-SNHG7, prognosis, prostate cancer
1. Introduction
Prostate cancer ranks the second leading cause of cancer-related death among American men, and is one of the most common malignant tumor in both the developed and developing countries.[1,2] Prostate cancer is a currently common disease in Chinese male. The incidence is increasing rapidly in urban area and the mortality is high in rural area.[3,4] Although the 5-year survival rates of prostate cancer steadily increase in the United States, the mortality of prostate cancer has been increasing every year globally, particularly in East Asia.[5] Despite of significant improvement in outcomes of patients with early-stage by surgical prostatectomy, radiotherapy, hormone therapy, or immunotherapy, the treatments for the advanced patients are still challenging.[6,7] High rates of metastasis and cancer-associated mortality are major cause of poor prognosis in prostate cancer patients.[8] Hence, there is an urgent need to find novel mechanisms involved in prostate cancer, which may provide new perspectives on management of prostate cancer patients.
LncRNAs are non-coding RNA transcripts that are longer than 200 nucleotides.[9] In recent years, mounting evidence has accumulated indicating that lncRNAs play important roles in various physiological and pathological processes, such as cell proliferation, apoptosis, differentiation, and the development of cancers.[10–12] Several studies have demonstrated lncRNAs as potential biomarkers for cancer detection, prognosis, and treatment.[13]
Long non-coding small nucleolar RNA host gene 7 (lncRNA SNHG7) is located on chromosome 9q34.3 in length of 984 bp. SNHG7 has been found to play the role of oncogene, and its dysregulation has been found to be associated with carcinogenesis and progression of several cancers, such as breast cancer,[14] lung cancer,[15] gastric cancer,[16] glioblastoma,[17] melanoma,[18] thyroid cancer,[19] and pancreatic cancer.[20] Previously, Han et al found that lncRNA SNHG7 was overexpressed in prostate cancer cell lines and tissues. LncRNA SNHG7 promoted prostate cancer migration and invasion by modulating EMT.[21] Qi et al found that SNHG7 regulated the cycle progression and acted as an oncogenic gene in the prostate cancer tumorigenesis via miR-503/Cyclin D1 pathway, revealing the vital role of lncRNA/microRNAs (miRNA)/messenger RNA (mRNA) axis in prostate cancer carcinogenesis.[22] However, the clinical significance of lncRNA SNHG7 in prostate cancer has not been investigated.
In the present study, we examined the expression of SNHG7 in prostate cancer tissues and in paired adjacent normal prostate tissues, and we further explored the clinical significance and prognostic value of SNHG7 in prostate cancer patients.
2. Materials and methods
2.1. Human prostate cancer tissue samples
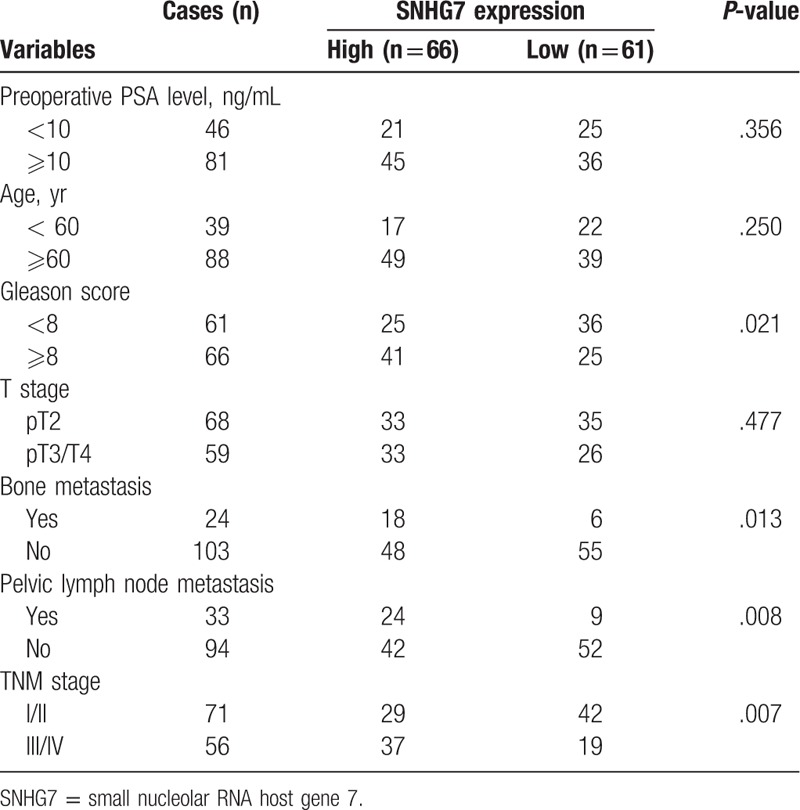
