Abstract
Cell cycle progression requires the coordination of cell growth, chromosome replication, and division. Consequently, a functional cell cycle must be coupled with metabolism. However, direct measurements of metabolome dynamics remained scarce, in particular in bacteria. Here, we describe an untargeted metabolomics approach with synchronized Caulobacter crescentus cells to monitor the relative abundance changes of ~400 putative metabolites as a function of the cell cycle. While the majority of metabolite pools remains homeostatic, ~14% respond to cell cycle progression. In particular, sulfur metabolism is redirected during the G1-S transition, and glutathione levels periodically change over the cell cycle with a peak in late S phase. A lack of glutathione perturbs cell size by uncoupling cell growth and division through dysregulation of KefB, a K+/H+ antiporter. Overall, we here describe the impact of the C. crescentus cell cycle progression on metabolism, and in turn relate glutathione and potassium homeostasis to timely cell division.
Introduction
Coordinating the cell division cycle with growth and metabolism is key to maintain homeostasis of all living organisms1–3. Not only is growth itself ultimately a metabolic challenge, but cells must ensure faithful production of viable daughter cells, despite changing environments or nutrient availability. The cell cycle provides an internal program that regulates the progressive execution of specific cellular processes such as replication, chromosome segregation, and cell division. However, as biomass formation and cell cycle progression can be decoupled4,5, their coordination in general, and with respect to metabolism in particular, is critical and subject to regulation. In eukaryotes, the cyclin-dependent-kinase (CDK) signaling cascade controls the cell cycle and gates central metabolism6,7. Additionally, growth regulators such as the molecular target of rapamycin (mTOR) complex sense metabolic cues and relay this information to cell cycle regulation8. Because defects or alterations of the regulatory links between metabolism and cell cycle manifests in disease, their elucidation gained substantial attention in eukaryotes6. Similarly, due to the ongoing effort to control bacterial growth, knowledge on the crosstalk between metabolism, growth, and the cell division cycle is important. Bacteria generally lack the CDK based cell cycle machinery, but employ other sophisticated regulatory cascades to drive their cell cycle9.
In the bacterial model organism for cell cycle control, Caulobacter crescentus, cell division produces a non-replicative, motile swarmer (G1) and a replication-competent, sessile stalked cell. To re-enter the replicative cycle, the swarmer cell differentiates into a stalked cell (S), a process during which it initiates chromosome replication10 and increases overall protein synthesis rates11. Coincident with ongoing replication, cells increase in mass and eventually initiate the division process (G2) (Fig. 1a). Analogous to eukaryotes, the metabolic state and available resources are critical parameters that guide bacteria along the path from cell cycle initiation to cell division12–18. For instance, in Bacillus subtilis carbon availability is signaled by UDP-glucose and controls cell size at the time of division14. Escherichia coli naturally alters cell size under different nutritional regimes, and mutants of central metabolism can display cell morphology defects19. While cell body length of C. crescentus is independent of carbon availability15, central metabolism is coupled to the cell cycle and cell division machinery, in which Z-ring formation is regulated by metabolic enzymes12,20. In addition, the signaling molecule c-di-GMP enables faithful cell division21, and (p)ppGpp coordinates a response to carbon or nitrogen limitation16–18. These studies exemplify that, like in their eukaryotic counterparts, metabolic cues are an integral part of cell cycle control in bacteria. However, the cell cycle machinery also entrains the metabolic reaction network to fuel varying metabolic demand7. This is because cell cycle progression globally affects bacterial physiology, as genome-wide expression studies describe large-scale transcriptional oscillations (up to ~1/3 of the entire gene set, including metabolic genes) that drive timely execution of cell cycle dependent processes and differentiation in C. crescentus22–25. Such fundamental reprogramming provokes questions on how the metabolic requirements change at distinct stages of the bacterial cell cycle, if and how resources are dynamically allocated, and how these fluctuations are met by adaptations of metabolic flux.
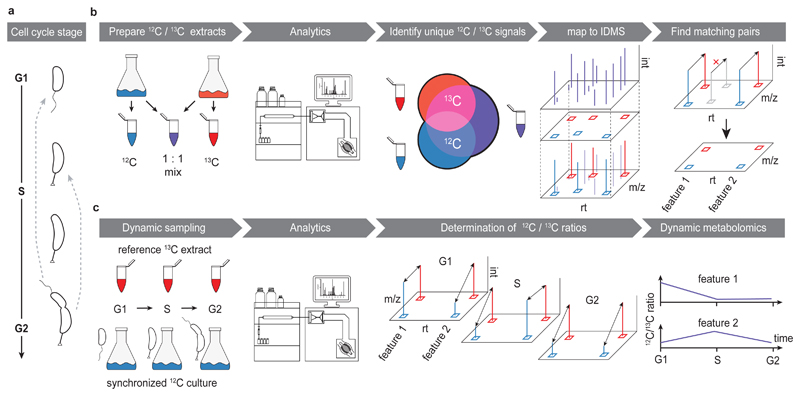
Figure 1. Untargeted dynamic metabolomics to monitor cell cycle progression.
(a) Schematics of the C. crescentus cell cycle and differentiation. Cell cycle progression is linked to clearly distinguishable changes in morphology. A motile swarmer cell (G1) sheds its flagellum, and develops into a stalked, proliferative cell that undergoes replication (S), and ultimately divides (G2) into new swarmer cell, while itself remaining a stalked cell that re-initiates S phase. (b) Building a non-targeted metabolite library. Production of naturally 12C - (blue) and highly uniformly (u) 13C -isotope enriched (red) C. crescentus extracts by growth on respectively labelled carbon sources to obtain a low- and high-molecular-weight metabolome. Accessible peaks from both extracts, as well as a mix thereof (12C/13C mix, purple), were subsequently detected using LC-HRMS that distinguishes mass shifts associated with isotope incorporation. Shared peaks between 12C and 13C samples were discarded as spectral noise. Unique peaks with a clear isotopic identity (u-12C, u-13C) were mapped to the peakmap of the mixed sample. Lastly, this peakmap was filtered by matching co-eluting peaks that were separated by a m/z shift explained by carbon labelling. This approach selected features of biological origin, and provided the extraction windows for the subsequent, isotope-dilution based metabolomics approach. (c) Dynamic metabolomics of the C. crescentus cell cycle. Synchronized cells growing on 12C glucose were followed throughout one cell cycle. For each time point, a 12C aliquot normalized to biomass was sampled and spiked with a constant amount of reference 13C enriched C. crescentus extract. Changes in relative pool sizes over time were then determined by calculating the peak area ratios of the u-12C relative to corresponding u-13C peak areas.
Here, we used untargeted metabolomics to monitor global temporal metabolite abundance profiles of the cell cycle progression in C. crescentus. We developed a workflow that combines various aspects of stable isotope dilution (SID) assisted liquid chromatography high resolution mass spectrometry (LC-HRMS) to precisely and accurately perform relative quantification of metabolites, to discriminate noise from signals of interest and to facilitate assignment of molecular identity26–28. We detect robust fluctuations of a set of metabolites throughout the cell cycle, including metabolites from central metabolic pathways, while also capturing novel potential (secondary) messengers involved in cell cycle control. While the quantitative changes of many primary metabolites are likely explained by varying metabolic needs for cell cycle dependent functions, they could indicate metabolites that are required for a functional cell cycle. To test this, we analyzed glutathione as one of the metabolites that fluctuates extensively throughout the cell cycle. Cells lacking glutathione display growth defects and aberrant cell size distributions. Our results indicate that glutathione deficiency results in activation of a potassium efflux system, which uncouples cellular growth from normal cell division.
Results
An isotope dilution-based workflow for untargeted metabolome profiling throughout the cell cycle
The here developed untargeted time-resolved metabolomics workflow starts with the SID-enabled chemoinformatic mining of cell extracts to build a library of detected metabolite features (Fig. 1b, for details see Supplementary Methods; the workflow can be retrieved from: https://pypi.org/project/idms-pairfinder-2/). In a second step, we used targeted peak extraction and SID-based quantification to study their intracellular dynamics during cell cycle progression in synchronized cultures (Fig. 1c). Two complementary LC-HRMS methods were applied to increase metabolite detection, i.e. broad coverage, with focus on polar compounds that prevail among intracellular metabolites29.
Applied to C. crescentus extracts, we detected on average (n=6 biological replicates) a total of ~5300 and ~5400 distinct LC-MS peaks (from molecules with a defined retention time [rt] and mass-to-charge [m/z]) unique to native (12C) or uniformly (u) 13C-enriched cells, respectively (Supplementary Fig. 1a). 12C and 13C peaks were matched based on congruent retention times and accurate m/z distances that conform to the theoretical patterns of 13C labeling incorporation. Subsequent isotopologue grouping resulted in ~920 feature-pairs, which we consider of biological origin (Supplementary Fig. 1b). To further reduce data redundancy resulting from multiple adduct formation during electrospray-ionization, we included a step of adduct grouping and curation. This deconvolution yielded 419 feature-pairs, representing putative metabolites (i.e. small molecules of biological origin). These molecules were then annotated by matching potential monoisotopic masses and further restricted by determined carbon-numbers against a genome-scale-reconstruction based metabolite list (see Supplementary Methods), complemented with the KEGG database30. In total, 211 features matched 277 metabolites. Of these, 123 features were unique (single database entries), while others corresponded to several metabolites, as it was expected due to the presence of isomers or overlapping masses that cannot be distinguished even at high mass accuracy31. Assigned putative metabolites showed excellent coverage of primary metabolism (Supplementary Fig. 1c, d). Approximately half (208 of 419) of the selected features could not be annotated, which is in agreement with current untargeted metabolomics experiments28,32. The generated library of C. crescentus metabolites, including unknowns, provided the basis for the following dynamic metabolomics approach.
To determine the effect of cell cycle progression on metabolism we employed a synchronization protocol yielding highly enriched (>95%) cultures of swarmer cells33. To avoid draining metabolism upon synchronization, we continuously supplemented growth media with glucose, and cultures readily retained exponential growth post-synchronization (Supplementary Fig. 2). Cell cycle synchrony was additionally validated by time-lapse microscopy (Supplementary Fig. 3). Because the synchronization procedure required cooling cultures, we included a control condition that mimicked this perturbation in non-synchronous populations. Differences in the metabolite abundance profiles of synchronized vs. non-synchronous controls are thus strongly associated with cell cycle dependency rather than the synchronization protocol itself, and were taken into account when determining the effect of cell cycle progression on metabolism.
Synchronized and non-synchronized cultures (n=3 biological replicates each) were then followed throughout one cell cycle, as judged by one doubling in biomass (doubling time 159 ± 5 and 147 ± 6 min respectively; Supplementary Fig. 2). We expect peak G1-S transition at ~20 min, peak S-phase at ~90 min, and pre-divisional cells after ~110 min based on previous studies10,23,34,35. Metabolomics samples were taken in ten minute time intervals and spiked with a 13C labelled C. crescentus metabolome extract as internal standard at each time point (Fig. 1c). As this reference extract was acquired from non-synchronized cells, each cell cycle stage is represented. Relative quantification of individual metabolites over time was based on the area ratios of u-12C signal relative to the u-13C isotope signal of the internal standard. This approach allowed quantification with high precision, as indicated by low coefficients of variations (CVs) between replicates compared to other spectral normalization approaches (Supplementary Fig. 4). Applying additional quality filters (see Methods), we ultimately obtained dynamic abundance profiles of ~400 putative metabolites throughout the cell cycle (Supplementary Data 1, 2; for metabolite annotation see Supplementary Table 1, 2).