A total of 127 prostate cancer tissues were collected from prostate cancer patients who underwent radical prostatectomy between April 2011 and March 2019 at the department of urology, Pudong New Area People's Hospital. All of the tissue were frozen and stored in liquid nitrogen until further research. None of the patients received preoperative chemotherapy or radiation. This study was performed with approval from the Ethics and Research Committees of Pudong New Area People's Hospital and all of the patients provided written informed consent. The clinical information of the 127 patients was summarized in Table 1.
Table 1.
Summary of clinicopathologic features of the patients with prostate cancer.
2.2. RNA extraction and real-time quantitative PCR
Total RNA in prostate cancer tissues and normal prostate tissues was extracted according to the instructions of TRIzol reagent (Invitrogen, Carlsbad, CA). After that, extracted RNA was reverse-transcribed to complementary deoxyribose nucleic acids using reverse Transcription Kit (TaKaRa Biotechnology Co., Ltd., Dalian, China). RT-qPCR was conducted on ABI 7500 system (Applied Biosystems, Foster City, CA) in strict accordance with SYBR Green Real-time PCR. Specific thermal cycle was as follows: 30 seconds at 95°C, 5 seconds at 95°C for a total of 40 cycles, and 35 seconds at 60°C. The upstream primer of lncRNA-SNHG7 was: 5′-GTTGGGGTGTTGGCATTCTTGTT-3′; the downstream primer was: 5′-TGGTCAGCCTGGTCACTCTGG-3′; the upstream primer of GAPDH was: 5′-ACCCAGAAGACTGTGGATGG-3′; the downstream primer of GAPDH was: 5′-TTCTAGACGGCAGGTCAGGT-3′. The relative expression of target gene was expressed in −ΔCt and 2−ΔΔCt.
2.3. Statistical analysis
Differences between groups were analyzed by the Student t test for continuous variables. The continuous data were described as mean ± standard deviation. Chi square tests were conducted to analyze associations between SNHG7 levels and clinicopathological factors. The Kaplan–Meier method was used to create survival curves and the log-rank test was used to determine statistical significance. A Cox proportional hazard analysis was used to evaluate the prognostic factors in univariate and multivariate analyses. Data were considered statistically significant when P < .05. Statistical Product and Service Solutions 19.0 Inc. (Chicago, IL) and Graph pad Prism 6.0 (San Diago, CA) were utilized for statistical analysis.
3. Results
3.1. SNHG7 expression is upregulated in prostate cancer tissues
To measure the expression status of SNHG7 in prostate cancer, real-time quantitative polymerase chain reaction (qRT-PCR) was conducted to quantify SNHG7 expression in prostate cancer tissues and paired adjacent normal prostatic tissues. Compared with paired adjacent normal prostatic tissues, SNHG7 expression was increased in prostate cancer tissues (5.81 ± 1.88 vs 1.00 ± 0.38, P < .001, shown in Fig. 1). The prostate cancer patients were divided into a high SNHG7 expression group (n = 66) and a low expression group (n = 61) according to the mean value of relative SNHG7 expression (6.622) in prostate cancer tissues as cut-off.
Figure 1.
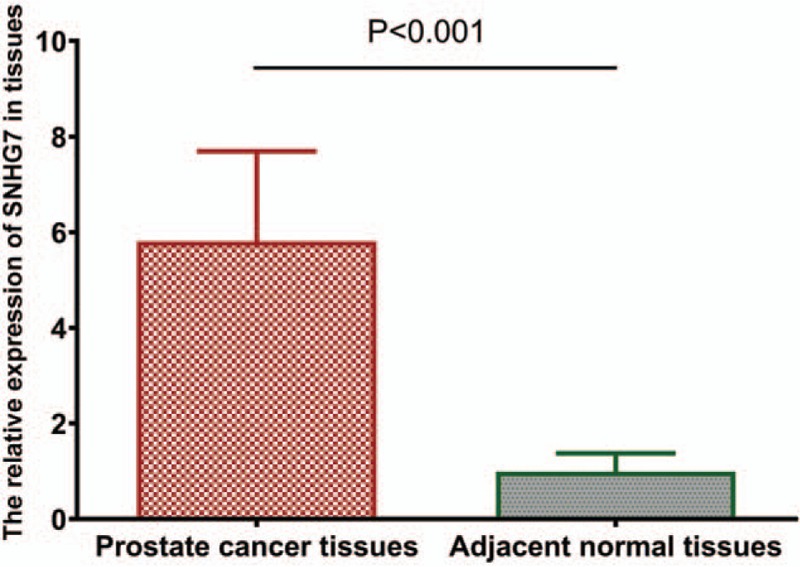
Compared with paired adjacent normal prostatic tissues, SNHG7 expression was increased in prostate cancer tissues (P < .001). SNHG7 = small nucleolar RNA host gene 7.
3.2. The associations between SNHG7 expression level and clinicopathological factors of prostate cancer patients
We then investigated associations between SNHG7 expression levels and clinicopathological factors in the prostate cancer patients. The results showed that increased SNHG7 expression correlated with Gleason score (P = .021), Bone metastasis (P = .013), Pelvic lymph node metastasis (P = .008), TNM stage (P = .007), but there were no significant associations between SNHG7 expression and age, preoperative PSA level, and T stage (all P > .05, shown in Table 1).
3.3. Prognostic value of SNHG7 in prostate cancer
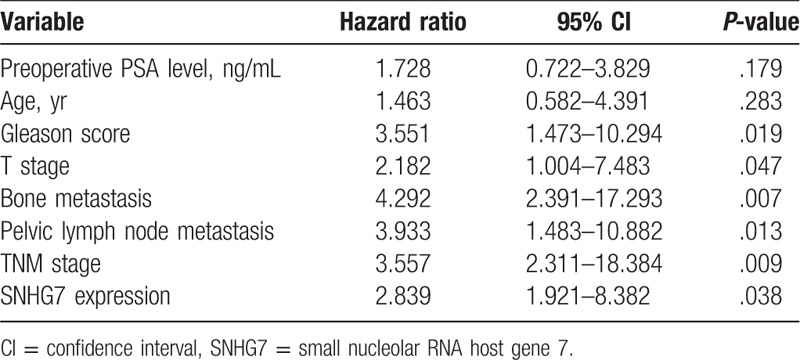
To explore the prognostic significance of SNHG7 in prostate cancer patients, the association between SNHG7 expression and overall survival were analyzed through log-rank test. All prostate cancer patients were divided into 2 subgroups according to the median SNHG7 expression. As shown in Figure 2, the prognosis of prostate cancer cases with high SNHG7 expression was poor than that of prostate cancer cases with low SNHG7 expression (P = .021). Multivariate Cox regression analyses were subsequently performed to determine the association of SNHG7 expression level and clinical prognostic factors in prostate cancer. The results revealed increased SNHG7 expression was independently associated with a poor prognosis of prostate cancer patients (HR = 2.839, 95% CI = 1.921–8.382, P = .038, shown in Table 2).
Figure 2.