The metabolome-wide response to cell cycle progression and differentiation in C. crescentus
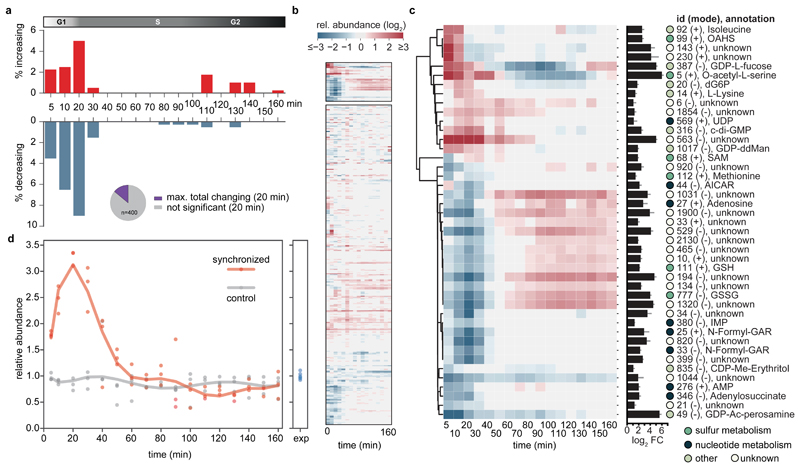
Comparative statistical analysis of synchronized vs. control samples at different time points of the cell cycle revealed that metabolite abundances remained largely homeostatic (Fig. 2a, Fig. 2b bottom, Supplementary Data 3). However, a discrete set of metabolites had a temporal response, with up to ~14% of metabolites significantly changing after 20 min (i.e. peak transition from G1/S or swarmer to stalked cell stage, Fig. 2a). The majority of these abundance profiles were characterized by an initial decrease, followed by peak levels during later stages of the cell cycle. Overall, cell cycle progression had a distinct impact on bacterial metabolism, while pool sizes remained largely constant despite the underlying substantial changes of biological function.
Figure 2. Global metabolic response to cell cycle progression.
(a) Fraction of metabolites over time (total of 400) that significantly increased (red) or decreased (blue) during cell cycle progression of synchronized compared to asynchronous C. crescentus cultures. Inlet pie chart shows the maximal cell cycle dependent response as observed at 20 min upon peak G1-S transition (~14 % of putative metabolites). (b, c) Metabolic signature of cell cycle progression. Cluster maps displaying the changes of metabolite abundances over time post synchronization. Hierarchical clustering (cosine distance matrix, single method agglomeration) of the relative abundance profiles of metabolites is shown (log2 fold-change of synchronized over control cultures). Overall, metabolism remains largely homeostatic as a function of a cell cycle (b, bottom). Instead, a fraction of metabolites strongly responded to cell cycle progression (b, top, enlarged in c). The bar chart depicts the maximal change of pool size (log2 fold change of minimum over maximum response). The circles next to compounds are colored according to the indicated metabolic identifiers. (d) Relative abundance profiles of mean c-di-GMP levels over time for synchronized (red) or control cultures (grey); n=3 biological replicates per condition. Data was normalized to indicated levels of exponentially ("exp") growing pre-cultures (n=6). Non-canonical abbreviations: OAHS, O-acetyl-L-homoserine; GDP-ddMan, GDP-4-dehydro-6-deoxy-D-mannose; CDP-Me-Erythritol, 4-CDP-2-C-methyl-D-erythritol; GDP-Ac-perosamine, GDP-N-acetyl-alpha-D-perosamine.
Next, we devised a stringent filter to identify metabolite abundance profiles with a highly robust response to cell cycle progression (Fig. 2b top, Fig. 2c). Due to the expected broadening of metabolic oscillations as a consequence of the internal age distribution (> 20 min) of the initially obtained swarmer populations in the G1-phase of the cell cycle34, we chose to retain those metabolites the concentrations of which differed significantly at two consecutive time-points and showed at least a twofold-change. The resulting 43 candidates were further subjected to higher level molecular identification including MS/MS validation (Supplementary Methods, Supplementary Tables 3-4, Supplementary Data 4, ~40% could be annotated with high confidence). Among these, we identified c-di-GMP, a known cell cycle dependent messenger that is peaking during the swarmer-to-stalked (i.e. G1-S phase) transition34. Indeed, c-di-GMP levels reached a maximum (> 3 times more compared to non-synchronized populations) after 20 min (Fig. 2d), indicating peak transition of G1 to S phase in agreement with previously published data on synchronized populations34. These findings validate that our untargeted approach uncovers metabolic cell cycle dependency. As expected, the amplitude is somewhat lower compared to single cell measurements (> 5 fold change)36 due to the broadening of the cell cycle dependent response at the population level (see above).
Hierarchical clustering of the metabolites that strongly correlated with cell cycle progression displayed the discrete metabolic footprint at the transition from swarmer to stalked cells and included metabolites from various pathways, such as nucleotide metabolism, cell-wall biosynthesis, as well as sulfur and amino acid metabolism (Fig. 2c).
Cell cycle dependent dynamics of biosynthetic pathways
In line with the expected periodic utilization of nucleotides throughout the cell cycle, we found an overrepresentation of precursors for de novo purine biosynthesis among cell cycle dependent pathways (Fig. 2c). The levels of AMP, adenylosuccinate as well as adenosine are low at the beginning of the cell cycle and gradually increase with a peak in S-phase. The pools of intermediates such as N-formylglycinamide ribonucleotide (N-formyl-GAR), aminoimidazole-carboxamide ribotide (AICAR), inosine monophosphate (IMP), markedly dropped sequentially at the G1-S phase transition (Supplementary Fig. 5). Because de novo purine synthesis is apparently not strictly differentially regulated by changes of transcription or translation levels with respect to cell cycle progression (Supplementary Fig. 6), their metabolite profiles provide examples of dynamic post-translational regulation. Notably, despite the temporal drop in respective precursor levels, the major purine pools ADP and ATP29,37, as well as guanylate pools largely maintained steady state throughout the cell cycle (Supplementary Fig. 5).
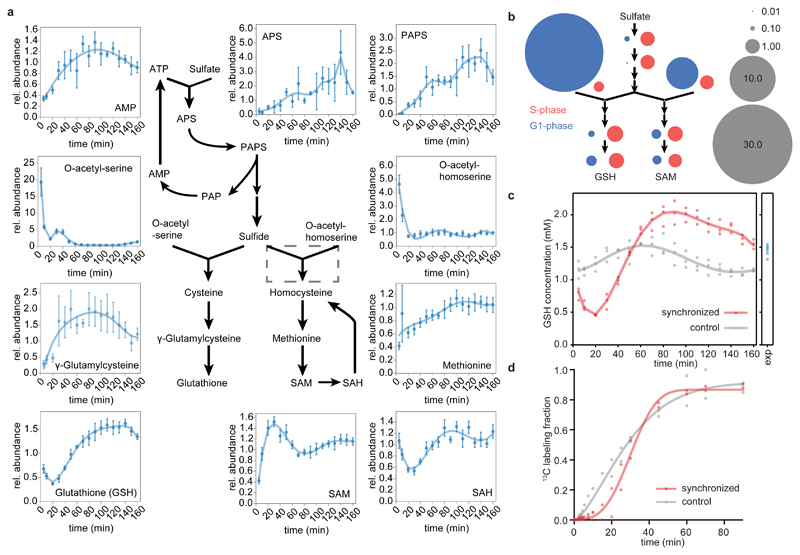
Another striking group of cell cycle-dependent metabolite changes concerns sulfur metabolism. Numerous pathway intermediates, including methionine, S-adenosylmethionine, glutathione, O-acetyl-serine and O-acetyl-homoserine, exhibited considerable concentration changes and are thus under temporal control (Fig. 2c, Fig. 3 a, b). Analysis of all detectable pathway intermediates suggest a block in sulfate assimilation to sulfide during the G1 phase, as indicated by low levels of the intermediates APS and PAPS, which is in sharp contrast to the accumulation of O-acetyl-serine/O-acetyl-homoserine (Fig. 3 a, b). This pattern of high versus low pathway intermediates within sulfur metabolism before the onset of G1 to S transition indicates a temporal block of sulfate reduction that would halt the sulfide-dependent synthesis of cysteine and homocysteine. Sulfate is initially assimilated by condensation with ATP, which is ultimately recycled to the adenylate pool as AMP (Fig. 3a, Supplementary Fig. 7). Thus, a low flux through sulfate assimilation could contribute to the observed drop in AMP levels. Indeed, during steady state, flux-balance-analysis (FBA) predicts that flux through sulfur assimilation pathway constitutes ~10 % of all reactions producing AMP (Supplementary Fig. 7; for details see Methods).
Figure 3. Cell cycle dependency of sulfur metabolism and glutathione.
(a) A simplified pathway representation of sulfate assimilation and biosynthesis of sulfur metabolites in C. crescentus based on homology mapping by KEGG30. Graphs display relative abundance profiles of respective pathway intermediates throughout one cell cycle and were calculated as the ratio of the response of synchronized vs. control samples per time point (mean values +/- SD; n=3 per condition; error propagation was applied). The dashed grey box indicates an ambiguous reaction with no predicted homolog in C. crescentus. (b) Schematic representation of cell cycle dependent block in sulfate assimilation and consequent accumulation of (homo-) cysteine precursors. Pathway layout relates to (a). Area of bubbles corresponds to the fold change of respective metabolite abundances at time points t=5 min (G1, blue) and t=90 min (S, red) over the median response of the time series; grey bubbles indicate scale. Note that APS, PAPS, and γ-glutamylcysteine were analyzed in targeted fashion (all are low abundance; APS, PAPS was below the detection limit in one replicate). For more information and corresponding transcriptional response of sulfur metabolism, see Supplementary Fig. 8. (c) Intracellular GSH concentration over time for synchronized (red) or control cultures (grey); n=3 biological replicates per condition. The time-point labelled "exp" indicates respective exponentially growing pre-cultures (n=6). (d) Fractional dynamic 13C labeling of glutathione in cell cycle synchronized (red) vs. control (grey) cells suggests attenuated de novo synthesis of glutathione early in the cell cycle (n=2 biological replicates).
Downstream of cysteine and homocysteine biosynthesis, we observed low γ-glutamylcysteine and methionine levels at the initial stages of the cell cycle (Fig. 3 a, b). This potential activation of sulfur assimilation would provide the capacity to rapidly metabolize the highly reactive intermediate sulfide to less harmful pathway intermediates. In consequence, it might represent temporal compartmentalization of pathway activity. Notably, temporal regulation of the sulfur metabolic network was also previously inferred from cell cycle dependent transcriptional changes22. The temporal mRNA expression and translational activity profiles of genes involved in sulfate uptake, assimilation, and synthesis of downstream sulfur metabolites cysteine and methionine, at the G1-S phase transition, align well with observed metabolic changes (Supplementary Fig. 8).
Additional anti-correlating major sulfur metabolites with cell cycle dependent abundance profiles were observed. The universal methyl-donor S-adenosylmethionine (SAM)38 accumulated at the G1-S transition, while its product S-adenosylhomocysteine (SAH) depleted simultaneously (Fig. 3a). Considering the observed low levels of sulfur pools during this phase, accumulation of SAM over SAH may suggest reduced methyl transferase activity during the transition to stalked cells. This hypothesis fits the temporal activation of SAM-dependent methylation of hemi-methylated DNA by CcrM that is set at mid- to late stages of the cell cycle23,39. Linked to cysteine biosynthesis, we also observed the production of another major cell cycle dependent sulfur metabolite, glutathione, which is discussed further below.
Glutathione levels vary throughout the cell cycle
The pool of glutathione, an evolutionarily widespread and major thiol in most bacteria40, displayed striking changes throughout the cell cycle (Fig. 3a,c). As a reduction equivalent, glutathione serves a variety of functions in cellular metabolism, modulates the cellular redox state, and has a regulatory role as a donor for post-translational protein modifications41,42. Glutathione is among the most abundant metabolites in bacteria29, and is highly abundant in C. crescentus with an intracellular concentration of >1 mM (Fig. 3c). Glutathione pools changed during cell cycle progression with an overall fluctuation by a factor of ~4.5 throughout a cell cycle (Fig. 3a, c). Initially the levels depleted to a minimum at the G1-S transition, followed by an increase in S-phase, reaching a peak in pre-divisional cells.
Varying metabolite concentrations do not necessarily allow inferences about pathway activity, as accumulation of a metabolite might both originate from its increased synthesis, or reduced consumption43. To determine metabolic rates by conventional fluxomics requires steady state conditions, which are not given during cell cycle progression7. Instead, we traced label incorporation of u-13C-glucose provided to synchronized versus non-synchronized cultures, and directly interpreted incorporation rates to estimate pathway activities43. This approach revealed temporal regulation of glutathione synthesis (Fig. 3d). Analogously to the dynamic changes of glutathione levels, labeling incorporation halts in G1, and spikes upon transition to S-phase. To rule out reduced flux through upstream reactions catabolizing u-13C-glucose, we confirmed that labeling dynamics of the precursors glutamate and O-acetyl-serine was largely independent of the cell cycle and had a substantially shorter half-life than glutathione (Supplementary Fig. 9).