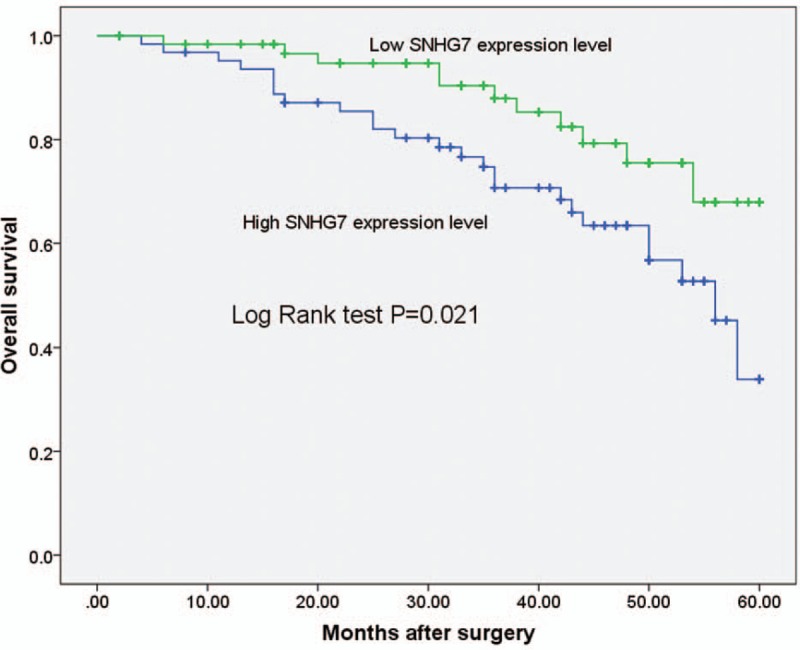
Log-rank test showed that the prognosis of prostate cancer cases with high SNHG7 expression was poor than that of prostate cancer cases with low SNHG7 expression (P = .021). SNHG7 = small nucleolar RNA host gene 7.
Table 2.
Multivariate analysis of overall survival in patients with prostate cancer.
4. Discussion
Prostate cancer is the second most common malignancy and a major leading cause of cancer death amongst men worldwide. Prostate cancer is a heterogeneous disease, where the majority of men experience an indolent form of the disease, while some experience a highly aggressive form of the disease that will metastasize.[1] The 5-year survival rate of patients with localized prostate cancer is ∼100%, due to the availability of effective treatments. However, certain patients experience progression to metastatic castrate-resistant prostate cancer, which is the final stage and has a poor prognosis, eventually resulting in mortality.[23–25] Therefore, identification of novel biomarker and a better understanding of the mechanism involved in the tumorigenesis and development of prostate cancer are instantly required.
LncRNAs, a class of noncoding RNAs with the length ranging from 200 nts to almost 100 kilobases, play vital roles in cancer development.[26] It has been certified that lncRNAs may be potential therapeutic targets and biomarkers for diagnosis and prognosis in many cancers.[27–30] Moreover, accumulating evidences showed that dysregulation of lncRNA expression attributes a tumor-suppressor or an oncogenic role to lncRNAs affecting the clinicopathological appearance, prognosis, and outcome of in the prostate cancer.[31]
LncRNA-SNHG7 (SNHG7) was a recently identified LncRNA, which was enhanced in tumor cells for tumor proliferation and survival. For example, Gao et al found that SNHG7 was significantly upregulated in breast cancer tissues when compared with normal tissues. Breast cancer cell lines showed higher levels of SNHG7 than normal breast epithelial cell line. The knockdown of SNHG7 by siRNA could remarkably repress breast cancer cell proliferation and invasion. Moreover, miRNA-381 was newly confirmed as a direct target of SNHG7 and it mediated the suppressing effects of SNHG7 on breast cancer cells.[14] She et al found that the expression levels of lncRNA-SNHG7 mRNA and protein obviously increased in lung cancer tissues compared to adjacent noncancerous tissues. lncRNA-SNHG7 promoted the proliferation, migration and invasion, and inhibited apoptosis of lung cancer cells by enhancing the FAIM2 expression, suggesting that lncRNA-SNHG7 as a key regulator of gene expression, maybe a promising therapeutic strategy for the treatment of lung cancer.[15] Wang et al found that the relative expression of SNHG7 was upregulated in 48 cases of gastric cancer tissues and 5 gastric cancer cell lines. The in vitro experiments showed that after SNHG7 expression was interfered, the proliferation of gastric cancer cells was inhibited with an increase in apoptotic rate and arrest of cell cycle in G1/G0 phase. Experiment on nude-mouse transplanted tumor model confirmed that after SNHG7 expression was interfered, in vivo tumor growth was inhibited, and the Western blotting assay revealed that regulation of p15 and p16 expressions constituted a part of the potential molecular mechanism.[16] Ren et al found that the expression of SNHG7 was significantly upregulated in glioblastoma tissues and cell lines compared with noncancerous brain tissues. Furthermore, we found that SNHG7 knockdown remarkably suppressed the proliferation, migration and invasion of A172 and U87 cells while inducing their apoptosis. Subsequently, we showed that SNHG7 knockdown significantly inhibited tumor growth and metastasis in vivo by using xenograft experiments in nude mice.[17] Wang et al found that SNHG7 expression level was higher in thyroid cancer samples than that in corresponding ones. The SNHG7 expression was associated with tumor size and TNM stage. Moreover, thyroid cancer cell proliferation was inhibited, and thyroid cancer cell apoptosis was induced after SNHG7 was knocked down in vitro. Moreover, the mRNA and protein expressions of brain-derived neurotrophic factor (BDNF) were downregulated after knockdown of SNHG7. Furthermore, the expression level of BDNF was positively related to the expression of SNHG7 in thyroid cancer tissues.[19] Cheng et al found that SNHG7 was overexpressed in both pancreatic cancer tissues and cell lines. High expression level of SNHG7 was correlated with the poor prognosis. SNHG7 knockdown inhibited the proliferation, migration, and invasion of pancreatic cancer cells. Moreover, SNHG7 was found to regulate the expression of ID4 via sponging miR-342-3p. Additionally, this finding was supported by in vivo experiments. The results indicated that SNHG7 as a potential target for clinical treatment of pancreatic cancer.[20] Xu et al found that that the expression of SNHG7 in esophageal cancer tissues and cells was significantly upregulated. After the si-SNHG7 intervention, the proliferation of esophageal cancer cells was inhibited, the apoptosis rate increased, and the cell cycle was blocked in G1-G0 phase. QRT-PCR and Western blot showed that, after the si-SNHG7 intervention, the expression of p15 and p16 increased significantly. These findings demonstrated that SNHG7 could partly promote the development of esophageal cancer by regulating the expression of p15 and p16.[32]
The role of lncRNA SNHG7 in prostate cancer has also been investigated. Han et al found that lncRNA SNHG7 was overexpressed in prostate cancer cell lines and tissues. LncRNA SNHG7 promoted prostate cancer migration and invasion by modulating EMT.[21] Qi et al found that SNHG7 regulated the cycle progression and acted as an oncogenic gene in the prostate cancer tumorigenesis via miR-503/Cyclin D1 pathway, revealing the vital role of lncRNA/miRNA/mRNA axis in prostate cancer carcinogenesis.[22] However, the clinical significance of lncRNA SNHG7 in prostate cancer has not been investigated. In the present study, to measure the expression status of SNHG7 in prostate cancer, qRT-PCR was conducted to quantify SNHG7 expression in prostate cancer tissues and paired adjacent normal prostatic tissues. Compared with paired adjacent normal prostatic tissues, SNHG7 expression was increased in prostate cancer tissues. We then investigated associations between SNHG7 expression levels and clinicopathological factors in the prostate cancer patients. The results showed that increased SNHG7 expression correlated with Gleason score, bone metastasis, pelvic lymph node metastasis, TNM stage, but there were no significant associations between SNHG7 expression and age, preoperative PSA level, and T stage. To explore the prognostic significance of SNHG7 in prostate cancer patients, the association between SNHG7 expression and overall survival were analyzed through log-rank test. All prostate cancer patients were divided into 2 subgroups according to the median SNHG7 expression. As shown in Figure 2, the prognosis of prostate cancer cases with high SNHG7 expression was poor than that of prostate cancer cases with low SNHG7 expression. Multivariate Cox regression analyses were subsequently performed to determine the association of SNHG7 expression level and clinical prognostic factors in prostate cancer. The results revealed increased SNHG7 expression was independently associated with a poor prognosis of prostate cancer patients.