In contrast to other major coenzymes such as PLP, NAD or CoA, we previously showed that in Escherichia coli the de novo synthesis of glutathione exceeds the requirement for dilution by growth44. Similarly, the observed turnover of glutathione in C. crescentus, and especially the amplitude of the initial decrease in glutathione pools cannot be explained by dilution by growth alone. Thus, additional sinks must exist. This is in line with the induction of genes involved in glutathione degradation and/or export (Supplementary Fig. 10), including as a major periplasmatic γ-glutamyltransferase (CCNA_01035) and a cysteine/glutathione transporter (cydC/CCNA_00798, cydD/CCNA_00799) at early stages of the cell cycle. In fact, degradation of glutathione at the G1-S transition would allow temporal reallocation of thiol groups, compensating for low sulfur pools. In terms of regulation, work with E. coli suggests that GSSG inhibits sulfate assimilation as it inactivates adenylyl-sulfate kinase45 (cysC/CCNA_01549) and PAPS reductase46 (cysH/CCNA_01179). Moreover, a negative feedback loop controls the two-step synthesis of GSH, because the activity of γ-glutamylcysteine synthetase (CCNA_003527) and glutathione synthetase (gshB/CCNA_00140) is reduced by GSH and GSSG respectively47. In accordance, overexpression of gshB from the xylX locus by full induction with xylose48 did not significantly alter glutathione levels (Supplementary Fig. 11). Overall, the observed drop of glutathione pools at the G1-S transition may set the stage for the following rapid increase of both sulfate assimilation and glutathione synthesis.
Lack of glutathione leads to uncoordinated cell division
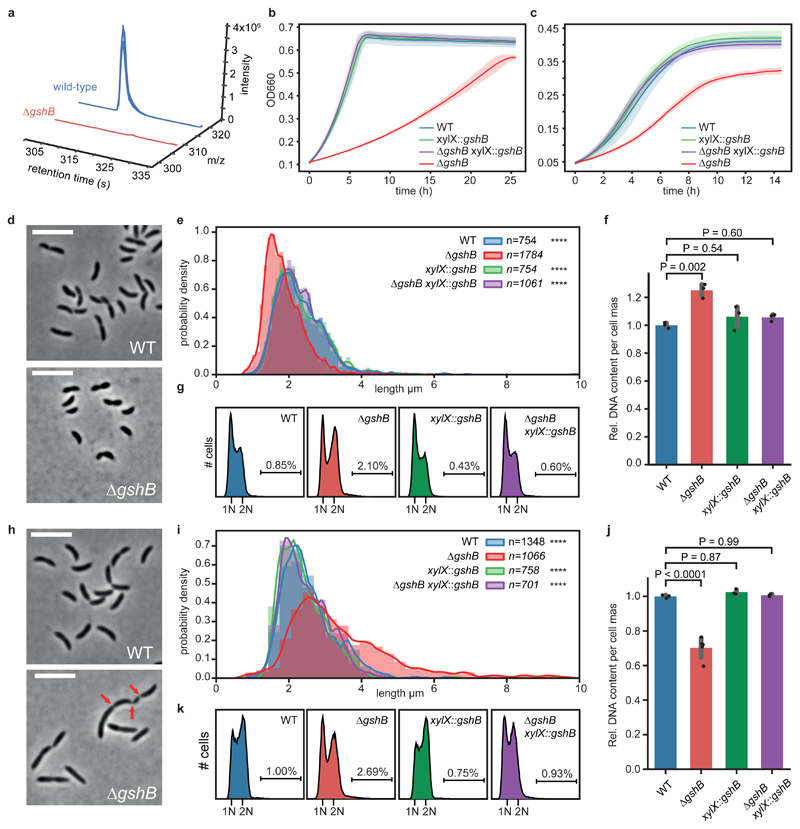
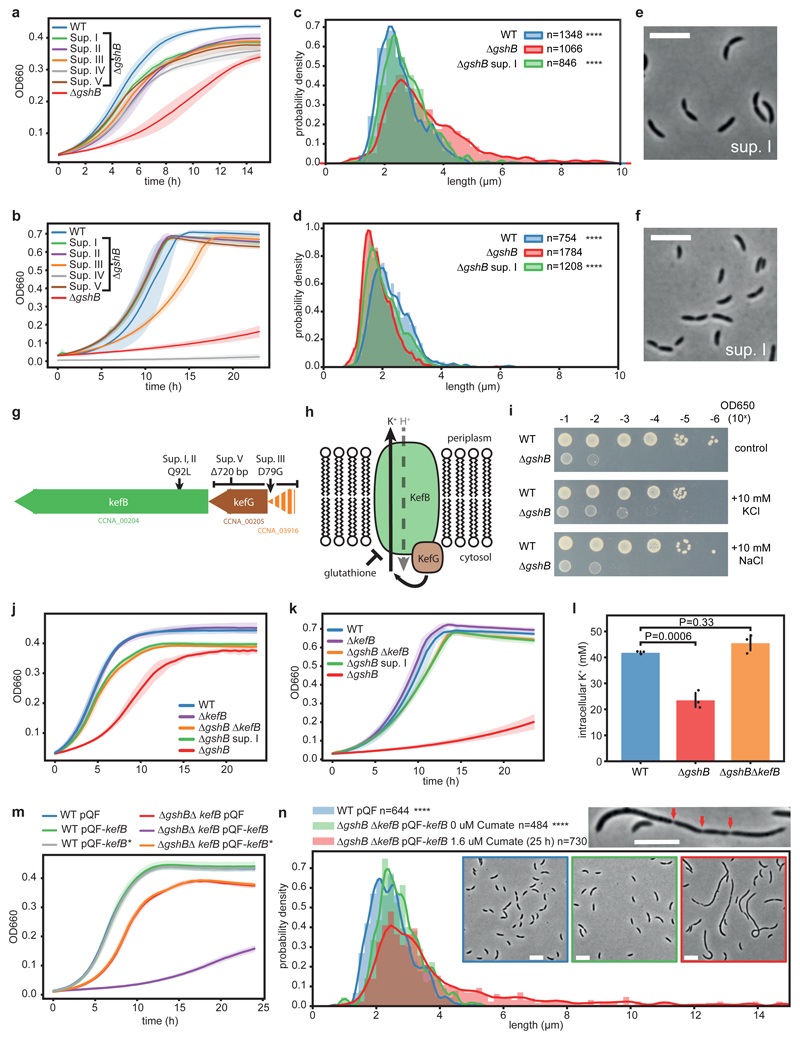
Glutathione is a highly abundant metabolite in the cell and is involved in a variety of functions29,42. In agreement, we determined high intracellular concentration of glutathione independent of growth conditions (Supplementary Fig. 12). Considering its pronounced cell cycle dependent quantitative changes (Fig. 3c), we wondered if glutathione itself is required for a faithful cell division cycle. To test this, we constructed a C. crescentus strain lacking the glutathione synthetase gene (gshB/CCNA_00140). In contrast to wild-type cells, a ΔgshB mutant had no detectable intracellular reduced or oxidized glutathione (Fig. 4a, Supplementary Fig. 13). The ΔgshB mutant displayed substantial growth defects on minimal and complex media, which could be complemented by a copy of gshB expressed in trans from the xylX locus (Fig 4b, c). Microscopic analyses revealed a perturbed cell-length distribution of ΔgshB cells grown on glucose-based minimal media (M2G) with cells being unusually short (Fig. 4d, e). Moreover, on average these cells accumulated more DNA per cell mass than the wild type (Fig. 4f, g), thus suggesting a defect in cell cycle progression. Surprisingly, the opposite phenotype was observed in complex growth media (PYE), as mutants were partly elongated and displayed heterogenous cell morphologies, including filamentous cells that occasionally displayed partial constrictions along their cell axis (Fig. 4h,i, Supplementary Fig. 14), indicating delayed or incomplete cell division. On average these cells showed significantly reduced DNA per cell mass (Fig. 4j). Importantly, the DNA profile of ΔgshB cells grown on PYE or M2G did not display a substantial fraction of cells with >2N (Fig. 4k). This suggests that rather than a loss of control of chromosome replication initiation, the ΔgshB mutant displays a defect in coordination of cell growth with division such that cells fail to divide at the usual cell length across the two media.
Figure 4. Loss of glutathione decouples growth from cell cycle progression.
(a) Knock-out strain of glutathione synthetase (ΔgshB) is devoid of reduced glutathione. Extracted ion chromatograms of reduced glutathione from ΔgshB cells (red traces, u-12C signals) vs. a spiked wild-type extract (blue traces, u-13C signals); n=3 biological replicates. For data on oxidized glutathione see Supplementary Fig. 13. (b, c) ΔgshB grown on M2G (b) and PYE (c) display growth defects; both were complemented by a copy of gshB (mean values +/- SD; n=3 biological replicates). (d, e) Reduced cell length of ΔgshB cells grown on M2G. Histograms of probability densities display cell length distributions; outlines show a fitted kernel estimate. Number of cells (n) per genotype and statistics (one-way ANOVA with a Kruskal-Wallis test of ΔgshB compared to controls; Dunn's multiple testing correction was applied) as indicated; n=4 biological replicates per genotype; ****P < 0.0001. (f) Relative DNA content per cell mass of M2G grown cells as estimated by flow cytometry. Mean values +/- SD of n=3 biological replicates. For statistical analysis, one-way ANOVA followed by multiple comparison analysis (MCA; Tukey) was performed. (g) Replication activity of M2G grown ΔgshB cells compared to controls from n=3 biological replicates per genotype as determined by flow cytometry. Chromosome equivalents (N) are depicted, and fractions of cells with more than 2N are indicated. (h, i) Mutant cells show aberrant cell morphology, increased cell body length and broadened distribution on complex media (PYE); data was reproduced with n=4 biological replicates per genotype. Data analysis, statistics like in (e); **** P < 0.0001. Red arrows indicate partial constrictions of a filamented cell. Cells larger >10 μm are not shown; examples are depicted in Supplementary Fig. 14. (j) Relative DNA content per cell mass of PYE grown cells. Mean values +/- SD of n=5 (ΔgshB) or n=3 (all other) biological replicates. Statistics like in (f) with one-way ANOVA followed by MCA (Tukey). (k) Replication activity of PYE grown biological replicates of ΔgshB cells (n=5) compared to controls (n=3 per genotype). For explanations, see (g). Cell body length was estimated using MicrobeJ78. Microscopy scale bar is 5 μm.
To test if the altered coupling between cell cycle progression and growth of PYE or M2G grown ΔgshB cells depends on the provided substrates or other media components, we tested different combinations of the respective growth media. Notably, only cells supported with glucose as the sole carbon source (M2G) produced unusually shortened cells. In contrast, supplementation with peptone/yeast-extract produced elongated cells independent of other media components such as glucose (Supplementary Fig. 15). These findings support the notion that the respective growth substrates determine the observed morphology defects, with peptone/yeast-extract displaying a dominant effect (Supplementary Fig. 15).
To further strengthen the link between glutathione and the observed defects of ΔgshB cells, we supplemented glutathione to the growth media. Notably, growth and morphology defects associated with gshB deletion could both be partially alleviated in a concentration dependent manner by extracellular glutathione supplementation (Supplementary Fig. 16a-c). Taken together, C. crescentus cells that are unable to maintain sufficient levels of glutathione are no longer able to properly couple growth to timely cell division.