In conclusions, this study showed that lncRNA-SNHG7 was overexpressed in prostate cancer tissues, and it might contributes to the development and progression of prostate cancer. Furthermore, the SNHG7 expression was associated with the prognosis of prostate cancer, suggesting a potential target for the treatment and prognosis of prostate cancer. Nevertheless, the underlying modulatory mechanism by which SNHG7 aggravates prostate cancer progression need to be further studied.
Author contributions
Data curation: Qier Xia, Jun Li.
Investigation: Qier Xia, Jun Li, Zhenyu Yang, Jinjun Tian.
Methodology: Qier Xia, Jun Li, Zhenyu Yang, Jinjun Tian.
Software: Qier Xia, Jun Li.
Writing – original draft: Qier Xia, Dingguo Zhang.
Footnotes
Abbreviations: lncRNA SNHG7 = long non-coding small nucleolar RNA host gene 7, lncRNAs = long noncoding RNAs, qRT-PCR = real-time quantitative polymerase chain reaction.
How to cite this article: Xia Q, Li J, Yang Z, Zhang D, Tian J, Gu B. Long non-coding RNA small nucleolar RNA host gene 7 expression level in prostate cancer tissues predicts the prognosis of patients with prostate cancer. Medicine. 2020;99:7(e18993).
The authors have no conflicts of interest to disclose.
References
- [1].Attard G, Parker C, Eeles RA, et al. Prostate cancer. Lancet 2016;387:70–82. [DOI] [PubMed] [Google Scholar]
- [2].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [3].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–32. [DOI] [PubMed] [Google Scholar]
- [4].Ye D, Zhu Y. Epidemiology of prostate cancer in China: an overview and clinical implication. Zhonghua Wai Ke Za Zhi 2015;53:249–52. [PubMed] [Google Scholar]
- [5].Hsing AW, Devesa SS. Trends and patterns of prostate cancer: what do they suggest? Epidemiol Rev 2001;23:3–13. [DOI] [PubMed] [Google Scholar]
- [6].Ostergren PB, Ternov KK, Jensen CFS, et al. Hormone naive metastatic prostate cancer: how to treat it? Arch Esp Urol 2019;72:192–202. [PubMed] [Google Scholar]
- [7].Zhang J, Wang L, You X, et al. Nanoparticle therapy for prostate cancer: overview and perspectives. Curr Top Med Chem 2019;19:57–73. [DOI] [PubMed] [Google Scholar]
- [8].Witte JS. Prostate cancer genomics: towards a new understanding. Nat Rev Genet 2009;10:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shi X, Sun M, Liu H, et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett 2013;339:159–66. [DOI] [PubMed] [Google Scholar]
- [10].Qadir MI, Bukhat S, Rasul S, et al. RNA therapeutics: Identification of novel targets leading to drug discovery. J Cell Biochem 2019;121:898–929. [DOI] [PubMed] [Google Scholar]
- [11].Chan JJ, Tay Y. Noncoding RNA: RNA regulatory networks in cancer. Int J Mol Sci 2018;19:E1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jandura A, Krause HM. The new RNA world: growing evidence for long noncoding RNA functionality. Trends Genet 2017;33:665–76. [DOI] [PubMed] [Google Scholar]
- [13].Zhang H, Chen Z, Wang X, et al. Long non-coding RNA: a new player in cancer. J Hematol Oncol 2013;6:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gao YT, Zhou YC. Long non-coding RNA (lncRNA) small nucleolar RNA host gene 7 (SNHG7) promotes breast cancer progression by sponging miRNA-381. Eur Rev Med Pharmacol Sci 2019;23:6588–95. [DOI] [PubMed] [Google Scholar]