Impaired cell division in glutathione-deficient cells depends on the potassium transporter KefB
To characterize the underlying defects of glutathione deficient strains, we took advantage of spontaneously emerging suppressors of the slow-growth phenotype of ΔgshB cells on PYE agar plates. Of the five suppressors analysed here (I-V), all reproducibly restored normal growth of the ΔgshB mutant in liquid PYE medium (Fig. 5a) and, with the exception of suppressor IV, also reduced the growth defect on M2G (Fig. 5b). Moreover, the suppressor strains showed restoration of morphology on both PYE and M2G (Fig. 5c-f, Supplementary Fig. 17). Additionally, we confirmed that DNA content per cell mass approached wild-type levels (Supplementary Fig. 18). Together, this suggested that the observed defects of ΔgshB cells were caused by the same mechanism. Whole-genome-resequencing of suppressor strains revealed mutations mapping to a predicted kefB operon for four of the five suppressors (Fig. 5g; Supplementary Data 5), while one suppressor (IV) had a mutation in a sulfate adenylyltransferase subunit, and is thus potentially impaired in sulfate assimilation (Fig. 5b; Supplementary Data 5). Suppressors I and II harboured a non-synonymous mutation (Q92L) at a highly conserved residue of KefB (CCNA_00204, Supplementary Fig. 19a, b), a predicted glutathione-dependent potassium efflux system (Fig. 5h). Suppressor V had a 720 bp deletion at the kefG locus (CCNA_00205, Supplementary Fig. 19c), encoding an ancillary protein that is required for full activity of KefB49 (Fig. 5h). Lastly, suppressor III resulted in a variant (D79G) of CCNA_3916, a hypothetical protein of unknown function that is encoded directly upstream of kefB and kefG (Fig. 5g).
Figure 5. Lack of glutathione leads to kefB-dependent cell division defects.
(a, b) Suppressors (I-V) improve growth of ΔgshB on PYE (a) and except for sup. IV, a sulfur assimilation mutant, see main text, on M2G (b). (c-f) Sup. I improves cell morphology of ΔgshB cells on PYE (c,e) and M2G (d,f). Corresponding data for all suppressors is shown in Supplementary Fig. 17. Distributions of wild-type and ΔgshB are from Fig. 4; for explanations, see corresponding caption. All data and statistics (one-way ANOVA with a Kruskal-Wallis test of ΔgshB compared to controls with Dunn's multiple testing correction) based on n=4 biological replicates; ****P < 0.0001. (g) ΔgshB suppressor strains have mutations around the kefB locus. (h) Schematic representation and regulation of the predicted KefB K+/H+ antiporter. (i) Serial dilutions of ΔgshB and wild-type cells grown on PYE alone, or supplemented with 10mM KCl, NaCl. The experiment was independently performed twice with similar results. (j, k) Deletion of kefB in a ΔgshB background (ΔgshBΔkefB) suppresses ΔgshB growth defects on PYE (j) and M2G (k). (l) Intracellular potassium concentrations of cells grown on PYE media as determined by ICP-MS. ΔgshB displays reduced intracellular K+ levels compared to wild-type and ΔgshBΔkefB cells. Mean values +/- SD of n=3 biological replicates are shown. Statistical analysis was done using a one-way ANOVA followed by multiple comparison analysis (Tukey). (m) Induction of kefB with cumate impairs growth on PYE in ΔgshBΔkefB cells complemented with wild-type kefB, while kefB-Q92L has no effect. All cultures were grown in presence of 100 μM cumate. (n) Modest induction of kefB by 1.6 μM cumate in ΔgshBΔkefB cells leads to increased cell body length and cell division defects on PYE media. For explanations, see Fig. 4e. Data from n=3 biological replicates. Statistics was done with one-way ANOVA with a Kruskal-Wallis test of induced vs. control cultures using Dunn’s multiple testing correction; ****P < 0.0001. Cells larger >15 μm are not plotted. Zoom-in displays a representative elongated cell; arrows indicate partial cell constrictions. All growth curves show mean values +/- SD of n=3 biological replicates; pQF, empty plasmid; pQF-kefB, wild-type kefB; pQF-kefB*, kefB-Q92L. Microscopy scale bar is 5 μm.
In E. coli, the two Kef transporters, KefB and KefC, are known to be subject to glutathione-dependent regulation50. Like E. coli, C. crescentus encodes two paralogs, kefB and kefC (CCNA_03611). In E. coli, glutathione represses Kef transporters by binding to their C-terminal KTN domains, whereas various electrophiles produce glutathione-derivatives that activate the system and compete with glutathione for binding50–52. Moreover, glutathione deficiency in E. coli perturbs potassium homeostasis by de-regulation of Kef proteins53. Consequently, we hypothesized that KefB is de-repressed in the absence of glutathione leading to increased potassium efflux and that the suppressor mutations would prevent overactivation of KefB. In agreement with the idea that potassium levels are decreased in a ΔgshB mutant and underlie the growth defect of this strain, we observed that supplementation of the medium with low levels of potassium improved growth of the ΔgshB mutant, while addition of sodium had no effect (Fig. 5i). In addition, deletion of kefB in a ΔgshB background largely suppressed the slow-growth phenotype of ΔgshB cells irrespective of the medium, phenocopying the kefB-Q92L suppressor (Fig. 5j, k), which likely represents a loss of function allele. As expected for glutathione-deficient strains54, ΔgshB ΔkefB cells remained sensitive to oxidative stress (Supplementary Fig. 20), supporting the idea that the cell cycle defect of the ΔgshB strain largely results from loss of glutathione-dependent gating of KefB activity.
To directly investigate the effect of glutathione deficiency on potassium levels, we performed elemental analysis with inductively coupled plasma mass spectrometry (ICP-MS). In agreement with the hypothesis that loss of glutathione affects potassium homeostasis, we found that ΔgshB cells suffered from a kefB-dependent reduction of the intracellular K+ concentration (~56% of wild-type levels; Fig. 5l, Supplementary Fig. 21). To further corroborate these results, kefB and kefB-Q92L or the kefB paralog kefC were expressed from a cumate-inducible promoter system55 in the ΔgshB ΔkefB background. Indeed, induction of kefB impaired growth of ΔgshB ΔkefB cells in a cumate concentration-dependent manner (Fig. 5m, Supplementary Fig. 22a), thus confirming that KefB is causing the growth defect of the ΔgshB mutant. Overexpression of kefB-Q92L in this background had no effect (Fig. 5m, Supplementary Fig. 22a), as it was the case for the native allele in the wild-type strain (Fig. 5m). Already a low induction of kefB led to a dramatic increase in cell body length on PYE, indicating a block of cell division (Fig. 5n). Moreover, elongated cells were partially constricted, indicative of incomplete cell division events (Fig. 5n). Together, this suggests a strong glutathione-based gating of the KefB transporter in C. crescentus. In contrast, even full induction of the homologous potassium efflux system kefC led to only a minor growth defect in a glutathione-deficient strain (Supplementary Fig. 22b). Notably, unlike kefB, overexpression of kefC also affected growth of the wild-type strain and resulted in a mild morphology defect resembling ΔgshB cells (Supplementary Fig. 23). Together, these data suggest less stringent regulation of KefC by glutathione in comparison with KefB. This is in agreement with the isolation of ΔgshB suppressor mutations affecting the KefB, but not the KefC system. Altogether, our results suggest that a loss of glutathione leads to dysregulation of a potassium transporter, which in turn uncouples growth from cell cycle progression.
Discussion
Bacterial cell cycle progression is predicted to involve major changes in metabolite pools based on temporal gene expression patterns observed at the transcript22,23,25 and protein56 level. We found that the abundances of most metabolites were largely independent of the cell cycle. This highlights the robustness of cellular metabolism against the intrinsic perturbations of cell cycle progression and differentiation, as most resources are effectively allocated to meet the varying metabolic needs. However, we identified a subset of metabolites with distinct dynamics throughout the cell cycle. Among these we observed intriguing parallels and conservation of temporal patterns to the eukaryotic cell cycle in yeast7 and mammalian cancer cells57, as well as the yeast metabolic cycle (YMC)58,59. Exemplarily, we observed sulfur metabolism in C. crescentus undergoes intracellular dynamics, and similarly shows cell cycle dependency in mammalian cells, and during the YMC. The underlying process that drives these and other fluctuations might well be the periodic changes in metabolic demand at different stages of the cell cycle and the accompanying differentiation process. Alternatively, it might represent temporal compartmentalization of potentially interfering cellular or metabolic processes60, especially in bacteria that generally lack organelles. This is in agreement with the observed temporal control of the bacterial sulfate assimilation pathway, which in plants is partly restricted to chloroplast61, probably due to highly reactive pathway intermediates62.
Because metabolism is not only serving the changing nutritional needs of the cell but may also drive cellular processes, a crucial question is how metabolic signals help coordinate the cell cycle and division program, especially under changing environmental conditions63. For instance, NAD(P)H-dependent proteins such as KidO or GdhZ link metabolism with the regulation of cell division and development in C. crescentus12,20. Other prime examples are molecules such as c-di-GMP or (p)ppGpp16–18,21,34. However, second messengers may not directly reflect a distinct metabolic state. Instead, a number of metabolites that show cell cycle dependency (including currently undescribed small molecules) could be metabolic signals for cell cycle control. Examples are purine metabolites (e.g. AMP, AICAR) or glutathione that due to their central metabolic role would serve as an excellent transmitter of the metabolic status41,64 and potential perturbations42. Among these, the cell cycle dependency of glutathione was especially striking, as pool size and synthesis rate are attenuated early in the cell cycle, but increase during S phase. Thus, elevated levels of glutathione and in turn reductive potential are provided during DNA synthesis, creating a protective environment for replicating cells. Likewise, yeast DNA synthesis is temporally confined to the more reductive phase of the YMC65. Notably also, C. crescentus displays thioredoxin (Trx1) peak expression during S phase66 and an overall redox shift during cell cycle progression67.
We found that glutathione-deficient strains show a distorted coordination of cellular growth with cell cycle progression, leading to impaired cell length distributions. The characteristic phenotypes of glutathione deficiency in C. crescentus are largely explained by a dysregulation of the glutathione-regulated potassium efflux system KefB. While the Kef systems have been well studied in E. coli49–52, their coupling to cell division in C. crescentus raises new questions on how ion homeostasis intersects with cell cycle control. Notably, kefB is annotated as a potential K+/H+ antiporter68, and its over-activation might increase proton influx. Indeed, the polymerization of the major prokaryotic cell division protein FtsZ is affected by pH and potassium concentration in vitro69, indicating their potential role for cell division control.
Morphology defects in C. crescentus related to the cell cycle include mutants with altered activity of the essential cell cycle regulators CtrA70, DnaA71 or mutants of the cell division machinery72–74. While deletion of gshB also affects cell size at time of division, initiation of additional rounds of replication was largely prohibited. Another intriguing characteristic of glutathione deficiency was that the apparent dependence on the provided substrates, producing either unusually short or elongated cells. Curiously, it was recently proposed that C. crescentus cell size is maintained by an adder mechanism, and modulation of a late cell cycle event after replication initiation75–77. Although a more detailed mechanistic understanding will be required, defects of this modulation could accordingly explain the aberrant cell-length distribution and DNA content relative to biomass of ΔgshB cells.
Overall, the here applied large-scale metabolomics approach uncovered cell cycle dependency of small molecules. A prominent example is glutathione which impacts cell size control. In conclusion, C. crescentus intersects metabolism with cell cycle progression, while effectively adjusting metabolism to fuel cell cycle and differentiation processes.
Methods
Strains, cultivation and reagents
The strain C. crescentus NA1000 was used throughout this study, and all strains constructed were in a NA1000 background (see Supplementary Table 7). A list of primers is stated in Supplementary Table 8. Bacteria were grown in batch cultures, and unless otherwise stated cultivated in Erlenmeyer shake flasks in Minitron incubators (Infors HT) with 150 rpm at 30°C and grown in minimal media (M2) supplemented with 0.2 % glucose (M2G) according to Ely et al79, which was modified by replacing MgCl2 with MgSO4. For strain construction and storage, peptone yeast extract (PYE)79 complex media was used. Antibiotics were used in the following concentrations (for liquid/solid media): kanamycin 5/20 μg/ml; streptomycin 5/5 μg/ml, gentamycin 1/5 μg/ml and tetracycline or oxytetracyclin, 1/5 μg/ml. Reagents, unless otherwise stated, were acquired from Sigma Aldrich. u-13C glucose (99%) was obtained from Cambridge Isotope Laboratories. LC solvents and additives were obtained in LC-MS grade. dH2O used for LC-MS analysis was from an ultrapure water system (ELGA Purelab).