- [15].She K, Huang J, Zhou H, et al. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep 2016;36:2673–80. [DOI] [PubMed] [Google Scholar]
- [16].Wang MW, Liu J, Liu Q, et al. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur Rev Med Pharmacol Sci 2017;21:4613–22. [PubMed] [Google Scholar]
- [17].Ren J, Yang Y, Xue J, et al. Long noncoding RNA SNHG7 promotes the progression and growth of glioblastoma via inhibition of miR-5095. Biochem Biophys Res Commun 2018;496:712–8. [DOI] [PubMed] [Google Scholar]
- [18].Zhang C, Zhu B, Li XB, et al. Long non-coding RNA SNHG7 promotes migration and invasion of melanoma via upregulating SOX4. Eur Rev Med Pharmacol Sci 2019;23:4828–34. [DOI] [PubMed] [Google Scholar]
- [19].Wang YH, Huo BL, Li C, et al. Knockdown of long noncoding RNA SNHG7 inhibits the proliferation and promotes apoptosis of thyroid cancer cells by downregulating BDNF. Eur Rev Med Pharmacol Sci 2019;23:4815–21. [DOI] [PubMed] [Google Scholar]
- [20].Cheng D, Fan J, Ma Y, et al. LncRNA SNHG7 promotes pancreatic cancer proliferation through ID4 by sponging miR-342-3p. Cell Biosci 2019;9:28. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [21].Han Y, Hu H, Zhou J. Knockdown of LncRNA SNHG7 inhibited epithelial-mesenchymal transition in prostate cancer though miR-324-3p/WNT2B axis in vitro. Pathol Res Pract 2019;215:152537. [DOI] [PubMed] [Google Scholar]
- [22].Qi H, Wen B, Wu Q, et al. Long noncoding RNA SNHG7 accelerates prostate cancer proliferation and cycle progression through cyclin D1 by sponging miR-503. Biomed Pharmacother 2018;102:326–32. [DOI] [PubMed] [Google Scholar]
- [23].Al-Salama ZT. Apalutamide: a review in non-metastatic castration-resistant prostate cancer. Drugs 2019;79:1591–8. [DOI] [PubMed] [Google Scholar]
- [24].Shenderov E, Isaacsson Velho P, Awan AH, et al. Genomic and clinical characterization of pulmonary-only metastatic prostate cancer: a unique molecular subtype. Prostate 2019;79:1572–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Battaglia A, De Meerleer G, Tosco L, et al. Novel insights into the management of oligometastatic prostate cancer: a comprehensive review. Eur Urol Oncol 2019;2:174–88. [DOI] [PubMed] [Google Scholar]
- [26].Li Z, Rana TM. Decoding the noncoding: prospective of lncRNA-mediated innate immune regulation. RNA Biol 2014;11:979–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gloss BS, Dinger ME. The specificity of long noncoding RNA expression. Biochim Biophys Acta 2016;1859:16–22. [DOI] [PubMed] [Google Scholar]
- [28].Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10:155–9. [DOI] [PubMed] [Google Scholar]
- [29].Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629–41. [DOI] [PubMed] [Google Scholar]
- [30].Whitehead J, Pandey GK, Kanduri C. Regulation of the mammalian epigenome by long noncoding RNAs. Biochim Biophys Acta 2009;1790:936–47. [DOI] [PubMed] [Google Scholar]
- [31].Mitobe Y, Takayama KI, Horie-Inoue K, et al. Prostate cancer-associated lncRNAs. Cancer Lett 2018;418:159–66. [DOI] [PubMed] [Google Scholar]
- [32].Xu LJ, Yu XJ, Wei B, et al. LncRNA SNHG7 promotes the proliferation of esophageal cancer cells and inhibits its apoptosis. Eur Rev Med Pharmacol Sci 2018;22:2653–61. [DOI] [PubMed] [Google Scholar]