Cell synchronization and preparation of asynchronous control cultures
C. crescentus was synchronized by isolation of swarmer cells by adapting a common large scale density gradient centrifugation protocol33. To this end, an overnight culture of C. crescentus cells in M2G was passaged to obtain fresh exponential pre-cultures. Subsequently, two 400 ml M2G cultures were inoculated in 2l Erlenmeyer shake flasks with an initial OD600 of ~5x10-4 and cells were then grown to an OD600 of ~0.6 and harvested by centrifugation (15 min, 4°C; 7000 g). Cell pellets were re-suspended in 80 ml M2G (4°C) and thoroughly mixed with pre-cooled (4°C) 40 ml Ludox colloidal silica (30 wt. % suspension, Sigma) that was pre-treated by passing through filter paper (MN615 1/4, Macherey-Nagel) and autoclaving. The mixture was aliquoted to 4x50 ml falcon tubes and was centrifuged (9700 g, 35 min, 4°C). The top fractions were aspirated and discarded, and the lower fractions corresponding to swarmer cells were isolated and pooled. Subsequently, the collected swarmer cells were aliquoted to two 50 ml falcon tubes and washed by mixing with precooled (4°C) M2G. Cells were pelleted (7650 g, 10 min, 4°C), the supernatant was discarded and again washed in 40 ml precooled (4°C) M2G. After another centrifugation step, the supernatant was removed. Cells were finally cell cycle released by re-suspension in pre-warmed (30°C) M2G to reach an initial OD600 of ~0.4 and cultivated in Erlenmeyer flasks situated in a shaking water bath. The duration of the whole synchronization procedure was typically ~100 min.
For preparation of asynchronous control cultures, 30 ml of respective pre-cultures were aliquoted and kept at 4°C for 15 min. Subsequently, 20 ml pre-cooled (4°C) M2G was added, mixed, and cells were kept on ice for additional ~75 min. Then, cells were pelleted (7650 g, 10 min, 4°C), the supernatant was discarded, and cells were re-suspended in pre-warmed (30°C) M2G to an OD600 of ~0.4 and cultivated as described above. Note that the comparison of synchronized cells to this (temperature-) control treatment was required as they showed a metabolic response to this perturbation (respective dynamic metabolome profiles are depicted in Supplementary Data 1 & 2; ~27% of metabolite pools were affected based on t-tests comparing cultures released after treatment with corresponding unperturbed pre-cultures).
Sample preparation for metabolome profiling
C. crescentus cells were sampled, quenched and extracted employing the fast-filtration method80. To this end, a culture volume corresponding to 1 ml OD600 of 1.0 was passed through a polyethersulfone filter (0.2 um, Sartorius Setim) placed on a vaccum-filtration apparatus. The medium was separated from cells and discarded as a flow through and cells were washed by quickly drawing 10 ml dH2O supplemented with 2 mM glucose through the filter. The filter situating bacterial cells was then immediately quenched in pre-cooled (-20°C) glass bottles (Schott) containing 8 ml quenching and extraction solution (60/20/20 % V/V acetonitrile/methanol/dH2O acidified to a final concentration of 0.1M formic acid). Cells were extracted for 10 min on ice with repeated treatment in a bath sonifier (Branson). The filter was removed, and the extract was frozen in liquid nitrogen and lyophilized overnight at -48°C. Finally, the tried extract was re-suspended in dH2O, centrifuged twice (10 000 g and 20 000 g, 10 min each, 4°C) and was stored at -80°C until further analysis.
Preparation of 13C enriched internal standard
C. crescentus was pre-grown for >10 generations on M2 with u-13C glucose (99%) in an Erlenmeyer flask and was diluted in fresh M2 with u-13C glucose (99%) in a stirred glass bottle (Schott) situated in a heated water bath (30°C) that was aerated with synthetic air containing 0.1% 13CO2 (Linde). Culture was grown for additional ~4 generations until OD600 reached ~1.0 and sampled by fast filtration as described above, except that 7 filters were pooled in 25 ml quenching solution. Final extracts were lyophilized, re-suspended in dH2O, centrifuged twice (10 000 g and 20 000 g, 10 min each, 4°C), pooled, and supernatant was then stored at -80°C until it was used as an internal 13C standard.
Sampling for dynamic metabolomics
Prior to synchronization, one sample of the pre-culture was obtained as a reference 12C extract, and one sample was spiked with 13C internal standard as described below to obtain a reference sample prior to treatment. Then, C. crescentus cells were cell cycle synchronized, incubated in Erlenmeyer flasks sitting in a shaking water bath and sampled at time points 5 min, 10 min, followed by 10 min intervals until 160 min after cell cycle release. At each time point, OD600 was determined and sampling volume for fast filtration was adjusted to yield a biomass corresponding to 1 ml of OD600 of 1.0. For quantification, an aliquot containing the same biomass of 13C internal standard was spiked into the respective quenching and extraction solution. Samples were further processed as described above and stored at -80°C until measurement.
Metabolite measurements
Nanoscale HPLC-HRMS was performed with an nLC-ultra (Eksigent) chromatography system hyphenated to a Q Exactive Plus (Thermo Scientific) Orbitrap mass spectrometer with a electrospray ionization probe (ESI). Separation was achieved by ion pairing with a reverse-phase C18 column (Dr. Maisch Reprosil-Gold 120, 3 μm, 100 × 0.1 mm, Morvay Analytik) as a stationary phase. Mobile phase consisted of solvent A with 230 μM tributylamine (TBA) in dH2O with 3% methanol (TBA was first dissolved in 230 μM aqueous acetic acid; pH was adjusted to 9.0 with ammonium hydroxide). The eluent solvent B (50/50 % V/V mix of methanol/2-propanol) was used for the following multi-step gradient: 3 min 0 % B; 35 min 12 % B; 36 min 90 % B; 48 min 90 % B; 49 min 0 % B; 60 min 0 % B. The Flow rate was 400 nl min-1. For analysis, bacterial extracts were diluted (1:5) in solvent A to a concentration of 100 ng ul-1; 1 μl was injected. ESI and mass spectrometer were operating in negative mode with a spray voltage of 2.1 kV without supplemental gas flows. The S-lens RF level was set to 50.0 and capillary temperature was 250°C. Mass acquisition was performed by data-independent Fourier transform mass spectrometry (FTMS) full scan mode (scan range m/z 100-1200) with a resolution of 70k (at m/z 200) and summing 3 micro-scans. For targeted tandem MS experiments, parallel reaction monitoring (PRM, resolution of 30k at m/z 200) was employed using pre-defined inclusion lists of parent retention time (rt) and m/z windows (isolation width was set to m/z 0.5). Metabolites were fragmented using higher energy collisional dissociations (HCDs) with a normalized stepped collision energy (NCE; 25, 30, 35). Data-independent FTMS scans were obtained in parallel (resolution of 30k at m/z 200).
Microbore UHPLC-HRMS was carried out employing a Dionex UltiMate 3000 chromatography (Thermo Scientific) coupled to a LTQ XL (Thermo Scientific) Orbitrap mass spectrometer with a heated electrospray ionization (HESI) probe. Separation was based on a modified protocol for hydrophilic interaction chromatography (HILIC) with a BEH amide column (130Å, 1.7 μm, 100 x 2.1 mm, Acquity)81. The mobile phase consisted of solvent C (95/5/5 % V/V of acetonitrile/methanol/dH2O) and the eluent solvent A (50 % V/V mix of acetonitrile and dH2O). Both solvents were buffered with 55 mM formic acid and 10 mM ammonium hydroxide. The gradient was: 1.5 min 85% C; 5.5 min 5% C; 7.5 min 5% C; 8.0 min 85% C; 10.0 min 85% C. For analysis, bacterial extracts were dried and concentrated in a vacuum concentrator (SpeedVac SVC100, Savant) and re-suspended in 85/15 % V/V of solvent C/A to a concentration of 2 μg ul-1; 20 μl were injected. HESI source and mass spectrometer were operating with an APCI vaporizer temperature of 400°C supported by sheath gas and aux gas flow of 50 and 20 respectively. The source voltage was 3.5 kV, the capillary temperature 275°C, the tube lens was set to 60.0. Mass spectrometer was acquiring in positive FTMS full scan mode with a resolution of 30k (at m/z 400) and a scan range of m/z 85-600. Targeted tandem MS experiments (resolution of 15k at m/z 400) were performed using MS/MS mass lists with preset parent rt and m/z values (isolation width was set to m/z 1.0). Metabolites were fragmented by collision-induced dissociation (CID) with a NCE of 25.0. Data-independent FTMS scans were obtained in parallel (resolution of 15k at m/z 400).
Untargeted, isotope assisted metabolite detection and dynamic metabolomics
The here described, semi-automated workflow was developed with the Python based framework eMZed282. A detailed description can be obtained from Supplementary Methods. Briefly, for each of the 6 sampled batches (i.e. three replicates of cell cycle synchronized and their respective asynchronous control cultures), a corresponding naturally labelled extract (12C), the u-13C enriched standard (13C) and mixture (mix) of both extracts were acquired with the two described, non-targeted LC-HRMS techniques. The data was converted to mzML files and was submitted to a workflow aiming at detection and isolation of extraction windows of u-12C, u-13C pairs corresponding to intracellular metabolites of C. crescentus. To this end, custom spectral processing and stable isotope assisted filtering was employed to build respective consensus tables containing the extraction windows (u-12C, u-13C pairs) of reproducibly detected metabolites for each applied LC-HRMS method.
For each measured batch, the respective consensus table was employed to build a batch-specific extraction table. To this end, retention times of all peakmaps were aligned to the reference peakmap, u-13C peaks were inspected in the batch-specific mixed sample, and if necessary, extraction windows were manually adjusted. u-12C extraction windows were shifted accordingly. Then, these tailored extraction tables were used for targeted extraction of respective unlabeled (12C) and fully labeled (13C) peak pairs of all detected metabolites throughout the time series of the batch. The targeted extraction algorithm allowed for minor retention time shifts occurring for data acquired by nano-scale LC-HRMS. Peak areas where then calculated using numerical integration with a trapezoid rule83. As 12C extracts and 13C internal standards were normalized to a constant biomass during sampling, relative pool size R of metabolite i at time point t was calculated as the ratio of peak areas AU12C over AU13C:
After processing of all samples, individual metabolites with high variability over time and between replicates were manually inspected and curated. As an additional quality criteria, peak pairs where the u-13C signal did not exceed the limit of quantification (LOQ, 10x signal to noise) were discarded from analysis, and only metabolite features that contained >75% u-13C signals per time-series were considered for analysis. Spectral noise levels were determined for each individual measurement adapted from Horn et al84 (for details see Supplementary Methods).
Absolute quantification of intracellular glutathione concentration
Glutathione was quantified by spiking increasing concentrations of naturally labelled reduced glutathione (p.a.) into the 13C enriched C. crescentus extract used as an internal standard throughout this paper. Data was analyzed as described above, a calibration curve was obtained by linear regression (R2=0.99), and glutathione concentration of the 13C C. crescentus extract (n=4 technical replicates) was determined. The intracellular glutathione concentration was estimated by spiking naturally labelled C. crescentus cultures with the quantified internal standard (see above), and assuming the following correlation factors: 1 ml of OD660=1.0 contains 1.4e9 cells and a single C. crescentus cell has an intracellular volume of 1.14e-15 l 34.
Data analysis, statistics, plotting
Data was analyzed using custom written Python 2.7 scripts. Plotting was performed using Python packages Matplotlib and Seaborn, as well as Graphpad Prism. Statistical analysis was performed using the Python packages Scipy and Statsmodels, and Graphpad Prism. Propagation of uncertainty was performed using the Uncertainties package. For estimation of metabolome-wide wide temporal response to cell-cycle progression, for each time-point, the abundances of all metabolites were compared by a two-sided t-test per metabolite of synchronous vs. control condition (from 3 biological replicates); p-values were corrected for multiple testing using the Benjamini-Hochberg procedure, with a FDR of 5%. To identify strong cell cycle regulated metabolites, respective abundances at all time-points of individual metabolites were compared using two-sided t-tests (synchronous vs. control response), and corrected for multiple testing as described above. Moreover, candidates significantly differed at least at two consecutive time-points from control, and a significant response between conditions exceeded a fold-change of 2. To compare DNA content per cell mass, as well as potassium concentration between different genotypes, one-way ANOVA was performed; the wild-type or ΔgshB was compared to controls as indicated. If deemed significant (p-value <0.05), multiple comparison analysis was performed using Dunnett or Tukey method as indicated, with a confidence interval of 95%. Difference of cell length distributions were evaluated using one-way ANOVA with a Kruskal-Wallis test where ΔgshB was compared to controls; if deemed significant (p-value <0.05), post-hoc multiple comparison analysis was performed with Dunn's multiple testing correction.
13C tracer labeling experiments
For labeling of synchronized and asynchronous control C. crescentus cultures, cells were obtained as described above, but after the final washing step, the cell pellet was re-suspended in pre-warmed (30°C) M2 medium with 0.2% (w/V) u-13C glucose (99%), which marked the starting point of the labeling experiment. The culture was subsequently cultivated in an Erlenmeyer flask situated in a shaking water bath (30°C), and samples were taken after 1, 2.5, 3.5, 4.5, 5.5, 7.5, 10, 15, 20, 25, 30, 45 min, followed by 15 min intervals until 165 min. Sampling was performed as described above, except that cells were washed with dH2O supplemented with 2 mM u-13C glucose.
Fractional carbon labelling was determined by targeted extraction of carbon isotopologue clusters of metabolites of interest, correction for natural labelling, followed by automated peak integration and calculation of fractional labeling of individual isotopologues and the average fractional labeling44,83.
Flux balance analysis
Flux balance analysis (FBA) of sulfate assimilation and other major AMP-dependent reactions was performed using OptFlux 3.2.6 software that was based on the E. coli model iAF126085. Constrains were defined by maximizing the objective function "R_Ec_biomass_iAF1260_core_59p81M" using parsimonious enzyme usage with a glucose uptake rate of 10 mM glucose per g cellular dry weight (CDW) and hour. Sulfate uptake rate was not limiting. The indicated E. coli model was employed as no curated metabolic model of C. crescentus has been published; however, both organisms employ homologous pathways for purine de novo synthesis and sulfate assimilation.
Plasmid and strain constructions
For construction of pNPTS-ΔCCNA_00140::Ω regions up- and downstream of CCNA_00140 were PCR-amplified from C. crescentus NA1000 gDNA with primer pairs 10946/11428 and 10945/11429, respectively. The Ω-cassette encoding Spec/Strep resistance was PCR-amplified from plasmid pGFPC-1 86 with primers 11426/11427. Upstream, downstream region and Ω-cassette PCR products were digested with EcoRI/KpnI, XbaI/BglII and BglII/KpnI, respectively, and cloned in pNPTS138 (M.R. Alley) digested with SpeI/EcoRI. For construction of pNPTStet-ΔkefB regions up- and downstream of CCNA_00204 were PCR-amplified from C. crescentus NA1000 gDNA with primer pairs 12527/12917 and 12525/12918, respectively, the resulting PCR products were joined by SOE-PCR using primer pair 12525/12527 and cloned in pNPTStet via EcoRI/XbaI. pNPTStet is a pNPTS138 derivative in which the kanamycin resistance cassette has been replaced by a tetracycline resistance cassette87. pNPTStet-ΔkefB::nptII was constructed by PCR amplification of nptII conferring kanamycin resistance from pAK40588 using primer pair 12528/12529 and cloning of the PCR product in pNPTStet-ΔkefB via BglII/KpnI.
Plasmid pPxylX-CCNA_00140 was constructed by PCR-amplifying CCNA_00140 from C. crescentus NA1000 gDNA with primers 11372/11373, digestion of the PCR product with NdeI/KpnI and cloning into pXGFPC-4 86 digested with the same enzymes. Plasmid pQF-kefB was constructed by PCR-amplifying CCNA_00204 (kefB) from C. crescentus NA1000 gDNA with primers 12530/12916, digestion of the PCR product with SpeI/KpnI and cloning into pQF55 digested with the same enzymes. This construct adds a C-terminal 3XFLAG tag to the kefB open reading frame. pQF-kefB* (encoding the Q92L amino acid substitution in KefB) was constructed in a similar way, but using C. crescentus strain AKS451 as a template for PCR. Plasmid pQF-kefC was constructed by PCR-amplifying CCNA_03611 (kefC) from C. crescentus NA1000 gDNA with primers 13892/13893, digestion of the PCR product with SpeI/KpnI and cloning into pQF digested with the same enzymes, adding a C-terminal 3XFLAG tag to the kefC open reading frame. All plasmid inserts were confirmed by Sanger sequencing (Microsynth, Balgach, Switzerland).
Strain AKS429, in which CCNA_00140 at the native locus is replaced by a Ω-cassette, but CCNA_00140 is expressed from the xylX locus, was constructed in two steps. An extra copy of CCNA_00140 was first introduced at the xylX locus of strain AKS75 by conjugation of plasmid pPxylX-CCNA_00140. Then, native CCNA_00140 in this strain was replaced by an Ω-cassette by conjugation of plasmid pNPTS-dCCNA_00140::Ω and selection on PYE/Spec/Strep containing 0.3% sucrose. Resulting colonies were confirmed to have lost the plasmid backbone by testing for kanamycin sensitivity. AKS439 was constructed by generalized transduction using a ΦCR30 lysate of AKS429 on AKS75 and selection on PYE/Gm, followed by purification of colonies on PYE. AKS441, AKS442 and AKS443 were constructed by generalized transduction using a ΦCR30 lysate of AKS429 on AKS75 and selection on PYE/Spec/Strep, followed by purification of colonies on PYE. For complementation, AKS498 was constructed by generalized transduction using a ΦCR30 lysate of AKS429 on AKS442.
Strain AKS522 was constructed by replacing CCNA_00204 (kefB) with nptII in AKS75 using conjugation of plasmid pNPTStet-ΔkefB::nptII and selection on PYE/Kan containing 0.3% sucrose. Resulting colonies were confirmed to have lost the plasmid backbone by testing for tetracycline sensitivity. AKS581 was constructed by generalized transduction using a ΦCR30 lysate of AKS429 on AKS522 and selection on PYE/Kan/Spec, followed by purification of colonies on PYE/Spec. Strains AKS582 and AKS585 were constructed by transformation of AKS581 and AKS75, respectively, with plasmid pQF and selection on PYE/Tet. Strains AKS583 and AKS586 were constructed by transformation of AKS581 and AKS75, respectively, with plasmid pQF-kefB and selection on PYE/Tet. Strain AKS584 was constructed by transformation of AKS581 with plasmid pQF-kefB* and selection on PYE/Tet. Strains AKS597 and AKS598 were constructed by transformation of AKS75 and AKS581, respectively, with plasmid pQF-kefC and selection on PYE/Tet.
Microscopy
For image acquisition, bacteria were fixed by addition of 0.3 ml culture to 0.7 ml ice-cold pure ethanol, cells were quickly vortexed and kept at -20°C. 4-5 μl of bacterial cells were loaded onto a thin pad of solidified 1% (w/V) agarose (GellyPhor®LE agarose in PBS, pH 7.4). After absorption of excess liquid, the agarose pads were transferred onto glass-bottom dishes (WillCo Wells, Amsterdam, Netherlands) and used immediately for imaging. Phase contrast images of C. crescentus were captured at room temperature using an Axio Observer Z1 inverted microscope (Carl Zeiss, Jena, Germany) equipped with a LD Plan-Neofluar 40x/0.6 or EC Plan-Neofluar 100x/1.30 oil immersion objective respectively and an AxioCamMR. Images were acquired using AxioVision software.
For time-lapse microscopy, freshly synchronized C. crescentus cells were re-suspended in M2G and seeded on M2G 1.5% agar pads. Time-lapse images were captured on an Axio Observer Z1 equipped with Plan-Apochromat 100x/1.40 oil immersion objective in 6 min intervals for 180 min at 30°C. The duration from re-suspension of synchronized cells to initial acquisition was ~15 min.
For growth on PYE, a single colony from PYE plates was used to inoculate 1 ml PYE in 15-ml tubes and grown for ~10 hours at 30°C and were diluted to OD650 of ~0.005 (ΔgshB) or ~0.0001 (all other strains) in 2.5 ml PYE, incubated o/n and sampled during exponential growth phase. For growth on M2G, a single colony of ΔgshB from a PYE plate was used to inoculate 2.5 ml of a M2G in 15-ml tubes and grown for ~18 hours (ΔgshB) or ~8 hours (all other strains) at 30°C. Cultures were diluted to OD650 of ~0.01 (ΔgshB) or ~0,001 (all other strains), incubated o/n and sampled during exponential growth phase. Note that for ΔgshB cultures pre-grown on PYE and transferred to M2G, an inoculum of filamented cells is present; sufficient growth on M2G must thus be ensured.
For kefB induction experiments, single colonies from PYE plates containing tetracycline were used to inoculate 2.5 ml liquid cultures in round bottom tubes, and grown o/n at 30°C. AKS585 and AKS583 were then diluted 1:50 in 2.5ml fresh PYE with tetracycline, and grown for 6 hours; AKS583 cultures were additionally diluted 1:200 in the same media, induced with 1.56 μM cumate and cultured for 25 hours. Cells were fixed and imaged as described above. For the kefC induction experiments, a single colony of AKS597 was used to inoculate 2.5ml fresh PYE with tetracycline, and grown for 12 hours. The culture was diluted 1:5000, split in two cultures as described above, and either induced with 100 μM cumate or mock treated, incubated o/n, and imaged during exponential growth.
To test the effect of media composition on ΔgshB morphology defects, M2 media (without glucose) was prepared and supplemented with 50x stocks of i) peptone (100 g/l) + yeast extract (50 g/l) to get "M2PYE" or ii) glucose (10%) to obtain "M2G". Similarly, PYE media was prepared alone ("PYE"), or with the addition of glucose from a 50x stock (10% glucose) to obtain "PYEG". Cultures were prepared, diluted and imaged as described above for PYE and M2G respectively.
Cell lengths were quantified with MicrobeJ78 (v5.12) using default settings for automated detection. Due to the highly heterogeneous morphology of glutathione deficient cells, detection results were manually inspected and curated.
Flow cytometry
For growth on PYE, a single colony from a PYE plates was used to inoculate 5 ml PYE in 15-ml glass tubes and grown overnight at 30°C in a drum roller at 140 rpm. Overnight cultures were diluted 40-fold (ΔgshB) or 80-fold (all other strains) in 5 ml PYE and grown for an additional 4.5 hours before sampling. For growth on M2G, a single colony from a PYE plate was used to inoculate 5 ml of a M2G/PYE mixture (95%/5%) in 15-ml glass tubes and grown overnight at 30°C in a drum roller at 140 rpm. Overnight cultures were diluted 20-fold (ΔgshB) or 80-fold (all other strains) in 5 ml M2G and grown for an additional 18 hours (ΔgshB) or 4.5 hours (all other strains) before sampling. For flow cytometry, 0.3 ml of cells culture were harvested and fixed by addition of 0.7 ml ice-cold, pure ethanol, quickly vortexed, and stored at -20°C until further processing. Fixed cells were pelleted and and resuspended in 0.5 ml FACS buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 50 mM sodium citrate, 0.01% TritonX-100) containing 2.5 μl RNaseA solution (Sigma, Cat# R6148) at room temperature for 30min. Cells were pelleted and resuspended in 1 ml FACS buffer containing 1.5 μl YO-PRO-1 iodide (Thermo Fisher Scientific, Cat# Y3603) at room temperature for 1h. Data were acquired using a FACS Canto II (BD Biosciences) with 50’000 events in the final gate and analyzed with FlowJo software. The gating strategy is outlined in Supplementary Fig. 24. DNA content per cell mass was calculated by dividing the mean fluorescent signal of the population by the mean FSC-H value of the population.
Growth curves
Strains were streaked on PYE plates and inoculated at 30°C until small colonies had formed (2-5 days depending on the strain). Single colonies were used to inoculate 175 μl medium (PYE or M2G) per well in a tissue culture-treated, flat-bottom 96-well polyststyrene microplate (Falcon, Cat# 353072). For kefB and kefC overexpression experiments, the indicated concentration of cumate was supplemented to the growth media. To test the effect of media composition on ΔgshB growth defects, respective media were prepared as described above to obtain PYE, PYEG, M2PYE and M2G respectively. Growth curves were recorded on a Synergy H4 Hybrid microplate reader (BioTek) or a Tecan Infinity M200 Pro spectrophotometer (Tecan) at 30°C with fast continuous shaking by reading the OD660 every 10-30 min. Wells containing medium only were used as blanks. To correct for differences in inoculum size, for each growth curve t0 was defined as the time point at which cultures had an OD660 of 0.045 (PYE) or 0.100 (M2G) (Fig. 4) or 0.03 (PYE and M2G) (Fig. 5a, b, j, k). For growth curves of strains carrying pQF, pQF-kefB, pQF-kefB* or pQF-kefC, pre-cultures were grown in 5 ml PYE containing oxytetracycline in glass tubes in a rotary wheel (30°C, 160 rpm) overnight and 5 μl were diluted back in 175 μl of fresh medium supplemented in addition with different cumate concentrations (added from a 1000X stock solution in 100% ethanol) in a tissue culture-treated, flat-bottom 96-well polyststyrene microplate (Falcon, Cat# 353072), and growth curves were recorded as described above. For growth curves with glutathione supplementation, exponentially growing o/n cultures of wild-type and ΔgshB were prepared. Cultures were diluted back to OD600 of ~0.05; each culture was aliquoted to 4 x 3 ml in 14 ml round bottom tubes, and reduced glutathione was added in the indicated concentrations from a freshly prepared 100 mM stock solution. Growth was monitored in semi-micro cuvettes (Bio-Greiner) using a Biophotometer Plus (Eppendorf) at OD600.
Dilution spotting
Single colonies from PYE plates were transferred to 4 ml PYE in 15-ml round bottom tubes and grown o/n at 30°C. PYE agar plates were freshly prepared; NaCl/KCl,reduced glutathione (prepared freshly, sterile filtered), or H2O2 (from freshly opened bottle kept at 4°C) were added from stock solutions (2M, 0.1M and 0.2M respectively) to 40 ml PYE at 43°C, mixed, and cast into 12 cm square plates. Cells were pelleted and re-suspended in dH2O and adjusted to OD650 or OD600 of 0.1. Serial dilutions were prepared in dH2O, and 4 μl of suspensions were spotted, air dried, and plates were incubated at 30°C for 3 days.
Sample preparation for ICP-MS
Purified H2O (≥18.2 MΩ cm, Millipore, Billerica, USA) was used throughout sample preparation. Briefly, a culture volume corresponding to 2 ml of OD600 of 1.0 from exponentially growing cultures in PYE media (OD600 ~ 0.2) was collected by centrifugation (10 min, 10000 g). The supernatant was removed and cells were re-suspended in 1 ml p.a. grade sodium phosphate buffer (12.3 mM NaH2PO4, 7.8 mM Na2HPO4). Cells were collected by centrifugation (3 min, 10000 g), supernatant was removed, and cells were re-suspended in 0.5 ml H2O. Cell suspension was incubated for 12 min at room temperature, and centrifuged (3 min, 10000 g). 0.45 ml of the supernatant was removed and stored at -20°C until analysis. The corresponding cell pellet was re-suspended in 0.45 ml water and extracted by 15 min incubation at 99°C, followed by three rapid freeze/thaw cycles using liquid nitrogen. The suspension was additionally lysed by indirect sonication (10 min, 100% amplitude, 0.8 cycle time) in a VialTweeter (HIFU, Hielscher, Teltow, Germany). Insoluble cell debris was removed by centrifugation (10 min, 20 000 g) and the supernatant containing lysed cells was stored at -20°C until analysis. To control for potassium contaminations during sample preparation, "mock" controls w/o cells were processed as described above. K contaminations contributed approximately 6% of K recovered from wild-type cells, and the average K levels of n=3 mock controls was subsequently subtracted from the K levels determined in respective samples.
Potassium quantification by ICP-MS
ICP reference standards of 6Li, K, Rb were purchased from Peak Performance and Inorganic Ventures and used to prepare tuning, calibration and sample solutions. Concentrated nitric acid (>65% wt; purified by double sub-boiling distillation), and purified water (≥18.2 MΩ cm, Millipore, Billerica, USA) were used for the sample preparation. The provided samples were diluted and internal standards were added. All the preparation steps were done gravimetrically using a Mettler Toledo AT400 balance. In order to quantify the K content, the samples were investigated using an (in-house modified) PerkinElmer Sciex ELAN 6100 DRC. The instrument is coupled to a quadrupole reaction cell in front of the analyzer quadrupole mass spectrometer. Ammonia (anhydrous, Praxair, Lot. 168434.002) was used as reaction gas in order to remove the 38ArH+ molecular ion and other molecular ions. Prior to analyzing the samples by ICP-MS, the control samples were diluted by a factor of 20 and the supernatant and lysed cell samples were diluted by a factor of 200 using a 1% HNO3 solution. 6Li and Rb were added as internal standards obtaining a concentration of 80 μg L-1 and 10 μg L-1, respectively. The solutions were introduced using a concentric nebulizer (PFA, Microflow, 100-1843) combined with a cyclonic spray chamber (quartz, AHF). The instrument was equipped with a quartz injector, quartz torch, sampler and skimmer made of nickel. A tuning solution containing 80 ppb of 6Li, 6 ppb of K, and 10 ppb of Rb was used to tune the instrument. Further instrumental parameters were adjusted as follows: nebulizer gas flow 0.92 L min-1, coolant gas flow 16 L min-1, auxiliary gas flow 0.75 L min-1, RF-power 1200 W. The dynamic bandpass tuning parameter was set to RPq = 0.4 and the cell gas flow to 0.64 L min-1. The isotopes 6Li, 39K, 41K, 85Rb were measured in the dynamic reaction cell (DRC) mode and the data was acquired in the peak-hopping mode at a dwell time of 100 ms and a settling time of 3 ms. 3 sweeps, 50 readings and 10 replicates resulted in a total measurement time of 3 minutes per sample.
An external calibration was carried out. Six calibration solutions were measured and a linear regression was conducted in order to fit the calibration curve. The limit of detection (LOD) for K was determined to be 63 ng L-1. Intracellular K concentration in C. crescentus cells was estimated as described above for glutathione. In order to validate the method and exclude any matrix effects, the samples were investigated applying a lower dilution factor, certain samples were spiked with a K ICP reference standard and a standard addition was carried out. Recoveries between 96% and 106% were obtained for all the spiked samples.
Whole genome sequencing
Single colonies of respective strains were used to inoculate PYE media and incubated o/n at 30°C. Genomic DNA was extracted using the MasterPure™ DNA purification kit. Dual indexed libraries were prepared from 100 ng purified genomic DNA using the TruSeq™ DNA Nano library preparation kit. Sequencing was performed on the Illumina NovaSeq6000 system (2x150 bp) at the Functional Genomics Center Zurich with a read depth of 5 million reads per library. Data was analyzed using breseq89 (version 0.33.2) assuming clonal populations and reference based alignment to predict mutations. Reference genome for alignment was C. crescentus NA1000 (NCBI reference sequence NC_011916.1).
Supplementary Material
Acknowledgements
We want to thank P. Christen (ETH Zurich) for technical support with LC-MS, J. v. Gienanth and C. Marulli (ETH Zurich) for experimental assistance in early stages of this project, and J. Bögli and S. Stefanova (FACS Core Facility, Biozentrum, University of Basel) for help with flow cytometry. This work was supported by the Swiss National Science Foundation (grant no. 31003A_173094) and ETH Zurich to J.A.V. and the Swiss National Science Foundation (grant no. 310030B_147090) and by an ERC Advanced Research Grant (grant no. 322809) to U.J.
Footnotes
Reporting Summary. Further information is available in the Nature Research Reporting Summary linked to this paper.
Data and code availability. The metabolomics dataset generated during this study is available through MetaboLights database entry linked to this paper (https://www.ebi.ac.uk/metabolights/). The computer code for the described “pairfinder” workflow is deposited at https://pypi.org/project/idms-pairfinder-2/. Other data that support the findings of this study are available from the corresponding author upon request
Author contributions
J.H. and J.A.V. conceived the project. J.H. synchronized cells. J.H. and F.M. prepared the metabolomics samples. J.H. performed LC-MS measurements. J.H. and P.K. developed the algorithm for metabolite detection, quantification and performed data analysis. A.K. and U.J. planned and performed genetic manipulations, growth experiments and FACS measurements. J.H., A.K. and M.M. performed microscopy and image analysis. T.V. and B.H. performed elemental analysis. J.H. and J.A.V. wrote the manuscript with input from all authors.
Competing interest statement
The authors declare no competing interest.
References
- 1.Cai L, Tu BP. Driving the cell cycle through metabolism. Annu Rev Cell Dev Bi. 2012;28:59–87. doi: 10.1146/annurev-cellbio-092910-154010. [DOI] [PubMed] [Google Scholar]
- 2.Wang JD, Levin PA. Metabolism, cell growth and the bacterial cell cycle. Nat Rev Microbiol. 2009;7:822–827. doi: 10.1038/nrmicro2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee IH, Finkel T. Metabolic regulation of the cell cycle. Curr Opin Cell Biol. 2013;25:724–729. doi: 10.1016/j.ceb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnston GC, Pringle JR, Hartwell LH. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1977;105:79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- 5.Boye E, Nordstrom K. Coupling the cell cycle to cell growth. EMBO Rep. 2003;4:757–760. doi: 10.1038/sj.embor.embor895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature. 2017;546:426–430. doi: 10.1038/nature22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ewald JC, Kuehne A, Zamboni N, Skotheim JM. The Yeast cyclin-dependent kinase routes carbon fluxes to fuel cell cycle progression. Mol Cell. 2016;62:532–545. doi: 10.1016/j.molcel.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saqcena M, et al. Amino acids and mTOR mediate distinct metabolic checkpoints in mammalian G1 cell cycle. Plos One. 2013;8:e74157. doi: 10.1371/journal.pone.0074157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Margolin W, Bernander R. How do prokaryotic cells cycle? Curr Biol. 2004;14:R768–R770. doi: 10.1016/j.cub.2004.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen RB, Wang SC, Shapiro L. A moving DNA replication factory in Caulobacter crescentus. Embo J. 2001;20:4952–4963. doi: 10.1093/emboj/20.17.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iba H, Fukuda A, Okada Y. Rate of major protein-synthesis during cell-cycle of Caulobacter-crescentus. J Bacteriol. 1978;135:647–655. doi: 10.1128/jb.135.2.647-655.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaufay F, et al. A NAD-dependent glutamate dehydrogenase coordinates metabolism with cell division in Caulobacter crescentus. Embo J. 2015;34:1786–1800. doi: 10.15252/embj.201490730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irnov I, et al. Crosstalk between the tricarboxylic acid cycle and peptidoglycan synthesis in Caulobacter crescentus through the homeostatic control of alpha-ketoglutarate. Plos Genet. 2017;13:1–27. doi: 10.1371/journal.pgen.1006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weart RB, et al. A metabolic sensor governing cell size in bacteria. Cell. 2007;130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campos M, et al. A constant size extension drives bacterial cell size homeostasis. Cell. 2014;159:1433–1446. doi: 10.1016/j.cell.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronneau S, Petit K, De Bolle X, Hallez R. Phosphotransferase-dependent accumulation of (p)ppGpp in response to glutamine deprivation in Caulobacter crescentus. Nat Commun. 2016;7 doi: 10.1038/ncomms11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiaverotti TA, Parker G, Gallant J, Agabian N. Conditions that trigger guanosine tetraphosphate accumulation in Caulobacter crescentus. J Bacteriol. 1981;145:1463–1465. doi: 10.1128/jb.145.3.1463-1465.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boutte CC, Crosson S. The complex logic of stringent response regulation in Caulobacter crescentus: starvation signalling in an oligotrophic environment. Mol Microbiol. 2011;80:695–714. doi: 10.1111/j.1365-2958.2011.07602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westfall CS, Levin PA. Comprehensive analysis of central carbon metabolism illuminates connections between nutrient availability, growth rate, and cell morphology in Escherichia coli. Plos Genet. 2018;14:e1007205. doi: 10.1371/journal.pgen.1007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radhakrishnan SK, Pritchard S, Viollier PH. Coupling prokaryotic cell fate and division control with a bifunctional and oscillating oxidoreductase homolog. Dev Cell. 2010;18:90–101. doi: 10.1016/j.devcel.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 21.Lori C, et al. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature. 2015;523:236–U278. doi: 10.1038/nature14473. [DOI] [PubMed] [Google Scholar]
- 22.Fang G, et al. Transcriptomic and phylogenetic analysis of a bacterial cell cycle reveals strong associations between gene co-expression and evolution. Bmc Genomics. 2013;14 doi: 10.1186/1471-2164-14-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laub MT, et al. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- 24.Schrader JM, et al. Dynamic translation regulation in Caulobacter cell cycle control. P Natl Acad Sci USA. 2016;113:E6859–E6867. doi: 10.1073/pnas.1614795113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou B, et al. The global regulatory architecture of transcription during the Caulobacter cell cycle. Plos Genet. 2015;11:1–17. doi: 10.1371/journal.pgen.1004831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L, et al. Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly 13C-labeled cell extracts as internal standards. Anal Biochem. 2005;336:164–171. doi: 10.1016/j.ab.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Bueschl C, et al. MetExtract II: A Software suite for stable isotope-assisted untargeted metabolomics. Anal Chem. 2017;89:9518–9526. doi: 10.1021/acs.analchem.7b02518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, et al. Peak annotation and verification engine for untargeted LC-MS metabolomics. Anal Chem. 2019;91:1838–1846. doi: 10.1021/acs.analchem.8b03132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett BD, et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kind T, Fiehn O. Metabolomic database annotations via query of elemental compositions: mass accuracy is insufficient even at less than 1 ppm. BMC Bioinformatics. 2006;7:234. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai ZJ, et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat Methods. 2018;15:53–+. doi: 10.1038/nmeth.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schrader JM, Shapiro L. Synchronization of Caulobacter crescentus for investigation of the bacterial cell cycle. Jove-J Vis Exp. 2015 doi: 10.3791/52633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abel S, et al. Bi-modal distribution of the second messenger c-di-GMP controls cell fate and asymmetry during the Caulobacter cell cycle. Plos Genet. 2013;9 doi: 10.1371/journal.pgen.1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 36.Christen M, et al. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science. 2010;328:1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen KF, Dandanell G, Hove-Jensen B, WillemoEs M. Nucleotides, nucleosides, and nucleobases. EcoSal Plus. 2008;3 doi: 10.1128/ecosalplus.3.6.2. [DOI] [PubMed] [Google Scholar]
- 38.Sekowska A, Kung HF, Danchin A. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J Mol Microbiol Biotechnol. 2000;2:145–177. [PubMed] [Google Scholar]
- 39.Gonzalez D, et al. The functions of DNA methylation by CcrM in Caulobacter crescentus: a global approach. Nucleic Acids Res. 2014;42:3720–3735. doi: 10.1093/nar/gkt1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masip L, Veeravalli K, Georgioui G. The many faces of glutathione in bacteria. Antioxid Redox Sign. 2006;8:753–762. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- 41.Kosower NS, Kosower EM. The glutathione status of cells. Int Rev Cytol. 1978;54:109–160. doi: 10.1016/s0074-7696(08)60166-7. [DOI] [PubMed] [Google Scholar]
- 42.Smirnova GV, Oktyabrsky ON. Glutathione in bacteria. Biochemistry-Moscow+ 2005;70:1199–1211. doi: 10.1007/s10541-005-0248-3. [DOI] [PubMed] [Google Scholar]
- 43.Buescher JM, et al. A roadmap for interpreting (13)C metabolite labeling patterns from cells. Curr Opin Biotechnol. 2015;34:189–201. doi: 10.1016/j.copbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartl J, Kiefer P, Meyer F, Vorholt JA. Longevity of major coenzymes allows minimal de novo synthesis in microorganisms. Nat Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2017.73. 17073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schriek U, Schwenn JD. Properties of the purified APS-kinase from Escherichia coli and Saccharomyces cerevisiae. Arch Microbiol. 1986;145:32–38. doi: 10.1007/BF00413024. [DOI] [PubMed] [Google Scholar]
- 46.Lillig CH, et al. Redox regulation of 3'-phosphoadenylylsulfate reductase from Escherichia coli by glutathione and glutaredoxins. J Biol Chem. 2003;278:22325–22330. doi: 10.1074/jbc.M302304200. [DOI] [PubMed] [Google Scholar]
- 47.Apontoweil P, Berends W. Glutathione Biosynthesis in Escherichia coli K-12 - properties of enzymes and regulation. Biochim Biophys Acta. 1975;399:1–9. doi: 10.1016/0304-4165(75)90205-6. [DOI] [PubMed] [Google Scholar]
- 48.Meisenzahl AC, Shapiro L, Jenal U. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J Bacteriol. 1997;179:592–600. doi: 10.1128/jb.179.3.592-600.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller S, et al. Identification of an ancillary protein, YabF, required for activity of the KefC glutathione-gated potassium efflux system in Escherichia coli. J Bacteriol. 2000;182:6536–6540. doi: 10.1128/jb.182.22.6536-6540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ness LS, Booth IR. Different foci for the regulation of the activity of the KefB and KefC glutathione-gated K+ efflux systems. J Biol Chem. 1999;274:9524–9530. doi: 10.1074/jbc.274.14.9524. [DOI] [PubMed] [Google Scholar]
- 51.Booth IR, Epstein W, Giffard PM, Rowland GC. Roles of the trkB and trkC gene products of Escherichia coli in K+ transport. Biochimie. 1985;67:83–89. doi: 10.1016/s0300-9084(85)80233-9. [DOI] [PubMed] [Google Scholar]
- 52.Roosild TP, et al. Mechanism of ligand-gated potassium efflux in bacterial pathogens. Proc Natl Acad Sci U S A. 2010;107:19784–19789. doi: 10.1073/pnas.1012716107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meury J, Kepes A. Glutathione and the gated potassium channels of Escherichia coli. Embo J. 1982;1:339–343. doi: 10.1002/j.1460-2075.1982.tb01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murata K, Kimura A. Overproduction of glutathione and its derivatives by genetically engineered microbial cells. Biotechnol Adv. 1990;8:59–96. doi: 10.1016/0734-9750(90)90005-v. [DOI] [PubMed] [Google Scholar]
- 55.Kaczmarczyk A, Vorholt JA, Francez-Charlot A. Cumate-inducible gene expression system for sphingomonads and other Alphaproteobacteria. Appl Environ Microbiol. 2013;79:6795–6802. doi: 10.1128/AEM.02296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grunenfelder B, et al. Proteomic analysis of the bacterial cell cycle. P Natl Acad Sci USA. 2001;98:4681–4686. doi: 10.1073/pnas.071538098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahn E, et al. Temporal fluxomics reveals oscillations in TCA cycle flux throughout the mammalian cell cycle. Mol Syst Biol. 2017;13 doi: 10.15252/msb.20177763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tu BP, et al. Cyclic changes in metabolic state during the life of a yeast cell. P Natl Acad Sci USA. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murray DB, Beckmann M, Kitano H. Regulation of yeast oscillatory dynamics. Proc Natl Acad Sci U S A. 2007;104:2241–2246. doi: 10.1073/pnas.0606677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: Temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 61.Bohrer AS, Takahashi H. Compartmentalization and regulation of sulfate Assimilation pathways in plants. Int Rev Cell Mol Biol. 2016;326:1–31. doi: 10.1016/bs.ircmb.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 62.Alam MT, et al. The self-inhibitory nature of metabolic networks and its alleviation through compartmentalization. Nat Commun. 2017;8 doi: 10.1038/ncomms16018. 16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monahan LG, et al. Coordinating bacterial cell division with nutrient availability: a role for glycolysis. Mbio. 2014;5 doi: 10.1128/mBio.00935-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25:1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Z, Odstrcil EA, Tu BP, McKnight SL. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science. 2007;316:1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- 66.Goemans CV, et al. An essential thioredoxin is involved in the control of the cell cycle in the bacterium Caulobacter crescentus. J Biol Chem. 2018;293:3839–3848. doi: 10.1074/jbc.RA117.001042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Narayanan S, Janakiraman B, Kumar L, Radhakrishnan SK. A cell cycle-controlled redox switch regulates the topoisomerase IV activity. Gene Dev. 2015;29:1175–1187. doi: 10.1101/gad.257030.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Masrati G, et al. Broad phylogenetic analysis of cation/proton antiporters reveals transport determinants. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06770-5. 4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Y, Bjornson K, Redick SD, Erickson HP. A rapid fluorescence assay for FtsZ assembly indicates cooperative assembly with a dimer nucleus. Biophys J. 2005;88:505–514. doi: 10.1529/biophysj.104.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quon KC, et al. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci U S A. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hottes AK, Shapiro L, McAdams HH. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol Microbiol. 2005;58:1340–1353. doi: 10.1111/j.1365-2958.2005.04912.x. [DOI] [PubMed] [Google Scholar]
- 72.Lariviere PJ, et al. FzlA, an essential regulator of FtsZ filament curvature, controls constriction rate during Caulobacter division. Mol Microbiol. 2018;107:180–197. doi: 10.1111/mmi.13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martin ME, Trimble MJ, Brun YV. Cell cycle-dependent abundance, stability and localization of FtsA and FtsQ in Caulobacter crescentus. Mol Microbiol. 2004;54:60–74. doi: 10.1111/j.1365-2958.2004.04251.x. [DOI] [PubMed] [Google Scholar]
- 74.Ohta N, et al. Identification, characterization, and chromosomal organization of cell division cycle genes in Caulobacter crescentus. J Bacteriol. 1997;179:2169–2180. doi: 10.1128/jb.179.7.2169-2180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Banerjee S, et al. Biphasic growth dynamics control cell division in Caulobacter crescentus. Nat Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2017.116. 17116. [DOI] [PubMed] [Google Scholar]
- 76.Si F, et al. Mechanistic origin of cell-size control and homeostasis in bacteria. Curr Biol. 2019;29:1760–1770 e1767. doi: 10.1016/j.cub.2019.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lambert A, et al. Constriction rate modulation can drive cell size control and homeostasis in C. crescentus. Iscience. 2018;4:180–+. doi: 10.1016/j.isci.2018.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ducret A, Quardokus EM, Brun YV. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.77. 16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 80.Bolten CJ, et al. Sampling for metabolome analysis of microorganisms. Anal Chem. 2007;79:3843–3849. doi: 10.1021/ac0623888. [DOI] [PubMed] [Google Scholar]
- 81.Mulleder M, Bluemlein K, Ralser M. A high-throughput method for the quantitative determination of free amino acids in Saccharomyces cerevisiae by hydrophilic interaction chromatography-Tandem Mass Spectrometry. Cold Spring Harb Protoc. 2017;2017 doi: 10.1101/pdb.prot089094. pdb prot089094. [DOI] [PubMed] [Google Scholar]
- 82.Kiefer P, Schmitt U, Vorholt JA. eMZed: an open source framework in Python for rapid and interactive development of LC/MS data analysis workflows. Bioinformatics. 2013;29:963–964. doi: 10.1093/bioinformatics/btt080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kiefer P, et al. DynaMet: a fully automated pipeline for dynamic LC-MS data. Anal Chem. 2015;87:9679–9686. doi: 10.1021/acs.analchem.5b01660. [DOI] [PubMed] [Google Scholar]
- 84.Horn DM, Zubarev RA, McLafferty FW. Automated reduction and interpretation of high resolution electrospray mass spectra of large molecules. Journal of the American Society for Mass Spectrometry. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 85.Feist AM, et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thanbichler M, Iniesta AA, Shapiro L. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 2007;35:e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaczmarczyk A, et al. Precise transcription timing by a second-messenger drives a bacterial G1/S cell cycle transition. bioRxiv. 2019 675330. [Google Scholar]
- 88.Kaczmarczyk A, Vorholt JA, Francez-Charlot A. Markerless gene deletion system for sphingomonads. Appl Environ Microbiol. 2012;78:3774–3777. doi: 10.1128/AEM.07347-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.